Abstract
We present a case of a patient who had a history of severe coronavirus disease (COVID-19) 4 months prior to this current presentation and, after a long asymptomatic period, subsequently tested positive for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) by a RNA PCR assay, after several interval negative SARS-CoV-2 RNA tests. We present this potential case of SARS-CoV-2 reinfection in order to incite discussion around differentiating persistent infection with intermittent viral shedding and reinfection, as well as to discuss evolving knowledge and approaches to the clinical management, follow-up molecular testing and treatment of COVID-19 reinfection.
Keywords: COVID-19, immunology, pneumonia (infectious disease)
Background
The COVID-19 pandemic continues to impose a formidable morbidity and mortality toll on almost every country in the world. As the pandemic progresses into its tenth month, cases of reinfection have been identified and mounting evidence shows that protective immunity after a first episode of infection may be short-lived, and that phenomenon may explain the potential for reoccurrence of the disease. Currently, limited data exist regarding SARS-CoV-2 reinfection as reported cases are very few. Thus, it is important to document cases of potential SARS-CoV-2 reinfection in order to elucidate the natural history of COVID-19 disease, to understand the risk factors which may make patients more susceptible to SARS-CoV-2 reinfection, and to discuss further clinical and therapeutic management. Furthermore, research to better understand the durability and breadth of natural immunity against SARS-CoV-2 is needed to better inform the diagnosis, management and prevention of COVID-19 reocurrence.
Case presentation
A 43-year-old Hispanic man with a past medical history of well-controlled type 2 diabetes mellitus, class 3 obesity, hypothyroidism and a history of COVID-19 (initially diagnosed in April 2020) 4 months prior to this current presentation presented to the hospital with dyspnoea, stridor and difficulty with managing his respiratory equipment at home. Of note, his initial hospitalisation in April 2020 for severe COVID-19 was complicated by chronic respiratory failure for which he had a tracheostomy placed following prolonged intubation for ongoing oxygen dependence and hypercoagulable state (elevated D-dimer), for which he remained on anticoagulation for a planned total duration of 3 months. On this admission, while in the emergency department, the patient tested positive for SARS-CoV-2 RNA via a nasal swab, which was newly positive, 3 months after his initial positive test and following four interval negative SARS-CoV-2 RNA tests.
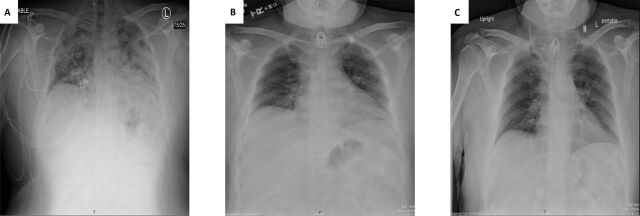
A review of his SARS-CoV-2 infection history is pertinent and showed that 4 months prior to this current presentation, in April 2020, he was initially hospitalised for 2 months with severe COVID-19. At the time he had outpatient testing for SARS-CoV-2 RNA that returned positive, after he developed and reported symptoms of fever, body aches and sore throat for 9 days. Five days after his initial positive SARS-CoV-2 RNA test, he experienced worsening shortness of breath which prompted him to seek hospital services. At that time, on requesting emergency medical services, they found him to be hypoxic with oxygen saturation around 80% on room air. On presentation to the emergency department his vital signs were notable for a temperature of 101.2°F, heart rate of 110 beats/min, respiratory rate of 30 breaths/min and oxygen saturation of 95% on a non-rebreather mask. He had a body mass index of 41.1 kg/m2. Laboratory data demonstrated a white blood cell count of 7800 cells/μL (neutrophils 78%, lymphocytes 17%, monocytes 5%), haemoglobin 14.8 g/dL and platelet count 204 000 cells/μL. Additionally, he had a bicarbonate level of 19 mmol/L and sodium level of 134 mmol/L, a normal procalcitonin of 0.17 ng/mL and raised high sensitivity C-reactive protein of 91.8 mg/L (<3 mg/L normal). A chest X-ray showed bilateral opacities (figure 1a).
Figure 1.
