Abstract
Background
Malignant gliomas including glioblastomas are characterized by a striking cellular heterogeneity, which includes a subpopulation of glioma cells that becomes highly resistant by integration into tumor microtube (TM)-connected multicellular networks.
Methods
A novel functional approach to detect, isolate, and characterize glioma cell subpopulations with respect to in vivo network integration is established, combining a dye staining method with intravital two-photon microscopy, Fluorescence-Activated Cell Sorting (FACS), molecular profiling, and gene reporter studies.
Results
Glioblastoma cells that are part of the TM-connected tumor network show activated neurodevelopmental and glioma progression gene expression pathways. Importantly, many of them revealed profiles indicative of increased cellular stemness, including high expression of nestin. TM-connected glioblastoma cells also had a higher potential for reinitiation of brain tumor growth. Long-term tracking of tumor cell nestin expression in vivo revealed a stronger TM network integration and higher radioresistance of the nestin-high subpopulation. Glioblastoma cells that were both nestin-high and network-integrated were particularly able to adapt to radiotherapy with increased TM formation.
Conclusion
Multiple stem-like features are strongly enriched in a fraction of network-integrated glioma cells, explaining their particular resilience.
Keywords: cancer stem cells, glioblastoma, glioma, networks, tumor microtubes
Key Points.
1. Glioma cells that are interconnected to a multicellular network show multiple stemness features.
2. Network-connected stem-like glioma cells are particularly malignant and radioresistant.
Importance of the Study.
Using a new technology to identify and isolate the subpopulation of glioma cells that connects to multicellular functional and resistant networks, we demonstrate that these tumor cells are characterized by specific molecular, cellular, and functional features that have all been associated with cancer stem cells. These results link the emerging field of communicating cancer cell networks (an area of “Cancer Neuroscience”) with the more established field of cancer heterogeneity/stemness, effectively contributing to a better understanding of both in Neuro-Oncology. The results of the study further support a specific role of the stemness marker nestin for tumor cell network biology and provide insights into molecular pathways involved in glioblastoma cell stemness and connectivity.
Malignant gliomas including glioblastomas are characterized by high resistance to all standard therapies.1–3 This is most likely attributable to a particularly resistant subpopulation of tumor cells.4–6 We have recently found that particularly treatment-resistant glioma cells extend long tubular membrane protrusions called tumor microtubes (TMs). Tumor cells use distinct neurodevelopmental pathways to extend those TMs in a dynamic fashion, invading and colonizing the brain, and interconnecting with them to one communicating multicellular network.7–11 Importantly, only about one half of the tumor cells is extending these interconnecting TMs and thereby becomes part of a multicellular glioma network, both in human and mouse diffuse astrocytomas and glioblastomas; the other half stays isolated.7 The TM-proficient and TM-connected glioma cell subpopulation is the one that resists the cytotoxic effects of radio- and chemotherapy and repairs surgical damage,12 thus constituting the resistant backbone of the disease. In humans, TM networks are especially prominent in prognostically unfavorable 1p/19q intact astrocytomas and glioblastomas and largely absent in oligodendrogliomas, and increase with tumor grade; moreover, TMs and TM networks serve also as the site of neuron–glioma synaptic input, again also demonstrated for patient material.8,9,13
Cellular stemness has previously been described for membrane tube-extending nonmalignant cells in the developing Drosophila testis.14 In light of the common concept that enhanced resistance against standard therapies is particularly found in glioma stem-like cells (named “CSCs” in this manuscript) which drive tumorigenesis and development similar to normal stem cells except following a strict hierarchy, and are key for tumor recurrence and poor patient prognosis,15–17 a more stem-like state of TM-connected glioma cells appears possible. This interrelation is so far undetermined but relevant for the field of Neuro-Oncology.
Here, using a newly developed methodology to isolate TM-connected vs TM-unconnected glioma cells, we show that TM-connected tumor cells are a functionally and molecularly distinct subpopulation characterized by high expression of markers indicative of cellular stemness, higher tumor-initiating properties, relative stability over time, and particular resistance and adaptability to cytotoxic stress. These findings provide an important link between hitherto unconnected key factors of morphological and molecular tumor cell heterogeneity in gliomas.
Methods
For additional information, see Supplementary Methods.
Cell Culture and Constructs
The human primary glioblastoma cell lines (S24, T269) were cultured as spheroids under stem-like conditions.7 Tumor cells were labeled with colors by lentivirus transduction for in vivo imaging, pLKO.1-puro-CMV-TurboGFP_shnon-target-vector (Sigma), LeGO-T2 vector (Addgene), and CX-nestinGFPas reporter plasmid (Addgene) were transduced as described.7 Serum-containing DMEM (Sigma) was used to induce the differentiation of S24-nestin reporter cells.
In Vivo Tumor Formation, Imaging, and Radiation Treatment
All in vivo experiments in this study were approved by the Regierungspräsidium Karlsruhe, Germany and compliant with the institutional laboratory animal research guidelines. Viable tumor cells in single-cell suspension were injected into the cortex of NMRI and NOD/SCID nude mice, which were previously implanted with chronic cranial window as described before.7 Following tumor formation, SR101 (Invitrogen) dissolved in saline solution was injected into the mice intraperitoneally at a dose of 0.12 mg per gram body weight. Three-dimensional images were acquired by multiphoton laser scanning microscopy (MPLSM) at the wavelengths of 850 nm (green fluorescent protein [GFP]) and 950 nm (tdTomato/SR101). For radiation treatment, a dose of 7 Gy was applied to mice at a rate of 3 Gy per minute for 3 continuous days.
Xenograft Isolation and Preparation
Mice were killed and perfused followed by sample isolation. Samples were either sequentially dehydrated and frozen under −80°C for immunostaining experiment or were mechanically digested into cell suspension with brain tumor dissociation kit and device (Miltenyi) for FACS sorting and subsequent sequencing analysis.
