Abstract
Background
T cell exhaustion compromises antitumor immunity, and a sustained elevation of co-inhibitory receptors is a hallmark of T cell exhaustion in solid tumors. Similarly, upregulation of co-inhibitory receptors has been reported in T cells in hematological cancers such as chronic lymphocytic leukemia (CLL). However, the role of CD160, a glycosylphosphatidylinositol-anchored protein, as one of these co-inhibitory receptors has been contradictory in T cell function. Therefore, we decided to elucidate how CD160 expression and/or co-expression with other co-inhibitory receptors influence T cell effector functions in patients with CLL.
Methods
We studied 56 patients with CLL and 25 age-matched and sex-matched healthy controls in this study. The expression of different co-inhibitory receptors was analyzed in T cells obtained from the peripheral blood or the bone marrow. Also, we quantified the properties of extracellular vesicles (EVs) in the plasma of patients with CLL versus healthy controls. Finally, we measured 29 different cytokines, chemokines or other biomarkers in the plasma specimens of patients with CLL and healthy controls.
Results
We found that CD160 was the most upregulated co-inhibitory receptor in patients with CLL. Its expression was associated with an exhausted T cell phenotype. CD160+CD8+ T cells were highly antigen-experienced/effector T cells, while CD160+CD4+ T cells were more heterogeneous. In particular, we identified EVs as a source of CD160 in the plasma of patients with CLL that can be taken up by T cells. Moreover, we observed a dominantly proinflammatory cytokine profile in the plasma of patients with CLL. In particular, interleukin-16 (IL-16) was highly elevated and correlated with the advanced clinical stage (Rai). Furthermore, we observed that the incubation of T cells with IL-16 results in the upregulation of CD160.
Conclusions
Our study provides a novel insight into the influence of CD160 expression/co-expression with other co-inhibitory receptors in T cell effector functions in patients with CLL. Besides, IL-16-mediated upregulation of CD160 expression in T cells highlights the importance of IL-16/CD160 as potential immunotherapy targets in patients with CLL. Therefore, our findings propose a significant role for CD160 in T cell exhaustion in patients with CLL.
Keywords: cytokines, T-lymphocytes, costimulatory and inhibitory T-cell receptors, B-lymphocytes, immunity, cellular
Background
Chronic lymphocytic leukemia (CLL), the most common leukemia among adults in Western countries, is identified by the clonal expansion of CD5+ mature B lymphocytes in the blood, bone marrow (BM), and secondary lymphoid organs.1 2 The disease fate is highly influenced by the advanced clinical stage (ie, Rai stage), the broad range of genetic factors and remarkable contributions from the tumor microenvironment.3 Malignant B cells (B-CLL) impair both innate and adaptive immune responses and thus increase susceptibility to infections.4 Several quantitative,5 phenotypic,6 and functional7 T cell alterations are reported in patients with CLL. Although CD8+ T cells play an important role against the tumor, the occurrence of T cell exhaustion compromises their effector functions. Exhausted CD8+ T cells are characterized by the loss of effector functions, altered epigenetic and transcriptional profiles, distinct metabolic style, and the inability to transition to memory T cells.8 Antigen persistence, soluble mediators, cytokines, and immunoregulatory cells modulate the severity and the pace of T cell exhaustion.9
The sustained upregulation of co-inhibitory receptors (co-IRs) is the hallmark of CD4+ and CD8+ T cell exhaustion in chronic viral infections and cancer.10 Likewise, it has been shown that T cells in CLL upregulate the expression of programmed cell death protein-1 (PD-1), CD160, and 2B4 (CD244),11 which results in their impairment (eg, proliferation, cytotoxicity, and cytolytic functions).12 13 Comparable alterations in the gene and protein expression in T cells of TCL1 transgenic CLL mouse model in which adopted T cells acquire the characteristic of T cell dysfunction on tumor antigen encounter have been reported.14
CD160 as one of these IRs belongs to glycosylphosphatidylinositol (GPI)-anchored glycoprotein of the immunoglobulin superfamily.15 It was first identified on the membrane of mice natural killer (NK) cells,16 mast cells,17 and B-CLL cells but not on normal B cells.18 CD160 can exist in three different forms such as the GPI-anchored, transmembrane (TM), and soluble forms. T cells mainly express the GPI but occasionally TM isoforms.19 The soluble isoform composed of the extracellular regions of CD160 that is susceptible to be cleaved from the cell membrane by a metalloprotease as CD160 shedding has been reported from NK20 and mast cells17 on activation and degranulation, respectively.
Human/mouse CD160 binds weakly to classical and non-classical major histocompatibility complex-I (MHC-I) molecules,21 triggering NK cell cytotoxicity22 and the release of proinflammatory cytokines. The interaction of CD160 with classical and non-classical MHC-I complexes inhibits binding of CD8 to α3 subunits of MHC class I, causing defect in MHC class I-dependent CD8+ T cell cytotoxicity.20 CD160 and B- T-lymphocyte attenuator (BTLA) interact with the cysteine-rich region of the extracellular domain of herpesvirus entry mediator (HVEM).23 24 The engagement of CD160 with the soluble HVEM has a costimulatory effect on human NK cell function.25 Paradoxically, CD160 interaction with HVEM receptor delivers an inhibitory signal to activated CD4+ T cells, resulting in diminished cytokine production and proliferation.26 However, the role of CD160 in CD8+ T cells is more complex. For example, CD160 exhibits an inhibitory function in viral infections such as HIV27 but a stimulatory property in Listeria monocytogenes infection28 and the allograft skin reaction.29 The various role of the CD160 molecule could be related to the expression of its corresponding ligands HVEM or MHC-I. For example, the interaction of CD160 with MHC-I induces a stimulatory signal,28 whereas the interaction with HVEM triggers an inhibitory signal.26 These discrepancies in the field warrant further investigations to better understand the role of CD160 in T cell function in chronic conditions such as cancer. Moreover, it is unclear how the soluble CD160 is transported out of immune cells and there is a possibility that extracellular vesicles (EVs) are involved in this process.
EVs are small endosomal-derived membrane microvesicles (50–100 nm) that are released extracellularly and act as intracellular communicators.30 EVs carry a complex of cargo proteins, lipids and nucleic acids to target cells. EVs express tetraspanins (CD9, CD63, CD81), cytoskeleton proteins, stress proteins, MHC, RNA, DNA, and glycolipids. Besides, EVs can carry co-inhibitory receptors and modulate the effector functions of different immune cells.31
In this study, we examined the expression of CD160 on the surface of both CD8+ and CD4+ T cells in 56 patients with CLL. We found CD160 upregulation was associated with an exhausted T cell phenotype. Notably, CD160+ CD8+ T cells were highly antigen-experienced/effector T cells, while CD160+ CD4+ T cells were more heterogeneous. The plasma cytokine profile of patients with CLL exhibited a proinflammatory phenotype. In particular, IL-16 was highly elevated in the plasma of patients with CLL and was correlated with the Rai stage. Finally, we show that isolated EVs from the plasma of patients with CLL can be a source of CD160.
Methods
Study population
The peripheral blood and bone marrow (BM) samples were collected from patients with B-CLL. We recruited 56 patients with CLL for the study (online supplemental table 1). The diagnosis was based on clinical, morphology, and immuno-phenotyping features. Peripheral blood from age-matched and sex-matched 25 healthy donors were obtained for comparison. The staging was done based on the clinical data using the Rai staging system reported elsewhere.32
jitc-2020-002189supp001.pdf (49.9KB, pdf)
Cell isolation and purification
The peripheral blood mononuclear cells (PBMCs) and BM mononuclear cells were isolated using Ficoll-Paque gradients (GE Healthcare). CD8+ or CD3+ T cells were isolated by negative selection using the Easysep isolation kits (Stem Cell Technologies) with a purity of >90% (online supplemental figure 1a, b). For effector T cell (CD8+CCR7-) isolation, CD8+ T cells were stained with phycoerythrin (PE)-conjugated anti-CCR7 antibody, followed by anti-PE-conjugated microbeads (Miltenyi) with a purity of >90% (online supplemental figure 1c). B-CLLs were isolated using the human B-CLL Cell Isolation Kit (Miltenyi) with a purity of >90% (online supplemental figure 1d).
jitc-2020-002189supp002.pdf (12.8MB, pdf)
Flow cytometry
The fluorochrome-conjugated antibodies were purchased from BD Biosciences, Thermo Fisher Scientific or Biolegend including human anti-CD3 (SK7), anti-CD4 (RPA-T4), anti-CD8 (RPA-T8), anti-CD5 (UCHT2), anti-CD19 (HIB19), anti-CD160 (BY55), anti-HVEM (94801), anti-2B4 (eBioDM244), anti-TIGIT (MBSA43), anti-PD1 (EH12.1), anti-BTLA (J168-540), anti-GAL-9 (9M1-3), anti-TIM-3 (7D3), anti-CD45RA (HL100), anti-CD45RO (UCHL1), anti-CD62L (DREG-56), anti-CCR7 (3D12), anti-CD27 (0323), anti-CD28 (CD28.2), anti-CD57 (NK-1), anti-TCR-(T10Bg.1A-31), anti-TCR-(B1), anti-CD16 (B73.1), anti-CD56 (B159), anti-CD25 (M-A251), anti-CD69 (FN50), anti-CD-38 (HIT2), anti-HLA-DR (LN3), anti-Glut-1 (202915), anti-CD137 (4B4-1), anti-CD154 (TRAp1), anti-CD122 (Mik-B3), anti-CD107a (H4A3), anti-IL-2 (MQ1-17H12), anti-TNF-(MAB11), anti-IFN-(45.B3), anti-Perforin (dG9), anti-Granzyme-B (GB11), anti-FOXP3 (150D/E4), anti-EOMES (WD1928), anti-T-bet (4B10), and anti-TCF-1 (7F11A10). The LIVE/DEAD kit (Life Technologies) was used to assess cell viability. The phosphorylation of STAT-5 was performed using a Phosflow kit (BD Biosciences) according to the manufacturer’s instruction. Stained cells were fixed in paraformaldehyde (PFA 4%) and data were acquired on a Fortessa-X20 or LSR Fortessa-SORP flow cytometry (BD Bioscience). Data were analyzed using FlowJo software (V.10.7.1). A representative gating strategy for CD160+ T cells is provided (online supplemental figure 1e).
Cell culture and ex vivo cytokine measurement
PBMCs or isolated CD8+ T cells were cultured and stimulated with soluble Purified NA/LE Mouse anti-human CD3 (UCHT1, 3 μg/mL) and anti-human CD28 (CD28.2, 1 μg/mL) or PMA/Ionomycin (Cell stimulation cocktail, BioLegend) in the presence of the protein transport inhibitor Brefeldin A (BD Biosciences) for 5 hours. After fixation and permeabilization (BD Cytofix/Cytoperm), intracellular cytokine staining was performed according to our previous methods.32 33 In some experiments, PBMCs were stimulated with anti-CD3/CD28 antibodies and treated with different cytokines such as IL-16 (R&D, 500 ng/mL) for 72 hours. Then, CD8+ T cells were subjected to flow cytometry and RT-PCR for the quantification of CD160. In long-term cultures, every 72 hours, fresh media and cytokine/stimulation cocktails were added. Also, isolated B-CLL cells (1×106) were cultured for 12 hours and then IL-16 levels were measured in the culture supernatant by the quantikine ELISA kit (R&D).
Proliferation assay
Isolated CD3+ T cells were labeled with the CFSE dye (Life Technologies) and then stimulated with the Dyna beads Human T-activator CD3/CD28 (Thermo Fisher Scientific) according to the manufacturer’s protocol and analyzed after 72 hours.
Image cytometry
After surface or intracytoplasmic staining (ICS), cells were fixed with PFA 4% and analyzed. More than 5000 images were collected for each panel after the appropriate compensations were applied using the Amnis ImageStream Mark II image cytometer (EMD Millipore) and images were analyzed by the IDEAS software as we have reported elsewhere.34–36 Only high-resolution and in-focus images were selected for further analysis.
