Abstract
Background
We and others have identified mutually exclusive molecular subgroups of meningiomas; however, the implications of this classification for clinical prognostication remain unclear. Integrated genomic and epigenomic analyses implicate unique oncogenic processes associated with each subgroup, suggesting the potential for divergent clinical courses. The aim of this study was to understand the associated clinical outcomes of each subgroup, as this could optimize treatment for patients.
Methods
We analyzed outcome data for 469 meningiomas of known molecular subgroup, including extent of resection, postoperative radiation, surveillance imaging, and time to recurrence, when applicable. Statistical relationships between outcome variables and subgroup were assessed. Features previously associated with recurrence were further investigated after stratification by subgroup. We used Kaplan–Meier analyses to compare progression-free survival, and identified factors significantly associated with recurrence using Cox proportional hazards modeling.
Results
Meningioma molecular subgroups exhibited divergent clinical courses at 2 years of follow-up, with several aggressive subgroups (NF2, PI3K, HH, tumor necrosis factor receptor–associated factor 7 [TRAF7]) recurring at an average rate of 22 times higher than others (KLF4, POLR2A, SMARCB1). PI3K-activated tumors recurred earlier than other subgroups but had intermediate long-term outcome. Among low-grade tumors, HH and TRAF7 meningiomas exhibited elevated recurrence compared with other subgroups. Recurrence of NF2 tumors was associated with male sex, high grade, and elevated Ki-67. Multivariate analysis identified molecular subgroup as an independent predictor of recurrence, along with grade and previous recurrence.
Conclusion
We describe distinct clinical outcomes and recurrence rates associated with meningioma molecular subgroups. Our findings emphasize the importance of genomic characterization to guide postoperative management decisions for meningiomas.
Keywords: meningioma genomics, molecular subgroups, precision medicine, tumor prognosis
Key Points.
In addition to distinct location, histological, transcriptional, and epigenetic profiles, meningioma molecular subgroups exhibit unique clinical courses.
Relative to other subgroups, PI3K-activated meningiomas recur earlier.
Among World Health Organization (WHO) grade I meningiomas, HH-activated and TRAF7 mutant tumors exhibit elevated recurrence rates.
Importance of the Study.
Previous studies have established 7 molecular subgroups of meningiomas, associated with distinct clinical, transcriptional, and epigenomic signatures. Though earlier investigations have identified molecular markers of meningioma aggressiveness, the relationship of subgroup with clinical outcome has not been comprehensively studied. Genomic findings now guide routine clinical management of gliomas and other brain tumors; however, meningiomas continue to rely on histologic markers that correlate poorly with underlying biology. Our results indicate divergent clinical courses among meningioma molecular subgroups, particularly within WHO grade I meningiomas. We identify the HH subgroup as an independent marker for meningioma aggressiveness, and find that PI3K tumors exhibit decreased time to recurrence. While we confirm the value of traditional markers of meningioma outcome, such as grade, extent of resection, and previous recurrence, we show that consideration of the molecular subgroup offers unique insights into the expected clinical course, and may explain the heterogeneity in outcome within these tumors.
Meningiomas are common central nervous system tumors that are primarily treated with surgical excision and, at times, radiation. The optimal use of radiation remains an area of intense study, as this treatment may itself lead to transformation of otherwise benign cells, as well as additional morbidities.1 Though there are ongoing clinical trials, there is currently no effective chemotherapy. Historically, clinicians have relied on histological features for classification into 3 pathological grades of increasing malignant potential. High-grade lesions (World Health Organization [WHO] grades II and III) represent approximately 20% of all cases and exhibit elevated recurrence rates and overall poor outcome.2 Given that the vast majority of lesions are WHO grade I, meningiomas are widely considered to be “benign” tumors. However, a subset of these low-grade meningiomas can have a more aggressive clinical course and, in certain instances, behave similar to higher-grade lesions.
Genomic studies have identified discrete meningioma subgroups, and further transcriptional and epigenomic characterization of these groups has revealed unique oncogenic processes associated with each.3–6 Biallelic loss of the neurofibromatosis 2 (NF2) gene, which is found in ~50% of all meningiomas, occasionally occurs with recurrent mutations in SWItch/sucrose nonfermentable–SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily B1 (SMARCB1). Somatic mutations in tumor necrosis factor receptor–associated factor 7 (TRAF7) are identified in half of the non-NF2 mutant meningiomas, and often co-occur either with activating mutations affecting the phosphatidylinositol-3 kinase (PI3K) pathway or with a recurrent Krüppel-like factor 4K409Q (KLF4K409Q) mutation. Together with other subgroups, characterized by Hedgehog (HH) activation and recurrent mutations in RNA polymerase II subunit A (POLR2A), these groups collectively account for more than 80% of all meningiomas. Large cohort analyses have identified statistical relationships between these subgroups and numerous clinical variables, including pathological grade, histology, and tumor location.7 However, associations between genomic subgroup and clinical outcome (including recurrence) have not been comprehensively investigated.
As our understanding of meningioma pathogenesis has advanced, interest has grown in elucidating the optimal features associated with long-term prognosis. The WHO grading paradigm was designed to help predict clinical course and currently serves as the primary metric that directs postoperative management, including imaging surveillance and need for adjuvant radiation. However, recent epigenomic studies indicate that tumors robustly cluster according to underlying genomic features, and not the histological markers that are widely utilized for stratification.4,8 This suggests that genomic drivers could activate distinct oncogenic processes, and consequently exhibit unique clinical courses and therapeutic susceptibilities. Indeed, recent changes to the WHO criteria for CNS tumor classification include molecular considerations for gliomas, ependymomas, and other common tumors, which has resulted in improved prognostic accuracy and clinical management.
In this study, we investigated associations between meningioma molecular subgroup and clinical outcome, with the goal of understanding differences in recurrence and optimal management of each subgroup. Leveraging a large cohort of previously characterized samples, we identified relationships with short- and long-term recurrence rates, identifying important prognostic associations particularly among WHO grade I lesions. Our findings demonstrate that individual subgroups exhibit distinct clinical courses, and thus may benefit from more tailored postoperative management strategies. Accordingly, we propose that the identification of the underlying molecular subgroup will enhance prognostic accuracy, and should be used routinely in meningioma management to improve outcome in these patients.
