Abstract
Background
The conditional reprogramming cell culture method was developed to facilitate growth of senescence-prone normal and neoplastic epithelial cells, and involves co-culture with irradiated fibroblasts and the addition of a small molecule Rho kinase (ROCK) inhibitor. The aim of this study was to determine whether this approach would facilitate the culture of compact low-grade gliomas.
Methods
We attempted to culture 4 pilocytic astrocytomas, 2 gangliogliomas, 2 myxopapillary ependymomas, 2 anaplastic gliomas, 2 difficult-to-classify low-grade neuroepithelial tumors, a desmoplastic infantile ganglioglioma, and an anaplastic pleomorphic xanthoastrocytoma using a modified conditional reprogramming cell culture approach.
Results
Conditional reprogramming resulted in robust increases in growth for a majority of these tumors, with fibroblast conditioned media and ROCK inhibition both required. Switching cultures to standard serum containing media, or serum-free neurosphere conditions, with or without ROCK inhibition, resulted in decreased proliferation and induction of senescence markers. Rho kinase inhibition and conditioned media both promoted Akt and Erk1/2 activation. Several cultures, including one derived from a NF1-associated pilocytic astrocytoma (JHH-NF1-PA1) and one from a BRAF p.V600E mutant anaplastic pleomorphic xanthoastrocytoma (JHH-PXA1), exhibited growth sufficient for preclinical testing in vitro. In addition, JHH-NF1-PA1 cells survived and migrated in larval zebrafish orthotopic xenografts, while JHH-PXA1 formed orthotopic xenografts in mice histopathologically similar to the tumor from which it was derived.
Conclusions
These studies highlight the potential for the conditional reprogramming cell culture method to promote the growth of glial and glioneuronal tumors in vitro, in some cases enabling the establishment of long-term culture and in vivo models.
Keywords: BRAFV600E, conditional reprogramming, low-grade glioma, NF1, enescence
Key Points.
Conditional reprogramming cell methods facilitate the culture of compact gliomas.
Preclinical testing was performed on new pilocytic astrocytoma and anaplastic pleomorphic xanthoastrocytoma models.
Importance of the Study.
Cell culture and xenograft models of pilocytic astrocytoma and other low-grade glial and glioneuronal tumors have been difficult to generate, but are critical for research into tumor biology and new treatments. Many of these tumors show relatively indolent growth, at least in part due to oncogene-induced senescence, which makes them difficult to study. Our study suggests that a modified conditional reprogramming cell technique using conditioned media from irradiated fibroblasts facilitates modeling of these tumors and will enable preclinical testing of potential therapies.
Many brain tumors, including low-grade gliomas and glioneuronal tumors in pediatric patients, as well as infiltrating isocitrate dehydrogenase (IDH) 1/2 mutant gliomas in adults are hard to model in the laboratory. Sanden et al1 attempted to establish cultures from 2 pediatric pilocytic astrocytoma (PA) patient samples, but cells could not be passaged for more than 3 generations. Kogiso et al2 injected 21 pediatric PA into the cerebellum of athymic nude mice, but no tumor xenografts formed. They also cultured 7 of these samples but failed to generate a cell line. Recently a new model was reported in which growth of a PA was maintained in culture, but required the introduction of SV40 large T antigen.3
One potential mechanism underlying difficulties in culturing and modeling pediatric low-grade glioma (pLGG) is oncogene-induced senescence. The majority of pLGGs harbor an alteration of the BRAF gene.4-6 Alterations in the BRAF locus which constitutively activate the kinase in human melanocytes induce senescence-associated growth arrest.7 Similar mechanisms limit the growth of both primary pLGG samples and models of these tumors.8-10 This may be due in part to a senescence-associated secretory phenotype.11 Another major driver of pLGG formation, loss of NF1, has also been associated with induction of senescence.12
Recently, a method resulting in conditionally reprogrammed cells (CRCs) was developed to facilitate growth of senescence-prone primary epithelia. Cells were co-cultured with irradiated 3T3 fibroblasts in the presence of Rho kinase (ROCK) inhibitor Y-27632. These conditions led to reversible blockage of senescence and increased proliferation.13 This method was subsequently applied to cultures of normal and neoplastic cells from human biospecimens, patient-derived tumor xenografts from outside the CNS, and murine tissues.14-18 Conditionally reprogrammed cells are reported to maintain signature genetic changes in a stable fashion, and provide a unique tool for preclinical research.17,19,20 Here, we report that CRC conditions facilitate the growth of pLGG and other circumscribed glial and glioneuronal tumors.
Materials and Methods
Tumor Dissociation and Cell Culturing
Human tumor specimens were collected at Johns Hopkins Hospital with local Institutional Review Board approval, and written informed consent was obtained from patients or their parents. Tissues were minced and digested with either papain dissociation system (Worthington, Lakewood, New Jersey) or TrypLE Express enzyme (ThermoFisher Scientific, Grand Island, New York), then filtered through a 70-μm Falcon cell strainer (ThermoFisher Scientific). Three different conditions were used to culture primary cells: Dulbecco’s modified Eagle’s medium (DMEM)/F-12 Nutrient Mixture (Ham) medium (1:1) supplemented with 10% heat-inactivated fetal bovine serum (ThermoFisher Scientific), designated FBS; FBS medium containing the 5 μM Y-27632 (Selleckchem, Houston, Texas) ROCK inhibitor, designated FBS + Y; and CRC medium. For CRC conditions, irradiated (30 Gy) 3T3 fibroblasts were cultured in F medium (3:1 [vol/vol] DMEM-F12, 10% heat-inactivated FBS, and 5 μg/mL insulin [ThermoFisher Scientific]), condition media were collected 48–72 hours later. CRC media were prepared fresh with F medium/conditioned medium (1:1 vol/vol) supplemented with 25 ng/mL hydrocortisone (MilliporeSigma, Burlington, Massachusetts), 8.4 ng/mL cholera toxin (MilliporeSigma), 10 ng/mL human recombinant epidermal growth factor (EGF, PeproTech, Rocky Hill, New Jersey), 5 μM Y-27632 (Selleckchem), 10 µg/mL gentamicin (ThermoFisher Scientific), 250 ng/mL Fungizone (ThermoFisher Scientific), and 100 µg/mL Penicillin–Streptomycin (Quality Biological, Gaithersburg, Maryland).
Pediatric glioma cell lines Res186 and Res259 provided by Dr. Chris Jones (Institute of Cancer Research, Sutton, UK) were maintained in DMEM/F12 Ham medium supplemented with 10% heat-inactivated FBS.21 DKFZ-BT66 cells provided by Dr. Till Milde (German Cancer Research Center, Heidelberg, Germany) were maintained in astrocyte growth medium (Lonza) with 1% doxycycline.3 All cell lines were verified to be Mycoplasma free by periodic PCR testing and subjected to routine cell line identity testing
The CellTiter-Blue cell viability assay kit (Promega, Madison, Wisconsin) was used to count viable cells, with cells seeded in 96-well plates at a density of 1000 cells per well. For drug treatments, cells in 96-well plates were cultured with different doses of Vinblastine, Carboplatin, or MEK162/binimetinib (Selleckchem). Vehicle (Dimethyl sulfoxide)-treated cells were used as controls and the cell survival fraction was calculated as percentage of control cells.
Apoptosis assays were performed using Muse Annexin V & Dead Cell reagent (MilliporeSigma) and a Muse flow cytometer (Millipore). Cell cycle analysis and Bromodeoxyuridine (BrdU) incorporation assays also utilized the Muse.
β-Galactosidase staining for senescence was performed according to the manufacturer’s instructions (#9680, Cell Signaling Technology).
