Abstract
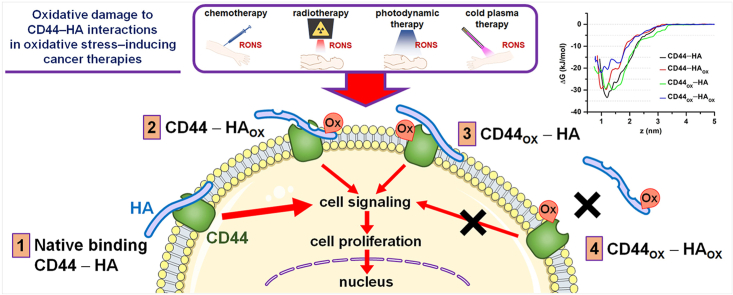
Multiple cancer therapies nowadays rely on oxidative stress to damage cancer cells. Here we investigated the biological and molecular effect of oxidative stress on the interaction between CD44 and hyaluronan (HA), as interrupting their binding can hinder cancer progression. Our experiments demonstrated that the oxidation of HA decreased its recognition by CD44, which was further enhanced when both CD44 and HA were oxidized. The reduction of CD44–HA binding negatively affected the proliferative state of cancer cells. Our multi-level atomistic simulations revealed that the binding free energy of HA to CD44 decreased upon oxidation. The effect of HA and CD44 oxidation on CD44–HA binding was similar, but when both HA and CD44 were oxidized, the effect was much larger, in agreement with our experiments. Hence, our experiments and computations support our hypothesis on the role of oxidation in the disturbance of CD44–HA interaction, which can lead to the inhibition of proliferative signaling pathways inside the tumor cell to induce cell death.
Keywords: Oxidative stress, Hyaluronan-CD44 interaction, Binding free energy, Cancer therapies, Cell proliferation
Graphical abstract
Highlights
-
•
Many cancer therapies rely on oxidative stress to damage cancer cells.
-
•
Reactive species oxidize both hyaluronan and CD44, lowering the binding free energy.
-
•
Low binding of CD44+ cancer cells to oxidized hyaluronan, lower when CD44 oxidizes.
-
•
Oxidation reduces cell proliferation of CD44+ cancer cells.
1. Introduction
In many types of cancer, malignant cells overexpress adhesion molecules to improve their interaction with the tumor environment and facilitate the spread of the disease. CD44 is the major cell adhesion receptor expressed in cancer and cancer stem cells, which aids cell-cell and cell-matrix interactions, proliferation, differentiation, invasion, and migration [1]. CD44 is the main membrane receptor for hyaluronan (HA), an essential component of the tumor extracellular matrix [2]. HA influences the biomechanical, physical and structural properties of tissues [3]. In cancer, there is evidence that both high- and low-molecular weight HA are abundant in the tumor microenvironment to favor tumor growth, reduce the immune surveillance and protect cancer cells from apoptosis [4]. Upon binding to CD44, HA activates signaling pathways that promote cancer cell proliferation, invasion and metastasis.
CD44 is present in multiple variant isoforms due to alternative mRNA splicing in several cancers, and its expression can be correlated with tumor subtypes [5]. Some CD44 variants can even participate in the reduction of intracellular reactive oxygen species (ROS) levels in cancer cells by coupling with the glutamate-cystine transporter xCT [6]. This mechanism of protection makes cancer cells resistant to chemo- and radiotherapy, as it is directly involved in the regulation of ROS defense and tumor progression. Nevertheless, all CD44 isoforms have a HA-binding domain in the N-terminal region of the extracellular domain, making HA the most specific ligand for the activation of the CD44 receptor [7]. It is clear that both the interaction of CD44 with HA and the availability of CD44 at the cell surface provide cancer cells with mechanisms of survival that perpetuate tumor growth. Modulating the levels of HA in the extracellular matrix and of CD44 at the cell surface in cancer cells, as well as their interaction, is thus of great importance for cancer therapy, to disrupt the signaling pathways that favor tumor progression [8].
The interaction of CD44 with HA can be disrupted by different methods. It is well documented that HA can be modified enzymatically via hyaluronidases, chemically by deacetylation, sulfation and oxidation [9,10], or broken down by gamma radiation [11]. HA degradation by hydroxyl (HO•) radicals and peroxynitrite (ONOO−) can cleave the glycosidic linkages of HA, and may even cause ring opening [10,12]. These modifications could disturb the CD44–HA interaction and hinder the crucial signaling events required for the survival and progression of cancer. This is particularly important in oncology, because radio-, chemo-, and photodynamic therapy −currently used to treat cancer− are known to increase the levels of reactive oxygen and nitrogen species (RONS) in cells to induce apoptosis [13,14]. For CD44, it has been reported that its structure and binding site can be altered by the reduction of disulfide bonds in the HA-binding groove [15]. Other methods to disrupt CD44–HA interaction are the use of small interfering RNA or blocking antibodies, which have shown antitumor effects in mice models and prevented post-chemotherapy tumor recurrence in breast cancer [[16], [17], [18]]. However, it is not well known how oxidative stress may affect CD44 receptors and their interaction with HA in cancer.
The aim of this study is to determine the interaction of HA with CD44 when exposed to oxidizing conditions, as well as its biological effect in cancer cells. We have performed functional experiments in three cancer cell lines to assess cell viability, proliferation, and binding to HA when delivering oxidative damage to CD44-expressing cells, HA or both. For this, we used a cold atmospheric plasma (CAP) source that generates exogenous RONS in ambient air. We have also carried out computer simulations to support the experiments, applying a combination of different simulation techniques, i.e., reactive and non-reactive molecular dynamics (MD) simulations.
2. Experimental section
2.1. Cell culture
Three CD44+ cancerous cell lines were used in this study: human glioblastoma (U87-MG), melanoma (A375), and colorectal cancer (HT29). All cell lines were cultured in Dulbecco's modified Eagle medium containing 10% fetal bovine serum (Gibco) and 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco). In addition, media was supplemented with l-glutamine (2 × 10−3 M for U87-MG and HT29 and 4 × 10−3 M for A375). Cells were cultured in a humidified environment with 5% CO2 at 37 °C. For all the experiments, 150,000/well of U87-MG, A375 and HT29 cells were seeded in 24-well plates the day prior to treatment to induce oxidative stress. After treatment, cells were detached with accutase at 37 °C, counted with Trypan Blue using the TC20 Automated Cell Counter (Biorad) and processed for the corresponding downstream experiments.
2.2. Quantification of endogenous HA
U87-MG, A375 (3 × 105 cells/T25 flask) and HT29 (1 × 106 cells/T25 flask) cell lines were cultured for 3 days, as indicated in section 2.1. After 3 days, an aliquot of the cell culture supernatant was removed and centrifuged at 1000×g to remove debris. Cells were washed with phosphate buffer saline (PBS) and lysed with Cell Lysis Buffer 2 (895347, R&D Systems) at room temperature (RT) for 30 min with gentle agitation. Debris was removed by centrifugation at 500×g for 5 min. The HA concentration was measured in cell lysates and culture supernatants using the Hyaluronan Quantikine ELISA kit (DHYAL0, R&D Systems), following the manufacturer's recommendations. Supernatants were 5-fold diluted and cell lysates were 10-fold diluted. To account for HA from fetal bovine serum in the culture media, the corresponding medium controls were used to determine the baseline concentration of HA. The optical density (OD) was recorded at 450 nm and 590 nm using the iMark microplate reader (Biorad). The background signal at 590 nm was subtracted from the absorbance at 450 nm and the HA concentration was calculated from a standard curve. The HA concentration is expressed as ng HA per million cells.
2.3. Exogenous RONS treatment
To test the effect of oxidative stress on CD44 and HA, we used a cold atmospheric plasma (CAP) device as an exogenous RONS source. CAP generates a rich mixture of RONS at body temperature by applying energy to a gas to induce partial ionization. The main biologically active components of CAP are the multiple RONS generated, which have medical applications for wound healing, disinfection, and cancer treatment [19]. Here we used a dielectric barrier discharge (DBD) CAP system (see Supporting Information (SI), section S.1, Fig. S1). Pulses were generated using a microsecond-pulsed power supply (Megaimpulse Ltd., St. Petersburg, Russia; 30 kV output, 1–1.5 μs rise time, and 2 μs pulse width). The short- and long-lived RONS produced by this device have been previously described [20]. Right before CAP treatment of cell cultures, the culture medium was removed. Untreated controls were sham-treated. A DBD electrode of 10 mm diameter for 24-well plates was used. For the treatment of plates coated with bovine serum albumin (BSA) and HA, a 3 mm diameter electrode was used. In both cases, the electrode was positioned 1 mm above the samples prior to treatment. The CAP pulse frequency was set at 50 or 100 Hz and samples were treated for 10 s each. Immediately after CAP treatment of cells, 500 μL of complete medium was added to the well and the cells were processed immediately. BSA- and HA-coated plates treated with CAP were immediately used.
2.4. Flow cytometry assays
Baseline CD44 expression was determined on the three cancer cell lines. Sham-treated cells were detached with accutase and washed with ice cold flow cytometry staining buffer (FACS) buffer (sheath buffer from BD supplemented with 0.1% BSA and 0.05% NaN3). Cell density was adjusted to 106 for all samples and controls. Cells were stained with CD44-PE (SAB4700183, Sigma) in the dark for 30 min at RT. As control, cells were labelled with the isotype IgG2b PE (12-4732-42, ThermoFisher Scientific). Unstained cells and acute T cell leukaemia Jurkat cells (lacking CD44 expression) were used as negative controls. All samples were washed with FACS buffer, centrifuged for 5 min at 500×g at RT, and resuspended in FACS buffer before measurement. Acquisition was done using the BD Accuri flow cytometer (BD). 10,000 events of the live population were collected and analyzed using the FlowJo software (FlowJo LLC, version 10).
