Abstract
Glioblastoma multiforme (GBM) is the most widespread and aggressive subtype of glioma in adult patients. Numerous long non-coding RNAs (lncRNAs) are deregulated or differentially expressed in GBM. These lncRNAs possess unique regulatory functions in GBM cells, ranging from high invasion/migration to recurrence. This review outlines the present status of specific involvement of lncRNAs in GBM pathogenesis, with a focus on their association with key molecular and cellular regulatory mechanisms. Also, we highlighted the potential of different novel RNA-based strategies that may be beneficial for therapeutic purposes.
Keywords: long non-coding RNAs, glioblastoma, blood-brain barrier, chemoresistance, therapeutics
Graphical abstract
GBM is invasive and accounts for 80% of all primary brain tumors. Most lncRNAs in glioma are aberrantly expressed and participate in the regulation of cellular processes and chemoresistance. Approaches such as RNAi, CRISPR-Cas9, NATs, and ASOs are used to target lncRNAs for their use in therapeutics
Introduction
The central dogma of molecular biology postulates the transmission of the genetic information from genomic DNA to messenger RNA (mRNA), then proteins.1 However, a large portion of genetic information that does not get translated into proteins raises a question about the possible roles of this non-coding portion of the genome in cellular functions. Later studies have highlighted the significance of these untranslated regions previously referred to as unused DNA responsible only for transcriptional noise.2,3 Most (~80%) of the mammalian genome consists of non-coding RNA (ncRNAs) genes, as annotated by ENCODE (Encyclopedia of DNA Elements) and FANTOM (Functional Annotation of the Mouse/Mammalian Genome). ncRNA falls into two sub-categories, based on the length of the transcripts; small ncRNAs are shorter than 200 nt and include transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small interfering RNAs (siRNAs), and microRNAs (miRNAs), whereas long ncRNAs (lncRNAs) are longer than 200 nt. They are structurally similar to mRNAs. lncRNA’s genes are transcribed by RNA polymerase II, capped at the 5′ end, polyadenylated at the 3′ end, and contain promoter region that can be spliced.2 lncRNAs comprise fewer exons (~2.8 exons) than do protein-coding genes and do not have an open reading frame (ORF). lncRNAs usually have a lower expression level but higher tissue-specificity than do most mRNAs.4,5 lncRNAs serve a dynamic role in modulating the expression of various genes at the transcriptional, post-transcriptional, translational, and epigenetic levels, but information on their specific mechanisms is limited.6
In the last few years, 127,802 transcripts from 56,946 lncRNAs genes have been cataloged in LNCipedia (https://lncipedia.org/), the largest lncRNA database. Some of these are aberrantly expressed and could be considered as potential therapeutic targets in clinical oncology and treatment of viral diseases.7, 8, 9
lncRNAs
Depending on the genomic location and with respect to neighboring protein-coding genes, lncRNAs can be sense, antisense, intronic, intergenic, and bidirectional. Sense and antisense are transcribed in the same or opposite direction of protein-coding gene strands. Bidirectional lncRNAs are transcribed in the opposite direction of their neighboring protein-coding gene (<1 kb away). Intronic lncRNAs are transcribed completely from the introns of the protein-coding gene, while intergenic lncRNAs are transcribed from different genomic locations.10 The activity of lncRNAs depends on its subcellular localization, allowing division of lncRNAs into nuclear or cytoplasmic subgroups.11 A gene transcriptional status can be regulated by transcriptional interference and chromatin remodeling acquired from lncRNA located in the nuclear region.12,13 In contrast, lncRNAs that are exported to the cytoplasm serve as mediators for RNA processing and can affect mRNA stability or regulate protein function.14
lncRNAs can interact with RNA, proteins, and DNA via different modes of molecular mechanisms such as signaling, decoy, guide, scaffold, and miRNA sponge. lncRNAs can function as a signaling molecule for any biological condition and are capable of regulating the expression of genes concerning a specific response; as a decoy molecule resulting in binding and sequestering of protein sequences, thereby regulating gene expression; as a guide, it can recruit RNA-binding protein to its target genes, either in trans or cis; and as a scaffold, lncRNAs function to keep proteins together and form a complex with specialized biological functions.15
lncRNAs in glioblastoma
Glioblastoma multiformes (GBMs) account for almost 80% of all primary brain tumors.16 In general, patients show a poor prognosis with a median survival of ~1 year.17 As of the 2016 edition of the World Health Organization classification, gliomas are classified based not only on histopathologic appearance but also on well-established molecular parameters.18 The incorporation of molecular features has most notably impacted the classification of astrocytic and oligodendroglial tumors, which are now grouped together as diffuse gliomas, based on the growth pattern, behavior, and shared isocitrate dehydrogenase (IDH) genetic status. Methylation profiling, a powerful method to evaluate one aspect of the epigenetic landscape of brain tumors, may be added to histologic and standard genetic approaches to classify brain tumors, possibly refining how brain tumors are classified in the future.19 The conventional treatment of GBM includes a combination of three different approaches: surgical resection, irradiation, or temozolomide (TMZ) chemotherapy.20 Most patients with glioma have a poor response to chemotherapy because of intrinsic and acquired resistance.17 Unfortunately, these treatments have not been able to significantly improve the survival rate because of complications from the treatment procedures or the inability to eliminate GBM invasiveness.20 To develop robust treatment options, it is crucial to develop a deep understanding of molecular and cellular aspects of glioma pathogenesis.
Main lncRNAs in gliomas
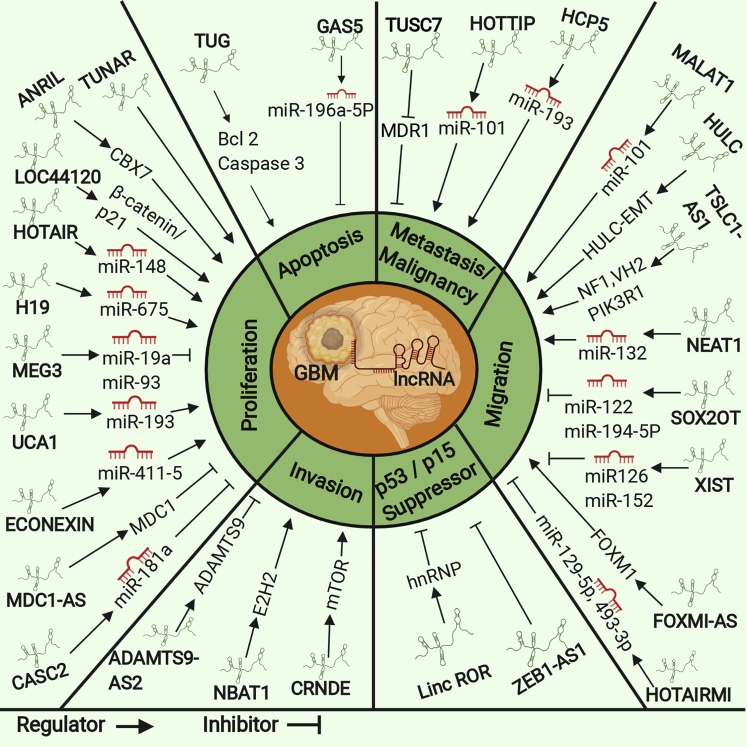
Most lncRNAs in glioma are aberrantly expressed and participate in the regulation of critical cellular processes21. lncRNAs can be organized based on their capabilities to regulate several characteristics and phenotypes as shown in detail in Figure 1 and Table 1.
Figure 1.
Representation of specific long non-coding RNAs that are well associated with glioma and function via targeting various miRNAs or genes/proteins and regulate various pathways such as proliferation, invasion, migration, apoptosis, and metastasis by acting as both regulators and inhibitors
Table 1.
Aberrant expression patterns of various long non-coding RNAs (lncRNAs) involved in glioblastoma
| LncRNAs | Expression pattern | Mechanism | Aberrant phenotype | References |
|---|---|---|---|---|
| HOTAIR | ↑ | interacts with the PRC2 complex | promotes cell cycle progression | 22, 23, 24 |
| HOTAIR/miR-326/FGF1 axis | promotes malignancy | |||
| enhances the tight junction by binding to miR-148b-3p as a ceRNA | decreases the blood-tumor barrier (BTB) permeability | |||
| HOTAIRMI | ↑ | acts as sponge for miR-129-5p and miR-493-3p | functions as ceRNA and promotes cell invasion and migration | 25,26 |
| associated with inflammation and T cell-mediated immune response initiation | ||||
| TUNAR | ↑ | positive regulator of neuronal development | involved in cancer progression, cell cycle conduct, and pluripotency | 27 |
| LincROR | acts as a tumor suppressor | promotes proliferation and enhances glioma stem cell forming ability | 28 | |
| negatively correlated with KLF4 | ||||
| H19 | ↑ | by reducing the miR-29a, it upregulates the VASH2 expression | promotes invasion and angiogenesis, along with tube formation | 29 |
| derives the miR-675, which in turn suppresses CDK6 directly | promotes proliferation and migration | |||
| CRNDE | ↑ | modulates the mTOR signaling pathway | promotes growth and invasion | 30, 31, 32 |
| attenuates the axis of miR-384/PIWIL4 /STAT3 | facilitates proliferation and invasion and inhibits apoptosis | |||
| protects Bcl-2 and Wnt2 by binding to miR-136-5p as a ceRNA | glioma cells migration and the capacity to invade are enhanced | |||
| XIST | ↑ | expression of ZO-2 and FOXC1 transcription factor as a miR-137 sponge are improved | decreases blood-tumor barrier permeability | 33, 34, 35 |
| forms silencing complex RISC with miR-152 induced by RNA | promotes angiogenesis, GSC proliferation, migration, and invasive capacities | |||
| NEAT1 | ↑ | mediates H3K27me3 in target promoters by binding to EZH2 | promotes growth and capacity to invade | 36 |
| TUG1 | ↑↓ | inhibits miR-144, which in turn reverses the effect of miR-144 on occludin, ZO-1, and claudin-5 | regulates blood-tumor barrier permeability | 37,38 |
| recruits polycomb to methylate locus-specific H3K27 histone | maintains stemness features of GSCs | |||
| MALAT1 | ↓↑ | negatively regulates miR-155 | suppresses invasion and proliferation | 39, 40, 41 |
| targets miR-203 | involved in chemoresistance | |||
| targets miR-140; interacts with nuclear factor YA | proliferation, blood-tumor barrier permeability, and chemosensitivity | |||
| targets miR-101 | induces proliferation, migration, autophagy, apoptosis, and correlates with survival | |||
| GAS5 | ↓ | the positive feedback loop of GAS5/miR-196a-5p/FOXO1 is formed | inhibits proliferation, migration, and invasion | 42 |
| CASC2 | ↓ | PTEN expression increases due to interaction with miR-181a | inhibit proliferations and increases TMZ sensitivity | 43 |
| NBAT-1 | ↓ | reduces the expression of the NRSF/REST neuronal-specific transcription factor | weakens proliferation, and neuroblastoma differentiation is increased | 44 |
| FOXM1-AS | ↑ | the smooth interaction of ALKBH5 and FOXM1 mRNA to demethylate FOXM1 mRNA | enhances reclamation and tumorigenesis of glioma stem-like cells | 45 |
| ECONEXIN | – | rise in TOP2A by wiping miR-411-5p | continues aggressive proliferation of glioma cells | 46 |
| SOX2OT | – | binds to miR-194-5p and miR-122, back-pedals SOX3 expression | boosts proliferation, migration, and invasion | 47 |
| HCP5 | – | forms a positive feedback loop of HCP5-miR-139-RUNX1 | causes proliferation, migration, and invasion | 48 |
| MEG3 | ↓ | acts in a p53-dependent manner; ceRNAs for miR-19a and miR-93 | inhibits glioma cell proliferation (in vitro) | 49,50 |
| UCA1 | ↑ | ceRNAs for miR-182 and miR-122 | promotes proliferation, invasion, and migration; modulates glycolysis and invasion of glioma | 51 |
| HOTTIP | ↓ | interacts via BRE, cycA and CDK2, p53 | promotes proliferation and apoptosis | 52 |
| HULC | ↑ | interacts with survivin, c-Myc, cyclin A/D1/E, p-Rb, Skp-1/2, CDK2/4 and EZH2, Bcl-2/Bax, caspase-3/8 | promotes proliferation invasion, migration, and angiogenesis | 53 |
| MDC1-AS | ↓ | upregulates antisense tumor-suppressing gene MDC1 | tumor suppressor; inhibits cell proliferation and cell cycle | 54 |
| TSLC1-AS1 | ↓ | _ | inhibits cell proliferation, migration, and invasion | 55 |
| ADAMTS9-AS2 | ↓ | modulates the protein-coding gene ADAMTS9 | suppresses cell migration | 56 |
| ZEB1-ASI | ↑ | _ | facilitates cell proliferation, migration, and invasion | 57 |
| TUSC 7 | ↓ | suppresses the levels of IC50, TMZ, and MDR1 expression and escalates TMZ cytotoxicity in U87TR cells | inhibits TMZ resistance and tumor malignancy | 58 |
| LOC441204 | ↑ | involved with the β-catenin/p21/cdk4 cascade | associated with cell proliferation and growth | 59 |
| ANRIL | ↑ | associated with the expression of CBX7 (chromo box homolog 7) | cell proliferation and migration | 60 |
↑, upregulation; ↓, downregulation.