PCR and Sequencing
For targeted sequencing studies, genomic DNA was isolated using the DNeasy Blood and Tissue Kit (QIAGEN, Germantown, Maryland) according to manufacturer’s instructions. For IDH1 sequencing, PCR product was purified using MinElute PCR Purification Kit (QIAGEN). The primers used for PCR amplification and sequencing were: forward 5′-AATGAGCTCTATATGCCATCACTG- 3′, reverse 5′-TTCATACCTTGCTTAATGGGTGT-3′.22 Sanger sequencing for IDH1 was performed at the Johns Hopkins Genetic Resources Core Facility. Polymerase chain reaction to test for BRAF p.V600E was performed as described by Huang et al.23
Next-generation sequencing (NGS) analysis of both surgical specimens and DNA extracted from CRC cultures was performed in the Johns Hopkins University Department of Pathology molecular diagnostics laboratory using standard clinical protocols. In brief, DNA was captured with Kapa Roche reagents and Integrated DNA technology probes, and sequenced using Illumina paired end technology. Analysis was performed using the human reference sequence genome assembly hg19 (NCBI build GRCh37) and an in-house variant caller (MDL VC 8.0) as well as Haplotyper Genome Analysis TK-3.3. Variants underwent further filtering with in-house algorithms and annotation using the COSMIC database v91, dbSNP v150, and Annovar (7042018) to confirm mutation status.
Quantitative Real-time PCR
Total RNA was isolated from cultured cells using RNeasy mini kit (QIAGEN), and cDNAs were produced using QuantiTect reverse transcription kit (QIAGEN). qRT-PCR was performed using PowerUp SYBR Green Master Mix (ThermoFisher Scientific). Primer sequences were NF1: forward 5′‐GTGGAATGGGTCCAGGC-3′, reverse 5′‐GACATTCCTTGTTG-3′; CDKN1B forward 5′‐ AAGAAGCCTGGCCTCAGAAG-3′, reverse 5′‐TTCATCAAGCAGTGATGTATCTGA-3′; HPRT1 forward 5′‐GTTATGGCGACCCGCAG-3′, reverse 5′‐ACCCTTTCCAAATCCTCAGC-3′. HPRT1 was used as the endogenous control. The relative fold changing was calculated based on the formula R = 2− (ΔCt sample − ΔCt control).
Immunofluorescence Staining
Cells were cultured on glass coverslips in 24-well plates then fixed with 4% paraformaldehyde for 15 minutes at room temperature (for glial fibrillary acidic protein [GFAP], Nestin, smooth muscle actin [SMA], and Vimentin) or 100% ice-cold methanol for 15 minutes at 4°C (for NG2, CD68, and α-tubulin). Primary antibodies used for immunofluorescence staining were: Nestin (#MAB5326, 1:1000, MilliporeSigma), GFAP (#3670, 1:300, Cell Signaling Technology, Danvers, Massachusetts), Vimentin (#5741, 1:100, Cell Signaling Technology), NG2 (14-6504-82, 1:50, eBioscience, San Diego, California), CD68 (#76437, 1:1000, Cell Signaling Technology), and α-tubulin (#3873, 1:1000, Cell Signaling Technology). Secondary antibodies used for immunofluorescence staining were: Fluorescein (FITC) AffiniPure Goat Anti-mouse IgG (115-095-205, 1:300, Jackson ImmunoResearch Laboratories, West Grove, Pennsylvania), Cy3 AffiniPure Donkey Anti-mouse IgG (H+L) (715-165-151, 1:500, Jackson ImmunoResearch Laboratories), and Cy3 AffiniPure Donkey Anti-rabbit IgG (H+L) (711-095-152, 1:500, Jackson ImmunoResearch Laboratories).
Western Blotting
Cells were lysed in RIPA lysis buffer supplemented with protease inhibitors (MilliporeSigma). Primary antibodies used for western blots were: p16 (sc-56330, 1:200, Santa Cruz), p21 (#2947, 1:1000, Cell Signaling Technology), p27 (sc-1641, 1:500, Santa Cruz Biotechnology), p53 (#9282, 1:1000, Cell Signaling Technology), p4E-BP1(Thr37/46) (#2855, 1:1000, Cell Signaling Technology), 4E-BP1 (#9644, 1:1000, Cell Signaling Technology), p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (#4370, 1:1000, Cell Signaling Technology), p42/44 (#9102, 1:1000, Cell Signaling Technology), pAKT(Ser473) (#4060, 1:1000, Cell Signaling Technology), Akt (#4691, 1:1000, Cell Signaling Technology), ATRX (sc-55584, 1:500, Santa Cruz Biotechnology), H3K27M (31-11175-00, 1:10000, RevMab Biosciences), PARP (#9542, 1:1000, Cell Signaling Technology), GFAP (# 12389, 1:500, Cell Signaling Technology), NF1 (A300-140A, 1:1000, BethyLaboratories), pRb (Ser807/811) (#8516, 1:1000, Cell Signaling Technology), Rb (#9309, 1:1000, Cell Signaling Technology), pS6 (Ser235/236) (#2211, 1:1000, Cell Signaling Technology), S6 (#2317, 1:1000, Cell Signaling Technology), and β-actin (sc-47778, 1:5000, Santa Cruz Biotechnology). Secondary antibody used for western blots were: anti-mouse IgG HRP-linked (#7076, 1:5000, Cell Signaling Technology) and anti-rabbit IgG HRP-linked (#7074, 1:5000, Cell Signaling Technology).
RNA Sequencing
Total RNA were isolated from 2 cell lines, JHH-NF1-PA1 and JHH-PXA1 cultured in CRC condition or 7 days after removal of Y-27632 or condition media by using RNeasy mini kit (QIAGEN). Illumine sequencing and data analysis were performed by Novogene Corporation Inc (Sacramento, California). Gene functions were annotated based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
Animal Xenografts
Murine and zebrafish studies were approved by the Johns Hopkins Institutional Animal Care and Use Committee. Intracranial xenografts in anesthetized animals (4- to 6-wk-old female athymic nude mice from Charles River) were established as previously described.24
The zebrafish background strain was “AB,” from the Zebrafish International Resource Center (ZIRC) and zebrafish xenotransplantation, largely followed a prior procedure.25 At 2 days post-fertilization (dpf), zebrafish embryos were dechorionated and anesthetized, and then mounted dorsally in 1% low-melting-point agarose (Fisher Scientific). Approximately 40–60 GFP-labeled cells were injected (Dagan PMI-100 microinjector) into the optic tectum. Afterward larvae were released into an incubator and maintained at 28.5°C overnight. At 1 day postinjection (dpi), larvae were screened for visible GFP+ cell mass at injection site via stereo fluorescence microscopy (Olympus SZX16, Center Valley, Pennsylvania). The localization of the GFP-expressing cells was monitored by confocal intravital microscopy (Olympus FV1000) at 1, 4, and 6 dpi, to observe dissemination over time.
Experimental Rigor and Statistical Analysis
At least 3 biological replicates were performed for all in vitro experiments and data were analyzed with 2-tailed Student’s t test. All data are presented as mean ± SD, and P <.05 was considered significant.