Mesoplex and ELISA assay
The plasma concentrations of interferon-γ (IFN-γ), interleukin (IL)-1β, IL-2, Il-4, Il-6, IL-10, IL-12p70, IL-13, and tumor necrosis factor-α (TNF-α) were measured using the V-plex plus Proinflammatory Panel 1 kit from Meso Scale Discovery (MSD). The plasma levels of granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-1α, IL-5, IL-7, IL-12/23 p40, IL-15, IL-16, IL-17A, TNF-β, and vascular endothelial growth factor (VEGF) were measured using the V-Plex Plus Cytokine Panel 1 (MSD). The concentrations of Eotaxin, MIP-1α, Eotaxin-3, TARC, IP-10, MIP-1β, IL-8, MCP-1, MDC, and MCP-4 were measured by the V-Plex Plus Chemokine Panel 1 kit (MSD). All assays were performed according to the manufacturer’s instructions. Plasma samples were diluted twofold for the proinflammatory and cytokine panels and fourfold for the chemokine panel. A total of 56 CLL samples and 20 healthy controls (HCs) were analyzed. Data were acquired on the V-plex Sector Imager 2400 plate reader. Analyte concentrations were extrapolated from a standard curve calculated using a four-parameter logistic fit using MSD Workbench V.3.0 software. The concentration of soluble CD160 in the plasma was detected by using ELISA kit (Sino Biological). The optical density was measured using a microplate reader (Synergy H1 Biotek) set to 450 nm and analyzed by Gen5 V.2.07 software.
EV isolation and uptake assay
Plasma samples were thawed and centrifuged at 16,000×g for 15 min at 4°C to remove debris. EVs were isolated using the exoEasy Maxi Kit (Qiagen) according to the manufacturer’s instruction. The protein concentration of isolated EV fraction was determined using the Pierce BCA protein assay kit (Thermo Fisher). Isolated EVs were quantified using the EXOCET Exosome Quantitation Assay kit (SBI, System Biosciences). For the uptake assay, EVs from patients with CLL were labeled with the CFSE dye (40 μM for 2 hours at 37°C as described).37 Unbound dye was removed using Exo-spin columns (Invitrogen). Next, PBMCs were co-cultured with the labeled EVs overnight followed by flow cytometry staining using the Image Stream analyzer.34
Western blotting
Isolated EVs (40 µg) solubilized in SDS-PAGE buffer were separated by electrophoresis on polyacrylamide gels (7% or 17%, depending on the molecular weight of the target protein). Blocking was done by 5% non-fat dry milk before probing overnight with the primary antibodies (CD9, CD63, CD81, CD160) (Thermo Fisher Scientific). Then blots were labeled with horseradish peroxidase (HRP)-conjugated secondary antibodies and transferred to polyvinylidene fluoride membranes for chemiluminescent protein detection (Thermo Fisher Scientific). Membranes were reprobed with loading controls (actin, GAPDH). Protein bands were quantified using Image Lab Software V.6.0.1 (Bio-Rad).
Gene expression analysis
The RNA was isolated from CD8+ T cells from HCs and patients with CLL using the Direct-zol RNA MicroPrep kit (Zymo Research). Normally, 100 ng of RNA was used for cDNA synthesis using the Quantitect Reverse Transcription Kit (Qiagen). RT-PCR was carried out using Quantitect and RT2 RT-PCR Kits (Qiagen) to measure the expression of CD160, NFKB, TCF-7, Perforin, granzyme-B, EOMES, and T-bet. Each sample was run in duplicate using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Beta-2-microglobulin was used as a reference gene and the relative fold change of the targeted genes was calculated by the ΔΔCT method.
Statistical analysis
Statistical analysis was performed using GraphPad Prism V.6 (GraphPad Software). For comparison, non-parametric Mann-Whitney U test or Wilcoxon signed-rank test was used for datasets that were non-paired or paired, respectively. Data were presented as means and SD of means (SD). Flow cytometry analysis and presentation of distributions were performed using SPICE V.638 downloaded from http://exon.niaid.nih.gov/spice. A comparison of distributions was performed using a Student’s t-test and a partial permutation test.
When more than two groups were compared, one-way analysis of variance (ANOVA) followed by Turkey’s test was used to compare the results. The expression level of targeted genes between groups was analyzed by one-way ANOVA followed by Tukey’s test.
Results
CLL influences the expression of co-inhibitory receptors on T cells
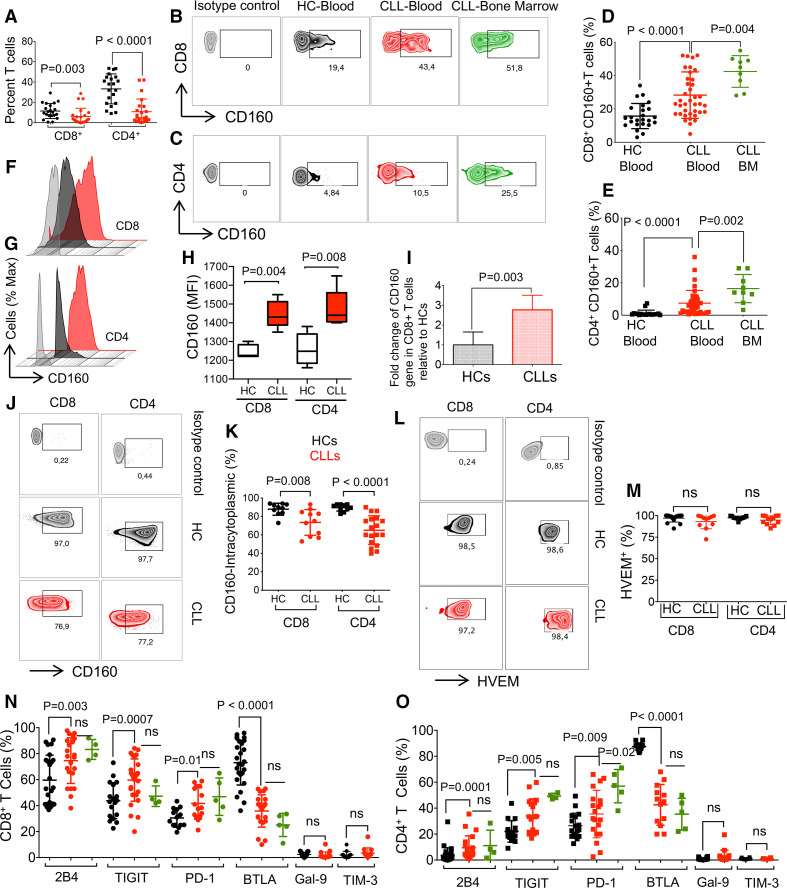
We found a significant decrease in percentages of both CD4+ and CD8+ T cells in PBMCs of patients with CLL compared with HCs (figure 1A). This difference was more prominent in CD4+ T cells as the CD4/CD8 ratio was reduced from 2.7 in HCs to 1.15 in CLLs (figure 1A). Unfortunately, we were unable to obtain the whole blood cell count from HCs; therefore, there is a possibility that a large population of malignant B cells in patients with CLL influences the T cell count in the peripheral blood. We observed significantly higher percentages of CD160 expressing cells among CD8+ and CD4+ T cells in patients with CLL compared with HCs (figure 1B, C and online supplemental figure 1e). Notably, the percentages of CD8+ and CD4+ T cells expressing CD160 were higher in the BM compared with the peripheral blood (figure 1B-E). However, we did not observe any difference in the percentages of CD160 expressing T cells in treatment naïve versus treated patients (online supplemental figure 1f). We also evaluated the influence of age on CD160 expression in CD8+ T cells in patients with CLL and found a higher proportion of CD160 expressing CD8+ T cells in 65–84 compared with the younger age group (online supplemental figure 1g). Since the mutation of the immunoglobulin heavy chain variable region gene (IgHV) status is considered as a prognostic factor in CLL,39 we compared the percentages of CD160 expressing CD8+ T cells in IgHV mutated versus unmutated patients. However, we did not observe any significant difference between the groups possibly due to a low sample size (online supplemental figure 1h). Once the fluorescence in situ hybridization (FISH) analysis was performed, we found significantly higher percentages of CD160 expressing cells among CD8+ T cells of Trisomy-12 subjects compared with those with Del13q (online supplemental figure 1i). Also, we measured the intensity of CD160 expression on T cells of patients with CLL and HCs, and found significantly higher expression levels of CD160 on both CD8+ and CD4+ T cells of patients with CLL compared with HCs (figure 1F, H). Then, we analyzed the expression of CD160 at the gene level and found a higher expression of CD160 mRNA in isolated CD8+ T cells of CLLs compared with HCs (figure 1I). Although percentages of both CD8+ and CD4+ T cells expressing intracytoplasmic CD160 were significantly higher than those expressing the surface CD160 expression in both patients with CLL and HCs (figure 1D, E and J, K), T cells expressing the intracytoplasmic CD160 had lower frequencies in patients with CLL compared with HCs (figure 1J, K). Also, we found that HVEM, the CD160 ligand, was highly expressed on the surface of T cells without any difference between CLLs and HCs (figure 1L, M). The same pattern was observed for percentages of B cells expressing HVEM (online supplemental figure 1j, k). Furthermore, we found significant increase in 2B4, TIGIT and PD-1 but the reduction of BTLA expressing CD8+ T cells in patients with CLL compared with HCs, however, the frequency of TIM-3 and Galectin-9 (Gal-9) expressing CD8+ T cells remained unchanged (figure 1N and online supplemental figure 1l). Of note, we did not find any difference in the frequency of CD8+ T cells expressing these co-inhibitory receptors in the peripheral blood versus the BM (figure 1N). Similar observations were made for CD4+ T cells expressing 2B4, TIGIT and PD-1 (figure 1O and online supplemental figure 1m). It is worth mentioning that the frequency of PD-1 expressing CD4+ T cells was significantly higher in the BM compared with the peripheral blood (figure 1O) but the frequencies of TIM-3 and Gal-9 expressing CD4+ T cells were low without any significant difference between patients with CLL and HCs (figure 1O). Overall, percentages of T cells expressing surface CD160 were significantly increased in patients with CLL compared with HCs.
Figure 1.
The expression of co-inhibitory receptors on T cells. (A) Cumulative data showing percentages of CD8+ and CD4+ T cells in peripheral blood mononuclear cells (PBMCs) of patients with chronic lymphocytic leukemia (CLL) versus healthy controls (HCs). (B) Representative flow cytometry plots of CD160+CD8+ T cells, and (C) CD160+CD4+ T cells in the blood and bone marrow (BM) of patients with CLL versus HC blood. (D) Cumulative data of percentages of CD160+ among CD8+ T cells, and (E) CD4+ T cells in CLL (blood and BM) versus HC blood. (F) Histogram plots of CD160 expression on CD8+, and (G) CD4+ T cells measured by the mean fluorescence intensity (MFI), and (H) cumulative data in CD8+/CD4+T cells of patients with CLL versus HCs. (I) Expression of CD160 mRNA level in CD8+ T cells of patients with CLL (PBMCs) relative to HCs. (J) Representative flow cytometry plots, and (K) cumulative data of percentages of CD8+ and CD4+ T cells expressing intracytoplasmic (ICS) CD160 in PBMCs of patients with CLL versus HCs. (L) Representative flow cytometry plots, and (M) cumulative data of percentages of HVEM+CD8+ and HVEM+CD4+ T cells in patients with CLL versus HCs. (N) Cumulative data of percentages of CD8+, and (O) CD4+ T cells expressing surface expression of 2B4, TIGIT, PD-1, BTLA, Galectin-9 (GAL-9) and TIM-3 on CD8+ T cells in patients with CLL (blood and BM) versus HC blood. Each dot represents data from a single patient with CLL or HC. Figure 1H and 1L from six human subjects/group. BTLA, B- and T-lymphocyte attenuator; HVEM, herpesvirus entry mediator; TIGIT, T cell immunoreceptor with Ig and ITIM domains; TIM-3, T-cell immunoglobin ad mucin-domain containing-3.
