Abstract
MEF2 and NKX2-5 transcription factors interact with each other in cardiogenesis and are necessary for normal heart formation. Despite evidence suggesting that these two transcription factors function synergistically and possibly through direct physical interactions, molecular mechanisms by which they interact are not clear. Here we determined the crystal structures of ternary complexes of MEF2 and NKX2-5 bound to myocardin enhancer DNA in two crystal forms. These crystal structures are the first example of human MADS-box/homeobox ternary complex structures involved in cardiogenesis. Our structures reveal two possible modes of interactions between MEF2 and NKX2-5: MEF2 and NKX bind to adjacent DNA sites to recognize DNA in cis; and MEF2 and NKX bind to different DNA strands to interact with each other in trans via a conserved protein-protein interface observed in both crystal forms. Disease related mutations are mapped to the observed protein-protein interface. Our structural studies provide a starting point to understand and further study the molecular mechanisms of the interactions between MEF2 and NKX2.5 and their roles in cardiogenesis.
Introduction
The MADS-box family transcription factor myocyte-enhancer factor 2 (MEF2) and the homeobox family transcription factor NKX2-5 play important roles in cardiogenesis [1–4] and have also been implicated in carcinogenesis [5–11]. Deletion of MEF2 or NKX2-5 causes embryonic death in mice with defects in heart development [12–14]. Mutations or misexpressions of MEF2 and NKX2-5 are frequently found in congenital heart disease and leukemia/lymphoma patients [10,15–24].
The functional synergy between MEF2 and NKX2-5 in cardiogenesis has long been postulated [25,26] based on experimental observations that MEF2 and NKX2-5 expression coincides with each other in cardiac muscle development, and the studies that suggests MADS-box protein and homeobox protein could interact with each other [27,28]. Co-immunoprecipitation and mammalian two-hybrid assays detected physical interactions between MEF2 and NKX2-5 in cells [29]. Chip-Seq studies show that MEF2 and NKX2-5 co-occupy active cardiac enhancer regions with other transcription factors such as GATA-4, TBX5 and SRF to initiate cardiac differentiation and maintain cardiac gene expression [30–33]. Furthermore, MEF2 and NKX2-5 have been shown as important factors in the reprogramming of fibroblasts into cardiomyocytes [34–38]. To further understand the molecular interactions between MEF2 and NKX2-5, we determined the co-crystal structures of ternary complexes of MEF2 and NKX2-5 bound to myocardin enhancer DNA in two crystal forms. Our structures reveal new insights into the interaction mechanisms between these two important transcription factors that are involved in the heart development.
Result and Discussion
The overall structure of the MEF2/NKX2-5/DNA complex
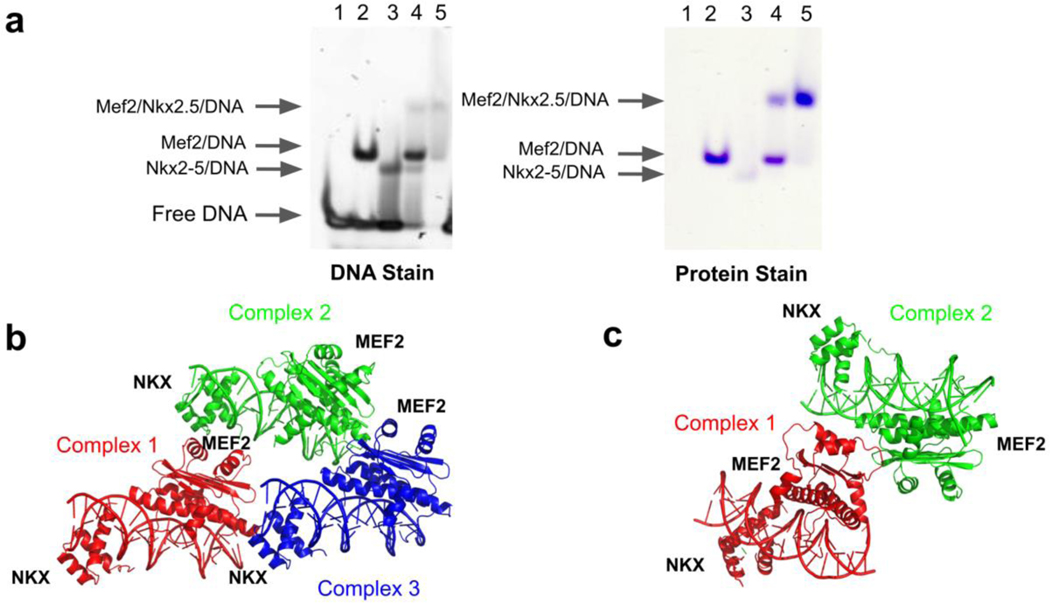
Before crystallography studies, we did electrophoretic mobility shift assays (EMSA) to study the ability of MEF2, NKX2-5 and DNA to form ternary complexes. The DNA in our EMSA and crystallization studies is designed based on the composite regulatory elements that regulate myocardin expression. Myocardin is a common transcriptional downstream target of MEF2 and NKX2-5 [31,39–41]. We analyzed Chip-seq peaks of MEF2 and NKX2-5 from literature in public Chip-seq databases [42], and found that many MEF2 and NKX2-5 binding sites were in the myocardin gene regulatory regions (Supplemental Fig. 1). We chose one composite regulatory element which contains adjacent MEF2 and NKX2-5 sites from myocardin enhancer (MyE) in our EMSA assays and crystallization studies. The EMSA results indicate that MEF2, NKX2-5 and myocardin enhancer DNA form stable ternary complexes (Fig. 1a). We used MEF2 and NKX2-5 protein constructs that contain the MEF2 MADS-box and MEF2 specific domain (residues 1–95) and the NKX2-5 homeobox domain (residues 137–197) (Supplemental Fig. 2) in the crystallization of MEF2/NKX2-5/DNA ternary complexes. We determined the MEF2/NKX2-5/DNA structures in two crystal forms, each from different MEF2 constructs: MEF2 Chimera/NKX2-5/DNA ternary complex at 2.1 Å resolution and MEF2B/NKX2-5/DNA at 2.9 Å resolution (Table 1). MEF2 Chimera is a MEF2 construct with the MADS-box domain from MEF2A and the MEF2 specific domain from MEF2B. MEF2 Chimera has over ninety percent amino acids identity as other MEF2 members (MEF2A-MEF2D) (Supplemental Fig. 2a), and is biochemically characterized as similar to wild-type MEF2 in previous studies [43]. The purpose of using MEF2 Chimera construct is to increase the chance of successful crystallization and better resolution. There are three sets of ternary complexes in the asymmetric unit of the MEF2 Chimera/NKX2-5/DNA crystal structure and two sets of ternary complexes in the asymmetric unit of the MEF2B/NKX2-5/DNA crystal structure (Fig. 1b, c). The overall folds of MEF2 and NKX2-5 are identical within the multiple copies in the asymmetric unit of the two crystal forms and also identical to their respective counterparts in previously published structures (Supplemental Fig. 3a, b) [44,45]. The structural features we present in this paper are conserved between the two crystal structures. Because MEF2 Chimera/NKX2-5/DNA crystals have better resolution, our structure analyses are based mainly on this structure unless indicated otherwise.
