Abstract
Computer-aided diagnosis has been extensively investigated for more rapid and accurate screening during the outbreak of COVID-19 epidemic. However, the challenge remains to distinguish COVID-19 in the complex scenario of multi-type pneumonia classification and improve the overall diagnostic performance. In this paper, we propose a novel periphery-aware COVID-19 diagnosis approach with contrastive representation enhancement to identify COVID-19 from influenza-A (H1N1) viral pneumonia, community acquired pneumonia (CAP), and healthy subjects using chest CT images. Our key contributions include: 1) an unsupervised Periphery-aware Spatial Prediction (PSP) task which is designed to introduce important spatial patterns into deep networks; 2) an adaptive Contrastive Representation Enhancement (CRE) mechanism which can effectively capture the intra-class similarity and inter-class difference of various types of pneumonia. We integrate PSP and CRE to obtain the representations which are highly discriminative in COVID-19 screening. We evaluate our approach comprehensively on our constructed large-scale dataset and two public datasets. Extensive experiments on both volume-level and slice-level CT images demonstrate the effectiveness of our proposed approach with PSP and CRE for COVID-19 diagnosis.
Keywords: Automated COVID-19 diagnosis, Chest CT images, Periphery-aware spatial prediction (PSP), Contrastive representation enhancement (CRE)
1. Introduction
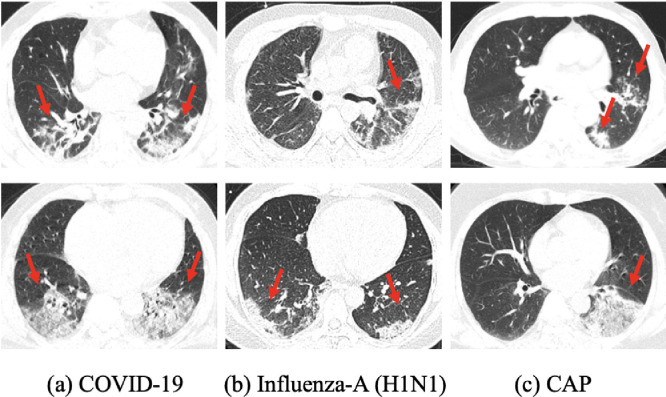
Coronavirus Disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is spreading quickly worldwide. To date (December 29th 2020), more than 79 million confirmed cases have been reported in over 200 countries and territories, with a mortality rate of 2.2% [1]. Considering the pandemic of COVID-19, early detection and treatment are of great importance to the slowdown of viral transmission and the control of the disease. Currently, the Reverse Transcription-Polymerase Chain Reaction (RT-PCR) testing remains the diagnostic gold standard of COVID-19. As a reliable complement to RT-PCR assay, thoracic computed tomography (CT) has also been recognized to be a major tool for clinical diagnosis in many hard-hit regions. As shown in Fig. 1 (a), some characteristic radiological patterns can be clearly detected in chest CT images, including ground-glass opacity (GGO), crazy-paving pattern, and later consolidation. In addition, bilateral, peripheral, and lower zone predominant distributions are mostly observed [2]. To assist radiologists in more efficient pneumonia screening during the outbreak of COVID-19, we aim to develop a deep learning based approach to automatically diagnose COVID-19 in complex diagnostic scenarios using chest CT images.
Fig. 1.
Some examples of (a) COVID-19, (b) influenza-A (H1N1), and (c) CAP cases in chest CT images. The red arrows indicate the locations of the main lesion regions. It can be seen that there are similar lesions among different types of pneumonia, which increases the challenge for the accurate diagnosis of COVID-19. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Deep learning approaches have demonstrated significant improvement in the field of medical image analysis. In fighting against COVID-19, these approaches have also been widely applied to the lung and infection region segmentation [3], [4], [5], [6], as well as the clinical diagnosis and assessment [7], [8], [9], [10]. The review articles [11], [12] have made thorough investigations on the advanced deep learning researches and techniques associated with COVID-19. However, there are still remaining issues to be addressed: 1) Some typical spatial patterns are valuable clinical cues in distinguishing COVID-19 from other categories of pneumonia. For instance, subpleural peripheral distributions are often observed in COVID-19 CT images, whereas peri-bronchial distributions are common in influenza pneumonia. However, there are merely a few researches that attempt to combine the domain knowledge of spatial patterns into deep learning networks for a more comprehensive COVID-19 diagnosis. 2) Despite a new disease, COVID-19 still has similar manifestations with various types of pneumonia, as illustrated in Fig. 1. For example, ground-glass opacity (GGO) is one of the characteristic CT findings of COVID-19, but it also occurs in other pneumonia such as adenovirus pneumonia, mycoplasmal pneumonia, and allergic pneumonia. It significantly increases the challenge of precise COVID-19 diagnostic performance. To address the first issue, it is critical to introduce the domain knowledge on specific spatial patterns of pneumonia into deep diagnosis networks explicitly. To address the second issue, it is important to learn more discriminative representations of various types of pneumonia that can capture the intra-class similarity and inter-class difference.
In this paper, we propose a novel COVID-19 diagnosis approach based on CNNs with spatial pattern prior knowledge and representation enhancement mechanism to distinguish COVID-19 in the complex scenario of multi-type pneumonia diagnosis. Based on the spatial patterns of COVID-19, we design an unsupervised Periphery-aware Spatial Prediction (PSP) to endow our pre-trained network with spatial location awareness. We then construct an adaptive joint learning framework that employs Contrastive Representation Enhancement (CRE) as an auxiliary task for a more discriminative and accurate COVID-19 diagnosis. In summary, the main contributions of this paper that significantly differ from other earlier works are the following three aspects: 1) We propose a novel diagnosis approach to efficiently diagnose COVID-19 from H1N1, CAP, and healthy cases. To the best of our knowledge, our approach first exploits the spatial pattern prior and representation enhancement on contrastive learning for further improving the diagnostic performance. 2) We integrate spatial patterns by introducing a Periphery-aware Spatial Prediction (PSP) task to pre-train the network without any human annotations. We also construct an adaptive Contrastive Representation Enhancement (CRE) mechanism by discriminating pneumonia-type guided positive and negative pairs in the latent space. 3) Extensive experiments and analyses are conducted on our constructed large-scale dataset and two public datasets. The quantitative and qualitative results demonstrate the superiority of our proposed model in COVID-19 diagnosis with both 3D and 2D CT data types.
The remainder of the paper is organized as follows. Section 2 reviews some related works. In Section 3, we describe in detail the periphery-aware COVID-19 diagnosis framework with contrastive representation enhancement mechanism. Section 4 discusses the experimental settings, results, comparisons, and analyses on our constructed CT dataset and two public datasets. Finally, conclusions and future work are given in Section 5.
2. Related work
2.1. Deep learning based COVID-19 screening
2.1.1. Categories of approach
During the outbreak of COVID-19, numerous deep learning approaches have been developed for COVID-19 screening in chest CT images. The recent works generally fall into two categories. The first category is to use the segmented infection regions in CT images for the COVID-19 diagnosis prediction. For example, Chen et al. [5] trained a UNet++ model to detect the suspicious lesions on three consecutive CT scans for COVID-19 diagnosis. Zhang et al. [13] and Jin et al. [6] both developed diagnosis systems following a combined “segmentation-classification” model pipeline. This category of research work requires a substantial number of segmentation annotations, which are, however, extremely difficult and time-consuming to acquire.
The second category uses 3D volume-level or 2D slice-level supervision to train a classification model to diagnose COVID-19 directly. Wang et al. [9] built a modified transfer-learning Inception model to distinguish COVID-19 from typical viral pneumonia. Ying et al. [10] developed a deep learning based system (DeepPneumonia) to detect COVID-19 patients from bacterial pneumonia and healthy controls. In addition, Wang et al. [3] proposed a 3D DeCoVNet to detect COVID-19, which took a CT volume with its lung mask as input. Huang et al. [14] proposed a belief function based CNN to produce more reliable and explainable diagnosis results. Since it is expensive to annotate the labels in a voxel-wise or pixel-wise manner, we follow the second category to diagnose COVID-19 from H1N1, CAP, and normal controls with only volume-level or slice-level supervision.
