Abstract
Cancer-associated cachexia is defined by loss of weight and muscle mass, and by the potential loss of adipose tissue, insulin resistance, and increased resting energy expenditure. Cachexia is most prevalent in pancreatic cancer, the 3rd leading cause of cancer-related deaths. While various factors interact to induce cachexia, the precise mechanisms underlying this clinical condition are not fully understood. Clinically relevant animal models of cachexia are needed given the lack of standard diagnostic methods or treatments for this condition. Described in this unit are in vitro and in vivo models used to study the role of macrophages in the induction of cachexia in pancreatic cancer. Included are procedures for isolating and culturing bone marrow-derived macrophages, harvesting tumor- and macrophage-derived conditioned medium, and for studying the effect of conditioned medium on C2C12 myotubes. Also described are procedures involving the use of an orthotopic model of pancreatic cancer, including a method for examining skeletal muscle atrophy in this model.
Keywords: Cancer Cachexia, Macrophages, Myotubes, Pancreatic Cancer, Skeletal Muscle
Introduction
Cancer-associated cachexia is a condition characterized by the loss of muscle mass, with or without adipose tissue loss, associated with hyper metabolism, a lower energy intake, and increased resting energy expenditure (Marceca et al., 2020). At present, up to 60% of cancer patients in the US exhibit a cachectic phenotype (Advani et al., 2018). In the clinical setting, a weight loss of 5% or more within 6 months, or greater than 2% in individuals with a Body Mass Index (BMI) less than 20 kg/m, confirms cachexia (Fearon et al., 2011). Cancer-associated cachexia is associated with 70% of pancreatic ductal adenocarcinoma PDAC (Pancreatic ductal adenocarcinoma), 60% of gastro-esophageal and head-neck cancers, and 40–50% of other malignancies (Marceca et al., 2020). Furthermore, cachexia increase susceptibility to the side effects and toxicities associated with chemotherapy, thereby negatively affecting the quality of life and survival. With PDAC, cachexia is the cause of death in a third of the patients (Henderson et al., 2018). Because of its complexity, there are currently no standard treatments for reversing cancer-associated cachexia. Appropriate, clinically relevant animal models of cachexia and PDAC are needed to address these issues. In vitro and in vivo studies have identified tumor necrosis factor-α, interleukins (e.g., IL-1, IL-6), interferon type II, microRNAs, toll-like receptors, and transforming growth factor-β superfamily of cytokines as substances involved in the induction of cachexia (Marceca et al., 2020). Inflammation generated by increased levels of inflammatory cytokines and increased proteolysis regulated by the ubiquitin-proteasome pathway are proposed to underlie the induction of cachexia (Advani et al., 2018). Like the inflammatory cytokines, immune cells contribute to the cachectic phenotype. For instance, a 100-fold expansion of myeloid-derived suppressor cells (MDSCs) is associated with increased energy expenditure and loss of tissue adiposity in tumor-bearing mice (Cuenca et al., 2014).
In contrast, macrophages exhibit both pro-and anti-inflammatory effects during muscle atrophy depending upon physiologic state. Macrophage depletion in healthy mice significantly delays muscle recovery, while the absence of macrophages in pancreatic tumor-bearing mice rescues the cachectic phenotype (Dumont & Frenette, 2010). Furthermore, macrophage/myotube co-cultures display a reduction in protein loss and myotube atrophy (Shukla et al., 2020b), whereas macrophage-derived conditioned medium induces myotrophy on co-incubation with pancreatic conditioned-CM. Hence, depending on the environment, macrophages alter myotube morphology and atrophy.
Described in this unit are the essential essays employed for the in vitro and in vivo assessment of skeletal muscle atrophy. The first protocol includes details for the preparation of pancreatic tumor cell- and macrophage-derived conditioned media (basic protocol 1). Described in Support Protocol 1 are the critical steps for the in vitro molecular evaluation of cachectic markers in the myotube model. Provided in Basic Protocol 2 and Support Protocol 2 are laboratory methods for evaluating the cachectic phenotype in pancreatic tumor-bearing mice.
Basic protocol 1: In vitro model of pancreatic tumor-induced cachexia using C2C12 cell lines (myotube model)
Summary
An immortalized myoblast cell line, C2C12 is established from murine satellite cells. This cell line is widely employed for studying myogenic regulation, muscle differentiation, regeneration, and muscle atrophy (Burattini et al., 2004; Ikeda et al., 2017; McMahon et al., 1994; Yaffe & Saxel, 1977). The C2C12 cells grow rapidly in culture, exit the cell cycle, and rapidly differentiate into muscle myotubes under low serum conditions. Also, the capacity of C2C12 to undergo step-wise differentiation from myoblast to myocyte and then to multinucleated myotubes, makes this cell line valuable for defining and examining factors affecting muscle growth and differentiation. Detailed below is a step-by-step, in vitro procedure for studying the cancer-induced cachexia phenotype using the mouse C2C12 muscle cell line and conditioned media from murine pancreatic cancer cell lines.
Materials
C2C12 cell line (purchased from ATCC, CRL-1772™)
Pancreatic Cancer cell lines:
KrasG12D; Trp53R172H/+; Pdx-1-Cre (KPC) mouse-derived pancreatic mouse cells (KPC1245 and KPC8069). Provided as gift from. Dr. David Tuveson (Cold Spring Harbor Laboratory, NY, USA) and Dr. Michael Hollingsworth (University of Nebraska Medical Center, NE, USA).
S2–013 (Pancreatic adenocarcinoma cell line derived from liver metastasis), obtained from Expasy (cat no: CVCL_B280).
L929 cells (ATCC, CCL-1)
Complete Dulbecco’s modified Eagle medium (DMEM) (Gibco Lot: 2120636)
Inactivated Horse Serum (Sigma-Aldrich, cat. No: H1270)
Fetal Bovine Serum (FBS) (Neuromics, cat. No: FBS002)
Trypsin (Corning, Lot: 30119014)
Dimethyl sulfoxide (DMSO) (Fisher Chemical, Lot: D139-1)
Insulin solution (Sigma-Aldrich, cat No: I0516)
Penicillin and Streptomycin (P/S) (Gibco, Lot: 2112692)
100 mm cell culture dishes (Thermo-Fisher Scientific)
IL-4 (Peprotech, cat no:214-14)
Collagen –coated plates (Corning, cat no: 354400)
Ficoll Plaque premium (GE Healthcare, cat no: GE17-5442-02)
Cell scraper (SL1250, Southern Labware, GA, USA)
Protocol Steps
Preparation of tumor-derived conditioned medium (CM) from pancreatic cancer cells lines
-
1Thaw and seed pancreatic cancer cell lines in T-75 flasks or 100 mm dishes with DMEM medium supplemented with 10% FBS and 1% penicillin and streptomycin (P/S) at 37°C to optimize growth.Note: The aim of this procedure is to evaluate the effect of conditioned medium-derived from three different tumor cell lines (KPC1245, KPC8069, and S2–013) on C2C12 myotubes. As these cell lines exhibit a similar doubling rate, the cell culture and collection of tumor-conditioned medium is the same for all three cell lines.
-
2At 70–80% confluence, wash the cells twice with a sterile 1x PBS, add 12ml of FBS-free DMEM media and incubate the cells at 37°C for 24 hours.70–80% cell confluence is attained 48hr after seeding the plate with 2×106 cells.
-
3
At the end of the incubation period collect the FBS-free medium and centrifuge at 300 × g for 5 minutes to remove any remaining cell components. Roughly, 15 ml CM is obtained from 100 mm cell culture dish. The medium can be stored in 15 or 50ml conical tubes to avoid repeated freze and thaw. Filter the CM through a 0.22um sterile filter and retain the filtered CM at −80 °C for future use.
Handling, maintenance and differentiation of mouse C2C12 muscle cells into myotubes
It is important to always passage and freeze the C2C12 cell lines when they reach 50–60% confluence. Do not allow the cells to grow over 60% confluency. Use 10% FBS and 1% P/S containing DMEM medium for C2C12 culture passaging and maintenance. Avoid prolonged incubation with trypsin (optimum 1 min) during the passaging process.
