Abstract
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Since the emergence of SARS-CoV-2 in the human population in late 2019, it has spread on an unprecedented scale worldwide leading to the first coronavirus pandemic. SARS-CoV-2 infection results in a wide range of clinical manifestations from asymptomatic to fatal cases. Although intensive research has been undertaken to increase understanding of the complex biology of SARS-CoV-2 infection, the detailed mechanisms underpinning the severe pathogenesis and interactions between the virus and the host immune response are not well understood. Thus, the development of appropriate animal models that recapitulate human clinical manifestations and immune responses against SARS-CoV-2 is crucial. Although many animal models are currently available for the study of SARS-CoV-2 infection, each has distinct advantages and disadvantages, and some models show variable results between and within species. Thus, we aim to discuss the different animal models, including mice, hamsters, ferrets, and non-human primates, employed for SARS-CoV-2 infection studies and outline their individual strengths and limitations for use in studies aimed at increasing understanding of coronavirus pathogenesis. Moreover, a significant advantage of these animal models is that they can be tailored, providing unique options specific to the scientific goals of each researcher.
Keywords: SARS-CoV-2, COVID-19, Animal models, Emerging infectious disease
INTRODUCTION
Coronaviruses are pleomorphic, enveloped, single-stranded, positive-sense RNA viruses belonging to the family Coronaviridae. Further classification under the Coronavirinae subfamily includes division into 4 genera, alpha, beta, gamma, and delta, based on phylogenetic relationships and genomic structures (1). In general, coronaviruses infect a wide range of hosts. The alpha and beta coronaviruses are reported to infect only mammals. On the other hand, gamma and delta coronaviruses infect birds, but can infect mammals to some extent (2).
Coronaviruses cause respiratory and intestinal infection in animals and humans. While most human coronaviruses induce self-limiting respiratory tract infections (HCoV-NL63, HCoV-229E, and HCoV-OC43), some can cause severe infections in the elderly, infants, and young children (2). In November 2002, there was an emergence of the severe acute respiratory syndrome coronavirus (SARS-CoV), followed by Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, and the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late December of 2019, all of which are beta-coronaviruses (3). Of these 3 coronavirus outbreaks, only SARS-CoV-2 has caused an ongoing pandemic, with over 100 million confirmed cases and a 2.1% case mortality rate as of February 2021 (4).
Although several SARS-CoV-2 vaccines have already been licensed, the host immune components involved in generating a successful immune response to this virus have not yet been fully elucidated. Thus, further understanding of the immunopathologies that arise from aberrant host immune responses against SARS-CoV-2 is needed. To address the remaining questions and further understanding the detailed mechanisms underlying disease pathology, suitable animal models that recapitulate the human immune response to SARS-CoV-2 infection are needed. Thus, this review aims to discuss the use of different animal models for pathological and immunological studies on SARS-CoV-2 in order to generate a better and more diverse understanding of viral pathogenesis and the host immune response.
RECEPTOR, VIRUS ENTRY, AND HOST IMMUNE RESPONSE
Previous studies on SARS-CoV lead to the discovery of the cell entry receptor, angiotensin-converting enzyme 2 (ACE2) (5,6). The ACE2 protein mediates several different functions including catalysis and amino acid transport, as well as acting as a viral receptor (7). ACE2 are expressed in many different cells of the body (8), however, ACE2 expression in the oral, nasal, and nasopharyngeal epithelium as well as on type II pneumocytes are believed to be important for SARS-CoV-2 transmission and respiratory manifestations (7). Due to the high nucleotide identity (79%) between SARS-CoV and SARS-CoV-2, ACE2 was hypothesized to be a common receptor for these viruses by computational studies (9,10,11), which was later proven by experimental studies (6,12). Cell entry is mediated by the SARS-CoV-2 spike (S) glycoprotein, which binds to the ACE2 receptor and subsequently activated via the transmembrane protease serine 2, a protease that operates as a cofactor enhancing SARS-CoV-2 entry by proteolysis of both viral S protein and the ACE2 receptor (13,14).
Studies of animal and human coronaviruses indicate that both the innate and adaptive immune responses are important mediators of host protection against these viruses. In general, innate immune cells express pathogen-recognition receptors, such as C-type lectin receptors, NOD-like receptors, RIG-I-like receptors, and TLRs, to recognize pathogen-associated molecular patterns (15). Coronaviruses are recognized by cytosolic and endosomal RNA sensors including RIG-I and TLR3, TLR7, and TLR8. Ligation of these cellular receptors results in activation of transcription factors including NF-κB and interferon regulatory factor 3 (IRF3), leading to expression of pro-inflammatory cytokines, chemokines, and type I IFN (16,17,18). Type I IFN activates JAKs and STAT via the IFNα/β receptor. JAKs phosphorylate STAT proteins, which then form a complex with IRF3 (19). These complexes stimulate the expression of IFN-stimulated genes, including antiviral proteins and important mediators of the innate immune response. However, most viruses have several strategies to antagonize the host's innate immune system by targeting viral sensors or blocking the downstream antiviral signaling molecules. For example, studies showed that non-structural proteins of coronaviruses impede the host innate immune response, and especially IFN signaling pathways (20,21,22,23,24). In addition, a recent study reported that SARS-CoV-2 induced delayed type I IFN response (21). The virus immune evasion provides a window for uncontrolled viral replication, leading to a surge of hyperactive host immune response (cytokine storm) later during infection, which eventually resulting in detrimental outcomes.
Adaptive immunity to SARS-CoV-2 infection involves T and B cell immune response and generation of antiviral neutralizing Abs (NAbs) (25). In general, after T cell activation, CD4+ and CD8+ T cells differentiate into effector T cells and cytotoxic T cells, respectively. CD4+ T cells function as a helper or effector producing cytokines and chemokines, while CD8+ T cells specifically kill virus-infected target cells. The B cell immune response against SARS-CoV-2 is characterized by the production of mainly IgG and IgM Ab. During SARS-CoV infection, seroconversion was detected as early as 4 days and generally within 10–16 days (25). In contrast, MERS-CoV patients were found to seroconvert 2–3 weeks post-onset of infection (26), and an absent or delayed Ab response was strongly associated with severe and fatal cases. Recent studies reported that SARS-CoV-2 infection induces very low levels of NAbs in mild disease while high neutralizing titers were only detected in severe cases of coronavirus disease 2019 (COVID-19) (27,28,29). Correlation of Ab levels with disease severity of COVID-19 had been reported from several studies, however, not yet fully elucidated (30,31,32). Furthermore, given that adaptive immune response is critical for the control of infection and future re-infection, however, several studies reported that many patients with severe SARS-CoV-2 infection develop lymphopenia (33,34). Although the mechanism behind is still an open question. Therefore, understanding the immune responses, proper innate and adaptive immune response, elicited after SARS-CoV-2 infection is critical for early recognition and control of coronavirus infection and replication within the host and protection against reinfection.
