Abstract
Zika virus (ZIKV) is a kind of flavivirus emerged in French Polynesia and Brazil, and has led to a worldwide public health concern since 2016. ZIKV infection causes various neurological conditions, which are associated with fetus brain development or peripheral and central nervous systems (PNS/CNS) functional problems. To date, no vaccine or any specific antiviral therapy against ZIKV infection are available. It urgently needs efforts to explore the underlying molecular mechanisms of ZIKV-induced neural pathogenesis. ZIKV favorably infects neural and glial cells specifically astrocytes, consequently dysregulating gene expression and pathways with impairment of process neural cells. In this study, we applied a model for ZIKV replication in mouse primary astrocytes (MPAs) and profiled temporal alterations in the host transcriptomes upon ZIKV infection. Among the RNA-sequencing data of 27,812 genes, we examined 710 genes were significantly differentially expressed by ZIKV, which lead to dysregulation of numerous functions including neurons development and migration, glial cells differentiation, myelinations, astrocytes projection, neurogenesis, and brain development, along with multiple pathways including Hippo signaling pathway, tight junction, PI3K-Akt signaling pathway, and focal adhesion. Furthermore, we confirmed the dysregulation of the selected genes in MPAs and human astroglioma U251 cells. We found that PTBP1, LIF, GHR, and PTBP3 were upregulated while EDNRB and MBP were downregulated upon ZIKV infection. The current study highlights the ZIKV-mediated potential genes associated with neurodevelopment or related diseases.
Author summary
Zika virus (ZIKV) infection causes serious neurological disorders of central and peripheral nervous system, and fetal brain development disorders including microcephaly. There are still uncovered explorations for the underlying molecular mechanism of ZIKV-infected pathogenesis. This study reveals a series of dysregulation of neuropathic genes mRNA and protein expression in mouse and human astrocytes upon ZIKV infection. As an ideal ZIKV infection model in mouse primary astrocytes (MPAs), RNA-seq was performed to profile transcriptome alteration by ZIKV infection. Bioinformatics analysis demonstrated the significant alterations of the 710 genes that were linked to glial cell differentiation and projection, neurogenesis and migration of neurons, myelination, as well as synaptic control. Among the top selected differentially expressed genes, such as PTBP1, LIF, GHR, PTBP3, EDNRB, and MBP, the mRNA and protein expressions were confirmed to identify the dysregulation of the transcriptome in MPAs upon ZIKV infection. Furthermore, ZIKV infection altered the mRNA and protein expression of these astrocytic genes involved in neurodevelopment in U251 cells following the analysis of the transcriptome. In conclusion, the alteration of astrocytic gene functions or associated-pathways suggest a novel clue of a mechanism involved in the ZIKV-induced neurodevelopment disorders.
Introduction
Zika virus (ZIKV) belongs to the Flaviviridae family, Flavivirus genus, and is transmitted to humans through Aedes mosquitoes bite and sexual intercourse [1–3]. ZIKV infection causes Zika fever that results in serious health issues for being potentially associated with fetus neurodevelopment disorders such as microcephaly and other central and peripheral nervous systems (CNS/PNS) neurological conditions including myelitis, encephalitis, polyneuritis, and demyelinating polyneuropathy [4–6]. ZIKV was isolated in the Zika forest of Uganda in 1947 from a monkey and the anti-ZIKV neutralizing antibodies in human serum were first reported in 1952 [7]. The first link between neurological diseases and ZIKV infection was described in the French Polynesia during the ZIKV outbreak in 2013–2014 [8,9]. In the near past between 2015 and 2016, an increase in the microcephaly prevalence reported in some areas of Brazil confirmed ZIKV transmission [10]. However, different approaches have been made to reveal the underlying links of microcephaly and ZIKV infection during pregnancy [11,12]. Additionally, ZIKV infection is also responsible for other neurological and brain abnormalities [10,11,13–15].
Differentiation failure or loss of the neuronal progenitor cell growth impairs neural development to causes microcephaly [16]. An increase in the fetal malformation cases upon ZIKV infection (congenital ZIKV syndrome), such as microcephaly and hydrocephaly, placental insufficiency in utero, stillbirth, and spontaneous abortion, indicating the capacity of ZIKV to readily infect the developing human brain, were reported as ZIKV infection deleterious consequences [17]. Most of these diseases are linked to the problems that occur during fetus brain development or CNS/PNS function [6]. To date, no antiviral therapy or vaccines are available for zika virus infection [18]. As a result, the relationship between neural disorders and ZIKV infection is of high concern.
Besides neural cells, glial cells such as microglia and astrocytes are the significant target cells for ZIKV infection in brain [19–22]. Glial cells are rich in astrocytes and found in the capillary’s vicinity as an important component of the blood-brain barrier (BBB). Thus, ZIKV could target astrocytes instantly after invading the CNS. Upregulation of pro-inflammatory molecules production upon Toll-like receptor (TLR) pathway activation in glial cells is a vital factor for neuro-inflammation which is associated with many other neurological disorders [23]. As such, ZIKV infection in glial cells may cause BBB leakage by producing pro-inflammatory molecules which leads to neuro-inflammation [24]. In addition, as astrocytes are exceedingly involved in synaptic and axonal guidance, other microglia’s coordination in postnatal synaptogenesis and neurons survival to death with genetic and functional alterations may result in various neurological disorders [25,26]. Astrocytes from different brain regions convert or help in-migration of new neuron through the rostral migratory stream organization during early developmental stages [25,27,28]. Based on this, the ZIKV infection in astrocytes is considered to be an important site in pathological neural disorders and brain abnormalities.
Emerging studies have been reported on virus-induced variations in the transcriptomic profiles of virus-treated cell lines and organisms [29,30]. Phenotypically, transcriptome profiling of ZIKV infection in mouse cortical tissues, human brain organoids, and neural progenitors have disclosed the dysregulation of many genes and pathways that are linked with cell death, DNA repair or replication, cell cycle, viral response, metabolism, and transcription [31–33]. The dysregulation of transcriptomes by ZIKV varies from Dengue virus (DENV) infection induced dysregulation [34]. ZIKV infection in embryonic mouse cortex and human neural progenitors dysregulate a number of known microcephaly-associated genes such as MCPH1 (Microcephalin), CENPF (Centromere Protein F), TBR2 (T-box brain protein 2), RBBP8 (RB binding protein 8), AXL (AXL receptor tyrosine kinase), and ASPM (Assembly factor for spindle microtubules) [31,35]. However, it remains unclear how such dysregulation may contribute to ZIKV-related pathophysiology in astrocytes.
This study evaluated the alteration of neuropathic genes mRNA and protein expression in mouse primary astrocytes (MPAs) and human astroglioma U251 cells upon ZIKV infection. We conducted an ideal ZIKV infection model in MPAs isolated from neonatal mice, followed by RNA-seq performance to profile ZIKV-induced transcriptome alterations. Briefly, we identified 710 significantly dysregulated genes from the RNA-seq of total of 27,812 genes upon ZIKV infection in MPAs, which were associated with astrocytic cell synaptic control, migration, cell growth and other brain development conditions under bioinformatic functional analyses. Subsequently, the most differentially dysregulated genes associated with neural function in MPAs were selected in ZIKV-infected MPAs. In detail, we selected top genes from the dysregulated pool and identified that LIF (Leukemia inhibitory factor), PTBP3 (Polypyrimidine tract binding protein 1), and GHR (Growth hormone receptor) were upregulated while EDNRB (Endothelin receptor type B) and MBP (Myelin basic protein) were downregulated upon ZIKV infection in both MPAs and U251 astrocytes. Another member from PTB family called PTBP1 plays a critical role in astrocyte-neuron conversion during neurodevelopment [27], was also detected along with PTBP3. Our findings systematically revealed dysregulation in a cluster of neural genes by ZIKV infection in astrocytes, which provides novel clues for the mechanism involved in ZIKV-associated neurodevelopment disorders. Datasets presented in this study could be a critical source to understand the molecular pathogenesis of ZIKV-induced abnormalities in fetal brain, and to develop an effective therapeutic approach against ZIKV infection and its consequences.
Materials and methods
Animal study
Wild type (WT) C57BL/6J mice were purchased from the Hubei Provincial Center for Disease Control and Prevention (Wuhan, Hubei, China). The mice were housed under specific pathogen-free (SPF) conditions in individually ventilated cages.
Ethics statement
The animal study was approved by the Institutional Review Board of the College of Life Sciences, Wuhan University and was conducted in accordance with the guidelines for the protection of animal subjects (permit number: WDSKY0201901).
Isolation of MPAs
Postnatal day 3 (P3) mice were sacrificed by decapitation for isolation of cortical astrocytes or also called MPAs. Cerebral cortices were carefully isolated and after processing to dissociate to a single cell suspension, and seeded in T75 tissue culture flask prior coated with Poly-D-lysine at a dose of 50 μg/ml, as previously reported [36]. After 7 to 8 days of incubation, confluent astrocytes monolayers were shaken on an orbital shaker for 40 minutes at 180 rpm to remove microglia cells, and then replaced culture medium following a second time shaking at 240 rpm for 6 hours to remove oligodendrocytes precursor cells (OPCs). Astrocyte monolayers were washed with phosphate buffer saline (PBS) and transferred into a new T75 cultural flask, and incubated for 12 to 14 days replacing fresh medium for every 2 to 3 days until usage [36]. The isolation of pure astrocytes and removal of other cells like microglia and OPCs was confirmed by a morphological overview of mix cortical cells at different time points after isolation and immunofluorescence microscopy against the astrocyte-specific marker GFAP [36].