-
4
Prior to C2C12 differentiation, seed 0.1×106 cells per well in 6-well collagen-coated plates with DMEM medium supplemented with 10% FBS and 1% P/S.
-
5
At 80% confluence, wash the 6-well plates with a sterile 1xPBS twice and add C2C12 differentiation 10ml medium (DMEM containing 2% inactivated horse serum and 1μg/ml insulin). At 80% confluence C2C12 differentiate uniformly into myotubes. These cells do not proliferate after they have differentiated. The differentiated myotubes are stable up to 7 days post-differentiation. The differentiation media is changed every 24 hours.
-
6
Change the differentiation media every 24 hours for 72 hour. Change the medium every 12 hours during the final 24 hours of the C2C12 differentiation process.
-
7Using a bright-field microscope, observe the morphological changes of C2C12 myoblasts towards their transformation into myotubes during the 72 hour differentiation process.Note: The C2C12 myoblasts gradually change shape from round to elongated, fiber-like morphology. At the end of the differentiation process the majority of the muscle cells differentiate into elongated muscle fibers that are termed muscle myotubes (Figure 1).
Figure 1. Pancreatic tumor-derived CM induces myotube atrophy.

(A) Representative bright-field photomicrographs display C2C12 myotubes after 24 hours of treatment with control medium, and tumor cell (KPC1245, KPC8069 and S2013 pancreatic cancer cell lines)-derived CM. The photomicrographs were taken with a Celigo imaging cytometer system; scale bar: 250μm. (B) Histogram plot represent myotube thickness (diameter) of control and tumor-derived CM-treated myotube groups at different time points. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, compared by one-way ANOVA with Dunnett’s multiple comparisons test.
Preparation of conditioned medium from M2 macrophages
Begin by preparing L929 cell-derived conditioned medium (L929-CM) for macrophage culture and harvest bone marrow-derived macrophages from 6–8 week old C57BL/6J mice as described elsewhere (Attri et al., 2018b). Briefly, aseptically isolate the femur and tibia from the mice and remove the soft tissue. Using a 21-gauge needle, flush from the bone marrow with 10 ml of serum-free DMEM medium from one end of the bone. Collect the flushed material in a 15 ml tube and wash with 10 ml complete DMEM (10% FBS). Centrifuge the cells at 500 × g for 5 minutes at room temperature and re-suspend the cells in fresh 6ml complete DMEM medium. Isolate monocytes using a Ficoll Paque gradient as previously described (Attri et al., 2018a). After monocyte isolation, determine the viability of the macrophage-precursor cells using trypan blue dye. Add 20μl of macrophage-precursor cell suspension to 20μl of trypan blue dye, using a hemocytometer to count the cells 10 μl of the mixture.
-
8
Following monocyte isolation, seed 4×106 the cells in a 100-mm cell culture dish containing 10 ml DMEM supplemented with 10% FBS and 20% L-929-CM. Subsequently incubate the cells for 24 hours in an incubator at 5% CO2, 20% O2 and 95% humidity.
-
9
Add an additional 10 ml of DMEM medium containing 10% FBS, and 20% L-929-CM to the plate and incubate the cells in the incubator for three days, during which time they must be monitored periodically for adherence and contamination. Cells should appear round and attached to the plate at the end of 3rd day. In case of contamination, discard the culture and isolate fresh batch of monocyte as mentions in step 7. If cells are not adhered to the plate, and medium is not contaminate, poor differentiation could be the underlying problem. Should that be the case, prepare a new batch of DMEM supplemented with L-929-CM and repeat monocyte isolation and culture steps as mentioned above.
-
10
Following the three-day incubation, replace the old medium with 10ml of freshly prepared DMEM (10% FBS+20% L929-conditioned medium). Incubate the cells for an additional three days. Every day monitor the cells for contamination. If cells are contaminated, discard the culture and isolate fresh batch monocyte as mentions in step 7.
-
11Harvest the macrophages at the end of the incubation period. Gently scrape the cells with sterile cell scraper in the presence of 1 ml trypsin, collect them in complete 10 ml of DMEM and centrifuge at 500 × g for 5 minutes at room temperature. Following centrifugation, remove the medium and add fresh 10 ml of DMEM medium and count the cells using hemocytometer. Next, re-seed the macrophages to a total of 1×106 cells/well in 2ml of DMEM medium containing 10% FBS and 20% L-929 cell-derived medium. Incubate the cells in the incubator at room temperature for overnight and monitor the cells the next day for adherence. If cells are not adhered, discard the culture. If cells are adhered and healthy proceed with next steps.Note: The L-929 cell secretes m-CSF that promotes the differentiation of macrophage progenitor cells into macrophages. Upon differentiation, macrophages appear elongated and resemble spindle shapes structure as described previously. To confirm the macrophage phenotype, perform cell surface staining using fluorescent labeled antibodies against CD11b and anti-F4/80 antibody and acquire he data through flow cytomer as previously
-
12
Once the macrophages are fully differentiated, add 20ng/ml IL-4 to the cells for 24 hours in the incubator at 37°C (Attri et al., 2018a; Attri et al., 2018b). In the presence of IL-4 macrophages express CD206, markers that are specific for M2-macrophages. The presence of this marker is verified using flow cytometric analysis. Subsequently, wash the polarized macrophages with 1X PBS and incubate the cells with serum-free DMEM media for 24 hours in the incubator at 37°C.
-
13On the following day, collect the supernatant and centrifuge at 300 × g for 5 minutes to remove the remaining cell components. Filter the medium through a 0.22um sterile filter and store the filtered medium at −80°C for future use. Macrophage CM is stable at −80°C for 6 months. To avoid repeyaed freeze thaw store them in 15 or 20ml aliquots.Note: Discard the cells as the study employs only macrophage-derived condition medium (Mac-CM) to determine the impact of the macrophage-secreted factor on muscle atrophy.
Pancreatic cancer cell and macrophage-derived conditioned medium treatment of differentiated C2C12 myotubes
-
14
To examine the effect of tumor-conditioned medium, incubate the differentiated C2C12 myotubes (step 4–7) with tumor-CM. Briefly, dilute tumor-CM at a 1:1 ratio with complete DMEM (containing 2% horse serum and 1%penicillin/streptomycin.). Likewise, to study the impact of Mac-CM on muscle atrophy, incubate the C2C12 myotubes with diluted 15 ml of Mac-CM (Mac-CM diluted with complete DMEM at a 1:1 ratio). Warm the media to 37°C prior to incubation with the myotubes.
-
15
Change the medium with diluted CM every 24 hours. The differentiated myotubes can be treated from 24 to 72 hours with CM.
-
16
Evaluate the morphological changes in myotubes by harvesting cells after at the 24, 48, and 72 hours (Fig. 1). The morphological changes in myotubes post macrophage-CM incubation is described elsewhere (Shukla et al., 2020b).
Data analysis
Morphological evaluation of C2C12 myotube wasting after CM treatment
-
17
The measurement of myotube thickness (fiber diameter) and evaluation of atrophy markers are used to define muscle wasting in the in vitro models. Morphological assessment of myotube determines the effect of tumor- or macrophage-CM on muscle morphology. Atrophic myotubes display a reduced diameter as compared to healthy myotubes.
-
18
To perform morphological analysis, use a light-transmitted microscope with a camera or an imaging-based system (e.g. Celigo imaging cytometer) to capture 5–10 bright-field images of control and CM-treated groups.
-
19
Use ImageJ software to determine the average myotube diameter. The average diameter length can be assessed from 200 myotubes in each group. Each myotube is measured at 3 points along its length. Data are analyzed by plotting the average diameter of the muscle fibers between control and tumor-CM-treated myotubes as demonstrated shown in Fig. 1.