FACS Sorting and Sphere-Forming Assay
Single-cell suspension was incubated with Calcein Violet 450 AM and TO-PRO®-3 Iodide (Invitrogen) before sorting. FACSAria cell sorting (BD Biosciences) was used to collect different populations according to diverse fluorescent intensities. YG586/15, tdTomato, and enhanced GFP channels were used to visualize SR101, tdTomato, and GFP signals, respectively.
For sphere-forming assay, the sorted cells were placed in 96-well plates (1 cell per well) and cultured under stem-like conditions. The spheres number was quantified after 30 days.
Validation of Nestin Reporter Expression
S24-nestin reporter cells either from culture or xenograft were processed with fixation, permeabilization, and subsequent staining with anti-nestin-Alexa Fluor 647 antibody (BD Biosciences) at 4°C. The overlap between GFP and nestin-Alexa Fluor 647 signals was identified by FACSCanto (BD Biosciences). Analysis was performed by FlowJo v9.0 software.
mRNA Profiling, Data Preprocessing, and Analysis
For bulk sequencing, samples were subjected to mRNA extraction (Qiagen) followed by dsDNA generation (Clontech), libraries preparation (NEB), sequencing in 50 bp single-end mode (Illumina), reads mapping, count data annotation, and group-wise comparison.
For single-cell sequencing, samples were prepared into single-cell gel beads (10× Genomics) followed by barcoded cDNA generation, sequencing (Illumina), alignment, quantification, and further clustering.
Ingenuity pathway analysis (IPA, Qiagen) and gene set enrichment analysis (GSEA) were performed to investigate diverse phenotypes reflected in the RNA-Seq gene list such as stem cell score and cell cycle.
Immunofluorescent Staining and Imaging
Samples (patient sections, xenografts, and cells) were subjected to standard protocols as previously described.7 Human nestin (Abcam), Musashi (Millipore), SOX2 (Cell Signaling), Ki67 (Abcam), HLA Class 1 ABC (Abcam), IDH1R132H (Dianove), and DAPI (Life Technologies) were selected for staining. The images of interest were recorded by confocal microscope (Leica).
Image Processing and Analysis
MPLSM images were captured with ZEISS ZEN software and subjected to Imaris or Fiji, for the purpose of removing unspecific background, identifying connectivity, counting TM and cell number, measuring signal intensity and mobility distance, cropping and generating illustrations in 3D or orthogonal view.
Statistical Analyses
All data with exception of RNA/scRNA Seq data (see above) were analyzed with GraphPad Prism 6 software. Mice were randomly allocated to control and irradiation groups. Sample size was designed based on our previous experimental experience. Quantitative data are presented as the mean ± SEM from at least three replicates. Sample sizes, P values, statistical tests, and error bars are described in the figure legends.
Results
A Novel Methodology for Intravital Selection of Network-Connected Glioma Cells
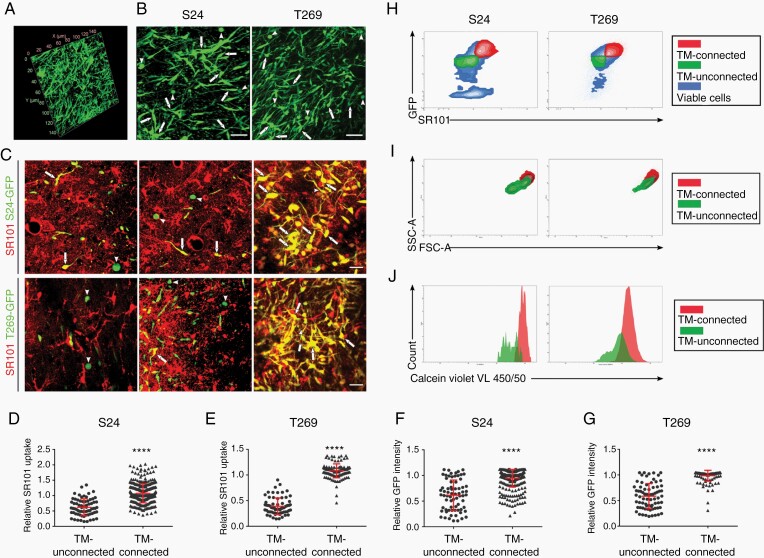
To characterize the specific cellular and molecular features of TM-connected vs -unconnected glioma cells, we first aimed to establish a methodology that allows to clearly distinguish these two principal tumor cell populations during glioma growth in the mouse brain. We have demonstrated before that glioma cells in TM-connected networks (Fig. 1A, human tumor sample) exchange gap junction-permeable small molecules with each other,7,18 similar to what is known from the functional syncytia of normal brain astrocytes. Those can take up an injected gap junction-permeable dye (sulforhodamine, SR101) from the circulation, which has been used for their in vivo labeling.19 We wondered whether a similarly straightforward intravital staining technology could also be used to label glioma cell networks.
Fig. 1.
TM-connected tumor cells exhibit distinct labeling characteristics. A, Representative image of TM-connected glioma cell network in a sample of a patient suffering from an IDH WT glioblastoma (140 × 140 × 60 µm volume; nestin staining). B, Representative images of TM-connected (arrows) and TM-unconnected (arrowheads) primary glioblastoma cells (GFP+, green) of two CSC lines, S24 and T269, during growth in the live mouse brain. Scale bars, 50 µm. C, Representative images of heterogeneous uptake of the fluorescent dye SR101 (red) by TM-connected (examples are shown with arrows) vs TM-unconnected (examples are shown with arrowheads) primary glioblastoma cells (GFP+, green) of both GSC lines during growth in the live mouse brain. SR101-positive normal astrocytes are GFP-negative and therefore red only. Scale bars, 25 µm. D-G, Quantification of SR101 and GFP fluorescence intensities measured in vivo. H, FACS analysis of SR101 and GFP signal intensities, allowing to differentiate tumor cell subpopulations enriched for TM-connected (SR101high, GFPhigh) and TM-unconnected (SR101low, GFPlow) tumor cells. Representative analysis of 3 independent mice. I and J, Morphology (I) and viability (J) of TM-connected vs -unconnected glioblastoma cells as analyzed by FACS, representative of 3 independent mice. In B-G, data obtained by in vivo MPLSM, two-tailed unpaired Student’s t test were used. ****P < .0001. Data are presented as mean ± SD of n = 3 independent samples.