Cell lines and culture
Mouse primary astrocytes (MPAs) were isolated from WT C57BL/6J mice brain cortices as described above [36]. Human astroglioma (U251), African green monkey kidney epithelial cells (Vero) (#CCL-81), Human lung cancer cell line (A549), and Aedes albopictus mosquito cell line (C6/36) (#CRL-1660) were purchased from ATCC (Manassas, VA, USA). All these cells were cultured in DMEM (Gibco; Grand Island, NY, USA) supplemented with 10% FBS (Gibco) with100 U/ml Penicillin/Streptomycin (Gibco) at 37°C and 5% CO2, except that C6/36 cells were maintained in RPMI1640 (Gibco) at 30°C and 5% CO2 [37].
Reagents
Rabbit anti- EDNRB (A2980), MBP (A11162), LIF (A1288), GHR (A14735) and β-actin antibodies were purchased from ABclonal Technology (Wuhan, China). Rabbit anti-PTBP1 (12582-1-AP) antibody was purchased from Proteintech Group (Rosemont, IL, USA). Rabbit anti-dsRNA [J2] (Ab01299) was purchased from Absolute Antibody (Wilton, UK). Rabbit anti-ZIKV-protein (GTX133312) was purchased from GeneTex (Irvine, CA, USA). Mouse anti-GFAP (E4L7M) was purchased from Cell Signaling Technology (Beverly, MA, USA).
Zika virus propagation and isolation
ZIKV Asian strain isolate z16006 (GenBank accession number, KU955589.1) obtained from Institute of Pathogenic Microbiology, Center for Disease Control and Prevention of Guangdong (Guangzhou, China) as previously reported [37]. ZIKV was propagated in C6/36 cell line. Briefly, C6/36 cells were cultured in RPMI1640 containing 10% FBS and 1% Penicillin/Streptomycin solution. Cells were infected with ZIKV at an MOI (Multiplicity of Infection) of 0.1 (diluted in 1 ml RPMI1640). After incubation for 2 h at 32°C, 5% CO2, 4 ml fresh medium was added to the culture and continued the incubation for additional 4 days. Cell supernatants were harvested, centrifuged and filtrated using a 0.45 μm filter. The stock was aliquoted in 1.5 ml Eppendorf tubes for single time thawing [37]. The determination of virus titer was performed by serial dilutions for the infectivity in Vero and A549 cells detected by standard plaque assay and further amplified by TCID50 (50% Tissue Culture Infective Dose) [38–40].
RNA-seq and bioinformatics analysis
MPAs were infected with ZIKV at MOI = 2 and harvested at 24 h p.i. The detailed methodology of the transcriptomic study is present in S1 Text. Briefly, the total RNA samples were extracted in TRIzol reagent and cDNAs were prepared. The library preparations (S1 Table) were sequenced on an Illumina Hiseq 2500 PE150 platform (Illumina; San Diego, CA, USA) in Novogene Bioinformatics Institute (Beijing, China). The raw reads of RNA-seq sequences are treated by the fastp (https://github.com/OpenGene/fastp). Clean reads are aligned to the hg38 genome with HISAT2 [41], duplicates are removed using Samtools [42]. Genes identified as rRNAs, and pseudogenes are removed. Transcript assembly is determined using StringTie and Ballgown is used to identify differentially expressed isoforms gene [43]. Then, the differential expression analysis is performed using DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html), using a fold change (FC) cutoff of 1.5 and padj-value cutoff of 0.05. Gene set enrichment analysis was done using QuSAGE [44]. The gene symbols were converted to ensemble IDs using BioDBnet (https://biodbnet-abcc.ncifcrf.gov/db/dbOrthoRes.php). For expression-based analysis the data were subjected to an online ImageGP tool for converting numerical data into heatmaps (http://www.ehbio.com/ImageGP/index.php/Home/Index/PHeatmap.html), and volcano plot (http://www.ehbio.com/ImageGP/index.php/Home/Index/Volcanoplot.html). The significantly differentially expressed genes selected were subjected to functional annotation analysis (gene ontology) using online ToppGene Suit (https://toppgene.cchmc.org/enrichment.jsp) and DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/summary.jsp) to find the most related KEGG pathways. The scripts (transcriptomic data) are available online in the NCBI Sequence Read Archive (SRA) database with the SRA (BioProject) accession number: PRJNA692303.
RNA quality and quantity, relative quantitative real-time PCR
Total cellular RNA was extracted from mock and ZIKV-infected MPAs and U251 cells using TRIzol reagent (Invitrogen; Carlsbad, CA, USA), following the manufacturer’s instructions. A detailed explanation of methodology is provided in S2 Text. Briefly, the quality of extracted RNA was assessed by spectrophotometry (NanoDrop; Thermo Scientific) (A260/A280) [45,46] and cDNAs were prepared using HiScript II Q Select RT SuperMix, according to the manufacturer’s instructions. (http://www.vazymebiotech.com/products_detail/productId=80.html). Relative quantitative Real-time polymerase chain reaction (RT-qPCR) analysis was performed on Light Cycler 480 (Roche, Basel, Switzerland) using SYBR Green Real-time PCR master mix (Bio-Rad; Hercules, CA, USA) as previously described [45,47]. Briefly, in a reaction mixture of 10 μl SYBR, 1 μl cDNA diluted template, 1 μl specific gene primers (0.5 μl forward and reverse primer, 10 μM each), and RNase-free water to complete 20 μl as a final volume. The amplification protocol used as follows: denaturation at 95°C for 30 s, 40 cycles of 95°C for 5 s, 55°C for 30 s, and 72°C for 30 s), and a final extension step. The melting curve was obtained by heating the amplicon from 65°C to 95°C at 0.5°C s−1. The relative expression level was based on 3 biological replicates mean at each time point using the 2−ΔΔCT approach [48]. GAPDH was used as an internal reference control in this study. Real-time PCR primers were designed by Primer Premier 5.0 (Premier Biosoft; Palo Alto, California, USA), and the sequences are provided in Table 1.
Table 1. Information of primers sequences for qPCR assay in this study.
| Primer name | Sequences (5’ to 3’) | |
|---|---|---|
| Human | Mouse | |
| qRT GAPDH F | ACCCAGAAGACTGTGGATGG | GGGATGATGGAGGACGTGAT |
| qRT GAPDH R | TCAGCTCAGGGATGACCTTG | CCAATACGGCCAAATCCGTT |
| qRT ZIKV NS5 F | GGTCAGCGTCCTCTCTAATAAACG | GGTCAGCGTCCTCTCTAATAAACG |
| qRT ZIKV NS5 R | GCACCCTAGTGTCCACTTTTTCC | GCACCCTAGTGTCCACTTTTTCC |
| qRT LIF F | ATCTGTCCATCCCAACAGCA | TGGAGCTGTATCGGATGGTC |
| qRT LIF R | ATCCTGGACAAGGGTGAGTG | GCATTGAGCTTGACCTGGAG |
| qRT EDNRB F | AGATGTGTAAGCTGGTGCCT | TTAGCCCTGTGTTCGTCACT |
| qRT EDNRB R | AACCACAGAGACCACCCAAA | GTTAAAGCTCTCGGGCTTGG |
| qRT GHR F | ACTCAGCAGCCCAGTGTTAT | AGCGAAGTCCTCCGTGTAAT |
| qRT GHR R | ATATGGGCAGCTTGGTGAGT | GGATCCTCTGAAGCTGGTGA |
| qRT MBP F | GAAGGCCAGAGACCAGGATT | ACTTGCCATCCATCCTGTGA |
| qRT MBP R | TGAATCCCTTGTGAGCCGAT | CCGGATCCCTGAGTTCTCTC |
| qRT PTBP1 F | AGGTCACCAACCTCCTGATG | TCTAAGTTTGGCACCGTCCT |
| qRT PTBP1 R | GGGTCACCGAGGTGTAGTAG | TGAGACTGGTGAGCTTGGAG |
| qRT PTBP3 F | CAGATGGCGGATGCAAATCA | CCATCGCATTTCCTCAAGCA |
| qRT PTBP3 R | TGGTCTTCTTGTCCCTCTCG | AATGGCCATCCTTCCACTGA |
qRT: quantitative Real-time PCR; F: forward; R: reverse.
Western blot
Cell lysates were extracted in cell lysis buffer as previously described [49] (https://www.novusbio.com/support/support-by-application/western-blot/protocol.html). The protein concentrations were determined by Bradford assay (Bio-Rad, Hercules, CA, USA), subjected to 12% SDS-PAGE gel and transferred onto a nitrocellulose (NC) membrane. The membrane was blocked with 5% nonfat dried skim milk and incubated with specific antibodies. Protein bands were detected by using enhanced chemiluminescence Luminescent Image Analyzer (Fujifilm LAS-4000, Fujifilm, Tokyo, Japan). β-actin was used as an internal control refers to the target genes [37] (http://docs.abcam.com/pdf/protocols/general-western-blot-protocol.pdf).
Bright-field microscopy
Morphological representation of U251 cells treated with ZIKV and Mock were observed for cytopathic effect (CPE) at 12, 24, 48, and 60 h p.i. using bright field Nikon TE-2000 inverted microscope (Tokyo, Japan). All images’ brightness and contrast were adjusted in PowerPoint 2016 [6].
Cell viability
Cell viability was determined using the Cell Counting Kit 8 (CCK8) according to the manufacturer’s instructions (Dojindo, Japan). ZIKV-treated cells for indicated times were provided with 500 μl of CCK8 solution in each well in 12-well plate and incubated for 30 minutes at 37°C. Cell injury absorbance was measured at 450 nm [6,47].