HOX transcript antisense intergenic RNA (HOTAIR)
HOTAIR is mapped at chromosome 12q13 and is 2,158 nt long.22,61 Recent reports suggest that it can impact histone methylation, resulting in the silencing of target genes.22 HOTAIR is highly expressed in GBM and can act as an independent prognostic factor.61 In gliomas, HOTAIR targets miRNA (miR)-148b-3p and miR-326.23 One study showed that HOTAIR is involved in the proliferation and tumorigenic potential of GBM by regulating the cells via the EZH2 and Wnt/β-catenin pathways.62 Moreover, HOTAIR can act as exosomal lncRNAs playing a significant role in the regulation of angiogenesis in glioma. For instance, patients with recurrent GBM showed increased serum levels of HOTAIR exosomal lncRNA that were associated with poor response to TMZ and worse prognosis.63
HOTAIR myeloid-specific 1 (HOTAIRM1)
HOTAIRM1 was first identified as a myeloid-specific regulator of the HOXA (homeobox A cluster) gene family and is located between the HOXA1 and HOXA2 loci. It regulates the transcription of specific genes during myeloid cell differentiation and maturation via chromosome remodeling.64 HOTAIRM1 showed an increased expression level especially in the high-grade astrocytomas and is involved in glioma development.25 Moreover, reports suggest that HOTAIRM1 has significant roles in glioma recurrence.26 A study by Liang et al.65 in 2019 observed that the overexpression of HOTAIRM1 was positively related to glioma grade progression. In U87 and LN229 cells, knockdown of HOTAIRM1 resulted in inhibition of glioma proliferation, invasion, epithelial-to-mesenchymal transition (EMT), cell migration, and enhanced sensitivity to TMZ in vitro. Furthermore, HOTAIRM1 functions as a ceRNA (competing endogenous RNA) to promote glioma cell invasion and migration by acting as a sponge for miR-129-5p and miR-495-3p. Additionally, a higher expression of HOTAIRM1 was associated with increased inflammation and initiation of a T cell-mediated immune response. lncRNA HOTAIRM1 was reported to promote glioma malignancy by forming a ceRNA network.
TCL1 upstream neural differentiation-associated RNA (TUNAR)
TUNAR is a conserved lncRNA found in vertebrates and is significantly expressed in the central nervous system (CNS).66 It is found to be a positive regulator of neuronal development and differentiation in humans, mice, and zebrafish, which indicates that downregulated TUNAR in brain tumors (GBM and lower-grade glioma [LGG]) is required to gain oncogenic characteristics.67 Some reports suggest that lncRNAs such as BCAR4 and TUNAR are found to be involved in cancer progression, cell cycle conduct, and pluripotency.27 A recent comprehensive study by Reon et al.68 analyzed the deregulated expression of three lncRNAs (TUNAR, LINC00152, LINC01476) and found that TUNAR and LINC01476 were highly upregulated in GBMs compared to normal brain tissue.
HOXA distal transcript antisense RNA (HOTTIP)
HOTTIP, an antisense transcript mapped at the distant end of the HOXA gene cluster, is related to the PRC2 and WDR5/MLL1 chromatin-modifying complexes and binds directly with WDR5. It is overexpressed in glioma tissues with high viability and hypoxia-treated glioma cells under hypoxic conditions. HOTTIP knockdown may inhibit hypoxia-induced EMT in glioma cells by increasing miR-101, which can regulate metastasis via modulation of ZEB1 expression. Based on these outcomes, HOTTIP may be a significant marker and therapeutic target in GBM.52
Large intergenic noncoding RNA regulator of reprogramming (LINC-ROR)
LINC ROR is 2.6 kb long, a cytoplasmic lncRNA located on chromosome 18q21.3, and is involved in the modulation of cellular reprogramming.28,69,70 Recently it was discovered that lncRNA-ROR mainly acts as an endogenous miRNA sponge in cancer cells to promote tumor development. Evidence reveals that LINC-ROR, p53, and caspase-3 are found to be upregulated in GBM patients.69 In GBM, LINC ROR is involved in the process of DNA damage, suppresses the expression of p53 by binding to hnRNP, and can regulate cell apoptosis.71 In another study, it was shown that upregulated expression of KRUPPEL-like factor 4 (KLF4) results in proliferation by enhancing glioma stem cell (GSC) formation.
H19 imprinted maternally expressed transcript (H19)
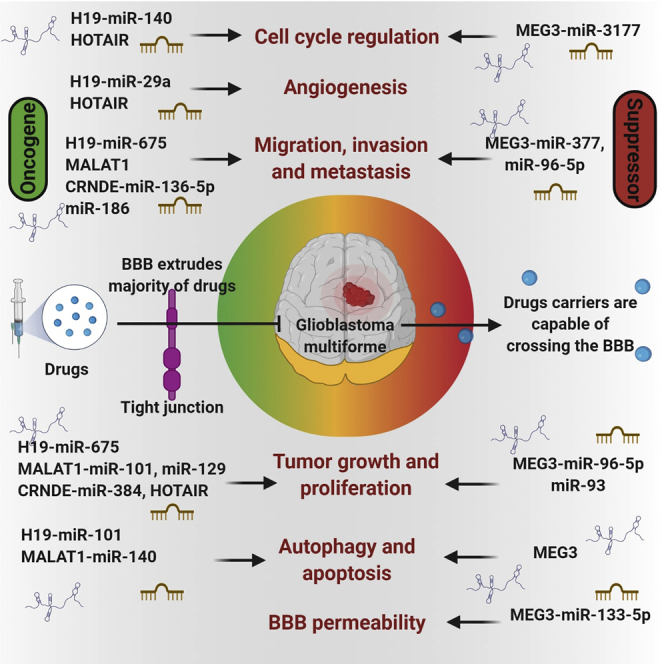
H19 mapped on the chromosome 11p15.5 and is 2.3 kb long.72,73 In glioma cells, binding of H19 with c-Myc oncogene results in the induction of tumorigenesis.73 Increasing evidence suggests that upregulated expression of H19 in GBM tissues is associated with GBM patient survival. It has been reported that miR-675, a miRNA derived by the first exon of H19, could potentially promote migration through CDK6, regulate glioma cell proliferation, and induce glioma cell invasion by targeting cadherin 13.72 Few other miRNAs that participate in H19-induced glioma tumorigeneses are miR-29a,51, miR-140,52, and miR-152.26 The depletion of H19 by siRNAs is associated with decreased glioma invasion.73 Higher expression of H19 results in angiogenesis, stemness of GBM cells in vitro, and invasion.74 Increased level of H19 lncRNA promotes invasion, angiogenesis, and stemness of glioblastoma cells.74 In the murine xenograft model, overexpression of H19 is associated with increased tumor growth. Suppressing H19 modulates tumorigenicity and stemness in U251 and U87 MG glioma cells.75 Upregulated H19 promotes TMZ resistance in U251 and M059J glioma cell lines as studied by Jia et al.76 The study has reported that the resistant cells have higher H19 expression than do non-resistant cells, and silencing of H19 resulted in downregulation of EMT markers and Wnt/β-catenin markers.58,76
Taurine upregulated gene 1 (TUG1)
TUG1 is nearly 7.1 kb long and resides on chromosome 22q12. It is highly upregulated in GSCs.77 The expression of the TUG1 is highly dependent on Notch1 activation in GSCs. In the cytoplasm, TUG1 functions to promote self-renewal by sponging miR-145, whereas in the nucleus it recruits polycomb repressive complex 1/2 (PRC1/2) to suppress the proliferation of genes by locus-specific methylation of histone H3K27 via YY1-binding activity.37 Studies conducted based on gain and loss of function showed that dysregulation of TUG1 affects apoptosis and cell proliferation.78
Colorectal neoplasia differentially expressed (CRNDE)
CRNDE is mapped on the 16th human chromosome and is 1,059 nt long.30,61 The higher expression of CRNDE was shown to play a regulatory role in glioma development.25 A microarray analysis-based study performed by Hu and colleagues26 revealed that CRNDE is involved in glioma recurrence, and higher expression of CRNDE contributed to cell apoptosis while it inhibited cell proliferation in glioma. CRNDE has many transcriptional isoforms because of alternative splicing.61 In glioma cells, CRDNE is regulated via the mTOR signaling pathway. The upregulation of CRNDE promotes tumor formation in a murine model and in the in vitro invasion system.30 Another role of this lncRNA includes the regulation of PIWIL4 mediating gene silencing, leading to the promotion of gliomagenesis via activation of STAT3 signaling.61
Mediator of DNA damage checkpoint protein 1 antisense (MDC1-AS)
MDC1-AS is an antisense transcript of tumor suppressor MDC1.54 However, its function is undetermined. MDC1-AS positively regulates the expression of MDC1 and inhibits the proliferation and cycle of U87MG and U251 cell lines when overexpressed.54 In glioma cells, MDC1-AS acts as a tumor suppressor by upregulating its antisense tumor-suppressing gene MDC1.54
Maternally expressed gene 3 (MEG3)
MEG3 is transcribed from the maternal allele.79,80 The lower expression of MEG3 has been found to behave as a tumor suppressor in glioma and is capable of activating both p53-dependent and p53-independent pathways.61 The overexpression of MEG3 inhibits cancer cell growth by hypomethylating MEG3.81 When MEG3 is overexpressed, the p53 level is elevated due to MDM2 downregulation.82 A study revealed that MEG3 may compete with miR-19a and miR-93 ceRNAs to inhibit the cellular growth of GBM.83,84 Also, increased MEG3 and Wnt/β-catenin expression in glioma cells was shown to inhibit cell growth and metastasis.49
X-inactive specific transcript (XIST)
XIST is involved in X chromosome inactivation in mammals to balance gene expression between sexes.61 In general, XIST knockout studies in glioblastoma cells revealed attenuated cell growth and uptake of glucose by sponging miR-126 via the IRSI/phosphatidylinositol 3-kinase (PI3K)/Akt pathway.85 XIST knockdown causes upregulation of miR-152, leading to the suppression of GSC proliferation, migration, invasion, and tumorigenic potential.33 XIST knockdown improved the brain-tumor barrier (BTB) permeability to repress angiogenesis in glioma.33 Mechanistically, XIST can regulate the expression of the transcription factor (TF) forkhead box C1 (FOXC1) and zonula occludens 2 (ZO-2).33 Gene expression analysis studies found that XIST is overexpressed in glioma endothelial cells.61 Consequently, a few other studies show how the behavior of XIST as ceRNAs suppress the action of miRNAs, thus conferring chemoresistance and oncogenic properties to glioma cells.34,61,86
Tumor suppressor in lung cancer-1 antisense-1 (TSLC1-AS1)
