Abstract
Background
Aberrant epidermal growth factor receptor (EGFR) activation is observed in over 50% of cases of adult glioblastoma (GBM). Nevertheless, EGFR antibodies are ineffective in clinical GBM treatment, suggesting the existence of redundant EGFR activation mechanisms. Whether circular RNA (circRNA) encodes a protein involved in EGFR-driven GBM remains unclear. We reported an unexpected mechanism in which circular EGFR RNA (circ-EGFR) encodes a novel EGFR variant to sustained EGFR activation.
Method
We used RNA-seq, Northern blot, and Sanger sequencing to confirm the existence of circ-EGFR. Antibodies and a liquid chromatograph tandem mass spectrometer were used to identify circ-EGFR protein products. Lentivirus-transfected stable cell lines were used to assess the biological functions of the novel protein in vitro and in vivo. Clinical implications of circ-EGFR were assessed using 97 pathologically diagnosed GBM patient samples.
Results
The infinite open reading frame (iORF) in circ-EGFR translated repeating amino acid sequences via rolling translation and programmed −1 ribosomal frameshifting (-1PRF) induced out-of-frame stop codon (OSC), forming a polymetric novel protein-complex, which we termed rolling-translated EGFR (rtEGFR). rtEGFR directly interacted with EGFR, maintained EGFR membrane localization and attenuated EGFR endocytosis and degradation. Importantly, circ-EGFR levels correlated with the EGFR signature and predicted the poor prognosis of GBM patients. Deprivation of rtEGFR in brain tumor-initiating cells (BTICs) attenuated tumorigenicity and enhanced the anti-GBM effect.
Conclusion
Our findings identified the endogenous rolling-translated protein and provided strong clinical evidence that targeting rtEGFR could improve the efficiency of EGFR-targeting therapies in GBM.
Keywords: CircRNA, EGFR, glioblastoma, iORF, rolling translation
Key Point.
1. Rolling-translated EGFR (rtEGFR) is a novel EGFR variant in glioblastoma translated from circular EGFR. rtEGFR sustains EGFR membrane localization via direct binding. rtEGFR deprivation sensitizes brain tumor-initiating cells for EGFR-targeting therapy.
Importance of the Study.
In this study, we reported that circ-EGFR encoded rolling-translated EGFR (rtEGFR), a polymetric protein complex that persistently activates oncogenic EGFR signaling by sustaining EGFR membrane localization. circ-EGFR deprivation suppressed tumorigenicity in brain tumor-initiating cells and enhanced the efficiency of nimotuzumab in glioblastoma (GBM) treatment, indicating an important role of rtEGFR in EGFR-driven GBM. Circ-EGFR is selectively expressed in brain tumor-initiating cells relative to normal astrocytes or neuro stem cells (NSC), which provides a wide therapeutic window for targeting rtEGFR in GBM with monoclonal antibodies or future small molecule inhibitors.
Activated oncogenic signaling stimulated by amplified or overexpressed genes is a hallmark of human cancers.1 EGFR amplification and/or mutation occur in 57% of primary glioblastoma (GBM) and is an established diagnostic marker as well as therapeutic target.2 However, many chemicals or antibodies targeting EGFR are not efficient at crossing the blood–brain barrier, which limits their efficacy.3 Additionally, redundant receptor tyrosine kinases and downstream molecules may bypass EGFR-targeting therapy, suggesting the complexity of EGFR activation.4
Vesicle transport provides a membrane platform for signaling-complex assembly and signal transduction termination.5 In GBM, EGFR is one of the most important clients of vesicle transport, and defective endocytic downregulation of EGFR alters its membrane localization and constitutive activation.6 Specifically, members of the vesicle-trafficking involved small GTPases are overexpressed in GBM and promoted tumor progression,7 and interacting proteins in vesicle trafficking also attenuated EGFR degradation,8,9 indicating the role of the EGFR endocytosis modulation network in GBM. Novel targets that control aberrant EGFR membrane localization in GBM may provide therapeutic opportunities for this lethal tumor.
CircRNAs are covalently closed, endogenous molecules in eukaryotes with important biological functions.10 CircRNAs largely exert their functions by acting as microRNAs or protein “sponges” or by acting as protein scaffolds.11 Surprisingly, the coding potential of circRNAs has recently been intensively studied.10 Given the covalently closed structure, open reading frames in circRNAs may also be translated through multiple rounds.12 Theoretically, the translation could go a maximum of three rounds (+0, +1, and +2 frame) if the nucleotides in the circRNA are not integral multiples of three. If stop codons are not engaged in all reading frames, an infinite open reading frame (iORF) is formed. Artificial circRNAs with iORF could translate long repeating proteins.13,14 Whether rolling translation exists endogenously in vivo remains largely unknown.
In this study, we found that circ-EGFR encoded a polymetric protein complex rtEGFR through iORF rolling-translation and the programmed −1 ribosomal frameshifting (−1PRF) induced out-of-frame stop codon (OSC) mechanism. rtEGFR sustained the aberrant activation of EGFR signaling in GBM. Because rtEGFR was not detectable in normal brain (NB), it may serve as a potential therapeutic target in GBM.
Materials and Methods
Human Cancer and Normal Tissues
All clinical samples were from the Department of Neurosurgery of the 1st Affiliated Hospital of Sun Yat-sen University. The approval of the Clinical Research Ethics Committee of the 1st Affiliated Hospital of Sun Yat-sen University were obtained.
Cell Lines
293T cell was purchased from ATCC (CRL-11268). U251, HS683, and SW1783 cells were kindly provided by Dr. Suyun Huang (VCU). Brain tumor-initiating cells (BTICs) were kindly supported by Dr. Jeremy N. Rich (UCSD). Normal human astrocytes (NHA) cells were purchased from Lonza and cultured in an AGM Bullet Kit™ (Lonza, Walkersville, MD) as recommended by the instructions.
RNA Sequencing
Total RNA was digestion with RNAase R (Epicentre Biotechnologies, Madison, WI), purified with RNeasy MinElute Cleanup Kit (Qiagen, Venlo, the Netherlands). After the rRNAs removed, RNAs was reversed into cDNA using random primers. The cDNA fragments were purified with a QiaQuick PCR extraction kit and ligated to Illumina sequencing adapters. Then the second-strand cDNA was digested with UNG (uracil-N-glycosylase), size-selected via agarose gel electrophoresis, PCR-amplified, and sequenced using an Illumina HiSeq TM 4000 by Gene Denovo Biotechnology Co. (Guangzhou, China).
Bioinformatics Analysis
20mers from both ends of the unmapped reads were extracted using in-house perl scripts and aligned to the reference genome (bowtie2, version 2.3.0) to locate unique anchor positions within splice sites. Anchor reads that aligned in the reverse orientation (head-to-tail) indicated circRNA splicing and were then subjected to find_circ (version 1.2, https://github.com/marvin-jens/find_circ/) to identify circRNAs. A candidate circRNA was called if it was supported by at least 2 unique back-spliced reads from at least one sample. The default parameters of edgeR were used, and differentially expressed genes (DEGs) were selected based on log2-fold changes ≥1 and q-values >.05.
