Graphical abstract
Keywords: Origin of life, Membrane, Abiogenesis, Compartment, Evolution
Abstract
The history of modern biochemistry started with the cellular theory of life. By putting aside the holistic protoplasmic theory, scientists of the XX century were able to advance the functional classification of cellular components significantly. The cell became the unit of the living. Current theories on the abiogenesis of life must account for a moment in evolution (chemical or biological) when this was not the case. Investigating the role of compartments and membranes along chemical and biotic evolution can lead a more generalised idea of living organisms that is fundamental to advance our efforts in astrobiology, origin of life and artificial life studies. Furthermore, it may provide insights in unexplained evolutionary features such as the lipid divide between Archaea and Eubacteria. By surveying our current understanding of the involvement of compartments in abiogenesis and evolution, the idea of cells as atomistic units of a general theory of biology will be discussed. The aim is not to undermine the validity of the cellular theory of life, but rather to elucidate possible biases with regards to cellularity and the origin of life. An open discussion in these regards could show the inherent limitations of non-cellular compartmentalization that may lead to the necessity of cellular structures to support complex life.
1. Introduction
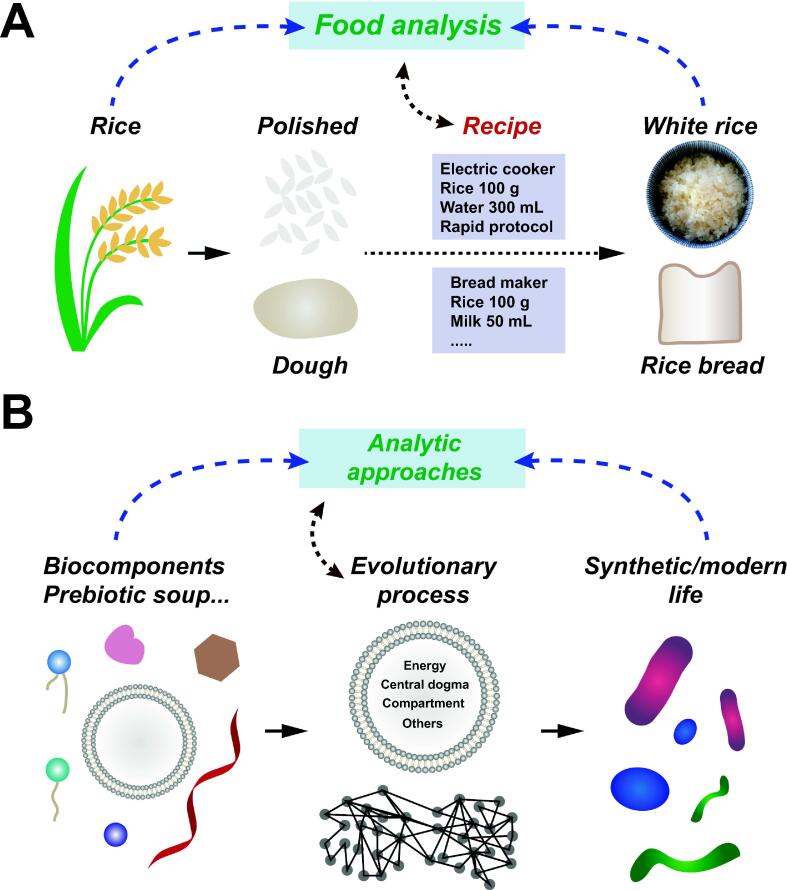
Studying biology today means studying the cell. These membrane-bound vessels are the constituents of all known forms of life and, for this reason, their emergence is a fundamental question in the origin of life. Historically, the cell theory of life emerged during the first decades of the XX century as the dominating framework in biology, with close ties to analytic and reductionist approaches. The analytic framework will be discussed by examining how the cell theory first emerged and its role in contemporary origins research. A focus of this examination will be the comparison to the constructive framework described by [1], to suggest how the universality of cells as units of life may be subject of future investigation (especially Chapter 2). Briefly, this framework aims at finding general principles governing life by constructing systems that capture some of their properties and by looking for the necessary and universal factors in such systems. Origins research is comparable to formulating a recipe for life from a list of ingredients. For a complex baked product such as bread, efforts spent analysing the end result are unlikely to give detailed information on the procedure that yielded it. The only way to understand how the properties of bread came about, is to reconstruct a recipe for it. As summarised in Fig. 1, previous approaches to this discipline excelled at the classification of parts and relationships between them. However, they also demonstrated an insufficient understanding of how these relationships and elements can emerge from a complex system. This understanding is key to universal biology and specifically verifies if life and cellularity are linked by chance or necessity. To explore these issues, Section 2 will lay out a brief history of the concept of cells as atoms of the living, Section 3 will address the advantages of compartmentalization for replicators generated abiogenetically (protobionts), Section 4 will discuss how and why such compartments would eventually evolve into cells. Section 5 questions cellularity as a necessity for the evolution of life in a general sense.
Fig. 1.
The constructive approach to biology is compared to the analytic one through this parallelism: in A it is depicted how, by processing raw ingredients (rice) with the appropriate recipe, one can successfully bake rice bread or prepare a portion of white rice. The analytic study of rice bread can provide a list of its components but, knowing nothing of the recipe beforehand, it will not inform how to make it or what makes it “bread”. Similarly, life's origin and early evolution are not understood by analysing modern forms of it but by formulating a recipe for its emergent behaviours and functions.
2. The cellular theory of life: a history
Biology in the XIX century was in the arduous process of trying to secularize itself. Living things were understood by two opposed philosophical frameworks: cartesian mechanism on one hand, vitalism on the other [2]. The latter ascribed “aliveness” to metaphysical properties of living things that set it distinctly apart from matter subject to Newtonian and thermodynamical laws. The former tried to explain organisms as complex and coordinated machinery, with a reductionist and materialistic outlook. The ideological pull away from vitalism and towards mechanism shaped biological discourse for centuries, eventually leading to the dominance of reductionism throughout the 20th century and beyond. These discussions aimed at atomising living things, aligning biology with physics and chemistry in having a fundamental object of study. Physiologists were spending most of their efforts identifying a common unit of the living, which could allow the generalisation of laws and observations among different clades and kingdoms of taxonomy.
The idea of cells as the basic functional units of organisms is univocally accepted as the foundation of modern biology, but that has not always been the case. Being limited by the technology at their disposal, biologists in the 1800s had little means to make sense of subcellular organization. Bladder-like enclosures were first described in Suberites by Robert Hooke [3]. Later findings identified the nucleus, the vacuole, and other subcellular structures, together with the striking dynamics of inter and intracellular transport. Different observations were brought together by Schleiden, Schwann, and, later, Virchow into the now accepted cellular theory of life. The theory postulates that all animals and plants are composed of at least one cell and that each cell is generated from a pre-existing one [4]. This first attempt at generalization was heavily criticized for putting too much emphasis on the housing of the fundamental constituent of living things [3]. Observations of membrane-like boundaries enveloping cells were not consistent, being limited by the microscopes available at the time, so the attention of influential investigators shifted to the cell’s contents. The substance variously described as “grey and viscous” or “granular with both viscous and liquid parts” or, again, as “contractile and complex in composition” received many definitions over the century [5]. Huxley, with his seminal lecture in 1869, finally shaped these different accounts in a protoplasmic theory of life that took traction among scientists of the time. By affirming that “a nucleated mass of protoplasm may be termed the structural unit of the human body” and further generalising this to all forms of life, the British biologist grounded all processes in the realm of biology on a common physical basis [6]. In his view, the differences between organisms are of degree, not kind. Life now had a physical embodiment -the protoplasm- which endowed every living thing with common properties. By endorsing the protoplasmic theory of life, physiologists and biologists could now achieve the mechanical comprehension of the living [3]. Even though the theory enjoyed a surge of popularity at the dawn of the XX century, modern academic curricula include cell biology, rather than protoplasm biology. The fall out of grace of the protoplasmic theory is an interesting topic in the history of life sciences that involved both ideological and technological shifts in the first half of the XX century. It was impossible to investigate the protoplasm without irreversibly changing its properties chemically, as it was a functional whole and alive as such. Huxley himself noted this apparent weakness in his lecture [6]. And yet, the theory he popularized was based on a dynamical organisation that was impervious to analytical techniques available at the time. This peculiarity stifled many reductionist approaches to the study of this unit of life and the debate between vitalists and mechanists with them. The inability of the protoplasm to advance secular thought in biology manifested in the heated debate that followed Huxley’s lecture, for example in the works of Beale [7]. His distinction of two types of matter (germinal and formed) in the protoplasm and of their role in organisms fully relied on the vitalist framework and stood in stark opposition with Huxley’s materialist view of the “vital force”. Most of their contemporaries maintained a more moderate perspective, recognising that the unique properties of the protoplasm were the result of its uniquely complex composition, but the term composition was not to be employed in the strict chemical sense. The endorsement of a protoplasmic unit of life was used by various investigators to justify their points of view, rather than to supply evidence that could rule out one of the two philosophies. For all the traction the theory gained in the mid-1800s, the chemical concept of protoplasm had shortcomings that inhibited any further reduction of the living to a physical phenomenon [2].
