Abstract
Background
Subphenotypes have been identified in patients with sepsis and ARDS and are associated with different outcomes and responses to therapies.
Research Question
Can unique subphenotypes be identified among critically ill patients with COVID-19?
Study Design and Methods
Using data from a multicenter cohort study that enrolled critically ill patients with COVID-19 from 67 hospitals across the United States, we randomly divided centers into discovery and replication cohorts. We used latent class analysis independently in each cohort to identify subphenotypes based on clinical and laboratory variables. We then analyzed the associations of subphenotypes with 28-day mortality.
Results
Latent class analysis identified four subphenotypes (SP) with consistent characteristics across the discovery (45 centers; n = 2,188) and replication (22 centers; n = 1,112) cohorts. SP1 was characterized by shock, acidemia, and multiorgan dysfunction, including acute kidney injury treated with renal replacement therapy. SP2 was characterized by high C-reactive protein, early need for mechanical ventilation, and the highest rate of ARDS. SP3 showed the highest burden of chronic diseases, whereas SP4 demonstrated limited chronic disease burden and mild physiologic abnormalities. Twenty-eight-day mortality in the discovery cohort ranged from 20.6% (SP4) to 52.9% (SP1). Mortality across subphenotypes remained different after adjustment for demographics, comorbidities, organ dysfunction and illness severity, regional and hospital factors. Compared with SP4, the relative risks were as follows: SP1, 1.67 (95% CI, 1.36-2.03); SP2, 1.39 (95% CI, 1.17-1.65); and SP3, 1.39 (95% CI, 1.15-1.67). Findings were similar in the replication cohort.
Interpretation
We identified four subphenotypes of COVID-19 critical illness with distinct patterns of clinical and laboratory characteristics, comorbidity burden, and mortality.
Key Words: coronavirus, COVID-19, latent class analysis, phenotypes, subphenotypes
Abbreviations: AKI, acute kidney injury; SP1, SP2, SP3, SP4, subphenotype 1, subphenotype 2, subphenotype 3, subphenotype 4; STOP-COVID, Study of Treatment and Outcomes in Critically Ill Patients With COVID-19
FOR EDITORIAL COMMENT, SEE PAGE 795
COVID-19, caused by the novel coronavirus SARS-CoV-2, results in a broad spectrum of disease manifestations, ranging from mild self-limited illness to life-threatening organ dysfunction.1 The presence of biologically distinct subgroups, or subphenotypes, has been hypothesized in critically ill patients with COVID-19 based on reports of differences in lung compliance, thrombotic complications, and inflammatory response.2, 3, 4, 5, 6, 7 However, to date, empiric evidence identifying and characterizing COVID-19 clinical subphenotypes is limited.
In other heterogeneous syndromes, such as sepsis, ARDS, and acute kidney injury (AKI), latent class analysis has been used to identify distinct subphenotypes based on clinical and biochemical characteristics.7, 8, 9, 10, 11, 12 These subphenotypes have been associated not only with clinical outcomes, but also with response to trial therapies such as high positive end-expiratory pressure for ARDS and vasopressin for AKI in retrospective analyses of clinical trials data.9 , 10 Furthermore, the subphenotype-defining characteristics in these studies yielded important insights into the heterogeneity of disease pathophysiologic features. Identifying subphenotypes in critically ill patients with COVID-19 may have a similar ability to elucidate pathophysiologic features, to predict outcomes, and to guide treatment selection.
We hypothesized that latent, currently uncharacterized, clinically and physiologically distinct subphenotypes exist within critically ill patients with COVID-19 and that they are associated with differences in the risk of important clinical outcomes. We therefore aimed to identify and replicate COVID-19 subphenotypes based on clinical and laboratory variables in a large, multicenter cohort of ICU patients. We then examined the associations of subphenotypes with 28-day mortality and other clinically relevant outcomes.
Methods
Patient Population
We used data from patients enrolled in the Study of the Treatment and Outcomes in Critically Ill Patients with COVID-19 (STOP-COVID), a multicenter cohort study conducted at 67 geographically diverse hospitals across the United States (e-Table 1).13 We included consecutive adults (≥ 18 years of age) with laboratory-confirmed SARS-CoV-2 infection admitted to participating ICUs between March 4 and April 13, 2020. Patients were followed up until the first incidence of the following: hospital discharge, death, or May 11, 2020 (the date on which the database for the current analysis was locked). Details of data collection for STOP-COVID are described elsewhere and are summarized e-Table 2.13 Clinical outcomes were recorded daily up to 14 days (defined in e-Table 2). Survival was tracked for a minimum of 28 days after ICU admission or until hospital discharge, whichever occurred first.
Statistical Analysis
Subphenotypes were identified using latent class analysis. Latent class analysis is a group of statistical methods that can be used to identify unobserved (latent) subphenotypes within a heterogeneous population by clustering on observed characteristics, such as laboratory variables.11 We randomly split enrolling centers into discovery and replication cohorts targeting a 2:1 overall patient ratio between the cohorts (e-Fig 1). We used 25 acute clinical and laboratory variables, selected a priori, as endogenous, class-defining variables (e-Table 3). Outcomes such as mortality were not used to define classes. We used ICU day 1 values for all class-defining variables included in the models.
Latent class analysis models containing between one and five classes were created sequentially in the discovery cohort. Selection of the most appropriate latent class model was determined using Akaike and Bayesian information criteria, the Vuong-Lo-Mendell-Rubin likelihood ratio test, probability of class assignment (class separation), entropy, class prevalence (favoring models with classes > 10% of the sample population), and clinical interpretability.14 , 15 After model selection in the discovery cohort, we repeated the same steps independently in the replication cohort. Further detailed description of the latent class methodology is contained in e-Appendix 1.
After class assignment, we examined the distributions of class-defining variables, baseline demographics, and clinical characteristics across the candidate subphenotypes using heatmaps. Heatmaps were created using Microsoft Excel for Mac version 16.38 (Microsoft). Baseline characteristics across the two cohorts and across subphenotypes were compared using descriptive statistics.
Survival analysis was performed using Kaplan-Meier estimation and restricted mean survival time.16 Poisson regression was used to compare individual 28-day mortality across subphenotypes.17 The interaction between subphenotype and the effect of steroids and anti-IL-6 therapies on mortality was also assessed. Models were adjusted for demographic features and comorbidities previously associated with mortality in COVID-19, US region, and hospital ICU bed capacity. Differences in the incidence of ARDS, thrombotic events, AKI, secondary infection, and new-onset congestive heart failure within 14 days after ICU admission were assessed using Cox proportional hazards regression, adjusted for reported risk factors for each outcome.18, 19, 20, 21, 22 To account for the competing risk of death, we computed cause-specific Cox proportional hazards and treated death as a censoring event.23
All statistical analyses were performed using Stata/IC version 15.1 software (StataCorp LP), unless otherwise specified. Additional details of the statistical methods used in this work can be found in e-Appendix 1 and e-Figure 1. All findings are reported in concordance with the Strengthening the Reporting of Observational Studies in Epidemiology guidelines (e-Table 4).
Results
Discovery and Replication Cohorts
Of the 67 centers in STOP-COVID, 45 centers (2,188 patients) were selected randomly for inclusion in the discovery cohort and 22 centers (1,112 patients) were included in the replication cohort. Table 1 compares patient demographics, clinical characteristics, US region, and missing data between the two cohorts. The replication cohort included a higher percentage of patients of Hispanic or Latino ethnicity, altered mental status on ICU day 1, and vasopressor use on ICU day 1 and also was characterized by slightly lower lymphocyte percentage, lower C-reactive protein levels, and higher D-dimer and ferritin values on ICU day 1. e-Table 5 shows a comparison of patient characteristics with and without missing values for several of these key variables missing in > 40% of patients. Standardized mean differences were all < 0.2, most were < 0.1, and a few of the observed differences seemed to be clinically meaningful. More than 70% of patients in the replication cohort were treated at centers in the Northeast United States, compared with approximately 50% in the discovery cohort, which had greater representation from the South, Midwest, and West regions.
Table 1.