Northern Blot
Ten micrograms of total RNA of each sample was separated in 1.5% formaldehyde agarose gels in MOPS buffer and transferred onto Hybond N+ membranes (Amersham) with 10× SSC. Then RNA was hybridized with DIG-labeled circ-EGFR probe or exon probe.
Plasmid Construction
All plasmids were synthesized by Generay Biotech (Shanghai, China) and were listed in Supplementary Table 1.
Stable Cell Line Generation
The lentivirus infected cell lines were selected with 1 μg/ml puromycin for 72 h. The sequences of the nucleotide siRNA targeting the splice sites of circ-EGFR and scramble siRNA were listed in Supplementary Table 2. All the shRNA sequences were obtained from Genepharma (Shanghai, China).
RNA Fluorescence In Situ Hybridization
Oligonucleotide-modified probe sequence for circ-EGFR was synthesized from Sangon Biotech (Shanghai, China). Then hybridization was performed at 37 °C overnight in a dark moist chamber. Images were acquired on ZEISS LSM 880 with Airyscan (Carl Zeiss, Jena, Germany).
EGFR Ubiquitylation
Cells were harvested for subsequent IP and IB experiments. The amounts of ubiquitin were corrected for the amount of immunoprecipitated EGFR, which was determined from parallel gels.
EGFR Degradation
Indicated cells were starved for 4 h in serum-free medium and incubated for the indicated times with 100 ng/ml EGF in the presence of 10 mg/ml cycloheximide. Cells were harvested on ice and the total protein were subjected to western blot analysis.
LC–MS/MS Analysis
Total proteins were collected and separated by SDS-Gel and the 30–40, 40–70, and 70–170 kD were isolated and subjected to digestion with sequencing-grade trypsin (Promega, Madison, WI). The digested peptides were analyzed by QExactive mass spectrometer (Thermo Fisher, Carlsbad, CA). Then the acquired spectra were analyzed with the SEQUEST HT algorithm.
CCK-8 Assay
Two thousand per well of indicated cells were seeded in 96-well plates. Viabilities of cells were measured every 24 h for 5 days.
Colony Formation Assay
Two thousand per well of indicated cells were seeded and incubated for 2 weeks with complete medium. The colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 10 min.
Limiting Dilution Assay
BITCs were seeded into 96-well plate at density of 5, 10, 20, 50, 100, or 200 cells per well and the frequency of spheres of each well was examined. Glioma sphere forming frequency was calculated using extreme limiting dilution analysis systerm (http://bioinf.wehi.edu.au/software/elda/).
Intracranial Injection
Six mice were grouped in each cage and randomly assigned to different experimental groups. We intracranially injected 1 × 105 cells in 5 μl PBS for each of the indicated BTICs or 5 × 105 cells for each of the indicated glioma cell into nude mice.
Statistical Analysis
Statistical analysis was carried out using GraphPad Prism version 8.0. Experimental data are represented as the average ± SD of a minimum of three biological replicates. The Student's two-tailed unpaired t test was used to determine statistical significance of in vitro experiments. The log-rank test or Gehan–Breslow–Wilcoxon test was used to determine the statistical differences of the survival data. All statistical tests were two-sided, and a P value of <.05 was considered statistically significant.
Data Availability
RNA-seq data from this study have been deposited into NCBI SRA: PRJNA525736.
More detailed methods are described in Supplementary data.
Results
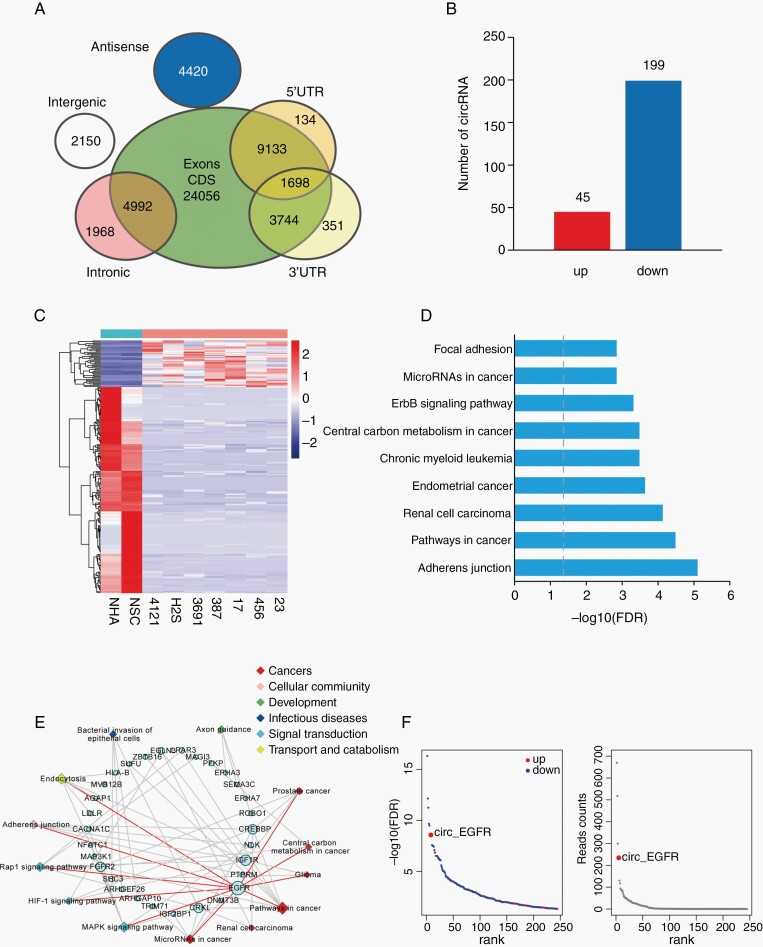
To detect differentially expressed circRNA (DEcRs) in GBM, we performed linear RNA depleted RNA-seq in a panel of BTICs. Neuro stem cells (NSCs) and NHAs were used as normal controls. A total of 52 646 circRNAs were identified, 14 531 of which were matched in circBase.15 We annotated these identified candidates using the ensemble database.16 Most identified circRNAs originated from protein-coding exons, and others aligned with introns, 5′-UTR, 3′-UTR, or antisense sequences of known genes (Fig. 1A). The majority of the identified circRNAs were less than 1500 nucleotides (nt) with a peak at 300–400 nt. Two hundred forty-four DEcRs (45 upregulated, 199 downregulated, Fig. 1B) were identified between BTICs and normal cells with a false discovery rate (FDR) < .05 and fold change > 2 (Fig. 1C). We next analyzed host genes of these DEcRs with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment (Fig. 1D, Supplementary Table 3). EGFR involved 10 of 13 KEGG enriched pathways, consistent with the notion that EGFR plays a key role during GBM tumorigenesis (Fig. 1E). Circ-EGFR expression ranked no. 7 among these DEcRs (Fig. 1F). In the top 10 DEcRs, circ-EGFR was the only one that was upregulated in GBM. Thus, we focused on circ-EGFR for the next step of our investigation.
Fig. 1.