2.1.2. Multi-type pneumonia classification
Confronting the challenge of distinguishing COVID-19 from other types of pneumonia with similar manifestations, several works have made great efforts to improve the COVID-19 diagnostic performance. Li et al. [4] developed a 3D COVNet based on ResNet50, which extracted both 2D local and 3D global features to classify COVID-19, CAP, and non-pneumonia. Ouyang et al. [15] proposed an attention module to focus on the infections and a sampling strategy to mitigate imbalanced infection sizes for COVID-19 and CAP diagnosis. Wang et al. [16] established a framework with prior-attention residual learning (PARL) blocks, where the attention maps were generated for COVID-19 and Interstitial Lung Disease identification. Xu et al. [17] designed a location-attention model to categorize COVID-19, Influenza-A viral pneumonia, and healthy cases, which took the relative distance-from-edge of segmented lesion candidates as an extra weight in a fully connected layer. Besides, Qian et al. [18] proposed a Multi-task Multi-slice Deep Learning System (M3Lung-Sys), which consisted of slice-level and patient-level classification networks for multi-class pneumonia screening.
Although these attempts have demonstrated their validity in COVID-19 screening, some drawbacks remain in clinical application. The spatial pattern, as an important basis for clinical diagnosis, has not been effectively integrated into the deep network. Additionally, the intra-class similarity and inter-class difference between various pneumonia need to be further explored.
2.2. Contrastive learning
Contrastive learning, as the name implies, aims to learn data representations by contrasting positive and negative samples. It is at the core of recent works on self-supervised learning, generally using a contrastive loss called the InfoNCE loss. Contrastive Predictive Coding (CPC) [19] was a pioneering work that first put forward the concept of the InfoNCE loss. It learned representations by predicting “future” information in a sequence. Moreover, instance-level discrimination has become a popular paradigm in recent contrastive learning works. It aimed to learn representations by imposing transformation invariances in the latent space. Two transformed images from the same image are regarded as the positives, which should be closer together than those from different images in the representation space. Several recent works, such as SimCLR [20] and MoCo [21], have followed this pipeline and achieved great empirical successes in self-supervised representation learning.
There are also some researches that have attempted to explore the potential of contrastive learning on COVID-19 screening. He et al. [22] proposed a Self-Trans approach, which integrated contrastive self-supervised learning and transfer learning to pre-train the networks. Wang et al. [23] proposed a redesigned Covid-Net and used contrastive learning to tackle the cross-site domain difference when diagnosing COVID-19 on heterogeneous CT datasets. Chen et al. [24] adopted a prototypical network for the few-shot COVID-19 diagnosis, which was pre-trained by the momentum contrastive learning method [21]. Li et al. [25] presented a contrastive multi-task CNN which can improve the generalization on unseen CT or X-ray samples for COVID-19 diagnosis. In our work, we fully exploit contrastive learning to obtain more discriminative representations between COVID-19 and other types of pneumonia, i.e. H1N1 and CAP, in chest CT images. We further introduce the pneumonia-type information into contrastive learning for better exploration of intra-class similarity and inter-class difference. Particularly, we adopt it as an auxiliary learning task that can effectively improve the classification performance of COVID-19 from other pneumonia.
3. Methodology
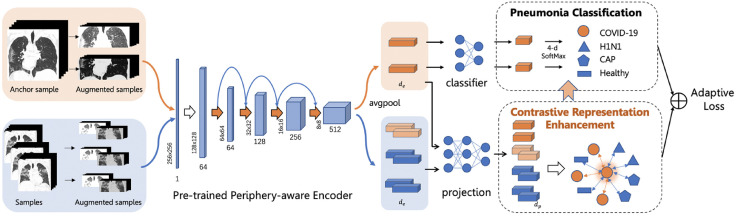
To make a more accurate diagnosis of COVID-19 from other pneumonia, we particularly construct a periphery-aware deep learning network with contrastive representation enhancement (CRE), as illustrated in Fig. 2 . It takes a CT sample as the input and outputs the pneumonia classification result in an end-to-end manner. We first perform the Periphery-aware Spatial Prediction (PSP) to obtain the pre-trained encoder network. Then the CRE, as an auxiliary task, is adaptively combined to the pneumonia classification for more discriminative representation learning. We utilize ResNet [26] as the backbone architecture since it has proven effective in the previous works.
Fig. 2.
An overview of our network architecture for COVID-19 diagnosis (best viewed in colour). Each augmented CT image is fed into the pre-trained periphery-aware encoder, generating representations in -dimension. A classifier is learned on top of the representations for pneumonia classification. Meanwhile, the representations are further enhanced in a contrastive learning manner, discriminating the positive (orange arrows) and negative (blue arrows) pairs after being mapped by the projection network into the -dimensional space. The enhanced representations can promote more precise diagnostic performance. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.1. Periphery-aware spatial prediction
To integrate the spatial patterns of infections into the pneumonia classification, we design an unsupervised Periphery-aware Spatial Prediction (PSP) to pre-train the encoder network without any human annotations. Given a CT image and its automatically generated boundary distance map , we train a neural network to solve the boundary distance prediction problem . We would like to obtain a feature encoder that is endowed with spatial location awareness by learning the PSP.
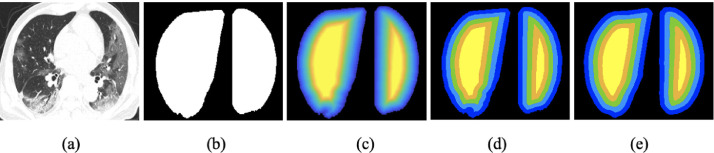
Inspired by the radiological signs that COVID-19 infections mostly appear in the subpleural area, the boundary distance map is created to represent the location information about whether a pixel belongs to the interior of the lung region as well as the distance to the region boundary. As shown in Fig. 3 , given a CT image, we first generate the segmented lung region mask by our segmentation algorithm without manual labeling, detailed in Section 4 (Preprocessing). Then, let denote the set of pixels on the region boundary, and is the set of pixels inside the lung region. For every pixel in the mask, we compute the truncated boundary distance as:
| (1) |
where is the relative Euclidean distance between the pixels and . It is divided by the segmented lung volume and then normalized to the interval . denotes the truncation threshold, which is the largest distance to represent. The distance is additionally weighted by the sign function to represent whether the pixel lies inside or outside the lung region. This prediction task can be trained with regression loss, as shown in Eq. (2).
| (2) |
Fig. 3.
An illustration of the boundary distance map (best viewed in colour). (a) CT image; (b) lung mask; (c) truncated boundary distance map; (d) truncated and quantized boundary distance map; and (e) our predicted boundary distance map. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Another feasible solution is to translate the dense map prediction problem to a set of pixel-wise binary classification tasks. We quantize the values in the pixel-wise map into uniform bins. To be specific, we encode the truncated distance of the pixel using a -dimensional binary vector as:
| (3) |
where is the distance value corresponding to the th bin. The continuous pixel-wise map is now converted into a set of binary pixel-wise maps by this one-hot encoding. A standard cross-entropy loss between the prediction and ground truth is used, as shown in Eq. (4).
| (4) |
It is discovered that the classification loss is more effective for the boundary distance prediction task [27]. In our experiments, we use and . We employ the UNet-style prediction network with an encoder-decoder architecture. We adopt ResNet as the encoder and construct the decoder as a mirrored version of the encoder by replacing the pooling layers with bilinear upsampling layers. A typical predicted result is shown in Fig. 3. After solving the PSP problem, we discard the decoder and take the pre-trained periphery-aware encoder for the downstream pneumonia classification.
3.2. Contrastive representation enhancement
To learn more discriminative representations of different pneumonia, we develop an adaptive Contrastive Representation Enhancement (CRE) as an auxiliary task for a precise COVID-19 diagnosis. Our novel network architecture with CRE is comprised of the following components.
-
•
A data augmentation function, , which transforms an input CT sample into a randomly augmented sample . We generate randomly augmented volumes from each input CT sample. To be concrete, we sequentially apply three augmentations for CT samples: 1) random cropping on the transverse plane followed by resizing back to the original resolution; 2) random cropping on the vertical section to a fixed depth ; and 3) random changes in brightness and contrast. We set in our model, and each augmented CT sample represents a different part of the original sample.
-
•
An encoder network, , which maps the augmented CT sample to a representation vector in the -dimensional latent space. The augmented CT samples of different pneumonia categories share the same encoder and generate pairs of representation vectors. In principle, any CNN architecture can be adopted as the encoder here, of which the outputs of the average pooling layer are used as the representation vectors. Then, the representations are normalized to the unit hypersphere.
-
•
A projection network, , which is used to map the representation vector to a relative low-dimension vector for the contrastive loss computation. A multi-layer perception (MLP) can be employed as the projection network. This vector is also normalized to the unit hypersphere, which enables the inner product to measure distances in the projection space. This network is only used for training the contrastive loss.