After CAP treatment, CD44 expression and binding to fluorescein-labelled HA was assessed. For CD44-PE staining, cells were processed as described above. For HA staining, cells were incubated with 30 μg/mL fluorescein-labelled HA (F1177, Sigma) or non-fluorescent HA (H5388, Sigma) as a negative control. Samples were incubated at 4 °C for 2 h, washed three times with FACS buffer and resuspended in FACS buffer for acquisition. In all cases, 5 μL 7-amino-actinomycin D (7AAD PC5.5) viability stain (Biolegend) was added to all tubes. Samples were acquired using the BD Accuri flow cytometer (BD). 10,000 events of the live population were collected and analyzed using the FlowJo software (FlowJo LLC, version 10). CD44 expression was calculated as the difference in mean fluorescence intensity (ΔMFI) between cells stained with anti-CD44 PE and the isotype control. For fluorescein-HA, the ΔMFI was calculated between cells stained with fluorescein-HA and the negative non-fluorescent HA control.
2.5. Preparation of HA-coated plates
96-well flat bottom plates (3599, Corning) were coated overnight at 37 °C with 50 μL of 200 μg/mL rooster comb HA (H5388, Sigma) for the enzyme-linked immunosorbent assay (ELISA) and HA-binding assays. HA from Streptococcus zooepidemicus (H9390, Sigma) was used for the proliferation assays. Plates were washed with PBS before use to remove aggregates. As negative controls, wells were coated with 2% BSA in PBS. The HA-coated plates were used for the HA binding, cell proliferation and ELISA assays.
2.6. HA-binding assay
To determine whether oxidative stress to cells affects their binding with HA, we performed a HA-binding assay. HA-coated plates were blocked with 0.2% BSA in PBS for 2 h at 37 °C. After CAP treatment, cells were detached with accutase, resuspended in 200 μL cell adhesion medium and counted. Immediately after, cells were transferred to the HA-coated plate. Samples were centrifuged for 2 min at 200×g to favor cell contact with the HA-coated plate and then incubated for 1 h at 4 °C. After cell binding, the plate was gently washed three times with cell adhesion medium to remove unbound cells and bright field images were collected with the Spark Cyto (Tecan). Confluence of bound cells in the HA-coated plate was analyzed using the Image Analyzer (Tecan).
2.7. Cell proliferation assay
We also determined whether the modified binding of cells to HA, due to oxidative stress to cells, affects cell proliferation. After CAP treatment, cells were detached with accutase, resuspended in 500 μL culture medium and counted. From the total number of cells collected, 2000 viable cells/well were seeded in the 96-well HA-coated plate and incubated in a humidified environment with 5% CO2 at 37 °C for 4 h. Afterwards, 20 μL/well of sterile resazurin (0.15 mg/mL, TCI) were added to assess the metabolic activity. The OD at wavelengths of 570 and 600 nm was measured every 2 h for a period of 24 h with the Spark Cyto (Tecan). To correct the absorbance values, the background OD at 600 nm was subtracted from the OD at 570 nm. In addition, cell confluency was monitored with the Spark Cyto every 24 h for up to 72 h after treatment.
2.8. CD44-Fc HA plate binding assays – ELISA
Finally, to determine whether oxidative damage to HA would affect its binding specifically with CD44 receptor (and not to alternative receptors binding HA in cells, for example), we used a human Fc chimera fusion protein (CD44-Fc). CAP-treated and untreated BSA- and HA-coated wells were blocked with 1% non-fat dry milk (Sigma) in PBS for 1 h at RT. 50 μL of human CD44-Fc chimera protein (3660-cd-050, R&D Systems) were added to each well at 10 μg/mL in PBST (phosphate buffer saline + 0.05% Tween-20, wash buffer). The plate was left to equilibrate at RT for 1 h. After washing, bound CD44-Fc was probed with 50 μL 1/130000 horseradish peroxidase (HRP)-conjugated anti-human Fc antibody (ab6759, Abcam) for 1 h at RT and in the dark. Plates were washed 5 times prior to developing the reaction. The colorimetric reaction of HRP was generated using the TMB (3,3′,5,5′-tetramethyl benzidine) substrate set (Biolegend) and quenched with the Stop Solution for TMB substrate (Biolegend). The OD was recorded at 450 nm and 590 nm using the iMark microplate reader (Biorad). The background signal at 590 nm was subtracted from the absorbance at 450 nm.
2.9. Statistical analysis
Statistical differences of the experimental data were analyzed using GraphPad Prism software. All experiments were done in triplicates. Results of experimental data are reported as mean ± S.D. In all cases, the post-hoc Dunnett's test was used to calculate the adjusted p value compared to untreated and sham-treated controls. The Shapiro-Wilk test of normality was applied. A p value less than or equal to 0.05 was considered statistically significant.
3. Computational section
3.1. Reactive MD simulations
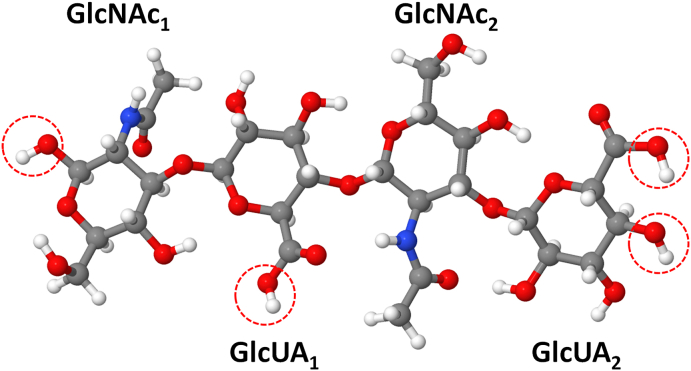
To study the interaction of RONS (i.e., atomic oxygen (O), hydroxyl (HO•), hydroperoxyl (HO2•), nitric oxide (•NO) and nitrogen dioxide (•NO2) radicals, hydrogen peroxide (H2O2) and ozone (O3) molecules as well as nitrite (NO2−) and nitrate (NO3−) ions) with HA, we applied reactive MD simulations based on the density functional-tight binding (DFTB) method [21,22]. The model system for HA in our DFTB-MD simulations consists of four monosaccharide molecules, i.e., two glucuronic acid (GlcUA) and two N-acetylglucosamine (GlcNAc) connected with both β(1,4) and β(1,3) glycosidic bonds (Fig. 1). This model system (called HA4) contains all possible bonds that are repeated in HA polysaccharide. In this way, we are able to study these bonds, i.e., their breaking and/or formation of new bonds taking place in HA upon interaction with RONS.
Fig. 1.
Schematic illustration of the HA4model system used in the DFTB-MD simulations. The system consists of two GlcNAc and two GlcUA units, connected with β(1,3) (between GlcUA1 and GlcNAc1, and between GlcUA2 and GlcNAc2) and with β(1,4) (between GlcNAc2 and GlcUA1) glycosidic links. The H-abstraction reactions occurring on OH groups in red dashed circles are not considered in the interaction of RONS with HA4, since these H atoms are used either to end up the truncated HA sugar chain or to protonate the GlcUA residues, which is not the case in real conditions. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Details about the preparation of the HA4 system are given in the SI, see section S.2.1 and Fig. S2 therein. Briefly, the HA4 structure was placed in a simulation box with periodic boundary conditions applied in all three directions. Next, the geometry of the HA4 was optimized followed by equilibration at 310 K in the canonical (i.e., NVT) ensemble (constant number of particles N, volume V and temperature T).
Subsequently, a single RONS species (e.g., an O atom) was randomly created around the structure with a minimum distance of 5 Å. This distance is greater than the cutoff radii used for non-bonded interactions in the DFTB method, so there was no initial interaction between the RONS species and HA4. Note that due to the absence of a water layer surrounding the HA4 (see section S.2.1 for more details), we protonated the GlcUA residues and added H atoms at the terminal residues, i.e., GlcNAc1 and GlcUA2 (see red circles in Fig. 1). Firstly, we performed 20 simulations for each impinging species to check whether they react with HA4, breaking or forming some bonds. This was done to save computation time and resources, as not all RONS react with the HA4 system. Indeed, our computations showed that only O and HO• radicals chemically react with HA and their reaction mechanism was found the same. Hence, to obtain more statistically valid results, we performed 180 additional simulations (in total 200 runs) for the interaction of only O atoms with the HA4 system (see section 4.4 for more details). It should be mentioned that other species might probably also react with HA4, but on longer time scales. They probably have larger energy barriers and therefore we did not see their reactions within the investigated time scale used. Thus, in our simulations we selected the faster reactions. Note that if an O atom reacted with the H atoms shown within the red circles of Fig. 1, we excluded this reaction and performed an additional MD simulation. All simulations were performed applying the DFTB + package, version 18.2 [23].