Comparison of Baseline Variables in the Discovery and Replication Cohorts
| Variable | Discovery Cohort | Replication Cohort | Missing | P Value |
|---|---|---|---|---|
| Baseline characteristics | ||||
| Age, y | 63 (52-71) | 62 (52-71) | 0 (0) | .607 |
| Male sex | 1,418 (64.8) | 702 (63·1) | 0 (0) | .342 |
| Race | ... | ... | 0 (0) | < .001 |
| White | 755 (34.5) | 466 (41.9) | ... | ... |
| Black | 783 (35.8) | 260 (23.4) | ... | ... |
| Other | 161 (7.4) | 94 (8.5) | ... | ... |
| Unknown | 489 (22.3) | 292 (26.3) | ... | ... |
| Latino ethnicity | 360 (16.5) | 318 (28.6) | 0 (0) | < .001 |
| BMI, kg/m2 | 30 (26-36) | 30 (27-36) | 384 (11.6) | .927 |
| Diabetes | 913 (41.7) | 445 (40.0) | 0 (0) | .346 |
| Hypertension | 1,363 (62.3) | 673 (60.5) | 0 (0) | .322 |
| Kidney function (eGFRa), mL/min/1.73 m2 | ... | ... | 0 (0) | .867 |
| ≥ 90 | 684 (31.3) | 365 (32.8) | ... | ... |
| 60-< 90 | 789 (36.1) | 388 (34.9) | ... | ... |
| 30-< 60 | 454 (20.7) | 229 (20.6) | ... | ... |
| 15-< 30 | 104 (4.8) | 56 (5.0) | ... | ... |
| < 15 or RRT | 157 (7.2) | 74 (6.7) | ... | ... |
| Coronary artery disease | 309 (14.1) | 148 (13.3) | 0 (0) | .523 |
| Congestive heart failure | 227 (10.4) | 101 (9.1) | 0 (0) | .241 |
| Atrial fibrillation or flutter | 164 (7.5) | 72 (6.5) | 0 (0) | .282 |
| COPD | 175 (8.0) | 103 (9.3) | 0 (0) | .216 |
| Asthma | 238 (10.9) | 109 (9.8) | 0 (0) | .341 |
| Chronic liver disease | 74 (3.4) | 38 (3.4) | 0 (0) | .958 |
| HIV/AIDS | 31 (1.4) | 17 (1.5) | 0 (0) | .800 |
| Active malignancy | 102 (4.7) | 60 (5.4) | 0 (0) | .356 |
| Solid organ transplantation | 69 (3.2) | 37 (3.3) | 0 (0) | .789 |
| Bone marrow transplantation | 4 (0.2) | 4 (0.4) | 0 (0) | .329 |
| Blood type A | 433 (19.8) | 250 (22.5) | 0 (0) | .071 |
| Smoking status | ... | ... | 0 (0) | .419 |
| Never smoker | 1,245 (5.9) | 610 (54.9) | ... | ... |
| Current or former | 648 (29.6) | 336 (30.2) | ... | ... |
| Unknown | 295 (13.5) | 166 (14.9) | ... | ... |
| Preadmission ACEI or ARB | 729 (33.3) | 394 (35.4) | 0 (0) | .226 |
| Preadmission anticoagulation | 225 (10.3) | 98 (8.8) | 0 (0) | .179 |
| Preadmission immunosuppressive medication | 236 (10.8) | 106 (9.5) | 5 (0.2) | .255 |
| Clinical and laboratory data | ... | ... | ... | ... |
| Coinfection on ICU day 1 | 513 (23.5) | 213 (19.2) | 2 (0.1) | .005 |
| Altered mental status on ICU day 1 | 437 (20.0) | 320 (28.8) | 1 (< 0.1) | < .001 |
| Tmax ICU day 1, °F | 100.5 (99.1-102.0) | 100.4 (99.0-101.9) | 3 (0.1) | .058 |
| HRmax ICU day 1 | 105 (91-120) | 103 (90-119) | 3 (0.1) | .165 |
| SBPmin ICU day 1, mm Hg | 96 (85-111) | 97 (86-110) | 4 (0.1) | .156 |
| RRmax ICU day 1 | 32 (26-38) | 30 (25-36) | 0 (0) | < .001 |
| Respiratory support and oxygenation on ICU day 1 | ... | ... | 0 (0) | .624 |
| Neither HFNC, NIPPV, nor MVb | 266 (12.2) | 155 (13.9) | ... | ... |
| BPV, CPAP, HFNC, or NRBc | 494 (22.6) | 226 (20.3) | ... | ... |
| Vent P to F ratio | ||||
| > 300 | 347 (15.9) | 175 (15.7) | ... | ... |
| > 200-≤ 300 | 187 (8.5) | 104 (9.4) | ... | ... |
| > 100-≤ 200 | 486 (22.2) | 242 (21.8) | ... | ... |
| ≤ 100 | 397 (18.1) | 203 (18.3) | ... | ... |
| ECMO | 11 (0.5) | 7 (0.6) | ... | ... |
| No. of vasoactive infusions on ICU day 1 | 1 (0-1) | 1 (0-1) | 1,069 (33.4) | .024 |
| 24-h UOP on ICU day 1, mL/d | 700 (300-1,168) | 722 (324-1,300) | 1,537 (46.6) | .145 |
| WBC count, K/mm3 | 8.1 (5.9-11.3) | 8.4 (6.0-11.8) | 188 (5.7) | .129 |
| Lymphocyte, % | 10.1 (6.3-15.4) | 9.5 (5.4-14.7) | 613 (18.6) | .005 |
| Hemoglobin, g/dL | 12.6 (11.1-14.1) | 12.6 (11.0-13.9) | 193 (5.9) | .205 |
| Platelet count, K/mm3 | 210 (160-269) | 215 (166-274) | 204 (6.2) | .249 |
| Creatinine, mg/dL | 1.10 (0.80-1.69) | 1.04 (0.80-1.70) | 157 (4.8) | .608 |
| Albumin, g/dL | 3.2 (2.8-3.6) | 3.2 (2.8-3.5) | 638 (19.3) | .607 |
| AST, U/L | 54 (36-85) | 55 (36-85) | 631 (19.1) | .733 |
| ALT, U/L | 35 (23-58) | 37 (23-61) | 609 (18.5) | .190 |
| Total bilirubin, mg/dL | 0.6 (0.4-0.8) | 0.6 (0.4-0.8) | 615 (18.6) | .102 |
| Lactate, mM | 1.5 (1.1-2.3) | 1.5 (1.1-2.2) | 1,246 (37.8) | .742 |
| CRP, mg/L | 162 (97-243) | 144 (76-232) | 1,316 (39.9) | < .001 |
| Arterial pH | 7.37 (7.30-7.43) | 7.37 (7.30-7.43) | 1,036 (31.4) | .897 |
| D-dimer, ng/mL | 1,209 (660-2,815) | 1,600 (822-3,935) | 1,646 (49.9) | < .001 |
| Ferritin, ng/mL | 1,015 (488-1,979) | 1,077 (550-2,261) | 1,451 (44.0) | .024 |
| Procalcitonin, ng/mL | 0.38 (0.15-1.22) | 0.40 (0.14-1.72) | 1,319 (40.0) | .381 |
| CK, U/L | 210 (98-565) | 210 (97-505) | 1,795 (54.4) | .546 |
| Center and region data | ||||
| Total centers | 45 | 22 | ... | ... |
| Total patients | 2,188 (66.3) | 1,112 (33.7) | ... | ... |
| US regiond | ... | ... | ... | ... |
| Northeast | 1,088 (49.7) | 807 (72.6) | ... | ... |
| South | 249 (11.4) | 90 (8.1) | ... | ... |
| Midwest | 618 (28.2) | 155 (13.9) | ... | ... |
| West | 233 (10.6) | 60 (5.4) | ... | ... |
Data are presented as No. (%) or median (interquartile range), unless otherwise indicated. The discovery and replication cohorts were constructed by randomly choosing centers from all available STOP-COVID sites with a target patient split of approximately 2:1 between discovery and replication. Differences between cohorts were compared using the Kruskal-Wallis test and the χ 2 test, as appropriate. ACEI = angiotensin converting enzyme inhibitor; ALT = alanine aminotransferase; ARB = angiotensin receptor blocker; AST = aspartate aminotransferase; BPV = bilevel pressure ventilation; CK = creatine kinase; CRP = C-reactive protein; ECMO = extracorporeal membrane oxygenation; eGFR = estimated glomerular filtration rate; HFNC = high-flow nasal cannula; HRmax = maximum heart rate; MV = mechanical ventilation; NIPPV = noninvasive positive pressure ventilation; NRB = nonrebreather mask; P to F = partial pressure of arterial oxygenation to fraction of inspired oxygenation; RRmax = maximum respiratory rate; RRT = renal replacement therapy; SBPmax = maximum systolic BP; STOP-COVID = Study of Treatment and Outcomes in Critically Ill Patients with COVID-19; Tmax = maximum temperature; UOP = urine output.
Via the Chronic Kidney Disease Epidemiology Collaboration equation.
Includes patients who received supplemental oxygen by nasal cannula administration.
Ninety-two percent of patients in this category (n = 662 of 720) were receiving respiratory support via HFNC or nonrebreather mask on ICU day 1. The remaining 8% (n = 58 of 720) were receiving BPV or CPAP.
Regions comprise the following US states (only states that contributed to the STOP-COVID database are listed): (1) Northeast: Connecticut, Washington, DC, Massachusetts, Maryland, New Jersey, New York, and Pennsylvania; (2) South: Alabama, Florida, Louisiana, North Caroline, Tennessee, Texas, and Virginia; (3) Midwest: Illinois, Indiana, Kentucky, Michigan, Minnesota, Missouri, Ohio, Oklahoma, and Wisconsin; and (4) West: Arizona, California, Colorado, Nevada, Oregon, and Washington.
Identification of COVID-19 Critical Illness Subphenotypes
In the discovery cohort, model fit statistics showed improving fit with increasing number of classes (Table 2 ). However, the five-class model showed a substantial increase in class assignment uncertainty (7.2%-11.1% across classes vs 0.7%-2.3% for the four-class model) and contained a relatively small class comprising 7.4% of patients. The four-class model included class proportions of between 11.2% and 39.7%, with high mean class probabilities (0.85-0.91 across four classes) (e-Fig 2). Investigator review of the four-class model suggested that differences in characteristics by class were clinically interpretable (see later). We then repeated model derivation steps in the replication cohort. The four-class model again showed improved model fit and reduced class uncertainty compared with models with fewer classes (Table 2), good representation across classes (13.0%-32.4%), and evident clinical interpretability (see later); a five-class model did not converge. We therefore maintained four-class models for each cohort, with classes labeled as COVID-19 subphenotypes 1 through 4 (SP1, SP2, SP3, and SP4).
Table 2.