Of course, this debate did not bring the whole field of physiology to a stop. In 1855, Nägeli’s observation of osmotic responsiveness in protoplasmic droplets and electrophysiological measurements of solute mobility strongly indicated the presence of a morphological and functional separation layer between protoplasm and environment [8]. Three possibilities for a protoplasm-membrane theory were proposed:
-
•
The protoplasm is separated by a surface layer typical of other colloidal systems
-
•
The membrane is composed of densely packed protoplasm
-
•
A compositionally distinct membrane endows semi-permeability and compartmentalization to the protoplasm
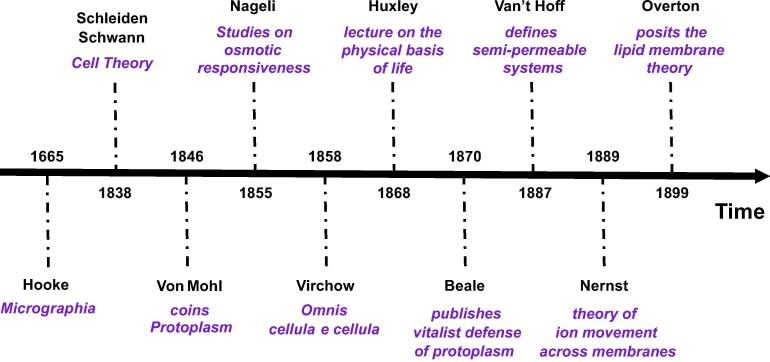
In Fig. 2 the salient developments in both cell theory and protoplasmic theory during the XIX century are chronologically listed for further reference.
Fig. 2.
Chronology of salient developments in the protoplasm and cell theory of life during the XIX century.
During the first half of the XX century, a gradual but inexorable shift from the first theory to the third took place. Thanks to the development of more powerful tools for measurement and observation, such as Langmuir troughs, more sophisticated electrodes for intracellular potential monitoring, and electron microscopy, an overwhelming amount of evidence undermined the accepted viewpoint in favour of a lipidic membrane as the cell-environment boundary [9]. Alongside these developments, the merging of biochemistry and enzymology started to shed light on the internal chemistry of cells, by characterizing the catalytic activity of purified enzymes and their organization in metabolic pathways [9]. Accusations of obscurantism were directed at the protoplasmic theory, reinforcing the process of association between protoplasm and vitalism. The reframed importance of the membrane as a structural element and the view of cells as chemical factories painted our view of the cell: a cell membrane compositionally distinct but functionally linked to an internal cytoplasm that contains the nucleus [4]. The cytoplasm was further subdivided in cytosol and corpuscular elements previously identified as different phases of a unitary protoplasm. As the existence of membrane channels, receptors, pumps were first postulated and then empirically verified [8], the centrality of membranes in biochemistry shaped a new conception of the cell. The membrane system became the locus of cell sensing and cellular metabolism, thanks to its ability to maintain concentration gradients. The cell could now be taken apart and its molecular components studied in isolation to understand the inner workings of the living unit. Reductionism and mechanism found strength in a theory that allowed structure–function relationships to be identified in vitro.
With the ability to analyse the cell, its overwhelming complexity was soon realised. As the new disciplines in cell biology produced ever-growing amounts of data, the simplistic view of the cell as a membrane-bound reaction vessel could not be sustained. During these first decades of the XXI century, holistic studies of biology have taken foot, thanks to data provided by omic disciplines and their synthesis in Systems Biology [10]. The recognition of reductionism as an insufficient tool in biology as well as a rejection of the machine-organism equivalence [11] are widespread. A more accurate picture of the intracellular medium as a crowded environment that is not comparable to ideal solutions is contributing to the reframing of cells as functional wholes. Techniques to investigate the physical state of the cytoplasm in vivo are being used to characterise quinary structures, the role of fractal kinetics in cell homeostasis [12] and the physical basis underlying the link between global oscillations in cytoplasm properties and metabolism [13].
The historical circumstances surrounding the formulation of protoplasmic and cell theories led to their antagonistic relationship. With our privileged position as future observers, this antagonism can easily be put aside, recognising that the two theories are not mutually exclusive. On the contrary, extensive use of the two is revealing itself instrumental in furthering our studies of cell biology. However, it is doubtful that current approaches will contribute to the identification of a valid physical basis of the living. Holism in the form of systems biology tends to devolve in the enumeration of components identified via the established reductionist framework [14]. This enumeration has been extended to interactions of components and its scale is enabling new statistical approaches but, as argued by [1], to understand life we need a different framework. Instead of looking at the one example of life currently available, features of known organisms could be replicated via emergence in de-novo constructed systems. This synthetic or constructive approach to biology is the most promising route to a universal biology yet, with the ability to indicate which features of life are the result of chance and which of necessity. The next sections are dedicated to the examination of cells and compartments in abiogenesis through this lens.
3. Compartmentalization and abiogenesis
The establishment of a common compositional identity of all living things was an essential achievement in the then new-born field of abiogenesis. Up until Pasteur’s confutation of spontaneous generation, it was commonly believed that life originates wherever it may. The fall of this concept, together with Lamarck’s and Darwin’s theories of evolution posed a whole new set of questions. All forms of life are evolutionary related and share a common morphology, thus life must have originated in the distant past and then differentiated in all the forms that populate the biosphere nowadays. Additionally, the protoplasmic paradigm states that life is based on common physicochemical principles, so nothing should prevent an all-knowing biochemist from replicating the event that gave rise to the original protoplasm. The quest for the synthesis of the protoplasm was, in fact, an early offshoot of the protoplasm theory, carried forward by pioneers such as Herrera in his experiments on plasmogeny [15]. In these first attempts we can find the core ideas of the constructive approach: the origin of life as viewed in plasmogeny addresses the subject very similarly to how a modern synthetic biologist would, albeit with much less refined tools. Parallel to that, the synthesis of urea from inorganic precursors by Wohler in 1828 initiated research on the abiogenesis of organic compounds and, with it, the complex interplay between chemistry and geology that is prebiotic chemical synthesis. A full discussion of the history of the field is beyond the scope of this section and is expertly reviewed in [16], but an introduction is needed in order to address the relevance of compartments in early life properly.
3.1. Evolvability in prebiotic replicators
Decades of development in origins research yielded two groups of theories, commonly described as information-first (or replication-first, genes-first, genetics-first) and metabolism-first [17]. To the first group belong theories that emphasise the emergence of self-replicating molecules from a prebiotic reaction mixture. As an extreme simplification of the central dogma of molecular biology, these self-replicators would act both as information storage molecules and catalysts for information transfer. The prime example of this is a replicase ribozyme that, once folded, could self-replicate using its complementary strand as template. These models focus on restricted inventories of compounds and polymeric replicators that would act in a very similar way as genetic replicators in modern organisms. The information encoded in the sequence of a polymer is interpreted via molecular recognition to copy it using freely available monomers. These protobionts would be absolute heterotrophs, relying on the environment for the supply of monomers or precursors. The most prominent member of the group is the RNA world [18] but other examples exist such as the amyloid world [19]. The second group focuses more on emergent phenomena in complex reaction mixtures and on how these could harvest geochemical gradients to persist. Wachtershauser’s iron-sulphur world is an exemplary case where the nature of heredity and replication is sidestepped, concerning only in the availability of energy gradients across physical barriers that would enable the growth of autocatalytic sets of molecules [20]. Kauffman’s theory of reflexively autocatalytic sets [21] and the more recent Graded Autocatalysis Replication Domain (GARD) model for autocatalytic growth of amphiphilic aggregates [22] deal more directly with the replication process, leaving the actual chemistry more abstract. The theory of autocatalytic sets as formulated by Kauffman states that, when a network grows to a certain size, catalytic closure is likely achieved. All members in a closed network have at least one reaction in their synthesis catalysed by a member of the network. Catalysed reactions lead to every member of the network from a maintained food set of molecules. Hordijk [21] provides an in-depth review of the theory’s development. GARD takes a similar point of view in addressing self-replication and focuses on self-assembled amphiphilic structures. For this reason, the model has been related to the “Lipid world hypothesis”. The key hypothesis of GARD is that the composition of an assembly can influence the entry/exit rates of monomers, a sort of catalysis (although the specific terminology has been disputed [23]). When kept out of equilibrium, simulations of such systems often lead to assembly with stable compositions (composomes) that could be the basis for information transfer.