ANIMALS NATURALLY INFECTED WITH SARS-CoV-2
Several animal species, such as companion animals, captive tigers and lions in New York zoo, and minks, have been reported positive for SARS-CoV-2 (35,36,37). Whereby, transmission to these animals were primarily due to closed contact with infected humans. Among these animals, cats including domestic cats, tigers and lions showed to be more susceptible to SARS-CoV-2. While sustained SARS-CoV-2 infections within domestic setting is implausible, however, environment within intensive farms with susceptible animals may pose higher probability of zoonotic or reverse zoonotic transmission. In April 2020, minks from 2 separate farms in the Netherlands showed respiratory and gastrointestinal disorders, mortality rates of 1.2% to 2.4%, and lesions of interstitial pneumonia (37,38). The outbreak was presumably introduced by infected employee with a subsequent pervasive transmission between animals in the farm. Auspiciously, direct impact of SARS-CoV-2 to animals only cause transient clinical infections, and most companion and captive animals showed less susceptibility, relatively weak viral replication and showed no strong evidence of sustained transmission from animal-to-humans or animals-to-animals. However, further research is warranted to definitively determine probable role of these animals to community transmission of SARS-CoV-2.
EXPERIMENTAL ANIMAL MODELS
Mouse model
The mouse (Mus musculus) has been adopted as a primary animal model for various virological and immunological investigations due to several advantages, including cost-effectiveness, ease of handling, and suitability for large-scale studies (39). However, wild-type and inbred mouse strains cannot be used for SARS-CoV-2 research due to the low affinity between the viral S protein and the murine ACE2 receptor, an interaction required to initiate virus infection (40). Advanced biotechnology has provided several excellent tools to resolve this issue. These include the generation of transgenic animals expressing human ACE2 (hACE2) and were used to elucidate by providing significant findings regarding SARS-CoV-2 pathogenesis.
Transgenic mouse strains expressing the hACE2 receptor have been developed using different promoters, allowing an avenue for precision research (41,42,43,44,45). Hepatocyte nuclear factor-3/forkhead homologue 4 (HFH-4/FOXJ) is a transcription factor with expression highly restricted in cells possessing motile cilia (39), hence, used as a promoter in developing HFH4-hACE2 transgenic mice. HFH4-hACE2 transgenic mice overexpress hACE2 under the control of the HFH4/FOXJ promoter and have been shown to be permissive to SARS-CoV-2 infection (37,38). Thus, using a ciliated cell-specific promoter, it was postulated that hACE2 will be expressed in the ciliated epithelial cells of the respiratory tract (46). HFH4-hACE2 mice exhibited only minimal weight loss during the course of infection; however, severe pneumonia was observed as characterized by extensive fibroplasia and alveolar necrosis. In addition, significant lymphopenia and neutrophilia in peripheral blood were also observed. The K18-hACE2 transgenic mouse (known as B6.Cg-Tg(K18-ACE2)2Prlmn/J), in which hACE2 expression is driven by the epithelial cell cytokeratin-18 promotor, was originally developed for SARS-CoV and subsequently evaluated for SARS-CoV-2 infection (42). Both the HFH4-hACE2 and the K18-hACE2 transgenic mice exhibited virus replication in the lungs, lung inflammation, and body weight loss after infection (47,48). Disease severity in these TG mouse models correlates with hACE2 expression levels, however, hACE2 is also highly expressed in the brain leading to neurological-related mortality (42,43,48). This finding is in contrast to COVID-19 in humans as central nervous system infection is rarely observed (49). Therefore, there are some issues with the use of hACE2 transgenic mice for SARS-CoV-2 pathogenesis and immunological studies. Further, another limitation with this mouse model has been its limited availability and difficulty obtaining it in a reasonable timeframe in certain countries during the 2020 COVID-19 pandemic. To circumvent this, researchers developed mouse-adapted SARS-CoV-2 strains, avoiding the need for hACE2 mice and permitting use of more readily available mouse strains (50,51). In addition, researchers have also transferred hACE2 into mouse lung cells using adenoviral vectors to rapidly produce mice susceptible to SARS-CoV-2 infection (52,53). For example, Hassan et al. (53) rendered mice susceptible to SARS-CoV-2 infection through the use of a recombinant human adenovirus type 5-expressing hACE2, which resulted in the expression of hACE2 in the lower respiratory tract of wild-type mice. In these mice, SARS-CoV-2 replication was found to be restricted to the respiratory tract where it caused the development of mild to moderate interstitial and perivascular inflammation. However, with this approach, there is the possibility that an antiviral immune response might be elicited against the adenoviral vector. In addition, adeno-associated virus (AAV)-mediated hACE2 expression (AAV-hACE2) has also been developed. This is another vector-based delivery system for the expression of hACE2 in the mouse respiratory tract (54). SARS-CoV-2-infected AAV-hACE2 mice had inflammatory pulmonary infiltrates and developed SARS-CoV-2 S-specific IgG and NAbs at 4–7 days post-infection (dpi). However, studies in the transduced hACE2 mouse model have generated inconsistent results, possibly due to non-uniform transduction of the lung epithelium leading to variable hACE2 expression in the mouse lung (55).
Currently, most of what is known with respect to viral pathogenesis and host immune responses has been derived from studies using mouse models. However, the genomic comparison has revealed significant variation between the human and murine immune systems, bringing the suitability of this animal model for the study of human physiology into debate (56,57). These disparities include differences in leukocyte subsets, TLRs, defensins, and Ab subsets, which may underlie the divergent immune response sometimes seen between the 2 species (39,58). To address this, Brumeanu et al. (59) developed a human immune system (HIS)-humanized animal model expressing hACE2. This HIS mouse model mimics the human immune system making it an appealing for the study of the mechanisms of human-like anti-viral immune responses (59,60). In fact, the HIS mouse model is readily infected with SARS-CoV-2 at doses as low as 2.8×103 pfu through the intranasal route, and importantly, recapitulates the main immunopathological events and immune responses seen in COVID-19 patients. In addition, the HIS mouse can also generate human IgM and IgG Abs that are specific for the SARS-CoV-2 RBD protein (59). Despite the limitations of humanized mice, this mouse model shows promise as a tool for studying SARS-CoV-2 pathogenesis and the resulting human immune response.