Methods
Genomic Characterization
This study’s methods were approved by Yale University’s institutional review board, which included ethical considerations. Informed consent was obtained from all study participants. Meningioma specimens were screened for driver mutations using targeted and/or whole-exome sequencing approaches that included the coding regions of NF2, TRAF7, SMARCB1, PIK3CA, PIK3R1, PRKAR1A, SMO, and SUFU, as well as the recurrent mutation sites POLR2AQ403K, L438_H439del, AKT1E17K, and KLF4K409Q. The molecular results and clinical features of this screen have been previously reported.3,4,7–9 Though other candidate driver mutations have been reported in meningioma (eg, BAP1, SMARCE1), the overall prevalence of these alterations is low, and they were not included in our analysis.7 In addition to this patient cohort, we also included clinical and outcomes data from 51 publicly reported meningiomas associated with the subgroups described below.10
Sample Selection and Subgroup Classification
Based on genomic results, all meningiomas were classified into one of 7 molecular subgroups, including “NF2,” “PI3K,” “KLF4,” “TRAF7,” “Hedgehog,” “SMARCB1,” and “POLR2A,” as previously defined.3,4 Meningiomas in the TRAF7 subgroup included all cases that harbored mutations in this gene but lacked reported driver mutations in the PI3K pathway or KLF4 based on targeted and/or whole-exome sequencing. It is important to point out that these meningiomas might harbor molecular alterations that activate the PI3K pathway, which were not detected based on our sequencing approaches. Of 22 samples in our cohort with recurrent SMARCB1 mutations, we identified a damaging coding variant in the gene NF2 in 13 cases; however, previous transcriptional and epigenetic studies suggest that these tumors form a distinct molecular subgroup.4 We did not include “mutation unknown” meningiomas, as it remains unclear if these lesions might be better classified by an existing subgroup. Samples in which clinical outcome data were not available were removed, as were cases known to have undergone radiation prior to resection. As a result of these filters, all cases known to have received chemotherapy prior to resection or recurrence were removed. Thus, the final analysis was performed using data from 469 meningiomas.
Clinical Data Acquisition
Data were collected by retrospective chart review. The preoperative variables used for analysis included tumor location, patient sex, and prior history of resection. The postoperative variables included extent of resection (EOR), WHO grade, histology, Ki-67 index, duration of radiographic follow-up, presence and timing of recurrence, and adjuvant therapies. Recurrence was defined as interval growth after resection based on surveillance imaging. Grade was classified as either low-grade (WHO I) or high-grade (WHO II/III). Location was defined as midline (M) or non-midline (NM), and skull base (SB) or non-skull base (NSB). Additionally, we further classified skull base samples as anterior fossa (AF), middle fossa (MF), or posterior fossa (PF), while non-skull base samples were further classified as anterior (ANT), posterior (POST), tentorial (TENT), or ventricular (VENT). Histologies that underwent analysis included the most common ones: meningothelial, secretory, fibrous, atypical, and transitional, with all other known histologies being classified as Other. We did not consider atypical features among grade I meningiomas in our analysis. EOR was defined as either gross total resection (GTR), which included Simpson grades I, II, and III, or subtotal resection (STR), which included Simpson grades IV and V. The EOR was determined based on imaging or, if unavailable, based on operative report. Ki-67 index was defined as either high (≥5%) or low (<5%), as this threshold has previously been shown to correlate with outcome.11 In the event that a sample lacked data for a particular variable, or it could not be classified according to the listed parameters, that sample was excluded from analyses involving the variable.
Statistical Analyses
Relationships between genomic subgroup and clinical features were assessed to confirm similarity of our cohort to previously reported datasets. For individual variables, chi-square tests were performed to identify significant relationships, with correction based on the number of conditions within each variable as noted. In some instances (as noted), specific subgroup/variable relationships were further interrogated via Fisher’s exact tests, with appropriate P-value correction based on the number of tests. Univariate associations with recurrence were assessed at 2 years, as previous reports indicate that this duration encompasses most cases of recurrent symptoms and/or progression.12 Similar to other variables, significant relationships between recurrence and clinical or molecular features were determined using Fisher’s exact tests, with correction based on the number of tests performed for a given variable. Subsequent univariate analyses of recurrence were performed after stratification by subgroup. As the intention of these analyses was to understand the covariate effects of subgroup on various features, we included recurrences at any time point to maximize availability of data. We performed one-way ANOVA tests for each feature to ensure no relationship existed between follow-up duration and either feature values or subgroup.
We calculated time to recurrence (TTR) as the elapsed time between tumor resection and radiographic discovery of recurrence, and comparisons between each subgroup and the remaining samples were performed using Student’s t-test. The analysis was repeated after stratification by EOR. Relationships between TTR and grade, EOR, location, and Ki-67 were also tested.
Multivariate determinants of clinical features were assessed by logistic regression using the generalized linear model. We included grade, location, sex, and molecular subgroup as covariates. Samples lacking data in one or more of these variables were excluded from this analysis.
Outcome Analyses
To understand the genomic and clinical factors associated with progression-free survival (PFS), we performed Kaplan–Meier analyses. Patients with recurrence-free imaging at the assessed period of study (2 and 5 y) or greater were censored. Significant differences of recurrence between the emerging more aggressive (NF2, PI3K, HH, TRAF7) and quiescent (SMARCB1, POLR2A, KLF4) subgroups were assessed using the log-rank test. We performed univariate regression analyses on available clinical data using Cox proportional hazards models, with each variable assessed individually as the dependent covariate. All nominal variables were converted into binary dummy variables for analysis. From these results, we selected non-subgroup variables for Cox multivariate analysis based on availability of data (requiring values in at least 80% of samples) and significance in univariate modeling (P < 0.05). Samples were filtered to include only those with complete data for the included variables (n = 383). Each genomic subgroup (represented as a binary variable) was sequentially used in a side-model to determine its association with recurrence as an independent factor among other covariates.
