Abstract
The development of germ cell tumors (GCTs) is a unique pathogenesis occurring at an early developmental stage during specification, migration or colonization of primordial germ cells (PGCs) in the genital ridge. Since driver mutations could not be identified so far, the involvement of the epigenetic machinery during the pathogenesis seems to play a crucial role. Currently, it is investigated whether epigenetic modifications occurring between the omnipotent two-cell stage and the pluripotent implanting PGCs might result in disturbances eventually leading to GCTs. Although progress in understanding epigenetic mechanisms during PGC development is ongoing, little is known about the complete picture of its involvement during GCT development and eventual classification into clinical subtypes. This review will shed light into the current knowledge of the complex epigenetic and molecular contribution during pathogenesis of GCTs by emphasizing on early developmental stages until arrival of late PGCs in the gonads. We questioned how misguided migrating and/or colonizing PGCs develop to either type I or type II GCTs. Additionally, we asked how pluripotency can be regulated during PGC development and which epigenetic changes contribute to GCT pathogenesis. We propose that SOX2 and SOX17 determine either embryonic stem cell-like (embryonal carcinoma) or PGC-like cell fate (seminoma). Finally, we suggest that factors secreted by the microenvironment, i.e. BMPs and BMP inhibiting molecules, dictate the fate decision of germ cell neoplasia in situ (into seminoma and embryonal carcinoma) and seminomas (into embryonal carcinoma or extraembryonic lineage), indicating an important role of the microenvironment on GCT plasticity.
Keywords: Germ cell tumor, Primordial germ cell, Epigenetic reprogramming, Plasticity, Microenvironment, BMP signaling, SOX2, SOX17
1. Classification of germ cell tumors (GCTs)
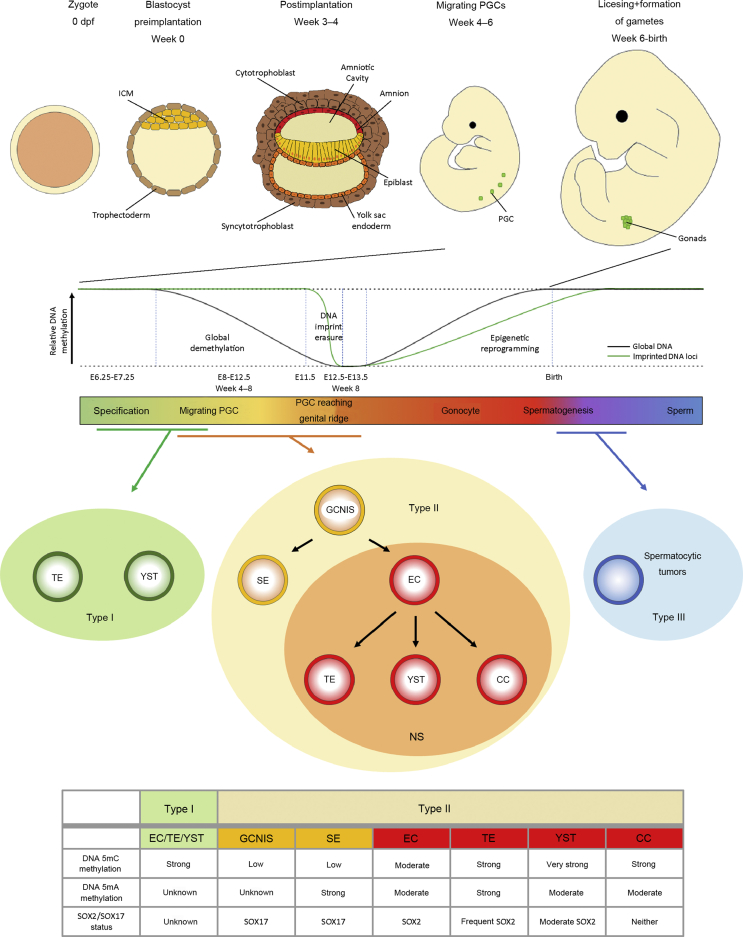
GCTs can be categorized into five common (I–V) and two new subtypes (0, IV) [1]. Type I and Type II can occur in both sexes and gonadal as well as extra-gonadal. Pediatric type I tumors consist of pure teratoma (TE) or pure yolk sac tumors (YSTs) and share cytogenetically gains on chromosomes 1q, 12p13 and 20q, as well as losses on 1p, 4 and 6q [2,3]. Type II tumors, which develop from primordial germ cells (PGCs), harbor mostly chromosome 12p aberrations (12p gain), while chromosome 9p alterations are characteristic in type III tumors (spermatocytic tumors) [4]. In males, type II GCTs can be divided into seminomas (SEs) (female: Dysgerminoma) and non-seminomas (NSs) (female: Non-dysgerminoma) (Fig. 1, lower panel). Embryonal carcinomas (ECs), the stem cell population of the NS, present a pluri- to totipotent character and can further differentiate into either YST or choriocarcinomas (CC), representing extraembryonic differentiation, or TE, which are known for their somatic differentiation into cells of all three germ layers. These subtypes can either develop as a single tumor entity or as a mixed tumor [5]. All type II testicular GCTs (TGCTs) are thought to develop from an arrested PGC, which acquired genetic and/or epigenetic aberrations (Fig. 1). This pre-invasive precursor lesion of SE and NS in postpubertal gonads is termed “germ cell neoplasia in situ” (GCNIS), formerly also known as “carcinoma in situ” (CIS) or “intratubular germ cell neoplasia unclassified” (IGCNU) [6]. Type III GCTs are spermatocytic tumors found in older men, and type IV are dermoid cysts of the ovaries, while type V comprises complete hydatidiform moles in the uterus [7]. Recently, Oosterhuis and Looijenga [1] proposed the classification of two additional as well as intermediate types: Type 0 (GCTs arising at the attachment site of conjoined twins) and type VI (GCTs derived from somatic cells).
Figure 1.
Development of GCs and pathogenesis of TGCTs. Upper panel: Development of the human embryo from the zygote (0 day post fertilization [dpf]) until Week 6 post fertilization when the PGC reach the gonads. Lower panel: DNA methylation events during PGC migration and arrival at the genital ridge and where different types pf TGCTs are thought to originate from. The table summarizes molecular and epigenetic findings from this review. Data from Refs. [1,48,49,52,58,74]. PGC, primordial germ cell; ICM, inner cell mass; SE, seminoma; EC, embryonal carcinoma; CC, choriocarcinoma; TE, teratoma; YST, yolk sac tumor; NS, non-seminoma; GCNIS, germ cell neoplasia in situ; GC, germ cell; TGCTs, testicular germ cell tumors.