Figure 1. MEF2 and NKX2-5 form ternary complexes on DNA.
(a) Electrophoretic mobility shift assays (EMSA) of MEF2, NKX2-5 binding to myocardin enhancer DNA element (MyE). The MyE DNA sequence, 5’- CACTATTTTAAGAAAGTGCTT-3’, contains adjacent MEF2 and NKX2-5 binding sites. The molar ratios of DNA, MEF2, and NKX2-5 are 1: 1: 1 in lane 4 and 1: 1: 2 in lane 5 (see method section for details). (b) Overall crystal structure of the MEF2 Chimera/NKX2-5/DNA ternary complex. Three sets of complexes in asymmetric units are colored in red, green and blue, respectively. (c) Overall crystal structure of the MEF2B/NKX2-5/DNA ternary complexes. Two sets of complexes in asymmetric units are colored in red and green, respectively.
Table 1:
Data collection and refinement statistics
| MEF2BWT/NKX2-5/DNA | MEF2ChimWT/NKX2-5/DNA | |
|---|---|---|
| Resolution range | 47.46 – 2.90 (3.00 – 2.90) | 49.28 – 2.10 (2.18 – 2.10) |
| Space group | P 21 21 21 | P 21 21 21 |
| Cell dimensions | ||
| a, b, c (Å) | 66.04 93.24 136.5 | 69.37 133.9 140.04 |
| α, β, γ (°) | 90 90 90 | 90 90 90 |
| Total reflections | 74630 (7279) | 499421 (47928) |
| Unique reflections | 18336 (1904) | 76673 (7471) |
| Multiplicity | 4.1 (3.8) | 6.5 (6.4) |
| Completeness (%) | 94.3 (99.6) | 99.7 (98.9) |
| Mean I/sigma(I) | 11.3 (3.0) | 12.9 (3.0) |
| Wilson B-factor | 60.64 | 35.73 |
| R-merge | 0.079 (0.580) | 0.077 (0.608) |
| R-meas | 0.089 (0.665) | 0.084 (0.661) |
| R-pim | 0.042 (0.319) | 0.033 (0.257) |
| CC1/2 | 0.997 (0.815) | 0.997 (0.854) |
| CC* | 0.999 (0.948) | 0.999 (0.960) |
| Reflections used in refinement | 18220 (1901) | 76582 (7468) |
| Reflections used for R-free | 878 (93) | 3802 (350) |
| R-work | 0.206 (0.320) | 0.188 (0.264) |
| R-free | 0.254 (0.357) | 0.225 (0.290) |
| CC (work) | 0.954 (0.845) | 0.965 (0.842) |
| CC (free) | 0.932 (0.778) | 0.968 (0.811) |
| Number of non-hydrogen atoms | 5706 | 8973 |
| macromolecules | 5703 | 8528 |
| solvent | 3 | 445 |
| Protein residues | 469 | 704 |
| RMS (bonds) | 0.012 | 0.015 |
| RMS (angles) | 1.55 | 2.05 |
| Average B-factor | 70.4 | 47.85 |
| macromolecules | 70.42 | 48.16 |
| solvent | 23.72 | 42.01 |
Values in parentheses are for the highest-resolution shell.
Protein-DNA interactions
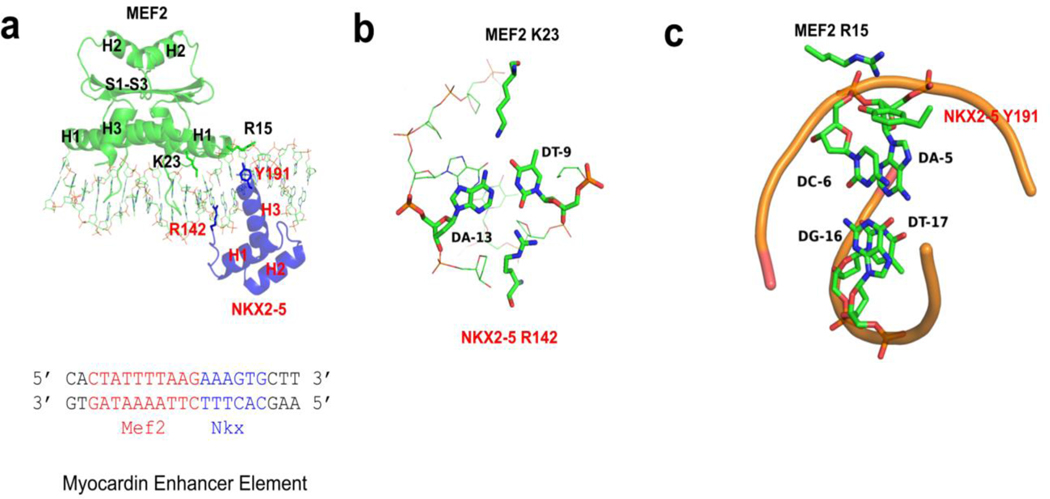
MEF2 and NKX2-5 interact with DNA from opposite faces of the DNA double helix in the ternary complex. Consistent with previous literature reports, MEF2 interacts mainly with DNA minor groove through helix H1 and the N terminal extension, and NKX2-5 interacts mainly with DNA major groove through helix H3 of the homeodomain (Fig. 2a, Supplemental Fig. 4). MEF2 and NKX2-5 bind to overlapping DNA regions at the junction of their binding sites. The N terminal extension from MEF2 and the N terminal extension from NKX2-5 interact with the same minor groove region (Figure 2a). Moreover, K23 of MEF2 and R142 of NKX2-5 interact with the same AT base pair from the major and minor groove, respectively (Figure 2b). Interestingly, the NKX2-5 R142C mutation, which is found in congenital heart defects patients, has reduced DNA affinity and diminished synergistic interaction with transcription partner MEF2, TBX5, and GATA4 in mouse models and biochemical assays [46–49]. There is a cation pi interaction between MEF2 R15 and NKX2-5 Y191, both residues are involved in DNA interaction: MEF2 R15 interacts with DNA phosphate backbone, and NKX2-5 Y191 interacts with bases A5’ and C6’ (Figure 2c). MEF2 R15 and NKX2-5 Y191 residues are conserved in MEF2 family proteins and NKX family proteins respectively [45,50], and residues Y191 are reported to be an important residue involving specific interaction between NKX2-5 and NK2 element [45]. There is no obvious DNA bending in our MEF2/NKX2-5/DNA structures as compared to published ternary structures involving MADS-box family members such as the yeast