Results
CRC Conditions Promote Growth of PA and Other Noninfiltrative Gliomas In Vitro
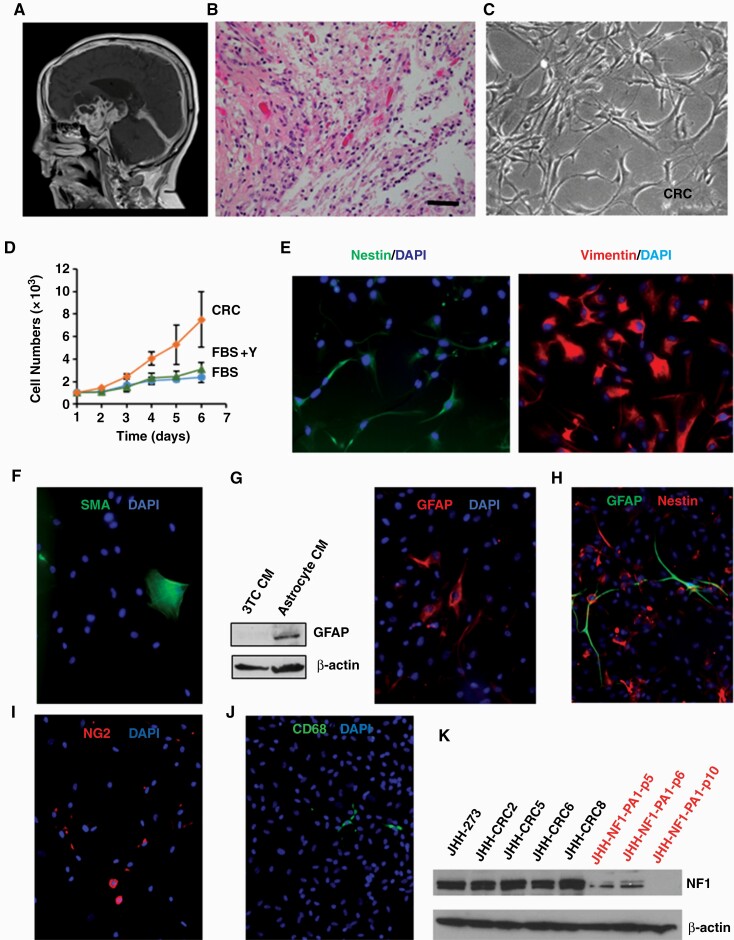
Previously, we attempted to culture 17 PAs and other noninfiltrative gliomas in high serum media, serum-free neurosphere conditions, or both methods, but failed to establish table cell lines (Supplementary Table 1). The first tumor cultured using the CRC method was a PA from the hypothalamus of a 14-year-old boy with neurofibromatosis type 1 (NF1). Both radiographic and histopathological features were typical of PA (Fig. 1A and B). An outline of the culture approach is shown in Supplementary Figure 1A; tumor cells were dissociated and grown in either media supplemented with high serum alone (FBS), the addition of ROCK inhibitor Y-27632 (FBS+Y), or CRC conditions. The appearance of cells was similar in the 3 conditions, with growth as a monolayer of cells showing a variable morphology and elongated processes (Fig. 1C; Supplementary Figure 1B). However, growth was most robust in CRC conditions, with several-fold more cells present as compared to those cultured in FBS alone or with ROCK inhibition (Fig. 1D; Table 1). Cells grown in CRC conditions were stable for up to 27 passages in terms of their appearance, with a doubling time of approximately 30 hours after 8 passages. We designated this stable culture JHH-NF1-PA1. Cell identity testing was performed at passages 10 and 20, with a short tandem repeat profile identical to the tumor from which it was derived (Supplementary Table 2).
Fig. 1.
Culturing of primary pilocytic astrocytoma cells. (A) Magnetic resonance imaging of tumor from which JHH-NF1-PA1 cells were derived. (B) Representative H&E image of JHH-NF1-PA1 primary tumor (×200, scale bar 50 microns). (C) Phase contrast image of JHH-NF1-PA1 cultures. (D) Growth of JHH-NF1-PA1 in CRC, FBS, or FBS + Y conditions. (E) Expression of nestin (left) and vimentin (right) in JHH-NF1-PA1. (F) Focal staining of SMA in JHH-NF1-PA1. (G) Increased GFAP in JHH-NF1-PA1 cultured in astrocyte condition media, and immunofluorescent staining of these cells. (H) Glial fibrillary acidic protein (green) and nestin (red) positive cells in JHH-CRC43. (I) NG2-positive cells in JHH-NF1-PA1. (J) Rare CD68-positive microglia in JHH-NF1-PA1. (K) Western blots showing loss of NF1 in CRC cultures. CRC, conditionally reprogrammed cells; FBS, fetal bovine serum; GFAP, glial fibrillary acidic protein; PA, pilocytic astrocytoma.
Table 1.
Circumscribed Gliomas and Glioneuronal Tumors
| Age/Gender | Diagnosis | Molecular Characteristics | Cell Growth | Passages CRC | |||
|---|---|---|---|---|---|---|---|
| FBS | FBS/Y | CRC | |||||
| CRC1 (JHH- NF1-PA1) | 14M | Pilocytic astrocytoma, Grade I | NF1-associated | ± | ++ | ++++ | >27 |
| CRC3 | 13M | Anaplastic ganglioglioma, Grade III | BRAF p.V600E | ++ | ++ | ++++ | >16 |
| CRC4 | 50F | Anaplastic glioma with ependymal features, Grade III | Negative for IDH1/2 mutations | ++ | ++ | ++++ | >15 |
| CRC10 | 14F | Ganglioglioma, Grade I | BRAF p.V600E | N/A | N/A | ++++ | >7 |
| CRC14 | 10M | Low-grade neuroepithelial tumor with ependymal/astroblastic features, Grade I/II | N/A | N/A | ++ | >4 | |
| CRC15 | 40F | Pilocytic astrocytoma with anaplastic features, Grade I | No growth | ||||
| CRC18 (JHH-PXA1) | 40M | Anaplastic PXA, Grade III | BRAF p.V600E | + | N/A | ++ | >20 |
| CRC19 | 66M | Myxopapillary ependymoma, Grade I | ± | N/A | + | >6 | |
| CRC21 | 41F | Myxopapillary ependymoma, Grade I | + | N/A | ++ | >7 | |
| CRC23 | 8F | Low-grade neuroepithelial tumor with features of DNET, Grade I | No mutations detected by NGS | + | N/A | ++ | >5 |
| CRC27 | 21F | Anaplastic glioma arising from pilocytic astrocytoma, Grade III/IV | H3K27M, ATRX, and NF1 mutations | N/A | N/A | + | >10 |
| CRC32 | 9M | Desmoplastic infantile ganglioglioma, Grade I | BRAF p.V600delinsDL, MET p.A320V | N/A | N/A | +++ | >17 |
| CRC33 | 14M | Pilocytic astrocytoma, Grade I | NF1-associated | N/A | N/A | ++ | >16 |
| CRC43 | 7F | Low-grade glioma with pilomyxoid features | BRCA2, RET | N/A | N/A | +++ | >4 |
Abbreviations: CRC, conditionally reprogrammed cell; FBS, fetal bovine serum; FBS/Y, FBS medium containing the 5 μM Y-27632 ROCK inhibitor.
Pilocytic astrocytomas generally express markers of glial differentiation such as GFAP, and are also often immunoreactive for vimentin and nestin.26,27 Immunofluorescence analysis showed that JHH-NF1-PA1 cells cultured in CRC media were strongly positive for vimentin, and more focally for the neural progenitor markers nestin and NG2, which are commonly expressed in gliomas including PA (Fig. 1E and data not shown).28 In contrast, immunostaining for the smooth muscle marker SMA highlighted only rare larger cells which lacked fine glial processes, suggesting that only a few myofibroblast-like cells were present (Fig. 1F). Glial fibrillary acidic protein was not expressed in this initial analysis, but after a short period of growth in astrocyte-conditioned medium, GFAP expression was detected in JHH-NF1-PA1 cells by both western blot and immunofluorescence staining (Fig. 1G).
