Abstract
For decades, cell biologists and cancer researchers have taken advantage of non-murine species to increase our understanding of the molecular processes that drive normal cell and tissue development, and when perturbed, cause cancer. The advent of whole-genome sequencing has revealed the high genetic homology of these organisms to humans. Seminal studies in non-murine organisms such as Drosophila melanogaster, Caenorhabditis elegans, and Danio rerio identified many of the signaling pathways involved in cancer. Studies in these organisms offer distinct advantages over mammalian cell or murine systems. Compared to murine models, these three species have shorter lifespans, are less resource intense, and are amenable to high-throughput drug and RNA interference screening to test a myriad of promising drugs against novel targets. In this review, we introduce species-specific breeding strategies, highlight the advantages of modeling brain tumors in each non-mammalian species, and underscore the successes attributed to scientific investigation using these models. We conclude with an optimistic proposal that discoveries in the fields of cancer research, and in particular neuro-oncology, may be expedited using these powerful screening tools and strategies.
Keywords: Drosophila, Zebrafish, Worms, C elegans, brain tumor, signaling pathways, high-throughput screening
Our understanding of the origins of human brain tumors has advanced considerably in the era of molecular biology and advanced genomics.1,2 Compared to other areas of cancer research, however, the field of neuro-oncology has seen less headway due to challenges such as: overcoming the blood-brain barrier (BBB), drug-efflux pumps, brain tumor location precluding gross total resection, the complex brain tumor micro-environment, and the paucity of reliable pre-clinical models.3,4
Murine models of human brain tumors are the workhorse of the modern neuro-oncology laboratory, and they have been instrumental in elucidating the biology of human brain tumors, identifying new therapeutic targets, and evaluating the effectiveness of novel treatments. While these models offer a powerful, reproducible, and sustainable system for studying human cancers, they also have several limitations. Chief among these include the reliance on immunodeficient mice, the use of human brain tumor implants or cell lines within a murine-based system, and the failure to recapitulate intratumoral heterogeneity within genetically engineered mice.3 Furthermore, mouse colonies are resource-intensive, requiring substantial cost and time investment, tedious experimental protocols, and lengthy ethical approval processes.3,5 Consequently, alternate model systems that enable high-throughput investigation are needed to ensure the rapid advancement of preclinical discoveries in the contemporary neuro-oncology era.
Simpler and more genetically tractable model organisms such as Drosophila melanogaster (Drosophila), Caenorhabditis elegans (C. elegans), and Danio rerio (Zebrafish) represent valuable alternatives to mouse models. In recent years, a resurgence of interest in non-murine modeling has led to the identification of new genes, therapeutic targets, and technologies for high-throughput genetic and chemical manipulation.
Advantages of Modeling Brain Tumors in Non-Mammalian Species
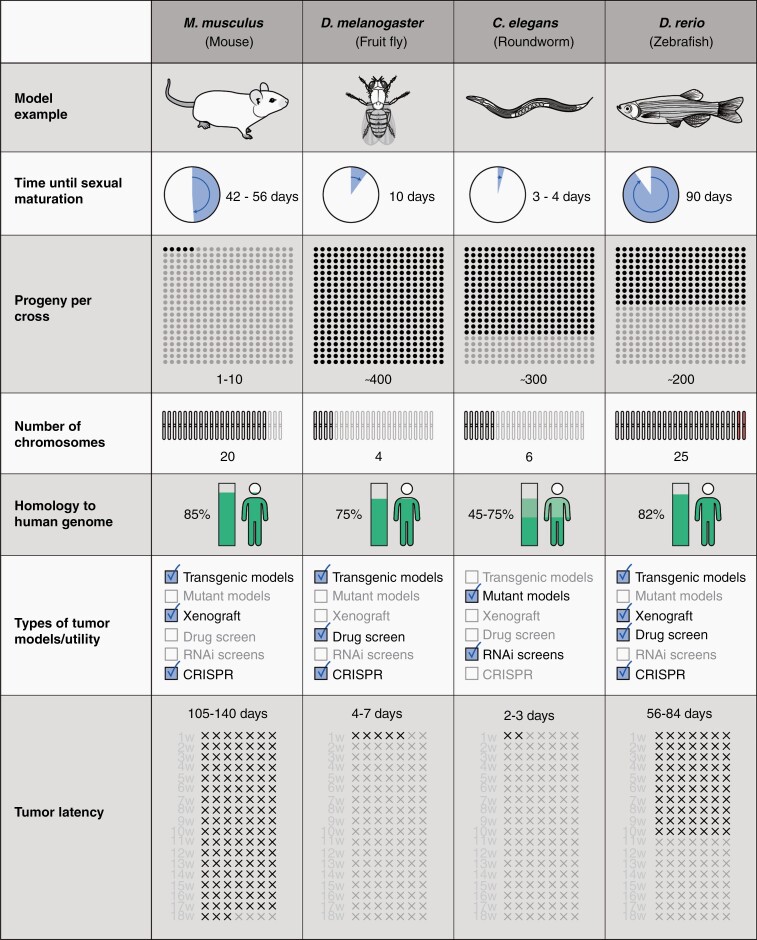
Multiple cancer models have been generated and validated using non-murine organisms. Notably, the use of non-murine species to model human brain tumors has several advantages over mammalian models (Fig. 1). The functional conservation of genes and pathways, coupled with powerful genetics and cell biology techniques makes D. melanogaster, C. elegans, and D. rerio invaluable in vivo systems to model human brain cancers.6–8 These organisms share a high degree of evolutionary conservation with the human genome.6–10 In addition, D. melanogaster and D. rerio share physiological characteristics with humans, including the development of discrete organ systems and representation of the BBB, making them advantageous for the generation of brain tumor models.7,11 Despite the differences in glial cell specification for the myelination process, there is considerable homology between the morphology, function, and progenitor cell development and differentiation of neural and glial cells of these species and vertebrates.12–14 These biological similarities enable investigation of the tumor and its microenvironment, while their simplicity facilitates tracking of tumor development and direct drug delivery.
Fig. 1.
Comparison of brain tumor models in different species.
Additionally, the maintenance of these organisms is comparatively inexpensive and less resource-intensive compared to complex animal models. Furthermore, D. melanogaster, C. elegans, and D. rerio all have high fecundity and undergo rapid sexual maturation.15 Not only does this expedite large-scale, high-throughput screening, it also allows for rapid genetic crosses and the development of transgenic models that are typically time-consuming to establish in mammalian models. Here, we describe these three model organisms and their feasibility in understanding the molecular underpinnings of various human brain tumors.
