Abstract
Abstract
Mycobacterium tuberculosis (Mtb) is one of the deadliest pathogens encountered by humanity. Over the decades, its characteristic membrane organization and composition have been understood. However, there is still limited structural information and mechanistic understanding of the constituent membrane proteins critical for drug discovery pipelines. Recent advances in single-particle cryo-electron microscopy and cryo-electron tomography have provided the much-needed impetus towards structure determination of several vital Mtb membrane proteins whose structures were inaccessible via X-ray crystallography and NMR. Important insights into membrane composition and organization have been gained via a combination of electron tomography and biochemical and biophysical assays. In addition, till the time of writing this review, 75 new structures of various Mtb proteins have been reported via single-particle cryo-EM. The information obtained from these structures has improved our understanding of the mechanisms of action of these proteins and the physiological pathways they are associated with. These structures have opened avenues for structure-based drug design and vaccine discovery programs that might help achieve global-TB control. This review describes the structural features of selected membrane proteins (type VII secretion systems, Rv1819c, Arabinosyltransferase, Fatty Acid Synthase, F-type ATP synthase, respiratory supercomplex, ClpP1P2 protease, ClpB disaggregase and SAM riboswitch), their involvement in physiological pathways, and possible use as a drug target.
Graphic Abstract
Tuberculosis is a deadly disease caused by Mycobacterium tuberculosis. The Cryo-EM and tomography have simplified the understanding of the mycobacterial membrane organization. Some proteins are located in the plasma membrane; some span the entire envelope, while some, like MspA, are located in the mycomembrane. Cryo-EM has made the study of such membrane proteins feasible.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00232-021-00179-w.
Keywords: Cryo-EM, Drug discovery, Membrane protein, Mycobacterium, Protein structure
Introduction
Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis (TB)—a disease that costs as many as 200,000 deaths per year worldwide, and almost a quarter of the world population is infected with it (Gaensbauer and Broadhurst 2019). Even after decades of research, there still is a lack of an effective short treatment course or a potent vaccine and better diagnostics. Due to non-compliance with the drug regimen, a high percentage of TB patients drop-out from the treatment program that lasts at least 6 months in general (Awofeso 2008; Churchyard 2018). This high-dropout rate has led to increased cases of TB relapse and drug resistance, making TB even more challenging to treat. Although new drugs have been introduced, none of them have proven to be completely effective. The only notable exception is the use of Pretomanid in combination with Bedaquiline (BDQ) and Linezolid for the treatment of a specific type of highly treatment-resistant TB of the lungs that has shown promise (Bigelow et al. 2020).
Given the current situation, the World Health Organization’s (WHO) ‘End the TB’ strategic program that aims to reduce TB incidences by 80% and deaths by 90% by the year 2030 seems rather ambitious (World Health Organization 2020). The ongoing COVID-19 pandemic has made the situation even more challenging (Grüber 2020) by severely disrupting the TB, Human Immunodeficiency Virus (HIV), and Malaria programs worldwide. In addition, according to recent estimates, COVID-19 has set back worldwide TB treatment efforts by almost a decade and reducing gains to control the disease (The Global Fund 2020). Under such circumstances, we are in urgent need of effective treatment for TB.
A deep understanding of the mechanism of action and atomic details of critical elements contributing to any pathogenic micro-organism's virulence is crucial for successful and effective drug development. The mycobacterial membrane enables the bacilli to survive in the hostile environment inside the host (Lin et al. 2002; Hahn et al. 2005; McClean 2012). The characteristic cell envelope, slow growth rate and unique metabolic features (e.g. glyoxalate pathway, diauxic growth, etc.) of Mtb make it an exciting but challenging system to study (Faller et al. 2004; Målen et al. 2011; Koul et al. 2014). The Mtb cell wall has a distinctive composition, and despite close resemblance with Gram-positive bacteria, its outer membrane contains a complex mixture of lipids such as mycolic acids that distinguishes it from other bacteria (Marrakchi et al. 2014). An extensive physicochemical and structural characterization of Mtb membrane samples could help us understand its composition. Further, various ‘omics’ studies have revealed information about constituent membrane proteins and the secretome of the pathogen. However, there is a large gap in the understanding of the Mtb membrane proteins from their structural point of view. This has been a significant bottleneck in deciphering the roles and mechanism of action of these proteins, which in turn has thwarted the discovery and development of novel therapeutics.
Cryo-electron microscopy (cryo-EM) is proving instrumental in breaking this bottleneck (Munir et al. 2021). Recent advances in cryo-EM have been the silver lining and have proven to be extremely helpful in unraveling the structures of key drug targets of various pathogens, including Mtb. A relatable example has been structure elucidation of various membrane proteins and drug targets of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in record time. This has advanced drug discovery and vaccine development efforts against SARS-CoV-2 worldwide (Jeong et al. 2020; Wrapp et al. 2020). Not surprisingly, cryo-EM has also helped determine structures of key Mtb proteins that are important for virulence and survival of the pathogen and are important targets for drug discovery and vaccine development programs. Furthermore, developments in cryo-electron tomographic (cryo-ET) techniques and cryo-EM have contributed a tremendous amount of structural knowledge about mycobacterial membrane organization and its constituent membrane proteins. This has proven instrumental in modern drug discovery and a better understanding of host–pathogen interactions (Sani et al. 2010b). So far, 75 cryo-EM structures of mycobacterial proteins have been deposited in the Protein data bank (PDB) (Table 1) (Burley et al. 2019). The majority of these structures are membrane proteins. However, very few of these have been tested as targets for drug discovery, while others are yet to be explored. In this review, we discuss the structures of some of the key Mtb proteins and protein complexes elucidated by EM. The list includes cryo-EM structures of type VII secretion systems, Rv1819c, Arabinosyltransferase, Fatty Acid Synthase (FAS), F-type ATP synthase, respiratory supercomplex, Caseinolytic proteases (Clp) P1P2 protease, ClpB disaggregase and S-adenosylmethionine (SAM) riboswitch proteins-protein complexes. These proteins-protein complexes are involved in distinct and vital cellular processes like transport, virulence, electron transport, etc., in Mtb and are promising candidates for drug development. These cryo-EM structures have opened new avenues for novel drug discovery programs. In addition, how recent advents in cryo-EM have helped us in gaining insights into the Mtb membrane organization is briefly discussed. Compiled structural information about mycobacterial membrane and membrane proteins deduced using cryo-EM would provide the researchers with a visual picture of reaction mechanisms associated with them and help design novel antituberculars.
Table 1.
List of mycobacterial membrane protein structures determined using cryo-EM and deposited in PDB (The titles are as deposited by respective authors)
| Sr. no | PDB code | Title of the PDB Entry | References |
|---|---|---|---|
| 1 | 5V93 | Cryo-EM structure of the 70S ribosome from M. tuberculosis bound with Capreomycin | Yang et al. (2017) |
| 2 | 5ZEP | M. Smegmatis hibernating state 70S ribosome structure | Mishra et al. (2018) |
| 3 | 6DZI | Cryo-EM Structure of M. Smegmatis 70S C(minus) ribosome 70S-MPY complex | Li et al. (2018a) |
| 4 | 5ZEB | M. Smegmatis P/P state 70S ribosome structure | Mishra et al. (2018) |
| 5 | 5V7Q | Cryo-EM structure of the large ribosomal subunit from M. tuberculosis bound with a potent linezolid analog | Yang et al. (2017) |
| 6 | 5ZET | M. smegmatis P/P state 50S ribosomal subunit | Mishra et al. (2018) |
| 7 | 6DZP | Cryo-EM Structure of M. smegmatis C(minus) 50S ribosomal subunit | Li et al. (2018a) |
| 8 | 6DZK | Cryo-EM Structure of M. smegmatis C(minus) 30S ribosomal subunit with MPY | Li et al. (2018a) |
| 9 | 5XYM | Large subunit of M. smegmatis ribosome | Li et al. (2018b) |
| 10 | 5XYU | Small subunit of M. smegmatis ribosome | Li et al. (2018b) |
| 11 | 5ZEU | M. smegmatis P/P state 30S ribosomal subunit | Mishra et al. (2018) |
| 12 | 5O61 | The complete structure of the M. smegmatis 70S ribosome | Hentschel et al. (2017) |
| 13 | 5O60 | 50S large ribosomal subunit from M. smegmatis | Hentschel et al. (2017) |
| 14 | 5O5J | 30S small ribosomal subunit from M. smegmatis | Hentschel et al. (2017) |
| 15 | 6EDT | M. tuberculosis RNAP open promoter complex with RbpA/CarD and AP3 promoter | Boyaci et al. (2019) |
| 16 | 6EE8 | M. tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter | Boyaci et al. (2019) |
| 17 | 6EEC | M. tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter captured by Corallopyronin | Boyaci et al. (2019) |
| 18 | 6C04 | M. tuberculosis RNAP Holo/RbpA/double fork DNA -closed clamp | Boyaci et al. (2018) |
| 19 | 6BZO | M. tuberculosis RNAP Holo/RbpA/Fidaxomicin/upstream fork DNA | Boyaci et al. (2018) |
| 20 | 6FBV | M. tuberculosis RNA polymerase in complex with Fidaxomicin | Lin et al. (2018) |
| 21 | 6HWH | Functional obligate respiratory supercomplex from M. smegmatis | Wiseman et al. (2018) |
| 22 | 6C06 | M. tuberculosis RNAP Holo/RbpA/Fidaxomicin | Boyaci et al. (2018) |
| 23 | 6C05 | M. tuberculosis RNAP Holo/RbpA in relaxed state | Boyaci et al. (2018) |
| 24 | 6M7J | M. tuberculosis RNAP with RbpA/us fork and Corallopyronin | Boyaci et al. (2019) |
| 25 | 5LZP | Binding of the C-terminal GQYL motif of the bacterial proteasome activator Bpa to the 20S proteasome | Bolten et al. (2016) |
| 26 | 7BVF | Cryo-EM structure of M. tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | Zhang et al. (2020b) |
| 27 | 6SGZ | M. smegmatis Structure of protomer 2 of the ESX-3 core complex | Famelis et al. (2019) |
| 28 | 6SGX | M. smegmatis Structure of protomer 1 of the ESX-3 core complex | Famelis et al. (2019) |
| 29 | 6SGW | M. smegmatis Structure of the ESX-3 core complex | Famelis et al. (2019) |
| 30 | 6BGL | Doubly PafE-capped 20S core particle in M. tuberculosis | Hu et al. (2018) |
| 31 | 6BGO | Singly PafE-capped 20S CP in M. tuberculosis | Hu et al. (2018) |
| 32 | 6EYD | M. smegmatis RNA polymerase Sigma-A holoenzyme | Kouba et al. (2019) |
| 33 | 6UMM | M. smegmatis ESX-3 translocon complex | Poweleit et al. (2019) |
| 34 | 6F6W | Structure of M. smegmatis RNA polymerase core | Kouba et al. (2019) |
| 35 | 7BVC | M. smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | Zhang et al. (2020b) |
| 36 | 7BVG | M. smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose | Zhang et al. (2020b) |
| 37 | 7BX8 | M. smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | Zhang et al. (2020a) |
| 38 | 7BWR | M. smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | Zhang et al. (2020a) |
| 39 | 6VGQ | ClpP1P2 complex from M. tuberculosis with GLF-CMK bound to ClpP1 | Vahidi et al. (2020) |
