Abstract
Objective
Liver glycogen levels are dynamic and highly regulated by nutrient availability as the levels decrease during fasting and are restored during the feeding cycle. However, feeding in the presence of fructose in water suppresses glycogen accumulation in the liver by upregulating the expression of the glucose-6-phosphatase catalytic subunit (G6pc) gene, although the exact mechanism is unknown. We generated liver-specific knockout MED13 mice that lacked the transcriptional Mediator complex kinase module to examine its effect on the transcriptional activation of inducible target gene expression, such as the ChREBP- and FOXO1-dependent control of the G6pc gene promoter.
Methods
The relative changes in liver expression of lipogenic and gluconeogenic genes as well as glycogen levels were examined in response to feeding standard low-fat laboratory chow supplemented with water or water containing sucrose or fructose in control (Med13fl/fl) and liver-specific MED13 knockout (MED13-LKO) mice.
Results
Although MED13 deficiency had no significant effect on constitutive gene expression, all the dietary inducible gene transcripts were significantly reduced despite the unchanged insulin sensitivity in the MED13-LKO mice compared to that in the control mice. G6pc gene transcription displayed the most significant difference between the Med13 fl/fl and MED13-LKO mice, particularly when fed fructose. Following fasting that depleted liver glycogen, feeding induced the restoration of glycogen levels except in the presence of fructose. MED13 deficiency rescued the glycogen accumulation defect in the presence of fructose. This resulted from the suppression of G6pc expression and thus G6PC enzymatic activity.
Among two transcriptional factors that regulate G6pc gene expression, FOXO1 binding to the G6pc promoter was not affected, whereas ChREBP binding was dramatically reduced in MED13-LKO hepatocytes. In addition, there was a marked suppression of FOXO1 and ChREBP-β transcriptional activities in MED13-LKO hepatocytes.
Conclusions
Taken together, our data suggest that the kinase module of the Mediator complex is necessary for the transcriptional activation of metabolic genes such as G6pc and has an important role in regulating glycogen levels in the liver through altering transcription factor binding and activity at the G6pc promoter.
Keywords: Mediator complex, Kinase module, Fructose, G6pc, Liver glycogen
Highlights
-
•
G6pc is upregulated with fructose supplementation through the ChREBP-dependent pathway.
-
•
Upregulation of G6pc suppresses glycogen synthesis in the liver.
-
•
Knockout of the Mediator complex MED13 subunit inhibits G6pc gene expression.
-
•
The Mediator complex kinase module is required for ChREBP-dependent activation of the G6pc gene.
1. Introduction
Different types of dietary carbohydrates have marked differential effects on metabolism and the development of obesity, diabetes, fatty liver, and cardiovascular diseases [[1], [2], [3]]. In particular, fructose is a more potent driver of de novo lipogenesis in the liver than glucose primarily through a greater induction of lipogenic gene expression through the activation of the transcription factors, sterol regulatory element-binding protein 1c (SREBP-1c) and carbohydrate response element-binding protein (ChREBP) [[4], [5], [6], [7]]. While both glucose and fructose are phosphorylated to fructose 6-phosphate (F6P) and fructose 1-phosphate (F1P), respectively, only glucokinase but not fructokinase (ketohexokinase) is highly sensitive to its end-product inhibition [[8], [9], [10]]. It was recently reported that at low doses, the major absorption and metabolism of dietary fructose occurs in the small intestines, whereas at high doses, fructose saturates intestinal metabolism resulting in excess fructose primarily going to and being metabolized by the liver [11,12]. The liver contains high levels of aldolase B that rapidly hydrolyzes F1P to dihydroacetone phosphate (DHAP) and glyceraldehyde [13]. The rapid phosphorylation of fructose results in depletion of ATP and subsequent activation of the AMP-dependent protein kinase (AMPK), nucleotide degradation, and uric acid formation [[14], [15], [16], [17]]. AMPK is an important energy sensor that suppresses the mechanistic target of rapamycin complex 1 (mTORC1), another important energy-sensing signaling node [18]. Because insulin induction of SREBP1c-mediated lipogenic gene expression depends on mTORC1, fructose induction of lipogenic gene likely results from a distinct pathway. Fructose was recently shown to induce the expression of the constitutively active ChREBP-β transcription factor and lipogenic gene expression through the nuclear translocation of ChREBP-α independent from mTORC1 [7,19]. Importantly, the catalytic subunit of glucose 6-phosphatase (G6pc) is a critical gluconeogenic gene and is also transactivated by ChREBP-β [20,21]. These findings suggest a pathophysiologic basis for the elevation of both de novo lipogenesis and increased hepatic glucose production (glycogenolysis and gluconeogenesis) that occurs in insulin-resistant states.
However, most eukaryotic transcription factors, including SREBP-1c and ChREBP-β, are unable to directly contact and activate RNA polymerase II (Pol II) required for mRNA transcription. The Mediator complex links these and the vast majority of regulated transcriptional factors and co-regulators with the basal transcriptional machinery [22,23]. The small or core mammalian Mediator complex contains 26 subunits whereas the large Mediator complex consists of the core Mediator without MED26 plus the kinase module composed of MED13, MED12, CDK8, and cyclin C (CCNC) [[24], [25], [26]]. Under normal physiologic conditions, the Mediator complex in the mouse liver undergoes a dynamic nutrient and hormonal-dependent interconversion between the large and small Mediator complex regulated through the mTORC1 pathway [27]. The relative contribution of the large and small Mediator complex to Pol II-dependent transcription has remained enigmatic and appears to result from complex interactions between combinatorial factors in a cell context-dependent manner [[28], [29], [30]]. In the present study, we used liver-specific MED13 knockout mice that resulted in the complete loss of the Mediator complex kinase module and generated the constitutive formation of the small/core Mediator. In these mice, we demonstrated that the kinase module is necessary for the transcriptional activation of G6pc expression and protects mice against some deleterious effects of fructose.
2. Materials and methods
2.1. Animals
All the animal experiments followed guidelines that were approved and in agreement with the Albert Einstein College of Medicine Institutional Animal Care and Use Committee (2016–0901). Med13fl/fl mice were kindly provided by Dr. Eric Olson and backcross-bred with C57BL/6J from Jackson Laboratories for 8 generations. MED13 liver-specific knockout mice (MED13-LKO) were generated by cross-breeding with C57BL/6J. Albumin-Cre (Alb-Cre) mice were purchased from Jackson Laboratories. For G6pc overexpression in the Med13fl/fl and MED13-LKO mice, AAV8-Tbg-G6pc was cloned and packaged (Vector Biolabs) for intravenous injection at 5 × 1011 GC/mouse. The mice had free access to food and water unless otherwise noted. All of the mice were housed in groups and under a facility equipped with a 12-h light/dark cycle.
Standard laboratory mouse chow (LabDiet #5053) that contained 25% protein, 13% fat, and 62% carbohydrates was provided for the mice. Prior to feeding experiments, the mice were trained for 3 days by removing food at 5 PM and feeding at 9 AM the next day (16-h fast overnight and 8-h feeding daytime). On the day of the experiment, the fasting group of mice was sacrificed in the morning at 11 AM (16-h fast) while the 9 AM fed group was also provided drinking water that contained 30% sucrose or fructose (w/v). The fed mice were sacrificed at 1 PM equal to 4 h of feeding. Then 30% sucrose or fructose water was prepared (w/v) and provided to the mice on the day of the experiment and not during the fasting/feeding training period.
For glucose, insulin, and pyruvate tolerance tests, mice were fasted overnight for 16 h (glucose tolerance test) or 6 h (insulin and pyruvate tolerance test) and tests were performed as previously described [31]. Blood glucose was determined by a Precision Xtra (Abbott) glucose-monitoring system, and plasma insulin levels were measured by an Ultra-Sensitive Mouse Insulin ELISA kit (Crystalchem). The respiratory exchange ratio was determined by indirect calorimetry using a Columbus Instruments Oxymax system as previously described [32].
2.2. Antibodies and immunoblotting
Antibodies against the following proteins were used in this study: MED13 (1:1000, Fisher PA5-35924 or 1:1000 Bethyl A301-278A), MED12 (1:1000, Abcam ab70842), CDK8 (1:1000, Abcam ab54561 or 1:1000, Bethyl A302-501A-M), CCNC (1:1000, Bethyl A301-989A), MED15 (1:1000, Bethyl A302-422A), nucleolin (1:1000, Cell Signaling 14574S), tubulin (1:1000, Cell Signaling 2144S), ChREBP (1:1000, Novus Biologicals NB400–135SS), phospho-S6 ribosomal protein (1:1000, Cell Signaling 4858P), S6 ribosomal protein (1:1000, Cell Signaling 2317S), phospho-AKT S473 (1:1000, Cell Signaling 4060), T308 (1:1000, Cell Signaling 4056), T450 (1:1000, Cell Signaling 12,178), total AKT (1:1000, Cell Signaling 9272), phospho GS (S641, 1:1000 Cell Signaling 3891), GS (1:1000 Cell Signaling 3893), phospho FOXO1 (S256, 1:1000 Cell Signaling 84,192), and FOXO1 (1:1000 Cell Signaling 2880). Total liver tissue lysates were generated using Pierce IP lysis buffer (87,787, Thermo Fisher Scientific) with freshly added 1 mM of dithiothreitol (DTT), protease inhibitors, and phosphatase inhibitors. Then, 3–8% Tris-acetate gels (Invitrogen) and nitrocellulose membranes (Invitrogen, iBlot gel transfer) were used to analyze protein samples. Western blotting signals were detected with either goat anti-rabbit or goat anti-mouse secondary antibodies (IRDye 800CW) from LI-COR. For a densitometric analysis, the relative band intensity was quantified by ImageJ and normalized to the loading control nucleolin or tubulin.
2.3. RNA isolation and quantitative PCR
Total RNA isolation from mouse livers and a quantitative PCR analysis were performed as previously described [33]. Briefly, an RNeasy purification kit (Qiagen) was used for RNA purification according to the manufacturer's protocol. A SuperScript VILO cDNA synthesis kit (Invitrogen) and PowerUp SYBR Green Master Mix (Applied Biosystems) were used for cDNA strand synthesis and a qPCR and StepOnePlus Real-Time PCR system (Applied Biosystems) was used for quantification. Samples were normalized to the Ppib gene to determine the relative mRNA levels. The primer sequences used in this study are listed in Table S1.
2.4. Nuclear extraction and glycerol gradient fractionation
All of the procedures were performed on ice in a 4 °C cold room. All the buffers contained a cocktail of protease inhibitors (Sigma Aldrich), phosphatase inhibitors (Sigma Aldrich), and 1 mM of DTT and were made immediately before use. Fresh mouse liver nuclear extracts were prepared as previously described with some modifications [34]. Briefly, the pooled liver extracts for each condition were loaded directly onto a glycerol gradient column generated by an auto mix gradient former (Jule) with 40% and 80% (w/v) glycerol in gradient buffer (20 mM of Hepes, 100 mM of NaCl, and 1.5 mM of MgCl2, pH 7.8) on top of a 200 μL cushion of 100% glycerol. Gradients containing samples or native marker protein standards that ran in parallel (Invitrogen) were centrifuged at 16,000×g for 16 h at 4 °C in a Beckman ultracentrifuge with SW55 Ti or VTi 65.2 rotors. Gradient fractions (50–100 μL) were collected from the top of the column and analyzed by Western blotting. Fractions taken from the protein standard column were resolved on NuPAGE 3–8% Tris-acetate native gel (Thermo Fisher Scientific) followed by silver staining and compared with the fractions from the lysate samples.
2.5. Chromatin immunoprecipitation (ChIP) assay
ChIP was performed as previously described [35]. Briefly, cross-linked mouse liver nuclear samples were sonicated in TE buffer containing protease inhibitor (Sigma Aldrich) and phosphatase inhibitor cocktail (Sigma Aldrich) on ice until the cross-linked chromatin DNA was sheared to an average length of ∼500 bp. The samples were precleaned with beads and aliquoted for incubation with anti-ChREBP (Novus Biologicals, NB400-135), anti-FOXO1 (Abcam, Ab39670), or anti-MED1 (Bethyl Laboratories, A300-793A) at 4 °C for 16 h. The immunoprecipitated DNA complex was incubated with proteinase-K (Sigma Aldrich) mix at 65 °C for 12 h, extracted with phenol-chloroform, and precipitated with ethanol. Next, 1% of starting chromatin input values or cycle threshold (Ct) values were used for normalization.
2.6. Histology, glycogen quantification, and G6PC activity assay
Liver tissues were fixed in formalin and subjected to standard H&E staining and PAS staining at the Albert Einstein College of Medicine Histology and Comparative Pathology Core. Glycogen levels in frozen liver tissues were measured by using a Glycogen assay kit II (Abcam, ab169558). For G6PC activity measurement, a G6PC activity assay kit (University of Buffalo BMR, E−120) was used.