-
•
A classifier network, , which classifies the representation vector to the pneumonia prediction . It is composed of the fully connected layer and the Softmax operation.
Given a minibatch of randomly sampled CT images and their pneumonia-type labels , we can generate a minibatch of samples after performing data augmentations, where and are two random augmented CT samples of , and .
In this work, our goal is to enhance the representation similarity within the same pneumonia, and simultaneously increase the inter-class difference between different types of pneumonia. Therefore, we introduce the information of pneumonia-type labels into contrastive learning. Different from the general definition of positive and negative pairs in contrastive self-supervised learning [20], [21], we redefine the positives as any two augmented CT samples from the same category, whereas the CT samples from different classes are considered as negative pairs. Assume that two samples and from the same class are considered as a positive pair, the contrastive loss is optimized when is similar to its positive sample and dissimilar to the other negative samples. Let be the index of an arbitrary augmented sample, the contrastive InfoNCE loss function [28] is defined as:
| (5) |
| (6) |
where stands for the representation vector of through the encoder and projection network , is an indicator function, and denotes a scalar temperature hyper-parameter. Samples which have the same label as sample (i.e., ) are the positives. is the total number of samples in a minibatch that have the same label . Besides, the inner product is used to measure the similarity between the normalized vectors and in the -dimensional space. Within the context of Eq. (6), the encoder is trained to maximize the similarity between positive samples and from the same class , while simultaneously minimizing the similarity between negative pairs. We compute the probability using a Softmax, and Eq. (5) sums the loss over all pairs of indices and . As a result, the encoder learns to discriminate positive and negative samples for enhancing the intra-class similarity and inter-class difference.
Finally, we learn the classifier to predict the pneumonia classification results using the standard cross-entropy loss , which is defined as:
| (7) |
| (8) |
where denotes the one-hot vector of ground truth label, and is predicted probability of the sample .
3.3. Adaptive joint training for COVID-19 diagnosis
Different from the previous works [20], [21], [28] which separate the contrastive learning and classification into two stages, we train the network by both loss functions, i.e., for the representation enhancement and for the pneumonia classification. To greatly improve the pneumonia classification accuracy by the enhanced representations from CRE, we particularly design a combined objective function with adaptive weights [29] to balance the two loss functions for better joint learning performance:
| (9) |
where and are utilized to learn the relative weights of the losses and , adaptively. We derive our joint loss function based on maximizing the Gaussian likelihood with homoscedastic uncertainty. The detailed derivation is explained in the Appendix section. Algorithm 1 summarizes our proposed diagnosis approach.
Algorithm 1.
Adaptive Joint Learning.
4. Experiments
4.1. Dataset
In our work, we particularly construct a large-scale chest CT dataset with two significant contributions. 1) Our dataset provides the annotations of both 3D volume-level and 2D slice-level CT samples for COVID-19 diagnosis research and development. 2) We specially select four classes of CT samples to construct our dataset, including COVID-19, Influenza-A (H1N1), CAP, and normal controls. These cases are extremely significant in both clinical applications and epidemiological studies. Generally, clinicians need not only to determine whether a subject contracts pneumonia, but also to further identify the type of pneumonia. Therefore, discriminating COVID-19 from CAP automatically offers reliable clues to assist radiologists in clinical diagnosis, as COVID-19 is highly infectious. The Novel Influenza-A H1N1 viral pneumonia in 2009 pandemic is also an infectious disease with high mortality. Our research of COVID-19 and H1N1 differentiation is significant, as it aims to help epidemiologists and clinicians to summarize historical experiences and obtain valuable references for the epidemic prevention in the future. Besides, we also conduct experiments on two public COVID-19 CT datasets, namely Covid-CT dataset [30] and CC-CCII dataset [13]. In the following sections, we introduce our constructed 3D and 2D CT datasets and the public datasets in detail.
4.1.1. In-house CT dataset
3D CT dataset. The protocol of this study was approved by the Ethics Committee of Shanghai Public Health Clinical Center, Fudan University. We searched and selected chest CT exams from the hospital Picture Archiving and Communication System (PACS) and finally constructed our dataset. This dataset comprises a total of 801 3D chest CT exams from 707 subjects, including 238 samples from 236 COVID-19 patients, 191 samples from 99 H1N1 patients, 122 samples from 122 CAP patients, and 250 normal controls with non-pneumonia. All chest CT exams are stored in the DICOM file format. The detailed statistics are summarized in Table 1 . Specifically, all the COVID-19 cases were acquired from Jan 20th to Feb 22nd, 2020. Every patient was confirmed with an RT-PCR testing kit, and the cases that had no manifestations on the chest CT images were excluded. The H1N1 cases were acquired from May 24th, 2009 to Jan 18th, 2010, which were confirmed according to the diagnostic criteria of the National Health Commission of the Peoples Republic of China. Besides, the CAP and healthy cases were randomly selected between Jan 3rd, 2019 and Jan 30th, 2020, and the CAP cases were confirmed positive by bacterial culture. To ensure the diversity of our dataset, we retained an at least three days gap between infected CT samples if they were taken from the same patient.
Table 1.
The statistics of our constructed dataset. Ages are reported as mean standard deviation.
| Category | COVID-19 | H1N1 | CAP | Healthy | Total |
|---|---|---|---|---|---|
| Patients | 236 | 99 | 122 | 250 | 707 |
| Sex (M/F) | 139/97 | 58/41 | 72/50 | 131/119 | 400/307 |
| Age | 5216 | 2915 | 4917 | 3212 | 4115 |
| 3D Exams | 238 | 191 | 122 | 250 | 801 |
| 2D Exams | 29,964 | 25,417 | 12,173 | 21,180 | 88,734 |
2D CT Dataset. In addition to the 3D volume-level labels obtained by clinical diagnosis reports, our constructed dataset particularly provides the annotations on 2D slice-level images. A slice image from three types of pneumonia is annotated with the corresponding CT volume’s category if it is determined to have infected lesions; otherwise, it is randomly selected as “Healthy”. The slices taken out of every three slices from healthy controls are annotated as “Healthy”. In this way, we obtain a total of 88,734 CT slices of the four classes, which is a substantial amount of CT data compared with all the existing public COVID-19 CT datasets. Moreover, the slice-level labels are annotated by five professional experts and supervised by two radiologists with clinical experience of more than five years. These valuable annotations of our dataset make it versatile for both 3D and 2D diagnosis research and development.
4.1.2. Covid-CT dataset
The Covid-CT dataset [30] is comprised of 349 CT scans for COVID-19 and 397 CT scans that are normal or contain other types of pneumonia. The COVID-19 scans are collected from 760 preprints posted from Jan 19th to Mar 25th, 2020. The non-COVID-19 scans are selected from the PubMed Central (PMC) search engine and a public medical image database named MedPix, which contains CT scans with various diseases. All images are stored in the PNG file format.
4.1.3. CC-CCII dataset
The CC-CCII dataset [13] contains 444,034 slices from 4,356 CT scans of 2,778 people, including 1,578 COVID-19 scans, 1,614 common pneumonia scans, and 1,164 normal control scans. Patients were randomly divided into the training set (80%), the validation set (10%), and the testing set (10%). Among all the CT slices, a section of 750 CT slices from 150 COVID-19 patients was manually segmented into the background, lung field, ground-glass opacity (GGO), and consolidation (CL). All images are stored in the PNG, JPG, and TIFF file formats.
4.2. Evaluation metrics
We adopt several metrics to measure the diagnostic performance of models thoroughly. To be concrete, we report the Sensitivity, Specificity, and Area Under Curve (AUC) for each class, as well as Accuracy, Macro-average AUC, and F1 score for overall comparison. Sensitivity and Specificity measure the proportion of positives and negatives that are correctly identified, respectively. Accuracy is the percentage of correct predictions among the total number of samples. F1 score is the harmonic mean of the precision and recall. We further present statistical analysis based on the independent two-sample -test. Besides, we visualize the Gradient-weighted Class Activation Mapping (Grad-CAM) [31], which identifies the infected regions that are most relevant to the predictions.