| 40 | 6VGK | ClpP1P2 complex from M. tuberculosis | Vahidi et al. (2020) |
| 41 | 6VGN | ClpP1P2 complex from M. tuberculosis bound to ADEP | Vahidi et al. (2020) |
| 42 | 7BVE | M. smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | Zhang et al. (2020b) |
| 43 | 3J83 | M. tuberculosis Heptameric EspB Rosetta model | Solomonson et al. (2015) |
| 44 | 5ZEY | M. smegmatis Trans-translation state 70S ribosome | Mishra et al. (2018) |
| 45 | 6SGY | Structure of EccB3 dimer from the ESX-3 core complex | Famelis et al. (2019) |
| 46 | 6GJC | Structure of M. tuberculosis Fatty Acid Synthase—I | Elad et al. (2018) |
| 47 | 4V8V | M. tuberculosis fatty acid synthase multienzyme complex | Ciccarelli et al. (2013) |
| 48 | 4V8W | M. tuberculosis fatty acid synthase multienzyme complex | Ciccarelli et al. (2013) |
| 49 | 6ED3 | M. tuberculosis ClpB in complex with AMPPNP | Yu et al. (2018) |
| 50 | 6PPU | Cryo-EM structure of AdnAB-AMPPNP-DNA complex | Jia et al. (2019) |
| 51 | 6DJU | M. tuberculosis ClpB in complex with ATPgammaS and casein, Conformer 1 | Yu et al. (2018) |
| 52 | 6DJV | M. tuberculosis ClpB in complex with ATPgammaS and casein, Conformer 2 | Yu et al. (2018) |
| 53 | 6PPR | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | Jia et al. (2019) |
| 54 | 6WBY | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 2 | Tan et al. (2020b) |
| 55 | 6WBX | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 1 | Tan et al. (2020b) |
| 56 | 6TQF | The structure of ABC transporter Rv1819c in AMPPNP bound state | Rempel et al. (2020) |
| 57 | 6TQE | The structure of ABC transporter Rv1819c without addition of substrate | Rempel et al. (2020) |
| 58 | 6UES | Apo SAM-IV Riboswitch | Zhang et al. (2019b) |
| 59 | 6PPJ | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP | Jia et al. (2019) |
| 60 | 4V8L | M. smegmatis Fatty Acid Synthase | Boehringer et al. (2013) |
| 61 | 6UET | SAM-bound SAM-IV riboswitch | Zhang et al. (2019b) |
| 62 | 6W98 | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | Tan et al. (2020b) |
| 63 | 6ADQ | Respiratory Complex CIII2CIV2SOD2 from M. smegmatis | Gong et al. (2018) |
| 64 | 2BYU | M.tuberculosis Acr1(Hsp 16.3) fitted with wheat sHSP dimer | Kennaway et al. (2005) |
| 65 | 6X0O | Cryo-EM structure of M. tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | Tan et al. (2020a) |
| 66 | 6XZC | CryoEM structure of the ring-shaped virulence factor EspB from M. tuberculosis | Korotkova et al. (2015) |
| 67 | 6LUM | Structure of M. smegmatis succinate dehydrogenase 2 | Not published |
| 68 | 7JG5 | Cryo-EM structure of bedaquiline-free M. smegmatis ATP synthase rotational state 1 | Guo et al. (2020a) |
| 69 | 7JG6 | Cryo-EM structure of bedaquiline-free M. smegmatis ATP synthase rotational state 2 (backbone model) | Guo et al. (2020a) |
| 70 | 7JG7 | Cryo-EM structure of bedaquiline-free M. smegmatis ATP synthase rotational state 3 (backbone model) | Guo et al. (2020a) |
| 71 | 7JG8 | Cryo-EM structure of bedaquiline-saturated M. smegmatis ATP synthase rotational state 1 (backbone model) | Guo et al. (2020a) |
| 72 | 7JG9 | Cryo-EM structure of bedaquiline-saturated M. smegmatis ATP synthase rotational state 2 (backbone model) | Guo et al. (2020a) |
| 73 | 7JGA | Cryo-EM structure of bedaquiline-saturated M. smegmatis ATP synthase rotational state 3 | Guo et al. (2020a) |
| 74 | 7JGB | Cryo-EM structure of bedaquiline-free M. smegmatis ATP synthase FO region | Guo et al. (2020a) |
| 75 | 7JGC | Cryo-EM structure of bedaquiline-saturated M. smegmatis ATP synthase FO region | Guo et al. (2020a) |
Cryo-Electron Tomography and Mycobacterial Membrane Organization
The mycobacterial cell wall is a unique structure that acts as a permeability barrier, and it is essential for survival, virulence, and pathogenesis (Barry Iii et al. 1998; Etienne et al. 2002). Over the decades, researchers have been trying to elucidate the mycobacterial membrane structure using transmission electron microscopy. However, sample preparation methods that involved fixing with various solvents and staining with heavy metal salts in different labs led to imaging of dehydrated membranes (Daffe and Draper 1997). The information obtained from these studies thus suffered from sample preparation artifacts along with lack of details (Paul and Beveridge 1992; Zuber et al. 2008). Advances in sample preparation methods, specially CEMOVIS (Al-Amoudi et al. 2004; Hoffmann et al. 2008) and cryo-EM of the mycobacterial capsular layer (Sani et al. 2010a), have allowed the visualization of fully hydrated and unstained mycobacterial membrane providing new structural insights into membrane organization. This unique combination of CEMOVIS and cryo-ET of plunge frozen membranes has provided an improved understanding of the structure of lipid bilayer and the outer membrane (Hoffmann et al. 2008).
Summing up the information available, the four-layered envelope of Mtb is about 75–80-nm-thick and can be described from the exterior to the interior of the cell (Daffé and Marrakchi 2019). A capsule layer of ~ 35 nm thickness is associated with ~ 8 nm outer membrane made up of mycolipids. The outer membrane is, in turn, linked to a ~ 35 nm thick cell wall primarily made up of the arabinogalactan layer tethered to the peptidoglycan layer (Alsteens et al. 2008). This is followed by a periplasmic space and ~ 7 nm thick granular layer tethered to the cell's plasma membrane (Lee et al. 2005), where membrane proteins are embedded (Besra and Chatterjee 1994). Interestingly, about thirteen enzymes are involved in synthesizing the arabinogalactan-peptidoglycan complex, which are unique to Mycobacteria (Lee et al. 2005; Crick and Brennan 2008) and hence are putative drug targets against Mtb.
While the understanding of Mtb cell envelops has improved due to their imaging in cryo-conditions, the resolutions achieved are still low. Moreover, it has been shown that lab-grown bacteria may have compositional heterogeneity in the membrane due to the kind of media used (Sani et al. 2010a) or the presence of detergents that may lead to capsule shedding (Hoffmann et al. 2008; Sani et al. 2010a). These problems may be overcome by cryo-EM of vitrified Mtb engulfed by macrophages. However, these are usually too thick to be vitrified properly. The recent developments in sample vitrification methods, high-pressure freezing, and preparation of thin lamella suitable for cryo-EM via FIB-SEM may make it possible (Gorelick et al. 2019; Ravelli et al. 2020). Besides, advances in cryo-ET and the establishment of unique Biosafety Level–3 cryo-EM facilities have raised the hopes of carrying out an exhaustive and detailed structural characterization of native Mtb envelop and membranes (Parvate 2018).
Cryo-EM Structures of Mtb Membrane Proteins
The majority of membrane proteins are recalcitrant to crystallization. Thus, several key Mtb membrane protein structures could not be determined by X-ray crystallography, severely impacting the progress of the Mtb drug discovery field. One of the key issues had been the requirement of milligram quantities of highly pure, compositionally, and conformationally homogenous protein sample for crystallization (Acharya and Lloyd 2005). Moreover, most Mtb membrane proteins express poorly in recombinant expression systems and require extensive optimization of expression and purification conditions (Korepanova et al. 2007). Unlike cytosolic proteins, membrane proteins are highly unstable when extracted from the lipid bilayer (Haltia and Freire 1995). Therefore, to mimic such an environment, detergents/lipids are added to the sample during membrane protein extraction and purification. Due to the presence of lipids/detergents, crystallization becomes more difficult. Technological advancements in cryo-EM, such as the development of direct electron detectors and stable microscopes, have proven beneficial for protein structure determination, especially for membrane proteins (Frank 2017). It overcomes the above two limitations as it requires less amount of sample and does not require crystallization (Renaud et al. 2018). By overcoming these two significant barriers, cryo-EM has accelerated the membrane protein structure determination.
Cryo-EM single-particle analysis technique has led to the structure determination of several essential Mtb proteins providing novel structural and mechanistic insights into their functions. We discuss some of the protein–protein complexes that are therapeutically important targets and whose structures were recently solved in a near-native environment due to current advances in the cryo-EM technique.
Structural Insights into Mycobacterial Secretion Systems and Transporters
ESX/ Type VII Secretion System
Bacterial cell membranes contain specific protein complexes used by the pathogenic bacteria to secrete virulence factors helping them to invade the host. Such secretion systems are classified into different types (e.g., Type I, Type II,…Type IX) based on their structure, composition, and function (Green and Mecsas 2016; Lasica et al. 2017). Mycobacteria consist of five paralogous ESX (ESX: ESAT-6 Secretion System, ESAT-6: 6 kDa early secretory antigenic target) secretion systems important for virulence, immune modulation, gene transfer, nutrient and metabolite uptake, and cell physiology of the bacterium (Bunduc et al. 2020). These systems release certain effectors, commonly known as virulence factors, that help the Mtb infect the host. It is proven that the mycobacterial cytosolic translocation is mediated via ESAT-6, a secretion product of the ESX-1 secretion system, and established a connection between the translocation of pathogenic mycobacterium and its virulence (Houben et al. 2012). Although the role of effector proteins from ESX-1, 3, and 5 systems have been traced, the lack of three-dimensional structure of the ESX system had limited the knowledge of its mechanism of operation until the structure of ESX-3 from M. smegmatis was solved at ~ 4 Å via EM (Famelis et al. 2019). During the limited supply of iron, ESX-3 is expressed (Rodriguez et al. 2002; Macikag et al. 2009) and is reported to be associated with various functions such as metal ion homeostasis (Serafini et al. 2009; Siegrist et al. 2009; Tinaztepe et al. 2016; Tufariello et al. 2016), repair of phagosomal damage, and phagosome maturation (Mehra et al. 2013; Portal-Celhay et al. 2016; Mittal et al. 2018).
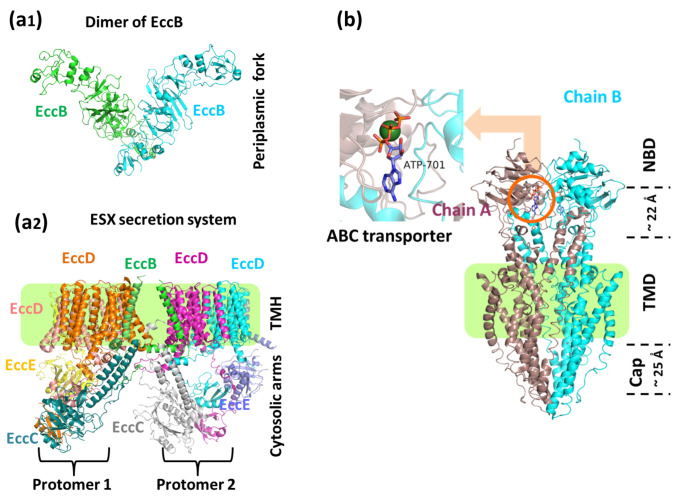
The ESX-3 secretion system consists of core components called 'ESC conserved component,' (Ecc) i.e., Ecc viz; EccB, EccC, EccD, EccE, and fifth component MycP. These components are responsible for releasing virulence factors EsxA and EsxB (Abdallah et al. 2006; Brodin et al. 2006; Ohol et al. 2010; Sloan Siegrist et al. 2014; Ates et al. 2015; Van Winden et al. 2016). The Ecc components together form a hexameric pore in its cell envelope. This pore releases virulence factors that help them infect the host and evade the actions of the host immune system. The membrane-anchored element ‘MycP’ possesses protease activity, while the fork domain of EccB is towards the periplasmic face (Fig. 1a1). The ESX-3 complex is made up of two protomers. Each protomer constitutes EccB3, EccC3, and EccE3 as a single copy and EccD3 in two copies of homo-dimers. The EccB3 from each protomer is attached to a transmembrane helix (TMH) that anchors it firmly within two protomers and holds the two protomers together with the help of the fork domain (Fig. 1a2). Together, these two protomers form the secretion pore with dimensions large enough to transport dimeric substrate proteins in folded conformation across its cell envelope. Another structure resolved at 3.7 Å, has elaborated how the pseudo-ATPase domain (reconstructed at ~ 10 Å) in the translocation gate is positioned in the EccC3 domain towards the cytoplasm (Poweleit et al. 2019).