Profiling of circular RNAs in brain tumor-initiating cells (BTICs), neuro stem cells (NSCs), and NHAs. (A). Venn plot showing the number of all circRNAs derived from different genomic regions. (B). The numbers of differentially expressed circRNAs (DEcRs) with false discovery rate (FDR) < .05 and fold change > 2 between cancerous and normal cells. (C). Heat map of all DEcRs. (D). The top 9 enrichment scores by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis among DEcRs derived host genes. X axis: enriched pathways, Y axis: −log10 FDR. (E). The network of 13 enriched pathways and host genes of DEcRs. Circular nodes represent host genes; rhombic nodes represent pathways. The size denotes the number of genes or pathways related to the nodes. (F). Left, −log10 FDR of DEcRs. The blue and red points represented downregulated and upregulated circRNAs. X axis: DEcRs, Y axis: −log10 FDR. Right, Reads count of DEcRs. X axis: DEcRs, Y axis: read counts.
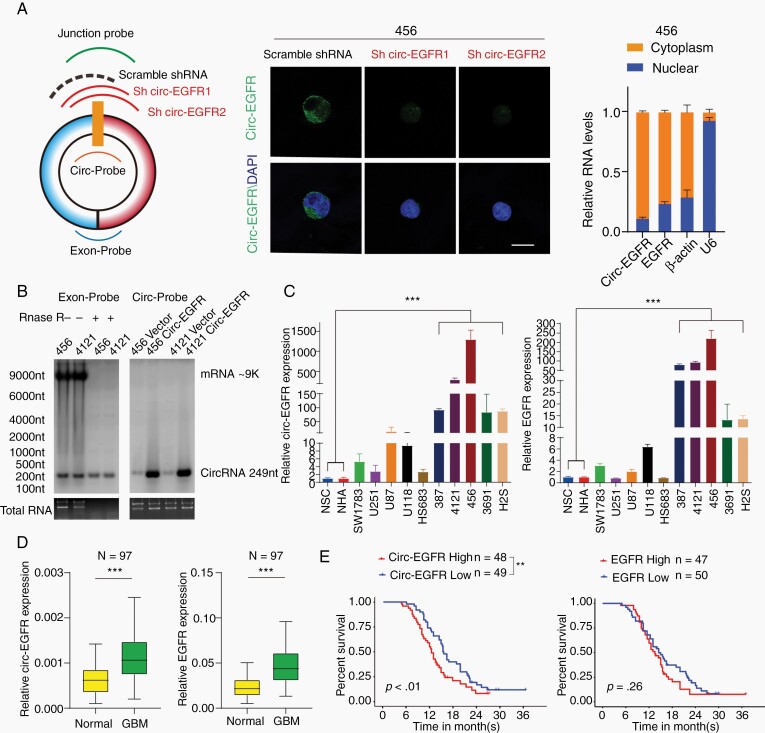
Inspection of the EGFR gene and circBase revealed that circ-EGFR (has_circ_0080229) was formed from exon 14 and 15 of EGFR (Supplementary Figure 1A, upper). Compared with EGFR mRNA, circ-EGFR had a longer half-life and was more resistant to RNase-R digestion17 (Supplementary Figure 1B). Using a junction-specific probe, circ-EGFR localization and specificity of 2 shRNA targeting circ-EGFR (referred as sh-1/sh-2 hereafter) were determined by fluorescence in situ hybridization (FISH) (Fig. 2A, left, middle). Cell fraction q-PCR further validated the cytoplasmic localization of circ-EGFR (Fig. 2A, right). By using an exon and junction probe, we validated the circ-EGFR expression and resistance to RNase-R compared with that of EGFR mRNA (Fig. 2B). Using a panel of cells including NSCs, NHA, SW1783, Hs683, U87, U251, U118 (glioma cell lines with different grade), 387, 4121, 456, 3691, and H2S (BTIC lines), we identified that circ-EGFR was more preferentially expressed in BTICs. The circ-EGFR expression level was approximately 50–1000 times higher in BTICs than in other cells, which is more striking differential expression than the linear EGFR mRNA level (Fig. 2C and Supplementary Figure 1C). We also determined circ-EGFR and EGFR mRNA levels in a cohort of 97 GBM samples and their paired adjacent normal brain (NB). Both circ-EGFR and EGFR mRNA were more highly expressed in GBM than in paired NB (Fig. 2D). In a recently reported circRNA database,13 circ-EGFR was also seen in in multiple cancers (Supplementary Figure 1D). Patients with higher circ-EGFR (higher than the median expression level) had worse prognosis than those with lower circ-EGFR (Fig. 2E, left). Moreover, circ-EGFR level was a better indicator to predict worse overall survival of GBM patients than EGFR mRNA (Fig. 2E, right). These data supported that circ-EGFR was overexpressed in BTICs/GBM and predicted worse overall survival of GBM patients.
Fig. 2.
Characterization of circ-EGFR in glioblastoma (GBM). (A). Left, circ-EGFR junction probe, 2 junction shRNAs and a scramble shRNA. Middle, fluorescence in situ hybridization (FISH) with junction probes was used to decide the subcellular localization of circ-EGFR in 456 brain tumor-initiating cells (BTICs). circ-EGFR shRNA1 and circ-EGFR shRNA2 were used to show the specificity. Scale bars, 20 μM. Right, cytoplasmic and nuclear fractions were isolated to determine circ-EGFR and linear EGFR. β-actin and U6 were used as cytoplasmic or nuclear markers. (B). Left, detection of linear EGFR mRNA and circ-EGFR with exon probe plus RNAse R treatment. Right, detection of circ-EGFR with junction probe. (C). Relative circ-EGFR (left) and linear EGFR (right) mRNA levels in indicated cell lines. (D). Relative circ-EGFR and linear EGFR mRNA levels of GBM and paired adjacent normal tissues in a cohort of 97 GBM patients. (E). Left, 97 patients in the cohort were divided into 2 groups according to relative circ-EGFR expression. The overall survival time of each group was calculated, Right, GBM patient overall survival based on linear EGFR mRNA expression in above cohort. Lines show the mean ± SD. *P < .05, **P < .01, ***P < .001. Data are representative of 2–3 experiments with similar results.