Support protocol 1: Molecular evaluation of cachectic markers in C2C12 myotubes using real-time PCR and immunoblotting
Summary
Muscle atrophy is characterized by an imbalance of muscle protein synthesis and catabolic processes (proteolysis). An up-regulation of muscle proteins associated with ubiquitin-proteasome pathways such as atrogin-1/MAFbx (muscle atrophy F-box protein) and MuRF1/TRIM63 (muscle-specific RING-Finger-1) indicates muscle atrophy. In addition, the elevated expression of selectin P (SELP), inhibin beta subunit A (INHBA), and Cathepsin L (CTSL) signal skeletal muscle atrophy. Moreover, atrophic muscles demonstrate decreased expression of myosin heavy chain (MyHC) ((Bodine et al., 2001; Gomes et al., 2001; Michaelis et al., 2017; Zhong et al., 2019)). Collectively, in vitro assays therefore employ the evaluation of MuRF1, INHBA, SELP, CTSL, and MyHC to determine muscle atrophy.
Materials
Qiagen RNA extraction kit (Qiagen, cat No: 74104)
SYBR Green PCR Master Mix (Thermo-Fisher Scientific, cat no: A25742)
Verso cDNA synthesis kit (Thermo-Fisher Scientific, cat no: AB1453A)
PCR QuantStudio system (Applied Biosystems)
Phosphate-buffered saline 1x (PBS) (NaCl, KCl, Na2HP04, KH2P04, dH20)
Phosphatase inhibitor cocktail (100x) (Cell Signaling Technology, cat no: 5870)
RIPA lysis buffer (Thermo-Fisher Scientific, cat no: 89900)
10% NuPAGE Bis-Tris protein gel (Thermo-Fisher Scientific, cat no: NP0301BOX)
NuPAGE MES SDS Running Buffer (Thermo-Fisher Scientific, cat no: NP00202)
Nitrocellulose transfer membrane (Fisher Scientific, cat no: IPVH00010)
Tris-base transfer buffer (tris base, glycine, dH20)
Tris-buffered saline (TBS) (tris base, NaCl, dH20)
Tween 20 (Sigma-Aldrich, cat no: P1379)
Nonfat dried milk
Normal Goat Serum (NGS) (Sigma-Aldrich, cat no: G9023)
NuPAGE LDS Sample Buffer (Thermo-Fisher Scientific, cat no: NP0008)
Bio-Rad protein standard ladder (Bio-Rad, cat no:1610376)
BCA protein assay kit (Thermo-Fisher Scientific, cat no: 23225)
Chemiluminescent Substrate Reagent Kit (Fisher Scientific, cat no: WP20005)
Bio-Rad Chemidoc imaging (Bio-Rad, cat no: 17001402)
Celigo Imaging Cytometer (Nexcelom Bioscience)
Corning 6-well collagen coated plates (Fisher Scientific, cat No: 08-772-69)
37°C, 5% CO2 humidified incubator (Thermo-Fisher Scientific, cat no: 3110)
Primer sequences:
Mouse MuRF1:
Forward primer - TGGCGATTGTCACAAAGTGG
Reverse primer - CCCTCTCTAGGCCACCGAGT
Mouse CTSL:
Forward primer - TCTCACGCTCAAGGCAATCA
Reverse primer - AAGCAAAATCCATCAGGCCTC
Mouse SELP:
Forward primer – CATCTGGTTCAGTGCTTTGATCT
Reverse primer - ACCCGTGAGTTATTCCATGAGT
Mouse INHBA:
Forward primer – TGAGAGGATTTCTGTTGGCAAG
Reverse primer - TGACATCGGGTCTCTTCTTCA
Mouse 18S RNA:
Forward primer – CGGACAGGATTGACAGATTG
Reverse primer - CAAATCGCTCCACCAACTAA
Western blot Antibodies:
Anti-mouse Myosin Heavy Chain (MyHC) antibody (R&D system, clone: MF20; dilution: 1/1000)
Anti-mouse MuRF1 antibody (Santa Cruz Biotechnology, clone: C11; dilution: 1/500)
Anti-mouse Atrogin-1 antibody (Thermo Fisher Scientific, Polyclonal: PA5–43915; dilution: 1/1000)
Anti-β-Actin antibody (Sigma-Aldrich, clone: AC-15; dilution: 1/5000)
Anti-mouse/anti-rabbit IgG HRP-conjugated antibody (dilution 1/10000) (Thermo Fisher Scientific cat no: 31460, 31430)
Protocol steps
Preparation of RNA and evaluation of cachectic markers in treated C2C12 myotubes
-
1
Extract the total RNA from the CM-treated C2C12 myotubes with the Qiagen RNA extraction kit per the manufacturer’s instructions.
-
2
For the gene expression assay, prepare cDNA using the Verso cDNA synthesis commercial kit. Use qPCR with a PCR QuantStudio system (Applied Biosystems) to assess the expression of different cachectic genes, including mouse MuRF1, INHBA, SELP, and CTSL. Use mouse 18S primers for internal controls. The primer sequences are provided in the material section of this unit. Use the ΔΔCt method as described previously (Livak & Schmittgen, 2001) to calculate and compare the cachectic gene expressions between the control and CM-treated groups. The following equation calculate the change in the expression of gene between the groups from real time-PCR data; ΔΔCt = ΔCt (treated sample) – ΔCt (untreated sample).
Preparation of protein and evaluation of cachectic markers in treated C2C12 myotubes
-
3
Extract the total protein with 40μl of RIPA cell lysis buffer supplemented with 1x phosphatase inhibitors from each control and CM-treated myotube group. 40μl of RIPA should be enough for the myotubes harvested from one well from 6-well plate.
-
4
Quantify the protein using a commercially available BCA-protein assay kit and the manufacturer’s instructions.
-
5
Perform an immunoblotting assay by loading 30μg of protein extract into 10% SDS-PAGE gel and transferring to a nitrocellulose membrane for 1hour. Block the membrane with 5% nonfat dry milk dissolved in 20 ml TBST for 1 hour at room temperature (RT).
-
6
Incubate the membrane overnight at 4°C with the anti-mouse antibodies including MuRF1, Atrogin-1, and Myosin Heavy Chain (MyHC) diluted in 10 ml total volume of TBST with 0.5% nonfat dried milk and 2% normal goat serum (NGS). Use mouse β-actin as a loading control. Incubate the membrane with HRP-conjugated secondary antibodies diluted in TBST with 0.5% nonfat dried milk and 1% NGS for 1 hour at RT. The clones and optimum dilution of each antibody are provided in the material section. After incubation with secondary antibody, wash the blot three times with 20 ml TBST and develop the blot using the ECL Chemiluminescent Substrate Reagent Kit. Changes in the cachectic markers post tumor-CM incubation are shown in Fig. 2D and in a previous study (Shukla et al., 2020a).
Figure 2. Pancreatic CM-derived medium up-regulates the expression of the cachectic genes in treated C2C12 myotubes.