2. The origin of GCTs—the PGC
It is generally believed that the precursor of GCTs arises during (early) germ cell (GC) development in the fetus (Fig. 1, upper panel). The murine development of GCs starts after a few divisions of the totipotent zygote, when cells can be divided into the pluripotent primitive ectoderm (inner cell mass [ICM]) as well as trophectoderm cells of blastocysts. After blastocyst implantation (E3.5 in mice), a part of the ICM starts differentiation towards somatic cell fates (epiblasts). PGC specification starts at E6.25–E7.25 in mice (human Week 4 post fertilization) from a few epiblasts located in the posterior proximal region of the embryo [1,[8], [9], [10]]. This induction of PGC specification occurs through signaling pathways from extraembryonic tissue, such as BMP2, BMP4, BMP8b and SMAD1 [11]. SMAD1 activity is dependent on SETDB1, which catalyzes H3K9 trimethylation leading to repression of negative SMAD1 regulators DPPA2, OTX2, and UTF1 [12]. Subsequently, expression of BLIMP1/PRDM1, the key factor of PGC specification, is triggered, causing the PGC precursors to escape the somatic differentiation program [1,8,10]. Similar to stem cells of the ICM, PGCs express multiple markers of pluripotency. However, they also act as precursors of the unipotent GC lineage, since their only fate is the production of gametogenic stem cells. Therefore, expression of somatic genes is considered pathologic [13]. Based on expression of chemokine receptors cKIT and CXCR4, PGCs migrate at E8 (human Week 4–5) along the developing hindgut towards the genital ridge [1,14]. The reset of genomic DNA methylation during migration of the PGCs to the gonadal ridge results in an open chromatin structure, alongside the re-expression of pluripotency genes Stella, Sox2, Oct4, and Nanog, as well as PRDM1-dependent expression of Dnd1, Nanos3, and Prdm14 [11]. Around E8.5, PRDM1 maintains the repression of the somatic program through complex formation with diverse histone modifiers, such as histone deacetylases (HDACs), as well as maintaining the unipotent cell lineage in a complex with PRMT5, a methyltransferase causing specific symmetrical demethylation of H2AR3 and H4R3 [15]. Lately, Mochizuki et al. [12] revealed a Prdm1-dependent enrichment of Hdac3 and deacetylation of H3 and H4 histones in murine epiblast-like cells, which is crucial for repression of somatic gene expression of Hoxa1, Dnmt3b, Crabp2, and Meis2. Upon E10.5–11.5, PGCs arrive at the genital ridge. These late PGCs undergo rapid demethylation of imprinted genes (E11.5–12.5) by active demethylation [16]. Subsequently, after embryonic sex determination, male PGCs go into a G1-phase mitotic arrest and are termed prospermatogonia or gonocytes, giving later rise to sperm cells. Gonocytes transact complete epigenetic reprogramming by re-establishing DNA methylation and histone marks, as well as sex-specific imprints [17]. The “licensing” process occurs until birth, which prepares the gonocytes for meiosis and further development into gametes [10,14,18].
3. Comparability of murine and human PGC development
The first major difference between rodent and human GC development is the formation of an egg cylinder (E5) or a bilaminar disc (Week 3–4), respectively [4,19]. Both structures consist of an epithelial monolayer, known as the epiblast, next to a layer of yolk-sac epithelium, called the hypoblast (Fig. 1) [4]. In humans, migration of PGCs occurs in Week 4–6 and arrives at the gonadal region at Week 6, while immature Sertoli cells encircle them [9]. Human and murine GCs display a highly similar expression profile of GC specifiers PRDM1/Prdm1 and TFAP2C/Tfap2c, GC factors NANOS3/Nanos3, DND1/Dnd1, DDX4/Ddx4, DAZL/Dazl and pluripotency factors OCT4/Oct4 and NANOG/Nanog [19]. In contrast, SOX17, which is involved in endodermal differentiation in early embryogenesis [20], is a key player in human PGCs, which lack SOX2 expression [21]. Factors as SOX17, PRDM1 and TFAP2C specify human PGC cell fate and, thereby, mediate establishment and maintenance of pluripotency [21] (Table 1).
Table 1.
Main findings of this review.
| Chapter | References |
|---|---|
| Comparability of murine and human (primordial) GC development | |
| The Phenotype differs: Egg cylinder in mice and bilaminar disc in human | [4,19] |
| Commonly expressed genes between the species: PRDM1/Prdm1, TFAP2C/Tfap2c, NANOS3/Nanos3, DND1/Dnd1, DDX4/Ddx4, DAZL/Dazl, OCT4/Oct4, NANOG/Nanog | [19] |
| Mouse specific genes: Prdm14, Sox2 | [21,22] |
| Human specific genes: SOX17, SOX15 | [21,24] |
| Development of type I GCTs | |
| PGC specification depends on WNT and BMP pathways | [27,28] |
| The surrounding tissue and extracellular matrix might influence BMP and WNT signaling pathways | this review |
| Misrouting of PGCs and failures in downregulating the pluripotency program could lead to type I GCTs | [7] |
| Development of type I GCT from PGCs in a short time-frame | [7] |
| The testicular dysgenesis syndrome and formation of GCNIS | |
| TDS results from disturbed hormonal microenvironmental factors during fetal development | [31,32] |
| TDS is related to TGCT development | [33] |
| Impaired Sertoli and Leydig cell function are not the only triggers leading to GCNIS formation | [33] |
| The PGC gene expression program in type II GCTs | |
| TFAP2C expression is associated with a PGC-like cell fate | [39] |
| TFAP2C in GCTs allows maintenance of a latent pluripotent state | this review |
| DAZL, DDX4, MAEL and TDRD12 are gonad-specifically activated in PGCs upon arrival at the genital ridge | [40] |
| DAZL deficiency leads to an increase in TE formation | [40] |
| SOX2 and SOX17 regulating GCT fate | |
| SOX2 and SOX17 share a common set of overlapping target genes, such as NANOG, OTX2, PIM1/2, PRDM14, DPP4, TDGF1, LIN28A, and TRIM71 | [45] |
| Epigenetic re-arrangements might contribute to SOX17 target gene accessibility including GC-fate and pluripotency genes in PGCs | this review |
| DNA-Methylation in GCTs | |
| Compared to SE and GCNIS, NS shows high de novo DNA methylation levels (high expression of DNMT3A and DNMT3L) | [[47], [48], [49]] |
| Active 5mC demethylation in TGCTs | [49] |
| The influence of microenvironment on TGCT pathogenesis | |
| Important interactions between SDF1/CXCL12 and CXCR4 as well as functioning Msx genes are involved in a correct migration during PGC development | [[62], [63], [64], [65],67,69] |
| A pro-inflammatory micromilieu (IL-1β, IL-6, TNF-α, CCL5, SDF-1/CXCL12, CXCL-13) might favor TGCT development | [68] |
| FOXA2 is identified as a key factor of differentiation of SE to NS (not EC) | [44,73] |
| Microenvironmental components and BMP-inhibiting factors could directly differentiate SE into EC | [22,44] |
| Colonization of the genital ridge is influenced by factors secreted by surrounding Sertoli-, Leydig-, and immune cells, such as chemokines and cytokines | this review |
EC, embryonal carcinoma; GC, germ cells; GCNIS, germ cell neoplasia in situ; GCTs, germ cell tumors; NS, non-seminoma; TDS, testicular dysgenesis syndrome; TE,teratoma; TGCT, testicular germ cell tumor; PGC, primordial germ cell; SE, seminoma.
Another differentially expressed gene is PR-domain containing protein 14 (Prdm14), which mediates murine PGC maintenance. By immunohistochemical staining, only minor expression levels of PRDM14 were found in human PGC-like cells [21,22]. Nevertheless, a significantly reduced efficacy of PGC-like cell derivation from PRDM14-deficient hESC (human embryonic stem cells) argues for a role of PRDM14 in human PGC development [23]. Interestingly, in human primordial germ cell-like cells (hPGCLC), the inducible depletion of SOX15, another member of the SOX family, resulted in increased mRNA expression of pluripotency factors PRDM14, VETNX, and NANOG [24]. This upregulation suggests for compensating pathways resulting in maintenance of a PGC-like state. Nevertheless, even though SOX15 seems replaceable, its inducible depletion in hPGCLCs also resulted in elevated SOX15 mRNA, indicating a negative feedback loop and a prominent role during PGC perpetuation [24].
4. Development of type I GCTs
Pediatric type I GCTs occur in children below the age of five years and often arise in extragonadal locations in midline structures, such as the sacrococcygeal, retroperitoneal, mediastinal, cervical, and intracranial region [1,25]. Although type I GCTs are most frequent in infants and children, a variant of immature TE is presenting in adults and highly malignant [7]. Epigenetically, type I GCTs may have either normal biparental genomic imprints or partially erased imprints, implicating an immature PGC origin [26].