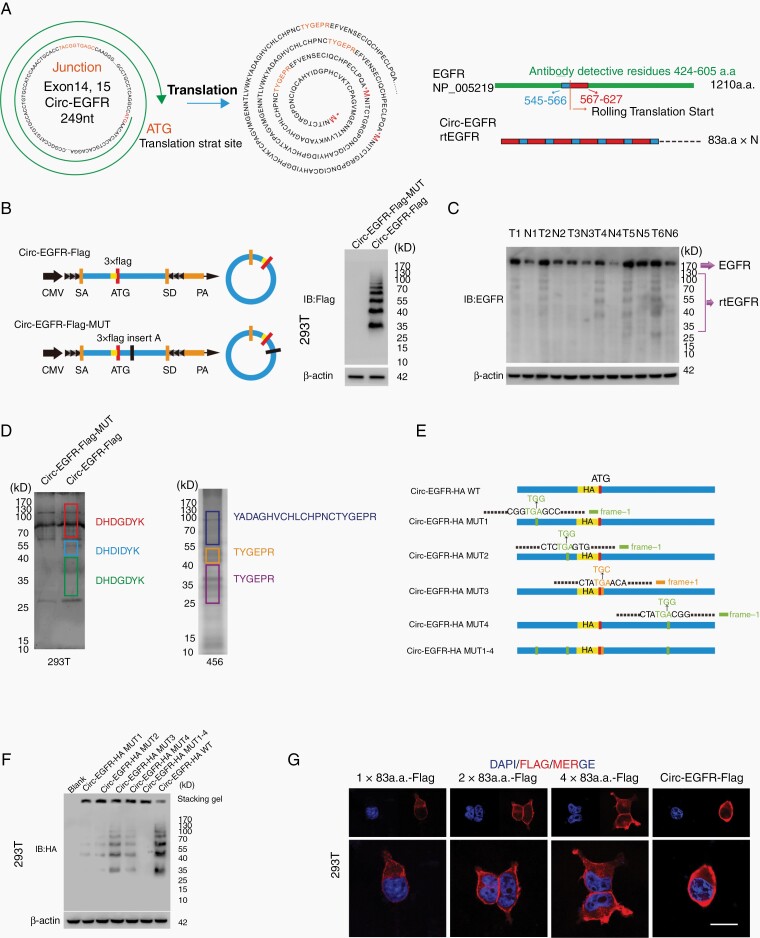
We have previously described several circRNA-encoded proteins in GBM.18,19 To explore whether circ-EGFR could also be translated, we first analyzed potential ORFs in circ-EGFR. Circ-EGFR contained an iORF (Fig. 3A, left) that started with “ATG” but had no in-frame stop-codon, and this iORF theoretically translated an endless protein composed by repeating 83 amino acid sequences. Of these 83 amino acids, 60 in the N-terminal sequence were identical to amino acids 567–627 of EGFR, and the remaining 22 amino acids in the C-terminus were identical to amino acids 545–566 of EGFR (Fig. 3A, right) due to the “rolling translation” of this iORF. To assess the translation potential of circ-EGFR, we transfected circ-EGFR or circ-EGFR with start codon ATG deletion (noATG) into 293T cells. A ribosome enrichment assay with the monosome (M), light polysome (L) and heavy polysome (H) is shown (Supplementary Figure 2A, upper). Circ-EGFR was mainly detected in M and L fractions instead of H fractions. In contrast, EGFR mRNA was mainly localized in H fractions. The noATG circ-EGFR reduced the ribosomal distribution of circ-EGFR but not EGFR mRNA, indicating that circ-EGFR could be translated (Supplementary Figure 2A, lower).
Fig. 3.
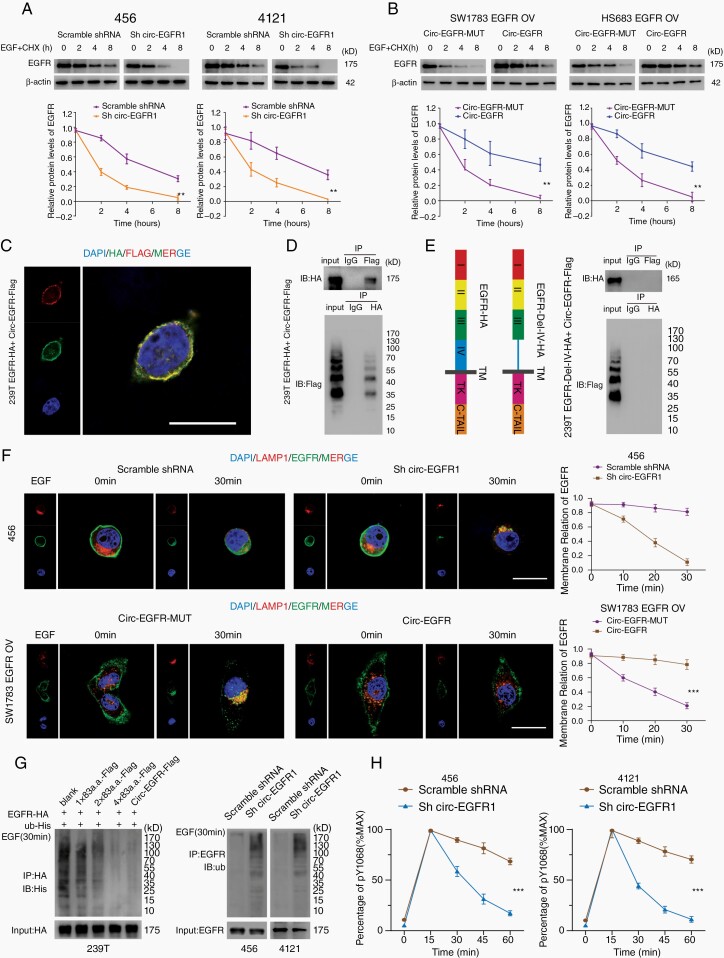
Circ-EGFR encoded rolling-translated EGFR (rtEGFR). (A). Upper, the putative iORF in circ-EGFR and the sequences. Lower, illustration of EGFR sequence and rtEGFR sequence. The antibody used in the study recognized 424–605 a.a. of EFGR. (B). Left, illustration of Circ-EGFR-Flag and Circ-EGFR-Flag-MUT. Right, rtEGFR expression was confirmed by immunoblotting (IB) using Flag antibody in Circ-EGFR-Flag and Circ-EGFR-Flag-MUT transfected 293T. (C). Endogenous rtEFGR expression was detected in paired glioblastoma (GBM) samples by using EGFR antibody. (D). Left, the differential gel bands 30–40 kD, 40–55 kD, and 70–170 kD from Circ-EGFR-Flag and Circ-EGFR-Flag-MUT transfected 293T cells were cut and subjected to LC–MS/MS separately. The identified 3xFlag sequences are shown. Right, the differential gel bands 30–40 kD, 40–55 kD, and 70–170 kD from 456 cells were cut and subjected to LC–MS/MS separately. rtEGFR junction-specific peptides are shown. (E). Illustration of circ-EGFR-HA-WT, circ-EGFR-HA-MUT1, circ-EGFR-HA-MUT2, circ-EGFR-HA-MUT3, circ-EGFR-HA-MUT4, and circ-EGFR-HA-MUT1-4 plasmids. Out of frame stop codons (OSC) are shown (green for −1 frame, orange for +1 frame). (F). Total protein from circ-EGFR-HA-WT, circ-EGFR-HA-MUT1, circ-EGFR-HA-MUT2, circ-EGFR-HA-MUT3, circ-EGFR-HA-MUT4 and circ-EGFR-HA-MUT1-4, circ-EGFR-HA-WT transfected 293T cells was evaluated by immunoblotting using HA antibody. Stacking gel was preserved and transferred to detected extra-large proteins. (G). 293T cells were transfected with 1×83a.a.-Flag, 2×83a.a.-Flag, 4×83a.a.-Flag or Circ-EGFR-Flag. Immunofluorescence using anti-Flag was performed. Scale bars, 20 μM. Lines show the mean ± SD. ***P < .001. Data are representative of 2–3 experiments with similar results.