Fig. 1.
a1 Structure of EccB dimer from the ESX-3 core complex showing fork architecture at the periplasm (PDB: 6SGY). a2 The ESX-3 core complex structure formed by EccC, EccD, and EccE subunits is arranged laterally. The pale green box is the trans-membrane region. The translocon pore is formed by the EccD subunit (PDB: 6SGW). b The structure of ABC transporter Rv1819c. In the inset, the binding site for the nucleotide (PDB: 6TQE) is shown. The substrate enters from the cap end of the transporter. The conversion of ATP to ADP leads to the import of the incoming substrate from the NBD end (Color figure online)
The structure of ESX-1 secretion associated protein B (EspB), a secretion protein was also predicted from a cryo-ET, with a resolution of about 30 Å from Mtb (Solomonson et al. 2015). It confers sevenfold symmetry with approximate dimensions of 100 Å × 80 Å cylinders with about 50 Å pore. It has been reported that EspB C-terminal domain (CTD) is processed by the membrane-bound protease MycP1P2 following secretion (McLaughlin et al. 2007; Xu et al. 2007; Ohol et al. 2010; Solomonson et al. 2013; Wagner et al. 2013). A visual snapshot of mycobacterial membrane organization using electron microscopy with proteomic confirmation of the presence of ESX-1-secreted proteins has been reported (Sani et al. 2010b). Moreover, an EM structure of ESX-1 secreted Mtb-EspB has been reconstructed at 3.37 Å resolution providing important structural and functional insights. The seven units of EspB form a heptameric cylinder with dimensions of 90 Å × 90 Å. The central channel pore is about 45 Å wide. The overall cylindrical arrangement of EspB indicates that it could facilitate the transport of ESX-1 substrates such as EsxA-EsxB or B form DNA molecules (Korotkova et al. 2015). The ESX-3 is proven to be indispensable for Zinc and iron metabolism in Mtb. BBH7, a benzyloxybenzylidene-hydrazine compound, has been identified as an inhibitor of the ESX system, which induces Zinc stress in Mtb, thereby severely inhibiting bacterial growth (Rybniker et al. 2014; Cole 2016). Combining the structural information with the knowledge of inhibitors like BBH7, highly specific inhibitors against ESX systems could be designed that may reduce the virulence of Mtb and thus could be an effective alternative antitubercular strategy.
Rv1819c, an ABC Transporter
The ATP-binding cassette transporters (ABC transporters) are probably one of the most studied superfamilies of the multi-subunit transporters, which transport a broad spectrum of molecules at the expense of ATP (Choudhuri et al. 2002). The presence of membrane-spanning domains (MSD) and the nucleotide-binding domains (NBD) is typical of ABC transporters. The ATPase activity is associated with the NBD, which is positioned towards the cytoplasm. ATP hydrolysis provides the energy required to transport substrates across membranes (De Rossi et al. 2006). The gene rv1819c was shown to be associated with the uptake/transport of cobalamin in Mycobacteria (Gopinath et al. 2013). However, the protein Rv1819c was related to the transport of antimicrobial peptides like Bleomycin (Domenech et al. 2009) and efflux of antibiotic ciprofloxacin when expressed in C. glutamicum, indicating its potential role in providing resistance through drug efflux (Mazando et al. 2013).
Recently, the structure of the Rv1819c homodimer in the nucleotide bound state was determined at 3.5 Å resolution using cryo-EM (Rempel et al. 2020). The electron density for detergent micelles, observed in the cryo-EM map, helped approximate membrane position around the protein. The structure also revealed the presence of an ABC exporter fold in Rv1819c. The characteristic features in this fold that distinguish it from the similar fold found in previously reported ABC transporters are the unusually large hydrophilic cavity and an additional TMH-0 towards the N-terminal (Korkhov et al. 2012; Choudhury et al. 2014; Lin et al. 2015). The cavity volume measures about 7,700 Å3 spanning almost the entire width of the lipid bilayer membrane. It is made up of two lateral chambers, larger across the membrane, while a smaller one acts as a junction between the larger chamber and the NBD (Fig. 1b). The structures also revealed a 17 amino acid residues long loop in TMH3, constricting between the two chambers near the membrane towards the cytoplasm. The cavity's inner side is lined by the network of polar and negatively charged amino acid residues. Such a lining could facilitate the exchange of hydrophilic molecules across the cavity, thus in and out of the membrane. Considering the cavity volume, it could facilitate the transport of peptides, too, as seen in transporters of Microcin-J25 export ATP-binding/permease protein McjD and the peptidase-containing ATP-binding cassette transporters (PCAT) (Choudhury et al. 2014; Lin et al. 2015). Further, the structure shows that the exporter fold cavity of the Rv1819c is big enough to accommodate about six cobalamin moieties. This indicates that there is no specific, stable binding site for cobalamin in the cavity. This hypothesis is strengthened with the observation that no substrate was eluted during the co-purification experiments of the Rc1819c. However, the presence of several antimicrobial peptides, including Bleomycin, was confirmed in the cavity. Since Mtb is a slow-growing pathogen, such a large cavity in a non-specific exporter could explain the uptake and accumulation of hydrophilic antibiotics inside the bacterial cell (Taber et al. 1987; Aller et al. 2009). In another study, it was observed that Rv1819c helped the Mtb cells in excreting out the antibiotics, serving as a salvage pathway for Mtb (Balganesh et al. 2010). It would be interesting to see if the Rc1819c could rescue the Mtb from antibiotic stress when treated with a specific inhibitor designed using current structural information. If it could do so, specific inhibitors against Rc1819c would prove to be effective antituberculars.
Proteins Involved in Mycobacterial Cell Wall Synthesis and Maintenance
Arabinosyltransferases: Members of Mycobacterial Cell Wall Synthesis Pathway
As described earlier, the unique cell wall of Mtb has a distinctive structure called mycolyl-arabinogalactan-peptidoglycan. It consists of three essential components: mycolic acids, arabinogalactan, and a cross-linking network of peptidoglycans (Alderwick et al. 2015) synthesized by various transferases and synthases unique to mycobacteria. Recently, cryo-EM enabled the structure determination of some of the key enzymes involved in cell wall biosynthesis. These structures hold particular importance in anti-TB drug development since most of the constituents associated with these enzymes are exclusive to Mtb and thus could act as excellent drug targets. One example is membrane-associated arabinosyltransferases EmbA, EmbB, EmbC enzymes that are members of the mycobacterial cell wall synthesis pathway against which the drug ethambutol (EMB) is considered to be active. EMB is a front-line antitubercular used to treat multi-drug resistant TB (MDR-TB) (Sreevatsan et al. 1997). Interestingly, mutations in these arabinosyltransferases result in resistance to EMB (Safi et al. 2008; Sun et al. 2018).
Structures of EmbA-EmbB and EmbC-EmbC complexes in different states, i.e., bound to their glycosyl donor and acceptor, EMB, were determined (Zhang et al. 2020b). These structures help in elucidating mechanisms of EmbA-EmbB and EmbC-EmbC complexes and mode of inhibition by EMB. EmbA and EmbB together form a heterodimer, while EmbC forms a homodimer for its activity. EmbB, a fifteen TMH spanning protein, catalyzes the transfer of arabinose from the donor decaprenyl-phosphate-arabinose (DPA) to its arabinosyl acceptor. On the cytoplasmic face of EmbB is an acyl-carrier protein (AcpM), which is suggested to be associated with the transferase activity. The DPA and di-arabinose bound structures help us understand the overall mechanism of catalysis. The EmbA-EmbB complex catalyzes the branching reaction, while the elongation reaction is catalyzed with the help of the EmbC dimer. In another report, the complex EmbB-EmbB’-AcpM2 in resting-state on binding to DPA gets activated, as shown in Fig. 2a (Zhang et al. 2020a). It leads to the transfer of arabinose from the donor DPA to its arabinosyl acceptor. EMB bound to EmbB structure explains the interaction of the drug with the active site in EmbB (Fig. 2a). The drug EMB competes for the binding sites for substrates in EmbB and EmbC (Goude et al. 2009). The 3.3 Å EM structures of EmbB (substrate and drug bound) provided crucial insights into the mode of action of substrate binding and presented a map of how resistance against ethambutol could have occurred due to mutations accumulated around the active site (Tan et al. 2020a).
Fig. 2.
a Mycobacterial Arabinosyltransferase complex. The complex is composed of two chains, each of EmbB and AcpM. On binding of DPA (red-green spheres), resting-state complex (PDB: 7BX8) is activated (PDB: 7BWR). The arabinosyltransferase is inhibited by ethambutol (Etmb: red) binding in the active site in EmbB (7BVC). b The barrel-shaped MsFAS is a hexamer with one monomer chain shown in cyan (PDB: 6GJC). Each chain is made up of 6 subunits, namely—acetyltransferase (AT), enoyl reductase (ER), dehydratase (DH), malonyl transacylase (MPT), ketoacyl reductase (KR), and ketoacyl synthase (KS) (Color figure online)
Similarly, arabinofuranosyltransferase D (AftD) is another class of transferase. The structure from M. abscessus at 2.9 Å resolution (Tan et al. 2020b) revealed similarities in overall architecture with EmBA, EmbB, and EmbC (Zhang et al. 2020b). The AftD structure is α-helix rich with sixteen TMHs and a cytoplasmic portion consisting of both α and β components. The cytoplasmic α/β folds are homologous to EmbC, three carbohydrate-binding modules (CBMs), and one structural β-sandwich domain. Besides, AftD has a conserved glycosyltransferase (type C) fold, which binds to complex arabinose glycans. The high-resolution structure of AftD enabled the identification of the active site for the transferase activity and highlighted the importance of Arg1389 residue.
Mycobacterial Fatty Acid Synthase
Mycolic acid is a significant and unique fatty acid found in the Mtb cell wall playing a vital role in pathogenicity (Marrakchi et al. 2014). The enzyme system that catalyzes the synthesis of fatty acid precursors of mycolic acids is known as the FAS complex. In Mtb, there are two types of FAS systems FAS1 and FAS2. FAS1 is found in other mycolic acid producers like certain Bifidobacteriales and Actinomycetales species and in Mtb, it is known to produce fatty acid precursors with a carbon chain length of C16-18 and C26. However, in M. smegmatis, FAS produces C24 fatty acids instead of C26 (Trivedi et al. 2004). FAS2 is involved in the elongation of products of FAS1 in Mycobacteria (Takayama et al. 2005). With the help of other enzymes, FAS2 converts C16-18 long products of FAS1 to C45-64 long meroacids, which are further converted to mycolic acids (Bhatt et al. 2007).