Stably GFP-expressing primary glioblastoma cell lines were implanted into the mouse brain, and their growth was monitored by in vivo two-photon microscopy over time. TM-connected and -unconnected glioma cells could be clearly distinguished by analysis of high-resolution three-dimensional microscopy datasets (Fig. 1B). TM-rich and interconnected glioma cells had a significantly higher SR101 uptake than those glioma cells without a detectable TM-based connection to other glioma cells (Fig. 1C-E). Furthermore, SR101 uptake was highest in the bulk/core tumor area, where TM-based tumor cell interconnectivity is most extensive (Supplementary Fig. 1A and B). Astrocytes were clearly distinguishable from tumor cells by their GFP negativity (Fig. 1C). Of note, tumor cells not connected to other tumor cells by TMs showed also significantly less GFP fluorescence intensity than TM-rich, connected tumor cells (Fig. 1C, F, and G). We took advantage of this observation and used it together with SR101 fluorescence intensity to separate TM-connected (SR101high, GFPhigh) from TM-unconnected glioma cells (SR101low, GFPlow) by FACS after brain removal and acute cell dissociation (Fig. 1H). Here, the glioma cell subpopulation enriched for TM-connected cells showed relatively higher values of forward and side scatter parameters in FACS analysis (Fig. 1I), compared to that enriched for TM-unconnected cells, and also larger calcein violet uptake (Fig. 1J). Together this argues for distinct differences in cellular properties, particularly a larger cell size and also a higher principle viability of TM-connected glioblastoma cells, when compared to the TM-unconnected ones.
TM Connectivity Is Associated With Tumorigenicity and Poor Survival
Next, we investigated the capability of sorted TM-connected (SR101high, GFPhigh) vs -unconnected (SR101low, GFPlow) glioma cells to successfully re-establish tumors in vivo. Despite reimplantation of very few initiating tumor cells (200-3000 cells/mouse), obvious tumorigenesis accompanied by poor survival was seen in the mice receiving SR101high/GFPhigh cells. By contrast, no animal mortality or detectable tumor growth was observed in the mice implanted with SR101low/GFPlow cells (Supplementary Figure 2A). Due to the low number of animals, this reimplantation study needs to be regarded as exploratory. Of note, the heterogeneity of glioma cells regarding their TM content and network integration was reconstituted by the re-implanted TM-connected population (Supplementary Figure 2B and C). No such experiments could be performed for mice receiving TM-unconnected glioma cells for their lack of tumorigenesis in mice. No significant differences could be observed with in vitro sphere formation experiments (Supplementary Figure 1C).
Collectively these findings hint toward an increased ability of TM network-integrated glioma cells to reinitiate tumor growth and give rise to a heterogeneous tumor cell population, two important features proposed for stem-like cancer cells.17
Network-Integrated Glioblastoma Cells Have Distinct Gene Expression Patterns
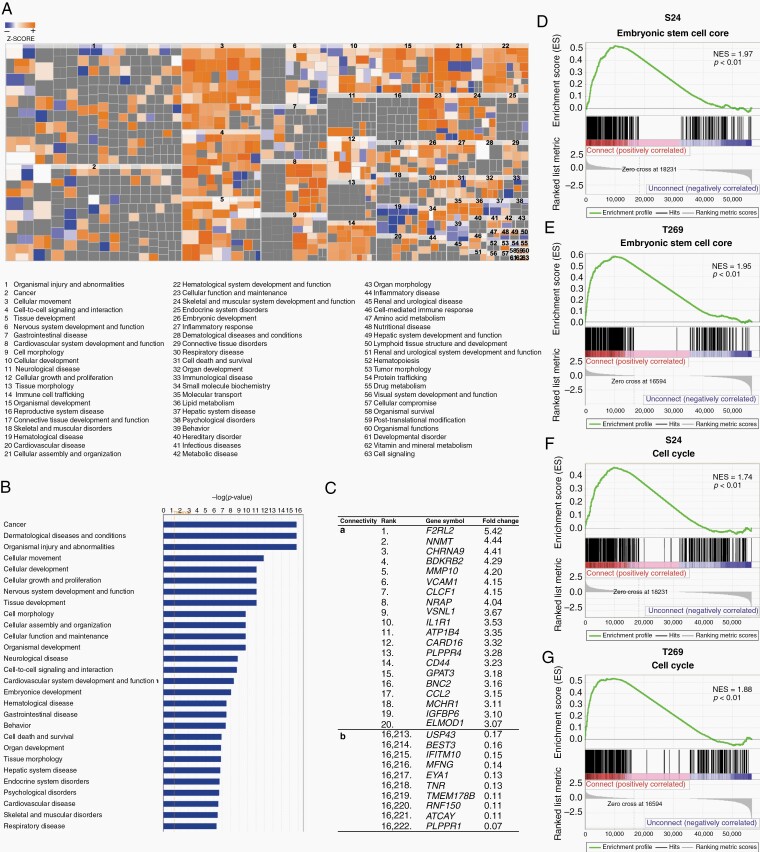
We next used the novel opportunity to separate TM-connected from -unconnected glioma cells by FACS (Fig. 1), and performed comparative bulk RNA sequencing of both principle tumor cell populations. Both populations showed distinct gene expression differences, and IPA revealed that biological functions including “cellular movement,” “cell-to-cell signaling and interaction,” “nervous system development and function,” “immune cell trafficking,” “molecular transport,” “formation of cellular protrusions,” and “microtubes dynamics” were predominately activated in the TM-connected group, corresponding to the known functions of TMs reported in our previous work,7,18 next to other genes associated with tumor progression (Fig. 2A-C, Supplementary Table 1, Supplementary Figure 3).