2.7. RNA-seq
Total isolated RNA samples were sent to Novogene Corporation for genome-wide mRNA sequencing and followed the company's standard protocol in generating 150 bp cDNA fragments, paired and two-ended reads, and 30, 000, 000 fragments/library construction for an Illumina machine. The raw sequencing reads were trimmed using Trim Galore with the stringency 5 option. Differential expression was performed with DESeq2 [36] using transcript-level counts estimated by Kallisto [37]. A gene ontology analysis was performed using the DAVID functional annotation tool [38].
The RNA-seq data are publicly available at the GEO repository (accession number: GSE165033).
2.8. Luciferase reporter assay
Luciferase reporter assay was performed as previously described [39]. Briefly, isolated primary hepatocytes from 12-week-old male Med13fl/fl or MED13-LKO mice were seeded and cultured at a density of 2 × 105 per well in 12-well collagen-coated plates in DMEM with 10% FBS. The same day, the Med13fl/fl cells were transduced with either adenovirus (Ad)-GFP or Ad-Cre. After an overnight culture, the primary hepatocytes were co-transfected with plasmids of pcDNA3 (vector), pcDNA-Foxo1, pGL4-G6pc-Luc, and Renilla-Luc (control) for FOXO1-induced G6pc promoter activity or pcDNA-Chrebp-β and pGL4-3xChoRE-Luc for ChREBP-β-induced ChoRE activity. The cells were then collected 24 h post-transfection for luciferase assays. The firefly luciferase activity of each sample was normalized by the renilla activity. The 3xChoRE sequence was 5′-AACATGCGCTGCAGGCATGTTCTTCCCATACCACGAGGTGGGAGGCTGTCCTAGACTAGTCACGTGTTCTACACGTGTT.
2.9. Statistical analysis
Data are presented as the mean ± SEM unless otherwise noted and compared between two groups using Student's t-test. A two-sided p < 0.05 was considered statistically significant. One-way ANOVA is also used as noted in the figure legends, followed by post hoc analysis for comparisons between individual groups. Identical letters in figures indicate datasets that are not statistically significant (p > 0.05).
3. Results
3.1. Liver-specific knockout of MED13 did not affect insulin sensitivity
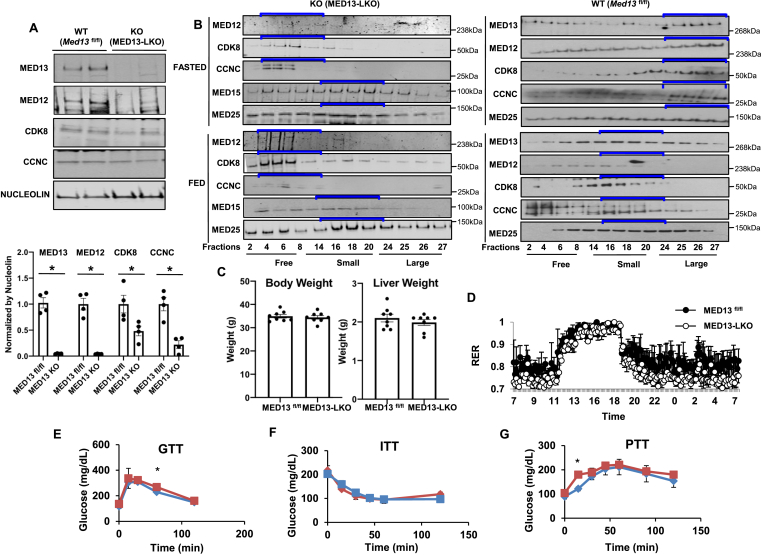
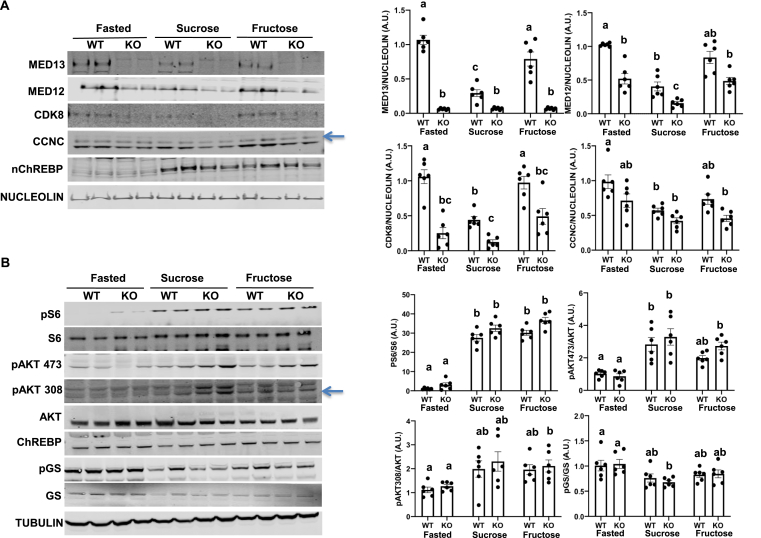
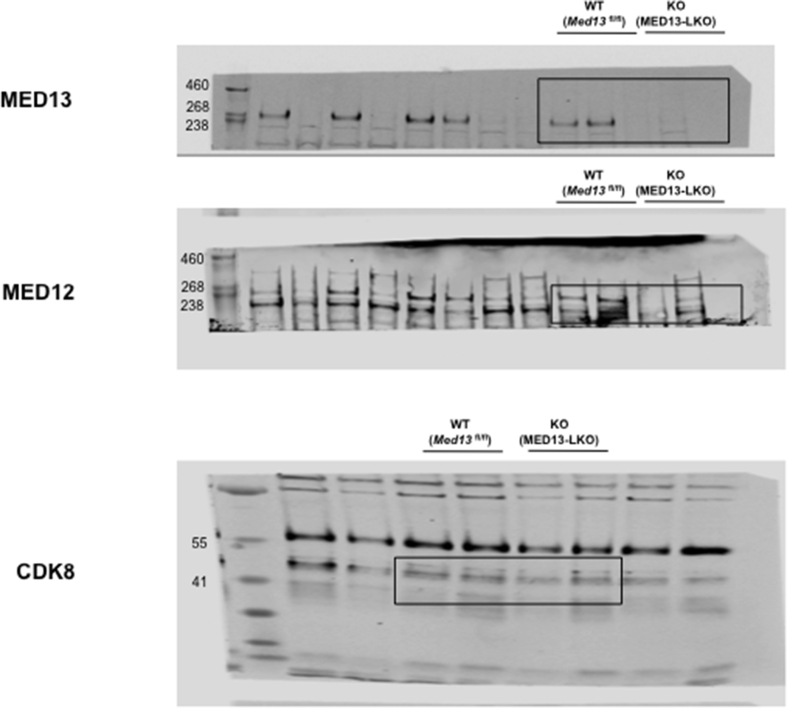
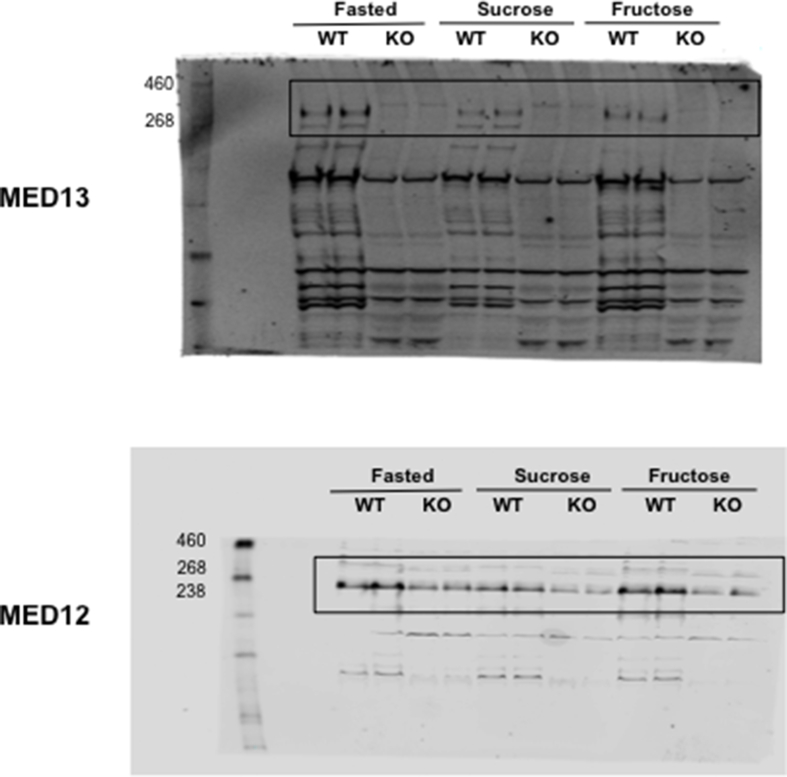
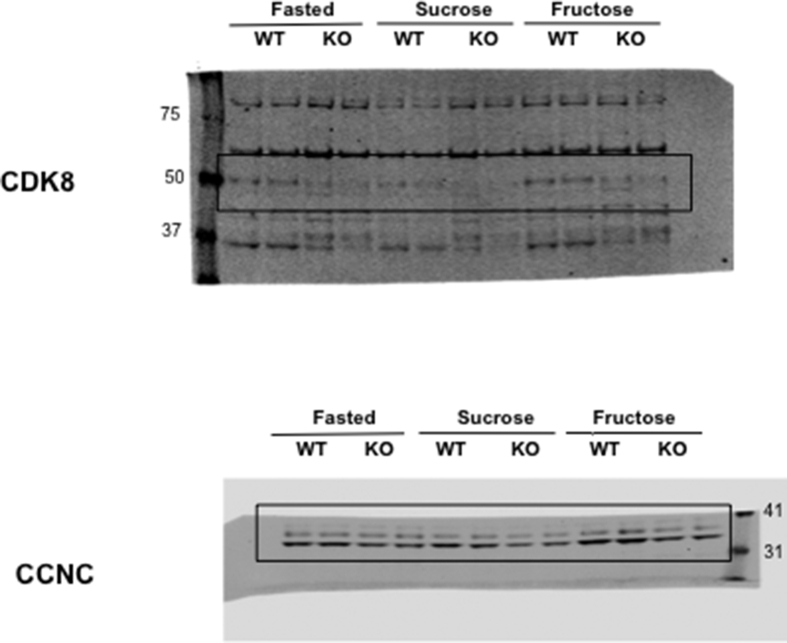
We recently reported that the kinase module subunits MED13, MED12, CDK8, and CCNC in the fed state dissociate from the large Mediator complex (∼1,200 kDa) to generate the small/core Mediator complex (∼600 kDa) with the subsequent degradation of the kinase module proteins [27]. Since MED13 directly links the kinase module to the core Mediator complex [40], loss of MED13 will essentially convert the large Mediator complex into the small Mediator complex. To understand the biological significance of fasting/feeding-regulated conversion of the Mediator complexes, we generated hepatocyte-selective MED13 knockout (MED13-LKO) mice by crossing Med13fl/fl mice with Alb-Cre mice. As expected, compared with controls (Med13fl/fl), the MED13-LKO mice displayed a near complete loss of MED13 and MED12 with an approximately 40% reduction in CDK8 and CCNC proteins (Figure 1A).
Figure 1.
Glucose metabolic phenotype of the MED13-LKO mice. Twelve-week-old male Med13fl/fl and MED13-LKO mice were fed normal chow (LabDiet #5053) ad libitum as described in Methods (n = 8 per group). A) Representative Western blotting images of liver nuclear proteins (5 separate experiments performed) and relative densitometric analysis of the mediator complex kinase module subunits. B) Glycerol gradient column separation and shift of kinase module subunits to the size that correlates with different complexes shown on gels. Fractions 2–8 represent free subunits, 14–20 represent small complexes, and 24–27 represent large complexes according to the size standards. Five livers of MED13-LKO (left panel) or Med13fl/fl (right panel) mice that were fasted or fed with normal chow and 30% sucrose containing water were pooled and the same amount of liver protein lysate was loaded onto each column as described in Methods. C) Body weight and liver weight were measured in 12-week-old Med13fl/fl and MED13-LKO mice after 4-h normal chow ad libitum feeding. D) Respiratory exchange ratio (RER) over a 24-h period. E) Glucose tolerance test, F) insulin tolerance test, and G) pyruvate tolerance test for Med13fl/fl (red) and MED13-LKO (blue) mice were performed as described in Methods (n = 8 per group). P values were determined by Student's t-test (p < 0.05). Values are the mean ± SEM.