Fit Statistics for Latent Class Models From One to Five Classes in the Discovery and Replication Cohorts
| No. of Classes | Log-Likelihood (Model) | df | AICa | BICa | N1 | N2 | N3 | N4 | N5 | Entropyb | Class Uncertaintyc | P Valued |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery cohort | ||||||||||||
| One | –16,694.32 | 52 | 33,492.64 | 33,788.56 | 2,188 (100) | ... | ... | ... | ... | —e | —e | —e |
| Two | –14,457.54 | 105 | 29,125.09 | 29,722.62 | 868 (39.7) | 1,320 (60.3) | ... | ... | ... | 0.791 | 3.2-4.8 | < .001 |
| Three | –13,702.23 | 158 | 27,720.47 | 28,619.61 | 682 (31.2) | 553 (25.3) | 953 (43.6) | ... | ... | 0.773 | 4.8-7.8 | < .001 |
| Four | –13,185.91 | 211 | 26,793.83 | 27,994.57 | 244 (11.2) | 600 (27.4) | 475 (21.7) | 869 (39.7) | ... | 0.771 | 0.7-2.3 | < .001 |
| Five | –12,806.37 | 264 | 26,140.73 | 27,643.09 | 393 (18.0) | 162 (7.4) | 452 (20.7) | 512 (23.4) | 669 (30.6) | 0.765 | 7.2-11.0 | < .001 |
| Replication cohort | ||||||||||||
| One | –8,338.35 | 52 | 16,780.71 | 17,041.43 | 1,112 (100) | ... | ... | ... | ... | —e | —e | —e |
| Two | –7,129.74 | 105 | 14,469.47 | 14,995.93 | 664 (59.7) | 448 (40.3) | ... | ... | ... | 0.801 | 1.7-2.5 | < .001 |
| Three | –6,692.71 | 158 | 13,701.43 | 14,493.63 | 431 (38.8) | 327 (29.4) | 354 (31.8) | ... | ... | 0.790 | 4.0-7.4 | < .001 |
| Four | –6,369.49 | 211 | 13,160.98 | 14,218.92 | 145 (13.0) | 360 (32.4) | 251 (22.6) | 356 (32.0) | ... | 0.791 | 0.7-2.3 | < .001 |
| Five | Model does not converge | |||||||||||
Data are presented as No. (%) or percentage, unless otherwise indicated. AIC = Akaike information criterion; BIC = Bayesian information criterion; df = degrees of freedom; N = number.
Criteria for model selection that estimate the relative distance of the fitted likelihood model to the unknown, true likelihood function that generated the data, while including a penalty for higher numbers of model parameters. Hence, a lower AIC and BIC suggest improved model fit, but may not yield a simpler (more parsimonious) model.
Measure of class separation. Values range from 0-1, with larger values indicating a greater degree of class separation.
Percentage of patients within each class with a marginal probability of belonging to their assigned class, defined as a probability of 0.45-0.55. After model fitting, probability of being assigned to each class was calculated for each patient, with the highest probability determining patient class assignment.
Vuong-Lo-Mendell-Rubin likelihood ratio test assesses whether the number of classes provides improved model fit compared with a model using one fewer class. The test is applicable only when comparing models with two or more classes.
For a model containing a single class, there is no class uncertainty and entropy cannot be defined. The P value corresponds to the comparison of models to model 1 (single class model) as the reference. Therefore, it was not appropriate to list a P value here.
Characteristics Distinguishing COVID-19 Subphenotypes
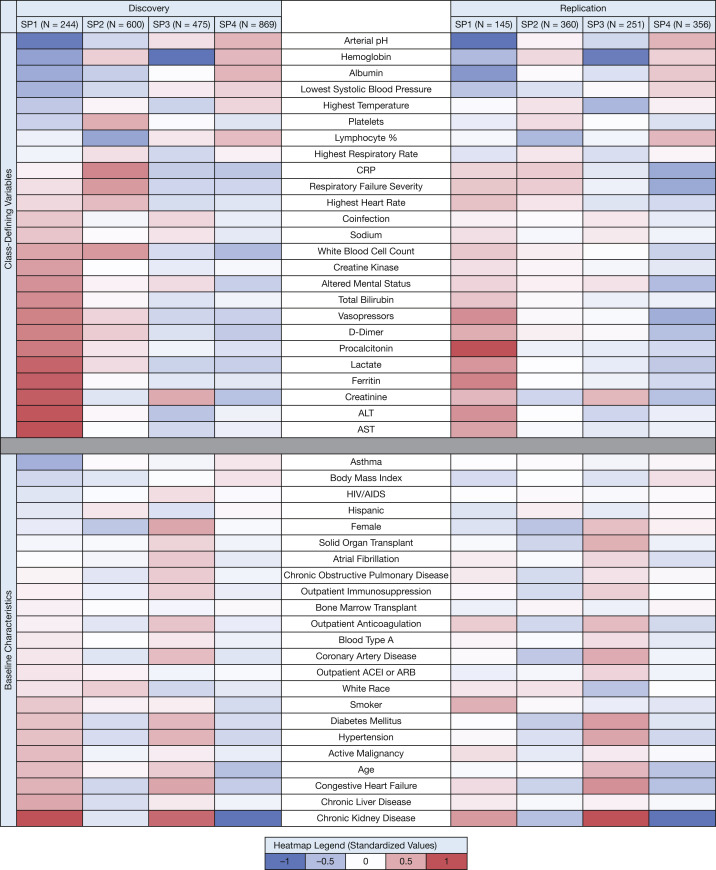
Patterns of class-defining variables and baseline characteristics across the four subphenotypes are shown in Figure 1 (see e-Tables 6 and 7 for nonstandardized numeric data). Patterns across subphenotypes were similar between the discovery and replication cohorts. For example, lactate, ferritin, and D-dimer levels were all highest in SP1, with progressively lower median values across SP2, SP3, and SP4. Across subphenotypes, the time from hospital admission to ICU admission was similar, although SP2 and SP4 had approximately 1 day more of symptoms before hospital admission.
Figure 1.
Heatmap displaying the standardized mean values for each variable across COVID-19 subphenotypes in the discovery and replication cohorts. The heatmap is divided into two sections: latent class-defining variables (top) and baseline characteristics that were not used to define class membership (bottom). Within each section, variables are ordered by standardized mean values, lowest to highest, among patients in the discovery cohort assigned to subphenotype 1 (SP1). The same order of variables is maintained in the replication cohort to facilitate comparison across cohorts. Within each cohort, variables were standardized by scaling to a mean = 0 and an SD = 1 and are represented graphically using a continuous color scale. A variable with value + 1 represents a mean value for that subphenotype, which is 1 SD more than the mean value for the entire cohort population. ACEI = angiotensin converting enzyme inhibitor; ALT = alanine aminotransferase; ARB = angiotensin receptor blocker; AST = aspartate aminotransferase; CRP = C-reactive protein.
SP1 was the least common subphenotype and was characterized by acidemia, elevated lactate, and the highest frequency of vasopressor use on ICU day 1. Acute organ dysfunction was more common in SP1 than in other SPs, including receipt of invasive mechanical ventilation, creatinine and transaminase elevation, and altered mental status. In addition, SP1 patients showed the highest elevations in D-dimer, ferritin, and procalcitonin.
SP2 patients showed the highest rates of acute respiratory failure requiring invasive mechanical ventilation on ICU day 1, even compared with SP1 patients (85%-90% across discovery and replication cohorts for SP2 vs 75%-83% in SP1). SP2 patients showed the highest C-reactive protein values, high maximum temperature, and highest WBC counts on ICU day 1.
SP3 patients were characterized by the highest chronic disease burden, with the highest rates of diabetes mellitus, hypertension, coronary artery disease, congestive heart failure, COPD, and solid organ transplantation, as well as the lowest admission hemoglobin concentration. Coinfections present on the day of ICU admission were as common as in SP1. Shock, acidemia, and acute organ dysfunction markers, such as elevated transaminases, were less common among SP3 patients than SP1 or SP2 patients.
SP4 patients showed relatively preserved indexes of organ function and demonstrated the lowest rates (38%-51%) of invasive mechanical ventilation on ICU day 1, although > 30% of SP4 patients in both cohorts required high-flow nasal canula or noninvasive positive pressure ventilation. SP4 patients received less vasopressor use and demonstrated normal or minimally elevated liver transaminases, lactate, and creatinine levels. SP4 patients showed the highest median BMI, but otherwise a relatively low chronic disease burden, especially compared with SP1 and SP3 patients.
Subphenotype distribution varied by region. Northeast centers showed the highest percentage of SP2 patients, centers in the South showed the highest percentage of SP3 patients, and centers in the Midwest and West showed the highest percentage of SP4 patients (e-Fig 3). Characteristics of patients across the subphenotypes in the discovery cohort were similar when the classes were defined using only data from those receiving mechanical ventilation on ICU day 1 (e-Fig 4). Compared with the subphenotypes derived using latent class modelling, a limited variable model containing fewer, commonly available laboratory and vital sign data did not predict class assignment reliably (e-Table 8).