(A) Chest X-ray on the initial presentation with severe COVID-19 (4 months prior to index hospitalisation). (B) Chest X-ray on the second hospitalisation when the patient developed shortness of breath and hypoxia with a positive SARS-CoV-2 RNA test after four interval negative SARS-CoV-2 RNA tests. (C) Chest x-ray on the third hospitalisation when the patient presented with shortness of breath and persistent SARS-CoV-2 RNA positivity.
He was admitted to the stepdown unit where he required intubation given the risk for respiratory fatigue. Thereafter, he was transferred to the medical intensive care unit (MICU) for ventilator dependence. With regard to his COVID-19 treatment, the patient initially received tocilizumab 800 mg intravenously once, a monoclonal antibody which targets interleukin-6 and a 5-day course of hydroxychloroquine (loading dose of 400 mg orally twice daily on day 1 followed by 200 mg orally daily for 4 days). He also received a 3-day course of methylprednisolone 40 mg administered intravenously every 6 hours.
In the MICU he had a complicated hospital course, including prolonged hypoxic respiratory failure and acute respiratory distress syndrome due to severe COVID-19 eventually transitioning to ventilation through a tracheostomy. He also required veno-venous extracorporeal membrane oxygenation for 1 month as well as vasopressor support with norepinephrine and vasopressin for shock, which was eventually titrated off. His hospital course was also complicated by methicillin-sensitive Staphylococcus aureus bacteraemia attributed to a central venous catheter infection, which was treated with a course of intravenous cefazolin. Other complications occurred including gastrointestinal bleeding requiring blood transfusions and cryoprecipitate, ventilator-associated pneumonia and renal failure requiring temporary continuous veno-venous haemofiltration. When he stabilised after a prolonged 2-month hospitalisation course, he was subsequently discharged to an acute care facility for rehabilitation. Prior to his discharge from the hospital, he had three serial negative nasopharyngeal swab SARS-CoV-2 PCR tests.
Two months later in early August 2020, he presented to the hospital again and was hospitalised for 1 day after he reported sudden-onset shortness of breath coincident with malfunctioning of his Oxymizer. He was placed on ventilation mask with FiO2 of 28% and 10 L/min oxygen flow (baseline of 5–8 L/min oxygen via tracheostomy mask at home). At that time, he denied fever, chills or cough. He had a temperature of 98.5°F, heart rate of 85 beats/min, respiratory rate of 20 breaths/min, blood pressure of 131/85 mmHg and oxygen saturation of 100%. On examination he was in no acute distress and noted to have a tracheostomy mask in place with tracheal secretions, which he was able to cough up. He had scattered rhonchi on pulmonary examination. He also had noted generalised oedema. The remainder of his examination was otherwise within normal limits. His nasopharyngeal SARS-CoV-2 RNA test was positive again. A chest X-ray showed infrahilar peribronchial cuffing, which may reflect sequela of viral aetiology per report (figure 1b). He was ultimately discharged home.
Approximately 2 weeks later in late August 2020 on the third and latest admission, the patient presented again with complaints of shortness of breath after his Oxymizer had broken and was hospitalised for 1 week. At home he reportedly had intermittent episodes of choking and shortness of breath, as well as stridor. He was again found to have a positive SARS-CoV-2 RNA test result in the hospital (which likely represented persistent shedding that began on the antecedent admission in early August). Of note, he had four interim negative tests prior to his most recent positive SARS-CoV-2 test. He denied much exposure to other people, except for his family members, including his wife and two children who were asymptomatic and had not recently been ill.
On this current presentation, he was initially afebrile with a temperature of 98.5°F, heart rate of 94 beats/min, respiratory rate of 24 breaths/min, blood pressure of 121/81 mmHg and oxygen saturation of 97% on FiO2 28% at 8 L/min via tracheostomy mask. On examination he was in no acute distress. He had decreased breath sounds in the right lower lobe and had a tracheostomy in place. The remainder of the examination was not remarkable. Initial chest X-ray showed no acute process. In the emergency department, he had a brief episode of desaturation to oxygen saturation of 84% attributed to mucous plugging, which temporarily required a non-rebreather mask and he quickly returned to his baseline oxygenation. This recurred 3 days into his hospitalisation for which he required manual Ambu bagging transiently. He was started on intravenous vancomycin and piperacillin-tazobactam empirically due to presumed pneumonia and admitted to the hospital.