In type I TE cell-intrinsic as well as niche-related factors have been recently suggested to be involved during the maintenance of the PGC phenotype. These factors are involved in the regulation of survival (DND1, KIT, KITLG, BAK1, and AKT1) or control of pluripotency and developmental programs (SOX17, BLIMP1, OCT4, SOX2, and NANOG) [1]. PGC specification depends on BMP and WNT signaling, which is active around E7.25. Hackett et al. [27] described a PRDM1-dependent upregulation of NR5A2 and ZFP296 in early human PGCs. Here, an activation (Brachyury [T] and Wnt3) or inhibition (T, Cdx2, Notum, and Dkk1) of the WNT signaling was observed in Nr5a2-deficient and Zfp296-deficient PGC-like cells (PGCLC), respectively. These data sets identified NR5A2 and ZFP296 as key players of WNT-modulated GC specification [27]. Regarding the BMP pathway, specifically in type I YSTs (compared to germinomas), SMAD6 and SMAD7 protein expression was highly elevated, as were several TGF-β-related genes, such as GSC, TGFB1/1, COL31A1, IGFBP3, TSC22D1, and JUNB [28]. This suggested that the surrounding tissue or the extracellular matrix (ECM) might have an influence on signaling pathways, like BMP or WNT, being crucial for PGC development processes. Hence, perturbations in these pathways might lead to formation of type I GCTs (Table 1).
Migrating PGCs are in a close cross-talk with their surrounding environment. To avoid anomalies occurring through misguided PGCs, apoptosis is induced as a result of KITLG (stem cell factor)-reduction and BAK1 (Bcl-2 homologous antagonist killer) increase [1,29]. If these apoptosis inducing factors are genetically or epigenetically altered, misrouted cells could survive and maintain their PGC phenotype. Microenvironmental factors possibly induce reprogramming in these mismigrated cells, resulting in uncontrolled proliferation, thereby giving rise to type I GCT [7]. Thus, misrouting and/or failures in shutting down the pluripotency program in PGCs might lead to development of type I GCTs.
5. The testicular dysgenesis syndrome and formation of GCNIS
TGCTs often coincide with other male reproductive disorders including impaired spermatogenesis, hypospadias and cryptorchidism. Skakkebeak et al. [30] suggested that all these clinical conditions could be symptoms of one main disorder, the so-called testicular dysgenesis syndrome (TDS). TDS is thought to be the result of a disturbed hormonal milieu during early fetal development, due to genetic effects and/or environmental factors during pregnancy, such as anti-androgens [31,32]. The micromilieu in the developing testis is needed for correct GC development and mainly established and maintained by Sertoli and Leydig cells [9]. It is, therefore, not surprising that histologic alterations in these cell types are often found in patients with TGCTs [33]. The TDS theory proposes genetic and environmental factors, like estrogen, estrogen-mimics, anti-androgens and other factors yet to be determined, to disturb Sertoli and Leydig cell function, avert proper GC differentiation, thereby leading to GCNIS and TGCTs [31]. Normally, the micromilieu harbors chemokines, which mediate migration direction and growth, but the micromilieu produced by aberrant somatic cells could create an environment favoring tumor growth. The exact mechanisms behind the signaling leading to secretion of these somatic chemokines are to date not completely understood. Sertoli cells express cytokine stromal-derived factor 1 (SDF-1/CXCL12) which is thought to contribute to testis development in guiding migration through its G-protein-coupled receptor CXCR4 on PGCs and is responsible for the maintenance of adult stem cell niches [34,35]. It was proposed that Sertoli and immune cells synthesizing SDF-1/CXCL12 in neoplastic testis could create a somatic niche allowing for cancer cell maintenance [35], especially as its receptor CXCR4 has been implicated in the metastatic progression of a range of tumor types [36]. Further, Szarek et al. [35] also found CCL17 in Sertoli cell cytoplasm only when GCNIS cells were present. CCL17 is a cytokine mainly expressed in the thymus and functions as a chemotactic factor for T-cells. It is regulated by the TNF-β superfamily, which has been linked to GCNIS formation [37]. Nevertheless, histologic alterations of Sertoli and Leydig cells were only found in about 25% of patients with TGCT in a study looking for histologic dysgenetic features [33] (Table 1).
In theory, SEs are the default pathway of GCNIS [38]. GCNIS that later develop into SEs are thought to arise from migratory PGCs during the global DNA demethylation phase, so shortly before arriving at the genital ridge and before coming into contact with Sertoli and Leydig cells. Therefore, TDS can strongly contribute to TGCT development, but other triggers besides impaired Sertoli and Leydig cell functions are possible. This is in accordance with the hypothesis by Skakkebaek et al. [31] and Sonne et al. [32] that these altered somatic cells from TDS are less responsible for aberrant GC differentiation, but rather display disturbed signaling, which fails to induce apoptosis in already existing GCNIS of prepubertal and adult testes.
6. The PGC gene expression program in type II GCTs
TFAP2C/AP-2γ was identified as a critical factor maintaining PGC cell fate in the PRDM1 signaling pathway. Knock-out of Tfap2c in mice led to a sterile phenotype, with PGC specification still intact up to E8.0 when the PGC signal (monitored by alkaline phosphatase staining) was lost before the start of migration [39]. Further, murine PGC-like cells lacking TFAP2C lost expression of Nanos3 and Dazl, but presented upregulation of Hoxa1, Hoxb1, and Brachyury/T indicating somatic differentiation. This was further cross-validated using TCam-2 cells as an in vitro model for PGCs. TFAP2C knock-down in TCam-2 cells showed upregulation of genes indicative of mesoderm differentiation (MYOD1, HAND1 and GATA2), while makers for ectoderm and endoderm remained unchanged, concluding TFAP2C involvement in the repression of a gene set inducing mesodermal differentiation [39]. TFAP2C is highly expressed in GCNIS and SE reflecting their PGC origin and indicating that TFAP2C is necessary to keep up PGC-like expression programs in these cell types, allowing maintenance of a latent pluripotent state.
Recently, Nicholls et al. [40] questioned whether a conserved transcriptional program would be initiated upon arrival in the genital ridge to differentiate GCs from their germ line precursors and somatic lineages. Specifically, based on transcriptome data, they identified the RNA-binding protein DAZL, DDX4, MAEL, and TDRD12 as being gonad-specifically activated after colonization. Further, the activation of DAZL-dependent GC developmental programs by diminution of pluripotency factors was elucidated. Here, Dazl-deficient mice continued to express the pluripotency factor Nanog to maintain a PGC-like state [40]. Interestingly, these mice spontaneously developed TE, which could arise from uncommitted gonadal PGCs. Moreover, upon reversion of gonadal sex of both XX and XY animals, the authors observed TE development in both sexes, though with a higher incidence in Dazl-deficient XY male mice. Therefore, not only was Dazl necessary for further development of the germ line, but, compared to ovaries, the testis served as a better TE-promoting microenvironment raising the importance of the tumor micromilieu [40]. In summary, the study by Nicholls et al. [40] provides a model for a DAZL-dependent germ line development. They proposed an additional developmental step after colonization, eventually determining the PGC fate. Thus, type II GCTs might also arise at a later PGC stage (Table 1).