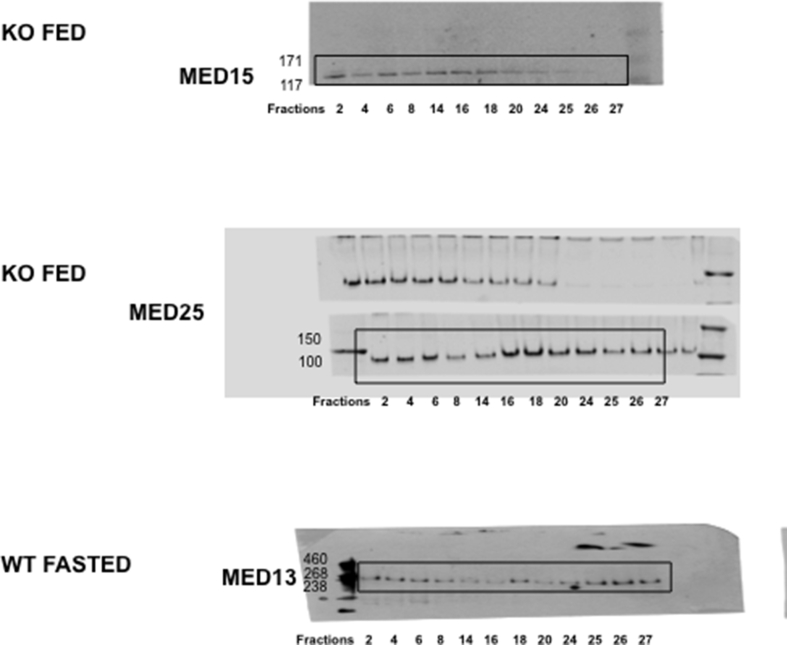
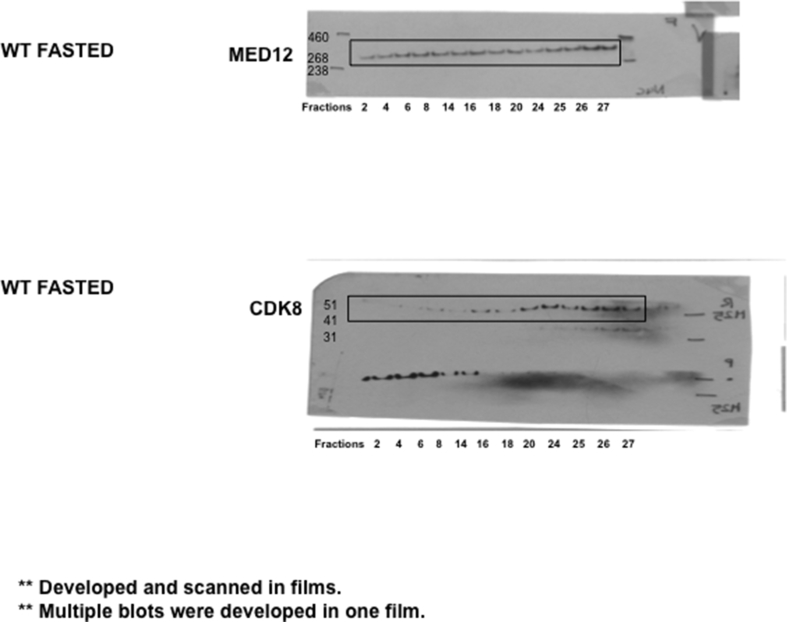
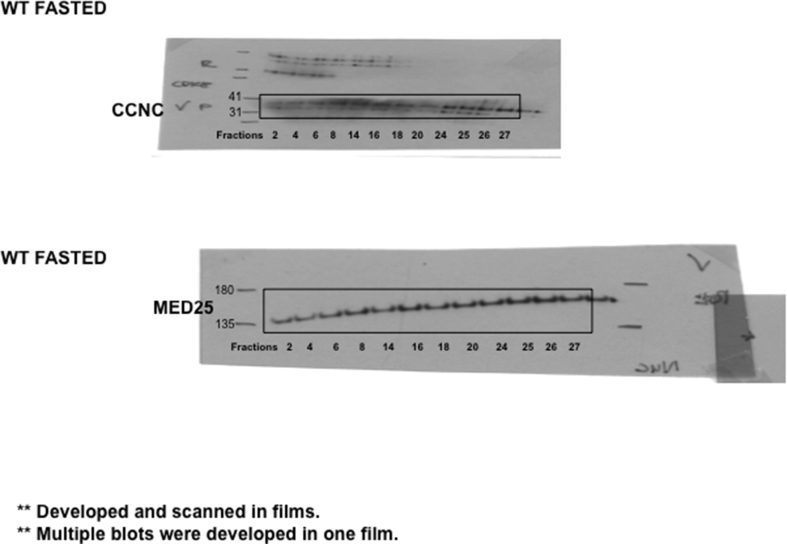
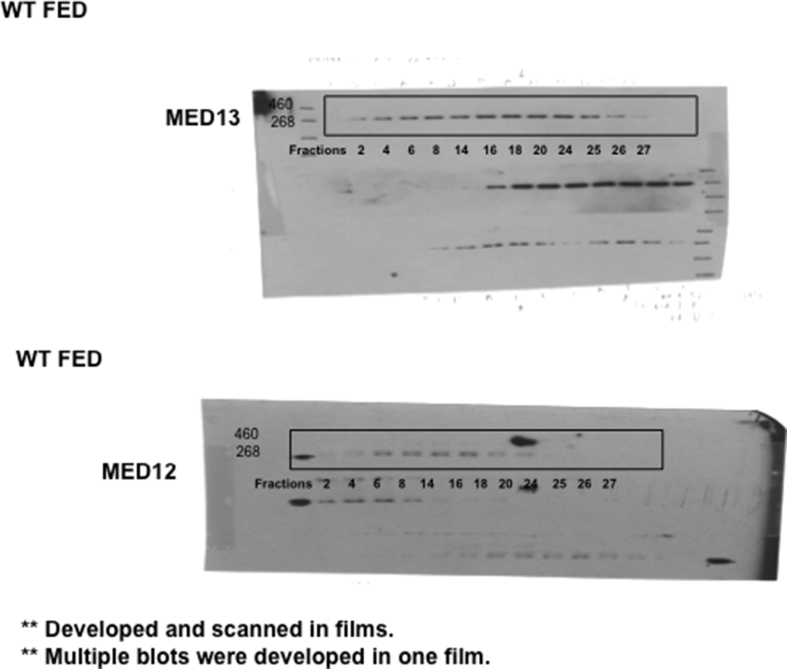
To determine the assembly state of the Mediator complex, we utilized glycerol gradient fractionation to separate different complexes based on the molecular size differences, the small Mediator complex (∼600 kDa) from the large Mediator complex (∼1.2 MDa). As we previously reported [27], in the fasted state, Mediator subunits in nuclear extracts of livers from control, in this case Med13fl/fl, mice migrated on glycerol gradients as a high molecular weight structure in the ∼1 MDa range (Figure 1B, right panel). However, a significant fraction of liver Mediator subunits migrated to the ∼600 kDa range following 4 h of feeding due to the dissociation of the kinase module. As expected, liver nuclear extracts of fasted MED13-LKO had a very low level of MED12 but the migration of CDK8 and CCNC to low molecular weight proteins range was indicative of their dissociation from MED12 (Figure 1B, left panel) and this was confirmed by co-immunoprecipitation in our previous study [27]. Despite the dissociation and loss of the kinase module subunits, the small/core Mediator complex remained intact as observed by MED15 and MED25 enrichment in the ∼600 kDa range. Moreover, MED13 knockout did not cause additional changes in the Mediator subunit assembly state following 4 h of feeding.
To determine whether liver MED13 deficiency and loss of the Mediator kinase module affected whole body metabolic homeostasis, we compared the changes in body weight, liver weight, respiratory exchange ratio (RER), glucose tolerance (GTT), insulin tolerance (ITT), and pyruvate tolerance (PTT) between the control Med13fl/fl and MED13-LKO mice (Figure 1C–G). In all of these metabolic assessments, there were no apparent differences between the Med13fl/fl and MED13-LKO mice.
3.2. Liver-specific knockout of MED13 significantly reduced inducible gene expression
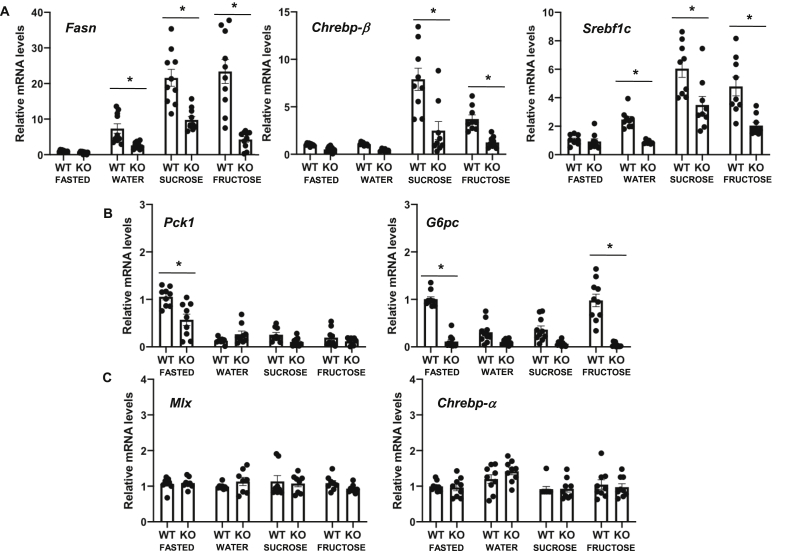
To explore the kinase module's role in regulating metabolic gene expression, we assessed the feeding induction of lipogenic gene expression using qRT-PCR (Figure 2). Fasn, Chrebp-β, and Srebf1c mRNA were induced in the control Med13fl/fl mice livers following 4 h of feeding with mouse chow only (Figure 2A). Supplementation of drinking water during the 4 h feeding period with sucrose or fructose further enhanced lipogenic gene expression. In contrast, the feeding induction of these genes with supplementation of sucrose and fructose was significantly suppressed in the MED13-LKO mice livers. In contrast to lipogenic genes that were induced in the fed state, the canonical fasting-induced gluconeogenic genes (Pck1 and G6pc) were elevated in the fasted state and suppressed following feeding with chow and when supplemented with sucrose in the control Med13fl/fl mice livers (Figure 2B). However, feeding with fructose suppressed Pck1 mRNA but not G6pc, which remained elevated (Figure 2B). The MED13-LKO livers suppressed G6pc mRNA even in the presence of fructose.
Figure 2.
Inducible gene expression was suppressed in MED13-LKO mice under different carbohydrate-containing feedings. FASTED for 16 h fasted with water, WATER for normal chow supplemented with water, SUCROSE for normal chow supplemented with 30% sucrose containing water, and FRUCTOSE for normal chow supplemented with 30% fructose containing water. A, B, and C) Real-time quantitative RT-PCR (qRT-PCR) of lipogenic genes (Fasn, Chrebp-β, and Srebf1c), gluconeogenic genes (G6pc and Pck1), and constitutively active and unaffected by nutrient signal genes (Mlx and Chrebp-α) performed in livers of the Med13fl/fl (WT) or MED13-LKO (KO) mice (n = 9 per group, 3 separate experiments were conducted). P values were determined by Student's t-test (p < 0.05). Values are the mean ± SEM.
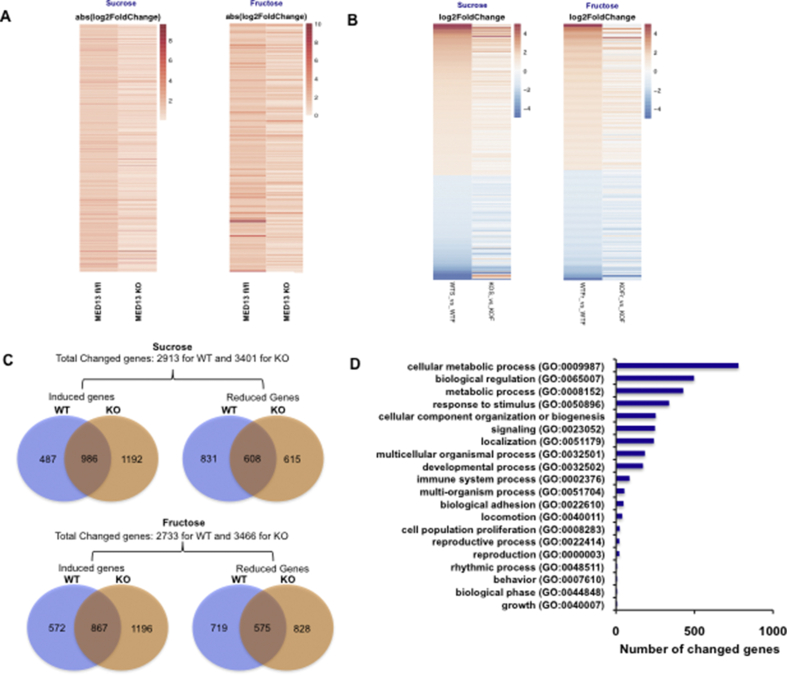
To determine whether MED13 deficiency affected non-nutrient regulated genes in the liver, Mlx and Chrebp-α mRNA expression levels were measured in the Med13fl/fl and MED13-LKO mice under these dietary conditions from the same mice (Figure 2C). There was no significant difference in the expression of these genes. Similarly, other constitutively expressed liver genes such as Lxr-α and Usf1 were unaffected by MED13 deficiency (Fig. S1). In addition, genes that regulate glycogen synthesis or breakdown, Gbe, Phkb, and Gs, showed no significant changes with changes in nutritional states or MED13 deficiency (Fig. S1). The relative effect of MED13 deficiency on nutrient-regulated liver genes but not constitutive gene expression was confirmed by genome-wide RNA-seq analyses for the sucrose- and fructose-fed mice (Fig. S2). Based on these data, we conclude that the kinase module primarily regulates inducible gene expression but is not essential for basal gene expression in the liver. Moreover, MED13 deficiency suppressed G6pc gene expression even in the fasting and fructose-fed conditions where G6pc expression levels should be highly induced.