Results
Association of Subgroup with Clinical and Therapeutic Variables
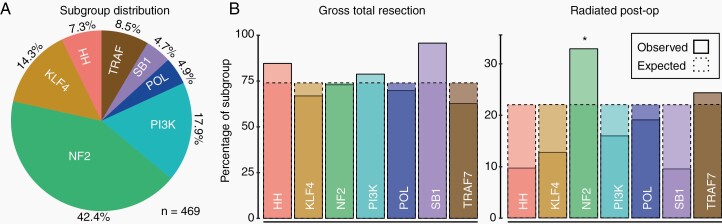
Based on availability of clinical outcomes data, a total of 469 meningiomas were used for the final analysis. The demographics of our cohort were consistent with previously reported large studies (Supplementary Table 1), and the distribution of subgroups among our samples was comparable to earlier cohorts,5–7,13,14 though with a disproportionate number of non-NF2 cases included (Figure 1A). As expected and previously reported, univariate analysis found that genomic subgroup was significantly associated with grade, location, histology, Ki-67 index, and sex7 (Supplementary Table 2). Similarly, further assessment with logistic regression indicated that subgroup was an independent predictor of skull base and midline location, cranial fossa location, histology, and Ki-67, when simultaneously considered with grade, sex, and/or location (Supplementary Table 3).
Fig. 1.
Subgroup relationships with clinical and therapeutic variables. (A) The distribution of genomic subgroups in our cohort is similar to previous reports, but with enrichment of non-NF2 cases. SB1: SMARCB1, POL: POLR2A, HH: Hedgehog, TRAF: TRAF7. (B) Plots of expected vs observed percentages for gross total resection (left) and use of postoperative radiation (right). significance was assessed using sample counts (not percentages).
With regard to surgical resection, we observed an overall GTR rate of 73.8%, which did not differ by grade (adj. P = 0.13). Several variables were found to be associated with GTR during univariate analysis, including non–skull base location (80.4%, vs 62.0% in SB; adj. P = 1.2 × 10−4), fibrous histology (92.1%, vs 72.8% in non-fibrous; adj. P = 0.045), and as expected, use of postoperative radiation (43.4%, vs 76.7% in non-radiated; adj. P = 2.9 × 10−8). EOR was found to be marginally associated with recurrence at 2 years after correction, with GTR and STR cases recurring 9.0% and 17.7% of the time, respectively (adj. P = 0.045). When we limited our analysis to low-grade meningiomas, the relationship was stronger, with GTR and STR cases recurring 4.4% and 17.5% of the time, respectively (adj. P = 4.5 × 10−3). Possibly due to insufficient power, we did not observe a difference in recurrence rate among the 54 high-grade meningiomas that underwent analysis. When investigating EOR and subgroup, a borderline association was found specifically among low-grade samples, which was driven by complete GTR rate among SMARCB1 mutant cases (18 of 18; adj. P = 0.056; Figure 1B). This relationship may be due to the enrichment of SMARCB1 meningiomas in parasagittal regions, as that location could render them more amenable to GTR.
Regarding postoperative management, we identified a significant relationship between molecular subgroup and use of radiation (adj. P = 4.3 × 10−3), with NF2 cases more commonly receiving this treatment after surgery (32.9%, vs 15.4% in non-NF2; adj. P = 6.5 × 10−4) (Figure 1B). However, this relationship was not significant when only low-grade samples were considered (18.6% vs 14.6% in non-NF2; adj. P = 1.00), suggesting that this effect is likely due to grade. Associations between subgroup and therapeutic variables are further summarized in Supplementary Table 4.
Univariate Relationships with Meningioma Recurrence
The average length of surveillance was 54.2 months, which was not significantly different among subgroups (adj. P = 0.22). The overall rate of recurrence at 2 years was 12.0%, including 7.4% of low-grade and 33.3% of high-grade lesions (adj. P = 7.3 × 10−7). Among selected histologies, we found a particularly high recurrence rate among atypical meningiomas (27.0%, 10/37; adj. P = 4.6 × 10−3), which are high-grade by definition. By contrast, there were no recurrences of transitional meningiomas at 2 years (0.0%; 0/46; adj. P = 0.057), though with inclusion of later time points, the overall rate was 7.6% (4/53). Further description of associations between 2-year recurrence and these features is provided in Supplementary Table 5.
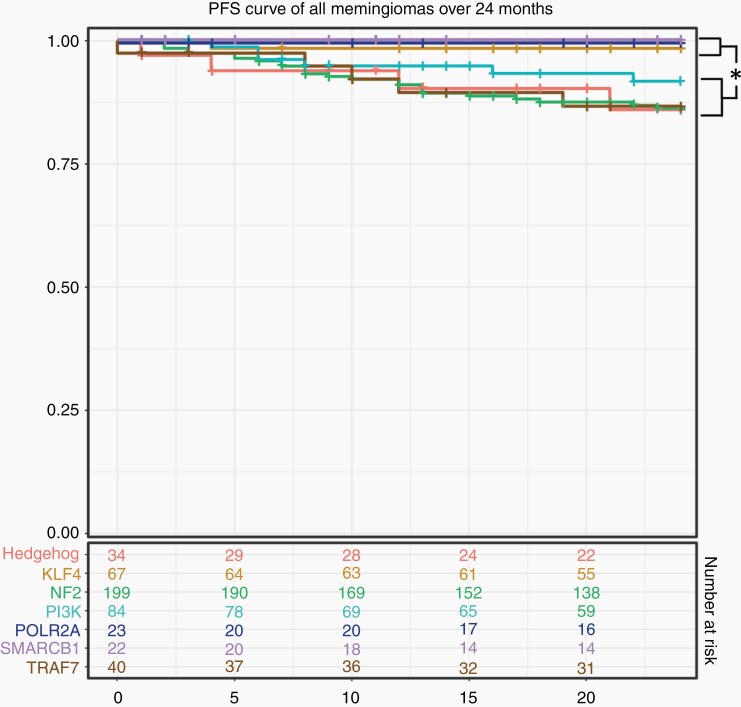
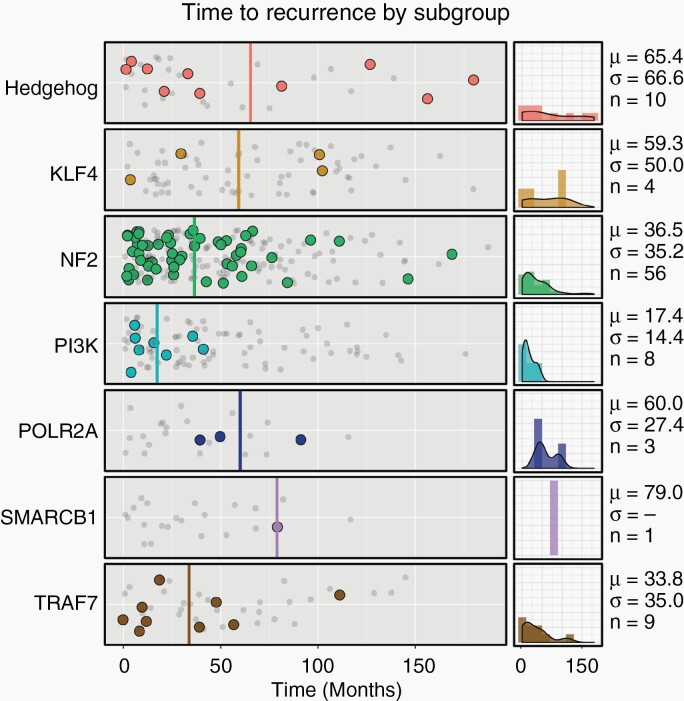