7. SOX2 and SOX17 regulating TGCT fate
Human GC development relies on SOX17, while in mice SOX2 is crucial during GC formation. Both SOX17 and SOX2 are able to bind OCT4 and represent transcription factors of the SOX family. They can regulate transcription either in complex with OCT4 or alone. The SOX2/OCT4 complex binds to the canonical DNA motif (CTTTGTCATGCAAAT) combining the SOX2 and OCT4 binding motifs, whereas the SOX17/OCT4 complex prefers the compressed motif (CTTTGTATGCAAAT) lacking a central cytosine [20]. While the canonical motif is located in enhancers of pluripotency genes, the compressed motif is found in regulatory regions of genes involved in endodermal differentiation. When comparing the DNA binding sites of SOX2 and SOX17 complexes with their respective binding motives, 50% of SOX2 sites contained the canonical motif (7.5% compressed motif). Interestingly, more than 40% of SOX17 sites contained the compressed-motif and 21.1% the canonical-motif [20]. This concludes that the SOX17/OCT4 complex is often bound to the canonical motif. The SOX2/OCT4 complex induces pluripotency in iPSCs and embryonic stem cells [41]. Even though PGCs lack SOX2, they present latent pluripotency while being unable to spontaneously differentiate [21,42]. Latent pluripotency is defined as a dormant or poised form of pluripotency that is thought to exist in PGCs. Cells are latent pluripotent if they express pluripotency transcription factors (OCT4, NANOG, and LIN28), but do not have the ability to differentiate like naïve pluripotent cells into all three germ layers [43]. Further SEs, highly similar to PGCs, express SOX17, whereas ECs, highly resembling ESCs, express SOX2 (Fig. 1, lower panel) [38]. The importance of SOX17 for SE phenotype was demonstrated by knock-down of SOX17 in SE cells (TCam-2), which resulted in downregulation of GC and pluripotency markers [21]. Further, SOX2 was not necessary in SE, but during reprogramming into EC. Upon xenotransplantation into the microenvironment of a murine flank, seminoma-like TCam-2 cells reprogrammed into an EC-like state, which was initiated upon inhibition of BMP signaling. Subsequently, SOX2 was upregulated (SOX17 downregulated) and NODAL signaling was induced (Fig. 2) [22]. Xenotransplanted SOX2 knock-out TCam-2 cells did not undergo this transition into an EC-like state but remained in an SE-like state, where gene expression closely resembled in vitro cultivated TCam-2 cells [44]. Jostes et al. [45] finally illuminated the different roles of SOX2 and SOX17 in TGCT fate. ChIP-seq data revealed that in TCam-2 cells, SOX17 also bound to both motifs (canonical and compressed), resulting in an overlap of SOX2 and SOX17 target genes. These target genes included factors involved in regulation of pluripotency and GC development like NANOG, OTX2, PIM1/2, PRDM14, DPPA4, TDGF1, LIN28A, and TRIM71. Therefore, pluripotency factors can be regulated by SOX17 as well as SOX2, indicating an overlap in their function as transcriptional regulators (Table 1). However, SOX2 also binds to genes upregulated in embryonic stem cells like GDF3, LEFTY2, SALL4, TP53, as well as SOX2 and OCT4 itself, creating a regulating loop, which was not found for SOX17 binding sites [45]. Further, comparing SOX17 binding sites in TCam-2 and differentiated ESC, only showed an overlap of 10%, where, out of all genes bound by SOX17 in TCam-2, 66% were only in TCam-2, but not found in the somatic lineages. PRDM1 and TFAP2C were found within these specific genes for TCam-2 [45], which are known to be transactivated by SOX17 during PGC specification [21,46], further demonstrating high similarity between SE and PGCs. The canonical motif was not found in hits of differentiated lineages, suggesting the canonical motif being specific for SE and possibly PGC. This concludes that DNA accessibility plays an important role in availability of target genes to SOX17 [45]. Considering the dramatic epigenetic changes, like DNA demethylation and histone modification during GC development, the accessibility of SOX17 to its target genes might be much easier in PGCs compared to somatic cells, making it possible for SOX17 to regulate factors of GC fate and pluripotency.
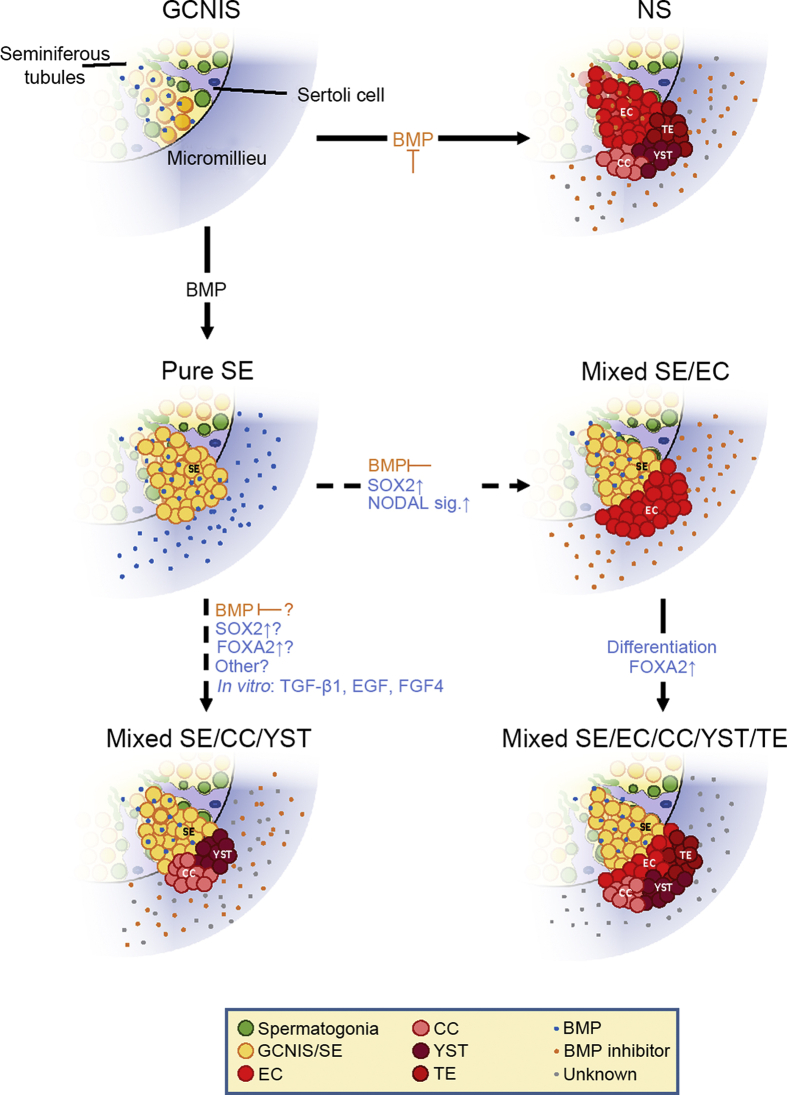
Figure 2.
Plasticity of SE cells—Transformation into EC and CC/YST. Depending on BMP activity GCNIS either develop into SE (BMP active) or EC (BMP inhibited) which further differentiates into TE, CC and YST. Additionally, to the commonly accepted TGCT developmental theory (Fig. 1), in vitro experiments indicated reprogramming of SE to EC [22] as well as directly into extraembryonal tumors without EC intermediate (SOX2) [44,75]. SE to EC transformation is initiated in vitro by BMP inhibition through the microenvironment. SOX2 upregulation and establishment of NODAL signaling give rise to mixed SE/EC. These can further differentiate into mixed SE/NS, eventually leading to mixed tumors with SE components while also containing extraembryonal proportions. Direct SE differentiation into extraembryonal-like tissue has been demonstrated in vitro and in vivo although the factors involved in the latter still remain unknown and have yet to be determined. GCNIS, germ cell neoplasia in situ; EC, embryonal carcinoma; SE, seminoma; NS, non-seminoma; CC, choriocarcinoma; TE, teratoma; YST, yolk sac tumor; NS, non-seminoma; GC, germ cell; TGCT, testicular germ cell tumor.
In summary, SOX2 and SOX17 share a common set of target genes, which are regulators of pluripotency (Fig. 1, lower panel). Additionally, SOX2 and SOX17 specifically regulate gene sets leading to either ESC-like cell fate (ECs) or PGC cell fate (SE). In addition, Jostes et al. [45] could identify genes bound by SOX2 in EC cell line 2102EP, but not in ESC, concluding that MLEC, PIM2, CD99L2, and APOBEC3F are genes which might be involved in GC tumorigenesis. The same applies to cancer related MYC and IGF1, which are bound by SOX17 only in TCam-2 cells.