Hamster model
The golden Syrian hamster (Mesocricetus auratus) has been used for many decades to study human-associated diseases and infections caused by other respiratory viruses such as SARS, adenovirus, and influenza virus (61). Analysis of the ACE2 gene showed that the Syrian hamster differs at only 4 out of 29 amino acids in the SARS-CoV-2 S-contacting regions of ACE2 in humans (62). In addition, previous animal studies showed that SARS-CoV, which also uses the ACE2 receptor for cell entry, can replicate robustly in Syrian hamsters (63). Thus, this suggested that the golden Syrian hamster would also be suitable as an experimental animal model for investigating SARS-CoV-2 infection. To this end, Sia et al. (64) recently reported that Syrian hamsters are susceptible to SARS-CoV-2 infection. Following inoculation, Syrian hamsters develop moderate interstitial pneumonia leading to transient mild to moderate disease. Infection with high titers of SARS-CoV-2 was also found to cause severe pathological lesions in the lungs of Syrian hamsters (63). In addition, induction of marked SARS-CoV-2 replication in the lungs, a strong inflammatory response with neutrophil infiltration, and edema were also observed (65). Boudewijns et al. (65) showed that the SARS-CoV-2-induced lung pathology observed in hamsters appears to be caused by the host immune response. Several studies showed significant evidences that the dysregulation of the innate immune system contributes to the clinical presentation of severe SARS-CoV-2 infection (66,67). The hyperactivity of the innate immune system, causing cytokine storm, has been postulated as pathological driver in severe SARS-CoV-2 infection (68,69). Furthermore, T cell immunosuppression due to adaptive immune dysfunction may increase the risk of SARS-CoV-2 systemic dissemination (70). Further, an immunological study in hamsters showed that the type I IFN response, and especially the STAT 2 signaling pathway, is key in the pathogenesis of SARS-CoV-2. A study in STAT2−/− hamsters showed that STAT2 signaling promotes severe lung injury caused by SARS-CoV-2, but restricts systemic viral dissemination (65). Furthermore, a study in RAG2 KO hamsters showed that the absence of functional B and/or T cells exacerbates pathogenesis at the early stage after SARS-CoV-2 exposure (71).
Studies showed that SARS-CoV-2-infected hamsters can mount a strong humoral immune response with the development of potent Nabs, which can confer protection from subsequent infection, within 14 days (63,72). Further, passive transfer of convalescent hamster sera has a protective effect and reduces viral loads in recipient hamsters after SARS-CoV-2 infection (62,63). It is noteworthy that NAb titers were higher in younger hamsters than in their aged counterparts (72), a direct contrast to what has been observed in COVID-19 patients where lower NAb titers were reported in younger COVID-19 patients (<50 years old) compared to that in elderly COVID-19 patients. Several cohort studies postulated that disease severity was associated with the production of higher NAb titers against SARS-CoV-2 (30-32), though this correlation needs further verification. However, a higher viral load may lead to more severe disease, but may also trigger the generation of a stronger Ab response late during infection due to increased levels of viral antigen.
Compared to other animal models Syrian golden hamsters exhibit a more consistent lung disease phenotype during SARS-CoV-2 infections. Despite the fact that this animal model recapitulates the main immunopathological events and immune responses observed in COVID-19 patients, Syrian hamsters do not succumb to SARS-CoV-2 infection, which does not match the human clinical profile. Recently, another hamster model, the roborovski dwarf hamster (Phodopus roborovskii), was found to be highly susceptible to SARS-CoV-2 (73). These animals also exhibited a rapid onset of severe clinical disease, mimicking the clinical and pathological outcomes observed in severe human COVID-19. However, mortality was not observed even in roborovski dwarf hamsters, outside of humane euthanasia required at a bodyweight loss of 15% (in contrast to the requirement at 25% body weight loss in the murine model).
While hamsters have the advantage of the ease of handling, this model is less frequently used due to the limited availability of research reagents (74), such as immunologic reagents for examining host immune responses, particularly for protein expression, and transgenic disease models which are required for a more complete evaluation of the host response. Nonetheless, hamsters exhibit viral pathogenesis and immune responses consistent with those seen in humans and are capable of transmitting the virus. Hence, this strengthens the argument for use of hamsters as an animal model for SARS-CoV-2 studies.
Ferret model
The ferret (Mustula putorius furo) has been well characterized as an animal model for infection with many respiratory viral pathogens due to their natural susceptibility to human respiratory viruses (75,76,77). In addition to the presence of the appropriate viral receptor, histo-anatomical features of the ferret respiratory tract are analogous to that of the human respiratory tract. These include the anatomical proportions of the upper and lower respiratory tract, glandular density in the bronchial wall, and the number of generations of terminal bronchioles (78,79,80,81). In addition, SARS-CoV-2 replicates efficiently in the respiratory tract of ferrets without prior adaptation (78,79). Unlike other small mammalian models, numerous clinical signs found in humans following SARS-CoV-2 infection are also present in ferrets after experimental intranasal inoculation. These include nasal discharge, wheezing, mucus build-up in the oropharynx, sneezing, and loose stool. Thus, this further supports the significance of this animal model for the investigation of viral transmission by measuring the capacity of SARS-CoV-2 to spread to naïve ferrets through direct contact or indirectly through respiratory droplets (78,79). Another advantage of the ferret model is its size. Compared to rodents their large size allows for more repeated blood sampling at volumes sufficient for immunological analyses throughout the course of infection (81). Thus, this increases the ability to monitor the progression of the host immune response.