3.3. Fructose suppressed liver glycogen accumulation in the fed state
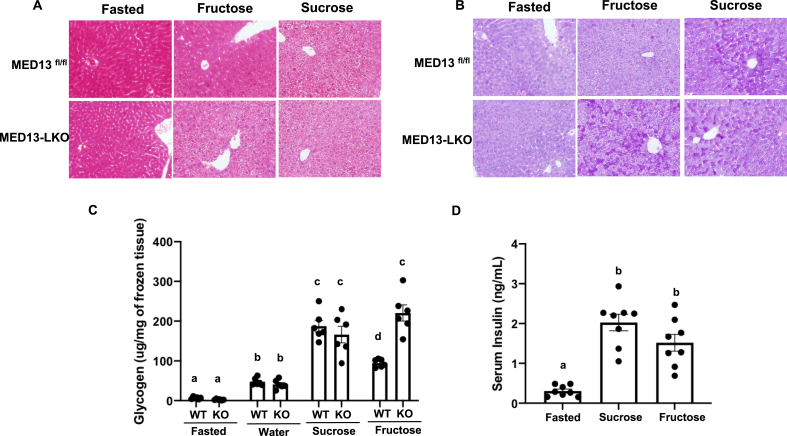
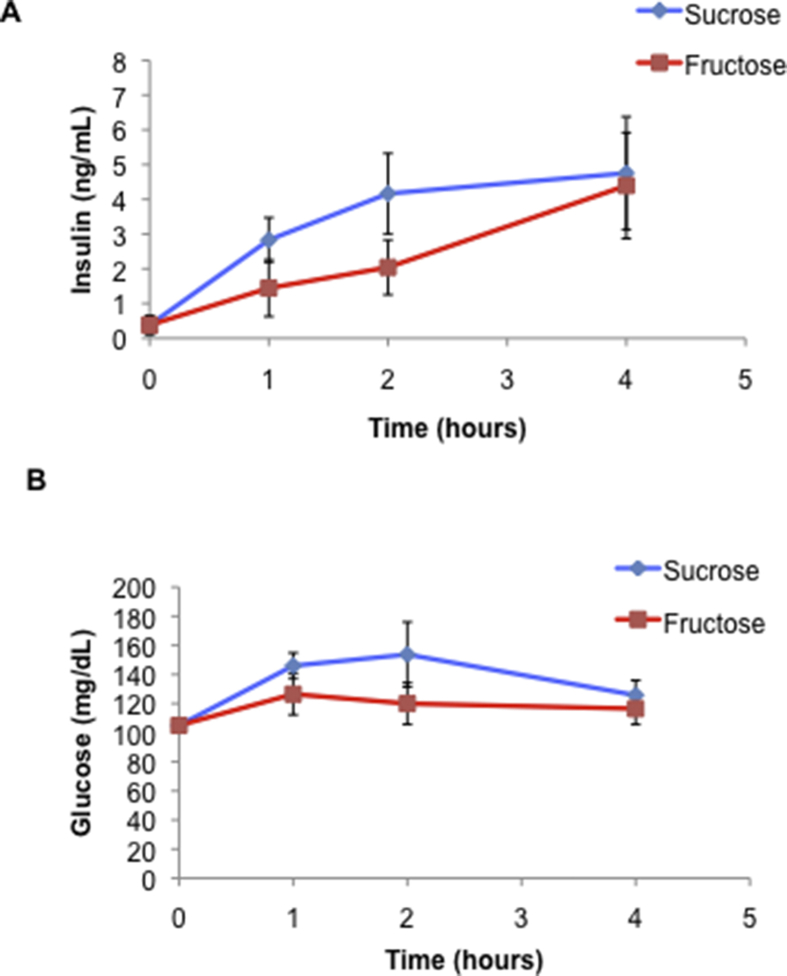
The hepatic production and release of glucose is a complex process that includes de novo formation of glucose by gluconeogenesis and breakdown of glycogen through glycogenolysis [41,42]. Previous studies also indicated that chronic fructose feeding results in elevated gluconeogenesis due to increased expression of G6pc, which encodes the catalytic subunit of G6PC (or G6Pase) [43,44]. Because G6Pase converts glucose 6-phosphate to glucose in the last step for both gluconeogenesis and glycogenolysis, we assessed the effect of acute fructose and MED13 deficiency on glycogen repletion. As expected, in the fasted state in the control Med13fl/fl and MED13-LKO mice, the glycogen levels were relatively low as assessed by H&E staining, periodic acid Schiff (PAS) labeling, and quantification of liver glycogen (Figure 3A–C). When the mice were fed with sucrose supplementation, there was a marked increase in liver glycogen content that was not significantly different between the control Med13fl/fl and MED13-LKO mice. However, supplementation with fructose in the drinking water resulted in a reduction in glycogen accumulation compared to sucrose supplementation in the control Med13fl/fl mice but that was not observed in the MED13-LKO mice livers. The reduction in the glycogen levels with fructose supplementation in the control Med13fl/fl mice was not a result of altered insulin secretion as there was no statistically significant difference in insulin levels (Figure 3D), although there was a trend of lower insulin levels in the fructose-supplemented mice during early periods of the post-prandial time course (Fig. S3A). The post-prandial rise in glucose also trended to be lower in the fructose-supplemented mice but this was not statistically significant (Fig. S3B).
Figure 3.
Glycogen accumulation was observed in MED13-LKO liver under fructose feeding. A) Representative H&E stained liver sections of Med13fl/fl and MED13-LKO after overnight fasting followed by chow feeding supplemented with 30% fructose containing water for 4 h. B) Representative periodic acid Schiff (PAS)-stained liver sections of the Med3fl/fl and MED13-LKO mice. C) Hepatic glycogen measurement from the Med13fl/fl and MED13-LKO mice under fasted or ad libitum fed supplemented with either water or water with 30% fructose or 30% sucrose conditions. D) Serum insulin levels of WT mice after feeding (n = 8 per group). Statistical significance was determined by one-way ANOVA followed by the Turkey-Kramer multiple comparisons test. Bars with the same letter are not significantly different.
3.4. Overexpression of G6PC restored fructose-caused reduction of glycogen accumulation in MED13-LKO mice
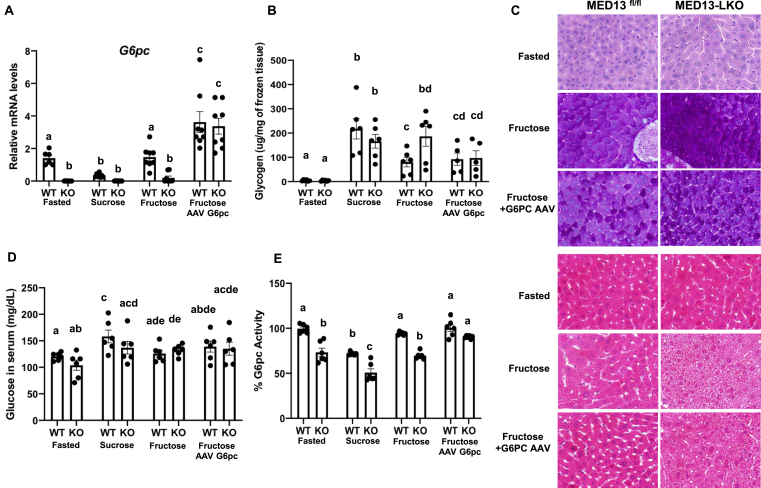
If the inability of fructose to suppress G6pc gene expression is the causative pathway preventing glycogen accumulation in the fed state, then constitutive G6pc expression should reverse the ability of the MED13-LKO to restore glycogen. To this end, we infected the control Med13fl/fl and MED13-LKO mice with adeno-associated viruses (AAV) (AAV-Tbg-GFP or AAV-Tbg-G6pc) to overexpress G6Pase specifically in hepatocytes under the control of the Tbg promoter, which is not regulated by nutrients. As shown in Figure 2, the G6pc mRNA level was elevated in the fasted state and suppressed when the control Med13fl/fl mice were fed with sucrose supplementation (Figure 4A). However, G6pc mRNA failed to decrease in the fructose-fed control mice whereas G6pc was suppressed in both the fasted and fructose-fed states in the MED13-LKO mice livers. In contrast, G6pc mRNA remained elevated approximately 2-fold over the fasted state in both the control Med13fl/fl and MED13-LKO mice following G6pc overexpression with fructose supplementation.
Figure 4.
Overexpression of G6pc decreased glycogen accumulation in MED13-LKO. A)G6pc gene expression in both Med13fl/fl and MED13-LKO mice liver was measured by qRT-PCR 10 days after AAV-Tbg-GFP or AAV-Tbg-G6pc injections. B) Hepatic glycogen measurement, C) H & E staining and PAS staining in liver sections and hepatic glycogen levels, D) changes in serum glucose levels, and E) % changes in G6pc activity (n = 6) were measured as described in Methods (n = 8 per group). Identical letters indicate values that are not statistically different from each other (p > 0.05). One-way ANOVA followed by the Turkey-Kramer multiple comparisons test.
Glycogen content increased in the sucrose-supplemented control Med13fl/fl mice and was suppressed in the fructose-supplemented mice whereas the MED13-LKO mice were able to accumulate glycogen following fructose supplementation (Figure 4B and C). In contrast, the fructose suppression of liver glycogen accumulation was not significantly different between the control Med13fl/fl and MED13-LKO mice when G6pc was overexpressed. As a control, serum glucose levels were measured that increased in the post-prandial states but were not significantly different in the Med13fl/fl and MED13-LKO mice whether fed with sucrose or fructose supplementation or by overexpression of G6pc (Figure 4D). G6Pase enzymatic activities were measured and the levels were in agreement with the G6pc mRNA levels (Figure 4E). These data demonstrate that constitutive overexpression of G6PC is sufficient to prevent liver glycogen restoration in the fed state.