4.3. Preprocessing
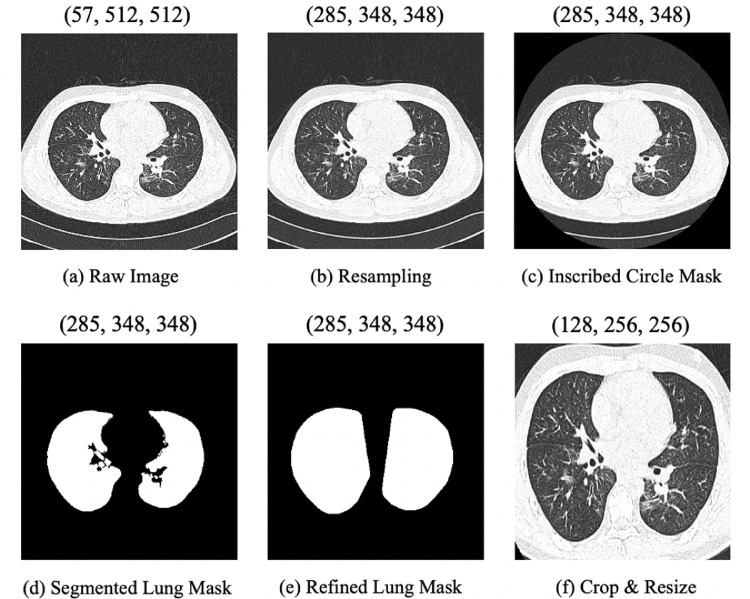
In this work, raw data is converted into Hounsfield Unit (HU), which is a standard quantitative scale for describing radiodensity. As Fig. 4 illustrates, our proposed preprocessing procedure is as follows. (a) Loading the raw CT volume. (b) Resampling the CT volume to the unified (1, 1, 1) mm spacing using the linear interpolation algorithm, since their original resolutions and slice thicknesses are different. (c) Applying an inscribed circle mask on each slice before the lung region segmentation, in order to make the following segmentation algorithm more resistant to “trays” on which the patient lays. (d) Extracting the preliminary lung mask from the CT volume based on the lung region segmentation algorithm [32]. The threshold is set to 320 in this work. (e) Computing a convex hull and performing dilation to get the refined lung mask, as some lesions attached to the outer wall of the lung are not included in the mask obtained in the previous step. (f) Generating a bounding box to crop the lung region based on the refined lung mask, removing most of the redundant background information. Then the volume is resized to 256256 on the transverse section and 128 in the depth-axis. Finally, we set the window width (WW) and the window level (WL) as 600 and -600, and linearly transform the CT volume to the interval for intensity normalization.
Fig. 4.
An example of H1N1 CT images in the preprocessing procedure. The tuple above each figure indicates the shape (D, H, W) of the preprocessed 3D volume in each step.
With the unsupervised lung region segmentation algorithm, the burden of human labeling can be largely reduced, and the majority of lung regions in CT images can be segmented accurately. However, the results may be incorrect in the two cases: 1) severe samples, and 2) lesions attached to the outer wall of the lung. For the second case, although we compute a convex hull and perform dilation to refine most imperfect segmented masks, a small part of inaccurate results still exists. In our work, the segmented mask from the unsupervised segmentation algorithm serves a dual purpose. 1) For the preprocessing, the segmented mask is used to generate the expanded bounding box of the region of interest. The region inside the bounding box is cropped as the input. In this step, the inaccurate mask does not affect the accuracy of bounding box localization. 2) For the PSP task, the segmented mask is used to generate the boundary distance map, as the label of boundary distance prediction. The experimental results show that the network can correctly predict the boundary distance map in most cases after learning PSP with a large number of samples. The network makes mistakes in a few stubborn cases which hold only a small proportion (about 4%) in our dataset.
4.4. Implementation details
For our 3D dataset, we use 3-fold cross validation. For our 2D dataset, we randomly divide it into training and testing sets with an approximate ratio of 2:1, comprising 59,413 and 29,321 slice images, respectively. In the training process, we adopt the random oversampling strategy [33] to mitigate the imbalance of training samples across different classes. The 3D ResNet18 and 2D ResNet50 models are adopted as backbone architectures for 3D and 2D datasets, respectively. The parameter is 512 for 3D ResNet18 and 2,048 for 2D ResNet50. The value of parameter is 128. We optimize the network using the Adam algorithm with a weight decay of . We present the detailed training configurations, including training data, input size, max training epoch, and training time of our models in Table 2 . The shape of input size is (N, C, D, H, W) for 3D data and (N, C, H, W) for 2D data, where N, C, D, H, W stand for the number of batch-size, channel, slice, height, and width, respectively. The initial learning rate is set to 0.001 and then divided by 10 at 30% and 80% of the total number of training epochs. Our networks are implemented in PyTorch and run on four NVIDIA GeForce GTX 1080 Ti GPUs. Our code will be publicly available at https://github.com/FDU-VTS/Periphery-aware-COVID.
Table 2.
The detailed training configurations, including training data, input size, max training epoch, and training time of our models.
| Methods | Data | Input Size | Epoch | Time |
|---|---|---|---|---|
| PSP pre-train (3D) | 571 | (8,1,64,256,256) | 40 | 6h |
| CRE classification (3D) | 571 | (8,1,64,256,256) | 300 | 10h |
| PSP pre-train (2D) | 59,413 | (64,1,256,256) | 40 | 18h |
| CRE classification (2D) | 59,413 | (64,1,256,256) | 100 | 8.5h |
4.5. Results on our 3D dataset
4.5.1. Comparison with existing methods
Firstly, we compare our diagnosis approach with several existing COVID-19 diagnosis methods [3], [4] based on 3D CT samples. The COVNet [4] takes CT slices as input, generates a series of 2D local features based on ResNet50, and combines them to 3D global features for COVID-19 diagnosis. The 3D DeCovNet [3] is composed of Stem, ResBlocks, and Progressive classifier, which inputs a CT volume with its lung mask and outputs the diagnosis probabilities. We retrain their approaches on our 3D dataset, and the related comparison results are presented in Table 3 .
Table 3.
The quantitative results on our 3D dataset, which are the mean values of 3-fold cross validation. We report Sensitivity (Sen.), Specificity (Spec.) and AUC for each class, as well as Accuracy, AUC, and F1 score for overall performance (unit:%). The values in brackets are 95% confidence intervals.
| Methods | COVID-19 |
H1N1 |
CAP |
Healthy |
Acc | AUC | F1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sen. | Spec. | AUC | Sen. | Spec. | AUC | Sen. | Spec. | AUC | Sen. | Spec. | AUC | ||||
| COVNet [4] | 87.00 | 79.89 | 92.18 | 91.08 | 97.88 | 98.63 | 65.57 | 88.51 | 86.59 | 33.94 | 90.76 | 79.25 | 68.16 [65.42, 70.79] | 89.39 | 66.66 |
| DeCovNet [3] | 85.73 | 92.03 | 95.14 | 98.94 | 99.67 | 99.89 | 67.22 | 96.32 | 91.43 | 82.04 | 91.11 | 94.53 | 84.89 [82.77, 86.89] | 95.44 | 83.94 |
| Ours (Res18+PSP) | 87.37 | 96.10 | 97.87 | 98.95 | 99.18 | 99.47 | 65.63 | 97.78 | 93.84 | 91.61 | 90.38 | 96.88 | 88.14 [86.14, 89.89] | 97.20 | 86.71 |
| Ours (Res18+CRE) | 90.73 | 96.45 | 98.20 | 98.95 | 99.34 | 99.84 | 66.40 | 98.82 | 94.16 | 95.60 | 92.02 | 96.75 | 90.51 [88.76, 92.13] | 97.42 | 88.95 |
| Ours (Res18+PSP+CRE) | 90.75 | 97.51 | 98.63 | 98.75 | 99.34 | 99.94 | 72.15 | 98.08 | 94.60 | 92.79 | 91.84 | 97.18 | 90.51 [88.76, 92.26] | 97.78 | 89.42 |
| Res18 (=2, =64) | 90.32 | 91.82 | 96.73 | 98.43 | 98.69 | 99.66 | 59.00 | 99.11 | 95.67 | 88.79 | 92.01 | 95.91 | 87.01 [85.14, 88.89] | 97.14 | 85.32 |
| Res18 (=1, =64) | 87.38 | 96.27 | 97.65 | 97.89 | 98.20 | 98.95 | 52.50 | 98.67 | 91.87 | 98.00 | 89.84 | 96.71 | 87.89 [86.02, 89.76] | 96.54 | 84.90 |
| Res18 (=1, =128) | 87.37 | 93.95 | 96.92 | 100.00 | 99.67 | 100.00 | 64.70 | 97.05 | 92.54 | 88.00 | 91.46 | 96.15 | 87.13 [85.14, 89.14] | 96.58 | 85.67 |
| Res18+CRE () | 90.32 | 93.96 | 98.00 | 98.95 | 98.36 | 99.79 | 62.38 | 98.08 | 95.16 | 89.98 | 92.92 | 97.12 | 88.01 [86.02, 89.89] | 97.70 | 86.12 |
| Res18+CRE () | 89.47 | 95.38 | 98.20 | 99.48 | 98.85 | 99.97 | 63.09 | 98.38 | 94.82 | 90.79 | 90.93 | 96.72 | 88.26 [86.39, 90.14] | 97.59 | 86.64 |
| Res18+CRE () | 90.32 | 94.49 | 97.90 | 99.48 | 99.01 | 99.86 | 63.05 | 98.53 | 94.75 | 90.41 | 91.65 | 96.67 | 88.39 [86.52, 90.14] | 97.47 | 86.76 |
| Res10+CRE | 91.58 | 93.96 | 98.28 | 99.47 | 98.85 | 99.94 | 65.61 | 98.53 | 95.77 | 89.21 | 92.92 | 96.98 | 88.76 [86.89, 90.51] | 97.65 | 87.36 |
| Res34+CRE | 91.16 | 90.41 | 96.36 | 99.47 | 98.69 | 99.80 | 58.23 | 97.79 | 92.41 | 82.41 | 92.73 | 96.13 | 85.39 [83.15, 87.39] | 96.40 | 83.62 |
Ours (Res18+PSP+CRE) is with =2, =64, the learned adaptive weight , and ResNet18 backbone.