Recently, Kim et al. (82) found evidence of reinfection with a heterologous variant of SAR-CoV-2 in ferrets, which occurred in a manner correlating with decreased NAbs titers . Thus, ferrets with high Nab titers were found to show attenuated virus titers in both the respiratory and digestive tracts following reinfection thereby leading to rapid viral clearance. This suggests Nabs play a critical role in cross-protection against heterologous SARS-CoV-2 reinfection. With the emergence of several virus variants, SARS-CoV-2 has been grouped into more than 6 different clusters (83). Thus, determining the potential for and consequence of reinfection with SARS-CoV-2 sequence variants is critical to further understand the development of protective immunity against this virus.
Although ferrets are outbred animals, they are considered specific-pathogen-free because they are bred under a controlled condition and raised in facilities following strict biosecurity protocols solely for medical research. Studies performed with ferrets, in contrast to non-human primates (NHPs), have the advantage of eliminated additional confounding factors that may affect the result of certain immunological or viral pathogenesis studies. Hence, they are considered a good animal model for assessing the efficacy of novel vaccines and therapeutic approaches (81). While ferrets provide usefulness and value as a comparative model in which to study the pathogenesis and transmission of human respiratory diseases, the greatest limitation of this model is the inadequate availability of ferret-specific immunological reagents. This impedes the in-depth determination of any immunological mechanisms underpinning viral disease transmission, protection, and immunopathology in this animal model. To overcome this limitation, researchers have employed a revolutionary approach using single-cell RNA (scRNA) sequencing to decipher the transcriptional signatures of immune cells and can be used to identify stochastic variations in gene expression within a single population (84,85,86). A study by Lee et al. (87) employed this scRNA technique and found an increase in response to IFN by certain subsets of natural killer cells and CD8+ T cells, suggesting increased expression of the IFN-responsive genes might have a beneficial effect on the rapid clearance of SARS-CoV-2.
Therefore, ferrets represent a useful animal model not only for viral transmission and pathogenesis studies but also for the investigation of the safety and efficacy of vaccines and therapeutic approaches for emerging viruses. Thus, given the important contribution of the ferret model to virus research, additional ferret-specific immunologic, genomic, and proteomic tools are currently being developed.
NHP model
The phylogenetic proximity of NHPs to humans makes these animals invaluable as models and as a result, they are widely recognized as ideal animal models for emerging and re-emerging diseases. Coronavirus studies, in particular, have greatly benefited from the use of NHPs as infection and disease models. NHPs, including rhesus macaques (Macaca mulatta) (88,89,90,91), cynomolgus macaques (Macaca fascicularis) (90,92), African baboons (Papio hamadryas) (89), and African green monkeys (AGMs) (Chlorocebus aethiops) (93) have all been demonstrated to be permissive for SARS-CoV-2 infection and to exhibit symptoms also observed in COVID-19 patients such as fever, diarrhea and clinical manifestations of pneumonitis.
Recently, Lu et al. (90) compared SARS-CoV-2 infection in 3 different non-human primates, rhesus macaque, cynomolgus macaque, and common marmoset. Of the 3 species of NHPs, only rhesus and cynomolgus macaques were permissive to SARS-CoV-2 and manifested symptoms of infection. Moreover, both macaque species showed detectable viral RNA in different extrapulmonary organs, including the urinary and digestive tracts, as well as in hilar and mesenteric lymph nodes. However, rhesus macaques showed more severe lung lesions than those in cynomolgus macaques. Although SARS-CoV-2 specific Abs were detectable as early as 4 dpi in the majority of the macaques used, adult and elderly rhesus macaques displayed higher virus-specific Ab titers than did young rhesus macaques. Rhesus and cynomolgus macaques also showed increased inflammatory cytokine expression, IL-1a, IL-8, IL-15, and MCP-1, including anti-inflammatory IL-10 that play a role in the underlying pathophysiology. In addition, transient lymphopenia was also observed, similar to what has been seen in humans (94,95), with the greatest decrease in cell number observed at 10–12 dpi.
A comprehensive study by Singh et al. (89), evaluated the fitness of the Indian rhesus macaque, African baboons, and common marmosets animal models for SARS-CoV-2 infection, and found that common marmosets suffered from a more mild form of infection. Compared to African baboons and Indian rhesus macaques, marmosets had accelerated clearance of viral RNA with only 1 of 6 infected marmosets positive at 6 dpi. Although African baboons and Indian rhesus macaques showed a similar degree of infection, African baboons, and especially older animals, develop a more severe lung pathology compared to Indian rhesus macaques. This is believed to be due to the persistence of SARS-CoV-2 in the lung compartment coupled with higher expression of several cytokines, chemokines, and inflammatory markers resulting in more severe inflammation in the lungs. Cytokine analysis of infected Indian rhesus macaques showed a significant elevation of IL-6, IFNα, IFNγ, IL-8, perforin, IP-10, MIP1α, and MIP1β in the broncho-alveolar lavage (BAL) fluid. In comparison, MIF, IL-6, C-reactive protein, and IP-10 were significantly increased in BAL fluid of African baboons. Moreover, IL1Ra, perforin IL-8, plasma IP-10, RANTES, and IFNα levels were also increased in BAL fluid and serum.
Similar to rhesus and cynomolgus macaques, AGMs recapitulate mild manifestations of human SARS-CoV-2 infection with a pronounced viral pneumonia. According to Woolsey et al. (93), a notable aspect of using AGMs is that they can develop virus-induced pneumonia, exhibited by pulmonary consolidation and hemorrhage along with increases in serum levels of IL-6, IFN-β, IL-10, and other pro-inflammatory cytokines after infection. Moreover, AGMs seroconverted past 5 dpi with IgG titers as high as 1:6,400.
Age-related differences in disease manifestation following SARS-CoV-2 infection were observed in both the macaque NHP model (rhesus and cynomolgus) and in baboons (89,90,93). Singh et al. (89) showed a more conspicuous age-related effect of SARS-CoV-2 infection in African baboons compared to Indian rhesus macaques. Furthermore, older African baboons developed more severe infections of longer duration. Hence, this model could warrant further development for the study of SARS-CoV-2 in elderly COVID-19 patients with co-morbidities, such as diabetes and heart disease. This implies that age has an effect and may be a factor for the more severe disease manifestations seen in elderly COVID-19 patients. Thus, older macaques/baboons may be a suitable NHP animal model for the study of age-dependent pathogenesis of SARS-CoV-2. The NHP genera show varied levels of SARS-CoV-2 infection and exhibit different degrees of pathological manifestation suggesting NHP species-dependent pathogenesis. Moreover, difference in the mounting of protective Abs were also observed. In a study of rhesus macaques, Singh et al. (89) showed that old rhesus macaques had lower Ab titers than young rhesus macaques, even though other studies demonstrated the opposite (90).