3.2. Non-reactive MD simulations
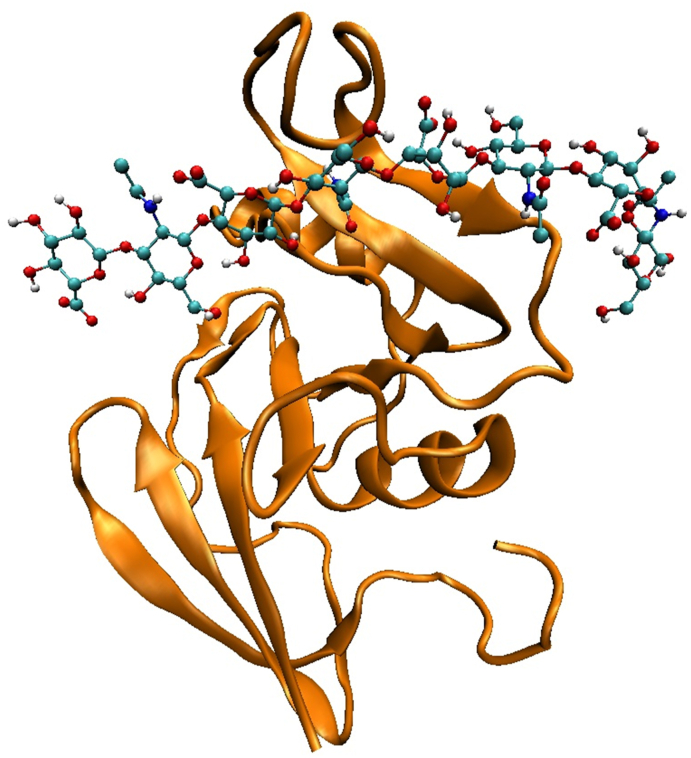
We performed non-reactive MD simulations to investigate the effect of oxidation on the interaction (i.e., binding free energy) of the HA sugar chain with its receptor protein CD44. For this purpose, we prepared four model systems, i.e., the native (CD44–HA8) system shown in Fig. 2 and its three oxidized forms. The latter include the systems with either oxidized HA8 (CD44–HA8-OX) or oxidized CD44 (CD44OX–HA8) or both (CD44OX–HA8-OX). These oxidized systems are called OX1, OX2 and OX3, respectively.
Fig. 2.
Schematic illustration of the native CD44–HA8system used in the non-reactive MD simulations. The suffix in the HA oligomer denotes the number of monosaccharides. CD44 (or more exactly, CD44-HABD) is displayed in cartoon view and HA8 is represented in CPK view.
Note that in our model systems, we used the Hyaluronan Binding Domain of CD44 (CD44-HABD) for the interaction with the HA8 and we call CD44-HABD as CD44 throughout the text, for simplicity. Moreover, we used the HA8 sugar octamer (i.e., HA containing eight sugar residues) in our simulations, as at least hexasaccharide (HA6) is required for high affinity interaction with CD44 on most cell backgrounds [[24], [25], [26]]. Thus, the chosen size of the HA (i.e., HA8) was sufficient for investigation of the binding properties. HA8 was also utilized in other MD simulation studies (see Ref. [27] and references therein).
Details about the preparation of the model systems are given in the SI (section S.2.2.1). Briefly, we prepared three replicas of each model system, placed in a simulation box and equilibrated with different initial velocities at 310 K and 1.0 bar. Thus, in total 12 structures were prepared. These model systems were used to calculate the secondary structure of the CD44 protein before and after oxidation, as well as to estimate the binding energies of the native, OX1, OX2 and OX3 systems without accounting for the entropic contribution (section S.2.2.1).
We also placed these complex systems in a bigger box enlarged in the z-direction, to calculate the free energy profiles (FEPs) of the native, OX1, OX2 and OX3 systems (see section S.2.2.1 and Fig. S3 therein, for more details). Specifically, we applied the umbrella sampling (US) method [28] to calculate the FEPs of the HA8/HA8-OX sugar octamer dissociating from the binding site of the CD44/CD44OX receptor protein. Initially, we performed pulling simulations, by slowly pulling the HA8/HA8-OX octamer along the z-axis. Slow pulling was needed to cause a minimum disturbance to the system during the detachment of HA8/HA8-OX from CD44/CD44OX. Details about the pulling simulations are given in section S.2.2.2 of the SI. After the pulling simulations, we extracted 40 windows (i.e., 40 starting structures for free energy calculations) along the z-axis, each separated by 0.10 ± 0.01 nm. Subsequently, we performed US simulations using the same parameters as employed in the equilibration runs (see SI; section S.2.2) to obtain a single FEP of HA8/HA8-OX dissociating from CD44/CD44OX. As we used three replicas for each model system (i.e., native, OX1, OX2 and OX3), we obtained the final FEP by averaging three individual FEPs for each of these systems (see section S.2.2.3 for more details). Thus, in total 40 US × 3 replica structures × 4 model systems = 480 US simulations were performed to obtain the FEPs.
All simulations were carried out using the GROMACS 5.1.2 package [29], employing the GROMOS 54A7 force field [30]. This force field version was chosen because it has been parametrized for an improved description of protein structure. The parameter set of oxidized residues (used in CD44OX of the oxidized systems) obtained from Ref. [31]. For description of the HA8 octamer, we used the GROMOS 56A6CARBO_R force field [32] supplemented with the uronate and N-acetylamino parameters [33,34]. Combination of the GROMOS 56A6CARBO_R and the GROMOS 54A7 force field versions was straightforward because non-bonded interactions are very similar, with exception of a few ionic interactions that were kept as in GROMOS 54A7. Information about the testing of the HA8 topology is given in the SI (section S.2.2.4 and Figs. S4–S6 therein).
We developed new GROMOS-type parameters for the HA8-OX structure, namely the modified N-acetylamino groups of GlcNAc residues. More information about the parametrization procedure and comparison of new parameters with the standard ones is given in the SI (sections S.2.2.5 and S.2.2.6 and Figs. S7–S11 therein). Briefly, we performed a fitting of standard potential energy functions to quantum chemistry data from the literature [35]. In this way, we obtained the parameters of newly formed bonds, angles and dihedrals in GlcNAc residues.
4. Results and discussion
4.1. Oxidation of HA decreases its recognition by CD44-Fc
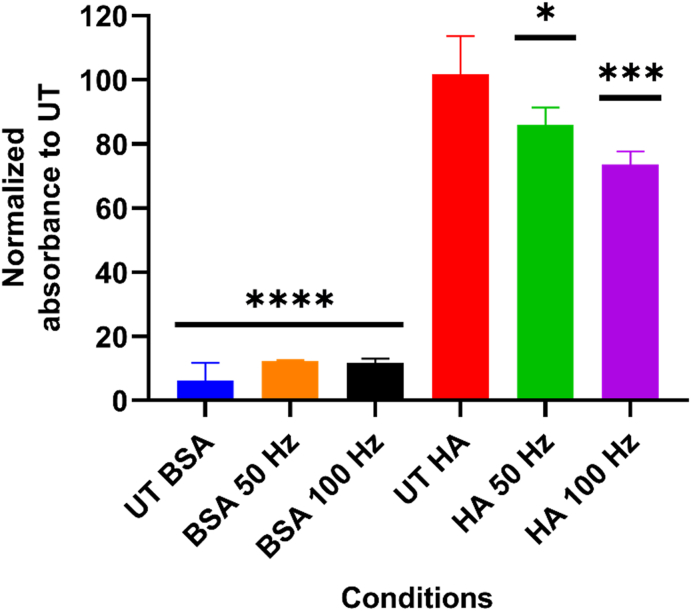
To determine the ability of exogenous RONS to alter the structure of HA and their effect on the CD44–HA interaction, we performed an ELISA test using a Fc chimera fusion protein CD44-Fc. As explained in section 2.3, we used a CAP device as an external source of RONS to oxidize HA deposited in plastic cell culture plates. After CAP treatment of HA-coated plates, the binding of CD44-Fc was reduced by approximately 15% (50 Hz, p ≤ 0.001) and 28% (100 Hz, p ≤ 0.0001), compared to the untreated (UT) control (Fig. 3). As expected, higher doses of RONS (i.e., CAP treatment at 100 Hz vs 50 Hz) increased the oxidative modifications in HA, most likely causing damage to the molecule chains. No significant binding was observed in the untreated and treated BSA-coated wells.
Fig. 3.
HA oxidation reduces its binding to CD44-Fc. HA-coated plates were treated with CAP to induce oxidation of HA molecules. A significant reduction in the binding levels was observed when compared to the untreated (UT) control. Wells coated only with 2% BSA (treated and untreated) served as negative controls. Results were analyzed using the Dunnett post hoc test. Mean ± S.D. * = p ≤ 0.01; *** = p ≤ 0.001; **** = p ≤ 0.0001.
It is known that oxidizing agents such as sodium periodate can modify the hydroxyl groups in HA, forming aldehyde groups [36]. Endogenous RONS produced by Fenton reaction in normal epidermal cells (H2O2 and O2•−) have also shown to partially degrade HA [37]. In addition, HA is highly susceptible to degradation by ONOO− but not by •NO radicals [38], as well as to changes in the tertiary structure of HA due to 1O2 [39]. These modifications fragment HA and reduce its molecular weight, affecting its interaction with CD44 receptors and interrupting the signal transduction.
The CAP source used here has been previously characterized [20], using the same treatment conditions as in this study. It produced a mixture of short- and long-lived RONS with small amounts of O/O3, •OH, •NO, and ONOO− and higher levels of NO3−, NO2− and H2O2 produced, whereas 1O2 and O2•− were undetectable. In addition, •OH generated from H2O2 in the presence of transition metal ions may degrade HA macromolecules [10,12]. Altogether, these results suggest that exogenous RONS alter the structure of HA, hindering its ability to bind with CD44-Fc.
4.2. Oxidative damage immediately decreases the ability of CD44+ cancer cells to bind HA
To evaluate whether the interruption of the CD44–HA binding upon oxidative stress is due to oxidation of HA, CD44, or both, we performed an HA binding assay using three CD44+ cancer cell lines under three different conditions: i) oxidizing CD44+ cells only; ii) oxidizing HA-coated plates only; and iii) oxidizing CD44+ cells and HA-coated plates.