Confocal microscopy
The confocal microscopy was performed according to the method previously reported [6]. Briefly, MPAs and U251 cells were seeded on 20-mm, and mock-infected or with ZIKV- infected for different time intervals. The cells were washed with PBS, fixed with 4% formaldehyde for 15 min and then permeabilized with 0.2% Triton X-100 for 5 min at room temperature (RT). After three times wash with PBS, the cells were blocked with 5% bovine serum albumin (BSA) for 1 h, subsequently incubated with specific primary antibodies overnight at 4°C. The cells were washed and incubated with FITC-conjugated or Cy3-conjugated goat anti-rabbit IgG, FITC-conjugated goat anti-mouse IgG (Proteintech Group) secondary antibodies for 1 h at RT. Finally, the cells were washed and mounted for Immunofluorescence (IF) assay by confocal laser scanning microscopy (Fluoview FV1000; Olympus, Tokyo, Japan) after nuclei staining with 4,6-diamidino-2-phenylindole solution (DAPI) [6,37].
Statistical analysis
All experiments were repeated at least three times. The results are presented as means ± SD unless stated otherwise. Statistical significance was determined by paired Student’s t-test using the Prism 8 software (GraphPad Software Inc., San Diego, CA, USA). A P ≤ 0.05 was considered statistically significant.
Results
The replication of ZIKV in MPAs
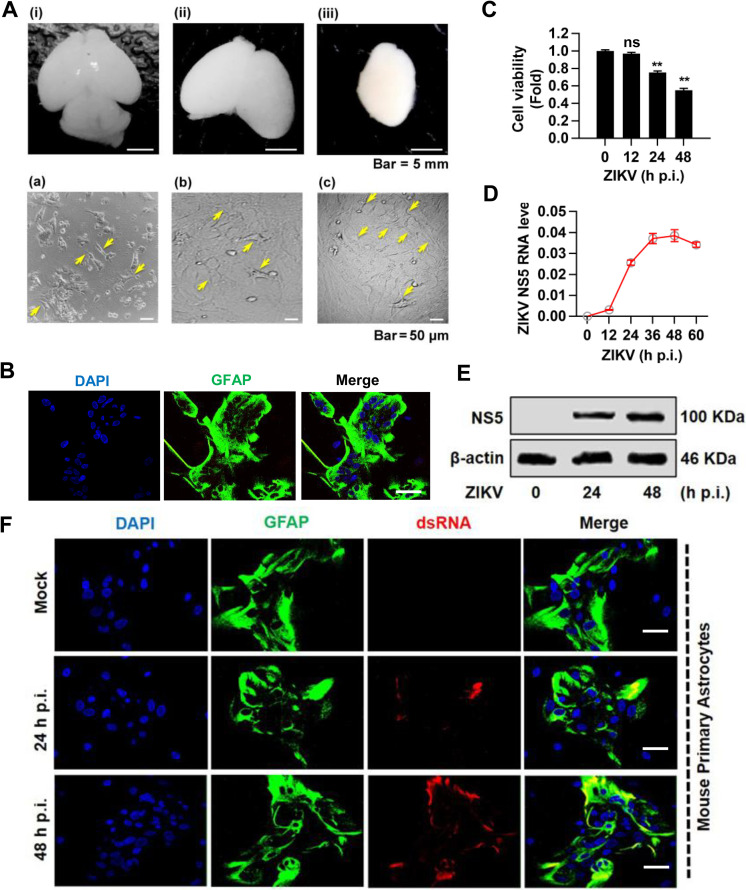
Considering astrocytes as one of the most significant target cells for ZIKV infection in the brain, we assessed the infection and replication of ZIKV in MPAs. Initially, we isolated and cultured MPAs from mouse brain cortex by standard protocol (Fig 1A). To confirm the purified isolation of MPAs, the cells were stained with astrocyte marker GFAP (glial fibrillary acidic protein). Immunofluorescence (IF) staining results revealed that nearly all isolated cells were GFAP positive (Fig 1B). We further evaluated ZIKV infection and replication in MPAs. The increasing cytopathic effect (CPE) was measured by cell viability assays (Fig 1C), ZIKV replication was determined by an obvious increase of viral NS5 RNA level detected by qPCR (Fig 1D), while expression of viral NS5 protein detected by Western blot (Fig 1E), suggesting a robust viral replication in MPAs upon ZIKV infection. Furthermore, IF analysis showed the ZIKV replication by the continuous increase of viral dsRNA in MPAs at 24 and 48 h p.i. (Fig 1F). Overall, these results illustrated that MPAs are susceptible to ZIKV infection and elicited well-observed replication, which allows MPAs as a kind of considerable model for ZIKV infection.
Fig 1. The isolation, culture, and identification of ZIKV infection in mouse primary astrocytes (MPAs).
(A) Postnatal (P3) mouse dissection for cortical astrocytes isolation. (i) Whole brain; (ii) Brain after cerebellum and olfactory bulbs removal; (iii) Cortices isolation by peeling off the meninges, a plate-like structure on the brain cortex, scale bar 5 = mm. Culture of mouse primary astrocytes from mouse brain cortex. (a) The yellow arrows indicate single cell astrocytes attached to the bottom of the culture flask and floating are the dead neurons in supernatant. (b) Astrocytes layer is forming and confluent after vigorous shaking to remove OPCs and Microglia cells. (c) Primary astrocytes layer shows a high density of the cells after 2 weeks of the first split, scale bar = 50 μm. (B) Identification of mouse primary astrocytes with rabbit anti-GFAP (green) marker and DAPI (blue) for nuclei by confocal microscopy, scale bar = 20 μm. (C-F) MPAs were mock-treated or infected with ZIKV (MOI = 2) for indicated times. Cell viability was measured by CCK8 assay (C), ZIKV viral NS5 mRNA (D) and NS5 protein expression (E) were determined by qPCR with GAPDH as an internal control and Western blot with β-actin as internal control, respectively, and viral dsRNA (a replication intermediate) was immunoprobed with dsRNA-antibody (red), along with GFAP (green) for astrocytes and DAPI (blue) for nuclei by Immunofluorescent (IF), scale bar = 20 μm (F). The underlying numerical data for “Fig 1C and 1D” is provided in the supporting file “S1 Data”.
Profile of mRNA expression in ZIKV-infected MPAs
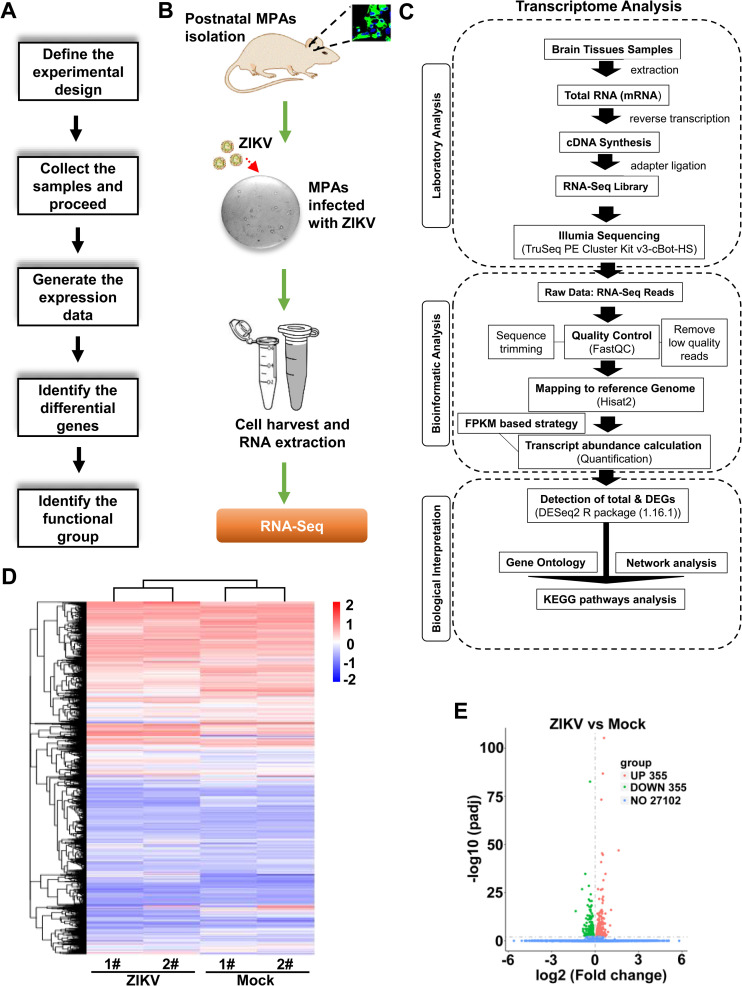
To explore the transcriptional differences by ZIKV infection in MPAs, RNA-Seq was performed and the transcriptional profile for altered genes was designed (Fig 2A). Briefly, we cultured MPAs isolated from the mouse brain cortices and then infected them with ZIKV, following cell harvest and RNA-Seq (Fig 2B). The transcriptome analysis was proceeded according to laboratory analysis, bioinformatic analysis, and biological interpretation (Fig 2C). In total 27,812 genes represented by a heatmap (Fig 2D), we identified 710 significantly dysregulated genes upon ZIKV infection in MPAs based on fold change with cut off value of 1.5 (S2 Table). Among these 710 genes, 355 genes were significantly up-regulated and 355 genes were down-regulated upon ZIKV infection in MPAs (Fig 2E). Thus, the data demonstrated that ZIKV infection altered transcriptome in MPAs.
Fig 2. Expression profile for altered mRNAs upon ZIKV infection in MPAs.