CD160+ T cells exhibit impaired effector functions but maintain their proliferative capacity in patients with CLL
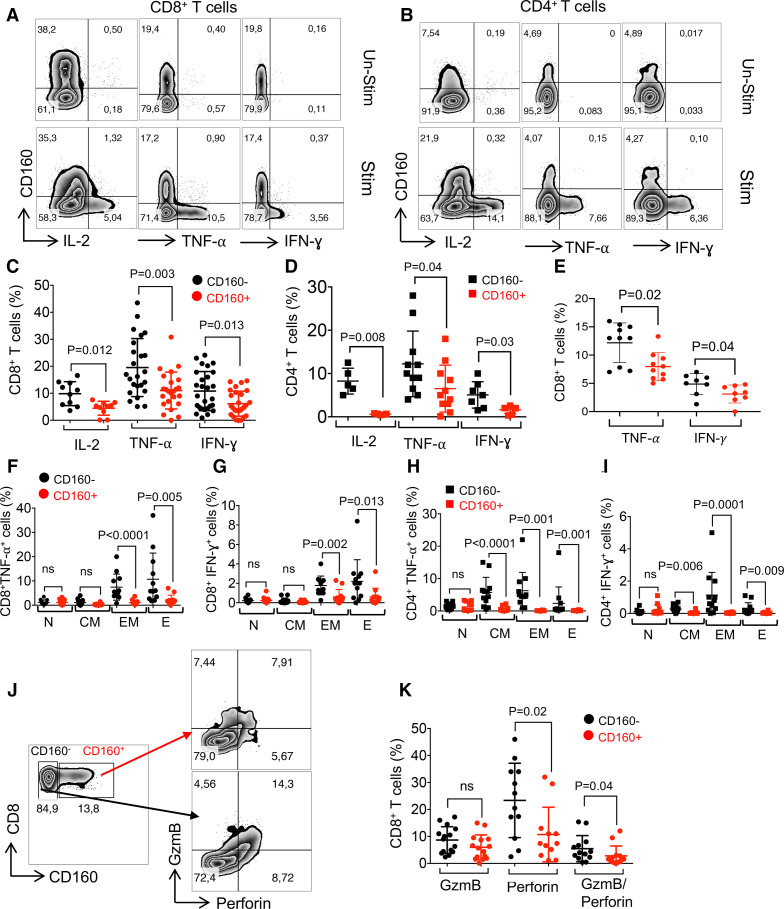
We found that CD160+ T cells had impaired production of IL-2, TNF-α and IFN-γ among CD8+ and CD4+ T cells in patients with CLL (figure 2A-D), which is in agreement with another report.12 We noted that CD8+CD160+ T cells compared with their CD160- counterparts in HCs exhibited the same phenotype as CLLs ((figure 2E and online supplemental figure 2a); however, the frequency of CD4+CD160+ T cells in HCs was very low to quantify (online supplemental figure 2b). Because of the reported differences in the cytokine production capacity of different T cell subsets,40 we measured TNF-α and IFN-γ production in CD160+ and CD160- in relation to their differentiation status (eg, naïve, central memory, effector memory and effector T cells). We found that CD160+ cells among either CD8+ or CD4+ T cells regardless of their differentiation status expressed significantly lower levels of cytokines compared with their CD16- counterparts (figure 2F-I and online supplemental figure 2c). Since the coordinated action of perforin41 and granzyme-B (GzmB)42 is essential for the optimal CD8+ T cell-mediated cytotoxicity, we analyzed perforin and GzmB co-expression in CD8+ T cells ex vivo. We found that significantly lower percentages of CD160+ compared with CD160- CD8+ T cells expressed perforin but this was not the case for GzmB. As such, significantly lower percentages of perforin/GzmB co-expressing cells were observed in CD160+ compared with CD160- CD8 +T cells in patients with CLL (figure 2J, K). In contrast, we detected higher mRNA for GzmB and perforin in CD8+ T cells of patients with CLL compared with HCs (online supplemental figure d, e). Higher mRNA for GzmB and perforin but lower protein expression may suggest that the genes are not efficiently translated into protein in CD8+CD160+ T cells in patients with CLL. Moreover, the ability of CD8+ T cells to degranulate in response to stimulation with anti-CD3/CD28 antibodies was assessed by CD107a expression (lysosomal-associated membrane protein I (LAMP-I)). We found that although CD160+ and CD160- CD8+ T cells expressed similar CD107a levels in the absence of stimulation, CD160+ CD8+ T cells exhibited impaired degranulation capacity following stimulation (figure 3A-D). Another feature of T cell exhaustion is the gradual loss of proliferation capacity.43 Thus, we assessed the proliferative capacity of isolated CD160+ versus CD160- T cells in response to stimulation with anti-CD3/CD28-coated microbeads for 72 hours in vitro. However, no significant difference was observed in the percentages of proliferated CD160+ versus CD160- in both CD8+ and CD4+ T cells (figure 3E-H). Because of the difference in proliferative capacities of T cell subsets,34 40 we measured the proliferative capacity of isolated effector T cells; however, both CD8+CD160+ and CD8+CD160- T cells exhibited similar proliferative capacity (online supplemental figure 2f-i). Similar observations were made for the Ki67 expression in CD160+ versus CD160- T cells (online supplemental figure 2j, k). Thus, our findings show that CD160+ T cells exhibit a dysfunctional phenotype in patients with CLL.
Figure 2.
Impaired cytokine production and cytolytic activity of CD160+ T cells in patients with chronic lymphocytic leukemia (CLL). (A) Representative flow cytometry plots of interleukin-2 (IL-2), tumor necrosis factor-α (TNF-α) and interferon-γ (IFN-γ) expression in CD8+CD160- and CD8+CD160+, and (B) CD4+CD160- versus CD4+CD160+ T cells. (C) Cumulative data of percentages of IL-2, TNF-α and IFN-γ expressing cells among CD8+CD160- and CD8+CD160+, and (D) CD4+CD160- and CD4+CD160+ T cells after 5 hours of in vitro stimulation with anti-CD3/CD28 antibodies. (E) Cumulative data of percentages of TNF-α and IFN-γ expressing cells among CD8+CD160- and CD8+CD160+ T cells in healthy controls (HCs). (F) Cumulative data of percentages of TNF-α, and (G) IFN-γ expressing cells in CD8+CD160- versus CD8+CD160+ T cells of patients with CLL defined as (N: naive; CM: central memory; EM: effector memory; E: effector). (H) Cumulative data of percentages of TNF-α, and (I) IFN-γ expressing cells in CD4+CD160- versus CD4+CD160+ T cells in different T cell subsets as shown. (J) Representative flow cytometry plots, and (K) cumulative data of percentages of granzyme-B (GzmB) and perforin expressing cells in CD8+CD160- versus CD8+CD160+ T cells in patients with CLL. For TNF-α and IFN-γ analysis, peripheral blood mononuclear cells (PBMCs) were stimulated with the anti-CD3 (3 μg/mL) and anti-CD28 (1 μg/mL) in the presence of protein transporter inhibitor (1:1000) for 5 hours. Each dot represents data from a single patient with CLL.
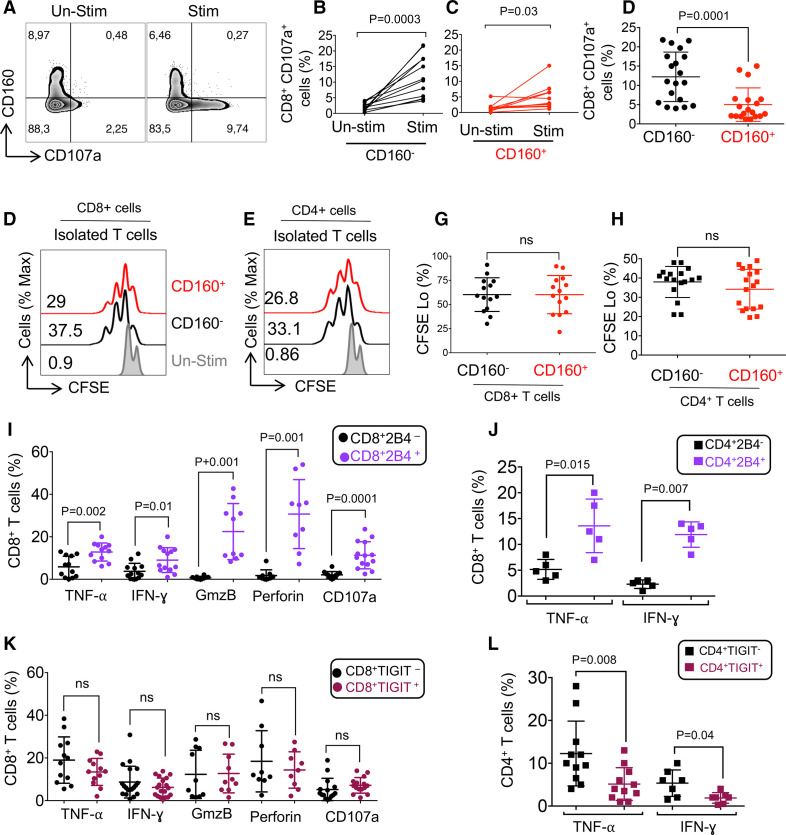
Figure 3.
The impact of CD160, 2B4 and TIGIT expression on T cell effector functions in patients with chronic lymphocytic leukemia (CLL). (A) Representative flow cytometry plots, and (B–D) cumulative data of percentages of CD107a expressing cells among CD8+CD160- and CD160+CD8+ T cells in unstimulated versus stimulated peripheral blood mononuclear cells (PBMCs) with anti-CD3/CD28 antibodies for 5 hours. (E) Representative flow cytometry plot of percentages of CFSElo (proliferated) CD8+CD160- and CD8+CD160+, and (F) CD4+CD160- and CD4+CD160+ T cells. (G) Cumulative data of percentages of CFSElo CD8+CD160- versus CD8+CD160+, and (H) CD4+CD160- versus CD4+CD160- T cells after stimulation of PBMCs from CLL patients with anti-CD3/CD28 antibodies for 3 days. (I) Cumulative data of percentages of tumor necrosis factor-α (TNF-α), interferon-γ (IFN-γ), granzyme-B (GzmB), perforin and CD107a expressing cells among 2B4-/2B4+CD8+ T cells. (J) Cumulative data showing percentages of TNF-α and IFN-γ expressing cells among 2B4-/2B4+ CD4+ T cells of patients with CLL. (K) Cumulative data showing percentages of TNF-α, IFN-γ, GzmB, perforin and CD107a expressing cells among TIGIT-/TIGIT+ CD8+ T cells. (L) Cumulative data showing percentages of TNF-α and IFN-γ expressing cells among TIGIT-/TIGIT+ CD4+ T cells of patients with CLL. For TNF-α and IFN-γ and CD107a analysis, PBMCs were stimulated with the anti-CD3 (3 μg/mL) and the anti-CD28 (1 μg/mL) antibody in the presence of protein transporter inhibitor (1:1000) for 5 hours. Each dot represents data from a single patient with CLL. TIGIT, T cell immunoreceptor with Ig and ITIM domains.