Drosophila melanogaster
The fruit fly, D. melanogaster, is the most impactful and robust biological model system to inform our understanding of modern genetics and development over the past 100 years. The remarkable genetic similarity between Drosophila and humans, with up to 75% of disease-related human genes having homologs in the fly genome, ensures that D. melanogaster biology will continue to accelerate our understanding of human diseases for years to come.7
D. melanogaster as a Model for Cancer
Several features of D. melanogaster have been exploited to investigate various hallmarks of cancer. Williams and colleagues used the single-hit model of cancer to investigate the relationship between neurofibromin and the Ras signaling pathway, leading to a greater understanding of tumor initiation.16–19 In colorectal cancer, Drosophila was paramount in the discovery that Rb can act as an oncogene through its repression of E2F1 resulting in the unsuppressed activity of the β-catenin signaling pathway.20 Additionally, the Drosophila CNS houses six types of glia which share developmental and functional characteristics with mammals.12 Although a recent entry into the field of oncology, the foundational genetic discoveries in D. melanogaster have been instrumental in our understanding of human brain tumors.21
Life Cycle
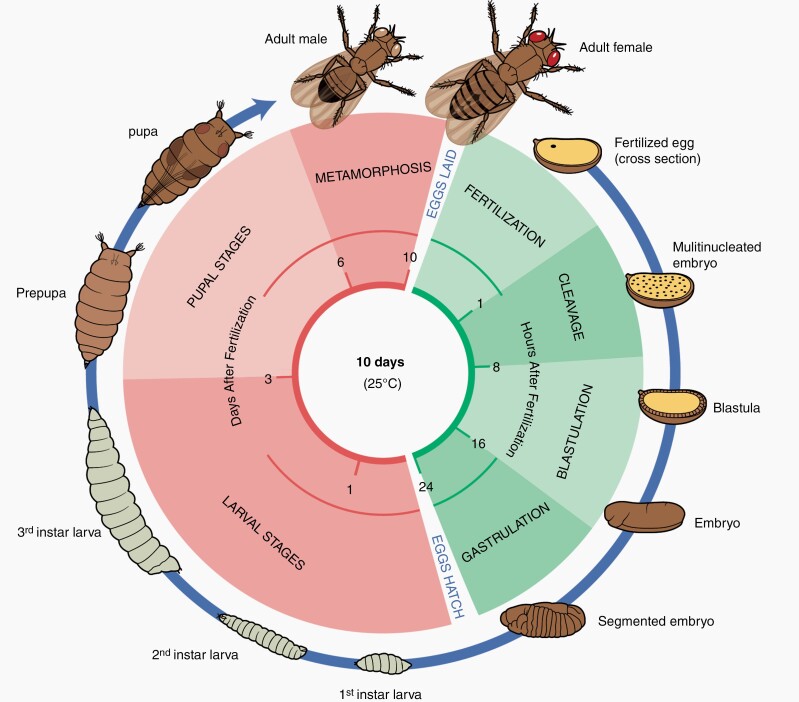
D. melanogaster has a considerably short life cycle, completing a revolution in approximately 10 days (Fig. 2). Drosophila has a total of three larval stages, called “instars,” each associated with distinct morphological and behavioral characteristics which progress through sequentially over 72 -96-hour period. At the end of the third instar, the larva migrates away from its source of food and initiates the process of pupariation. During the pupal stage, organism-wide structural degradation (histolysis) and tissue changes trigger metamorphosis into the adult fly, which forms one week later at the time of eclosion.22
Fig. 2.
D. melanogaster life cycle. Drosophila completes a life cycle revolution in approximately 10 days at 25°C. Upon fertilization, the embryo completes initial cleavage, blastulation, gastrulation, and hatches into its larval form in 24 h. This is followed by three larval stages, instars, each associated with distinct morphological and behavioral characteristics. Under optimal growth conditions, the larva migrates away (wandering 3rd instar) from its source of food and initiates pupariation. As a pupa, the larva undergoes histolysis which triggers metamorphosis into the adult fly.
Breeding and Genetic Manipulation Strategies
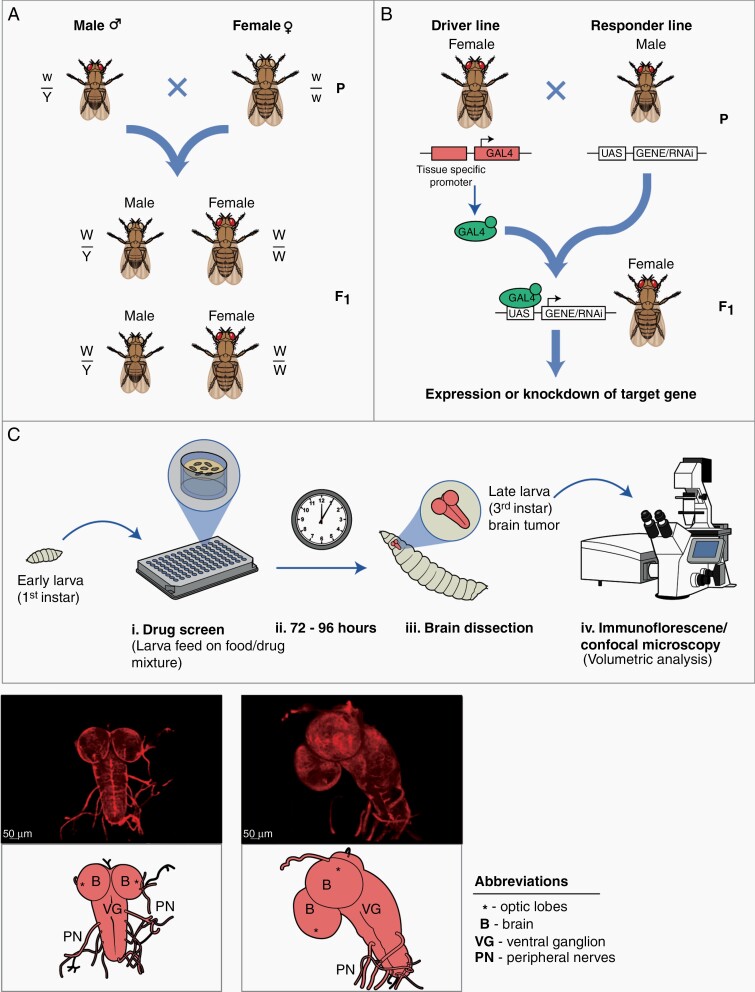
As in all breeding strategies, flies must be selected for the intended phenotypic markers that correspond to the underlying genotype of interest. Mating schemes must, therefore, be carefully designed to ensure that the intended progeny is produced with the correct genotype, often after a series of sequential crosses (Fig. 3A).15,22
Fig. 3.
The utility of Drosophila in brain tumor research. (A) Illustrative cross between a male and female Drosophila on the heterosomal 1st/X chromosome for mutant recessive allele w (white eyes) and dominant wild-type allele W (red eyes). Mendelian inheritance is observed. F1 progeny maintain a 1:1 proportionality of dominant and recessive sex-linked alleles yielding 50% heterozygous and 50% hemizygous recessive. (B) Simplified transgenic cross between a tissue specific Gal4 expressing construct and a UAS expressing construct upstream of the gene of interest. In the F1 progeny, Gal4 binds and activates the UAS enhancer to transcribe downstream genes. Expression of the gene of interest will reflect the tissue specific expression of Gal4. (C) Drug/small molecule screening platform. (i) Early 1st instar larvae are placed into 96 well plate with larval food mixed with a chemical compound of interest. (ii) Larvae consume drug-food mixture over 72–96 h and enter late 3rd instar stage. (iii) Late 3rd instar larvae undergo central nervous system extraction. (iv) Volumetric analysis/drug response can be microscopically evaluated. (D) Non-transformed confocal microscopy (above) and representative illustration (below) of wild-type Drosophila central nervous system with fluorescent UAS-red fluorescent protein reporter. (E) Transformed transgenic (DP110CAAX and dEGFRλ) glioma model as imaged by confocal microscopy (above) with representative illustration (below). Note hypertrophic brain and ventral ganglion reflecting hyperproliferative Drosophila glia.
