Abstract
The majority of non-melanoma skin cancer (NMSC) is cutaneous basal cell carcinoma (BCC) or squamous cell carcinoma (SCC), which are also called keratinocyte carcinomas, as both of them originate from keratinocytes. The incidence of keratinocyte carcinomas is over 5 million per year in the US, three-fold higher than the total incidence of all other types of cancer combined. While there are several reports on gene expression profiling of BCC and SCC, there are significant variations in the reported gene expression changes in different studies. One reason is that tumor-adjacent normal skin specimens were not included in many studies as matched controls. Furthermore, while numerous studies of skin stem cells in mouse models have been reported, their relevance to human skin cancer remains unknown. In this report, we analyzed gene expression profiles of paired specimens of keratinocyte carcinomas with their matched normal skin tissues as the control. Among several novel findings, we discovered a significant number of zinc finger encoding genes up-regulated in human BCC. In BCC, a novel link was found between hedgehog signaling, Wnt signaling, and the cilium. While the SCC cancer-stem-cell gene signature is shared between human and mouse SCCs, the hair follicle stem-cell signature of mice was not highly represented in human SCC. Differential gene expression (DEG) in human BCC shares gene signature with both bulge and epidermal stem cells. We have also determined that human BCCs and SCCs have distinct gene expression patterns, and some of them are not fully reflected in current mouse models.
Keywords: Basal cell carcinoma, Gli1, Hedgehog signaling, PTCH1, Squamous cell carcinoma
Introduction
Keratinocyte carcinomas, which encompass cutaneous squamous cell carcinoma (SCC) and basal cell carcinoma (BCC), are the most common malignancies in Caucasians, with continually increasing incidence worldwide.1 UV irradiation via excessive sun exposure is the most significant risk factor for both SCC2,3 and BCC,4,5 and most gene mutations have UV signatures. Additionally, organ transplantation patients have a significantly higher incidence of SCC (>100 times than the general population) and an elevated incidence of BCC (>10 times more than the general population).6, 7, 8 Although SCC and BCC both originate from keratinocytes, they are very distinct in several aspects. While BCC is known to be caused by uncontrolled activated hedgehog signaling, SCC has alterations in many genes, including Notch gene families, RAS and its downstream effectors, and Cyclin-Dependent Kinase Inhibitor 2A (CDKN2A). Mutations of TP53 are found in both BCC and SCC, and those mutations show an UV irradiation signature.8,9
The link between hedgehog signaling and BCC came from studies of a rare genetic disorder of the skin, basal cell nevus syndrome, initially through genetic linkage analysis followed by direct mutation analysis of gene Patched 1 (PTCH1).10,11 Further studies revealed frequent mutations of the hedgehog signaling molecules in sporadic BCC, including PTCH1 and Smoothened (SMO).12, 13, 14 Inactivated mutations of the hedgehog ligand-receptor, PTCH1, or gain-of-function mutations of SMO all lead to constitutive activation of the hedgehog pathway. Currently, two SMO antagonists, vismodegib15, 16, 17 and sonidegib,18, 19, 20 have been approved to treat locally advanced and metastatic BCCs. Despite the fact that almost all BCCs have activated hedgehog signaling through gene mutations of PTCH1 and SMO, less than 40% of tumors in sporadic BCCs are responsive to SMO antagonists, and the underlying mechanisms are currently unknown. It is hoped that the transcriptome of BCC may reveal why some BCCs are quite responsive to SMO antagonists while others are nonresponsive.
In contrast to BCC, treatment of SCC with chemotherapy or radiation often yields low and unsatisfactory responses. There are several clinical trials with promising results in SCC,21, 22, 23, 24 including EGFR inhibitor-based trials and immune therapy using PD-1 or PD-L1 antibodies. One major challenge for such trials is identifying the right patient population. Further analyses of gene expression profiling in individual SCC tumors may identify biomarkers for SCC treatment.
Thus, transcriptome data on BCC and SCC are currently badly needed, both for understanding the biology of keratinocyte carcinomas and for designing strategies for effective treatment. While several groups have reported gene expression profiles of human BCCs and SCCs, only a few studies have compared gene expression between cancerous skin specimens and corresponding adjacent normal skin using microarray technology.25, 26, 27, 28, 29 To our knowledge, no RNA-seq data were generated from non-melanoma skin tumors and the matched adjacent normal specimens in The Cancer Genome Atlas (TCGA) database. A previous systematic comparison of several microarray-based gene expression analyses did not reveal consensus genes, and individual differences contributed significantly to this issue.30 In addition, tumor contents vary significantly. Comparing the transcriptome data of BCC and SCC will enable us to understand the differences and the similarities between BCCs and SCCs.
Several mouse models have been generated to study skin stem cell biology and the cancer biology of SCC and BCC. While these model systems provide an insightful understanding of skin cancer biology, the relevance of these studies to human tumors is currently unclear. Genetic lineage tracing in the mouse model has generated a wealth of information about the specific subpopulations of cells that contribute to tissue homeostasis and cancer, particularly in intestine, skin, and bone marrow.31, 32, 33, 34, 35, 36, 37 In the skin, there are several stem cell pools, including hair follicle bulge and inter-follicular epidermal stem cells. It is proposed that BCC preferentially arises from stem cells within a hair follicle or a region within the inter-follicular epidermis called touch dome epithelia.5,38, 39, 40 As predicted from the mouse models, squamous cell carcinomas (SCCs), on the other hand, can arise from inter-follicular epidermal stem cells, hair follicle stem cells, or both.41, 42, 43 Currently, it is not known whether the gene signatures generated in mice match those in the corresponding human tumor type.
In this study, we analyzed the transcriptomes of 25 paired BCCs and 10 paired SCCs. Through comparison of tumor tissues with the adjacent normal skin tissues, we identified tumor-specific gene expression alterations as well as signaling networks in BCC and SCC. We also compared BCC with SCC by their genome-wide transcriptome patterns and corresponding biological functions. Furthermore, we assessed the enrichment of gene expression signatures from mouse models in human cancer specimens.
Materials and methods
Human specimens and tissue processing
The Institutional Review Board approved this study at Indiana University School of Medicine, Indianapolis, IN. Each patient provided written informed consent. Skin specimens (cancerous and adjacent normal skin tissues) were collected in the operating room, quick-frozen in liquid nitrogen, and stored in an 80゜C freezer. For tissue processing, each specimen was divided into three portions: one for haemotoxylin and eosin (H&E) staining, one for RNA extraction, and one for backup. The portion for H&E staining was first fixed in formalin for 24 h, then processed for paraffin embedding in a Leica TP1020 tissue processor. Paraffin-embedded tissues were sectioned at 5 μm for H&E staining, which was used to determine the tumor contents in the specimens using Image J software, and only tumor tissues with high tumor content (>30% of the tissue area as shown in H&E images) proceeded to gene expression analysis. Matched normal specimens were collected to contain epidermis (Supplementary Fig. 1).