First, the basal expression of CD44 in the three cancer cell types was determined by flow cytometry (Fig. S12) following dual staining with an anti-CD44 PE monoclonal antibody and a live-dead stain. Detailed gating strategies for analysis are provided in Fig. S13. Glioblastoma (U87-MG) and melanoma (A375) showed a pronounced expression of CD44 at the cell surface (CD44high), compared to colorectal adenocarcinoma (HT29, CD44low). In the same way, the basal expression of HA was determined by ELISA (Fig. S14). U87-MG produced more HA than A375, as measured in the cell culture supernatants and cell lysates. This is in agreement with the literature, as HA is the main component of the brain extracellular matrix and high HA levels are expected in glioblastoma [40,41]. It is possible that both cell lines (and especially U87-MG) retain HA at the cell surface, forming a pericellular matrix that could contribute to their proliferation and locomotion [42]. We did not find detectable levels of HA in HT29 samples.
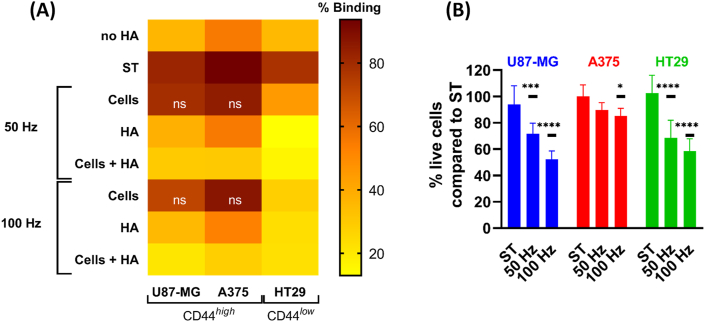
The binding assay demonstrated that sham-treated U87-MG and A375 cells presented high levels of binding, followed by HT29 (Fig. 4A). As expected, wells coated with BSA only (no HA) showed low levels of cell binding (Fig. 4A). Oxidation of CD44+ cancer cells did not reduce the binding to HA-coated plates with either treatment (50 and 100 Hz; p > 0.05). It is possible that the abundance of CD44 receptors at the cell surface could compensate for the oxidative damage induced to some of the CD44 molecules, or that the oxidative damage inflicted was not sufficient to alter the CD44 structure and decrease the binding. To date, changes in the HA-binding groove of CD44 have been only reported when CD44 was reduced with tris(2-carboxyethyl)phosphine hydrochloride (TCEP.HCl) [15], but the effect of RONS in CD44 is not yet reported. Alternatively, it is possible that the oxidation of CD44+ cancer cells affected other molecules at the cell surface. As HA is produced and secreted by U87-MG cells, and in lesser extent by A375, HA anchored to CD44 could react with the RONS delivered and partly quench oxidative damage. If that was the case, this could explain the low effect of oxidative damage in binding when only CD44high U87-MG and A375 cancer cells were treated.
Fig. 4.
Oxidation of HA significantly reduces the binding of CD44+cancer cells. (A) CAP treatment of cancer cells (causing oxidation of surface receptors such as CD44) had little or no effect on their binding to HA-coated plates. However, oxidation of HA significantly reduced the binding of CD44high (U87-MG and A375) and CD44low (HT29) cells to the HA-coated plate. CAP treatment of both HA and CD44+ cells further reduced the binding. Wells coated only with BSA (no HA) served as negative controls. Mean values plotted express the percentage of binding of CD44+ cells to HA-coated plates for each condition. p ≤ 0.0001 for all samples compared to their corresponding sham-treated (ST) control, except when “ns” (not significant) is indicated. (B) Live cell count was done after CAP treatment, before transferring cells to the HA-coated plate. The percentage of live cells compared to the ST control is shown. Mean ± S.D. * = p ≤ 0.05; *** = p ≤ 0.001; **** = p ≤ 0.0001. In all cases, results were analyzed using the Dunnett post hoc test.
In contrast, the RONS effect on HA significantly reduced the binding of all CD44+ cancer cell lines to the treated HA-coated plate (p ≤ 0.0001, Fig. 4A). These results are in agreement with our ELISA results (Fig. 3), supporting that the oxidative damage in HA hinders its binding to CD44. Further oxidation of both HA and CD44+ cells completely abolished the CD44–HA binding to levels comparable to those in wells without HA, suggesting that the oxidative damage inflicted in both CD44 and HA molecules is required to fully inhibit their interaction.
In all treatment conditions, the effect was more evident in the CD44low HT29 cell line. This could be due to the intrinsic low expression of CD44 at the cell surface, the main receptor for HA. Cancer cells also express RHAMM (receptor for hyaluronic acid mediated motility; HMMR; CD168) [43], a receptor with invasive functions similar to CD44, when CD44 is absent [44]. As cancer cells were immediately challenged to HA-plates after CAP treatment and analyzed only a couple of hours after, it is unlikely that this time was sufficient to allow the expression of RHAMM to overcome the lack of CD44 at the cell surface. In addition, the lack of HA production by HT29 cells could contribute towards a more pronounced effect of RONS on cell surface molecules such as CD44.
As oxidative damage can also affect the cell viability, the latter was assessed in all cells before transference into the HA-coated plates. As expected, the exogenous RONS reduced the number of viable cells in all cell lines, which was directly proportional to the dose of RONS, as determined by the CAP treatment frequency (Fig. 4B). However, none of the treatments decreased the percentage of viable cells below 50% in all the cell lines. Most of the CD44+ cancer cells exposed to the CAP treatments preserved their ability to bind native HA-coated plates (approx. 10% reduction for U87-MG and A375, and 50% reduction for HT29, compared to ST).
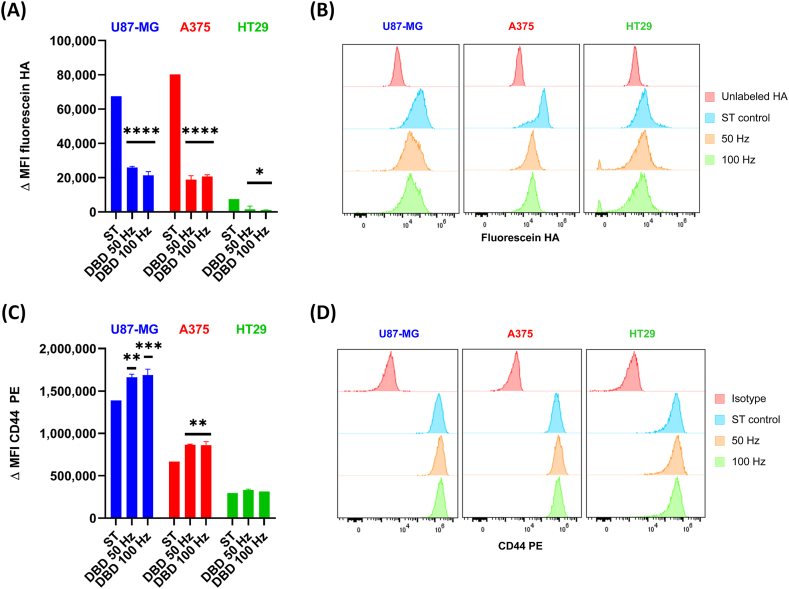
To further determine if the reduction in binding was specifically due to modifications to CD44 receptors in CD44+ cells and not due to other receptors present at the cell membrane, we performed a flow cytometry analysis. Cells were labelled with CD44 PE or fluorescein-HA and counterstained with live-dead stain immediately after CAP treatment. When we analyzed the live population of fluorescein-HA labelled CD44+ cells, we observed a highly statistically significant difference in the ΔMFI of CAP-treated cells compared to the ST controls for U87-MG and A375 cells (p ≤ 0.0001) and HT29 cells (p ≤ 0.05) (Fig. 5A and B). These results show a significant reduction in binding of CD44+ cells to HA due to the oxidative damage of CD44 receptors in cancer cells.
Fig. 5.
Oxidation of CD44+cells affects their binding to HA and availability at the cell surface.(A) The binding of fluorescein-HA to CD44+ cells treated with CAP was analyzed by flow cytometry. The results show a decrease in binding immediately after CAP treatment in all cell lines. (B) Representative histograms of each treatment condition per cell line. (C) CAP treatment increased the ΔMFI of CD44 PE in U87-MG and A375 cells, but not in HT29. (D) Representative histograms of each treatment condition per cell line. A and B: mean ΔMFI ±S.D. * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001; **** = p ≤ 0.0001 when compared to their corresponding sham-treated (ST) control. Results were analyzed using the Dunnett post hoc test.
Compared to the HA binding assay results, the flow cytometry data indicated a more dramatic consequence of the oxidation of CD44+ cells. It is known that the way HA is presented to cells (whether HA is soluble or immobilized in a surface) affects the ability of CD44 to bind HA in different cell lines [45,46]. The consequence is that many cell lines can only bind immobilized but not soluble HA, which could explain the differences we observed between the flow cytometry and HA binding assays. Nevertheless, the result of both experiments suggests alterations to the HA-binding site of CD44, which limit its binding to HA.
When analyzing CD44 expression, we observed an increase in the ΔMFI of CD44 PE in CAP-treated cells compared to the ST control for U87-MG and A375 (p ≤ 0.01 and 0.001), and a trend to increase for HT29, albeit it was not statistically significant (Fig. 5C and D). CD44 binds to a minimum of six HA sugar residues and its binding capacity depends on the formation of CD44 aggregates, a process that can be hindered if oligomerization is inhibited [24]. The increased number of CD44 molecules identified by the anti-CD44 PE monoclonal antibody at the cell surface after CAP treatment could be due to variations in the packing density of CD44 in lipids rafts, or due to the interaction of CD44 with other cell membrane receptors that might increase the detectability of CD44 by flow cytometry. In some cells, it has been reported that the oxidative stress induced by H2O2 results in the overexpression of CD44 at the cell surface of cancer cells, a process that involves a fine dysregulation of microRNAs [47]. It is possible that this occurs as a response to oxidative stress, as some CD44 isoform variants have been implicated in the regulation of the defense against ROS [6]. However, we have not determined the isoforms expressed in the CD44+ cell lines before and after treatment, and this possibility should be explored further to determine the role of isoform variants in the response to oxidative stress. Nevertheless, it is worth mentioning that the expression of CD44 on the cell surface does not compel them to bind HA [45].