Perhaps one of the greatest strengths of D. melanogaster as a model is the capacity to interrogate the function of any gene through forward or reverse genetics, typically achieved through large-scale unbiased screens to identify genetic aberrations that augment the cancer process in question.23 A useful technique to manipulate gene expression in Drosophila is the FLP recombinase/FLP recognition target (FLP/FRT) system, which has been widely used to either activate or inactivate genes or cause somatic recombination in homologous chromosomes.24 Genes flanked by FRT are excised by the FLP enzyme. To add additional control, FLP can be placed under a regulatory element or it can be indirectly coupled to the genomic enhancer using the Gal4/Upstream Activating Sequence (UAS) system. Pagliarini and Xu used the expression of activated Ras (UAS- RasV12) to induce benign tumor growth in the larval brain, visualized using a fluorescent tag.25,26
One of the most common D. melanogaster ectopic expression systems is the yeast-derived Gal4/UAS system, which has generated a myriad of transgenic Drosophila lines (Fig. 3B).27 Since Gal4 binds exclusively to UAS, only target genes bearing a fused UAS will be activated. The spatiotemporal expression pattern is additionally regulated through the addition of silencer genes like Gal80 (which represses Gal4 when co-expressed) as well as through the use of combination strategies such as clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9).
Drug Screening and Discovery Strategies
One of the most promising uses of D. melanogaster to model brain tumors is its recent emergence as a tool for drug discovery and high-throughput drug screening.28,29 Provided that drug targets are evolutionarily conserved, Drosophila drug screening platforms can be used as proof of principle in vivo systems to screen panels of putative antineoplastic drugs.30 A schematic of the experimental workflow we use is shown (Fig. 3C). Using Drosophila brain tumor models as a drug-screening tool has advantages over in vitro drug screens, namely the ability to witness novel whole-system drug effects, trialing multiple drug-delivery methods, and to help delineate anti-neoplastic effects versus toxic effects of candidate compounds.
Brain Tumor Models
Glioma Models
Because of the high degree of conservation between the human and Drosophila homologs of phosphoinositide-3 kinase (PI3K) and epidermal growth factor receptor (EGFR), Read and colleagues developed the repoGal4-glial specific model of constitutively active, overexpressed, PI3K/EGFR (DP110CAAX and dEGFRλ).31 This model recapitulates proliferation and disordered growth signaling resulting in neoplastic glia of the bilateral lobes and ventral nerve cord that is detectable as early as the first instar stage of development (Fig. 3D-E). As a foundational model of glioma, multiple versions and adaptations of the Read et al. repo-Gal4 driven PI3K/EGFR model was generated to examine specific malignant processes including, but not limited to: ion channels,32 tumor metabolism,33 potentially therapeutic pathometabolic pathways,34 and signaling pathway convergence between WNT β-catenin and EGF.35 We previously used a Gal4/UAS transgenic line that overexpresses the fly homologs of human PI3K and EGFR in glial cells to model glioma in Drosophila.33 In this study, Drosophila with overexpressed PTEN-induced kinase 1 (PINK1) localized to glial cells resulted in a reduced proliferative phenotype versus controls, indicating a novel role of PINK1 as a suppressor of glioma growth.33
Activating genetic alterations in the B-Raf proto-oncogene (BRAF) is common in low-grade gliomas (LGGs), so we generated a BRAF Drosophila LGG model by expressing constitutively active Raf (RafGOF) in the glia cells.32 FGFR-TACC (transforming acidic coiled-coil containing protein) gene fusions are frequent in human cancers and are found in 3% of GBM cases.36 We generated a Drosophila model by expressing patient-derived FGFR3-TACC3 in the glia cells.37 In RafGOF-, FGFR3-TACC3-, and DP110CAAX; dEGFRλ-driven Drosophila gliomas, we showed that all tumors develop tissue stiffening, a physical hallmark of human gliomas,38 and that the mechanosensitive ion channel Piezo increases tumor tissue stiffness and proliferation.32
Finally, Drosophila has also been used to model glial migration and invasion. Through overexpression of orthologs of PDGFR/VEGFR, FGFR1, and insulin-receptor genes Pvr, htl, and Inr respectively, Witte and colleagues were able to model increased migratory and neoplastic glia in the Drosophila brain.39
Medulloblastoma Models
Current models of medulloblastoma in Drosophila use transgenic lines that induce abnormal embryologic development in pathways such as Sonic Hedgehog (SHH) and WNT.40 These transgenic Drosophila are used in the context of loss-of-function or gain-of-function screens, epigenetic discovery, and large dataset bioinformatic analyses.21,40,41
Caenorhabditis elegans
C. elegans is a free-living soil nematode first described by Sydney Brenner as a simple metazoan model.42 It is a transparent, self-fertilizing hermaphrodite with a short life cycle and a simple genome, consisting of only five pairs of autosomes and one pair of sex chromosomes. The body of the worm consists of 959 somatic cells and lacks a respiratory and circulatory system, but has a nervous system comprised of 302 neurons and fifty-six glial cells.13,43 It feeds on bacteria such as Escherichia coli (E. coli) and can be cultivated in liquid culture or on agar plates, which makes it easy and inexpensive to grow in large numbers.42,44 Since its introduction for biological research, it has been extensively utilized as a model organism for neural development.
C. elegans as a Model for Cancer
In 1998, C. elegans became the first metazoan to have a completely sequenced genome,10 where approximately 52.6% of human protein-coding genes have known orthologs in C. elegans,45 and 40–75% of human disease genes have been predicted to have homologs in the C. elegans genome.46 In addition, many biological processes such as cell signaling, apoptosis, cell polarity, metabolism, and aging, are conserved between human and C. elegans.47
Indeed, the use of C. elegans as a model organism in cancer research has yielded important insights into biological processes known to be dysregulated in cancer, such as cell cycle progression, invasion and migration, growth factor signaling, apoptosis, and genome instability, among others.48,49 As one example, C. elegans was used to demonstrate that apoptosis was a genetically programmed suicide process, which helped elucidate the in vivo function of the anti-apoptotic gene BCL2 in development and cancer.50
Life Cycle
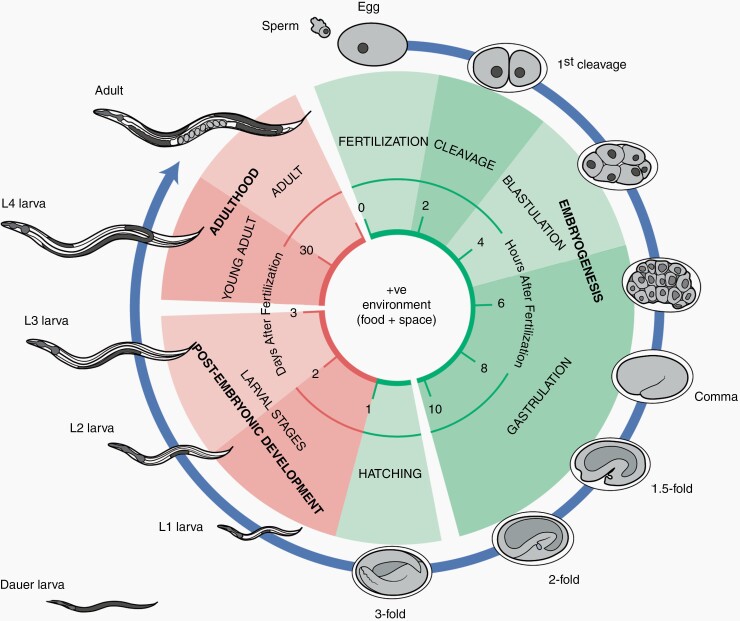
The life cycle of C. elegans is composed of a period of embryogenesis, followed by post-embryonic development consisting of four larval stages (~3 days), and then finally, adulthood (Fig. 4). The hermaphrodite embryo hatches with approximately 558 nuclei,43,51 and by the end of embryogenesis, the main body plan of the worm is established. In unfavorable growth circumstances (e.g., high population density, shortage of food, or high temperature), the worm may enter a non-developmental hibernation phase known as Dauer (L2d), which allows the worm to live up to 4 months in harsh conditions.43,51
Fig. 4.