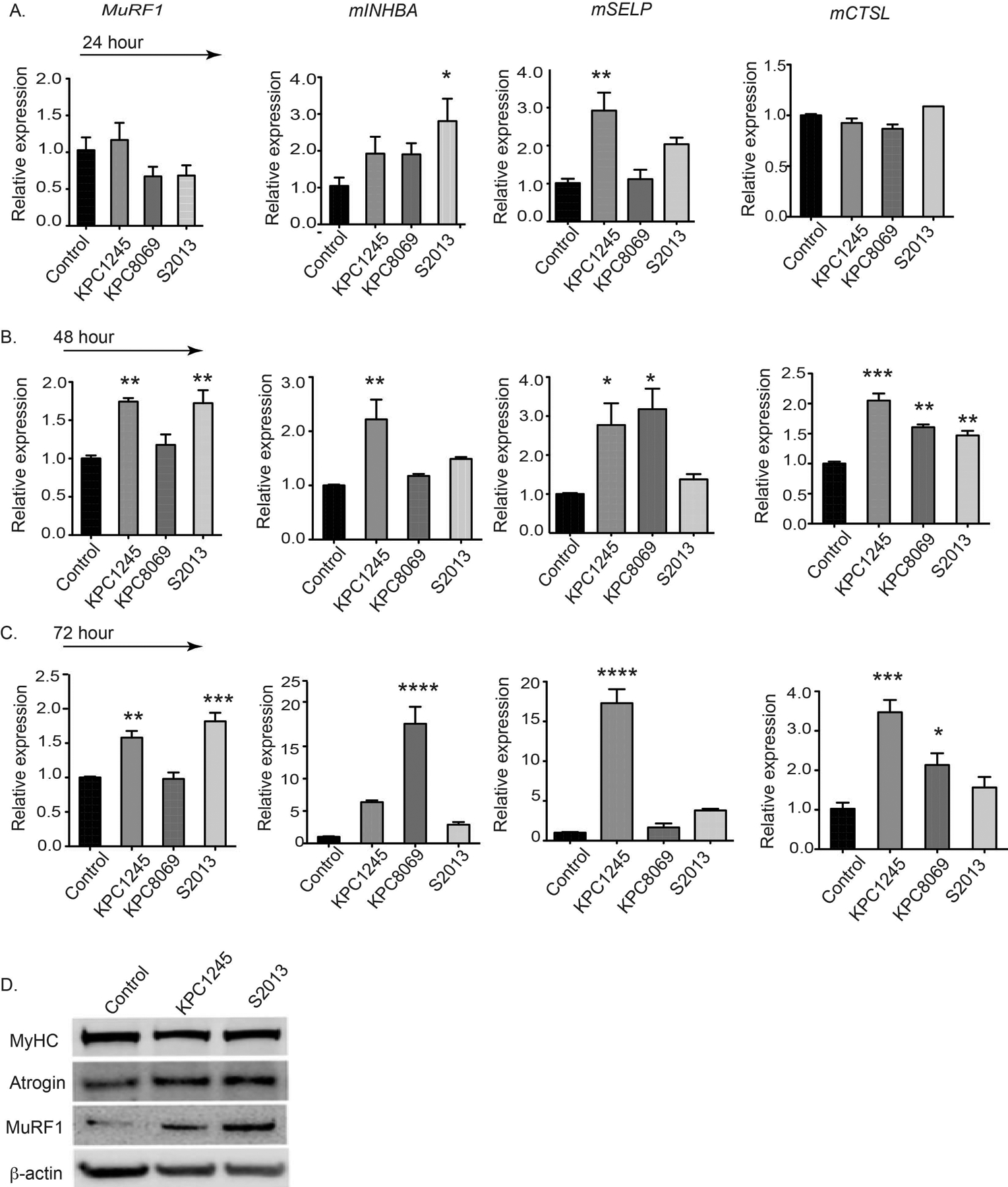
(A-C) histogram demonstrates mRNA expression levels of MuRF1, INHBA, SELP, CTSL, in C2C12-derived myotubes treated with control and tumor-derived CM at different time-points.The (D) immunoblot represents the protein levels of myosin heavy chain (MyHC), Atrogin-1 and MuRF1 in control and tumor CM-treated myotubes after 48 hours of treatment. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, compared by one-way ANOVA with Dunnett’s multiple comparisons test.
Basic protocol 2: In vivo model to study cachectic phenotype in pancreatic tumor-bearing mice
Summary
Orthotopic models of pancreatic cancer mimic the critical hallmarks of human cancer cachexia in a rapid and accurate manner (Michaelis et al., 2017; Talbert et al., 2019). In contrast to the long-term KPC GEMM model, the KPC allograft orthotopic model of PDAC displays a median survival of approximately one month after tumor implantation, with detection of tumor lesions as early as 1-week post-implantation. In the past, the KPC allograft orthotopic model of pancreatic cancer was used for studying the cachectic phenotype in skeletal muscle and adipose tissue (Shukla et al., 2015; Shukla et al., 2014). Detailed in the protocol below are the methods for tumor implantation in mice and as well as the identification of cachectic parameters in mice (Shukla et al., 2020a).
Materials
KrasG12D; Trp53R172H/+; Pdx-1-Cre (KPC) mouse-derived pancreatic mouse cells (KPC8069) (These cell lines were prepared in house from the spontaneous KPC model of PDAC)
C57BL/6J mice (Jackson Lab)
Clodronate liposomes (Cat no: SKU# CLD-8909, Encapsula NanoSciences, Brentwood, TN
Grip Strength meter (Columbus Instruments, OH, USA)
Rotarod apparatus, Rotamex-5 (Columbus Instruments, OH, USA)
Lunar PIXImus densitometer (GE Medical-Lumar, Madison, WI)
Electronic weighing scale
Ketamine/Xylazine (UNMC pharmacy)
Isoflurane anesthesia chamber with oxygen supply (SKU: 93805107, Midmark Animal Health)
Sterile surgical instruments – scissors, forceps, pincers
Absorbable and non-absorbable surgical sutures (B. Braun)
70% ethanol
Betadine solution (Santa Cruz animal health, cat no: sc-359867)
27G needle syringe (BD syringe)
Heating pads and heating laps (SoftHeat, Comparative Medicine, UNMC supply)
Electric trimmer (Wahl, Amazon)
Paper towels
Aluminum foil
10% formalin (Cat no: SF100-20, Thermo Fisher Scientific,)
Hematoxylin and Eosin (Cat no: HHS32, Sigma-Aldrich,)
Liquid nitrogen
Eppendorf 1.5 tubes (Fisher Scientific cat no: 05-408-129)
Complete Dulbecco’s modified Eagle medium (DMEM); (Gibco Lot: 2120636)
Fetal Bovine Serum (FBS); (Neuromics, cat. no: FBS002)
Trypsin (Corning, Lot: 30119014)
Penicillin and Streptomycin (P/S) (Gibco, Lot: 2112692)
100mm cell culture dishes (Thermo-Fisher Scientific)
Protocol steps
Establishment of orthotopic cachectic mouse model of pancreatic cancer and macrophage depletion
-
1
A day prior to orthotopic implantation of tumor cells, use weighing scale to measure and record the body weight of all 6-week old C57BL/6J mice.
-
2
Anesthetize the mice the following day with i.p ketamine (100–200mg/kg)/xylazine (5–16 mg/kg) as per IACUC approved protocol.
-
3
Once the mice are anesthetized, carefully shave the left abdominal side with electric trimmer and disinfect the skin with a 70% ethanol and betadine solution.
-
4
Using sterile surgical micro-scissors, make a small incision in the upper left abdomen and carefully expose the pancreas with the aid of sterile surgical pincers.
-
5
Culture healthy KPC8069 cells, as described in the protocol 1.
-
6
Inject into the splenic lobe of the pancreas freshly prepared KPC8069 cells in 1X PBS (5×104/30μl) using a sterile 27G needle syringe. Use a 6–0 mm absorbable suture to close the peritoneal incision and a non-absorbable suture or clips to close the skin incision.
-
7
Inject each animal subcutaneously with 100μl of sterile saline (0.9% NaCl) solution. Place the mice into a clean cage with an electric heating pad set at 38–40°C until they have fully recovered from the anesthesia.
-
8
Once they have recovered, inject the mice s.c. with an analgesic such as buprenorphine (Buprenex, 0.05–0.01mg/kg) every 12 hours for the next 3 days as approved by the institutional IACUC.
-
9
Visually monitor the mice daily for the next 7 days for the presence of adverse consequences of surgery such as open wounds or bleeding. Re-suture skin if necessary. Carefully remove the skin suture or clips 7–10 days post-surgery. The appearance of cachectic parameters typically begins a week after surgery.
-
10To examine the impact of macrophage depletion in pancreatic tumor-bearing mice, deplete the macrophages with clodronate liposomes (100μl) with intravenous injections three times a week. The clodronate liposome injections should begin three days prior to the implantation of the pancreatic tumor. Confirm macrophage depletion by collecting blood from the submandibular vein, performing immunofluorescence staining to detect macrophage markers (CD11b+ F4/80+) (Shukla et al., 2020b).Note: All protocols involving mice must be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC). Mice must be housed and handled per institutional regulations and approved IACUC protocols.
Data analysis
Evaluation of body weight of the cachectic mice model
-
11
A week after surgery begin measuring and recording the body weights of all control, healthy mice (without tumor) and tumor-bearing mice using a calibrated electronic weighing scale.