Taken together, these results demonstrate that RONS immediately decrease the ability of CD44+ cancer cells to bind HA. This effect is not due to the reduction of cell viability, but to modifications to CD44 receptor, that is further abrogated when HA is also oxidized.
4.3. The oxidative damage to CD44 and HA affects the proliferation of cancer cells
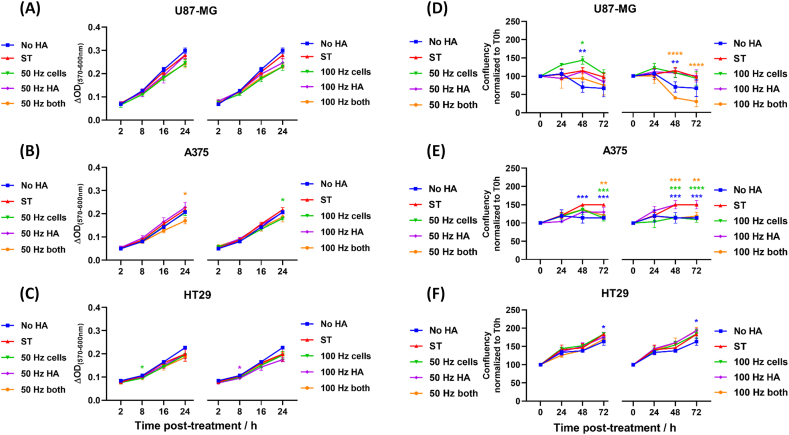
After demonstrating that exogenous RONS immediately reduced the binding of CD44-Fc and CD44+ cancer cells to HA, we evaluated whether this effect could translate into alterations on the proliferative and metabolic activity of CD44+ cells. We treated the CD44+ cells at the same three conditions as previously used: i) oxidizing CD44+ cells only; ii) oxidizing HA-coated plates only; and iii) oxidizing CD44+ cells and HA-coated plates. Endotoxin-free HA was used for the cell proliferation assays. After treatment, cells were incubated with resazurin, a cell viability reagent that is used to measure the metabolic capacity of cells and their corresponding proliferative state. Viable cells reduce resazurin into resorufin, which results in a signal that is measured by colorimetry. In addition, cell confluence was measured for 72 h post-treatment to monitor cell growth, which represents the percentage of the well surface covered by adherent cells.
Exogenous RONS affected the metabolic activity of the CD44high U87-MG cell line in a similar way when HA, cells or both were treated (Fig. 6A). The metabolic activity of CD44 high A375 and CD44low HT29 was reduced when both HA and cells were treated (Fig. 6B and C; p ≤ 0.05 for A375 for 50 Hz - both), although in most cases the difference to the sham-treated control was small 24 h post-treatment.
Fig. 6.
The disruption of CD44–HA binding altered the metabolic and proliferative state of cancer cells. Metabolic activity of (A) U87-MG, (B) A375 and (C) HT29 cancer cells challenged to HA-coated plates was assessed with resazurin for 24 h after treatment. Cell, HA-coated plates or both were treated with CAP (A-C: 50 Hz left; 100 Hz right). Metabolic activity was assessed with resazurin for 24 h after treatment. Wells coated only with BSA (no HA) served as negative controls. ΔOD = optical density corrected for background. Cell proliferation was assessed by measuring the confluence of the cultures over 72 h for (D) U87-MG, (E) A375 and (F) HT29 cancer cells. (D–F) Data is expressed as confluency normalized to T = 0 h. Statistically significant differences were found between the sham-treated (ST) control and corresponding treated samples. Mean ± S.D. * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001; **** = p ≤ 0.0001. In all cases, data were analyzed using the Dunnett post hoc test.
When looking at the confluence of the cell cultures, we observed that cell proliferation of U87-MG was enhanced when cells were treated with 50 Hz CAP. In contrast, it significantly decreased when both cells and HA were treated with 100 Hz CAP (Fig. 6D, p ≤ 0.0001). Similarly, A375 cells failed to proliferate over time after CAP treatments, especially when using CAP at 100 Hz (Fig. 6E, p ≤ 0.0001 for 100 Hz - cells; p ≤ 0.01 for 100 Hz - both). HT29 cell proliferation was not significantly affected by CAP when monitored for 72 h post-treatment (Fig. 6F). This is particularly interesting, as CAP treatments significantly disrupted the binding of HT29 to HA-coated plates, as shown above. It is possible that HT29 cells exposed to oxidative stress overcame the lack of CD44 at the cell surface over time by enhancing CD44 or RHAMM expression. Further studies to determine the role of other HA receptors in cancer cells in the context of oxidative stress could bring light to this issue.
CD44 is the main receptor for HA, able to induce proliferation, migration, and invasion in cells in a HA-dependent manner, as it activates cell survival anti-apoptotic proteins [48]. Another HA receptor RHAMM is mostly involved in HA-mediated motility [48]. It has been suggested that CD44 binds to HA with five times greater affinity than RHAMM [49], thus we believe that our results are a consequence of the disruption of HA–CD44 interaction. Note that the binding of HA–CD44 can promote the association of CD44 with EGFR in the intracellular compartment, as well as induce EGFR phosphorylation [50]. However, these events take place upon interaction of CD44 with HA, and EGFR does not interact directly with HA. Our experiments demonstrate that the ability of CD44+ cells to proliferate in a HA-coated plate is reduced upon exposure to oxidative damage, even more so after both cells and HA-coated plates are treated. As the disruption of HA–CD44 leads to a reduction in proliferation, the downstream signaling pathways affected by the treatment could include EGFR. However, a first HA–CD44 interaction would be needed to lead to this downstream event.
The disruption of CD44–HA interaction has been suggested as an alternative method to target cancers, to reduce tumor progression and metastasis [8]. Our results demonstrate that RONS alter not only HA but also cell surface receptors such as CD44, that could interrupt the CD44–HA binding and reduce cell proliferation. To understand the molecular level mechanisms of RONS-induced modifications on both HA and CD44 and its subsequent effect on CD44–HA binding, we performed in silico analyses.
4.4. RONS induces oxidative modifications in the HA structure
We performed reactive DFTB-MD simulations to study the interaction of RONS (i.e., O, •OH, HO2•, H2O2, O3, •NO, •NO2, NO2− and NO3−) with HA4, i.e., HA containing four monomer sugars (see Fig. 1). We mainly focused on the bond breaking and formation processes taking place in HA4. Our simulation results showed that, during the investigated time scale, HO2•, H2O2, O3, •NO, •NO2, NO2− and NO3− species do not cause any bond dissociation and formation in the HA4 sugar molecule. The fact that no reaction is observed does not necessarily indicate that it is thermodynamically unfavorable. Certain reactions may be associated with larger energy barriers, meaning that they might still occur given enough time. Species that do not react only have non-bonded interactions with the HA4 structure. HO2•, H2O2, NO2− and NO3− formed weak attractive interaction with HA4 (see Fig. S15), whereas •NO and •NO2 showed weak repulsive interaction, and O3 showed both, i.e., sometimes weak attractive and sometimes weak repulsive interaction, depending on what side (i.e., which oxygen atom) of O3 approaches the structure. On the other hand, we did observe bond breaking and formation processes upon interaction of O atoms and •OH radicals with HA4. The reaction mechanisms of these species are very similar, i.e., they both abstract a H-atom from the HA4 molecule, forming an HO• radical (in the case of the O atom) or a water molecule (in the case of the HO• radical). In other words, the O atom acts as two HO• radicals. Therefore, we only considered the interaction of O atoms in our further reactive MD simulations, to obtain more statistically valid results on the reaction mechanisms.
We observed many reaction mechanisms upon impact of an O atom on the HA4 molecule and all of these reactions are initiated by H-abstraction from different positions of the structure (see Table S1 and Fig. S16). From the observed 34 reaction mechanisms, most of them (i.e., reactions 1–30) lead to the addition of an extra O atom in the system. In other words, the O atom abstracts a H atom from the structure, forming an HO• radical, and this radical reacts back with the structure, creating a stable alcohol group. Other reaction mechanisms of O atom interaction with HA4 result in either the formation of a C O bond in the structure (reaction 32) or a breakage of the C–C bond (reactions 31, 33 and 34). The last two reaction mechanisms (reactions 33 and 34), where the oxidation of GlcUA leads to its ring opening, forming two aldehyde groups, were also observed in experiments with periodate oxidation [51]. The ring-opening in GlcUA by HO• radicals was also found earlier [52]. In general, it was stated that the ring-opening by oxidation of HA can further lead to a degradation of this polymeric chain, thereby lowering its molecular weight [12,36]. HA degradation by ROS (mainly HO• radicals) can also be due to dissociation of glycosidic bonds, thereby leading to fragmentations in the HA chain structure [9,10]. Short fragments of HA (i.e., HA oligomers) can themselves serve as blockers of the interaction of high molecular weight HA (HMW-HA) with CD44, thereby being effective in cancer treatment [53]. Blocking of HA–CD44 interaction by short HA oligomers helped to remove HMW-HA from the surface of both human (HaCaT) and mouse keratinocytes [26,54], to displace HMW-HA from cell surface CD44 [55] and to significantly inhibit the formation of pericellular matrix by ovarian cancer cells [56]. Moreover, exogenously added short HA oligomers have shown the antigrowth effects in lung, mammary, and colon carcinoma cell lines (see Ref. [53] and references therein). In our simulations, we did not observe the glycosidic bond cleavage, and hence the fragmentation, by O atom impact. However, we only studied single impacts of O atoms on the HA surface, and the glycosidic bond breakage can probably occur during consecutive impacts of O atoms on the modified/oxidized surface of HA (i.e., during high level of oxidation). The formation of OH groups in the HA structure was observed in our simulations mainly in GlcNAc compared to GlcUA residues, i.e., 65% vs 35%, respectively (see Table S1). This is in agreement with experiments from literature, where it was mentioned that the GlcNAc moiety is attacked more specifically by ROS (mainly HO• radicals) rather than GlcUA [10]. Overall, these oxidative modifications in HA can lead to HA degradation, which may disturb the CD44–HA interaction and hinder the crucial signaling events required for the survival and progression of cancer.