C. elegans life cycle. C. elegans has a temperature-dependent life cycle of 3–4 days at 20–25°C, and a short lifespan of approximately 2–3 weeks. After fertilization and first cleavage, the embryo undergoes blastulation and gastrulation, which takes 24 h to complete. By the end, the main body of the worm is developed, and the egg is ready to hatch. During the first larval (L1) stage, lasting approximately 12 h, the nervous system, reproductive system, and the digestive tract start to develop. Under favorable conditions and ambient temperature of 20°C, the worm progresses through larval stages L2-L4, and finally into adulthood. However, under unfavorable conditions, such as temperature extremes, high population density, and food scarcity, the L2 larva may choose to enter the Dauer stage (L2d) and enter a non-developmental phase, which allows them to survive up to 4 months under harsh environmental conditions.
Breeding Strategies
C. elegans follows an androdioecious breeding system where hermaphrodites can either self-fertilize or mate with males. A single hermaphrodite can produce approximately 300 progeny by self-fertilization, and over 1000 progeny when mated with a male. Sex determination is based upon the number of X-chromosomes, with hermaphrodites possessing two (X/X), whereas males have only one (X/O). While it is true that all offspring are genetically similar to each other, it is important to note that only 0.1–0.2% of progeny are males.43
Modeling Cancer in C. elegans
C. elegans lacks a complex segmented brain similar to the vertebrates. Instead, its nervous system is comprised of a bundle of neurons and glial cells, with just fifty-eight of the neurons residing in the head.52 In addition, four of the cephalic glia wrap their processes around the nerve ring, akin to how the BBB isolates neurons in the vertebrate brain.13 Although brain tumor models in C. elegans do not exist in the traditional sense, it is possible to model the conserved driver pathways to study the effects on different hallmarks of cancer.
One of the defining characteristics of cancer is cell cycle dysregulation causing uncontrolled cellular proliferation. In C. elegans, the Ras/mitogen-activated protein kinase (MAPK) pathway plays a role in regulating cell proliferation, differentiation, and various cell fate decisions. The insulin growth factor-1 (IGF-1) dependent activation of the MAPK cascade induces vulval development and generates a hyperproliferative multivulva (Muv) phenotype.53,54 Ablation of the daf-2 (IGF1R) gene can suppress the Muv phenotype of let-60 (Kirsten Rat Sarcoma [KRAS]) gain-of-function (gf),54 as well as hypersensitivity to irradiation-induced apoptosis in the germline of let-60(gf) mutants.53 In addition, daf-18 (Phosphatase and tensin homolog [PTEN]) negatively regulates MAPK signaling to control vulval development as well as neuroblast cell divisions in the worm.55 The high degree of functional conservation of these pathways in C. elegans provides opportunities for exploiting the powerful genetics of this organism to understand mechanisms by which Ras and PTEN collaborate to control cell fate decisions.
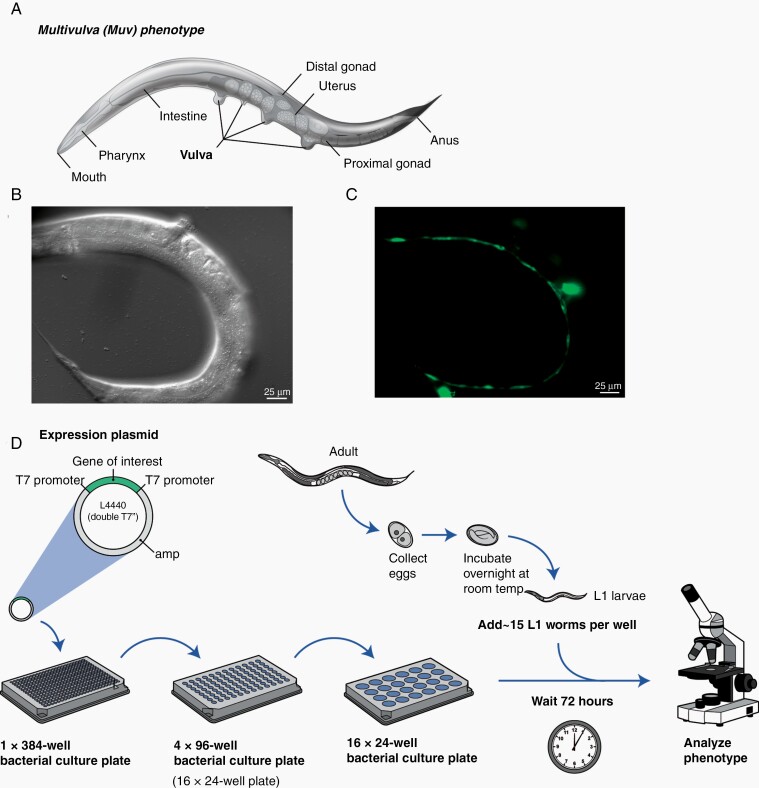
In glioblastoma (GBM), PTEN loss occurs in around 40–50% of primary cases,4 whereas receptor tyrosine kinase/Ras/PI3K pathway is activated in 88% of all gliomas.1,4 In the worm, the Ras signaling pathway regulates vulva cell specification. Increased activity of Ras signaling causes ectopic induction of vulva precursor cell fates that result in a Muv phenotype56 (Fig. 5A). Our lab has generated a let-60 (KRAS); daf-18 (PTEN) double mutant model of glioma, which enhances the Muv phenotype of let-60(gf) to approximately 100%. We also introduced a cdh-3::GFP transgene, which is expressed in the vulva,57 into the daf-18(lf) and let-60(gf) double mutant strain to easily distinguish the Muv phenotype by fluorescence microscopy (Fig. 5B-C). The combination of activated let-60 (KRAS) and loss of daf-18 (PTEN) offers a genetically tractable animal model to study synergy between the most commonly altered signaling pathways in gliomas.
Fig. 5.
C. elegans in brain tumor research. (A, B) KRAS; PTEN (let-60; daf-18) double mutation in C. elegans results in a strain that exhibits a nearly 100% penetrant multivulva phenotype. (C) The KRAS; PTEN double mutant strain also expresses a cdh-3::GFP reporter in the vulva cells as well as the nervous system. (D) Experimental scheme for RNAi screen. First, distinct E. coli strains expressing dsRNA to each individual gene in the worm are grown in overnight cultures, then induced to activate expression of the dsRNAs, and then cultured onto 24-well agar solid media plates. Next, ~30 L1 worms are plated onto each well containing a different dsRNA-expressing E. coli strain and cultivated until the worms reach adulthood. Finally, the phenotype is analyzed for the suppression of the multivulva phenotype.