Four subgroups had appreciable rates of uncensored recurrence at 2 years, including HH (17.4%, 4/23), NF2 (16.8%, 26/155), PI3K (9.5%, 6/63), and TRAF7 (14.7%, 5/34), while among the remaining subgroups (KLF4, POLR2A, SMARCB1) (n = 75), only a single recurrence was observed. The former group (HH/NF2/PI3K/TRAF7) thus recurred at a rate 21.9 times higher than the latter during this time period (17.2 times higher when considering only low-grade samples), leading to a significant difference in survival curves (Figure 2; adj. P = 8.7 × 10−4). At 60 months, HH, NF2, and TRAF7 meningiomas continued to exhibit elevated rates of recurrence (35.3%, 43.7%, and 36.4%, respectively). By contrast, most PI3K tumor recurrences occurred within the first 24 months (75.0%; 6 of 8), resulting in an intermediate rate at later time points (Supplementary Table 6). Indeed, the average TTR of PI3K tumors was 17.4 months, which differed significantly from an average of 40.0 months among all non-PI3K lesions (adj. P = 7.5 × 10−3; Figure 3). The significant relationship of TTR and PI3K persisted when only GTR and only low-grade samples were considered (adj. P = 0.037 and 0.042, respectively), and we did not observe an independent relationship between TTR and grade (adj. P = 0.19), EOR (adj. P = 0.51), Ki-67 index (adj. P = 0.32), or skull base location (adj. P = 0.70). Thus, activating mutations in the PI3K pathway is a predictor of early TTR, independent of other prognostic clinical variables.
Fig. 2.
Progression free survival by genomic subgroup. Recurrence rates of HH, NF2, TRAF7, and PI3K meningiomas were elevated relative to other subgroups. Survival curves show significant deviation of these subgroups from others (log-rank P-value = 8.7 × 10−4).
Fig. 3.
Timing of meningioma recurrence. Shown is the time to recurrence (solid dots) or last recurrence-free scan (gray, decreased opacity) for each subgroup in months. The average time to recurrence of PI3K meningiomas was significantly less than other tumors (P = 7.5 × 10−3). Averages for each subgroup are shown in solid lines. Density plots of each subgroup’s recurrences are shown on the right, along with the mean (μ), standard deviation (σ), and number of recurrences (n).
At later time points, we observed several recurrences among POLR2A meningiomas (n = 3; range 39 to 91 mo), suggesting that these tumors tend to recur relatively later. Though noticeably higher than the NF2 and PI3K subgroups (with a mean of 60.0 mo), the TTR for POLR2A did not differ significantly overall from other subgroups, likely owing to the limited number of POLR2A samples and wide distribution of recurrence timing in other subgroups. Collectively, these results suggest that PI3K tumors tend to recur earlier, POLR2A recur later, and HH, NF2, and TRAF7 meningiomas have steady rates of increased recurrence over time.
The PI3K and HH subgroups are associated with multiple distinct driver genes, and it is possible that events in specific genes may stratify prognosis of these groups. The most common variants in the PI3K subgroup include recurrent mutations in AKTE17K, PIK3CAE545K, and PIK3CAH1047R, and we observed a similar rate of recurrence between AKT1 (8.8%, 5/57) and PIK3CA (11.1%, 3/27) mutants in our cohort (P = 0.71). In HH meningiomas, the most frequently identified events include oncogenic SMOL412F or SMOW535L mutations, or biallelic loss of SUFU. Though 66% of SUFU meningiomas recurred (2/3; vs 23.1% of SMO mutants, 6/26), we were underpowered to establish a statistical relationship in this subgroup (P = 0.18).
Previous studies have reported relationships with other genomic features and outcome, including deletions or loss of heterozygosity of chromosome 1p (chr1p_Loss), as well as mutations in the promoter of the gene telomerase reverse transcriptase (TERT) in samples undergoing malignant progression.8,15–19 Among meningiomas with DNA copy number information available (n = 123), we identified chr1p_Loss in 23.6% of cases. This feature was almost entirely limited to samples in the NF2 subgroup (82.8%; adj. P = 2.6 × 10−4) and trended toward enrichment in higher grade (43.8%, vs 20.8% in low-grade cases; adj. P = 0.077). Among our samples, we did not observe a convincing relationship of chr1p_Loss with elevated recurrence (17.2%, vs 9.6% recurrence among chromosome 1p wild-type; adj. P = 0.33), and this feature did not stratify outcome when only NF2 mutant cases were considered (16.7%, vs 11.8% recurrence among chromosome 1p wild-type; adj. P = 0.72). We identified TERT promoter mutations in 10 out of 338 screened samples in our larger cohort (including meningiomas outside of the included genomic subgroups). All of these cases were high-grade (7 WHO grade II and 3 grade III), and among those with outcomes data available (n = 2), both recurred.