Models for the origin of life based on catalytic networks have encountered resistance when proposed for two main reasons: their apparent lack of heredity and, thus, evolvability and the lack of experimental results on the matter [17], [24], [25]. Given the abstract nature of the models and their requirement for complex reaction environments, experimental work on the subject is mainly carried out in silico [26]. There is a somewhat diffident attitude towards results obtained by computer simulation over ones coming from wet-lab settings, probably because such contributions are a relatively new addition to a field otherwise dominated by chemistry. The other criticism generally levied at metabolism-first theories is on the evolvability of collections of molecules since, in known organisms, evolution operates primarily on the sequence of a specific polymer (Deoxyribonucleic acid or DNA). As noted in [26], every division event partitions genetic material and all other cellular components (cytoplasm, organelles…) between the daughter cells. Inheritance involves distribution of information digitally stored in DNA sequences and analogically stored in the concentrations of molecules present in different regions of the parent cell. Metabolism-first self-replicators just use analogic information transfer during replication. Inheritance for GARD simulations was assessed in [23], finding that simulations involving two kinds of interacting molecules behave very closely to models for the distribution of two alleles for a gene in population genetics. Inheritance is a necessary but not sufficient requirement for evolution. To evolve, the information transfer process must have non-zero error rates. Since there is no example of a purely analogue replicator, their ability to evolve cannot be taken for granted given compositional inheritance. The replicating units for different models, an autocatalytic (RAF) set for Kauffman’s framework and composomes for GARD, have been demonstrated to be theoretically equivalent [26], so no distinctions will be made in discussing their evolvability. Simulations comparing the variability within composomes and the quasispecies model introduced by Eigen [27] shows that composomes behave like quasispecies, thus making them targets for selection [28]. Based on the nature of the compositional space explored through variation, evolution can be open-ended or not: if the space can be exhaustively navigated by an arbitrary population, evolution is not open-ended, it is limited by the nature of the evolving units. Some extensions of GARD seem to indicate that these scenarios encompass open-ended evolution as well [29].
As models and scenarios are refined through time, the distinction between information-based and metabolism-based approaches is blurred. For example, the “molecular biologist’s dream” posited that an ancestral RNA-polymerase ribozyme used copies of itself as template in the first self-replicating systems [30]. Finding such a ribozyme proved difficult, given requirements on processivity and accuracy needed to avoid the error catastrophe [31]. This induced the expansion of the theory to accommodate alternative modes of polymer growth and intermediate steps where molecular ecologies would have produced the complex machinery needed for self-replication [32]. Sets of RNA oligomers that collectively replicate are very similar to mutually catalytic sets and have shown intricate evolutionary dynamics in previous experiments [33]. Although classically collocated in the information-first one, to which set the non-naïve RNA world should belong is not immediate: the distinction usually implied with metabolism-first and information-first is not meaningful given the state of the art. Additionally, the meaning of information in biology has changed since the first inception of these theories, justifying a revision of the usual nomenclature [34]. Since the main feature that sets the two views apart is the nature of inheritance, the next discussions will use analogue versus digital replicators to indicate the two groups [35]. This way, it is apparent that these are not mutually exclusive theories, and they can concern different scenarios or different moments in the development of prebiotic replicators. Once replicators started populating their environments, they had to thrive and diversify according to the respective evolutionary dynamics. Digital replicators are subject to variation through random mutation on sites of the polymer, while analogue ones can evolve according to pathways outlined in [36]. All species belonging to a catalytic network are overrepresented in terms of concentration in a given milieu thanks to catalytic focusing [37]. For this reason, all non-catalysed reactions that involve a member of the net are more likely. According to [36], a net of mutually catalytic reactions has inheritable units (cores) represented by strongly connected autocatalytic cycles, while species catalysed by cores but not part themselves of autocatalytic cycles are subject to fluctuations and represent the periphery of the net. Cores can be thought of as the genotype of the network, while the periphery is an expression of the cores, a kind of phenotype. Through rare chance, new catalytic species may emerge from rare non-catalysed reactions, fixating new members in the net and altering the core-periphery partition. Additionally, the net may lose some of its structures or members due to stochastic dilution of species.
One universal feature of evolvable replicators is the presence of two separate timescales of variation: processes that allow for the persistence and growth of the replicator must be faster than ones leading to its variation. In cells, this is evident in the rates of metabolic reactions, orders of magnitudes larger than a cell cycle and mutations. Reactions that lead to inheritable change are in most cases non catalysed and lead to cell degeneration if favoured. The same considerations hold for ancestral replicators. The theory regarding digital replicators has solid foundations in Eigen’s treatment [32]. Less is known on the robustness of analogue replicators’ evolvability. The basics of inheritance and variation are recent advancements of the theory so it is likely that soon work will be dedicated to this issue. Clarifying what ranges of parameters allow for evolvability while still accounting for robustness to environmental changes and parasitism are essential steps to inform wet lab realizations of these hypothetical systems.
3.2. Compartments yield more robust replicators
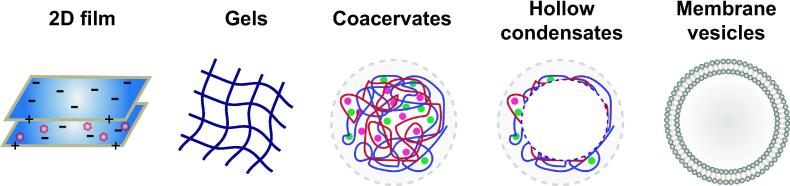
A growing population of replicators will encounter several limitations in free solution: catalysis can require interactions between many molecules and low concentrations of species in bulk volumes could be a limiting factor. For digital replicators, parasites are quick to emerge from mutants that lose catalytic activity but retain template properties, and usually they grow at faster rates than the actual replicator [38]. Similarly, in analogue replicators side reactions may drain the network, hindering its survival. Other than parasitism, the stability of the replicators’ basic constituents in solution would also be a limiting factor. Amino acids and other biomolecules readily racemize when dissolved in water, effectively reducing the availability of food. The presence of compartments or environments with low water activity slows down this process, endowing an additional selective advantage to these systems [39]. One of the simplest mechanisms for replicators to escape being overrun by parasites is their collocation in a spatially defined setting. An illustrative example of this effect has been reported by [40], where simulations of a hypercycle [32] that would normally be sensitive to parasites, gained resistance by segregating the various steps of the cycle in spiral patterns that emerged in spatially discrete simulation. A number of such systems have supposedly played a role in the origin of life, the most relevant of which are schematically depicted in Fig. 3 and further described here.
Fig. 3.
Different modes of compartmentalization may have been involved in the evolution of life. Ordered from left to right based on their capacity to limit exchanges with the environment: 2D films, like adsorbed nucleic acids on clay or lipid lamellar phases; organic hydrogels such as those formed by high concentrations of nucleic acids; coacervates that phase separate from the bulk above critical concentrations of charged polymers; hollow coacervates, obtained from the previous assemblies under specific conditions; lipid membranes closed in a spherical shape, such as vesicles or liposomes.
