Summary
Enhanced stemness in colorectal cancer has been reported and it contributes to aggressive progression, but the underlying mechanisms remain unclear. Here we report a Wnt ligand, Dickkopf-2 (DKK2) is essential for developing colorectal cancer stemness. Genetic depletion of DKK2 in intestinal epithelial or stem cells reduced tumorigenesis and expression of the stem cell marker genes including LGR5 in a model of colitis-associated cancer. Sequential mutations in APC, KRAS, TP53, and SMAD4 genes in colonic organoids revealed a significant increase of DKK2 expression by APC knockout and further increased by additional KRAS and TP53 mutations. Moreover, DKK2 activates proto-oncogene tyrosine-protein kinse Src followed by increased LGR5 expressing cells in colorectal cancer through degradation of HNF4α1 protein. These findings suggest that DKK2 is required for colonic epithelial cells to enhance LGR5 expression during the progression of colorectal cancer.
Subject areas: Cell Biology, Stem Cells Research, Cancer
Graphical abstract
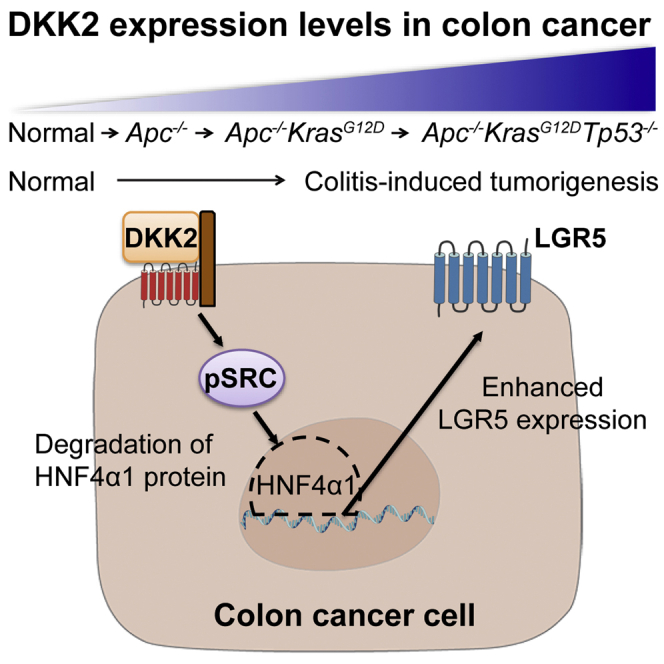
Highlights
-
•
APC, KRAS, and TP53 mutations induce DKK2 expression in murine colon cancer
-
•
DKK2 increases Src phosphorylation in colon cancer cells
-
•
Activated Src leads to degradation of HNF4α1 protein
-
•
This DKK2 downstream signaling enhances LGR5 expression in colon cancer
Cell Biology; Stem Cells Research; Cancer
Introduction
Colorectal cancer is the third most common cancer in the world and the second leading cause of cancer related death (Bray et al., 2018). About 80% of colorectal cancer patients have mutations in the adenomatous polyposis coli (APC) gene, a negative regulator of canonical Wnt signaling (Phelps et al., 2009). APC mutations in stem cells initiate intestinal tumorigenesis (Barker et al., 2009). Barker et al. has identified LGR5 as an intestinal stem cell marker gene (Barker et al., 2007). LGR5 positive intestinal stem cell-specific APC conditional knockout mice showed spontaneous tumor formation whereas APC deletion in non-stem cells was rarely tumorigenic (Barker et al., 2009). Lineage tracing studies further confirmed LGR5 positive stem cells as cancer stem cells in the gut (Schepers et al., 2012). Additional oncogenic mutations in KRAS, TP53 and SMAD4 in epithelial cells promote colorectal carcinogenesis (Vogelstein et al., 1988). Briefly, oncogenic alterations in intestinal stem cells lead to initiation and progression of colorectal cancer.
Recent advances in organoid culture techniques allowed in vitro primary intestinal epithelial cell culture (Sato et al., 2009; Drost and Clevers, 2018). Mutations in APC, KRAS, TP53 and SMAD4 in human intestinal organoids using CRISPR methods provided a better understanding of carcinogenesis in colorectal cancer (Matano et al., 2015). Depletion of LGR5 expressing cancer stem cells in murine primary or metastatic colon cancer cells carrying those mutations have shown the necessity of cancer stem cells in metastatic progression (de Sousa e Melo et al., 2017). However, underlying mechanisms of how LGR5 expressing cancer stem cells arise and are maintained are still elusive.
In this study, we introduce DKK2 as an essential factor for LGR5 expression in colon cancer. We showed increased DKK2 expression in colon tumor epithelial cells. Using colon cancer organoids, we showed a significant increase of DKK2 transcription by triple mutations in APC, KRAS, and TP53 genes. Gain and loss of function studies using normal or cancer organoids revealed a feedforward feedback regulation of DKK2 expression. Mechanistically, we have identified that DKK2 activates proto-oncogene tyrosine-protein kinse Src (c-Src), which then results in HNF4α1 protein degradation followed by enhanced LGR5 expression in colon cancer. Our findings provide a better understanding of the formation of LGR5 expressing cancer stem cells during colorectal cancer progression.