As can be apparently seen, our method “Ours (Res18+PSP+CRE)” significantly outperforms these existing methods (p<0.001) on the 3D CT dataset. For COVID-19 diagnosis, our method retains a strong classification performance with 90.75% Sen., 97.51% Spec., and 98.63% AUC. It indicates that the predictions of our model can provide assistance with high reliability and practicality for radiologists in clinical COVID-19 diagnosis. Besides, our method achieves superior overall performance in the complex scenario of multi-type pneumonia classification. Specifically, our model consistently surpasses the performance of other methods on all the overall evaluation metrics, reaching 90.51% Acc, 97.78% AUC, and 89.42% F1 score. For the diagnostic performance of the other specific classes, our approach greatly improves the CAP diagnostic results by a large margin. It also gains the high sensitivity and specificity over 91% as well as the AUC over 97% on H1N1 and healthy cases. These results show the superiority of our approach for the precise diagnosis of COVID-19 and other categories of pneumonia.
4.5.2. Analysis on periphery-aware spatial prediction (PSP)
We evaluate the impact of the proposed PSP by comparing our model “Ours (Res18+PSP)” with the baseline model “Res18 (=2, =64)”. This baseline model is trained to predict pneumonia classification, in which both PSP and CRE are discarded. It keeps the same augmentation parameters (i.e., =2 and =64) as our model, where denotes the number of augmented samples that are generated from one input sample, and denotes the number of slices in each augmented 3D sample. As shown in Table 3, “Ours (Res18+PSP)” achieves significant improvements on the AUC of COVID-19 and all three overall metrics than the baseline. It also reaches satisfactory diagnosis results in other categories. The effectiveness of our proposed PSP will be further demonstrated by comprehensive experiments on our 2D dataset.
4.5.3. Analysis on contrastive representation enhancement (CRE)
We investigate the effectiveness of the proposed CRE by comparing our model “Ours (Res18+CRE)” with three baselines on our 3D dataset. We discard the CRE and train two additional models (“Res18 (=1, =64)” and “Res18 (=1, =128)”) to predict pneumonia classification as the baselines. To eliminate the specificity of data augmentation which is designed for the contrastive learning framework, the baselines are applied by three different augmentation settings (i.e., (=2, =64), (=1, =64), and (=1, =128)). It is observed from the 6th to 8th rows of Table 3 that the variance of diagnostic performance of the three baselines with different settings is small. It indicates that the success of our proposed CRE owes little to the two data augmentation operations for each sample. Our proposed approach with CRE (“Ours (Res18+CRE)”) further improves the pneumonia diagnosis performance over all the baselines (p0.001). The results demonstrate that our proposed CRE can effectively capture the intra-class similarity and inter-class difference by contrasting the pneumonia-type guided positive and negative samples. The enhanced representations are more discriminative to greatly improve the multi-type pneumonia classification.
4.5.4. Analysis on adaptive joint training
As illustrated in Eq. (9), we design a joint training loss with adaptive weights, and , to combine the contrastive loss and cross-entropy loss. During the training process, the two learnable weights are also optimized along with the network’s parameters. Taking the average value of 3-fold cross validation results, we get the learned weights and . We normalize them to the interval [0,1] and obtain and . To analyze the validity of the learned weights, we use the grid search to compare the performance of different set weights. Let the joint training loss be balanced by fixed weights and , it can be expressed as:
| (10) |
We implement three tests with the configurations of , respectively. Note that when , our whole framework degenerates to the baseline that is trained with only the cross-entropy loss. The results of different weights in the loss function are illustrated in the 9th to 11th rows of Table 3. “Ours (Res18+CRE)” with the adaptive learned weights ( and ) consistently outperforms all three methods by 2%3% improvements on Acc and F1 score. It also achieves a comparable AUC score with marginal differences (0.05%0.28%). The results indicate that it is effective to learn the weights adaptively for balancing the contrastive loss and cross-entropy loss in our diagnosis approach.
4.5.5. Analysis on backbone architectures
To show the impact of backbone models, we run experiments with 3D ResNet10, ResNet18, as well as ResNet34 [26], and the results are shown in the last two rows of Table 3. It can be seen that 3D ResNet10 and ResNet34 achieve inferior overall performance than ResNet18. Besides, both ResNet10 and ResNet34 get high sensitivity but extremely low specificity in COVID-19 diagnosis, which indicates that they tend to make more false positive predictions of COVID-19. In contrast, our ResNet18 model achieves a better comprehensive performance on AUC for COVID-19. Finally, we adopt the best-performing 3D ResNet18 model as the backbone architecture.
4.5.6. Visual explanation
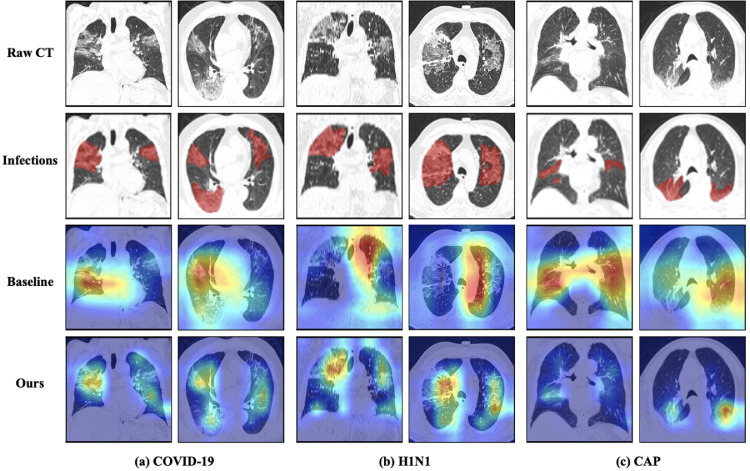
We exploit a model explanation technique, namely Gradient-weighted Class Activation Mapping (Grad-CAM) [31], to show the attention maps for pneumonia classification. In Fig. 5 , we present some typical attention maps obtained by our proposed model trained in one fold. For a fair comparison, we also show the attention results of the baseline model trained in the same fold. It can be seen in the 3rd row that although the pneumonia predictions are correct, the attention maps of the baseline just roughly indicate the suspicious area, and even largely focus on irrelevant regions rather than the infections. However, our approach greatly improves the localization ability of suspicious lesions. As shown in the attention maps of the 4th row, most infection areas can be located precisely, and the irrelevant regions are excluded. These highlighted areas in attention maps are in close proximity to the annotation results of infections, which demonstrates the reliability and interpretability of the diagnosis results from our model. Therefore, the attention maps can be possibly used as the basis to derive the COVID-19 diagnosis in clinical practice.
Fig. 5.
The visualization results on COVID-19, H1N1, CAP cases in both axial view and coronal view. We show the raw CT images (1st row) and the annotation results of pneumonia infections (2nd row). We visualize the attention results of the baseline model (3rd row) and our proposed model (4th row).