The first mycobacterial FAS1 structure was reported at a global resolution of 7.5 Å from M. smegmatis, succeeded by a 3.3 Å structure from Mtb, determined using single-particle cryo-EM (Boehringer et al. 2013; Elad et al. 2018). The FAS1 complex structure appears like a barrel of ~ 25 nm diameter. Even though the resolution is not high, almost all the helices were resolved in the density map. Although separate β-strands were not visible, their presence could be confidently marked due to flattened density clouds corresponding to β-sheets in the MsFAS model. At the same time, these features could be characterized in Mtb FAS1. However, a crucial difference in the reactive pocket of fungal FAS1 and MsFAS is visible. MsFAS at the opening of the reactive pocket is broader than that observed in fungal FAS1 (Gipson et al. 2010). These differences in the cleft size of fungal and mycobacterial FAS1 could enable Mtb FAS1 to produce C26 fatty acids.
The multifunctional, mycobacterial FAS1 is a 2MDa complex consisting of α6 homo-hexamers (Fig. 2b). Each protomer is a product of six subunits named acetyltransferase (AT), enoyl reductase (ER), dehydratase (DH), malonyl transacylase (MPT), ketoacyl reductase (KR), and ketoacyl synthase (KS), assuming respective function as indicated by its name (Fig. 2b). Although considered homologous, M. smegmatis FAS1 (MsFAS) is 900 amino acid residues shorter than the fungal FAS1. This difference could be attributed to the presence of the C-terminal phosphopantetheinyl transferase domain in the fungal FAS. This structural data, particularly regions that are different from fungal FAS, can be used for virtual screening and identification of drug candidates that could target mycolate synthesis.
Electron Transport Chain Proteins are Important Therapeutic Targets
F-type ATP Synthase
The mycobacterial F-type ATP synthase is an essential enzyme among its respiratory chain protein members, which provides ATPs crucial for survival. Encoded by unc operon, the ATP synthase consists of six subunits. F1 subunit crystal structure had been reported (Zhang et al. 2019a). However, the complete enzyme structure was lacking, which was recently reported from M. smegmatis using cryo-EM (Fig. 3a) (Guo et al. 2020a).
Fig. 3.
The complete F-type ATPase complex. a F0 is the membrane-bound region that mainly consists of a C-ring. The F1 is the peripheral region where ATP is synthesized (PDB: 7JG5). b SF0 regions with and without BDQ (dark red) bound to it. c BDQ binds in two sites. The leading site lies between ‘a’ and ‘c’ subunits, while the lagging site lies only on the ‘c’ subunit (Color figure online)
The structure of mycobacterial F-type ATP synthase holds particular importance since the recently approved antitubercular drug BDQ specifically inhibits mycobacterial F-type ATP synthase (Haagsma et al. 2009). It was speculated that it binds either to the c subunit of the enzyme or targets ε subunit (Preiss et al. 2015; Kundu et al. 2016). However, the recent cryo-EM structure describes the full-length enzyme in different drug bound states while resolving all of its subunits. The EM structure remarkably reveals that there are in all 7 BDQ binding sites at F0 region (Fig. 3b) in the ATP synthase: 5 BDQ molecules exclusively bind to the c chains of the ring called the ‘lagging site’ (Fig. 3c) while the remaining 2 interact with the subunit a and subunit c, called the ‘leading site’ (Fig. 3c). These findings have ruled out the proposed interaction of the BDQ with the ε subunit and have elucidated the overall mechanism and biochemistry of the drug action (Guo et al. 2020b). A recent report also indicates that the drug BDQ can target human mitochondrial ATPase (Luo et al. 2020). This has posed serious concerns regarding the treatment of Multi/Extremely drug resistant (M/XDR) TB. Thus, the availability of the complete structure of mycobacterial ATPase is a significant advancement in designing highly specific inhibitors.
Mycobacterial Respiratory Supercomplex
Several membrane-bound multi-protein enzyme complexes together form the respiratory chain in Mtb (Gong et al. 2018). Popularly termed as respiratory supercomplexes, which help transfer electrons from reduced nicotinamide adenine dinucleotide (NADH) to molecular oxygen (O2), thus linking this electron transfer to transmembrane proton translocation (Roy and Wani 2020). The net result is the generation of a proton gradient that drives the ATP production and transport across the membrane (Rao et al. 2008).
The M. smegmatis respiratory supercomplex’s (Fig. 4a1) structure was determined at 3.3–3.5 Å resolutions with its crucial cofactors like heme clearly visible in the electron density map (Fig. 4a2) (Wiseman et al. 2018). The complex is primarily made up of two sub-complexes, complex III and complex IV. The dimer of complex III lies in the middle and is flanked by individual complex IV subunits from both sides. The center for oxygen reduction lies in complex IV, while the quinol site is present in complex III. The electron transfer between these two sub-complexes is facilitated via a cytochrome subunit bridged between complex III and IV. The complex of dimeric CIII–CIV arrangement is C2 symmetric with dimensions of 200 × 70 × 120 Å. Such symmetry is the first of its kind observed among respiratory supercomplexes so far. At a global resolution of 3.5 Å, all the prosthetic groups of respiratory supercomplex, menaquinone, and heme molecules could be resolved and built. The two menaquinone molecules could be observed in the conserved quinone binding site located in the CIII complex. The complex CIII comprises of QcrA, QcrB, and QcrC subunits, while the complex CIV consists of CtaC, CtaD, CtaE, and CtaF subunits. When the supercomplex is in an open state, the cc head of the QcrC is bound to only complex CIV, while in the closed state, the head domain is associated with both CIII and CIV complexes (Gong et al. 2018).
Fig. 4.
Structure of respiratory supercomplex (PDB: 6HWH). a1 The supercomplex is composed of two subcomplexes and some unidentified peptides (grey). Complex IV (blue) consists of CtaC, CtaD, CtaE, and CtaF units. Complex III is composed of QcrA, QcrB, and QcrC domains. Open QcrC is in orange, while closed conformation is in red. a2 The electron from heme bL is transferred to heme bH rather than to FeS. The cardiolipins (CDL) are anchored to the two Arg residues near the menaquinone binding site in QcrB and the helix of QcrC that embeds in the membrane. The heme cofactors are shown in purple and the cardiolipin in green. b The MtClpP1P2 complex (PDB: 6VGK). The heptamer ClpP1 (cyan) ring is placed below the heptameric ClpP2 (maroon) ring, but each monomer of ClpP1 interacts with its ClpP2 counterpart. c The ClpB is a hexamer of six protomers. From the top view, the core of the free ClpB appears like a tunnel (PDB: 6ED3). On the binding of nucleotides (ADP: blue spheres, ATPγS brown spheres) and the substrate—casein (red), the size of the tunnel shrinks due to the movement of protomers (e.g., P1), and the overall structure becomes more compact with the movement of each monomer subunit (PDB: 6DJU) (Color figure online)
Mycobacterial bc-type CIII supercomplex appears similar to the dimeric association, as seen in bacterial bc1-type and mitochondrial CIII; however, certain subtle structural differences are present (Xia et al. 1997). Q203, an imidazopyridine amide, is a candidate reported to be an effective inhibitor of cytochrome bc1-aa3 complex, binds explicitly to QcrB of the supercomplex (Lee et al. 2020). Since complex CIII is an essential ROS production site, it is a crucial target to produce lethal stress in mycobacteria (Zhao and Drlica 2014). The superoxide dismutase (SodC) enzyme is known to protect the electron transport chain (ETC) of mycobacteria from this ROS stress when phagocytized by human macrophages, thus helping in the survival of the bacterium inside the host lungs (D’Orazio et al. 2001). It has been found that the SodC is integral to the respiratory supercomplex situated at the periplasmic face. It is probably involved in scavenging the ROS produced by the host, providing a robust defense mechanism to Mtb (Bhattacharya 2015). In summary, the cryo-EM structure of a CIII–CIV respiratory supercomplex from M. smegmatis illustrates how an electron transfer pathway occurs in complex CIII and reaches the final oxygen reduction step in the complex CIV, a new branch of electron transfer for energy transduction in association with superoxide dismutase domain acting as the detoxing agent of ROS.
Structural Insights into Key Mycobacterial Proteases
ClpP1P2 Protease
Proteostasis is a necessary process for maintaining overall protein turnover in the cell using various synthesis and lysis pathways. Specific proteolytic machines in assistance with AAA + unfoldase (AAA: ATPases Associated with diverse cellular Activities) partner recognize certain substrate proteins and mediate their unfolding and further carry out the proteolytic activity (Sauer and Baker 2011). Among such proteases, the ClpP system (Caseinolytic proteases P) is crucial as it helps maintain homeostasis and assist virulence in several bacteria. Particularly in Mtb, the ClpP1P2 protease system is of major importance and indispensable for bacterial growth and virulence (Frees et al. 2014; Bhandari et al. 2018), making it an attractive drug target. Unlike other bacteria, Mtb contains two genes, clpP1 and clpP2, encoding for MtClpP1 and MtClpP2 proteins, respectively (Personne et al. 2013; Alhuwaider and Dougan 2017).
The structures of MtClpP1P2 were recently solved in two conformations; an apo form, which is a compact conformation, and cyclic acyldepsipeptides (ADEPs) bound conformation, which is relatively more relaxed and ordered at the N-terminus. The overall ClpP complex (single ClpP protein 200 residues long) appears like a barrel of about 83 Å height with a central channel formed by two rings stacked one upon another (Fig. 4b). The products of gene clp1 and clp2, i.e., proteins Clp1 and Clp2, form a heptameric ring. The ClpP2 heptamer ring is stacked above that of the ClpP1 ring coaxially to produce a barrel-like structure. In the bacterial ClpP complex, a ‘handle’ region made up of a helix and a β-sheet in each protomer, mediates the interactions within the rings (Liu et al. 2014). The handle region helps the catalytic triad Asp-His-Ser maintain the active site's geometry for proper binding of the substrate polypeptide (Szyk and Maurizi 2006). The 3.0 Å resolution structure of the apo form of mycobacterial ClpP complex suggests that the β-strands in the handle region are disordered, leading to the distorted catalytic triad. This explains the inactivity of the enzyme system in the apo form. On binding with the ADEP, there is no change in the overall architecture of the 'handle' domain (Vahidi et al. 2020).
According to one report, the chloromethyl ketone derivative of compound Bortezomib specifically inhibits the MtClpP1P2 system, imparting cytotoxic effects on Mtb (Moreira et al. 2017). In the same report, it was shown that the inhibitor does not act on human proteases. This structural information of the MtClpP1P2 system could be exploited to design inhibitors highly specific to the protease active site and could be further developed as novel antitubercular. Candidates like Lassomycin, Rufomycin bind to the ClpC1- an ATP unfoldase, thereby affecting the ATP-dependent protein unfolding from proteolysis, ultimately showing a bactericidal effect on active and latent TB infections (Dougan et al. 2020).