In a second pLGG culture, CRC43, at day 19, the majority of cells were vimentin and/or nestin positive, while a significant number also expressed GFAP. As in JHH-NF1-PA1, SMA-positive cells were larger, lacked fine glial processes, and represented a small fraction of the culture. At day 29 in CRC conditions, we were able to undertake a more extensive analysis of cellular differentiation in CRC43. The culture remained largely fibrillary and glial in morphology, with mainly nestin-positive and scattered GFAP-positive cells showing long, fine processes (Fig. 1H). A significant number of NG2-positive glial-appearing cells were also present (Fig. 1I). In contrast, cells expressing the myofibroblast marker SMA or the microglial/macrophage marker CD68 were much less common and lacked long, fine glial processes (Fig. 1J). CRC27, a high-grade tumor with H3 p.K27M mutation that arose from a PA, also largely contained cells expressing vimentin and nestin or GFAP. Thus, the bulk of these cultures are composed of cells with glial or neural progenitor phenotypes, with only a minor component of inflammatory cells or fibroblasts.
We cultured a total of 14 glial and glioneuronal tumors with circumscribed growth patterns (Table 1). This included additional PAs, hard to classify low-grade neuroepithelial tumors from pediatric patients, gangliogliomas of various grades, an anaplastic PXA, and a high-grade glioma with a H3 p.K27M mutation arising from a PA diagnosed 10 years previously which also harbored H3 p.K27M, ATRX, and NF1 mutations. In all cases for which multiple conditions were tested, CRC media promoted enhanced growth as compared to FBS or FBS with the ROCK inhibitor Y-27632 (Table 1).
Genetic Stability in CRC Cultures
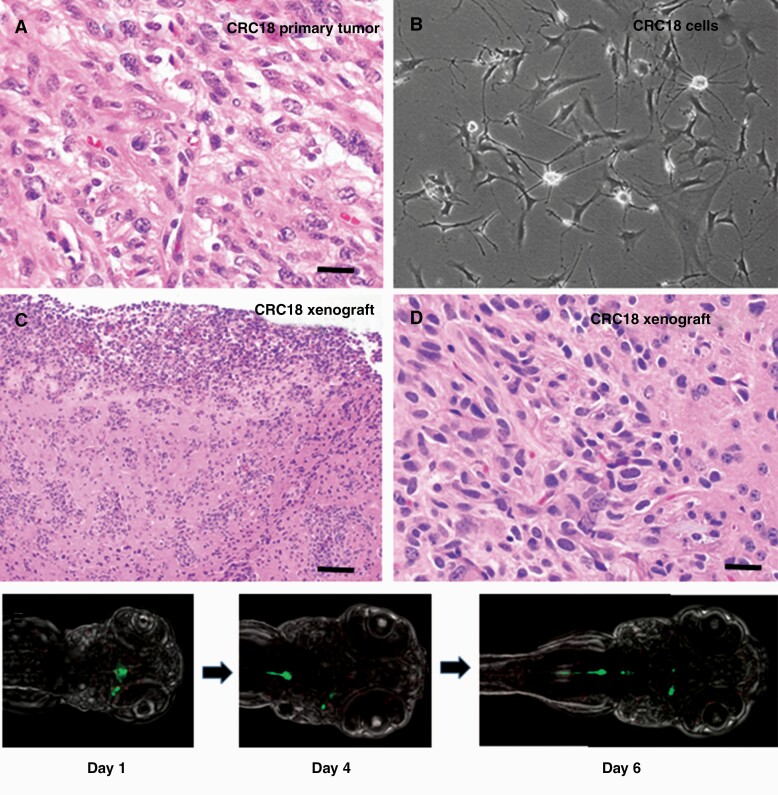
For tumors with signature genetic changes in the clinical specimen, cultured cells were analyzed to confirm the presence of similar molecular alterations. Short tandem repeat profiling was also performed on both primary tumor material and cell cultures, with confirmation of identity in all cases. JHH-NF1-PA1 was analyzed by western blot in order to confirm loss of NF1 protein. Only a faint NF1 band was identified in the first few passages, and after passage 8 no NF1 protein was present (Fig. 1K; Supplementary Figure 1C). Reduced NF1 expression was also confirmed at the mRNA level (Supplementary Figure 1D). No alterations known to be associated with aggressive PA growth, such as ATRX or CDKN2A loss, were identified by NGS analysis in either the JHH-NF1-PA1 patient sample or CRC cells. A BRAF p.V600E mutation was present in both the primary anaplastic PXA specimen and the culture derived from it (CRC18/JHH-PXA1, Fig. 2A and B) as detected by pyrosequencing and PCR analysis. The BRAF p.V600E alteration present in the CRC3 surgical specimen was also confirmed by PCR in the cultures. However, while BRAF p.V600E was present in the low-grade ganglioglioma giving rise to CRC10, we could not detect it in the cultured cells. In CRC27, loss of NF1 and ATRX protein, as well as the presence of mutant H3 p.K27M, was confirmed by western blot (Supplementary Figure 1E), while in CRC43, the BRCA2 mutation was confirmed by Sanger sequencing.
Fig. 2.
Xenografts of JHH-CRC cultures. (A) Representative H&E-stained image of JHH-CRC18 primary tumor (×400, scale bar 25 microns). (B) Phase contrast image of JHH-PXA1 (CRC18) culture. (C, D) Representative H&E-stained images of JHH-PXA1 mouse xenografts (×100, ×400, scale bars 100 microns, 25 microns). (E) Confocal images showing migration of JHH-NF1-PA1 cells (green) in zebrafish xenografts, taken at ×15 on days 1, 4, and 6 postinjection. CRC, conditionally reprogrammed cells; PA, pilocytic astrocytoma.
To more broadly assess the genetic stability of CRC cultures as compared to the tumors from which they were derived, NGS data from the surgical sample and longer-term cultures (at least 15 passages) were compared in detail for JHH-NF1-PA1, CRC32, and CRC33. We did not identify any new copy number alterations or clearly pathogenic mutations in the cultures. With respect to mutations/single nucleotide polymorphisms (SNPs) of no known clinical significance, almost all alterations were detected in both the surgical specimen and DNA from CRC cells, with only minimal changes in Variant Allele Frequency in the range of 1%–21%. A total of 6 alterations, all of no known clinical significance, were present only in the surgical specimen or in the CRC cells (supplementary Table 3).
Growth of Xenografts Derived From CRC Cells in Mice and Fish
In a subset of cultures, intracranial xenografts were attempted. Cells from JHH-NF1-PA1 were injected into the flank (n = 6) and cortex (n = 7) of athymic (nude) mice. However, animals remained asymptomatic over 12 months and no tumor cells were identified microscopically. In contrast, JHH-PXA1 (CRC18) formed orthotopic tumors in both mice injected, and microscopic analysis revealed atypical, somewhat epithelioid glial cells in the brain and leptomeninges morphologically similar to those in some regions of the surgical specimen (Fig. 2C and D).
We also evaluated if JHH-NF1-PA1 cells could survive and migrate in the brains of larval zebrafish prior to maturation of their adaptive immune system. Larvae were injected with approximately 40–60 GFP-tagged cells into the midline in the optic tectum at 2 dpf. The GFP-expressing tumor cells survived over 6 days in 23 of the 25 larvae grown at 28°C. In some cases, the cells migrated caudally within the developing spinal cord (Fig. 2E).