This lncRNA is the antisense transcript of a tumor suppressor.55 TSLC1-AS1 expression is downregulated in tumor specimens55 and acts as a TSLC1 expression mediator.55 TSLC1-AS1’s overexpression results in TSLC1 upregulation and inhibits proliferation, migration, and invasion of U87 cells.55 This regulation may require a new modulator instead of promoter hypermethylation.55
Highly upregulated in liver cancer (HULC)
HULC is a lncRNA located on chromosome 6p24.3.53 Upregulation of HULC promotes proliferation and colony formation of glioma cells.53 The underlying molecular mechanisms of HULC overexpression in glioma remain undetermined. In the case of hepatocellular carcinoma (HCC) cells, by sequestering miR-200a-3p, the increase in HULC gene expression and activity was associated with improved EMT-related parameters.53 Therefore, the HULC-EMT pathway could cause cell migration and invasion.
Nuclear paraspeckle assembly transcript 1 (NEAT1)
The lncRNA oncogene NEAT1 is reportedly involved in promoting glioma development by suppressing miR-132.87 This indirectly promotes SOX2 expression involved in tumor formation of glioma.87 NEAT1 knockdown could potentially inhibit glioma cell proliferation and metastasis. The differential expression of NEAT1 in astrocytomas and oligodendrogliomas was associated with lineage specificity and terminal differentiation of glial cells.25 Moreover, NEAT1 expression also relates to overall survival and may act as a prognostic candidate for LGG patients. Upregulated expression of NEAT1 was related to poor outcomes in LGG patients and promoted proliferation, invasion, and migration.88 Interestingly, few studies have shown a NEAT1 knockdown inhibitory effect on cell viability and invasion in gliomas.87 It is known to upregulate the c-Met oncogene expression via miR-449b-5p, which negatively regulates c-Met expression levels. A recent study demonstrated that NEAT1 is probably upregulated by the oncogenic epidermal growth factor receptor (EGFR) pathway where it acts as a scaffold molecule, promoting GBM tumorigenesis by recruiting histone-modifying enzyme.87
ADAM metallopeptidase with thrombospondin type 1 motif, 9 antisense RNA 2 (ADAMTS9-AS2)
ADAMTS9-AS2 is the antisense partner lncRNA of the protein-coding ADAMTS9 gene, which is a well-recognized tumor suppressor.56 ADAMTS9-AS2 expression level is regulated by promoter hypermethylation and histone modification.56 One study indicated that the high methylation level is responsible for the modulation of its inactivation in glioma cells.56 Also, this study highlighted the ability of ADAMTS9-AS2 to suppress cell migration by modulating the protein-coding gene ADAMTS9.56 Reportedly, ADAMTS9-AS2’s overexpression inhibits the migration and invasion abilities of T98G cells, which allows increased tumor progression. ADAMTS-AS2 was shown to be downregulated during tumorigenesis of glioma.56
ZEB antisense RNA 1 (ZEB1-AS1)
ZEB1-AS1 upregulation is correlated with tumor progression. ZEB1-AS1 has a significant effect on glioma cellular proliferation, migration, and invasion.57 Recent reports have shown that the knockout of ZEB1-AS1 attenuated tumor growth in the glioma murine model.57 Thus, it is a tumor-promoting lncRNA in glioma cells. Similarly, in other loss-of-function studies, ZEB1-AS1 was seen to effectively promote colorectal cancer cell proliferation, partially by suppressing p15 gene expression.57
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)
MALAT1, or nuclear-enriched transcript 2 (NEAT2), is located on chromosome 11q13.1.39 MALAT1 silencing leads to increased apoptosis of glioma cells and promotes TMZ resistance by miR-203 suppression and TS function activation.89 The downregulation of MALAT1 reduced levels of TMZ resistance in U251 and U87 cell lines by regulating ZEB1 expression.39 ZEB1 is synthesized by EMT, whose role has been recently identified in MDR. Furthermore, Cai et al.90 showed that MALAT1 shares crosstalk with TMZ resistance, and upregulated expression of MALAT1 was found both in vitro and in vivo in glioma and was shown to direct binding with miR-101 to decrease miR-101 expression to regulate TMZ resistance. However, decreased expression of MALAT1 in cells resulted in overexpression of miR-101 followed by decreased levels of MGMT and glycogen synthase kinase-3β (GSK3B), indicating that both of these proteins are involved in the mechanism of TMZ resistance.58,90
Growth arrest-specific 5 (GAS5)
GAS5 is mapped on chromosome 1q25 and can inhibit tumor growth in gliomas.42 One study revealed that miR-196a-5p is overexpressed in glioblastoma tissues. GAS5 targets miR-196a-5p, which results in improved FOXO1 expression and a high transcriptional rate of PID1 and MIIP genes. When FOXO1 is upregulated, it causes an increase in GAS5 transcription. This results in the formation of a positive regulatory feedback loop. The GAS5/miR-196a-5p/FOXO1 axis suppresses GSC proliferation, migration, invasion, and apoptosis. These outcomes demonstrate that GAS5 is a prominent target for glioma treatment.42
Cancer susceptibility candidate 2 (CASC2)
CASC2 is located on chromosome 10q26. Alternative splicing yields five CASC2 exons, generating three lncRNAs: CASC2a, CASC2b, and CASC2c.67 Exogenous CACS2 was reported to inhibit the proliferation of glioma cells and amplify cell proliferation repression induced by TMZ. The regulation of sensitivity to TMZ chemotherapy by modulating PTEN is due to the CASC2 and miR-181a interactions.43 Correlation studies have found that CASC2 positively correlates with PTEN and shares an inverse correlation with miR-181a in glioma tissues resistant to TMZ. Subsequently, miR-181a was inversely related to PTEN. Data revealed direct competitive binding with miR-181a between CASC2 and PTEN mRNA. This indicates that glioma cells are sensitized to TMZ as the miR-181a expression is inhibited by CASC2 directly targeting glioma cells.43
Neuroblastoma associated transcript-1 (NBAT-1)
NBAT-1 is mapped on the 6p22 chromosome. It controls the expression of genes involved in the proliferation and invasion of cells by EZH2 interaction epigenetically. Studies demonstrate that during upregulation of NBAT-1 target genes, cell proliferation and invasion are affected in both EZH2 and NBAT-1 knockdown cells. These genes act as a potential target of repression by NBAT-1/EZH2-mediated H3K27me3. NBAT-1 also acts as a risk-assessing biomarker for neuroblastoma tumors. It also has a role in cellular proliferation and invasion. NBAT-1’s role in neuronal cell differentiation is linked to mechanisms and networks of genes involved in the development of neuroblastoma tumors and their progression. NBAT-1, along with SOX9 and NRSF/REST downstream effectors, might be potential targets for the development of therapeutics.44
Forkhead box M1 antisense (FOXM1-AS)
FOXM1-AS is a single lncRNA involved in the interaction between nascent transcripts ALKBH5 and FOXM1. LOC100507424 (FOXM1-A) is a lncRNA found on the 12th chromosome (chr12: 2945982–2968961, GRCh37/hg19), is transcribed in the direction opposite of FOXM1, and is 457 nt long, which are complementary to the last exon of FOXM1 mRNA.45 Its removal increases the m6A modification of FOXM1 precursor (pre-)mRNA. Furthermore, FOXM1-AS knockdown reduced the expression of FLAG-FOXM1 in a similar pattern to ALKBH5 knockdown, which decreased both the activity of FOXM1 3 UTR luciferase and collaboration of HuR and FOXM1 RNA. Therefore, FOXM1-AS vitalizes FOXM1 expression and aids in maintaining GSCs.45
ECONEXIN
The ECONEXIN/C130071C03Rik expression seems to be regulated indirectly by Tp53 and Nf1. It shows sequence homology of 80% or more to their promoter region.46 ECONEXIN is predominantly located in the cytoplasm; it has two miR-411-5p binding sites. ECONEXIN’s binding to the AGO2 protein and miR-411-5p, which is a part of the RNA complex for silencing, indicates that it plays a role in miRNA regulation through the mechanism of silencing complex induced by RNA. Lack of ECONEXIN in cells causes an inclined miR-411-5p level. Thus, it regulates miR-411-5p through its role as a miRNA sponge for entrapping miR-411-5p, resulting in the upregulation of TOP2A upon glioma formation.46
SOX2OT
SOX2OT is a lncRNA located on chromosome 3q26.3 (Chr3q26.3).47 SOX2OT elimination suppresses cellular proliferation, migration, and invasion, escalating GSCs apoptosis. It also affects apoptosis via the upregulation of miR-194-5p and miR-122 expression, which can downregulate SOX3 expression by SOX2OT, leading to the activation of TDGF-1 expression. This affects GSC biology via the JAK/STAT pathway. The feedback loop of SOX2OT-miR-194-5p/miR-122-SOX3-TDGF-1 plays a vital role in modulating the biology of GSCs.47
Histocompatibility leukocyte antigen (HLA) complex P5 (HCP5)
HCP5 is located on chromosome 6p21.3. HCP5 knockdown restricts cell proliferation, migration, and invasion and promotes apoptosis in glioma cells.48 HCP5 regulates the malignancy of glioma cells by binding to miR-139, which functions as a tumor suppressor. HCP5’s elimination also downregulates the RUNX1 TF, which can promote the expression of AEG-1 involved in a sequence of oncogenic effects in glioma cells.48 RUNX1 plays a part in miR-139-mediated inhibition of malignancy in glioma cells. HCP5 downregulates miR-139 to upregulate RUNX1. Also, interestingly, being primarily expressed in immune system cells, lncRNA HCP5 may have a vital role in autoimmunity.
