Abstract
Colorectal cancer is a leading cause of cancer deaths. Most colorectal cancer patients eventually develop chemoresistance to the current standard-of-care therapies. Here, we used patient-derived colorectal cancer organoids to demonstrate that resistant tumor cells undergo significant chromatin changes in response to oxaliplatin treatment. Integrated transcriptomic and chromatin accessibility analyses using ATAC-Seq and RNA-Seq identified a group of genes associated with significantly increased chromatin accessibility and upregulated gene expression. CRISPR/Cas9 silencing of fibroblast growth factor receptor 1 (FGFR1) and oxytocin receptor (OXTR) helped overcome oxaliplatin resistance. Similarly, treatment with oxaliplatin in combination with an FGFR1 inhibitor (PD166866) or an antagonist of OXTR (L-368,899) suppressed chemoresistant organoids. However, oxaliplatin treatment did not activate either FGFR1 or OXTR expression in another resistant organoid, suggesting that chromatin accessibility changes are patient-specific. The use of patient-derived cancer organoids in combination with transcriptomic and chromatin profiling may lead to precision treatments to overcome chemoresistance in colorectal cancer.
Keywords: Chromatin accessibility, Drug screening, Patient-derived organoids, Personalized medicine, Target discovery
Introduction
Colorectal cancer (CRC) is the third most common cancer type in both men and women in the United States. It is estimated that more than 145,600 new cases of CRC were diagnosed and approximately 51,020 deaths occurred in 2019.1,2 In the US, 140,250 people were diagnosed with CRC, and 50,630 patients died from CRC in 2018.2 The 5-year survival rate of CRC patients drops from 90% in the early stage to less than 15% in the late stages.3 Although surgical resection for curative intent has improved over the past decade, the 5-year survival rate has not significantly increased in part due to the fact that most patients are diagnosed with late-stage disease.4,5 Additionally, many CRC patients will develop metastases or chemotherapy resistance in advanced CRC.5, 6, 7 The median overall survival of CRC patients treated with oxaliplatin, one of the standard drugs for the treatment of advanced CRC cases, is less than 1 year mainly due to drug resistance7, 8, 9 Therefore, it is pivotal to discover effective therapeutic treatments to circumvent drug resistance for CRC patients.
Patient-derived cell lines and patient-derived xenograft (PDX) models have been used for drug screening, although each has its limitations. Long-passaged cell lines often lose some of their original properties while PDX models are expensive and time-consuming to develop.10 Recently, patient-derived organoids (PDOs) have emerged as models for diseases and personalized drug testing.11, 12, 13, 14, 15 PDOs recapitulate many properties of the primary tumor, including the patient's unique genetic background and intrinsic tumor heterogeneity, and exhibit drug responses that correlate well with patient outcomes.16, 17, 18, 19
Epigenetic alterations, including histone modifications and DNA methylation, have been shown to contribute to CRC chemoresistance.20 For instance, the expression of thymidylate synthetase can be epigenetically elevated to promote CRC resistance to 5-FU, and silencing the epigenetically-mediated upregulation of thymidylate synthetase with a HDAC inhibitor reverses the resistance.21, 22, 23, 24, 25 Additionally, UGT1A1 silencing by DNA methylation (which occurs in 82% of primary CRCs) and ABC transporter gene silencing by histone deacetylation affect the pharmacokinetic profile of irinotecan, a first-line treatment for colorectal cancer.26, 27, 28 Further, hyper-methylation has been shown to contribute to cisplatin resistance.29
In this study, we developed metastatic patient-derived CRC organoids for personalized drug testing. These PDOs were found to have different sensitivities to frontline CRC drugs. Integrated chromatin accessibility and transcriptomic profiling using ATAC-Seq and RNA-Seq identified genes associated with treatment-induced chromatin alterations, particularly in more resistant organoids. Notably, we identified fibroblast growth factor 1 (FGFR1)30 and oxytocin receptor (OXTR)31 as potential therapeutic targets. Silencing of FGFR1 or OXTR by CRISPR/Cas9 or small molecule inhibitors synergized with oxaliplatin to overcome resistance to oxaliplatin. However, FGFR1 or OXTR upregulation was not consistent among patient organoids, suggesting that drug-resistant pathways may be personalized.
Materials and methods
Patient-derived organoid culture
Tumor samples from metastatic CRC patients were collected under a Duke IRB approved protocol (Pro00002435) at Duke University Hospital. All participants provided written informed consent to participate in the study. Tumor samples were chopped into 5 mm pieces and washed with PBS several times. The tumor fragments were incubated in digestion buffer (Dulbecco's modified Eagle medium with 2.5% fetal bovine serum, penicillin/streptomycin [Invitrogen], 75 U/mL collagenase type IX [Sigma], 125 μg/mL dispase type II [Invitrogen]) for 60 min at 37 °C. The supernatant was collected in a 50 mL Falcon tube, centrifuged at 1000RPM for 5 min, and then washed with PBS repeatedly. Isolated cancer cells were counted using a hemocytometer. Single cells were embedded in ice cold Matrigel (Corning Life Sciences) and seeded in 24-well plates. Matrigel was polymerized for 10 min at 37 °C. Basal culture medium was supplemented with a combination of growth factors as previously described.13
Drug sensitivity assays
CRC240, CRC159, CRC344, and CRC119 organoids were enzymatically dissociated using Accumax (Sigma), passed through a 40 μm cell strainer (Falcon), and seeded into 96-well plates pre-coated with Matrigel (Corning Life Science) at densities between 500 and 1000 organoids/well with conditioned media. Three replicates were used for each drug concentration. After 24 h of incubation at 37 °C, organoids were treated for 6 days at different drug concentrations to determine the IC50 values. Drug responses were determined by measuring ATP levels using CellTiter-Glo 3D Luminescent Cell Viability Assay (Promega, USA) on day 7, and IC50 values were calculated for each cell line using a nonlinear regression model in GraphPad Prism software (La Jolla, CA, USA).
CRISPR/Cas9 genomic editing
All-in-one CRISPR/Cas9-gRNA plasmids (pLentiCRISPR-v2) were purchased from GenScript. Plasmids were extracted using Qiagen Plasmid Maxi Kit. HEK293T cells were transfected with the plasmids to package lentiviruses using TransIT®- LT1 Transfection Reagent (Mirus Bio) according to the manufacturer's instructions. The collected lentiviruses were used to infect organoid cultures to silence genes of interest. Puromycin (2 μg/mL, Thermo Fisher Scientific) was added to the cell culture medium for selection.
Quantitative RT-PCR
Total RNA was extracted using the RNeasy Kit (Qiagen) according to the manual. cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen). PCR reactions were prepared using the QuantiFast SYBR Green PCR Kit (Qiagen). RT-qPCR was performed using the Applied Biosystems StepOnePlus™ Real-Time PCR System in a two-step cycling protocol, with a denaturation step at 95 °C and a combined annealing/extension step at 60 °C. RT-qPCR measurements represent the average of three independent experiments normalized to GAPDH expression. The primers listed in Table 2 were purchased from Integrated DNA Technologies.
Table 2.