Results
DKK2 is highly expressed in colorectal cancer epithelium and facilitates tumorigenesis
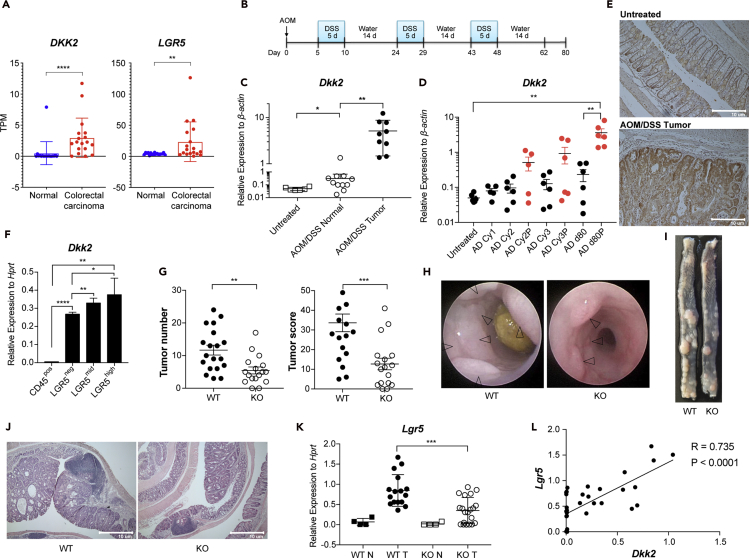
DKK2 expression is increased about 50-fold in colorectal cancer patients compared to healthy normal controls and polyp-specific DKK2 expression in APC mutant (Apcmin/+) mice has been reported by in situ hybridization (Matsui et al., 2009; Gregorieff et al., 2005). We have confirmed the elevation of DKK2 in colorectal cancer using colorectal cancer patient samples and a murine colitis-induced tumor model. Transcriptional increase of DKK2 was observed in human colorectal carcinoma RNA sequencing (RNA-seq) data along with increased expression of the stem cell marker gene, LGR5 (Figure 1A). DKK2 expression was increased about 100-fold in tumor tissues compared to untreated controls in the azoxymethane and dextran sodium sulfate (AOM/DSS)-treated colitis-induced tumor model (Figures 1B and 1C). Polyp-specific increase of DKK2 expression was observed in a time course experiment (Figure 1D). Immunohistochemistry analysis for DKK2 showed epithelial expression of DKK2 in murine colitis-induced tumors (Figure 1E). To confirm the pattern of DKK2 expression, we sorted epithelial cells from colitis-induced tumors based on epithelial marker gene (EpCAM) and stem cell marker gene expression (LGR5) using Lgr5CreERT2-IRES-EGFP reporter mice (Figures S1A and S1B). The sorted LGR5high, LGR5mid, and LGR5neg populations represent intestinal stem cell populations, transit amplifying cell and Paneth cell populations and differentiated epithelial cell populations, respectively (Sato et al., 2009). Increased expression of DKK2 was shown along with LGR5 expression (Figure 1F). These data demonstrate a significant increase of DKK2 expression and its pattern in colonic epithelium in the colitis-induced tumor model.
Figure 1.
DKK2 expression in colon cancers and reduced colitis-induced tumorigenesis in DKK2 deficient mice
(A) Expression of DKK2 and LGR5 in human colorectal carcinoma and adjacent normal tissues was analyzed using previously deposited RNA-seq data (GSE50760, n = 18 each group, TPM; Transcripts per million).
(B) A schematic diagram of azoxymethane/dextran sodium sulfate (AOM/DSS) treatment schedule.
(C) Quantitative PCR analysis of DKK2 expression in mouse colon tissues or polyps upon AOM/DSS treatment. Each symbol indicates an individual tissue sample.
(D) Expression kinetics of DKK2 in the AOM/DSS-treated wild type C57BL/6 colon tissues measured by quantitative PCR (AD; AOM/DSS, Cy1, 2 and 3; the first, second and third DSS cycle, P; polyps, d80; day 80, Red; polyps, Black; normal tissues). Each dot represents an individual tissue sample.
(E) Representative DKK2 immunohistochemistry in AOM/DSS-induced tumors. Scale bar = 10 μm.
(F) Quantitative analysis of DKK2 expression in the sorted tumor cells based on EGFP signal from AOM/DSS treated Lgr5-EGFP reporter mice.
(G) Tumor number and score by colonoscopy in wild type (WT, n = 19) or VillinCre-Dkk2fl/fl knockout (KO, n = 17) mice on day 80. n represents the number of independent mice.
(H–J) Representative of colonoscopy, macroscopic view and histology on day 80.
(K) Quantitative gene expression analysis of LGR5 in tumor tissues after AOM/DSS treatment. WT N: wild type normal colon, n = 4, WT T: wild type tumor, n = 16, KO N: Dkk2 knockout normal colon, n = 4, KO T: Dkk2 knockout tumor, n = 20. n represents the number of tumor tissues.
(L) Correlation of DKK2 and LGR5 relative expression to Hprt in tumor samples by Pearson r test. Each dot represents an individual tissue sample. n.s, not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; two-tailed Welch's ttest. Error bars indicate mean ± s.d. Results are representative of three independent experiments.
Next, in order to investigate the roles of DKK2 in intestinal tumorigenesis, we generated intestinal epithelium-specific DKK2 conditional knockout VillinCre-Dkk2fl/fl mice. Eighty days after AOM/DSS treatment, the number and size of polyps were recorded by colonoscopy. DKK2 knockout in the intestinal epithelium showed about 60% reduction of tumor growth (Figures 1G–1J and S1C). Interestingly, DKK2 knockout does not alter proliferation of colitis-induced tumor cells (Figures S1D and S1E). Rather, the expression of stem cell marker gene LGR5 was reduced in the absence of DKK2 and there was a correlation of expression between DKK2 and LGR5 during colitis-induced tumor development (Figures 1K and 1L). These findings suggest that DKK2 contributes to intestinal tumorigenesis and the elevation of LGR5 expression in colorectal cancer.