Urothelial carcinoma-associated 1 (UCA1)
UCA1, a lncRNA mapped on chromosome 19p13.12, is a globally identified oncogenic gene in a variety of malignant tumors.51 It was primarily diagnosed with bladder cancer. UCA1 modulates the biological growth of glioblastoma cells by negatively mediating miR-193a expression, which negatively modulates CDK6 by directly targeting its 3′ UTR. However, it was discovered that CDK6 stimulates PI3K/AKT and MAPK cascades, along with the Notch signaling pathway. UCA1 along with miR-193 may impact these signal cascades via regulating the expression of CDK6. UCA1 silently represses glioblastoma cell (U-118 MG and A172) potential and weakens migration and cell invasion. However, it aids apoptotic progress through miR-193a-mediated CDK6 silencing, while inhibiting the PI3K/AKT, MAPK, and Notch pathways.51 This proves that the lncRNA UCA1 could be a valuable therapeutic target.
Tumor suppressor candidate 7 (TUSC7)
TUSC7 is an antisense lncRNA that has four exons mapped at 3q13.3.91 According to Pasic et al.,92 it was primarily detected in osteosarcoma. Recently, a few studies reported that low TUSC7 expression significantly suppresses tumor growth. TUSC7 expression was interconnected negatively with TMZ resistance.58 Its overexpression reduces levels of half-maximal inhibitory concentration (IC50) of TMZ and MDR1 expression, and it escalates TMZ cytotoxicity in U87TR cells. Up-modulation of miR-10a improves MDR1 expression and IC50 of TMZ, increases cell viability and division, and restricts U87TR cell apoptosis. Furthermore, TUSC7 inhibits the resistance of TMZ in U87TR cells through miR-10a interaction. TUSC7 downregulation in glioma worked as a prognostic biological marker for glioma patients and restricted malignant behavior.91
LOC441204
Lin and colleagues59 investigated the role of LOC441204 in brain tumor samples using oligonucleotide microarray analysis. The study reported an increased expression of LOC441204 that was positively associated with tumor size and grade. Evidence indicates that the diminution of LOC441204 in T98 human glioma cells led to a considerable decrease in proliferative ability and amplified β-catenin degradation. Moreover, LOC441204 accelerated the cell proliferation ability of glioma cell lines via a mechanism involving the β-catenin/p21/cdk4 cascade. Therefore, these results strongly support the fact that LOC441204 is a novel oncogene and may serve as a promising cell growth marker for glioma progression.
Antisense non-coding RNA in the INK4 locus (ANRIL)
Paul et al.60 investigated the role of oncogenic lncRNA ANRIL concerning glioma biology. In general, lncRNA ANRIL transcribes antisense to the INK4b-ARF-INK4a locus that encodes CDKN2B and CDKN2, the two cyclin-dependent kinase inhibitors, as well as ARF that binds to MDM2 and results in its degradation, thereby activating p53. Knocking out ANRIL in LN229 and T98G glioma cell lines inhibited colony formation and cell proliferation. In general, ANRIL is upregulated in glioma cell lines; however, ANRIL is co-deleted with a homozygous deletion of the INK4b/ARF/INK4a locus in GBMs. This is positively associated with the upregulation of all genes present in the locus, i.e., ANRIL, CDKN2A, and CDKN2B. Thus, the lack of ANRIL-mediated suppression of the INK4b-ARFINK4a locus is possibly due to the lower expression of CBX7 (chromo box homolog 7), a co-repressor molecule in GBMs.
lncRNAs involved in chemoresistance
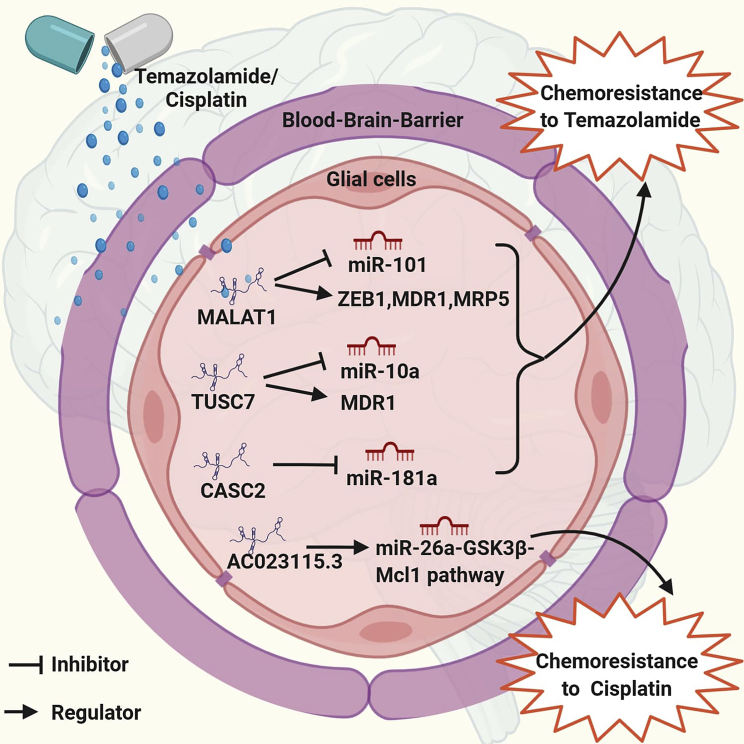
Zhang et al.93 developed TMZ (Temodar), an oral chemotherapeutic drug for clinical use in glioma with the best therapeutic efficacy against patients with high-grade gliomas. A substantial proportion of patients suffering from GBM have inherent or acquired TMZ resistance that significantly deters the clinical outcome.90,94 Since chemoresistance is the main hurdle to a better outcome of GBM chemotherapy, the mechanism of this resistance needs to be understood, and we have to identify a unique therapeutic target. Our current understanding is that MALAT1 boosts the chemoresistance by suppressing the miR-101 pathway of signaling via direct GBM cell binding.90 Another lncRNA AC023115.3 competes with endogenous RNA for miR-26a and constricts the miR-26a inhibitory effect on GSK3β. It promotes Mcl1 deterioration by inducing a GSK3β increase and autophagy decline. Furthermore, AC023115.3 enhances the glioma cell’s chemosensitivity to cisplatin by regulating the miR-26a-GSK3β-Mcl1 pathway. These data indicate that the AC023115.3-miR-26a-GSK3β signaling axis plays a vital role in the reduction of the chemoresistance of gliomas.94
Recently, lncRNAs have been identified as playing a role in chemotherapy resistance in multiple cancers95 (Figure 2). lncRNA RP11-838N2.4 can strengthen the in vivo and in vitro cytotoxic effects of TMZ on GBM cells. lncRNA RP11-838N2.4 plays the role of an endogenous sponge, curbing the function of miR-10a via conserved sequences and strengthening the expression of EphA8, which enhances the rate of cell apoptosis. This leads to intensification of the sensitivity of GBM cells to TMZ. Transforming growth factor β (TGF-β) activity is impeded by lncRNA RP11-838N2.4, which is miR-10a-independent. lncRNA RP11- 838N2.4 characterization might aid in approaches for heightening TMZ efficacy.95
Figure 2.
Illustration of how the mechanism of chemoresistance in glioma to temozolomide (TMZ) and cisplatin with the involvement of lncRNAs, CASC2, MALAT1, and TUSC7 promotes chemoresistance to TMZ by suppressing the miRNAs while AC023115.3 mainly targets the mcl1 pathway and enhances glioma cell chemosensitivity to cisplatin
Partially studied lncRNAs in glioma
Chen et al.96 used a Cox regression analysis and identified four independent lncRNA biomarkers named AK098425, AL833059, AK056155, and CR613436. These were positively associated with the clinical outcome of patients with GBM. Later, functional analysis revealed that the genes co-expressed with these four lncRNAs tended to be clustered most significantly in immune-related biological processes in Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Therefore, understanding the molecular mechanisms of these novel lncRNAs biomarkers during the tumorigenesis and malignant progression of gliomas may provide novel diagnostic, prognostic, and therapeutic targets for gliomas.96
A study by Dong and colleagues97 used lncRNA gene network analysis to identify ASLNC22381 and ASLNC20819 lncRNAs that exhibited significantly higher expression in growth factor-related insulin growth factor (IGF)-1 pathway, leading to recurrence and malignant progression of GBM. Furthermore, the differential expression of lncRNAs in glioma compared to normal brain tissue may have a regulatory function in the PPAR signaling pathways that may be involved in the pathogenesis of GBM.
Studies conducted by Zhang et al.25 on differential lncRNA expression signatures revealed that different lncRNA expression profiles may be potentially involved in distinguishing between astrocytic and oligodendroglial tumors, as well as between grades of the gliomas. For example, LOC400043 was found to be upregulated with increasing malignancy grades while a dramatic increase in the expression level of LOC400043 was reported to play an important role in gliomagenesis.
RFPL1S was reported to function as a regulator of cell-cycle progression, and it was reported as antisense of the coding gene RFPL1 and found to play an essential role in the post-transcriptional regulation of the RFPL1. Furthermore, the differential expression of DLX6-AS between astrocytomas and oligodendrogliomas was displayed to be associated with lineage specificity and terminal differentiation of glial cells.25
A clinical-based investigation performed by Hegi and colleagues98 reported that prominin-1 lncRNA (or CD133) is a predictor of poor survival in patients treated with chemoradiotherapy. Interestingly, its expression was associated with resistance and it maintains a self-renewal signature. This indicates that a tumor stem cell phenotype is involved in the escape of glioblastoma from chemoradiotherapy. Moreover, multivariate analysis revealed the two major clusters characterized by EGFR and HOX (homeobox) gene expression. These are two autonomous prognostic factors for O-6-methylguanine-DNA methyltransferase (MGMT) methylation status and are associated with cell adhesion, innate immune response, and tumor vascularization.