Covariates Associated with Subgroup and Recurrence
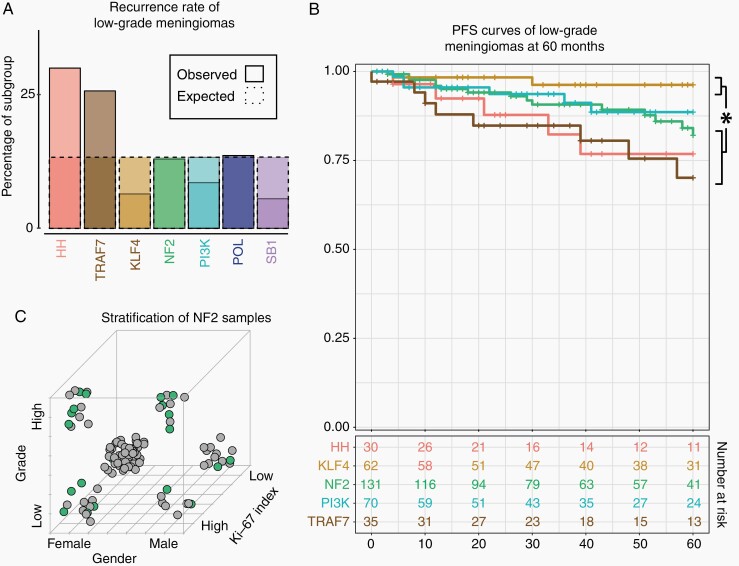
Postoperative management of low-grade meningiomas can be clinically challenging, as it is difficult to predict the minority of these cases that will ultimately exhibit poor outcome. To understand patterns associated with their recurrence, we performed additional analyses specific to low-grade samples. In addition to lesions that underwent STR (discussed above), we found elevated recurrence among the HH and TRAF7 samples (30.0% and 25.7% respectively; Figure 4A; adj. P = 0.01). We did not identify relationships with recurrence between location or histology among low-grade meningiomas, nor among other subgroups (including NF2). Survival analysis over 60 months indicated a difference in PFS curves of the KLF4 subgroup with NF2 (log-rank P = 0.035), HH (log-rank P = 0.011) and TRAF7 (log-rank P = 1.7 × 10–3) samples, with the PI3K subgroup exhibiting intermediate survival (Figure 4B).
Fig. 4.
Effect of subgroup on recurrence rates stratified by covariates. (A) Samples classified as HH or TRAF7 were more likely to recur relative to other subgroups (adj. P = 0.01). (B) Among low-grade samples, subgroups exhibited distinct PFS curves, with KLF4 meningiomas exhibiting better prognosis than NF2 (log-rank P = 0.035), TRAF7 (log-rank P = 1.7 × 10−3) and HH (log-rank P = 0.016) meningiomas. Subgroups with > 10 samples available for analysis at 60 months are included. (C) Among NF2 mutant samples, recurrence was enriched among High-Grade, High Ki-67, and Male patients. Samples falling within each corner of the displayed cube are associated with the corresponding values for these features. Solid green circles indicate recurrences, while gray circles indicate samples that did not recur.
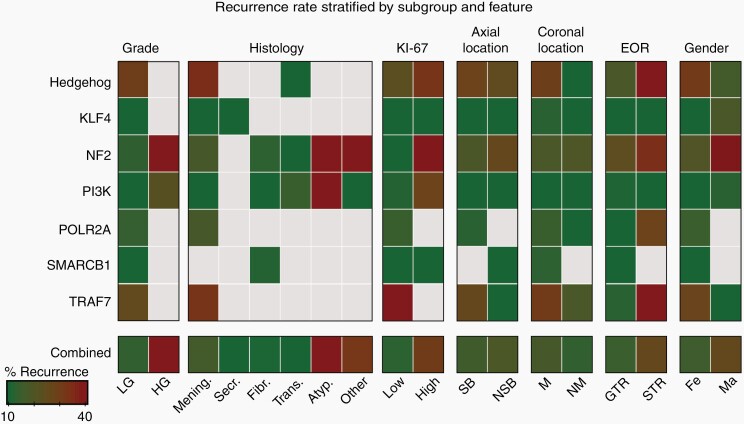
Further analysis revealed additional variables that stratified outcome among various subgroups. Recurrence among NF2 samples, for instance, was associated with features traditionally associated with meningioma aggressiveness, including high-grade (60.7%, vs 13.0% in low-grade; adj. P = 8.7 × 10–11), high Ki-67 (39.5%, vs 7.8% in Low Ki-67; adj. P = 1.4 × 10−4), male (39.0% vs 20.2% in female; adj. P = 0.022) and atypical (60.0% vs 20.6% in non-atypical; adj. P = 1.5 × 10−4) samples (Figure 4C). Transitional histology was associated with a low rate in NF2 meningiomas (9.1%, vs 36.7% in non-transitional; adj. P = 0.011). Among all cases, recurrence in HH meningiomas showed a borderline association with EOR (60.0% vs 18.5% in STR and GTR samples respectively, adj. P = 0.085), though we were underpowered to conclusively detect a relationship (Figure 5). A previous study noted elevated recurrence of olfactory groove meningiomas in the anterior skull base, a location enriched for HH-activated samples.20 Among our meningiomas in this region (n = 35; including 34.3% HH), we indeed found that most recurrences occurred among HH-mutant samples (57.1%; 4 of 7).
Fig. 5.
Stratification of recurrence rate by subgroup and feature. Shown is a heatmap of recurrence rates stratified by subgroup and feature. Boxes that represent less than 4 samples are grayed out. LG: Low-Grade, HG: High-Grade, Mening: Meningothelial, Secr: Secretory, Fibr: Fibrous, Trans: Transitional, Atyp: Atypical, SB: Skull Base, NSB: Non-Skull Base, M: Midline, NM: Non-Midline, GTR: Gross Total Resection, STR: Sub-Total Resection, Fe: Female, Ma: Male.
