Abstract
Background and aims
The Hepatitis E virus hijacks the endosomal system for its release. These structures are highly dependent on cholesterol. Hence, this study investigates the impact of HEV on cholesterol-metabolism, the effect of intracellular cholesterol content on HEV-release and the potential of cholesterol-modulators to serve as antivirals.
Methods
Intracellular cholesterol-content of cells was modulated and impacts on HEV were monitored using qPCR, Western blot, microscopy, virus-titration and density-gradient centrifugation. Blood-lipids and HEV-RNA were routinely quantified in chronically infected patients during follow-up visits.
Results
In HEV-infected cells, decreased levels of cholesterol are found. In patients, HEV infection decreases serum-lipid concentrations. Importantly, statin treatment herein increases viral titers. Similarly, reduction of intracellular cholesterol via simvastatin treatment increases viral release in vitro. On the contrary, elevating intracellular cholesterol via LDL or 25-hydroxycholesterol strongly reduces viral release due to enhanced lysosomal degradation of HEV. Drug-induced elevation of intracellular cholesterol via fenofibrate or PSC833 impairs HEV release via the same mechanism.
Conclusions
This study analyses the crosstalk between HEV and intracellular cholesterol. The results highlight the importance of an intact cholesterol homeostasis for HEV-release and thereby identify a potential target for antiviral strategies. Especially fenofibrate is considered a promising novel antiviral against HEV. Beyond this, the study may help clinicians evaluating co-treatments of HEV-infected patients with statins, as this may be counter indicated.
Keywords: Antiviral, Cholesterol, HEV, Lipids, Lysosomes
Abbreviations used in this paper: 25-HC, 25-hydroxycholesterol; CI, confidence interval; DMEM, Dulbecco’s modified Eagle’s medium; eHEV, quasi-enveloped hepatitis E virus; EC50, half maximal effective concentration; FGF19, fibroblast growth factor 19; HDL, high-density lipoprotein; HEV, hepatitis E virus; LAMP2, lysosome-associated membrane protein 2; LDL, low-density lipoprotein; MVB, multivesicular body; PCSK9, proprotein convertase subtilisin/kexin type 9; qPCR, quantitative polymerase chain reaction; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; TCID50, half maximal tissue culture infective dose
Graphical abstract
Summary.
A hepatitis E virus (HEV) infection alters cholesterol homeostasis in vitro and in vivo. Reduced cholesterol levels enhance viral release, yet an increase induces lysosomal HEV degradation. Concordantly, cholesterol-modulating drugs such as fenofibrate and PSC833 were identified as novel antivirals against HEV making use of this mechanism.
The hepatitis E virus (HEV) is a hepatotropic (+) single-strand RNA virus and the sole member of the Hepeviridae family. Its human pathogenic forms are restricted to genotypes 1–4 and 7 within the genus Orthohepevirus A. Genotypes 3, 4, and 7 display a zoonotic potential also infecting species such as swine or camelids.1 Infection occurs via the fecal-oral route, conventionally via contaminated water, whereas food-borne or transfusion-related transmissions represent an increasing risk in industrialized countries.2,3 The virus is endemic in large parts of the developing world, yet it displays major threats to the developed world for previously mentioned reasons.4,5 Over the past years, infections were observed to reach as high as 20 million cases per year. This results in up to 44,000 HEV-related fatalities annually.6,7 Among high-risk groups, pregnant women can be found with mortality rates reaching as high as 25%–30%8 or immunosuppressed patients tending to develop chronic infections.9 In all cases, chronification can be observed for infections with genotype 3, although course of disease is milder when compared with genotype 1 or 2, which frequently display fulminant outcomes.10,11 As of now, treatment options are restricted to either ribavirin, where development of drug-resistance can occur,12 or pegylated interferon. Both treatment options can cause severe side effects.13
HEV hijacks multivesicular bodies (MVBs) for release of quasi-enveloped HEV (eHEV) viral particles.14 The viral pORF3 protein leads to a tumor susceptibility gene 101 (tsg101)–dependent tethering of capsids to the ESCRT (endosomal sorting complex required for transport)15 via its viral late-domain (PSAP-motif).16 Thus, an impact of cellular lipids on HEV release appears likely, since successful maturation and gain of functionality of these structures require an intact orchestration of membrane composition.17 Especially cholesterol has important functions during invagination of MVBs’ intraluminal vesicles18 by means of inducing membrane curvature and forming lipid microdomains. Therefore, cholesterol may also play a crucial role in the morphogenesis and release of eHEV, displaying the only described route of viral egress, besides its implications for viral entry.19 Cholesterol also is important for determining the fate of endosomal vesicle trafficking.20 This affects the release of viruses in the form of quasi-enveloped particles, whose release depends on the endosomal system.
Cholesterol and lipid metabolism per se are of central interest in the context of treating clinical symptoms caused by an HEV infection. As reported, there are several, yet ambiguous, changes in biomarkers related to cholesterol or its derivatives,21, 22, 23 which may also pose the need of treatment (eg, hepatitis-induced hypercholesterolemia or cholestasis). Several different examples of drugs impacting systematic distribution of lipids are available as prescription drugs or subject of clinical studies, among which are (1) fibrates (eg, gemfibrozil or fenofibrate), lowering triglycerides via activation of respective catabolism24; (2) statins (eg, simvastatin), inhibiting the rate-limiting step in cholesterol-biosynthesis25; (3) PSC-833, lowering sterol-efflux from cells26; (4) NGM282, a fibroblast growth factor 19 (FGF19) analogue, inhibiting primary bile acid synthesis27; (5) alirocumab or evolocumab, monoclonal antibodies triggering increased low-density lipoprotein (LDL) uptake28; or (6) avasimibe, a cholesterol storage inhibitor.29 By acting on the cholesterol homeostasis in cells, these compounds inevitably act on the endolysosomal system and are of great interest when it comes to modulation of endosomal fate.
This study aims to investigate the impact of HEV on cholesterol metabolism and to analyze impacts of intracellular cholesterol level on the HEV life cycle. The virus strongly relies on the endosomal system for its release, specifically on MVBs. These structures, in turn, are highly dependent on intracellular cholesterol homeostasis for intact functionality and maturation. Thus, a focus was set on whether altering the content of cellular cholesterol can be used as a new approach for an antiviral treatment. Insights into this novel field help broadening both the understanding of the viral life cycle and of drugs being potential candidates for an off-label use against the viral hepatitis.
Results
HEV Modulates Cholesterol Homeostasis In Vitro and In Vivo
To study the impact of an HEV infection on the intracellular cholesterol content, a quantitative filipin stain of cholesterol in infected cells compared with uninfected cells was performed. Herein, the viral infection caused reduced amounts of intracellular cholesterol (Figure 1A and B). In line with this, gene expression profiling of HEV-infected cells revealed that HEV is also capable of inducing a dysregulation in gene expression of cholesterol metabolism–related genes as compared with uninfected cells (Figure 1C). Quantification of selected transcripts by quantitative polymerase chain reaction (qPCR) confirmed that HEV modulates key enzymes of cholesterol metabolism and transport that cause a decrease on the intracellular cholesterol content (Figure 1D).
Figure 1.
HEV dysregulates cholesterol in vitro and in vivo. (A) Representative filipin stain of cholesterol (purple) and pORF2 (green) in uninfected and infected cells; scale bar = 20 μm. (B) Quantification of A; filipin intensity depicted as corrected total cell fluorescence (CTCF) per cell; unpaired t test with Holm-Sidak correction. (C) Gene expression profiling of persistently HEV-infected A549 cells vs uninfected A549/D3 cells. (D) qPCR validation of C, fold-change messenger RNA (mRNA) infected vs uninfected; unpaired t test with Holm-Sidak correction. (E–H) Comparison blood lipids in patients at t–1, t0, and t+1; t–1 vs t0 (triglycerides [TG] = 29 pairs / total cholesterol [TC] = 29 pairs / LDL = 29 pairs / HDL = 28 pairs); t0 vs t+1 (TG= 20 pairs / TC = 20 pairs / LDL = 21 pairs / HDL = 21 pairs); paired, nonparametric t test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
To correlate these findings from in vitro experiments with an in vivo situation, serum-lipids of chronically infected patients were analyzed with respect to triglycerides, total cholesterol, LDL, and high-density lipoprotein (HDL). Comparing serum lipids before infection (t–1), at first positive testing (t0), and at the first follow-up (t+1) revealed that there were significantly lowered concentrations of triglycerides, total cholesterol, and LDL levels (Figure 1E–G) once an HEV infection occurred in the patient. No changes were induced with respect to HDL levels (Figure 1H).