The simplest forms of spatial localisation are imposed on models by embedding them in a bidimensional lattice, emulating the effect of an adsorbing surface. Many such materials may have played a role in early chemical evolution, such as pyrite, silicates like montmorillonite, liquid interfaces, or porous rock channels to name a few [41], [42]. The reduction in dimensionality awards some resistance to parasitism, but diffusion is still allowed on the surface and through adsorption/desorption equilibria. A more functional spatial organisation for replicators is compartmentalization. Entities that contain spatially distinct populations of replicators confine the effect of parasites to the compartments where they emerge, greatly reducing their impact. In silico simulations of systems with limited diffusion (e.g. surfaces or porous materials) and protocellular ones highlighted the key advantage of the latter. The presence of a boundary that limits diffusion among compartments greatly increases the replicators’ survivability [43]. Hydrogels and lipidic aggregates can show such behaviour by slowing the diffusion of species hosted in them, even though they lack a neat boundary. Another essential role of compartmentalization is its ability to maintain concentration gradients between a continuous phase and an enclosed one. A protobiont can exploit these gradients through transport mechanisms to maintain an active metabolism, for example, by importing food and excreting waste. Two prebiotically-plausible systems have been subject to extensive research until now: coacervates and lipid vesicles.
Coacervates are organic molecular aggregates that are readily formed in polyelectrolytes solutions. At a critical concentration, polymers of opposite charge start interacting favourably leading to liquid–liquid phase separation of polymer-rich droplets. Oparin was among the first to propose the relevance of coacervation to the origin of life since coacervates would be readily formed in a hypothetical prebiotic soup. Coacervate droplets have demonstrated the ability to retain macromolecules with catalytic activity such as enzymes [44] and ribozymes [45] and to maintain concentration gradients [46]. Recent advances have shown that coacervation takes place even in the presence of low molecular weight poly Lysine when combined with nucleotides [47]. Additionally, it has been reported that the ratio of positive to negative polymer can induce shape transitions in the polymer-rich phase, from droplets to hollow condensates [48]. The presence of an aqueous lumen hints at the possible establishment of transport mechanisms and of different environments within the same particle, making them more chemically versatile. Coacervates are ideal candidates for a messy origin of life, where the environment was populated by a highly heterogeneous mix of compounds since they can harness this heterogeneity to their advantage. By combining them with lipids, it was shown that the droplet’s surface can support the formation of a lipid bilayer that would constitute a second permeability barrier [49]. Even though coacervates are a promising avenue for prebiotic encapsulation, by far the most studied systems are lipid vesicles. These compartments are bound by a lipid bilayer, similar to a simplified cell membrane. The main distinction between the two is the absence of proteins and the relative homogeneity in composition: where a cell may have hundreds of different amphiphiles in the same membrane, liposomes and vesicles are often made up of few components. For their relevance in cell biology, lipids have been extensively characterised and mechanical or chemical properties of several membrane compositions are readily available in the literature. In origin of life studies, single chain amphiphiles, mainly fatty acids, are preferred since they are easier to synthetise abiotically. Their disadvantage is the sensitivity of the vesicles thus formed to pH, ions concentrations and temperature, all variables difficult to account for in most origin of life scenarios. Furthermore, their high critical vesicular concentration prevents their formation unless a large supply of amphiphiles is available. These constraints have been softened by studying the effect of heterogeneous membrane composition, including ones that combine fatty acids with alcohols, amines, or phosphates [50]. These results highlight the stabilization of the bilayer by heterogeneous compositions. Other advantages of lipid vesicles as compartments are their osmotic responsiveness, the over-encapsulation of macromolecular solutes and their ability to spontaneously deform according to the phase behaviour of the membrane-forming lipids:
-
•
Osmotic imbalance across a lipidic membrane increases tension, which has been shown to drive membrane growth. If the compartment hosts an active metabolism, this may fuel the appropriate osmotic gradient and lead to compartment growth proportional to metabolic activity. This is a minimal way to link compartment multiplication with the processivity of an internal metabolism [51].
-
•
Luisi et al. have first reported that when incapsulating macromolecules in vesicles by spontaneous swelling of lipid films, the distribution of encapsulated proteins does not follow the expected Poissonian. Compartments are populated according to an exponential distribution, resulting in higher frequencies of empty compartments, but also in an over-representation of vesicles with high protein concentration. This has an important impact for the use of vesicles containing complex reaction mixtures, for example in bioreactor applications. The over-representation of vesicles with high contents of macromolecules increases co-encapsulation likelihood even at relatively low concentrations of essential components [52].
-
•
Lipids are packed in monolayers according to their molecular geometry. Bulky hydrophilic or hydrophobic groups can influence the overall curvature of the monolayer and stabilise different geometries. Local variations of curvature have been used to stabilize pores [53], adhesion patches [54] and to induce vesicle division via budding [55]. Most of these studies used phospholipids, but these principles of molecular packing and geometry applies to any self-assembled structure. Spontaneous shape deformations of compartments would lay the basis for a primitive cell cycle.
A third class of relevant cell-like enclosures are porous mineral precipitates. These have an important role in hypothesis on the origin of life in hydrothermal sites on the ocean floor [56], [57]. The newly established field of chemobrionics [58] focuses on the patterns and structures that arise in heterogeneous systems such as chemical gardens. Vents like black smokers and chimneys found at Lost City naturally establish pH gradients across the precipitate walls that constitute them. These gradients may have fuelled the first steps in chemical evolution towards replicators and the cell-sized chambers of the structure would have acted as a built-in form of compartmentalization. Studies in chemobrionics are now focussing on the structural properties of precipitate structures, such as ion fluxes [59] or morphology of precipitates in flow systems [60], opening up the possibility of systems chemistry approaches to the study of mineral precipitate compartments.
Different ways of compartmentalising prebiotic replicators may have participated in chemical evolution towards the first protocells [41], with possible intermediate forms such as membrane-bound coacervates [42]. An important aspect to underline is that in no case compartments were merely ways of establishing boundaries with the environment. They likely played a role in metabolism and communication from the beginning [61]. For example, a recent proposal for the origin of life in terrestrial environments that relies on hydration-dehydration cycles posits that lipid membranes and progenote-like lipid gel phases alternated in the key steps of chemical evolution [62].
4. Membranes and evolution: LUCA and the membrane divide
The simplistic replicators described in the previous section still had a long way to go before resembling an actual cell. The evolution from replicators to reproducers as described by [63], and included among the major transitions in evolution [64], is still on the fringes of origins of life discourse since crucial steps before and after are still up for debate. Strong evidence suggests that this transition required profound modifications of the replicators’ compartment boundary, in the direction of reduced permeability. The encapsulation of replicators in compartments ensures genotype-phenotype linking. Compartment division combined with replication of inheritable material appear to be required for Darwinian evolution, namely the inheritance of phenotypes along distinct lineages. In experiments on RNA replication by the Qβ replicase, evolution towards increasing affinity for the template occurred only once the reaction mix was encapsulated in water droplets [65]. Additionally, according to the stochastic corrector model, the transition to cellularity led to the organization of genetic material in chromosomes, that would ensure the inheritance of all genetic traits hosted in the compartment [66], [67]. The random redistribution of genes separated in different molecules lowers the likelihood of faithful inheritance upon cell division. More reliable information transmission would be selected for by concatenation of separate genes into chromosomes. At first, the boundary needed to be permeable, to avoid the starvation of the hosted replicator. Permeability plays an important role in non-enzymatic RNA replication experiments, where the diffusion of RNA oligomers from the environment to the protocell allows for the replication of long stretches of mixed-sequence RNA [68]. After generations, the membrane became gradually less permeable by coevolving with transport mechanisms. The alternative hypothesis would require the emergence of replicators and transporters together which is not plausible without selective pressure towards control of exchanges with the environment. This theme of progressive loss of compartment permeability in the cellularization of protobionts is invoked in numerous theories. For example, various modes of compartmentalization are sometimes put in a hierarchy according to their permeability (and cell-like appearance), where the more permeable ones are relevant to older stages of the origin of life [41]. There are also indications that the incorporation of compounds that lower permeability in lipid membranes would award them with selective advantages. It was shown by [69] that phospholipid-fatty acids mixed membranes tend to grow at the expense of pure fatty acids ones. This is due to the low diffusion rate of phospholipids from one membrane to another and ultimately leads to higher growth and division rates of less permeable compartments. Lower permeability is also the driver of metabolic pathway enzymatization according to the progressive sequestration hypothesis [70]. Starting from pathways fuelled by freely exchanged materials, there will be a selective pressure for the catalytic formation of the most depleted substrates as the boundary becomes less permeable. Once these catalysts are established, the pressure moves upstream, leading to the progressive enzymatization of the whole pathway. The model is further subdivided in pathway retention or pathway innovation for autotrophic and heterotrophic metabolisms respectively and is an alternative to the patchwork model [71].