List of primers used in RT-qPCR.
| FGFR1-F | AACCTGACCACAGAATTGGAGGCT |
|---|---|
| FGFR1-R | ATGCTGCCGTACTCATTCTCCACA |
| FGFR2-F | TGATGGACTTCCTTATGTCCGCGT |
| FGFR2-R | AGCGTCCTCTTCTGTGACATTGGT |
| FGFR3-F | ACCAATGTGTCTTTCGAGGATGCG |
| FGFR3-R | AGAGCACGCAGCTTGTCACATAGA |
| FGFR4-F | ATGGAACTGGTGTGCTCAAGAAGC |
| FGFR4-R | TTCACATGTCCTCCGACCAACACA |
| OXTR-F | CCTTCATCGTGTGCTGGACG |
| OXTR-R | CTAGGAGCAGAGCACTTATG |
| GAPDH-F | TCGACAGTCAGCCGCATCTTCTTT |
| GAPDH-R | GCCCAATACGACCAAATCCGTTGA |
Western blotting
Cells were lysed in Radioimmunoprecipitation assay buffer (Thermo Fisher Scientific) with protease and phosphatase inhibitor cocktail (Roche). Cell lysate was subjected to a standard Bio-Rad western blotting workflow using Mini-PROTEAN® TGX Stain-Free™ Precast Gels and Trans-Blot® Turbo™ Transfer System. The following primary antibodies and dilutions were used: FGFR1 (#9740), FGFR2 (#11835), pFGFR (#3471), β-Tubulin (#2128) antibodies (Cell Signaling Technology), OXTR (Abcam) and FGFR3 (sc-390423), FGFR4 (sc-136988) (Santa Cruz Biotechnology). All antibodies were used at the 1:1000 ratio. Protein bands were processed using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) or Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Science) followed by visualization in a ChemiDoc™ Touch Imaging System (Bio-Rad). Images were edited in Image Lab™ Software (Bio-Rad).
RNA-Seq and ATAC-Seq analysis
Organoids were dissociated into single cells using Accumax (Sigma). 50,000 viable cells were collected for ATAC-Seq preparation as described previously.32 RNA-Seq was performed on dissociated organoids samples. RNA-Seq libraries were generated using the Kapa Stranded RNA-Seq kit. Triplicates of samples were collected for sequencing, and sequencing experiments were performed at the Duke Center for Genomic and Computational Biology sequencing core facility. Sequence files of RNA-Seq were aligned to human genome hg19 using Hisat2.33 Sequence files of ATAC-Seq were aligned to human genome hg19 using bowtie2,34 and MACS2 was utilized to call open chromatin peaks.35 DESeq236 and DiffBind (https://doi.org/10.18129/B9.bioc.DiffBind)37 were used for differential analysis of RNA-Seq and ATAC-Seq, respectively. The open chromatin peaks and differential peaks from ATAC-Seq were annotated to nearby genes using Homer.38 Triplicates of samples were merged for plotting heatmaps of chromatin accessibility and reads coverage on identified peak regions by Deeptools.39
Integrative analysis of ATAC-Seq and RNA-Seq were performed in R. Gene Set Enrichment Analysis (GSEA) was performed using the GSEA tool (http://software.broadinstitute.org/gsea/index.jsp) developed by Broad Institute.40 The cancer modules curated by Sagel et al41 were applied to the discovery of cancer-associated genes. The Drug Gene Interaction Database (DGIdb) was used to identify druggable gene targets.42
Predictions of transcriptional factor binding site
Putative binding transcriptional factors (TFs) were predicted using PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)43 on human genome, and only TFs with dissimilarity lower than 1% were reported. The associated binding motifs of TFs were collected from MotifMap (http://motifmap.ics.uci.edu)44 using human genome hg19.
Statistical analysis
The displayed data are presented as mean ± SEM. Statistical comparisons were made using unpaired two-tailed Student's t-test and one-way ANOVA with Tukey's post hoc test in GraphPad Prism to calculate significance. Differences were considered significant at P < 0.05.
Results
Drug responses in patient-derived CRC organoids
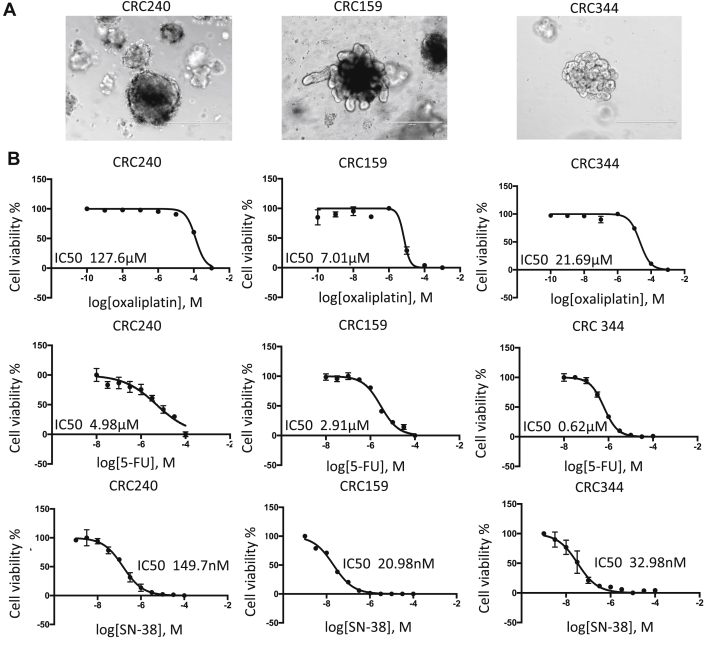
To generate PDOs of CRC, CRC samples were obtained from patients undergoing resection of their metastatic CRC at Duke University under an IRB-approved protocol. CRC119, CRC159, and CRC240 were derived from CRC metastasizing to the liver, and CRC344 was derived from CRC metastasizing to the omentum. Patient demographics are described in Table 1. Tissues were dissociated and subsequently cultured as tumor organoids according to an established protocol.13 The CRC organoids derived from three different patients (CRC240, CRC159, CRC344) varied in morphology under the same culture conditions (Fig. 1A). Organoids were enzymatically dissociated and seeded into 96-well plates at a density of 500–1000 organoids per well. After 24 h, organoids were treated with three standard chemotherapy drugs: 5-fluorouracil (5-FU), oxaliplatin, and SN-38 (the active metabolite of irinotecan). The drugs were applied over a logarithmic range of concentrations to measure the IC50 values, which are shown in Fig. 1B. The IC50 values of oxaliplatin were 127.6 μM, 7.01 μM, and 21.69 μM in CRC240, CRC159, and CRC344, respectively, suggesting that CRC240 organoids are particularly resistant to oxaliplatin (Fig. S1A). In comparison, the IC50 values of 5-FU were 4.98 μM, 2.91 μM, and 0.62 μM, and the IC50 values of SN38 were 149.7 nM, 20.98 nM, and 32.98 nM in respective organoids.
Table 1.