Differential effects of 2B4 and TIGIT expression on cytokine production and cytolytic activity of T cells in patients with CLL
We also evaluated the effect of other highly expressed co-inhibitory receptors such as 2B4 and TIGIT on T cell effector functions in patients with CLL. In contrast to CD160 expression, 2B4 expression was associated with a greater cytokine production ability of both CD8+ and CD4+ T cells in patients with CLL (figure 3I, J, and online supplemental figure 2i), which is consistent with a previous report.34 While the expression of TIGIT had no significant impact on TNF-α and IFN-γ production in CD8+ T cells, it dampened TNF-α and IFN-γ production in CD4+ T cells (figure 3K, L, and online supplemental figure 3m). Of note, the co-expression of GmzB/perforin was negligible in 2B4- CD8+ T cells (online supplemental figure 2n). However, the expression of TIGIT did not affect the expression of GzmB/perforin in CD8+ T cells in patients with CLL (figure 3K and online supplemental figure 2o). Also, we found a significantly higher frequency of cytolytic molecules and CD107a expressing cells among 2B4+ CD8+ T cells compared with their negative counterparts (figure 3I and online supplemental figure 2p). Finally, we found a higher frequency of CD107a expressing cells among 2B4+CD8+ versus TIGIT+CD8+ T cells after stimulation with anti-CD3/CD28 antibodies for 5 hours (online supplemental figure 2p). Overall, the expression of 2B4 was associated with enhanced T cell effector functions in patients with CLL but this was not the case for TIGIT.
Co-expression of CD160 with other co-inhibitory receptors results in a more impaired CD8+ T cell phenotype in patients with CLL
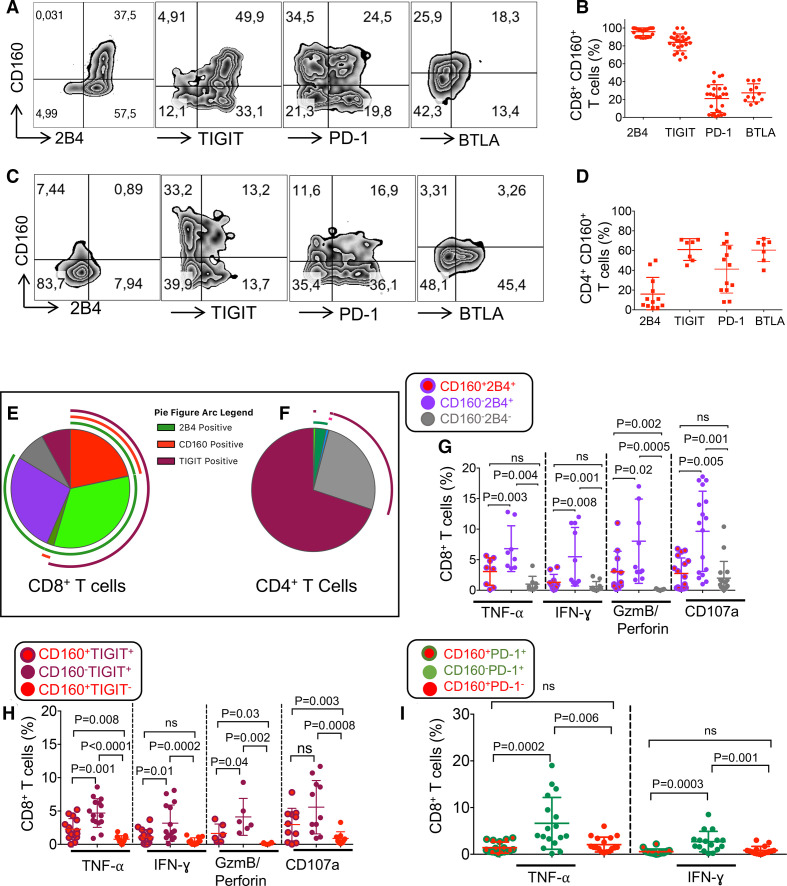
Since the co-expression of multiple co-inhibitory receptors dictates a more impaired T cell phenotype,10 we analyzed the co-expression of CD160 with other co-inhibitory receptors. We found that CD160 was highly co-expressed with 2B4 and TIGIT but to a lesser extent with PD-1, and BTLA in CD8+ T cells (figure 4A, B). Conversely, CD160 was moderately co-expressed with TIGIT, PD-1 and BTLA but even lower with 2B4 on CD4+ T cells (figure 4C, D). Besides, we analyzed the simultaneous co-expression of 2B4, CD160, and TIGIT using the SPICE software. We found that most of CD8+ T cells expressed one or two of these co-inhibitory receptors but 205% of CD8+ T cells co-expressed CD160, 2B4, and TIGIT (red-colored sector on the pie chart and the bar graph) (figure 4E). In contrast, the majority of CD4+ T cells expressed only TIGIT (gray-colored sector on the pie chart and bar graph, or none of these co-inhibitory receptors (purple-colored sector colored on the pie chart and bar graph) (figure 4F). We found about 50% of CD8+ and 10% of CD4+ T cells co-expressed 2B4 and TIGIT (online supplemental figure 3a, b).
Figure 4.
Co-expression of CD160 with other co-inhibitory receptors on T cells in patients with lymphocytic leukemia (CLL). (A) Representative flow cytometry plots, and (B) cumulative data showing percentages of CD160 co-expression with 2B4, TIGIT, PD-1, and BTLA in CD8+ T cells. (C) Representative flow cytometry plots, and (D) cumulative data showing percentages of CD160 co-expression with 2B4, TIGIT, PD-1, and BTLA in CD4+ T cells. (E) The pie chart showing percentages of CD8+ T cells co-expressing CD160, 2B4, and TIGIT, simultaneously. (F) The pie chart showing percentages of CD4+ T cells co-expressing CD160, 2B4, and TIGIT, simultaneously. (G) Cumulative data showing percentages of tumor necrosis factor-α (TNF-α), interferon-γ (IFN-γ), granzyme-B (GzmB), perforin and CD107a expressing cells among CD160+2B4+/CD160-2B4+/CD160-2B4- CD8+ T cells. (H) Cumulative data showing percentages of TNF-α, IFN-γ, GzmB, perforin and CD107a expressing cells among CD160+TIGIT+/CD160-TIGIT+/CD160+TIGIT- CD8+ T cells. (I) Cumulative data showing percentages of TNF-α, IFN-γ expressing cells among CD160+PD-1+/CD160-PD-1+/CD160+PD-1- CD8+ T cells. Each dot represents data from a single patient with CLL. BTLA, B- and T-lymphocyte attenuator; PD-1, programmed cell death protein-1; TIGIT, T cell immunoreceptor with Ig and ITIM domains.
We also analyzed the effect of CD160 co-expression with 2B4, TIGIT, and PD-1 on CD8+ T cell effector functions. Co-expression of CD160 with 2B4 and TIGIT corresponded with impaired cytokine production (TNF-α and IFN-γ), lower levels of cytolytic molecules (GzmB and perforin) expression, and reduced degranulation (CD107a) capacity (figure 4G, H. The reduced effector functions in CD160-/2B4- and CD160-/TIGIT- CD8+ T cells could be explained by their exclusion from the effector T cell pool. Although the frequency of CD160+PD-1+CD8+ T cell subset was low, these cells exhibited impaired cytokine production (figure 4I), which is consistent with a report.44 These observations indicate that co-expression of CD160 with 2B4, TIGIT, and PD-1 dictates an impaired CD8+ T cell phenotype in patients with CLL (figure 4G-I).
CD160 expression is associated with different stages of T cell differentiation
To determine the differentiation status of CD160+ T cells, we stained them with CD45RA and CCR7. We observed that not only the majority of CD8+ T cells but also CD160+CD8+ T cells were effector T cells (TEFF) in patients with CLL (online supplemental figure 3c-f). This was not the case for CD4+ T cells, while total CD4+ T cells were enriched with TEFF cells (online supplemental figure 3g, h), CD160+CD4+ T cells were heterogeneous (online supplemental figure 3i, j). When we compared CD8+ T cell subsets in patients with CLL versus HCs, we found significantly higher percentages of TEFF but lower naïve, CM, and EM in patients with CLL (online supplemental figure 3d). The same pattern was the case for CD4+ T cells except for percentages of naïve T cells remained unchanged in HCs versus patients with CLL (online supplemental figure 3h). Also, we observed significantly higher replicative senescence in CD8+CD160+ T cells compared with their negative counterparts as evidenced by a lower CD28 but higher CD57 and PD-1 expression43 (online supplemental figure 3k-n).
Next, we analyzed the expression of CD160 in different generations of antigen-experienced CD8+ T cells as follows: naïve (CD27+CD28+CD45RA+CCR7+CD57-), Ag experienced level-1 (CD27+ CD28+CD45RA-CCR7+CD57-), Ag experienced level-2 (CD27+CD28+CD45RA-CCR7-CD57+/-), Ag experienced level-3 (CD27+CD28-CD45RA+/-CCR7-CD57+/-), and Ag experienced level-4 (CD27-CD28-CD45RA-CCR7-CD57+).45 46 We found an increase in the expression of CD160 on CD8+ T cells as the level of antigen experience progressed (online supplemental figure 3o, p). This was further confirmed by a higher expression of CD137, as a marker of antigen-experienced T cells,47 on CD160+CD8+ T cells (online supplemental figure 3q, r). Thus, CD160+CD8+ T cells have a characteristic of an antigen-experienced effector phenotype with very few senescent cells.
Differential expression of exhaustion transcriptional factors and IL-2 signaling in CD160+ T cells in patients with CLL
T-bet and EOMES are T-box family transcription factors, whose intricate equilibrium delineates and maintains different subsets of exhausted CD8+ T cells.48 Tcf1 is another transcription factor related to preserving the effector function of exhausted T cells.49 We found no change in the expression of EOMES at the gene level but T-bet (TBX21) was significantly upregulated in CD8+ T cells from patients with CLL compared with HCs. However, the expression of the Tcf1 gene was downregulated in CD8+ T cells of patients with CLL versus HCs (online supplemental figure 4a-c). At the protein level, we found no change in EOMES and T-bet expressing CD8+ T cells but significantly higher percentages of CD160+CD4+ T cells expressed EOMES compared with CD4+CD160- T cells (online supplemental figure d-f).
Also, we measured the expression of IL-2 receptors, CD25 (IL-2Rα), and CD122 (IL-2Rβ) on CD160+ T cells. We found that CD122 was significantly higher in CD4+CD160+ but not in CD8+CD160+ T cells (online supplemental figure 4g, h). On the other hand, CD25 was highly expressed on both CD4+ and CD8+ T cells expressing CD160 (online supplemental figure 3i, j). Considering the higher IL-2 receptor expression in CD160+ T cells, we measured the signal transduction in response to IL-2 stimulation in vitro. We treated PBMCs from CLL patients with or without IL-2 and phosphorylation of STAT-5 was measured by Phospho-flow. We found that CD160+ T cells had higher baseline phosphorylation of STAT-5 compared with CD160- population and also after stimulation with IL-2 (online supplemental figure 4k-n).