RNAi Screening and Potential Target Discovery Strategies in C. elegans
Extensive RNAi screens have been performed in a variety of C. elegans mutants, which have generated a large repository of functional data to help understand detailed gene function in more complex organisms.58 RNAi screens can be performed by feeding the worms E. coli expressing double-stranded RNA (dsRNA) that targets a specific C. elegans gene then visualizing effects on numerous phenotypes, such as Muv (Fig. 5D).58 The availability of reagents and the ease of performing the whole organism RNAi screens have made C. elegans an attractive model for modeling human diseases.59–61 Our lab recently performed a whole-genome siRNA screen of the daf-18(lf); let-60(gf) double mutants, which uncovered several genes that potently suppress their Muv phenotype (Shahzad et al., unpublished data). Not only do these targets have human orthologs, but these genes are also dysregulated in GBM, indicating the utility of using C. elegans to identify potential therapeutic targets.
C. elegans is also useful for small molecule discovery. Drug screens involving target validation can be performed through agar-based assays with live worms to visualize morphology or locomotion.48,62 Alternatively, high throughput drug screens can be performed using either a drug sprayer or a microplate reader, which can use both live or immobilized worms.48,63 Despite certain limitations such as intestinal drug uptake inefficiency, C. elegans is an economic and rapid system in the drug discovery pipeline before testing candidates in the complex and costly vertebrate model systems.
Danio rerio
Zebrafish (D. rerio) has emerged as a robust model for studying vertebrate development and disease, investigating physiological effects, as well as for high throughput drug and genetic screens. Zebrafish are a small tropical aquatic species that shares considerable genetic homology with humans.15 Approximately 71.4% of human genes have at least one zebrafish orthologue and 82% of genes associated with human diseases have a zebrafish counterpart.9 Major organs including the heart, pancreas, liver, and gastrointestinal tract are developmentally and functionally comparable to humans.11 In addition, mechanisms of glial and neural development as well as their functions are highly conserved.14 Lastly, genetic manipulation to create transgenic zebrafish models is efficient, underscoring their potential for modeling human brain tumors.15,64
D. rerio as a Model for Cancer
Zebrafish models of cancer have led to numerous discoveries over the last decade.11 The transparent embryos and larvae permit visualization of tumor initiation and progression with high-resolution microscopy techniques.65 Using lineage-specific transgenic models or xenotransplantation of patient-derived tumor cells, zebrafish provide many unique opportunities to investigate the etiology of brain tumor progression with efficiency and precision.66
Life Cycle
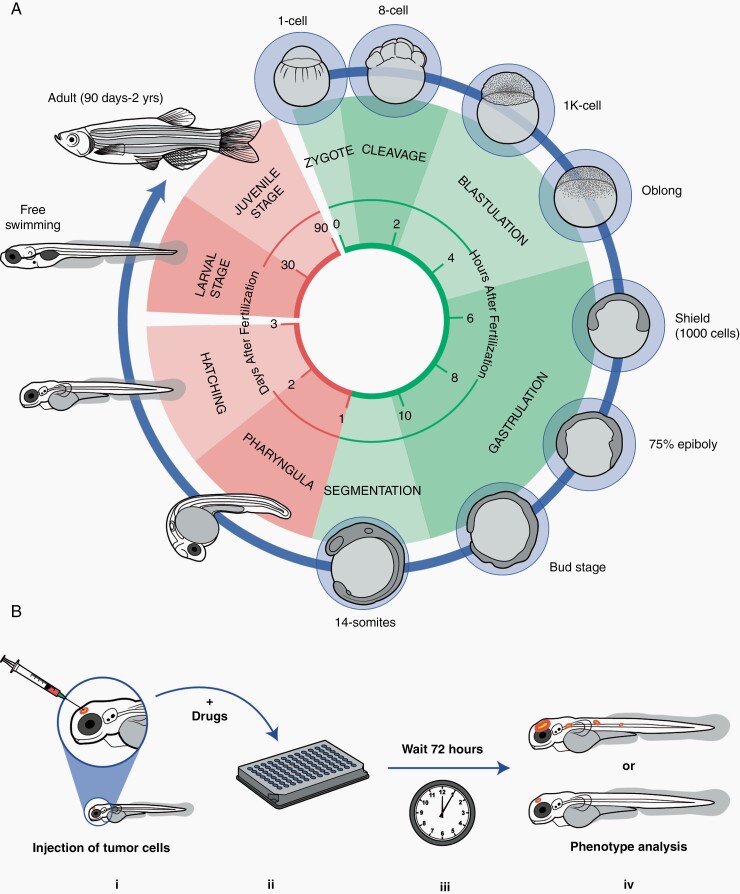
Zebrafish embryos are generated through ex-utero fertilization. Within 30 minutes, a single cell embryo has begun to develop over a large yolk cell, and by 6 hours, gastrulation is underway65 (Fig. 6A). By 3–4 days, hatching of the larvae occurs and embryogenesis comes to an end, marked by the protruding mouth stage.67 Larval development lasts approximately 6 weeks, as the animal undergoes pigmentation, fin growth, and scale development, resulting in the juvenile morphology, followed by sexual maturation that is completed by approximately 3 months.67 Normalization of the zebrafish life cycle, particularly in the laboratory setting, is highly dependent on external factors including temperature, hypoxia, diet, and fish density.67 The ability to regulate these factors provides an additional layer of control over developmental progression.67,68
Fig. 6.
Life cycle and high-throughput drug screening of patient-derived xenografts in D. rerio. (A) The zebrafish life cycle comprises four major stages, mainly the embryo, larva, juvenile, and adult stages. The first 24 h post fertilization (green) is comprised of five stages – zygote, cleavage, blastulation, gastrulation, and segmentation. The pharyngula phase encompasses the subsequent 24 h of development leading towards hatching. Zebrafish hatch into larva by 3 days post fertilization (dpf), and by 7dpf, the fry begin developing specialized structures such as the swim bladder and are capable of motility. The larvae continue to grow reaching the juvenile stage at 30 days and proceed to adulthood (sexual maturity) by 90dpf. (B) Schematic of the workflow for high-throughput drug screening of xenograft models of brain tumors. (i) Orthotopic injection of patient-derived tumor cells into the brain of a zebrafish embryo at 2dpf.89 (ii) Incubation of zebrafish larvae in a 96-well plate format with a drug or small molecule library. (iii) Monitored culture of the larvae for fixed incubation lengths. (iv) Analysis of phenotype (e.g., tumor growth inhibition or decreased metastasis) using imaging techniques.