Clinical characteristics of the four colorectal cancer patients.
| ID | Gender | Histology | Grade | Microsatellite Status | KRAS | BRAF | Primary | Metastatic Site |
|---|---|---|---|---|---|---|---|---|
| CRC344 | M | Adenocarcinoma | poorly differentiated | MSS | mutated | WT | colon | omentum |
| CRC240 | F | Adenocarcinoma | poorly differentiated | MSS | WT | WT | colon | liver |
| CRC159 | F | Adenocarcinoma | moderately differentiated | MSI | WT | mutated | colon | liver |
| CRC119 | F | Adenocarcinoma | moderately differentiated | MSS | mutated | WT | colon | liver |
Figure 1.
Chemo-sensitivity of CRC 240, CRC344, and CRC159 PDOs. (A) Bright-field images of colorectal cancer organoids. From left to right: CRC240, CRC159, and CRC344. Scale bar = 400 μm. (B) Drug sensitivities to oxaliplatin, 5-FU, and SN-38 were assessed in CRC240, CRC159, and CRC344 organoids. Organoids were exposed to chemotherapy for 6 days, and cell viability was assessed by CellTiter-Glo 3D cell viability assay. The IC50 values were calculated by a nonlinear regression model in GraphPad Prism. Error bars represents the standard error of the mean.
Chromatin and transcriptional profiling
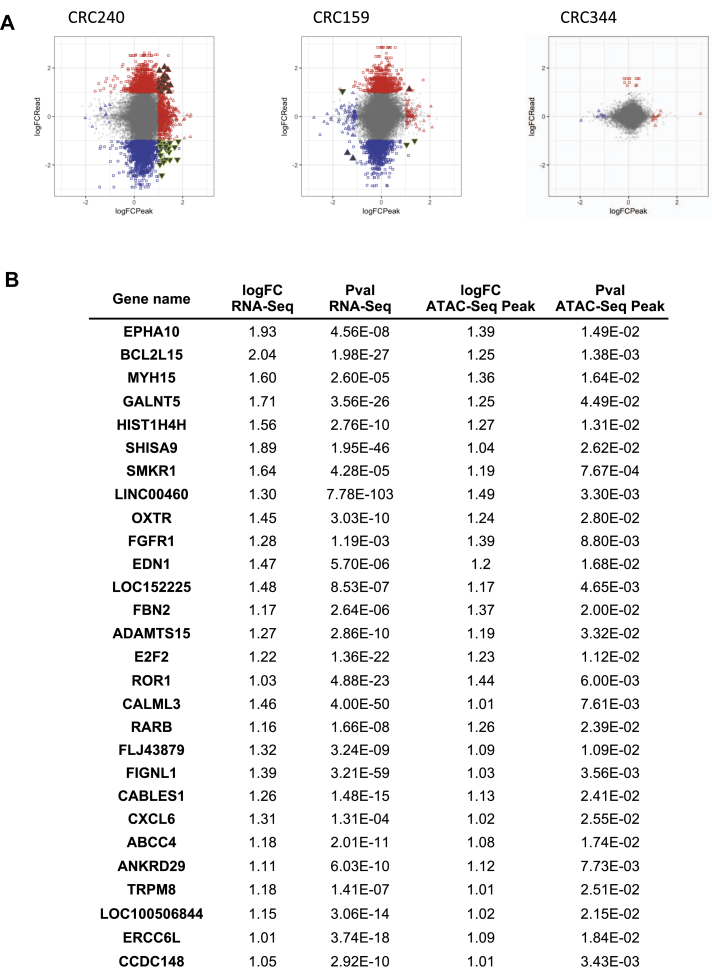
We next used ATAC-Seq and RNA-Seq to profile the chromatin accessibility and transcriptome of CRC organoids comparing 10-day oxaliplatin vs. DMSO (control) treatment (Fig. S2A). In CRC 240 organoids, which were the most resistant to oxaliplatin, 1493 genes were differentially expressed after 10 days of oxaliplatin treatment according to RNA-Seq (Fig. S2B). According to ATAC-Seq (Fig. S2C), 893 chromatin accessibility peaks were significantly altered compared to the DMSO control (Fig. S2D). In comparison, fewer genes and ATAC-Seq peaks were altered by oxaliplatin treatment in CRC159 and CRC344 organoids (Fig. S2B-D).
As CRC240 organoids were most resistant to oxaliplatin and displayed more alterations in chromatin accessibility and gene expression than the other organoids, we further integrated the differential analyses of ATAC-Seq and RNA-Seq to identify genes associated with both chromatin accessibility and gene expression changes in CRC240 (Fig. 2A, filled triangles). Twenty-eight genes experienced consistent changes in chromatin opening and upregulation of expression in response to oxaliplatin treatment (Fig. 2A, filled up-pointing red triangles, and Fig. 2B). Hence, the chromatin accessibility changes for these genes may play a role in CRC240 resistance to oxaliplatin.
Figure 2.
Transcriptomic and chromatin accessibility profiling of oxaliplatin-treated organoids. (A) Integration of ATAC-Seq and RNA-Seq. The differential analyses of ATAC-Seq and RNA-Seq were performed by using DESeq2 and DiffBind respectively. The differential genes and peaks were filtered by P-values (p-value < 0.05). Red color represents increase changes (logFC >1) for expression (square) or chromatin accessibility (triangle). Blue color represents decreased changes (logFC < −1) for expression (square) or chromatin accessibility (triangle). The filled triangles represent both ATAC-Seq peaks and RNA-Seq expression significantly altered (P < 0.05). (B) Top ranked genes that display both increased chromatin accessibility nearby and increased gene expression in oxaliplatin-resistant CRC240.
Upregulation of drug targetable genes FGFR1 and OXTR
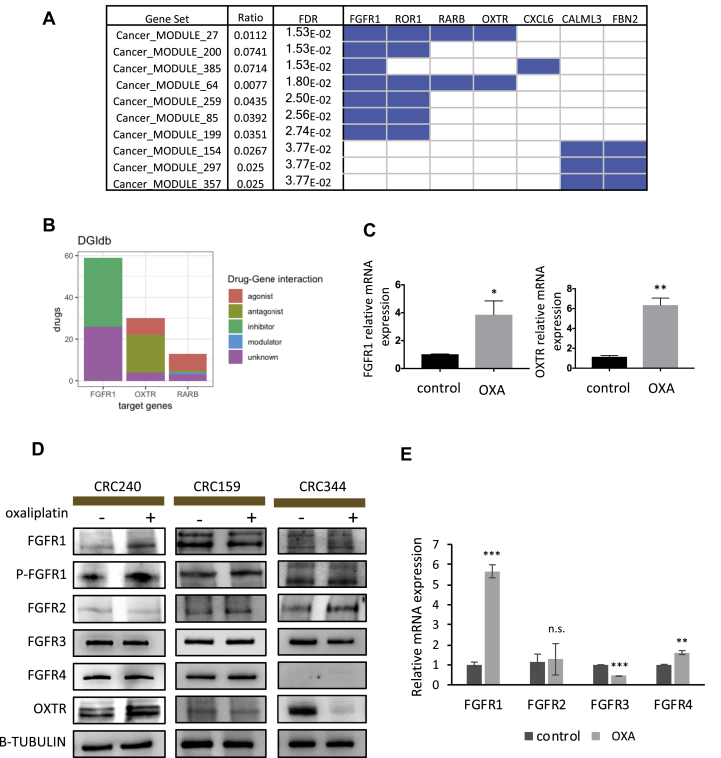
To identify potential therapeutic targets to overcome oxaliplatin resistance from the list of 28 genes, we first performed Gene Set Enrichment Analysis (GSEA) based on cancer modules curated by the Broad institute (Fig. 3A). FGFR1, ROR1, RARB, OXTR, and CXCL6 are the top five genes enriched in the cancer modules, and only FGFR1, RARB, and OXTR are known targets of FDA-approved drugs (Fig. 3B). While RARB has been extensively discussed in terms of its epigenetic roles in CRCs,45, 46, 47, 48 FGFR1 and OXTR are relatively new to CRC treatment.