Investigations
On the most recent admission, laboratory investigations included white blood cell count of 7600 cells/μL (61% neutrophils, 25% lymphocytes, 8% monocytes, 5% eosinophils), haemoglobin of 10.6 g/dL and a platelet count of 333 000 cells/μL. Creatinine was 1.2 mg/dL with an estimated glomerular filtration rate of >60 mL/min/1.73 m2. Glucose was 135 mg/dL, procalcitonin was <0.06 ng/mL and C-reactive protein was elevated to 17.2 mg/L. D-dimer was normal at 0.38 mg/L. SARS-CoV-2 IgG antibody in the blood was elevated to 268 AU/mL (<15 AU/mL). A chest X-ray showed interval development of patchy opacities in the right lung likely representing aspiration (figure 1c).
The Infectious Diseases service was consulted to adjudicate the significance of the positive SARS-CoV-2 RNA test results and trend and to give management recommendations. Serial SARS-CoV-2 testing platforms and cycle threshold (Ct) values are shown in table 1.
Table 1.
SARS-CoV-2 testing and cycle threshold (Ct) values over a 154-day timeframe
| Date | Test result | Source of specimen | SARS-CoV-2 cycle threshold (Ct) |
Testing platform |
| Day 1: 2 April 2020 | Positive | Nasopharynx-oropharynx | ORF1a 15.49, E 16.08 | Mayo Clinic Roche C6800* |
| Day 8: 9 April 2020 | Positive | Nasopharynx | N1 24.6, N2 23.8 | Cepheid GeneXpert† |
| Day 17: 18 April 2020 | Positive | Nasopharynx | N1 29.2, N2 30.0 | Yale CDC laboratory developed test‡ |
| Day 43: 14 May 2020 | Negative | Nasopharynx | Not applicable | Panther TMA§ |
| Day 46: 17 May 2020 | Positive | Nasopharynx | N2 40, E 0.0 | Cepheid GeneXpert† |
| Day 49: 20 May 2020 | Negative | Nasopharynx | Not applicable | Yale CDC laboratory developed test‡ |
| Day 51: 22 May 2020 | Negative | Nasopharynx | Not applicable | Panther TMA§ |
| Day 56: 27 May 2020 | Negative | Nasopharynx | Not applicable | Thermo Fisher¶ |
| Day 108: 17 July 2020 | Negative | Nasopharynx | Not applicable | Cepheid GeneXpert† |
| Day 129: 6 August 2020 | Positive | Nasopharynx | N2 38.4, E 35.4 | Cepheid GeneXpert† |
| Day 144: 20 August 2020 | Positive | Nasopharynx | N2 43.1, E 0 | Cepheid GeneXpert† |
| Day 148: 24 August 2020** | Negative | Nasopharynx | Not applicable | Panther TMA§ |
| Day 154: 30 August 2020 | Negative | Nasopharynx | Not applicable | Cepheid GeneXpert† |
*Mayo Clinic Laboratories, Rochester, Minnesota, USA
†Cepheid, Sunnyvale, California, USA.
‡Yale Virology Lab, New Haven, Connecticut, USA.
§Hologic, San Diego, California, USA.
¶Thermo Fisher Scientific, Waltham, Massachusetts, USA.
**Received remdesivir on 24–30 August 2020.
E, SARS-CoV-2 E gene; N1, SARS-CoV-2 Nucleocapsid gene 1; N2, SARS-CoV-2 Nucleocapsid gene 2; ORF1a, SARS-CoV-2 Open Reading frame 1a gene.
Differential diagnosis
We present a case of a patient with initial severe COVID-19 infection with multiple complications including multiorgan failure who improved after a prolonged hospital course and was discharged to a rehabilitation facility. He presented 4 months later with mild respiratory symptoms and new positive diagnostic testing for SARS-CoV-2 after many interval negative tests, suggesting that this could be attributed to SARS-CoV-2 reinfection. Following his initial SARS-CoV-2 testing, the subsequent Ct values increased, representing a decreased SARS-CoV-2 viral load, as noted in table 1. This finding could be attributable to treatments received or represent the natural history of virus clearance over time. Subsequently (4 months later), he had detectable SARS-CoV-2 IgG antibodies in the blood. Together, the finding of a new positive SARS-CoV-2 RNA test, mild clinical symptoms, in conjunction with positive serum IgG antibodies led us to postulate that the milder clinical syndrome on the most recent presentation could be attributable to a relatively lower viral load as well as the presence of humoral immunity developed in response to the prior infection. However, a definitive diagnosis would require more information with regard to molecular testing/sequencing, including comparing viral sequences from his initial syndrome with his current isolate, which was not performed as samples from his first admission were not retained.