DKK2 depletion modulates stemness and differentiation of colitis-induced tumor cells
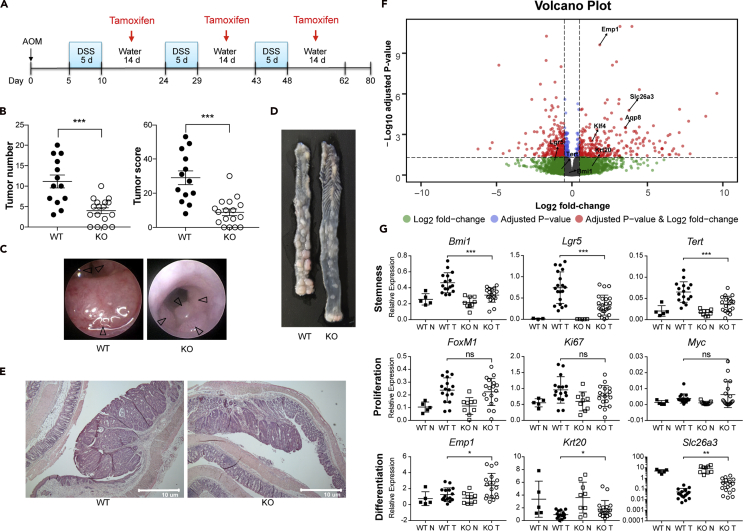
Using tamoxifen-inducible intestinal stem cell-specific DKK2 knockout (Lgr5CreERT2-IRES-EGFP-Dkk2fl/fl) mice, we have confirmed that DKK2 expression in LGR5 positive cells and its daughter cells promotes colitis-induced tumorigenesis (Figures 2A–2E). The correlation between DKK2 and LGR5 expression was confirmed as well and those results were not a consequence of the functional loss of one LGR5 promoter allele in the Lgr5CreERT2-IRES-EGFP-Dkk2fl/fl mice (Figures S1F and S1G).
Figure 2.
DKK2 regulates stemness and differentiation of colon cancer cells
(A) A schematic diagram of AOM/DSS treatment schedule with tamoxifen administration in order to knockout DKK2 in LGR5 positive cells. 2 mg of tamoxifen was administered by intra-peritoneal injection every other day from day 7 after DSS water was removed.
(B) Tumor number and score by colonoscopy in wild type (WT, n = 13) or Lgr5CreERT2-Dkk2fl/fl knockout (KO, n = 17) mice on day 80.
(C–E) Representative view of colonoscopy, macroscopic view and histology at the same time.
(F) Volcano plot generated by RNA-seq analysis for AOM/DSS-induced tumors from wild type and Lgr5CreERT2-Dkk2fl/fl knockout mice. Wald test, Lgr5; p = 0.004, adjust p = 0.078, Tert; p = 0.070, adjust p = 0.336, Bmi1; p = 0.290, adjust p = 0.639, Aqp8; p = 1.07 × 10−6, adjust p = 0.003, Emp1; p = 6.01 × 10−14, adjust p = 2.39E-10, Krt20; p = 0.025, adjust p = 0.206, Klf4; p = 2.99 × 10−5, adjust p = 0.003, Slc26a3; p = 3.32 × 10−8, adjust p = 1.65 × 10−5.
(G) Quantitative gene expression analysis of AOM/DSS-induced tumors in stem cell maker genes (Bmi1, Lgr5 and Tert), proliferation marker genes (FoxM1, Ki67 and Myc) and differentiation marker genes (Emp1, Krt20 and Slc26a3). WT N: wild type normal colon, WT T: wild type tumor, KO N: Dkk2 knockout normal colon, KO T: Dkk2 knockout tumor. Each symbol represents an individual tissue sample. n.s, not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; two-tailed Welch's ttest. Error bars indicate mean ± s.d.
As an unbiased way to assess the roles of DKK2 in intestinal tumorigenesis, we performed a bulk RNA-seq analysis on colitis-induced polyps from the AOM/DSS-treated Lgr5CreERT2-IRES-EGFP-Dkk2fl/fl mice. Consistent with the previous VillinCre-Dkk2fl/fl mice data, the expression of LGR5 was decreased in the absence of DKK2 (Figure 2F). Quiescent stem cell marker genes, BMI1 and TERT, were reduced as well whereas the differentiation marker genes, EMP1, KRT20 and SLC26A3 were increased and it has been confirmed with quantitative PCR analysis (Figure 2G). Again, expression of proliferation marker genes such as KI67 was unaltered in DKK2-deficient polyps (Figure 2G). These results imply that DKK2 regulates stemness and differentiation of epithelial cells in a colitis-induced tumor model.
DKK2 enhances Lgr5 expression in colonic epithelial cells
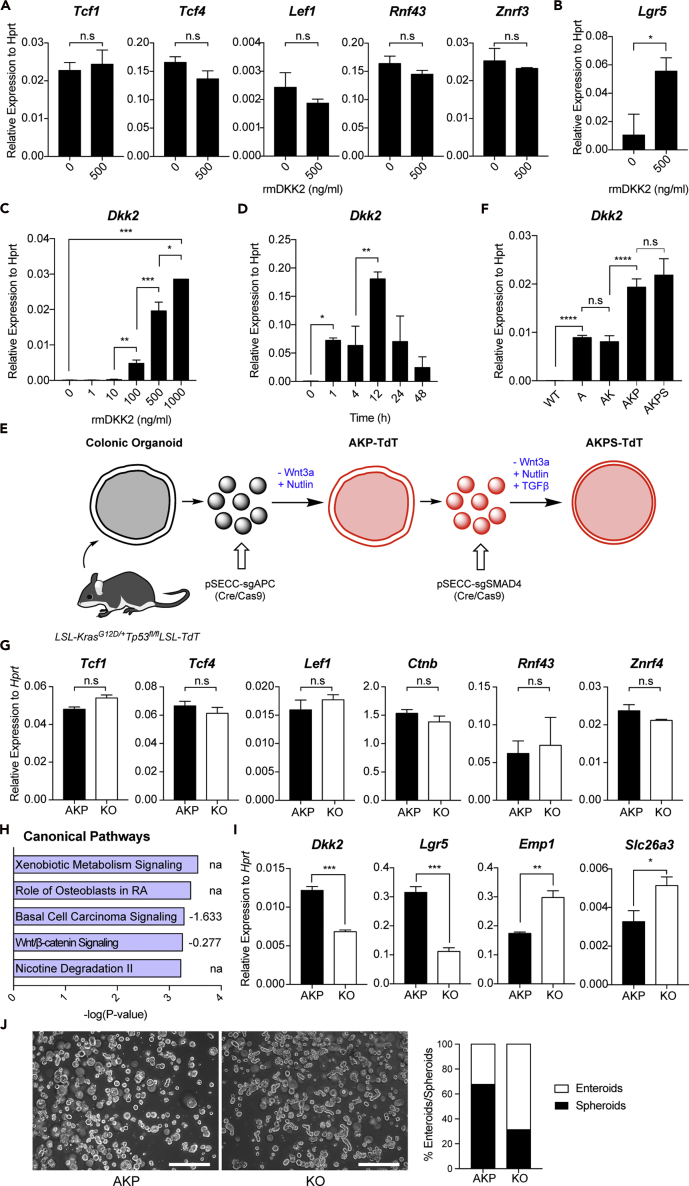
In the canonical Wnt signaling pathway, DKK2 has been introduced as a Wnt antagonist, binding to the common Wnt receptors, LRP5 and LRP6 (Niehrs, 2006). The following studies have reported the agonistic roles of DKK2 in the canonical Wnt pathway in the absence of another Wnt receptor Kremen2 (Mao and Niehrs, 2003). To test the direct effects of DKK2 on colonic epithelial cells, we performed normal colonic organoid culture in the presence of mouse recombinant DKK2 protein. Notably, expression of the canonical Wnt target genes was unchanged in the cultured organoids after recombinant DKK2 treatment (Figure 3A). Meanwhile, LGR5 expression was increased about 3-fold compared to the untreated controls (Figure 3B). In addition, recombinant DKK2 treatment further increased DKK2 expression in organoids in a dose- and time-dependent manner indicating the feedforward regulation of its expression (Figures 3C and 3D).