Association of COVID-19 Subphenotypes With 28-Day Mortality
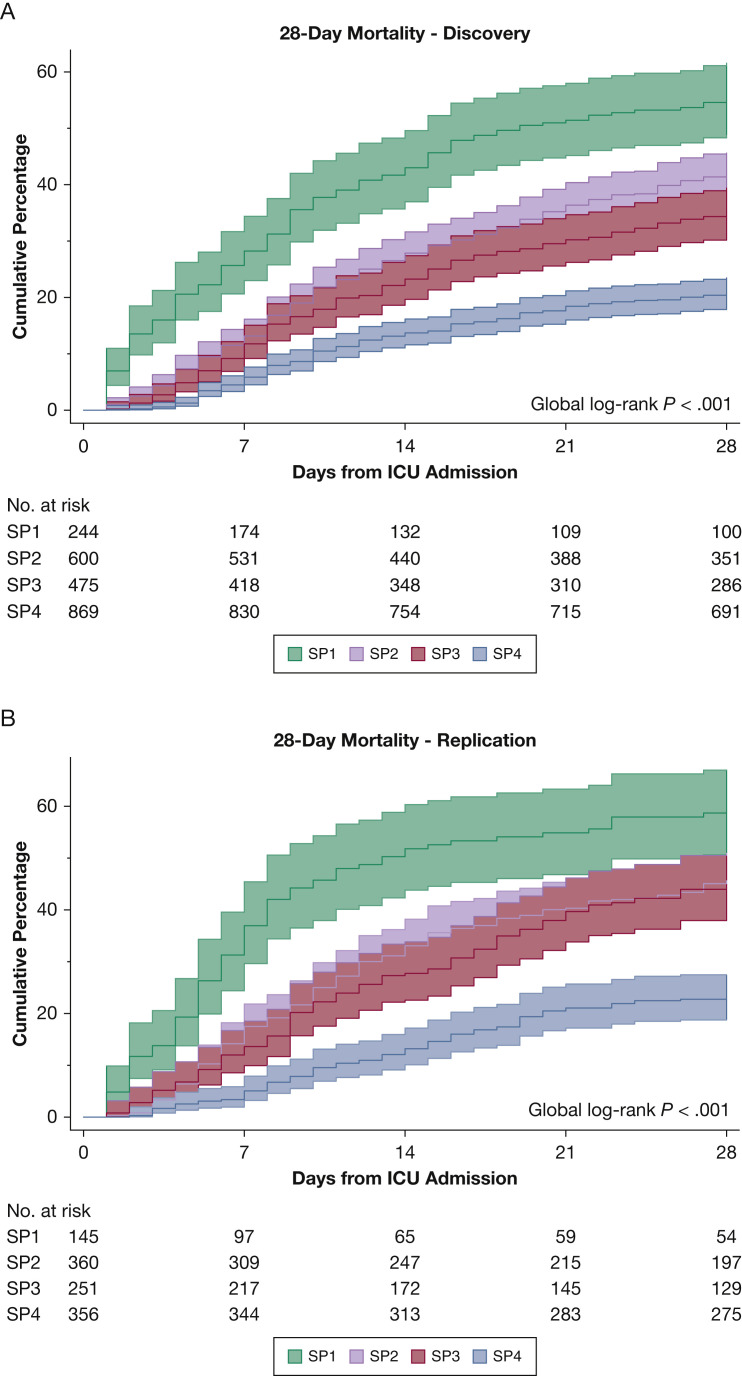
Overall, 28-day mortality was 32.7% in the discovery cohort and 38.9% in the replication cohort. In both cohorts, mortality was highest in SP1 patients, lowest in SP4 patients, and intermediate in SP2 and SP3 patients (P < .001, global log-rank test in both cohorts) (Fig 2 ). Compared with SP4 patients, patients in the SP1, SP2, and SP3 groups showed a significantly higher risk of mortality after adjustment for demographics, comorbidities, organ dysfunction and illness severity, and regional and hospital factors (Table 3 ). Similarly, restricted mean survival times all were significantly shorter in SP1, SP2, and SP3 groups compared with the SP4 group (e-Table 9). Additional pairwise comparisons using adjusted models are shown in e-Table 10.
Figure 2.
A, B, Graphs showing the cumulative incidence of mortality in the discovery (A) and replication (B) cohorts stratified by subphenotype. No patients were censored before mortality determination at 28 days. Patients discharged alive before 28 days were assumed to be alive at day 28. Numbers of at-risk individuals are displayed in the corresponding table, and 95% CIs are shown for each survival curve. SP1, SP2, SP3, SP4 = subphenotype 1, subphenotype 2, subphenotype 3, subphenotype 4.
Table 3.
Unadjusted and adjusted mortality analysis, stratified by subphenotype, in the Discovery and Replication cohorts
| Mortality | ||||
|---|---|---|---|---|
| Discovery |
Replication |
|||
| Unadjusted | Adjusted | Unadjusted | Adjusted | |
| SP1 | 2·57 (2·15-3·06) | 1.67 (1.36-2.04) | 2·49 (1·96-3·16) | 1.67 (1.28-2.17) |
| SP2 | 2·01 (1·71-2·37) | 1.39 (1.17-1.65) | 2·00 (1·60-2·50) | 1.37 (1.09-1.73) |
| SP3 | 1·61 (1·35-1·94) | 1.39 (1.15-1.67) | 1·86 (1·46-2·36) | 1.49 (1.16-1.91) |
| SP4 | 1 (ref) | 1 (ref) | 1 (ref) | 1 (ref) |
Data are shown as relative risk (95% CI). Associations of SP1, SP2, and SP3 with mortality are compared with the reference of SP4. Models adjusted for age, sex, race and ethnicity, hypertension, diabetes, COPD, end-stage kidney disease, active malignancy, chronic liver disease, illness severity, and organ dysfunction on ICU day 1 (respiratory support and oxygenation, platelet count, altered mental status, and creatinine), hospital ICU bed capacity, and US region. BMI, vasopressor support, and liver transaminases were missing in a nontrivial number of patients (11.6%-33.4%) (Table 1). Models also adjusting for these variables in patients with nonmissing data showed minimal change in relative risks compared with those reported above. SP1, SP2, SP3, SP4 = subphenotype 1, subphenotype 2, subphenotype 3, subphenotype 4.
Association of COVID-19 Subphenotypes With 14-Day Clinical Outcomes
Although the unadjusted rates of invasive mechanical ventilation and ARDS were highest in the SP1 and SP2 groups in both cohorts (Table 4 ), only the SP2 group showed an increased ARDS incidence after adjustment (discovery: adjusted hazard ratio, 1.63 [95% CI, 1.49-1.78]; replication: adjusted hazard ratio, 1.53 [95% CI, 1.35-1.75]) (e-Table 11).18 SP1 and SP2 were associated significantly with a higher risk of thrombosis, even after adjustment for known VTE risk factors (e-Table 11).20
Table 4.
Clinical Outcomes, Stratified by Subphenotype, in the Discovery and Replication Cohorts
| Variable | Subphenotype |
P Value | |||
|---|---|---|---|---|---|
| SP1 | SP2 | SP3 | SP4 | ||
| Discovery cohort | |||||
| Patients | 244 (11.2) | 600 (27.4) | 475 (21.7) | 869 (39.7) | ... |
| Mortality | |||||
| Mortality at 28 d | 129 (52.9) | 249 (41.5) | 158 (33.3) | 179 (20.6) | < .001 |
| Clinical outcomes | |||||
| Mechanical ventilation | 181 (85.0) | 572 (95.7) | 295 (74.5) | 635 (73.5) | < .001 |
| Time from hospital admission to invasive mechanical ventilation (d) | 8 (3-12) | 5 (1-9) | 6 (2-10) | 4 (1-8) | < .001 |
| ARDS | 165 (67.6) | 542 (90.5) | 323 (68.1) | 609 (70.1) | < .001 |
| AKI | |||||
| AKI | 152 (71.4) | 374 (62.5) | 216 (54.5) | 356 (41.2) | < .001 |
| Stage 3 | 64 (30.0) | 158 (26.4) | 76 (19.2) | 121 (14.0) | < .001 |
| Thrombosis | 28 (11.5) | 81 (13.5) | 31 (6.5) | 66 (7.6) | < .001 |
| New-onset CHF | 13 (5.3) | 32 (5.3) | 6 (1.3) | 20 (2.3) | < .001 |
| Secondary Infection | 75 (30.7) | 208 (34.7) | 138 (29.1) | 244 (28.1) | .053 |
| Replication cohort | |||||
| Patients | 145 (13.0) | 360 (32·4) | 251 (22·6) | 356 (32·0) | ... |
| Mortality | |||||
| Mortality at 28 d | 82 (56.6) | 164 (45·6) | 106 (42·2) | 81 (22·8) | < .001 |
| Clinical outcomes | |||||
| Mechanical ventilation | 124 (92.5) | 338 (94·2) | 176 (81·9) | 253 (71·5) | < .001 |
| Time from hospital admission to invasive mechanical ventilation | 8 (3-12) | 5 (1-9) | 5.5 (2-10) | 4 (1-7) | < .001 |
| ARDS | 113 (77.9) | 309 (85.8) | 177 (71.1) | 241 (68.3) | < .001 |
| AKI | |||||
| No AKI | 101 (75.4) | 214 (59.6) | 136 (63.3) | 126 (35.6) | < .001 |
| Stage 3 RRT | 42 (31.3) | 87 (24.2) | 60 (27.9) | 29 (8.2) | < .001 |
| Thrombosis | 17 (11.7) | 61 (16.9) | 30 (12.0) | 34 (9.6) | .026 |
| New-onset CHF | 10 (6.9) | 17 (4.7) | 10 (4.0) | 7 (2.0) | < .001 |
| Secondary infection | 49 (33.8) | 121 (33.6) | 78 (31.3) | 94 (26.6) | .183 |
Data are presented as No. (%) or median (interquartile range), unless otherwise indicated. Within each cohort, differences across subphenotypes were compared using the χ 2 test. AKI = acute kidney injury; CHF = congestive heart failure; RRT = renal replacement therapy; SP1, SP2, SP3, SP4 = subphenotype 1, subphenotype 2, subphenotype 3, subphenotype 4.