4.6. Results on our 2D dataset
4.6.1. Low-data classification
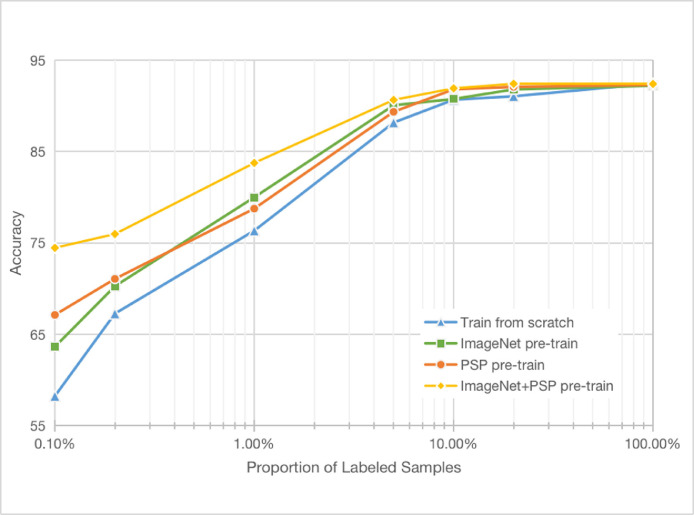
We conduct low-data classification experiments to investigate the impact of our PSP as an unsupervised pre-training scheme. We start by evaluating the performance of a supervised network, which is trained from scratch. Concretely, we train a separate network on each subset with various proportions of labeled data from 0.1% to 100%, and then evaluate each model's performance on our entire testing set. The detailed numbers of 2D training images corresponding to each proportion are presented in Table 4 . As can be seen in Fig. 6 (blue curve), the model trained from scratch tends to overfit more severely with decreasing amounts of data. When the proportion of labeled data decreases from 100% to 0.1%, the accuracy drops from 92.47% to 58.24%.
Table 4.
The detailed numbers of 2D training images corresponding to each proportion.
| Proportion | 0.1% | 0.2% | 1% | 5% | 10% | 20% | 100% |
|---|---|---|---|---|---|---|---|
| Number | 64 | 127 | 657 | 2,968 | 6,308 | 13,065 | 59,413 |
Fig. 6.
The diagnosis accuracy as a function of the proportion of labeled CT examples on our 2D dataset (best viewed in colour).
We next evaluate our pre-trained periphery-aware model on the same data proportions. Here, we pre-train the feature encoder on our entire unlabeled training set, and then learn the classifier and fine-tune the encoder using a subset of labeled images. Figure 6 (orange curve) shows the results. In contrast to the model trained from scratch (blue curve), learning the majority of the model’s parameters with PSP greatly alleviates the degradation in diagnosis accuracy as the amount of labeled data decreases. Specifically, our pre-trained model's accuracy in the low-data regime (only 0.1% of labeled CT samples) is 67.13%, outperforming the model trained from scratch by nearly 9%, while retaining a high accuracy (92.3%) in the high-data regime including all CT labels.
We compare our approach with the ImageNet pre-trained model, which is a commonly used model pre-trained by a large-scale natural image dataset [34]. Figure 6 shows that our PSP pre-trained model even surpasses the ImageNet pre-trained model (green line) in the regimes with 0.1%, 0.2%, 10%, 20% proportions of data, and achieves a comparable performance with 1% and 5% data. These results demonstrate that our unsupervised PSP pre-training method effectively introduces the important spatial pattern prior, which largely improves the diagnosis performance in most regimes, especially in the low-data regime.
To further improve the diagnostic performance in the low-data regime, we train the PSP task on the ImageNet pre-trained model and then fine-tune the “ImageNet+PSP” pre-trained model. As can be seen from the yellow curve in Fig. 6, it significantly boosts the diagnosis accuracy compared with other models when the number of images decreases. It even represents a 16% improvement over the model trained from scratch in the 0.1% data regime. The results suggest that the diagnostic performance can be further improved by the combination of ImageNet and PSP pre-training.
4.6.2. Analysis on 1% data
Since it is difficult to acquire a large number of annotated medical images in many real-world scenarios, we further show the effect of our PSP and CRE on 1% CT samples of our 2D dataset, as illustrated in Table 5 . Compared with the ResNet50 trained from scratch (“Res50”), the PSP pre-trained model “Res50+PSP” clearly improves the performance on both COVID-19 diagnosis and overall classification (p<0.001). The model “Res50+Mask” indicates that we degenerate our PSP pre-training task to the general lung segmentation task, in which the network is trained to predict lung region masks obtained by our unsupervised segmentation algorithm. It can be observed that the network pre-trained by our proposed PSP task (“Res50+PSP”) achieves better overall performance than the lung segmentation task (“Res50+Mask”) with 2% improvements on both Acc and F1 metrics. In COVID-19 diagnosis, “Res50+PSP” even improves the sensitivity and the AUC score by over 11% and 1.5%. The results confirm the significance of PSP with the boundary distance prediction and indicate the effectiveness of PSP in multi-type pneumonia classification and COVID-19 diagnosis.
Table 5.
The quantitative results on our 2D dataset with the proportion of 1% CT samples. We report Sensitivity (Sen.), Specificity (Spec.) and AUC for each class, as well as Accuracy, AUC, and F1 score for overall performance (unit:%). The values in brackets are 95% confidence intervals.
| Methods | COVID-19 |
H1N1 |
CAP |
Healthy |
Acc | AUC | F1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sen. | Spec. | AUC | Sen. | Spec. | AUC | Sen. | Spec. | AUC | Sen. | Spec. | AUC | ||||
| Res50 | 82.63 | 86.65 | 92.52 | 95.94 | 93.82 | 98.79 | 68.59 | 94.57 | 92.18 | 49.78 | 92.64 | 85.79 | 76.36 [75.95, 76.78] | 92.32 | 73.77 |
| Res50+PSP | 85.77 | 86.66 | 92.80 | 96.25 | 94.50 | 98.71 | 67.98 | 95.98 | 92.77 | 55.40 | 93.59 | 84.18 | 78.77 [78.42, 79.17] | 92.12 | 76.48 |
| Res50+Mask | 74.45 | 88.82 | 91.33 | 97.03 | 93.04 | 98.62 | 59.17 | 97.34 | 92.76 | 65.77 | 88.70 | 86.77 | 76.70 [76.33, 77.14] | 92.37 | 74.77 |
| Res50+ImageNet | 84.19 | 88.59 | 93.57 | 98.65 | 93.68 | 98.97 | 62.81 | 97.64 | 94.73 | 62.48 | 92.35 | 88.32 | 80.00 [79.64, 80.39] | 93.90 | 77.79 |
| Res50+ImageNet+PSP | 87.22 | 92.56 | 95.51 | 97.95 | 95.38 | 99.26 | 61.61 | 97.58 | 94.05 | 75.17 | 92.23 | 92.80 | 83.75 [83.42, 84.11] | 95.41 | 81.27 |
| Res50+CRE | 83.57 | 86.48 | 92.20 | 97.69 | 95.00 | 99.00 | 60.76 | 96.55 | 92.31 | 56.02 | 91.10 | 86.27 | 77.63 [77.23, 78.04] | 92.44 | 74.98 |
| Res50+PSP+CRE | 85.32 | 86.97 | 92.11 | 98.66 | 95.93 | 99.17 | 67.76 | 96.85 | 93.90 | 55.84 | 91.80 | 87.36 | 79.40 [79.02, 79.82] | 93.14 | 77.26 |
| Res50+ImageNet+CRE | 86.67 | 87.53 | 94.01 | 94.95 | 97.12 | 99.12 | 75.81 | 95.66 | 95.19 | 59.34 | 93.32 | 88.62 | 80.76 [80.41, 81.16] | 94.24 | 79.10 |
| Res50+ImageNet+PSP+CRE | 85.79 | 91.43 | 96.13 | 97.40 | 94.55 | 98.84 | 77.45 | 96.80 | 95.84 | 69.75 | 95.24 | 93.49 | 84.00 [83.64, 84.36] | 96.07 | 82.74 |
Then, we evaluate the importance of our CRE. For each initialization, (i.e., random initialization, PSP, ImageNet, ImageNet+PSP), incorporating our CRE can improve the performance on all the overall metrics, as shown in the last four rows of Table 5. Our method “Res50+ImageNet+PSP+CRE” reaches the best result with 84.00% Acc, 96.07% AUC, and 82.74% F1 score. These results validate that our proposed approach is effective to utilize the spatial pattern information and contrastive representations for a better COVID-19 diagnosis on the 2D CT dataset.