Figure 3.
DKK2 enhances Lgr5 expression in intestinal epithelial cells without affecting expression of the canonical Wnt target genes
Normal colonic organoids were cultured in the presence or absence of recombinant DKK2 for 3 days (rmDKK2: Recombinant mouse DKK2 protein).
(A and B) Quantitative gene expression analyses of the canonical Wnt target genes (A) and Lgr5 (B).
(C and D) Dkk2 expression after rmDKK2 treatment in a dose- (C) and time-dependent (D) manner.
(E) A schematic diagram of colon cancer organoid development. Apc, Kras and Tp53 triple (AKP) and plus Smad4 quadruple (AKPS) organoids are shown. Colonic organoids were generated from LSL-KrasG12D/+Tp53fl/flLSL-TdT mice. AKP triple mutations were acquired by adenoviral transduction of pSECC plasmids carrying Cre, Cas9 and guide RNA targeting Apc to isolated single organoid cells. Mutant tumor organoids were selected during culture in the absence of Wnt3a, EGF and Nutlin recombinant proteins for Apc, Kras and Tp53 mutations, respectively. Additional adenoviral transduction and recombinant TGFβ selection were performed to incorporate Smad4 mutations in order to generate AKPS organoids. LSL: LoxP-Stop-Loxp, TdT: Tandem dimer Tomato.
(F) Dkk2 expression in wild type (WT) colonic organoids and colon cancer organoids carrying Apc mutation (A), Apc and Kras mutations (AK), Apc, Kras and Tp53 mutations (AKP) and Apc, Kras, Tp53 and Smad4 mutations (AKPS).
(G) Expression of the canonical Wnt target genes in Dkk2 knockout AKP (KO) organoids.
(H) Ingenuity pathway analysis of colitis-induced tumor RNA-seq data suggested canonical pathways by Dkk2 knockout. Numbers indicate activation Z score.
(I) Expression of Dkk2, Lgr5, Emp1 and Slc26a3 in KO organoids.
(J) Representative pictures and the percentiles of spheroid versus enteroids in control and KO organoids. Cystic organoids were counted as spheroids while budding shape organoids were counted as enteroids. 20 independent images were analyzed in each group. Scale bar = 50 μm. n.s, not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.001; two-tailed Welch's ttest. Error bars indicate mean ± s.d.
Next, we questioned whether our in vitro findings are reproducible in tumor settings. In order to understand the expression conditions of DKK2 during colorectal cancer development, we generated colon cancer organoids carrying various combinations of mutations in APC, KRAS, TP53, and SMAD4 genes that are commonly observed during colorectal cancer progression (Vogelstein et al., 2013) (Figure 3E). Accumulating mutations in those genes resulted in the elevation of LGR5 expression in colonic organoids (Figure S2A). Expression of DKK2 was detected in Apc knockout (A) and Apc knockout KrasG12D (AK) mutant organoids (Figure 3F). It was further increased in Apc knockout KrasG12D and Tp53 knockout triple mutant (AKP) organoids and this expression level was maintained in quadruple mutant (AKPS) organoids with additional knockout in the SMAD4 gene (Figure 3F). Given that information, AKP organoids were used for the following experiments.
The time course expression of DKK2 and LGR5 in AKP organoids was analyzed for 10 days after single isolated cells were embedded in 3-dimensional matrigel culture. DKK2 expression increased and reached a plateau at day 8 (Figure S2B). LGR5 expression increased at day 4-6 when spheroids formed then decreased about two-fold and was maintained at day 8-10 when enteroid formation was mostly observed (Figure S2C). Based on these results, we decided to perform transcriptome analysis at day 8.
We then developed DKK2 knockout AKP (KO) organoids using the CRISPR technique. To rule out unwanted results by Cas9-driven random mutations on DKK2, we cloned DKK2 knockout AKP organoids through single cell sorting and sequencing (Figure S2D). The identified clones could only express truncated DKK2 protein (Figure S2E). While the CRISPR technique does not alter target gene transcription, we observed more than 50% reduction in DKK2 expression after knockout, indicating that the positive feedback regulation in DKK2 expression was abolished as well (Figure S2F). In accordance with the above in vitro data using DKK2-treated normal colonic organoids, DKK2 knockout in AKP colon cancer organoids resulted in unchanged expression of the canonical Wnt target genes (Figure 3G). Confocal microscopy and flow cytometry analysis of CTNNB1 in AKP organoids showed that DKK2 knockout did not alter cellular localization and protein levels of CTNNB1 (Figures S3A and S3B). Moreover, ingenuity pathway analysis of the RNA-seq data from colitis-induced polyps showed marginal reduction of the canonical Wnt signaling pathway activity in the absence of DKK2 (Figure 3H). These results imply the possibility of the presence of non-canonical DKK2 signaling.