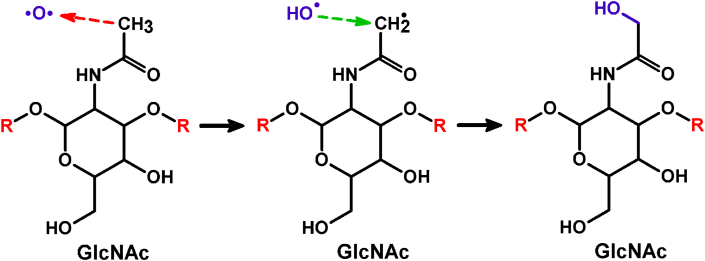
The most often observed reaction mechanism (i.e., in 29% of the cases, see Table S1) is illustrated in Fig. 7.
Fig. 7.
Reaction mechanism of an O atom with the N-acetylamino group of GlcNAc, leading to the formation of an additional OH group in the system. The H-abstraction and OH addition reactions are indicated by red and green dashed arrows, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In this figure, the O atom abstracts a H atom from the N-acetylamino group of the GlcNAc residue (red dashed arrow), leading to the formation of two radicals, i.e., CH2• and •OH. Subsequently, these two radicals bind with each other (green dashed arrow), forming a new OH group in the system.
The formation of the OH groups in the HA sugar chain can influence the binding of HA to its receptor CD44, which may lead to a disturbance of proliferative signaling pathways inside the cells, affecting the cell growth. Therefore, in our non-reactive MD simulations we studied the effect of this most frequent oxidation of HA (i.e., formation of OH in the N-acetylamino groups of the GlcNAc residues), as well as oxidation of CD44, on the binding of HA with CD44 receptor protein.
4.5. Oxidation of both HA and CD44 reduces the CD44–HA binding free energy
As mentioned above, the oxidation of HA can affect its binding to CD44 receptor protein. Moreover, this oxidation can also take place in CD44 itself, modifying its specific residues. Thus, studying the effect of CD44 oxidation on binding with native or oxidized HA is highly relevant. These simulations can provide insight into the molecular-level mechanism of the suppressive effect of oxidation on CD44–HA binding, observed in our experiments. For this purpose, we investigated the effect of oxidation on the binding free energies of the native, OX1, OX2 and OX3 systems.
For the creation of CD44OX used in the OX2 and OX3 systems, we modified specific amino acids in the protein to their oxidized forms, based on literature [[57], [58], [59], [60]] and the analysis of the solvent accessible surface area (SASA). Details about the chosen amino acids and the SASA results are given in section S.4.2 of the SI (Table S2).
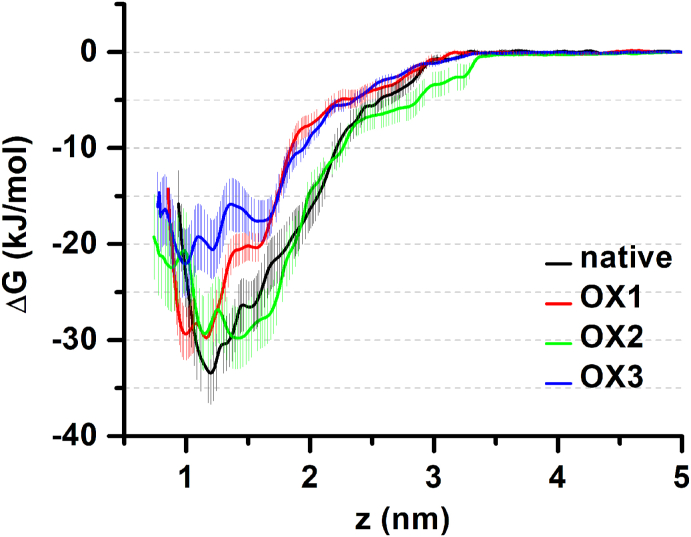
It is clear from Fig. 8 that oxidation of either HA8 (in the case of OX1) or CD44 (in the case of OX2) decreases the binding free energy (i.e., it becomes more positive, see Table 1), thereby making the interaction weaker and increasing the probability of HA dissociation from the binding groove of CD44. Indeed, the dissociation free energy (which is equal to the binding free energy) decreases by 3.6 kJ/mol in both the OX1 and OX2 systems, in comparison with the native system (see Table 1).
Fig. 8.
Calculated free energy profiles of the native (CD44–HA8), OX1 (CD44–HA8-OX), OX2 (CD44OX–HA8) and OX3 (CD44OX–HA8-OX) systems. The uncertainties associated with sampling are shaded accordingly.
Table 1.
Calculated binding free energies ΔG for the native, OX1, OX2 and OX3 systems. ΔΔG denotes the loss of energy compared to the native case.
| System | ΔG (kJ/mol) | ΔΔG (kJ/mol) |
|---|---|---|
| native (CD44–HA8) | −33.4 ± 3.3 | – |
| OX1 (CD44–HA8-OX) | −29.8 ± 2.6 | 3.6 |
| OX2 (CD44OX–HA8) | −29.8 ± 3.2 | 3.6 |
| OX3 (CD44OX–HA8-OX) | −22.1 ± 3.6 | 11.3 |
The effect of oxidation is most clearly visible in the case of the OX3 system, where the binding energy decreases to −22.1 kJ/mol (i.e., a drop by 11.3 kJ/mol compared to the native system), see Table 1. Thus, oxidation of both HA8 and CD44 leads to a much weaker CD44–HA interaction. These results are in qualitative agreement with our experimental results, where oxidation of HA or CD44 or both interrupts the binding of HA to its receptor CD44, thereby disturbing the proliferative signaling pathways inside the cancer cells and reducing their metabolic activity.
To understand the reason of the oxidation effect on the binding free energy, we performed secondary structure analysis of the CD44/CD44OX structure in all systems to obtain detailed information on the conformational changes. The results showed that there is almost no change in α-helix and β-sheet conformations after oxidation (see Table S5). This is most likely attributed to a stabilizing effect of HA, as it was suggested that the presence of HA in the binding site stabilizes CD44 (more specifically its B-form) [[61], [62], [63]]. Note that in the B-conformation of CD44 (which was used in this study), the side chain of Arg41 is flipped towards the bound HA ligand, forming hydrogen bonds. Analysis of the secondary structure also revealed a decrease of the coil and turn structures by 1–2%, and a rise in the bend conformation by 2–3%. This is most probably due to the breakage of the disulfide bonds between Cys77–Cys97 and Cys28–Cys129 residues, which results in a reduced stability of the protein structure and hence lower binding to HA. Indeed, Kellett-Clarke et al. reported that the disulfide bridge between Cys77–Cys97 residues stabilizes the HA binding groove and a cleavage of this bond over the other disulfide bonds results in inhibition of the CD44–HA interaction [15].
We also estimated the binding energies of the native, OX1, OX2 and OX3 systems, applying the g_mmpbsa tool of GROMACS, using the molecular mechanics Poisson−Boltzmann surface area (MMPBSA) method for calculation of the binding energy components. Although this tool cannot calculate the entropic contribution of the binding free energy (which was accounted for using the US simulations, see above), it is useful to compare the relative binding energies, e.g., between the native and OX3 systems, and more importantly to find out the energy contribution of each residue to the total binding energy. Due to the dynamic nature of the interaction, it is difficult to study the residues involved in the interaction and therefore this tool is suitable for estimation of the energy contribution per individual residue before and after oxidation, which helps to understand the effect of oxidation on the binding free energy, as shown in Fig. 8 above. The obtained results show the decreasing trend in binding energy (i.e., it becomes more positive) upon oxidation (see Table S6). Note that the energy values are positive, but as stated earlier, the entropic contribution to the binding free energy is neglected in this method. Still, the energy trend observed is in line with the trend of the binding free energy calculated with the US method (see Table 1).
Comparison of the energy contributions of the important residues (Table S7) also shows an overall decrease of the binding energy, which is clearly seen between the native (no oxidation) and OX3 (oxidation of both the protein and ligand) systems, except for a few residues that have lower energies than the others. We focus here on the residues Arg41, Cys77 and Cys97 that have higher absolute energy values and they play a crucial role in binding [15,61]. The binding energy of Arg41 decreases upon oxidation, but it is still negative, meaning that this residue exhibits attraction with HA8/HA8-OX. The binding energies of Cys77 and Cys97 residues also decrease, and become even positive after oxidation to cysteic acid (i.e., in OX2 and OX3 systems). This indicates that the interaction becomes repulsive after oxidation.