Breeding & Genetic Manipulation Strategies
Zebrafish are a cost-effective model, as a single breeding pair can lay up to 200–300 eggs/week and requires little space and maintenance. Uniquely, they exhibit sexual dimorphism whereby their sex is not obviously determined by heteromorphic chromosomes, but rather is highly dependent on environmental cues including temperature, food, and rearing density.68 Peak sexual maturity is maintained between six months to one year of age, during which zebrafish can continuously spawn at an optimal breeding frequency of every 10 days.69 A secondary breeding method involves mixing female spawned eggs with male sperm that has been treated with ultraviolet radiation to produce haploid zebrafish with or without the desired mutation and can be used for identifying candidate mutant phenotypes and genetic interactions in cancer.70
Additionally, DNA, RNA, and/or protein can be micro-injected into externally fertilized embryos for transient or permanent manipulations and is a simple yet robust method for functional genetic analysis in an intact vertebrate system.11,15,71 Furthermore, with the emergence of CRISPR/Cas9 technologies, researchers can readily generate targeted loss-of-function or gain-of-function alleles to recapitulate disease-specific alterations.72
Drug Screening and Drug Discovery Strategies
Using zebrafish models of cancer, several high throughput drug screens have been performed to streamline chemotherapeutic discovery.73,74 Drug screening is extremely efficient given that zebrafish can absorb soluble chemical compounds directly from water, and screening can be performed in multi-well format (Figure 6B).11 For example, White and colleagues utilized mitf:BRAF(V600E);tp53-/- zebrafish to identify dihydroorotate dehydrogenase as an effective compound in abrogating melanoma growth, a finding that was translatable in vitro to human cells and to mouse models.73
Brain Tumor Models
Over the last decade, zebrafish has emerged as an effective system for modeling human brain cancers.66,75–77 Because of the transparent nature of zebrafish embryos and larvae, tumors are monitored in real-time to assess tumor cell infiltration and the microenvironment, key aspects of glioma biology.78–80 Another important consideration for modeling CNS tumors in zebrafish is the integrity of the BBB, which is critical to drug testing methodologies.70,81 Jeong et al. performed the extensive functional analysis to validate the BBB in zebrafish as robust compared to higher vertebrates.70 Interestingly, Fleming et al. have shown that the BBB does not completely form until 15 days post fertilization, which allows for experimentation with and without the BBB, if desired.81 Lastly, the development of the cortex and cerebellum have been well studied in zebrafish. Key transcriptional regulators have been identified and a diversity of tissue specific transgenic promoters are available, which enables modeling cancers of known cellular origin.66,75–77
Genetic Models
Multiple genetic models of human brain cancer have been generated using zebrafish that differ based on their location, potential cell of origin, and methodology. Jung et al. were the first to demonstrate that cerebellar specific gliomagenesis was possible in zebrafish by utilizing a ptf1-driven activated AKT.76 Subsequently, it was found that somatic inactivation of Rb leads to medulloblastoma-like tumors arising in the zebrafish cerebellum that have been classified as primitive neuroectodermal tumors based on their location and histopathological features.82
Similarly, Ju and colleagues validated a role for oncogenic Ras signaling in gliomagenesis by expressing human KRASG12V under the control of the zebrafish gfap and krt5 promoters, resulting in CNS tumors and neoplasms resembling nerve sheath tumors, respectively.66 Using these models, the MAPK/ERK kinase (MEK) inhibitor U0126 was identified as an inhibitor of abnormal phenotypes associated with KRAS over-expression,66 revealing a novel platform for chemical screens for inhibitors of Ras signaling in glioma and beyond.
Mayrhofer et al. also developed a robust model of GBM, using both somatic and germ-line Gal4/UAS approaches to express Harvey Rat Sarcoma viral oncogene homolog (HRAS)V12 from a zic4 enhancer, leading to the development of lesions in the telencephalon, ventricles, and diencephalon.83 In the future, it will be interesting to utilize zebrafish transgenic and mutant models like those described to explore tumor initiation, clonal expansion, and invasion within an endogenous tumor microenvironment.
Transplant Models
Transplantation of human brain tumor cells into zebrafish is an easy and effective strategy for direct visualization of tumor cell engraftment, growth, and metastasis. Eden et al. utilized a retrovirus strategy to over-express Erb-B2 Receptor Tyrosine Kinase 2 - Red Fluorescent Protein (ERBB2-RFP) in neural stem cells isolated from mice followed by orthotopic xenotransplantation into zebrafish larvae.84 Resulting tumors recapitulated major phenotypic and histological markers including pleomorphic cell morphology, high mitotic rate (Ki67+ve), and neovascularization. Importantly, the metastatic potential seen in mice was retained in a proportion of the fish, providing a novel and high-throughput platform to study drivers of metastasis in vivo.84 More recently, GBM patient-derived xenografts (PDXs) have been characterized using zebrafish hosts.85–87 Neurospheres engrafted into the midbrain of zebrafish embryos were able to recapitulate the heterogeneity of GBM tumors, including Sox2+ tumor stem cells,85 and models like this have been utilized for high-throughput testing of common chemotherapy agents to discern factors involved in drug resistance and metastasis.85–87 For example, Zeng et al. were able to identify a novel nitrogen-based DNA crosslinking small chemical called TNB, that traversed the BBB and inhibited xenograft establishment.86 Similarly, validation of targets identified in vitro, such as MutT homolog 1 (MTH1) inhibition, could be validated in vivo using zebrafish PDX models.88
Models of ependymoma (EPRTBDN-RFP, EPEPHB2-RFP) and choroid plexus carcinoma (Tp53flx/flx;Rbflx/flx;PTENflx/flx;CRE-RFP) xenotransplantation have also been generated using zebrafish.84 Ependymoma xenografts had histological similarities to patient biopsies, including pseudorosettes, while choroid plexus carcinoma models expressed the highly specific transthyretin receptor.84 Altogether, xenograft approaches in zebrafish reveal conserved tumor cell dynamics compared to what is seen in rodents and the future can be harnessed for large-scale drug screening approaches in vivo.
Limitations of Non-Mammalian Species
It is important to note a few limitations of using non-mammalian species to model brain tumors. The first is the lack of a complex segmented nervous system similar to humans, which may prevent accurate modeling of the environmental niche during xenotransplantation.23,43,64 However, the lack of complex anatomy also provides a clean canvas for investigating aspects of sufficiency and necessity, as well as the tumor cell of origin.67 The second disadvantage is the duplication of genes within the genome of non-mammalian species. Although there is much homology with humans, there is also redundancy due to genetic duplication during evolutionary divergence. This has resulted in the sub-functionalization of genes and can complicate gene discovery or transgenic modeling.11,23,44 Furthermore, the smaller size of these species may pose technical challenges when performing intricate experimental procedures. Nonetheless, the potential to conduct experiments at a large scale in a cost-effective manner is tantamount to their success as tools for oncological investigation.
Conclusions and Future Directions
With the high evolutionary conservation of human genes in model organisms like D. melanogaster, C. elegans, and D. rerio, coupled with their advanced forward and reverse genetics methods, it is now easier than ever to construct meaningful models of multigenic diseases like cancer. Worms, flies, and zebrafish offer rapid and reliable screening tools to probe and query the underlying genetic alterations of a variety of human brain tumors and can function as a powerful drug development pipelines. Importantly, it is cost effective and quick to engineer multiple cancer-relevant mutations into these animals to identify genetic vulnerabilities by screening methods that can be validated in murine models that more accurately mimic human brain cancers. The study of neuro-oncology in these organisms is a rapidly evolving field that has already demonstrated great promise. Future work using these organisms to develop in vivo brain tumor models will add powerful arsenal to the established murine and cell-based systems for elucidating the intricacies of these neoplasms and will add a strong foundation to the drug discovery pipeline and pre-clinical testing. It is our strong belief that information derived from such studies in non-mammalian species will continue to yield better understanding of targetable mechanisms that can be rapidly translated to shorten the journey towards more effective therapies for patients with brain tumors.
Funding
This study was supported by grants from the Canadian Institutes of Health Research (PJT-155967, PJT-153104 and GSD-140274), Meagan’s Hug (Meagan Bebenek Foundation), b.r.a.i.nchild and the Wiley Fund.