To validate whether this iORF was actively translated by “rolling-translation” in vivo, we generated a 3xFlag-tagged-circ-EGFR vector (circ-EGFR-Flag). 3xFlag-tag was added in front of the “ATG” to ensure the reading frame was intact. In contrast, an adenine was added inside the iORF to compromise the in-frame reading (circ-EGFR-Flag-Mut) (Fig. 3B, left). We transfected these vectors into 293T cells and performed immunoblotting (IB) by using an anti-Flag antibody. We did not detect any band in circ-EGFR-Flag-Mut vector-transfected 293T cells, indicating that the compromised iORF could not be translated. We also did not detect an extreme large “endless” protein in circ-EGFR-Flag vector-transfected 293T cells. Instead, several “ladder-shaped” bands were observed (Fig. 3B, right). We termed these “ladder-shaped” bands as rolling-translated-EGFR (rtEGFR). To further validate that rtEGFR existed endogenously, we performed IB in NSCs and 456 and 4121 BTICs by using an anti-EGFR antibody. The antigen of this commercial antibody was generated near the 424–605 amino acid sequence of EGFR, which ensured that both EGFR and rtEGFR could be recognized. EGFR and rtEGFR were detected in 456 and 4121 BTICs, while only EGFR was detected in NSCs (Supplementary Figure 2B, left). In contrast, antibody against EGFR outside these domains could not detect rtEGFR in these cells (Supplementary Figure 2B, right). rtEGFR was also identified in 6 paired GBM samples, and cancerous tissues had higher expression levels of rtEGFR than adjacent NB (Fig. 3C). Furthermore, mass spectra (MS) identified 3xFlag-tag sequences from different molecular weight sites in circ-EGFR-Flag-transfected 293T cells (Fig. 3D, left; Supplementary Figure 3A), indicating that rtEGFR was “rolling-translated.” MS also identified part of these 83 amino acid sequences and the circular junction sequences of different molecular weights endogenously in 456 BTIC (Fig. 3D, right; Supplementary Figure 3B).
The most obvious rtEGFR bands were around ~35 kD, 40 kD, 55 kD, 70 kD, etc., which were not integer multiples of the 83 amino acid sequences (~9 kD). To determine whether these “ladder-shaped” rtEGFRs were formed by translation–termination or due to protein purification, we further investigated circ-EGFR sequences. There were 4 out-of-frame stop codon (OSC) “TGAs” in circ-EGFR (Fig. 3E, left).20,21 Viruses, such as HIV-1, can use a mechanism called “programmed -1 ribosomal frameshifting” (-1PRF) to recode Gag-pol from Gag-coding mRNA.22 We hypothesized that -1PRF could also cause ribosomes to encounter an OSC and terminate the iORF in circ-EGFR. To validate this, we generated several synonymous mutated circ-EGFR, in which the OSCs “TGAs” were compromised (Fig. 3E). We found that mutation of all 4 “TGAs” in circ-EGFR (circ-EGFR-mut1-4) could not generate the “ladder-shaped” rtEGFR in 293T cells. Furthermore, compromising the first 2 “TGAs” in circ-EGFR (circ-EGFR-mut1 and circ-EGFR-mut2) also significantly attenuated the “ladder-shaped” bands, while mutation of the last 2 “TGA” (circ-EGFR-mut3 and circ-EGFR-mut4) had lower efficiency to do so (Fig. 3F). Importantly, when we transferred the stacking gel together with the resolving gel, the extremely large “endless” protein was increased in circ-EGFR mut compared with that from circ-EGFR (Fig. 3F). The first 2 “TGA”-induced terminations could compose rtEGFR through 144, 227, 310… amino acid or 153, 236, 319… amino acid repeating sequences, respectively, which is consistent with the rtEGFR molecular weight in IB (Fig. 3F).
As rtEGFR was composed by repeating the core 83 amino acid sequences, we next determined cellular localization of rtEGFR, as well as 1×, 2× and 4× 83 amino acid sequences. IB of the above constructs indicated that only circ-EGFR could generate rtEGFR, further discrediting the notion that “ladder-shaped” bands formed due to protein purification (4× 83 amino acid sequences did not show “ladder-shaped” bands, Supplementary Figure 4A). Using a Flag antibody, we determined the cellular localization of all of the above proteins in 293T cells (Fig. 3G, Supplementary Figure 4B) and showed that these 83 amino acid sequences preferred to stay on the cell membrane.
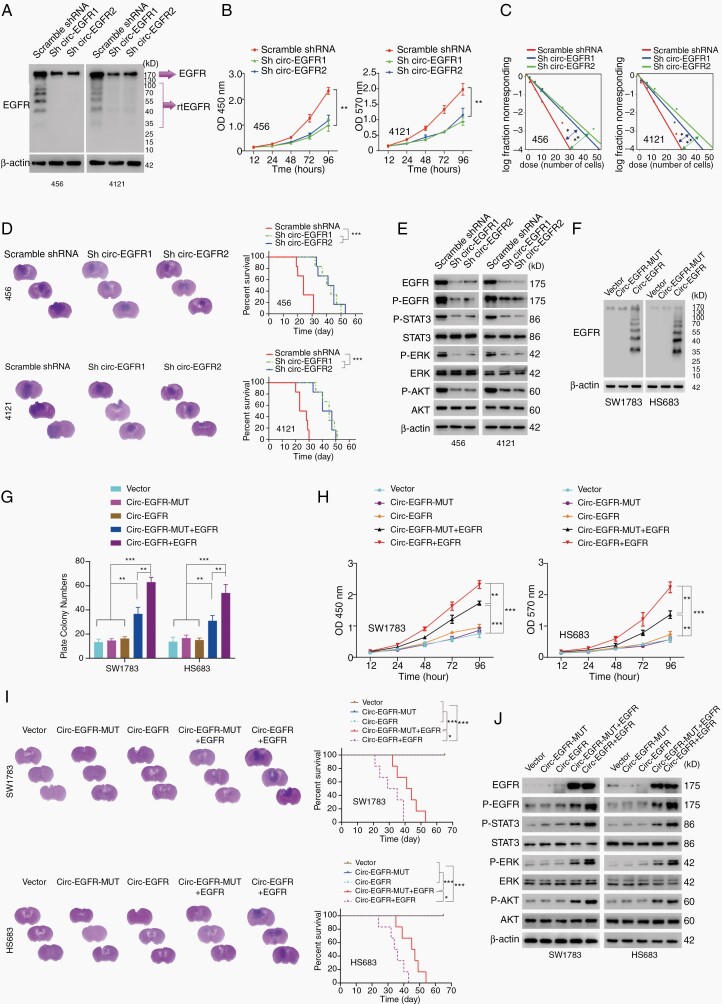
Given the discovery of rtEGFR in BTICs, we next explored its unknown functions. We generated circ-EGFR stable knockdown (KD) 456 and 4121 BTICs with sh-1 and sh-2 as mentioned previously. Circ-EGFR KD did not affect EGFR mRNA levels in either BTIC (Supplementary Figure 5A). In contrast, both rtEGFR and EGFR protein levels were attenuated in 456 and 4121 BTICs (Fig. 4A). As EGFR plays critical roles in BTICs,3,23 we tested several biological characters in 456 and 4121 BTICs with stable circ-EGFR KD. Both modified BTICs exhibited impaired cell proliferation (Fig. 4B), inhibited limited dilution assay (LDA) results (Fig. 4C, Supplementary Figure 5B) and decreased tumorigenicity in vivo (Fig. 4D, left). Mice bearing BTICs with stable circ-EGFR KD exhibited longer overall survival than control groups (Fig. 4D, right). Consistent with downregulated EGFR protein levels, p-EGFR (Y1068), downstream p-STAT3, p-AKT(S308) and p-ERK levels were inhibited in BTICs with circ-EGFR KD (Fig. 4E, Supplementary Figure 5C). The stemness markers, including C-Myc, Nestin, Oct-4, and SOX-2, were also decreased markedly in these modified BTICs, while the expression of differentiation markers Oligo2, GFAP, and β3-tublin was elevated (Supplementary Figure 5D). We also performed RNA-seq in 456 with stable circ-EGFR KD. KEGG pathway enrichment and differential analysis indicated that EGFR signaling, as well as EGFR downstream effectors, were attenuated dramatically after circ-EGFR knockdown (Supplementary Figure 5E).