Figure 3.
Confirmation of drug-associated genes FGFR1 and OXTR in CRC organoids. (A) Gene set enrichment analysis of top genes identified by integrated analysis in Fig. 2B. Cancer modules curated by the Broad institute were applied for the enrichment analysis. Blue color in module-gene matrix indicates the significantly enriched cancer modules and the associate gene hits from the top gene list. (B) Bar diagram of drug-gene interactions. The known drugs targeting the identified cancer associated genes are classified into groups annotated by DGIdb based on the targeting mechanisms. Among these drugs, 33 drugs are inhibitors targeting FGFR1, 18 drugs are antagonists targeting OXTR, and one antagonist targets RARB. (C) Left: RT-qPCR showed mRNA expression of FGFR1 in CRC240 control (DMSO) organoids compared to oxaliplatin treatment. Right: RT-qPCR showed mRNA expression of OXTR in CRC240 control (DMSO) organoids compared to oxaliplatin treatment. Organoids were exposed to the IC50 concentration of oxaliplatin, and RNA was isolated after incubation. Expression levels are given relative to the housekeeping gene GAPDH. Data were mean ± SEM (n = 3) and the statistical significance was assessed by unpaired two-tailed student's t-test. *P < 0.05, **P < 0.01. (D) Protein expression was assessed by Western blot using antibodies to FGFR1, phospho-FGFR1, FGFR2, FGFR3, FGFR4, OXTR, and beta-tubulin in CRC240, CRC159, and CRC344 organoids with DMSO or oxaliplatin treatment. Immunoblots between 90KD to 120KD are different isoforms and glycosylated fibroblast growth factor receptors. (E) RT-qPCR showed mRNA expression of FGFR1, FGFR2, FGFR3, and FGFR4 in CRC240 control (DMSO) organoids compared to oxaliplatin treatment. Organoids were exposed to the IC50 concentration of oxaliplatin, and RNA was isolated after incubation. Expression levels are given relative to the housekeeping gene GAPDH. Data were mean ± SEM (n = 3) and the statistical significance was assessed by unpaired two-tailed student's t-test. *P < 0.05; **P < 0.01; ***P < 0.01; n.s., not significant.
We analyzed mRNA levels by RT-qPCR and protein expression by Western blot to validate FGFR1 and OXTR expression in PDOs. Consistent with RNA-Seq, RT-qPCR showed that oxaliplatin treatments increase FGFR1 and OXTR mRNA expression in CRC240 organoids but not in CRC159 and CRC344 (Fig. 3C and Fig. S3A). According to Western blot, among the three patient-derived organoids, only CRC240 showed elevated FGFR1 and OXTR protein levels after oxaliplatin treatment. In contrast, CRC344 displayed a decrease in OXTR protein expression level after oxaliplatin treatment (Fig. 3D). Among the four FGFR family members (FGFR1-4), FGFR1 is the only receptor that showed elevated mRNA and protein levels in CRC240 in response to oxaliplatin treatment (Fig. 3D and E). Phosphorylation of FGFR1 also increased in CRC240, indicating more active FGFR1 signaling in response to oxaliplatin treatment (Fig. 3D). Combing the validation results from RT-qPCR and western blots, these also confirm the alternations of FGFR1 and OXTR in response to oxaliplatin from the integrative analysis of RNA-Seq and ATAC-Seq (Fig. 2B, Fig. S3B and C).
In order to validate these findings, a patient-derived CRC organoid CRC119 was derived from another oxaliplatin-resistant patient (Fig. S3D) to investigate whether oxaliplatin-induced FGFR1 and OXTR upregulation is specific to CRC240. The IC50 of oxaliplatin in CRC119 was 104 μM (Fig. S3E), similar to that in CRC240 (127.6 μM) and higher than the IC50s of CRC159 and CRC344 (Fig. 1B). However, levels of FGFR1-4 and phosphor-FGFR1 in CRC119 did not change significantly in response to oxaliplatin treatment, while the expression of OXTR slightly decreased (Fig. S3F). The differences between CRC240 and CRC119 suggest that the response and resistance to oxaliplatin may occur through patient-specific mechanisms.
Inhibition of FGFR1/OXTR enhances the effect of oxaliplatin
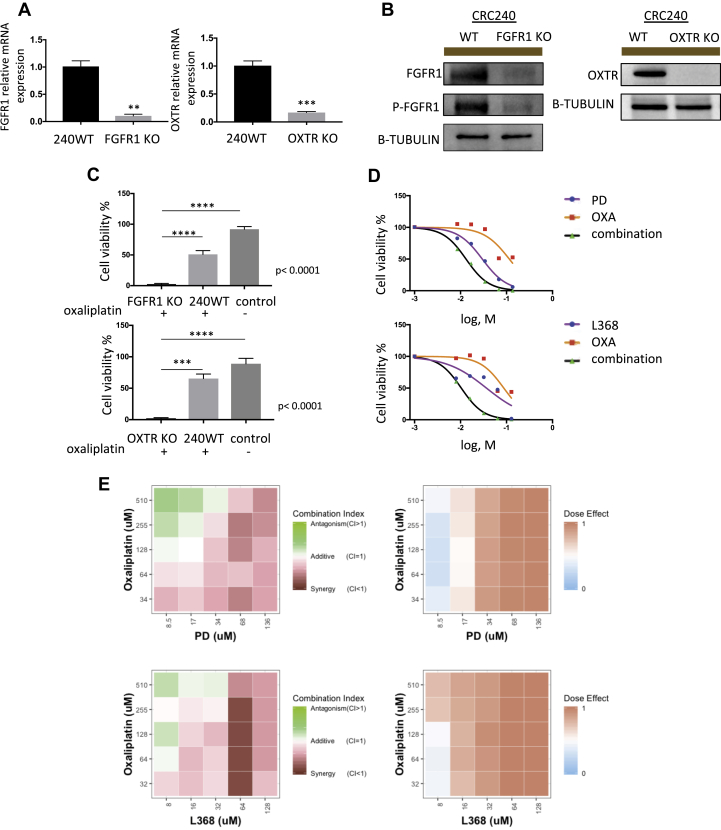
We examined whether targeting FGFR1/OXTR could sensitize CRC240 to oxaliplatin. We first used CRISPR-Cas9 to silence FGFR1 and OXTR separately in PDOs. Single organoids with either FGFR1 or OXTR knockout were clonally expanded after Puromycin selection. mRNA and protein levels of FGFR1 and OXTR were reduced significantly in these knockout organoids (Fig. 4A and B). With oxaliplatin treatment, FGFR1 or OXTR knockout organoids exhibited significantly reduced proliferation rates compared with wild-type CRC240 organoids (Fig. 4C). Therefore, genetic silencing of FGFR1 or OXTR seemed to synergize with oxaliplatin in resistant CRC240 organoids.