ClpB—an ATP-Driven Disaggregase
Like the ClpP system described earlier, another protease system, known as ClpB disaggregase, functions in association with heat-shock proteins. In bacteria, ClpB is a ring-shaped ATP-driven disaggregase belonging to the family of Hsp100 chaperones (Doyle et al. 2013). Under stress conditions, the ClpB, in association with other chaperons and accessory proteins, helps specific proteins refold from an aggregated state, thus helping the cell survive and overcome the stress (Mogk 1999; Doyle and Wickner 2009; Lupoli et al. 2016). Like Hsp104, ClpB utilizes ATP hydrolysis energy, and thus, it is known as 'protein disaggregase,' where it carries polypeptides through its central pore (Weibezahn et al. 2004). The cryo-EM structure of Mtb ClpB was determined at ~ 3.8–3.9 Å, in different conformations (AMPPNP bound conformer, open and closed state, AMPPNP- Adenylyl imidodiphosphate) (Yu et al. 2018). The structure indicates that ClpB is a hexamer arranged in a ring (Fig. 4c). Each protomer consists of three domains: an N-terminal domain (NTD), and NBD1 and NBD2 as two AAA+ type nucleotide domains that carry out ATP assisted peptide translocation (Gates et al. 2017). Thus, each hexamer accommodates 12 NBDs. Eukaryotic Hsp104 and mycobacterial ClpB belong to the same family (Doyle et al. 2013). Still, there are key differences, especially in the NBD regions of these two proteins. The Mtb ClpB has both NBD1 and NBD2 moving in an upward direction. However, in Hsp104, the two NBDs move in opposite directions, i.e., if NBD1 moves upwards, then the NBD2 orients itself downwards. This movement creates a temporary gap in the Hsp104 ring, which is absent in ClpB. This structural difference could probably reflect a different mode of ATP hydrolysis in ClpB and Hsp104. Further experiments, however, are warranted to confirm this observation. ClpB is crucial for Mtb survival inside the host (Lupoli et al. 2018). The insights gained from the ClpB structure could help in developing it into a novel anti-TB drug target.
Cryo-EM Structures that Elucidate Basic Cellular Processes in Mtb
Apart from the proteins mentioned above, a few other crucial structures were reported recently. These structures are essential considering their druggability but are more significant in terms of bacterial physiology and have provided us with a more in-depth understanding of mycobacteria's underlying cellular processes. The list includes Mtb and M. smegmatis ribosomes, small heat-shock protein Acr1, proteasome assembly and its bacterial proteasome activator (Bpa), nuclease AdnAB motor, RNA polymerase, and few others (Table 1). The structure of α-crystallin related protein 1 is among the first few mycobacterial protein structures determined using cryo-EM (Kennaway et al. 2005).
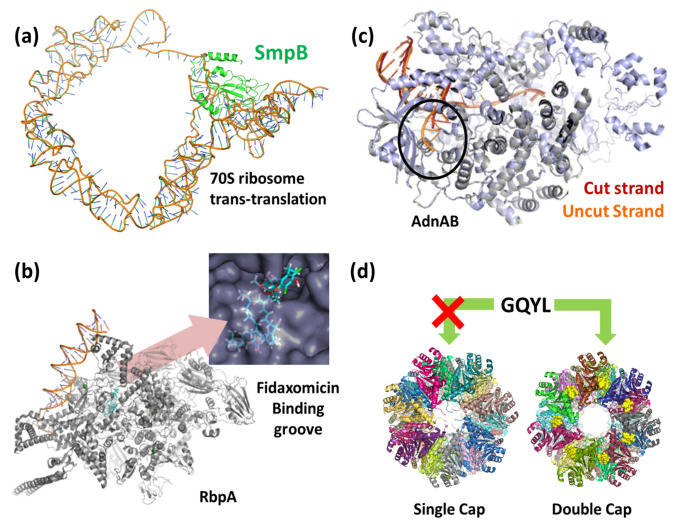
The cryo-EM structures of ribosomes describe how Mtb remodels its ribosomes in Zinc-depleted conditions (Li et al. 2018a). The 70S structure has underlined the differences between Mtb and other bacterial ribosomes, especially the variations in lengths of specific rRNA helices (Li et al. 2018b). The 70S structure now enables us to understand the movement between specific ribosomal proteins, handle, small subunit, and other regions/domains of the complete ribosome assembly (Yang et al. 2017). These structures offer detailed information about the transcription occurring at the ribosome. Additionally, two non-ribosomal proteins were detected in these structures. These two proteins could be some transcription regulators; however, these are yet to be identified. The EM reconstructions of the M. smegmatis 70S ribosomes in the hibernating state, trans-translating state (Fig. 5a), and the peptidyl tRNA bound state at the resolutions 4.1 Å, 12.5 Å, and 3.4 Å, respectively, highlight the new role of bS1 protein in 70S protection during hibernation in mycobacteria. With these structures' help, we now have a complete snapshot of H54a-bS1 interactions that prevent subunit dissociation and degradation (Mishra et al. 2018).
Fig. 5.
a The overall architecture of tmRNA and SmpB in the 70S bound state—the trans-translation. The SmpB is a small protein (green) that helps in the stabilization of tmRNA at A and P-tRNA binding sites (PDB: 5ZEY). b The open clamp structure of the Mtb RNAP bound to Fidaxomicin (in cyan), which stabilizes the clamp in a fully open position without DNA in the active site cleft (PDB: 6BZO). The Fidaxomicin binds in the narrow gap between the clamp and the RNAP, as shown in the inset. c Overlapping structure of AdnAB helicase-nuclease before (PDB: 6PPR) and after (PDB: 6PPU) cleavage DNA-complex. Pre-cleavage DNA is shown in orange, while post-cleavage is in red. d PafE-doubly capped (PDB: 6BGL), and PafE-singly capped (PDB: 6BGO) 20S core particle in Mtb. The GQYL motif (yellow spheres) can be seen in the doubly capped CP complex. This motif enables PafE to mount on the α ring, thus allowing the substrate to enter (Color figure online)
Since, using cryo-EM, protein structures could be captured in various intermediate steps of transcription, they improve the understanding of the underlying conformational transitions. The available EM structures of mycobacterial RNA polymerase in core and holoenzyme containing σA—a principal sigma factor form have filled this gap (Kouba et al. 2019). These structures depict the steps where the promoter partially melts within the RNA polymerase cleft. This leads to the restriction of DNA at the RNA polymerase active site. The structures of RNA polymerase-promoter DNA complex intermediates elaborate on how and why clamp opening is essential to open a passage for the entry of single-stranded DNA into the active pocket, including fork-loop 2 and switch 2 structures in promoter melting (Boyaci et al. 2019). This RNA polymerase is arrested by antibiotic Fidaxomicin, in an open state, i.e., Fidaxomicin on binding in the cleft of RNA polymerase when in an open state (Fig. 5b) halts the transcription by stopping the promoter from interacting with the RNA polymerase (Boyaci et al. 2018; Lin et al. 2018). Alongside transcription regulation, cryo-EM also sheds light on other molecular machines such as those involved in homologous recombination. AdnAB is a helicase-nuclease present in mycobacteria. It is a dimeric complex with a C-terminal nuclease domain that creates cleavage of 5′ strand DNA. The structures of AdnB in the absence of DNA and complex with forked duplex DNAs before and after cleavage of 5′ ssDNA describe the process of DNA double-strand break resection in M. smegmatis (Fig. 5c) (Jia et al. 2019).
Proteasomes are the assemblies of proteins that perform regulated proteolysis at times required by the cell (Becker and Darwin 2017). For example, the mycobacterial proteasome is activated by proteasome accessory factor E (PafE) (Fig. 5d). This activation promotes proteolysis through proteasome, helping in the virulence of Mtb in the human host. PafE binds to proteasome via glycine–glutamine–tyrosine–leucine (GQYL) motif at its C terminus, independent of ATP. This binding stimulates proteolysis (Hu et al. 2018). Like PafE, Bpa is another molecule that activates the 20S proteasome with the GQYL motif's help (Bolten et al. 2016).
S-Adenosylmethionine IV Riboswitch RNA
In bacteria, specific RNAs can regulate downstream gene expression when bound to cellular metabolites, especially in response to a change in those particular metabolites' concentration. Such RNAs are known as riboswitches. Even though riboswitch RNA does not fall into the membrane protein category, its structure holds particular importance. Despite its small size of 40 kDa, a relatively high-resolution EM structure was determined. These metabolite-binding riboswitches are highly specific but non-coding in nature. The metabolite S-adenosylmethionine (SAM), when acting as a ligand for riboswitches, regulates sulfur metabolism in bacteria (Roth and Breaker 2009).
SAM acts as a cofactor in several methylation reactions, wherein it particularly recognizes seven types of riboswitches like SAM-I, SAM-II… to SAM VI, and SAM-I/IV (Mirihana Arachchilage et al. 2018). SAM-I, SAM-I/IV, and SAM-IV are commonly found SAM riboswitches; crystal structures are already available for them except SAM-IV (Montange and Batey 2006; Gilbert et al. 2008; Lu et al. 2008; Trausch et al. 2014; Huang and Lilley 2018). Recently, the SAM-IV riboswitch cryo-EM structures in both apo and SAM-bound forms were successfully determined at an overall resolution of 3.7 Å and 4.1 Å, respectively (Zhang et al. 2019b). SAM-IV riboswitch comprises of P1, P2a, P2b, P3, and P5 as five helices with two 'pseudoknot' (PK) folds, PK-1 and PK-2. The comparison of SAM-IV structure with that of SAM-I marks critical differences between these two structures. The SAM-IV contains a pseudoknot PK2, which is not seen in SAM-I but lacks the P4 helix present in SAM-I. The kink-turn motif in SAM-I and IV contains six and four nucleotides, respectively (Montange and Batey 2006). It can be noticed that the SAM-I/IV riboswitch contains the P4 helix but lacks PK1 pseudoknot. However, the P4 helix is present in the SAM-IV riboswitch. Despite all these significant structural differences among these three classes of riboswitches (SAM-I, SAM-I/IV, and SAM-IV), the overall core topology at the SAM binding site in these riboswitches remains the same.
Additional Cryo-EM Structures that could be Novel Drug Targets
Despite the recent progress in structure determination of Mtb proteins, there are still several proteins that remain to be explored not just for drug discovery purposes but also to delineate the mechanism of action of crucial enzymes involved in different metabolic processes.
Phenolphthiocerol/phthiocerol polyketide synthase subunit E (PpsE), a type I polyketide synthase, is involved in the elongation of C22-24 fatty acids by adding malonyl-CoA and methylmalonyl-CoA extender units. This elongation step yields phthiocerol derivatives (Astarie-Dequeker et al. 2009). In Mtb, PpsE is proven to be associated with the ESX-1 secretion system (Augenstreich et al. 2017), thus makes it an exciting target. The msl3 gene product, mycolipanoate synthase, is a homodimer. It is a part of the polyacylated trehalose synthesis pathway. Mutations in this gene lead to cell aggregation (Dubey et al., 2002).
Another candidate protein, Lnt, is 93 kDa in size and catalyzes the N-acylation of the N-terminal cysteine of apolipoprotein (Tschumi et al. 2009). Specific inhibitors of Lnt could be designed based on the structural information to study the after-effects of tri-acetylation inhibition in Mtb cell morphology. Apart from such membrane proteins, other relatively smaller proteins like iron import ATP-binding protein, Serine/Threonine-protein kinase, Protoheme IX farnesyltransferase should be targeted for structure determination since these hold great potential as novel drug targets (Nalamolu et al. 2019).