CRC-like Conditions Only Transiently Promote the Growth of Primary IDH Mutant Tumors
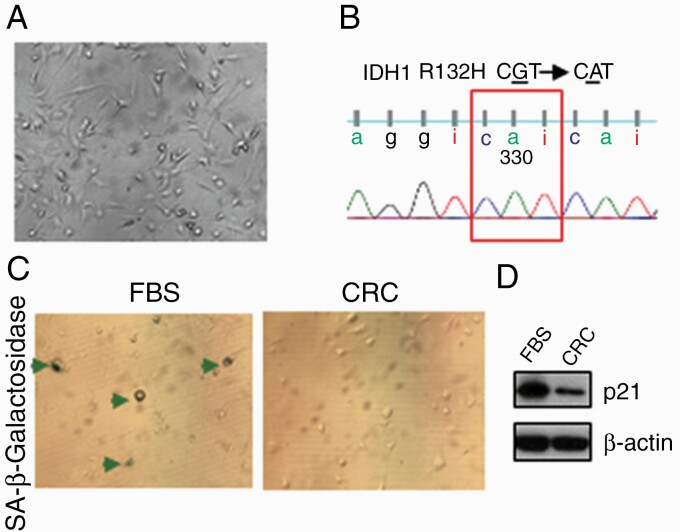
JHH-273 is a patient-derived IDH1 mutant anaplastic astrocytoma serial xenograft model.29 Cells from the JHH-273 flank xenograft do not grow in vitro using serum or serum-free medium with growth factors. We successfully cultured JHH-273 derived cells in CRC conditions for up to 8 passages and detected the IDH1 p.R132H (G/A) mutation by Sanger sequencing (Fig. 3A and B). In contrast, cells cultured in parallel in FBS media exhibited significant cell senescence by β-galactosidase staining and increased expression of p21 on western blots (Fig. 3C). This suggests that CRC conditions might also facilitate the growth of IDH mutant gliomas. However, while 8 cell cultures derived from primary IDH1 p.R132H mutant gliomas survived for several passages, they grew poorly and were negative for the mutation by Sanger sequencing.
Fig. 3.
Culturing of JHH-273 in CRC conditions. (A) Phase contrast image of JHH-273 CRC cells. (B) The IDH1 mutation was present in cultured cells. (C) Induction of senescence in FBS evidenced by acidic β-galactosidase staining of JHH-273 (arrows), as well as increased p21 expression in (D). CRC, conditionally reprogrammed cells; FBS, fetal bovine serum; IDH, isocitrate dehydrogenase.
CRC Conditions Reversibly Modulate Cell Proliferation and Survival
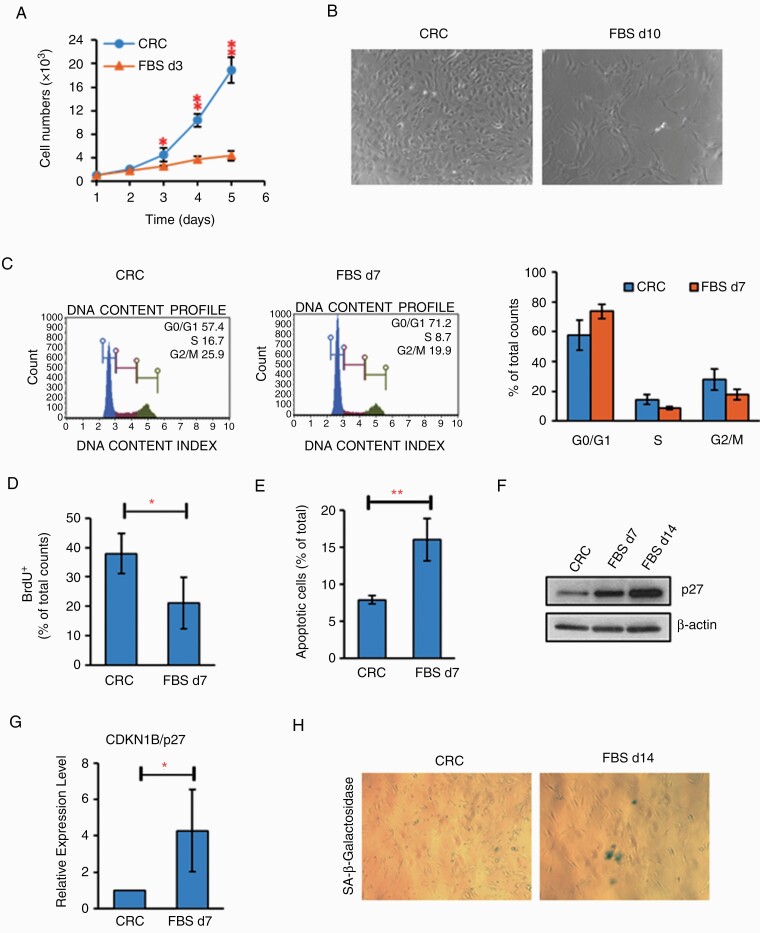
To determine if conditional reprogramming of brain tumor cells was reversible, we switched the JHH-NF1-PA1 culture to both standard FBS medium and serum-free neurosphere medium supplemented with EGF and fibroblast growth factor (FGF). As shown in Fig. 4A and B, growth of JHH-NF1-PA1 rapidly decreased over several days in FBS medium, suggesting that ongoing CRC conditions were required. Cell cycle analysis exhibited a decreased S-phase population, further confirmed by BrdU incorporation assay (Fig. 4C and D). Cells grown in serum also exhibited an increase of Annexin V-positive apoptotic cells (Fig. 4E) as well as signs of senescence with increased expression of p27 and acidic β-galactosidase-positive cells (Fig. 4F–H). Growth inhibition, apoptotic induction, and induction of senescence were also noted when the CRC2, CRC3, and CRC4 cultures were shifted to high serum growth conditions.
Fig. 4.
Switching JHH-NF1-PA1 cultures to FBS condition induced apoptosis, senescence and inhibited cell proliferation. (A, B) Slow growth of JHH-NF1-PA1 cells switched to FBS conditions. (C) After 7 days in FBS conditions, JHH-NF1-PA1 cells accumulated in G1 phase. (D, E) Decreased proliferation (BrdU+ cells) and increased apoptosis (Annexin V assay) after the switch to FBS conditions. (F, G) Levels of p27 protein and mRNA also increased when JHH-NF1-PA1 was cultured in FBS. (H) Induction of acidic β-galactosidase-positive cells in FBS (*P < .05, **P < .01). BrdU, bromodeoxyuridine; FBS, fetal bovine serum; PA, pilocytic astrocytoma.
Serum-free conditions with added EGF and FGF growth factors also failed to support robust growth. After switching JHH-NF1-PA1 to this serum-free neural stem cell (NSC) media, the cells stopped growing immediately. Addition of a ROCK inhibitor to the NSC did not increase growth (Supplementary Figure 2A and B). As was true for FBS conditions, these cells showed increased apoptosis. Over 60% of cells were Annexin V positive, and a significant increase was seen in cleaved PARP (Supplementary Figure 2C–E). Induction of p27 was also noted (Supplementary Figure 2E). Similar results were obtained with CRC2, CRC3, and CRC4 cultures.