Despite the similarity between NHP ACE2 and human ACE2, the NHP animal model does have several limitations. NHPs do not recapitulate severe human disease and no mortality was observed in this model. In addition, variability in the innate and adaptive immune responses against SARS-CoV-2 infection has been reported for NHPs (90,96). It should be noted that there are several species and subspecies of NHPs, which may result in significant variation in the level of viral replication and clinical diseases. Additional disadvantages of using NHPs include significant expenses, limited numbers of animals, and the tight regulation of their use due to ethical reasons (97).
The use of animal models (Table 1) has allowed deducing experimental results into the context of human infection (Fig. 1) (98,99). Furthermore, the use of mammalian models of SARS-CoV-2 infection to evaluate both pathogenicity and transmissibility has resulted in a fuller understanding of the capacity of the virus to cause disease.
Table 1. General animal models of SARS-CoV-2.
| Variables | Description | ||
|---|---|---|---|
| Mouse | |||
| Advantage | Cost-effective, ease of handling, and suitable for large scale studies (47). | ||
| Limitations | Wild-type mice not permissive to SARS-CoV-2 infection (47). | ||
| Transgenic and transduced mice | |||
| Advantage | Permissive to SARS-CoV-2 infection (54,55). | ||
| Virus replicates in the lungs causing lung inflammation. | |||
| Bodyweight loss after infection (54,55). | |||
| Limitations | Limited availability in a timely manner. | ||
| Neurological-related mortality (50,51,54). | |||
| Possibility of antiviral immune response against the vector used. | |||
| Variable hACE2 expression in the mouse lungs due to non-uniform transduction (55). | |||
| Humanized animal model expressing hACE2 | |||
| Advantage | Recapitulates the main human immunopathological events and generates human IgM and IgG SARS-CoV-2 RBD-specific Abs (59). | ||
| Limitations | |||
| Hamster | |||
| Golden syrian hamster | |||
| Advantage | High similarity between the SARS-CoV-2 spike-contacting regions of human ACE2 and hamster ACE2 (62). | ||
| Susceptible to SARS-CoV-2 infection (64). | |||
| More consistent lung disease phenotype compared to other animal models against SARS-CoV-2 infections (64). | |||
| Limitations | Limited availability of immunological reagents (74). | ||
| Lack of mortality. | |||
| Roborovski dwarf hamsters | |||
| Advantage | Highly susceptible to SARS-CoV-2 infection (73). | ||
| Developed a rapid onset of severe clinical disease mimicking the outcomes of severe COVID-19 cases. | |||
| Limitations | Real mortality was not observed. | ||
| Ferret | |||
| Advantage | Histo-anatomical features of the ferret respiratory tract can reproduce condition similar to the human respiratory tract (78,79,80,81). | ||
| SARS-CoV-2 replicate efficiently in the respiratory tract of ferrets without prior adaptation (78,79). | |||
| Suitable for transmission study (78,79). | |||
| Apt size allows repeated blood sampling at volumes (81). | |||
| Limitations | Inadequate availability of ferret-specific immunological reagents. | ||
| NHP | |||
| Advantage | Phylogenetic proximity with humans. | ||
| Permissive for SARS-CoV-2 infection. | |||
| Similar symptoms observed in mild to moderate COVID-19. | |||
| Develop virus-induced pneumonia. | |||
| Suitable animal model for studying SARS-CoV-2 in elderly COVID-19 patients. | |||
| Limitations | Does not recapitulate severe human disease and lack of mortality. | ||
| Variable disease outcome, clinical signs, and immune responses dependent on NHP species (90,96). | |||
| Expensive, NHP studies are limited to a small number of animals and are highly regulated due to ethical reasons (97). | |||
RBD, receptor-binding domain.
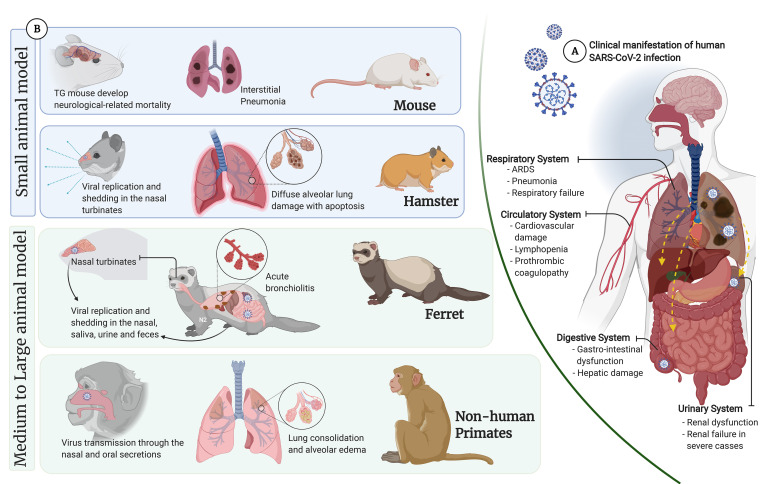
Figure 1. (A) Overview of human clinical manifestation of SARS-CoV-2 infection: showing common clinical features observed in hospitalized patients (98,99). Pneumonia, a major clinical feature in COVID-19 patients, complicated by ARDS leading to respiratory failure. Reports on human autopsies reported that the lung generally appeared congested and edematous, with different stages of diffuse alveolar damage characterized by fibrosis of the interstitium, desquamation of pneumocytes, hyaline membrane formation with inflammatory cell infiltration. In addition, direct assault to other organs by disseminated SARS-CoV-2 and immune pathogenesis were also observed such as cardiovascular complications, hypercoagulopathy, substantial reduction of lymphocytes, gastrointestinal dysfunction, and impaired liver function, and renal injury. (B) Overview of SARS-CoV-2 manifestations in different animal models. (created with BioRender.com).
ARDS, acute respiratory distress syndrome; TG, transgenic.