Figure 4.
Inhibition of fibroblast growth factor 1 (FGFR1) and oxytocin receptor (OXTR) reduce tumor growth. (A) RT-qPCR measurement validated the efficiency of FGFR1 and OXTR knockout by CRISPR-Cas9 editing in CRC240 organoids. Left: RT-qPCR measurement of FGFR1 mRNA levels in either wild-type or FGFR1 knockout CRC240 organoids. Right: RT-qPCR measurement of OXTR mRNA levels in either wild-type or OXTR knockout CRC240 organoids. Expression was normalized to GAPDH. Data represent mean ± SEM (n = 3), and the statistical significance was assessed by unpaired two-tailed student's t-test. **P < 0.01, ***P < 0.001. (B) Western blot analysis of knockout efficiency of either FGFR1 or OXTR in CRC240 organoids. Beta-tubulin was used as an internal control. (C) Cell viability of either FGFR1 (top) or OXTR (bottom) knockout organoids after oxaliplatin treatment. FGFR1 knockout or OXTR knockout CRC240 organoids were treated with IC50 of oxaliplatin, and wild-type CRC240 organoids were treated with same IC50 for comparison. Cell viability of wild-type CRC240 organoids without oxaliplatin treatment were measured as control. Data represent mean ± SEM (n = 3). One-way ANOVA with Tukey's post hoc test was performed. ***P < 0.001, ****P < 0.0001. (D) Dose-response curves of CRC240 organoids treated with monotherapy or a combination therapy. Top: Combination treatment with oxaliplatin (OXA) and PD166866 (PD). Bottom: Combination treatment with oxaliplatin (OXA) and L-368,899 (L368). Organoids were treated with a series of six different drug doses of oxaliplatin (OXA), PD166866 (PD), and L-368,899 (L368) or a combination of both agents for 6 days. Then, cell viability was measured via CellTiter-Glo 3D cell viability assay (Promega). Purple (PD or L368) line, orange (OXA) line, and black (PD/OXA or L368/OXA combination therapy) line represent the dose-response curves. (E) Combination index of combination treatments. Top: Combination treatment of oxaliplatin (OXA) and PD166866 (PD). Bottom: Combination treatment with oxaliplatin (OXA) and L-368,899 (L368). Left: 5 × 5 dose matrix of combination index. Right: 5 × 5 dose matrix of dose effect. CRC240 organoids were treated with increasing concentrations of oxaliplatin, and PD166866, or L-368,899 or co for 6 days in conditional medium. Combination index (CI) was calculated using CompuSyn software. Additive area was selected by CI between 0.9 and 1.1. CI ≥ 1.1 indicates antagonism; <0.9 indicates synergism. Red and green color indicate synergism and antagonism in CI matrix. Dose effect represents fraction of cells killed by drug treatment. Dark red indicates 100% killing, while light blue indicates 0% killing.
We subsequently targeted FGFR1 and OXTR pharmacologically. We first measured the IC50s of the FGFR1-specific inhibitor PD166866 (PD) and non-peptide oxytocin receptor antagonist L368,899 (L368) (Fig. S4A). We then treated organoids with oxaliplatin in combination with PD or L368. Dose-response curves indicated that CRC240 organoids were more sensitive to combination therapy than monotherapy (Fig. 4D), which was not observed in CRC159, CRC344, or CRC119 organoids (Fig. S4B). A 5 × 5 combination dose-response screen of PD and L368 with oxaliplatin was performed to characterize the effects of combination treatments. Combination index heat maps demonstrated synergism between PD/L368 and oxaliplatin that at the majority of doses tested in CRC240 organoids (Fig. 4E, Fig. S4C).
Discussion
Emerging evidence suggests that human cancer organoids provide a versatile pre-clinical platform by maintaining patient-specific molecular and histopathologic phenotypes.16, 17, 18, 19,49, 50, 51, 52 In this study, patient-derived CRC organoids were used to test sensitivity to frontline CRC chemotherapy drugs. Integrated chromatin and transcriptomic profiling of CRC organoids identified altered chromatin regions and gene expression associated with the response to chemotherapy in resistant tumor cells. Among them, FGFR1 and OXTR were computationally predicted as druggable targets associated with the oxaliplatin-resistant CRC240 organoids. Pharmacological inhibition and genetic silencing of FGFR1 or OXTR synergized with oxaliplatin treatment in these organoids. Interestingly, neither FGFR1 nor OXTR was upregulated in CRC119 organoids from another oxaliplatin-resistant patient, suggesting that chemoresistance pathways may be highly personalized.
Cancer drug resistance is typically associated with genetic mutations and clonal evolution. However, this study suggests that, in resistant clones, chromatin accessibility changes may play a role in protecting these cells in response to treatment. Among the many genes that have altered expression levels, genes associated with altered chromatin accessibility regions may play a more lasting role. By focusing on those genes, we were able to narrow down the list to identify top gene candidates. However, the fact that FGFR1 and OXTR were not upregulated in another patient-derived resistant organoid suggests that there is not a uniform target for overcoming oxaliplatin resistance, thus combination regimens may have to be personalized.
FGFR1 amplification was reported to promote breast cancer resistance to 4-hydroxytamoxifen53 and is a potential therapeutic target in squamous cell lung carcinoma.54,55 Lower expression of OXTR was reported to promote breast cancer,56 and OXTR is associated with prostate cancer metastasis by mediating cancer cell migration.57 Despite those reports, the mechanism of FGFR1 and OXTR in CRC chemotherapy resistance remains to be elucidated. Potential upstream factors could be predicted based on sequences of open-chromatin regions and binding motifs of transcriptional factors (TFs) (Fig. S5 and 6, Table 3). Our analysis suggests that the genomic regions of FGFR1 and OXTR share some common putative TF binding sites as well as other TF sites unique to each peak (Fig. S5, Table 3), which could be investigated to understand the resistance mechanisms. Large-scale integrated epigenetic/transcriptomic profiling might reveal additional potential targets to treat chemotherapy resistance. Continued profiling of drug responses from patient-derived organoids may identify new biomarkers and targets for future precision medicine to treat drug-resistant cancer.
Table 3.