CRC Conditions Promote Akt and Erk Activity
The mechanism(s) by which CRC conditions promote in vitro growth are not well understood. Suprynowicz et al30 demonstrated that in some conditionally reprogrammed epithelial cells, Akt phosphorylation is decreased, while β-catenin activation, expression of WNT targets such as Axin2, and mTOR signaling are all increased. In our PA1 and PXA1 glioma cells grown in CRC conditions, we found that Akt phosphorylation increased as compared to cultures lacking ROCK inhibitor or conditioned media (Supplementary Figure 3A). In contrast, markers of mTOR signaling including phosphorylated 4E-BP1 and S6 protein levels did not change, nor did mRNA levels of WNT targets such as Axin2 (Supplementary Figure 3B and data not shown). Another study suggested that a natural p53 isoform contributes to conditional reprogramming and long-term proliferation of primary epithelial cells, and that overall p53 protein levels are increased.31 While conditioned media removal seemed to reduce p53 protein levels to a degree, overall we did not see a clear association between p53 and CRC conditions in the 2 lines tested (Supplementary Figure 3A). Other cell cycle proteins such as Rb, p21 and p27 were also not affected by removal of ROCK inhibitor or conditioned media, however, both seemed to promote Erk1/2 phosphorylation (Supplementary Figure 3A and B).
To determine how the ROCK inhibitor and conditioned media affect gene expression within tumor cells more broadly, either Y-27632 or conditioned media were removed from JHH-NF1-PA1 and JHH-PXA1 and RNAseq data were compared to cells grown in complete CRC media. In both of these tumor cultures, removal of conditioned media resulted in more prominent alterations than removal of the ROCK inhibitor (Supplementary Figure 4A and data not shown). KEGG pathway analysis of these data identified reductions in TNF signaling which could potentially impact Akt activity following removal of conditioned media in both JHH-NF1-PA1 and JHH-PXA1 cells (Supplementary Figure 4B), as well as removal of ROCK inhibitor from PXA1 cells. KEGG analysis also suggested that removal of conditioned media from both PA1 and PXA1 cells, as well as ROCK inhibitor from PXA1 cells could inhibit Akt and Erk via reductions in cAMP (Supplementary Figure 4C).
CRC Cell Lines Can Be Used for Therapeutic Testing
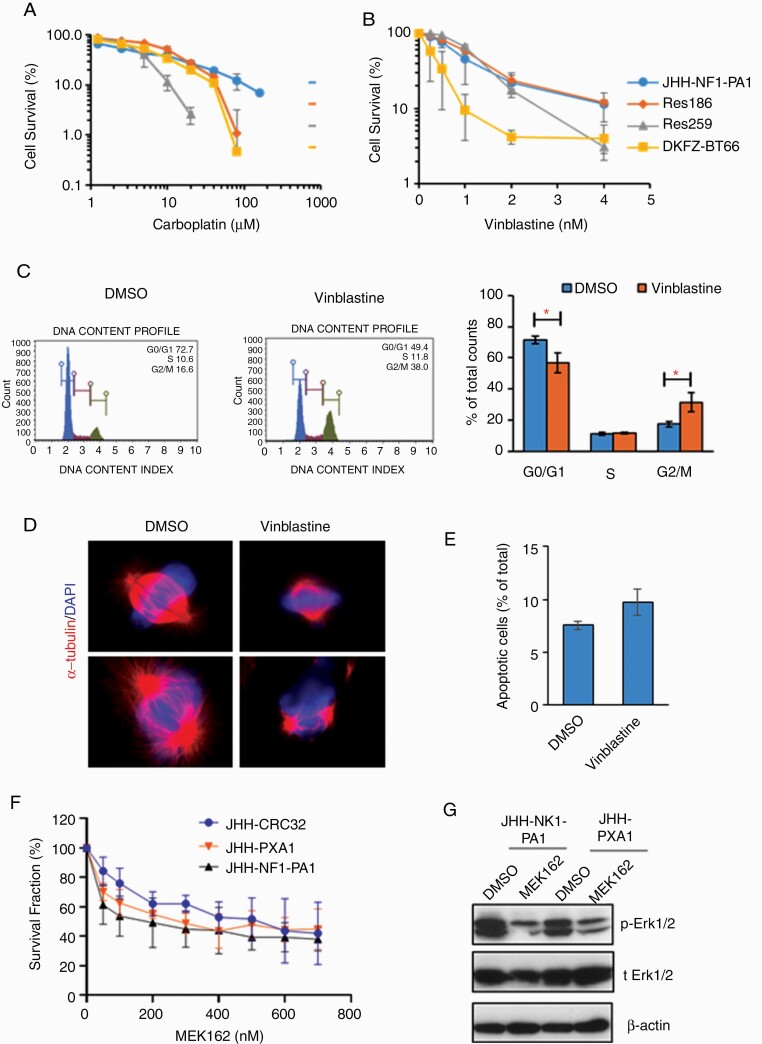
JHH-NF1-PA1 was derived from a patient treated with carboplatin, bevacizumab, and everolimus prior to surgical removal of the recurrent tumor used to generate the line. JHH-NF1-PA1 cells were more resistant to carboplatin than 3 other PLGG cell lines—Res186, Res259, and DKFZ-BT66—perhaps due to this prior clinical exposure (Fig. 5A). The response to vinblastine, an agent sometimes used to treat pLGG but not in this patient, was more promising. JHH-NF1-PA1 cells responded to the drug in the nM range, as did the other 3 pLGG cell lines (Fig. 5B). Vinblastine binds to β-tubulin and inhibits mitotic spindle formation, and after 6 hours in 2 nM vinblastine, JHH-NF1-PA1 cells were at least partially arrested in G2/M phase by cell cycle analysis (Fig. 5C). Immunostaining for α-tubulin detected few normal spindles as compared to vehicle treated control cells (Fig. 5D). The treatment did not significantly induce cell apoptosis (Fig. 5E).
Fig. 5.
Chemosensitivity of CRC cultures. (A, B) Survival fraction of JHH-NF1-PA1 and other pLGG cell lines (Res186, Res259, and BT66) after 5-d treatment with carboplatin or vinblastine. (C) JHH-NF1-PA1 cells treated with vinblastine for 6 h accumulate in G2/M. (D) Immunofluorescent staining of α-tubulin in JHH-NF1-PA1 cells treated with vinblastine for 6 h shows abnormal spindles. (E) Minimal increase in Annexin V+ apoptotic cells after vinblastine treatment. (F) Decreased numbers of JHH-NF1-PA1, JHH-PXA1, and JHH-CRC32 cells after 5-d treatment with MEK162. (G) Inhibition of Erk1/2 phosphorylation by MEK162. CRC, conditionally reprogrammed cells; PA, pilocytic astrocytoma; pLGG, pediatric low-grade glioma. *P < .05.
We also tested the effects of the MAPK pathway inhibitor MEK162 (binimetinib) in 2 cultures with genetic alterations activating BRAF, as well as the JHH-NF1-PA1 cells. JHH-PXA1, JHH-NF1-PA1, and the culture derived from a desmoplastic infantile ganglioglioma (JHH-CRC32) showed growth inhibition over 5 days (Fig. 5F). Inhibition of MAPK signaling by MEK162, as evidenced by suppression of Erk phosphorylation, was documented by western blot analysis (Fig. 5G).
Discussion
The CRC technique was previously mainly used to facilitate the growth of benign and neoplastic epithelial cells.13-17,19 Here, we harnessed this approach to develop experimental models for low-grade gliomas and other neuroepithelial tumors which have been historically difficult to propagate in standard serum or serum-free conditions. In studies by the Schlegel group, where this approach was discovered, cells were grown on an irradiated feeder layer of 3T3 cells.13 In our modified approach, conditioned media from irradiated 3T3 cells was used in place of feeder cells. Overall, we found that compact glial and glioneuronal tumors, most of which were low grade, grew better in CRC conditions than in standard high serum or serum-free media.
Pilocytic astrocytomas, including several which arose in patients with neurofibromatosis, represent one tumor type which benefitted from this new approach. Prior studies generally relied on genetically engineered mouse models for NF1-associated PA research.32-35 The human NF1-associated PA culture we describe here, JHH-NF1-PA1, provides an additional model system with which to study the biology of these tumors. A second NF1-associated PA culture, JHH-CRC33, was useful for some initial low passage studies.