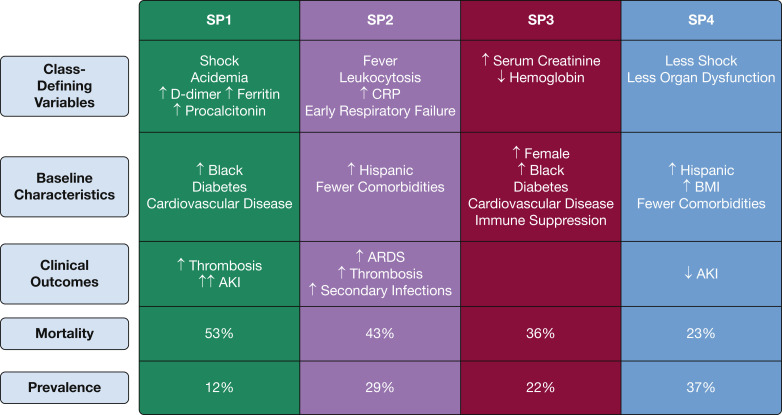
The rates of AKI and AKI requiring renal replacement therapy were higher in SP1, SP2, and SP3 compared with SP4 (Table 4, e-Table 11). Secondary infections occurring after ICU admission were common across all subphenotypes, but only patients with SP2 experienced secondary infections at a significantly higher rate than those with SP4, after adjustment for age, coinfection status at ICU admission, and immunosuppression (e-Table 11). Key distinguishing characteristics of each subphenotype, overall prevalence, and mortality risk are shown in Figure 3 .
Figure 3.
Diagram showing summary of class-defining variables, baseline characteristics, and clinical outcomes for each subphenotype of COVID-19. Overall cumulative 28-day mortality and the class prevalence, across both cohorts, are shown as percentages. AKI = acute kidney injury; CRP = C-reactive protein. SP1, SP2, SP3, SP4 = subphenotype 1, subphenotype 2, subphenotype 3, subphenotype 4.
Interaction Between COVID-19 Subphenotypes With Antiinflammatory and Antimicrobial Therapies
Corticosteroid use varied significantly across subphenotypes in both the discovery and replication cohorts (e-Table 12), with the greatest use in SP1 and SP2 patients compared with SP3 and SP4 patients. Steroid use was associated with significantly increased mortality risk in SP2, SP3, and SP4 groups in the discovery cohort, but not in the replication cohorts (e-Table 13). Anti-IL-6 therapy was used less frequently than corticosteroids across all subpheontypes (e-Table 12). Receipt of anti-IL-6 therapy was not associated mortality within any subphenotype in both the discovery and replication cohorts (e-Table 13). Antimicrobial therapy was used commonly across all subphenotypes on ICU day 1 and was not associated with mortality risk within any subphenotype in either the discovery or replication cohorts (e-Table 13).
Discussion
In our study of more than 3,000 critically ill patients with COVID-19, we identified four clinical subphenotypes with distinct physiologic and laboratory characteristics, unique clustering across demographic and baseline patient characteristics, and clinically meaningful differences in mortality and acute organ dysfunction. Our findings lend empiric support to the hypothesized and clinically observed heterogeneity of COVID-19 critical illness. These subphenotypes were identified using standard clinical and laboratory data obtained during the first day of ICU admission and may provide a useful framework for understanding COVID-19 critical illness pathophysiologic features and prognosis. In addition, these findings may help to identify patient subgroups most likely to benefit from emerging therapeutics.
In our study, we reliably assigned patients with COVID-19 to one of four subphenotypes based on readily available clinical and physiologic characteristics. Our findings have several similarities with a previous study that used clustering techniques to identify four subgroups in a broad population of hospitalized patients with sepsis based on demographic characteristics, clinical signs, and laboratory variables.8 In that study, patients with sepsis were classified into one of four subgroups: a first subgroup characterized by shock (δ phenotype), a second subgroup characterized by high C-reactive protein and pulmonary dysfunction (γ), a third subgroup characterized by chronic illness and a high prevalence of kidney dysfunction (β), and a fourth subgroup characterized by relatively limited physiologic derangements (α). This pattern has several similarities to the COVID-19 subphenotypes we identified (SP1, SP2, SP3, and SP4, respectively) (Fig 3). This is not unexpected, given that most critically ill patients with COVID-19 meet criteria for sepsis.
Notably, however, the COVID-19 subphenotypes were characterized by a more defined relationship between chronic and acute conditions. SP1 patients, who had severe acute physiologic changes, also were typified by the high burden of comorbidities and elevated initial serum creatinine seen in SP3 patients; in contradistinction, the δ sepsis subphenotype was defined more by shock, with notably fewer comorbidities and lower creatinine levels than the β phenotype. In addition, the δ sepsis phenotype showed the highest Pao 2, whereas patients with COVID-19 demonstrating the SP1 phenotype included a high proportion with hypoxemia and respiratory failure, second only to the SP2 group in severity. These differences, as well as a substantially higher mortality in our cohort, could be explained by our inclusion of only patients admitted to the ICU, although some dissimilarities are likely because of the uniformity of pulmonary sepsis source in the present cohorts as well as the unique characteristics of the novel SARS-CoV-2 virus, which may differ even from other respiratory viral illnesses.24
Like prior latent class analyses in critical care, we focused heavily on acute clinical and laboratory variables to define subgroups.10 , 25 As noted (see Methods), we considered demographics and chronic medical conditions to be exogenous, non-class-defining variables, rather than features of acute COVID-19 illness. However, we found that these characteristics occurring before acute illness showed remarkably consistent patterns of association with subphenotypes across the discovery and replication cohorts. Comorbidities were more prevalent in SP1 and SP3 groups compared with SP2 and SP4 groups. Black race prevalence showed overlap with these comorbidities, with higher representation in SP1 and SP3 groups. This finding is consistent with other large US cohort studies of COVID-19 and with observed health disparities within the United States.26 Not surprisingly, SP3 was seen most commonly in the South, the US region with the largest Black population.27 In contrast, Hispanic patients were more likely to belong to SP2 and SP4 groups across both discovery and replication cohorts, which is somewhat surprising, given that Hispanic populations are known to carry an increased burden of chronic medical diseases compared with non-Hispanic White patients.28
Beyond providing a construct for understanding COVID-19 heterogeneity and outcome risk, the clinical subphenotypes we identified may be the result of distinct underlying disease mechanisms and may help to inform treatment decisions. Prior studies of ARDS, AKI, and sepsis identified both simple and complex molecular signatures that defined patient subgroups while elucidating associated pathophysiologic processes.9 , 10 , 29 , 30 These studies also suggested that such molecular subphenotypes may affect response to therapies, identifying potential beneficial effects in certain subgroups even when overall trial results were negative.31 , 32 We did not have biospecimens to test molecular markers in patients in the STOP-COVID population, but small cohort studies already have suggested the presence of molecular endotypes within the population with COVID-19. For example, Sinha et al33 applied previously defined biomarker-based ARDS subphenotypes to 39 patients with COVID-19, distinguishing patients between hypoinflammatory and hyperinflammatory subgroups. In addition, Mathew et al34 identified immunophenotypes based on lymphocyte activation that were linked to clinical outcomes in 125 patients hospitalized with COVID-19. Informed by such deep molecular phenotyping analyses, further studies in large cohorts with COVID-19 could determine whether the clinical subphenotypes we identified are associated with distinct molecular characteristics, potentially identifying patients for targeted treatments. Dexamethasone, for example, improves survival in patients hospitalized with COVID-19, but seems to exert significant effect differences across levels of respiratory support.35 In our secondary analysis, we did not find an association of steroid treatment with improved survival, and in several subphenotypes, steroid treatment was associated with an increased mortality risk, findings that we believe are likely the result of confounding by indication in the early pandemic experience. Neither IL-6 receptor antagonists nor early antimicrobial therapy were associated with a significant effect across subphenotypes in either the discovery or replication cohorts. We hypothesize that clinical and molecular subphenotypes characterized by immune activation may help to target more precisely patients who are responsive to suppression of inflammation, while avoiding the risks of immune suppression and excess viral replication that could lead to harm in other patient subgroups.
Our study has several notable strengths. First, latent class models were generated independently in two patient populations totaling 3,300 patients at 67 centers across the United States. The subphenotype similarities between discovery and replication cohorts, two randomly selected sets of STOP-COVID centers, lend external validity to our findings. Further strengthening the validity of the identified classes, we found consistent subphenotype associations with demographics, medical comorbidities, and clinical outcomes, although these were not used to define classes. Furthermore, the similarities in the timing of disease onset relative to hospital and ICU admission across subphenotypes, provide evidence that subphenotypes describe truly separate classes, rather than the same disease at different time points. Ascertainment of similar subphenotypes when analysis is limited to patients receiving mechanical ventilated on ICU day 1 also supports this conclusion. In addition, our study population was diverse, both geographically and demographically, which increases the generalizability of our findings and suggests that our subphenotype classifications may be applicable to other cohorts with COVID-19. Our study’s large sample size may have enhanced our ability to detect more clinically distinguishable subphenotypes than in studies of ARDS and AKI and the same number of classes as a study of > 20,000 sepsis patients.8, 9, 10
Our study has several important limitations. First, the STOP-COVID study from which our data were drawn was an observational study; although data were collected manually and systematically, they were limited by what was available in the medical record. Substantial missingness of some laboratory variables could have impacted our findings, despite the use of full-information maximum likelihood and good balance of characteristics between those with and without missing values. Despite the challenges of creating a parsimonious model to assign patients to four unique classes, further study to develop class-assigning models with fewer, reliably available variables is important to facilitate application. Alternative clustering methods, such as consensus k-means clustering, also could be used to validate our findings further.
An additional limitation was lack of data on circulating novel molecular markers, such as soluble tumor necrosis factor receptor 1, IL-6, and IL-8, which have been described as important class-defining variables in other critical illness latent class models.9 , 10 As noted, follow-up studies in cohorts with biospecimens will be key to associating clinical with molecular subphenotypes. The true incidence of some clinical outcomes, such as ARDS, may differ from the reported incidence because of misclassification. For example, we were unable to phenotype ARDS using direct chest radiography review and expert consensus. Similarly, our data lacked detailed information of respiratory physiologic features, such as respiratory system compliance, or quantitative radiographic information, including extent of lung consolidation. The absence of detailed biomarker and physiologic variables, as well as substantial missingness of variables such as ferritin and D-dimer, likely contributed to the inability to predict class assignment using fewer, more commonly available clinical variables. However, our findings regarding subphenotype association with such outcomes were consistent with the associations we observed for outcomes less likely to have been misclassified, including AKI and mortality.