Fig. 2.
Gene expression profiles of network-integrated glioma cells reflect known biological TM functions and a stemness signature. A, Heat map of biological functions from ingenuity pathway analysis (IPA) comparing TM-connected (SR101high, GFPhigh) and -unconnected (SR101low, GFPlow) S24 tumor cells. RNA-Seq data are obtained from triplicate samples. A highly positive Z-score indicates relative activation in the TM-connected population (red), a highly negative Z-score indicated relative deactivation (blue). B, Bar plot of the most-activated pathways in TM-connected cells compared to the TM-unconnected group shown by IPA. C, Top differentially expressed genes between TM-connected vs -unconnected glioma cell subpopulations. The top 20 upregulated (a) and top 10 downregulated (b) genes of TM-connected cells are shown. Fold change of the mean normalized value for each gene is shown. For the full dataset, see Supplementary Table 1. D-G, Gene set enrichment analysis of RNA-Seq data indicates that genes associated with mouse and human embryonic stem cells (Wong ESC Core) and cell cycle (Reactome cell cycle) are enriched in the TM-connected subpopulation. NES, normalized enrichment score. P values are calculated based on 1000 permutations by the GSEA algorithm and were not adjusted for multiple comparisons.
Significant enrichment of gene signatures associated with embryonic stem cell states20 was found in the TM-connected group when compared to the TM-unconnected group (Fig. 2D and E). Furthermore, significant differences between the groups were detected for GSEA of cell cycle-associated genes (Fig. 2F and G). Together these data suggested that functional TM connectivity in gliomas is linked to a distinct gene expression pattern of this tumor cell subpopulation that is consistent with TM proficiency and cellular stemness.17
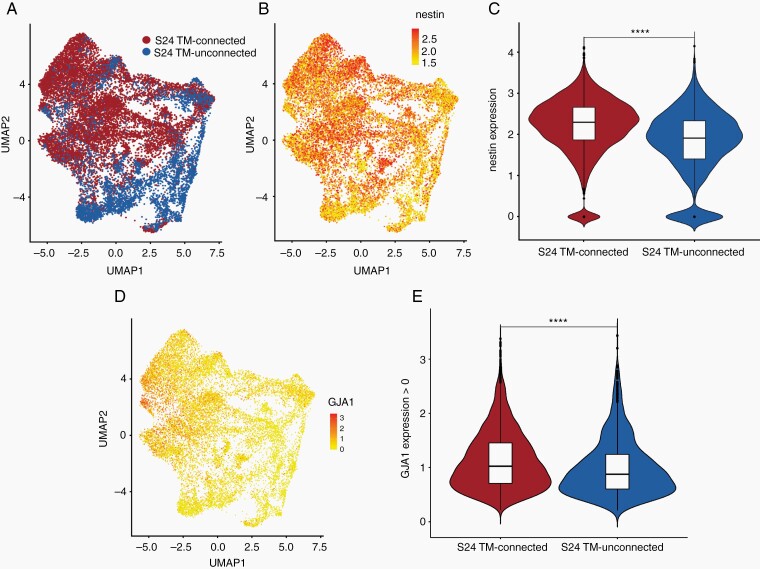
For deeper insights into the two principal glioblastoma cell populations defined by TM connectivity, single-cell RNA-Seq analysis of TM-connected vs TM-unconnected tumor cells was performed. UMAP (Uniform Manifold Approximation and Projection) analysis revealed that TM-connected and TM-unconnected glioblastoma cells show only minor overlap and different cellular subpopulations (Fig. 3A). To examine the expression level of stemness markers in these populations, we compared the expression levels of nestin in those cell clusters, one of the best-established markers of stemness15,17 and therapy resistance16,21 in malignant gliomas. RNA expression patterns of nestin correlated with the distribution of TM-connected tumor cell clusters (Fig. 3B), and overall mean nestin expression was significantly higher in TM-connected glioblastoma cells (Fig. 3C), which is well in line with their higher stemness signatures reported above. Finally, the principle connexin for connecting TMs to multicellular networks in gliomas, Cx43,7 was significantly higher expressed in TM-connected glioblastoma cells (Fig. 3D and E), confirming the cellular identity of the cells analyzed.
Fig. 3.
Single-cell transcriptome analysis of TM-connected and TM-unconnected tumor cells. A, UMAP (Uniform Manifold Approximation and Projection) visualization of TM-connected (red) and TM-unconnected (blue) S24 glioblastoma cells. B, Expression of nestin on UMAP from A. C, Violine blot showing the level of nestin expression in TM-connected and TM-unconnected S24 glioblastoma cells. D, UMAP visualization of single tumor cell GJA1 (Cx43) expression patterns. E, Violin blot of the expression of GJA1 (Cx43) in TM-connected and TM-unconnected S24 glioblastoma cells which show any detectable gene expression. ****P < .001.