The other consideration was intermittent viral shedding which can either be a true clinical phenomenon or a laboratory artefact, as it can occur with the use of multiple tests (as occurred in this patient) with different detection thresholds and/or those that are not standardised or comparable to each other. However, even with this consideration, it seemed highly unlikely, as he had four interval negative SARS-CoV-2 RNA tests using four different testing platforms prior to the new positive test approximately 4 months later (table 1). SARS-CoV-2 cultures, which would provide information on the viability of the detected virus, were not performed as this is not available at our institution.
Regarding the aetiology of his respiratory decompensation, he likely had multiple factors contributing to his episodic respiratory decompensations: mucus plugging or aspiration events, possible bacterial pneumonia (antibiotics were discontinued quickly when this was thought to be less likely given the absence of fevers, purulent sputum and normal procalcitonin) and SARS-CoV-2 reinfection.
Treatment
Due to concern for SARS-CoV-2 reinfection, his risk factors for adverse outcomes and recurrent hospitalisation for respiratory decompensation, he was started on remdesivir with tocilizumab or placebo given once (administered via a clinical trial that was ongoing at our institution at the time). He eventually received an 8-day course of remdesivir treatment, which was discontinued when he was discharged. During the hospitalisation he had a bronchoscopy and tracheostomy exchange and was noted to have findings of granulation tissue and stenosis at the distal end of the tracheostomy leading to severe airway obstruction. This was managed with a 3-day course of intravenous methylprednisolone 40 mg every 6 hours.
Outcome and follow-up
The patient clinically improved during the hospital stay. His oxygen requirements were weaned back to his baseline prior to discharge and he was discharged from the hospital to his home with home healthcare services.
Discussion
We present a case of a patient with severe COVID-19 4 months prior to presentation who developed new positive detection of SARS-CoV-2 RNA after several interval negative SARS-CoV-2 RNA tests. This case highlights SARS-CoV-2 reinfection in patients with a prior diagnosis of COVID-19 and adds to the scarce literature on this occurrence.
As the COVID-19 pandemic has evolved, emerging reports have shown that SARS-CoV-2 reinfection is possible, such that positive SARS-CoV-2 RNA testing over a long period of time does not necessarily indicate persistent viral shedding from prior COVID-19 infection. We propose features to help distinguish between SARS-CoV-2 RNA reinfection and persistent viral shedding (table 2). To date there are no known tissue reservoirs other than the lungs that are associated with recrudescent or persistent disease over long periods of time for COVID-19. Moreover, sustained viral shedding detected via positive SARS-CoV-2 RNA testing in patients previously infected with SARS-CoV-2 does not necessarily correlate with infectivity supported by organism viability studies such as cultures.1 2 While viral shedding via the respiratory tract has been detected up to 63 days after symptom onset and some studies have documented 41 severe cases of COVID-19 with a median viral shedding period of 31 days,1 these are outliers as most individuals clear the virus in the first 2 weeks following symptom onset.3 One factor that impacts the duration of viral shedding is immunocompromised states such as in transplant recipients, who exhibit relatively prolonged SARS-CoV-2 viral shedding times compared with controls.1
Table 2.