Predicted Transcriptional Factor Binding Sites on ATAC-Seq peaks of FGFR1 and OXTR.
| Gene | Peak Coordinate | Factor name | Start position | End position | Dissimilarity | String | RE equally | RE query |
|---|---|---|---|---|---|---|---|---|
| FGFR1 | chr8_38299794_38300181 | TCF-4E [T02878] | 16 | 22 | 0 | AGCAAAG | 0.02368 | 0.02196 |
| FGFR1 | chr8_38299794_38300181 | C/EBPbeta [T00581] | 17 | 20 | 0 | GCAA | 3.03125 | 2.83287 |
| FGFR1 | chr8_38299794_38300181 | C/EBPbeta [T00581] | 76 | 79 | 0 | TTGT | 3.03125 | 2.83287 |
| FGFR1 | chr8_38299794_38300181 | C/EBPbeta [T00581] | 286 | 289 | 0 | GCAA | 3.03125 | 2.83287 |
| FGFR1 | chr8_38299794_38300181 | AP-2alphaA [T00035] | 174 | 179 | 0.226186 | CCAGGC | 0.18945 | 0.23994 |
| FGFR1 | chr8_38299794_38300181 | YY1 [T00915] | 187 | 190 | 0 | CCAT | 1.51562 | 1.50405 |
| FGFR1 | chr8_38299794_38300181 | YY1 [T00915] | 239 | 242 | 0 | ATGG | 1.51562 | 1.50405 |
| FGFR1 | chr8_38299794_38300181 | c-Ets-1 [T00112] | 218 | 224 | 0 | CTTCCTG | 0.04736 | 0.04972 |
| FGFR1 | chr8_38299794_38300181 | GR-beta [T01920] | 238 | 242 | 0.840383 | AATGG | 1.51562 | 1.25141 |
| FGFR1 | chr8_38299794_38300181 | GR-beta [T01920] | 248 | 252 | 0.840383 | TCATT | 1.51562 | 1.25141 |
| FGFR1 | chr8_38299794_38300181 | GR-beta [T01920] | 363 | 367 | 0 | AATGT | 0.75781 | 0.587 |
| FGFR1 | chr8_38299794_38300181 | GR-alpha [T00337] | 38 | 42 | 0 | CCTGT | 1.51562 | 1.50405 |
| FGFR1 | chr8_38299794_38300181 | GR-alpha [T00337] | 207 | 211 | 0 | CCTGT | 1.51562 | 1.50405 |
| FGFR1 | chr8_38299794_38300181 | GR-alpha [T00337] | 290 | 294 | 0.207689 | AGAGG | 1.51562 | 1.50405 |
| FGFR1 | chr8_38299794_38300181 | GR-alpha [T00337] | 383 | 387 | 0.207689 | CCTCT | 1.51562 | 1.50405 |
| FGFR1 | chr8_38299794_38300181 | ER-alpha [T00261] | 327 | 331 | 0 | TGACC | 0.37891 | 0.39927 |
| FGFR1 | chr8_38299794_38300181 | ER-alpha [T00261] | 341 | 345 | 0 | GGTCA | 0.37891 | 0.39927 |
| FGFR1 | chr8_38299794_38300181 | TFII-I [T00824] | 241 | 246 | 0 | GGAAAG | 0.28418 | 0.29328 |
| FGFR1 | chr8_38299794_38300181 | HNF-1A [T00368] | 78 | 85 | 0.287765 | GTTAAAGT | 0.04736 | 0.03641 |
| FGFR1 | chr8_38299794_38300181 | FOXP3 [T04280] | 75 | 80 | 0 | GTTGTT | 0.28418 | 0.27002 |
| FGFR1 | chr8_38299794_38300181 | XBP-1 [T00902] | 30 | 35 | 0 | CGTCAT | 0.18945 | 0.17638 |
| OXTR | chr3_8887435_8887795 | TFII-I [T00824] | 0 | 5 | 0 | CTTTCC | 0.2644 | 0.24722 |
| OXTR | chr3_8887435_8887795 | TFII-I [T00824] | 37 | 42 | 0 | CTTTCC | 0.2644 | 0.24722 |
| OXTR | chr3_8887435_8887795 | TFII-I [T00824] | 259 | 264 | 0 | CTGTCC | 0.2644 | 0.24722 |
| OXTR | chr3_8887435_8887795 | YY1 [T00915] | 41 | 44 | 0 | CCAT | 1.41016 | 1.37888 |
| OXTR | chr3_8887435_8887795 | YY1 [T00915] | 159 | 162 | 0 | CCAT | 1.41016 | 1.37888 |
| OXTR | chr3_8887435_8887795 | YY1 [T00915] | 182 | 185 | 0 | ATGG | 1.41016 | 1.37888 |
| OXTR | chr3_8887435_8887795 | YY1 [T00915] | 193 | 196 | 0 | ATGG | 1.41016 | 1.37888 |
| OXTR | chr3_8887435_8887795 | AP-1 [T00029] | 54 | 62 | 0.436196 | TCTGAGTCA | 0.01653 | 0.01807 |
| OXTR | chr3_8887435_8887795 | AP-1 [T00029] | 104 | 112 | 0.401835 | TGACTCACT | 0.01653 | 0.01807 |
| OXTR | chr3_8887435_8887795 | c-Jun [T00133] | 56 | 62 | 0 | TGAGTCA | 0.02203 | 0.02329 |
| OXTR | chr3_8887435_8887795 | c-Jun [T00133] | 104 | 110 | 0 | TGACTCA | 0.02203 | 0.02329 |
| OXTR | chr3_8887435_8887795 | C/EBPbeta [T00581] | 66 | 69 | 0 | TTGT | 2.82031 | 3.08444 |
| OXTR | chr3_8887435_8887795 | C/EBPbeta [T00581] | 96 | 99 | 0 | TTGT | 2.82031 | 3.08444 |
| OXTR | chr3_8887435_8887795 | C/EBPbeta [T00581] | 117 | 120 | 0 | ACAA | 2.82031 | 3.08444 |
| OXTR | chr3_8887435_8887795 | C/EBPbeta [T00581] | 133 | 136 | 0 | GCAA | 2.82031 | 3.08444 |
| OXTR | chr3_8887435_8887795 | C/EBPbeta [T00581] | 152 | 155 | 0 | TTGT | 2.82031 | 3.08444 |
| OXTR | chr3_8887435_8887795 | C/EBPbeta [T00581] | 178 | 181 | 0 | ACAA | 2.82031 | 3.08444 |
| OXTR | chr3_8887435_8887795 | C/EBPbeta [T00581] | 238 | 241 | 0 | TTGC | 2.82031 | 3.08444 |
| OXTR | chr3_8887435_8887795 | C/EBPbeta [T00581] | 345 | 348 | 0 | ACAA | 2.82031 | 3.08444 |
| OXTR | chr3_8887435_8887795 | GR-beta [T01920] | 74 | 78 | 0.840383 | TCATT | 1.41016 | 1.91809 |
| OXTR | chr3_8887435_8887795 | GR-beta [T01920] | 82 | 86 | 0.840383 | AATGA | 1.41016 | 1.91809 |
| OXTR | chr3_8887435_8887795 | GR-beta [T01920] | 174 | 178 | 0.840383 | AATGA | 1.41016 | 1.91809 |
| OXTR | chr3_8887435_8887795 | GR-beta [T01920] | 181 | 185 | 0.840383 | AATGG | 1.41016 | 1.91809 |
| OXTR | chr3_8887435_8887795 | GR-beta [T01920] | 187 | 191 | 0 | ACATT | 0.70508 | 1.05515 |
| OXTR | chr3_8887435_8887795 | FOXP3 [T04280] | 116 | 121 | 0 | GACAAC | 0.2644 | 0.27362 |
| OXTR | chr3_8887435_8887795 | ER-alpha [T00261] | 143 | 147 | 0 | TGACC | 0.35254 | 0.30462 |
| OXTR | chr3_8887435_8887795 | LEF-1 [T02905] | 150 | 157 | 0.641865 | CTTTGTTC | 0.01102 | 0.01241 |
| OXTR | chr3_8887435_8887795 | IRF-2 [T01491] | 108 | 113 | 0 | TCACTT | 0.08813 | 0.10791 |
| OXTR | chr3_8887435_8887795 | IRF-2 [T01491] | 232 | 237 | 0 | TCACTT | 0.08813 | 0.10791 |
| OXTR | chr3_8887435_8887795 | IRF-2 [T01491] | 304 | 309 | 0 | AAGTGA | 0.08813 | 0.10791 |
| OXTR | chr3_8887435_8887795 | GR-alpha [T00337] | 30 | 34 | 0 | CCTGT | 1.41016 | 1.41143 |
| OXTR | chr3_8887435_8887795 | GR-alpha [T00337] | 163 | 167 | 0.207689 | CCTTT | 1.41016 | 1.41143 |
| OXTR | chr3_8887435_8887795 | GR-alpha [T00337] | 323 | 327 | 0 | ATAGG | 1.41016 | 1.41143 |
| OXTR | chr3_8887435_8887795 | GR-alpha [T00337] | 348 | 352 | 0 | ATAGG | 1.41016 | 1.41143 |
| OXTR | chr3_8887435_8887795 | TFIID [T00820] | 313 | 319 | 0 | TTTTCTA | 0.1983 | 0.35789 |
| OXTR | chr3_8887435_8887795 | c-Ets-1 [T00112] | 264 | 270 | 0 | CAGGAAG | 0.04407 | 0.0403 |
| OXTR | chr3_8887435_8887795 | SRY [T00997] | 150 | 158 | 0 | CTTTGTTCC | 0.00551 | 0.00768 |
| OXTR | chr3_8887435_8887795 | PR B [T00696] | 90 | 96 | 0 | GACTGTT | 0.0661 | 0.07627 |
| OXTR | chr3_8887435_8887795 | PR A [T01661] | 90 | 96 | 0 | GACTGTT | 0.0661 | 0.07627 |