It is also possible that therapies were applied differentially across subphenotypes and represent a source of residual confounding in mortality and other outcomes analyses. Secondary analyses of emerging COVID-19 clinical trials may be particularly useful to address this limitation. Our study also is limited to patients admitted to the ICU, themselves a subset of patients infected with SARS-CoV-2. Finally, our study lacks data on the association of subphenotypes with clinically relevant long-term outcomes like functional status, quality of life, and complications occurring after discharge.
Interpretation
Overall, our results provide empirical evidence that COVID-19 critical illness is characterized by distinct clinical subphenotypes. These findings pave the way for future studies using molecular data aimed at further clarifying whether these subphenotypes reflect differences in underlying pathophysiologic processes. In addition, differences in therapy responses could be tested across subphenotypes to target COVID-19 treatments more effectively, both in retrospective analyses of emerging clinical trials and in stratified designs for future trials of novel COVID-19 therapies.
Take-home Points.
Study Question: Can unique subphenotypes be identified among critically ill patients with COVID-19?
Results: Based on clinical and laboratory characteristics assessed within 24 h of ICU admission, we identified four clinically distinct subphenotypes of COVID-19, with significant differences in mortality at 28 days, as well as clinical outcomes, including ARDS, acute kidney injury, and thrombotic events, at 14 days.
Interpretation: To the best of our knowledge, this is the first study to comprehensively investigate the existence of distinct subphenotypes of COVID-19 in a large, geographically and demographically diverse population of critically ill patients and to replicate the results in a second independent population.
Acknowledgments
Author contributions: C. R. V. is the guarantor of this manuscript and takes responsibility for the content of the manuscript, including integrity of the data and analysis. C. R. V. contributed to the concept, design, and data collection; conducted the analyses; and drafted the manuscript. S. G. and D. E. L. contributed to concept and design, data collection, and critical revisions of the manuscript and designed the parent STOP-COVID study. T. A. M. contributed to concept and design, data collection, statistical analysis, and critical revisions of the manuscript. M. G. S. S. contributed to concept and design and statistical analysis and assisted in drafting and critical revisions of the manuscript. M. R., J. H., W. Y., D. N. H., J. P. R., and S. J. S. contributed to concept and design, statistical analysis, and critical revisions of the manuscript.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: D. E. L. has received research funding from BioPorto. S. G. is the scientific coordinator for GlaxoSmithKline’s ASCEND trial. None declared (C. R. V., T. A. M., M. R., J. H., W. Y., D. N. H., J. P. R., S. J. S., M. G. S. S.).
Role of sponsors: No representatives from external funding agencies or groups had any role in the development of this research or manuscript preparation.
∗ STOP-COVID Investigators: Baylor College of Medicine: Carl P. Walther (PI), and Samaya J. Anumudu; Baylor University Medical Center: Justin Arunthamakun (PI), Kathleen F. Kopecky, Gregory P. Milligan, Peter A. McCullough, and Thuy-Duyen Nguyen; Beth Israel Deaconess Medical Center: Shahzad Shaefi (PI), Megan L. Krajewski, Sidharth Shankar, Ameeka Pannu, and Juan D. Valencia; Boston Medical Center: Sushrut S. Waikar (PI) and Zoe A. Kibbelaar; Cook County Health: Ambarish M. Athavale (PI), Peter Hart, Shristi Upadhyay, Ishaan Vohra, and Ajiboye Oyintayo; Cooper University Health Care: Adam Green (PI), Jean-Sebastien Rachoin, Christa A. Schorr, and Lisa Shea; Duke University Medical Center: Daniel L. Edmonston (PI) and Christopher L. Mosher; Hackensack Meridian Health Mountainside Medical Center: Alexandre M. Shehata (PI), Zaza Cohen, Valerie Allusson, Gabriela Bambrick-Santoyo, Noor ul aain Bhatti, Bijal Mehta, and Aquino Williams; Hackensack Meridian Health Hackensack University Medical Center: Samantha K. Brenner (PI), Patricia Walters, Ronaldo C. Go, and Keith M. Rose; Icahn School of Medicine at Mount Sinai: Lili Chan (PI), Kusum S. Mathews (PI), Steven G. Coca, Deena R. Altman, Aparna Saha, Howard Soh, Huei Hsun Wen, Sonali Bose, Emily A. Leven, Jing G. Wang, Gohar Mosoyan, Girish N. Nadkarni, Pattharawin Pattharanitima, and Emily J. Gallagher; Indiana University School of Medicine/Indiana University Health: Allon N. Friedman (PI), John Guirguis, Rajat Kapoor, Christopher Meshberger, and Katherine J. Kelly; Johns Hopkins Hospital: Chirag R. Parikh (PI), Brian T. Garibaldi, Celia P. Corona-Villalobos, Yumeng Wen, Steven Menez, Rubab F. Malik, Carmen Elena Cervantes, and Samir C. Gautam; Kings County Hospital Center: Mary C. Mallappallil (PI), Jie Ouyang, Sabu John, Ernie Yap, Yohannes Melaku, Ibrahim Mohamed, Siddhartha Bajracharya, Isha Puri, Mariah Thaxton, Jyotsna Bhattacharya, John Wagner, and Leon Boudourakis; Loma Linda University: H. Bryant Nguyen (PI) and Afshin Ahoubim; Mayo Clinic: Kianoush Kashani (PI) and Shahrzad Tehranian; Mayo Clinic, Arizona: Leslie F. Thomas (PI) and Dheeraj Reddy Sirganagari; Mayo Clinic, Florida: Pramod K. Guru (PI); Medical College of Wisconsin: Yan Zhou, (PI) Paul A. Bergl, Jesus Rodriguez, Jatan A. Shah, and Mrigank S. Gupta; MedStar Georgetown University Hospital: Princy N. Kumar (PI), Deepa G. Lazarous, and Seble G. Kassaye; Montefiore Medical Center/Albert Einstein College of Medicine: Michal L. Melamed (PI), Tanya S. Johns. Ryan Mocerino, Kalyan Prudhvi, Denzel Zhu, Rebecca V. Levy, Yorg Azzi, Molly Fisher, Milagros Yunes, Kaltrina Sedaliu, Ladan Golestaneh, Maureen Brogan, Neelja Kumar, Michael Chang, and Jyotsana Thakkar; New York-Presbyterian Queens Hospital: Ritesh Raichoudhury (PI), Akshay Athreya, and Mohamed Farag; New York-Presbyterian/Weill Cornell Medical Center: Edward J. Schenck (PI), Soo Jung Cho, Maria Plataki, Sergio L. Alvarez-Mulett, Luis G. Gomez-Escobar, Di Pan, Stefi Lee, Jamuna Krishnan, and William Whalen; New York University Langone Hospital: David Charytan (PI), Ashley Macina, Sobaata Chaudhry, Benjamin Wu, and Frank Modersitzki; Northwestern Memorial Hospital: Northwestern University Feinberg School of Medicine—Anand Srivastava (PI), Alexander S. Leidner, Carlos Martinez, Jacqueline M. Kruser, Richard G. Wunderink, and Alexander J. Hodakowski; Ochsner Medical Center: Juan Carlos Q. Velez (PI), Eboni G. Price-Haywood, Luis A. Matute-Trochez, Anna E. Hasty, and Muner M. B. Mohamed; Oregon Health and Science University Hospital: Rupali S. Avasare (PI) and David Zonies (PI); Partners Healthcare (Brigham and Women’s Hospital, Brigham and Women’s Faulkner Hospital, Massachusetts General Hospital, and Newton Wellesley Hospital): David E. Leaf (PI), Shruti Gupta (PI), Meghan E. Sise, Erik T. Newman, Samah Abu Omar, Kapil K. Pokharel, Shreyak Sharma, Harkarandeep Singh, Simon Correa, Tanveer Shaukat, Omer Kamal, Wei Wang, Heather Yang, Jeffery O. Boateng, Meghan Lee, Ian A. Strohbehn, Jiahua Li, and Ariel L. Mueller; ProMedica Health System: Roberta E. Redfern (PI), Nicholas S. Cairl, Gabriel Naimy, Abeer Abu-Saif, Danyell Hall, and Laura Bickley; Renown Health: Chris Rowan (PI) and Farah Madhani-Lovely (PI); Rush University Medical Center: Vasil Peev (PI), Jochen Reiser, John J. Byun, Andrew Vissing, Esha M. Kapania, Zoe Post, Nilam P. Patel, and Joy-Marie Hermes; Rutgers/New Jersey Medical School: Anne K. Sutherland (PI), Amee Patrawalla, Diana G. Finkel, Barbara A. Danek, Sowminya Arikapudi, Jeffrey M. Paer, Peter Cangialosi, and Mark Liotta; Rutgers/Robert Wood Johnson Medical School: Jared Radbel (PI), Sonika Puri, Jag Sunderram, Matthew T. Scharf, Ayesha Ahmed, Ilya Berim, and Jayanth S. Vatson; Stanford Healthcare (Stanford University School of Medicine): Shuchi Anand (PI), Joseph E. Levitt, and Pablo Garcia; Temple University Hospital: Suzanne M. Boyle (PI) and Rui Song; Thomas Jefferson University Hospital: Jingjing Zhang (PI), Sang Hoon Woo, Xiaoying Deng, Goni Katz-Greenberg, and Katharine