CONCLUSION
The emergence of the novel SARS-CoV-2 pathogen has resulted in the need for the development of an appropriate animal model to aid in the understanding of disease causation, transmission, and pathogenesis, as well as to elucidate the details of the host immune response. Perhaps the most perplexing aspect of SARS-CoV-2 infection is the wide range of disease severity in humans, which can vary from asymptomatic or mild presentation to severe and fatal disease. Developing animal models that can reflect such diverse clinical manifestations is remarkably important, albeit arduous. While many SARS-CoV-2 infection models have been developed and investigated thus far, including mice, hamsters, ferrets, and NHPs, a large variation in results has been obtained both between and within species.
While standard mouse strains are not susceptible to natural SARS-CoV-2 infection, several approaches to generate mouse strains susceptible to SARS-CoV-2 have been developed. Unfortunately, engineered mice exhibit uneven hACE2 transduction, and ectopic hACE2 expression results in changes in viral tissue tropism. In addition to the mouse model, hamsters have proven to be a useful animal model for SARS-CoV-2 infection due to their development of clinical signs of illness, including lung damage similar to that seen in humans. While hamsters do offer several advantages, a caveat with this species is the scarcity of research reagents for immunological studies compared to those available for mice.
Several medium to large size animals are closer anatomically and physiologically to humans, having ACE2 receptors similar to human ACE2, and manifest many clinical similarities to human infection. With respect to ferrets, not only are they susceptible to SARS-CoV-2, they can spread the virus with high transmission rates via both direct contact and through airborne transmission. In addition, several studies have indicated that non-human primate animal models are both permissive to SARS-CoV-2 and are suitable for pathology studies. Moreover, a notable variation of infection was found with respect to the age of infected NHPs, with aged animals succumbing to infection and showing more severe symptoms and pathology. In addition, different NHP species exhibit heterogeneous responses to SARS-CoV-2 infection.
The use of an appropriate animal model can markedly accelerate understanding of the immunopathogenesis of novel pathogens such as SARS-CoV-2. Although numerous animal models are available, however, currently, no single existing animal model can really recapitulate the totality of human SARS-CoV-2 infection. Several possible reasons may explain why experimental animal models only reflect moderate lung pathology and disease manifestation. These include, route and method of virus inoculation, absence nor not been rigorously explored effects of co-morbidities and other risk factors associated with high-case fatality such as advancement in the age of the animal, obesity, diabetes, cardiovascular disease and immunocompromised state. Incorporation of these features may predispose existing animal models to reflect pathological states showing clinical manifestation similar to those observed in humans with severe COVID-19. Nevertheless, each animal model has advantages and disadvantages, and importantly, the value of different animal models is that they can be tailored, providing different options in order to achieve the scientific goals of the researcher.
ACKNOWLEDGEMENTS
This work was supported by the National Research Council of Science & Technology (NST) grant by the Korea government (MSIT) (No. CAP-20-01-KRIBB).
Abbreviations
- AAV
adeno-associated virus
- AAV-hACE2
adeno-associated virus-mediated hACE2 expression
- ACE2
angiotensin-converting enzyme 2
- AGM
African green monkey
- BAL
broncho-alveolar lavage
- COVID-19
coronavirus disease 2019
- dpi
days post-infection
- hACE2
human angiotensin-converting enzyme 2
- HFH-4/FOXJ
hepatocyte nuclear factor-3/forkhead homologue 4
- HIS
human immune system
- IRF3
interferon regulatory factor 3
- MERS-CoV
Middle East respiratory syndrome coronavirus
- NAb
neutralizing antibody
- NHP
non-human primate
- S
spike
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- scRNA
single-cell RNA
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Casel MAB, Rollon RG, Choi YK.
- Writing - original draft: Casel MAB, Rollon RG.
- Writing - review & editing: Casel MAB, Rollon RG, Choi YK.
References
- 1.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV) [Internet] [accessed on 20 January 2021]. Available at https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-mers#tab=tab_1.
- 4.World Health Organization. Who coronavirus disease (COVID-19) dashboard [Internet] [accessed on 1 February 2021]. Available at https://covid19.who.int/
- 5.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medina-Enríquez MM, Lopez-León S, Carlos-Escalante JA, Aponte-Torres Z, Cuapio A, Wegman-Ostrosky T. ACE2: the molecular doorway to SARS-CoV-2. Cell Biosci. 2020;10:148. doi: 10.1186/s13578-020-00519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, Zhong W, Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 11.Zou J, Yin J, Fang L, Yang M, Wang T, Wu W, Bellucci MA, Zhang P. Computational prediction of mutational effects on SARS-CoV-2 binding by relative free energy calculations. J Chem Inf Model. 2020;60:5794–5802. doi: 10.1021/acs.jcim.0c00679. [DOI] [PubMed] [Google Scholar]
- 12.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tartey S, Takeuchi O. Pathogen recognition and Toll-like receptor targeted therapeutics in innate immune cells. Int Rev Immunol. 2017;36:57–73. doi: 10.1080/08830185.2016.1261318. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 17.Krieg AM. AIMing 2 defend against intracellular pathogens. Nat Immunol. 2010;11:367–369. doi: 10.1038/ni0510-367. [DOI] [PubMed] [Google Scholar]
- 18.Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front Immunol. 2020;11:1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao L, Li J, Cui X, Jia N, Wei J, Xia L, Wang H, Zhou Y, Wang Q, Liu X, et al. Distribution of Haemaphysalis longicornis and associated pathogens: analysis of pooled data from a China field survey and global published data. Lancet Planet Health. 2020;4:e320–e329. doi: 10.1016/S2542-5196(20)30145-5. [DOI] [PubMed] [Google Scholar]
- 23.Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsueh PR, Huang LM, Chen PJ, Kao CL, Yang PC. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corman VM, Albarrak AM, Omrani AS, Albarrak MM, Farah ME, Almasri M, Muth D, Sieberg A, Meyer B, Assiri AM, et al. Viral shedding and antibody response in 37 patients with middle east respiratory syndrome coronavirus infection. Clin Infect Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legros V, Denolly S, Vogrig M, Boson B, Siret E, Rigaill J, Pillet S, Grattard F, Gonzalo S, Verhoeven P, et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol. 2021;18:318–327. doi: 10.1038/s41423-020-00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henss L, Scholz T, von Rhein C, Wieters I, Borgans F, Eberhardt FJ, Zacharowski K, Ciesek S, Rohde G, Vehreschild M. Analysis of humoral immune responses in SARS-CoV-2 infected patients. J Infect Dis. 2020;223:56–61. doi: 10.1093/infdis/jiaa680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau EH, Tsang OT, Hui DS, Kwan MY, Chan WH, Chiu SS, Ko RL, Chan KH, Cheng SM, Perera RA, et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat Commun. 2021;12:63. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luchsinger LL, Ransegnola BP, Jin DK, Muecksch F, Weisblum Y, Bao W, George PJ, Rodriguez M, Tricoche N, Schmidt F, et al. Serological assays estimate highly variable SARS-CoV-2 neutralizing antibody activity in recovered COVID-19 patients. J Clin Microbiol. 2020;58:e02005-20. doi: 10.1128/JCM.02005-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muecksch F, Wise H, Batchelor B, Squires M, Semple E, Richardson C, McGuire J, Cleary S, Furrie E, Greig N, et al. Longitudinal analysis of clinical serology assay performance and neutralising antibody levels in COVID19 convalescents. J Infect Dis. 2021;223:389–398. doi: 10.1093/infdis/jiaa659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu F, Liu M, Wang A, Lu L, Wang Q, Gu C, Chen J, Wu Y, Xia S, Ling Y, et al. Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in shanghai, china. JAMA Intern Med. 2020;180:1356–1362. doi: 10.1001/jamainternmed.2020.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, Damoraki G, Gkavogianni T, Adami ME, Katsaounou P, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sit TH, Brackman CJ, Ip SM, Tam KW, Law PY, To EM, Yu VY, Sims LD, Tsang DN, Chu DK, et al. Infection of dogs with SARS-CoV-2. Nature. 2020;586:776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Animal and Plant Health Inspection Service (US) USDA statement on the confirmation of COVID-19 in a tiger in New York [Internet] [accessed on 3 April 2021]. Available at https://www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2020/NY-zoo-covid-19.