Cryo-EM and Vaccine Development
‘Structural vaccinology’ is a rational approach for vaccine development. It is primarily based on determining the atomic structure of the antigen or antigen–antibody complex, mainly against viruses (Anasir and Poh 2019). The cryo-EM technique facilitated the structure determination of giant viruses like the herpes simplex virus for vaccine development. (Dai and Zhou 2018). Based on a similar approach, vaccines could be developed against different viruses. Recent EM structures of the African swine fever virus (ASFV)—one of the most complex DNA viruses could help in a more in-depth understanding of the virus's structure–function relationship (Gallagher and Harris 2020). Such progress should aid vaccine development and could form the basis of a new era of ‘Structural Vaccinology.’ During the recent SARS-CoV-2 pandemic, several structures have been deposited that have proven instrumental in research and development on drug discovery and vaccine development for SARS-CoV-2 (Walsh et al. 2020). Similarly, in TB's case, structural information from the above discussed Mtb proteins could be utilized to develop an effective treatment/vaccine against all TB types towards worldwide TB control by 2030 as per WHO’s goals.
At present, 14 TB vaccines are at different stages of clinical trials, while the live, attenuated Mycobacterium bovis bacille Calmette-Guérin (BCG) vaccine is still the most widely used (Schrager et al. 2020). With the amalgamation of available cryo-EM information and additional experiments to understand mycobacterial membrane using EM and tomography techniques, improved and effective vaccine alternatives to the classical BCG vaccine may be an achievable goal.
Concluding Remarks
As mentioned earlier, membrane protein structure determination is a cumbersome process since membrane proteins are difficult to crystallize. However, in the past decade, cryo-EM technology has taken a giant leap forward, making these limitations almost invisible. Since 2005, about 75 EM structures have been deposited (as updated on July 2020) from mycobacteria in the PDB. We have briefly discussed key membrane proteins that are therapeutically important. These structures include protein candidates from various biological pathways. Some are associated with cell growth and metabolic pathways, while others are associated with virulence, defense, and survival strategies. The snapshots provided by cryo-EM structures depicting proteins in various stages of their activity help us establish underlying mechanisms with structural evidence. Several structures of ribosomes help us visualize different steps in transcription. Not only membrane protein structures, we now have snapshots of membrane organization, which would prove instrumental in the expansion of knowledge of material transport across the mycomembrane, especially when mechanistic insights into the functioning of secretion systems and transporters have started to emerge, thanks to cryo-EM.
In recent years, we have witnessed a tremendous increase in structure determination using cryo-EM. Compared to the past two decades, we see exponential growth in the number of membrane protein structures determined using EM in the past three years (Supplementary Table S1). These membrane proteins are associated with crucial pathways involved in bacterial metabolism and/or virulence; at the same time, they are non-native to human physiology. Hence, biosynthesis or metabolic pathways associated with these proteins could serve as a goldmine of novel and effective anti-TB drug targets.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors are thankful to the Department of Biotechnology, India, for funding and Research Associate fellowship to ADB and the Netherlands Organisation for Scientific Research (NWO) in the framework of the National Roadmap NEMI Project Number 184.034.014.
Abbreviations
- AAA
ATPases associated with diverse cellular activities
- ABC
ATP-binding cassette
- AcpM
Acyl-carrier protein M, Acr1: alpha-crystallin-related protein 1
- Acr1
Alpha-crystallin-related protein 1
- ADEP
Acyldepsipeptide
- Adn
ATP-dependent nuclease
- AftD
Arabinofuranosyltransferase D
- AMPPNP
Adenylyl imidodiphosphate
- BBH
Benzyloxybenzylidene-hydrazine
- BDQ
Bedaquiline
- Bpa
Bacterial proteasome activator
- CBM
Carbohydrate-binding module
- CDL
Cardiolipin
- CIII2CIV2SOD2
CIII and CIV subunits of respiratory supercomeplex bound to superoxide dismutase in C2, symmetry
- Clp
Caseinolytic proteases
- Cryo-EM
Cryo-Electron Microscopy
- Cryo-ET
Cryo-Electron Tomography
- Cta
Subunits of aa3-type cytochrome c oxidase
- CTD
C-terminal domain
- DPA
Decaprenyl-phosphate-arabinose
- Ecc
ESC conserved component
- EMB
Ethambutol
- ESAT-6
6 KDa early secretory antigenic target
- Esp
ESX-1 secretion-associated protein
- ESX
ESAT-6
- ETC
Electron transport chain
- FAS
Fatty Acid Synthase
- Galf
Galactofuranosyl
- GLF-CMK
Z-Gly-Leu-Phe-chloromethyl ketone
- GQYL
Glycine–glutamine–tyrosine–leucine
- HIV
Human immunodeficiency virus
- HSP
Heat shock protein
- MPY
Mycobacterial-specific protein Y
- MSD
Membrane spanning domain
- Mtb
Mycobacterium tuberculosis
- M/XDR
Multi/extremely drug resistant
- NBD
Nucleotide-binding domain
- PafE
Proteasome accessory factor E
- PCAT
Peptidase-containing ATP-binding cassette transporters
- PDB
Protein data bank
- PK
Pseudoknot
- PpsE
Phenolphthiocerol/phthiocerol polyketide synthase subunit E
- RbpA
RNA polymerase-binding protein A
- RNAP
RNA polymerase
- ROS
Reactive oxygen species
- SAM
S-Adenosylmethionine
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- Sr. No.
Serial Number
- TB
Tuberculosis
- TMH
Trans-membrane helix/helices
- WHO
World Health Organization
Author Contributions
The manuscript was conceptualized, designed, and written by ADB, PJP, and JK.
Funding
This work was funded by a Centre of Excellence Grant BT/PR15450/COE/34/46/2016 from the Department of Biotechnology, India. ADB gratefully acknowledges the financial support from the DBT-RA Program in Biotechnology and Life Sciences (India).
Declarations
Conflict of interest
PJP is co-founder and shareholder of www.cryosol-world.com.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abdallah AM, Verboom T, Hannes F, et al. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol. 2006 doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- Acharya KR, Lloyd MD. The advantages and limitations of protein crystal structures. Trends Pharmacol Sci. 2005;26:10–14. doi: 10.1016/j.tips.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Al-Amoudi A, Chang J-J, Leforestier A, et al. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderwick LJ, Harrison J, Lloyd GS, Birch HL. The mycobacterial cell wall—peptidoglycan and arabinogalactan. Cold Spring Harb Perspect Med. 2015 doi: 10.1101/cshperspect.a021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhuwaider AAH, Dougan DA. AAA+ machines of protein destruction in mycobacteria. Front Mol Biosci. 2017;4:49. doi: 10.3389/fmolb.2017.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aller SG, Yu J, Ward A, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science (80–) 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsteens D, Verbelen C, Dague E, et al. Organization of the mycobacterial cell wall: a nanoscale view. Pflügers Arch J Physiol. 2008;456:117–125. doi: 10.1007/s00424-007-0386-0. [DOI] [PubMed] [Google Scholar]
- Anasir MI, Poh CL. Structural vaccinology for viral vaccine design. Front Microbiol. 2019;10:738. doi: 10.3389/fmicb.2019.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astarie-Dequeker C, Le Guyader L, Malaga W, et al. Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog. 2009 doi: 10.1371/journal.ppat.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates LS, Ummels R, Commandeur S, et al. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet. 2015 doi: 10.1371/journal.pgen.1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augenstreich J, Arbues A, Simeone R, et al. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol. 2017;19:e12726. doi: 10.1111/cmi.12726. [DOI] [PubMed] [Google Scholar]
- Awofeso N. Anti-tuberculosis medication side-effects constitute major factor for poor adherence to tuberculosis treatment. Bull World Health Organ. 2008;86:B–D. doi: 10.2471/BLT.07.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balganesh M, Kuruppath S, Marcel N, et al. Rv1218c, an ABC transporter of Mycobacterium tuberculosis with implications in drug discovery. Antimicrob Agents Chemother. 2010;54:5167–5172. doi: 10.1128/AAC.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry Iii CE, Lee RE, Mdluli K, et al. Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- Becker SH, Darwin KH. Bacterial proteasomes: mechanistic and functional insights. Microbiol Mol Biol Rev. 2017;81(1):e00036–e116. doi: 10.1128/MMBR.00036-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besra GS, Chatterjee D (1994) Lipids and carbohydrates of Mycobacterium tuberculosis. In: Tuberculosis. American Society of Microbiology, Washington, pp 285–306
- Bhandari V, Wong KS, Zhou JL, et al. The role of ClpP protease in bacterial pathogenesis and human diseases. ACS Chem Biol. 2018;13:1413–1425. doi: 10.1021/acschembio.8b00124. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S (2015) Reactive oxygen species and cellular defense system. In: Free radicals in human health and disease. Springer, Cham, pp 17–29
- Bhatt A, Molle V, Besra GS, et al. The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development. Mol Microbiol. 2007;64:1442–1454. doi: 10.1111/j.1365-2958.2007.05761.x. [DOI] [PubMed] [Google Scholar]
- Bigelow KM, Tasneen R, Chang YS, et al. Preserved efficacy and reduced toxicity with intermittent linezolid dosing in combination with bedaquiline and pretomanid in a murine tuberculosis model. Antimicrob Agents Chemother. 2020;64(10):e01178–e1220. doi: 10.1128/AAC.01178-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D, Ban N, Leibundgut M. 7.5-Å cryo-EM structure of the mycobacterial fatty acid synthase. J Mol Biol. 2013;425:841–849. doi: 10.1016/j.jmb.2012.12.021. [DOI] [PubMed] [Google Scholar]
- Bolten M, Delley CL, Leibundgut M, et al. Structural analysis of the bacterial proteasome activator Bpa in complex with the 20S proteasome. Structure. 2016;24:2138–2151. doi: 10.1016/j.str.2016.10.008. [DOI] [PubMed] [Google Scholar]
- Boyaci H, Chen J, Lilic M, et al. Fidaxomicin jams Mycobacterium tuberculosis RNA polymerase motions needed for initiation via RbpA contacts. Elife. 2018;7:e34823. doi: 10.7554/eLife.34823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyaci H, Chen J, Jansen R, et al. Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding. Nature. 2019;565:382–385. doi: 10.1038/s41586-018-0840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P, Majlessi L, Marsollier L, et al. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun. 2006 doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunduc CM, Bitter W, Houben ENG. Structure and function of the mycobacterial type VII secretion systems. Annu Rev Microbiol. 2020;74:315–335. doi: 10.1146/annurev-micro-012420-081657. [DOI] [PubMed] [Google Scholar]
- Burley SK, Berman HM, Bhikadiya C, et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri BS, Bhakta S, Barik R, et al. Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. Biochem J. 2002 doi: 10.1042/BJ20020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury HG, Tong Z, Mathavan I, et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1320506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchyard GJ. A stratified approach to tuberculosis treatment. Nat Med. 2018;24:1639–1641. doi: 10.1038/s41591-018-0244-y. [DOI] [PubMed] [Google Scholar]
- Ciccarelli L, Connell SR, Enderle M, et al. Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex. Structure. 2013;21:1251–1257. doi: 10.1016/j.str.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Cole ST. Inhibiting Mycobacterium tuberculosis within and without. Philos Trans R Soc B Biol Sci. 2016;371:20150506. doi: 10.1098/rstb.2015.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick DC, Brennan PJ (2008) Biosynthesis of the arabinogalactan-peptidoglycan complex of mycobacterium tuberculosis. In: The mycobacterial cell envelope, pp 25–40 [DOI] [PubMed]