Lin et al.99 predicted the role of potential oncogenic lncRNAs such as CARD8-AS1 and PWAR6. These are dynamically expressed lncRNAs and are regulated by grade-specific TFs. These TFs show higher expression with increasing grades of glioma. Some TF regulators such as SPI1 and PLAU play a critical function in glioma and are associated with tissue remodeling and wound repair. Moreover, CARD8-AS1 and PWAR6 have a significant role in the process of the metastatic potential of glioma cell lines in vitro, in DNA repair, the EMT, regulating immune responses, and patient survival. Therefore, these oncogenic lncRNAs can serve as promising candidates for prognostic biomarkers in glioma.99
Li and colleagues100 have evaluated the role of lncRNAs in GBM that showed epigenetic dysregulation in promoter regions, most of which were hypomethylated. Their study identified six lncRNAs of note; one of them, PRRT3-AS1, was among the most hypermethylated lncRNAs and maybe a potential biomarker of GBM prognosis. Furthermore, it was revealed that the aberrant methylation of either hypermethylated or hypomethylated lncRNAs may result in the disruption of some pathways such as mTOR signaling, apoptosis, ErbB signaling, and FoxO signaling pathways.
Studies conducted by Matjašič et al.101 examined the expression level of several lncRNAs and certain lncRNA-associated miRNAs, which reported that there is a significant change in the expression between the analyzed glioma subtypes and grades. For example, expression levels of seven lncRNAs (7SL, EGO-A, HOTAIR, JPX, MEG3, RNCR3, and ZFAS1) and four miRNAs (miR-770, miR-125b, miR-124a, and miR-196a) showed major association with overall survival considering only the expression of astrocytoma, primary GBMs, and oligodendroglioma.
lncRNA 7SL is downregulated in astrocytic tumors and upregulated in oligodendroglioma and oligoastrocytoma. This difference may be an additional genetic parameter for differentiating these subtypes from each other; upregulated expression of EGO-A (eosinophil granule ontogeny isoform A) was observed in all glioma subtypes, except secondary GBMs. EGO-A host gene ITPR1 (inositol triphosphate receptor type 1) is encoded near to a transcriptional regulator, EGR-1 (early growth response 1), that is responsible for the induction of mitosis, cell differentiation, and growth. The expression of the EGR-1 gene in glioma cells is induced by overexpression of the EGFR gene and platelet-derived GFR (PDGFR) gene.101,102
RNCR3 (retinal noncoding RNA3) is overexpressed in the brain and acts as a sponge for miR-124a. RNCR3 is a precursor for miR-124a and both are downregulated in primary and secondary GBMs. A lower expression of RNCR3 was correlated with a lower survival probability.101
Zottel et al.58 have reviewed a few lncRNAs that are involved in resistance to TMZ in glioblastoma including RP11-838N2.4, NC-TALC, NONHSAT163779, ADAMTS9 antisense RNA2 (ADAMTS9-AS2), P73 antisense RNA 1T non-protein-coding (TP73-AS1), AC003092.1, and SET binding factor 2 antisense RNA 1 (lncSBF2-AS1).
Recently, Cheng and colleagues103 discussed the function of exosomal lncRNAs in glioma progression and clinical applications. Exosomes are small endosomes derived from extracellular vesicles (EVs) of 40–150 nm in diameter. These are secreted by different cells in the body and almost from all body fluids, including urine, blood, saliva, breast milk, and cerebrospinal fluid (CSF).104 Some of the exosomal lncRNAs, such as lncRNA-AHIF and lncRNA-ATB, were reported to be involved in invasion and cell proliferation in gliomas; lincRNA-POU3F3 and lincRNA-CCAT2 played a significant role in the regulation of angiogenesis in gliomas, and lncRNA-SBF2-AS1 enhances chemoresistance to TMZ. Alternatively, few other exosomal lncRNAs such as circATP8B4 and lncRNA-AHIF were reported to promote cell radioresistance and invasion. Furthermore, the exosomal lncRNAs may have a promising clinical function in glioma diagnosis because of their distinctive characteristics such as small size, the ability to circulate freely or package into exosomes, their double-layer phospholipid membrane acting as a seal to protect ncRNAs from the ribonuclease-mediated degradation, and their ability to cross the blood-brain barrier (BBB), thereby suggesting their role as potential biomarkers. For instance, patients with recurrent GBM showed increased serum levels of SBF2-AS1 exosomal lncRNAs that were associated with poor response to TMZ therapy and worse prognosis.63
Lin et al.88 have identified six aberrantly expressed lncRNAs of prognostic significance with their ceRNA networks against LGGs. Based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) RNA sequencing (RNA-seq) databases, six differentially expressed lncRNAs between LGG and normal brain tissue were screened, i.e., AC021739.2, AL031722.1, AL354740.1, FGD5-AS1 (FGD5 antisense RNA1), LINC00844. and NEAT1. These were associated with overall survival and may act as prognostic candidates for LGG patients. For instance, FGD5-AS1 was found to be a prospective therapeutic target for LGG; in contrast, upregulated expression of LINC00844 showed a favorable prognosis, whereas information on the remaining lncRNAs (AC021739.2, AL031722.1, and AL354740.1) is limited. Furthermore, based on the connectivity map (CMap) dataset, six putative drugs (irinotecan, camptothecin, mitoxantrone, azacitidine, mestranol, and enilconazole) were used for glioma treatment that potentially targeted glioma-associated pathways, such as the JAK-STAT signaling pathway, the TGF-β signaling pathway, the HIF-1 signaling pathway, and the mitogen-activated protein kinase (MAPK) signaling pathway.
A microarray analysis-based study performed by Hu and colleagues26 revealed some differentially expressed lncRNAs in recurrent gliomas relative to primary glioma. For example, some lncRNAs such as AC016745.3, XLOC_001711, and RP11-128A17.1 have significant roles in glioma recurrence. Furthermore, the report suggests that mRNA in the lncRNA-mRNA pair may be the target of lncRNA. For example, higher expression of FOXD4L1 (forkhead box protein D4-like 1) correspond to increased expression of enhancer lncRNA AC016745.3 that is involved in increasing the expression of genes that lead to the maintenance of proliferative neural precursors. Decreased expression of mRNA RPRM (reprimo) in pituitary tumors is related to decreased lncRNA XLOC_001711 expression that acts as a tumor suppressor. Another highly expressed sense mRNA, MEIS2 (myeloid ecotropic insertion site 2), corresponds to increased expression of antisense lncRNA RP11-128A17.1, thereby promoting cell proliferation in human neuroblastoma cell lines.
Therapeutic approaches targeting lncRNAs in glioma
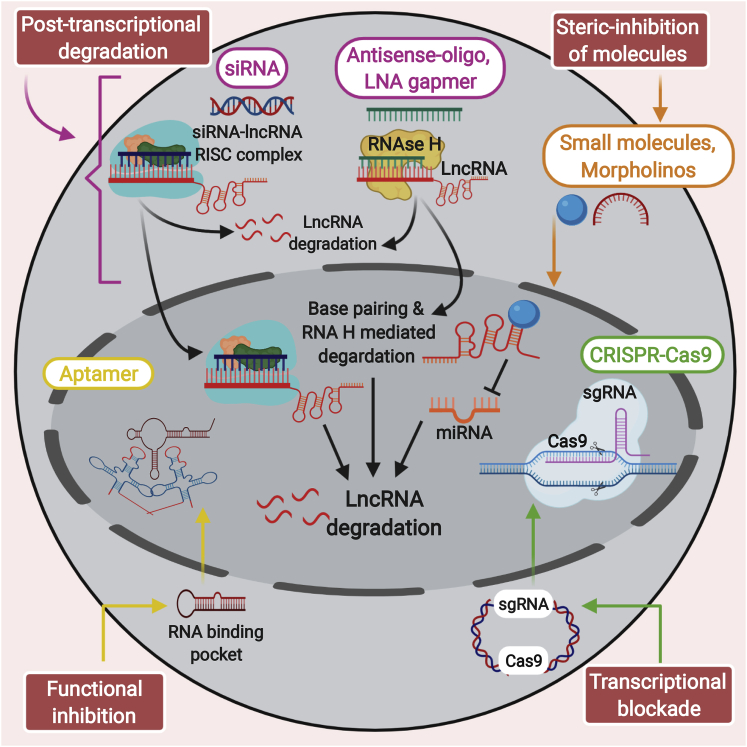
lncRNAs have a significant role at various stages of tumor onset and progression in GBM. Therefore, targeting both nuclear and cytoplasmic lncRNAs may allow us to discover additional therapeutic applications (Figure 3).
Figure 3.
Depiction of the most widely recognized approaches to focus on the RNA of interest
To post-transcriptionally disintegrate the desired RNAs, RNA interference recruits the RISC complex to post-transcriptionally disintegrate the target RNAs; antisense oligonucleotides (ASOs) degrade the RNA of interest through an RNase H-dependent mechanism; CRISPR-Cas9 is used to accomplish transcriptional silencing; and small molecules such as morpholinos and tailored ASOs are used to hinder interactions between the lncRNA and the protein.