RNA extraction
Total RNAs were extracted using TRI@ reagent.44 For transcriptomic analysis, the RNA quality in each specimen was assured by the Agilent Bioanalyzer profiles before moving on to cDNA synthesis and library construction (service provided by the Broad Institute).
RNA-seq analysis
More than 95% of samples had total raw reads over 65 million (see Supplementary Table 1 for details). The raw sequencing data were processed with one of the most popular RNA-seq workflows, Tophat-HTSeq. The sequencing reads were aligned to the human genome (hg19) by Tophat.45 The mean percentage of mapped reads reached about 81%, with an average of 51 million reads aligned in pairs. We then used HTSeq-count46 to summarize transcript counts from uniquely mapped reads based on the reference transcriptome profile (Homo_sapiens.GRCh37.73.gtf). The gene-level counts were transformed by a base-2 logarithmic scale on read counts per million (CPM) for normalization. Those genes with average CPM less than 1.0 for all four groups of samples, BCC normal/tumor, and SCC normal/tumor, were removed from the differentiation analysis. The paired t-test was performed between tumors and matched normal specimens for BCC and SCC. All p-values were modified by multiple testing correction via a false discovery rate (FDR) estimation. Differentially expressed genes (DEGs) were identified according to the cutoffs chosen, |log2FC| > 1 and FDR < 0.01 for BCC, or |log2FC| > 1 and FDR < 0.05 for SCC.
Real-time PCR
Real-time quantitative PCR analyses were performed according to a previously published procedure.47 The CT values were analyzed in Microsoft Excel using the comparative CT (ΔΔCT) method as described by the manufacturer (Applied Biosystems, Foster City, CA). The amount of target (2−ΔΔCT) in the tumor was calculated by normalization to an endogenous reference (Gapdh for mice and GAPDH for humans), and relative to the value of the adjacent normal skin specimen. All TaqMan primers and probes were purchased from Applied Biosystems Inc. For each target gene, four pairs of tumors, either BCC or SCC, and matched adjacent normal skin were used for gene expression analyses to validate the RNA-sequencing data.
GO and KEGG pathway analysis
The webserver, DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/home.jsp), was used for GO and KEGG pathway analysis. We compared the occurrences of GO terms and KEGG pathways in the foreground gene, e.g., DEG up-regulated in BCCs, to the background, e.g., all expressed genes in our RNA-seq dataset. The significantly over-represented GO terms and KEGG pathways were determined by the cutoff of FDR-adjusted P < 0.05.
Statistical enrichment analysis
The statistical significance of an overlap between two gene sets was evaluated by the fold enrichment (F.E.) and the p-value. p-value was calculated based on a hypergeometric model.
Results
Identification of differentially expressed genes (DEGs) in BCC and SCC
We selected 35 paired specimens, with tumor samples and the adjacent normal skin tissues, from 126 skin cancer patients of the Indiana University Dermatology Clinics. The rationale for selecting patient samples instead of using all specimens is two-fold. First, we want to reduce individual differences among patients, for which we performed a paired comparison between each tumor specimen and the surrounding matched normal skin. Second, because we want to reduce the noise of gene expression from non-cancerous cells, we used ImageJ analyses of H&E images to determine the tumor contents in the specimens (see Method). We exclude those specimens that do not have matched normal skin, or the adjacent skin tissue has cancer cells from the study. We anticipate that if tumor content is less than 30%, the influence of non-cancerous cells will be significant. Only specimens with tumor content >30% were used for further analyses. As shown in Fig. S1, all skin specimens included epidermis and dermis. The normal tissue was the skin tissue adjacent to the tumor but without any tumor cells (as assessed by H&E staining). These 35 paired human tumor specimens included 25 paired BCCs and 10 paired SCCs. RNA-seq technology was adopted to examine the gene expression profiles of all 70 specimens.
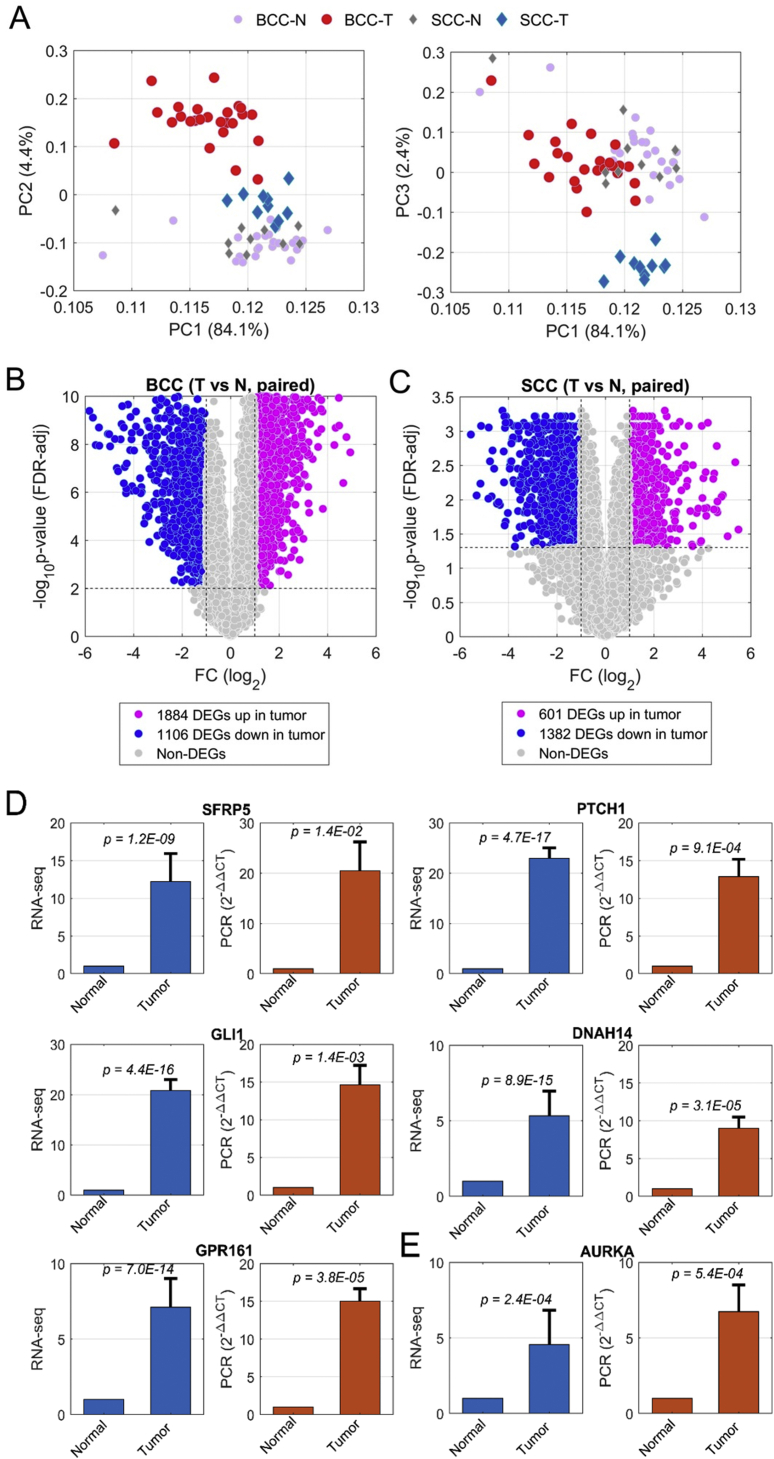
First, we performed several quality-control analyses on RNA-seq data of 35 pairs of specimens. Principal component analysis (PCA) was used to evaluate the relationship between these 70 samples (Fig. 1A). The first three principal components (PCs) together explain about 91% of the total variance. The PCA plots (Fig. 1A) show substantial similarity between most normal samples, regardless of whether they were gathered from BCC or SCC patients. Significant differences between tumor and normal samples were observed in the comparisons of PC1 and PC2, or PC1 and PC3.