MCM1/Matα2/DNA and the human SRF/SAP-1/DNA ternary structures (Supplemental Fig. 5) [28,51,52]. There is no protein-protein interaction involving secondary structural elements between MEF2 and NKX2-5 in our crystal structures, which is in contrast to the yeast MCM1/Matα2/DNA and the human SRF/SAP-1/DNA ternary structures, in which there are direct protein-protein interactions between strand S2 of the MADS-box domain and another beta strand from co-factors (Supplemental Fig. 5) [51–53]. As the protein constructs in our crystallization studies only contain the MADS-box and MEF2 specific domain of MEF2 and homeobox domain of NKX2-5, we could not rule out the possibility that other parts in the full length MEF2 and NKX2-5 proteins may interact with each other in this cis-mode.
Figure 2. Protein and DNA interaction features of MEF2/NKX2-5/DNA structure.
(a) MEF2 chimera/NKX2-5/DNA ternary complex structure. DNA interacting residues of interests (MEF2 R15 and K23, NKX2-5 R142 and Y191) are shown as sticks. The sequence of the DNA in the crystal is shown below, with the MEF2 binding site colored in red and the NKX2-5 binding site colored in blue. H1-H3: helix 1–3; S1-S3: beta strand 1–3. (b) MEF2 K23 and NKX2-5 R142 interact with the same AT base pair from the major and minor groove, respectively. (c) Cation ion and pi interaction between MEF2 R15 and NKX2-5 Y191. MEF2 R15 interacts with phosphate backbone of A-5’. NKX2-5 Y191 interacts with the bases of A-5’ and C-6’.
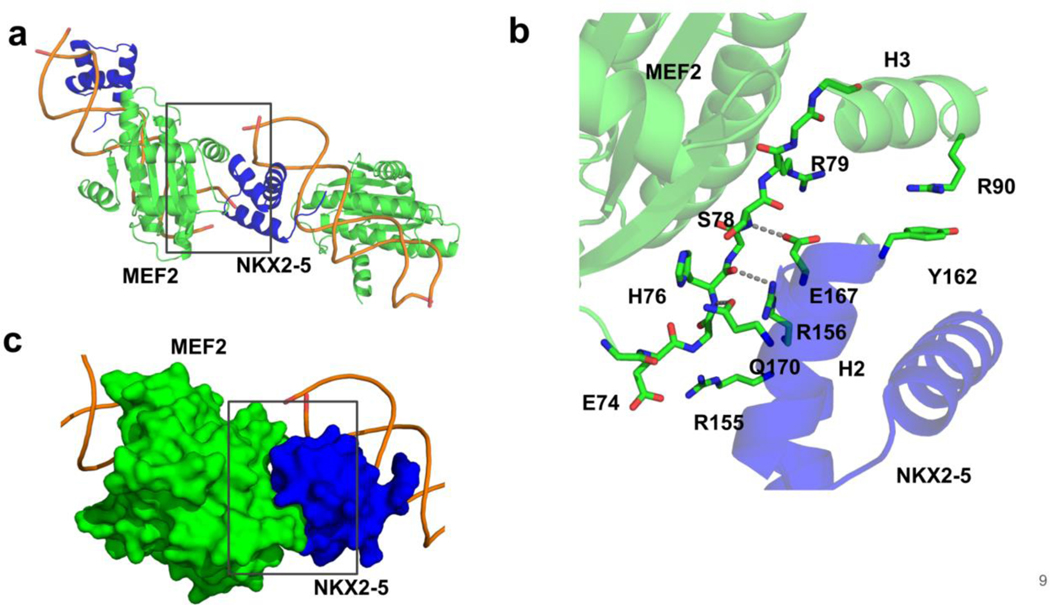
Protein-Protein Interaction Interface
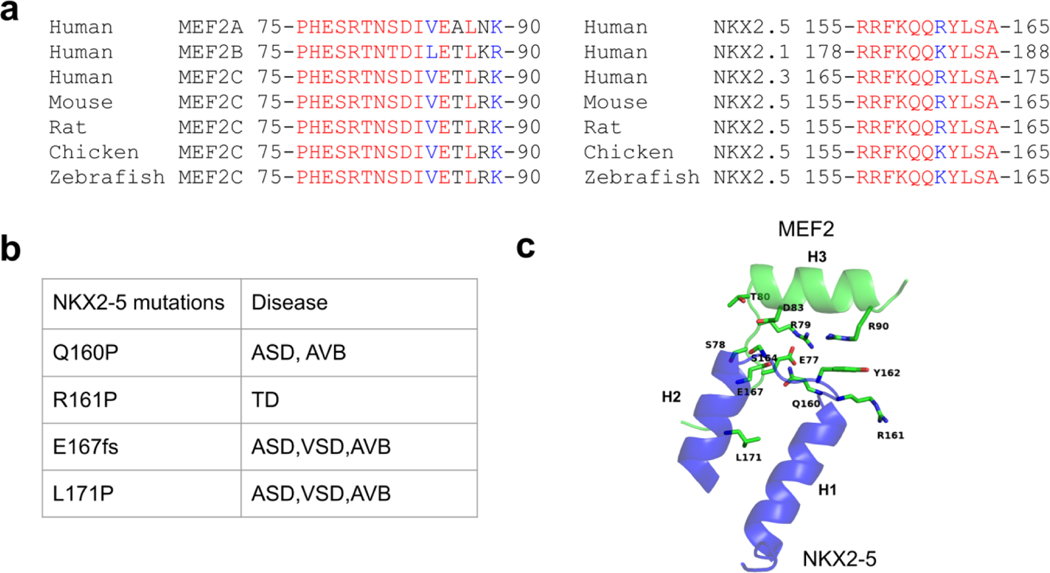
In the crystal structures, we noticed a protein-protein interaction interface (with buried surface area around 359.6 Å2) between a MEF2 dimer bound to one DNA duplex and a NKX2-5 molecule bound to another DNA duplex in symmetry related complexes. We refer to this as the trans interaction mode. This protein-protein interaction interface is conserved in both the MEF2 Chimera/NKX2-5/DNA crystal structure and the MEFB/NKX2-5/DNA crystal structure. In this interface, the MEF2 specific domain strand S3 and helix H3 interact with helix H1, loop1 and helix H2 from NKX2-5 homeobox domain (Figure 3a). This interface shows remarkable chemical and shape complementarity: charge-charge interactions between MEF2 E74 and NKX2-5 R155, and MEF2 R79 and NKX2-5 E167; hydrogen bonding interactions between the main chain of MEF2 H76, S78, and the side chains of NKX2-5 R156, E167, and Q170; and a cation-pi interaction between MEF2 R90 and NKX2-5 Y162 (Figure 3b, c). Residues in this protein-protein interaction interface are evolutionarily conserved (Figure 4a). Mutations associated with heart disease such as NKX2-5 Q160P and L171P are mapped in this interface (Figure 4b, c) [17,54–57], these disease mutations are likely disrupting MEF2 and NKX2-5 interaction. These analyses suggest that the MEF2 and NKX2-5 interaction interfaces observed in our structures are likely to be functionally important. This interface also harbors residues which has been reported to be subject to post-translational modifications in literature, for example, MEF2 T80 and NKX2-5 and NKX2-5 S164 could be modified by phosphorylation [58–60]. Further studies are needed to test if these disease mutations and post-translational modifications impact the interaction between MEF2 and NKX2-5.
Figure 3. The conserved MEF2 and NKX2-5 protein-protein interaction interface in two crystal forms.
(a) Direct protein-protein interaction interface between MEF2 and NKX2-5 from symmetry related complexes. (b) Detailed interactions of residues involved in the protein-protein interaction interface. Dashed line indicates hydrogen bonds. (c) MEF2 and NKX2-5 protein-protein interaction interface represented in surface mode. H3: MEF2 helix 3. H2: NKX2-5 helix 2.
Figure 4. MEF2 and NKX2-5 protein-protein interaction interface is evolutionary conserved.