-
12
Tare the scale between measurement of each mouse. Place the animal on the scale and note the steady weight value.
-
13
Weigh each mouse three times/week at approximately the same time of day. Compare the time-course body weight plots for each group at the conclusion of the study.
Assessment of forelimb grip strength
Cachectic patients exhibit progressive body-weight loss and functional impairment of skeletal muscle. In tumor-bearing mice, the assessment of forelimb grip strength measures deteriorating muscle function. Utilizing the grip strength meter, assess the peak amount of force that mice apply in grasping the pull bar with their forelimbs.
-
14
Set up the grip strength meter to measure the maximum pull force in kilogram-force (KGF). Tare the meter to zero between each measurement.
-
15
Acclimatize the mice for 15–20 minutes before measuring the grip strength of each animal by grabbing the base of the animal’s tail and placing it between the thumb and index finger. Once the animal is immobilized, place only its front paws over the pull grid of the apparatus and wait until the mouse grabs the grid as shown in Fig 4. Next, pull the animal towards in the opposite direction of the meter until the mouse drops from the pull grid. Be sure to always maintain the animal’s body in a horizontal position during the measurement process.
-
16
Record the peak full force in KGF for three independent measurements for each animal and calculate the average value comparison with the subsequent analysis.
-
17
Measure the grip strength of each group of mice twice per week at approximately the same time of the day. Compare the time-course grip strength plots for each group at the conclusion of the study.
Figure 4. Pancreatic tumor-bearing mice display cachectic phenotype.

The (A-E) histograms display cachectic parameters in healthy and tumor-bearing mice. Total body weight (A), carcass weight (B), muscle weight (C), fat weight (D), heart weight (E) are shown for healthy controls and tumor-bearing mice. N=9 (control mice), n=6 (pancreatic tumor-bearing mice), * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, unpaired student’s t test.
Rotarod testing of motor coordination
In addition to weight loss, cancer cachexia is associated with a decrease in motor skills and attention span. This suggests a critical link between hypothalamic inflammation and regulation of locomotor activity in cancer cachexia. In the hypothalamus, the arcuate nuclei (ARC) region contains two distinct populations of neurons that regulate energy balance. While one group of neurons produces α-MSH to induces anorexia, another expresses agouti-related protein (AgRP) and neuropeptide-Y (NPY), which stimulate appetite. Binding of α-MSH to type-3 melanocortin receptors (MC3-R), and the type-4 melanocortin receptors (MC4-R) induces anorexia. Studies indicate a critical association between melanocortin activation and locomotor activity, with MC4-R null mice being resistant to inflammation-induced decrease in physical activity (wheel running)(Marks et al., 2001). Assessment of locomotor activity is essential for determining the cachectic phenotype in tumor-bearing mice. To this end, the rotarod performance test is utilized to assess locomotor activity in tumor-bearing mice (Broberger et al., 1998; Cone, 2005; Grossberg et al., 2010; Murphy et al., 2012).
-
18
Place a soft padded or paper cushion in each exercise line where mice will fall in the Rotamex-5 apparatus. Establish an exercise program with an initial spinning speed of 3 rpm and acceleration of 1 rpm every 10 seconds. Use the same parameters throughout the entire study.
-
19
Acclimatize the mice for approximately 15–20 minutes before initiating the test. For the assessment, place each mouse on the rotarod facing away from the direction of the rotor’s spin. Here mouse would try to maintain the grip and move forward to stay standing. It is recommended during the first exposure the animal be allowed to become familiar with the rotating rod for a 1 minute period (3 rpm speed) prior to recording data.
-
20
Activate the rotarod testing program and record the latency of fall in seconds for each animal. Record three independent measurements for each subject, allowing 10 minutes of rest between each test. Use the average of the three values for the subsequent analysis.
-
21
Perform rotarod testing of each group of mice twice each week at approximately the same time of the day. Compare the time-course latency of fall plots for each group at the conclusion of the study.
Post-Necropsy processing and measurements of cachectic parameters in tumor- bearing mice
-
22Euthanize the mice at the end of the study period and prepare them for further post-necropsy processing, including organ weight measurements and appropriate tissue preservation for further morphological and molecular analysis. The following post-necropsy measurements are performed for evaluating the cancer-induced cachectic phenotype:Bodyweight – Immediately weigh each mouse after death. Do not collect any blood or tissue before bodyweight measurement.Carcass weight – The carcass weight represents the animal’s weight after removal of the tumor. To determine this value, weigh the animal after removing the entire tumor, including any visible tumor nodules on the peritoneal wall the near-site of injection.Heart weight– After removing the mouse heart, wash it thoroughly with 1X PBS or saline to remove remaining blood. Use paper tissue to dry the organ before weighing.Muscle weight– Using surgical scissors, remove the skin form the hind legs and cut all muscles from the leg including, gastrocnemius, tibialis anterior and quadriceps. Determine the weight of each muscle. Freeze a fraction of muscle tissue in liquid nitrogen and store at −80 °C. Fix the remaining portion of the muscle tissue in 10% formalin for immunohistochemical analysis.Fat weight– Carefully remove and weigh the white abdominal fat. As was done with the skeletal muscles, process the fatty tissue for further molecular and immunohistochemical analysis.
-
23
Tumor weight and volume– weigh the tumor tissue on an electronic scale. For tumor volume measurement, determine the length and the width of the tumor by using a caliper. Store the tumor tissue for further molecular and immunohistochemical analysis, as described above. Calculate the tumor volume using the following formula V = (W2 xL)/2, where W is the tumor’s width and L is tumor’s length.
Support protocol 2: Evaluation of cachectic markers in the skeletal muscle of tumor-bearing mice
Summary
As with the C2C12 myotube model, evaluation of muscle morphology and cachectic markers is important for quantifying the extent of muscle wasting in the tumor-bearing mice. The protocol below describes the procedure for assessing cachectic markers in gastrocnemius muscle obtained from pancreatic tumor-bearing mice.
Materials
10% formalin (Sigma Aldrich, HT501128-4L)
Hematoxylin and Eosin staining kit (Abcam, cat no: 245880)
Eppendorf 1.5 tubes (Fisher Scientific cat no: 05-408-129)
Hand-operated tissue homogenizer (BTlab system.com, cat no: BT704)
Penicillin and Streptomycin (P/S) (Gibso, Lot: 2112692)
100mm cell culture dishes (Thermo-Fisher Scientific)
Qiagen RNA extraction kit (Qiagen, cat no: 74104)
SYBR Green PCR Master Mix (Thermo-Fisher Scientific, cat no: A25742)
Verso cDNA synthesis kit (Thermo-Fisher Scientific, cat no: AB1453A)
PCR QuantStudio system (Applied Biosystems)
Phosphate-buffered saline 1x (PBS) (NaCl, KCl, Na2HP04, KH2P04, dH20)
Saline 0.9%
Phosphatase inhibitor cocktail (100x) (Cell Signaling Technology, cat no: 5870)
RIPA lysis buffer (Thermo-Fisher Scientific, cat no: 89900)
10% NuPAGE Bis-Tris protein gel (Thermo-Fisher Scientific, cat no: NP0301BOX)
NuPAGE MES SDS Running Buffer (Thermo-Fisher Scientific, cat no: NP00202)
Nitrocellulose transfer membrane (Fisher Scientific, cat no: IPVH00010)
Tris-base transfer buffer (Tris base, glycine, dH20)
Tris-buffered saline (TBS) (Tris base, NaCl, dH20)
Tween 20 (Sigma-Aldrich, cat no: P1379)
Nonfat dried milk
Normal Goat Serum (NGS) (Sigma-Aldrich, cat no: G9023)
NuPAGE LDS Sample Buffer (Thermo-Fisher Scientific, cat no: NP0008)
Bio-Rad protein standard ladder (Bio-Rad, cat no: 1610376)
BCA protein assay kit (Thermo-Fisher Scientific, cat no: 23225)
Chemiluminescent Substrate Reagent Kit (Fisher Scientific, cat no: WP20005)
Bio-Rad Chemidoc imaging system (Bio-Rad, cat no: 17001402)
37°C, 5% CO2 humidified incubator (Thermo-Fisher Scientific, cat no: 3110)
Protocol steps
Morphological evaluation of cachectic mouse muscles
-
1
To assess the degree of muscle wasting in tumor-bearing mice, fix the freshly harvested muscles (gastrocnemius, tibialis anterior) with 10% formalin and embed in paraffin blocks. Cut 5mm thick sections using a microtome.