Figure 1.
Differentially expressed genes (DEGs) identified in human BCC and SCC by comparison between tumor and adjacent normal skin. (A) shows principal component analysis (PCA) of BCC (PC2, left) and SCC (PC3, right) (including both tumors and matched adjacent normal skin). (B) and (C) show volcano plots for BCC (B) and SCC (C), where the purple dots represent the DEGs up-regulated and blue dots being those down-regulated. (D) and (E) show gene expression profiles achieved by RNA-seq (blue bars in the left panel) and validated by real-time PCR (brown bars in the right panel) of several DEG examples for BCC (D) and SCC (E).
Furthermore, notable disparities between SCC and BCC in the human tumor specimens confirm that gene expression profiles differ between the two cancer types. Such differences were also reflected by the correlations on the fold-change (FC) of tumor vs. normal among 35 patients. Higher correlation coefficients were observed within either the BCC or the SCC group, whereas the correlations across BCC and SCC were lower (Fig. S2). These results also indicate the high quality of our RNA-seq data.
Next, differential expression was analyzed by paired comparison between normal and tumor specimens (either SCC or BCC), regardless of gender identity alongside other patient information. Differentially expressed genes (DEGs) were identified based on defined cutoff values. For BCC, if one gene had an amplitude of fold change (FC) larger than 1 (in log scale with base 2, corresponding to linear 2-fold) and P-value less than 0.01 after multiple test correction via False Discovery Rate (FDR) estimation, the gene was identified as DEG (Fig. 1B). Considering the sample size for SCC, which is only 40% of BCC numbers, we chose cutoffs, |log2FC| > 1 and FDR-adjusted P-value < 0.05, to mark the DEGs (Fig. 1C). In summary, we found 1884 up-regulated and 1106 down-regulated genes in BCC tumors in comparison with matched normal specimens (Fig. 1B and Table S2). In SCC tumors, on the other hand, we found 601 up-regulated and 1382 down-regulated DEGs (Fig. 1C and Table S2). Several genes of interest, some of which are implicated in the Wnt or the hedgehog signaling pathway, were selected to validate gene expression for both BCC and SCC by analysis of real-time PCR results (Fig. 1D). For example, Secreted Frizzled Related Protein 5 (SFRP5) is a protein involved in the Wnt signaling pathway that was highly expressed in BCC tumors, as shown in both RNA-seq and real-time PCR (Fig. 1D).
Similarly, the pattern of hedgehog-signaling target gene expression (e.g., PTCH1) indicates elevated hedgehog signaling in BCC. GLI Family Zinc Finger 1 (GLI1), a critical transcription factor (TF) in hedgehog signaling, was significantly up-regulated in BCC (Fig. 1D). Elevated gene expression in BCC tumor was also observed on cilium assembly genes, such as ciliary Dynein Axonemal Heavy Chain 14 (DNAH14), associated with microtubule motor activity and G Protein-Coupled Receptor 161 (GPR161) (Fig. 1D). In SCC, elevated expression of Aurora Kinase A (AURKA), which is involved in the cell cycle,48 was observed (Fig. 1E). These gene expression changes were further confirmed by real-time PCR (Fig. 1D and E).
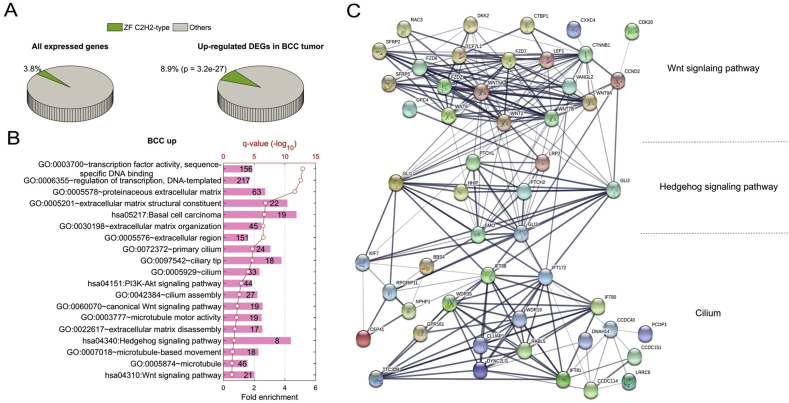
One significant discovery in our analysis is the elevated expression of a significant number of C2H2-type Zinc finger encoding genes. C2H2-type Zinc Finger (ZF) proteins form a large transcription-factor family across the whole genome.49 In this study, 628 out of 16382 expressed genes (3.8%) belong to C2H2-type ZF (Fig. 2A). Surprisingly, about one-quarter of C2H2-ZF (168) proteins were found in the group of 1884 elevated DEGs in BCC tumors. For instance, GLI1, which contains C2H2-ZF domains, was up-regulated in BCC (Fig. 1D). The ratio of 8.9% was much higher than its proportion in all expressed genes (3.8%), reflected by 2.3-fold enrichment (F.E.) with p-value 3.2 × 10−27 (Fig. 2A and Fig. S3A). When we focused on 156 and 217 up-regulated transcription factors in BCC implicated in transcription activity associated with sequence-specific DNA binding (GO:0003700) and regulation of transcription (GO:0006355) as seen in Fig. 2B, we found that 97 (62.2%, p = 1.7 × 10−98) and 148 (68.2%, p = 1.8 × 10−161) TFs have C2H2-ZF domains (Fig. S3A), indicating a critical role of C2H2-ZF transcription regulation in BCC. In contrast, only 16 out of 1106 (1.4%) repressed genes in BCC have a C2H2-type domain (Fig. S3A and B). Significant enrichment of C2H2-type ZF protein was not observed in neither up- nor down-regulation of SCC (Fig. S3A and B), suggesting a BCC-specific phenotype associated with regulation of these C2H2-ZF proteins for BCC tumor formation. It is known that C2H2-type ZF proteins directly bind specific DNA sequences in the genome, involving skin homeostasis regulation. Structural analyses indicate that the majority of the C2H2-type zinc finger molecules up-regulated in BCC contain a KRAB domain.50 Previous work indicates that KRAB domain proteins are critical for the regulation of embryonic development, cell differentiation, and cell cycle regulation. The exact mechanisms for selective up-regulation of KRAB domain-containing molecules in BCC are currently unknown.