These data indicate that an HEV infection modulates cholesterol homeostasis in vitro, leading from dysregulated gene expression of related genes to reduced intracellular cholesterol levels in infected cells. Similarly, HEV-dependent lipid modulation could be observed in infected patients resulting in lowered serum lipid concentrations of cholesterol and triglycerides.
Increased Intracellular Cholesterol Inhibits HEV
The data described previously indicate that the cholesterol content is decreased in HEV replicating cells. To further characterize the crosstalk between HEV replication and intracellular cholesterol levels, the latter was elevated by a 48-hour treatment with LDL or 25-hydroxycholesterol (25-HC). Vice versa, decrease of intracellular cholesterol was achieved by treating cells with simvastatin for 48 hours. Treatment efficiency and cytotoxicity or cytostaticity was controlled via filipin-stain (Figure 2A and B) or viability assays (Figure 2D and E), respectively. As the use of 100 μM 25-HC caused major cytotoxicity and cytostaticity, it was not used for further analyses but the Western blot.
Figure 2.
Intracellular cholesterol is increased via LDL or 25-HC treatment and decreased via simvastatin treatment. (A) Representative immunofluorescent filipin stain of cholesterol (purple) and pORF2 (green) upon treatment in persistently HEV-infected A549 cells; scale bar = 20 μm. (B) Quantification of A; filipin intensity depicted as CTCF per cell. (C) qPCR evaluation of transcriptional effects of treatment with 25-HC in Huh-7 or A549/D3 cells, values referred to respective untreated control. (D) Cytotoxicity in persistently HEV-infected cells treated with different compounds as determined via lactate dehydrogenase assay; untreated cells were set to 100%, changes in amount of intact cells are depicted as % of untreated control. (E) Cytostaticity in persistently HEV-infected cells treated with different compounds as determined via PrestoBlue assay; untreated cells were set to 100%, changes in metabolic activity are depicted as % of untreated control. Unpaired t test with Holm-Sidak correction for all panels. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. DMSO, dimethyl sulfoxide.
Suitability of A549/D3 cells for cholesterol modulation was demonstrated by comparing effects of 25-HC treatment on gene expression of certain, cholesterol-related genes in comparison with Huh-7 cells after 24 hours (Figure 2C). In persistently HEV-infected A549 cells, both increase and decrease of intracellular cholesterol level led to a dose-dependent reduction in the amount of the HEV capsid protein (pORF2) (Figure 3A–D) and a reductive trend in intracellular HEV transcripts (Figure 3E). This went along with a reduction in release of viral RNA and infectious viral particles for LDL or 25-HC treatment (Figure 3F and G) and a shift in particle density (Figure 3H). Interestingly, treatment with simvastatin led to a significant increase of released infectious viral particles (Figure 3G) with no changes in density of eHEV (Figure 3H).
Figure 3.
HEV release is inhibited by LDL and 25-HC but induced by simvastatin. (A) Representative Western blot of pORF2 and GAPDH; neg = uninfected A549/D3 cells; black arrows indicate pORF2 bands. (B–D) Quantification of pORF2-signals in A; fold-change compared with untreated group; unpaired t test with Holm-Sidak correction. (E) Fold-change of intracellular HEV transcripts as determined by RT-qPCR; unpaired t test with Holm-Sidak correction. (F) Fold-change of extracellular HEV RNA as determined by RT-qPCR; unpaired t test with Holm-Sidak correction. (G) Fold-change of released infectious viral particles as determined by end-point dilution assay; unpaired t test with Holm-Sidak correction. (H) HEV RNA in fractions of density-gradients as determined by RT-qPCR; depicted as % of whole genomes in gradient. (I) HEV titer in chronically infected patients with or without statin treatment; n = 42 patients, 129 measured values (without = 74 values; with = 55 values); Mann-Whitney-test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
As simvastatin induced viral release in vitro, viral titers of chronically infected patients were compared with respect to patients receiving a statin treatment. Strikingly, treatment with this class of cholesterol-lowering drugs led to significantly elevated viral loads in respective patient sera (Figure 3I), which goes in line with in vitro findings. No statin-dependent changes in blood lipids could be observed (Figure 4A–D).
Figure 4.
Statin treatment does not reduce blood-lipids in chronically HEV-infected patients. (A–D) Serum-lipid concentration in chronically infected patients grouped as treated or untreated with statins; n = 42 patients; TG (without = 53 values; with = 45 values), TC (without = 52 values; with = 44 values), LDL (without = 56 values; with = 45 values), HDL (without = 52 values; with = 44 values). Mann-Whitney test.
These data indicate that increased cellular cholesterol content significantly reduces HEV both intracellularly and extracellularly with formation of a new population of released eHEV. Opposing to this, low cholesterol content of cells significantly reduces intracellular viral content, which is accompanied by an increase in viral release. The, patient data argue for a counter indication of statins in the context of an HEV infection.
Excess of Cholesterol Induces Lysosomal Degradation of pORF2
As HEV seems to be affected negatively by elevated levels of intracellular cholesterol, the question arose whether this could be due to a degradative process or to an impaired release. Therefore, infected cells were subjected to confocal laser scanning microscopy to visualize subcellular localization and to further evaluate the amount of pORF2 after modulation of cholesterol levels for 48 hours (Figure 5A).
Figure 5.
High cholesterol induces lysosomal localization of HEV. (A) Representative immunofluorescent stain of DAPI (blue), pORF2 (green), and LAMP2 (red) upon treatment in persistently HEV-infected A549 cells; scale bar = 20 μm; zoom = magnified section indicated with white square in merge. (B) Quantification of A; pORF2 intensity depicted as CTCF per cell. (C) Quantification of A; thresholded Mander’s overlap coefficient (tMOC) = red signal being present in green areas per cell. Unpaired t test with Holm-Sidak correction for all panels; ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
Here, the viral capsid protein accumulates in dot-like structures, once there is more intracellular cholesterol (Figure 5A). Further, reduced protein amounts were detected (Figure 5B). These cholesterol-induced, dot-like structures co-localized with the lysosomal marker lysosome-associated membrane protein 2 (LAMP2), meaning that these clustered structures represent pORF2 being present inside of lysosomes. Notably, this increase in cholesterol-induced lysosomal localization of pORF2 is further represented by calculation of the thresholded Mander’s overlap coefficient between the viral protein and LAMP2 (Figure 5C). In contrast to this, cells with lowered cholesterol content, as induced by simvastatin treatment, showed a decrease in intracellular pORF2 amount (Figure 5B), which was independent from dot formation and the co-localization with LAMP2 (Figure 5C). To verify a cholesterol-induced lysosomal degradation of pORF2, lysosomal protease inhibition was implemented by using leupeptin during the last 24 hours of cholesterol modulation (Figure 6A). Applying leupeptin generally leads to a slight increase in intracellular pORF2 (Figure 6D and E). Within cholesterol modulation, the pORF2 amount was not reduced by 25-HC or LDL any longer, although being present in lysosomes. However, simvastatin-induced pORF2 reduction was not reverted (Figure 6A–C).
Figure 6.
Inhibition of lysosomal degradation rescues cholesterol-induced pORF2 reduction. (A) Representative immunofluorescent stain of DAPI (blue), pORF2 (green), and LAMP2 (red) upon treatment in persistently HEV-infected A549 cells; scale bar = 20 μm; zoom = magnified section indicated with white square in merge. (B) Quantification of A; pORF2 intensity depicted as CTCF per cell. (C) Quantification of A; tMOC = red signal being present in green areas per cell. (D) Representative Western blot of pORF2 and GAPDH of leupeptin-treated cells; neg = uninfected A549/D3 cells; black arrows indicate pORF2 bands. (E) Quantification of pORF2-signals in D; fold-change compared with untreated group. Unpaired t test with Holm-Sidak correction for all panels; ∗∗P < .01, ∗∗∗P < .001.
These data imply that the intracellular HEV load is reduced once intracellular cholesterol is elevated. This reduction is due to a targeting of pORF2 into lysosomes, where it is degraded. Thus, less viral particles are being released. Low intracellular cholesterol, however, does not induce pORF2 degradation but leads to an increased viral release.