-
2
Deparaffinize and rehydrate the muscle sections in a sequential order by placing the slides in the following order; 100% xylazine (3 minutes), 100% Xylazine (3minutes), 95% ethanol (5 minutes), 75% ethanol (5 minutes), 50% ethanol (5 minutes) and water. Perform the Hematoxylin and Eosin staining as per manufacturer instructions (Abcam, cat no: 245880). After staining, remove excess xylene and mount the section with a few drops of mounting medium. Place the coverslips and image the slide using a bright field microscope. Take approximately 5–10 bright-field images across the muscle section for each mouse.
-
3
Use Image J software to determine the average cross-sectional area from 5 independent bright-field images for each group of mice. Use at least 3 to 5 biological replicates for calculating the value for each group. Compare the plots of the muscle average cross-sectional area for each experimental group. Using these approaches we have demonstrated the a decrease in muscle cross sectional area in the gastrocnemius muscle of the KPC-tumor bearing mice as compare to healthy mice (Dasgupta et al., 2020).
Molecular evaluation of cachectic markers in skeletal muscle by RT-PCR and western blot
-
4
Extract from the muscle samples of each mouse group the total RNA from the gastrocnemius muscle using the Qiagen RNA extraction kit and the protein content with 40 μl RIPA cell lysis buffer supplemented with 1X of protease inhibitors cocktail. from the muscle samples of each mice group.
-
5
For the gene expression assay, prepare cDNA using the Verso cDNA synthesis kit as per the manufacturer instructions. Perform qPCR with the PCR QuantStudio system (Applied Biosystems) to assess the gene expression of MuRF1, Atrogin-1, INHBA, SELP, and CTSL (primers list in method section). Use the mouse 18S RNA marker for the internal control.
-
6
Use the ΔΔCt method as described previously (11846609) to calculate and compare gene expression levels between the control and tumor-bearing mice.
-
7
For evaluation of cachectic markers at the protein level, prepare the protein extract from the skeletal muscle using RIPA lysis buffer. First, prepare the muscle tissue homogenate by adding a small volume of liquid nitrogen and crushing the tissue with a mortar and pestle. Afterwards, add RIPA lysis buffer and extract the protein. For this, add RIPA Buffer to the homogenate (an approximately 60μl to 30 mg crushed tissue) and place it on ice for 30 minutes. Following incubation centrifuge the sample at 16000 g for 15 minutes at 4 C. Transfer the supernatant into fresh tubes and discard the pellet.
-
8
Quantify total protein using the BCA protein assay. Perform the immunoblotting assay by loading 30μgs of protein extract into 10% SDS-PAGE gel, transferring the gel to a nitrocellulose membrane for 1 hour. Block the membrane by placing it in 5% nonfat dried milk dissolved in TBST for 1 hour at room temperature (RT).
-
9
Incubate the membrane overnight at 4°C with the anti-mouse cachectic antibodies including, MurF1, Atrogin-1, and Myosin Heavy Chain (MyHC) diluted in TBST with 0.5% nonfat dried milk and 2% normal goat serum (NGS). Use either β-actin or GAPDH as a housekeeping gene. Then incubate the membrane for 1 hour at RT with HRP-conjugated or fluorescence-conjugated secondary antibodies diluted in TBST with 0.5% nonfat dried milk and 1% NGS. Shown in the material section are the clone and optimum dilution of each antibody used in the immunoblot assay.
Statistical analyses
Perform statistical analysis and create data plots using GraphPad Prism software. Be sure to use appropriate statistical tests for multiple comparisons.
COMMENTARY:
Background Information
Pancreatic cancer is a lethal disease with a 5-year survival of less than 10%. Pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic cancer (80–90%). Pancreatic ductal adenocarcinoma arises from preinvasive lesions, such as intraductal papillary mucinous neoplasia (IPMNs), and pancreatic intraepithelial neoplasia (PanINs) (Matthaei et al., 2013). Pancreatic intraepithelial neoplasia are categorized into four grades- PanIN-1A, PanIN-1B, PanIN-2, and PanIN-3. While PanIN-1 lesions display minimal atypia and are present in up to 40% of noncancerous pancreas, PanIN-2 lesions exhibit moderate atypia. The PanIN-3 lesions are highly atypic, detected in 30–50% of the pancreas with infiltrating ductal carcinoma, and are more closely associated with invasive carcinoma (Hruban et al., 2007). In most cases, PDAC arises from successive progression in PanINs, which accumulate genetic alterations at each successive lesion. The earliest genetic events in pancreatic cancer development, which include activation of KRAS oncogene by single point mutations in codon 12, occur in PanIN-1. Subsequently, PanIN-2 harbors inactivated tumor suppressor gene p16. With PanIN-3 there is accumulation of additional alterations in TP53, SMAD4, which translates into full-blown PDAC. The preclinical models in immunocompetent mice have been employed extensively for studying different aspects of cancer, including its development, progression, metastasis, resistance, dormancy, and cachexia (Galuschka et al., 2017). Genetically Engineered Mouse Models (GEMM) models have for decades been used to study pancreatic cancer biology. The GEMM models of pancreatic cancer are generated through the incorporation of specific pancreatic cancer-associated genetic mutations in KRAS, P16, TP53, and SMAD4. KrasG12D; Trp53R172H/+; Pdx-1-Cre (KPC) is one of the best-characterized and widely used GEMM models of PDAC. It expresses an oncogenic KRASG12D mutation and gain-of-function TP53R172H mutation under a pancreas-specific Pdx1-Cre driver (Hingorani et al., 2005; Hoffman, 2015). Typically, the pancreatic tumor appears in 10-week old KPC mice, with the median survival rate being five months. The KPC exhibit the key characteristic feature of cachexia. In end-point studies (mice reaching IACUC-approved end-points), KPC mice exhibit a significant reduction in overall weight that correlates with reduced hind limb tibialis anterior and gastrocnemius muscle mass as compared to littermate controls. In addition, reduced body weight correlates with reduced white adipose tissue mass (Talbert et al., 2019). Tumor allograft models of PDAC are now preferred over GEMM models because the allograft animals provide fast, efficient, and reproducible modeling of the cancer. For tumor allograft models, multiple syngeneic cell lines are generated from primary and metastatic PDAC GEMM tumors. These syngeneic cell lines are surgically implanted, either orthotopically or heterotopically, into immunocompetent mice. Thus, these orthotopic models provide a suitable platform that not only mimic the hallmark of primary human pancreatic tumors, but also display key characteristic features of cancer cachexia. Numerous investigators have utilized the KPC allograft orthotopic model of pancreatic cancer to study the cachectic phenotype in the skeletal muscle and adipose tissue (Dasgupta et al., 2020; Shukla et al., 2020a).