TM-Positive Glioma Cells Express Higher Stem Cell Markers in Humans and Mice
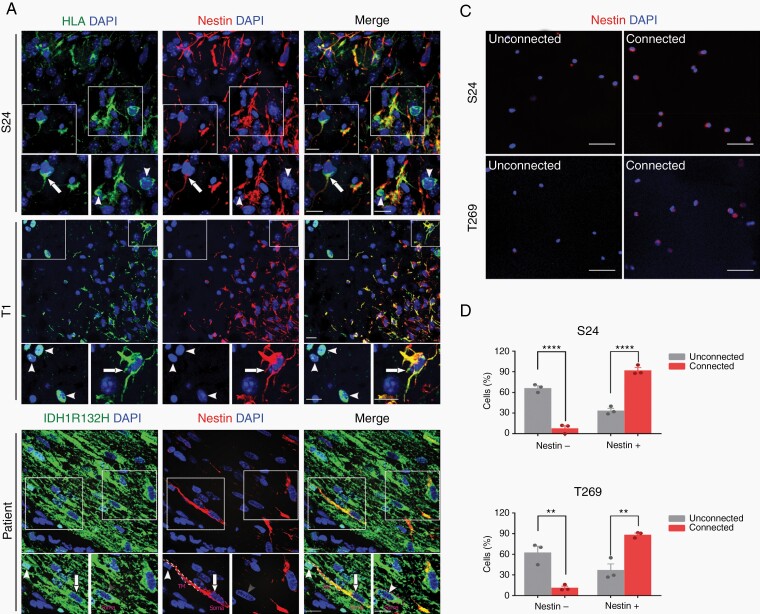
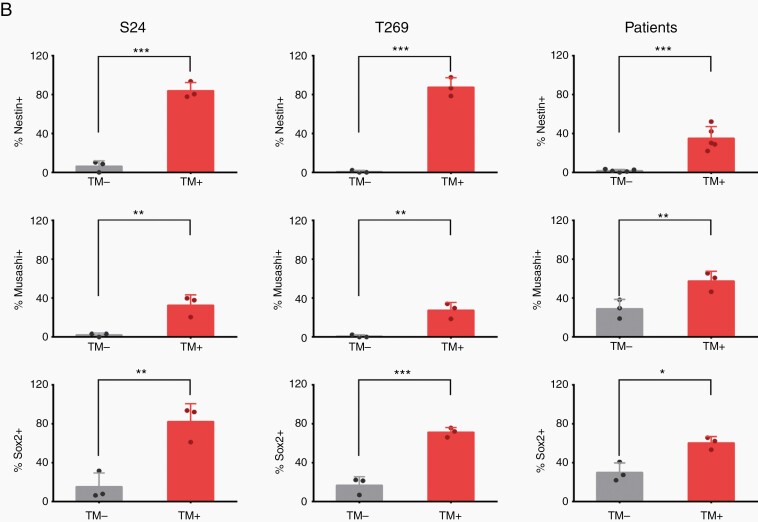
To verify that TM-positive glioma cells in tumor tissue from glioma patients and mouse models show gene expression patterns indicative of cellular stemness, protein expression of nestin, and also the putative stem cell markers Musashi and Sox2 was determined by immunohistochemistry (IHC). The extent of nestin protein expression was positively associated with TM formation in tumor cells (Fig. 4A and B), and Musashi and Sox2 protein expression was also significantly increased in tumor cells with detectable TMs in the sections analyzed (Fig. 4B and Supplementary Figure 4). Finally, increased nestin protein expression was also confirmed in the TM-connected FACS-sorted tumor cell population according to SR101 and GFP positivity (Fig. 4C and D).
Fig. 4.
TM formation is associated with expression of nestin and other stem cell markers in glioma tissue. A, Representative nestin immunofluorescence images in glioma cells from implantation xenografts derived from primary tumor cell lines (S24, T1) and patient samples of IDH-mutated anaplastic astrocytoma or glioblastoma. HLA and highly tumor cell-specific IDH1R132H stainings (green) were used to clearly identify tumor cells in mouse and human samples: Nestin (red), DAPI (blue). Exemplary, TM-positive glioma cells are marked with arrows, TM-negative cells with arrowheads. Scale bars, 50 µm. B, Percentage of tumor cells with identifiable TMs in histological sections (TM+) vs those tumor cells where no TMs were identified (TM−) with respect to detectable immunohistochemical expression of nestin, Musashi and Sox2. C, Representative images of nestin immunofluorescence staining of FACS-sorted, TM-connected (SR101high, GFPhigh) vs -unconnected (SR101low, GFPlow) tumor cells from CSC xenograft glioma models. Please note that after FACS sorting, cells were directly stained without preculture and are thus always rounded and isolated, so TM proficiency/interconnectivity cannot be analyzed. Nestin (red), DAPI (blue). D, Quantification of detectable nestin protein expression of both groups (shown in C). Two-tailed unpaired Student’s t test (B) and two-way ANOVA with Tukey’s multiple comparisons test (D) were used. **P < .01, ***P < .001, ****P < .0001. Data are presented as the mean ± SD (B) or mean ± SEM (D) of n = 3-5 independent samples.
In Vivo Nestin Reporter Positivity Reflects Distinct Tumor Cell States
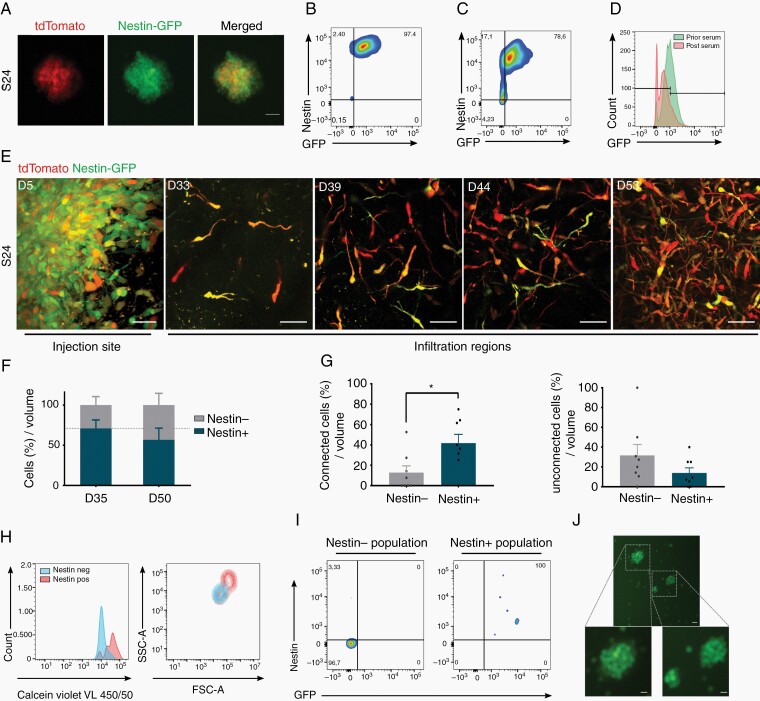
To dynamically monitor cellular stemness in relation to TM network properties in vivo, tdTomato-expressing primary glioblastoma cells were transduced with a reporter vector where GFP expression is driven by the nestin promoter (Fig. 5A). Initially, these tumor cells were strictly kept under stem-like condition in vitro, leading to a very high level of nestin expression indicated by high GFP positivity (Fig. 5B). After 1 week of serum-containing culture which leads to a less stem-like cellular state of glioblastoma cells (Supplementary Figure 5),22 a reduction of GFP intensity was observed (Fig. 5C and D), reconfirming the use of GFP expression intensity as a marker for distinct stem-like cellular states of glioma cells.