Cholesterol-Modulating Drugs Act Antivirally Against HEV
The previously presented findings of this study imply that an HEV infection results in reduced intracellular cholesterol levels, therefore guaranteeing efficient viral release. Disruption of this effect via artificially increasing intracellular cholesterol levels of infected cells led to a reduction in intracellular pORF2 amount and concordantly to reduced viral release. This effect was observed to be due to a cholesterol-induced lysosomal degradation of pORF2. Thus, different drugs targeting cholesterol metabolism and distribution were tested for their antiviral capacity against HEV. The aim was to induce lysosomal degradation of the virus based on a drug-related elevation of intracellular cholesterol.
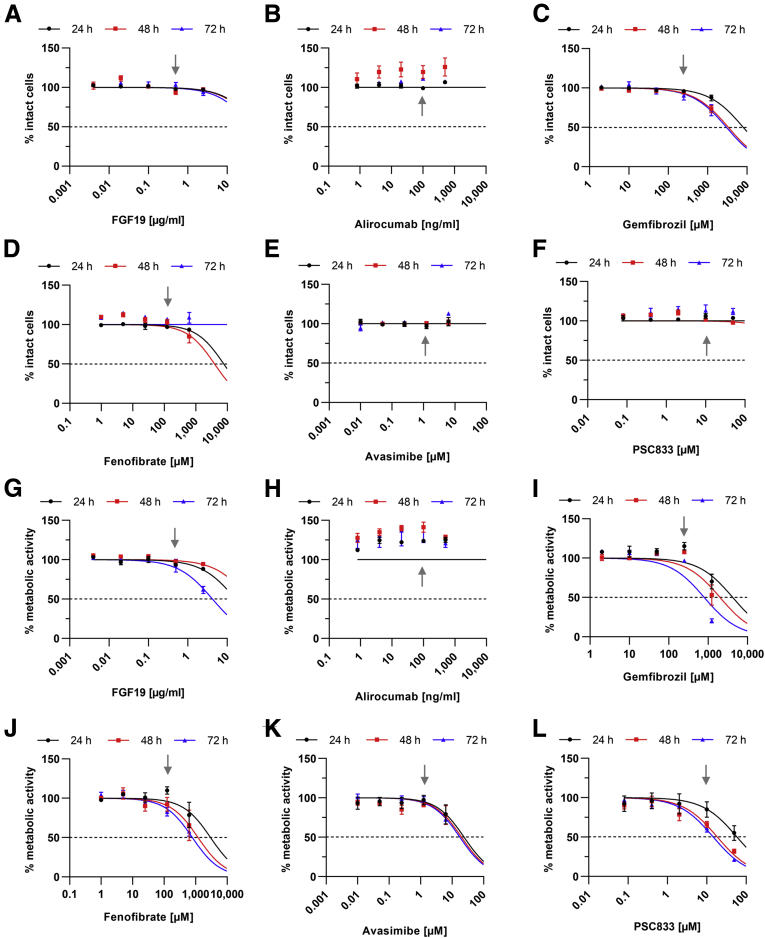
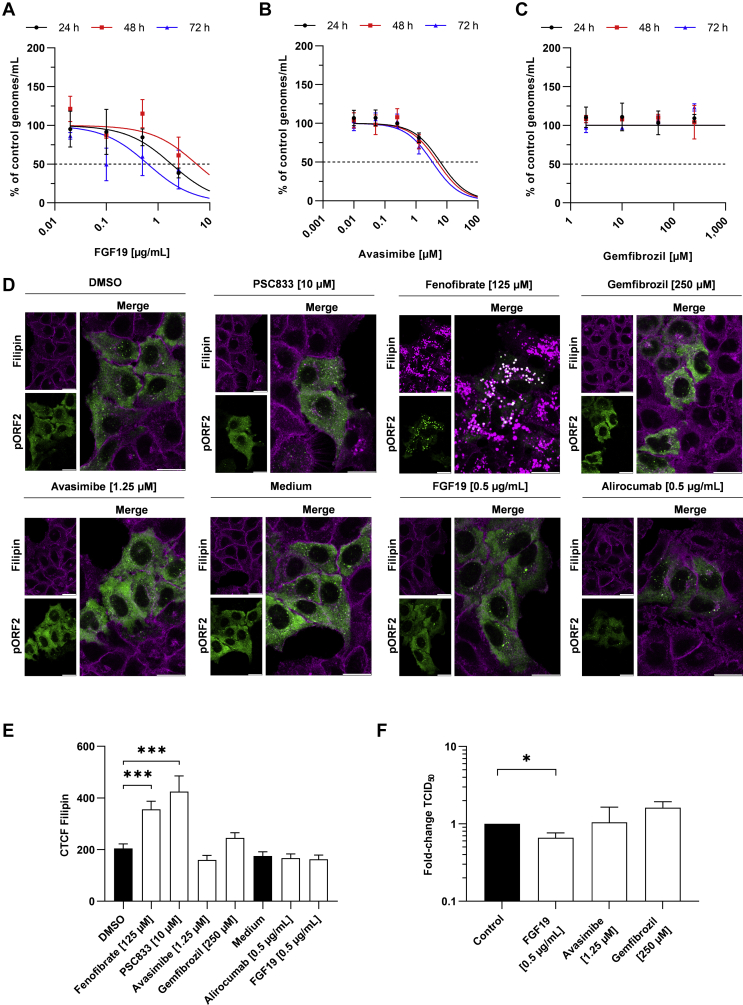
Assessment of the cell viability indicated that none of the tested concentration of the compounds led to increased cell death, yet some compounds led to slight cytostatic effects (Figure 7) at very high concentrations, which thereafter were excluded from antiviral testing. Determination of the half maximal effective concentration (EC50) via qPCR indicated that Avasimibe (EC50 = 3.4 μM at 72 hours; 95% confidence interval [CI], 1.84–8.5 μM) and FGF19 (EC50 = 0.64 μg/mL at 72 hours; 95% CI, 0.08–4.27 μg/mL) displayed low potentials to inhibit viral release. Gemfibrozil did not display an antiviral effect (Figure 8A–C and F). Alirocumab (EC50 = 155 ng/mL at 72 hours; 95% CI, 34.94–810.4 ng/mL) inhibited HEV mildly, while both fenofibrate (EC50 = 71.24 μM at 72 hours; 95% CI, 34.52–154.3 μM) and PSC833 (EC50 = 12.47 μM at 72 hours; 95% CI, 4.95–48.0 μM) showed strong antiviral effects (Figure 9A–C). This reduction was in line with Western blot analyses detecting intracellular pORF2 protein (Figure 9D and E). Importantly, the inhibitory capacity of each drug correlated well with its ability or inability to increase intracellular cholesterol after 24 hours (Figure 8D–F). Especially treatment with fenofibrate led to a heavy increase and accumulation of cholesterol inside the cells. Notably, A549/D3 cells behaved similarly as Huh-7 cells with respect to intracellular cholesterol being modulated by fenofibrate or PSC833 for 24 hours (Figure 10).
Figure 7.
Cholesterol-modulating drugs are not cytotoxic at concentrations showing inhibition of HEV. (A–F) Dose-response curve of persistently HEV-infected cells treated with different compounds as determined via lactate dehydrogenase assay; untreated cells were set to 100%, changes in amount of intact cells are depicted as % of untreated control. Gray arrows indicate highest concentrations used for subsequent experiments. (G–L) Dose-response curve of persistently HEV-infected cells treated with different compounds as determined via PrestoBlue assay; untreated cells were set to 100%, changes in metabolic activity are depicted as % of untreated control. Gray arrows indicate highest concentrations used for subsequent experiments.
Figure 8.
Antiviral effect of drugs is accompanied by increased intracellular cholesterol. (A–C) Dose-response curve of viral RNA being released by treated persistently HEV-infected cells as determined via RT-qPCR; untreated cells were set to 100%, changes in RNA amount are depicted as % of untreated control; basis for calculation of EC50. (D) Representative immunofluorescent filipin stain of cholesterol (purple) and pORF2 (green) in untreated or treated persistently HEV-infected A549 cells; scale bar = 20 μm. (E) Quantification of F; Cholesterol intensity depicted as CTCF per cell. (F) Fold change of released infectious viral particles as determined by end-point dilution assay. Unpaired t test with Holm-Sidak correction for all panels; ∗P < .05, ∗∗∗P < .001.
Figure 9.