Senter; Tulane Medical Center: Moh’d A. Sharshir (PI) and Vadym V. Rusnak; United Health Services Hospitals: Muhammad Imran Ali; University of Colorado Anschutz Medical Campus: Anip Bansal (PI), Amber S. Podoll, Michel Chonchol, Sunita Sharma, and Ellen L. Burnham; University Hospitals Cleveland Medical Center: Arash Rashidi (PI) and Rana Hejal; University of Alabama-Birmingham Hospital: Eric Judd (PI), Laura Latta, and Ashita Tolwani; University of California, Davis Medical Center: Timothy E. Albertson (PI) and Jason Y. Adams; University of California, Los Angeles Medical Center: (Ronald Reagan-UCLA Medical Center) Steven Y. Chang (PI), Rebecca M. Beutler, and (UCLA Medical Center, Santa Monica) Carl E. Schulze; University of California, San Diego Medical Center: Etienne Macedo (PI) and Harin Rhee; University of California, San Francisco Medical Center: Kathleen D. Liu (PI) and Vasantha K. Jotwani; University of Chicago Medical Center: Jay L. Koyner (PI) and Alissa Kunczt; University of Florida Health, Gainesville: Chintan V. Shah (PI); University of Florida Health, Jacksonville: Vishal Jaikaransingh (PI); University of Illinois Hospital and Health Sciences System: Stephanie M. Toth-Manikowski (PI), Min J. Joo (PI), and James P. Lash; University of Kentucky Medical Center: Javier A. Neyra (PI), Nourhan Chaaban, Yahya Ahmad, and Madona Elias; University Medical Center of Southern Nevada: Rajany Dy (PI), Alfredo Iardino, Elizabeth H. Au, and Jill H. Sharma; University of Miami Health System: Marie Anne Sosa (PI), Sabrina Taldone, Gabriel Contreras, David De La Zerda, and Hayley B. Gershengorn; University of Michigan: Salim S. Hayek (PI), Pennelope Blakely, Hanna Berlin, Tariq U. Azam, Husam Shadid, Michael Pan, Patrick O’ Hayer, Chelsea Meloche, Rafey Feroze, Rayan Kaakati, Danny Perry, Abbas Bitar, Elizabeth Anderson, Kishan J. Padalia, John P. Donnelly, and Andrew J. Admon; University of North Carolina School of Medicine: Jennifer E. Flythe (PI), Matthew J. Tugman, and Emily H. Chang; University of Oklahoma Health Sciences Center: Brent R. Brown (PI); University of Pennsylvania Health System: Amanda K. Leonberg-Yoo (PI), Ryan C. Spiardi, Todd A. Miano, Meaghan S. Roche, and Charles R. Vasquez; University of Pittsburgh Medical Center: Amar D. Bansal (PI), Natalie C. Ernecoff, Sanjana Kapoor, Siddharth Verma, and Huiwen Chen; University of Tennessee Health Science Center and Memphis VA Medical Center/Methodist University Hospital: Csaba P. Kovesdy (PI), Miklos Z. Molnar (PI), and Ambreen Azhar; University of Texas Southwestern Medical Center and Parkland Health and Hospital System: S. Susan Hedayati (PI), Mridula V. Nadamuni, Shani Shastri, and Duwayne L. Willett; University of Vermont Larner College of Medicine: Samuel A. P. Short; University of Virginia Health System: Amanda D. Renaghan (PI) and Kyle B. Enfield; University of Washington Medical Center: Pavan K. Bhatraju (PI) and A. Bilal Malik; Vanderbilt University Medical Center: Matthew W. Semler; Washington University in St. Louis/Barnes Jewish Hospital: Anitha Vijayan (PI), Christina Mariyam Joy, Tingting Li, Seth Goldberg, and Patricia F. Kao; Wellforce Health System: (Lowell General Hospital) Greg L. Schumaker (PI), (Tufts Medical Center) Nitender Goyal (PI), Anthony J. Faugno, Greg L. Schumaker, Caroline M. Hsu, Asma Tariq, Leah Meyer, Ravi K. Kshirsagar, Daniel E. Weiner, and Aju Jose; Westchester Medical Center: Marta Christov (PI), Jennifer Griffiths, Sanjeev Gupta, Aromma Kapoor, and Savneek Chugh; and Yale School of Medicine: Perry Wilson, (PI) Tanima Arora, and Ugochukwu Ugwuowo. PI = site principal investigator.
Additional information: The e-Appendix, e-Figures, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
Drs Vasquez and Gupta contributed equally to this manuscript. Drs Leaf and Shashaty contributed equally to this manuscript.
FUNDING/SUPPORT: The authors are funded by the following grants from the National Institutes of Health: T32-DK007785 (C. R. V.), K23-DK118198 (S. J. S.), R01HL144566 (D. E. L.), R01DK125786 (D. E. L.), and R01-DK111638 (M. G. S. S.).
Contributor Information
STOP-COVID Investigators:
Carl P. Walther, Samaya J. Anumudu, Justin Arunthamakun, Kathleen F. Kopecky, Gregory P. Milligan, Peter A. McCullough, Thuy-Duyen Nguyen, Shahzad Shaefi, Megan L. Krajewski, Sidharth Shankar, Ameeka Pannu, Juan D. Valencia, Sushrut S. Waikar, Zoe A. Kibbelaar, Ambarish M. Athavale, Peter Hart, Shristi Upadhyay, Ishaan Vohra, Ajiboye Oyintayo, Adam Green, Jean-Sebastien Rachoin, Christa A. Schorr, Lisa Shea, Daniel L. Edmonston, Christopher L. Mosher, Alexandre M. Shehata, Zaza Cohen, Valerie Allusson, Gabriela Bambrick-Santoyo, Noor ul aain Bhatti, Bijal Mehta, Aquino Williams, Samantha K. Brenner, Patricia Walters, Ronaldo C. Go, Keith M. Rose, Lili Chan, Kusum S. Mathews, Steven G. Coca, Deena R. Altman, Aparna Saha, Howard Soh, Huei Hsun Wen, Sonali Bose, Emily A. Leven, Jing G. Wang, Gohar Mosoyan, Girish N. Nadkarni, Pattharawin Pattharanitima, Emily J. Gallagher, Allon N. Friedman, John Guirguis, Rajat Kapoor, Christopher Meshberger, Katherine J. Kelly, Chirag R. Parikh, Brian T. Garibaldi, Celia P. Corona-Villalobos, Yumeng Wen, Steven Menez, Rubab F. Malik, Carmen Elena Cervantes, Samir C. Gautam, Mary C. Mallappallil, Jie Ouyang, Sabu John, Ernie Yap, Yohannes Melaku, Ibrahim Mohamed, Siddhartha Bajracharya, Isha Puri, Mariah Thaxton, Jyotsna Bhattacharya, John Wagner, Leon Boudourakis, H. Bryant Nguyen, Afshin Ahoubim, Kianoush Kashani, Shahrzad Tehranian, Leslie F. Thomas, Dheeraj Reddy Sirganagari, Pramod K. Guru, Yan Zhou, Paul A. Bergl, Jesus Rodriguez, Jatan A. Shah, Mrigank S. Gupta, Princy N. Kumar, Deepa G. Lazarous, Seble G. Kassaye, Michal L. Melamed, Tanya S. Johns, Ryan Mocerino, Kalyan Prudhvi, Denzel Zhu, Rebecca V. Levy, Yorg Azzi, Molly Fisher, Milagros Yunes, Kaltrina Sedaliu, Ladan Golestaneh, Maureen Brogan, Neelja Kumar, Michael Chang, Jyotsana Thakkar, Ritesh Raichoudhury, Akshay Athreya, Mohamed Farag, Edward J. Schenck, Soo Jung Cho, Maria Plataki, Sergio L. Alvarez-Mulett, Luis G. Gomez-Escobar, Di Pan, Stefi Lee, Jamuna Krishnan, William Whalen, David Charytan, Ashley Macina, Sobaata Chaudhry, Benjamin Wu, Frank Modersitzki, Anand Srivastava, Alexander S. Leidner, Carlos Martinez, Jacqueline M. Kruser, Richard G. Wunderink, Alexander J. Hodakowski, Juan Carlos Q. Velez, Eboni G. Price-Haywood, Luis A. Matute-Trochez, Anna E. Hasty, Muner M.B. Mohamed, Rupali S. Avasare, David Zonies, David E. Leaf, Shruti Gupta, Meghan E. Sise, Erik T. Newman, Samah Abu Omar, Kapil K. Pokharel, Shreyak Sharma, Harkarandeep Singh, Simon Correa, Tanveer Shaukat, Omer Kamal, Wei Wang, Heather Yang, Jeffery O. Boateng, Meghan Lee, Ian A. Strohbehn, Jiahua Li, Ariel L. Mueller, Roberta E. Redfern, Nicholas S. Cairl, Gabriel Naimy, Abeer Abu-Saif, Danyell Hall, Laura Bickley, Chris Rowan, Farah Madhani-Lovely, Vasil Peev, Jochen Reiser, John J. Byun, Andrew Vissing, Esha M. Kapania, Zoe Post, Nilam P. Patel, Joy-Marie Hermes, Anne K. Sutherland, Amee Patrawalla, Diana G. Finkel, Barbara A. Danek, Sowminya Arikapudi, Jeffrey M. Paer, Peter Cangialosi, Mark Liotta, Jared Radbel, Sonika Puri, Jag Sunderram, Matthew T. Scharf, Ayesha Ahmed, Ilya Berim, Jayanth S. Vatson, Shuchi Anand, Joseph E. Levitt, Pablo Garcia, Suzanne M. Boyle, Rui Song, Jingjing Zhang, Sang Hoon Woo, Xiaoying Deng, Goni Katz-Greenberg, Katharine Senter, Moh’d A. Sharshir, Vadym V. Rusnak, Muhammad Imran Ali, Anip Bansal, Amber S. Podoll, Michel Chonchol, Sunita Sharma, Ellen L. Burnham, Arash Rashidi, Rana Hejal, Eric Judd, Laura Latta, Ashita Tolwani, Timothy E. Albertson, Jason Y. Adams, Steven Y. Chang, Rebecca M. Beutler, Carl E. Schulze, Etienne Macedo, Harin Rhee, Kathleen D. Liu, Vasantha K. Jotwani, Jay L. Koyner, Alissa Kunczt, Chintan V. Shah, Vishal Jaikaransingh, Stephanie M. Toth-Manikowski, Min J. Joo, James P. Lash, Javier A. Neyra, Nourhan Chaaban, Yahya Ahmad, Madona Elias, Rajany Dy, Alfredo Iardino, Elizabeth H. Au, Jill H. Sharma, Marie Anne Sosa, Sabrina Taldone, Gabriel Contreras, David De La Zerda, Hayley B. Gershengorn, Salim S. Hayek, Pennelope Blakely, Hanna Berlin, Tariq U. Azam, Husam Shadid, Michael Pan, Patrick O’ Hayer, Chelsea Meloche, Rafey Feroze, Rayan Kaakati, Danny Perry, Abbas Bitar, Elizabeth Anderson, Kishan J. Padalia, John P. Donnelly, Andrew J. Admon, Jennifer E. Flythe, Matthew J. Tugman, Emily H. Chang, Brent R. Brown, Amanda K. Leonberg-Yoo, Ryan C. Spiardi, Todd A. Miano, Meaghan S. Roche, Charles R. Vasquez, Amar D. Bansal, Natalie C. Ernecoff, Sanjana Kapoor, Siddharth Verma, Huiwen Chen, Csaba P. Kovesdy, Miklos Z. Molnar, Ambreen Azhar, S. Susan Hedayati, Mridula V. Nadamuni, Shani Shastri, Duwayne L. Willett, Samuel A.P. Short, Amanda D. Renaghan, Kyle B. Enfield, Pavan K. Bhatraju, A. Bilal Malik, Matthew W. Semler, Anitha Vijayan, Christina Mariyam Joy, Tingting Li, Seth Goldberg, Patricia F. Kao, Greg L. Schumaker, Nitender Goyal, Anthony J. Faugno, Greg L. Schumaker, Caroline M. Hsu, Asma Tariq, Leah Meyer, Ravi K. Kshirsagar, Daniel E. Weiner, Aju Jose, Marta Christov, Jennifer Griffiths, Sanjeev Gupta, Aromma Kapoor, Savneek Chugh, Perry Wilson, Tanima Arora, and Ugochukwu Ugwuowo