Post-transcriptional degradation
Post-transcriptional degradation is the most widely recognized method used to focus on the desired RNA. The development of the RNA-RNA or RNA-DNA duplex is a profoundly explicit procedure that has permitted specialists to examine the capabilities of oligonucleotide-based therapeutics. These therapeutics offer higher efficacy, improved stability, and reduced off-target effects compared to traditional treatment options.105,106
There are two significant methodologies utilized. One is RNA interference (RNAi): in this methodology, siRNAs stimulate dicer action in the cytoplasm and recruit the RISC (RNA-induced silencing complex) to post-transcriptionally disintegrate the desired RNAs. This strategy has been utilized to target mRNAs in various neurotic conditions, such as malignancies, neurological ailments, and metabolic disorders.107 The other methodology is antisense oligonucleotides (ASOs), which are promising therapeutics that target and degrade the RNA of interest through the RNase H-dependent mechanism. The binding of ASOs to the desired RNA can restrain or change the expression of the gene via a steric blockade, alternating the splicing mechanism, and degradation. For instance, intravenous treatment with ASOs targeting TUG1 coupled with a drug delivery system induces GSC differentiation and efficiently represses GSC growth in vivo.37 The improvement of locked nucleic acids (LNAs) and S-constrained ethyl (cEt) alterations represent a portion of the advances in ASOs with improved strength and have promising pharmacokinetic profiles for different malignant growth-related and neurological sicknesses.108, 109, 110
Another subset of ncRNAs is natural antisense RNAs (NATs), which coincide with protein-coding genes and are transcribed in the reverse direction. Upregulated expression of NATs in malignant growth results in the suppression of the related tumor suppressor genes.111 The inhibition of NATs via ASOs, referred to as “antagoNATs,” may increase the expression of tumor suppressor genes. AntagoNATs are linear, synthetically altered LNAs, or different ASOs.111,112 TUG1 is targeted with ASOs intravenously mix with a medication delivery system that results in the induction of differentiation in GSCs and in vivo growth suppression of tumors.37 To protect ASOs, chemical modification at the ends is performed with the addition of 2′-O-methyl and LNA at both the 5′ and 3′ ends that protects it from degradation and helps in the maintenance of constant ASO levels. A preclinical trial demonstrated that MALAT1 ASO, when administered subcutaneously in the MMTV-PyMT mouse model of luminal B breast malignancy, leads to the separation of early tumors with an almost 80% decrease in metastasis compared to non-specific ASOs.113 These investigations showed that MALAT1 ASO may be developed as a possible restorative for metastatic ailment in a few disease types, yet further assessment is required.114 The contemporary and clinically progressed oligonucleotides, peptide nucleic acids (PNAs), are exclusively nucleic acids linked with a neutral-charged peptide spine and are thermally steady in the tumor microenvironment. These are altered in vivo for their utilization in “anti-ncRNA treatment.”115 The PNAs-based technique might be effective in solid tumors, such as gliomas.116
Steric blockade
lncRNA genes can be regulated by the steric shunt of the promoter or by utilizing genome-altering strategies such as CRISPR-Cas9.117 In this methodology, a transcriptional repressor is combined with dead-Cas9, and this complex is guided by guide RNAs to a particular gene promoter to accomplish transcriptional silencing.118 The altering of genome, epigenome, or RNA intervened by CRISPR-Cas9 is a promising methodology for targeting lncRNAs expressed in glioma and could be profitable over regular quality treatment approaches.119
Steric inhibition
The interactions between the lncRNA and the protein can be hindered by utilizing small molecules such as morpholinos and tailored ASOs, which are incapable of using the RNA degradation pathway. These can bring about loss of function by specifically binding to the RNA and blocking the RNA and the protein boundary.120 lncRNAs are capable of forming stable secondary and tertiary structures that can be precisely mapped using SHAPE and PARIS, which are RNA structure determination assays.121, 122, 123 Small molecule inhibitors for bromodomain and extra-terminal (BET) proteins decrease the level of several oncogenes involved in the succession of GBM. For instance, BET domain proteins diminish the expression of HOTAIR, and the utilization of BET inhibitors can re-establish the tumor growth brought about by oncogenic lncRNAs.124
Conclusions
Glioblastoma is one of the most lethal brain diseases. Clinical advances in surgery and radiotherapy have proved to be useful; however, the development of non-invasive methods is still a prerequisite for truly great treatment options. Within the last few years, ncRNAs have gained a fresh impetus in the field of glioma research and could be utilized as a promising tool for targeted therapy. lncRNAs are known to participate in various cellular regulatory pathways that modulate the process of tumorigenesis. Numerous deregulated lncRNAs have been associated with GBM and are involved in almost all of the features of this malignancy, including cell multiplication, movement, angiogenesis, stemness, tumor relapse, severity, and chemoresistance. The cutting-edge biochemical and molecular methodologies have unequivocally helped in elucidating how lncRNAs control and regulate various functions. Although it is promising to find novel diagnostics and therapeutics to improve glioma survival using lncRNAs, to date, the attributes of most of these ncRNAs are indefinite. The distribution of drugs across the intact BBB into the brain is a critical factor to consider during the development of valuable therapies. The therapeutics based on lncRNAs are restricted to only in vivo application because of low stability and poor drug uptake. Thus, future investigations need to give more consideration to recognizing cell-to-cell heterogeneity of lncRNAs in gliomas that will help in the development of novel RNA-based methodology to treat such a malignancy and bring new trust in patients with GBM.
Acknowledgments
This work was supported by the Science and Engineering Research Board under its ECRA scheme (SERB file no. ECR/2018/000059) and the Indian Council of Medical Research (ICMR) file no. 5/13/10/2019/NCD-III (to A.K.P.), and the Department of Biotechnology (DBT) under its Ramalingaswami Fellowship (no. BT/RLF/Re-entry/24/2014) and the Science and Engineering Research Board under its ECRA scheme (SERB file no. ECR/2016/001519) (to M.G.). This work was also supported by a Department of Science and Technology (DST) international grant (DST-RBFR; file no. DST/INT/RUS/RSF/P-29) (to A.K.P.), a Junior Research Fellowship (DST) (to B.Y.), and RSF grant no. 19-44-02027 (МНК-DST(2018)) (to M.P.).
Author contributions
A.K.P. conceived the idea of this review. A.K.P., B.Y., and S.P. wrote the manuscript and presented the concepts in the manuscript. A.K.P., M.G., B.Y., Y.R., M.P., A.G., and S.P. designed and created the figures, revised the paper, and made final corrections. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.CRICKCentral F. Dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Kung J.T.Y., Colognori D., Lee J.T. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong X.L., Jing J.X., Fang Z.M., Bannu-Kuken, Miao H.J., Song H.Y., Zhong H., Lu Y., Liu S.M. Efficacy and safety analysis of gastrodin injection combined with anti-anxiety drug for treatment of climacteric hypertension. Int. J. Clin. Exp. Med. 2016;9:12120–12124. [Google Scholar]
- 4.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pal S., Garg M., Pandey A.K. Deciphering the mounting complexity of the p53 regulatory network in correlation to long non-coding RNAs (lncRNAs) in ovarian cancer. Cells. 2020;9:527. doi: 10.3390/cells9030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q. LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y., Wang P., Wang Y., Ma X., Zhi H., Zhou D., Li X., Fang Y., Shen W., Xu Y. Lnc2Cancer v2.0: updated database of experimentally supported long non-coding RNAs in human cancers. Nucleic Acids Res. 2019;47(D1):D1028–D1033. doi: 10.1093/nar/gky1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volders P.J., Anckaert J., Verheggen K., Nuytens J., Martens L., Mestdagh P., Vandesompele J. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019;47(D1):D135–D139. doi: 10.1093/nar/gky1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L.L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Jain A.K., Xi Y., McCarthy R., Allton K., Akdemir K.C., Patel L.R., Aronow B., Lin C., Li W., Yang L., Barton M.C. lncPRESS1 is a p53-regulated lncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol. Cell. 2016;64:967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y., Merkurjev D., Zhang J., Ohgi K., Song X. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin A., Li C., Xing Z., Hu Q., Liang K., Han L., Wang C., Hawke D.H., Wang S., Zhang Y. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat. Cell Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartonicek N., Maag J.L.V., Dinger M.E. Long noncoding RNAs in cancer: Mechanisms of action and technological advancements. Mol. Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanif F., Muzaffar K., Perveen K., Malhi S.M., Simjee ShU. Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac. J. Cancer Prev. 2017;18:3–9. doi: 10.22034/APJCP.2017.18.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Y., Zhou G., Li M., Hu D., Zhang L., Liu P., Lin K. Long noncoding RNA DANCR mediates cisplatin resistance in glioma cells via activating AXL/PI3K/Akt/NF-κB signaling pathway. Neurochem. Int. 2018;118:233–241. doi: 10.1016/j.neuint.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 19.Capper D., Jones D.T.W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D.E. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X.Q., Sun S., Lam K.F., Kiang K.M.Y., Pu J.K.S., Ho A.S.W., Lui W.M., Fung C.F., Wong T.S., Leung G.K.K. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol. Dis. 2013;58:123–131. doi: 10.1016/j.nbd.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Cruickshanks N., Pahuski M., Yuan F., Dutta A., Schiff D., Purow B., Abounader R. Noncoding RNAs in glioblastoma. In: De Vleeschouwer S., editor. chapter 6. Codon Publications; 2017. https://exonpublications.com/index.php/exon/article/view/128/151 (Gliobastoma). [PubMed] [Google Scholar]
- 22.Sa L., Li Y., Zhao L., Liu Y., Wang P., Liu L., Li Z., Ma J., Cai H., Xue Y. The role of HOTAIR/miR-148b-3p/USF1 on regulating the permeability of BTB. Front. Mol. Neurosci. 2017;10:194. doi: 10.3389/fnmol.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Ke J., Yao Y.L., Zheng J., Wang P., Liu Y.H., Ma J., Li Z., Liu X.B., Li Z.Q., Wang Z.H., Xue Y.X. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6:21934–21949. doi: 10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K., Sun X., Zhou X., Han L., Chen L., Shi Z., Zhang A., Ye M., Wang Q., Liu C. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6:537–546. doi: 10.18632/oncotarget.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., Sun S., Pu J.K.S., Tsang A.C.O., Lee D., Man V.O.Y., Lui W.M., Wong S.T.S., Leung G.K.K. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y., Wu J.-J., Lin X.-B., Bao Y., Chen Z.-H., Zhang C.-R., Cai Z., Zhou J.-Y., Ding M.-H., Wu X.-J. Differential lncRNA expression profiles in recurrent gliomas compared with primary gliomas identified by microarray analysis. Int. J. Clin. Exp. Med. 2015;8:5033–5043. [PMC free article] [PubMed] [Google Scholar]