4.7. Results on the Covid-CT dataset
4.7.1. Comparison with existing methods
We analyze the effectiveness of our proposed approach on the public Covid-CT dataset, and the results are shown in Table 6 . Note that our PSP pre-training task is unable to be validated in this dataset because the PNG image format cannot provide HU values for the pre-segmentation of lung regions during pre-training. Our unsupervised PSP perfectly deals with the DICOM file which is extensively adopted in real-world clinical applications. Therefore, we only transfer the ImageNet pre-trained weights to improve the diagnostic results, as the scale of this dataset is relatively small. Firstly, we compare our model “ResNet50+ImageNet+CRE” with the existing COVID-19 diagnosis methods [9], [14], [22], [23].
Table 6.
The comparison results of our proposed approach and existing methods on the Covid-CT dataset. We report Accuracy, AUC, and F1 score for performance comparison.
| Methods | Acc | AUC | F1 |
|---|---|---|---|
| DensNet169 [22] | 0.83 | 0.87 | 0.81 |
| DenseNet169 (self-trans) [22] | 0.83 | 0.90 | 0.84 |
| Redesigned Covid-Net [23] | 0.79 | 0.85 | 0.79 |
| M-Inception [9] | 0.81 | 0.88 | 0.82 |
| Evidential Covid-Net [14] | 0.81 | 0.87 | 0.81 |
| Ours (Res50+ImageNet+CRE (=0.38)) | 0.84 | 0.90 | 0.86 |
For a fair comparison, we retrained the DenseNet169 (self-trans) model with same settings, except for the pre-training on the additional LUNA dataset.
Notably, in the Covid-CT dataset, COVID-19 is defined as negative (label = 0) and non-COVID-19 is positive (label = 1). It is observed from Table 6 that our method “Ours (Res50+ImageNet+CRE (=0.38))” clearly outperforms all these existing methods on the three metrics. Acc, AUC, and F1 score are 0.84, 0.90, and 0.86. Compared with DenseNet169 (self-trans) [22] and Redesigned COVID-Net [23] which also exploit contrastive learning in COVID-19 diagnosis, our CRE demonstrates better diagnostic performance, improving Acc and AUC by 1% and 2%, respectively. These results show the superiority of our method with CRE for COVID-19 and non-COVID-19 diagnosis.
4.7.2. Ablation study
We also conduct an ablation study on the Covid-CT dataset, as shown in Table 7 . With random initialization, our model “Res50+CRE (=0.37)” significantly outperforms the baseline model “Res50 (=1)” by 2% Acc, 11% AUC, and 2% F1 score. When pre-trained on ImageNet, our model “Res50+ImageNet+CRE (=0.38)” also achieves the improvements of 2%∼3% on the three metrics, compared with the baselines “Res50+ImageNet (=1)” and “Res50+ImageNet (=2)”. We can also observe that our designed joint training strategy can learn better adaptive loss weights (=0.38), compared with the models with other fixed weights (=0.25,0.50,0.75). As for the network architecture, the ResNet50 is verified to be a preferable backbone architecture, compared with the other commonly used networks such as ResNet34, ResNet101, DenseNet121, and DenseNet169.
Table 7.
The results of ablation study on the Covid-CT dataset.
| Methods | Acc | AUC | F1 |
|---|---|---|---|
| Res50 (m=1) | 0.65 | 0.67 | 0.70 |
| Res50+ImageNet (=1) | 0.82 | 0.88 | 0.83 |
| Res50+ImageNet (=2) | 0.80 | 0.88 | 0.83 |
| Res50+ImageNet+CRE () | 0.82 | 0.89 | 0.83 |
| Res50+ImageNet+CRE () | 0.80 | 0.89 | 0.82 |
| Res50+ImageNet+CRE () | 0.80 | 0.88 | 0.83 |
| Res34+ImageNet+CRE | 0.81 | 0.87 | 0.83 |
| Res101+ImageNet+CRE | 0.78 | 0.86 | 0.80 |
| Dense121+ImageNet+CRE | 0.84 | 0.88 | 0.85 |
| Dense169+ImageNet+CRE | 0.79 | 0.88 | 0.80 |
| Res50+CRE () | 0.67 | 0.78 | 0.72 |
| Res50+ImageNet+CRE () | 0.84 | 0.90 | 0.86 |
4.8. Results on the CC-CCII dataset
To train our model “Res50+ImageNet+PSP+CRE” on the CC-CCII dataset, we first use 2D ImageNet pre-trained ResNet50 to perform a 2D PSP task with a small number of labeled lung masks (750 masks). Then, we initialize the 3D ResNet50 model with the inflated 2D weights pre-trained on PSP [35]. Finally, we train the 3D model with CRE on the entire dataset.
Table 8 shows the results of our method and other existing approaches on the CC-CCII dataset. “Binary cls” denotes the classification between non-COVID-19 and COVID-19; and “Ternary cls” is the classification among normal controls, COVID-19, and common pneumonia (CP). For “Binary cls”, our model achieves 98.18% Acc, 97.75% Sen., 98.44% Spec., and 99.29% AUC, which surpasses the other approaches by a large margin. For “Ternary cls”, our model obtains the improvements of 4.24% on Acc and 1.37% on AUC compared with Zhang et al.’s in [13]. Both binary classification task and ternary classification task demonstrate the superiority of our method on the CC-CCII dataset.
Table 8.
The comparison results on the CC-CCII dataset (unit:%).
5. Conclusion and future work
In this work, we propose a periphery-aware COVID-19 diagnosis approach with contrastive representation enhancement, which detects COVID-19 in complex scenarios of multi-type pneumonia using chest CT images. Particularly, we design an unsupervised Periphery-aware Spatial Prediction (PSP) pre-training task, which can effectively introduce the important spatial pattern prior to networks. It is confirmed that the model pre-trained by our proposed PSP task even outperforms the fully-supervised ImageNet pre-trained model in the low-data regime. The design of the unsupervised PSP pre-training task provides a new paradigm to inject location information into neural networks. To further obtain more discriminative representations, we build a joint learning framework that integrates Contrastive Representation Enhancement (CRE) as an adaptive auxiliary task to pneumonia classification. Our CRE extends the general contrastive learning, which can capture the intra-class similarity and inter-class difference for a more precise multi-type pneumonia diagnosis. We also construct a large-scale COVID-19 dataset with four categories for the fine-grained differential diagnosis. The effectiveness and clinical interpretation of our proposed approach have been verified on our in-house 3D and 2D CT datasets as well as the public Covid-CT and CC-CCII datasets. In the future, we will further explore the potential of our proposed CRE and PSP in multiple lesion diagnosis tasks, including lesion detection and segmentation. Contrastive learning assists the network to learn lesion-specific features in images or volumes, making the network a stronger feature extractor. As our proposed PSP embeds location prior about COVID-19, it can also be extended to location-sensitive tasks such as lesion localization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by National Natural Science Foundation of China (No. 61976057); Science and Technology Commission of Shanghai Municipality (No. 20511100800, No. 20511101103, No. 20511101203, No. 20DZ1100205, No. 19DZ2205700, No. 2021SHZDZX0103); and Project (No. AWS15J005). The authors would like to thank Jiawang Cao (for his help in data processing), Nan Zhang, Huifeng Zhang, and Weifeng Jin (for their help in language and writing assistance).
Biographies
Junlin Hou received the B.S. degree in Mathematics and Applied Mathematics from Shanghai University, Shanghai, China, in 2018. She is currently a PhD student in School of Computer Science, Fudan University, Shanghai, China. She is a member of Shanghai Key Lab of Intelligent Information Processing in School of Computer Science. Her research interest is medical image analysis, including self-supervised learning and lung diseases detection.
Jilan Xu received the B.S. degree in Computer Science from Shanghai University, Shanghai, China, in 2018. He is currently a master student in School of Computer Science, Fudan University, Shanghai, China. He is a member of Shanghai Key Lab of Intelligent Information Processing in School of Computer Science. His research interest is medical image analysis, including weaklysupervised learning and histopathology image analysis.
Longquan Jiang received the B.S. degree in Computer Science and Technology from East China University of Science and Technology, Shanghai, China, in 2010, and the M.S. degree in Computer Science from Fudan University, Shanghai, China, in 2013. His research interests include medical image analysis, pattern recognition, and machine learning.
Shanshan Du received the B.S. degree in Computer Science and Technology from Yangtze University, Hubei, China, in 2006, and the M.S. degree in Computer Science from Fudan University, Shanghai, China, in 2015. She is currently a PhD student in School of Information Science and Technology, Fudan University, Shanghai, China. Her research interests include computer version, multimedia information analysis and processing, machine learning and medical image analysis.