Drugs modulating cholesterol are effective against HEV release. (A-C) Dose-response curve of viral RNA being released by treated, persistently HEV-infected cells as determined via RT-qPCR; untreated cells set to 100%, changes in RNA amount depicted as % of control. (D) Representative Western blot of pORF2 and GAPDH; neg = uninfected A549/D3 cells; black arrows indicate pORF2 bands. (E) Quantification of pORF2-signals in D; fold change compared with untreated group. (F) Fold change of released infectious viral particles as determined by end-point dilution assay. (G) HEV RNA in fractions of density gradients as determined by RT-qPCR; % of whole genomes in gradient. Unpaired t test with Holm-Sidak correction for all panels; ∗∗P < .01, ∗∗∗∗P < .0001.
Figure 10.
Fenofibrate and PSC833 induce similar changes in cholesterol in A549/D3 and Huh-7 cells. (A) Representative immunofluorescent filipin stain of cholesterol (purple) in A549/D3 and Huh-7 cells; scale bar = 20 μm. (B) Quantification of A; Cholesterol intensity depicted as CTCF per cell. Unpaired t test with Holm-Sidak correction for all panels; ∗P < .05, ∗∗∗P < .001, ∗∗∗∗P < .0001.
To correlate changes in extracellular viral RNA with actual infectious viral particles, half maximal tissue culture infective dose (TCID50) values of the highest tolerated concentrations were compared after 72 hours of treatment (Figure 9F). Again, alirocumab showed a mild inhibition of viral release. Fenofibrate and PSC833 drastically inhibited viral release. This was accompanied by a change in eHEV density (Figure 9G).
In summary, these data suggest that alirocumab inhibits HEV release to some extent. Drastic reductions in viral release going along with increased intracellular cholesterol levels were observed for both fenofibrate and PSC833. Further, the aforementioned phenotype of eHEV displaying a shift in density to denser fractions upon LDL or 25-HC treatment of infected cells could be mimicked by applying fenofibrate or PSC833.
Effective Drug Candidates Induce Lysosomal Degradation of pORF2
Whether the drug-induced reduction in viral release is due to lysosomal degradation of pORF2, similar to cells with a high cholesterol-content (Figure 3), was addressed by confocal laser scanning microscopy analyses after 48 hours of treatment.
Here, treatment with fenofibrate, PSC833 or alirocumab led to the same, dot-like accumulation of pORF2 (Figure 11A) and to a significant decrease of its amount (Figure 11B). This accumulation of pORF2 was comparable to the once induced by LDL or 25-HC. Again, these structures could be found to colocalize with the lysosomal marker LAMP2 (Figure 11A). Just like the direct increase of intracellular cholesterol, the drug-induced elevation led to an increase of pORF2 being found inside of lysosomes as evaluated by calculation of thresholded Mander’s overlap coefficient (Figure 11C). Blocking of lysosomal degradation via leupeptin treatment during the last 24 hours of drug treatment rescued pORF2 amounts from being reduced (Figure 12). Notably, both PSC833 and fenofibrate impacted lysosomal morphology. Compounds being inefficient against HEV, namely gemfibrozil and FGF19, did not show such reducing activities with respect to the pORF2 amount and similarly did not show increased lysosomal localization (Figure 13).
Figure 11.
Antiviral effect of fenofibrate, PSC833, and alirocumab due to lysosomal localization of HEV. (A) Representative immunofluorescent stain of DAPI (blue), pORF2 (green), and LAMP2 (red) upon treatment in persistently HEV-infected A549 cells; scale bar = 20 μm; zoom = magnified section indicated with white square in merge. (B) Quantification of A; pORF2 intensity depicted as CTCF per cell. (C) Quantification of A; tMOC = red signal being present in green areas per cell. Unpaired t test with Holm-Sidak correction for all panels; ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
Figure 12.
Inhibition of lysosomal degradation rescues drug-induced, cholesterol-dependent pORF2 reduction. (A) Representative immunofluorescent stain of DAPI (blue), pORF2 (green), and LAMP2 (red) upon treatment in persistently HEV-infected A549 cells; scale bar = 20 μm; zoom = magnified section indicated with white square in merge. (B) Quantification of A; pORF2 intensity depicted as CTCF per cell. (C) Quantification of A; tMOC = red signal being present in green areas per cell. Unpaired t test with Holm-Sidak correction for all panels; ∗P < .05, ∗∗P < .01.
Figure 13.
FGF19, avasimibe, or gemfibrozil treatment is not accompanied by lysosomal localization of HEV. (A) Representative immunofluorescent stain of DAPI (blue), pORF2 (green), and LAMP2 (red) upon treatment in persistently HEV-infected A549 cells; scale bar = 20 μm; zoom = magnified section indicated with white square in merge. (B) Quantification of A; pORF2 intensity depicted as CTCF per cell. (C) Quantification of A; tMOC = red signal being present in green areas per cell. Unpaired t test with Holm-Sidak correction for all panels; ∗∗∗∗P < .0001.
Applying these findings to a broader context indicates that especially PSC833 and fenofibrate are capable of inducing cholesterol-dependent pORF2 degradation via lysosomes leading to an antiviral effect. This mimics effects observed for 25-HC and LDL, which is in line with our initial aim of treatment. Major findings regarding antiviral activity of compounds are summarized in Table 1.
Table 1.
Summary of Cholesterol- and Drug-Based Effects on HEV
| Intracellular cholesterol (CTCF) | Intracellular pORF2 (CTCF) | Extracellular viral particles (TCID50) | Lysosomal localization (tMOC) | |
|---|---|---|---|---|
| LDL (600 μg/mL) | ↑↑ | ↓↓↓ | ↓ | ↑↑↑↑ |
| 25-HC (25 μM) | ↑↑ | ↓↓↓↓ | ↓↓↓↓ | ↑↑ |
| Simvastatin (5 μM) | ↓ | ↓ | ↑ | ↓ |
| Fenofibrate (125 μM) | ↑↑↑ | ↓↓↓↓ | ↓↓↓↓ | ↑↑ |
| PSC833 (10 μM) | ↑↑↑ | ↓↓↓ | ↓↓↓↓ | ↑↑↑↑ |
| Alirocumab (0.5 μg/mL) | — | ↓↓ | ↓↓ | ↑↑ |
| Gemfibrozil (250 μM) | — | — | — | — |
| Avasimibe (1.25 μM) | — | — | — | — |
| FGF19 (0.5 μg/mL) | — | — | ↓ | — |
NOTE. Arrows indicate P value; upward arrow = increase; downward arrow = decrease. ↑P < .05, ↑↑P < .01, ↑↑↑P < .001, ↑↑↑↑P < .0001.
25-HC, 25-hydroxycholesterol; CTCF, corrected total cell fluorescence; HEV, hepatitis E virus; LDL, low-density lipoprotein; TCID50, half maximal tissue culture infective dose; tMOC, thresholded Mender’s overlap coefficient.
Discussion
The endosomal system, especially trafficking via late endosomes through multivesicular bodies, plays a central role in the life cycle of a variety of viruses. Among these is HEV. These organelles are particularly sensitive for changes in cholesterol content. Thus, this study aimed to analyze the impact of changes in cholesterol and lipid content on HEV and to use these findings as an antiviral strategy.
The initial finding of this study identified that an HEV infection causes a reduction in the amount of intracellular cholesterol in cell culture. This reduction goes along and may be based on changes in the transcriptomic profile infected cells display in comparison with uninfected cells. Here, various genes being important for maintaining cholesterol homeostasis and regulating cholesterol trafficking are dysregulated. Evidence for an HEV-induced modulation of cholesterol levels can also be found in an in vivo situation. Upon infection, patients’ blood-lipids are significantly reduced. Thus, a modulation of lipid metabolism being induced by the virus is evident, which is common not only for hepatotropic viruses.30, 31, 32 A lack of changes in HDL levels may indicate that the virus induces not a general change in lipid-metabolism, but rather a targeted change.
As HEV induces a reduction in intracellular cholesterol, the question arose whether a disturbed cholesterol homeostasis could affect the viral life cycle. Thus, cholesterol was modulated using LDL or 25-HC as inducers of intracellular cholesterol and simvastatin was used to induce a reduction in intracellular cholesterol. Efficient reductions in intracellular viral content were achieved by both reducing and elevating cholesterol levels. Surprisingly, the cause for this observation is of completely different nature when comparing low-cholesterol with high-cholesterol situations. Low cholesterol content was accompanied by less viral content intracellularly because more infectious viral particles are released upon statin treatment. This, in terms, is contradictory to situations found for other viruses such as the Hepatitis B virus.33 However, reductions in HEV under high cholesterol concentrations was caused by lysosomal degradation of pORF2, ultimately leading to an overall reduction of viral egress. This may be explained by an inhibition of proper endosomal trafficking and subsequent endolysosomal fusion, once cholesterol is present in excess.20 Such mechanism has previously been described (eg, for the influenza A virus).34 Importantly, chronically infected patients treated with statins showed significantly higher viral loads compared with chronically infected patients without statin administration. This is directly comparable with the effect observed in vitro and displays a key finding with major importance for clinicians caring for HEV-infected patients. While statins may help regulating lipid homeostasis, it poses a high risk of inducing viral spread in the liver and body. Summarizing these aspects implies the following 2 statements: (1) low intracellular cholesterol levels are beneficial for HEV, inducing its release from infected cells, and (2) high intracellular cholesterol levels are detrimental for HEV release because viral content is degraded in lysosomes in a cholesterol-dependent manner.