- D’Orazio M, Folcarelli S, Mariani F, et al. Lipid modification of the Cu,Zn superoxide dismutase from Mycobacterium tuberculosis. Biochem J. 2001 doi: 10.1042/0264-6021:3590017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffé M, Marrakchi H (2019) Unraveling the structure of the mycobacterial envelope. In: Gram-positive pathogens. ASM Press, Washington, pp 1087–1095
- Daffe M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol. 1997;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- Dai X, Zhou ZH. Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science (80–) 2018;360(6384):eaao7298. doi: 10.1126/science.aao7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rossi E, Aínsa JA, Riccardi G. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol Rev. 2006;30:36–52. doi: 10.1111/j.1574-6976.2005.00002.x. [DOI] [PubMed] [Google Scholar]
- Domenech P, Kobayashi H, Levier K, et al. BacA, an ABC transporter involved in maintenance of chronic murine infections with Mycobacterium tuberculosis. J Bacteriol. 2009 doi: 10.1128/JB.01132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Alver R, Turgay K. Exploring a potential Achilles heel of Mycobacterium tuberculosis: defining the ClpC1 interactome. FEBS J. 2020;288(1):95–98. doi: 10.1111/febs.15430. [DOI] [PubMed] [Google Scholar]
- Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Doyle SM, Genest O, Wickner S. Protein rescue from aggregates by powerful molecular chaperone machines. Nat Rev Mol Cell Biol. 2013;14(10):617–629. doi: 10.1038/nrm3660. [DOI] [PubMed] [Google Scholar]
- Dubey VS, Sirakova TD, Kolattukudy PE. Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol Microbiol. 2002;45:1451–1459. doi: 10.1046/j.1365-2958.2002.03119.x. [DOI] [PubMed] [Google Scholar]
- Elad N, Baron S, Peleg Y, et al. Structure of Type-I Mycobacterium tuberculosis fatty acid synthase at 3.3 Åresolution. Nat Commun. 2018;9:1–6. doi: 10.1038/s41467-018-06440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne G, Villeneuve C, Billman-Jacobe H, et al. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of s. Microbiology. 2002;148:3089–3100. doi: 10.1099/00221287-148-10-3089. [DOI] [PubMed] [Google Scholar]
- Faller M, Niederweis M, Schulz GE. The structure of a mycobacterial outer-membrane channel. Science (80–) 2004;303:1189–1192. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- Famelis N, Rivera-Calzada A, Degliesposti G, et al. Architecture of the mycobacterial type VII secretion system. Nature. 2019;576:321–325. doi: 10.1038/s41586-019-1633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. Advances in the field of single-particle cryo-electron microscopy over the last decade. Nat Protoc. 2017;12:209–212. doi: 10.1038/nprot.2017.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frees D, Gerth U, Ingmer H. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int J Med Microbiol. 2014;304(2):142–149. doi: 10.1016/j.ijmm.2013.11.009. [DOI] [PubMed] [Google Scholar]
- Gaensbauer J, Broadhurst R. Recent innovations in diagnosis and treatment of pediatric tuberculosis. Curr Infect Dis Rep. 2019;21:4. doi: 10.1007/s11908-019-0662-0. [DOI] [PubMed] [Google Scholar]
- Gallagher JR, Harris AK. Cryo-EM cools down swine fever. J Biol Chem. 2020;295:13–14. doi: 10.1074/jbc.H119.012169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates SN, Yokom AL, Lin J, et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science (80–) 2017;357:273–279. doi: 10.1126/science.aan1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- Gipson P, Mills DJ, Wouts R, et al. Direct structural insight into the substrate-shuttling mechanism of yeast fatty acid synthase by electron cryomicroscopy. Proc Natl Acad Sci USA. 2010;107:9164–9169. doi: 10.1073/pnas.0913547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gloabal Fund (2020) Global Fund Survey: majority of HIV, TB and Malaria programs face disruptions as a result of COVID-19. https://www.theglobalfund.org/en/covid-19/news/2020-06-17-global-fund-survey-majority-of-hiv-tb-and-malaria-programs-face-disruptions-as-a-result-of-covid-19/
- Gong H, Li J, Xu A, et al. An electron transfer path connects subunits of a mycobacterial respiratory supercomplex. Science (80–) 2018 doi: 10.1126/science.aat8923. [DOI] [PubMed] [Google Scholar]
- Gopinath K, Moosa A, Mizrahi V, Warner DF. Vitamin B12 metabolism in Mycobacterium tuberculosis. Fut Microbiol. 2013;8(11):1405–1418. doi: 10.2217/fmb.13.113. [DOI] [PubMed] [Google Scholar]
- Gorelick S, Buckley G, Gervinskas G, et al. PIE-scope, integrated cryo-correlative light and FIB/SEM microscopy. Elife. 2019;8:e45919. doi: 10.7554/eLife.45919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goude R, Amin AG, Chatterjee D, Parish T. The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2009;53:4138–4146. doi: 10.1128/AAC.00162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ER, Mecsas J. Bacterial secretion systems: an overview. Microbiol Spectr. 2016 doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüber G. Introduction: Novel insights into TB research and drug discovery. Prog Biophys Mol Biol. 2020;152:2–5. doi: 10.1016/j.pbiomolbio.2020.02.003. [DOI] [PubMed] [Google Scholar]
- Guo H, Courbon GM, Bueler SA, et al. Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline. Nature. 2020;589(7840):143–147. doi: 10.1038/s41586-020-3004-3. [DOI] [PubMed] [Google Scholar]
- Guo H, Courbon GM, Bueler SA, et al (2020a) Structure of mycobacterial ATP synthase with the TB drug bedaquiline. bioRxiv [DOI] [PubMed]
- Haagsma AC, Abdillahi-Ibrahim R, Wagner MJ, et al. Selectivity of TMC207 towards mycobacterial ATP synthase compared with that towards the eukaryotic homologue. Antimicrob Agents Chemother. 2009;53:1290–1292. doi: 10.1128/AAC.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M-Y, Raman S, Anaya M, Husson RN. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J Bacteriol. 2005;187:7062–7071. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltia T, Freire E. Forces and factors that contribute to the structural stability of membrane proteins. Biochim Biophys Acta (BBA) 1995;1241:295–322. doi: 10.1016/0304-4157(94)00161-6. [DOI] [PubMed] [Google Scholar]
- Hentschel J, Burnside C, Mignot I, et al. The complete structure of the Mycobacterium smegmatis 70S ribosome. Cell Rep. 2017;20:149–160. doi: 10.1016/j.celrep.2017.06.029. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Leis A, Niederweis M, et al. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben D, Demangel C, Van Ingen J, et al. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol. 2012;14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- Huang L, Lilley DMJ. Structure and ligand binding of the SAM-V riboswitch. Nucleic Acids Res. 2018;46:6869–6879. doi: 10.1093/nar/gky520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Jastrab JB, Zhang S, et al. Proteasome substrate capture and gate opening by the accessory factor PafE from Mycobacterium tuberculosis. J Biol Chem. 2018;293:4713–4723. doi: 10.1074/jbc.RA117.001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong GU, Song H, Yoon GY, et al. Therapeutic strategies against COVID-19 and structural characterization of SARS-CoV-2: a review. Front Microbiol. 2020;11:1723. doi: 10.3389/fmicb.2020.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Unciuleac MC, Xue C, et al. Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection. Proc Natl Acad Sci USA. 2019;116:24507–24516. doi: 10.1073/pnas.1913546116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennaway CK, Benesch JLP, Gohlke U, et al. Dodecameric structure of the small heat shock protein Acr1 from Mycobacterium tuberculosis. J Biol Chem. 2005;280:33419–33425. doi: 10.1074/jbc.M504263200. [DOI] [PubMed] [Google Scholar]
- Korepanova A, Moore JD, Nguyen HB, et al. Expression of membrane proteins from Mycobacterium tuberculosis in Escherichia coli as fusions with maltose binding protein. Protein Expr Purif. 2007;53:24–30. doi: 10.1016/j.pep.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhov VM, Mireku SA, Locher KP. Structure of AMP-PNP-bound vitamin B 12 transporter BtuCD–F. Nature. 2012;490:367–372. doi: 10.1038/nature11442. [DOI] [PubMed] [Google Scholar]
- Korotkova N, Piton J, Wagner JM, et al. Structure of EspB, a secreted substrate of the ESX-1 secretion system of Mycobacterium tuberculosis. J Struct Biol. 2015;191:236–244. doi: 10.1016/j.jsb.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouba T, Pospíšil J, Hnilicová J, et al. The core and holoenzyme forms of RNA polymerase from Mycobacterium smegmatis. J Bacteriol. 2019;201:e00583–e618. doi: 10.1128/JB.00583-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A, Vranckx L, Dhar N, et al. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat Commun. 2014;5:3369. doi: 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S, Biukovic G, Grüber G, Dick T. Bedaquiline targets the ε subunit of mycobacterial F-ATP synthase. Antimicrob Agents Chemother. 2016;60:6977–6979. doi: 10.1128/AAC.01291-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasica AM, Ksiazek M, Madej M, Potempa J. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front Cell Infect Microbiol. 2017;7:215. doi: 10.3389/fcimb.2017.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee REB, Li W, Chatterjee D, Lee RE. Rapid structural characterization of the arabinogalactan and lipoarabinomannan in live mycobacterial cells using 2D and 3D HR-MAS NMR: structural changes in the arabinan due to ethambutol treatment and gene mutation are observed. Glycobiology. 2005;15:139–151. doi: 10.1093/glycob/cwh150. [DOI] [PubMed] [Google Scholar]
- Lee BS, Sviriaeva E, Pethe K. Targeting the cytochrome oxidases for drug development in mycobacteria. Prog Biophys Mol Biol. 2020;152:45–54. doi: 10.1016/j.pbiomolbio.2020.02.001. [DOI] [PubMed] [Google Scholar]
- Lin J, Huang S, Zhang Q. Outer membrane proteins: key players for bacterial adaptation in host niches. Microbes Infect. 2002;4:325–331. doi: 10.1016/s1286-4579(02)01545-9. [DOI] [PubMed] [Google Scholar]
- Lin DYW, Huang S, Chen J. Crystal structures of a polypeptide processing and secretion transporter. Nature. 2015 doi: 10.1038/nature14623. [DOI] [PubMed] [Google Scholar]
- Lin W, Das K, Degen D, et al. Structural basis of transcription inhibition by fidaxomicin (lipiarmycin A3) Mol Cell. 2018;70:60–71. doi: 10.1016/j.molcel.2018.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Ologbenla A, Houry WA. Dynamics of the ClpP serine protease: a model for self-compartmentalized proteases. Crit Rev Biochem Mol Biol. 2014;49:400–412. doi: 10.3109/10409238.2014.925421. [DOI] [PubMed] [Google Scholar]
- Li Z, Ge X, Zhang Y, et al. Cryo-EM structure of Mycobacterium smegmatis ribosome reveals two unidentified ribosomal proteins close to the functional centers. Protein Cell. 2018;9:384–388. doi: 10.1007/s13238-017-0456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sharma MR, Koripella RK, et al. Zinc depletion induces ribosome hibernation in mycobacteria. Proc Natl Acad Sci USA. 2018;115:8191–8196. doi: 10.1073/pnas.1804555115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Zhou W, Patel H, et al. Bedaquiline inhibits the yeast and human mitochondrial ATP synthases. Commun Biol. 2020;3:1–10. doi: 10.1038/s42003-020-01173-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupoli TJ, Fay A, Adura C, et al. Reconstitution of a Mycobacterium tuberculosis proteostasis network highlights essential cofactor interactions with chaperone DnaK. Proc Natl Acad Sci USA. 2016;113:E7947–E7956. doi: 10.1073/pnas.1617644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupoli TJ, Vaubourgeix J, Burns-Huang K, Gold B. Targeting the proteostasis network for mycobacterial drug discovery. ACS Infect Dis. 2018;4:478–498. doi: 10.1021/acsinfecdis.7b00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Smith AM, Fuchs RT, et al. Crystal structures of the SAM-III/S MK riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol. 2008;15:1076. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macikag A, Piazza A, Riccardi G, Milano A. Transcriptional analysis of ESAT-6 cluster 3 in Mycobacterium smegmatis. BMC Microbiol. 2009;9:48. doi: 10.1186/1471-2180-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Målen H, De Souza GA, Pathak S, et al. Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiol. 2011;11:18. doi: 10.1186/1471-2180-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrakchi H, Lanéelle MA, Daffé M. Mycolic acids: Structures, biosynthesis, and beyond. Chem Biol. 2014;21(1):67–85. doi: 10.1016/j.chembiol.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Mazando S, Zimba M, Zimudzi C, et al. Proposed contribution of Rv1819c an efflux pump (ABC protein) from Mycobacterium tuberculosis to drug efflux in Corynebacterium glutamicum. J Med Biomed Sci. 2013;4(2):28–36. [Google Scholar]