Although the canonical Wnt target genes were unaffected by DKK2 knockout, reduced Lgr5 expression and enhanced differentiation of intestinal epithelial cells are still observed in AKP organoids (Figures 3I and 3J). LGR5 expression was reduced by random mutations in the DKK2 gene and the knockout clones showed about three-fold reduction in LGR5 expression compared to the control AKP organoids at day 8 (Figures S2G and S2H). Also, elevation of differentiation marker genes such as Emp1 and Slc26a3 was confirmed (Figure 3I). Decreased spheroid/enteroid ratios in morphology reflect reduced stemness in KO organoids as well (Rodriguez-Colman et al., 2017) (Figures 3J and S4A). These data confirmed the necessity of DKK2 in the stemness of colorectal cancer represented by enhanced LGR5 expression and the underlying mechanisms may not be through the canonical Wnt signaling pathway.
DKK2-driven loss of HNF4α1 protein enhances LGR5 expression in colon cancer cells
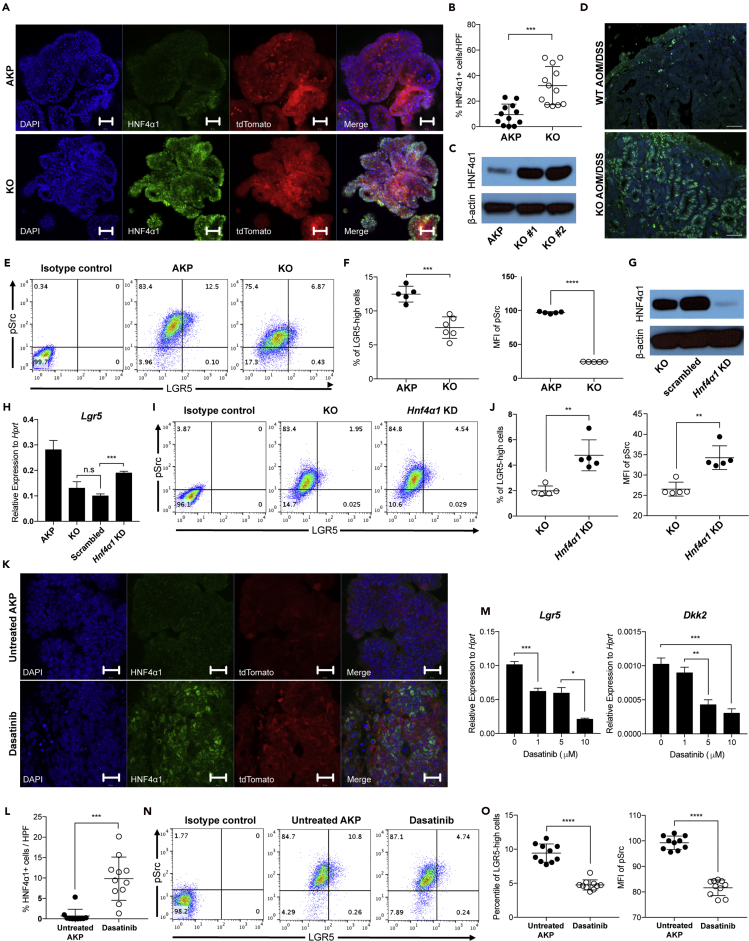
Ingenuity pathway analysis (IPA) of the RNA-seq data from colitis-induced polyps suggested a transcription factor, hepatocyte nuclear factor 4-alpha (HNF4A) as an upstream regulator that is activated in the absence of DKK2 (Figure S5A). HNF4A is a nuclear receptor protein which is crucial for development of the gut (Chen et al., 2019). VillinCreERT2-Hnf4afl/fl conditional knockout mice showed destabilization of intestinal epithelium following increased expression of LGR5 upon tamoxifen administration (Cattin et al., 2009). Chellappa et al. have reported the existence of two Hnf4a isoforms—Hnf4α1 and Hnf4α7—in the gut epithelium and have discovered the loss of the HNF4α1 isoform in AOM/DSS-driven intestinal tumor cells (Chellappa et al., 2016). Furthermore, inhibition of the HNF4α1 isoform using shRNA in the Caco2/15 human colon cell line showed reduced ileum villi cells signature and colon differentiated cell signature by gene set enrichment analysis (Babeu et al., 2018). Briefly, loss of HNF4A in colorectal cancer impairs epithelial differentiation.
We tested whether HNF4A is involved in DKK2 downstream signaling in colorectal cancer using AKP and KO organoids. Quantitative gene expression analysis showed that two isoforms of HNF4A, HNF4α1 and HNF4α7, were unchanged in the absence of DKK2 (Figure S5B). On the other hand, the protein level of HNF4α1 was significantly increased in the nucleus of KO organoids compared to AKP organoids (Figures 4A–4C, S5C, and S5D). This has also been observed in DKK2 knockout colitis-induced polyps in vivo (Figure 4D). The modulation of HNF4α1 protein but not HNF4α1 RNA expression is consistent with a previous report that post-translational modification of HNF4α1 isoform by c-Src leads to proteasomal degradation of HNF4α1 in human colorectal cancer (Chellappa et al., 2012a). We then performed flow cytometry analysis of c-Src phosphorylation and LGR5 expression in colon cancer organoids. DKK2 knockout in AKP organoids resulted in dramatic reduction of c-Src phosphorylation as well as LGR5 expression followed by decreased numbers of cells expressing high levels of LGR5 (Figures 4E and 4F). Those results indicate that DKK2 is necessary for c-Src-induced proteasomal degradation of HNF4α1, which contributes to LGR5 expression in colon cancer.
Figure 4.
Loss of HNF4α1 protein mediates DKK2-driven LGR5 expression in colon cancer organoids through proto-oncogene tyrosine-protein kinse Src (c-Src) phosphorylation
(A) Confocal microscopy of HNF4α1 staining in AKP organoids carrying the tdTomato reporter gene. Scale bar = 50 μm. KO; Dkk2 knockout AKP organoids.