Altogether, the oxidation of both HA8 and CD44 overall decreases the binding energy contribution of the residues, except for a few of them, which is negligible. This in turn affects the overall binding affinity of HA8 to CD44, as observed above (see Fig. 8 and Table 1). The drop in binding affinity will affect the signaling pathways in cancer cells, eventually reducing their proliferation. In our experiments, we observed a lower effect of CD44 oxidation on the CD44–HA binding, compared to that of HA (see Fig. 4 above). In our simulations, we observed the same values of the binding energy for HA and CD44 oxidation, although the errors are slightly different (cf. OX1 and OX2 in Table 1). As is clear from Fig. 8, the error bars of OX2 are close to that of the native system, whereas the errors of OX1 are relatively small (cf. minima of OX1 and OX2 in Fig. 8). In this sense, the effect of OX1 (i.e., HA oxidation) is probably more pronounced than that of OX2 (i.e., CD44 oxidation). It should also be mentioned that we used only the most frequently observed oxidative modification, to create HA8-OX and to study its interaction with CD44. It is likely that other oxidations (with low percentages of occurrence) can also play a role in CD44–HA binding. As we discussed in section 4.4, some of the reaction mechanisms obtained by our reactive MD simulations (i.e., reactions 33 and 34, see Table S1) result in a ring opening and formation of dialdehyde, which can further lead to a degradation of HA [36]. Destruction of HA most likely affects its interaction with CD44, thereby causing a reduction of CD44–HA binding observed in our experiments.
Thus, in general our computational results are in qualitative agreement with our experimental results, and show that oxidation of HA or CD44 or both of them induces alterations in their structures, leading to a disturbance of CD44–HA interaction. As a result, cancer cells express a reduced metabolic activity, which prevents their growth. The harmful effect of oxidation was more pronounced when both HA and CD44 are oxidized, as observed both in our in silico and in vitro experiments.
5. Conclusions
Using various experimental and computational methods, we demonstrate that oxidation of not only HA but also CD44 has a significant detrimental effect on the CD44–HA interaction, which reduces the proliferative state of CD44+ cancer cells. This effect is enhanced when both CD44 and HA are oxidized. Our reactive MD simulations show that the most frequent oxidation of HA leads to the formation of alcohol groups in the GlcNAc residues. Our non-reactive MD simulations reveal that oxidation of both HA and CD44 leads to a drop of the binding free energy of HA to CD44, thereby disturbing their interaction. As shown in our experiments, this reduction in binding is not due to a decreased viability of cancer cells exposed to RONS, but to the changes in the structure of CD44 at the cell surface. Our in vitro and in silico experiments are in qualitative agreement. The drop in binding energy will affect the signaling pathways in cancer cells, ultimately reducing their proliferation. Hence, this could be one of the underlying mechanisms of cancer therapies based on oxidative stress.
Funding statement
This work was financially supported by the Research Foundation – Flanders (FWO) [grant number 1200219N].
Declaration of competing interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Acknowledgements
The authors acknowledge the Turing HPC infrastructure at the CalcUA core facility of the University of Antwerp (UA), a division of the Flemish Supercomputer Center VSC, funded by the Hercules Foundation, the Flemish Government (department EWI) and the UA, where all computational work was performed.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101968.
Contributor Information
Maksudbek Yusupov, Email: maksudbek.yusupov@uantwerpen.be.
Angela Privat-Maldonado, Email: angela.privatmaldonado@uantwerpen.be.
Annemie Bogaerts, Email: annemie.bogaerts@uantwerpen.be.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Thapa R., Wilson G.D. The importance of CD44 as a stem cell biomarker and therapeutic target in cancer. Stem Cell. Int. 2016;2016:2087204. doi: 10.1155/2016/2087204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misra S., Hascall V.C., Markwald R.R., Ghatak S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front. Immunol. 2015;6:201. doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monslow J., Govindaraju P., Pure E. Hyaluronan - a functional and structural sweet spot in the tissue microenvironment. Front. Immunol. 2015;6:231. doi: 10.3389/fimmu.2015.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu M., Tolg C., Turley E. Dissecting the dual nature of hyaluronan in the tumor microenvironment. Front. Immunol. 2019;10:947. doi: 10.3389/fimmu.2019.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senbanjo L.T., Chellaiah M.A. CD44: a multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front Cell Dev Biol. 2017;5:18. doi: 10.3389/fcell.2017.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishimoto T., Nagano O., Yae T., Tamada M., Motohara T., Oshima H., Oshima M., Ikeda T., Asaba R., Yagi H., et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Canc. Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 7.Chen C., Zhao S., Karnad A., Freeman J.W. The biology and role of CD44 in cancer progression: therapeutic implications. J. Hematol. Oncol. 2018;11:1–23. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misra S., Heldin P., Hascall V.C., Karamanos N.K., Skandalis S.S., Markwald R.R., Ghatak S. Hyaluronan-CD44 interactions as potential targets for cancer therapy. FEBS J. 2011;278:1429–1443. doi: 10.1111/j.1742-4658.2011.08071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soltes L., Mendichi R., Kogan G., Schiller J., Stankovska M., Arnhold J. Degradative action of reactive oxygen species on hyaluronan. Biomacromolecules. 2006;7:659–668. doi: 10.1021/bm050867v. [DOI] [PubMed] [Google Scholar]
- 10.Soltes L., Kogan G., Stankovska M., Mendichi R., Rychly J., Schiller J., Gemeiner P. Degradation of high-molar-mass hyaluronan and characterization of fragments. Biomacromolecules. 2007;8:2697–2705. doi: 10.1021/bm070309b. [DOI] [PubMed] [Google Scholar]
- 11.Daar E., King L., Nisbet A., Thorpe R.B., Bradley D.A. Viscosity changes in hyaluronic acid: irradiation and rheological studies. Appl. Radiat. Isot. 2010;68:746–750. doi: 10.1016/j.apradiso.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 12.Cowman M.K. vol. 74. Elsevier; 2017. Hyaluronan and hyaluronan fragments; pp. 1–59. (Advances in Carbohydrate Chemistry and Biochemistry). [DOI] [PubMed] [Google Scholar]
- 13.Yang H., Villani R.M., Wang H., Simpson M.J., Roberts M.S., Tang M., Liang X. The role of cellular reactive oxygen species in cancer chemotherapy. J. Exp. Clin. Canc. Res. 2018;37:266. doi: 10.1186/s13046-018-0909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou Z., Chang H., Li H., Wang S. Induction of reactive oxygen species: an emerging approach for cancer therapy. Apoptosis. 2017;22:1321–1335. doi: 10.1007/s10495-017-1424-9. [DOI] [PubMed] [Google Scholar]
- 15.Kellett-Clarke H., Stegmann M., Barclay A.N., Metcalfe C. CD44 binding to hyaluronic acid is redox regulated by a labile disulfide bond in the hyaluronic acid binding site. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marangoni E., Lecomte N., Durand L., de Pinieux G., Decaudin D., Chomienne C., Smadja-Joffe F., Poupon M.F. CD44 targeting reduces tumour growth and prevents post-chemotherapy relapse of human breast cancers xenografts. Br. J. Canc. 2009;100:918–922. doi: 10.1038/sj.bjc.6604953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin L., Hope K.J., Zhai Q., Smadja-Joffe F., Dick J.E. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat. Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 18.Subramaniam V., Vincent I.R., Gilakjan M., Jothy S. Suppression of human colon cancer tumors in nude mice by siRNA CD44 gene therapy. Exp. Mol. Pathol. 2007;83:332–340. doi: 10.1016/j.yexmp.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Privat-Maldonado A., Schmidt A., Lin A., Weltmann K.-D., Wende K., Bogaerts A., Bekeschus S. ROS from physical plasmas: redox chemistry for biomedical therapy. Oxid Med Cell Longev. 2019;2019:29. doi: 10.1155/2019/9062098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin A., Gorbanev Y., De Backer J., Van Loenhout J., Van Boxem W., Lemière F., Cos P., Dewilde S., Smits E., Bogaerts A. Non-thermal plasma as a unique delivery system of short-lived reactive oxygen and nitrogen species for immunogenic cell death in melanoma cells. Advanced Science. 2019;6:1802062. doi: 10.1002/advs.201802062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elstner M., Porezag D., Jungnickel G., Elsner J., Haugk M., Frauenheim T., Suhai S., Seifert G. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys. Rev. B. 1998;58:7260. [Google Scholar]
- 22.Gaus M., Goez A., Elstner M. Parametrization and benchmark of DFTB3 for organic molecules. J. Chem. Theor. Comput. 2013;9:338–354. doi: 10.1021/ct300849w. [DOI] [PubMed] [Google Scholar]
- 23.Hourahine B., Aradi B., Blum V., Bonafé F., Buccheri A., Camacho C., Cevallos C., Deshaye M.Y., Dumitrică T., Dominguez A., et al. DFTB+, a software package for efficient approximate density functional theory based atomistic simulations. J. Chem. Phys. 2020;152:124101. doi: 10.1063/1.5143190. [DOI] [PubMed] [Google Scholar]