Proposed features for distinguishing SARS-CoV-2 reinfection from prolonged SARS-CoV-2 viral shedding
| SARS-CoV-2 reinfection | Prolonged SARS-CoV-2 viral shedding | |
| Sequencing | Confirmed RNA sequencing with evidence of distinct SARS-CoV-2 viral strains at distinct episodes would provide a definitive diagnosis of SARS-CoV-2 reinfection | RNA sequencing showing the same SARS-CoV-2 viral strains at distinct periods could suggest prolonged SARS-CoV-2 viral shedding rather than reinfection; unless clinical suspicion is high for SARS-CoV-2 reinfection, further evaluation would be needed to assess if the patient has been reinfected with the same SARS-CoV-2 strain |
| Cycle threshold (Ct) values | Variability in SARS-CoV-2 Ct detection has been documented in cases of SARS-CoV-2 reinfection, with some cases with increased or decreased Ct values on reinfection episode (with Ct varying from 16.6 to 36.85)8 | Likely to have high Ct values suggesting low level viral shedding; however, low Ct values may be observed in immunocompromised patients who have decreased ability to clear the virus |
| Viral culture | Likely to be positive | Could be positive or negative (latter representing non-viable virus detected via SARS-CoV-2 PCR tests) |
| Timing of repeat positive testing | Variable; time between positive SARS-CoV-2 RNA PCR in documented cases of SARS-CoV-2 reinfection ranges from 48 to 142 days8 | Variable |
| Antibody testing in blood | Could be positive or negative* | Could be positive or negative |
| Host characteristics | Variable; the majority of documented cases of SARS-CoV-2 infections noted in immunocompetent hosts (with the exception of one patient taking inhaled corticosteroids)8 | Variable; notably, immunocompromised hosts are more likely to have prolonged episodes of SARS-CoV-2 viral shedding |
| Symptomatic vs. asymptomatic presentation | Variable symptoms; may have milder (including asymptomatic presentations) or worsening symptoms as noted in prior cases of SARS-CoV-2 reinfection8 | Predicted to have asymptomatic presentation if due to prolonged SARS-CoV-2 viral shedding |
*SARS-CoV-2 re-infections may occur more commonly in individuals with waning immunity after their first SARS-CoV-2 infection.
Recent reports have shown that SARS-CoV-2 reinfection with distinct virological strains can occur. Such cases of SARS-CoV-2 positivity after recovery from a prior diagnosis of COVID-19 have emerged from China, Hong Kong and Vietnam.4 5 In the Hong Kong case, whole genome sequencing was performed on respiratory samples from two separate COVID-19 episodes in the patient—initially when the patient was symptomatic with cough, sputum production, fever and headache and subsequently during the second episode when the patient was asymptomatic 142 days later. Testing showed two separate SARS-CoV-2 viral strains (with the first viral genome related to USA or England strains circulating around April 2020 while the second viral genome was related to strains isolated in Switzerland and England in August 2020).4
According to the Centers for Disease Control and Prevention (CDC) guidance, a positive SARS-CoV-2 PCR test within 90 days of an initial infection may represent sustained viral shedding rather than reinfection.6 Therefore, based on the guidance of the CDC, those who are asymptomatic during the 90-day period do not need to be retested for COVID-19.6 However, for previously infected individuals in this 90-day timeframe who develop respiratory symptoms and without an alternative aetiology identified on evaluation, it may be reasonable to evaluate for SARS-CoV-2 reinfection.6 Such individuals should be considered for retesting for SARS-CoV-2 infection within the 90-day timeframe.6 In this scenario, pre-emptive isolation is warranted, especially if the individual reports epidemiological risk and certainly if there is a new positive test that could suggest infectiousness.6
Interestingly, persistent SARS-CoV-2 viral shedding has been noted despite seroconversion, and SARS-CoV-2 virus has been cultured after development of antibodies to the virus (table 2).7 8 For instance, another study found that 14.5% of convalescent patients had noted re-detection of SARS-CoV-2 PCR RNA in the time of follow-up (minimum time to follow-up of 14 days).9 Additionally, studies have shown that the presence of antibodies did not correlate with a rapid decline in viral shedding.10 These studies suggest that measured antibody responses may not represent true neutralising antibody levels; studies that focus on identifying and isolating the specific antibodies that neutralise the virus are ongoing. This approach has informed emergence of monoclonal antibody candidates that are being evaluated in clinical trials, having been isolated from individuals who have recovered from COVID-19 and using viral or pseudoviral neutralisation tests.