Figure 2.
Up-regulated DEGs in BCC. (A) shows a 3-D pie chart for the ratio of C2H2-ZF proteins in all expressed genes (left) or DEGs up-regulated in BCC tumor (right). (B) shows top Gene Ontology (GO) terms and KEGG pathways significantly enriched in up-regulated DEGs in BCC. (C) shows a diagram of protein–protein interactions (PPIs) of DEGs implicated in the Wnt signaling pathway, Hedgehog signaling pathway, and Cilium components.
Novel signaling networks in BCC and SCC
We used DAVID51 to perform functional enrichment analysis on different groups of DEGs. Several Gene Ontology (GO) functions and KEGG pathways were found to be directly associated with the up-regulation of 1884 DEGs in BCC tumors (Fig. 2B). It is not surprising that 19 out of 46 genes annotated in the pathway of basal cell carcinoma were activated in our human BCC cohort, including known hedgehog target genes GLI1, Hedgehog Interacting Protein (HHIP), and PTCH1. Our analyses also uncovered several new alterations in BCCs. First, many pathways associated with transcription activity were up-regulated in BCC tumors, including transcription regulation (GO:0003700 and GO:0006355, see Fig. 2B). Also, expression of genes encoding extracellular matrix (GO:0005578, GO:0005576, GO:0005201, and GO:0022617) and developmental signaling pathways (has04340, hsa 04310, and GO:0060070) was elevated in BCC (see Fig. 2B). Many prominent up-regulated DEGs (Fig. 1B and D) were implicated in the Wnt signaling pathway, the Hedgehog signaling pathway, and cilium. It is known that cilium is a critical structure for hedgehog and Wnt signaling.52,53 For example, cilium is required for signal transduction from Smoothened to Gli transcription factors, and lack of cilium prevents hedgehog signaling.54,55 When examining the connections among these three pathways in BCC, we found that hedgehog signaling plays a central role in terms of protein–protein interaction (PPI) network on up-regulated DEGs. Hedgehog signaling is connected with Wnt signaling and cilium components at multiple points, whereas Wnt signaling has little connection with cilium components (Fig. 2C). For example, another C2H2-type ZF protein, GLI Family Zinc Finger 2 (GLI2), involved in hedgehog signaling, interacts with the Wnt pathway through PPI with Cyclin D2 (CCND2), while being connected with other molecules in Cilium, including RPGR Interacting Protein 1-Like (RPGRIP1L), Kinesin Family Member 7 (KIF7), IFT family genes. This functional analysis of up-regulated DEGs in BCC further confirms the existence of the hedgehog-Wnt signaling network and the significance of cilium for hedgehog signaling.
Transcriptome comparison between BCC and SCC
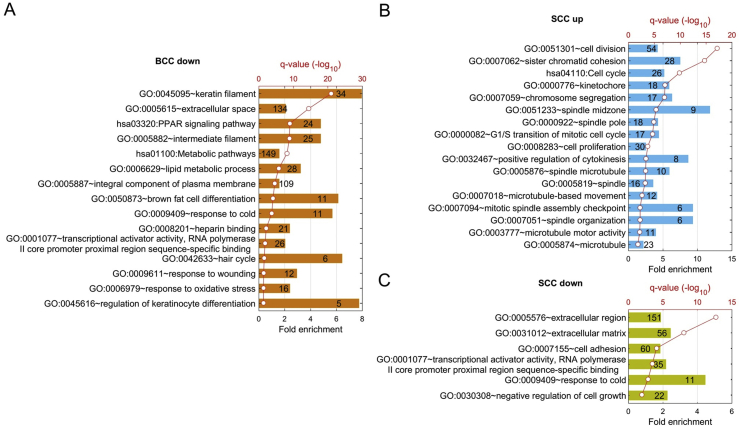
Through a paired comparison between BCC and SCC tumors and normal specimens, we generated a list of DEGs that are either up-regulated or down-regulated. The different sets of DEGs may reflect commonalities between BCC and SCC, or tumor-specific changes and their corresponding biological functions. For example, as shown in Fig. 3A, 1106 BCC down-regulated genes were significantly over-represented in genes encoding for proteins for keratin filament (GO:0045095), epithelial morphology (GO:0005615, GO:0005887, and GO:0005882), keratinocyte differentiation (GO:0045615), hair follicle (GO:0042633), and reduced lipid metabolism (such as lipid metabolic process GO:0006629), suggesting reduced cell differentiation in BCC (see Fig. 3A). In SCC, significantly up-regulated genes encode proteins for cell division (GO:0051301, GO:0007062), cell cycle (hsa 04110), spindle organization (GO:0051233, GO:0000922 and GO0005819), and microtubule-related biological processing (GO:0000776, GO:0007059 and GO:0032467) (Fig. 3B). Consistently, genes associated with negative regulation of cell growth (GO:0030308), cell adhesion (GO:0007155), RNA polymerase II core promoter proximal region sequence-specific binding activity (GO:0001077), or extracellular matrix (GO:0005576, GO:0031012) were notably suppressed in SCC (Fig. 3C). This finding is consistent with increased cell proliferation in SCC.
Figure 3.
Functional enrichment analysis of different sets of DEGs. (A) shows top GO terms and KEGG pathways from down-regulated genes in BCC; (B) shows pathways from up-regulated genes in SCC, and (C) shows pathways from down-regulated genes in SCC.
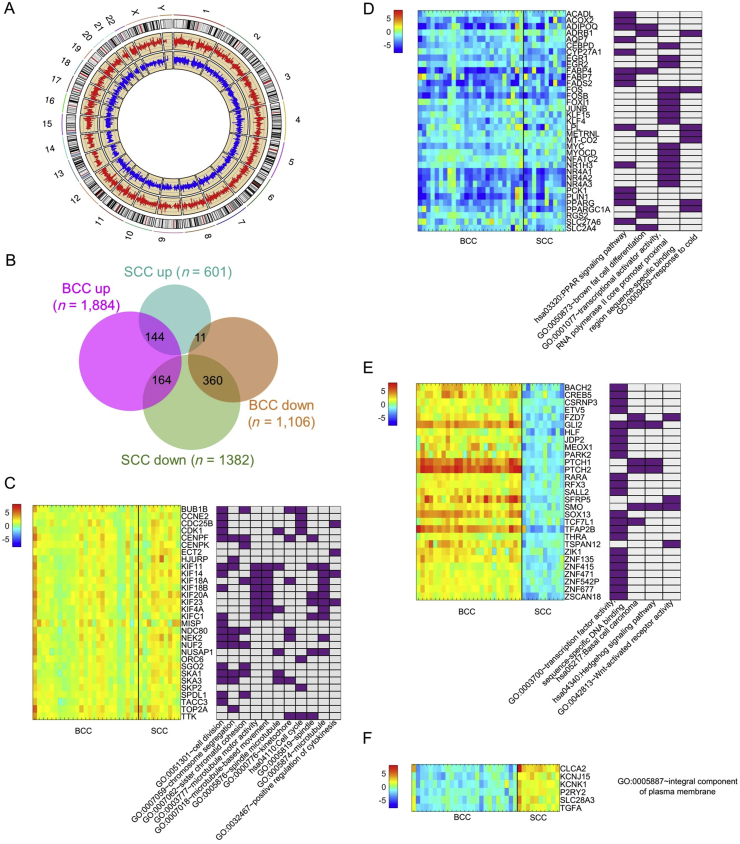
BCC and SCC had distinct transcriptome patterns in general (Fig. 4A). The Circos plot56 in Fig. 4A displays the average FCs between tumors and matched normal specimens for BCC (red) and SCC (blue). DEGs identified from BCC were quite different from those of SCC. However, we did identify 679 genes as shared DEGs between both BCC and SCC (Fig. S4) regardless of the direction of gene expression change. Among them, 144 DEGs were elevated and 360 DEGs were suppressed in both BCC and SCC (Fig. 4B). These overlaps were statistically significant, in that they had fold enrichments of 2.1 and 3.9 compared with random selection, with P = 1.1 × 10−18 and 2.4 × 10−128, respectively. Only 11 genes were down-regulated in BCC but up-regulated in SCC. The fold enrichment was very low, 0.27, with P = 9.0 × 10−9. It thus appears that there are more overlaps between BCC and SCC among up-regulated than among down-regulated genes.