The knowledge about HEV being affected negatively by cholesterol was used to set up a novel strategy of antiviral treatment against HEV: using cholesterol-modulating drugs to induce lysosomal pORF2 degradation. The proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitor alirocumab showed some antiviral activity, yet lacked lysosome-based phenotypes observed for LDL or 25-HC. Notably, treatment of cells with alirocumab did not lead to the expected increase in intracellular cholesterol explaining the lack in antiviral activity. As an inhibition of PCSK9 results in increased LDL uptake,35 a certain extracellular concentration of LDL is mandatory for it to work as intended. Thus, application in an organism could perhaps improve alirocumab’s impact on HEV.
In our screening, most promising candidates were the PPARα (peroxisome proliferator-activated receptor alpha) agonist fenofibrate and the P-glycoprotein efflux transporter inhibitor PSC833. Both inhibited viral release drastically, which correlated with a strong increase in intracellular cholesterol and finally lysosomal degradation of pORF2. This can be deduced from PSC833 inhibiting the oxysterol exporter ABCA126 and fenofibrate directly acting on lysosomal lipid and cholesterol content.24,36 Slight increases in intracellular pORF2 under PSC833 treatment may be explained by secondary effects being induced by the compound, leading to a lysosomal trapping without degradation, which may display the need of careful titration during treatment. Another aspect accompanying drug-induced HEV inhibition are changes in the density of eHEV, which was observed for direct increases in intracellular cholesterol. Hence, the aim of modulating cholesterol content to induce lysosomal degradation of pORF2 was perfectly achieved. Notably, the underlying hypothesis is further backed up by fenofibrate being discussed to display antiviral effects against SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2).37 This highlights that this antiviral strategy could be of universal use.
Conclusion
As of now, this study is among the first approaches of antiviral screenings targeting the lipid metabolism of host cells. HEV is herein observed to be degraded lysosomally in a cholesterol-dependent manner, which can be efficiently induced by treatment with fenofibrate or PSC833. These findings correlate with lipid alterations induced upon HEV infection in patients, which also revealed that statins are counter indicated during the viral infection. Hereby, a milestone is set for a better understanding of metabolic host processes, which can be directly modulated to induce antiviral effects. Not only is this a strategy to fight HEV, but also bears a huge potential to tackle various viruses making use of the endolysosomal system.
Material and Methods
Cell Culture
Cell lines used included Huh-7,38 A549/D3,39 and persistently HEV-infected A549 cells40 representing parental A549 cells initially infected with HEV genotype 3c isolate 47832c. These cells were chosen as cell culture model as they maintain viral infection while being passaged or cryopreserved, yielding a stable infection, which avoids establishing an infection at each experiment. This feature is superior to other, hepatocyte-based systems and yields high titers and HEV infection efficiency. Cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) (supplemented with 2 mM L-glutamine, 100 μg/mL streptomycin, 100 U/mL penicillin, and 10% v/v fetal bovine serum; FBS.S 0615, Bio & Sell GmbH, Feucht, Germany) at 37°C with 95% relative humidity and 5% CO2.
Treatments with LDL (360-10-0.1; Lee Biosolutions, Maryland Heights, MO), 25-HC (11097; Cayman Chemicals, Ann Arbor, MI) and simvastatin (sc-200829B; Santa Cruz Biotechnology, Heidelberg, Germany) were performed in serum-free DMEM (D6546-500ML; Sigma Aldrich, Schnelldorf, Germany). Fully supplemented DMEM was used for treatment with gemfibrozil, fenofibrate, avasimibe, PSC833, FGF19, and alirocumab. A final concentration of 200 μM leupeptin (L2884-1mg; Sigma-Aldrich, St Louis, MO) over 24 hours was used to inhibit lysosomal degradation.
Viability Assays and Determination of EC50
Viability assays were performed using PrestoBlue Assay (A-13261; Life Technologies, Carlsbad, CA) and lactate dehydrogenase assay (MK401; Clontech, Mountain View, CA), as described previously.41 Serial dilutions of compounds were applied (5 steps, 1:5 ratio) ranging from 2 to 1250 μM (gemfibrozil; sc-204764; Santa Cruz Biotechnology, Heidelberg, Germany), from 1 to 625 μM (fenofibrate; sc-204751; Santa Cruz Biotechnology), from 0.01 to 6.25 μM (avasimibe; sc-364315; Santa Cruz Biotechnology), from 0.08 to 50 μM (PSC-833; sc-361298; Santa Cruz Biotechnology), from 0.004 to 2.5 μg/mL (FGF19; 100-32; PeproTech, Hamburg, Germany), or from 0.8 to 500 ng/mL (alirocumab; TAB-719; Creative Biolabs, Shirley, NY) and incubated on cells over 24 hours, 48 hours, and 72 hours.
Determination of EC50 was achieved by using similar experimental settings as previous, with the highest concentration being spared for all compounds but alirocumab. Subsequently, release of viral RNA was monitored using reverse-transcription qPCR (RT-qPCR). Fitting of graphs (dose-response curve) was performed via GraphPad Prism 8.0 (GraphPad Software, San Diego, CA).
Reverse-Transcription qPCR
Intracellular RNA was isolated, reverse transcribed into complementary DNA and analyzed via Maxima SYBR-Green qPCR Kit (K0221; Thermo Fisher Scientific, Braunschweig, Germany) as described previously.41 Primers targeted HEV ORF2 or 60S ribosomal protein L27 (RPL27) as reference gene. For analysis of host genes, primers targeting cholesterol 25-hydroxylase (CH25H); ATP-binding cassette transport A1 (ABCA1); PCSK9; apolipoprotein E (APOE); 25-hydroxycholesterol 7-alpha-hydroxylase (CYP7B1); nuclear receptor subfamily 1, group H, member 4 (NR1H4); and 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase (HMGCR) (sequences see Table 2) were used. Calculation of fold-change values was achieved by the ΔΔCT method.42
Table 2.
Primer Sequences Used for qPCR Analyses
| Name | Sequence | Supplier |
|---|---|---|
| HEV ORF2-FWD | 5′-GGT GGT TTC TGG GGT GAC-3′ | biomers.net |
| HEV ORF2-REV | 5′-AGG GGT TGG TTG GAT GA-3′ | biomers.net |
| RPL27-FWD | 5′-AAA GCT GTC ATC GTG AAG AAC -3′ | biomers.net |
| RPL27-REV | 5′-GCT GCT ACT TTG CGG GGG TAG-3′ | biomers.net |
| CH25H-FWD | 5′-GGT CCT GGA TAT CCT GTG CTC C-3′ | GATC Eurofins |
| CH25H-REV | 5′-GAG TAG CAG GCA GAA CAG GAT GTG G-3′ | GATC Eurofins |
| ABCA1-FWD | 5′-GCT CGC CTG TTC TCA GAT GC-3′ | GATC Eurofins |
| ABCA1-REV | 5′-GGA GAA TGA CAT CAG CCC TCA GC-3′ | GATC Eurofins |
| PCSK9-FWD | 5′-GCC AGG ACA GCA ACC TCT CC-3′ | GATC Eurofins |
| PCSK9-REV | 5′-TTC AGC ACC ACC ACG TAG GTG C-3′ | GATC Eurofins |
| APOE-FWD | 5′-CCA ATC ACA GGC AGG AAG ATG AAG G-3′ | GATC Eurofins |
| APOE-REV | 5′-AGA CAG TGT CTG CAC CCA GC-3′ | GATC Eurofins |
| CYP7B1-FWD | 5′-TTG GCT TCC TTA TCT TGG AGT GGT CC-3′ | GATC Eurofins |
| CYP7B1-REV | 5′-CTG CAT CAT GCT TTC CAA GAG TAT GTC C-3′ | GATC Eurofins |
| NR1H4-FWD | 5′-CAT GCG AAG AAA GTG TCA AGA GTG TCG-3 | GATC Eurofins |
| NR1H4-REV | 5′-CCT GCA TGA CTT TGT TGT CGA GG-3′ | GATC Eurofins |
| HMGCR-FWD | 5′-CCT TAG TGG CTG AAA CAG ATA CCC-3′ | GATC Eurofins |
| HMGCR-REV | 5′-CTG GAT GAT CTC AGC ATC ACT AAG G-3′ | GATC Eurofins |
qPCR, quantitative polymerase chain reaction.