Authors contribution
Conception and design: K.L. Tung, K.Y. Chen, X. Shen.
Development of methodology: K.L. Tung, K.Y. Chen, G.E. Crawford, D.S. Hsu, X. Shen.
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.):
K.L. Tung, K.Y. Chen, M. Negrete, T. Chen, A. Safi, L. Song, A.A. Aljamal, G.E. Crawford, D.S. Hsu.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): K.L. Tung, K.Y. Chen, L. Song, A.A. Aljamal.
Writing, review, and/or revision of the manuscript: K.L. Tung, K.Y. Chen, G.E. Crawford, S. Ding, D.S. Hsu, X. Shen.
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): K.L. Tung, K.Y. Chen, M. Negrete, T. Chen.
Study supervision: X. Shen.
Conflict of Interests
The authors declare no potential conflicts of interest.
Acknowledgements
We thank Duke Center for Genomic and Computational Biology sequencing core facility for research support. This work was supported by NIH NCI U01 CA217514 and U01 CA214300. We would also like to thank Erdem Altunel, and Wayne Glover from Dr. David S. Hsu's lab for their assistance and helpful discussions. The authors would like to gratefully acknowledge all study participants for their contributions.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2019.10.012.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA A Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. 2019. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA A Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Facts & Figures 2019. American Cancer Society; 2019. [Google Scholar]
- 4.Manfredi S., Lepage C., Hatem C., Coatmeur O., Faivre J., Bouvier A.M. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244(2):254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang G.J., Rodriguez-Bigas M.A., Skibber J.M., Moyer V.A. Lymph node evaluation and survival after curative resection of colon cancer: systematic review. J Natl Cancer Inst. 2007;99(6):433–441. doi: 10.1093/jnci/djk092. [DOI] [PubMed] [Google Scholar]
- 6.Dashwood R.H. Early detection and prevention of colorectal cancer (review) Oncol Rep. 1999;6(2):277–281. [PubMed] [Google Scholar]
- 7.Longley D.B., Johnston P.G. Molecular mechanisms of drug resistance. J Pathol. 2005;205(2):275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 8.Comella P., Casaretti R., Sandomenico C., Avallone A., Franco L. Role of oxaliplatin in the treatment of colorectal cancer. Ther Clin Risk Manag. 2009;5(1):229–238. doi: 10.2147/tcrm.s3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegel R., Desantis C., Jemal A. Colorectal cancer statistics. CA A Cancer J Clin. 2014;64(2):104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 10.Seruga B., Ocana A., Amir E., Tannock I.F. Failures in phase III: causes and consequences. Clin Cancer Res. 2015;21(20):4552–4560. doi: 10.1158/1078-0432.CCR-15-0124. [DOI] [PubMed] [Google Scholar]
- 11.Miranda C.C., Fernandes T.G., Diogo M.M., Cabral J.M.S. Towards multi-organoid systems for drug screening applications. Bioengineering. 2018;5(3) doi: 10.3390/bioengineering5030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clevers H. Modeling development and disease with organoids. Cell. 2016;165(7):1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 13.Sato T., Stange D.E., Ferrante M. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 14.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194) doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 15.Bu P., Wang L., Chen K.Y. A miR-34a-Numb feedforward loop triggered by inflammation regulates asymmetric stem cell division in intestine and colon cancer. Cell Stem Cell. 2016;18(2):189–202. doi: 10.1016/j.stem.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buzzelli J.N., Ouaret D., Brown G., Allen P.D., Muschel R.J. Colorectal cancer liver metastases organoids retain characteristics of original tumor and acquire chemotherapy resistance. Stem Cell Res. 2018;27:109–120. doi: 10.1016/j.scr.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weeber F., Ooft S.N., Dijkstra K.K., Voest E.E. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem Biol. 2017;24(9):1092–1100. doi: 10.1016/j.chembiol.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Skardal A., Devarasetty M., Rodman C., Atala A., Soker S. Liver-tumor hybrid organoids for modeling tumor growth and drug response in vitro. Ann Biomed Eng. 2015;43(10):2361–2373. doi: 10.1007/s10439-015-1298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlachogiannis G., Hedayat S., Vatsiou A. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crea F., Nobili S., Paolicchi E. Epigenetics and chemoresistance in colorectal cancer: an opportunity for treatment tailoring and novel therapeutic strategies. Drug Resist Updates. 2011;14(6):280–296. doi: 10.1016/j.drup.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Glaser K.B., Staver M.J., Waring J.F., Stender J., Ulrich R.G., Davidsen S.K. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2(2):151–163. [PubMed] [Google Scholar]
- 22.Flis S., Gnyszka A., Flis K., Splawinski J. MS275 enhances cytotoxicity induced by 5-fluorouracil in the colorectal cancer cells. Eur J Pharmacol. 2010;627(1–3):26–32. doi: 10.1016/j.ejphar.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 23.Fazzone W., Wilson P.M., Labonte M.J., Lenz H.J., Ladner R.D. Histone deacetylase inhibitors suppress thymidylate synthase gene expression and synergize with the fluoropyrimidines in colon cancer cells. Int J Cancer. 2009;125(2):463–473. doi: 10.1002/ijc.24403. [DOI] [PubMed] [Google Scholar]
- 24.Tumber A., Collins L.S., Petersen K.D. The histone deacetylase inhibitor PXD101 synergises with 5-fluorouracil to inhibit colon cancer cell growth in vitro and in vivo. Cancer Chemother Pharmacol. 2007;60(2):275–283. doi: 10.1007/s00280-006-0374-7. [DOI] [PubMed] [Google Scholar]
- 25.Lee J.H., Park J.H., Jung Y. Histone deacetylase inhibitor enhances 5-fluorouracil cytotoxicity by down-regulating thymidylate synthase in human cancer cells. Mol Cancer Ther. 2006;5(12):3085–3095. doi: 10.1158/1535-7163.MCT-06-0419. [DOI] [PubMed] [Google Scholar]