Fig. 4.
Biological functions of circ-EGFR in glioma cell lines. (A). 456 and 4121 brain tumor-initiating cells (BTICs) were transfected with circ-EGFR shRNA1, 2 or scramble shRNA. (B). Cell proliferation of indicated cells was determined by CCK8. (C) In vitro extreme limiting dilution assays (LDAs) in indicated cells. (D) 456 and 4121 BTICs with indicated modification were intracranially injected into nude mice (1 × 105 cells, per mice, 6 mice per group). Representative HE staining of brain sections in each experimental group is shown. Survival analysis was calculated by Kaplan–Meier curve. (E). The expression of the downstream signaling pathway of EGFR was determined by IB in 456 and 4121 BTICs with indicated modifications. (F). SW1783 and Hs683 glioma cells were transfected with circ-EGFR-MUT, circ-EGFR, or empty vector. The rolling-translated EGFR (rtEGFR) expression was confirmed by immunoblotting. (G). Plate colony formation of vector, circ-EGFR-MUT, circ-EGFR, circ-EGFR-MUT+EGFR, and circ-EGFR+EGFR transfected SW1783 and Hs683. (H). Cell proliferation of vector, circ-EGFR-MUT, circ-EGFR, circ-EGFR-MUT+EGFR, and circ-EGFR+EGFR transfected SW1783 and Hs683. (I). SW1783 and Hs683 cells with vector, circ-EGFR-MUT, circ-EGFR, circ-EGFR-MUT+EGFR, and circ-EGFR+EGFR overexpression were intracranially injected into nude mice (1 × 105 per mice, 6 mice per group). Representative HE staining of brain sections in each experimental group is shown. Survival analysis was calculated by Kaplan–Meier curve. (J) The expression of EGFR downstream signaling was determined by Western blotting in vector, circ-EGFR-MUT, circ-EGFR, circ-EGFR-MUT+EGFR, and circ-EGFR+EGFR transfected SW1783 and Hs683. Lines show the mean ± SD. *P < .05, **P < .01, ***P < .001. Data are representative of 2–3 experiments with similar results.
To further investigate whether circ-EGFR or rtEGFR exerted the above-described functions, we generated circ-EGFR overexpression (OV) SW1783 and Hs683 anaplastic astrocytoma cells. Circ-EGFR and circ-EGFR Mut OV did not alter EGFR mRNA levels in either SW1783 or Hs683 cells (Supplementary Figure 5F). Only circ-EGFR, instead of circ-EGFR-Mut, translated rtEGFR in the above 2 cell lines (Fig. 4F). Compared with circ-EGFR-Mut plus EGFR, circ-EGFR plus EGFR promoted plated colony formation, cell proliferation, in vivo tumorigenesis and shortened the overall survival of mice administered SW1783 and Hs683 cells, indicating circ-EGFR had a synergistic effect with EGFR (Fig. 4G–J, Supplementary Figure 5G). Mechanistically, compared to circ-EGFR Mut plus EGFR, circ-EGFR plus EGFR enhanced the expression of EGFR, p-EGFR, p-STAT3, p-AKT, and p-ERK (Fig. 4J, Supplementary Figure 5H). EGFR-Y5F kinase-dead mutants24 could not activate EGFR downstream signaling, indicating rtEGFR mainly promote EGFR activation (Supplementary Figure 5I).
As circ-EGFR did not affect EGFR mRNA, we then tested the half-life of EGFR in 456 and 4121 BTICs with circ-EGFR stable KD. Compared with the control, circ-EGFR KD reduced the EGFR half-life in both BTICs (Fig. 5A, Supplementary Figure 6A). In contrast, the half-life of EGFR in SW1783 and Hs683 cells was much longer with circ-EGFR and EGFR co-OV (Fig. 5B, Supplementary Figure 6B). rtEGFR repeatedly covered the amino acid sequence 567–627 of EGFR. These amino acids belong to part of extracellular domain IV and cross membrane domain, which is essential for the formation of homolog dimers of EGFR.25 We hypothesized that rtEGFR would also interact with EGFR through extracellular domain IV. We coexpressed EGFR-HA and circ-EGFR-Flag vector in 293T cells, and colocalization of EGFR/rtEGFR was observed on the cell membrane (Fig. 5C). Immunoprecipitation (IP) further confirmed that circ-EGFR-Flag and EGFR-HA interacted with each other; and 1×, 2×, and 4× amino acid sequences also interacted with EGFR (Fig. 5D, Supplementary Figure 6C). Next, we constructed an EGFR vector in which extracellular domain IV was deleted (EGFR-Del-IV-HA) (Fig. 5E, left). EGFR-Del-IV-HA abolished the interaction of EGFR to rtEGFR, indicating that extracellular domain IV of EGFR was required for EGFR/rtEGFR interaction (Fig. 5E, right). To address which amino acid decided rtEGFR-EGFR interaction, we noticed that I545, I556, 1562, and 1592 in EGFR Domain IV were critical for EGFR multimeration.26 These 4 amino acids were all localized in rtEGFR (I3, I14, I20, and V50 in rtEGFR). We mutated these amino acids in circ-EGFR (circ-EGFR-4 mut KKRE-Flag, I3K, I14K, I20R, and V50E) and performed co-IP with EGFR-HA. Mutation of these amino acids abolished rtEGFR and EGFR interaction, validated the critical sites for rtEGFR/EGFR binding (Supplementary Figure 6D).
Fig. 5.