Rui Feng received the B.S. degree in Industrial Automatic from Harbin Engineering University, Haerbin, China, in 1994, the M.S. degree in Industrial Automatic from Northeastern University, Shenyang, China, in 1997, and the Ph.D. degree in Control Theory and Engineering from Shanghai Jiaotong University, Shanghai, China, in 2003. In 2003, He joined Department of Computer Science and Engineering (now School of Computer Science), Fudan University as an Assistant Professor, and then become Associate Professor and Full Professor. His research interests include medical image analysis, intelligent video analysis, and machine learning.
Yuejie Zhang received the B.S. degree in Computer Software, the M.S. degree in Computer Application, and the Ph.D. degree in Computer Software and Theory from Northeastern University, Shenyang, China, in 1994, 1997 and 1999, respectively. She was a Postdoctoral Researcher at Fudan University, Shanghai, China, from 1999 to 2001. In 2001, she joined Department of Computer Science and Engineering (now School of Computer Science), Fudan University as an Assistant Professor, and then become Associate Professor and Full Professor. Her research interests include medical image analysis, multimedia/cross-media information analysis, processing, and retrieval, and machine learning.
Fei Shan received the B.S. degree in Imaging Medical from the Nantong University, Nantong, China, in 2001, the M.S. degree and the Ph.D. degree in Imaging and Nuclear Medicine from the Fudan University, Shanghai, China in 2004 and 2009 respectively. He is currently Chief physician of Radiology Department in Shanghai Public Health Clinical Center in Shanghai, China. He has published more than 35 refereed articles in many premier radiological and artificial intelligence journals and conferences such as Eur Respir J, American Journal of Roentgenology, IEEE Transactions on Medical Imaging and so on. His research interests include the chest imaging diagnosis, especially the differential diagnosis of pulmonary nodules, and the artificial intelligence imaging of lung diseases.
Xiangyang Xue received the BS, MS, and PhD degrees in communication engineering from Xidian University, Xian, China, in 1989, 1992, and 1995, respectively. He is currently a professor of Computer Science in Fudan University, Shanghai, China. His research interests include computer vision, image analysis, and machine learning.
Appendix A
Derivation of our joint loss
Our joint loss is comprised of the InfoNCE loss in the contrastive representation enhancement and the cross-entropy loss in the pneumonia classification task. From [29], the log likelihood of the classification task can be written as:
| (11) |
where is the cross-entropy loss of . Here, we derive the log likelihood of InfoNCE loss in detail and obtain our joint loss with adaptive weights.
The InfoNCE loss can be formulated as the instance-level classification objective using the Softmax criterion. Given samples and their features, for a sample with feature , the probability of it being recognized as th example is:
| (12) |
We adapt the likelihood to squash a scaled version of the model output with a positive scalar :
| (13) |
The log likelihood can then be written as:
| (14) |
In maximum likelihood inference, we maximize the log likelihood of the network, which equals to minimize:
| (15) |
where is written for the contrastive InfoNCE loss. The second term is approximately equal to when .
In the case of multiple network outputs, the likelihood is defined to factorize over the outputs. We define as the sufficient statistics and obtain the following likelihood:
| (16) |
with the model outputs stand for classification and contrastive learning, respectively. Therefore, our joint loss is given as:
| (17) |
References
- 1.WHO, COVID-19 weekly epidemiological update, (https://www.who.int/publications/m/item/weekly-epidemiological-update---29-december-2020. December 29th, 2020.
- 2.Wong H.Y.F., Lam H.Y.S., Fong A.H.-T., Leung S.T., Chin T.W.-Y., Lo C.S.Y., et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020;296(2):E72–E78. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., et al. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans. Med. Imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 4.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020;296:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., et al. Ai-assisted ct imaging analysis for COVID-19 screening: building and deploying a medical ai system in four weeks. MedRxiv. 2020 doi: 10.1016/j.asoc.2020.106897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z., Xiao Y., Li Y., Zhang J., Lu F., Hou M., et al. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest x-rays. Pattern Recognit. 2020;110:107613. doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shorfuzzaman M., Hossain M.S. MetaCOVID: a siamese neural network framework with contrastive loss for n-shot diagnosis of COVID-19 patients. Pattern Recognit. 2020:107700. doi: 10.1016/j.patcog.2020.107700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., et al. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19) Eur. Radiol. 2021:1–9. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. MedRxiv. 2020 doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2987975. 1–1. [DOI] [PubMed] [Google Scholar]
- 12.Mohamadou Y., Halidou A., Kapen P.T. A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID-19. Appl. Intell. 2020;50:3913–3925. doi: 10.1007/s10489-020-01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., et al. Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.L. Huang, S. Ruan, T. Denoeux, COVID-19 classification with deep neural network and belief functions, arXiv preprint arXiv:2101.06958 (2021).
- 15.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., et al. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39(8):2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 16.Wang J., Bao Y., Wen Y., Lu H., Luo H., Xiang Y., et al. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imaging. 2020;39(8):2572–2583. doi: 10.1109/TMI.2020.2994908. [DOI] [PubMed] [Google Scholar]
- 17.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian X., Fu H., Shi W., Chen T., Fu Y., Shan F., et al. M Lung-sys: a deep learning system for multi-class lung pneumonia screening from CT imaging. IEEE J. Biomed. Health Inf. 2020;24(12):3539–3550. doi: 10.1109/JBHI.2020.3030853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A.v. d. Oord, Y. Li, O. Vinyals, Representation learning with contrastive predictive coding, arXiv preprint arXiv:1807.03748 (2018).
- 20.Chen T., Kornblith S., Norouzi M., Hinton G. International Conference on Machine Learning. 2020. A simple framework for contrastive learning of visual representations; pp. 1597–1607. [Google Scholar]
- 21.He K., Fan H., Wu Y., Xie S., Girshick R. 2020 IEEE Conference on Computer Vision and Pattern Recognition. 2020. Momentum contrast for unsupervised visual representation learning; pp. 9726–9735. [Google Scholar]
- 22.He X., Yang X., Zhang S., Zhao J., Zhang Y., Xing E., et al. Sample-efficient deep learning for COVID-19 diagnosis based on ct scans. MedRxiv. 2020 [Google Scholar]
- 23.Wang Z., Liu Q., Dou Q. Contrastive cross-site learning with redesigned net for COVID-19 CT classification. IEEE J. Biomed. Health Inf. 2020;24(10):2806–2813. doi: 10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X., Yao L., Zhou T., Dong J., Zhang Y. Momentum contrastive learning for few-shot COVID-19 diagnosis from chest CT images. Pattern Recognit. 2021;113:107826. doi: 10.1016/j.patcog.2021.107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Zhao G., Tao Y., Zhai P., Chen H., He H., et al. Multi-task contrastive learning for automatic CT and x-ray diagnosis of COVID-19. Pattern Recognit. 2021;114:107848. doi: 10.1016/j.patcog.2021.107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He K., Zhang X., Ren S., Sun J. 2016 IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 27.Hayder Z., He X., Salzmann M. 2017 IEEE Conference on Computer Vision and Pattern Recognition. 2017. Boundary-aware instance segmentation; pp. 587–595. [Google Scholar]
- 28.Khosla P., Teterwak P., Wang C., Sarna A., Tian Y., Isola P., Maschinot A., Liu C., Krishnan D. Annual Conference on Neural Information Processing Systems 2020. 2020. Supervised contrastive learning. [Google Scholar]
- 29.Kendall A., Gal Y., Cipolla R. 2018 IEEE Conference on Computer Vision and Pattern Recognition. 2018. Multi-task learning using uncertainty to weigh losses for scene geometry and semantics; pp. 7482–7491. [Google Scholar]
- 30.J. Zhao, Y. Zhang, X. He, P. Xie, Covid-CT-dataset: a CT scan dataset about COVID-19, arXiv preprint arXiv:2003.13865 (2020).
- 31.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. 2017 IEEE International Conference on Computer Vision. 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 32.Liao F., Liang M., Li Z., Hu X., Song S. Evaluate the malignancy of pulmonary nodules using the 3-D deep leaky noisy-or network. IEEE Trans. Neural Netw. Learn. Syst. 2019;30(11):3484–3495. doi: 10.1109/TNNLS.2019.2892409. [DOI] [PubMed] [Google Scholar]
- 33.F. He, Y. Ma, Imbalanced learning: foundations, algorithms, and applications, (2013) 45.
- 34.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE Conference on Computer Vision and Pattern Recognition. Ieee; 2009. ImageNet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 35.Carreira J., Zisserman A. 2017 IEEE Conference on Computer Vision and Pattern Recognition. 2017. Quo vadis, action recognition? A new model and the kinetics dataset; pp. 6299–6308. [Google Scholar]