Further research is needed to elucidate the breadth and durability of protection conferred by natural immunity to SARS-CoV-2. A literature review has found that immune responses to SARS-CoV-2 infection are similar to that of SARS-CoV-1 and Middle East Respiratory Syndrome (MERS) coronavirus.11 Most infected individuals have detectable seroconversion within 10–14 days following onset of symptoms,12 although other studies have demonstrated seroconversion as early as 7 days.11 Antibody detection may also vary by disease severity with undetectable or lower SARS-CoV-2 antibodies detected in mild cases compared with severe cases.11 An analysis of 173 patients in China with acute SARS-CoV-2 respiratory syndromes with chest CT imaging abnormalities showed a median seroconversion time of 11–14 days.13 In another cohort study of COVID-19 patients, SARS-CoV-2 seroconversion was noted in all patients by days 17–19 after symptom onset.10
In general, severely ill patients have higher SARS-CoV-2 IgG blood levels than non-severe cases within 7–14 days after symptom onset; however, no difference in antibody titres existed between these groups at 15–21 days in one study.10 Other studies have shown that neutralising antibody titres start to wane within a timeframe of 1–2 months, with a steeper decline in patients with mild disease compared with severe cases, and may be a plausible explanation for the risk of reinfection although it remains to be definitively identified as a correlate of protection against reinfection.4 Furthermore, one could hypothesise that, if patients with severe disease develop more robust antibody levels, their duration of protection against reinfection and resulting severity of disease, if it does occur, may be muted. Future observations would certainly shed more light on this if this hypothesis holds true. The role of the presence or absence of antibodies after initial infection in survivors of a first episode of COVID-19 and its role in mitigating the risk of SARS-CoV-2 reinfection is not clearly defined. It is plausible, however, that waning immunity or absence of antibodies after the first episode of SARS CoV-2 infection may make one more susceptible to reinfection.
Ultimately, understanding the clinical course and immune responses to SARS-CoV-2, as well as mechanisms and patterns of reinfection, are essential and have important implications for testing, treatment and prevention of SARS-CoV-2 reinfection.
Learning points.
SARS-CoV-2 reinfection does occur and mild reinfection may follow initial severe disease.
There are differences in the timing, robustness and durability of immune responses to SARS-CoV-2 infection between patients with mild and severe COVID-19.
It is important to evaluate and test patients with a history of COVID-19 who develop new symptoms suggestive of SARS-CoV-2 reinfection, given the need to evaluate them for retreatment and to prevent disease transmission.
Footnotes
Contributors: JT, AS-M and OO have contributed individually and as a group to the writing of this case report with regard to the planning, conduct, reporting, conception and design, acquisition of data or analysis and interpretation of data.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Patient consent for publication: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Widders A, Broom A, Broom J. SARS-CoV-2: the viral shedding vs infectivity dilemma. Infect Dis Health 2020;25:210–5. vol.. 10.1016/j.idh.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu J, Peng J, Xiong Q, et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine 2020;59:102960. 10.1016/j.ebiom.2020.102960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. He X, EHY L, Wu P. Temporal dynamics in viral shedding and transmissibility of COVID-19 [published correction appears in Nat Med. 2020 Sep;26(9):1491-1493]. Nat Med 2020;26:672–5. [DOI] [PubMed] [Google Scholar]
- 4. To KK-W, Hung IF-N, Ip JD, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis 2020:ciaa1275. 10.1093/cid/ciaa1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thach PN, Anh NT, Hang NV, et al. Two recovered COVID-19 cases testing positive again for SARS-CoV-2 after discharge in Hanoi, Vietnam. Arch Clin Med Case Rep 2020;4:766–73. [Google Scholar]
- 6. Centers for Disease Control and Prevention . Duration of isolation and precautions for adults with COVID-19, August 16, 2020. [Google Scholar]
- 7. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9 https://doi.org/ 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 8. European Centre for Disease Prevention and Control . Re-infection with SARS-CoV-2: considerations for public health response. Threat Assessment Brief; 21 September 2020.
- 9. An J, Liao X, Xiao T, et al. Clinical characteristics of recovered COVID-19 patients with re-detectable positive RNA test. Ann Transl Med 2020;8:1084–20044222. 10.21037/atm-20-5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Long Q-X, Liu B-Z, Deng H-J, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020;26:845–8. 10.1038/s41591-020-0897-1 [DOI] [PubMed] [Google Scholar]
- 11. Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol 2020;101:791–7. 10.1099/jgv.0.001439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang AT, Garcia-Carreras B, Hitchings MDT, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun 2020;11:4704. 10.1038/s41467-020-18450-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis 2020;71:2027–34. 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]