Membrane permeability also determined in large part how protocells harvested energy from their environment. The universality of proton gradient use by the ATP synthase complex and the use of sodium ions by some acetogens for the same purpose can be explained by evolutionary considerations on ion permeability during the evolution of cellularity in alkaline vents. Amphiphiles involved in the first forms of compartmentalization were unlikely to support a strong coupling between gradient and phosphorylation due to their permeability, so in [72] it was proposed that at this evolutionary stage sodium was used instead of protons. Cellularity in geochemically sustained autotrophic origins of life (for example in the iron-sulphur world or abiogenesis in alkaline vents) is also a central tenant in some of the most complete narrations of the origin of life published thus far [73]. These theories include a particular evolutionary step between protocells and the first divergence between eubacteria and archaea, the progenote stage, where intercellular exchange of materials was commonplace. All cells in the progenote evolved collectively because of the extensive horizontal gene transfer that supposedly took place. According to [74] this stage of pre-Darwinian evolution allowed for the fixation of the universal genetic code and the progressive reduction of gene sharing through led to the so-called Darwinian transition, where cells could behave as distinct entities and eventually develop in separate species. [73] extends the non-free-living state after the divergence point from common ancestry. This would mean that there was no last universal common ancestor, rather a common progenote that split into diverging populations. The evidence in support of common ancestry is not contradicted, just the idea that there was a single lineage of organisms that originated all domains of life [75]. Non-free-living ancestors of Archaea and Eubacteria are incorporated in theories on the lipid divide, accounting for the fact that eukaryotes and eubacteria have membranes made of sn‐glycerol‐3‐phosphate lipids, while members of archaea mainly use sn‐glycerol‐1‐phosphate isomers. Additionally, archaea most often have membranes made of isoprenoid lipids arranged in a monolayer, but more detailed analysis revealed that they retain the ability to synthetise phospholipids [76]. To complicate things, Eukaryotes are phylogenetically closest to Archaea than Eubacteria, but their membranes are composed of bacterial-type phospholipids. The branch point of Eukaryotes and the incongruence in membrane composition are stimulating a large number of studies, as summarised in [77]. According to the progenote theory, the divide emerged because eubacteria and archaea originated from two separate populations and independently gained the ability to thrive as free-living cells. The same paper also outlines how the endosymbiosis of an eubacterium in an archaeon led to the first eukaryotic cell being formed and how lipids from the endosymbiont ended up replacing the original membrane. It should be noted that this theory is highly disputed. Other models account for the lipid divide via biophysical arguments, such as the lower stability of racemic membranes [78] or the heightened activity of membrane-bound proteins in enantiopure membranes [79]. Furthermore, the idea of a progenote that evolved via non-Darwinian dynamics is far from validated, and many phylogenetic approaches indicate its implausibility [80]. All in all, little is known of the LUCA (last universal common ancestor) and how it evolved. Phylogenetic approaches are biased by the limited number of archaea genomes at our disposal and, although this number is rapidly increasing, it is a factor that strongly limits these techniques [76]. Archaea is a relatively new addition to the tree of life, and radical changes in the state of the art on the matter are quite frequent (e.g. the discovery of Asgardarchaeota superphylum). Omic techniques are accelerating progress and approaches combining multiple structural homologies in their criterion for monophyly have been able to circumvent some of the shortcomings of past efforts [81]. This field is rapidly changing and, even though we are far from having a solid picture of the evolutionary history that led to prokaryotes, we may soon be able to infer the history of cellularization and successive major evolutionary transitions with greater confidence.
In this section, two approaches to the study of cellularity have been presented. The first one starts with hypothetical scenarios and builds experimental systems around such hypothesis to draw conclusions. The second is more interested in reading the leftover evidence in modern organisms and reconstructing the phylogeny of life on Earth. If we want to rewind the tape of evolution both ways of proceeding are needed, but only the first one will yield generalisable statements on biology.
5. Non-cellular life and its’ limits
What discussed thus far indicates the foundational importance of cellularity and especially of membrane-bound compartments in the evolution of living things. Mitigating parasitism, regulating exchanges of food and waste, enabling the use of gradients to drive cellular metabolism are all evolutionary advantages of compartmentalised systems. Additionally, as reviewed in sections 3 and 4, once compartments are established, a selective pressure towards less permeable membrane compositions is naturally instated, allowing tighter control over the exchanges between protobiont and environment. This suggests that the physical basis of life is indeed the cell, in a universal sense.
Biology is in a unique position among all sciences when it comes to universal principles. Since all living things characterised so far are monophyletically related, extrapolating general features from the examples we can study is an arduous task. Additionally, contrary to what happened with computer science, where the general theory was already available when the first computing machines were built, biology does not have general theories to base deduction upon [74]. This is a contributing factor to the elusive nature of a comprehensive definition of life. Attempts at it are abundant and relevant, but fundamentally operational [82]: such definitions are very useful framing devices but limited as such. None of the currently available lists of properties of living things has the predictive power to inform astrobiology or synthetic cell studies in an unbiased way. Such efforts are conditioned by what is currently available to observation and would benefit greatly from general principles [83]. This is not to say that a definition is necessary to further scientific understanding in these fields. As noted by Szostak for the origin of life, it is sufficient to know the beginning and end points, not where the (often arbitrary) line between the inanimate and the living stands [84]. But, even though the definition of life does not bear much relevance for its origin, the reciprocal may not be as valid. Through the study of the origin of life, we are narrowing down on the features that lead to open-ended evolution and the increase in complexity associated with major evolutionary transitions [85]. At the same time, studies inspired by origins of life scenarios are expanding the semantic field usually involved in definitions of life, bringing new ideas to the table [86]. The current focus of origins research is to find out how life originated, here it is advocated that it could instead focus on how life originates. The question of the origin of life as it happened roughly 4.5 billion years ago on Earth is answered by providing a chronology of subsequent evolutionary changes from relevant prebiotic chemistry to relevant protobiotic systems to relevant cellular ancestors. It is corroborated by fossil and geochemical records and phylogenetic studies that confine the plausibility of proposed theories. But this specificity is also a shortcoming if such an answer is not used to extrapolate alternative scenarios and developmental pathways for life. Most of the criticism levied at certain theories or models has been related to their prebiotic plausibility, a notion that is inextricably linked to the many contingencies of the one origin of life we know of. By softening this constraint, origins studies could define how life develops in a general sense, yielding an inclusive origin of life. As depicted in Fig. 4, a general tree of life would have the tree-like branching we associate to evolution nowadays, but also net-like components and even completely different trunks that were not populated during our origin of life.
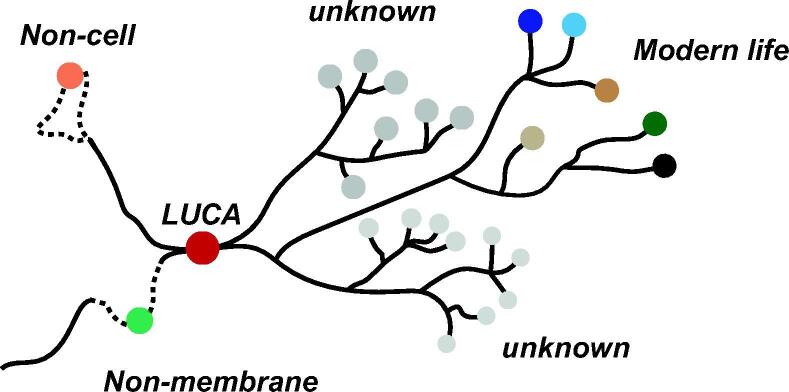
Fig. 4.
Sketch for a universal tree of life. The central branch originates at the Last Universal Common Ancestor and yields many leaves, including but not limited to the three surviving domains. Side paths connecting different branches can indicate horizontal gene transfer and endosymbiotic events. The basis of the tree has two additional hypothetical paths for evolution: non-cellular life, like the communal progenote described previously, and non-membrane life, where evolution leads to compartments that were not bound by impermeable lipid membranes. The viability and complexity of such branches can be investigated via the constructive approach and could also indicate whether we would still classify these as domains of life.