(a) Sequence alignment of MEF2 and NKX2-5 interface across species. Identical amino acids are colored in red. Amino acids with strong similar properties are colored in blue. (b) NKX2-5 disease related mutations in the interface region in literature. ASD, Atrial Septal Defect; AVB: atrioventricular block; VSD, Ventricular Septal Defect; TD: thyroid dysgenesis. fs: frameshift mutation. (c) Disease relevant residues of NKX2-5 are shown as sticks in the MEF2 and NKX2-5 interaction interface. Residues from MEF2 which are involving interactions with these NKX2-5 residues are shown as sticks and labelled in green. Although NKX2-5 L171 is not involved in direct contact with MEF2, the L171P mutation may impact NKX2-5 and MEF2 interaction as proline is considered to be an alpha helix breaker.
Our study indicates that the MEF2 specific domain could be an important interaction interface for cofactor interactions. This novel cofactor interaction interface is distinct from the classical MEF2 cofactor binding groove formed by the MEF2 specific domain helix H2 and strand S1-S3 [44,61,62]. Our structures could explain previous observations in literature that MEF2C VLL65-67ASR mutants that were unable to bind histone deacetylase (HDAC4) could interact with the bHLH family member myogenin [63]. According to our model, the VLL65-67ASR mutations disrupt classical MEF2 co-factor interaction groove which is responsible for class IIa HDAC interaction but not for myogenin interaction; myogenin interacts with MEF2 through MEF2 specific domain, as mutations (E77V/S78N/R79Q/T80A or N73I/E74A/H76L) in MEF2 specific domain disrupts MEF2 and myogenin interaction and synergistic activation of target genes [64].
However, we were unable to detect MEF2 and NKX2-5 interaction in vitro using two DNA oligomers with one containing a MEF2 binding site and the other containing a NKX2-5 binding site, and the MEF2 and NKX2-5 protein fragments used in our crystallography study (data not shown). One possibility is that the stable interaction by this interface requires full-length MEF2 or NKX2-5 proteins wherein other parts of MEF2 and NKX2-5 may contribute to the overall interaction stability. Another possibility is that the interaction inferred by this interface is a weak and transient interaction in solution but would be further stabilized by other proteins in cellular contexts. Our preliminary data shows that the interaction of DNA with either or both MEF2C and NKX2-5 is important for their mutual interactions, as nuclease treatment disrupts the interaction between MEF2C and NKX2-5 in pull-down assays (Supplemental Fig. 6).
In conclusion, our studies suggest that MEF2 and NKX2-5 could interact with each other through at least two different modes: by binding adjacent and overlapping DNA regions in cis on the same DNA strands and interactions in trans across different DNA strands through a conserved protein-protein interaction interface. The latter could also have implications for long-range chromatin interactions (e.g., enhancer-promoter interaction) mediated by transcription machinery containing MEF2 and NKX2-5 proteins. This model of long range chromatin interactions is consistent with the observations that NKX2-5 and MEF2 are found in super-enhancers [65,66]. Further functional and genome structural studies are needed to test the biological roles of this interface in cell and animal models, and the crystal structures presented in this study provide a foundation for these studies.
Materials and Methods
Protein purification
MEF2 Chimera (residues 1–95) and MEF2B (residues 1–93) which contain MADS-box domain (residues 1–57) and MEF2 specific domain (residues 58–95) were purified as previously described [43]. The final storage buffer for MEF2B WT, MEF2 Chimera was as follows: 10 mM HEPES (pH 7.5), 200 mM NaCl, 0.5 mM EDTA, and 0.5 mM TCEP.