- 26.Gagnon J.F., Bernard O., Villeneuve L., Tetu B., Guillemette C. Irinotecan inactivation is modulated by epigenetic silencing of UGT1A1 in colon cancer. Clin Cancer Res. 2006;12(6):1850–1858. doi: 10.1158/1078-0432.CCR-05-2130. [DOI] [PubMed] [Google Scholar]
- 27.Hauswald S., Duque-Afonso J., Wagner M.M. Histone deacetylase inhibitors induce a very broad, pleiotropic anticancer drug resistance phenotype in acute myeloid leukemia cells by modulation of multiple ABC transporter genes. Clin Cancer Res. 2009;15(11):3705–3715. doi: 10.1158/1078-0432.CCR-08-2048. [DOI] [PubMed] [Google Scholar]
- 28.Innocenti F., Kroetz D.L., Schuetz E. Comprehensive pharmacogenetic analysis of irinotecan neutropenia and pharmacokinetics. J Clin Oncol: Off J Am Soc Clin Oncol. 2009;27(16):2604–2614. doi: 10.1200/JCO.2008.20.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang X., Monitto C.L., Demokan S. Identification of hypermethylated genes associated with cisplatin resistance in human cancers. Cancer Res. 2010;70(7):2870–2879. doi: 10.1158/0008-5472.CAN-09-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen D.L., Wang Z.Q., Zeng Z.L. Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology. 2014;60(2):598–609. doi: 10.1002/hep.27118. [DOI] [PubMed] [Google Scholar]
- 31.Cassoni P., Sapino A., Marrocco T., Chini B., Bussolati G. Oxytocin and oxytocin receptors in cancer cells and proliferation. J Neuroendocrinol. 2004;16(4):362–364. doi: 10.1111/j.0953-8194.2004.01165.x. [DOI] [PubMed] [Google Scholar]
- 32.Corces M.R., Trevino A.E., Hamilton E.G. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14(10):959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu T. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol. 2014;1150:81–95. doi: 10.1007/978-1-4939-0512-6_4. [DOI] [PubMed] [Google Scholar]
- 36.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12) doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross-Innes C.S., Stark R., Teschendorff A.E. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinz S., Benner C., Spann N. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramirez F., Ryan D.P., Gruning B. Deep tools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A., Tamayo P., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segal E., Friedman N., Koller D., Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36(10):1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 42.Cotto K.C., Wagner A.H., Feng Y.Y. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46(D1):D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farre D., Roset R., Huerta M. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31(13):3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daily K., Patel V.R., Rigor P., Xie X., Baldi P. MotifMap: integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinf. 2011;12 doi: 10.1186/1471-2105-12-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tommasi S., Pinto R., Petriella D. Oncosuppressor methylation: a possible key role in colon metastatic progression. J Cell Physiol. 2011;226(7):1934–1939. doi: 10.1002/jcp.22524. [DOI] [PubMed] [Google Scholar]
- 46.Youssef E.M., Estecio M.R., Issa J.P. Methylation and regulation of expression of different retinoic acid receptor beta isoforms in human colon cancer. Cancer Biol Ther. 2004;3(1):82–86. doi: 10.4161/cbt.3.1.591. [DOI] [PubMed] [Google Scholar]
- 47.Stewart L.V., Thomas M.L. Retinoids differentially regulate the proliferation of colon cancer cell lines. Exp Cell Res. 1997;233(2):321–329. doi: 10.1006/excr.1997.3569. [DOI] [PubMed] [Google Scholar]
- 48.Selaru F.M., David S., Meltzer S.J., Hamilton J.P. Epigenetic events in gastrointestinal cancer. Am J Gastroenterol. 2009;104(8):1910–1912. doi: 10.1038/ajg.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romero-Calvo I., Weber C.R., Ray M. Human organoids share structural and genetic features with primary pancreatic adenocarcinoma tumors. Mol Cancer Res. 2019;17(1):70–83. doi: 10.1158/1541-7786.MCR-18-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagle P.W., Plukker J.T.M., Muijs C.T., van Luijk P., Coppes R.P. Patient-derived tumor organoids for prediction of cancer treatment response. Semin Cancer Biol. 2018;53:258–264. doi: 10.1016/j.semcancer.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Mazzocchi A.R., Rajan S.A.P., Votanopoulos K.I., Hall A.R., Skardal A. In vitro patient-derived 3D mesothelioma tumor organoids facilitate patient-centric therapeutic screening. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-21200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walsh A.J., Cook R.S., Sanders M.E., Arteaga C.L., Skala M.C. Drug response in organoids generated from frozen primary tumor tissues. Sci Rep. 2016;6 doi: 10.1038/srep18889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner N., Pearson A., Sharpe R. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70(5):2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malchers F., Ercanoglu M., Schutte D. Mechanisms of primary drug resistance in FGFR1-amplified lung cancer. Clin Cancer Res. 2017;23(18):5527–5536. doi: 10.1158/1078-0432.CCR-17-0478. [DOI] [PubMed] [Google Scholar]
- 55.Heist R.S., Mino-Kenudson M., Sequist L.V. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7(12):1775–1780. doi: 10.1097/JTO.0b013e31826aed28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ariana M., Pornour M., Mehr S.S. Preventive effects of oxytocin and oxytocin receptor in breast cancer pathogenesis. Personalized Med. 2019;16(1):25–34. doi: 10.2217/pme-2018-0009. [DOI] [PubMed] [Google Scholar]
- 57.Zhong M., Boseman M.L., Millena A.C., Khan S.A. Oxytocin induces the migration of prostate cancer cells: involvement of the Gi-coupled signaling pathway. Mol Cancer Res. 2010;8(8):1164–1172. doi: 10.1158/1541-7786.MCR-09-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.