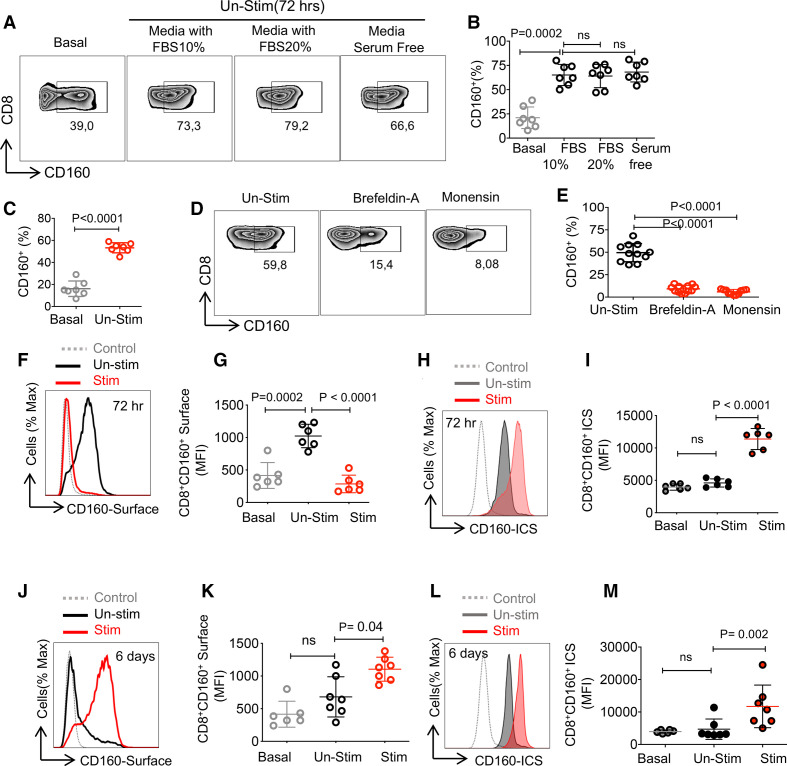
Prolonged T cell stimulation upregulates surface CD160 expression on T cells
To understand the mechanism underlying CD160 upregulation on T cell surface in patients with CLL, we analyzed the effect of T cell stimulation on CD160 surface and intracytoplasmic expression. Interestingly, we observed that culture of PBMCs (72 hours) from patients with CLL in the absence of any stimulation resulted in the expansion of CD160 expressing CD8+ and CD4+ T cells, respectively (figure 5A, B, and online supplemental figure 5a, b). To rule out the external source of CD160 such as NK cells,20 we cultured isolated CD8+ T cells and again the same pattern was observed (figure 5C). We found that this was unrelated to the fetal bovine serum (FBS) in the culture media (figure 5A, B, and online supplemental figure 5a, b). These observations suggested that the intracellular CD160 may translocate to the surface of T cells when cultured in vitro. To confirm this, Brefeldin A (endoplasmic reticulum (ER) to Golgi inhibitor) and Monensin (Golgi to plasma membrane pathway inhibitor) were added to the culture media for 16 hours, which resulted in a significant reduction of cells expressing CD160 compared with non-treated T cells (figure 5D, E, and online supplemental figure 5c, d). These observations suggested that CD160 gets transported to the surface via the ER–Golgi–plasma membrane pathway.
Figure 5.
Prolonged T cell stimulation upregulates CD160 expression. (A) Representative flow cytometry plots, and (B) cumulative data showing percentages of CD8+ T cells expressing CD160 at the baseline, in the presence of 10%, 20% (fetal bovine serum (FBS)) or in the absence of FBS after 72 hours in vitro culture of total peripheral blood mononuclear cells (PBMCs) from patients with chronic lymphocytic leukemia (CLL). (C) Cumulative data showing percentages of isolated CD8+ T cells expressing CD160 at the baseline and after 72 hours in vitro culture. (D) Representative flow cytometry plots, and (E) cumulative data of percentages of CD8+ T cells expressing CD160 in the absence or presence of Brefeldin A and Monensin after 6-hour culture of peripheral blood mononuclear cells (PBMCs). (F) Representative histogram, and (G) cumulative data of the mean fluorescence intensity (MFI) of surface CD160 expression on CD8+ T cells stimulated with anti-CD3/CD28 antibodies (stim) versus unstimulated (Un-stim) for 72 hours. (H) Representative histogram, and (I) cumulative data of intracytoplasmic CD160 expression in CD8+ T cells unstimulated versus stimulated with anti-CD3/CD28 for 72 hours. (J) Representative histogram, and (K) cumulative data of MFI for the surface expression of CD160 in CD8+ T cells, unstimulated versus stimulated with anti-CD3/CD28 after 6 days of in vitro culture. (L) Representative histogram, and (M) cumulative data of MFI for the intracytoplasmic expression of CD160 in CD8+ T cells, unstimulated versus stimulated with anti-CD3/CD28 after 6 days of in vitro culture. Each dot represents data from a single study subject. ICS, intracytoplasmic staining.
Moreover, we observed that on stimulation of PBMCs with anti-CD3/CD28 antibody (for 72 hours), the surface CD160 expression was significantly decreased, while intracytoplasmic expression was increased in both CD8+ (figure 5F-I) and CD4+ T cells (online supplemental figure 5e-h). However, the prolonged stimulation with anti-CD3/CD28 antibody (for 6 days) resulted in a significant upregulation of surface CD160 on CD8+ (figure 5J, K) and CD4+ T cells (online supplemental figure 5i, j). Similarly, the intracytoplasmic expression of CD160 was increased in stimulated CD8+ cells with anti-CD3/CD28 antibodies (figure 5L, M) and CD4+ T cells (online supplemental figure k, l).
To test the possibility of CD160 shedding from T cells, isolated CD8+ T cells were stimulated with anti-CD3/CD28 antibodies, IL-2 or IL-15 for 72 hours. Interestingly, CD160 protein was undetectable in cell culture supernatants as measured by western blotting (using an anti-CD160 antibody (Clone: BY55) (online supplemental figure 5m). Notably, we detected significantly lower levels of CD160 in the plasma of patients with CLL compared with HCs (online supplemental figure 5n). These observations suggest that constant antigenic stimulation may result in the upregulation of CD160 in T cells of patients with CLL.
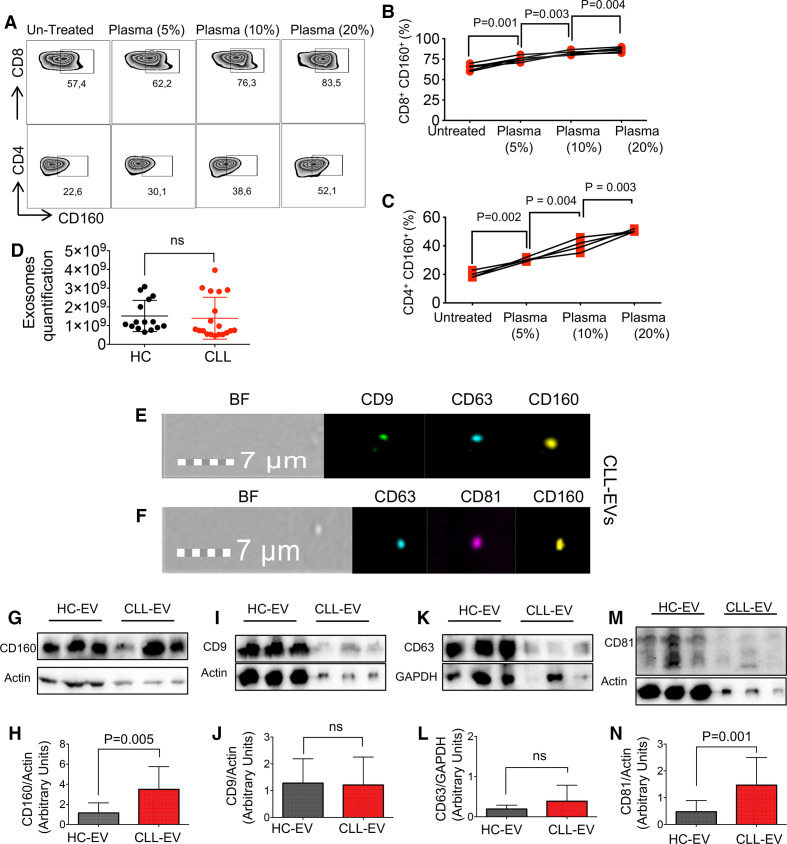
Plasma-derived EVs contain CD160 in patients with CLL
To determine the role of soluble mediators in CD160 expression on T cells, we added 5%, 10%, and 20% of plasma from CLL or HCs to unstimulated PBMCs from patients with CLL and HCs for 72 hours. Interestingly, we observed the upregulation of CD160 on both CD8+ and CD4+ T cells in a dose-dependent manner (figure 6A-C). To better understand the mechanism, we isolated EVs from the plasma of patients with CLL and HCs. In contrast to a previous report,50 using the Exocet ELISA method we did not observe any difference in the quantity of EVs between patients with CLL and HCs (figure 6D). We found that EVs, as characterized by CD9, CD81, and CD63 markers, contained CD160 (figure 6E, F). This was further examined by western blotting (figure 6G-N). Due to the differential expression of actin in HC and CLL samples in western blot, we normalized the amount of CD9, CD81, CD63, and CD160 based on the actin expression and identified that CD160 expression was higher in CLL EVs compared with HCs (figure 6G, H). While CD9 and CD63 expressions were remained unchanged (figure 6I-L), CD81 expression in EVs from patients with CLL was higher than HCs (figure 6M, N). We further performed the EV uptake assay by labeling isolated EVs from CCL patients with the CFSE dye and then adding them into PBMCs from HCs overnight. We then investigated the uptake of EVs in T cells by using the ImageStream analysis. As shown in online supplemental figure 6a, we observed the uptake of EVs by T cells. These data suggest that plasma-derived EVs of patients with CLL contain CD160 and could be taken up by T cells which may influence CD160 levels in T cells of patients with CLL.
Figure 6.
Plasma-derived extracellular vesicles (EVs) contain CD160. (A) Representative flow cytometry plots of CD160 expression in CD8+ and CD4+ T cells untreated versus treated with the plasma from patients with chronic lymphocytic leukemia (CCL) (using 5%, 10% and 20% plasma) after 72 hours of in vitro culture. (B) Cumulative data of percentages of CD160 expressing cells among CD8+, and (C) CD4+ T cells either untreated or treated with indicated plasma concentrations after 72 hours. (D) Quantification of EV numbers isolated from the plasma of CLLs versus healthy controls (HCs) by Exocet ELISA kit. (E) ImageStream plots of plasma-derived EVs showing the expression of CD9, CD63 and CD160, bright field (BF). (F) ImageStream plots of plasma-derived EVs showing expression of CD63, CD81 and CD160. (G) Representative western blot (WB) images of plasma-derived EVs from HCs and patients with CLL depicting CD160 presence. (H) Cumulative data showing normalized arbitrary units of CD160/actin in plasma-derived EVs in HCs versus patients with CLL. (I) Representative WB images of plasma-derived EVs depicting CD9 expression, and (J) cumulative data showing normalized arbitrary units of CD9/actin in plasma-derived EVs in HCs versus patients with CLL. (K) Representative WB images of plasma-derived EVs depicting CD63 expression, and (L) cumulative data showing normalized arbitrary units of CD63/GAPDH in plasma-derived EVs in HCs versus patients with CLL. (M) Representative WB images of plasma-derived EVs depicting CD81 expression, and (N) cumulative data showing normalized arbitrary units of CD160/actin in plasma-derived EVs in HCs versus patients with CLL. Actin was used as a loading control to normalize protein amounts of CD81, CD9, and CD160, and GAPDH was used as a loading control to normalize protein amount of CD63. Each dot/band represents data from a subject.