Fig. 5.
A reporter system for dynamic intravital tracking of nestin expression. A, S24-tdTomato cells cultured under stemness conditions in vitro express GFP driven by the nestin promoter. B-D, Relationship of GFP signal and in vitro nestin expression of tumor cells as determined by antibody staining. FACS analysis of tumor cells cultured under stemness condition (B), or after growth under differentiating serum-containing culture conditions for 1 week (C and D). E, Representative in vivo MPLSM images of S24 nestin reporter cells growing in the mouse brain in a tumor microregion over a period of 53 days. Nestin-GFP is shown in green, tdTomato is shown in red. Scale bars, 50 µm. F, Quantification of the relative change of tumor cell numbers in the same tumor microregions from Day 35 to Day 50, separately for the nestin+ (green and yellowish) and nestin− (red) tumor cell subpopulation. Dotted line represents the relative percentage value of nestin+ at Day 35. G, Relative fractions of connected nestin+ vs -nestin− subpopulations, and of unconnected nestin+ vs -nestin− subpopulations within tumor cells per volume at Day 35. H, Viability and morphology of nestin+ vs nestin− tumor cells at Day 57 of in vivo growth, analyzed by FACS. I, Relationship of GFP signal and in vivo nestin expression of tumor cells as determined by antibody staining. FACS analysis of GFP+ vs GFP− glioma cells from xenograft. J, Glioma cells that were nestin− (GFP−) after isolation from the mouse brain express GFP after re-exposition to stem-like culture conditions in vitro for 2 weeks. Scale bars, 20 µm. In F-H, one-way ANOVA with Tukey’s multiple comparisons test or two-way ANOVA with Sidak’s multiple comparisons test were used. *P < .05. Data presented as mean ± SEM of n = 7-12 tumor regions from 2-3 animals.
After implantation of these cells into the mouse brain, single tumor cells in distinct tumor regions could be dynamically tracked over time by repetitive in vivo two-photon microscopy (Fig. 5E). In the days after injection, nestin reporter intensity remained at high levels and was detectable in the vast majority of tumor cells. This pattern changed when the tumor started to grow and to infiltrate the brain; at this time point, an increasing fraction of glioblastoma cells fell with the GFP signal below the detection threshold (referred to as nestin reporter-negative [nestin−] cells vs clearly GFP-positive [nestin+] cells) (Fig. 5E and F). TM connectivity was strongly associated with nestin reporter positivity in vivo (Fig. 5G), confirming the immunostaining experiments of Fig. 4. After tumor removal and FACS analysis of GFP-positive brain tumor cells, in vivo nestin− cells showed lower values in forward and side scatter parameters, and less calcein violet uptake than the in vivo nestin+ population (Fig. 5H). For validation of the in vivo nestin reporter system, including the transduction stability of tumor cells, a strong overlap between nestin reporter GFP signal and nestin expression measured by immunofluorescence in glioblastoma cells separated from the mouse brain was demonstrated (Fig. 5I), and regain of GFP fluorescence after their exposure to 2 weeks of stem-like conditioned culture in vitro (Fig. 5J).
The nestin+ population demonstrated a trend to higher sphere-forming potential than nestin− cells (Supplementary Figure 6A and B). Of note, Ki67 expression was particularly evident in the nestin+ tumor cell population (Supplementary Figure 6C), which is a feature that has recently been associated with cancer cell stemness5,23–25 but can also speak for a cycling cancer cell subpopulation within the nestin+ cells.5 Interestingly, TMs had been associated with tumor cell proliferation in gliomas before.7,26 Overall, nestin+ glioblastoma cells were more static than nestin− ones, and some could be tracked over many weeks, including those with cell divisions (Supplementary Results; Supplementary Figure 7).