Figure 4.
Cross-comparison of gene expression profiles between BCC and SCC. (A) shows the Circos plot of average fold change (FC) between tumor and normal specimens for BCC (red) and SCC (blue), respectively. (B) shows the Venn diagram of numbers of DEGs in BCC and SCC and the numbers of DEGs that overlap. (C–F) shows selected DEGs and their associated GO functions. (C) shows up-regulated DEGs in BCC and SCC; (D) shows down-regulated DEGs in both BCC and SCC; (E) shows DEGs up-regulated in BCC but down-regulated in SCC, and (F) shows DEGs down-regulated in BCC but up-regulated in SCC.
We also found distinct functions for genes with similar expression patterns in both BCC and SCC. For example, we found that genes up-regulated in both BCC and SCC tumors tend to be involved in cell division, cell cycle, chromosome segregation, and cytokinesis regulation, e.g., Cyclin-Dependent Kinase 1 (CDK1), TTK Protein Kinase (TTK) (Fig. 4C). In contrast, genes suppressed in both BCC and SCC, e.g., Peroxisome Proliferator-Activated Receptor Gamma (PPARG), are associated with metabolism (Fig. 4D). As expected, some BCC-specific genes, e.g., GLI2, PTCH1/2, Frizzled Class Receptor 7 (FZD7), were high in BCC but suppressed in SCC (Fig. 4E). Of the 11 genes down-regulated in BCC but up-regulated in SCC, six are integral components of plasma membrane proteins (Fig. 4F), whose roles in the development of BCC and SCC are not yet understood.
The relevance of mouse model-generated data to human BCCs and SCCs
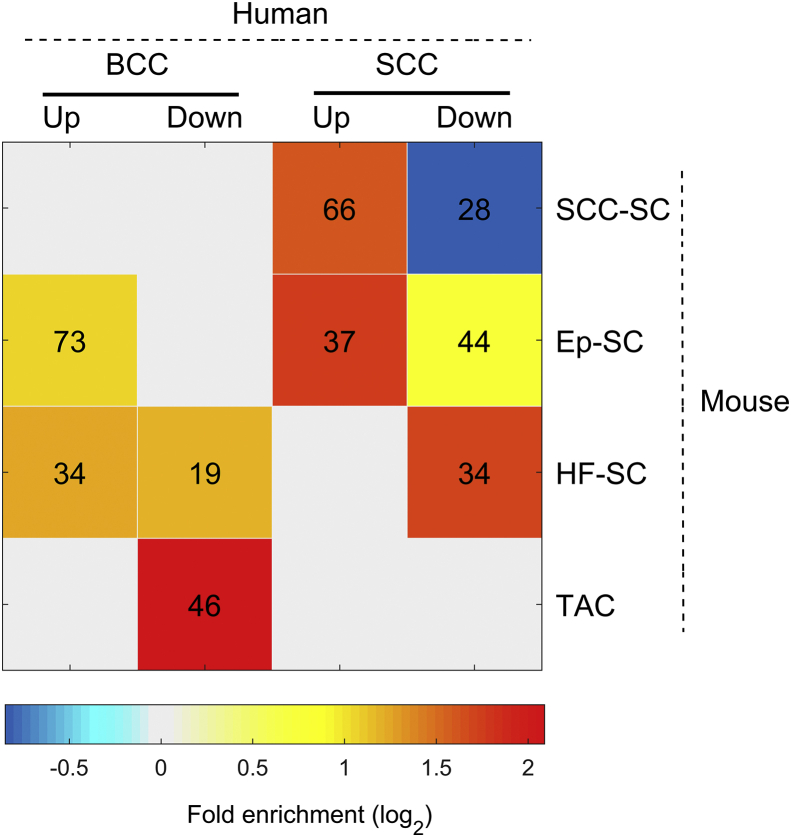
Different mouse models of SCC and BCC are widely used for studying stem cell, cancer stem cell, and cancer biology.57, 58, 59, 60, 61, 62, 63, 64, 65, 66 To determine the relevance of established mouse models to human skin cancer, we first determined whether the mouse SCC stem cell (SCC-SC) signature genes are highly represented in the human SCC.66 We found that 598 mouse SCC-SC genes with human orthologs were expressed in our human tumor or normal tissues. Of those orthologs, 66 (11.0%) were recognized as up-regulated DEGs in human SCC specimens (Fig. 5 and Table S2). In contrast, only 3.7% of all 16,382 expressed genes were up-regulated in SCC (F.E. = 3.0 and P = 6.0 × 10−16), indicating that the mouse SCC-SC gene signature was indeed enriched in human SCCs. Examples of SCC stem cell marker include Sex Determining Region Y-Box 2 (SOX2), BUB1 Mitotic Checkpoint Serine/Threonine Kinase (BUB1), and Cyclin E1 (CCNE1). The function of these SCC-SC genes includes cell division and sister chromatid cohesion. On the other hand, very few mouse SCC-SC genes were down-regulated in human SCCs: only 28 (4.7% of) SCC-SC signature genes were present in 1382 down-regulated SCC DEGs, which is significantly under-represented (F.E. = 0.56 and P = 1.0 × 10−4, Fig. 5). These results suggest that human SCC does share some cancer-stem-cell gene signature with its mouse counterpart.
Figure 5.
Presence of mouse signature genes in human BCC/SCC DEGs. These include signature genes for SCC stem cell gene signature (SCC-SC), epidermal stem cell gene signature (Ep-SC), hair-follicle-stem-cell gene signature (HF-SC), and transient amplifying cell gene signature (TAC). Only significant (a number marked P < 0.01) overlap were colored by the fold enrichment (in log scale with base 2 shown at the bottom). The numeral indicates the number of mouse genes present in human DEGs.