Extracellular, viral RNA or RNA from density gradient fractions were quantified using LightMix Modular Hepatitis E Virus Kit (53-0638-96; TIB MolBiol, Berlin, Germany) in combination with LightCycler Multiplex RNA Virus Master (6754155001; Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions. Prior to RT-qPCR, samples were diluted 1:5 in ddH2O supplemented with 0.1% w/v DEPC.
All RT-qPCR experiments were performed using the LightCycler 480 Instrument II (Roche Diagnostics) and analyzed using the LightCycler480 Software (v1.5.1; Roche Diagnostics).
Density Gradient Centrifugation
Discontinuous density gradients were prepared using 10%–60% w/v iodixanol in ddH2O (OptiPrep; 1114542; Progen Biotechnik, Heidelberg, Germany). In total, 6 layers of iodixanol with decreasing density (steps of 10% iodixanol) were layered above one another with each layer matching a volume of 300 μL. Subsequently, 200 μL of cell culture supernatant of treated or untreated cells were layered on top of the density gradient. Isopycnic centrifugation was performed using a TLS-55 rotor (Beckman Coulter, Brea, CA) over 4 hours at 255,000 g at 4°C. Fractionation occurred from top to bottom with fraction volumes equaling 200 μL. Viral RNA being present in each fraction was analyzed via RT-qPCR. Calculation of percentages of HEV RNA being present in each fraction was achieved by dividing the number of genomes per fraction by the total sum of genomes present in the respective total gradient and subsequent multiplication with 100.
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis and Western Blot
Western blot samples were prepared, subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis, and blotting and detection of primary antibodies were performed as described previously.41 Detection of the viral capsid protein was achieved using a polyclonal rabbit-α-pORF2 antibody (HCD3K129; raised against aa112-608 of recombinant pORF2 protein). Detection of GAPDH (glyceraldehyde 3-phosphate dehydrogenase) (monoclonal mouse anti-GAPDH, sc-32233; Santa Cruz Biotechnology) served as reference protein. Primary antibodies were detected using either polyclonal donkey anti-mouse IgG IRDye 680RD (926-32222; LI-COR Biosciences, Lincoln, NE) or polyclonal donkey anti-rabbit IgG IRDye 800CW (926-32213; LI-COR Biosciences) secondary antibodies. Membranes were imaged using the LI-COR Odyssey infrared imager (LI-COR Biosciences), and band intensities were measured with the ImageStudio Lite software (v5.2; LI-COR Biosciences). Each signal of pORF2 was divided by the GAPDH signal and subsequently referred to the respective experimental control to yield a fold-change value.
Immunofluorescence Microscopy
Treated, ethanol-fixed cells were subjected to immunofluorescence stain under conditions as described previously.41 pORF2 was detected using HCD3K129 (see Western blot). LAMP2 was detected using a monoclonal mouse anti-CD107b (555803; BD Biosciences, San Jose, CA). Secondary antibodies were labeled with either AlexaFluor488 (A21206; Invitrogen, Carlsbad, CA) or AlexaFluor546 (A10036; Invitrogen). Staining cholesterol via filipin III (F4767-1MG; Sigma-Aldrich, Schnelldorf, Germany) was performed on formaldehyde-fixed cells as described previously.43 For imaging of stained, Mowiol-mounted cells, a confocal laser scanning microscope Leica TCS SP8 System with a DMi8 microscope (Leica, Wetzlar, Germany) was used. Images were captured with a 100× magnification oil immersion objective (numerical aperture = 1.4) at room temperature. Image acquisition and analysis was performed with the LAS X Control Software or FIJI.44 A minimum of 7 cells were analyzed for thresholded Mender’s overlap coefficient or corrected total cell fluorescence.
Determination of TCID50
HEV-permissive A549/D3 cells were infected using a serial dilution of cell culture supernatant (7 steps, 1:5 ratio) in 6 replicates for 96 hours. Fixation and blocking were performed similar to immunofluorescence microscopy. Incubation of HCD3K129 was performed overnight at 4°C. Horseradish peroxidase–coupled donkey-α-rabbit IgG (NA934; GE Healthcare, Chicago, IL) served as secondary antibody and subsequent stain was performed using 3-amino-9-ethylcarbazol (30 mM Na-acetate, 12 mM acetic acid, 0.05% w/v 3-amino-9-ethylcarbazol, 0.01% H2O2). The resulting TCID50 was calculated as described previously.45
Patient Data
Lipid levels and HEV viral loads from 42 patients were assessed retrospectively. The group included 33 female and 9 male patients with age varying from 24 to 78 years. All patients received renal transplants with 7 patients receiving a co-transplant along with the kidney. Treatment with immunosuppressants was implemented for all patients (tacrolimus = 35 patients, cyclosporin A = 2 patients, belatacept = 3 patients, azathioprin = 1 patient) and all but 1 patient received a mycophenolate mofetil treatment. Of the analyzed patients, 10 were diagnosed with Diabetes mellitus and 3 were tested positive for a different chronic hepatic infection (hepatitis B or C). Values from each patient were available from a range of 2–37 individual appointments during routine follow-up visits. Retrospective assessment of data from patients with chronic hepatitis E after renal transplantation was approved by the local institutional review board of the ethics committee of Charité Universitätsmedizin Berlin, Germany (approval number EA1/249/16). All patients gave their written informed consent.
Positive testing of an HEV infection included determination of IgG/IgM levels detecting the viral capsid protein or testing for viral genomes in serum via PCR. Exact viral titers were quantified via qPCR, whereas blood lipids were quantified via standard laboratory procedures. Time-periods, where an HEV-targeted therapy was applied (ribavirin or sofosbuvir) were excluded from analysis. Outliers were identified using the ROUT method (Q = 1%).
Microarray Gene Expression Profiling and Analysis
Global gene expression profiling was performed using SurePrint G3 Human Gene Expression 8x60K Microarray Kit (G4851C; Agilent One Color Microarray Technology; Agilent Technologies, Santa Clara, CA). A total of 100 ng of total RNA was used for amplification, which was extracted using RNeasy Mini Kit (217004; QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. Microarray results were extracted using Agilent Feature Extraction Version 11.0.1.1 and analyzed using Genespring 14.9.1 GX software (Agilent Technologies). Genespring 14.9.1 GX software was used for fold-change calculation and for the analysis of pathways of KEGG (Kyoto Encyclopedia of Genes and Genomes.46 Data are deposited at Gene Expression Omnibus (GSE157820; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157820).
Statistical Analyses
All in vitro data presented in this study are depicted as mean value ± SEM. Statistical analyses for these were performed using GraphPad Prism 8.0 using nonpaired t tests, unless stated otherwise. Each dataset that is presented as fold-change value is referred to its suitable, experimental control group. As the control groups were arbitrarily set as 1, a standard deviation for the control groups cannot be reported, as the standardization of the measured values (relative to the control group) was performed for each of the independent assays. Therefore, measurements for the treatment groups in each assay were dependent (matched). N = x displays the number of independent experiments. Unless stated otherwise, experiments are based on biological replicates with n = 3. If no mathematical significance could be determined in in vitro studies, no annotation was made.
Acknowledgments
The authors thank Gert Carra, Robin Oliver Murra, and Sandra Weickhardt for their superb technical support. Further, they thank Daniela Bender, Marie-Luise Herrlein, Catrina Spengler, Tobias Zahn, Michael Basic, Catarina Sabino, and Patrycja Dudek for their topic-related discussions and establishment of methods. Many thanks further go to Reimar Johne (Federal Institute for Risk Assessment, Germany) for providing cells and virus.