In light of these considerations, the subject of compartmentalization and cellularity as basis for life can be assessed. It is not uncommon to find sentences such as “all life is cellular” in the literature. Of course, these short-hand expressions are not meant to include acellular stages in the life cycle of some organisms [87] and really mean all known life is cellular, but they nonetheless show how the cell theory is used to ground arguments in contemporary biological discourse. That life is cellular is self-evident at this point, what can be challenged in these statements is the implicit assumption that life has to be cellular. For instance, simulations conducted by [88] do not specify any form of compartmentalization, rather complex behaviour emerges in a spatially heterogeneous chemical system due to the combination of diffusion and degradation rates used. The patches of chemicals can be followed as individuals for the course of the simulation, showing that individuation is not synonymous with the presence of a membrane. Furthermore, the presence of parasitic relationships between chemical species can lead to the emergence of complex behaviour rather than the disruption of homeostasis. Within some parameters’ subspaces, it even results in a mutualism that allows for the survival of the replicators only when the parasite is present [89]. A recent work has also shown that the benefits imparted by compartments to simple replicators can stem from spatial segregation in turbulent flow [90]. These findings suggest that forms of compartmentalization can appear in a homogeneous solvent, softening the constraints for replicator sustainability to any condition that limits diffusion, rather than requiring material partitioning of the environment. Cellularity is not a fundamental requirement of self-replicating units but, as discussed in sections 2 and 3, it enables further evolutionary transitions after it emerges. Through these considerations, it appears that replicators can emerge and evolve without cells, but there may be an evolutionary threshold that can only be surpassed after the acquisition of endogenous boundaries. This line of thought may inspire future research in what such limits in complexity might be, or how they may be circumvented. An interesting study in this direction is reported in [91]. In this simulation, cellularity and metabolism are orthogonal properties of hypothetical protobionts. Rather than implementing compartmentalization as a fixed condition, it is linked to a variable permeability degree that can evolve through generations. Environments with different food material and energy availability deeply affect the evolutionary outcome of simulations. With scarce resources, cellularity is beneficial and co-evolves with a proficient metabolism, whereas when resources are abundant and freely available, there is no pressure to develop less permeable boundaries and efficient metabolisms.
The studies here presented indicate a way of generalising our ideas on the evolution of life. Besides contributing to universal biology, questioning the necessity of compartments and cells could inspire experiments to assess the limitations of acellular systems. In silico results could be corroborated by wet lab counterparts, taking inspiration by ancillary fields such as techniques in enzyme immobilization and interface-rich systems used in chemical engineering [92], [93].
6. Conclusions
The widespread acceptance of the cell theory of life opened a new age of biology. Today we are witnessing a similar process, with the reconceptualization of living things as systems. Systems biology is expanding biological knowledge in ever new directions with the integrated use of omic data and techniques. This systemic view together with the constructive approach are influencing origins research, with the birth of systems chemistry and the recent proposal for a “systems protobiolgy” [26] as a way forward for the field. The integration of approaches and theories in recent times helped the unification of previously separate and often antagonized theories [94] and is a trend that will with no doubt help develop a generalised theory for the origin of life. With these optics the idea of cells as atomic units of the living has been examined, in lieu of updated theories on abiogenesis and evolution. Cellularity appears to be facultative for the emergence and evolution of replicators, while it gains a more profound importance at the transition to reproducers. Once reproducers emerge, selective pressure can drive the impermeabilization of the boundary, leading to cell membranes with sensing and transport faculties. The existence of boundaries that lead to strong individuation lays the basis for the further complexification that characterizes the domains of life, such as endosymbiotic relationships, multicellularity, and modularity. Questioning the necessity of compartments, cells and membranes is a fruitful endeavour in the current biological landscape, with potential repercussions on astrobiology and evolutionary biology, and will reframe many questions in the origins field, eventually yielding generalisable answers. The lack of clear individuation and endogenous barriers (such as in the inhabitants of hydrothermal vents [73]) would put many concepts that are now part of our basic understanding of the living in a new light. For example, autopoiesis [95] would not apply to the systems just mentioned but we would argue they still belong in the tree of life, given their ability to reproduce and evolve. This would subdivide the domains of life in autopoietic and non-autopoietic entities.
CRediT authorship contribution statement
Adriano Caliari: Writing - original draft. Jian Xu: Conceptualization, Visualization, Supervision, Writing - review & editing. Tetsuya Yomo: Conceptualization, Supervision, Writing - review & editing, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Dr Martin M. Hanczyc for helpful comments on this manuscript. We also thank Dr Doron Lancet for insightful discussions on the GARD model. This work was supported by the National Key R&D Program of China, Synthetic Biology Research (2019YFA0904500); MOE International Joint Laboratory of Trustworthy Software at East China Normal University.
Contributor Information
Jian Xu, Email: xujian@sei.ecnu.edu.cn.
Tetsuya Yomo, Email: tetsuyayomo@gmail.com.
References
- 1.Kaneko K. Springer; 2006. Life: an introduction to complex systems biology. [Google Scholar]
- 2.Allen G.E. Mechanism, vitalism and organicism in late nineteenth and twentieth-century biology: the importance of historical context. Stud Hist Philos Biol Biomed Sci. 2005;36(2):261–283. doi: 10.1016/j.shpsc.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Welch G.R., Clegg J.S. From protoplasmic theory to cellular systems biology: a 150-year reflection. Am J Physiol Cell Physiol. 2010;298(6):C1280–C1290. doi: 10.1152/ajpcell.00016.2010. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds A. The cell's journey: from metaphorical to literal factory. Endeavour. 2007;31(2):65–70. doi: 10.1016/j.endeavour.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Welch G.R., Clegg J.S. Cell versus protoplasm: revisionist history. Cell Biol Int. 2012;36(7):643–647. doi: 10.1042/CBI20120128. [DOI] [PubMed] [Google Scholar]
- 6.Huxley T.H., Harry Houdini Collection (Library of Congress) Macmillan and Co.; Macmillan Co; London New York: 1902. Lectures and essays; p. 128. [Google Scholar]
- 7.Geison G.L. The protoplasmic theory of life and the vitalist-mechanist debate. Isis. 1969;60(3):273–292. doi: 10.1086/350498. [DOI] [PubMed] [Google Scholar]
- 8.At Kleinzeller. Exploring the cell membrane: conceptual developments. 1995;xiv:359 p.. [Google Scholar]
- 9.Yeagle P. xii. Elsevier/AP; Amsterdam; Boston: 2016. (The membranes of cells). [Google Scholar]
- 10.Kollek R., Petersen I., Döring M., Brüninghaus A. Back into future: the systems biology to come. Contextual Syst Biol. 2015:283–301. [Google Scholar]
- 11.Nicholson D.J. Organisms not equal machines. Stud Hist Philos Biol Biomed Sci. 2013;44(4 Pt B):669–678. doi: 10.1016/j.shpsc.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Aon M.A., Cortassa S. Function of metabolic and organelle networks in crowded and organized media. Front Physiol. 2014;5:523. doi: 10.3389/fphys.2014.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagatolli L., Stock R. The cell as a gel: material for a conceptual discussion. Physiol Mini Rev. 2016;9(5):38–49. [Google Scholar]
- 14.Kaneko K. The challenges facing systemic approaches in biology: an interview with Kunihiko Kaneko. Front Physiol. 2011;2:93. doi: 10.3389/fphys.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negrón-Mendoza A. Alfonso L. Herrera: a Mexican pioneer in the study of chemical evolution. J Biol Phys. 1995;20(1–4):11–15. [Google Scholar]
- 16.Lazcano A. Historical development of origins research. Cold Spring Harb Perspect Biol. 2010;2(11) doi: 10.1101/cshperspect.a002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazcano A. What is life? A brief historical overview. Chem Biodivers. 2008;5(1):1–15. doi: 10.1002/cbdv.200890001. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert W. Origin of life: the RNA world. Nature. 1986;319(6055) 618 618. [Google Scholar]