In addition, we analyzed the expression of inter-follicular epidermal stem cell (303 Ep-SC genes) and hair follicle stem cell (124 HF-SC genes) signatures in human SCCs. We found that only Ep-SC genes were over-represented in up-regulated SCC DEGs (F.E. = 3.3 and P = 1.0 × 10−10) (Fig. 5). The functions of these 37 activated Ep-SC genes, e.g., ITGA and LAMA family genes, include hemidesmosome assembly, ECM-receptor interaction, and focal adhesion. ITGA6 (CD49f) is a known marker for epidermal stem cells.67 We also noted that Ep-SC and HF-SC signatures were significantly over-represented in down-regulated human SCC DEGs (F.E. = 1.7 and 3.3, P = 1.3 × 10−4 and 3.1 × 10−10, respectively). These genes encode proteins in extracellular space or are associated with tissue homeostasis.
Finally, we also surveyed the overlap between these mouse signature genes and DEGs in human BCC specimens. As expected, the mouse SCC-SC gene signature was not significantly over-represented in BCC DEGs (Fig. 5 and Table S2). We did notice the co-existence of both Ep-SC and HF-SC signature genes in the up-regulated BCC DEGs (F.E. = 2.1 and 2.4, P = 2.7 × 10−10 and 6.0 × 10−7, respectively). These results are consistent with previous findings in mouse models that BCCs can arise from different stem cell pools,38,39,42,58 particularly HF-SC.40 Further KEGG pathway analysis showed that the HF-SC genes activated in human BCCs were enriched in genes encoding proteins in the extracellular matrix or involved in cell adhesion, ECM-receptor interaction, and protein digestion and absorption. However, 73 Ep-SC genes over-represented in up-regulated human BCC DEGs encode proteins essential for positive regulation of cell proliferation and cell migration. Some of them are associated with the PI3K-Akt signaling pathway, the Ras signaling pathway, or transcriptional dysregulation in cancer, which is consistent with previous analyses of human BCCs.25 Some of the mouse HF-SC genes were also enriched in the human BCC down-regulated DEG group (F.E. = 2.3 and P = 4.1 × 10−4), especially HF-SC genes encoding for proteins in the extracellular matrix.
Transit-amplifying cell (TAC) progenitors work during intermediate stages in tissue regeneration and have been known to regulate HF morphogenetic growth.68 When we compared mouse TAC signature with human SCC and BCC DEGs, 46 out of 160 TAC genes were found significantly over-represented in suppressed gene sets of human BCC (F.E. = 4.3 and P = 9.2 × 10−18) but not represented in SCC DEGs. These down-regulated genes encode intermediate filament proteins, proteins involved in structural molecule activity, or hair follicle morphogenesis, or were implicated in the pathway of tight junction, e.g., KRT and CLDN families.
Taken together, our findings indicate that some mouse signature genes are highly represented in human tumors, including elevated expression of SCC SC genes and Ep-SC in human SCC, as well as up-regulation of Ep-SC, HF-SC, and TAC genes in human BCC. However, several mouse signature genes are not represented in human tumors, e.g., the low representation of HF-SC and TAC genes in human SCC. For SCC-SC signature genes, not all are up-regulated in human SCC, like the Sox 9 gene. While the exact reasons for the discrepancies between mouse and human tumors are not known, we suggest that current mouse models may not recapture all the features of human tumors, and thus additional mouse models for BCC and SCC are in greatly needed for future human relevance studies.
In summary, we have discovered many novel gene expression features in human BCC and SCC. We discovered high representation of C2H2-type zinc finger molecules and several new pathway links in human BCC. We found quite a few shared genes up-regulated in both BCC and SCC as well as distinct gene expression signatures in these two tumor types. Another significant finding in our studies is that many gene expression signatures in mouse models are not well-represented in human tumors, such as the low representation of mouse HF-SC in human SCC. It appears that development of mouse models for SCC and BCC is necessary for use in relevance studies.
Discussion
Our comparative analyses of the transcriptome profiles of tumors and matched normal tissues identified many gene sets representing human BCC and SCC signatures, including both those reported previously and novel players. Specifically, we discovered a significant number of C2H2-type Zinc Finger molecules highly represented in human BCC. While Gli molecules are known downstream transcription factors for the hedgehog signaling pathway, the exact function of other zinc finger molecules identified in this study for stem cells remains to be demonstrated using novel mouse models.
While our data are consistent with several previously published results,25,26,28,30 we had several novel findings, including novel players and signaling networks in BCC. There are many possible reasons that we uncovered these new features of BCC and SCC. First, we used matched tumor-normal tissues to identify tumor-specific gene expression, which reduced individual differences observed using random normal skin tissues. Second, we selected tissues with high tumor contents (>30% by H&E) to reduce the influence of gene expression from normal cells.
We performed a comprehensive comparison among human BCC DEGs, SCC DEGs, and mouse skin signature genes, e.g., mouse SCC-SC, Ep-SC, HF-SC, and TAC, in order to identify shared and distinct gene signatures between mouse and human models. Our results detected a substantial quantity of mouse SCC-SC signature69 in human SCC, indicating the similarity of cancer-stem-cell gene signatures between human SCC and mouse SCC. This relevance suggests that mouse models of SCC stem cell biology may help us understand human SCCs. Meanwhile, many well-studied mouse skin SCC stem cell genes were not differentially expressed in the human skin SCC specimens. We noticed that not all cancer stem cell markers generated in mouse models were highly expressed in the human SCCs. For example, CD44 is a cancer stem cell marker for SCC in the mouse model that was not highly expressed in human SCC. We suspect that this finding may explain the failed clinical trial with the CD44 antibody bivatuzumab in SCC,70,71 as human SCC does not have a high level of CD44.
A substantial portion of Ep-SC signature genes generated in the mouse model (but not HF-SC signature genes), were highly represented in human SCC. Our data are consistent with the general conclusion from a recent publication that epidermal stem cells, not HF-SC, give rise to SCCs.72 In BCC, on the other hand, both Ep-SC and HF-SC gene signatures of mice were enriched in human BCC. We also noticed that many HF-SC genes for metabolism, BMP, and Jak-STAT3 signaling were not enriched in human BCC. These results indicate that cancer cells are not merely an expansion of tissue stem cells. Instead, cancer cells have undergone significant alterations in metabolism and signaling. For example, BMP2 is known to be important for regulation of proliferation and differentiation in the hair follicle stem cells,68 and high levels of BMP2 promote cell differentiation.73 In human BCC, BMP2 expression was not significantly altered, suggesting that BMP regulatory mechanisms may be hijacked in the cancer cells.
Data availability
Both raw and analyzed data of RNA-seq have been deposited in the GEO database with the accession number GSE125285. The list of DEGs is available as Supplementary Table 2 in the paper.
Conflict of Interests
The authors state no conflict of interest.