(A) A basic workflow diagram for the differential gene expression profile starting from experimental design to using bioinformatic tools. (B) Sketch diagram of main experiments from isolation of MPAs to RNA-Seq. (C) Flow sheath of transcriptome analysis of MPAs upon ZIKV infection. (D) Hierarchical clustering Heatmap of a distinguishable mRNA expression profiling in mouse primary astrocyte cells of ZIKV-infected group relative to Mock group (each group, n = 2). (E) Volcano analysis for a profile of mRNAs expressions of ZIKV-infected relative to Mock group. Red dots illustrate up-regulated genes, green dots represent down-regulated genes, while blue dots show insignificant genes. The underlying numerical data for “Fig 2D and 2E” is provided in the supporting file “S1 Data”.
Functional categorization of the differentially expressed genes by ZIKV in MPAs
Transcriptome analyses provide a better understanding of how the genome is transformed into functional proteins. ZIKV-treated human brain organoids, neural progenitors, and mouse cortical tissues including primary astrocytes exposed numerous altered gene ontologies or functions and pathways associated with cell death, metabolism, transcription, DNA replication and repair, cell cycle, and viral responses [50]. Since the transcriptome alteration was a conspicuous molecular phenotype of ZIKV infection in MPAs, we further showed the top 30 dysregulated basic functions of the related genes, and assigned the altered genes ontologies to three classes: biological process (BP, red color), cellular component (CC, dark gray color), and molecular function (MF, blue color) as reported previously [51], with 30 functional terms and genes in the annotation (Fig 3A). The dot plot also showed the enrichment score values of the top 30 most significant enrichment terms (Fig 3B), suggesting that ZIKV induces a change in the expression of genes associated with neurogenesis, neuron differentiation, development, migration, and maturation, astrocytes projection, and brain development in infected cells.
Fig 3. Functional categorization of the differentially expressed genes (DEGs) in ZIKV-infected MPAs.
(A) The DEGs were assigned to three gene ontology classes: biological process (BP, red color), cellular component (CC, dark grey color), and molecular function (MF, blue color), with top 30 functional terms. (B) The dot plot displays the enrichment score values of the top 30 most significant enrichment terms. (C) Pathway enrichment (top 20) involved in significant genes reported in ZIKV-infected relative to Mock mouse primary astrocyte cells from RNA-seq analysis. KEGG was used as the pathway database. Enrichment scores are based on (A). (D) Top 20 enriched KEGG pathways among the annotated DEGs in 2 groups comparisons. The Y-axis represents KEGG pathways. Low padj values are shown in red, and high padj values are depicted in blue. Pathways with padj values less than 0.05 are significantly enriched. The size of the spot reflects the number of DEGs, and the color of the spot corresponds to different padj-value ranges. The underlying numerical data for “Fig 3A, 3B, 3C, and 3D” is provided in the supporting file “S1 Data”.
Furthermore, we showed the pathway enrichment results (top 20) involved in significant genes between mock and ZIKV-infected MPAs from RNA-seq analysis. KEGG was used as the pathway database and enrichment scores was depicted (Fig 3C). The comparisons of top 20 enriched KEGG pathways among the annotated differentially expressed genes in 2 groups were shown (Fig 3D), which reveals that ZIKV infection was related to multiple pathways including Hippo signaling pathway, focal adhesion, PI3K-Akt signaling pathway, and tight junction. Thus, the functional categorization is associated with brain development, and neurogenesis from differentially expressed genes by ZIKV in MPAs.
ZIKV infection dysregulate neural genes expression in MPAs
Bioinformatic analysis of RNA-seq data illustrated that ZIKV can potentially dysregulate the astrocyte genes. We summarized the top 10 genes that were significantly differentially expressed (up- and down- regulated) among the 710 genes, which were significantly dysregulated in ZIKV-infected MPAs at 24 h p.i. compared to mock-treated cells. The comparisons of gene expression between two groups are presented by fold change (Table 2). It is known that these selected genes were directly or indirectly involved in neurological conditions or brain development [52,53], which is consistent with the fact that ZIKV infection dysregulates a number of known genes in the embryonic mouse cortex and human neural progenitors that are associated with neurodevelopment or neural disorders [31].
Table 2. The enrichment of top 10 genes in ZIKV infected MPAs.
| List of most variable differentially expressed genes from RNA-seq data. (FC ≥ 1.5, P < 0.05) | |||||||
|---|---|---|---|---|---|---|---|
| Expression | NCBI ID | Gene ID | Description | ZIKV | Mock | log2(V/M) | P-values |
| Up-regulated | 14528 | ENSMUSG00000037580 | GTP cyclohydrolase 1 | 411.0951 | 255.2346 | 0.686851214 | 2.80E-05 |
| 16948 | ENSMUSG00000024529 | lysyl oxidase | 1955.848 | 1278.814 | 0.611777167 | 1.96E-14 | |
| 14600 | ENSMUSG00000055737 | growth hormone receptor | 971.2079 | 693.0997 | 0.485122031 | 1.40E-05 | |
| 16878 | ENSMUSG00000034394 | leukemia inhibitory factor | 821.1017 | 591.2651 | 0.475197313 | 6.50E-05 | |
| 19878 | ENSMUSG00000020580 | Rho-associated coiled-coil containing protein kinase 2 | 5737.706 | 4195.744 | 0.45306051 | 7.90E-19 | |
| 17294 | ENSMUSG00000051855 | mesoderm specific transcript | 1454.704 | 1083.248 | 0.427033195 | 1.43E-06 | |
| 20393 | ENSMUSG00000019970 | serum/glucocorticoid regulated | 1304.022 | 974.9833 | 0.418896285 | 9.53E-06 | |
| 230257 | ENSMUSG00000028382 | polypyrimidine tract binding protein 3 | 1069.396 | 808.1253 | 0.40267405 | 9.09E-05 | |
| 66234 | ENSMUSG00000031604 | methylsterol monooxygenase 1 | 2787.93 | 2108.884 | 0.400968214 | 2.97E-09 | |
| 19205 | ENSMUSG00000006498 | polypyrimidine tract binding protein 1 | 4517.417 | 4254.308 | 0.085671 | 0.084453 | |
| Down-regulated | 17196 | ENSMUSG00000041607 | myelin basic protein | 621.6153 | 945.9696 | -0.607703751 | 1.71E-07 |
| 50913 | ENSMUSG00000039830 | oligodendrocyte transcription | 1866.69 | 2697.02 | -0.53178784 | 2.98E-16 | |
| 17172 | ENSMUSG00000020052 | achaete-scute family bHLH transcription factor 1 | 321.8875 | 465.8106 | -0.53166219 | 0.000573 | |
| 17762 | ENSMUSG00000018411 | microtubule-associated protein tau | 685.4954 | 958.5018 | -0.483424535 | 3.92E-06 | |
| 18823 | ENSMUSG00000031425 | proteolipid protein (myelin) 1 | 549.6362 | 767.9758 | -0.482498168 | 3.89E-05 | |
| 13618 | ENSMUSG00000022122 | endothelin receptor type | 5027.983 | 6649.826 | -0.40246941 | 6.08E-21 | |
| 18128 | ENSMUSG00000026923 | notch 1 | 3787.42 | 4951.807 | -0.387878131 | 1.88E-15 | |
| 23805 | ENSMUSG00000020135 | adenomatosis polyposis coli 2 | 2954.399 | 3839.891 | -0.378530646 | 1.38E-12 | |
| 20847 | ENSMUSG00000040033 | signal transducer and activator of transcription 2 | 2639.62 | 3375.84 | -0.354998851 | 5.01E-10 | |
| 12266 | ENSMUSG00000024164 | complement component 3 | 204812 | 261725.1 | -0.353735272 | 6.43E-87 | |
ENSMUXXXX, ensemble gene IDs; V, MPAs with ZIKV infection; M, MPAs with mock infection; FC, fold change
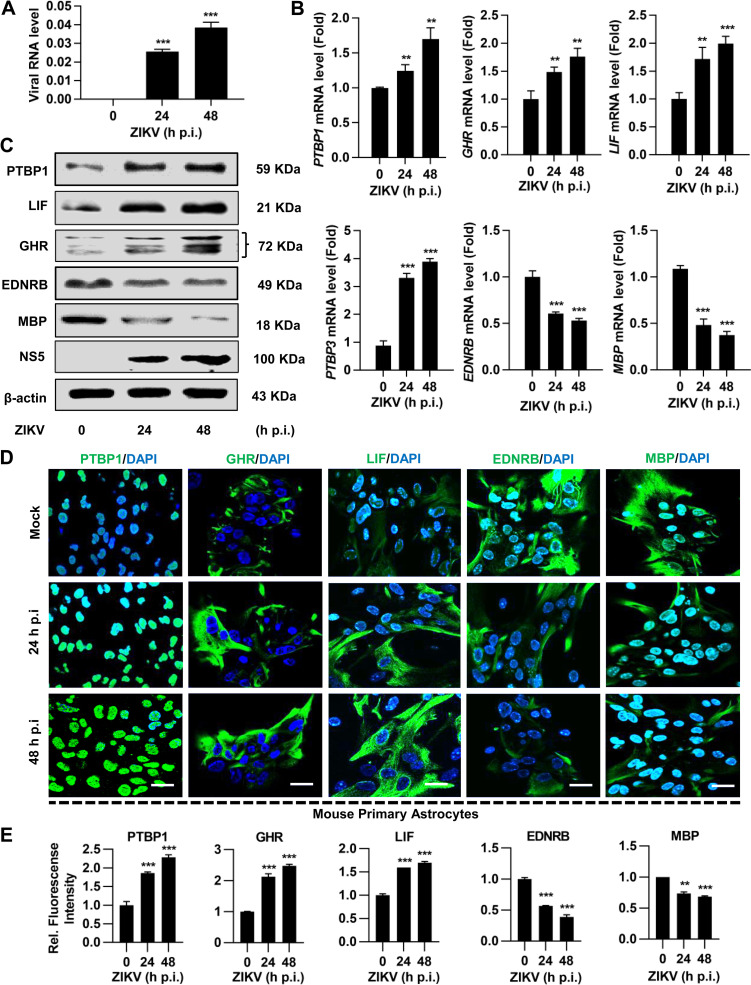
Since ZIKV infection is responsible for causing differential gene expression in MPAs, we further selected six cellular genes that were significantly dysregulated by ZIKV infection to validate the RNA-seq results. ZIKV RNA (Fig 4A) and protein (Fig 4C) expression levels for NS5 were detected to confirm the significant infectivity and replication at 24 and 48 h p.i. We determined the differential gene expression in MPAs by qPCR. Our results showed that mRNA levels of PTBP1, LIF, PTBP3, and GHR were significantly upregulated, while EDNRB and MBP were downregulated upon ZIKV infection at indicated time (Fig 4B). The similar results were also observed for the protein expression of PTBP1, LIF, GHR, EDNRB, and MBP genes (Fig 4C). Furthermore, IF assay (Fig 4D) along with the intensity of illustrated gene expression (Fig 4E) confirmed the upregulation and downregulation of indicated gene expression in ZIKV-infected MPAs, respectively. These results suggested that ZIKV infection potentially dysregulated neural genes in MPAs, which may lead to neurodevelopmental disorders.