Importantly, drug sensitivity testing of JHH-NF1-PA1 paralleled prior in vivo responses in the patient from which it was derived. These cells originated from an optic pathway glioma that progressed after treatment with carboplatin and everolimus as single agents. Our results recapitulated the carboplatin resistance and revealed sensitivity to vinblastine. We previously reported that this tumor was resistant to everolimus.36 These initial studies support the utility of CRC cultures such as this for preclinical therapeutic testing.
Additional new models of rare tumor types reported in this study include an anaplastic ganglioglioma with BRAF p.V600E mutation, an anaplastic PXA with BRAF p.V600E mutation, a desmoplastic infantile ganglioglioma with BRAF and MET mutations, an anaplastic glioma with H3 p.K27M, ATRX, and NF1 mutations that initially presented as a PA and then recurred after many years, and a low-grade glioma with pilomyxoid features and mutations in BRCA2 and RET. The BRAF-altered anaplastic PXA and desmoplastic infantile ganglioglioma cultures were both treated with the MAPK pathway inhibitor MEK162 (binimetinib), and this clinically active compound inhibited their growth in the low nanomolar range.37,38 Prior studies have shown in vivo inhibitory effects of MEK162 in murine glioblastoma xenografts, suggesting that it can penetrate into the brain.39 These results suggest that such inhibitors could be beneficial in patients harboring gliomas with similar features.
The mechanisms by which ROCK inhibition and fibroblast conditioned media promote glioma growth are not entirely clear. Unlike prior studies in epithelial cells, our results in CNS tumors did not reveal alterations in p53, WNT, or mTOR activity.30,31 However, we did find that CRC conditions promoted Akt1 and Erk1/2 phosphorylation, and broader analysis of gene expression using RNA sequencing provided additional support for changes in these pathways. Importantly, molecular comparisons between primary tumor samples and CRC cultures indicated that growth was not due to the accumulation of new genetic drivers in vitro. These results suggest that mechanisms through which CRC conditions enhance ex vivo growth may vary depending upon the tissue of origin and underlying mechanisms.
Our studies also revealed that a subset of CRC cultures could be used to generate xenografts in mice or zebrafish. The anaplastic pleomorphic xanthoastrocytoma-derived JHH-PXA1 cells formed orthotopic xenografts which microscopically resembled the tumors from which they were derived in athymic mice. These neoplastic masses took 9–12 months to form, a time course consistent with that of another recently described PXA xenograft model.2 In contrast, JHH-NF1-PA1 did not establish either flank or intracranial xenografts in athymic mice. Interestingly, optic glioma cells from the murine NF1 model generated glioma-like lesions in immunocompetent wild-type mice, but not in athymic nude mice, providing a possible explanation.40 Recently, Pan et al41 reported that T-cell-microglia interaction is required to establish a microenvironment for NF1 low-grade glioma growth in athymic nude mice. Our group and others have recently developed methods to xenograft human tumor cells in immunocompetent mice, which might provide a more appropriate microenvironment for establishing pLGG xenografts.42,43
Zebrafish xenografts represent an additional rapid and cost-effective in vivo model system for tumor xenografts. The models can be established with fewer cells, and the adaptive immune system is not yet established in early embryonic or larval stages. The short experiment timeframes and convenience of tracking tumor cells in translucent larvae make zebrafish a powerful tool for drug screening.44-46 Zebrafish are typically raised at 28°C, which can slow the growth of human tumor cells. Eden et al47 developed a method to adapt both fish and cells to 34°C for orthotopic xenograft of murine pediatric brain tumors. More recently, Yan et al48 adapted immune-deficient adult zebrafish to 37°C and shown that this facilitates successful intraperitoneal engraftment of numerous non-CNS cancer types.
In summary, CRC conditions facilitate growth of a subset of pLGG and other brain tumors which have not been amenable to other culture methods. Our mechanistic studies indicate that CRC conditions enhance proliferation pathways while suppressing apoptosis and the induction of senescence in a reversible fashion. Novel models generated using these techniques will provide opportunities for in vitro and in vivo drug screening, mechanistic studies, and other research on low-grade gliomas associated with NF1 or driven by sporadic mutations such as BRAF p.V600E.
Supplementary Material
Funding
This work was supported by the Imagine An Answer To Kids’ Brain Cancer Foundation, and in part by the Pilocytic/Pilomyxoid Fund (including Lauren’s First and Goal and Stick It to Brain Tumors), Department of Defense grant W81XWH-18-1-0496, and Alex’s Lemonade Stand Foundation.
Conflict of interest statement. None declared.
Authorship statement. Planning of study: M.Y., E.R., F.R., and C.G.E.; sample and data collection and analysis: M.Y., D.W., L.R., E.B., M.G., A.C., E.J., J.B., J.R., J.M., L.C., L.J., E.R., F.R., and C.G.E.; and editing of manuscript: M.Y., D.W., A.C., E.J., J.M., L.C., L.J., E.R., F.R., and C.G.E.
References
- 1. Sandén E, Eberstål S, Visse E, Siesjö P, Darabi A. A standardized and reproducible protocol for serum-free monolayer culturing of primary paediatric brain tumours to be utilized for therapeutic assays. Sci Rep. 2015;5:12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kogiso M, Qi L, Lindsay H, et al. . Xenotransplantation of pediatric low grade gliomas confirms the enrichment of BRAF V600E mutation and preservation of CDKN2A deletion in a novel orthotopic xenograft mouse model of progressive pleomorphic xanthoastrocytoma. Oncotarget. 2017;8(50):87455–87471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Selt F, Hohloch J, Hielscher T, et al. . Establishment and application of a novel patient-derived KIAA1549:BRAF-driven pediatric pilocytic astrocytoma model for preclinical drug testing. Oncotarget. 2017;8(7):11460–11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodriguez FJ, Lim KS, Bowers D, Eberhart CG. Pathological and molecular advances in pediatric low-grade astrocytoma. Annu Rev Pathol. 2013;8:361–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumar R, Liu APY, Orr BA, Northcott PA, Robinson GW. Advances in the classification of pediatric brain tumors through DNA methylation profiling: from research tool to frontline diagnostic. Cancer. 2018;124(21):4168–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ellison DW, Hawkins C, Jones DTW, et al. . cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAFV600E mutation. Acta Neuropathol. 2019;137(4): 683–687. [DOI] [PubMed] [Google Scholar]
- 7. Michaloglou C, Vredeveld LC, Soengas MS, et al. . BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. [DOI] [PubMed] [Google Scholar]
- 8. Raabe EH, Lim KS, Kim JM, et al. . BRAF activation induces transformation and then senescence in human neural stem cells: a pilocytic astrocytoma model. Clin Cancer Res. 2011;17(11):3590–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jacob K, Quang-Khuong DA, Jones DT, et al. . Genetic aberrations leading to MAPK pathway activation mediate oncogene-induced senescence in sporadic pilocytic astrocytomas. Clin Cancer Res. 2011;17(14):4650–4660. [DOI] [PubMed] [Google Scholar]
- 10. Sievert AJ, Lang SS, Boucher KL, et al. . Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci USA. 2013;110(15):5957–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buhl JL, Selt F, Hielscher T, et al. . The senescence-associated secretory phenotype mediates oncogene-induced senescence in pediatric pilocytic astrocytoma. Clin Cancer Res. 2019;25(6):1851–1866. [DOI] [PubMed] [Google Scholar]