Acknowledgements
This research is generously supported by Riley Children's Foundation (J.X.) and AGA Foundation (J.X.). We acknowledge support from the IU Simon Cancer Center (grant number P30CA082709), the Purdue University Center for Cancer Research (grant number P30CA023168), and the Walther Cancer Foundation.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2019.10.004.
Contributor Information
Jingwu Xie, Email: jinxie@iu.edu.
Jiali Han, Email: jialhan@iu.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplemental Figure 1. H&E images of typical skin cancer specimens and their adjacent normal skin tissues. Bar=100μm. Supplemental Figure 2. Correlations of fold change (FC) of the tumor and matched normal among all 35 samples. Supplemental Figure 3. (A) Numbers of DEGs and C2H2-type ZF proteins in different groups of DEGs. (B) Percentage of C2H2-type ZF proteins in each set of genes of interest. The dashed line represents the percentage of C2H2-type ZF proteins in all expressed genes in our study. Supplemental Figure 4. Heatmap of DEGs recognized in both BCC and SCC groups.
RNA-seq statistics on 70 paired normal and tumor specimens from 25 BCC and 10 SCC patients. The table lists essential information on each sample, including raw reads, reads aligned in pairs, and mapping ratio.
List of DEGs in either BCC or SCC or both, and their corresponding gene annotation for mouse SCC-SC, Ep-SC, HF-SC, and TAC, which were selected from previous publications. The genes encoding Zinc fingers C2H2-type binding proteins are indicated in the table.
References
- 1.Nehal K.S., Bichakjian C.K. Update on keratinocyte carcinomas. N Engl J Med. 2018;379(4):363–374. doi: 10.1056/NEJMra1708701. [DOI] [PubMed] [Google Scholar]
- 2.Johnson T.M., Rowe D.E., Nelson B.R., Swanson N.A. Squamous cell carcinoma of the skin (excluding lip and oral mucosa) J Am Acad Dermatol. 1992;26(3 Pt 2):467–484. doi: 10.1016/0190-9622(92)70074-p. [DOI] [PubMed] [Google Scholar]
- 3.Green A.C., McBride P. Squamous cell carcinoma of the skin (non-metastatic) BMJ Clin Evid. 2014;2014:e1709. [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova A., Wang G., Dolorito J.A., Kc S., Libove E., Epstein E.H., Jr. Vitamin D3 produced by skin exposure to UVR inhibits murine basal cell carcinoma carcinogenesis. J Investig Dermatol. 2017;137(12):2613–2619. doi: 10.1016/j.jid.2017.05.037. [DOI] [PubMed] [Google Scholar]
- 5.Epstein E.H., Jr. Skin cancer: basal cell carcinoma--pay your money, take your choice. Nat Rev Clin Oncol. 2013;10(9):489–490. doi: 10.1038/nrclinonc.2013.133. [DOI] [PubMed] [Google Scholar]
- 6.Wells J.L., 3rd, Shirai K. Systemic therapy for squamous cell carcinoma of the skin in organ transplant recipients. Am J Clin Oncol. 2012;35(5):498–503. doi: 10.1097/COC.0b013e318201a3ef. [DOI] [PubMed] [Google Scholar]
- 7.Perez H.C., Benavides X., Perez J.S. Basic aspects of the pathogenesis and prevention of non-melanoma skin cancer in solid organ transplant recipients: a review. Int J Dermatol. 2017;56(4):370–378. doi: 10.1111/ijd.13409. [DOI] [PubMed] [Google Scholar]
- 8.Hussein M.R. Ultraviolet radiation and skin cancer: molecular mechanisms. J Cutan Pathol. 2005;32(3):191–205. doi: 10.1111/j.0303-6987.2005.00281.x. [DOI] [PubMed] [Google Scholar]
- 9.Jayaraman S.S., Rayhan D.J., Hazany S., Kolodney M.S. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J Investig Dermatol. 2014;134(1):213–220. doi: 10.1038/jid.2013.276. [DOI] [PubMed] [Google Scholar]
- 10.Johnson R.L., Rothman A.L., Xie J. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272(5268):1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 11.Hahn H., Wicking C., Zaphiropoulous P.G. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 12.Hahn H., Christiansen J., Wicking C. A mammalian patched homolog is expressed in target tissues of sonic hedgehog and maps to a region associated with developmental abnormalities. J Biol Chem. 1996;271(21):12125–12128. doi: 10.1074/jbc.271.21.12125. [DOI] [PubMed] [Google Scholar]
- 13.Gailani M.R., Stahle-Backdahl M., Leffell D.J. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet. 1996;14(1):78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- 14.Aszterbaum M., Rothman A., Johnson R.L. Identification of mutations in the human PATCHED gene in sporadic basal cell carcinomas and in patients with the basal cell nevus syndrome. J Investig Dermatol. 1998;110(6):885–888. doi: 10.1046/j.1523-1747.1998.00222.x. [DOI] [PubMed] [Google Scholar]
- 15.Sekulic A., Migden M.R., Oro A.E. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366(23):2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekulic A., Migden M.R., Basset-Seguin N. Long-term safety and efficacy of vismodegib in patients with advanced basal cell carcinoma: final update of the pivotal ERIVANCE BCC study. BMC Cancer. 2017;17(1):e332. doi: 10.1186/s12885-017-3286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekulic A., Migden M.R., Lewis K. Pivotal ERIVANCE basal cell carcinoma (BCC) study: 12-month update of efficacy and safety of vismodegib in advanced BCC. J Am Acad Dermatol. 2015;72(6):1021–1026. doi: 10.1016/j.jaad.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Migden M.R., Guminski A., Gutzmer R. Treatment with two different doses of sonidegib in patients with locally advanced or metastatic basal cell carcinoma (BOLT): a multicentre, randomised, double-blind phase 2 trial. Lancet Oncol. 2015;16(6):716–728. doi: 10.1016/S1470-2045(15)70100-2. [DOI] [PubMed] [Google Scholar]
- 19.Lear J.T., Migden M.R., Lewis K.D. Long-term efficacy and safety of sonidegib in patients with locally advanced and metastatic basal cell carcinoma: 30-month analysis of the randomized phase 2 BOLT study. J Eur Acad Dermatol Venereol. 2018;32(3):372–381. doi: 10.1111/jdv.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dummer R., Guminski A., Gutzmer R. The 12-month analysis from Basal Cell Carcinoma Outcomes with LDE225 Treatment (BOLT): a phase II, randomized, double-blind study of sonidegib in patients with advanced basal cell carcinoma. J Am Acad Dermatol. 2016;75(1):113–125. doi: 10.1016/j.jaad.2016.02.1226. [DOI] [PubMed] [Google Scholar]
- 21.Magrini S.M., Buglione M., Corvo R. Cetuximab and radiotherapy versus cisplatin and radiotherapy for locally advanced head and neck cancer: a randomized phase II trial. J Clin Oncol. 2016;34(5):427–435. doi: 10.1200/JCO.2015.63.1671. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham T.J., Tabacchi M., Eliane J.P. Randomized trial of calcipotriol combined with 5-fluorouracil for skin cancer precursor immunotherapy. J Clin Investig. 2017;127(1):106–116. doi: 10.1172/JCI89820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevenson M.L., Wang C.Q., Abikhair M. Expression of programmed cell death ligand in cutaneous squamous cell carcinoma and treatment of locally advanced disease with pembrolizumab. JAMA Dermatol. 2017;153(4):299–303. doi: 10.1001/jamadermatol.2016.5118. [DOI] [PubMed] [Google Scholar]