3.5. MED13 deficiency did not inhibit proximal insulin signaling
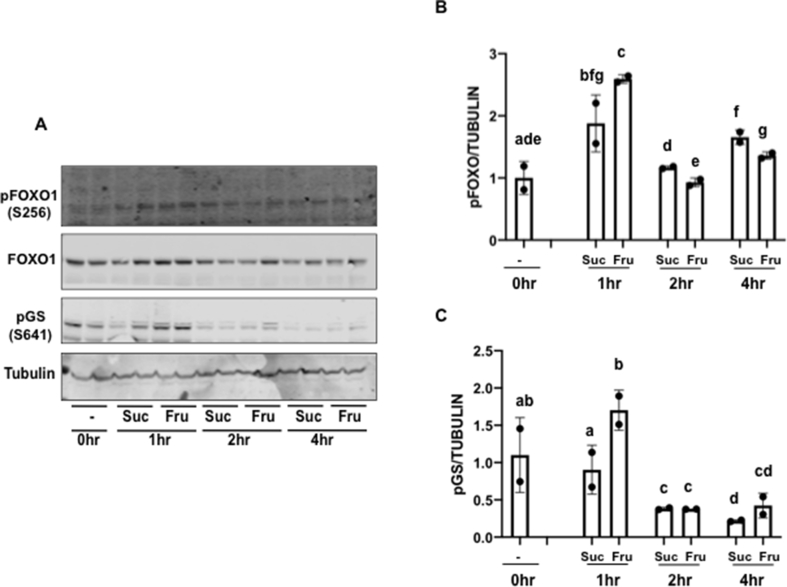
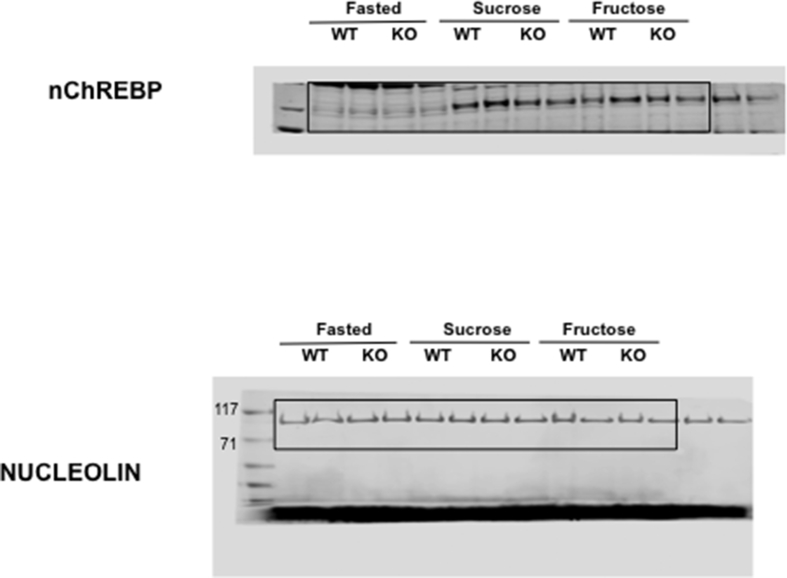
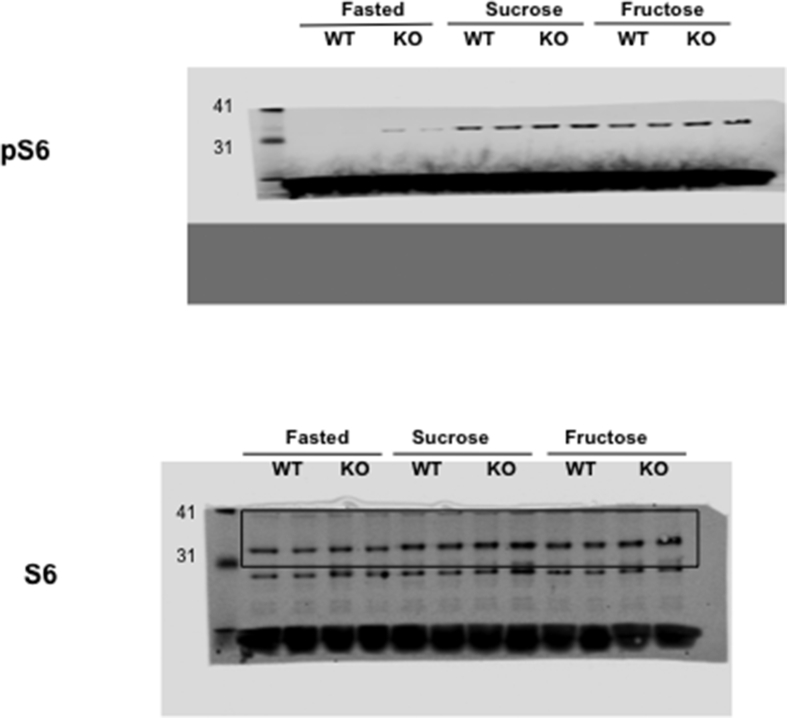
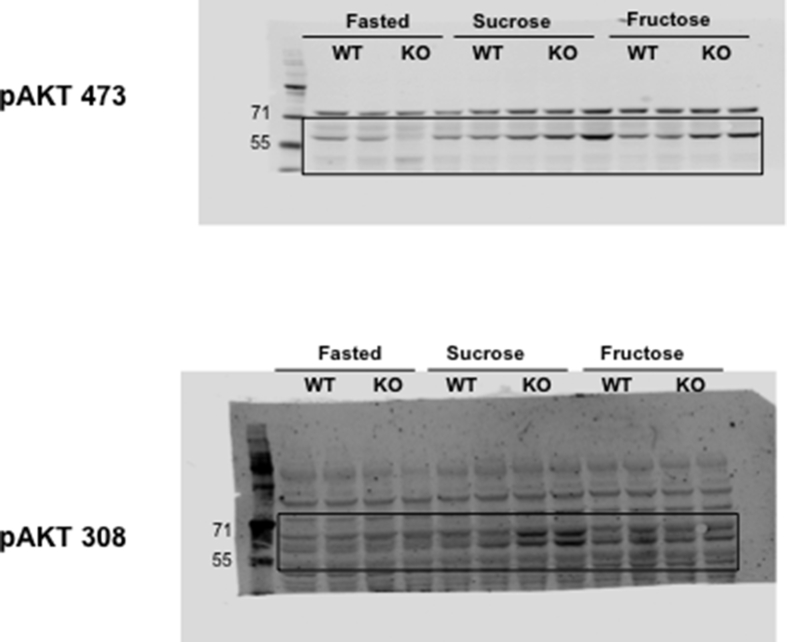
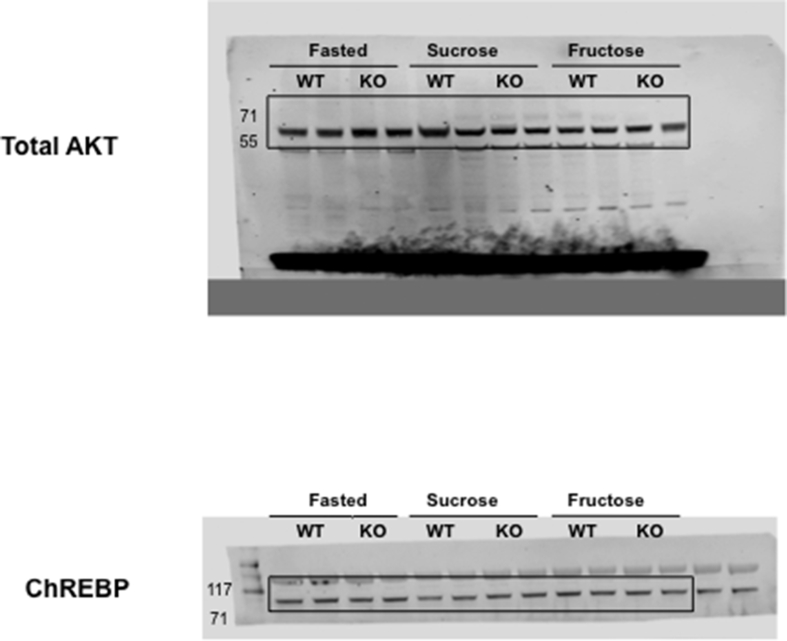
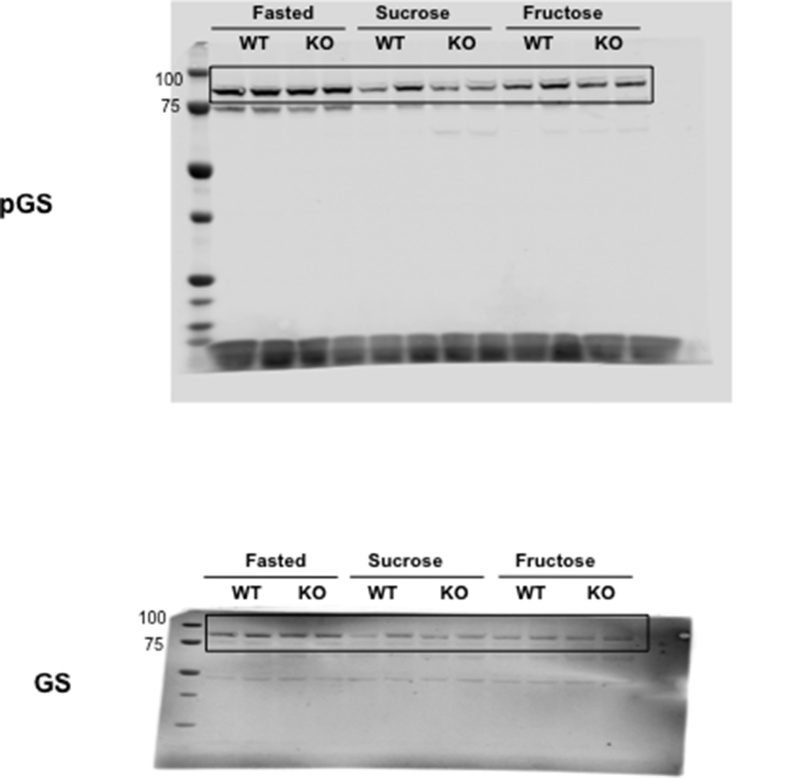
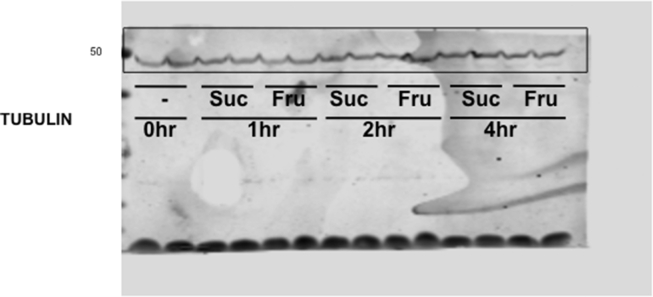
The Mediator complex kinase module subunits (MED13, MED12, CDK8, and CCNC) undergo degradation in the fed state [27,33]. Consistent with these findings, Western blotting analyses of the kinase module subunits displayed a reduction in these proteins in the control Med13fl/fl mice following feeding with chow plus sucrose compared to fasted mice livers (Figure 5A). In contrast, when the mice were fed with fructose-supplemented water, there was no significant decrease in the kinase module subunit proteins. The transcription factor ChREBP is known to undergo nuclear translocation from the cytoplasm in the fed state [19], and we observed a similar extent of ChREBP nuclear translocation in both the sucrose and fructose-supplemented fed mice. Moreover, MED13 deficiency had no significant effect on ChREBP nuclear translocation (Figure 5A), with no apparent change in the total levels of ChREBP protein (Figure 5B). Our previous studies suggested that kinase module protein reduction depends on activation of the mTORC1 pathway [27,33]. There was no significant difference in S6, AKT, or glycogen synthase (GS) phosphorylation states in the control Med13fl/fl mice induced by either sucrose or fructose-supplemented feeding (Figure 5B). Consistent with previous reports [63,65], feeding resulted in a transient increase in FOXO1 phosphorylation but with a slower and more sustained decrease in GS phosphorylation (Figs. S5A–C). However, there was no significant difference between the effect of sucrose or fructose supplementation, consistent with the similar levels of insulin secretion (Figure 3 and Fig. S3).
Figure 5.
Fructose supplementation reduced insulin signaling and suppressed the downregulation of the Mediator kinase module. A) Western blotting and relative densitometric analysis of hepatic nuclear MED13, MED12, CDK8, CCNC, nuclear ChREBP, and nucleolin loading control. B) Western blotting and relative densitometric of total liver including nuclear and cytosolic fractions (n = 6 per group, representative duplicate from 3 separate experiments shown on gels). P values were determined by Student's t-test (p < 0.05). Values are the mean ± SEM. Identical letters indicate values that are not statistically different from each other (p > 0.05). Multiple sample comparisons were determined by one-way ANOVA followed by the Turkey-Kramer multiple comparisons test.