- 19.Maury C.P.J. Amyloid and the origin of life: self-replicating catalytic amyloids as prebiotic informational and protometabolic entities. Cell Mol Life Sci. 2018;75(9):1499–1507. doi: 10.1007/s00018-018-2797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wachtershauser G. Groundworks for an evolutionary biochemistry: the iron-sulphur world. Prog Biophys Mol Biol. 1992;58(2):85–201. doi: 10.1016/0079-6107(92)90022-x. [DOI] [PubMed] [Google Scholar]
- 21.Hordijk W. A History of autocatalytic sets. Biol Theory. 2019;14(4):224–246. [Google Scholar]
- 22.Segre D., Ben-Eli D., Deamer D.W., Lancet D. The lipid world. Orig Life Evol Biosph. 2001;31(1–2):119–145. doi: 10.1023/a:1006746807104. [DOI] [PubMed] [Google Scholar]
- 23.Wu M., Higgs P.G. Compositional inheritance: comparison of self-assembly and catalysis. Orig Life Evol Biosph. 2008;38(5):399–418. doi: 10.1007/s11084-008-9143-4. [DOI] [PubMed] [Google Scholar]
- 24.Lancet D., Segre D., Kahana A. Twenty years of “lipid world”: a fertile partnership with David Deamer. Life (Basel) 2019;9(4):77. doi: 10.3390/life9040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasas V., Szathmary E., Santos M. Lack of evolvability in self-sustaining autocatalytic networks constraints metabolism-first scenarios for the origin of life. Proc Natl Acad Sci U S A. 2010;107(4):1470–1475. doi: 10.1073/pnas.0912628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lancet D., Zidovetzki R., Markovitch O. Systems protobiology: origin of life in lipid catalytic networks. J R Soc Interface. 2018;15(144):20180159. doi: 10.1098/rsif.2018.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eigen M., McCaskill J., Schuster P. vol. 75. J. Wiley & Sons; New York: 1989. (The molecular quasispecies). [Google Scholar]
- 28.Gross R., Fouxon I., Lancet D., Markovitch O. Quasispecies in population of compositional assemblies. BMC Evol Biol. 2014;14:265. doi: 10.1186/s12862-014-0265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markovitch O., Sorek D., Lui L.T., Lancet D., Krasnogor N. Is there an optimal level of open-endedness in prebiotic evolution? Orig Life Evol Biosph. 2012;42(5):469–474. doi: 10.1007/s11084-012-9309-y. [DOI] [PubMed] [Google Scholar]
- 30.Joyce GF, Orgel LE (1999) Prospects for understanding the origin of the RNA world. Cold Spring Harbor Monograph Series pp. 3749–3478.
- 31.Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58(10):465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 32.Higgs P.G., Lehman N. The RNA World: molecular cooperation at the origins of life. Nat Rev Genet. 2015;16(1):7–17. doi: 10.1038/nrg3841. [DOI] [PubMed] [Google Scholar]
- 33.Vaidya N., Manapat M.L., Chen I.A., Xulvi-Brunet R., Hayden E.J. Spontaneous network formation among cooperative RNA replicators. Nature. 2012;491(7422):72–77. doi: 10.1038/nature11549. [DOI] [PubMed] [Google Scholar]
- 34.Ball P. The problems of biological information. Philos Trans A Math Phys Eng Sci. 2016;374(2063):20150072. doi: 10.1098/rsta.2015.0072. [DOI] [PubMed] [Google Scholar]
- 35.Szathmáry E. Chemes, genes, memes: a revised classification of replicators. Lect Mathem Life Sci. 1999;26:1–10. [Google Scholar]
- 36.Vasas V., Fernando C., Santos M., Kauffman S., Szathmary E. Evolution before genes. Biol Direct. 2012;7(1) doi: 10.1186/1745-6150-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bagley R.J., Farmer J.D. Los Alamos National Lab; NM (USA): 1990. Spontaneous emergence of a metabolism. [Google Scholar]
- 38.Bansho Y., Furubayashi T., Ichihashi N., Yomo T. Host–parasite oscillation dynamics and evolution in a compartmentalized RNA replication system. Proc Natl Acad Sci U S A. 2016;113(15):4045–4050. doi: 10.1073/pnas.1524404113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toxvaerd S. A prerequisite for life. J Theor Biol. 2019;474:48–51. doi: 10.1016/j.jtbi.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Boerlijst M.C., Hogeweg P. Spiral wave structure in pre-biotic evolution: hypercycles stable against parasites. Physica D. 1991;48(1):17–28. [Google Scholar]
- 41.Monnard P.A., Walde P. Current ideas about prebiological compartmentalization. Life (Basel) 2015;5(2):1239–1263. doi: 10.3390/life5021239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monnard P.A. Taming prebiotic chemistry: the role of heterogeneous and interfacial catalysis in the emergence of a prebiotic catalytic/information polymer system. Life (Basel) 2016;6(4):40. doi: 10.3390/life6040040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah V., de Bouter J., Pauli Q., Tupper A.S., Higgs P.G. Survival of RNA replicators is much easier in protocells than in surface-based, spatial systems. Life (Basel) 2019;9(3):65. doi: 10.3390/life9030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokolova E., Spruijt E., Hansen M.M.K., Dubuc E., Groen J. Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proc Natl Acad Sci U S A. 2013;110(29):11692–11697. doi: 10.1073/pnas.1222321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drobot B., Iglesias-Artola J.M., Le Vay K., Mayr V., Kar M. Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nat Commun. 2018;9(1):3643. doi: 10.1038/s41467-018-06072-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vieregg J.R., Tang T.Y.D. Polynucleotides in cellular mimics: coacervates and lipid vesicles. Curr Opin Colloid Interface Sci. 2016;26:50–57. [Google Scholar]
- 47.Cakmak F.P., Choi S., Meyer M.O., Bevilacqua P.C., Keating C.D. Prebiotically-relevant low polyion multivalency can improve functionality of membraneless compartments. Nat Commun. 2020;11(1):5949. doi: 10.1038/s41467-020-19775-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alshareedah I., Moosa M.M., Raju M., Potoyan D.A., Banerjee P.R. Phase transition of RNA−protein complexes into ordered hollow condensates. Proc Natl Acad Sci U S A. 2020;117(27):15650–15658. doi: 10.1073/pnas.1922365117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dora Tang T.Y., Rohaida Che Hak C., Thompson A.J., Kuimova M.K., Williams D.S. Fatty acid membrane assembly on coacervate microdroplets as a step towards a hybrid protocell model. Nat Chem. 2014;6(6):527–533. doi: 10.1038/nchem.1921. [DOI] [PubMed] [Google Scholar]
- 50.Deamer D. The role of lipid membranes in life's origin. Life (Basel) 2017;7(1):5. doi: 10.3390/life7010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen I.A., Roberts R.W., Szostak J.W. The emergence of competition between model protocells. Science. 2004;305(5689):1474–1476. doi: 10.1126/science.1100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luisi P.L., Allegretti M., Pereira De Souza T., Steiniger F., Fahr A. Spontaneous protein crowding in liposomes: a new vista for the origin of cellular metabolism. ChemBioChem. 2010;11(14):1989–1992. doi: 10.1002/cbic.201000381. [DOI] [PubMed] [Google Scholar]
- 53.Sakuma Y., Taniguchi T., Imai M. Pore formation in a binary giant vesicle induced by cone-shaped lipids. Biophys J. 2010;99(2):472–479. doi: 10.1016/j.bpj.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakuma Y., Imai M., Yanagisawa M., Komura S. Adhesion of binary giant vesicles containing negative spontaneous curvature lipids induced by phase separation. Eur Phys J E Soft Matter. 2008;25(4):403–413. doi: 10.1140/epje/i2007-10307-0. [DOI] [PubMed] [Google Scholar]
- 55.Urakami N., Jimbo T., Sakuma Y., Imai M. Molecular mechanism of vesicle division induced by coupling between lipid geometry and membrane curvatures. Soft Matter. 2018;14(16):3018–3027. doi: 10.1039/c7sm02188g. [DOI] [PubMed] [Google Scholar]
- 56.Russell M.J., Martin M. The rocky roots of the acetyl-CoA pathway. Trends Biochem Sci. 2004;29(7):358–363. doi: 10.1016/j.tibs.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 57.Russell M.J., Ponce A. Six 'Must-Have' minerals for life's emergence: olivine, pyrrhotite, bridgmanite, serpentine, fougerite and mackinawite. Life (Basel) 2020;10(11):291. doi: 10.3390/life10110291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barge L.M., Cardoso S.S.S., Cartwright J.H.E., Cooper G.J., Cronin L. From chemical gardens to chemobrionics. Chem Rev. 2015;115(16):8652–8703. doi: 10.1021/acs.chemrev.5b00014. [DOI] [PubMed] [Google Scholar]
- 59.Ding Y., Cartwright J.H.E., Cardoso S.S.S. Intrinsic concentration cycles and high ion fluxes in self-assembled precipitate membranes. Interface Focus. 2019;9(6) doi: 10.1098/rsfs.2019.0064. 20190064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding Y., Batista B., Steinbock O., Cartwright J.H.E., Cardoso S.S.S. Wavy membranes and the growth rate of a planar chemical garden: Enhanced diffusion and bioenergetics. Proc Natl Acad Sci U S A. 2016;113(33):9182–9186. doi: 10.1073/pnas.1607828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanczyc M.M., Monnard P.A. Primordial membranes: more than simple container boundaries. Curr Opin Chem Biol. 2017;40:78–86. doi: 10.1016/j.cbpa.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 62.Damer B., Deamer D. The hot spring hypothesis for anorigin of life. Astrobiology. 2020;20(4):429–452. doi: 10.1089/ast.2019.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szathmáry E, Santos M, Fernando C (2005) Evolutionary potential and requirements for minimal protocells. In: Walde P, editor. Prebiotic Chemistry. Berlin, Heidelberg: Springer Berlin Heidelberg. pp. 167–211.