Rolling-translated EGFR (rtEGFR) sustained EGFR membrane localization and prevented EGFR degradation. (A). Upper panel, 456 and 4121 brain tumor-initiating cells (BTICs) with indicated modifications were treated with EGF and cycloheximide. Total lysates at the indicated time points were collected, and indicated proteins were evaluated by immunoblotting. Lower panel, semi-quantification of EGFR protein levels in above immunoblotting. (B). Upper panel, SW1783 and Hs683 glioma cells with indicated modifications were treated with EGF and cycloheximide. Total lysates at the indicated time points were collected, and indicated proteins were determined by immunoblotting. Lower panel, semi-quantification of EGFR protein levels in above immunoblotting. (C). EGFR-HA and circ-EGFR-Flag vector were transfected into 239T cells, and immunofluorescence was performed using anti-Flag and anti-HA antibodies. Scale bar, 20 μm. (D). Mutual interaction of EGFA-HA and circ-EGFR-Flag was determined by IP in 293T cells. (E). Left, illustration of EGFR-HA and EGFR-Del-IV-HA plasmids. Right panel, in vivo interaction of EGFR-HA and circEGFR-Flag was detected by IP in 293T cells. (F). Immunofluorescence using anti-EGFR (green) and anti-lamp1 (red) was performed to show EGFR cellular localization after EGF stimulation in 456 BTICs or SW1783 with indicated modifications. Scale bars, 20 μM. (G) Left, HA-tagged-EGFR and His-tagged-Ub were cotransfected with 1×83a.a.-flag, 2×83a.a.-flag, 4×83a.a.-flag or circ-EGFR-Flag in 293T cells. IP was performed, followed by IB using the indicated antibodies. Right, Ub levels were detected in 456 and 4121 BTICs with circ-EGFR knockdown and their control cells, followed by IP using the indicated antibodies. (H). Quantitation of EGFR phosphorylation time courses, normalized by signal at 15 min. Lines show the mean ± SD. *P < .05, **P < .01, ***P < .001. Data are representative of 2–3 experiments with similar results.
EGFR level is dynamically regulated through endocytosis and follows degradation.27 As rtEGFR directly interacts with EGFR on the cell membrane, we next explored whether rtEGFR reduced EGFR endocytosis. We found that 456 BTICs with circ-EGFR stable KD exhibited enhanced endocytosis of EGFR. In contrast, circ-EGFR OV in SW1783 cells prolonged EGFR membrane localization (Fig. 5F). Endocytic EGFR was sorted by early endosome, either recycled to membrane via the recycling compartment or ubiquitinated and degraded in lysosomes.28 Given that rtEGFR enhanced EGFR stability, we suspected that rtEGFR also decreased EGFR ubiquitination. Indeed, rtEGFR, as well as the 1×, 2×, and 4× amino acid sequences, inhibited EGFR ubiquitination in 293T cells (Fig. 5G, left). Among these proteins, rtEGFR had the strongest effect. In 456 and 4121 BTICs with circ-EGFR stable KD, EGFR ubiquitination was markedly promoted (Fig. 5G, right). Using an EGFR-Y1068 kinase activity assay, we also showed that circ-EGFR stable KD in both BTICs decreased p-EGFR activity (Fig. 5H).
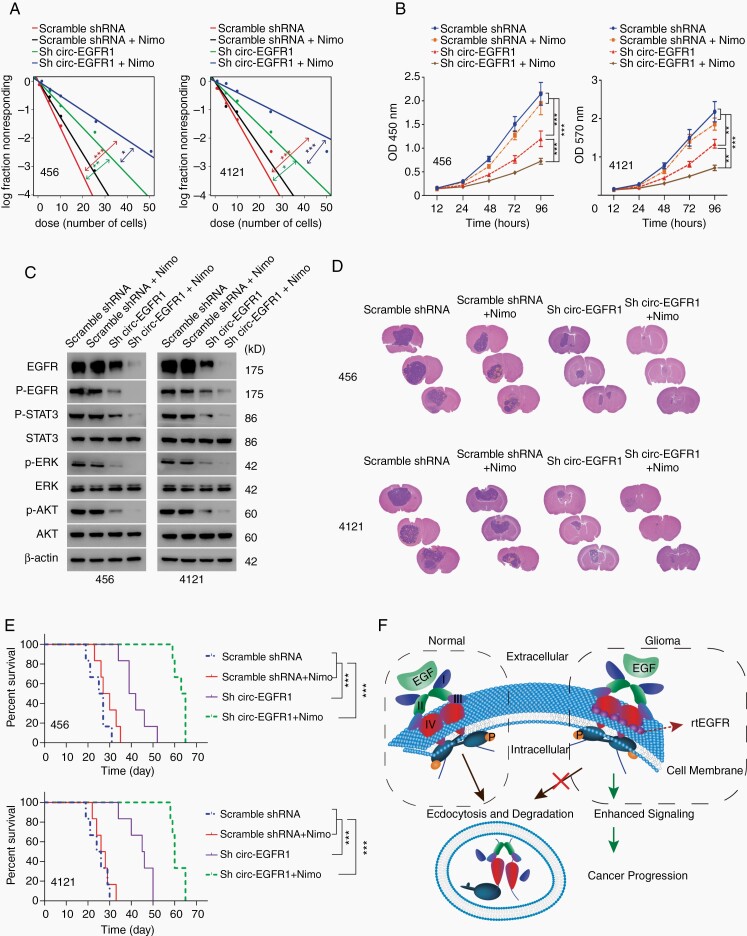
We next explored whether targeting rtEGFR had clinical implications. Nimotuzumab, a clinically used EGFR antibody, did not show the desired effect in GBM treatment29 (Fig. 6A–C). Nimotuzumab treatment only partially inhibited p-EGFR, p-AKT, p-STAT3, and p-ERK, supporting redundant activation mechanisms for EGFR signaling (Fig. 6C, Supplementary Figure 6E).30 In sharp contrast, circ-EGFR KD plus nimotuzumab drastically inhibited EGFR signaling activation (Fig. 6C). In mouse xenografts intracranially planted with 456 and 4121 BTICs, nimotuzumab did not showed satisfactory results and had limited survival benefits compared with control groups. However, in combination with circ-EGFR KD, nimotuzumab dramatically inhibited EGFR signaling activation and prolonged overall survival, demonstrating the promising clinical implication of targeting rtEGFR in clinical GBM (Fig. 6D and E; Supplementary Figure 6F).
Fig. 6.
Circ-EGFR knockdown enhanced the therapeutic effect of nimotuzumab in glioblastoma (GBM). (A). Neurosphere-forming capability in indicated cells. (B). Cell proliferation of the indicated groups was determined by CCK8. (C). The expression of EGFR downstream signaling was determined in each group. (D). Representative HE staining in each experimental group are shown. (E). Survival analysis was conducted with the Kaplan–Meier curve in indicated groups. (F). Illustration of circ-EGFR function. Normally, after activation by EGF, the ubiquitylated EGFR was followed by endocytosis and degradation. In brain tumor cells where rolling-translated EGFR (rtEGFR) was abundantly expressed, rtEGFR formed a complex with EGFR and prevented its endocytosis. This disrupted the normal downregulation of the EGFR, extended it signaling lifespan and promoted tumorigenesis. Lines show the mean ± SD. *P < .05, **P < .01, ***P < .001. Data are representative of 2–3 experiments with similar results.
Discussion
A subset of circRNAs were found to be upregulated in cancer, indicating that circRNAs were functionally diverse and that their clinical implications remained to be investigated.31EGFR pre-mRNA formed several circRNAs. Among these circRNAs, circ-EGFR had the highest differential expression between BTICs and normal cells. Mmu_circ_0002861, which is the homolog of circ-EGFR in mice, was reported to promote granulosa cell proliferation by modulating miR-125a-3p.32 Circ-EGFR was conserved between human and mouse.33 Their identical nucleotide sequences and similar biological functions implied that rtEGFR also exert a similar function in mice, which could be further validated by circRNA-specific KO.