8. DNA methylation in TGCTs
8.1. De novo 5-methylcytosine (5mC) DNA methylation in TGCTs
Considering the higher DNA methylation content in NS compared to SE and GCNIS, it is rather likely that reprogramming from latent into stem cell like pluripotency is coincident with epigenetic reprogramming. The establishment of de novo methylation is carried out by DNA methyltransferases DNMT3A and DNMT3B. The establishment of parental methylation imprints requires further DNMT3L, which has no methyltransferase activity itself but stimulates the activity of DNMT3A and DNMT3B through complex formation [47]. Expression of DNMT3B was highly elevated in ECs compared to SE [47] (Table 1).
Immunohistochemical analysis revealed lower 5mC content in GCNIS and SE compared to NS (Fig. 1, lower panel), whereas no significant difference in DNMT1 expression was reported [48]. During the transition from SE to EC, shown in xenotransplants of TCam-2 cells into mice, DNMT3 and DNMT3L were strongly upregulated resulting in higher 5mC content suggesting de novo methylation. Moreover, 5-hydroxymethylcytosine (5hmC) levels were also increased, indicating active DNA demethylation processes [49]. Netto et al. [48] proposed a model where the higher methylation content of NS compared to GCNIS and SE is attributed to de novo methylation of GCNIS mediated by DNMT3A and DNMT3L rather than developmentally acquired CpG island methylation from PGCs. This model supports the linear development from GCNIS to SE and further to NS as well as the direct transition from GCNIS to NS but does not support GCNIS deriving from late PGCs (after Week 8/E13.5) which underwent epigenetic re-programming.
The difference in methylation status has also been promising for the classification of different TGCT subtypes as they show distinct methylation patterns independent from location, gender, or genetic changes [[50], [51], [52]] (Fig. 1). A recent study identified five gene promoters (CRIPTO, HOXA9, MGMT, RASSF1A and SCGB3A1) by which it was possible to distinguish between SE and NS as well as the NS subtypes or even between pure and mixed types [52]. Further, methylation at tumor suppressor genes is associated with the development of malignancies from GCNIS, which is also true for EC as well as SE, which are mainly hypomethylated, but show some DNA-methylation at these genes [53].
8.2. N6-adenosine methylation (6mA) DNA methylation in TGCTs
In addition to the well-known cysteine methylation, 6mA might play a role during epigenetic reprogramming of GCs. In general, RNA and DNA adenosine methylation is mediated by epigenetic writers (e.g. METTL3, METTL14), erasers (e.g. ALKBH5, FTO), and readers (YTHDC1-2, YTHDF1-3) [54].
Recently, in several TGCT cell lines, METTL3, ALKBH5, YTHDC1, YTHDF1/2, and HNRNPC have been identified as fundamental factors from the epigenetic machinery mediating 6mA modifications [55]. 6mA modifications were mostly found in the cytoplasm of TGCT tissue and cell lines (SE and NS) suggesting 6mA presence on RNA but not DNA level, which was verified by mass spectrometry analysis of DNA from TGCT cell lines. As 6mA was not found on DNA in TGCT (except for potential 6mA methylation on GCNIS DNA), the authors concluded that TGCTs do not use 6mA DNA modifications to regulate transcription. They further found differentiation associated with changes in RNA 6mA levels [55]. In vitro differentiation of TCam-2 cells (using murine fibroblast conditioned medium) to mixed NS led to a strong increase of RNA 6mA levels, while differentiation of EC cell lines NCCIT and NT2/D1 induced by retinoic acid only led to a mild increase. This is in compliance with results from Batista et al. [56] using Mettl3 knock-out murine ESCs. These knock-out cells presented increased self-renewal ability and were unable to differentiate upon directed in vitro differentiation protocols. Therefore, increased 6mA RNA levels were associated with differentiation.
Subtypes of GCNIS vary in expression of 6mA writer VIRMA which is part of the methylation complex consisting of METTL3, METTL14, VIRMA and WTAP [57], and the reader YTHDF3. An in silico analysis of TCGA-data revealed VIRMA and YTHDF3 as the most commonly altered modifiers/readers of 6mA (mostly transcript upregulations) in nearly half of the available TGCT samples [58]. Interestingly, the data showed a strong correlation in the upregulation of both enzymes. In a patient tissue study performed by Lobo et al. [58], SE indicated higher VIRMA and YTHDF3 mRNA levels and higher VIRMA protein expression (as well as 6mA levels) compared to NS (Fig. 1, lower panel). In this study 6mA was found predominantly nuclear and only in 29.1% of the tumor samples in the cytoplasm [58], which stands in contrast to the observations made by Nettersheim et al. [55]. Lobo et al. [58] further found nuclear VIRMA and cytoplasmic YTHDF3 immunostaining in tissue samples. There was no significant association found between VIRMA and YTHDF3, indicating no correlation between the coincident alteration rate from the in silico analysis. From a developmental point of view, SEs are considered the default pathway for GCNIS [38] and reprogramming of GCNIS results in NS. As SEs show high levels of 6mA, Lobo et al. [58] proposed, based on the experiments by Wang et al. [59] showing reduced stemness upon Mettl3 and Mettl14 knock-down (i.e. decrease in 6mA), that 6mA might contribute to the maintenance of the SE phenotype, which was surprising regarding the high 6mA levels, which were associated with differentiation as stated by Nettersheim et al. [55], and Batista et al. [56].
Future studies on deciphering the RNA and DNA 6mA methylation landscape and functional experiments including knock-outs of different complex components might give insight in the complex regulation of 6mA modifiers and further regulatory capacity of 6mA itself.
9. The influence of microenvironment on TGCT pathogenesis
When removed from their microenvironment, PGCs undergo reversion to pluripotent embryonic GCs in vitro, which differ from embryonic stem cells in their variable methylation at imprinting control centers [60]. However PGC-like cells, derived from embryonic stem cells in vitro can differentiate into gametes if placed in the microenvironment at the gonadal niche [61]. This concludes that the microenvironment might play a crucial role in the transformation of PGCs to GCNIS.
It has been proven that SDF-1/CXCL12 and its receptor CXCR4 are crucial during colonization of PGCs [62]. By following PGC development in SDF-1-deficient mice, the authors reported not only a delayed migration, but also a decreased number of PGCs in the gonads [62]. Murine embryos injected with morpholino antisense for either SDF-1/CXCL12 or CXCR4 also represented a disrupted and scattered migratory phenotype [63]. Later, the crucial interaction between SDF-1 and CXCR4 was validated in CXCR4-deficient mice having misrouted PGCs along the mesentery and the hindgut around E10.5 instead of the genital ridge [64]. Additionally, in transgenic zebrafish, reticulons (RTNs) have been described to directly interact with CXCR4, thereby also regulating migration of PGCs [65]. Clinically, positive CXCR4 expression was documented in TGCTs. In detail, not only was CXCR4 expressed in SE and EC, but also in extragonadal primary GCTs [66], again indicating the influence of SDF-1/CXCR4 interaction during colonization. Interestingly, based on expression pattern of murine sdf-1β, Stebler et al. [67] inferred that migration was not solely dependent on SDF-1/CXCL12, but rather by other chemokines and cytokines. In a model of rat experimental autoimmune orchitis (EAO), the later pathway was recently acknowledged to activate Galectin-1 (Gal-1) and TNF-α secretion by Sertoli cells. This inflammatory response eventually leads to onset of further cytokines, such as IL-1β and IL-6 [68]. This pro-inflammatory environment consisting of cytokines, such as IL-1β, IL-6, TNF-α, and chemokines, such as CCL5, SDF-1/CXCL12, and CXCL-13, has been reported in GCNIS and SE [69], rising the thought of a pro-inflammatory micromilieu being favorable during the development of TGCTs (Table 1).