A proinflammatory cytokine profile in the plasma of patients with CLL
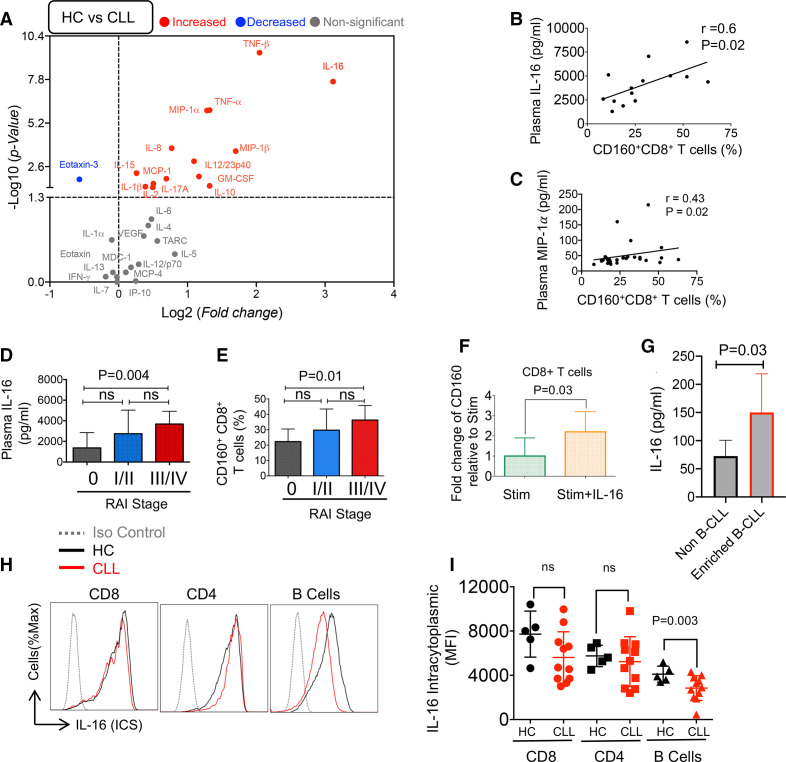
Several studies have shown the impact of cytokines and chemokines on the modulation of co-inhibitory receptors and T cell effector functions.51 For example, IL-10 has been reported to upregulate CD160 in CD8+ T cells.28
Plasma specimens from 37 patients with CLL and 20 HCs were subjected to multiplex assay. Z-scores were calculated for each cytokine and chemokine in each patient/HC and individual results are shown as a heatmap diagram for better comparison (online supplemental figure 6b, c). Plasma levels of TNF-β IL-16, TNF-α, MIP-1α, MIP-1β (CCL4), IL-12/23p40, GM-CSF, IL-10, IL-8, IL-15, MCP-1, IL-1β, IL-17A and IL-2 were significantly elevated in CLL compared with HC samples (figure 7A). The Eotaxin-3 was significantly lower in patients with CLL (figure 7A) but the other analyzed cytokines including IL-1α, IL-4, IL-5, IL-6, IL-7, IL12/23p70, IL-13, IFN-γ, VEGF, MCP-4 (CCL13), MDC-1, TARC (CCL17), IP-10, and Eotaxin were similar in both groups (figure 7A). Among these cytokines, IL-16, TNF-α, TNF-β, MIP-1α and MIP-1β (CCL4) showed the highest deviation between CLL and HCs (p<0.0001) followed by IL-8 (p=0.0002). We did not find any significant difference in levels of IFN-γ, TARC, IL-5, and IL-6, as it has been reported.52 MIP-1α and IL-16 concentration (pg/mL) showed a positive correlation with percentages of CD160+CD8+ T cells (figure 7B, C). In summary, our data indicate a more proinflammatory milieu in patients with CLL.
Figure 7.
The plasma cytokines/chemokines in the plasma of patients with chronic lymphocytic leukemia (CLL) versus healthy controls (HCs). (A) The volcano plot illustrating the magnitude and significance of differences in cytokine/chemokines plasma concentrations (measured by the Mesoplex assay) in patients with CLL versus HCs. (B) Scatter plot of the correlation between percentages of CD160+CD8+ T cells in peripheral blood mononuclear cells (PBMCs) with the interleukin-16 (IL-16), and (C) MIP-1α concentrations in the plasma of patients with CLL. (D) Cumulative data showing IL-16 concentrations in the plasma of patients with CLL in low (0), intermediate (I/II), and high (III/IV) Rai stages, 16, 24 and 7 patients/group, respectively. (E) Cumulative data of percentages of CD160+ expressing cells among CD8+ T cells in PBMCs of patients with CLL in low (0), intermediate (I/II), and high (III/IV) Rai stages, 9, 20 and 6 patients/group, respectively. (F) Fold regulation of CD160 gene in CD8+ T cells stimulated with the anti-CD3/CD28 antibodies in the absence or presence of rh-IL-16 (500 ng/mL) for 72 hours relative to stimulated as quantified by qPCR from seven human subjects/group. (G) Cumulative data of IL-16 production in cell culture supernatants of isolated B-CLL versus non-B-CLLs after 12 hours’ culture as detected by ELISA from four patients. (H) Representative flow cytometry plots, and (I) cumulative data of intracytoplasmic IL-16 expression in CD8+, CD4+, and B cells of patients with CLL versus HCs. MIP-1α, macrophage inflammatory protein-1 alpha.
B-CLLs are a major source of elevated IL-16 in the plasma of patients with CLL and correlated to a high RAI stage (III/IV)
Plasma cytokine levels were stratified based on the individual Rai stage, percentages of CD160+CD8+ T cells, and the lymphocyte count. Although we did not find any correlation between the measured cytokine levels and lymphocyte counts, we observed that IL-16 was higher in the high-risk Rai staging system (III/IV) compared with the low-risk group (0) without any difference between low/intermediate and intermediate/high-risk patients (figure 7D). Interestingly, CD160 expressing CD8+ T cells were higher in the high risk (Rai stage: III/IV) compared with the low-risk group (Rai stage: 0) (figure 7E). Because of the dramatically elevated levels of IL-16 in the plasma of patients with CLL, and a positive correlation with the CD160 expressing CD8+ T cells, we were curious about the possible role of IL-16 in CD160 upregulation. Therefore, we treated PBMCs with different concentrations of IL-16 (50, 100, 250, and 500 pg/mL) and stimulated them with anti-CD3/CD28 antibodies for 3 days. Then we measured the surface expression of CD160. We noted that the addition of recombinant IL-16 (500 pg/mL) increased the gene expression of CD160 in CD8+ T cells after 72 hours (figure 7F). Moreover, we measured the level of IL-16 in the culture supernatant of malignant B cells when cultured without any stimulation for 12 hours. We found that malignant B cells secreted IL-16 when compared with non-B-CLL cells (figure 7G). Also, we assessed the viability of non-B-CLL versus B-CLL after 12 hours’ culture, which indicated cell viability of >95% (online supplemental figure 6d). Finally, we observed a significantly lower intracytoplasmic IL-16 in B cells of patients with CLL (figure 7H, I), which suggests B-CLL may serve as a source of IL-16.
Discussion
In this study, we found significantly higher percentages of CD160, 2B4, TIGIT, and PD-1 expressing T cells in patients with CLL compared with HCs. However, a lower proportion of BTLA expressing T cells was observed, which can be explained by a decrease in naïve T cell population in patients with CLL.40
Although the frequency of CD160+CD8+ T cells was lower than 2B4+CD8+ and TIGIT+CD8+ T cells in patients with CLL, CD160 was associated with a prominent T cell impairment. Notably, we found that the intensity of CD160 expression was significantly higher in both T cell subsets in patients with CLL compared with HCs. The role of CD160 as a co-inhibitory molecule in the induction of T cell dysfunction has been reported in chronic viral infections such as HIV,27 hepatitis virus C (HCV),53 Epstein-Barr virus (EBV), cytomegalovirus (CMV),44 and in pancreatic cancer.54 Likewise, the inhibitory nature of CD160 in T cells has been reported in the context of autoimmunity.55 In contrast, some studies have shown a stimulatory role for CD160 in CD8+ T cells in mucosal immunity and skin allograft in a mouse model.28 29
In our cohort, we did not find any association between the IgHV mutation and the upregulation of CD160 on T cells; however, the Trisomy-12 subjects were enriched with CD160+CD8+ T cells. These findings provide a novel and unreported, to our knowledge, insight into the relationship between the Trisomy-12 and CD160 expression. Our further characterization of CD160+ T cells in CLL revealed that while CD160+CD8+ T cells were mainly TEFF, CD160+CD4+ T cells were phenotypically heterogeneous and scattered through different T cell subsets. We also observed a higher expression of T-bet in CD8+ T cells of patients with CLL, which supports terminal differentiation of TEFF cells.56 However, a lower expression of Tcf1 mRNA in CD8+ T cells in patients with CLL may stem from a systemic inflammatory signal that suppresses Tcf1 expression in primed CD8+ T cells or a lower number of naïve CD8+ T cells in patients with CLL. Although high EOMES expression is reported to be correlated with severe CD8+ T cell exhaustion, we did not find a high mRNA level for EOMES in CD8+ T cells of patients with CLL. This can be explained by the complex reciprocity between IRs, T-bet, and EOMES in exhausted T cells.57 In addition, we found that CD160+ T cells in patients with CLL had an impaired degranulation capacity as reported for mast cells.17 This might be related to an impaired immunologic synapse formation with defective vesicle trafficking,7 which can decrease the cytotoxic ability of CD160+CD8+ T cells in patients with CLL.
Similar to an exhausted phenotype, CD160+ T cells exhibited an impaired cytokine production ability but surprisingly maintained their proliferative capability. This is in contrast with a report showing that CD160 blockade was associated with the restoration of virus-specific CD8+ T cell proliferative capacity.27 This discrepancy might be explained by the different status of CD160+ T cells in a viral infection versus CLL. Interestingly, CD160 was always co-expressed with 2B4 and highly co-expressed with TIGIT on CD8+ T cells of patients with CLL. We found that 2B4+CD8+ T cells had a greater cytokine production ability, perforin/GzmB expression, and degranulation capacity compared with 2B4- CD8+ T cells in patients with CLL. Interestingly, 2B4-CD8+ T cells were almost devoid of perforin and GzmB expression. This demonstrates an important role for 2B4 in the cytolytic property of T cells in CLL. However, the co-expression of CD160 with 2B4 diminished their T cell effector functions. These findings indicate the dictated inhibitory role of CD160 on 2B4 expressing CD8+ T cells in CLLs. Moreover, CD160 was highly co-expressed with TIGIT on CD8+ T cells in CLLs. Despite previous reports on the inhibitory role of TIGIT on tumor-infiltrating CD8+ T cells in follicular lymphoma58 and multiple myeloma,59 this was not the case for CLL. Although TIGIT expression had no effects on CD8+ T cells, it was associated with impaired cytokine production ability of CD4+ T cells in patients with CLL. Despite previous reports on the important role of TIM-3 and Gal-9 interactions in T cell exhaustion in hematological and non-hematological cancers,60 61 we observed a very low expression level of these co-inhibitory receptor/ligand in T cells of patients with CLL.
Despite the recognition of some tumor-associated antigens in CLL by CD8+ T cells, identification of these cells is complex and might be inefficient due to the heterogeneity of tumor-associated antigens.62 To overcome this limitation, we used an alternative immunophenotyping approach that enabled us to confirm that CD160+CD8+ T cells were highly antigen-experienced. Adoptive T cell transfer studies have shown an enhanced antitumor efficacy in less antigen-experienced T cells.63 This observation may explain impaired effector functions of CD160+CD8+ T cells in patients with CLL as they appeared to be highly antigen-experienced.
Another potential mechanism for dysfunctional CD160+CD8+ T cells in CLLs may be related to their high IL-2 and IL-15-dependent signaling which can restrict their survival and effector functions as reported for TIM-3+ T cells.51 We noted spontaneous upregulation of CD160 on rested T cells ex vivo, as reported elsewhere.19 Our further studies revealed that CD160 trafficking occurs via the ER/Golgi to the plasma membrane. This is in agreement with the GPI anchored protein trafficking that has been reported for CD59.64 GPI-anchored proteins are synthesized in the ER and then selectively packaged into the coat protein complex II (COPII) vesicles and delivered via Golgi to the membrane in a clustered form partitioning in lipid rafts.65 However, on short-term TCR-dependent stimulation (anti-CD3/CD28) of T cells, we noted decreased surface but increased intracellular CD160, which is in contrast to one study26 but consistent with another study.19 Our further observations showed increased surface CD160 expression following prolonged T cell activation (6 days) which is in agreement with the concept of chronic antigenic stimulation and the upregulation of co-inhibitory receptors.66
Intriguingly, for the very first time, we detected a high CD160 content in plasma-derived EVs of patients with CLL. Moreover, we found that CD160+ EVs can be taken up by T cells, and therefore, there is a possibility to propose that CD160+ EVs serve as a potential source of CD160. However, it is unclear whether uptake of CD160+EVs influences T cell effector functions. The inhibitory signal of PDL-1+EVs with T cells expressing PD-1 has been reported.67 Therefore, how these CD160+EVs impact the interaction of T cells with CD160 ligands is unknown and merits further investigation.