3.6. MED13 deficiency altered ChREBP binding to the G6pc promoter
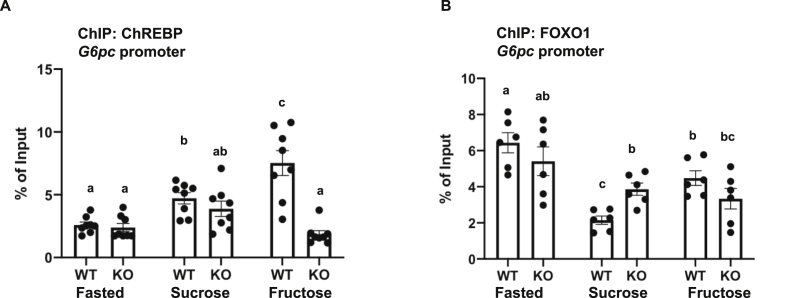
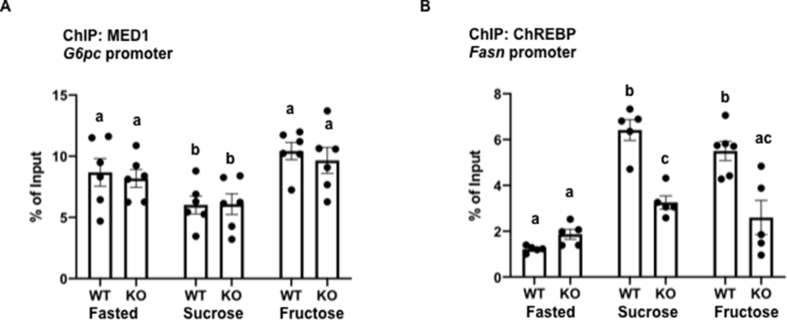
In the fasted state, G6pc expression is transactivated by FOXO1, and in the fed state, FOXO1 is inactivated by acetylation and nuclear-exported due to AKT-dependent phosphorylation [45]. The G6pc gene also contains a ChREBP-responsive cis-DNA element and fructose is a potent activator of ChREBP, which has been reported to be the basis for the elevated activation of G6pc expression by fructose [7]. To examine the basis for MED13 regulation of G6pc, we first performed chromatin immuno-precipitation coupled with quantitative PCR (ChIP-qPCR) to determine FOXO1 and ChREBP binding on the G6pc gene promoter (Figure 6A and B). In the fasted state, there was a relatively low level of ChREBP binding to G6pc, which was not different between the control Med13fl/fl and MED13-LKO mice livers (Figure 6A). Surprisingly, in the control livers, ChREBP binding increased following feeding with sucrose supplementation, although G6pc gene expression was substantially suppressed. Similarly, the control mice fed with fructose supplementation also displayed increased ChREBP binding, but more importantly, fructose supplementation did not induce ChREBP binding to the G6pc promoter in the MED13-LKO mice, suggesting that the interaction of ChREBP with the G6pc gene promoter is MED13-dependent. In contrast, the binding of FOXO1 to the G6pc gene in the fasted state was not disrupted in the MED13-LKO mice (Figure 6B). Moreover, under sucrose supplementation, FOXO1 binding to the G6pc gene promoter was downregulated in both the Med13fl/fl and MED13-LKO mice.
Figure 6.
Binding of ChREBP and FOXO1 transfactors on G6pc promoter in MED13-LKO mice. ChIP-qPCR was performed to analyze the enrichment for transfactors in the G6pc promoter region. A) ChREBP-specific antibody was used to pull down sonicated liver chromatin of the Med13fl/fl and MED13-LKO mice (n = 6). Precipitated DNA enrichment is shown as a percentage of input DNA, with average Ct values of 31.2 (Fasted WT), 29.65 (Fasted KO), 30.38 (Sucrose WT), 29.3 (Sucrose KO), and 30.0 (Fructose WT). B) FOXO1-specific antibody was used to pull down sonicated liver chromatin of the Med13fl/fl and MED13-LKO mice (n = 6). Precipitated DNA enrichment is shown as a percentage of input DNA, with average Ct values of 32.4 (Fasted WT), 31.5 (Fasted KO), 33.8 (Sucrose WT), 31.9 (Sucrose KO), 34.1 (Fructose WT), and 31.8 (Fructose KO). Non-specific IgG was used as a control, which displayed Ct values > 38 or were undetectable. A promoter set of the non-specific upstream region was also used as a control, which showed Ct value > 38 or was undetectable. P values were determined by one-way ANOVA followed by the Turkey-Kramer multiple comparisons test. Error bars indicate the mean ± SEM. Bars with different letters are significantly different from each other (p < 0.05).
To confirm that the Mediator complex remains able to bind to the G6pc promoter in either the presence or absence of MED13, we also performed ChIP-qPCR using a well-tested antibody against the core Mediator subunit MED1 in the control and MED13-LKO mice. As shown in Fig. S4A, MED13 deficiency had no effect on MED1 occupancy on the G6pc promoter. MED1 occupancy was somewhat reduced in the fed state with sucrose but not in the fed state with fructose, indicating that Mediator binding was not the primary reason for the nutrient regulation of G6pc gene expression. As an additional control, we also assessed ChREBP binding to the Fasn promoter (Fig. S4B). As expected, ChREBP occupancy increased in the sucrose- and fructose-fed state in the control mice that was reduced in the MED13-LKO mice livers.
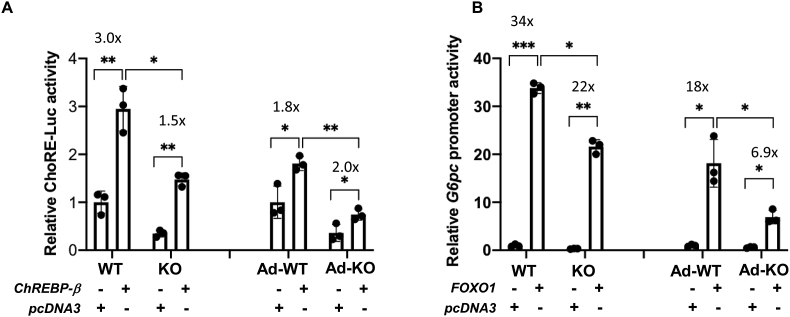
Although DNA binding of regulatory factors is an important component of gene expression, this does not directly address functional transcriptional activity. Therefore, we next isolated primary hepatocytes from the control and MED13-LKO mice and co-transfected them with expression plasmids for ChREBP-β and a synthetic ChoRE-luciferase reporter, which only contains three copies of ChoRE (Figure 7A). As shown in the left bars, both the basal and ChREBP-β-induced ChoRE promoter activity of the ChREBP reporter was significantly suppressed by MED13 deficiency. To eliminate any potential development effects in the MED13-LKO hepatocytes, we also isolated primary hepatocytes from the Med13fl/fl mice, which were then infected with adenovirus expressing GFP (Ad-GFP) or Cre (Ad-Cre) with the expression plasmids for ChREBP-β and the ChoRE-luciferase reporter (Figure 7A, right bars). We again observed a significant suppression of basal and ChREBP-β-stimulated transcriptional activity in the acute MED13 knockout hepatocytes. Similarly, examination of the FOXO1 transcriptional activity using the proximal G6pc promoter also demonstrated a reduced ability of FOXO1 to transactivate G6pc reporter in the MED13 knockout hepatocytes (Figure 7B). In this case, there was no significant change in basal reporter activity only on the FOXO1-induced transcriptional activation.
Figure 7.
Luciferase reporter assay of ChREBP and FOXO1 transfactors in MED13-LKO mice. Isolated primary hepatocytes from adenovirus (Ad)-GFP (Ad-WT) or Ad-Cre (Ad-KO) transduced Med13fl/fl (WT) or Med13fl/flAlb-Cre (KO) mice were used. A) Luciferase reporter assays for the effects of ChREBP-β on ChoRE activity in either Med13fl/fl (WT and Ad-WT) or Med13-LKO (KO and Ad-KO) as indicated. B) Luciferase reporter assays for the effects of FOXO1 on G6pc promoter activity in indicated cells, either from Med13fl/fl (WT and Ad-WT) or Med13-LKO (KO and Ad-KO). P values were determined by Student's t-test (p < 0.05). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and n = 3. The values above each of the error bars indicate the fold stimulation.
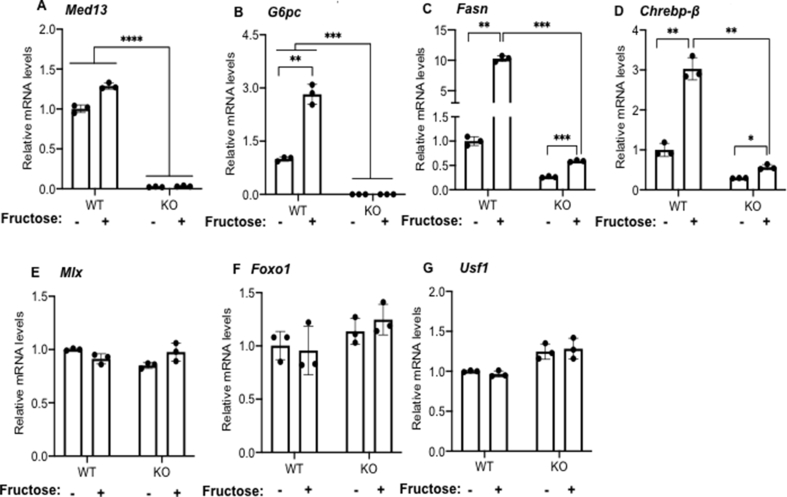
To further examine whether the fructose regulation of G6pc expression was cell autonomous and independent of systemic metabolic signaling, we isolated primary hepatocytes from the control and MED13-LKO mice (Fig. S6A). Fructose treatment alone increased the expression of G6pc, Fasn, and Chrebp-β mRNA in the control cells compared to primary hepatocytes maintained under regular tissue culture conditions (Figs. S6B–D). Importantly and consistent with the effect of fructose supplementation in vivo (Figure 2), in the MED13-deficient hepatocytes, both the basal and fructose-stimulated increase in G6pc, Fasn and, Chrebp-β mRNA was markedly suppressed. In contrast, constitutively expressed genes Mlx, Foxo1, and Usf1 that are not nutrient-regulated invivo were neither affected by fructose treatment nor suppressed in the MED13 knockout hepatocytes (Figs. S6E–G). Together, these data support the hypothesis that the kinase module of the Mediator complex plays an important regulator role in controlling transcription factor DNA occupancy and transcriptional activity in liver hepatocytes that are i) transfactor-specific, ii) nutrient-specific, and iii) target gene-specific.
4. Discussion
The integration of gene transcription is coordinated by multiple regulator factors of which the Mediator plays an essential role [46,47]. In particular, elements of the Mediator kinase module have been observed to play various roles in a tissue/cell-type context-dependent manner. For example, CDK8 was found to function both as a proto-oncogene and tumor suppressor [[48], [49], [50]]. In Drosophila and mice, CDK8 was also observed to regulate nuclear SREBP-1c degradation and independently CCNC was reported to regulate mitochondrial fission [51,52]. Deletion of MED13 in mouse cardiomyocytes enhanced obesity in response to a high-fat diet and exacerbated metabolic syndrome due to the inhibition of a cardiomyocyte secretory factor [53]. In contrast, MED13 deficiency in skeletal muscle increased insulin sensitivity and protected mice against high-fat diet-induced hepatic steatosis [54].
In the liver we recently observed that the large Mediator complex predominantly exists in the fasted state but that a substantial proportion converts to the small/core Mediator complex in the fed state due to both dissociation and subsequent degradation of the kinase module subunits, MED13, MED12, CDK8, and CCNC [27,33]. Of the approximately 12,000 protein-encoding genes expressed in the liver, approximately 3000 undergo regulated changes between the fasted and fed states [55]. In particular, among the most dynamically modulated genes during the fasted/fed transitions are those regulating the gluconeogenic and lipogenic pathways. In this regard, several studies have demonstrated that dietary fructose has significant patho-metabolic properties and may be an important contributor to the development of fasting hyperglycemia, insulin resistance, obesity, and non-alcoholic fatty liver disease [56]. However, the mechanisms of fructose action and downstream physiologic responses are highly complex and poorly understood. For example, fructose has been reported to induce [57] and inhibit beta cell insulin secretion [[58], [59], [60]]. In this regard, we have observed a trend for fructose to reduce insulin secretion compared to sucrose but this was not statistically significant and in any case, insulin levels were within the post-prandial physiologic range with no deleterious effect on proximal insulin-signaling events.
As shown in Fig. S4, we observed no significant difference in the upregulation of blood glucose levels and serum insulin levels in the mice that were under the sucrose water or fructose water supplementation conditions in all of the post-prandial time points measured. Consistent with several recent studies [6,7,61], fructose feeding also prevented the post-prandial decline in G6pc expression. Under the combination of a high-fat and high-fructose diet, chronic feeding elevated G6pc levels through transactivation by ChREBP-β [6]. CHREBP-α is activated in the fed state, which in turn transactivates ChREBP-β that is an important transfactor regulating lipogenic gene expression [62]. Although the G6pc gene is typically suppressed in the fed state, its distal promoter contains a ChREBP-binding element and can be transactivated by ChREBP transcription factors [7]. Our data indicate that the acute effect of fructose to prevent the suppression of G6pc is responsible for the inhibition of glycogen accumulation during the fasted to fed transition.
It should be noted that two previous studies also examined the effect of post-prandial fructose on liver glycogen repletion. Conlee et al. [64] provided gastric-infused glucose and fructose to rats following fasting but only observed a small increase of glycogen repletion to approximately 20% of normal liver glycogen levels in both cases. Similarly, Sato et al. [63] also used intragastric-delivered fructose in fasted mice and observed glycogen restoration to relatively normal fed liver glycogen levels. However, in this study, fructose prevented the decline in G6pc mRNA that occurs following feeding but with a reduction in G6PC activity. Other than the different methodologies used in these previous studies, the basis for the differences with our findings remains to be determined.