NKX2-5 homeobox domain (residues 138–197) with C193S was cloned into pET28 vector for crystallization [45]. The protein was expressed as sumo fusion protein with both 6x His-tag and sumo tag at its N terminal in E.coli BL21(DE3) pLysS cells. Protein expression was induced in 1L 2XYT medium with 0.5 mM IPTG at 22 °C overnight (16 to 20 hours). Protein was initially purified by QIAGEN Ni-NTA agarose. The His-tag and sumo-tag was cleaved by Ulp-1 enzyme, and the protein was further purified by heparin Fastflow column (GE Healthcare) with buffer A containing 20 mM HEPES (pH 7.0), 0.5 mM EDTA, and 0.5 mM TCEP and buffer B containing all the components in buffer A and 1.5 M NaCl. Protein peak fractions were pooled together and subject to final Superdex 75 (GE Healthcare) size exclusion column purification with buffer as follows: 10 mM HEPES (pH 7.5), 200 mM NaCl, and 0.5 mM TCEP.
DNA purification
The DNA used in crystallization for MEF2 Chimera/NKX2-5/DNA ternary complex is 5′ CACTATTTTAAGAAAGTGCTT 3′ and its complementary strand 5′ AAGCACTTTCTTAAAATAGTG 3′. The DNA used in crystallization for MEF2B/NKX2-5/DNA is 5′ CCACTATTTTAAGAAAGTGCTT 3′ and its complementary strand 5′ AAGCACTTTCTTAAAATAGTGG 3′. DNA was purchased from Integrated DNA Technologies (Coralville, IA) at 1-μmol scale in the crude and desalted form. The crude DNA was dissolved in a 10 mM NaOH and purified by a Mono Q cation-exchange column (GE Healthcare) on FPLC (GE Healthcare) as previously described [67]. Complementary DNA strands were annealed at 95 °C in the annealing buffer (100 mM NaCl, 5 mM HEPES pH 7.6) in PCR machine (Eppendorf Mastercycler Personal 5332 Thermal Cycler) for 2 min and cool to room temperature on bench for 1 hour.
Electrophoresis mobility shift assay (EMSA)
EMSA was performed in 20 mM HEPES (pH 7.6), 250 mM NaCl, 1 mM DTT, 12% glycerol in a 10 μl volume. The final concentration of DNA was kept at 10 μM. The final concentration of MEF2 Chim WT (residues 1–95) and MEF2B (residues 1–93) was kept at 10 μM. The final NKX2-5 (residues 138–197) concentration was kept at 10 μM (DNA: MEF2: NKX2-5 molar ratio 1:1:1) or 20 μM (DNA: MEF2: NKX2-5 molar ratio 1:1:2). The binding reactions were analyzed on a 4%–20% (w/v) acrylamide gradient native gel in TBE and stained with Sybr Safe DNA Dye (Thermo Fisher Scientific).
Crystallization and structure determination
MEF2, NKX2-5 and DNA were mixed at a molar ratio of 1:1:1.2, and the final protein concentration in the mixture was around 10 to 15 mg/mL. Sitting drop crystal trays were set up by a crystallization robot (Crystal Gryphon from Art Robbins Instruments) at 18°C, in which 0.4 μL protein complex and 0.4 μL mother liquor were mixed. MEF2 Chimera/NKX2-5/DNA crystals appeared within three weeks with rod shape in crystallization buffer [0.15 M DL-Malic acid pH 7.0, 20% Polyethylene glycol (PEG) 3350]. Crystals were harvested, cryoprotected in the crystallization buffer with increased PEG concentration to 35% and flash frozen in liquid nitrogen. MEF2B/NKX2-5/DNA crystals appeared within three weeks with needle or plate shape in crystallization buffer [100 mM HEPES pH 7.0, 18% PEG 2000]. Crystals were harvested, cryoprotected in the crystallization buffer with 30% PEG 400 as cryoprotectant. Data were collected at Advanced Photon Source (APS Chicago) beamline 23 ID-B. Crystal diffraction data were processed with iMosflm and initial space group assignment by pointless in CCP4 suite [68–70]. The MEF2 Chimera/NKX2-5/DNA and MEF2B/NKX2-5/DNA ternary complex structures were determined by molecular replacement with Phaser in CCP4 suite using MEF2/DNA complex (PDB: 1N6J) and NKX2-5/DNA complex (PDB: 3RKQ) as partial search models [44,45,71]. Model building was done in Coot and refinement was done in Refmac5, Phenix refine, and PDB_REDO [72–76]. Composite omit maps were generated by “Composite omit map” tool in Phenix suite [77–79]. Crystallographic and refinement statistics table (Table 1) was generated by utility tools in Phenix suite [73].
Protein sequence alignment was performed with the Clustal Omega [80] and visualized with ESPript 3.0 [81], and protein and DNA interaction plots are generated with the DNAproDB tool [82].
Chip-seq data analysis
MEF2C and NKX2-5 Chip-seq data was retrieved from ChIP-Atlas database [42] and visualized in IGV browser [83].
Co-immunoprecipitation (Co-IP) and Immunoblotting
HEK293T cells were transfected with the indicated expression plasmids for 48 hours. Whole cell lysates were prepared with NP40 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 5 mM EDTA) supplemented with 20 mM β-glycerophosphate and 1 mM sodium orthovanadate. Whole cell lysates were sonicated, centrifuged and pre-cleared with protein A/G agarose for 1 hour. Pre-cleared samples were then incubated with the indicated antibody-conjugated agarose overnight at 4°C. The agarose beads were washed extensively, and samples were eluted by boiling at 95°C for 10 min. Precipitated proteins were analyzed by SDS gel electrophoresis and immunoblotting.
Immunoblotting was performed using the indicated primary antibodies (1:1000 dilution) and IRDye800-conjugated secondary antibodies (1:10,000 dilution, LI-COR). Proteins were visualized by Odyssey infrared imaging system (LI-COR).
Supplementary Material
Acknowledgements
The work was supported by the National Institutes of Health (R01AI113009, 5U54DK107981, and R21HG010528 to L.C. and R35GM130376 to R.R.).