CRediT Authorship Contributions
Mirco Glitscher (Conceptualization: Equal; Formal analysis: Lead; Investigation: Lead; Methodology: Equal; Visualization: Lead; Writing – original draft: Lead)
David Heiler Martin (Formal analysis: Equal; Investigation: Equal; Writing – review & editing: Supporting)
Kathrin Woytinek (Formal analysis: Equal; Investigation: Equal)
Benjamin Schmidt (Formal analysis: Supporting; Investigation: Supporting) Denna Tabari, Master (Formal analysis: Equal; Writing – review & editing: Supporting)
Catharina Scholl (Formal analysis: Equal; Writing – review & editing: Supporting) Julia Stingl (Resources: Supporting)
Evelyn Seelow (Formal analysis: Supporting; Investigation: Supporting; Resources: Supporting)
Mira Choi (Investigation: Supporting; Resources: Equal; Writing – review & editing: Supporting)
Eberhard Hildt (Conceptualization: Lead; Funding acquisition: Lead; Methodology: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of Interest The authors disclose no conflicts.
Funding Eberhard Hildt obtained a grant by the LOEWE Center DRUID (Novel Drug Targets against Poverty-Related and Neglected Tropical Infectious Diseases). The sponsors had no role in study design, collection, analysis and interpretation of data, the writing of the report or in the decision to submit the article for publication.
Supplementary Material
References
- 1.Purdy M.A., Harrison T.J., Jameel S., Meng X.-J., Okamoto H., van der Poel W.H.M., Smith D.B. Ictv Report Consortium. ICTV virus taxonomy profile: hepeviridae. J Gen Virol. 2017;98:2645–2646. doi: 10.1099/jgv.0.000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boxall E., Herborn A., Kochethu G., Pratt G., Adams D., Ijaz S., Teo C.-G. Transfusion-transmitted hepatitis E in a ‘nonhyperendemic’ country. Transfus Med. 2006;16:79–83. doi: 10.1111/j.1365-3148.2006.00652.x. [DOI] [PubMed] [Google Scholar]
- 3.Ricci A., Allende A., Bolton D., Chemaly M., Davies R., Fernandez Escamez P.S., Herman L., Koutsoumanis K., Lindqvist R., Nørrung B., Robertson L., Ru G., Sanaa M., Simmons M., Skandamis P., Snary E., Speybroeck N., Ter Kuile B., Threlfall J., Wahlström H., Di Bartolo I., Johne R., Pavio N., Rutjes S., van der Poel W., Vasickova P., Hempen M., Messens W., Rizzi V., Latronico F., Girone R. Public health risks associated with hepatitis E virus (HEV) as a food-borne pathogen. EFSA J. 2017;15:e4886. doi: 10.2903/j.efsa.2017.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minuk G.Y., Sun A., Sun D.F., Uhanova J., Nicolle L.E., Larke B., Giulivi A. Serological evidence of hepatitis E virus infection in an indigenous North American population. Can J Gastroenterol. 2007;21:439–442. doi: 10.1155/2007/289059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Echevarría J.M., González J.E., Lewis-Ximenez L.L., Dos Santos D.R.L., Munné M.S., Pinto M.A., Pujol F.H., Rodríguez-Lay L.A. Hepatitis E virus infection in Latin America: a review. J Med Virol. 2013;85:1037–1045. doi: 10.1002/jmv.23526. [DOI] [PubMed] [Google Scholar]
- 6.Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V., Abraham J., Adair T., Aggarwal R., Ahn S.Y., AlMazroa M.A., Alvarado M., Anderson H.R., Anderson L.M., Andrews K.G., Atkinson C., Baddour L.M., Barker-Collo S., Bartels D.H., Bell M.L., Benjamin E.J., Bennett D., Bhalla K., Bikbov B., Bin Abdulhak A., Birbeck G., Blyth F., Bolliger I., Boufous S., Bucello C., Burch M., Burney P., Carapetis J., Chen H., Chou D., Chugh S.S., Coffeng L.E., Colan S.D., Colquhoun S., Colson K.E., Condon J., Connor M.D., Cooper L.T., Corriere M., Cortinovis M., Courville de Vaccaro K., Couser W., Cowie B.C., Criqui M.H., Cross M., Dabhadkar K.C., Dahodwala N., De Leo D., Degenhardt L., Delossantos A., Denenberg J., Des Jarlais D.C., Dharmaratne S.D., Dorsey E.R., Driscoll T., Duber H., Ebel B., Erwin P.J., Espindola P., Ezzati M., Feigin V., Flaxman A.D., Forouzanfar M.H., Fowkes F.G.R., Franklin R., Fransen M., Freeman M.K., Gabriel S.E., Gakidou E., Gaspari F., Gillum R.F., Gonzalez-Medina D., Halasa Y.A., Harin D., Harrison J.E., Havmoeller R., Hay R.J., Hoen B., Hotez P.J., Hoy D., Jacobsen K.H., James S.L., Jasrasaria R., Jayaraman S., Johns N., Karthikeyan G., Kassebaum N., Keren A., Khoo J.-P., Knowlton L.M., Kobusingye O., Koranteng A., Krishnamurthi R., Lipnick M., Lipshultz S.E., Ohno S.L., Mabweijano J., MacIntyre M.F., Mallinger L., March L., Marks G.B., Marks R., Matsumori A., Matzopoulos R., Mayosi B.M., McAnulty J.H., McDermott M.M., McGrath J., Memish Z.A., Mensah G.A., Merriman T.R., Michaud C., Miller M., Miller T.R., Mock C., Mocumbi A.O., Mokdad A.A., Moran A., Mulholland K., Nair M.N., Naldi L., Venkat Narayan K.M., Nasseri K., Norman P., O’Donnell M., Omer SZb, Ortblad K., Osborne R., Ozgediz D., Pahari B., Pandian J.D., Panozo Rivero A., Perez Padilla R., Perez-Ruiz F., Perico N., Phillips D., Pierce K., Pope C.A., 3rd, Porrini E., Pourmalek F., Raju M., Ranganathan D., Rehm J.T., Rein D.B., Remuzzi G., Rivara F.P., Roberts T., Rodriguez de León F., Rosenfeld L.C., Rushton L., Sacco R.L., Salomon J.A., Sampson U., Sanman E., Schwebel D.C., Segui-Gomez M., Shepard D.S., Singh D., Singleton J., Sliwa K., Smith E., Steeer A., Taylor J.A., Thomas B., Tleyjeh I.M., Towbin J.A., Truelsen T., Undurraga E.A., Venkatsubramanian N., Vijayakumar L., Vos T., Wagner G.R., Wang M., Wang W., Watt K., Weinstock M.A., Weintraub R., Wikinson J.D., Woof A.D., Wulf S., Yeh P.-H., Yip P., Zabetian A., Zheng Z.-J., Lopez A.D., Murray C.J.L. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rein D.B., Stevens G.A., Theaker J., Wittenborn J.S., Wiersma S.T. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology. 2012;55:988–997. doi: 10.1002/hep.25505. [DOI] [PubMed] [Google Scholar]
- 8.Patra S., Kumar A., Trivedi S.S., Puri M., Sarin S.K. Maternal and fetal outcomes in pregnant women with acute hepatitis E virus infection. Ann Intern Med. 2007;147:28–33. doi: 10.7326/0003-4819-147-1-200707030-00005. [DOI] [PubMed] [Google Scholar]
- 9.McPherson S., Elsharkawy A.M., Ankcorn M., Ijaz S., Powell J., Rowe I., Tedder R., Andrews P.A. Summary of the British Transplantation Society UK Guidelines for Hepatitis E and Solid Organ Transplantation. Transplantation. 2018;102:15–20. doi: 10.1097/TP.0000000000001908. [DOI] [PubMed] [Google Scholar]
- 10.Kamar N., Izopet J., Dalton H.R. Chronic hepatitis e virus infection and treatment. J Clin Exp Hepatol. 2013;3:134–140. doi: 10.1016/j.jceh.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouilly J., Chen Q., Siewiera J., Cartron G., Levy C., Dubois M., Al-Daccak R., Izopet J., Jabrane-Ferrat N., El Costa H. Genotype specific pathogenicity of hepatitis E virus at the human maternal-fetal interface. Nat Commun. 2018;9:4748. doi: 10.1038/s41467-018-07200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debing Y., Ramière C., Dallmeier K., Piorkowski G., Trabaud M.-A., Lebossé F., Scholtès C., Roche M., Legras-Lachuer C., de Lamballerie X., André P., Neyts J. Hepatitis E virus mutations associated with ribavirin treatment failure result in altered viral fitness and ribavirin sensitivity. J Hepatol. 2016;65:499–508. doi: 10.1016/j.jhep.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Kamar N., Abravanel F., Garrouste C., Cardeau-Desangles I., Mansuy J.M., Weclawiak H., Izopet J., Rostaing L. Three-month pegylated interferon-alpha-2a therapy for chronic hepatitis E virus infection in a haemodialysis patient. Nephrol Dial Transplant. 2010;25:2792–2795. doi: 10.1093/ndt/gfq282. [DOI] [PubMed] [Google Scholar]