(B) Statistical analysis of HNF4α1 positive cells per high power field (HPF) in (A) using ImageJ. Each dot represents a single image of HNF4α1 staining.
(C) Quantification of HNF4α1 in AKP organoids by Western blot analysis. HNF4α1 = 52 kDa, β-actin = 42 kDa.
(D) Representative images of HNF4α1 staining in AOM/DSS-induced tumors from wild type (WT AOM/DSS) and VillinCre-Dkk2fl/fl conditional knockout (KO AOM/DSS) mice. Scale bar = 1 μm.
(E) Flow cytometry analysis of c-Src phosphorylation (pSrc) and LGR5 expression in KO organoids.
(F) Statistic analysis of the percent of cells expressing high levels of LGR5 (LGR5-high: LGR5 and pSrc double-positive) and the mean fluorescence intensity of pSrc in (E).
(G) Quantification of HNF4α1 protein in the HNF4α1 knockdown in DKK2 KO AKP organoids (HNF4α1 KD) by Western blot analysis.
(H) Quantitative gene expression analysis of LGR5 in HNF4α1 KD oragnoids.
(I) Flow cytometry analysis of LGR5 expression and c-Src phosphorylation (pSrc) in HNF4α1 KD.
(J) Statistic analysis of the percent of LGR5-high cells and the mean fluorescence intensity of pSrc in HNF4α1 KD.
(K) Confocal microscopy of HNF4α1 staining in AKP organoids carrying the tdTomato reporter gene in the presence or absence of 5 μM of c-Src inhibitor, dasatinib for 48 hr. Scale bar = 20 μm.
(L) Statistic analysis of HNF4α1 positive cells per high power field (HPF) in (K). Each symbol represents a single image of HNF4α1 staining.
(M) Quantitative gene expression analysis of LGR5 and DKK2 in dasatinib treated AKP organoids at 24 hr in a dose-dependent manner.
(N) Flow cytometry analysis of c-Src phosphorylation (pSrc) and LGR5 expression in AKP organoids after 5 μM of dasatinib treatment for 24 hr.
(O) Statistic analysis of LGR5-high cells and pSrc MFI in AKP organoids in (N). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; two-tailed Welch's ttest. Error bars indicate mean ± s.d. Results are representative of three independent experiments.
To validate the inhibitory effects of HNF4α1 in LGR5 expression in colon cancer, we performed a loss of function study targeting the HNF4α1 gene in KO organoids where the protein level of HNF4α1 was increased by DKK2 knockout (Figure 4G). Transcriptional expression of LGR5 was recovered by CRISPR-induced random loss of function mutations in the HNF4α1 gene in KO organoids (Figure 4H). The percentile of cells expressing high levels of LGR5 in KO organoids was also increased by HNF4α1 knockdown (Figures 4I and 4J). Those data confirmed that protein depletion of HNF4α1 by c-Src is a key step in DKK2 downstream signaling for LGR5 expression.
c-Src-induced HNF4α1 phosphorylation has been reported as an underlying mechanism of its degradation in human colorectal cancer cells (Chellappa et al., 2012b). It has been reported that activation of c-Src is sufficient to drive stem cell expansion in intestinal tumorigenesis (Cordero et al., 2014). To validate whether c-Src is the downstream target of DKK2 in the process of HNF4α1 protein degradation, we utilized a well-known c-Src inhibitor, dasatinib in AKP organoid culture. Inhibition of c-Src activation using dasatinib showed increased HNF4α1 protein in the nucleus of AKP organoids similar to what has been shown in KO organoids (Figures 4K and 4L). Reduced transcription of LGR5 and DKK2 was observed in dasatinib-treated AKP organoids in a dose-dependent manner (Figure 4M). Flow cytometry analysis of c-Src phosphorylation and LGR5 expression further confirmed that c-Src activation is critical to elevate the number of cells expressing high levels of LGR5 in AKP organoids (Figures 4N and 4O). Following the loss of function study using dasatinib, we performed a gain of function study using recombinant DKK2 protein and normal colonic organoids, which have marginal expression of DKK2. Recombinant DKK2 protein treatment in normal colonic organoid culture increased c-Src phosphorylation about two-fold compared to untreated controls (Figures S6A and S6B). The percentile of LGR5 positive cells in the recombinant DKK2-treated organoids was increased about two-fold as well (Figure S6C). Taken together, our findings suggest DKK2 as an essential factor for the stemness of colorectal cancer represented by enhanced LGR5 expression.
Discussion
Increased expression of the intestinal stem cell marker gene LGR5 is commonly observed in colorectal cancers (Leushacke and Barker, 2012; Szkandera et al., 2015; Li et al., 2016). Recent studies have shown that depletion of LGR5 expressing cells suppressed metastatic progression of colon cancer in murine model systems (de Sousa e Melo et al., 2017; Fumagalli et al., 2020). Here, we report that DKK2 is required for enhanced LGR5 expression in colorectal cancer. We have confirmed an increased expression of DKK2 in colorectal cancer and identified its positive feedback regulation. DKK2 facilitates c-Src activation followed by proteasomal degradation of the transcription factor HNF4α1. Loss of HNF4α1 elevated LGR5-expressing cancer stem cell populations. These alterations by DKK2 eventually promote colorectal cancer development.
DKK2 is known as a Wnt antagonist in the presence of the receptor Kremen-2 (Wu et al., 2000). Marginal expression of Kremen-2 in colon cancer cells potentiates the agonistic roles of DKK2 in the gut epithelium (Uhlén et al., 2015). Abnormal proliferation of the gut epithelium is often caused by alterations in the β-catenin-induced Wnt canonical pathway (Shtutman et al., 1999). However, proliferation of cancer cells was unchanged in the absence of DKK2 in colitis-induced polyps and colon cancer organoids. We think this is the consequence of functional loss of APC in both colitis-induced tumors and our APC knockout cancer organoids (Fillippo et al., 1998). It permanently activates β-catenin such that the Wnt agonistic or antagonistic functions of DKK2 are minimized in this context. Ingenuity pathway analysis of our RNA-seq data also showed marginal reduction of Wnt/β-catenin signaling in the DKK2 knockout polyps whereas basal carcinoma signaling was significantly reduced. Expression of the canonical Wnt target genes was unaltered by DKK2 knockout in APC knockout cancer organoids. Cellular localization and the protein quantity of β-catenin in cancer organoids were unaffected by DKK2 knockout as well. Collectively, DKK2 effects on the Wnt canonical signaling pathway were not apparent in colonic epithelial cells in the context of functional loss of APC such as colitis-induced tumors.