Association of Subgroup with Progression-Free Survival
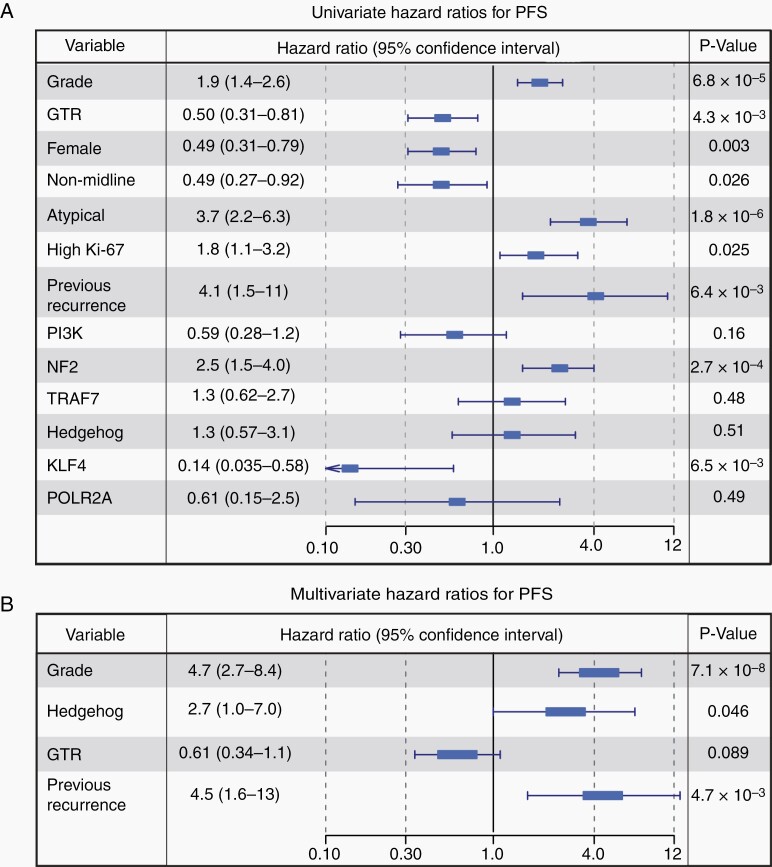
To understand the effects of genomic subgroup and other covariates on long-term outcome, we performed regression analyses using Cox proportional hazards modeling. Univariate analysis was conducted on all available data fields, and statistical associations were identified with grade (P = 6.8 × 10−5), GTR (P = 64.3 × 10−3), female sex (P = 3.0 × 10−3), NM location (P = 0.026), atypical histology (P = 1.8 × 10−6), high Ki-67 (P = 0.025), and previous recurrence (P = 6.4 × 10−3). Among subgroups, NF2 meningiomas were negatively associated with PFS (P = 2.7 × 10−4), while KLF4 meningiomas were positively associated (P = 6.5 × 10−3) (Figure 6A). We selected factors for multivariate regression using univariate results and availability of data (see Methods). This analysis identified clinical grade (P = 7.1 × 10−8) and previous recurrence (P = 4.7 × 10−3) as independent significant covariates (Figure 6B). Among genomic subgroups, we found that NF2 samples were not observed to be an independent factor associated with PFS (P = 0.30), consistent with dependence of this factor on grade and other associated aggressive features. By contrast, a marginal association was identified in the HH subgroup (P = 0.046), while KLF4 mutations were found to be a protective variable (P = 0.045).
Fig. 6.
Proportional hazards modeling. (A) Univariate proportional hazards modeling identified low-grade , GTR, female sex, NM location, non-atypical histology, low Ki-67, and primary lesions as protective factors. Among subgroups, KLF4 meningiomas were positively associated with PFS, while NF2 lesions were negatively associated. The 95% confidence interval of the hazard ratio is labeled. Clinical features that were not significant are omitted. (B) Further analysis with Cox multivariate regression identified grade, previous recurrence, and Hedgehog subgroup as significant independent factors associated with recurrence.
Discussion
While it is well established that WHO grades II and III meningiomas have aggressive clinical courses, poor outcomes may also occur in a subset of low-grade lesions for less clear reasons. In this report, we investigated the role of genomic drivers in PFS and other clinical-outcome related variables, as this feature defines transcriptional and epigenetic subgroups of meningioma, and thus may represent distinct pathways of disease. Using a large cohort of well-annotated samples, we identified unique clinical courses associated with mutually exclusive molecular subgroups in meningioma, thus providing unambiguous genomic criteria to identify patients who may benefit from closer follow-up or potentially earlier adjuvant treatments following surgical resection, such as radiation. Indeed, our study suggests genomic factors may explain why some WHO grade I meningiomas clinically behave more aggressive than others, particularly with regards to recurrence, and provides cautious insight into classifying some low-grade meningiomas as more aggressive or quiescent.
Samples in the KLF4 and SMARCB1 subgroups rarely recurred, suggesting that avoidance of more aggressive adjuvant treatment, such as radiation, could be the most reasonable approach for these meningiomas. SMARCB1 meningiomas were associated with high Ki-67 index, a marker for elevated recurrence in our cohort; however, they underwent GTR in almost all cases, which may explain their relatively improved PFS. We have previously reported recurrent SMARCB1 mutations to be an important factor in the formation of de novo high-grade meningiomas,8 as opposed to the progression of low to high grade meningiomas, which typically acquire TERT promoter mutations. Because our current cohort consisted primarily of low-grade lesions, our conclusions with regards to SMARCB1 are limited to these cases, and further study is needed in high-grade tumors. Similarly, POLR2A tumors also had a low recurrence rate at 2 years, though at later time points, several recurrences were observed. Our results thus suggest a trend for later recurrence; however, additional confirmatory studies with a larger number of POLR2A samples are needed.
By contrast, other molecular subgroups were associated with more aggressive clinical courses, and may benefit from closer follow-up and consideration of early adjuvant postoperative radiotherapy. PI3K meningiomas exhibited a significantly shorter time to recurrence than other subgroups, though overall recurrence at 5 years was moderate. Indeed, we observed minimal recurrence of PI3K meningiomas after 24 months, despite mean follow-up time of 55.9 months. Our results are consistent with a previous cohort of AKT1 mutant meningiomas that showed a trend toward decreased time to recurrence among skull-base lesions (P = 0.094).21 Tumors of this subgroup tend to localize along the sphenoid wing, and may encase critical neurovascular structures. As a result, there is often residual tumor left in place, which should be closely monitored if found to harbor PI3K activation. Upfront adjuvant radiotherapy in this subset could also be considered, but more understanding is needed.