Further, also the ECM is a crucial surrounding environment during migration of PGCs. Besides integrins and glycoproteins (e.g. Fibronectin, Laminin, and Type IV Collagen) [70], Msx genes have been recently correlated to PGC cell migration [71]. Similar to the observations found in SDF-1/CXCL12 or CXCR4 mutants, Sun et al. [71] demonstrated a delayed PGC migration with subsequent colonization in the mesentery in Msx1/2-deficient mice. Moreover, since Msx1/2 is initially expressed in the distal end of the primitive streak, the authors questioned, whether a conditional knock-out of mesoderm-specific Msx1,2 would lead to similar observations. Indeed, these knock-out mutants (Mesp1-Cre;Msx1,2) displayed a similar phenotype compared to Msx1/2-deficient mice, concluding a non-autonomous cascade being involved upon loss of Msx1,2 [71].
Another major role of the microenvironment is its capacity of cellular reprogramming leading to phenotypic plasticity during the development of GCTs. Xenotransplantation of TCam-2 cells (SE) into the murine flank led to reprogramming into ECs (Fig. 2) [22]. Molecularly, the somatic microenvironment caused inhibition of BMP signaling, thereby, inducing NODAL. Upregulation of SOX2 induced expression of NODAL co-factors CRIPTO and LEFTY1/2, thereby establishing NODAL-signaling, which is thought to mediate EC reprogramming [72]. Xenotransplantation of SOX2-deficient cells mainly retained the SE phenotype, but a proportion of cells differentiated into CC- or YST-like cells [44]. Similarly, in vitro differentiation of TCam-2 cells using TGF-β1, EGF and FGF4 supplemented medium, resulted in mixed NS lineages without EC intermediate (no SOX2 upregulation) (Fig. 2) [70]. These differentiated TCam-2 cells presented a roundish giant multinucleated phenotype, indicating differentiation into extraembryonic lineage. Members of the Hippo pathway YAP1 and TEAD4, triggers of extraembryonic cell fate found in TCam-2, were thought to drive the extraembryonic differentiation [70]. Even though SOX2-deficient TCam-2 xenografts represented an SE phenotype, the authors observed demarcated regions in the tumor, which clearly induced differentiation [73]. In this cluster, FOXA2 pathway was the crucial network inducing differentiation. Indeed, while the previously stated murine xenotransplanted SOX2-deficient TCam-2 cells were positive for differentiation markers FOXA2, AFP, and EOMES, the SOX2/FOXA2 double knock-out TCam-2 cells were negative for these differentiation markers and remained positive for SE and pluripotency markers, such as OCT4, SOX17, TFAP2C, and BLIMP1 [73]. Since FOXA2 is absent in most of TGCT cell lines and tissues, the authors concluded FOXA2 being induced only during the late differentiation of SOX2-deficient TCam-2 into NS lineage. Since reprogramming could start as soon as GCTs get into contact with the microenvironment outside of the seminiferous tubules, we hypothesize that BMP-inhibitory factors, as well as factors yet to be determined (chemokines, environmental toxins, hormone mimics, etc.) might lead to direct differentiation/re-programming of SE to extra-embryonal tumors CC or YST (Fig. 2).
Summarizing, so far not enough is known about the interaction between the microenvironment and PGCs and how disturbances could result in misguided or miscolonized PGCs eventually leading to TGCTs. Nevertheless, it seems that it is rather colonization than migration being influenced by factors, such as chemokines and cytokines, which are secreted by surrounding Sertoli, Leydig, and immune cells or the ECM.
10. Conclusion
Latest research determining the specific and redundant roles of SOX2 and SOX17 gave insight in the SOX17 and SOX2 driven regulation of pluripotency in SE/PGCs and ECs, respectively [45] (Fig. 1). Further analyses of the genes found to be regulated by either SOX17 or SOX2, such as MYC and IGF1 in SE and MLEC, PIM2, CD99L2 and APOBEC3F might give new insights into tumor pathogenesis.
Based on our literature revision, we conclude that in TGCTs a higher cytosine methylation status is correlated with a more differentiated TGCT subtype (Fig. 1). This makes it possible to distinguish between SE and NS as well as the NS subtypes or even between pure and mixed types [52].
Adenosine methylation was only recently identified in GCTs. Further research is needed to fully understand the function of 6mA, which might differ if mediated by various modifiers. In general, it could be shown that 6mA is associated with pluripotency as well as differentiation, and therefore surely plays a role in GCT tumorigenesis.
It is generally accepted that the microenvironment is an important factor during TGCT formation. TDS certainly has an effect on the prevalence of TGCT but is most likely not the first step in line to tumor development. The TGCT microenvironment consists mostly of Sertoli, Leydig, and immune cells and the ECM, which can secrete pro-inflammatory chemokines, cytokines and interleukins, leading to the activation of signaling pathways, such as BMP, and eventually maintenance of pluripotency and/or reprogramming of TGCTs. Vice versa, inhibition of BMP signaling pathway can reprogram SEs to ECs via SOX2-dependent maintenance of pluripotency. Specifically, the interaction between SDF-1/CXCL12 and CXCR4 seems crucial during the migration, as well as colonization of PGCs giving rise to the development of TGCTs due to misguided and altered PGCs. In addition, we propose a mechanism for the development of mixed SE/NS GCTs (Fig. 2).
In conclusion, the switch of regular PGC development to different types of TGCTs is a process occurring in the short time-frame between murine E6.25-E13.5 and human Week 4–8. Nevertheless, many questions on the pathogenesis of GCTs remain and further research should be performed on the epigenetic and microenvironmental causes to GCT formation.
Author contributions
Study concept and design: Daniel Nettersheim.
Data acquisition: Melanie R. Müller, Margaretha A. Skowron.
Data analysis: Melanie R. Müller, Margaretha A. Skowron.
Drafting of manuscript: Melanie R. Müller, Margaretha A. Skowron, Peter Albers.
Critical revision of the manuscript: Daniel Nettersheim, Peter Albers.
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Second Military Medical University.