Supplementary Data
References
- 1.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 2.Gattinoni L., Chiumello D., Caironi P., et al. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med. 2020;46(6):1099–1102. doi: 10.1007/s00134-020-06033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X., Ma X. Acute respiratory failure in COVID-19: is it “typical” ARDS? Crit Care. 2020;24(1):198. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marini J.J., Gattinoni L. Management of COVID-19 respiratory distress. JAMA. 2020;323(22):2329–2330. doi: 10.1001/jama.2020.6825. [DOI] [PubMed] [Google Scholar]
- 5.Spyropoulos A.C., Weitz J.I. Hospitalized COVID-19 patients and venous thromboembolism: a perfect storm. Circulation. 2020;142(2):129–132. doi: 10.1161/CIRCULATIONAHA.120.048020. [DOI] [PubMed] [Google Scholar]
- 6.Cardone M., Yano M., Rosenberg A.S., Puig M. Lessons learned to date on COVID-19 hyperinflammatory syndrome: considerations for interventions to mitigate SARS-CoV-2 viral infection and detrimental hyperinflammation. Front Immunol. 2020;11:1131. doi: 10.3389/fimmu.2020.01131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy K., Sinha P., O’Kane C.M., Gordon A.C., Calfee C.S., McAuley D.F. Subphenotypes in critical care: translation into clinical practice. Lancet Respir Med. 2020;8(6):631–643. doi: 10.1016/S2213-2600(20)30124-7. [DOI] [PubMed] [Google Scholar]
- 8.Seymour C.W., Kennedy J.N., Wang S., et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA. 2019;321(20):2003–2017. doi: 10.1001/jama.2019.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calfee C.S., Delucchi K., Parsons P.E., et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2(8):611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatraju P.K., Zelnick L.R., Herting J., et al. Identification of acute kidney injury subphenotypes with differing molecular signatures and responses to vasopressin therapy. Am J Respir Crit Care Med. 2019;199(7):863–872. doi: 10.1164/rccm.201807-1346OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagenaars J.A., McCutcheon A.L. Cambridge University Press; 2002. Applied Latent Class Analysis. [Google Scholar]
- 12.Reilly J.P., Bellamy S., Shashaty M.G., et al. Heterogeneous phenotypes of acute respiratory distress syndrome after major trauma. Ann Am Thorac Soc. 2014;11(5):728–736. doi: 10.1513/AnnalsATS.201308-280OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta S., Hayek S.S., Wang W., et al. Factors associated with death in critically ill patients with coronavirus disease 2019 in the US. JAMA Intern Med. 2020;180(11):1436–1447. doi: 10.1001/jamainternmed.2020.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nylund K.L., Asparouhov T., Muthén B.O. Deciding on the number of classes in latent class analysis and growth mixture modeling: a Monte Carlo simulation study. Struct Equ Modeling. 2007;14(4):535–569. [Google Scholar]
- 15.Lo Y., Mendell N.R., Rubin D.B. Testing the number of components in a normal mixture. Biometrika. 2001;88(3):767–778. [Google Scholar]
- 16.Harhay M.O., Porcher R., Cantu E., et al. An alternative approach for the analysis of time-to-event and survival outcomes in pulmonary medicine. Am J Respir Crit Care Med. 2018;198(5):684–687. doi: 10.1164/rccm.201801-0189LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou G. A modified Poisson regression approach to prospective studies with binary data. Am J Epidemiol. 2004;159(7):702–706. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
- 18.Thompson B.T., Chambers R.C., Liu K.D. Acute respiratory distress syndrome. N Engl J Med. 2017;377(6):562–572. doi: 10.1056/NEJMra1608077. [DOI] [PubMed] [Google Scholar]
- 19.Hoste E.A.J., Kellum J.A., Selby N.M., et al. Global epidemiology and outcomes of acute kidney injury. Nat Rev Nephrol. 2018;14(10):607–625. doi: 10.1038/s41581-018-0052-0. [DOI] [PubMed] [Google Scholar]
- 20.Wolberg A.S., Rosendaal F.R., Weitz J.I., et al. Venous thrombosis. Nat Rev Dis Primers. 2015;1:15006. doi: 10.1038/nrdp.2015.6. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Acelas A.L., de Abreu Almeida M., Engelman B., Canon-Montanez W. Risk factors for health care-associated infection in hospitalized adults: systematic review and meta-analysis. Am J Infect Control. 2017;45(12):e149–e156. doi: 10.1016/j.ajic.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Bui A.L., Horwich T.B., Fonarow G.C. Epidemiology and risk profile of heart failure. Nat Rev Cardiol. 2011;8(1):30–41. doi: 10.1038/nrcardio.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu J.Y., Roy J.A., Xie D., et al. Statistical methods for cohort studies of CKD: survival analysis in the setting of competing risks. Clin J Am Soc Nephrol. 2017;12(7):1181–1189. doi: 10.2215/CJN.10301016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen E., Koopmans M., Go U., et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis. 2020;20(9):e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinha P., Delucchi K.L., McAuley D.F., O’Kane C.M., Matthay M.A., Calfee C.S. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: a secondary analysis of randomised controlled trials. Lancet Respir Med. 2020;8(3):247–257. doi: 10.1016/S2213-2600(19)30369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price-Haywood E.G., Burton J., Fort D., Seoane L. Hospitalization and mortality among Black patients and white patients with Covid-19. N Engl J Med. 2020;382(26):2534–2543. doi: 10.1056/NEJMsa2011686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.United States Census Bureau 2018 Population estimates by age, sex, race and Hispanic origin. 2019. United States Census Bureau website. https://www.census.gov/newsroom/press-kits/2019/detailed-estimates.html
- 28.Centers for Disease Control and Prevention Racial and ethnic disparities in heart disease. 2019. Centers for Disease Control and Prevention website. https://www.cdc.gov/nchs/hus/spotlight/2019-heart-disease-disparities.htm
- 29.Scicluna B.P., van Vught L.A., Zwinderman A.H., et al. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med. 2017;5(10):816–826. doi: 10.1016/S2213-2600(17)30294-1. [DOI] [PubMed] [Google Scholar]
- 30.Davenport E.E., Burnham K.L., Radhakrishnan J., et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. 2016;4(4):259–271. doi: 10.1016/S2213-2600(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calfee C.S., Delucchi K.L., Sinha P., et al. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med. 2018;6(9):691–698. doi: 10.1016/S2213-2600(18)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Famous K.R., Delucchi K., Ware L.B., et al. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med. 2017;195(3):331–338. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinha P., Calfee C.S., Cherian S., et al. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with COVID-19: a prospective observational study. Lancet Respir Med. 2020;8(12):1209–1218. doi: 10.1016/S2213-2600(20)30366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathew D., Giles J.R., Baxter A.E., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508):eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.RECOVERY Collaborative Group. Horby P., Lim W.S., et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2020;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.