Fig 4. ZIKV infection dysregulates certain astrocytic genes in MPAs.
MPAs were infected with ZIKV at MOI of 2 for 0, 24, and 48 h, respectively. (A) ZIKV NS5 RNA was determined by qPCR analysis in MPAs. (B) The mRNA expressions of PTBP1, GHR, LIF, PTBP3, EDNRB, and MBP were determined by qPCR. The data represent the relative expression of target genes normalized to GAPDH as a reference internal control. (C) The protein expressions of PTBP1, LIF, GHR, EDNRB, and MBP were determined by Western blot analyses. The β-actin was selected as an internal control. (D and E) Confocal microscopy images from MPAs stained with PTBP1, GHR, LIF, EDNRB, and MBP (green) for target genes and DAPI (blue) for nuclei, scale bar = 20 μm (D). Relative protein expression to control is quantified using Image J software (E). Data are shown as fold change from mock control. All the experiments were performed in triplicate, non-significant (ns); P < 0.05 (*); P < 0.01 (**); P < 0.001 (***). Student’s t-test. The underlying numerical data for “Fig 4A, 4B, and 4E” is provided in the supporting file “S1 Data”.
Validation of DEGs in ZIKV-infected MPAs
To access the validation of DEGs in ZIKV-infected MPAs, the comparison of the top 6 selected differentially expressed genes between qRT-PCR and RNA-Seq was performed. In the ZIKV-infected MPAs for 24 h, qRT-PCR analysis displayed the up-regulation of PTBP1 (Fig 5A), GHR (Fig 5B), LIF (Fig 5C), and PTBP3 (Fig 5D), while the down-regulation of EDNRB (Fig 5E) and MBP (Fig 5F), revealing that the selected genes expression profiles were accordant with obtained results from RNA-Seq (Fig 5A–5F). Therefore, the data illustrated that the methodology in the study concluded the accuracy and reproducibility.
Fig 5. Validation of astrocytic altered genes upon ZIKV infection.
MPAs were infected with ZIKV at MOI of 2 for 0 and 24 h, respectively. (A-F) The expression patterns of top 6 shortlisted genes: PTBP1 (A), GHR (B), LIF (C), PTBP3 (D), EDNRB (E), and MBP (F), were measured by RNA-seq and qRT-PCR (qPCR), respectively. GAPDH was used as an internal control in qRT-PCR. The individual bars represent the qPCR data as the means ± SD, n = 3. The underlying numerical data for “Fig 5A–5F” is provided in the supporting file “S1 Data”.
ZIKV infection dysregulate neural genes expression in human astroglioma U251 cells
Given the fact that MPAs were susceptible to ZIKV infection and ZIKV dysregulated several critical neural genes expression, such alterations by ZIKV infection were also evaluated in human U251 cells. As compared to mock-treated U251 cells, ZIKV-infected U251 cells displayed a visible cytopathic effect (CPE) in a time-dependent fashion (S1 Fig), and the increasing CPE was measured by cell viability assays (Fig 6A). The qPCR results revealed that ZIKV RNA level continuously increased from 0 to 36 h p.i., whereas slightly declined between 36 and 48 h p.i. (Fig 6B), suggesting U251 cells were susceptible to ZIKV infection. In parallel, we also observed a robust expression of viral dsRNA upon ZIKV infection in U251 at 12 and 24 h p.i. (Fig 6C). Furthermore, we evaluated the differential gene expression of indicated genes in ZIKV-infected U251 cells. Our results illustrated that PTBP1, LIF, PTBP3, and GHR mRNA levels were upregulated while EDNRB and MBP mRNA levels were downregulated in ZIKV-infected U251 cells at 12 and 24 h p.i. (Fig 6D). Similarly, we also observed the dysregulated protein expression of the indicated genes by Western blot assay (Fig 6E) and IF assay (Fig 6F), along with the intensity of illustrated genes expression (Fig 6G). Notably, ZIKV infection dysregulated astrocytic gene expression leading to brain abnormality in human U251 cells as similar to MPAs (Fig 7), which suggests ZIKV-infected MPAs as a compatible infection model in the exploration of neural disorders.
Fig 6. ZIKV infection differentially alters certain astrocytic genes in U251 cells.
U251 cells were infected with ZIKV at MOI of 2 for indicated periods. (A) Cell viability was measured by the CCK-8 assay. Data represent as fold change compared to mock-treated cells. (B) ZIKV NS5 RNA was determined by qPCR with GAPDH as an internal control, and (C) viral dsRNA was immunoprobed with dsRNA-antibody (red), along with GFAP (green) for astrocytes and DAPI (blue) for nuclei by Immunofluorescent (IF), scale bar = 20 μm. (D and E) The mRNA expressions of PTBP1, GHR, LIF, PTBP3, EDNRB, and MBP were determined by qPCR (D) and protein expressions of PTBP1, GHR, LIF, EDNRB, and MBP was determined by Western blot analyses (E), respectively. In the qPCR analysis, the data represent the relative expression of target genes normalized to GAPDH as a reference internal control. In Western blot analysis, the β-actin was selected as an internal control. (F and G) Confocal microscopy images from MPAs stained with PTBP1, GHR, LIF, EDNRB, and MBP (green) for target genes and DAPI (blue) for nuclei, scale bar = 20 μm (F). Relative protein expression to control is quantified using Image J software (G). Data are shown as fold change from mock control. All the experiments were performed in triplicate, non-significant (ns); P < 0.05 (*); P < 0.01 (**); P < 0.001 (***). Student’s t-test. The underlying numerical data for “Fig 6A, 6B, 6D, and 6G” is provided in supporting file “S1 Data”.
Fig 7. A mechanism underlying the brain-related neurological disorders upon ZIKV infection.
The neurotropic ZIKV favorably infects and replicates in astrocytes of the fetal brain cerebral cortex. ZIKV can alter the astrocytic genes including PTBP1, LIF, PTBP3, GHR, EDNRB, and MBP, which are involved in various functions leading to neurological conditions such as microcephaly and brain development impairment. The numerical data used in all figures are included in S1 Data.
Discussion
An increase of defects in neurodevelopmental has been associated with ZIKV, a flavivirus mainly transmitted by mosquitoes and sexual transmission [54,55]. Current studies have revealed that ZIKV can infect human neurospheres, neural progenitor cells, and brain organoids [31,56–58]. However, the exact mechanisms of infection and specifically which cell populations in the nervous system respond to ZIKV and contribute to brain developmental disorders is still unclear. In current study, we used the RNA sequencing to analyze the astrocytic mRNAs that are dysregulated upon ZIKV infection. Utilizing different bioinformatic analysis coupled with gene expression profiling, our findings suggest that ZIKV can potentially alter the astrocytic gene expression, which is associated with brain development and neurological conditions [18]. Almost half or more of the brain cells are represented by glial cells (in specific parts up to 90%) and glial cell development is critical for normal brain size and function [59]. Astrocytes are the abundant and first line of the brain glia cells that are infected or respond to ZIKV infection [24]. Several studies have been reported on the association of ZIKV with brain development or associated diseases but yet there are no potential therapeutic options to overcome indicated neurological conditions [18]. For a developing brain, astrocytes are the drivers and initiators of ZIKV infection. Therefore, this virus not only infects brain cells and its precursors but also can penetrate the placental barrier and causes different neurological disorders linked to PNS and CNS [6].
Several studies reported the gene expression profiles in ZIKV-induced human and mouse neuronal cells [60–62], however, the association between the changes in astrocytic mRNAs expression and neurodegeneration or brain development is not described. In this study, we aimed to figure out the underlying link between the ZIKV and brain astrocyte cells. Initially, we constructed a ZIKV infection model in MPAs by isolating cortical astrocytes from the postnatal mouse brain, following ZIKV infection for indicated time points. ZIKV infection was confirmed in the MPAs and human astroglioma U251 cells, indicating these two kinds of astrocytes susceptible to ZIKV infection and considerable infection model in the exploration between ZIKV and astrocytes. At present, human neural progenitor cells [63], brain organoids [56,57,64] are the platform to study zika virus-associated microcephaly and embryonic brain development in vitro. Primary cell culture is increasingly being a major tool in virus and host interaction, which provides an excellent model for the study of normal physiology of the cells when exposed to virus infection [65]. Our study identified MPAs along with U251 cells behaving as a considerable tool to explore the relationship of ZIKV and host, which is consistent with evidence that ZIKV can target human glial cells [66,67].