Conflict of interest statement. The authors have no financial or conflicts of interest to disclose.
References
- 1. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network . The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janbazian L, Karamchandani J, Das S. Mouse models of glioblastoma: lessons learned and questions to be answered. J Neurooncol. 2014;118(1):1–8. [DOI] [PubMed] [Google Scholar]
- 4. Agnihotri S, Burrell KE, Wolf A, et al. Glioblastoma, a brief review of history, molecular genetics, animal models and novel therapeutic strategies. Arch Immunol Ther Exp (Warsz). 2013;61(1):25–41. [DOI] [PubMed] [Google Scholar]
- 5. Chen L, Zhang Y, Yang J, Hagan JP, Li M. Vertebrate animal models of glioma: understanding the mechanisms and developing new therapies. Biochim Biophys Acta. 2013;1836(1):158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barbazuk WB, Korf I, Kadavi C, et al. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000;10(9):1351–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6(1):9–23. [DOI] [PubMed] [Google Scholar]
- 8. Blumenthal T, Evans D, Link CD, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417(6891):851–854. [DOI] [PubMed] [Google Scholar]
- 9. Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. The C. Elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–2018. [DOI] [PubMed] [Google Scholar]
- 11. Huiting LN, Laroche F, Feng H. The zebrafish as a tool to cancer drug discovery. Austin J Pharmacol Ther. 2015;3(2):1069. [PMC free article] [PubMed] [Google Scholar]
- 12. Yildirim K, Petri J, Kottmeier R, Klämbt C. Drosophila glia: few cell types and many conserved functions. Glia. 2019;67(1):5–26. [DOI] [PubMed] [Google Scholar]
- 13. Oikonomou G, Shaham S. The glia of Caenorhabditis elegans. Glia. 2011;59(9):1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lyons DA, Talbot WS. Glial cell development and function in zebrafish. Cold Spring Harb Perspect Biol. 2015;7(2):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kegelman TP, Hu B, Emdad L, Das SK, Sarkar D, Fisher PB.. In Vivo Modeling of Malignant Glioma: The Road to Effective Therapy. Vol 121. 1st ed. Oxford, UK: Elsevier Inc.; 2014. [DOI] [PubMed] [Google Scholar]
- 16. Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293(5538):2251–2256. [DOI] [PubMed] [Google Scholar]
- 17. Hannan F, Ho I, Tong JJ, Zhu Y, Nurnberg P, Zhong Y. Effect of neurofibromatosis type I mutations on a novel pathway for adenylyl cyclase activation requiring neurofibromin and Ras. Hum Mol Genet. 2006;15(7):1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yager J, Richards S, Hekmat-Scafe DS, et al. Control of Drosophila perineurial glial growth by interacting neurotransmitter-mediated signaling pathways. Proc Natl Acad Sci U S A. 2001;98(18):10445–10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lavery W, Hall V, Yager JC, Rottgers A, Wells MC, Stern M. Phosphatidylinositol 3-kinase and Akt nonautonomously promote perineurial glial growth in Drosophila peripheral nerves. J Neurosci. 2007;27(2):279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morris EJ, Ji JY, Yang F, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455(7212):552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Northcott PA, Korshunov A, Witt H, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ashburner M, Roote J. Maintenance of a Drosophila laboratory: general procedures. CSH Protoc. 2007;2007:pdb.ip35. [DOI] [PubMed] [Google Scholar]
- 23. St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3(3):176–188. [DOI] [PubMed] [Google Scholar]
- 24. Weasner BM, Zhu J, Kumar JP. FLPing genes on and off in drosophila. Methods Mol Biol. 2017;1642:195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003;302(5648):1227–1231. [DOI] [PubMed] [Google Scholar]
- 26. Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463(7280):545–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ornitz DM, Moreadith RW, Leder P. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci U S A. 1991;88(3):698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Willoughby LF, Schlosser T, Manning SA, et al. An in vivo large-scale chemical screening platform using Drosophila for anti-cancer drug discovery. Dis Model Mech. 2013;6(2):521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sonoshita M, Scopton AP, Ung PMU, et al. A whole-animal platform to advance a clinical kinase inhibitor into new disease space. Nat Chem Biol. 2018;14(3):291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gladstone M, Frederick B, Zheng D, Edwards A, Yoon P, Stickel S. A translation inhibitor identified in a Drosophila screen enhances the effect of ionizing radiation and taxol in mammalian models of cancer. Dis Model Mech. 2012;5(3):342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Read R, Cavenee W, FB F, Thomas J. A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 2009;5(2):e1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X, Wanggou S, Bodalia A, et al. A feedforward mechanism mediated by mechanosensitive ion channel PIEZO1 and tissue mechanics promotes glioma aggression. Neuron. 2018;100(4):799–815.e7. [DOI] [PubMed] [Google Scholar]
- 33. Agnihotri S, Golbourn B, Huang X, Remke M, Younger S, Cairns R. PINK1 Is a negative regulator of growth and the warburg effect in glioblastoma. Cancer Res. 2016;76(16):4708–4719. [DOI] [PubMed] [Google Scholar]
- 34. Chi KC, Tsai WC, Wu CL, Lin TY, Hueng DY. An adult drosophila glioma model for studying pathometabolic pathways of gliomagenesis. Mol Neurobiol. 2019;56(6):4589–4599. [DOI] [PubMed] [Google Scholar]
- 35. Hu T, Li C. Convergence between Wnt-β-catenin and EGFR signaling in cancer. Mol Cancer. 2010;9:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frattini V, Pagnotta SM, Tala, et al. A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature. 2018;553(7687):222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miroshnikova YA, Mouw JK, Barnes JM, et al. Tissue mechanics promote IDH1-dependent HIF1α-tenascin C feedback to regulate glioblastoma aggression. Nat Cell Biol. 2016;18(12):1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Witte HT, Jeibmann A, Klämbt C, Paulus W. Modeling glioma growth and invasion in Drosophila melanogaster. Neoplasia. 2009;11(9):882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monje M, Beachy PA, Fisher PG. Hedgehogs, flies, Wnts and MYCs: the time has come for many things in medulloblastoma. J Clin Oncol. 2011;29(11):1395–1398. [DOI] [PubMed] [Google Scholar]
- 41. Yi J, Wu J. Epigenetic regulation in medulloblastoma. Mol Cell Neurosci. 2018;87:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jorgensen EM, Mango SE. The art and design of genetic screens: caenorhabditis elegans. Nat Rev Genet. 2002;3(5):356–369. [DOI] [PubMed] [Google Scholar]
- 44. Kyriakakis E, Markaki M, Tavernarakis N. Caenorhabditis elegans as a model for cancer research. Mol Cell Oncol. 2015;2(2):e975027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim W, Underwood RS, Greenwald I, Shaye DD. OrthoList 2: a new comparative genomic analysis of human and Caenorhabditis elegans Genes. Genetics. 2018;210(2):445–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Silverman GA, Luke CJ, Bhatia SR, et al. Modeling molecular and cellular aspects of human disease using the nematode Caenorhabditis elegans. Pediatr Res. 2009;65(1):10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Apfeld J, Alper S. What can we learn about human disease from the nematode C. elegans? Methods Mol Biol. 2018;1706:53–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carretero M, Solis GM, Petrascheck M. C. elegans as model for drug discovery. Curr Top Med Chem. 2017;17(18):2067–2076. [DOI] [PubMed] [Google Scholar]