Understanding Results
Displayed on Figure 1 is C2C12 myotube morphology following CM treatment. As is evident from myotube diameter assessment, the CM from various tumor cell lines induces muscle atrophy. The data on Figure 2 illustrate the upregulation of cachexia-associated markers in the C2C12 following the KPC-8069-derived CM treatment, confirming myotube atrophy. While the appearance of few a cachectic markers begin within 24 hours, significant up-regulation of all essential genes (INHBA, SLEP, CTSL, and MuRF1) is confirmed only after the persistent presence of CM at 72 hours. Furthermore, while MurF1 and atrogin display significant upregulation, MyHC protein is attenuated in the CM-treated myotubes. These data validate pancreatic tumor-induced myotube atrophy in the in vitro assay. The in vivo study makes possible a post-necropsy evaluation of the cachectic phenotype (muscles, fat, heart, and carcass weight) in different groups of mice (Figure 3). The results reveal a significant decrease in total body weight, carcass weight, muscle, and fat weight in tumor-bearing mice as compared to healthy, non-tumor bearing controls. Furthermore, these data indicate a moderate reduction in heart weight tumor mice as compared to their control counterparts. Taken together these findings reveal that the KPC-derived pancreatic cancer cell line induces a cancer-specific cachexia phenotype including loss in whole body weight, carcass weight, muscle strength, and fat weight. Taking these models, we have demonstrated the impact of macrophage depletion on restoring cachectic phenotype in pancreatic tumor bearing mice (Shukla et al, 2020b). In our study, clodronate liposome treatment significantly attenuated circulating macrophage and in parallel attenuated muscle wasting in pancreatic tumor-bearing mice. In our study, KPC-tumor mice displayed a significant reduction in in gastrocnemius muscle and epididymal fat weight compare to healthy non-tumor bearing mice. Further, tumor-bearing mice displayed significantly attenuated grip strength and restricted mobility in rotarod measurement. Contrastingly, macrophage depleted mice restored muscle and epididymal fat weight with parallel increased in grip strength and mobility as compared to tumor-bearing mice. Together, these models are suitable for the investigation of molecular mechanism that underlie muscle wasting in pancreatic tumor-bearing mice model. In parallel, these models can be utilized for the assessment of the impact of chemo- or immunotherapies in PDAC models.
Figure 3. Grip strength meter for measuring grip strength in mice.
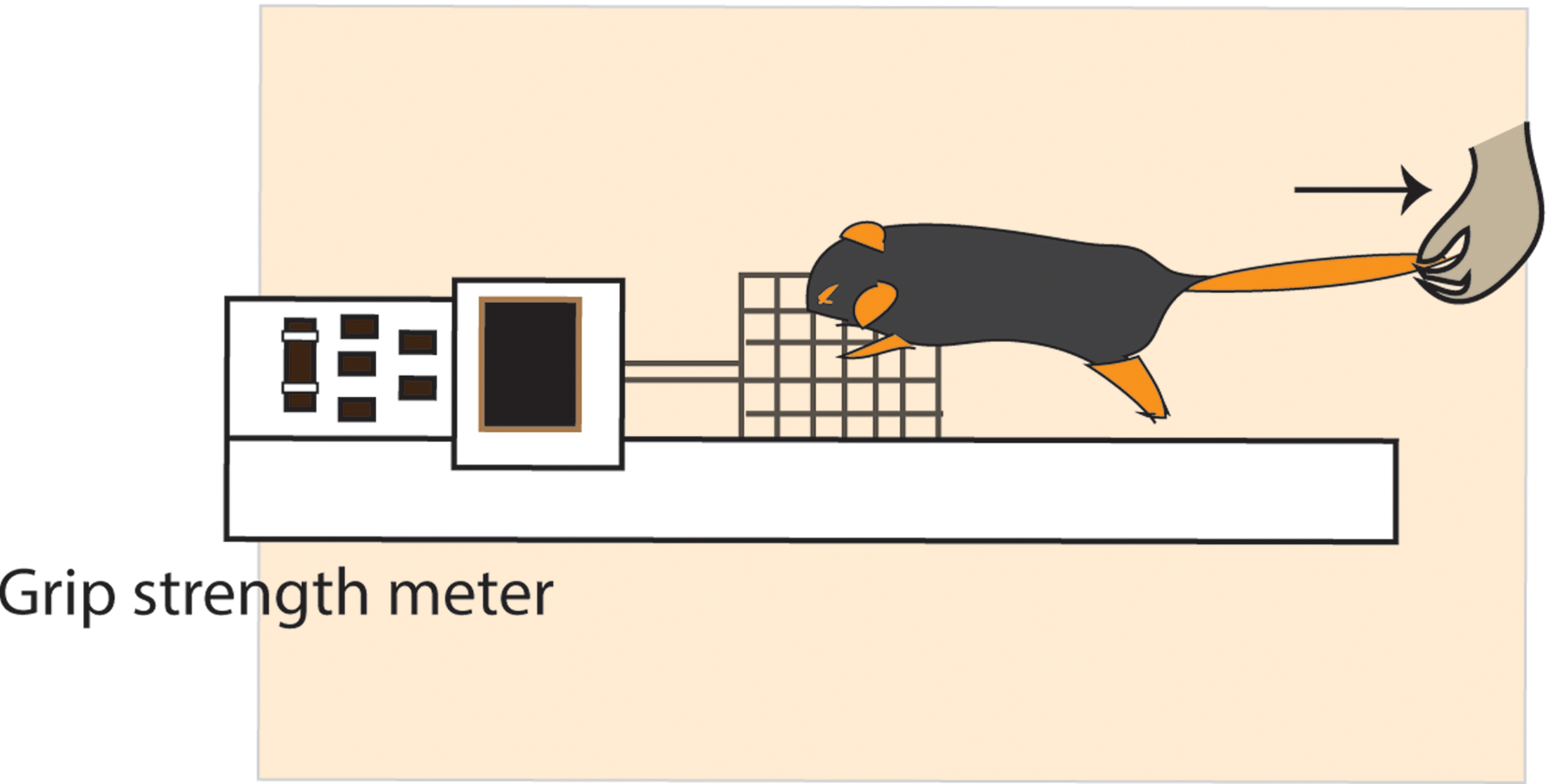
Pictorial representation of grip strength measurement of healthy or cachectic mice using grip strength meter.
Critical Parameter and Troubleshooting
There are several critical issues to considered when conducting the in vivo experiments. It is essential to ensure that the pancreatic cancer cell lines used for orthotopic implantation are in suitable condition and pathogen-free. It is highly recommended that growth of the cancer cells begins at least a week or two before the implantation to allow for ample time to observe cell behavior in the culture dishes, including their growth rate, viability, and morphology. The cells should be discarded, and another frozen vial seeded if any abnormalities are noted in morphology, growth kinetics, or health status. Preparation of the cancer cells is a critical step for ensuring the successful execution of the in vivo experiment, which itself is highly costly, time-consuming, and labor-intensive. Often, there is a delay or rejection of a tumor cell line if the tumor cells are stressed and have questionable viability at the time of implantation. It is also critical to confirm the establishment of tumors after implantation. In most cases, there is a 100% tumor take rate after implantation of murine pancreatic cancer cell lines. For macrophage depletion, it is important to confirm the systemic reduction of macrophages in peripheral blood isolated from submandibular capillaries before the end of the study. It is always important to make certain of the presence of tumors in the mice before initiating any procedure for evaluating the cachectic parameters. Discard all animals which fail to demonstrate tumor growth as their inclusion with complicate interpretation of findings. Evaluate the tumor appearance by palpating the abdominal part of the implanted mice one week after the tumor implantation. For tissue processing at the end of the study, rapid tissue freezing and timely formalin-fixation are critical as delays in these procedures can alter tissue morphology and architecture.
Special attention needs to be devoted to performing the grip strength, rotarod, and densitometer testing of tumor-bearing mice and for analyzing the results. For grip strength evaluation, it is crucial that every measurement be performed with proper positioning of the animal’s body on the metal mesh of the meter, being especially careful not to allow the mouse to grab the meter with their back legs. The animal’s body must always be maintained in a horizontal position during the measurement process. Such consistency will ensure accurate measurements and reliable, reproducible results. For rotarod testing, the mice must be carefully monitored while they are performing the test. In our experience, the rotarod apparatus sometimes fails to record the time of fall in the software program. Diligent observation of the animal’s performance will ensure detection of these sporadic failures, with manual recording of the time of fall saving labor and time.
Time Considerations
In contrast to the in vitro C2C12 assay, in vivo modelling and evaluation of cancer cachexia requires much more time. The in vitro preparation of the cancer cell lines for orthotopic implantation, which includes evaluation of the health status of the cells, could require up to 1 week to complete. The amount of time needed for the surgical implantation of pancreatic tumor cells line depends on the number of mice and groups and on the number of people involved in executing the procedure. Approximately 6 hours are needed for three people to complete surgery for 30 mice. The evaluation of body weight, forelimb grip strength measurement and motor coordination (rotarod test) each require about 60–90 minutes/mouse to complete. Post-necropsy processing and measurements depend on the number of mice and investigators, but can require a whole day of work. The time for morphological and molecular evaluation of the cachectic mice muscles using RT PCR and western blot analysis is usually completed in 2–3 days. Overall, an in vivo study typically requires to 45 days to execute all of the assays described in this unit in association with a 30-day study.