- 12. Larribere L, Wu H, Novak D, et al. . NF1 loss induces senescence during human melanocyte differentiation in an iPSC-based model. Pigment Cell Melanoma Res. 2015;28(4):407–416. [DOI] [PubMed] [Google Scholar]
- 13. Liu X, Ory V, Chapman S, et al. . ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180(2):599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dakic A, DiVito K, Fang S, et al. . ROCK inhibitor reduces Myc-induced apoptosis and mediates immortalization of human keratinocytes. Oncotarget. 2016;7(41):66740–66753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Timofeeva OA, Palechor-Ceron N, Li G, et al. . Conditionally reprogrammed normal and primary tumor prostate epithelial cells: a novel patient-derived cell model for studies of human prostate cancer. Oncotarget. 2017;8(14):22741–22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gentzsch M, Boyles SE, Cheluvaraju C, et al. . Pharmacological rescue of conditionally reprogrammed cystic fibrosis bronchial epithelial cells. Am J Respir Cell Mol Biol. 2017;56(5):568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu X, Krawczyk E, Suprynowicz FA, et al. . Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat Protoc. 2017;12(2):439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee HS, Lee JS, Lee J, et al. . Establishment of pancreatic cancer cell lines with endoscopic ultrasound-guided biopsy via conditionally reprogrammed cell culture. Cancer Med. 2019;8(7):3339–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Correa BRS, Hu J, Penalva LOF, et al. . Patient-derived conditionally reprogrammed cells maintain intra-tumor genetic heterogeneity. Sci Rep. 2018;8(1):4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mahajan AS, Sugita BM, Duttargi AN, et al. . Genomic comparison of early-passage conditionally reprogrammed breast cancer cells to their corresponding primary tumors. PLoS ONE. 2017;12(10):e0186190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bax DA, Little SE, Gaspar N, et al. . Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PLoS ONE. 2009;4(4):e5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li J, Zhang H, Wang L, et al. . Comparative study of IDH1 mutations in gliomas by high resolution melting analysis, immunohistochemistry and direct DNA sequencing. Mol Med Rep. 2015;12(3):4376–4381. [DOI] [PubMed] [Google Scholar]
- 23. Huang T, Zhuge J, Zhang WW. Sensitive detection of BRAF V600E mutation by Amplification Refractory Mutation System (ARMS)-PCR. Biomark Res. 2013;1(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mao XG, Hütt-Cabezas M, Orr BA, et al. . LIN28A facilitates the transformation of human neural stem cells and promotes glioblastoma tumorigenesis through a pro-invasive genetic program. Oncotarget. 2013;4(7):1050–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teng Y, Xie X, Walker S, White DT, Mumm JS, Cowell JK. Evaluating human cancer cell metastasis in zebrafish. BMC Cancer. 2013;13:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tchoghandjian A, Fernandez C, Colin C, et al. . Pilocytic astrocytoma of the optic pathway: a tumour deriving from radial glia cells with a specific gene signature. Brain. 2009;132(Pt 6):1523–1535. [DOI] [PubMed] [Google Scholar]
- 27. Martelli C, Iavarone F, D’Angelo L, et al. . Integrated proteomic platforms for the comparative characterization of medulloblastoma and pilocytic astrocytoma pediatric brain tumors: a preliminary study. Mol Biosyst. 2015;11(6):1668–1683. [DOI] [PubMed] [Google Scholar]
- 28. Shoshan Y, Nishiyama A, Chang A, et al. . Expression of oligodendrocyte progenitor cell antigens by gliomas: implications for the histogenesis of brain tumors. Proc Natl Acad Sci USA. 1999;96(18):10361–10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Borodovsky A, Meeker AK, Kirkness EF, et al. . A model of a patient-derived IDH1 mutant anaplastic astrocytoma with alternative lengthening of telomeres. J Neurooncol. 2015;121(3):479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suprynowicz FA, Kamonjoh CM, Krawczyk E, et al. . Conditional cell reprogramming involves non-canonical β-catenin activation and mTOR-mediated inactivation of Akt. PLoS ONE. 2017;12(7):e0180897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mondal AM, Zhou H, Horikawa I, et al. . Δ133p53α, a natural p53 isoform, contributes to conditional reprogramming and long-term proliferation of primary epithelial cells. Cell Death Dis. 2018;9(7):750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Freret ME, Gutmann DH. Insights into optic pathway glioma vision loss from mouse models of neurofibromatosis type 1. J Neurosci Res. 2019;97(1):45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khatua S, Gutmann DH, Packer RJ. Neurofibromatosis type 1 and optic pathway glioma: molecular interplay and therapeutic insights. Pediatr Blood Cancer. 2018;65(3):1–7. [DOI] [PubMed] [Google Scholar]
- 34. Bajenaru ML, Hernandez MR, Perry A, et al. . Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res. 2003;63(24):8573–8577. [PubMed] [Google Scholar]
- 35. Zhu Y, Harada T, Liu L, et al. . Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development. 2005;132(24):5577–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Poore B, Yuan M, Arnold A, et al. . mTORC1 inhibition in pediatric low-grade glioma depletes glutathione and therapeutically synergizes with carboplatin. Neuro Oncol. 2019;21(2):252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Herpen CML, Agarwala SS, Hauschild A, et al. . Biomarker results from a phase II study of MEK1/2 inhibitor binimetinib (MEK162) in patients with advanced NRAS- or BRAF-mutated melanoma. Oncotarget. 2019;10(19):1850–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dummer R, Ascierto PA, Gogas HJ, et al. . Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2018;19(10):1315–1327. [DOI] [PubMed] [Google Scholar]
- 39. Narayan RS, Gasol A, Slangen PLG, et al. . Identification of MEK162 as a radiosensitizer for the treatment of glioblastoma. Mol Cancer Ther. 2018;17(2):347–354. [DOI] [PubMed] [Google Scholar]
- 40. Chen YH, McGowan LD, Cimino PJ, et al. . Mouse low-grade gliomas contain cancer stem cells with unique molecular and functional properties. Cell Rep. 2015;10(11):1899–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pan Y, Xiong M, Chen R, et al. . Athymic mice reveal a requirement for T-cell-microglia interactions in establishing a microenvironment supportive of Nf1 low-grade glioma growth. Genes Dev. 2018;32(7-8):491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Semenkow S, Li S, Kahlert UD, et al. . An immunocompetent mouse model of human glioblastoma. Oncotarget. 2017;8(37):61072–61082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Basel MT, Narayanan S, Ganta C, et al. . Developing a xenograft human tumor model in immunocompetent mice. Cancer Lett. 2018;412:256–263. [DOI] [PubMed] [Google Scholar]
- 44. Wertman J, Veinotte CJ, Dellaire G, Berman JN. The zebrafish xenograft platform: evolution of a novel cancer model and preclinical screening tool. Adv Exp Med Biol. 2016;916:289–314. [DOI] [PubMed] [Google Scholar]
- 45. Brown HK, Schiavone K, Tazzyman S, Heymann D, Chico TJ. Zebrafish xenograft models of cancer and metastasis for drug discovery. Expert Opin Drug Discov. 2017;12(4):379–389. [DOI] [PubMed] [Google Scholar]
- 46. Asnaghi L, White DT, Key N, et al. . ACVR1C/SMAD2 signaling promotes invasion and growth in retinoblastoma. Oncogene. 2019;38(12):2056–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eden CJ, Ju B, Murugesan M, et al. . Orthotopic models of pediatric brain tumors in zebrafish. Oncogene. 2015;34(13):1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yan C, Brunson DC, Tang Q, et al. . Visualizing engrafted human cancer and therapy responses in immunodeficient zebrafish. Cell. 2019;177(7):1903–1914 e1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.