- 27.Bouckenheimer J., Fauque P., Lecellier C.H., Bruno C., Commes T., Lemaître J.M., De Vos J., Assou S. Differential long non-coding RNA expression profiles in human oocytes and cumulus cells. Sci. Rep. 2018;8:2202. doi: 10.1038/s41598-018-20727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao H., Wang T., Zhang P., Shang M., Gao Z., Yang F., Liu R. linc-ROR regulates apoptosis in esophageal squamous cell carcinoma via modulation of p53 ubiquitination by targeting miR-204-5p/MDM2. J. Cell. Physiol. 2020;235:2325–2335. doi: 10.1002/jcp.29139. [DOI] [PubMed] [Google Scholar]
- 29.Jia P., Cai H., Liu X., Chen J., Ma J., Wang P., Liu Y., Zheng J., Xue Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381:359–369. doi: 10.1016/j.canlet.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Wang Y., Li J., Zhang Y., Yin H., Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 31.Zheng J., Liu X., Wang P., Xue Y., Ma J., Qu C., Liu Y. CRNDE promotes malignant progression of Glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol. Ther. 2016;24:1199–1215. doi: 10.1038/mt.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Li D.X., Fei X.R., Dong Y.F., Cheng C.D., Yang Y., Deng X.F., Huang H.L., Niu W.X., Zhou C.X., Xia C.Y., Niu C.S. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget. 2017;8:88163–88178. doi: 10.18632/oncotarget.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu H., Xue Y., Wang P., Liu X., Ma J., Zheng J., Li Z., Li Z., Cai H., Liu Y. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis. 2017;6:e303. doi: 10.1038/oncsis.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Z., Li Z., Ma K., Li X., Tian N., Duan J., Xiao X., Wang Y. Long non-coding RNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. J. Cancer. 2017;8:4106–4116. doi: 10.7150/jca.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao Y., Ma J., Xue Y., Wang P., Li Z., Liu J., Chen L., Xi Z., Teng H., Wang Z. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 36.Chen Q., Cai J., Wang Q., Wang Y., Liu M., Yang J., Zhou J., Kang C., Li M., Jiang C. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/β-catenin pathway by scaffolding EZH2. Clin. Cancer Res. 2018;24:684–695. doi: 10.1158/1078-0432.CCR-17-0605. [DOI] [PubMed] [Google Scholar]
- 37.Katsushima K., Natsume A., Ohka F., Shinjo K., Hatanaka A., Ichimura N., Sato S., Takahashi S., Kimura H., Totoki Y. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016;7:13616. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai H., Xue Y., Wang P., Wang Z., Li Z., Hu Y., Li Z., Shang X., Liu Y. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget. 2015;6:19759–19779. doi: 10.18632/oncotarget.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Yuan X., Yan D., Li D., Guan F., Dong Y., Wang H., Liu X., Yang B. Long non-coding RNA MALAT1 decreases the sensitivity of resistant glioblastoma cell lines to temozolomide. Cell. Physiol. Biochem. 2017;42:1192–1201. doi: 10.1159/000478917. [DOI] [PubMed] [Google Scholar]
- 40.Cao S., Wang Y., Li J., Lv M., Niu H., Tian Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am. J. Cancer Res. 2016;6:2561–2574. [PMC free article] [PubMed] [Google Scholar]
- 41.Han Y., Wu Z., Wu T., Huang Y., Cheng Z., Li X., Sun T., Xie X., Zhou Y., Du Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7:e2123. doi: 10.1038/cddis.2015.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X., Liu Y., Zheng J., Liu X., Chen J., Liu L., Wang P., Xue Y. GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1605–1617. doi: 10.1016/j.bbamcr.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 43.Liao Y., Shen L., Zhao H., Liu Q., Fu J., Guo Y., Peng R., Cheng L. lncRNA CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J. Cell. Biochem. 2017;118:1889–1899. doi: 10.1002/jcb.25910. [DOI] [PubMed] [Google Scholar]
- 44.Pandey G.K., Mitra S., Subhash S., Hertwig F., Kanduri M., Mishra K., Fransson S., Ganeshram A., Mondal T., Bandaru S. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 45.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bögler O. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deguchi S., Katsushima K., Hatanaka A., Shinjo K., Ohka F., Wakabayashi T., Zong H., Natsume A., Kondo Y. Oncogenic effects of evolutionarily conserved noncoding RNA ECONEXIN on gliomagenesis. Oncogene. 2017;36:4629–4640. doi: 10.1038/onc.2017.88. [DOI] [PubMed] [Google Scholar]
- 47.Su R., Cao S., Ma J., Liu Y., Liu X., Zheng J., Chen J., Liu L., Cai H., Li Z. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol. Cancer. 2017;16:171. doi: 10.1186/s12943-017-0737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teng H., Wang P., Xue Y., Liu X., Ma J., Cai H., Xi Z., Li Z., Liu Y. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of glioma cells. Mol. Ther. 2016;24:1806–1822. doi: 10.1038/mt.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Cui X., Sun D., Shen B., Wang X. MEG-3-mediated Wnt/β-catenin signaling pathway controls the inhibition of tunicamycin-mediated viability in glioblastoma. Oncol. Lett. 2018;16:2797–2804. doi: 10.3892/ol.2018.9048. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Ma B., Gao Z., Lou J., Zhang H., Yuan Z., Wu Q., Li X., Zhang B. Long non-coding RNA MEG3 contributes to cisplatin-induced apoptosis via inhibition of autophagy in human glioma cells. Mol. Med. Rep. 2017;16:2946–2952. doi: 10.3892/mmr.2017.6897. [DOI] [PubMed] [Google Scholar]
- 51.Xin H., Liu N., Xu X., Zhang J., Li Y., Ma Y., Li G., Liang J. Knockdown of lncRNA-UCA1 inhibits cell viability and migration of human glioma cells by miR-193a-mediated downregulation of CDK6. J. Cell. Biochem. 2019;120:15157–15169. doi: 10.1002/jcb.28777. [DOI] [PubMed] [Google Scholar]
- 52.Zhang S., Wang W., Liu G., Xie S., Li Q., Li Y., Lin Z. Long non-coding RNA HOTTIP promotes hypoxia-induced epithelial-mesenchymal transition of malignant glioma by regulating the miR-101/ZEB1 axis. Biomed. Pharmacother. 2017;95:711–720. doi: 10.1016/j.biopha.2017.08.133. [DOI] [PubMed] [Google Scholar]
- 53.Yan H., Tian R., Zhang M., Wu J., Ding M., He J. High expression of long noncoding RNA HULC is a poor predictor of prognosis and regulates cell proliferation in glioma. OncoTargets Ther. 2016;10:113–120. doi: 10.2147/OTT.S124614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yue H., Zhu J., Xie S., Li F., Xu Q. MDC1-AS, an antisense long noncoding RNA, regulates cell proliferation of glioma. Biomed. Pharmacother. 2016;81:203–209. doi: 10.1016/j.biopha.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 55.Qin X., Yao J., Geng P., Fu X., Xue J., Zhang Z. lncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int. J. Clin. Exp. Pathol. 2014;7:3065–3072. [PMC free article] [PubMed] [Google Scholar]
- 56.Yao J., Zhou B., Zhang J., Geng P., Liu K., Zhu Y., Zhu W. A new tumor suppressor lncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. 2014;35:7935–7944. doi: 10.1007/s13277-014-1949-2. [DOI] [PubMed] [Google Scholar]
- 57.Wei N., Wei H., Zhang H. Long non-coding RNA ZEB1-AS1 promotes glioma cell proliferation, migration and invasion through regulating miR-577. Eur. Rev. Med. Pharmacol. Sci. 2018;22:3085–3093. doi: 10.26355/eurrev_201805_15068. [DOI] [PubMed] [Google Scholar]
- 58.Zottel A., Šamec N., Videtič Paska A., Jovčevska I. Coding of glioblastoma progression and therapy resistance through long noncoding RNAs. Cancers (Basel) 2020;12:1842. doi: 10.3390/cancers12071842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin T.K., Chang C.N., Tsai C.S., Huang Y.C., Lu Y.J., Chen W.J., Lin Y.H., Chung I.H., Lin K.H. The long non-coding RNA LOC441204 enhances cell growth in human glioma. Sci. Rep. 2017;7:5603. doi: 10.1038/s41598-017-05688-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul Y., Thomas S., Patil V., Kumar N., Mondal B., Hegde A.S., Arivazhagan A., Santosh V., Mahalingam K., Somasundaram K. Genetic landscape of long noncoding RNA (lncRNAs) in glioblastoma: Identification of complex lncRNA regulatory networks and clinically relevant lncRNAs in glioblastoma. Oncotarget. 2018;9:29548–29564. doi: 10.18632/oncotarget.25434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng T., Li L., Zhou Y., Gao L. Exploring long noncoding RNAs in glioblastoma: Regulatory mechanisms and clinical potentials. Int. J. Genomics. 2018;2018:2895958. doi: 10.1155/2018/2895958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou X., Ren Y., Zhang J., Zhang C., Zhang K., Han L., Kong L., Wei J., Chen L., Yang J. HOTAIR is a therapeutic target in glioblastoma. Oncotarget. 2015;6:8353–8365. doi: 10.18632/oncotarget.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y., Zhang M., Zhou F. Biological functions and clinical applications of exosomal long non-coding RNAs in cancer. J. Cell. Mol. Med. 2020;24:11656–11666. doi: 10.1111/jcmm.15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X.Q.D., Dostie J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2017;45:1091–1104. doi: 10.1093/nar/gkw966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang Q., Li X., Guan G., Xu X., Chen C., Cheng P. Long non-coding RNA, HOTAIRM1, promotes glioma malignancy by forming a ceRNA network. Aging. 2019;11:6805–6838. doi: 10.18632/aging.102205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin N., Chang K.Y., Li Z., Gates K., Rana Z.A., Dang J., Zhang D., Han T., Yang C.S., Cunningham T.J. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reon B.J., Anaya J., Zhang Y., Mandell J., Purow B., Abounader R., Dutta A. Expression of lncRNAs in low-grade gliomas and glioblastoma multiforme: an in silico analysis. PLoS Med. 2016;13:e1002192. doi: 10.1371/journal.pmed.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toraih E.A., El-Wazir A., Hussein M.H., Khashana M.S., Matter A., Fawzy M.S., Hosny S. Expression of long intergenic non-coding RNA, regulator of reprogramming, and its prognostic value in patients with glioblastoma. Int. J. Biol. Markers. 2019;34:69–79. doi: 10.1177/1724600818814459. [DOI] [PubMed] [Google Scholar]
- 70.Chen W., Wang H., Liu Y., Xu W., Ling C., Li Y., Liu J., Chen M., Zhang Y., Chen B. linc-RoR promotes proliferation, migration, and invasion via the Hippo/YAP pathway in pancreatic cancer cells. J. Cell. Biochem. 2020;121:632–641. doi: 10.1002/jcb.29308. [DOI] [PubMed] [Google Scholar]
- 71.Ge H., Liu J., Liu F., Sun Y., Yang R. Long non-coding RNA ROR mitigates cobalt chloride-induced hypoxia injury through regulation of miR-145. Artif. Cells Nanomed. Biotechnol. 2019;47:2221–2229. doi: 10.1080/21691401.2019.1620759. [DOI] [PubMed] [Google Scholar]