Functional and endogenous “rolling-translated” product has not previously been reported. Infinite GFP-ORF was used to show that circRNA translated extremely long repeating sequences in vivo.14 A multiple tandem Flag-tag-iORF in circRNA was also constructed to verify the “rolling translation” in eukaryotic cells.13 These reports provided strong evidence that “rolling translation” is an efficient way to produce proteins. But due to the lack of endogenous examples, whether iORF had certain functions remained elusive. More recently, circRtn4 was reported to generate “multimer” products through “infinite translation”.34 However, circRtn4 was also artificially generated, as several intron enhancers were added. Our discovery advanced the understanding of “rolling translation” in several ways. First, we indicated that “rolling translation” existed in vivo. Second, we showed that “rolling-translated” circRNAs could also be terminated. Third, we demonstrated that the “rolling-translated” product had a biological function. The cross-membrane amino acid sequence of rtEGFR could act like a “screw washer” to interact and reinforce EGFR membrane localization, while further structure-based studies are required (Fig. 6F).
Efforts aimed at targeting EGFR in combination with other approaches have been proven to be ineffective for GBM treatments.3 Among the plausible mechanisms, multiple EGFR ligands, redundant downstream signaling, multiple RTKs, and feedback activation have been attributed to therapeutic resistance.2,3 We described an unrevealed mechanism of aberrant EGFR activation in GBM, which is at least in part responsible for EGFR-targeting therapy resistance. The unique expression level of circ-EGFR may provide a wide therapeutic window for targeting BTICs without systematic toxicity. This study not only advanced our understanding of the molecular basis underlying an unknown circRNA translation model but also suggested a novel targeting approach for treating malignant GBM.
Supplementary Material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Acknowledgments
This study was approved by the IRB of the First Affiliated Hospital of Sun Yat-sen University.
Data availability
Raw sequencing and processed RNA Seq data from this study have been deposited into NCBI SRA: PRJNA525736.
Funding
This work was supported in part by the National Key Research and Development Program of China (2016YFA0503000 to N.Z.), the Natural Science Outstanding Youth Foundation of China (81822033 to N.Z.), the National Natural Science Foundation of China (81772683 to N.Z., 81873763 to Z.X.), the Guangdong Basic and Applied Basic Research Foundation (2020A1515010274 to Y.L.), and the Science and Technology Program Key Project of Guangzhou (201603010006 to Z.X.).
Conflict of interest statement. There are no potential conflicts of interest to disclose.
Author contributions. Conceptualization, N.Z.; Data production, analysis, and investigation, Y.L., Z.L., M.Z., H.Z., X. W., J.Z., F.X., N.H., X.Y., R. Z., L. Y., and Z. X.; Writing, review, and editing, N.Z., Z.X., Y.L., and Z.L.; Supervision, N.Z.; Funding acquisition, N.Z., L.Y., and Z.X.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 2. Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15(5):302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Westphal M, Maire CL, Lamszus K. EGFR as a target for glioblastoma treatment: an unfulfilled promise. CNS Drugs. 2017;31(9): 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. An Z, Aksoy O, Zheng T, Fan QW, Weiss WA. Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies. Oncogene. 2018;37(12):1561–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stasyk T, Huber LA. Spatio-temporal parameters of endosomal signaling in cancer: implications for new treatment options. J Cell Biochem. 2016;117(4):836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zahonero C, Sánchez-Gómez P. EGFR-dependent mechanisms in glioblastoma: towards a better therapeutic strategy. Cell Mol Life Sci. 2014;71(18):3465–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim JK, Lee SY, Park CW, et al. Rab3a promotes brain tumor initiation and progression. Mol Biol Rep. 2014;41(9):5903–5911. [DOI] [PubMed] [Google Scholar]
- 8. Portela M, Segura-Collar B, Argudo I, et al. Oncogenic dependence of glioma cells on kish/TMEM167A regulation of vesicular trafficking. Glia. 2019;67(2):404–417. [DOI] [PubMed] [Google Scholar]
- 9. Wang W, Zhang H, Liu S, et al. Internalized CD44s splice isoform attenuates EGFR degradation by targeting Rab7A. Proc Natl Acad Sci U S A. 2017;114(31):8366–8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. [DOI] [PubMed] [Google Scholar]
- 11. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. [DOI] [PubMed] [Google Scholar]
- 12. AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci U S A. 2014;111(40):14542–14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abe N, Matsumoto K, Nishihara M, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perriman R, Ares M Jr. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA. 1998;4(9):1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zerbino DR, Achuthan P, Akanni W, et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xia X, Li X, Li F, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. [DOI] [PubMed] [Google Scholar]
- 20. Ketteler R. On programmed ribosomal frameshifting: the alternative proteomes. Front Genet. 2012;3:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tse H, Cai JJ, Tsoi HW, Lam EP, Yuen KY. Natural selection retains overrepresented out-of-frame stop codons against frameshift peptides in prokaryotes. BMC Genomics. 2010;11:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X, Xuan Y, Han Y, et al. Regulation of HIV-1 gag-pol expression by shiftless, an inhibitor of programmed -1 ribosomal frameshifting. Cell. 2019; 176(3):625–635 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frattini V, Trifonov V, Chan JM, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45(10):1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deng W, Poppleton H, Yasuda S, et al. Optimal lysophosphatidic acid-induced DNA synthesis and cell migration but not survival require intact autophosphorylation sites of the epidermal growth factor receptor. J Biol Chem. 2004;279(46):47871–47880. [DOI] [PubMed] [Google Scholar]
- 25. Arkhipov A, Shan Y, Das R, et al. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152(3):557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Y, Bharill S, Karandur D, et al. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. Elife. 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274(5295): 2086–2089. [DOI] [PubMed] [Google Scholar]
- 28. Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24(1):26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Solomón MT, Selva JC, Figueredo J, et al. Radiotherapy plus nimotuzumab or placebo in the treatment of high grade glioma patients: results from a randomized, double blind trial. BMC Cancer. 2013;13:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eskilsson E, Røsland GV, Solecki G, et al. EGFR heterogeneity and implications for therapeutic intervention in glioblastoma. Neuro Oncol. 2018;20(6):743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019; 176(4):869–881 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jia W, Xu B, Wu J. Circular RNA expression profiles of mouse ovaries during postnatal development and the function of circular RNA epidermal growth factor receptor in granulosa cells. Metabolism. 2018;85:192–204. [DOI] [PubMed] [Google Scholar]
- 33. Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. [DOI] [PubMed] [Google Scholar]
- 34. Mo D, Li X, Raabe CA, et al. A universal approach to investigate circRNA protein coding function. Sci Rep. 2019;9(1):11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data from this study have been deposited into NCBI SRA: PRJNA525736.
More detailed methods are described in Supplementary data.
Raw sequencing and processed RNA Seq data from this study have been deposited into NCBI SRA: PRJNA525736.