- 37.Hobbs EC, Reid TJ. Animals and SARS-CoV-2: species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound Emerg Dis. 2020 doi: 10.1111/tbed.13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude Munnink BB, Hakze-van der Honing RW, Gerhards N, Tolsma P, Bouwstra R, Sikkema R, et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020;25:2001005. doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 40.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang XH, Deng W, Tong Z, Liu YX, Zhang LF, Zhu H, Gao H, Huang L, Liu YL, Ma CM, et al. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp Med. 2007;57:450–459. [PubMed] [Google Scholar]
- 42.McCray PB, Jr, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tseng CT, Huang C, Newman P, Wang N, Narayanan K, Watts DM, Makino S, Packard MM, Zaki SR, Chan TS, et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Netland J, Meyerholz DK, Moore S, Cassell M, Perlman S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol. 2008;82:7264–7275. doi: 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshikawa N, Yoshikawa T, Hill T, Huang C, Watts DM, Makino S, Milligan G, Chan T, Peters CJ, Tseng CT. Differential virological and immunological outcome of severe acute respiratory syndrome coronavirus infection in susceptible and resistant transgenic mice expressing human angiotensin-converting enzyme 2. J Virol. 2009;83:5451–5465. doi: 10.1128/JVI.02272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menachery VD, Yount BL, Jr, Sims AC, Debbink K, Agnihothram SS, Gralinski LE, Graham RL, Scobey T, Plante JA, Royal SR, et al. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 48.Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, Li Q, Zhang L, Zhu Y, Si HR, et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helms J, Kremer S, Merdji H, Clere-Jehl R, Schenck M, Kummerlen C, Collange O, Boulay C, Fafi-Kremer S, Ohana M, et al. Neurologic features in severe SARS-CoV-2 infection. N Engl J Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ, Wang Y, Teng Y, Zhao Z, Cui Y, et al. Rapid adaptation of SARS-CoV-2 in BALB/c mice: novel mouse model for vaccine efficacy. bioRxiv. 2020 doi: 10.1101/2020.05.02.073411. [DOI] [Google Scholar]
- 51.Dinnon KH, 3rd, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Jr, Hou YJ, Adams LE, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yun SM, Park SJ, Kim YI, Park SW, Yu MA, Kwon HI, Kim EH, Yu KM, Jeong HW, Ryou J, et al. Genetic and pathogenic diversity of severe fever with thrombocytopenia syndrome virus (SFTSV) in South Korea. JCI Insight. 2020;5:e129531. doi: 10.1172/jci.insight.129531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, McCune BT, Fox JM, Chen RE, Alsoussi WB, et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rathnasinghe R, Strohmeier S, Amanat F, Gillespie VL, Krammer F, García-Sastre A, Coughlan L, Schotsaert M, Uccellini MB. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg Microbes Infect. 2020;9:2433–2445. doi: 10.1080/22221751.2020.1838955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, Wakamatsu E, Benoist C, Koller D, Regev A, et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc Natl Acad Sci U S A. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ernst W. Humanized mice in infectious diseases. Comp Immunol Microbiol Infect Dis. 2016;49:29–38. doi: 10.1016/j.cimid.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Brumeanu TD, Vir P, Shashikumar S, Karim AF, Kar S, Chung KK, Pratt KP, Casares S. A Human-Immune-System mouse model for COVID-19 research (DRAGA mouse: HLA-A2.HLA-DR4.Rag1KO.IL-2Rγc KO.NOD) bioRxiv. 2021 doi: 10.1101/2020.08.19.251249. [DOI] [Google Scholar]
- 60.Tu W, Zheng J. Application of humanized mice in immunological research. Methods Mol Biol. 2016;1371:157–176. doi: 10.1007/978-1-4939-3139-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miao J, Chard LS, Wang Z, Wang Y. Syrian hamster as an animal model for the study on infectious diseases. Front Immunol. 2019;10:2329. doi: 10.3389/fimmu.2019.02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, Chan WM, Fan Z, Tsoi HW, Wen L, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sia SF, Yan LM, Chin AW, Fung K, Choy KT, Wong AY, Kaewpreedee P, Perera RA, Poon LL, Nicholls JM, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boudewijns R, Thibaut HJ, Kaptein SJ, Li R, Vergote V, Seldeslachts L, De Keyzer C, Bervoets L, Sharma S, Van Weyenbergh J, et al. STAT2 signaling as double-edged sword restricting viral dissemination but driving severe pneumonia in SARS-CoV-2 infected hamsters. bioRxiv. 2020 doi: 10.1101/2020.04.23.056838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, Liu X, Xie L, Li J, Ye J, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang D, Guo R, Lei L, Liu H, Wang Y, Wang Y, Qian H, Dai T, Zhang T, Lai Y, et al. Frontline science: COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J Leukoc Biol. 2021;109:13–22. doi: 10.1002/JLB.4HI0720-470R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ HLH Across Speciality Collaboration, UK. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vardhana SA, Wolchok JD. The many faces of the anti-COVID immune response. J Exp Med. 2020;217:e20200678. doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brocato RL, Principe LM, Kim RK, Zeng X, Williams JA, Liu Y, Li R, Smith JM, Golden JW, Gangemi D, et al. Disruption of adaptive immunity enhances disease in sars-cov-2-infected syrian hamsters. J Virol. 