NF2, HH, and TRAF7 meningiomas exhibited a persistently high recurrence rate relative to other subgroups, that did not taper by 5 years of surveillance. The elevated recurrence in NF2 lesions depended upon co-occurrence of several clinical features, including high-grade, male sex, and high Ki-67 index, suggesting that loss of NF2 is insufficient to drive clinical aggressiveness. Among low-grade lesions, NF2 meningiomas did not show elevated recurrence compared with other subgroups, and within this group we found that female patients with low Ki-67 index had an overall recurrence rate of only 5.8%. Recurrence of HH meningiomas, on the other hand, was independent of other covariates, with the possible exception of EOR, which trended toward significance. Indeed, we found that this was the only subgroup to be significantly associated with recurrence when grade, EOR, and previous recurrence were considered. Given that HH tumors typically arise along the anterior medial skull base and can be associated with hyperostosis, the potential benefit of aggressive drilling to decrease recurrence should be further investigated (though risk of cerebrospinal leak must be considered). While HH meningiomas make up only a fraction of cases found in this region, they accounted for a majority of recurrences, similar to a previous report.9 This suggests utility in profiling anterior skull base meningiomas for the commonly occurring SMOW535L and SMOL412F mutations, which could guide follow-up and adjuvant treatment strategies.
The typical co-occurrence of TRAF7 mutations with KLF4K409Q or PI3K activation suggests that they alone may be insufficient to drive meningioma formation. However, we identified a subset of TRAF7 mutant meningiomas that lacked alterations in previously implicated genes, which may suggest unknown mechanisms of activating PI3K or involvement of different oncogenic pathways altogether. Further molecular studies are needed to elucidate the exact genomic alterations within this TRAF7 subgroup, which clearly shows a difference in clinical prognosis and thus warrants attention.
The Simpson grading of EOR has long been used to guide clinical expectations after resection of grade I meningiomas, and indeed, our results support the relevancy of EOR in recurrence of these tumors. By contrast, univariate analysis did not suggest EOR was a significant factor in predicting recurrence in high-grade lesions. When we stratified our cohort by molecular subgroup, we found evidence suggesting that GTR may play a protective role in recurrence of HH meningiomas (adj. P = 0.085), though this result missed significance. Additionally, we observed that patients with SMARCB1 mutant meningiomas underwent GTR in almost all cases, and rarely recurred despite elevated Ki-67 index. Though relationships between EOR and recurrence of other subgroups were less convincing, it is possible that an expanded cohort may elucidate additional associations between subgroup, EOR, and recurrence.
Previous molecular studies have provided additional insights into prognostic stratification, which are complementary to the findings in this paper. The association of copy number events with outcome, for instance, has been extensively studied, with numerous reports identifying chr1p_Loss as a negative prognostic indicator.8,15,16 In our cohort, this feature was almost entirely limited to samples in the NF2 subgroup, though we were unable to demonstrate that chr1p_Loss stratifies outcome among this subgroup. Furthermore, we assessed TERT promoter mutations in a subset of samples, and our results are consistent with previous studies that associate these events with tumors undergoing malignant progression. Though we were underpowered to draw significant conclusions in our cohort of samples, previous independent studies have convincingly demonstrated the relevancy of this event for transformation.8,17,19 Using unbiased clustering approaches, multiple studies have demonstrated the prognostic value of DNA methylation.10,22–24 Though subgroup was simultaneously considered in a single study,10 the number of non-NF2 cases considered was limited. Finally, we and others have reported additional genomic, transcriptional, and epigenetic features that are associated with meningioma aggressiveness and recurrence, including DMD deletions,25,26BAP1 mutations,27 activation of the PRC2/EZH2, FOXM1, and Homeobox signaling pathways.8,26,28,29 However, we did not have sufficient samples with these confirmed alterations to be assessed in our current study.
Based on current evidence, several molecular features are thus predictive of long-term outcome of meningiomas, and in the absence of established practice guidelines, this could become a source of confusion about the use of this data in clinical management. Though rigorous evaluation through prospective studies and multi-specialty dialogue is needed to finalize a consensus recommendation, we believe the use of molecular subgroups offers advantages over other reported prognostic correlates. In a majority of cases, subgroup can be unambiguously determined using Sanger sequencing, a low-cost test that requires minimal training or bioinformatics expertise. However, this approach would not be sufficient to identify loss of tumor suppressors, such as NF2. In cases where Sanger is undiagnostic, low-coverage whole-exome sequencing is typically sufficient and can be performed at a favorable cost to epigenetic profiling. Furthermore, the mutual-exclusivity of meningioma genomic subgroups, as well as their direct relationship with gene expression and enhancer clustering, suggests that driver mutations represent unique routes to oncogenesis. The discovery of unique recurrence profiles associated with each subgroup is therefore unsurprising, as the underlying biology is distinct. Clinical genotyping could accordingly provide both prognostic and therapeutic benefits, as individual subgroups may respond to precision medications targeting specific downstream dependencies.
There are several limitations to our study, which could be addressed with future investigations. The mean follow-up among our cases was 54.2 months, and we were unable to perform rigorous recurrence analysis for some subgroups at 5 years due to data attrition. Though we did not observe any SMARCB1 recurrences among the limited data that was available at this time-point (n = 5), we did observe 2 POLR2A recurrences (of 9 with available data). It is unclear if this indicates a trend for later recurrence of these tumors or an artifact of limited sample size, but we continue to prospectively follow these patients. Additionally, as mentioned, it is possible that meningiomas in the TRAF7 subgroup might harbor molecular alterations that activate the PI3K pathway, which were not detected in our study. However, the TRAF7 and PI3K mutant tumors showed increased and earlier recurrence, respectively, suggesting that these low grade tumors act more aggressively. Further understanding will be needed to elucidate whether the PI3K pathway, or yet another contributing mechanism, is responsible. Further, our study is limited to the assessment of genomic and clinical data and does not simultaneously consider epigenetic or transcriptional factors that may play a role in outcome. However, previous reports suggest that meningioma genomic drivers segregate into distinct gene expression and enhancer clusters,4 and most non-NF2 samples are thought to fall into a single methylation subclass.10 Thus, in both cases it is unlikely that consideration of these variables as cofactors would alter our results.
In conclusion, our analysis reveals distinct clinical courses in meningioma according to genomic subgroup, suggesting utility in routine mutational profiling of these tumors to aid in postoperative management decisions. More specifically, we identified more quiescent and aggressive genomic subgroups, with the latter group experiencing recurrence at a rate 22 times higher than the former. In most cases, subgroup can be determined in a cost-effective and unambiguous manner, and this information could provide particular value in distinguishing low-grade lesions that may benefit from closer follow-up and consideration of adjuvant treatment. As several recent studies have identified convincing molecular correlates with meningioma outcomes, consensus is needed in determining the appropriate integration of these features into clinical practice. With development of an optimal stratification paradigm, an accompanying prospective clinical trial will be an essential step in leveraging these molecular discoveries to improve the care of meningioma patients.