Footnotes
Data availability
Coordinates and structure factors have been deposited in the PDB with accession numbers 6WC2 (MEF2 Chimera/NKX2-5/DNA complex) and 6WC5 (MEF2B/NKX2-5/DNA complex).
References
- [1].Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP, Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants, Development. 119 (1993) 419–431. [DOI] [PubMed] [Google Scholar]
- [2].Duprey P, Lesens C, Control of skeletal muscle-specific transcription: involvement of paired homeodomain and MADS domain transcription factors, Int. J. Dev. Biol 38 (1994) 591–604. [PubMed] [Google Scholar]
- [3].Shore P, Sharrocks AD, The MADS-box family of transcription factors, Eur. J. Biochem 229 (1995) 1–13. [DOI] [PubMed] [Google Scholar]
- [4].Potthoff MJ, Olson EN, MEF2: a central regulator of diverse developmental programs, Development. 134 (2007) 4131–4140. [DOI] [PubMed] [Google Scholar]
- [5].Nagel S, Kaufmann M, Drexler HG, MacLeod RAF, The Cardiac Homeobox Gene NKX2-5 Is Deregulated by Juxtaposition with BCL11B in Pediatric T-ALL Cell Lines via a Novel t(5;14)(q35.1;q32.2), Cancer Res. 63 (2003) 5329–5334. [PubMed] [Google Scholar]
- [6].Nagel S, Meyer C, Quentmeier H, Kaufmann M, Drexler HG, MacLeod RAF, MEF2C is activated by multiple mechanisms in a subset of T-acute lymphoblastic leukemia cell lines, Leukemia. 22 (2008) 600–607. [DOI] [PubMed] [Google Scholar]
- [7].Su X, Della-Valle V, Delabesse E, Azgui Z, Berger R, Merle-Béral H, Bernard OA, Nguyen-Khac F, Transcriptional activation of the cardiac homeobox gene CSX1/NKX2-5 in a B-cell chronic lymphoproliferative disorder, Haematologica. 93 (2008) 1081–1085. [DOI] [PubMed] [Google Scholar]
- [8].Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, Jackman S, Krzywinski M, Scott DW, Trinh DL, Tamura-Wells J, Li S, Firme MR, Rogic S, Griffith M, Chan S, Yakovenko O, Meyer IM, Zhao EY, Smailus D, Moksa M, Chittaranjan S, Rimsza L, Brooks-Wilson A, Spinelli JJ, Ben-Neriah S, Meissner B, Woolcock B, Boyle M, McDonald H, Tam A, Zhao Y, Delaney A, Zeng T, Tse K, Butterfield Y, Birol I, Holt R, Schein J, Horsman DE, Moore R, Jones SJM, Connors JM, Hirst M, Gascoyne RD, Marra MA, Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma, Nature. 476 (2011) 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, Vuerhard M, Buijs-Gladdines J, Kooi C, Klous P, van Vlierberghe P, Ferrando AA, Cayuela JM, Verhaaf B, Beverloo HB, Horstmann M, de Haas V, Wiekmeijer A-S, Pike-Overzet K, Staal FJT, de Laat W, Soulier J, Sigaux F, Meijerink JPP, Integrated Transcript and Genome Analyses Reveal NKX2-1 and MEF2C as Potential Oncogenes in T Cell Acute Lymphoblastic Leukemia, Cancer Cell. 19 (2011) 484–497. [DOI] [PubMed] [Google Scholar]
- [10].Pon JR, Marra MA, MEF2 transcription factors: developmental regulators and emerging cancer genes, Oncotarget. 7 (2016) 2297–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brescia P, Schneider C, Holmes AB, Shen Q, Hussein S, Pasqualucci L, Basso K, Dalla-Favera R, MEF2B Instructs Germinal Center Development and Acts as an Oncogene in B Cell Lymphomagenesis, Cancer Cell. 34 (2018) 453–465.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lin Q, Schwarz J, Bucana C, Olson EN, Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C, Science. 276 (1997) 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP, Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene NKX2-5, Genes Dev. 9 (1995) 1654–1666. [DOI] [PubMed] [Google Scholar]
- [14].Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S, The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development, Development. 126 (1999) 1269–1280. [DOI] [PubMed] [Google Scholar]
- [15].Bruneau BG, Transcriptional regulation of vertebrate cardiac morphogenesis, Circ. Res 90 (2002) 509–519. [DOI] [PubMed] [Google Scholar]
- [16].Bruneau BG, Signaling and transcriptional networks in heart development and regeneration, Cold Spring Harb. Perspect. Biol 5 (2013) a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schott J-J, Woodrow Benson D, Basson CT, Pease W, Michael Silberbach G, Moak JP, Maron BJ, Seidman CE, Seidman JG, Congenital heart disease caused by mutations in the transcription factor NKX2-5, Science. 281 (1998) 108–111. [DOI] [PubMed] [Google Scholar]
- [18].Chung I-M, Rajakumar G, Genetics of Congenital Heart Defects: The NKX2-5 Gene, a Key Player, Genes. 7 (2016). 10.3390/genes7020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Reamon-Buettner SM, Hecker H, Spanel-Borowski K, Craatz S, Kuenzel E, Borlak J, Novel NKX2-5 Mutations in Diseased Heart Tissues of Patients with Cardiac Malformations, Am. J. Pathol 164 (2004) 2117–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG, Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage, Cell. 151 (2012) 206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Qiao X-H, Wang F, Zhang X-L, Huang R-T, Xue S, Wang J, Qiu X-B, Liu X-Y, Yang Y-Q, MEF2C loss-of-function mutation contributes to congenital heart defects, Int. J. Med. Sci 14 (2017) 1143–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nagel S, Venturini L, Meyer C, Kaufmann M, Scherr M, Drexler HG, Macleod RA, Transcriptional deregulation of oncogenic myocyte enhancer factor 2C in T-cell acute lymphoblastic leukemia, Leuk. Lymphoma 52 (2011) 290–297. [DOI] [PubMed] [Google Scholar]
- [23].Rajasingh J, Bord E, Hamada H, Lambers E, Qin G, Losordo DW, Kishore R, STAT3-dependent mouse embryonic stem cell differentiation into cardiomyocytes: analysis of molecular signaling and therapeutic efficacy of cardiomyocyte precommitted mES transplantation in a mouse model of myocardial infarction, Circ. Res 101 (2007) 910–918. [DOI] [PubMed] [Google Scholar]
- [24].Cante-Barrett K, Pieters R, Meijerink JP, Myocyte enhancer factor 2C in hematopoiesis and leukemia, Oncogene. 33 (2014) 403–410. [DOI] [PubMed] [Google Scholar]
- [25].Olson EN, Perry M, Schulz RA, Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors, Dev. Biol 172 (1995) 2–14. [DOI] [PubMed] [Google Scholar]