We showed that DKK2 increased LGR5 expression in colon cancer cells. In the homeostatic condition, intestinal stem cells are primed with Wnt ligands such as Wnt3a for LGR5 expression (Clevers, 2013). In colorectal cancer, a recent study has shown that a transcription factor, GATA6 increases LGR5 expression through repressing BMP4 signaling (Whissell et al., 2014). GATA6 directly bound to the LGR5 promoter in this report. DKK2 knockout in AKP organoids increased BMP4 transcription whereas GATA6 expression was unchanged (Figure S5E). Future studies could investigate the molecular mechanisms of DKK2 and HNF4α1 in BMP4-mediated or direct regulation of LGR5 expression in colorectal cancer.
c-Src activation is commonly observed in aggressive colon cancers and is involved in cell adhesion, invasion, and migration of cancer cells which are essential for metastasis (Lieu and Kopetz, 2010; Cordero et al., 2014; Chen et al., 2014). The underlying mechanisms of c-Src activation in colon cancer cells are still unclear. Our results showed significant reduction of c-Src phosphorylation at Y416 in DKK2 knockout colon cancer organoids even in the presence of APC, KRAS, and TP53 mutations. Gain of function study using recombinant DKK2 further confirmed that DKK2 activates c-Src in normal colonic epithelial cells as well. These findings propose DKK2 as a key upstream regulator of c-Src activation in colonic epithelial cells during tumorigenesis. Further studies to identify the DKK2 receptors in c-Src activation will provide better molecular understanding of the DKK2 downstream signaling pathways.
In this study, we have shown that DKK2 is required for the elevated expression of LGR5 in colon cancer cells through c-Src activation and depletion of HNF4α1. Based on our findings, new therapeutic approaches targeting DKK2 would be beneficial against colorectal cancers, particularly to control LGR5 expressing cancer stem cells.
Limitations of the study
Increased expression of DKK2 is shown in the colitis-induced tumor model and cancer organoids with mutations in APC, KRAS, TP53, and SMAD4 genes. Based on the pattern of DKK2 expression, this study focused on the roles of DKK2 in colon cancer cells. Further study is needed to understand the roles of DKK2 in the homeostatic condition. In this study, we observed decreased expression of LGR5 by DKK2 knockout in colitis-induced polyps and cancer organoids. LGR5 is expressed in most stem cells including crypt base columnar stem cells and quiescent stem cells and paneth cells as well. A technical limitation of our data is that we cannot provide information about the sub-types of LGR5 expressing stem cells affected by DKK2 knockout.
Resource availability
Lead contact
Further requests for resources should be directed to and will be fulfilled by the lead contact, Alfred L. M. Bothwell (alfred.bothwell@yale.edu).
Materials availability
This study did not generate new materials.
Data and code availability
The accession number for the RNA sequencing data reported in this study is GSE157535.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by National Institutes of Health USA, RO1CA168670 (awarded to A.L.M.B).The authors would like to thank GouzelTokmulina and ZuzanaTobiasova for the FACS sorting.
Author contributions
J.S. and A.L.M.B. designed and conducted the experiments, analyzed the data and wrote the manuscript. J.J. performed immunofluorescence and confocal microscopy and Western blot analysis. J.C. and J.L. performed and analyzed RNA-seq experiments. R.K.D. cloned gRNA/Cas9 plasmids targeting DKK2 exon1 and HNF4α1 exon1. J.B. and O.Y. developed colon tumor organoids. J.H. contributed to RNA-seq and quantitative PCR analysis. S.E.M. supported in vivo experiments. M.C.A.V. was involved in colonoscopy. W.K., J.K., and J.C. generated recombinant mouse DKK2 protein and polyclonal anti-DKK2 antibody. W.T. and D.W. developed DKK2-floxed mice. H.N.B., R.X.M. and X.L. contributed to immunohistochemistry. J.S. performed all other experiments.
Declaration of interests
The authors declare no competing interests exist.
Published: May 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102411.