- McClean S. Eight stranded $β$-barrel and related outer membrane proteins: role in bacterial pathogenesis. Protein Pept Lett. 2012;19:1013–1025. doi: 10.2174/092986612802762688. [DOI] [PubMed] [Google Scholar]
- McLaughlin B, Chon JS, MacGurn JA, et al. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 2007 doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Zahra A, Thompson V, et al. Mycobacterium tuberculosis type VII secreted effector EsxH targets host ESCRT to impair trafficking. PLoS Pathog. 2013 doi: 10.1371/journal.ppat.1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirihana Arachchilage G, Sherlock ME, Weinberg Z, Breaker RR. SAM-VI RNAs selectively bind S-adenosylmethionine and exhibit similarities to SAM-III riboswitches. RNA Biol. 2018 doi: 10.1080/15476286.2017.1399232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Ahmed T, Tyagi A, et al. Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA. Sci Rep. 2018;8:1–12. doi: 10.1038/s41598-018-31850-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal E, Skowyra ML, Uwase G, et al. Mycobacterium tuberculosis type VII secretion system effectors differentially impact the ESCRT endomembrane damage response. MBio. 2018 doi: 10.1128/mBio.01765-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999 doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006 doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- Moreira W, Santhanakrishnan S, Dymock BW, Dick T. Bortezomib warhead-switch confers dual activity against mycobacterial caseinolytic protease and proteasome and selectivity against human proteasome. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir A, Wilson MT, Hardwick SW, et al. Using cryo-EM to understand antimycobacterial resistance in the catalase-peroxidase (KatG) from Mycobacterium tuberculosis. Structure. 2021 doi: 10.1016/j.str.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalamolu RM, Pasala C, Katari SK, Amineni U. Discovery of common putative drug targets and vaccine candidates for Mycobacterium tuberculosis sp. J Drug Deliv Ther. 2019;9:67–71. [Google Scholar]
- Ohol YM, Goetz DH, Chan K, et al. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe. 2010 doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvate AD (2018) An approach to perform Cryo-EM analysis of glutaraldehyde fixed BSL-3 viruses in BSL-1/2 conditions. Dissertation, Purdue University
- Paul TR, Beveridge TJ. Reevaluation of envelope profiles and cytoplasmic ultrastructure of mycobacteria processed by conventional embedding and freeze-substitution protocols. J Bacteriol. 1992;174:6508–6517. doi: 10.1128/jb.174.20.6508-6517.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personne Y, Brown AC, Schuessler DL, Parish T. Mycobacterium tuberculosis ClpP proteases are co-transcribed but exhibit different substrate specificities. PLoS ONE. 2013 doi: 10.1371/journal.pone.0060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal-Celhay C, Tufariello JM, Srivastava S, et al. Mycobacterium tuberculosis EsxH inhibits ESCRT-dependent CD4+ T-cell activation. Nat Microbiol. 2016 doi: 10.1038/nmicrobiol.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poweleit N, Czudnochowski N, Nakagawa R, et al. A large inner membrane pore defines the ESX translocon. bioRxiv. 2019 doi: 10.1101/800169. [DOI] [Google Scholar]
- Preiss L, Langer JD, Yildiz Ö, et al. Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline. Sci Adv. 2015 doi: 10.1126/sciadv.1500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SPS, Alonso S, Rand L, et al. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli RBG, Nijpels FJT, Henderikx RJM, et al. Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification. Nat Commun. 2020;11:1–9. doi: 10.1038/s41467-020-16392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel S, Gati C, Nijland M, et al. A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds. Nature. 2020;580:409–412. doi: 10.1038/s41586-020-2072-8. [DOI] [PubMed] [Google Scholar]
- Renaud J-P, Chari A, Ciferri C, et al. Cryo-EM in drug discovery: achievements, limitations and prospects. Nat Rev Drug Discov. 2018;17:471–492. doi: 10.1038/nrd.2018.77. [DOI] [PubMed] [Google Scholar]
- Rodriguez GM, Voskuil MI, Gold B, et al. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy KK, Wani MA. Emerging opportunities of exploiting mycobacterial electron transport chain pathway for drug-resistant tuberculosis drug discovery. Expert Opin Drug Discov. 2020;15:231–241. doi: 10.1080/17460441.2020.1696771. [DOI] [PubMed] [Google Scholar]
- Rybniker J, Chen JM, Sala C, et al. Anticytolytic screen identifies inhibitors of mycobacterial virulence protein secretion. Cell Host Microbe. 2014;16:538–548. doi: 10.1016/j.chom.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Safi H, Sayers B, Hazbón MH, Alland D. Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob Agents Chemother. 2008;52:2027–2034. doi: 10.1128/AAC.01486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M, Houben ENG, Geurtsen J, et al. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6:e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M, Houben ENG, Geurtsen J, et al. Direct visualization by Cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010 doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Schrager LK, Vekemens J, Drager N, et al. The status of tuberculosis vaccine development. Lancet Infect Dis. 2020;20:e28–e37. doi: 10.1016/S1473-3099(19)30625-5. [DOI] [PubMed] [Google Scholar]
- Serafini A, Boldrin F, Palù G, Manganelli R. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol. 2009 doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Unnikrishnan M, McConnell MJ, et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci USA. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan Siegrist M, Steigedal M, Ahmad R, et al. Mycobacterial Esx-3 requires multiple components for iron acquisition. MBio. 2014 doi: 10.1128/mBio.01073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomonson M, Huesgen PF, Wasney GA, et al. Structure of the mycosin-1 protease from the mycobacterial ESX-1 protein type VII secretion system. J Biol Chem. 2013 doi: 10.1074/jbc.M113.462036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomonson M, Setiaputra D, Makepeace KAT, et al. Structure of EspB from the ESX-1 type VII secretion system and insights into its export mechanism. Structure. 2015;23:571–583. doi: 10.1016/j.str.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Sreevatsan S, Stockbauer KE, Pan XI, et al. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob Agents Chemother. 1997;41:1677–1681. doi: 10.1128/aac.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Xiao T, Liu H, et al. Mutations within embCAB are associated with variable level of ethambutol resistance in Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother. 2018;62(1):e01279–e1317. doi: 10.1128/AAC.01279-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Maurizi MR. Crystal structure at 1.9 Åof E. coli ClpP with a peptide covalently bound at the active site. J Struct Biol. 2006;156:165–174. doi: 10.1016/j.jsb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Taber HW, Mueller JP, Miller PF, Arrow AS. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev. 1987;51:439. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Wang C, Besra GS. Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev. 2005;18:81–101. doi: 10.1128/CMR.18.1.81-101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YZ, Rodrigues J, Keener JE, et al. Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-17202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YZ, Zhang L, Rodrigues J, et al. Cryo-EM structures and regulation of arabinofuranosyltransferase AftD from Mycobacteria. Mol Cell. 2020;78(4):683–699.e11. doi: 10.1016/j.molcel.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinaztepe E, Wei JR, Raynowska J, et al. Role of metal-dependent regulation of ESX-3 secretion in intracellular survival of Mycobacterium tuberculosis. Infect Immun. 2016 doi: 10.1128/IAI.00197-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch JJ, Xu Z, Edwards AL, et al. Structural basis for diversity in the SAM clan of riboswitches. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1312918111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi OA, Arora P, Sridharan V, et al. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- Tschumi A, Nai C, Auchli Y, et al. Identification of apolipoprotein N-acyltransferase (Lnt) in mycobacteria. J Biol Chem. 2009;284:27146–27156. doi: 10.1074/jbc.M109.022715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020) Tuberculosis. https://www.who.int/news-room/fact-sheets/detail/tuberculosis
- Tufariello JAM, Chapman JR, Kerantzas CA, et al. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc Natl Acad Sci USA. 2016 doi: 10.1073/pnas.1523321113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahidi S, Ripstein ZA, Juravsky JB, et al. An allosteric switch regulates Mycobacterium tuberculosis ClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR. Proc Natl Acad Sci USA. 2020;117:5895–5906. doi: 10.1073/pnas.1921630117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Winden VJC, Ummels R, Piersma SR, et al. Mycosins are required for the stabilization of the ESX-1 and ESX-5 type VII secretion membrane complexes. MBio. 2016 doi: 10.1128/mBio.01471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JM, Evans TJ, Chen J, et al. Understanding specificity of the mycosin proteases in ESX/type VII secretion by structural and functional analysis. J Struct Biol. 2013;184:115–128. doi: 10.1016/j.jsb.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MA, Grimes JM, Stuart DI. Diamond light source: contributions to SARS-CoV-2 biology and therapeutics. Biochem Biophys Res Commun. 2020;538:40–46. doi: 10.1016/j.bbrc.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Wiseman B, Nitharwal RG, Fedotovskaya O, et al. Structure of a functional obligate complex III2IV2 respiratory supercomplex from Mycobacterium smegmatis. Nat Struct Mol Biol. 2018 doi: 10.1038/s41594-018-0160-3. [DOI] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (80–) 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Yu CA, Kim H, et al. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science (80–) 1997 doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Laine O, Masciocchi M, et al. A unique Mycobacterium ESX-1 protein co-secretes with CFP-10/ESAT-6 and is necessary for inhibiting phagosome maturation. Mol Microbiol. 2007;66:787–800. doi: 10.1111/j.1365-2958.2007.05959.x. [DOI] [PubMed] [Google Scholar]
- Yang K, Chang J-Y, Cui Z, et al. Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis. Nucleic Acids Res. 2017;45:10884–10894. doi: 10.1093/nar/gkx785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lupoli TJ, Kovach A, et al. ATP hydrolysis-coupled peptide translocation mechanism of Mycobacterium tuberculosis ClpB. Proc Natl Acad Sci USA. 2018;115:E9560–E9569. doi: 10.1073/pnas.1810648115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Li S, Kappel K, et al. Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 Å resolution. Nat Commun. 2019;10:1–6. doi: 10.1038/s41467-019-13494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AT, Montgomery MG, Leslie AGW, et al. The structure of the catalytic domain of the ATP synthase from Mycobacterium smegmatis is a target for developing antitubercular drugs. Proc Natl Acad Sci USA. 2019;116:4206–4211. doi: 10.1073/pnas.1817615116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhao Y, Gao Y, et al. Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science (80–) 2020;368:1211–1219. doi: 10.1126/science.aba9102. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhao Y, Gao R, et al. Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2. Protein Cell. 2020 doi: 10.1007/s13238-020-00726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Drlica K. Reactive oxygen species and the bacterial response to lethal stress. Curr Opin Microbiol. 2014;21:1–6. doi: 10.1016/j.mib.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber B, Chami M, Houssin C, et al. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol. 2008 doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.