Nevertheless, at the molecular level, our data indicate that ChREBP occupies the G6pc promoter in the fed state with both sucrose and fructose supplementation. However, G6pc gene expression remains elevated only when fructose is present in the diet. Previously using FOXO1 knockout mice, fructose feeding was reported to induce G6pc gene expression independent of FOXO1 [7]. Consistent with these data, in both sucrose- and fructose-supplemented feeding, FOXO1 binding to the G6pc promoter was similarly reduced. An important difference between sucrose- and fructose-supplemented feeding is the molecular state of the Mediator complex. Feeding with sucrose shifts the large Mediator complex present in the fasted state to that of the small/core Mediator complex. However, in the presence of fructose, the Mediator complex primarily remains as the large Mediator complex. Assuming that the large Mediator complex is the major driver for the induction of gluconeogenic gene expression whereas the small/core Mediator is the major driver for lipogenic gene expression, this would account for ChREBP's ability to drive G6pc gene expression in the presence of fructose. Moreover, this would also account for the effect of MED13 deficiency to suppress G6pc gene expression in the fasted, sucrose-, and fructose-fed states. As the Mediator continuously undergoes dynamic oligomeric state reorganization, it is likely that this reflects many of the gene and cell context effects of Mediator complex-dependent gene expression. Future studies are needed to identify the specific mechanisms responsible for this unusual form of transcription specificity in the liver.
Significance statement
The Mediator is a multisubunit protein complex that links transcription factors to RNA polymerase II and undergoes dynamic state reorganization between the small or core Mediator complex (26 subunits) and the large Mediator complex that also contains the kinase module subunits (MED13, MED12, CDK8, and CCNC). In the liver, we found that deletion of the MED13 subunit resulted in the constitutive formation of the small Mediator and the downregulation of nutrient-inducible gene expression. Fructose suppression of glycogen synthesis occurred due to increased G6pc expression and MED13 deficiency reversed the detrimental effect of fructose by inhibiting G6pc. These data provide direct evidence for the kinase module as a regulator of inducible gene expression and preventing fructose-mediated metabolic dysregulation in the liver.
Acknowledgments
We thank Drs. Leonela Amoasii and Eric Olson for generously providing the Med13fl/fl mice used in this study. We thank Dr. Mark Herman for providing the Chrebp alpha and beta plasmids. This work was supported by grants from the National Institutes of Health DK110063, DK098439, DK117417, and DK020541. DYY was supported by a T32 post-doctoral training award AG023475 and Einstein Marion Bessin Liver Research Center Pilot Grant DK041296.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101227.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
List of mouse oligonucleotide primer sequences for qRT-PCR.
Fig. s1.
Gene expression of nutrient-unresponsive genes. The Med13fl/fl control mice (WT) and MED13-LKO (KO) mice were fasted for 16 h overnight and sacrificed (fasted) or fed with water or fed with normal chow with supplementation of 30% sucrose or fructose water (sucrose, fructose). Real-time quantitative RT-PCR (qRT-PCR) analysis of glycogen synthesis or breakdown pathway-regulating genes (Gbe, Phkb, and Gs) and nutrient-unresponsive genes (Lxr-α and Usf1) was performed in livers of the Med13fl/fl or MED13-LKO mice (n = 4 per group for glycogen-related genes and n = 3 for nutrient-unresponsive genes). No statistically significant differences between samples were found.
Fig. s2.
Heatmap of RNA-seq reporting changes in hepatic genes in the Med13fl/fland MED13-LKO mice under fed states. A) Absolute of log2foldchange in gene expression for the Med13fl/fl and MED13-LKO mice under feeding with 30% sucrose or fructose water supplementation compared with fasted liver samples. Genes with |log2foldchange| > 1 and padj (adjusted p value) < 0.05 in the MED13fl/fl mice were selected as rows of the heatmap, and the intensity of the red color represents the magnitude of fold changes in either the up or down direction. B) Log2foldchange in gene expression for the Med13fl/fl and MED13-LKO mice under feeding with 30% sucrose or fructose water supplementation compared with fasted liver samples. Genes with |log2foldchange| > 1 and padj (adjusted p value) < 0.05 in the Med13fl/fl mice were selected as rows of the heatmap, and the intensity of the red color represents the magnitude of fold changes in either the up or down direction. C) Feeding induced and reduced gene populations in the Med13fl/fl (blue) and MED13-LKO (orange) mice with sucrose or fructose water supplementation. Genes with padj <0.05 were counted. D) Gene ontology analysis of both upregulated and downregulated genes in the MED13-LKO mice compared to the Med13fl/fl mice under fructose water supplementation. Genes with |log2foldchange| > 1 and padj (adjusted p value) < 0.05 were used as an input list to the DAVID functional annotation tool, and all of the genes detected in the experiment were used as background. Bar graph shows log10(padj) and the numbers beside the bars show the number of input genes in the GO term.
Fig. s3.
Circulating glucose and insulin levels under different carbohydrates containing water supplementations and at different post-prandial time points. Twelve-week-old C57BL/6J mice were fasted overnight and fed with normal chow supplemented with 30% sucrose or fructose in their drinking water. A) Blood glucose was measured at each post-prandial time point as indicated. B) Serum insulin was measured at each post-prandial time point as indicated. P values were determined by Student's t-test (p < 0.05). Values are the mean ± SEM.
Fig. s4.
Binding of MED1 on G6pc promoter and ChREBP transfactors on Fasn promoter in the MED13fl/fl(WT) and MED13-LKO (KO) mice. ChIP-qPCR was performed to analyze the enrichment of the core mediator subunit MED1 on the G6pc promoter region and ChREBP transfactors on the Fasn promoter region. A) Anti-MED1 and B) anti-ChREBP were used to pull down sonicated liver chromatin of the Med13fl/fl and MED13-LKO mice (n = 4). Precipitated DNA enrichment is shown as a percentage of input DNA. P values were determined by one-way ANOVA followed by the Turkey-Kramer multiple comparisons test. Error bars indicate the mean ± SEM. Bars with different letters are significantly different from each other (p < 0.05).
Fig. s5.
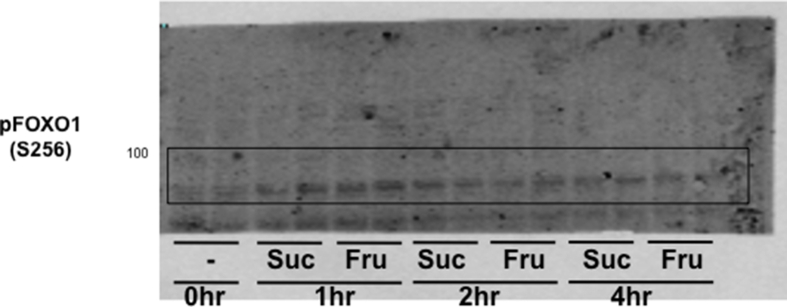
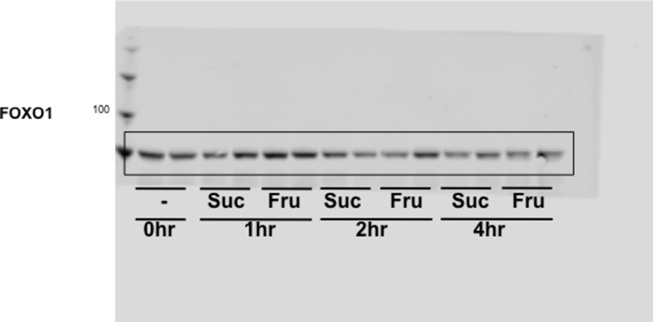
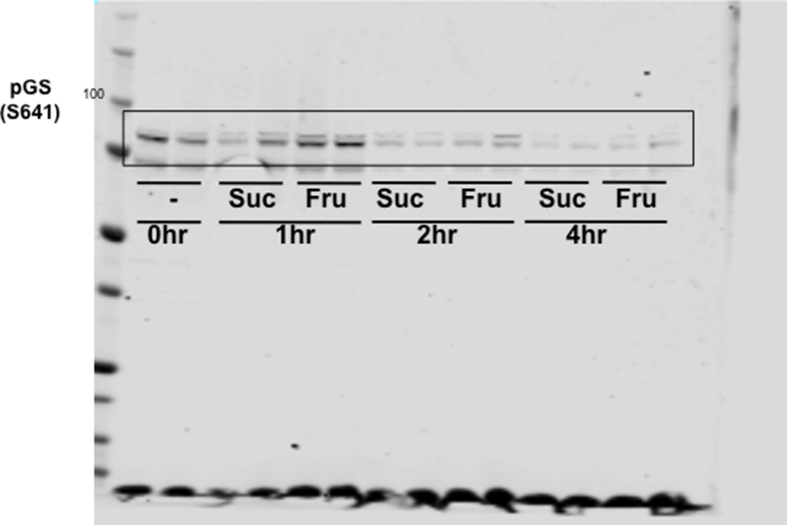
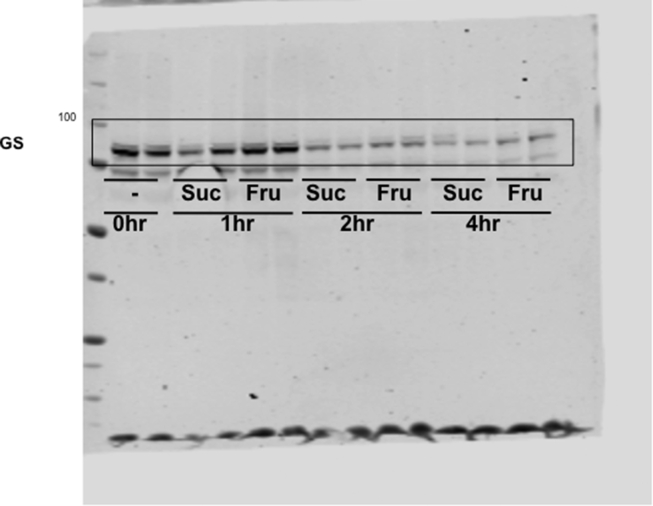
FOXO1 and GS phosphorylation levels under different carbohydrates containing water at different feeding times. Twelve-week-old C57BL/6J mice were fasted overnight and fed with normal chow supplemented with 30% sucrose or fructose in their drinking water. A) Representative Western blotting images of phosphorylated FOXO1 (S256), total FOXO1, phosphorylated GS (S641), and tubulin from two independent mouse livers. B) Relative densitometric analysis of pFOXO1/tubulin and C) pGS/tubulin is presented. P values were determined by one-way ANOVA followed by the Turkey-Kramer multiple comparisons test. Error bars indicate the mean ± SD. Bars with different letters are significantly different from each other (p < 0.05).
Fig. s6.
Fructose induction of endogenous gene expression in primary hepatocytes isolated from the control and MED13-LKO mice. Primary mouse hepatocytes were isolated from the WT (Med13fl/fl) or KO (Med13-LKO) mice. The cells were cultured on collagen-coated plates in DMEM without glucose (−) or with 45 mM of fructose (+) for 5 h. Cellular RNA was isolated and subjected to qRT-PCR for A)Med13, B)G6pc, C)Fasn, D)Chrebp-β, E)Mlx, F)Foxo1, and G)Usf1 mRNA. P values were determined by Student's t-test (p < 0.05). Values are the mean ± SD.
Fig. s7.
Fig. s8.
Fig.s9.
Fig. s10.
Fig. s11.
Fig. s12.
Fig. s13.
Fig. s14.
Fig. s15.
Fig. s16.
Fig. s17.
Fig. s18.
Fig. s19.
Fig. s20.
Fig. s21.
Fig. s22.
Fig. s23.
Fig. s24.
Fig. s25.
Fig. s26.
Fig. s27.
Fig. s28.
Fig. s29.
Fig. s30.
Fig. s31.
Fig. s32.
Fig. s33.