Cytokines and chemokines are other factors that can negatively influence T cell effector functions.68 We observed a significant increase in proinflammatory cytokines (IL-1β, TNF-α) and Th1-type cytokines (IL-2, TNF-α, TNF-β, IL-12/23p40, GM-CSF) in patients with CLL. However, Th2-type cytokines, IL-12/p70, IL-1, and IFN-γ were remained unchanged inconsistent with other studies.52 The dominancy of a proinflammatory cytokine profile in our study could be explained by the characteristics of our cohort that include mainly patients that were treatment naïve rather than treated (64.9% vs 35%) and in the early stages (low/intermediate) of the disease (online supplemental figure 6e). In our study, patients with CLL had elevated plasma IL-10 which is consistent with a previous study.69 IL-10 prolongs B-CLL survival and reduces the generation of effector CD4+ and CD8+ T cells.70
We also observed the elevation of chemokines71 such as MIP-1α, MIP-1β, MCP-1, and IL-8 that can contribute to the survival of neoplastic B cells by maintaining various anti-apoptotic mechanisms.71 72 For instance, MIP-1α is secreted by B-CLL cells and induces the recruitment of macrophage-lineage cells promoting the initiation of the leukemia niche.73 74 Interestingly, we observed a positive relationship between percentages of CD160+CD8+ T cells and the plasma MIP-1α plasma concentrations. Although we were unable to investigate the source/role of these elevated chemokines, there is evidence that B-CLL cells constitutively express IL-875 and they exhibit immunosuppressive on CD8+ T cells in cancer.76 Finally, we found a dramatic rise in IL-16 levels in the plasma of patients with CLL compared with HCs, which is consistent with another study.77 IL-16 has been identified as a chemotactic factor for CD4+ T cells that binds to CD4 and CD9 receptors on T cells.78 Similarly, in multiple myeloma, a strong expression of IL-16 in the BM that prolongs the survival of malignant cells has been reported.79 Although we were unable to identify the main source of IL-16, our observations suggest that B-CLL cells could be considered as one of the potential sources of this cytokine in patients with CLL. Importantly, we found a significant correlation between IL-16 levels with the cancer stage. This suggests that the plasma IL-16 could be used as a potential prognostic marker in CLL. However, further investigation on larger cohorts is required for the validity of our finding. Moreover, we found a positive correlation between the plasma IL-16 concentrations and percentages of CD160+CD8+ T cells, which suggests an intricate mechanism that calls for further investigations. Although our study provided a novel insight into the role of co-inhibitory receptors, in particular, CD160 in CLL, further studies in larger cohorts are warranted.
We are aware of multiple study limitations such as the lack of IgHV mutation results and the FISH analysis data for all of our patients. Also, the differential proportion of T cell subpopulations (eg, native, memory) in patients with CLL versus HCs may influence our data regarding the expression of T-bet, EOMES, and Tcf1 expression in T cells. Obtaining the BM aspirate from HCs was not possible in our studies, therefore, comparing immunological changes in the BM of HCs versus CLLs may assist in understanding the role of CD160 in the BM. Unfortunately, we were unable to obtain the whole blood cell count from HCs, therefore, there is a possibility that a large population of malignant B cells in patients with CLL influences the T cell count reported in PBMCs of patients with CLL.
Conclusions
Our data highlight the important role of CD160 in T cell exhaustion in patients with CLL. In particular, the co-expression of CD160 with other co-inhibitory receptors dictates more impaired T cell effector function. Also, the abundance of IL-16 in the plasma of patients with CLL and its impact on the upregulation of CD160 provide a novel insight into the mechanism underlying CD160 overexpression in patients with CLL. The correlation of the plasma IL-16 levels with the Rai stage suggests that IL-16 could be used as a prognostic biomarker in patients with CLL. Therefore, further studies on larger cohorts are warranted to determine whether targeting IL-16 and/or CD160 would have clinical implications in hematological cancers. Taken together, our findings provide a novel insight into the inhibitory role of CD160 alone and a synergistic inhibitory effect when co-expressed with other co-inhibitory receptors on T cells in patients with CLL.
Acknowledgments
We thank all the volunteers who supported this study by donating their samples and dedicating their time, and the staff at the CLL clinic and medical laboratory at the Cross-Cancer Institute (CCI) for sample collection. We also would like to thank the flow core facility of the Faculty of Medicine and Dentistry, at the University of Alberta for supporting the study.
Footnotes
Contributors: NB designed and performed most of the study, analyzed the data, designed the figures and wrote the first draft of the manuscript. IO contributed to extracellular vesicle (EV)-related experiments. OO performed and analyzed western blotting. LX performed and analyzed Image Stream data. AFontainefo, NC-K, LML, MH and AFagarasanu as clinicians provided the blood and bone-marrow aspirates. JB, a hemato-oncologist, provided advice, ACP as a hemato-oncologist provided most of the blood and bone-marrow aspirates, advised and assisted with the clinical data analysis. SE conceptualized the study, assisted in the experimental design, secured fund and resources, supervised the study and wrote the manuscript.
Funding: This study was supported by the Canadian Institute for Health Research (CIHR) through a Foundation Grant and a New Investigator Award (both to SE). Nevertheless, the funding bodies had no role in the design of the study, data collection, analysis, and interpretation of data.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1.Seifert M, Sellmann L, Bloehdorn J, et al. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J Exp Med 2012;209:2183–98. 10.1084/jem.20120833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scarfò L, Ferreri AJM, Ghia P. Chronic lymphocytic leukaemia. Crit Rev Oncol Hematol 2016;104:169–82. 10.1016/j.critrevonc.2016.06.003 [DOI] [PubMed] [Google Scholar]
- 3.Arruga F, Gyau BB, Iannello A, et al. Immune response dysfunction in chronic lymphocytic leukemia: dissecting molecular mechanisms and microenvironmental conditions. Int J Mol Sci 2020;21. 10.3390/ijms21051825. [Epub ahead of print: 06 Mar 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christopoulos P, Pfeifer D, Bartholomé K, et al. Definition and characterization of the systemic T-cell dysregulation in untreated indolent B-cell lymphoma and very early CLL. Blood 2011;117:3836–46. 10.1182/blood-2010-07-299321 [DOI] [PubMed] [Google Scholar]
- 5.Porakishvili N, Roschupkina T, Kalber T, et al. Expansion of CD4+ T cells with a cytotoxic phenotype in patients with B-chronic lymphocytic leukaemia (B-CLL). Clin Exp Immunol 2001;126:29–36. 10.1046/j.1365-2249.2001.01639.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsay AG, Clear AJ, Fatah R, et al. Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: establishing a reversible immune evasion mechanism in human cancer. Blood 2012;120:1412–21. 10.1182/blood-2012-02-411678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramsay AG, Johnson AJ, Lee AM, et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest 2008;118:2427–37. 10.1172/JCI35017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okoye IS, Houghton M, Tyrrell L, et al. Coinhibitory Receptor Expression and Immune Checkpoint Blockade: Maintaining a Balance in CD8+ T Cell Responses to Chronic Viral Infections and Cancer. Front Immunol 2017;8:1215. 10.3389/fimmu.2017.01215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wherry EJ. T cell exhaustion. Nat Immunol 2011;12:492–9 https://www.nature.com/articles/ni.2035 10.1038/ni.2035 [DOI] [PubMed] [Google Scholar]
- 10.McLane LM, Abdel-Hakeem MS, Wherry EJ. Cd8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol 2019;37:457–95. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- 11.Palma M, Gentilcore G, Heimersson K, et al. T cells in chronic lymphocytic leukemia display dysregulated expression of immune checkpoints and activation markers. Haematologica 2017;102:562–72. 10.3324/haematol.2016.151100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riches JC, Davies JK, McClanahan F, et al. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood 2013;121:1612–21. 10.1182/blood-2012-09-457531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanna BS, Roessner PM, Yazdanparast H, et al. Control of chronic lymphocytic leukemia development by clonally-expanded CD8+ T-cells that undergo functional exhaustion in secondary lymphoid tissues. Leukemia 2019;33:625–37. 10.1038/s41375-018-0250-6 [DOI] [PubMed] [Google Scholar]
- 14.Gassner FJ, Zaborsky N, Catakovic K, et al. Chronic lymphocytic leukaemia induces an exhausted T cell phenotype in the TCL1 transgenic mouse model. Br J Haematol 2015;170:515–22. 10.1111/bjh.13467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maïza H, Leca G, Mansur IG, et al. A novel 80-kD cell surface structure identifies human circulating lymphocytes with natural killer activity. J Exp Med 1993;178:1121–6. 10.1084/jem.178.3.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsujimura K, Obata Y, Matsudaira Y, et al. Characterization of murine CD160+ CD8+ T lymphocytes. Immunol Lett 2006;106:48–56. 10.1016/j.imlet.2006.04.006 [DOI] [PubMed] [Google Scholar]
- 17.Ortonne N, Ram-Wolff C, Giustiniani J, et al. Human and mouse mast cells express and secrete the GPI-anchored isoform of CD160. J Invest Dermatol 2011;131:916–24. 10.1038/jid.2010.412 [DOI] [PubMed] [Google Scholar]
- 18.Farren TW, Giustiniani J, Liu F-T, et al. Differential and tumor-specific expression of CD160 in B-cell malignancies. Blood 2011;118:2174–83. 10.1182/blood-2011-02-334326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Far M, Pellerin C, Pilote L, et al. CD160 isoforms and regulation of CD4 and CD8 T-cell responses. J Transl Med 2014;12:217. 10.1186/s12967-014-0217-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giustiniani J, Marie-Cardine A, Bensussan A. A soluble form of the MHC class I-specific CD160 receptor is released from human activated NK lymphocytes and inhibits cell-mediated cytotoxicity. J Immunol 2007;178:1293–300. 10.4049/jimmunol.178.3.1293 [DOI] [PubMed] [Google Scholar]
- 21.Maeda M, Carpenito C, Russell RC, et al. Murine CD160, Ig-like receptor on NK cells and NKT cells, recognizes classical and nonclassical MHC class I and regulates NK cell activation. J Immunol 2005;175:4426–32. 10.4049/jimmunol.175.7.4426 [DOI] [PubMed] [Google Scholar]
- 22.Tu TC, Brown NK, Kim T-J, et al. CD160 is essential for NK-mediated IFN-γ production. J Exp Med 2015;212:415–29. 10.1084/jem.20131601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Barbosa JI, Schneider P, Weigert A, et al. HVEM, a cosignaling molecular switch, and its interactions with BTLA, CD160 and light. Cell Mol Immunol 2019;16:679–82. 10.1038/s41423-019-0241-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kojima R, Kajikawa M, Shiroishi M, et al. Molecular basis for herpesvirus entry mediator recognition by the human immune inhibitory receptor CD160 and its relationship to the cosignaling molecules BTLA and light. J Mol Biol 2011;413:762–72. 10.1016/j.jmb.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 25.Šedý JR, Bjordahl RL, Bekiaris V, et al. CD160 activation by herpesvirus entry mediator augments inflammatory cytokine production and cytolytic function by NK cells. J Immunol 2013;191:828–36. 10.4049/jimmunol.1300894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai G, Anumanthan A, Brown JA, et al. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat Immunol 2008;9:176–85. 10.1038/ni1554 [DOI] [PubMed] [Google Scholar]
- 27.Peretz Y, He Z, Shi Y, et al. CD160 and PD-1 co-expression on HIV-specific CD8 T cells defines a subset with advanced dysfunction. PLoS Pathog 2012;8:e1002840. 10.1371/journal.ppat.1002840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan CL, Peluso MJ, Drijvers JM, et al. CD160 Stimulates CD8+ T Cell Responses and Is Required for Optimal Protective Immunity to Listeria monocytogenes. Immunohorizons 2018;2:238–50. 10.4049/immunohorizons.1800039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Del Rio M-L, Bravo Moral AM, Fernandez-Renedo C, et al. Modulation of cytotoxic responses by targeting CD160 prolongs skin graft survival across major histocompatibility class I barrier. Transl Res 2017;181:83–95. 10.1016/j.trsl.2016.09.004 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Liu Y, Liu H, et al. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci 2019;9:19. 10.1186/s13578-019-0282-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whiteside TL. Exosomes carrying immunoinhibitory proteins and their role in cancer. Clin Exp Immunol 2017;189:259–67 https://onlinelibrary.wiley.com/doi/abs/ 10.1111/cei.12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.www.cancer.ca . Stages of chronic lymphocytic leukemia (CLL) - Canadian Cancer Society [Internet]. Available: https://www.cancer.ca:443/en/cancer-information/cancer-type/leukemia-chronic-lymphocytic-cll/staging/?region=on [Accessed cited 2021 Feb 18].