- 72.Li C., Lei B., Huang S., Zheng M., Liu Z., Li Z., Deng Y. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am. J. Transl. Res. 2015;7:1747–1764. [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang P., Wang P., Sun X., Yuan Z., Zhan R., Ma X., Li W. Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy. OncoTargets Ther. 2016;9:3501–3509. doi: 10.2147/OTT.S96278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang X., Yan Y., Hu M., Chen X., Wang Y., Dai Y., Wu D., Wang Y., Zhuang Z., Xia H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 2016;124:129–136. doi: 10.3171/2014.12.JNS1426. [DOI] [PubMed] [Google Scholar]
- 75.Li W., Jiang P., Sun X., Xu S., Ma X., Zhan R. Suppressing H19 modulates tumorigenicity and stemness in U251 and U87MG glioma cells. Cell. Mol. Neurobiol. 2016;36:1219–1227. doi: 10.1007/s10571-015-0320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jia L., Tian Y., Chen Y., Zhang G. The silencing of lncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/β-Catenin pathway. OncoTargets Ther. 2018;11:313–321. doi: 10.2147/OTT.S154339. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Park J.Y., Lee J.E., Park J.B., Yoo H., Lee S.-H., Kim J.H. Roles of long non-coding RNAs on tumorigenesis and glioma development. Brain Tumor Res. Treat. 2014;2:1–6. doi: 10.14791/btrt.2014.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li J., Zhang M., An G., Ma Q. lncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp. Biol. Med. (Maywood) 2016;241:644–649. doi: 10.1177/1535370215622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miyoshi N., Wagatsuma H., Wakana S., Shiroishi T., Nomura M., Aisaka K., Kohda T., Surani M.A., Kaneko-Ishino T., Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- 80.Zhang X., Rice K., Wang Y., Chen W., Zhong Y., Nakayama Y., Zhou Y., Klibanski A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matjasic A., Popovic M., Matos B., Glavac D. Expression of LOC285758, a potential long non-coding biomarker, is methylation-dependent and correlates with glioma malignancy grade. Radiol. Oncol. 2017;51:331–341. doi: 10.1515/raon-2017-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D.L., Gejman R., Ansell P.J., Zhao J., Weng C., Klibanski A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 83.Zhang S., Guo W. Long non-coding RNA MEG3 suppresses the growth of glioma cells by regulating the miR-96-5p/MTSS1 signaling pathway. Mol. Med. Rep. 2019;20:4215–4225. doi: 10.3892/mmr.2019.10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang L., Liang X., Li Y. Long non-coding RNA MEG3 inhibits cell growth of gliomas by targeting miR-93 and inactivating PI3K/AKT pathway. Oncol. Rep. 2017;38:2408–2416. doi: 10.3892/or.2017.5871. [DOI] [PubMed] [Google Scholar]
- 85.Cheng Z., Luo C., Guo Z. lncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J. Cell. Biochem. 2020;121:2170–2183. doi: 10.1002/jcb.29440. [DOI] [PubMed] [Google Scholar]
- 86.Du P., Zhao H., Peng R., Liu Q., Yuan J., Peng G., Liao Y. lncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci. Rep. 2017;37 doi: 10.1042/BSR20170696. BSR20170696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou K., Zhang C., Yao H., Zhang X., Zhou Y., Che Y., Huang Y. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol. Cancer. 2018;17:105. doi: 10.1186/s12943-018-0849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin J.Z., Lin N., Zhao W.J. Identification and validation of a six-lncRNA prognostic signature with its ceRNA networks and candidate drugs in lower-grade gliomas. Genomics. 2020;112:2990–3002. doi: 10.1016/j.ygeno.2020.05.016. [DOI] [PubMed] [Google Scholar]
- 89.Chen W., Xu X.K., Li J.L., Kong K.K., Li H., Chen C., He J., Wang F., Li P., Ge X.S., Li F.C. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8:22783–22799. doi: 10.18632/oncotarget.15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cai T., Liu Y., Xiao J. Long noncoding RNA MALAT1 knockdown reverses chemoresistance to temozolomide via promoting microRNA-101 in glioblastoma. Cancer Med. 2018;7:1404–1415. doi: 10.1002/cam4.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shang C., Tang W., Pan C., Hu X., Hong Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother. Pharmacol. 2018;81:671–678. doi: 10.1007/s00280-018-3522-y. [DOI] [PubMed] [Google Scholar]
- 92.Pasic I., Shlien A., Durbin A.D., Stavropoulos D.J., Baskin B., Ray P.N. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160–171. doi: 10.1158/0008-5472.CAN-09-1902. [DOI] [PubMed] [Google Scholar]
- 93.Zhang J., Stevens M.F.G., Bradshaw T.D. Temozolomide: mechanisms of action, repair and resistance. Curr. Mol. Pharmacol. 2012;5:102. doi: 10.2174/1874467211205010102. [DOI] [PubMed] [Google Scholar]
- 94.Ma B., Yuan Z., Zhang L., Lv P., Yang T., Gao J., Pan N., Wu Q., Lou J., Han C., Zhang B. Long non-coding RNA AC023115.3 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1393–1404. doi: 10.1016/j.bbamcr.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 95.Liu Y., Xu N., Liu B., Huang Y., Zeng H., Yang Z., He Z., Guo H. Long noncoding RNA RP11-838N2.4 enhances the cytotoxic effects of temozolomide by inhibiting the functions of miR-10a in glioblastoma cell lines. Oncotarget. 2016;7:43835–43851. doi: 10.18632/oncotarget.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen G., Cao Y., Zhang L., Ma H., Shen C., Zhao J. Analysis of long non-coding RNA expression profiles identifies novel lncRNA biomarkers in the tumorigenesis and malignant progression of gliomas. Oncotarget. 2017;8:67744–67753. doi: 10.18632/oncotarget.18832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Han L., Zhang K., Shi Z., Zhang J., Zhu J., Zhu S., Zhang A., Jia Z., Wang G., Yu S. lncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int. J. Oncol. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 98.Murat A., Migliavacca E., Gorlia T., Lambiv W.L., Shay T., Hamou M.F., de Tribolet N., Regli L., Wick W., Kouwenhoven M.C.M. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 99.Lin X., Jiang T., Bai J., Li J., Wang T., Xiao J., Tian Y., Jin X., Shao T., Xu J. Characterization of transcriptome transition associates long noncoding RNAs with glioma progression. Mol. Ther. Nucleic Acids. 2018;13:620–632. doi: 10.1016/j.omtn.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ji J., Zhao L., Zhao X., Li Q., An Y., Li L., Li D. Genome-wide DNA methylation regulation analysis of long non-coding RNAs in glioblastoma. Int. J. Mol. Med. 2020;46:224–238. doi: 10.3892/ijmm.2020.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matjašič A., Tajnik M., Boštjančič E., Popović M., Matos B., Glavač D. Identifying novel glioma-associated noncoding RNAs by their expression profiles. Int. J. Genomics. 2017;2017:2312318. doi: 10.1155/2017/2312318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kaufmann K., Thiel G. Epidermal growth factor and platelet-derived growth factor induce expression of Egr-1, a zinc finger transcription factor, in human malignant glioma cells. J. Neurol. Sci. 2001;189:83–91. doi: 10.1016/s0022-510x(01)00562-7. [DOI] [PubMed] [Google Scholar]
- 103.Cheng J., Meng J., Zhu L., Peng Y. Exosomal noncoding RNAs in glioma: biological functions and potential clinical applications. Mol. Cancer. 2020;19:66. doi: 10.1186/s12943-020-01189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang L., Gu Y., Du Y., Liu J. Exosomes: diagnostic biomarkers and therapeutic delivery vehicles for cancer. Mol. Pharm. 2019;16:3333–3349. doi: 10.1021/acs.molpharmaceut.9b00409. [DOI] [PubMed] [Google Scholar]
- 105.Khvorova A., Watts J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stein C.A., Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 2017;25:1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Petrocca F., Altschuler G., Tan S.M., Mendillo M.L., Yan H., Jerry D.J., Kung A.L., Hide W., Ince T.A., Lieberman J. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24:182–196. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Obika S., Nanbu D., Hari Y., Morio K.I., In Y., Ishida T., Imanishi T. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Tetrahedron Lett. 1997;38:8735–8738. [Google Scholar]
- 109.Burel S.A., Han S.R., Lee H.S., Norris D.A., Lee B.S., Machemer T., Park S.Y., Zhou T., He G., Kim Y. Preclinical evaluation of the toxicological effects of a novel constrained ethyl modified antisense compound targeting signal transducer and activator of transcription 3 in mice and cynomolgus monkeys. Nucleic Acid Ther. 2013;23:213–227. doi: 10.1089/nat.2013.0422. [DOI] [PubMed] [Google Scholar]
- 110.Pandey S.K., Wheeler T.M., Justice S.L., Kim A., Younis H.S., Gattis D., Jauvin D., Puymirat J., Swayze E.E., Freier S.M. Identification and characterization of modified antisense oligonucleotides targeting DMPK in mice and nonhuman primates for the treatment of myotonic dystrophy type 1. J. Pharmacol. Exp. Ther. 2015;355:329–340. doi: 10.1124/jpet.115.226969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Modarresi F., Faghihi M.A., Lopez-Toledano M.A., Fatemi R.P., Magistri M., Brothers S.P., van der Brug M.P., Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J. Molecular biology: Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 113.Arun G., Diermeier S., Akerman M., Chang K.C., Wilkinson J.E., Hearn S., Kim Y., MacLeod A.R., Krainer A.R., Norton L. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mendell J.T. Targeting a long noncoding RNA in breast cancer. N. Engl. J. Med. 2017;374:2287–2289. doi: 10.1056/NEJMcibr1603785. [DOI] [PubMed] [Google Scholar]
- 115.Cheng C.J., Bahal R., Babar I.A., Pincus Z., Barrera F., Liu C., Svoronos A., Braddock D.T., Glazer P.M., Engelman D.M. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Özeş A.R., Wang Y., Zong X., Fang F., Pilrose J., Nephew K.P. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci. Rep. 2017;7:894. doi: 10.1038/s41598-017-00966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thakore P.I., D’Ippolito A.M., Song L., Safi A., Shivakumar N.K., Kabadi A.M., Reddy T.E., Crawford G.E., Gersbach C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang H.X., Li M., Lee C.M., Chakraborty S., Kim H.W., Bao G., Leong K.W. CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev. 2017;117:9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 120.Kloosterman W.P., Lagendijk A.K., Ketting R.F., Moulton J.D., Plasterk R.H.A. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pegueroles C., Gabaldón T. Secondary structure impacts patterns of selection in human lncRNAs. BMC Biol. 2016;14:60. doi: 10.1186/s12915-016-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lu Z., Zhang Q.C., Lee B., Flynn R.A., Smith M.A., Robinson J.T., Davidovich C., Gooding A.R., Goodrich K.J., Mattick J.S. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016;165:1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Novikova I.V., Dharap A., Hennelly S.P., Sanbonmatsu K.Y. 3S: Shotgun secondary structure determination of long non-coding RNAs. Methods. 2013;63:170–177. doi: 10.1016/j.ymeth.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 124.Pastori C., Kapranov P., Penas C., Peschansky V., Volmar C.H., Sarkaria J.N., Bregy A., Komotar R., St Laurent G., Ayad N.G., Wahlestedt C. The bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc. Natl. Acad. Sci. USA. 2015;112:8326–8331. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]