References
- 1.Oosterhuis J.W., Looijenga L.H.J. Human germ cell tumours from a developmental perspective. Nat Rev Canc. 2019;19:522–537. doi: 10.1038/s41568-019-0178-9. [DOI] [PubMed] [Google Scholar]
- 2.Palmer R.D., Foster N.A., Vowler S.L., Roberts I., Thornton C.M., Hale J.P. Malignant germ cell tumours of childhood: new associations of genomic imbalance. Br J Canc. 2007;96:667–676. doi: 10.1038/sj.bjc.6603602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fonseca A., Frazier A.L., Shaikh F. Germ cell tumors in adolescents and young adults. J Oncol Pract. 2019;15:433–441. doi: 10.1200/JOP.19.00190. [DOI] [PubMed] [Google Scholar]
- 4.Baraban E.G., Cooper K. Pathogenesis of testicular germ cell neoplasia: a conceptual approach. Adv Anat Pathol. 2019;26:241–245. doi: 10.1097/PAP.0000000000000233. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez-Vargas H., Sincic N., Ouzounova M., Herceg Z. Epigenetic signatures in stem cells and cancer stem cells. Epigenomics. 2009;1:261–280. doi: 10.2217/epi.09.19. [DOI] [PubMed] [Google Scholar]
- 6.Berney D.M., Looijenga L.H.J., Idrees M., Oosterhuis J.W., Rajpert-De Meyts E., Ulbright T.M. Germ cell neoplasia in situ (GCNIS): evolution of the current nomenclature for testicular pre-invasive germ cell malignancy. Histopathology. 2016;69:7–10. doi: 10.1111/his.12958. [DOI] [PubMed] [Google Scholar]
- 7.Oosterhuis J.W., Looijenga L.H.J. Testicular germ-cell tumours in a broader perspective. Nat Rev Canc. 2005;5:210–222. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi K., Sousa Lopes SMC de, Surani M.A. Germ cell specification in mice. Science. 2007;316:394–396. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- 9.Kristensen D.G., Skakkebæk N.E., Rajpert-De Meyts E., Almstrup K. Epigenetic features of testicular germ cell tumours in relation to epigenetic characteristics of foetal germ cells. Int J Dev Biol. 2013;57:309–317. doi: 10.1387/ijdb.130142ka. [DOI] [PubMed] [Google Scholar]
- 10.Suresh P.K. Mechanisms of pluripotency and epigenetic reprogramming in primordial germ cells: lessons for the conversion of other cell types into the stem cell lineage. Turk J Biol. 2015;39:187–193. [Google Scholar]
- 11.Kurimoto K., Yabuta Y., Ohinata Y., Shigeta M., Yamanaka K., Saitou M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mochizuki K., Tando Y., Sekinaka T., Otsuka K., Hayashi Y., Kobayashi H. SETDB1 is essential for mouse primordial germ cell fate determination by ensuring BMP signaling. Development. 2018;145:dev164160. doi: 10.1242/dev.164160. [DOI] [PubMed] [Google Scholar]
- 13.Killian J.K., Dorssers L.C.J., Trabert B., Gillis A.J.M., Cook M.B., Wang Y. Imprints and DPPA3 are bypassed during pluripotency- and differentiation-coupled methylation reprogramming in testicular germ cell tumors. Genome Res. 2016;26:1490–1504. doi: 10.1101/gr.201293.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson B.E., Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat Rev Mol Cell Biol. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckert D., Biermann K., Nettersheim D., Gillis A.J.M., Steger K., Jäck H.-M. Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev Biol. 2008;8:106. doi: 10.1186/1471-213X-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felici M de. Nuclear reprogramming in mouse primordial germ cells: epigenetic contribution. Stem Cell Int. 2011;2011:425863. doi: 10.4061/2011/425863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki H., Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y.C., Nicholls P.K., Soh Y.Q., Daniele J.R., Junker J.P., van Oudenaarden A. Licensing of primordial germ cells for gametogenesis depends on genital ridge signaling. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi T., Surani M.A. On the origin of the human germline. Development. 2018;145:dev150433. doi: 10.1242/dev.150433. [DOI] [PubMed] [Google Scholar]
- 20.Aksoy I., Jauch R., Chen J., Dyla M., Divakar U., Bogu G.K. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 2013;32:938–953. doi: 10.1038/emboj.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irie N., Weinberger L., Tang W.W.C., Kobayashi T., Viukov S., Manor Y.S. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nettersheim D., Jostes S., Sharma R., Schneider S., Hofmann A., Ferreira H.J. BMP inhibition in seminomas initiates acquisition of pluripotency via NODAL signaling resulting in reprogramming to an embryonal carcinoma. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sybirna A., Tang W.W.C., Pierson Smela M., Dietmann S., Gruhn W.H., Brosh R. A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat Commun. 2020;11 doi: 10.1038/s41467-020-15042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson Smela M., Sybirna A., Wong F.C.K., Surani M.A. Testing the role of SOX15 in human primordial germ cell fate. Wellcome Open Res. 2019;4:122. doi: 10.12688/wellcomeopenres.15381.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierce J.L., Frazier A.L., Amatruda J.F. Pediatric germ cell tumors: a developmental perspective. Adv Urol. 2018;2018:9059382. doi: 10.1155/2018/9059382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murrell A. Genomic imprinting and cancer: from primordial germ cells to somatic cells. Sci World J. 2006;6:1888–1910. doi: 10.1100/tsw.2006.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackett J.A., Huang Y., Günesdogan U., Gretarsson K.A., Kobayashi T., Surani M.A. Tracing the transitions from pluripotency to germ cell fate with CRISPR screening. Nat Commun. 2018;9:4292. doi: 10.1038/s41467-018-06230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fustino N., Rakheja D., Ateek C.S., Neumann J.C., Amatruda J.F. Bone morphogenetic protein signalling activity distinguishes histological subsets of paediatric germ cell tumours. Int J Androl. 2011;34:e218–e233. doi: 10.1111/j.1365-2605.2011.01186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bejarano I., Rodríguez A.B., Pariente J.A. Apoptosis is a demanding selective tool during the development of fetal male germ cells. Front Cell Dev Biol. 2018;6 doi: 10.3389/fcell.2018.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skakkebaek N.E., Holm M., Hoei-Hansen C., Jørgensen N., Rajpert-De Meyts E. Association between testicular dysgenesis syndrome (TDS) and testicular neoplasia: evidence from 20 adult patients with signs of maldevelopment of the testis. APMIS. 2003;111:1–11. doi: 10.1034/j.1600-0463.2003.11101031.x. [DOI] [PubMed] [Google Scholar]
- 31.Skakkebaek N.E., Rajpert-De Meyts E., Main K.M. Testicular dysgenesis syndrome: an increasingly common developmental disorder with environmental aspects. Hum Reprod. 2001;16:972–978. doi: 10.1093/humrep/16.5.972. [DOI] [PubMed] [Google Scholar]
- 32.Sonne S.B., Kristensen D.M., Novotny G.W., Olesen I.A., Nielsen J.E., Skakkebaek N.E. Testicular dysgenesis syndrome and the origin of carcinoma in situ testis. Int J Androl. 2008;31:275–287. doi: 10.1111/j.1365-2605.2007.00855.x. [DOI] [PubMed] [Google Scholar]
- 33.Hoei-Hansen C.E., Holm M., Rajpert-De Meyts E., Skakkebaek N.E. Histological evidence of testicular dysgenesis in contralateral biopsies from 218 patients with testicular germ cell cancer. J Pathol. 2003;200:370–374. doi: 10.1002/path.1372. [DOI] [PubMed] [Google Scholar]
- 34.Chen C., Ouyang W., Grigura V., Zhou Q., Carnes K., Lim H. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–1034. doi: 10.1038/nature03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szarek M., Bergmann M., Konrad L., Schuppe H.-C., Kliesch S., Hedger M.P. Activin A target genes are differentially expressed between normal and neoplastic adult human testes: clues to gonocyte fate choice. Andrology. 2019;7:31–41. doi: 10.1111/andr.12553. [DOI] [PubMed] [Google Scholar]
- 36.Dorsam R.T., Gutkind J.S. G-protein-coupled receptors and cancer. Nat Rev Canc. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 37.Young J.C., Jaiprakash A., Mithraprabhu S., Itman C., Kitazawa R., Looijenga L.H.J. TCam-2 seminoma cell line exhibits characteristic foetal germ cell responses to TGF-beta ligands and retinoic acid. Int J Androl. 2011;34:e204–e217. doi: 10.1111/j.1365-2605.2011.01170.x. [DOI] [PubMed] [Google Scholar]
- 38.de Jong J., Stoop H., Gillis A.J., van Gurp R.J., van de Geijn G.J., Boer Md. Differential expression of SOX17 and SOX2 in germ cells and stem cells has biological and clinical implications. J Pathol. 2008;215:21–30. doi: 10.1002/path.2332. [DOI] [PubMed] [Google Scholar]