- 33.Elahi S, Ertelt JM, Kinder JM, et al. Immunosuppressive CD71+ erythroid cells compromise neonatal host defence against infection. Nature 2013;504:158–62 https://www.nature.com/articles/nature12675 10.1038/nature12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shahbaz S, Dunsmore G, Koleva P, et al. Galectin-9 and vista expression define terminally exhausted T cells in HIV-1 infection. J Immunol 2020;204:ji1901481–91. 10.4049/jimmunol.1901481 [DOI] [PubMed] [Google Scholar]
- 35.Dunsmore G, Bozorgmehr N, Delyea C, et al. Erythroid Suppressor Cells Compromise Neonatal Immune Response against Bordetella pertussis. J Immunol 2017;199:2081-2095. 10.4049/jimmunol.1700742 [DOI] [PubMed] [Google Scholar]
- 36.Namdar A, Dunsmore G, Shahbaz S. CD71+ erythroid cells exacerbate HIV-1 susceptibility, mediate trans-infection. and Harbor Infective Viral Particles. mBio [Internet] 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morales-Kastresana A, Telford B, Musich TA, et al. Labeling extracellular vesicles for nanoscale flow cytometry. Sci Rep 2017;7:1878. 10.1038/s41598-017-01731-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roederer M, Nozzi JL, Nason MC. Spice: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 2011;79:167–74. 10.1002/cyto.a.21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yun X, Zhang Y, Wang X. Recent progress of prognostic biomarkers and risk scoring systems in chronic lymphocytic leukemia. Biomark Res 2020;8:40. 10.1186/s40364-020-00222-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuertes Marraco SA, Neubert NJ, Verdeil G, et al. Inhibitory receptors beyond T cell exhaustion. Front Immunol 2015;6:310. 10.3389/fimmu.2015.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Podack ER, Young JD, Cohn ZA. Isolation and biochemical and functional characterization of perforin 1 from cytolytic T-cell granules. Proc Natl Acad Sci U S A 1985;82:8629–33. 10.1073/pnas.82.24.8629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol 2003;3:361–70. 10.1038/nri1083 [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y, Shao Q, Peng G. Exhaustion and senescence: two crucial dysfunctional states of T cells in the tumor microenvironment. Cell Mol Immunol 2020;17:27–35. 10.1038/s41423-019-0344-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viganò S, Banga R, Bellanger F, et al. CD160-associated CD8 T-cell functional impairment is independent of PD-1 expression. PLoS Pathog 2014;10:e1004380. 10.1371/journal.ppat.1004380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero P, Zippelius A, Kurth I, et al. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J Immunol 2007;178:4112–9. 10.4049/jimmunol.178.7.4112 [DOI] [PubMed] [Google Scholar]
- 46.Walker EB, Haley D, Petrausch U, et al. Phenotype and functional characterization of long-term gp100-specific memory CD8+ T cells in disease-free melanoma patients before and after boosting immunization. Clin Cancer Res 2008;14:5270–83. 10.1158/1078-0432.CCR-08-0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elahi S, Dinges WL, Lejarcegui N, et al. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med 2011;17:989–95. 10.1038/nm.2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lazarevic V, Glimcher LH, Lord GM. T-Bet: a bridge between innate and adaptive immunity. Nat Rev Immunol 2013;13:777–89. 10.1038/nri3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z, Ji Z, Ngiow SF, et al. TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity 2019;51:840–55. 10.1016/j.immuni.2019.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeh Y-Y, Ozer HG, Lehman AM, et al. Characterization of CLL exosomes reveals a distinct microRNA signature and enhanced secretion by activation of BCR signaling. Blood 2015;125:3297–305. 10.1182/blood-2014-12-618470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z-Z, Grote DM, Ziesmer SC, et al. Il-12 upregulates Tim-3 expression and induces T cell exhaustion in patients with follicular B cell non-Hodgkin lymphoma. J Clin Invest 2012;122:1271–82. 10.1172/JCI59806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan X-J, Dozmorov I, Li W, et al. Identification of outcome-correlated cytokine clusters in chronic lymphocytic leukemia. Blood 2011;118:5201–10. 10.1182/blood-2011-03-342436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bengsch B, Seigel B, Ruhl M, et al. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog 2010;6:e1000947. 10.1371/journal.ppat.1000947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S, Zhang W, Liu K, et al. CD160 expression on CD8+ T cells is associated with active effector responses but limited activation potential in pancreatic cancer. Cancer Immunol Immunother 2020;69:789–97. 10.1007/s00262-020-02500-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He W, Wang B, Li Q, et al. Aberrant expressions of co-stimulatory and Co-inhibitory molecules in autoimmune diseases. Front Immunol 2019;10:261. 10.3389/fimmu.2019.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reiser J, Banerjee A. Effector, Memory, and Dysfunctional CD8+ T Cell Fates in the Antitumor Immune Response [Internet]. Journal of Immunology Research. Hindawi 2016;2016:e8941260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia B, Zhao C, Rakszawski KL, et al. Eomes+T-betlow CD8+ T Cells Are Functionally Impaired and Are Associated with Poor Clinical Outcome in Patients with Acute Myeloid Leukemia. Cancer Res 2019;79:1635–45. 10.1158/0008-5472.CAN-18-3107 [DOI] [PubMed] [Google Scholar]
- 58.Josefsson SE, Huse K, Kolstad A, et al. T cells expressing checkpoint receptor TIGIT are enriched in follicular lymphoma tumors and characterized by reversible suppression of T-cell receptor signaling. Clin Cancer Res 2018;24:870–81. 10.1158/1078-0432.CCR-17-2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guillerey C, Harjunpää H, Carrié N, et al. TIGIT immune checkpoint blockade restores CD8+ T-cell immunity against multiple myeloma. Blood 2018;132:1689–94 https://pubmed.ncbi.nlm.nih.gov/29986909/ 10.1182/blood-2018-01-825265 [DOI] [PubMed] [Google Scholar]
- 60.Tan J, Yu Z, Huang J, et al. Increased PD-1+Tim-3+ exhausted T cells in bone marrow may influence the clinical outcome of patients with AML. Biomark Res 2020;8:6. 10.1186/s40364-020-0185-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okoye I, Xu L, Motamedi M, et al. Galectin-9 expression defines exhausted T cells and impaired cytotoxic NK cells in patients with virus-associated solid tumors. J Immunother Cancer 2020;8:e001849 https://jitc.bmj.com/lookup/doi/ 10.1136/jitc-2020-001849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krackhardt AM, Witzens M, Harig S, et al. Identification of tumor-associated antigens in chronic lymphocytic leukemia by SEREX. Blood 2002;100:2123–31. 10.1182/blood-2002-02-0513 [DOI] [PubMed] [Google Scholar]
- 63.Klebanoff CA, Gattinoni L, Restifo NP. Sorting through subsets: which T-cell populations mediate highly effective adoptive immunotherapy? J Immunother 2012;35:651–60. 10.1097/CJI.0b013e31827806e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bonnon C, Wendeler MW, Paccaud J-P, et al. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J Cell Sci 2010;123:1705–15 https://jcs.biologists.org/content/123/10/1705 10.1242/jcs.062950 [DOI] [PubMed] [Google Scholar]
- 65.Sato K, Nakano A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett 2007;581:2076–82 https://pubmed.ncbi.nlm.nih.gov/17316621/ 10.1016/j.febslet.2007.01.091 [DOI] [PubMed] [Google Scholar]
- 66.L A, De S HP. Inhibitory Receptor Expression Depends More Dominantly on Differentiation and Activation than “Exhaustion” of Human CD8 T Cells [Internet] Frontiers in immunology. Front Immunol 2013;4 https://pubmed.ncbi.nlm.nih.gov/24391639/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie F, Xu M, Lu J, et al. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol Cancer 2019;18:146. 10.1186/s12943-019-1074-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sabzevary-Ghahfarokhi M, Shirzad H, Rafieian-Kopaei M, et al. The role of inflammatory cytokines in creating T cell exhaustion in cancer. Cancer Biother Radiopharm 2018;33:267–73. 10.1089/cbr.2018.2449 [DOI] [PubMed] [Google Scholar]
- 69.Alhakeem SS, McKenna MK, Oben KZ, et al. Chronic lymphocytic Leukemia–Derived IL-10 suppresses antitumor immunity. J.i. 2018;200:4180–9 https://www.jimmunol.org/content/200/12/4180 10.4049/jimmunol.1800241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kitabayashi A, Hirokawa M, Miura AB. The role of interleukin-10 (IL-10) in chronic B-lymphocytic leukemia: IL-10 prevents leukemic cells from apoptotic cell death. Int J Hematol 1995;62:99–106. 10.1016/0925-5710(95)00395-9 [DOI] [PubMed] [Google Scholar]
- 71.Schröttner P, Leick M, Burger M. The role of chemokines in B cell chronic lymphocytic leukaemia: pathophysiological aspects and clinical impact. Ann Hematol 2010;89:437–46. 10.1007/s00277-009-0876-6 [DOI] [PubMed] [Google Scholar]
- 72.Ghamlouch H, Ouled-Haddou H, Damaj G, et al. A combination of cytokines rescues highly purified leukemic CLL B-cells from spontaneous apoptosis in vitro. PLoS One 2013;8:e60370. 10.1371/journal.pone.0060370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baba T, Mukaida N. Role of macrophage inflammatory protein (MIP)-1α/CCL3 in leukemogenesis. Mol Cell Oncol [Internet] 2014;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schulz A, Toedt G, Zenz T, et al. Inflammatory cytokines and signaling pathways are associated with survival of primary chronic lymphocytic leukemia cells in vitro: a dominant role of CCL2. Haematologica 2011;96:408–16. 10.3324/haematol.2010.031377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wierda WG, Johnson MM, Do K-A, et al. Plasma interleukin 8 level predicts for survival in chronic lymphocytic leukaemia. Br J Haematol 2003;120:452–6. 10.1046/j.1365-2141.2003.04118.x [DOI] [PubMed] [Google Scholar]
- 76.Somasundaram A, Cillo A, Oliveri L. Il-6, IL-8 drive LAG3/PD1 immune suppression on effector and naïve, peripheral blood CD8+ T cells in cancer patients. J Immunol 2019;202:195–9. [Google Scholar]
- 77.Bellomo G, Allegra A, Alonci A, et al. Serum levels of interleukin-16 in lymphoid malignancies. Leuk Lymphoma 2007;48:1225–7. 10.1080/10428190701268767 [DOI] [PubMed] [Google Scholar]
- 78.Cruikshank WW, Kornfeld H, Center DM. Interleukin-16. J Leukoc Biol 2000;67:757–66 https://jlb.onlinelibrary.wiley.com/doi/abs/ 10.1002/jlb.67.6.757 [DOI] [PubMed] [Google Scholar]
- 79.Atanackovic D, Hildebrandt Y, Templin J, et al. Role of interleukin 16 in multiple myeloma. J Natl Cancer Inst 2012;104:1005–20. 10.1093/jnci/djs257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-002189supp001.pdf (49.9KB, pdf)
jitc-2020-002189supp002.pdf (12.8MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.