Nestin Expression, TM Networks, and Radiation Resistance
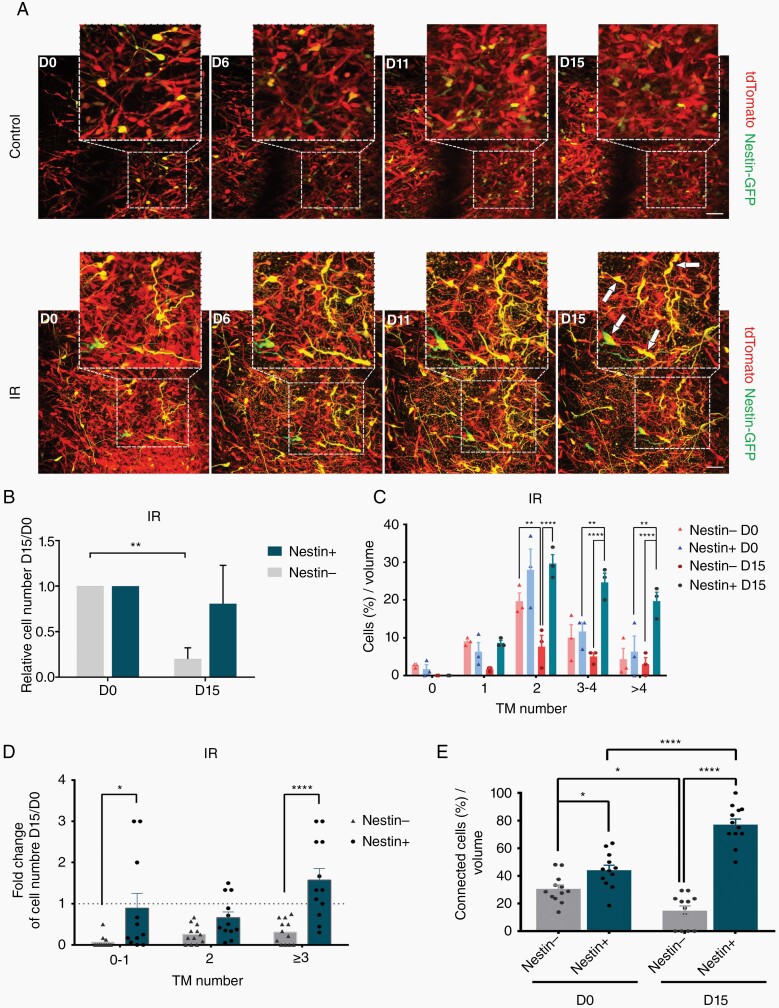
To investigate how standard therapy differentially affects the various glioma cell subpopulations, a mouse equivalent dose to human radiotherapy was applied. First, nestin− cells largely died over the next days, compared to nestin+ cells where the vast majority survived until Day 15 after irradiation, leading to a relative increase of the nestin+ population (Fig. 6A and B and Supplementary Figure 6D). Moreover, only nestin+ tumor cells collectively increased their TM number after irradiation, which has been described as a mechanism of adaptive resistance before7; the nestin− cells failed to execute this cellular response to radiotherapy (Fig. 6C). Of note, nestin+ cells survived better with or without tumor network integration, making glioma cell nestin positivity to a factor of radioresistance that can be independent of the network integration of tumor cells (Fig. 6D). In effect, the combination of both cellular/molecular mechanisms of radioresistance leads to an enrichment of nestin+ TM network-integrated glioma cells after radiotherapy over time (Fig. 6E). In summary, these data speak for nestin+ glioblastoma cells harboring a higher capacity for survival and for cellular plasticity after irradiation damage, with multiple relations to the glioma network.
Fig. 6.
Response to radiotherapy correlates with in vivo nestin expression and network connectivity. A, Representative MPLSM images of dynamic, morphological, and gene expression changes of nestin reporter S24 glioblastoma cells in control (upper panel) and irradiated (3 × 7 Gy at Days 0-2; lower panel) tumors in the live mouse brain. Examples of nestin+/TM+ cells that survive radiation are marked with arrows. Scale bars, 100 µm. B, Relative cell numbers of nestin+ and nestin− S24 glioma cells in the same tumor microregion over 15 days vs Day 0 before radiation, respectively. C and D, Relative fractions of nestin+ vs nestin− tumor cells with TMs of 0, 1, 2, 3-4, and >4 per volume prior and post-irradiation (C), and relative change overview of nestin+ vs nestin− subpopulations with TMs of 0-1, 2, and ≥3 after radiation (D). E, Relative fractions of TM-connected nestin+ vs nestin− tumor cell subpopulations per brain volume prior and post-irradiation. In B-E, one-way ANOVA with Tukey’s multiple comparisons test or two-way ANOVA with Sidak’s multiple comparisons test were used. *P < .05, **P < .01, ***P < .001, ****P < .0001. Data are presented as mean ± SEM of n = 12 tumor regions from 3 mice per group.
Discussion
Tumor heterogeneity, including the likely existence of a resistant stem-like glioma cell subpopulation, and diffuse tumor cell invasion leading to efficient brain colonization have been regarded as the main reasons for inevitable recurrence of malignant gliomas after therapy.7,15–17,27 Our study unveiled that the subpopulation of brain-colonizing glioblastoma cells that are part of a functionally connected tumor cell network are strongly enriched for stem-like cellular features. Together these data provide the first link between the emerging fields of tumor cell network connectivity and tumor cell heterogeneity in gliomas, which will help to optimally identify those tumor cells that are both of particular importance for tumor progression and later constitute the resistant backbone of the disease.
Intra- and intertumoral heterogeneity has been found in many tumor entities, including gliomas. While we are just at the beginning of understanding its functional role, single-cell RNA sequencing has revealed that distinct glioma cell subpopulations exist and can be separated from each other in glioblastoma, but also IDH (isocitrate dehydrogenase)-mutant lower-grade astrocytoma, oligodendroglioma, and diffuse intrinsic pontine glioma.6,28–31 In these studies, glioma cell subpopulations with a stemness signature were reproducibly detected. Moreover, earlier studies had found that small subpopulation of glioma cells might be capable of accelerate growth of the entire tumor.32 The results of the current study suggest that those molecularly defined stem-like cancer cell states are related to distinct network connectivity patterns of glioma cell subpopulations.
Interestingly, the typical diffuse brain infiltration of incurable gliomas seems to share features with the migration of stem and progenitor cells during central nervous system (CNS) development, particularly radial glia cells or nonmalignant CNS stem cells during damage repair.33 Our study adds to the concept by showing that tumor cells bearing TMs which are the known subpopulation that is capable of efficient brain invasion7,26 are characterized by molecular and functional signs of stemness. The in vitro sphere formation ability of TM-connected and -unconnected tumor cells did not markedly differ, which might be attributable to the fact that purified populations just reflect temporary cellular states, and also that in vivo stemness features in the brain can differ from those that are measurable in in vitro growth assays.34
It remains to be seen how many of the TM-connected and nestin+ glioma cells possess true stem-like states and capabilities, and whether more than one distinct stem cell subpopulations can be found within the population of TM-connected tumor cells. Finally, while we made experimental use of the fact that unconnected glioma cells showed lower GFP expression than connected ones, the biological basis for this observation is not clear and needs to be determined, as the identity of the very rare tumor cell populations that are SR101-positive but GFP low, and vice versa.