2020;94:e01683-20. doi: 10.1128/JVI.01683-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Osterrieder N, Bertzbach LD, Dietert K, Abdelgawad A, Vladimirova D, Kunec D, Hoffmann D, Beer M, Gruber AD, Trimpert J. Age-dependent progression of SARS-CoV-2 infection in syrian hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trimpert J, Vladimirova D, Dietert K, Abdelgawad A, Kunec D, Dökel S, Voss A, Gruber AD, Bertzbach LD, Osterrieder N. The roborovski dwarf hamster is a highly susceptible model for a rapid and fatal course of Sars-CoV-2 infection. Cell Reports. 2020;33:108488. doi: 10.1016/j.celrep.2020.108488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cleary SJ, Pitchford SC, Amison RT, Carrington R, Robaina Cabrera CL, Magnen M, Looney MR, Gray E, Page CP. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br J Pharmacol. 2020;177:4851–4865. doi: 10.1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chu YK, Ali GD, Jia F, Li Q, Kelvin D, Couch RC, Harrod KS, Hutt JA, Cameron C, Weiss SR, et al. The SARS-CoV ferret model in an infection-challenge study. Virology. 2008;374:151–163. doi: 10.1016/j.virol.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 2013;341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- 77.Stittelaar KJ, de Waal L, van Amerongen G, Veldhuis Kroeze EJ, Fraaij PL, van Baalen CA, van Kampen JJ, van der Vries E, Osterhaus AD, de Swart RL. Ferrets as a novel animal model for studying human respiratory syncytial virus infections in immunocompetent and immunocompromised hosts. Viruses. 2016;8:168. doi: 10.3390/v8060168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang JH, Kim EJ, Lee S, Casel MAB, et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson-Delaney CA, Orosz SE. Ferret respiratory system: clinical anatomy, physiology, and disease. Vet Clin North Am Exot Anim Pract. 2011;14:357–367. vii. doi: 10.1016/j.cvex.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 81.Enkirch T, von Messling V. Ferret models of viral pathogenesis. Virology. 2015;479-480:259–270. doi: 10.1016/j.virol.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim YI, Kim SM, Park SJ, Kim EH, Yu KM, Chang JH, Kim EJ, Casel MAB, Rollon R, Jang SG, et al. Critical role of neutralizing antibody for sars-cov-2 reinfection and transmission. Emerg Microbes Infect. 2021;10:152–160. doi: 10.1080/22221751.2021.1872352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.GISAID. Pandemic coronavirus causing COVID-19 [Internet] [accessed on 14 January 2021]. Available at https://www.epicov.org/epi3/frontend#264948.
- 84.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18:35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 85.Stubbington MJ, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358:58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, Fan X, Xia P, Fu JL, Wang SY, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 87.Lee JS, Koh JY, Yi K, Kim YI, Park SJ, Kim EH, Kim SM, Park SH, Ju YS, Choi YK, et al. Single-cell transcriptome of bronchoalveolar lavage fluid reveals dynamic change of macrophages during SARS-CoV-2 infection in ferrets. bioRxiv. 2020 doi: 10.1101/2020.11.18.388280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu P, Qi F, Xu Y, Li F, Liu P, Liu J, Bao L, Deng W, Gao H, Xiang Z, et al. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh DK, Singh B, Ganatra SR, Gazi M, Cole J, Thippeshappa R, Alfson KJ, Clemmons E, Gonzalez O, Escobedo R, et al. Responses to acute infection with SARS-CoV-2 in the lungs of rhesus macaques, baboons and marmosets. Nat Microbiol. 2021;6:73–86. doi: 10.1038/s41564-020-00841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu S, Zhao Y, Yu W, Yang Y, Gao J, Wang J, Kuang D, Yang M, Yang J, Ma C, et al. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct Target Ther. 2020;5:157. doi: 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bao L, Deng W, Gao H, Xiao C, Liu J, Xue J, Lv Q, Liu J, Yu P, Xu Y, et al. Lack of reinfection in rhesus macaques infected with SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.03.13.990226. [DOI] [Google Scholar]
- 92.Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den Brand J, Okba NM, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Woolsey C, Borisevich V, Prasad AN, Agans KN, Deer DJ, Dobias NS, Heymann JC, Foster SL, Levine CB, Medina L, et al. Establishment of an African green monkey model for COVID-19 and protection against re-infection. Nat Immunol. 2021;22:86–98. doi: 10.1038/s41590-020-00835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu R, Wang Y, Li J, Han H, Xia Z, Liu F, Wu K, Yang L, Liu X, Zhu C. Decreased T cell populations contribute to the increased severity of COVID-19. Clin Chim Acta. 2020;508:110–114. doi: 10.1016/j.cca.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, Ivison GT, Ranganath T, Vergara R, Hollis T, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Scientific Committee on Health, Environmental and Emerging Risks (SCHEER) The need for non-human primates in biomedical research, production and testing of products and devices. Brussels: European Union; 2009. [Google Scholar]
- 98.Maiese A, Manetti AC, La Russa R, Di Paolo M, Turillazzi E, Frati P, Fineschi V. Autopsy findings in COVID-19-related deaths: a literature review. Forensic Sci Med Pathol. 2020:1–18. doi: 10.1007/s12024-020-00310-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mohanty SK, Satapathy A, Naidu MM, Mukhopadhyay S, Sharma S, Barton LM, Stroberg E, Duval EJ, Pradhan D, Tzankov A, et al. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and coronavirus disease 19 (COVID-19) - anatomic pathology perspective on current knowledge. Diagn Pathol. 2020;15:103. doi: 10.1186/s13000-020-01017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]