Grant Support
This study was supported by NCI-SPORE P50 CA127297 Career Development Award to KM.
Footnotes
Disclosure of Potential conflict of interest
The authors declare no potential conflicts of interest.
References
- Advani SM, Advani PG, VonVille HM, Jafri SH. (2018). Pharmacological management of cachexia in adult cancer patients: a systematic review of clinical trials. BMC Cancer, 18, 1174. doi: 10.1186/s12885-018-5080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attri KS, Mehla K, Shukla SK, Singh PK. (2018a). Microscale Gene Expression Analysis of Tumor-Associated Macrophages. Sci Rep, 8, 2408. doi: 10.1038/s41598-018-20820-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attri KS, Mehla K, Singh PK. (2018b). Evaluation of Macrophage Polarization in Pancreatic Cancer Microenvironment Under Hypoxia. Methods Mol Biol, 1742, 265–276. doi: 10.1007/978-1-4939-7665-2_23. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, … Glass DJ. (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science, 294, 1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Broberger C, Johansen J, Johansson C, Schalling M, Hokfelt T (1998). The neuropeptide Y/agouti gene-related protein (AGRP) brain circuitry in normal, anorectic, and monosodium glutamate-treated mice. Proc Natl Acad Sci U S A, 95, 15043–15048. doi: 10.1073/pnas.95.25.15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burattini S, Ferri P, Battistelli M, Curci R, Luchetti F, Falcieri E (2004). C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur J Histochem, 48, 223–233. [PubMed] [Google Scholar]
- Cone RD. (2005). Anatomy and regulation of the central melanocortin system. Nat Neurosci, 8, 571–578. doi: 10.1038/nn1455. [DOI] [PubMed] [Google Scholar]
- Cuenca AG, Cuenca AL, Winfield RD, Joiner DN, Gentile L, Delano MJ, … Moldawer LL. (2014). Novel role for tumor-induced expansion of myeloid-derived cells in cancer cachexia. J Immunol, 192, 6111–6119. doi: 10.4049/jimmunol.1302895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A, Shukla SK, Vernucci E, King RJ, Abrego J, Mulder SE, … Singh PK. (2020). SIRT1-NOX4 signaling axis regulates cancer cachexia. J Exp Med, 217, doi: 10.1084/jem.20190745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont N, Frenette J (2010). Macrophages protect against muscle atrophy and promote muscle recovery in vivo and in vitro: a mechanism partly dependent on the insulin-like growth factor-1 signaling molecule. Am J Pathol, 176, 2228–2235. doi: 10.2353/ajpath.2010.090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon K, Strasser F, Anker SD, Bosaeus I, Bruera E, Fainsinger RL, … Baracos VE. (2011). Definition and classification of cancer cachexia: an international consensus. Lancet Oncol, 12, 489–495. doi: 10.1016/S1470-2045(10)70218-7. [DOI] [PubMed] [Google Scholar]
- Galuschka C, Proynova R, Roth B, Augustin HG, Muller-Decker K (2017). Models in Translational Oncology: A Public Resource Database for Preclinical Cancer Research. Cancer Res, 77, 2557–2563. doi: 10.1158/0008-5472.CAN-16-3099. [DOI] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. (2001). Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A, 98, 14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossberg AJ, Scarlett JM, Marks DL. (2010). Hypothalamic mechanisms in cachexia. Physiol Behav, 100, 478–489. doi: 10.1016/j.physbeh.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson SE, Makhijani N, Mace TA. (2018). Pancreatic Cancer-Induced Cachexia and Relevant Mouse Models. Pancreas, 47, 937–945. doi: 10.1097/MPA.0000000000001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, … Tuveson DA. (2005). Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell, 7, 469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Hoffman RM. (2015). Patient-derived orthotopic xenografts: better mimic of metastasis than subcutaneous xenografts. Nat Rev Cancer, 15, 451–452. doi: 10.1038/nrc3972. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Maitra A, Kern SE, Goggins M (2007). Precursors to pancreatic cancer. Gastroenterol Clin North Am, 36, 831–849, vi. doi: 10.1016/j.gtc.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ito A, Imada R, Sato M, Kawabe Y, Kamihira M (2017). In vitro drug testing based on contractile activity of C2C12 cells in an epigenetic drug model. Sci Rep, 7, 44570. doi: 10.1038/srep44570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marceca GP, Londhe P, Calore F (2020). Management of Cancer Cachexia: Attempting to Develop New Pharmacological Agents for New Effective Therapeutic Options. Front Oncol, 10, 298. doi: 10.3389/fonc.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks DL, Ling N, Cone RD. (2001). Role of the central melanocortin system in cachexia. Cancer Res, 61, 1432–1438. [PubMed] [Google Scholar]
- Matthaei H, Dal Molin M, Maitra A (2013). Identification and analysis of precursors to invasive pancreatic cancer. Methods Mol Biol, 980, 1–12. doi: 10.1007/978-1-62703-287-2_1. [DOI] [PubMed] [Google Scholar]
- McMahon DK, Anderson PA, Nassar R, Bunting JB, Saba Z, Oakeley AE, Malouf NN. (1994). C2C12 cells: biophysical, biochemical, and immunocytochemical properties. Am J Physiol, 266, C1795–1802. doi: 10.1152/ajpcell.1994.266.6.C1795. [DOI] [PubMed] [Google Scholar]
- Michaelis KA, Zhu X, Burfeind KG, Krasnow SM, Levasseur PR, Morgan TK, Marks DL. (2017). Establishment and characterization of a novel murine model of pancreatic cancer cachexia. J Cachexia Sarcopenia Muscle, 8, 824–838. doi: 10.1002/jcsm.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KT, Chee A, Trieu J, Naim T, Lynch GS. (2012). Importance of functional and metabolic impairments in the characterization of the C-26 murine model of cancer cachexia. Dis Model Mech, 5, 533–545. doi: 10.1242/dmm.008839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SK, Dasgupta A, Mehla K, Gunda V, Vernucci E, Souchek J, … Singh PK. (2015). Silibinin-mediated metabolic reprogramming attenuates pancreatic cancer-induced cachexia and tumor growth. Oncotarget, 6, 41146–41161. doi: 10.18632/oncotarget.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SK, Gebregiworgis T, Purohit V, Chaika NV, Gunda V, Radhakrishnan P, … Singh PK. (2014). Metabolic reprogramming induced by ketone bodies diminishes pancreatic cancer cachexia. Cancer Metab, 2, 18. doi: 10.1186/2049-3002-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SK, Markov SD, Attri KS, Vernucci E, King RJ, Dasgupta A, … Mehla K. (2020a). Macrophages potentiate STAT3 signaling in skeletal muscles and regulate pancreatic cancer cachexia. Cancer Lett, 484, 29–39. doi: 10.1016/j.canlet.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SK, Markov SD, Attri KS, Vernucci E, King RJ, Dasgupta A, … Mehla K. (2020b). Macrophages potentiate STAT3 signaling in skeletal muscles and regulate pancreatic cancer cachexia. Cancer Lett, doi: 10.1016/j.canlet.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert EE, Cuitino MC, Ladner KJ, Rajasekerea PV, Siebert M, Shakya R, … Guttridge DC. (2019). Modeling Human Cancer-induced Cachexia. Cell Rep, 28, 1612–1622 e1614. doi: 10.1016/j.celrep.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Saxel O (1977). Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature, 270, 725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- Zhong X, Pons M, Poirier C, Jiang Y, Liu J, Sandusky GE, … Zimmers TA. (2019). The systemic activin response to pancreatic cancer: implications for effective cancer cachexia therapy. J Cachexia Sarcopenia Muscle, 10, 1083–1101. doi: 10.1002/jcsm.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]