- 49. Jolliffe AK, Derry WB. The TP53 signaling network in mammals and worms. Brief Funct Genomics. 2013;12(2):129–141. [DOI] [PubMed] [Google Scholar]
- 50. Horvitz HR. Worms, life, and death (Nobel lecture). Chembiochem. 2003;4(8):697–711. [DOI] [PubMed] [Google Scholar]
- 51. Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100(1):64–119. [DOI] [PubMed] [Google Scholar]
- 52. White J, Southgate E, Thomson J, Brenner F. The structure of the nervous system of the nematode caenorhabditis elegans. Philos Trans R Soc L B Biol Sci. 1986;314:1–340. [DOI] [PubMed] [Google Scholar]
- 53. Perrin AJ, Gunda M, Yu B, et al. Noncanonical control of C. elegans germline apoptosis by the insulin/IGF-1 and Ras/MAPK signaling pathways. Cell Death Differ. 2013;20(1):97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Battu G, Hoier EF, Hajnal A. The C. elegans G-protein-coupled receptor SRA-13 inhibits RAS/MAPK signalling during olfaction and vulval development. Development. 2003;130(12):2567–2577. [DOI] [PubMed] [Google Scholar]
- 55. Nakdimon I, Walser M, Fröhli E, Hajnal A. PTEN negatively regulates MAPK signaling during Caenorhabditis elegans vulval development. PLoS Genet. 2012;8(8):e1002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Beitel GJ, Clark SG, Horvitz HR. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990;348(6301):503–509. [DOI] [PubMed] [Google Scholar]
- 57. Pettitt J, Wood WB, Plasterk RH. cdh-3, a gene encoding a member of the cadherin superfamily, functions in epithelial cell morphogenesis in Caenorhabditis elegans. Development. 1996;122(12):4149–4157. [DOI] [PubMed] [Google Scholar]
- 58. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. [DOI] [PubMed] [Google Scholar]
- 59. Jansen IE, Ye H, Heetveld S, et al. Discovery and functional prioritization of Parkinson’s disease candidate genes from large-scale whole exome sequencing. Genome Biol. 2017;18(1):1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu H, Dowdle JA, Khurshid S, et al. Discovery of stromal regulatory networks that suppress ras-sensitized epithelial cell proliferation. Dev Cell. 2017;41(4):392–407.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Otten C, Knox J, Boulday G, et al. Systematic pharmacological screens uncover novel pathways involved in cerebral cavernous malformations. EMBO Mol Med. 2018;10(10):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmeisser K, Fardghassemi Y, Parker JA. A rapid chemical-genetic screen utilizing impaired movement phenotypes in C. elegans: input into genetics of neurodevelopmental disorders. Exp Neurol. 2017;293:101–114. [DOI] [PubMed] [Google Scholar]
- 63. Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38(8):896–903. [DOI] [PubMed] [Google Scholar]
- 64. Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122(7):2337–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ablain J, Zon LI. Of fish and men: using zebrafish to fight human diseases Julien. Trends Cell Biol. 2013;23(12):584–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ju B, Chen W, Orr BA, et al. Oncogenic KRAS promotes malignant brain tumors in zebrafish. Mol Cancer. 2015;14:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 2009;238(12):2975–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Varga ZM. Aquaculture and husbandry at the zebrafish international resource center. Methods Cell Biol. 2011;104:453–478. [DOI] [PubMed] [Google Scholar]
- 69. Niimi A, LaHam Q. Influence of breeding time interval on egg number, mortality, and hatching of zebra fish Brachydanio rerio. Can J Zool. 1974;52:515–517. [DOI] [PubMed] [Google Scholar]
- 70. Jeong J-Y, Kwon H-B, Ahn J-C, Kang D, Kwon J. Functional and developmental analysis of the blood–brain barrier in zebrafish. Brain Res Bull. 2008;75:619–628. [DOI] [PubMed] [Google Scholar]
- 71. Zhao S, Huang J, Ye J. A fresh look at zebrafish from the perspective of cancer research. J Exp Clin Cancer Res. 2015;34:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Luo JJ, Bian WP, Liu Y, et al. CRISPR/Cas9-based genome engineering of zebrafish using a seamless integration strategy. FASEB J. 2018;32(9):5132–5142. [DOI] [PubMed] [Google Scholar]
- 73. White RM, Cech J, Ratanasirintrawoot S, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471(7339):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu NA, Jiang H, Ben-Shlomo A, et al. Targeting zebrafish and murine pituitary corticotroph tumors with a cyclin-dependent kinase (CDK) inhibitor. Proc Natl Acad Sci U S A. 2011;108(20):8414–8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shin J, Padmanabhan A, de Groh ED, et al. Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Dis Model Mech. 2012;5(6):881–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jung IH, Leem GL, Jung DE, et al. Glioma is formed by active Akt1 alone and promoted by active Rac1 in transgenic zebrafish. Neuro Oncol. 2013;15(3):290–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ju B, Chen W, Spitsbergen JM, et al. Activation of Sonic hedgehog signaling in neural progenitor cells promotes glioma development in the zebrafish optic pathway. Oncogenesis. 2014;3:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Eden CJ, Ju B, Murugesan M, et al. Orthotopic models of pediatric brain tumors in zebrafish. Oncogene. 2015;34(13):1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. White RM, Sessa A, Burke C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2(2):183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yan C, Yang Q, Do D, Brunson DC, Langenau DM. Adult immune compromised zebrafish for xenograft cell transplantation studies. EBioMedicine. 2019;47:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fleming A, Diekmann H, Goldsmith P. Functional characterisation of the maturation of the blood-brain barrier in larval zebrafish. PLoS One. 2013;8(10):e77548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shim J, Choi JH, Park MH, et al. Development of zebrafish medulloblastoma-like PNET model by TALEN-mediated somatic gene inactivation. Oncotarget. 2017;8(33):55280–55297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mayrhofer M, Gourain V, Reischl M, et al. A novel brain tumor model in zebrafish reveals the role of YAP activation in MAPK- and PI3K-induced malignant growth. Dis Model Mech. 2017;10(1):15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Eden C, Ju B, Murugesan M, et al. Orthotopic models of pediatric brain tumors in zebrafish. Oncogene. 2014;34:1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Welker AM, Jaros BD, Puduvalli VK, Imitola J, Kaur B, Beattie CE. Standardized orthotopic xenografts in zebrafish reveal glioma cell-line-specific characteristics and tumor cell heterogeneity. Dis Model Mech. 2016;9(2):199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zeng A, Ye T, Cao D, et al. Identify a blood-brain barrier penetrating drug-TNB using zebrafish orthotopic glioblastoma xenograft model. Sci Rep. 2017;7(1):14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pudelko L, Edwards S, Balan M, et al. An orthotopic glioblastoma animal model suitable for high-throughput screenings. Neuro Oncol. 2018;20(11):1475–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pudelko L, Rouhi P, Sanjiv K, et al. Glioblastoma and glioblastoma stem cells are dependent on functional MTH1. Oncotarget. 2017;8(49):84671–84684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Casey M, Modzelewska K, Anderson D, et al. Transplantation of Zebrafish pediatric brain tumors into immune-competent hosts for long-term study of tumor cell behavior and drug response. J Vis Exp. 2017;17(123):e55712. [DOI] [PMC free article] [PubMed] [Google Scholar]