Supplemental information
References
- Babeu J.P., Jones C., Geha S., Carrier J.C., Boudreau F. P1 promoter-driven HNF4alpha isoforms are specifically repressed by beta-catenin signaling in colorectal cancer cells. J.Cell Sci. 2018;131 doi: 10.1242/jcs.214734. [DOI] [PubMed] [Google Scholar]
- Barker N., Ridgway R.A., Van Es J.H., Van De Wetering M., Begthel H., Van Den Born M., Danenberg E., Clarke A.R., Sansom O.J., Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Barker N., Van Es J.H., Kuipers J., Kujala P., Van Den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Cattin A.L., Le Beyec J., Barreau F., Saint-Just S., Houllier A., Gonzalez F.J., Robine S., Pincon-Raymond M., Cardot P., Lacasa M., Ribeiro A. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol.Cell. Biol. 2009;29:6294–6308. doi: 10.1128/MCB.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappa K., Deol P., Evans J.R., Vuong L.M., Chen G., Briancon N., Bolotin E., Lytle C., Nair M.G., Sladek F.M. Opposing roles of nuclear receptor HNF4alpha isoforms in colitis and colitis-associated colon cancer. Elife. 2016;5 doi: 10.7554/eLife.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappa K., Jankova L., Schnabl J.M., Pan S., Brelivet Y., Fung C.L., Chan C., Dent O.F., Clarke S.J., Robertson G.R., Sladek F.M. Src tyrosine kinase phosphorylation of nuclear receptor HNF4alpha correlates with isoform-specific loss of HNF4alpha in human colon cancer. Proc. Natl. Acad. Sci. U S A. 2012;109:2302–2307. doi: 10.1073/pnas.1106799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappa K., Jankova L., Schnabl J.M., Pan S., Brelivet Y., Fung C.L.S., Chan C., Dent O.F., Clarke S.J., Robertson G.R., Sladek F.M. Src tyrosine kinase phosphorylation of nuclear receptor HNF4 correlates with isoform-specific loss of HNF4 in human colon cancer. Proc.Natl.Acad.Sci. U S A. 2012;109:2302–2307. doi: 10.1073/pnas.1106799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Elfiky A., Han M., Chen C., Saif M.W. The role of Src in colon cancer and its therapeutic implications. Clin. Colorectal Cancer. 2014;13:5–13. doi: 10.1016/j.clcc.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Chen L., Toke N.H., Luo S., Vasoya R.P., Aita R., Parthasarathy A., Tsai Y.H., Spence J.R., Verzi M.P. HNF4 factors control chromatin accessibility and are redundantly required for maturation of the fetal intestine. Development. 2019;146:dev179432. doi: 10.1242/dev.179432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Cordero J.B., Ridgway R.A., Valeri N., Nixon C., Frame M.C., Muller W.J., VIDAL M., SANSOM O.J. c-Src drives intestinal regeneration and transformation. EMBO J. 2014;33:1474–1491. doi: 10.1002/embj.201387454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa e Melo F., Kurtova A.V., Harnoss J.M., Kljavin N., Hoeck J.D., Hung J., Anderson J.E., Storm E.E., Modrusan Z., Koeppen H. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- Drost J., Clevers H. Organoids in cancer research. Nat. Rev. Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- Fillippo C.D., Carderni G., Bazzicalupo M., Briani C., Giannini A., Fazi M., Dolaral P. Mutations of the Apc gene in experimental colorectal carcinogenesis induced by azoxymethane in F344 rats. Br. J. Cancer. 1998;77:2148–2151. doi: 10.1038/bjc.1998.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli A., Oost K.C., Kester L., Morgner J., Bornes L., Bruens L., Spaargaren L., Azkanaz M., Schelfhorst T., Beerling E. Plasticity of Lgr5-negative cancer cells drives metastasis in colorectal cancer. Cell Stem Cell. 2020;26:569–578.e7. doi: 10.1016/j.stem.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A., Pinto D., Begthel H., Destree O., Kielman M., Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Leushacke M., Barker N. Lgr5 and Lgr6 as markers to study adult stem cell roles in self-renewal and cancer. Oncogene. 2012;31:3009–3022. doi: 10.1038/onc.2011.479. [DOI] [PubMed] [Google Scholar]
- Li X.B., Yang G., Zhu L., Tang Y.L., Zhang C., Ju Z., Yang X., Teng Y. Gastric Lgr5(+) stem cells are the cellular origin of invasive intestinal-type gastric cancer in mice. Cell Res. 2016;26:838–849. doi: 10.1038/cr.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu C., Kopetz S. The SRC family of protein tyrosine kinases: a new and promising target for colorectal cancer therapy. Clin. Colorectal Cancer. 2010;9:89–94. doi: 10.3816/CCC.2010.n.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B., Niehrs C. Kremen2 modulates Dickkopf2 activity during Wnt/lRP6 signaling. Gene. 2003;302:179–183. doi: 10.1016/s0378-1119(02)01106-x. [DOI] [PubMed] [Google Scholar]
- Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- Matsui A., Yamaguchi T., Maekawa S., Miyazaki C., Takano S., Uetake T., Inoue T., Otaka M., Otsuka H., Sato T. DICKKOPF-4 and -2 genes are upregulated in human colorectal cancer. Cancer Sci. 2009;100:1923–1930. doi: 10.1111/j.1349-7006.2009.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- Phelps R.A., Broadbent T.J., Stafforini D.M., Jones D.A. New perspectives on APC control of cell fate and proliferation in colorectal cancer. Cell Cycle. 2009;8:2549–2556. doi: 10.4161/cc.8.16.9278. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Colman M.J., Schewe M., Meerlo M., Stigter E., Gerrits J., Pras-Raves M., Sacchetti A., Hornsveld M., Oost K.C., Snippert H.J. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., Van De Wetering M., Barker N., Stange D.E., Van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schepers A.G., Snippert H.J., Stange D.E., Van Den Born M., Van Es J.H., Van De Wetering M., Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- Shtutman M., Zhurinsky J., Simcha I., Albanese C., D’amico M., Pestell R., Ben-Ze’ev A. The cyclin D1 gene is a target of the b-catenin LEF-1 pathway. Proc. Natl. Acad. Sci. U S A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkandera J., Herzog S., Pichler M., Stiegelbauer V., Stotz M., Schaberl-Moser R., Samonigg H., Asslaber M., Lax S., Leitner G. LGR5 rs17109924 is a predictive genetic biomarker for time to recurrence in patients with colon cancer treated with 5-fluorouracil-based adjuvant chemotherapy. Pharmacogenomics J. 2015;15:391–396. doi: 10.1038/tpj.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Fearon E.R., Hamilton S.R., Kern S.E., Preisinger A.C., Leppert M., Nakamura Y., White R., Smits A.M., Bos J.L. Genetic alteration during colorectal-tumor development. N. Engl. J. Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., JR., Kinzler K.W. Cancer genome landscape. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whissell G., Montagni E., Martinelli P., Hernando-Momblona X., Sevillano M., Jung P., Cortina C., Calon A., Abuli A., Castells A. The transcription factor GATA6 enables self-renewal of colon adenoma stem cells by repressing BMP gene expression. Nat.Cell Biol. 2014;16:695–707. doi: 10.1038/ncb2992. [DOI] [PubMed] [Google Scholar]
- Wu W., Glinka A., Delius H., Niehrs C. Mutual antagonism between dickkopf1 and dickkopf2 regulates Wnt:β-catenin signalling. Curr. Biol. 2000;10:1611–1614. doi: 10.1016/s0960-9822(00)00868-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA sequencing data reported in this study is GSE157535.