- 64.Szathmary E., Smith J.M. The major evolutionary transitions. Nature. 1995;374(6519):227–232. doi: 10.1038/374227a0. [DOI] [PubMed] [Google Scholar]
- 65.Mizuuchi R., Ichihashi N. Sustainable replication and coevolution of cooperative RNAs in an artificial cell-like system. Nat Ecol Evol. 2018;2(10):1654–1660. doi: 10.1038/s41559-018-0650-z. [DOI] [PubMed] [Google Scholar]
- 66.Grey D., Hutson V., Szathmary E. A re-exam ination of the stochastic corrector model. Proc R Soc Lond B. 1995;262(1363):29–35. [Google Scholar]
- 67.Kaneko K., Yomo T. On a kinetic origin of heredity: minority control in a replicating system with mutually catalytic molecules. J Theor Biol. 2002;214(4):563–576. doi: 10.1006/jtbi.2001.2481. [DOI] [PubMed] [Google Scholar]
- 68.O'Flaherty D.K., Kamat N.P., Mirza F.N., Li L., Prywes N., Szostak J.W. Copying of mixed-sequence RNA templates inside model protocells. J Am Chem Soc. 2018;140(15):5171–5178. doi: 10.1021/jacs.8b00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Budin I., Szostak J.W. Physical effects underlying the transition from primitive to modern cell membranes. Proc Natl Acad Sci U S A. 2011;108(13):5249–5254. doi: 10.1073/pnas.1100498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szathmary E. Coevolution of metabolic networks and membranes: the scenario of progressive sequestration. Philos Trans R Soc Lond B Biol Sci. 2007;362(1486):1781–1787. doi: 10.1098/rstb.2007.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jensen R.A. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 72.Lane N., Martin W.F. The origin of membrane bioenergetics. Cell. 2012;151(7):1406–1416. doi: 10.1016/j.cell.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 73.Martin W., Russell M.J. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci. 2003;358(1429):59–83. doi: 10.1098/rstb.2002.1183. discussion 83-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goldenfeld N., Biancalani T., Jafarpour F. Universal biology and the statistical mechanics of early life. Philos Trans A Math Phys Eng Sci. 2017;375(2109) doi: 10.1098/rsta.2016.0341. 20160341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Velasco J. Universal common ancestry, LUCA, and the Tree of Life: three distinct hypotheses about the evolution of life. Biol Philos. 2018;33:31. [Google Scholar]
- 76.Baker B.J., De Anda V., Seitz K.W., Dombrowski N., Santoro A.E. Diversity, ecology and evolution of Archaea. Nat Microbiol. 2020;5(7):887–900. doi: 10.1038/s41564-020-0715-z. [DOI] [PubMed] [Google Scholar]
- 77.Eme L., Spang A., Lombard J., Stairs C.W., Ettema T.J.G. Archaea and the origin of eukaryotes. Nat Rev Microbiol. 2017;15(12):711–723. doi: 10.1038/nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- 78.Koga Y. Early evolution of membrane lipids: how did the lipid divide occur? J Mol Evol. 2011;72(3):274–282. doi: 10.1007/s00239-011-9428-5. [DOI] [PubMed] [Google Scholar]
- 79.Sojo V. Why the lipid divide? membrane proteins as drivers of the split between the lipids of the three domains of life. BioEssays. 2019;41(5) doi: 10.1002/bies.201800251. [DOI] [PubMed] [Google Scholar]
- 80.Becerra A., Delaye L., Islas S., Lazcano A. The very early stages of biological evolution and the nature of the last common ancestor of the three major cell domains. Annu Rev Ecol Evol Syst. 2007;38:361–379. [Google Scholar]
- 81.Goldman A.D., Baross J.A., Samudrala R. The enzymatic and metabolic capabilities of early life. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0039912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bich L., Green S. Is defining life pointless? operational definitions at the frontiers of biology. Synthese. 2018;195:3919–3946. [Google Scholar]
- 83.Dick S.J. Cambridge University Press. ix; Cambridge: 2015. The Impact of Discovering Life Beyond Earth; p. 356. [Google Scholar]
- 84.Szostak J.W. Attempts to define life do not help to understand the origin of life. J Biomol Struct Dyn. 2012;29(4):599–600. doi: 10.1080/073911012010524998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szathmary E. Toward major evolutionary transitions theory 2.0. Proc Natl Acad Sci U S A. 2015;112(33):10104–10111. doi: 10.1073/pnas.1421398112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Egbert M.D., Barandiaran X.E., Di Paolo E.A. Behavioral metabolution: the adaptive and evolutionary potential of metabolism-based chemotaxis. Artif Life. 2012;18(1):1–25. doi: 10.1162/artl_a_00047. [DOI] [PubMed] [Google Scholar]
- 87.Vallverdu J., Castro O., Mayne R., Talanov M., Levin M. Slime mould: the fundamental mechanisms of biological cognition. Biosystems. 2018;165:57–70. doi: 10.1016/j.biosystems.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 88.Froese T, Virgo N, Ikegami T. Life as a process of open-ended becoming: Analysis of a minimal model; 2011. pp. 250–257.
- 89.Virgo N, Froese T, Ikegami T. The positive role of parasites in the origins of life. 2013 IEEE Symposium on Artificial Life.
- 90.Krieger M.S., Sinai S., Nowak M.A. Turbulent coherent structures and early life below the Kolmogorov scale. Nat Commun. 2020;11(1):2192. doi: 10.1038/s41467-020-15780-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takagi Y.A., Nguyen D.H., Wexler T.B., Goldman A.D. The coevolution of cellularity and metabolism following the origin of life. J Mol Evol. 2020;88(7):598–617. doi: 10.1007/s00239-020-09961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Serrano-Luginbühl S., Ruiz-Mirazo K., Ostaszewski R., Gallou F., Walde P. Soft and dispersed interface-rich aqueous systems that promote and guide chemical reactions. Nat Rev Chem. 2018;2:306–327. [Google Scholar]
- 93.Kuchler A., Yoshimoto M., Luginbuhl S., Mavelli F., Walde P. Enzymatic reactions in confined environments. Nat Nanotechnol. 2016;11(5):409–420. doi: 10.1038/nnano.2016.54. [DOI] [PubMed] [Google Scholar]
- 94.Preiner M., Asche S., Becker S., Betts H.C., Boniface A. The future of origin of life research: bridging decades-old divisions. Life (Basel) 2020;10(3):20. doi: 10.3390/life10030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luisi P.L. Autopoiesis: a review and a reappraisal. Naturwissenschaften. 2003;90(2):49–59. doi: 10.1007/s00114-002-0389-9. [DOI] [PubMed] [Google Scholar]