Funding
This work was supported by the Gregory M. Kiez and Mehmet Kutman Foundation, Connecticut Brain Tumor Alliance, T32GM007205, and F30CA213666.
Conflict of interest statement.
The authors declare no conflicts of interest associated with this manuscript.
Authorship statement.
Study Design: MWY, MG and JM. Data Collection: MWY, DFM, TG, CL, DD, JDM, AZ, AS, ET, KO, FI, MP, JB, MP, TA, AH, KB, TK, MNP, NA, MK, and EZE. Data Analysis: MWY, DFM, LJ, EZE, MG, and JM. Manuscript Drafting: MWY and JM. Genomics Expertise: MG. Supervision of Clinical Correlations: JM.
Supplementary Material
References
- 1. Day SE, Halasz LM. Radiation therapy for WHO grade I meningioma. Chin Clin Oncol. 2017;6(Suppl 1):S4. [DOI] [PubMed] [Google Scholar]
- 2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 3. Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clark VE, Harmancı AS, Bai H, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48(10):1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45(3):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016;18(5):649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Youngblood MW, Duran D, Montejo JD, et al. Correlations between genomic subgroup and clinical features in a cohort of more than 3000 meningiomas [published online ahead of print October 25, 2019]. J Neurosurg. 2019:1–10. doi: 10.3171/2019.8.JNS191266 [DOI] [PubMed] [Google Scholar]
- 8. Harmancı AS, Youngblood MW, Clark VE, et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat Commun. 2017;8:14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boetto J, Bielle F, Sanson M, Peyre M, Kalamarides M. SMO mutation status defines a distinct and frequent molecular subgroup in olfactory groove meningiomas. Neuro Oncol. 2017;19(3):345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682–694. [DOI] [PubMed] [Google Scholar]
- 11. Takahashi JA, Ueba T, Hashimoto N, Nakashima Y, Katsuki N. The combination of mitotic and Ki-67 indices as a useful method for predicting short-term recurrence of meningiomas. Surg Neurol. 2004; 61(2):149–155. [DOI] [PubMed] [Google Scholar]
- 12. Gupta PK, Sastry Kolluri VR, Das S, Chandra Mouli BA, Narayana Swamy KS, Das BS. Recurrences in meningioma after surgery. Acta Neurochir (Wien). 1989;100(3-4):104–107. [DOI] [PubMed] [Google Scholar]
- 13. Strickland MR, Gill CM, Nayyar N, et al. Targeted sequencing of SMO and AKT1 in anterior skull base meningiomas. J Neurosurg. 2017;127(2):438–444. [DOI] [PubMed] [Google Scholar]
- 14. Yuzawa S, Nishihara H, Yamaguchi S, et al. Clinical impact of targeted amplicon sequencing for meningioma as a practical clinical-sequencing system. Mod Pathol. 2016;29(7):708–716. [DOI] [PubMed] [Google Scholar]
- 15. Sulman EP, Dumanski JP, White PS, et al. Identification of a consistent region of allelic loss on 1p32 in meningiomas: correlation with increased morbidity. Cancer Res. 1998;58(15):3226–3230. [PubMed] [Google Scholar]
- 16. Bello MJ, Pestaña A, Rey JA, et al. Allelic loss at 1 p is associated with tumor progression of meningiomas. Genes, Chromosomes and Cancer. 1994; 9(4):296–298. [DOI] [PubMed] [Google Scholar]
- 17. Goutagny S, Nault JC, Mallet M, Henin D, Rossi JZ, Kalamarides M. High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol. 2014;24(2):184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spiegl-Kreinecker S, Lötsch D, Neumayer K, et al. TERT promoter mutations are associated with poor prognosis and cell immortalization in meningioma. Neuro Oncol. 2018;20(12):1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sahm F, Schrimpf D, Olar A, et al. TERT promoter mutations and risk of recurrence in meningioma. J Nat Cancer Inst. 2015;108(5):djv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mirimanoff RO, Dosoretz DE, Linggood RM, Ojemann RG, Martuza RL. Meningioma: analysis of recurrence and progression following neurosurgical resection. J Neurosurg. 1985;62(1):18–24. [DOI] [PubMed] [Google Scholar]
- 21. Yesilöz Ü, Kirches E, Hartmann C, et al. Frequent AKT1E17K mutations in skull base meningiomas are associated with mTOR and ERK1/2 activation and reduced time to tumor recurrence. Neuro Oncol. 2017;19(8):1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kishida Y, Natsume A, Kondo Y, et al. Epigenetic subclassification of meningiomas based on genome-wide DNA methylation analyses. Carcinogenesis. 2012;33(2):436–441. [DOI] [PubMed] [Google Scholar]
- 23. Olar A, Wani KM, Wilson CD, et al. Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol. 2017;133(3):431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nassiri F, Mamatjan Y, Suppiah S, et al. ; International Consortium on Meningiomas . DNA methylation profiling to predict recurrence risk in meningioma: development and validation of a nomogram to optimize clinical management. Neuro Oncol. 2019;21(7):901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Juratli TA, McCabe D, Nayyar N, et al. DMD genomic deletions characterize a subset of progressive/higher-grade meningiomas with poor outcome. Acta Neuropathol. 2018;136(5):779–792. [DOI] [PubMed] [Google Scholar]
- 26. Paramasivam N, Hübschmann D, Toprak UH, et al. Mutational patterns and regulatory ne2rks in epigenetic subgroups of meningioma. Acta Neuropathol. 2019;138(2):295–308. [DOI] [PubMed] [Google Scholar]
- 27. Shankar GM, Abedalthagafi M, Vaubel RA, et al. Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro Oncol. 2017;19(4):535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Collord G, Tarpey P, Kurbatova N, et al. An integrated genomic analysis of anaplastic meningioma identifies prognostic molecular signatures. Sci Rep. 2018;8(1):13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vasudevan HN, Braunstein SE, Phillips JJ, et al. Comprehensive molecular profiling identifies FOXM1 as a key transcription factor for meningioma proliferation. Cell Rep. 2018;22(13):3672–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.