We evaluated the ZIKV-induced MPAs for transcriptomics by RNA-seq and illustrated a huge number of astrocytic genes altered by ZIKV infection. Dysregulation of the neuronal genes, such as RBBP8, ASPM, AXL, TBR2, MCPH1, and CENPF upon ZIKV infection may cause different neurological or brain development related disorders [35]. By applying the bioinformatic tools, our RNA-seq data showed 27,812 genes alteration from MPAs upon ZIK infection, and around 355 genes were significantly dysregulated from each group of the differentiated the up- and down-regulated genes. Cellular function of the astrocytic genes, including modulation of stress response, remyelination, cell growth dysregulation, inhibition of differentiation, and modulation of stress response, have been identified in C6/36, Vero cells, human neural stem cells, and mouse glial cells [68,69]. The bioinformatic analysis revealed that ZIKV infection could interrupt neurogenesis, neuron differentiation, development, migration, and maturation, astrocytes projection, and brain development, as well as multiply pathways, including Hippo signaling pathway, focal adhesion, PI3K-Akt signaling pathway, and tight junction. To make the RNA-seq results more authentic, we further evaluated 6 genes associated with neurological disorders or brain development were significantly dysregulated upon ZIKV infection. Our results demonstrated that the expressions of PTBP1, PTBP3, LIF, and GHR were upregulated while EDNRB and MBP were downregulated in both ZIKV-infected MPAs and U251 cells, implying a potential dysfunction in ZIKV-induced neural disorders.
Among the selected differentially expressed genes, the RNA binding PTBP1 and PTBP3 from PTB family, which are responsible for the conversion of astrocytes into neurons [27], neurodevelopment including control of neuronal progenitor cells (NPCs) and embryonic stem cells (ESCs) splicing, and control of neural differentiation timing [70], are naturally decreased during neural progenitors’ differentiation [71] and give rise to multiple neuronal cell lineage in the brain [70,72]. On this account, our results suggest that the ZIKV-induced upregulation of the PTBP1 and PTBP3 levels in astrocytes may decrease or inhibit the conversion to neurons and eventually lead to various neurological conditions or make neurological infections more venerable.
LIF is a pleiotropic cytokine that promotes neurogenesis in fetus cerebrum [73,74] and autoimmune diseases treatment [75]. It is involved in neuroprotection, axonal regeneration, and preventing demyelination [76]. In addition, it induces neuronal progenitors [73] and acts as a stimulator for astrocytes differentiator [28]. Various studies reported that neurons and astrocytes are produced from the same precursor of CNS [28]. Based on these reports, our findings suggest that LIF works as a stimulator for astrocytes differentiation, so the upregulation of LIF may lead to the production of more astrocytes and fewer neurons, which results in different brain development defects.
We also reported the ZIKV-induced downregulation of EDNRB and MBP genes expression in astrocytes. EDNRB promotes reactive astrogliosis and helps in repairing brain injuries (remyelination) [77,78], while MBP releases from a sheath which plays a vital role in compacting myelin and protection of myelin sheath from degradation by myelinotoxic factors (proteinases) produced during multiple sclerosis [79], revealing that these two genes are essential housekeeping factors for normal myelin sheath control. Based on the previous studies, our results suggest that the downregulation of these genes might be the cause of neurological or brain disorders related to demyelination. Moreover, our results are consistent with the fact that the depletion or low EDNRB expression leads to defective B cell differentiation or intrinsic lymphoid defects [80], similar to that MBP reduction causes neurodegenerative disorders like microcephaly [81]. GHR promotes normal human postnatal growth and its deficiency or mutation may lead to utero and severe postnatal growth failure, intellectual impairment, microcephaly, and sensorineural deafness [82]. Additionally, upregulation of GHR levels have been reported to increase the risk of cancer in general and regulate key cellular functions, such as metastasis, apoptosis, proliferation, and differentiation [83]. The disruption of GHR also cause Laron syndrome (microcephalic syndrome) which not only affects the brain but also total somatic dwarfism [84,85]. In current study, the upregulation of GHR suggests that the alteration of this gene by ZIKV may lead to neurological impairments causing reported diseases, which further indicate that MBP and GHR are directly involved in the brain development and microcephaly.
Actually, there are some limitations in the study and some considerations should be taken into account when interpreting the findings. Firstly, the model we selected is the primary cells with some disadvantages, including limited potential for self-renewal and differentiation, which needs a further platform, such as human neural progenitor cells or brain organoids when studying ZIKV-associated microcephaly or embryonic brain development in vitro. Secondly, there should be more ZIKV strains included to access the astrocytic gene alteration to uncover the common rules of ZIKV-associated neurodevelopment. Thirdly, the dysfunctional genes associated with ZIKV-induced neurodevelopment are identified with observable expression regardless of poorly expressed genes, which may contribute to the uncertainty of findings. Even so, we verified a considerable model that ZIKV alters the transcriptome and discovered potential genes associated with neurodevelopment in MPAs, which requires an advanced model (e.g., human neural progenitor cells or brain organoids) and viral scope of ZIKV for biological functional exploration of neurodevelopment with genetic approaches (e.g., CRISPR-Cas9 or RNAi) in the future study.
In conclusion, the present study revealed ZIKV-infected MPAs as a compatible infection model for the exploration of ZIKV and astrocytes in neural disorders. In addition, we systematically exposed dysregulation in a cluster of neural genes by ZIKV infection in astrocytes, which provides novel clues for the mechanism involved in ZIKV-associated neurodevelopment or brain development impairment, and also lay the foundation for an approach to mechanistic research to develop potential vaccines or treatment against ZIKV infection to guard innumerable children and mothers exposed to the unfolding epidemics.
Supporting information
U251 cells were infected with ZIKV at an MOI of 2 for indicated periods. The cytopathic effect (CPE) was displayed in U251 cells infected with ZIKV (bottom) compared to mock (top) with bright-field microscopy, scale bar = 50 μm.
(TIF)
(DOCX)
(DOCX)
(XLSX)
(XLS)
Excel spreadsheet in separate sheets contains the underlying numerical data for Figure panels 1C, 1D, 2D, 2E, 3A & 3B, 3C & 3D, 4A, 4B, 4E, 5A-5F, 6A, 6B, 6D, 6G.
(XLSX)
Acknowledgments
We thank Dr. Jie Jin (Biorun Biosciences Co., Ltd, Wuhan, China), and Dr. Suliman Khan (the Second Affiliated Hospital of Zhengzhou University, Zhengzhou, China) for the technical support in bioinformatic tools and software.
Data Availability
All the relevant data are within the Revised Manuscript and Supporting Information files. The datasets generated from RNA-Seq raw reads data can be obtained from NCBI under BioProject accession number: PRJNA692303.