- [26].Molkentin JD, Firulli AB, Black BL, Martin JF, Hustad CM, Copeland N, Jenkins N, Lyons G, Olson EN, MEF2B is a potent transactivator expressed in early myogenic lineages, Mol. Cell. Biol 16 (1996) 3814–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Herskowitz I, A regulatory hierarchy for cell specialization in yeast, Nature. 342 (1989) 749–757. [DOI] [PubMed] [Google Scholar]
- [28].Tan S, Richmond TJ, Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex, Nature. 391 (1998) 660–666. [DOI] [PubMed] [Google Scholar]
- [29].Vincentz JW, Barnes RM, Firulli BA, Conway SJ, Firulli AB, Cooperative interaction of Nkx2.5 and Mef2c transcription factors during heart development, Dev. Dyn 237 (2008) 3809–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL, GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium, Mol. Cell. Biol 28 (2008) 5420–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].He A, Kong SW, Ma Q, Pu WT, Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart, Proceedings of the National Academy of Sciences. 108 (2011) 5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I, Sperling SR, The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs, PLoS Genet. 7 (2011) e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MAH, He D, Small EM, Mileikovsky M, Nagy A, Holloway AK, Pollard KS, Müller CW, Bruneau BG, Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis, Cell. 164 (2016) 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sepulveda JL, Vlahopoulos S, Iyer D, Belaguli N, Schwartz RJ, Combinatorial Expression of GATA4, NKX2-5, and Serum Response Factor Directs Early Cardiac Gene Activity, J. Biol. Chem 277 (2002) 25775–25782. [DOI] [PubMed] [Google Scholar]
- [35].Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D, Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors, Cell. 142 (2010) 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, Christoforou N, Epstein JA, Gearhart JD, Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success, J. Mol. Cell. Cardiol 60 (2013) 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Song K, Nam Y-J, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, Hill JA, Bassel-Duby R, Olson EN, Heart repair by reprogramming non-myocytes with cardiac transcription factors, Nature. 485 (2012) 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang L, Liu Z, Yin C, Asfour H, Chen O, Li Y, Bursac N, Liu J, Qian L, Stoichiometry of Gata4, Mef2c, and Tbx5 Influences the Efficiency and Quality of Induced Cardiac Myocyte Reprogramming, Circ. Res (2015). https://www.ahajournals.org/doi/abs/10.1161/circresaha.116.305547 (accessed August 15, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ueyama T, Kasahara H, Ishiwata T, Nie Q, Izumo S, Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis, Mol. Cell. Biol 23 (2003) 9222–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN, Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development, Development. 133 (2006) 4245–4256. [DOI] [PubMed] [Google Scholar]
- [41].Pagiatakis C, Gordon JW, Ehyai S, McDermott JC, A novel RhoA/ROCK-CPI-17-MEF2C signaling pathway regulates vascular smooth muscle cell gene expression, J. Biol. Chem 287 (2012) 8361–8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Oki S, Ohta T, Shioi G, Hatanaka H, Ogasawara O, Okuda Y, Kawaji H, Nakaki R, Sese J, Meno C, ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data, EMBO Rep. 19 (2018). 10.15252/embr.201846255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lei X, Kou Y, Fu Y, Rajashekar N, Shi H, Wu F, Xu J, Luo Y, Chen L, The Cancer Mutation D83V Induces an α-Helix to β-Strand Conformation Switch in MEF2B, J. Mol. Biol 430 (2018) 1157–1172. [DOI] [PubMed] [Google Scholar]
- [44].Han A, Pan F, Stroud JC, Youn H-D, Liu JO, Chen L, Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2, Nature. 422 (2003) 730–734. [DOI] [PubMed] [Google Scholar]
- [45].Pradhan L, Genis C, Scone P, Weinberg EO, Kasahara H, Nam H-J, Crystal structure of the human NKX2.5 homeodomain in complex with DNA target, Biochemistry. 51 (2012) 6312–6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gutierrez-Roelens I, Sluysmans T, Gewillig M, Devriendt K, Vikkula M, Progressive AV-block and anomalous venous return among cardiac anomalies associated with two novel missense mutations in the CSX/NKX2-5 gene, Hum. Mutat 20 (2002) 75–76. [DOI] [PubMed] [Google Scholar]
- [47].Kasahara H, Benson DW, Biochemical analyses of eight NKX2.5 homeodomain missense mutations causing atrioventricular block and cardiac anomalies, Cardiovasc. Res 64 (2004) 40–51. [DOI] [PubMed] [Google Scholar]
- [48].Zakariyah AF, Rajgara RF, Veinot JP, Skerjanc IS, Burgon PG, Congenital heart defect causing mutation in Nkx2.5 displays in vivo functional deficit, J. Mol. Cell. Cardiol 105 (2017) 89–98. [DOI] [PubMed] [Google Scholar]
- [49].Zakariyah AF, Rajgara RF, Horner E, Cattin M-E, Blais A, Skerjanc IS, Burgon PG, In Vitro Modeling of Congenital Heart Defects Associated with an NKX2-5 Mutation Revealed a Dysregulation in BMP/Notch-Mediated Signaling, Stem Cells. 36 (2018) 514–526. [DOI] [PubMed] [Google Scholar]
- [50].Santelli E, Richmond TJ, Crystal structure of MEF2A core bound to DNA at 1.5 A resolution, J. Mol. Biol 297 (2000) 437–449. [DOI] [PubMed] [Google Scholar]
- [51].Hassler M, Richmond TJ, The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex, EMBO J. 20 (2001) 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mo Y, Ho W, Johnston K, Marmorstein R, Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex, J. Mol. Biol 314 (2001) 495–506. [DOI] [PubMed] [Google Scholar]
- [53].Tan S, Richmond TJ, Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex, Nature. 391 (1998) 660–666. [DOI] [PubMed] [Google Scholar]
- [54].Inga A, Reamon-Buettner SM, Borlak J, Resnick MA, Functional dissection of sequence-specific NKX2-5 DNA binding domain mutations associated with human heart septation defects using a yeast-based system, Hum. Mol. Genet 14 (2005) 1965–1975. [DOI] [PubMed] [Google Scholar]