In conclusion, this study links the emerging fields of tumor cell networks and cellular heterogeneity in gliomas for the first time. By demonstrating that TM-connected glioma cells harbor multiple features previously associated with (tumor) cell stemness, a novel explanation for the remarkable ability of stem-like cancer cells for survival under cytotoxic stress, self-repair, and general tumor progression is provided.4,5,15–17 In consequence, this study adds another core feature to cancer stem-like cells in glioma: network connectivity. All in all, the subpopulation of glioma cells that is both TM network-integrated and nestinhigh emerges as the most relevant tumor cell subpopulation that should be studied with priority in the future. Targeting of this subpopulation holds the promise to effectively tackle tumor progression and resistance in incurable gliomas.
Supplementary Material
Acknowledgments
We thank A. Berghoff, M. Osswald, C. Felblinger, S. Schmitt, T. Rubner, K. Damir, M. Brom, D. Lemke, K. Gunkel, as well as the DKFZ Genomics and Proteomics, Omics IT and Data Management, and the animal core facilities for support and assistance. We thank A. Hotz-Wagenblatt for support with the analysis of RNA-Seq data, and F. Sahm for providing sections from patients. We thank the scOpen Lab at German Cancer Research Center in Heidelberg for support with the single-cell analysis.
Funding
This work was supported by a grant from the German Research Foundation (DFG, SFB1389). Single-cell RNA sequencing was supported by a DKFZ-HIPO grant (K32) to T.K., W.W., and F.W.
Conflict of interest statement. F.W. and W.W. report the patent (WO2017020982A1) “Agents for use in the treatment of glioma.” F.W. is co-founder of DC Europa Ltd. (a company trading under the name Divide & Conquer), which is developing new medicines for the treatment of glioma. GSK, Boehringer, Genentech, Roche and Divide, and Conquer have provided research funding to F.W.’s lab under research collaboration agreements. G.S. is an employee of Carl Zeiss Microscopy GmbH.
Authorship statement. R.X. and F.W. designed the study and interpreted the data. R.X. performed most experiments, wrote the manuscript, and contributed to all aspects of the study under the guidance of F.W. T.K. participated in the single-cell studies. T.K., D.C.H., and L.H. performed bioinformatic analysis of RNA-Seq data. J.G., L.H., and G.S. performed in vivo two-photon experiments. V.V. and M.R. offered Ki67 staining and imaging assistance. M.S. supervised bioinformatics analyses and performed data interpretation. W.W. provided conceptual input, performed data interpretation, and supported RNA-Seq experiments. F.W. directed the whole study, critically commented, and corrected the manuscript. All authors approved the content of the manuscript.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group . Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Wick W, Platten M. Understanding and targeting alkylator resistance in glioblastoma. Cancer Discov. 2014;4(10):1120–1122. [DOI] [PubMed] [Google Scholar]
- 3. Osuka S, Van Meir EG. Overcoming therapeutic resistance in glioblastoma: the way forward. J Clin Invest. 2017;127(2):415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–334. [DOI] [PubMed] [Google Scholar]
- 5. Lan X, Jörg DJ, Cavalli FMG, et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549(7671):227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osswald M, Jung E, Sahm F, et al. Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528( 7580):93–98. [DOI] [PubMed] [Google Scholar]
- 8. Venkataramani V, Tanev DI, Strahle C, et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature. 2019;573(7775):532–538. [DOI] [PubMed] [Google Scholar]
- 9. Venkatesh HS, Morishita W, Geraghty AC, et al. Electrical and synaptic integration of glioma into neural circuits. Nature. 2019;573(7775):539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Linkous A, Balamatsias D, Snuderl M, et al. Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 2019;26(12):3203–3211.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winkler F, Wick W. Harmful networks in the brain and beyond. Science. 2018;359(6380):1100–1101. [DOI] [PubMed] [Google Scholar]
- 12. Weil S, Osswald M, Solecki G, et al. Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro Oncol. 2017;19(10):1316–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gritsenko PG, Atlasy N, Dieteren CEJ, et al. p120-Catenin-dependent collective brain infiltration by glioma cell networks. Nat Cell Biol. 2020;22(1):97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inaba M, Buszczak M, Yamashita YM. Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature. 2015;523(7560):329–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- 16. Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 18. Osswald M, Solecki G, Wick W, Winkler F. A malignant cellular network in gliomas: potential clinical implications. Neuro Oncol. 2016;18(4):479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nimmerjahn A, Kirchhoff F, Kerr JN, Helmchen F. Sulforhodamine 101 as a specific marker of astroglia in the neocortex in vivo. Nat Methods. 2004;1(1):31–37. [DOI] [PubMed] [Google Scholar]
- 20. Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2(4):333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Charles N, Ozawa T, Squatrito M, et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010;6(2):141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 23. van Oijen MG, Medema RH, Slootweg PJ, Rijksen G. Positivity of the proliferation marker Ki-67 in noncycling cells. Am J Clin Pathol. 1998;110(1):24–31. [DOI] [PubMed] [Google Scholar]
- 24. Boral D, Vishnoi M, Liu HN, et al. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat Commun. 2017;8(1):196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cidado J, Wong HY, Rosen DM, et al. Ki-67 is required for maintenance of cancer stem cells but not cell proliferation. Oncotarget. 2016;7(5):6281–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jung E, Osswald M, Blaes J, et al. Tweety-homologue 1 drives brain colonization of gliomas. J Neurosci. 2017;37(29):6837–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu Z, Khan MA, Weiler M, et al. Targeting self-renewal in high-grade brain tumors leads to loss of brain tumor stem cells and prolonged survival. Cell Stem Cell. 2014;15(2):185–198. [DOI] [PubMed] [Google Scholar]
- 28. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Venteicher AS, Tirosh I, Hebert C, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017;355(6332):eaai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tirosh I, Venteicher AS, Hebert C, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539(7628):309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Filbin MG, Tirosh I, Hovestadt V, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018;360(6386):331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eskilsson E, Røsland GV, Solecki G, et al. EGFR heterogeneity and implications for therapeutic intervention in glioblastoma. Neuro Oncol. 2018;20(6):743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15(7): 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8(5):486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.