Funding Statement
This work was supported by National Natural Science Foundation of China (81730061 to JW; 32070148 and 31800147 to ZL) (https://isisn.nsfc.gov.cn/egrantweb/), Guangdong Basic and Applied Basic Research Foundation (2019A1515011073 to ZL).(https://pro.gdstc.gov.cn/egrantweb/), Open Research Fund Program of the State Key Laboratory of Virology of China (2021KF001 to ZL) (http://klv.whu.edu.cn/), and National Mega Project on Major Infectious Disease Prevention under Grant (2017ZX10103005 to FL) (https://isisn.nsfc.gov.cn/egrantweb/). The funders play no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Foy BD, Kobylinski KC, Foy JLC, Blitvich BJ, da Rosa AT, Haddow AD, et al. Probable non–vector-borne transmission of Zika virus, Colorado, USA. Emerging infectious diseases. 2011;17(5):880. 10.3201/eid1705.101939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Althaus CL, Low N. How relevant is sexual transmission of Zika virus? PLoS medicine. 2016;13(10):e1002157. 10.1371/journal.pmed.1002157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreira J, Peixoto TM, Siqueira AMd, Lamas CC. Sexually acquired Zika virus: a systematic review. Clinical Microbiology and Infection. 2017;23(5):296–305. 10.1016/j.cmi.2016.12.027 [DOI] [PubMed] [Google Scholar]
- 4.Parra B, Lizarazo J, Jiménez-Arango JA, Zea-Vera AF, González-Manrique G, Vargas J, et al. Guillain–Barré syndrome associated with Zika virus infection in Colombia. New England Journal of Medicine. 2016;375(16):1513–23. 10.1056/NEJMoa1605564 [DOI] [PubMed] [Google Scholar]
- 5.de Araújo TVB, Rodrigues LC, de Alencar Ximenes RA, de Barros Miranda-Filho D, Montarroyos UR, de Melo APL, et al. Association between Zika virus infection and microcephaly in Brazil, January to May, 2016: preliminary report of a case-control study. The Lancet Infectious Diseases. 2016;16(12):1356–63. 10.1016/S1473-3099(16)30318-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sher AA, Glover KK, Coombs KM. Zika virus infection disrupts astrocytic proteins involved in synapse control and axon guidance. Frontiers in microbiology. 2019;10:596. 10.3389/fmicb.2019.00596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ojha CR, Rodriguez M, Karuppan MKM, Lapierre J, Kashanchi F, El-Hage N. Toll-like receptor 3 regulates Zika virus infection and associated host inflammatory response in primary human astrocytes. PloS one. 2019;14(2):e0208543. 10.1371/journal.pone.0208543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao-Lormeau V-M, Blake A, Mons S, Lastère S, Roche C, Vanhomwegen J, et al. Guillain-Barré Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. The Lancet. 2016;387(10027):1531–9. 10.1016/S0140-6736(16)00562-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oehler E, Watrin L, Larre P, Leparc-Goffart I, Lastere S, Valour F, et al. Zika virus infection complicated by Guillain-Barre syndrome–case report, French Polynesia, December 2013. Eurosurveillance. 2014;19(9):20720. 10.2807/1560-7917.es2014.19.9.20720 [DOI] [PubMed] [Google Scholar]
- 10.de Oliveira WK, Cortez-Escalante J, De Oliveira WTGH, Carmo GMId, Henriques CMP, Coelho GE, et al. Increase in reported prevalence of microcephaly in infants born to women living in areas with confirmed Zika virus transmission during the first trimester of pregnancy—Brazil, 2015. Morbidity and Mortality Weekly Report. 2016;65(9):242–7. 10.15585/mmwr.mm6509e2 [DOI] [PubMed] [Google Scholar]
- 11.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika virus and birth defects—reviewing the evidence for causality. New England Journal of Medicine. 2016;374(20):1981–7. 10.1056/NEJMsr1604338 [DOI] [PubMed] [Google Scholar]
- 12.Schuler-Faccini L, Ribeiro EM, Feitosa IM, Horovitz DD, Cavalcanti DP, Pessoa A, et al. Possible association between Zika virus infection and microcephaly—Brazil, 2015. Morbidity and Mortality Weekly Report. 2016;65(3):59–62. 10.15585/mmwr.mm6503e2 [DOI] [PubMed] [Google Scholar]
- 13.Ventura CV, Maia M, Bravo-Filho V, Góis AL, Belfort R. Zika virus in Brazil and macular atrophy in a child with microcephaly. The Lancet. 2016;387(10015):228. [DOI] [PubMed] [Google Scholar]
- 14.Aragao MdFV, van der Linden V, Brainer-Lima AM, Coeli RR, Rocha MA, da Silva PS, et al. Clinical features and neuroimaging (CT and MRI) findings in presumed Zika virus related congenital infection and microcephaly: retrospective case series study. Bmj. 2016;353:i1901. 10.1136/bmj.i1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell K, Oliver SE, Lewis L, Barfield WD, Cragan J, Meaney-Delman D, et al. Update: interim guidance for the evaluation and management of infants with possible congenital Zika virus infection—United States, August 2016. Morbidity and Mortality Weekly Report. 2016;65(33):870–8. 10.15585/mmwr.mm6533e2 [DOI] [PubMed] [Google Scholar]
- 16.O’Driscoll M, Jeggo PA. The role of the DNA damage response pathways in brain development and microcephaly: insight from human disorders. DNA repair. 2008;7(7):1039–50. 10.1016/j.dnarep.2008.03.018 [DOI] [PubMed] [Google Scholar]
- 17.Jaenisch T, Rosenberger KD, Brito C, Brady O, Brasil P, Marques ET. Risk of microcephaly after Zika virus infection in Brazil, 2015 to 2016. Bulletin of the World Health Organization. 2017;95(3):191. 10.2471/BLT.16.178608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fauci AS, Morens DM. Zika virus in the Americas—yet another arbovirus threat. New England journal of medicine. 2016;374(7):601–4. [DOI] [PubMed] [Google Scholar]
- 19.Miner JJ, Diamond MS. Understanding how Zika virus enters and infects neural target cells. Cell stem cell. 2016;18(5):559–60. 10.1016/j.stem.2016.04.009 [DOI] [PubMed] [Google Scholar]
- 20.van den Pol AN, Mao G, Yang Y, Ornaghi S, Davis JN. Zika virus targeting in the developing brain. Journal of Neuroscience. 2017;37(8):2161–75. 10.1523/JNEUROSCI.3124-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muffat J, Li Y, Omer A, Durbin A, Bosch I, Bakiasi G, et al. Human induced pluripotent stem cell-derived glial cells and neural progenitors display divergent responses to Zika and dengue infections. Proceedings of the National Academy of Sciences. 2018;115(27):7117–22. 10.1073/pnas.1719266115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H, Liu B, Yves TD, He Y, Wang S, Tang H, et al. Zika virus induces autophagy in human umbilical vein endothelial cells. Viruses. 2018;10(5):259. 10.3390/v10050259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marinelli C, Di Liddo R, Facci L, Bertalot T, Conconi MT, Zusso M, et al. Ligand engagement of Toll-like receptors regulates their expression in cortical microglia and astrocytes. Journal of neuroinflammation. 2015;12(1):1–20. 10.1186/s12974-015-0458-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao Q, Herrlinger S, Yang S-L, Lai F, Moore JM, Brindley MA, et al. Zika virus infection disrupts neurovascular development and results in postnatal microcephaly with brain damage. Development. 2016;143(22):4127–36. 10.1242/dev.143768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Perez O, Quiñones-Hinojosa A. Role of astrocytes as neural stem cells in the adult brain. Journal of stem cells. 2012;7(3):181. doi: jsc.2012.7.3.181 [PMC free article] [PubMed] [Google Scholar]
- 26.Reemst K, Noctor SC, Lucassen PJ, Hol EM. The indispensable roles of microglia and astrocytes during brain development. Frontiers in human neuroscience. 2016;10:566. 10.3389/fnhum.2016.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian H, Kang X, Hu J, Zhang D, Liang Z, Meng F, et al. Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature. 2020;582(7813):550–6. 10.1038/s41586-020-2388-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koblar SA, Turnley AM, Classon BJ, Reid K, Ware CB, Cheema SS, et al. Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proceedings of the National Academy of Sciences. 1998;95(6):3178–81. 10.1073/pnas.95.6.3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vester D, Rapp E, Gade D, Genzel Y, Reichl U. Quantitative analysis of cellular proteome alterations in human influenza A virus-infected mammalian cell lines. Proteomics. 2009;9(12):3316–27. Epub 2009/06/09. 10.1002/pmic.200800893 . [DOI] [PubMed] [Google Scholar]
- 30.Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445(7125):319–23. Epub 2007/01/19. 10.1038/nature05495 . [DOI] [PubMed] [Google Scholar]
- 31.Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell stem cell. 2016;18(5):587–90. 10.1016/j.stem.2016.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X, Dong X, Li S-H, Zhou Y-P, Rayner S, Xia H-M, et al. Proteomic analysis of zika virus infected primary human fetal neural progenitors suggests a role for doublecortin in the pathological consequences of infection in the cortex. Frontiers in microbiology. 2018;9:1067. 10.3389/fmicb.2018.01067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scaturro P, Stukalov A, Haas DA, Cortese M, Draganova K, Płaszczyca A, et al. An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature. 2018;561(7722):253–7. 10.1038/s41586-018-0484-5 [DOI] [PubMed] [Google Scholar]
- 34.Zhang F, Hammack C, Ogden SC, Cheng Y, Lee EM, Wen Z, et al. Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic acids research. 2016;44(18):8610–20. 10.1093/nar/gkw765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, et al. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell stem cell. 2016;19(1):120–6. 10.1016/j.stem.2016.04.017 [DOI] [PubMed] [Google Scholar]
- 36.Schildge S, Bohrer C, Beck K, Schachtrup C. Isolation and culture of mouse cortical astrocytes. JoVE (Journal of Visualized Experiments). 2013;(71):e50079. 10.3791/50079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Li G, De W, Luo Z, Pan P, Tian M, et al. Zika virus infection induces host inflammatory responses by facilitating NLRP3 inflammasome assembly and interleukin-1β secretion. Nature Communications. 2018;9(1):106. 10.1038/s41467-017-02645-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen B, Wang X, Zhao W, Wu J. Klotho inhibits growth and promotes apoptosis in human lung cancer cell line A549. Journal of Experimental & Clinical Cancer Research. 2010;29(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolay A, Castilho LR, Reichl U, Genzel Y. Propagation of Brazilian Zika virus strains in static and suspension cultures using Vero and BHK cells. Vaccine. 2018;36(22):3140–5. 10.1016/j.vaccine.2017.03.018 [DOI] [PubMed] [Google Scholar]
- 40.Vicenti I, Boccuto A, Giannini A, Dragoni F, Saladini F, Zazzi M. Comparative analysis of different cell systems for Zika virus (ZIKV) propagation and evaluation of anti-ZIKV compounds in vitro. Virus research. 2018;244:64–70. 10.1016/j.virusres.2017.11.003 [DOI] [PubMed] [Google Scholar]