- 39.Weber S., Eckert D., Nettersheim D., Gillis A.J., Schäfer S., Kuckenberg P. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod. 2010;82:214–223. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- 40.Nicholls P.K., Schorle H., Naqvi S., Hu Y.C., Fan Y., Carmell M.A. Mammalian germ cells are determined after PGC colonization of the nascent gonad. Proc Natl Acad Sci U S A. 2019;116:25677–25687. doi: 10.1073/pnas.1910733116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 42.Nettersheim D., Jostes S., Schneider S., Schorle H. Elucidating human male germ cell development by studying germ cell cancer. Reproduction. 2016;152:R101–R113. doi: 10.1530/REP-16-0114. [DOI] [PubMed] [Google Scholar]
- 43.Leitch H.G., Smith A. The mammalian germline as a pluripotency cycle. Development. 2013;140:2495–2501. doi: 10.1242/dev.091603. [DOI] [PubMed] [Google Scholar]
- 44.Nettersheim D., Heimsoeth A., Jostes S., Schneider S., Fellermeyer M., Hofmann A. SOX2 is essential for in vivo reprogramming of seminoma-like TCam-2 cells to an embryonal carcinoma-like fate. Oncotarget. 2016;7:47095–47110. doi: 10.18632/oncotarget.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jostes S.V., Fellermeyer M., Arévalo L., Merges G.E., Kristiansen G., Nettersheim D. Unique and redundant roles of SOX2 and SOX17 in regulating the germ cell tumour fate. Int J Canc. 2019;146:1592–1605. doi: 10.1002/ijc.32714. [DOI] [PubMed] [Google Scholar]
- 46.Kojima Y., Sasaki K., Yokobayashi S., Sakai Y., Nakamura T., Yabuta Y. Evolutionarily distinctive transcriptional and signaling programs drive human germ cell lineage specification from pluripotent stem cells. Cell Stem Cell. 2017;21:517–532.e5. doi: 10.1016/j.stem.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 47.Lobo J., Henrique R., Jerónimo C. The role of DNA/histone modifying enzymes and chromatin remodeling complexes in testicular germ cell tumors. Cancers (Basel) 2018;11:E6. doi: 10.3390/cancers11010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Netto G.J., Nakai Y., Nakayama M., Jadallah S., Toubaji A., Nonomura N. Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol. 2008;21:1337–1344. doi: 10.1038/modpathol.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nettersheim D., Heukamp L.C., Fronhoffs F., Grewe M.J., Haas N., Waha A. Analysis of TET expression/activity and 5mC oxidation during normal and malignant germ cell development. PloS One. 2013;8 doi: 10.1371/journal.pone.0082881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smiraglia D.J., Szymanska J., Kraggerud S.M., Lothe R.A., Peltomäki P., Plass C. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene. 2002;21:3909–3916. doi: 10.1038/sj.onc.1205488. [DOI] [PubMed] [Google Scholar]
- 51.Lind G.E., Skotheim R.I., Lothe R.A. The epigenome of testicular germ cell tumors. APMIS. 2007;115:1147–1160. doi: 10.1111/j.1600-0463.2007.apm_660.xml.x. [DOI] [PubMed] [Google Scholar]
- 52.Costa A.L., Moreira-Barbosa C., Lobo J., Vilela-Salgueiro B., Cantante M., Guimarães R. DNA methylation profiling as a tool for testicular germ cell tumors subtyping. Epigenomics. 2018;10:1511–1523. doi: 10.2217/epi-2018-0034. [DOI] [PubMed] [Google Scholar]
- 53.Lobo J., Gillis A.J.M., Jerónimo C., Henrique R., Looijenga L.H.J. Human germ cell tumors are developmental cancers: impact of epigenetics on pathobiology and clinic. Int J Mol Sci. 2019;20:E258. doi: 10.3390/ijms20020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji P., Wang X., Xie N., Li Y. N6-Methyladenosine in RNA and DNA: an epitranscriptomic and epigenetic player implicated in determination of stem cell fate. Stem Cell Int. 2018;2018:3256524. doi: 10.1155/2018/3256524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nettersheim D., Berger D., Jostes S., Kristiansen G., Lochnit G., Schorle H. N6-Methyladenosine detected in RNA of testicular germ cell tumors is controlled by METTL3, ALKBH5, YTHDC1/F1/F2, and HNRNPC as writers, erasers, and readers. Andrology. 2019;7:498–506. doi: 10.1111/andr.12612. [DOI] [PubMed] [Google Scholar]
- 56.Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lobo J., Costa A.L., Cantante M., Guimarães R., Lopes P., Antunes L. m6A RNA modification and its writer/reader VIRMA/YTHDF3 in testicular germ cell tumors: a role in seminoma phenotype maintenance. J Transl Med. 2019;17 doi: 10.1186/s12967-019-1837-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oliveros-Etter M., Li Z., Nee K., Hosohama L., Hargan-Calvopina J., Lee S.A. PGC reversion to pluripotency involves erasure of DNA methylation from imprinting control centers followed by locus-specific re-methylation. Stem Cell Reports. 2015;5:337–349. doi: 10.1016/j.stemcr.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 62.Ara T., Nakamura Y., Egawa T., Sugiyama T., Abe K., Kishimoto T. Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1) Proc Natl Acad Sci U S A. 2003;100:5319–5323. doi: 10.1073/pnas.0730719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doitsidou M., Reichman-Fried M., Stebler J., Köprunner M., Dörries J., Meyer D. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–659. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 64.Molyneaux K.A., Zinszner H., Kunwar P.S., Schaible K., Stebler J., Sunshine M.J. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–4286. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- 65.Li H., Liang R., Lu Y., Wang M., Li Z. RTN3 regulates the expression level of chemokine receptor CXCR4 and is required for migration of primordial germ cells. Int J Mol Sci. 2016;17:382. doi: 10.3390/ijms17040382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilbert D.C., Chandler I., McIntyre A., Goddard N.C., Gabe R., Huddart R.A. Clinical and biological significance of CXCL12 and CXCR4 expression in adult testes and germ cell tumours of adults and adolescents. J Pathol. 2009;217:94–102. doi: 10.1002/path.2436. [DOI] [PubMed] [Google Scholar]
- 67.Stebler J., Spieler D., Slanchev K., Molyneaux K.A., Richter U., Cojocaru V. Primordial germ cell migration in the chick and mouse embryo: the role of the chemokine SDF-1/CXCL12. Dev Biol. 2004;272:351–361. doi: 10.1016/j.ydbio.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Lei T., Moos S., Klug J., Aslani F., Bhushan S., Wahle E. Galectin-1 enhances TNFα-induced inflammatory responses in Sertoli cells through activation of MAPK signalling. Sci Rep. 2018;8:3741. doi: 10.1038/s41598-018-22135-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klein B., Haggeney T., Fietz D., Indumathy S., Loveland K.L., Hedger M. Specific immune cell and cytokine characteristics of human testicular germ cell neoplasia. Hum Reprod. 2016;31:2192–2202. doi: 10.1093/humrep/dew211. [DOI] [PubMed] [Google Scholar]
- 70.Alvarez-Buylla A., Merchant-Larios H. Mouse primordial germ cells use fibronectin as a substrate for migration. Exp Cell Res. 1986;165:362–368. doi: 10.1016/0014-4827(86)90590-2. [DOI] [PubMed] [Google Scholar]
- 71.Sun X., Park C.B., Deng W., Potter S.S., Dey S.K. Uterine inactivation of muscle segment homeobox (Msx) genes alters epithelial cell junction proteins during embryo implantation. Faseb J. 2016;30:1425–1435. doi: 10.1096/fj.15-282798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spiller C.M., Feng C.-W., Jackson A., Gillis A.J.M., Rolland A.D., Looijenga L.H.J. Endogenous Nodal signaling regulates germ cell potency during mammalian testis development. Development. 2012;139:4123–4132. doi: 10.1242/dev.083006. [DOI] [PubMed] [Google Scholar]
- 73.Nettersheim D., Vadder S., Jostes S., Heimsoeth A., Schorle H. TCam-2 cells deficient for SOX2 and FOXA2 are blocked in differentiation and maintain a seminoma-like cell fate in vivo. Cancers (Basel) 2019;11:E728. doi: 10.3390/cancers11050728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nonaka D. Differential expression of SOX2 and SOX17 in testicular germ cell tumors. Am J Clin Pathol. 2009;131:731–736. doi: 10.1309/AJCP7MNCNBCRN8NO. [DOI] [PubMed] [Google Scholar]
- 75.Nettersheim D., Gillis A.J., Looijenga L.H., Schorle H. TGF-β1, EGF and FGF4 synergistically induce differentiation of the seminoma cell line TCam-2 into a cell type resembling mixed non-seminoma. Int J Androl. 2011;34:e189–e203. doi: 10.1111/j.1365-2605.2011.01172.x. [DOI] [PubMed] [Google Scholar]