- [55].van Engelen K, Mommersteeg MTM, Baars MJH, Lam J, Ilgun A, van Trotsenburg ASP, Smets AMJB, Christoffels VM, Mulder BJM, Postma AV, The Ambiguous Role of NKX2-5 Mutations in Thyroid Dysgenesis, PLoS One. 7 (2012) e52685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Abou Hassan OK, Fahed AC, Batrawi M, Arabi M, Refaat MM, DePalma SR, Seidman JG, Seidman CE, Bitar FF, Nemer GM, NKX2-5 mutations in an inbred consanguineous population: genetic and phenotypic diversity, Sci. Rep 5 (2015) 8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kalayinia S, Ghasemi S, Mahdieh N, A comprehensive in silico analysis, distribution and frequency of human NKX2-5 mutations; A critical gene in congenital heart disease, J Cardiovasc Thorac Res. 11 (2019) 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kasahara H, Izumo S, Identification of the in vivo casein kinase II phosphorylation site within the homeodomain of the cardiac tisue-specifying homeobox gene product Csx/Nkx2.5, Mol. Cell. Biol 19 (1999) 526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E, PhosphoSitePlus, 2014: mutations, PTMs and recalibrations, Nucleic Acids Res. 43 (2015) D512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Al Madhoun AS, Mehta V, Li G, Figeys D, Wiper-Bergeron N, Skerjanc IS, Skeletal myosin light chain kinase regulates skeletal myogenesis by phosphorylation of MEF2C, EMBO J. 30 (2011) 2477–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].He J, Ye J, Cai Y, Riquelme C, Liu JO, Liu X, Han A, Chen L, Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly, Nucleic Acids Res. 39 (2011) 4464–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cardoso AC, Pereira AHM, Ambrosio ALB, Consonni SR, Rocha de Oliveira R, Bajgelman MC, Dias SMG, Franchini KG, FAK Forms a Complex with MEF2 to Couple Biomechanical Signaling to Transcription in Cardiomyocytes, Structure. 24 (2016) 1301–1310. [DOI] [PubMed] [Google Scholar]
- [63].Molkentin JD, Black BL, Martin JF, Olson EN, Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C, Mol. Cell. Biol 16 (1996) 2627–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Molkentin JD, Black BL, Martin JF, Olson EN, Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins, Cell. 83 (1995) 1125–1136. [DOI] [PubMed] [Google Scholar]
- [65].Ounzain S, Pedrazzini T, Super-enhancer lncs to cardiovascular development and disease, Biochim. Biophys. Acta 1863 (2016) 1953–1960. [DOI] [PubMed] [Google Scholar]
- [66].Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA, Super-enhancers in the control of cell identity and disease, Cell. 155 (2013) 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wu Y, Dey R, Han A, Jayathilaka N, Philips M, Ye J, Chen L, Structure of the MADS-box/MEF2 domain of MEF2A bound to DNA and its implication for myocardin recruitment, J. Mol. Biol 397 (2010) 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Collaborative Computational Project, Number 4, The CCP4 suite: programs for protein crystallography, Acta Crystallogr. D Biol. Crystallogr 50 (1994) 760–763. [DOI] [PubMed] [Google Scholar]
- [69].Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW, iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM, Acta Crystallogr. D Biol. Crystallogr 67 (2011) 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Potterton E, Briggs P, Turkenburg M, Dodson E, A graphical user interface to the CCP4 program suite, Acta Crystallogr. D Biol. Crystallogr 59 (2003) 1131–1137. [DOI] [PubMed] [Google Scholar]
- [71].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software, J. Appl. Crystallogr 40 (2007) 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Emsley P, Cowtan K, Coot: model-building tools for molecular graphics, Acta Crystallogr. D Biol. Crystallogr 60 (2004) 2126–2132. [DOI] [PubMed] [Google Scholar]
- [73].Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr. D Biol. Crystallogr 66 (2010) 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD, Towards automated crystallographic structure refinement with phenix.refine, Acta Crystallogr. D Biol. Crystallogr 68 (2012) 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Joosten RP, Long F, Murshudov GN, Perrakis A, The PDB_REDO server for macromolecular structure model optimization, IUCrJ. 1 (2014) 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Joosten RP, Salzemann J, Bloch V, Stockinger H, Berglund A-C, Blanchet C, Bongcam-Rudloff E, Combet C, Da Costa AL, Deleage G, Diarena M, Fabbretti R, Fettahi G, Flegel V, Gisel A, Kasam V, Kervinen T, Korpelainen E, Mattila K, Pagni M, Reichstadt M, Breton V, Tickle IJ, Vriend G, PDB_REDO: automated re-refinement of X-ray structure models in the PDB, J. Appl. Crystallogr 42 (2009) 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Hodel A, Kim S-H, Brünger AT, Model bias in macromolecular crystal structures, Acta Crystallogr. A 48 (1992) 851–858. [Google Scholar]
- [78].Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Adams PD, Read RJ, Zwart PH, Hung L-W, Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias, Acta Crystallogr. D Biol. Crystallogr 64 (2008) 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Afonine PV, Moriarty NW, Mustyakimov M, Sobolev OV, Terwilliger TC, Turk D, Urzhumtsev A, Adams PD, FEM: feature-enhanced map, Acta Crystallogr. D Biol. Crystallogr 71 (2015) 646–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega, Mol. Syst. Biol 7 (2011) 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Robert X, Gouet P, Deciphering key features in protein structures with the new ENDscript server, Nucleic Acids Res. 42 (2014) W320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sagendorf JM, Markarian N, Berman HM, Rohs R, DNAproDB: an expanded database and web-based tool for structural analysis of DNA–protein complexes, Nucleic Acids Res. 48 (2020) D277–D287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Thorvaldsdóttir H, Robinson JT, Mesirov JP, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration, Brief. Bioinform 14 (2013) 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.