- 41.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature methods. 2015;12(4):357–60. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature biotechnology. 2015;33(3):290–5. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaari G, Bolen CR, Thakar J, Kleinstein SH. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic acids research. 2013;41(18):e170–e. 10.1093/nar/gkt660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE Guidelines: M inimum I nformation for Publication of Q uantitative Real-Time PCR E xperiments. Oxford University Press; 2009. [DOI] [PubMed] [Google Scholar]
- 46.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nature protocols. 2006;1(3):1559–82. 10.1038/nprot.2006.236 [DOI] [PubMed] [Google Scholar]
- 47.Luo Z, Su R, Wang W, Liang Y, Zeng X, Shereen MA, et al. EV71 infection induces neurodegeneration via activating TLR7 signaling and IL-6 production. PLoS pathogens. 2019;15(11):e1008142. 10.1371/journal.ppat.1008142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 49.Wang W, Hu D, Wu C, Feng Y, Li A, Liu W, et al. STING promotes NLRP3 localization in ER and facilitates NLRP3 deubiquitination to activate the inflammasome upon HSV-1 infection. PLoS pathogens. 2020;16(3):e1008335. 10.1371/journal.ppat.1008335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ming G-l, Tang H, Song H. Advances in Zika virus research: stem cell models, challenges, and opportunities. Cell stem cell. 2016;19(6):690–702. 10.1016/j.stem.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dedhia M, Kohetuk K, Crusio WE, Delprato A. Introducing high school students to the Gene Ontology classification system. F1000Research. 2019;8. 10.12688/f1000research.17047.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cairns DM, Boorgu DSSK, Levin M, Kaplan DL. Niclosamide rescues microcephaly in a humanized in vivo model of Zika infection using human induced neural stem cells. Biology open. 2018;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu B, Chen X. The Distinct Signaling Regulatory Roles in the Cortical Atrophy and Cerebellar Apoptosis of Newborn Nbn-Deficient Mice. Cellular and molecular neurobiology. 2013;33(8):1043–53. 10.1007/s10571-013-9971-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy M. Zika virus was transmitted by sexual contact in Texas, health officials report. BMJ: British Medical Journal (Online). 2016;352. 10.1136/bmj.i720 [DOI] [PubMed] [Google Scholar]
- 55.Musso D, Roche C, Robin E, Nhan T, Teissier A, Cao-Lormeau V-M. Potential sexual transmission of Zika virus. Emerging infectious diseases. 2015;21(2):359. 10.3201/eid2102.141363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcez PP, Loiola EC, da Costa RM, Higa LM, Trindade P, Delvecchio R, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–8. 10.1126/science.aaf6116 [DOI] [PubMed] [Google Scholar]
- 57.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–54. 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dang J, Tiwari SK, Lichinchi G, Qin Y, Patil VS, Eroshkin AM, et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell stem cell. 2016;19(2):258–65. 10.1016/j.stem.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan M, Luan R. Research progress of the causal link between Zika virus and microcephaly. Global Health Journal. 2018;2(4):11–8. [Google Scholar]
- 60.Quintens R. Convergence and divergence between the transcriptional responses to Zika virus infection and prenatal irradiation. Cell death & disease. 2017;8(3):e2672. 10.1038/cddis.2017.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rolfe AJ, Bosco DB, Wang J, Nowakowski RS, Fan J, Ren Y. Bioinformatic analysis reveals the expression of unique transcriptomic signatures in Zika virus infected human neural stem cells. Cell & bioscience. 2016;6(1):42. 10.1186/s13578-016-0110-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kozak RA, Majer A, Biondi MJ, Medina SJ, Goneau LW, Sajesh BV, et al. MicroRNA and mRNA dysregulation in astrocytes infected with Zika virus. Viruses. 2017;9(10):297. 10.3390/v9100297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghouzzi VE, Bianchi FT, Molineris I, Mounce BC, Berto GE, Rak M, et al. ZIKA virus elicits P53 activation and genotoxic stress in human neural progenitors similar to mutations involved in severe forms of genetic microcephaly. Cell Death Dis. 2016;7(10):e2440. Epub 2016/10/28. 10.1038/cddis.2016.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watanabe M, Buth JE, Vishlaghi N, de la Torre-Ubieta L, Taxidis J, Khakh BS, et al. Self-organized cerebral organoids with human-specific features predict effective drugs to combat Zika virus infection. Cell reports. 2017;21(2):517–32. 10.1016/j.celrep.2017.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, et al. Zika virus targets different primary human placental cells, suggesting two routes for vertical transmission. Cell host & microbe. 2016;20(2):155–66. 10.1016/j.chom.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meertens L, Labeau A, Dejarnac O, Cipriani S, Sinigaglia L, Bonnet-Madin L, et al. Axl Mediates ZIKA Virus Entry in Human Glial Cells and Modulates Innate Immune Responses. Cell Rep. 2017;18(2):324–33. Epub 2017/01/12. 10.1016/j.celrep.2016.12.045 . [DOI] [PubMed] [Google Scholar]
- 67.Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, et al. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proceedings of the National Academy of Sciences. 2016;113(50):14408–13. 10.1073/pnas.1618029113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Devhare P, Meyer K, Steele R, Ray RB, Ray R. Zika virus infection dysregulates human neural stem cell growth and inhibits differentiation into neuroprogenitor cells. Cell death & disease. 2017;8(10):e3106–e. 10.1038/cddis.2017.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hou S, Kumar A, Xu Z, Airo AM, Stryapunina I, Wong CP, et al. Zika virus hijacks stress granule proteins and modulates the host stress response. Journal of virology. 2017;91(16). 10.1128/JVI.00474-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng S, Gray EE, Chawla G, Porse BT, O’dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nature neuroscience. 2012;15(3):381–8. 10.1038/nn.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu J, Qian H, Xue Y, Fu X-D. PTB /nPTB: master regulators of neuronal fate in mammals. Biophysics reports. 2018;4(4):204–14. 10.1007/s41048-018-0066-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Critical reviews in biochemistry and molecular biology. 2012;47(4):360–78. 10.3109/10409238.2012.691456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simamura E, Shimada H, Higashi N, Uchishiba M, Otani H, Hatta T. Maternal leukemia inhibitory factor (LIF) promotes fetal neurogenesis via a LIF-ACTH-LIF signaling relay pathway. Endocrinology. 2010;151(4):1853–62. 10.1210/en.2009-0985 [DOI] [PubMed] [Google Scholar]
- 74.Simamura E, Shimada H, Shoji H, Otani H, Hatta T. Effects of melanocortins on fetal development. Congenital anomalies. 2011;51(2):47–54. 10.1111/j.1741-4520.2011.00316.x [DOI] [PubMed] [Google Scholar]
- 75.Tsukada T, Simamura E, Shimada H, Arai T, Higashi N, Akai T, et al. The suppression of maternal–fetal leukemia inhibitory factor signal relay pathway by maternal immune activation impairs brain development in mice. PloS one. 2015;10(6):e0129011. 10.1371/journal.pone.0129011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Slaets H, Hendriks JJ, Stinissen P, Kilpatrick TJ, Hellings N. Therapeutic potential of LIF in multiple sclerosis. Trends in molecular medicine. 2010;16(11):493–500. 10.1016/j.molmed.2010.08.007 [DOI] [PubMed] [Google Scholar]
- 77.Hammond TR, McEllin B, Morton PD, Raymond M, Dupree J, Gallo V. Endothelin-B receptor activation in astrocytes regulates the rate of oligodendrocyte regeneration during remyelination. Cell reports. 2015;13(10):2090–7. 10.1016/j.celrep.2015.11.002 [DOI] [PubMed] [Google Scholar]
- 78.Hammond TR, Gadea A, Dupree J, Kerninon C, Nait-Oumesmar B, Aguirre A, et al. Astrocyte-derived endothelin-1 inhibits remyelination through notch activation. Neuron. 2014;81(3):588–602. 10.1016/j.neuron.2013.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Latronico T, Branà MT, Gramegna P, Fasano A, Di Bari G, Liuzzi GM. Inhibition of myelin-cleaving poteolytic activities by interferon-beta in rat astrocyte cultures. Comparative analysis between gelatinases and calpain-II. PLoS One. 2013;8(2):e49656. Epub 2013/02/08. 10.1371/journal.pone.0049656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacLean GA, McEldoon J, Huang J, Allred J, Canver MC, Orkin SH. Downregulation of Endothelin Receptor B Contributes to Defective B Cell Lymphopoiesis in Trisomy 21 Pluripotent Stem Cells. Scientific Reports. 2018;8(1):8001. 10.1038/s41598-018-26123-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dupuis N, Fafouri A, Bayot A, Kumar M, Lecharpentier T, Ball G, et al. Dymeclin deficiency causes postnatal microcephaly, hypomyelination and reticulum-to-Golgi trafficking defects in mice and humans. Human molecular genetics. 2015;24(10):2771–83. 10.1093/hmg/ddv038 [DOI] [PubMed] [Google Scholar]
- 82.Hwa V. STAT5B deficiency: impacts on human growth and immunity. Growth Hormone & IGF Research. 2016;28:16–20. 10.1016/j.ghir.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subramani R, Lopez-Valdez R, Salcido A, Boopalan T, Arumugam A, Nandy S, et al. Growth hormone receptor inhibition decreases the growth and metastasis of pancreatic ductal adenocarcinoma. Experimental & Molecular Medicine. 2014;46(10):e117–e. 10.1038/emm.2014.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laron Z. Laron syndrome (primary growth hormone resistance or insensitivity): the personal experience 1958–2003. The Journal of Clinical Endocrinology & Metabolism. 2004;89(3):1031–44. 10.1210/jc.2003-031033 [DOI] [PubMed] [Google Scholar]
- 85.Zhou Y, Xu BC, Maheshwari HG, He L, Reed M, Lozykowski M, et al. A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse). Proceedings of the National Academy of Sciences. 1997;94(24):13215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
U251 cells were infected with ZIKV at an MOI of 2 for indicated periods. The cytopathic effect (CPE) was displayed in U251 cells infected with ZIKV (bottom) compared to mock (top) with bright-field microscopy, scale bar = 50 μm.
(TIF)
(DOCX)
(DOCX)
(XLSX)
(XLS)
Excel spreadsheet in separate sheets contains the underlying numerical data for Figure panels 1C, 1D, 2D, 2E, 3A & 3B, 3C & 3D, 4A, 4B, 4E, 5A-5F, 6A, 6B, 6D, 6G.
(XLSX)
Data Availability Statement
All the relevant data are within the Revised Manuscript and Supporting Information files. The datasets generated from RNA-Seq raw reads data can be obtained from NCBI under BioProject accession number: PRJNA692303.