- 14.Nagashima S., Takahashi M., Kobayashi T., Nishizawa T., Nishiyama T., Primadharsini P.P., Okamoto H. Characterization of the quasi-enveloped hepatitis E virus particles released by the cellular exosomal pathway. J Virol. 2017;91 doi: 10.1128/JVI.00822-17. e00822-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagashima S., Takahashi M., Jirintai S., Tanaka T., Nishizawa T., Yasuda J., Okamoto H. Tumour susceptibility gene 101 and the vacuolar protein sorting pathway are required for the release of hepatitis E virions. J Gen Virol. 2011;92:2838–2848. doi: 10.1099/vir.0.035378-0. [DOI] [PubMed] [Google Scholar]
- 16.Nagashima S., Takahashi M., Jirintai, Tanaka T., Yamada K., Nishizawa T., Okamoto H. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J Gen Virol. 2011;92:269–278. doi: 10.1099/vir.0.025791-0. [DOI] [PubMed] [Google Scholar]
- 17.Babst M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr Opin Cell Biol. 2011;23:452–457. doi: 10.1016/j.ceb.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Möbius W., van Donselaar E., Ohno-Iwashita Y., Shimada Y., Heijnen H.F.G., Slot J.W., Geuze H.J. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic. 2003;4:222–231. doi: 10.1034/j.1600-0854.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 19.Holla P., Ahmad I., Ahmed Z., Jameel S. Hepatitis E virus enters liver cells through a dynamin-2, clathrin and membrane cholesterol-dependent pathway. Traffic. 2015;16:398–416. doi: 10.1111/tra.12260. [DOI] [PubMed] [Google Scholar]
- 20.Sobo K., Le Blanc I., Luyet P.-P., Fivaz M., Ferguson C., Parton R.G., Gruenberg J., van der Goot F.G. Late endosomal cholesterol accumulation leads to impaired intra-endosomal trafficking. PLoS One. 2007;2:e851. doi: 10.1371/journal.pone.0000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taneja S., Ahmad I., Sen S., Kumar S., Arora R., Gupta V.K., Aggarwal R., Narayanasamy K., Reddy V.S., Jameel S. Plasma peptidome profiling of acute hepatitis E patients by MALDI-TOF/TOF. Proteome Sci. 2011;9:5. doi: 10.1186/1477-5956-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L., Yesupriya A., Chang M.-H., Teshale E., Teo C.-G. Apolipoprotein E and protection against hepatitis E viral infection in American non-Hispanic Blacks. Hepatology. 2015;62:1346–1352. doi: 10.1002/hep.27938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel I., Ching Companioni R., Bansal R., Vyas N., Catalano C., Aron J., Walfish A. Acute hepatitis E presenting with clinical feature of autoimmune hepatitis. J Community Hosp Intern Med Perspect. 2016;6:33342. doi: 10.3402/jchimp.v6.33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staels B., Dallongeville J., Auwerx J., Schoonjans K., Leitersdorf E., Fruchart J.C. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation. 1998;98:2088–2093. doi: 10.1161/01.cir.98.19.2088. [DOI] [PubMed] [Google Scholar]
- 25.Endo A. The discovery and development of HMG-CoA reductase inhibitors. J Lipid Res. 1992;33:1569–1582. [PubMed] [Google Scholar]
- 26.Nagao K., Maeda M., Mañucat N.B., Ueda K. Cyclosporine A and PSC833 inhibit ABCA1 function via direct binding. Biochim Biophys Acta. 2013;1831:398–406. doi: 10.1016/j.bbalip.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Hirschfield G.M., Chazouillères O., Drenth J.P., Thorburn D., Harrison S.A., Landis C.S., Mayo M.J., Muir A.J., Trotter J.F., Leeming D.J., Karsdal M.A., Jaros M.J., Ling L., Kim K.H., Rossi S.J., Somaratne R.M., DePaoli A.M., Beuers U. Effect of NGM282, an FGF19 analogue, in primary sclerosing cholangitis: a multicenter, randomized, double-blind, placebo-controlled phase II trial. J Hepatol. 2019;70:483–493. doi: 10.1016/j.jhep.2018.10.035. [DOI] [PubMed] [Google Scholar]
- 28.McDonagh M., Peterson K., Holzhammer B., Fazio S. A systematic review of PCSK9 inhibitors alirocumab and evolocumab. J Manag Care Spec Pharm. 2016;22:641–653q. doi: 10.18553/jmcp.2016.22.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llaverías G., Laguna J.C., Alegret M. Pharmacology of the AC AT inhibitor avasimibe (CI-1011) Cardiovasc Drug Rev. 2003;21:33–50. [PubMed] [Google Scholar]
- 30.Fabris C., Federico E., Soardo G., Falleti E., Pirisi M. Blood lipids of patients with chronic hepatitis: differences related to viral etiology. Clin Chim Acta. 1997;261:159–165. doi: 10.1016/s0009-8981(97)06532-7. [DOI] [PubMed] [Google Scholar]
- 31.Lo J. Dyslipidemia and lipid management in HIV-infected patients. Curr Opin Endocrinol Diabetes Obes. 2011;18:144–147. doi: 10.1097/MED.0b013e328344556e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quaye O., Amuzu B.G., Adadey S.M., Tagoe E.A. Effect of hepatitis B virus (HBV) infection on lipid profile in Ghanaian patients. Virology (Auckl) 2019;10 doi: 10.1177/1178122X19827606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bader T., Korba B. Simvastatin potentiates the anti-hepatitis B virus activity of FDA-approved nucleoside analogue inhibitors in vitro. Antiviral Res. 2010;86:241–245. doi: 10.1016/j.antiviral.2010.02.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kühnl A., Musiol A., Heitzig N., Johnson D.E., Ehrhardt C., Grewal T., Gerke V., Ludwig S., Rescher U. Late endosomal/lysosomal cholesterol accumulation is a host cell-protective mechanism inhibiting endosomal escape of influenza A virus. mBio. 2018;9:e01345–e01348. doi: 10.1128/mBio.01345-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagace T.A., Curtis D.E., Garuti R., McNutt M.C., Park S.W., Prather H.B., Anderson N.N., Ho Y.K., Hammer R.E., Horton J.D. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price S., Hinton R., Mitchell F., Hall D., Grasso P., Blane G., Bridges J. Time and dose study on the response of rats to the hypolipidaemic drug fenofibrate. Toxicology. 1986;41:169–191. doi: 10.1016/0300-483x(86)90198-8. [DOI] [PubMed] [Google Scholar]
- 37.Ehrlich A, Uhl S, Ioannidis K, Hofree M, tenOever BR, Nahmias Y. The SARS-CoV-2 transcriptional metabolic signature in lung epithelium. Cell Metabolism. 10.2139/ssrn.3650499. [DOI]
- 38.Nakabayashi H., Taketa K., Miyano K., Yamane T., Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 39.Schemmerer M., Apelt S., Trojnar E., Ulrich R.G., Wenzel J.J., Johne R. Enhanced replication of hepatitis E virus strain 47832c in an A549-derived subclonal cell line. Viruses. 2016;8:267. doi: 10.3390/v8100267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johne R., Reetz J., Ulrich R.G., Machnowska P., Sachsenröder J., Nickel P., Hofmann J. An ORF1-rearranged hepatitis E virus derived from a chronically infected patient efficiently replicates in cell culture. J Viral Hepat. 2014;21:447–456. doi: 10.1111/jvh.12157. [DOI] [PubMed] [Google Scholar]
- 41.Glitscher M., Himmelsbach K., Woytinek K., Johne R., Reuter A., Spiric J., Schwaben L., Grünweller A., Hildt E. Inhibition of hepatitis E virus spread by the natural compound silvestrol. Viruses. 2018;10:301. doi: 10.3390/v10060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Stoeck I.K., Lee J.-Y., Tabata K., Romero-Brey I., Paul D., Schult P., Lohmann V., Kaderali L., Bartenschlager R. Hepatitis C virus replication depends on endosomal cholesterol homeostasis. J Virol. 2018;92 doi: 10.1128/JVI.01196-17. e01196-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J Virol. 2016;5:85–86. doi: 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.