- 24.Sleeman J., Rudy W., Hofmann M., Moll J., Herrlich P., Ponta H. Regulated clustering of variant CD44 proteins increases their hyaluronate binding capacity. J. Cell Biol. 1996;135:1139–1150. doi: 10.1083/jcb.135.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahmann J.D., O'Brien R., Werner J.M., Heinegård D., Ladbury J.E., Campbell I.D., Day A.J. Localization and characterization of the hyaluronan-binding site on the link module from human TSG-6. Structure. 2000;8:763–774. doi: 10.1016/s0969-2126(00)00163-5. [DOI] [PubMed] [Google Scholar]
- 26.Tammi R., MacCallum D., Hascall V.C., Pienimäki J.-P., Hyttinen M., Tammi M. Hyaluronan bound to CD44 on keratinocytes is displaced by hyaluronan decasaccharides and not hexasaccharides. J. Biol. Chem. 1998;273:28878–28888. doi: 10.1074/jbc.273.44.28878. [DOI] [PubMed] [Google Scholar]
- 27.Vuorio J., Vattulainen I., Martinez-Seara H. Atomistic fingerprint of hyaluronan–CD44 binding. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kästner J. Umbrella sampling. Wiley Interdiscipl. Rev.: Comput. Mol. Sci. 2011;1:932–942. [Google Scholar]
- 29.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 30.Schmid N., Eichenberger A.P., Choutko A., Riniker S., Winger M., Mark A.E., van Gunsteren W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011;40:843–856. doi: 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- 31.Petrov D., Margreitter C., Grandits M., Oostenbrink C., Zagrovic B. A systematic framework for molecular dynamics simulations of protein post-translational modifications. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plazinski W., Lonardi A., Hünenberger P.H. Revision of the GROMOS 56A6CARBO force field: improving the description of ring‐conformational equilibria in hexopyranose‐based carbohydrates chains. J. Comput. Chem. 2016;37:354–365. doi: 10.1002/jcc.24229. [DOI] [PubMed] [Google Scholar]
- 33.Panczyk K., Gaweda K., Drach M., Plazinski W. Extension of the GROMOS 56a6CARBO/CARBO_R force field for charged, protonated, and esterified uronates. J. Phys. Chem. B. 2018;122:3696–3710. doi: 10.1021/acs.jpcb.7b11548. [DOI] [PubMed] [Google Scholar]
- 34.Oborský P. ETH Zurich; 2018. Conformational Properties of Specific Biomolecular Systems Investigated Using Molecular Dynamics Simulations. [Google Scholar]
- 35.Maris A. On the conformational equilibrium of glycolamide: a free jet millimetre-wave spectroscopy and computational study. Phys. Chem. Chem. Phys. 2004;6:2611–2616. [Google Scholar]
- 36.Pandit A.H., Mazumdar N., Ahmad S. Periodate oxidized hyaluronic acid-based hydrogel scaffolds for tissue engineering applications. Int. J. Biol. Macromol. 2019;137:853–869. doi: 10.1016/j.ijbiomac.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Agren U.M., Tammi R.H., Tammi M.I. Reactive oxygen species contribute to epidermal hyaluronan catabolism in human skin organ culture. Free Radic. Biol. Med. 1997;23:996–1001. doi: 10.1016/s0891-5849(97)00098-1. [DOI] [PubMed] [Google Scholar]
- 38.Duan J., Kasper D.L. Oxidative depolymerization of polysaccharides by reactive oxygen/nitrogen species. Glycobiology. 2011;21:401–409. doi: 10.1093/glycob/cwq171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andley U.P., Chakrabarti B. Role of singlet oxygen in the degradation of hyaluronic acid. Biochem. Biophys. Res. Commun. 1983;115:894–901. doi: 10.1016/s0006-291x(83)80019-9. [DOI] [PubMed] [Google Scholar]
- 40.Stern, R. Association between cancer and “acid mucopolysaccharides”: an old concept comes of age, finally. In Proceedings of Semin Cancer Biol; pp. 238-243. [DOI] [PubMed]
- 41.Hartheimer J.S., Park S., Rao S.S., Kim Y. Targeting hyaluronan interactions for glioblastoma stem cell therapy. Canc. Microenviron. 2019;12:47–56. doi: 10.1007/s12307-019-00224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evanko S.P., Tammi M.I., Tammi R.H., Wight T.N. Hyaluronan-dependent pericellular matrix. Adv. Drug Deliv. Rev. 2007;59:1351–1365. doi: 10.1016/j.addr.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mele V., Sokol L., Kölzer V.H., Pfaff D., Muraro M.G., Keller I., Stefan Z., Centeno I., Terracciano L.M., Dawson H., et al. The hyaluronan-mediated motility receptor RHAMM promotes growth, invasiveness and dissemination of colorectal cancer. Oncotarget. 2017;8:70617–70629. doi: 10.18632/oncotarget.19904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nedvetzki S., Gonen E., Assayag N., Reich R., Williams R.O., Thurmond R.L., Huang J.-F., Neudecker B.A., Wang F.-S., Turley E.A., et al. RHAMM, a receptor for hyaluronan-mediated motility, compensates for CD44 in inflamed CD44-knockout mice: a different interpretation of redundancy. Proc. Natl. Acad. Sci. U. S. A. 2004;101:18081–18086. doi: 10.1073/pnas.0407378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lesley J., Hyman R., Kincade P.W. CD44 and its interaction with extracellular matrix. Adv. Immunol. 1993;54:271–335. doi: 10.1016/s0065-2776(08)60537-4. [DOI] [PubMed] [Google Scholar]
- 46.Amorim S., da Costa D.S., Freitas D., Reis C.A., Reis R.L., Pashkuleva I., Pires R.A. Molecular weight of surface immobilized hyaluronic acid influences CD44-mediated binding of gastric cancer cells. Sci. Rep. 2018;8:16058. doi: 10.1038/s41598-018-34445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishimoto T., Sugihara H., Watanabe M., Sawayama H., Iwatsuki M., Baba Y., Okabe H., Hidaka K., Yokoyama N., Miyake K., et al. Macrophage-derived reactive oxygen species suppress miR-328 targeting CD44 in cancer cells and promote redox adaptation. Carcinogenesis. 2014;35:1003–1011. doi: 10.1093/carcin/bgt402. [DOI] [PubMed] [Google Scholar]
- 48.Misra S., Hascall V.C., Markwald R.R., Ghatak S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front. Immunol. 2015;6:201. doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherman L., Sleeman J., Herrlich P., Ponta H. Hyaluronate receptors: key players in growth, differentiation, migration and tumor progression. Curr. Opin. Cell Biol. 1994;6:726–733. doi: 10.1016/0955-0674(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 50.Wang S.J., Bourguignon L.Y.W. Hyaluronan and the interaction between CD44 and epidermal growth factor receptor in oncogenic signaling and chemotherapy resistance in head and neck cancer. Arch. Otolaryngol. 2006;132:771–778. doi: 10.1001/archotol.132.7.771. [DOI] [PubMed] [Google Scholar]
- 51.Kristiansen K.A., Dalheim M.Ø., Christensen B.E. Periodate oxidation and macromolecular compaction of hyaluronan. Pure Appl. Chem. 2013;85:1893–1900. doi: 10.1351/pac-con-13-01-05. [DOI] [Google Scholar]
- 52.Hawkins C.L., Davies M.J. Direct detection and identification of radicals generated during the hydroxyl radical-induced degradation of hyaluronic acid and related materials. Free Radic. Biol. Med. 1996;21:275–290. doi: 10.1016/0891-5849(96)00042-1. [DOI] [PubMed] [Google Scholar]
- 53.Skandalis S.S., Gialeli C., Theocharis A.D., Karamanos N.K. Advances and advantages of nanomedicine in the pharmacological targeting of hyaluronan-CD44 interactions and signaling in cancer. Adv. Canc. Res. 2014;123:277–317. doi: 10.1016/B978-0-12-800092-2.00011-3. [DOI] [PubMed] [Google Scholar]
- 54.Pasonen-Seppänen S., Hyttinen J.M., Rilla K., Jokela T., Noble P.W., Tammi M., Tammi R. Role of CD44 in the organization of keratinocyte pericellular hyaluronan. Histochem. Cell Biol. 2012;137:107–120. doi: 10.1007/s00418-011-0883-2. [DOI] [PubMed] [Google Scholar]
- 55.Teriete P., Banerji S., Noble M., Blundell C.D., Wright A.J., Pickford A.R., Lowe E., Mahoney D.J., Tammi M.I., Kahmann J.D. Structure of the regulatory hyaluronan binding domain in the inflammatory leukocyte homing receptor CD44. Mol. Cell. 2004;13:483–496. doi: 10.1016/s1097-2765(04)00080-2. [DOI] [PubMed] [Google Scholar]
- 56.Ween M.P., Hummitzsch K., Rodgers R.J., Oehler M.K., Ricciardelli C. Versican induces a pro-metastatic ovarian cancer cell behavior which can be inhibited by small hyaluronan oligosaccharides. Clin. Exp. Metastasis. 2011;28:113–125. doi: 10.1007/s10585-010-9363-7. [DOI] [PubMed] [Google Scholar]
- 57.Xu G., Chance M.R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 58.Takai E., Kitamura T., Kuwabara J., Ikawa S., Yoshizawa S., Shiraki K., Kawasaki H., Arakawa R., Kitano K. Chemical modification of amino acids by atmospheric-pressure cold plasma in aqueous solution. J. Phys. Appl. Phys. 2014;47:285403. [Google Scholar]
- 59.Zhou R., Zhou R., Zhuang J., Zong Z., Zhang X., Liu D., Bazaka K., Ostrikov K. Interaction of atmospheric-pressure air microplasmas with amino acids as fundamental processes in aqueous solution. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruno G., Heusler T., Lackmann J.-W., Von Woedtke T., Weltmann K.-D., Wende K. Cold physical plasma-induced oxidation of cysteine yields reactive sulfur species (RSS) Clin. Plasma Med. 2019;14:100083. [Google Scholar]
- 61.Banerji S., Wright A.J., Noble M., Mahoney D.J., Campbell I.D., Day A.J., Jackson D.G. Structures of the Cd44–hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 2007;14:234–239. doi: 10.1038/nsmb1201. [DOI] [PubMed] [Google Scholar]
- 62.Jamison F.W., II, Foster T.J., Barker J.A., Hills R.D., Jr., Guvench O. Mechanism of binding site conformational switching in the CD44–hyaluronan protein–carbohydrate binding interaction. J. Mol. Biol. 2011;406:631–647. doi: 10.1016/j.jmb.2010.12.040. [DOI] [PubMed] [Google Scholar]
- 63.Favreau A.J., Faller C.E., Guvench O. CD44 receptor unfolding enhances binding by freeing basic amino acids to contact carbohydrate ligand. Biophys. J. 2013;105:1217–1226. doi: 10.1016/j.bpj.2013.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.