References
- 1.Loomba R., Sanyal A.J. The global NAFLD epidemic. Nature Reviews. Gastroenterology & Hepatology. 2013;10:686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 2.Howard B.V., Wylie-Rosett J. Sugar and cardiovascular disease: a statement for healthcare professionals from the committee on nutrition of the council on nutrition, physical activity, and metabolism of the American heart association. Circulation. 2002;106:523–527. doi: 10.1161/01.cir.0000019552.77778.04. [DOI] [PubMed] [Google Scholar]
- 3.Aller E.E., Abete I., Astrup A., Martinez J.A., van Baak M.A. Starches, sugars and obesity. Nutrients. 2011;3:341–369. doi: 10.3390/nu3030341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iizuka K., Bruick R.K., Liang G., Horton J.D., Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linden A.G., Li S., Choi H.Y., Fang F., Fukasawa M., Uyeda K. Interplay between ChREBP and SREBP-1c coordinates postprandial glycolysis and lipogenesis in livers of mice. The Journal of Lipid Research. 2018;59:475–487. doi: 10.1194/jlr.M081836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Softic S., Gupta M.K., Wang G.X., Fujisaka S., O'Neill B.T., Rao T.N. Divergent effects of glucose and fructose on hepatic lipogenesis and insulin signaling. Journal of Clinical Investigation. 2017;127:4059–4074. doi: 10.1172/JCI94585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim M.S., Krawczyk S.A., Doridot L., Fowler A.J., Wang J.X., Trauger S.A. ChREBP regulates fructose-induced glucose production independently of insulin signaling. Journal of Clinical Investigation. 2016;126:4372–4386. doi: 10.1172/JCI81993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Schaftingen E. A protein from rat liver confers to glucokinase the property of being antagonistically regulated by fructose 6-phosphate and fructose 1-phosphate. European Journal of Biochemistry. 1989;179:179–184. doi: 10.1111/j.1432-1033.1989.tb14538.x. [DOI] [PubMed] [Google Scholar]
- 9.Detheux M., Vandercammen A., Van Schaftingen E. Effectors of the regulatory protein acting on liver glucokinase: a kinetic investigation. European Journal of Biochemistry. 1991;200:553–561. doi: 10.1111/j.1432-1033.1991.tb16218.x. [DOI] [PubMed] [Google Scholar]
- 10.Choi J.M., Seo M.H., Kyeong H.H., Kim E., Kim H.S. Molecular basis for the role of glucokinase regulatory protein as the allosteric switch for glucokinase. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10171–10176. doi: 10.1073/pnas.1300457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang C., Hui S., Lu W., Cowan A.J., Morscher R.J., Lee G. The small intestine converts dietary fructose into glucose and organic acids. Cell Metabolism. 2018;27:351–361 e353. doi: 10.1016/j.cmet.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen T., Abdelmalek M.F., Sullivan S., Nadeau K.J., Green M., Roncal C. Fructose and sugar: a major mediator of non-alcoholic fatty liver disease. Journal of Hepatology. 2018;68:1063–1075. doi: 10.1016/j.jhep.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox T.M. Aldolase B and fructose intolerance. The FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 1994;8:62–71. doi: 10.1096/fasebj.8.1.8299892. [DOI] [PubMed] [Google Scholar]
- 14.Van den Berghe G. Fructose: metabolism and short-term effects on carbohydrate and purine metabolic pathways. Progress in Biochemical Pharmacology. 1986;21:1–32. [PubMed] [Google Scholar]
- 15.Brosh S., Boer P., Sperling O. Effects of fructose on synthesis and degradation of purine nucleotides in isolated rat hepatocytes. Biochimica et Biophysica Acta. 1982;717:459–464. doi: 10.1016/0304-4165(82)90288-4. [DOI] [PubMed] [Google Scholar]
- 16.Mayes P.A. Intermediary metabolism of fructose. American Journal of Clinical Nutrition. 1993;58:754S–765S. doi: 10.1093/ajcn/58.5.754S. [DOI] [PubMed] [Google Scholar]
- 17.Rho Y.H., Zhu Y., Choi H.K. The epidemiology of uric acid and fructose. Seminars in Nephrology. 2011;31:410–419. doi: 10.1016/j.semnephrol.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J., Ji J., Yan X.H. Cross-talk between AMPK and mTOR in regulating energy balance. Critical Reviews in Food Science and Nutrition. 2012;52:373–381. doi: 10.1080/10408398.2010.500245. [DOI] [PubMed] [Google Scholar]
- 19.Kawaguchi T., Takenoshita M., Kabashima T., Uyeda K. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13710–13715. doi: 10.1073/pnas.231370798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L., Robinson L.N., Towle H.C. ChREBP∗Mlx is the principal mediator of glucose-induced gene expression in the liver. Journal of Biological Chemistry. 2006;281:28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 21.Petrie J.L., Al-Oanzi Z.H., Arden C., Tudhope S.J., Mann J., Kieswich J. Glucose induces protein targeting to glycogen in hepatocytes by fructose 2,6-bisphosphate-mediated recruitment of MondoA to the promoter. Molecular and Cellular Biology. 2013;33:725–738. doi: 10.1128/MCB.01576-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson C.M., Koleske A.J., Chao D.M., Young R.A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y.J., Bjorklund S., Li Y., Sayre M.H., Kornberg R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 24.Dotson M.R., Yuan C.X., Roeder R.G., Myers L.C., Gustafsson C.M., Jiang Y.W. Structural organization of yeast and mammalian mediator complexes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., Ranish J.A., Aebersold R., Hahn S. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. Journal of Biological Chemistry. 2001;276:7169–7175. [PubMed] [Google Scholar]
- 26.Malik S., Baek H.J., Wu W., Roeder R.G. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Molecular and Cellular Biology. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Youn D.Y., Xiaoli A.M., Kwon H., Yang F., Pessin J.E. The subunit assembly state of the Mediator complex is nutrient-regulated and is dysregulated in a genetic model of insulin resistance and obesity. Journal of Biological Chemistry. 2019;294:9076–9083. doi: 10.1074/jbc.RA119.007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemet J., Jelicic B., Rubelj I., Sopta M. The two faces of Cdk8, a positive/negative regulator of transcription. Biochimie. 2014;97:22–27. doi: 10.1016/j.biochi.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Mo X., Kowenz-Leutz E., Xu H., Leutz A. Ras induces mediator complex exchange on C/EBP beta. Molecular Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y., Kung C., Fishburn J., Ansari A.Z., Shokat K.M., Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Molecular and Cellular Biology. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y., Tarabra E., Yang G.S., Vaitheesvaran B., Palacios G., Kurland I.J. Alteration of de novo glucose production contributes to fasting hypoglycaemia in Fyn deficient mice. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shemesh A., Wang Y., Yang Y., Yang G.S., Johnson D.E., Backer J.M. Suppression of mTORC1 activation in acid-alpha-glucosidase-deficient cells and mice is ameliorated by leucine supplementation. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2014;307:R1251–R1259. doi: 10.1152/ajpregu.00212.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng D., Youn D.Y., Zhao X., Gao Y., Quinn W.J., Xiaoli A.M. mTORC1 down-regulates cyclin-dependent kinase 8 (CDK8) and cyclin C (CycC) PLoS One. 2015;10 doi: 10.1371/journal.pone.0126240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagata T., Redman R.S., Lakshman R. Isolation of intact nuclei of high purity from mouse liver. Analytical Biochemistry. 2010;398:178–184. doi: 10.1016/j.ab.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Abdulla A., Yi Z., Hsu F.N., Xiaoli A.M., Zhao X., Yang E.S. Regulation of lipogenic gene expression by lysine-specific histone demethylase-1 (LSD1) Journal of Biological Chemistry. 2014;289:29937–29947. doi: 10.1074/jbc.M114.573659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 38.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiaoli A.M., Song Z., Yang F. Lipogenic SREBP-1a/c transcription factors activate expression of the iron regulator hepcidin, revealing cross-talk between lipid and iron metabolisms. Journal of Biological Chemistry. 2019;294:12743–12753. doi: 10.1074/jbc.RA119.009644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knuesel M.T., Meyer K.D., Donner A.J., Espinosa J.M., Taatjes D.J. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Molecular and Cellular Biology. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersen M.C., Vatner D.F., Shulman G.I. Regulation of hepatic glucose metabolism in health and disease. Nature Reviews. Endocrinology. 2017;13:572–587. doi: 10.1038/nrendo.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharabi K., Tavares C.D., Rines A.K., Puigserver P. Molecular pathophysiology of hepatic glucose production. Molecular Aspects of Medicine. 2015;46:21–33. doi: 10.1016/j.mam.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ter Horst K.W., Serlie M.J. Fructose consumption, lipogenesis, and non-alcoholic fatty liver disease. Nutrients. 2017;9 doi: 10.3390/nu9090981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hannou S.A., Haslam D.E., McKeown N.M., Herman M.A. Fructose metabolism and metabolic disease. Journal of Clinical Investigation. 2018;128:545–555. doi: 10.1172/JCI96702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Der Heide L.P., Hoekman M.F., Smidt M.P. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochemical Journal. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poss Z.C., Ebmeier C.C., Taatjes D.J. The Mediator complex and transcription regulation. Critical Reviews in Biochemistry and Molecular Biology. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kornberg R.D. Mediator and the mechanism of transcriptional activation. Trends in Biochemical Sciences. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 48.Firestein R., Bass A.J., Kim S.Y., Dunn I.F., Silver S.J., Guney I. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim M.Y., Han S.I., Lim S.C. Roles of cyclin-dependent kinase 8 and beta-catenin in the oncogenesis and progression of gastric adenocarcinoma. International Journal of Oncology. 2011;38:1375–1383. doi: 10.3892/ijo.2011.948. [DOI] [PubMed] [Google Scholar]
- 50.Gu W., Wang C., Li W., Hsu F.N., Tian L., Zhou J. Tumor-suppressive effects of CDK8 in endometrial cancer cells. Cell Cycle. 2013;12:987–999. doi: 10.4161/cc.24003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K., Yan R., Cooper K.F., Strich R. Cyclin C mediates stress-induced mitochondrial fission and apoptosis. Molecular Biology of the Cell. 2015;26:1030–1043. doi: 10.1091/mbc.E14-08-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao X., Feng D., Wang Q., Abdulla A., Xie X.J., Zhou J. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. Journal of Clinical Investigation. 2012;122:2417–2427. doi: 10.1172/JCI61462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baskin K.K., Grueter C.E., Kusminski C.M., Holland W.L., Bookout A.L., Satapati S. MED13-dependent signaling from the heart confers leanness by enhancing metabolism in adipose tissue and liver. EMBO Molecular Medicine. 2014;6:1610–1621. doi: 10.15252/emmm.201404218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amoasii L., Holland W., Sanchez-Ortiz E., K Baskin K., Pearson M., Burgess S.C. A MED13-dependent skeletal muscle gene program controls systemic glucose homeostasis and hepatic metabolism. Genes & Development. 2016;30:434–446. doi: 10.1101/gad.273128.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chi Y., Youn D.Y., Xiaoli A.M., Liu L., Pessin J.B., Yang F. Regulation of gene expression during the fasting-feeding cycle of the liver displays mouse strain specificity. Journal of Biological Chemistry. 2020;295:4809–4821. doi: 10.1074/jbc.RA119.012349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basciano H., Federico L., Adeli K. Fructose, insulin resistance, and metabolic dyslipidemia. Nutrition and Metabolism. 2005;2:5. doi: 10.1186/1743-7075-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kyriazis G.A., Soundarapandian M.M., Tyrberg B. Sweet taste receptor signaling in beta cells mediates fructose-induced potentiation of glucose-stimulated insulin secretion. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E524–E532. doi: 10.1073/pnas.1115183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grant A.M., Christie M.R., Ashcroft S.J. Insulin release from human pancreatic islets in vitro. Diabetologia. 1980;19:114–117. doi: 10.1007/BF00421856. [DOI] [PubMed] [Google Scholar]
- 59.Curry D.L. Effects of mannose and fructose on the synthesis and secretion of insulin. Pancreas. 1989;4:2–9. doi: 10.1097/00006676-198902000-00002. [DOI] [PubMed] [Google Scholar]
- 60.Elliott S.S., Keim N.L., Stern J.S., Teff K., Havel P.J. Fructose, weight gain, and the insulin resistance syndrome. American Journal of Clinical Nutrition. 2002;76:911–922. doi: 10.1093/ajcn/76.5.911. [DOI] [PubMed] [Google Scholar]
- 61.Cui X.L., Soteropoulos P., Tolias P., Ferraris R.P. Fructose-responsive genes in the small intestine of neonatal rats. Physiological Genomics. 2004;18:206–217. doi: 10.1152/physiolgenomics.00056.2004. [DOI] [PubMed] [Google Scholar]
- 62.Abdul-Wahed A., Guilmeau S., Postic C. Sweet sixteenth for ChREBP: established roles and future goals. Cell Metabolism. 2017;26:324–341. doi: 10.1016/j.cmet.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 63.Sato T., Watanabe Y., Nishimura Y., Inoue M., Morita A., Miura S. Acute fructose intake suppresses fasting-induced hepatic gluconeogenesis through the AKT-FoxO1 pathway. Biochemistry and Biophysics Report. 2019;18:100638. doi: 10.1016/j.bbrep.2019.100638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conlee R.K., Lawler R.M., Ross P.E. Effects of glucose or fructose feeding on glycogen repletion in muscle and liver after exercise or fasting. Annals of Nutrition and Metabolism. 1987;31:126–132. doi: 10.1159/000177259. [DOI] [PubMed] [Google Scholar]
- 65.MacAulay K., Doble B.W., Patel S., Hansotia T., Sinclair E.M., Drucker D.J. Glycogen synthase kinase 3alpha-specific regulation of murine hepatic glycogen metabolism. Cell Metabolism. 2007;6(4):329–337. doi: 10.1016/j.cmet.2007.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of mouse oligonucleotide primer sequences for qRT-PCR.