- 24.Migden M.R., Rischin D., Schmults C.D. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med. 2018;379(4):341–351. doi: 10.1056/NEJMoa1805131. [DOI] [PubMed] [Google Scholar]
- 25.Bonilla X., Parmentier L., King B. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48(4):398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 26.Chitsazzadeh V., Coarfa C., Drummond J.A. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun. 2016;7:e12601. doi: 10.1038/ncomms12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asplund A., Gry Bjorklund M., Sundquist C. Expression profiling of microdissected cell populations selected from basal cells in normal epidermis and basal cell carcinoma. Br J Dermatol. 2008;158(3):527–538. doi: 10.1111/j.1365-2133.2007.08418.x. [DOI] [PubMed] [Google Scholar]
- 28.Jee B.A., Lim H., Kwon S.M. Molecular classification of basal cell carcinoma of skin by gene expression profiling. Mol Carcinog. 2015;54(12):1605–1612. doi: 10.1002/mc.22233. [DOI] [PubMed] [Google Scholar]
- 29.Wenzel J., Tomiuk S., Zahn S. Transcriptional profiling identifies an interferon-associated host immune response in invasive squamous cell carcinoma of the skin. Int J Cancer. 2008;123(11):2605–2615. doi: 10.1002/ijc.23799. [DOI] [PubMed] [Google Scholar]
- 30.Van Haren R., Feldman D., Sinha A.A. Systematic comparison of nonmelanoma skin cancer microarray datasets reveals lack of consensus genes. Br J Dermatol. 2009;161(6):1278–1287. doi: 10.1111/j.1365-2133.2009.09338.x. [DOI] [PubMed] [Google Scholar]
- 31.Tumbar T., Guasch G., Greco V. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kretzschmar K., Watt F.M. Markers of epidermal stem cell subpopulations in adult mammalian skin. Cold Spring Harb Perspect Med. 2014;4(10):e013631. doi: 10.1101/cshperspect.a013631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watt F.M. Stem cell fate and patterning in mammalian epidermis. Curr Opin Genet Dev. 2001;11(4):410–417. doi: 10.1016/s0959-437x(00)00211-2. [DOI] [PubMed] [Google Scholar]
- 34.Morris R.J., Liu Y., Marles L. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22(4):411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 35.Clevers H. Searching for adult stem cells in the intestine. EMBO Mol Med. 2009;1(5):255–259. doi: 10.1002/emmm.200900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weissman I.L. The E. Donnall Thomas lecture: normal and neoplastic stem cells. Biol Blood Marrow Transplant. 2008;14(8):849–858. doi: 10.1016/j.bbmt.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapidot T., Sirard C., Vormoor J. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 38.Grachtchouk M., Pero J., Yang S.H. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J Clin Investig. 2011;121(5):1768–1781. doi: 10.1172/JCI46307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G.Y., Wang J., Mancianti M.L., Epstein E.H., Jr. Basal cell carcinomas arise from hair follicle stem cells in Ptch1(+/-) mice. Cancer Cell. 2011;19(1):114–124. doi: 10.1016/j.ccr.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson S.C., Eberl M., Vagnozzi A.N. Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches. Cell Stem Cell. 2015;16(4):400–412. doi: 10.1016/j.stem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapouge G., Youssef K.K., Vokaer B. Identifying the cellular origin of squamous skin tumors. Proc Natl Acad Sci U S A. 2011;108(18):7431–7436. doi: 10.1073/pnas.1012720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanpain C. Tracing the cellular origin of cancer. Nat Cell Biol. 2013;15(2):126–134. doi: 10.1038/ncb2657. [DOI] [PubMed] [Google Scholar]
- 43.Ge Y., Gomez N.C., Adam R.C. Stem cell lineage infidelity drives wound repair and cancer. Cell. 2017;169(4):636–650. doi: 10.1016/j.cell.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan Q., Gu D., Liu H. Defective TGF-beta signaling in bone marrow-derived cells prevents hedgehog-induced skin tumors. Cancer Res. 2014;74(2):471–483. doi: 10.1158/0008-5472.CAN-13-2134-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu D., Liu H., Su G.H. Combining hedgehog signaling inhibition with focal irradiation on reduction of pancreatic cancer metastasis. Mol Cancer Ther. 2013;12(6):1038–1048. doi: 10.1158/1535-7163.MCT-12-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikonova A.S., Astsaturov I., Serebriiskii I.G., Dunbrack R.L., Jr., Golemis E.A. Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci. 2013;70(4):661–687. doi: 10.1007/s00018-012-1073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razin S.V., Borunova V.V., Maksimenko O.G., Kantidze O.L. Cys2His2 zinc finger protein family: classification, functions, and major members. Biochemistry (Mosc) 2012;77(3):217–226. doi: 10.1134/S0006297912030017. [DOI] [PubMed] [Google Scholar]
- 50.Mackeh R., Marr A.K., Fadda A., Kino T. C2H2-Type zinc finger proteins: evolutionarily old and new partners of the nuclear hormone receptors. Nucl Recept Signal. 2018;15 doi: 10.1177/1550762918801071. e1550762918801071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 52.Singla V., Reiter J.F. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313(5787):629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 53.Balmer S., Dussert A., Collu G.M., Benitez E., Iomini C., Mlodzik M. Components of intraflagellar transport complex a function independently of the cilium to regulate canonical Wnt signaling in Drosophila. Dev Cell. 2015;34(6):705–718. doi: 10.1016/j.devcel.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider L., Clement C.A., Teilmann S.C. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15(20):1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 55.Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., Reiter J.F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437(7061):1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 56.Krzywinski M., Schein J., Birol I. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nitzki F., Becker M., Frommhold A., Schulz-Schaeffer W., Hahn H. Patched knockout mouse models of Basal cell carcinoma. J skin Canc. 2012;2012:e907543. doi: 10.1155/2012/907543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Youssef K.K., Van Keymeulen A., Lapouge G. Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol. 2010;12(3):299–305. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- 59.Fan Q., Gu D., He M. Tumor shrinkage by cyclopamine tartrate through inhibiting hedgehog signaling. Chin J Canc. 2011;30(7):472–481. doi: 10.5732/cjc.011.10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen B., Trang V., Lee A. Posaconazole, a second-generation triazole antifungal drug, inhibits the hedgehog signaling pathway and progression of basal cell carcinoma. Mol Cancer Ther. 2016;15(5):866–876. doi: 10.1158/1535-7163.MCT-15-0729-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fontenete S., Perez-Moreno M. Isolation of cancer stem cells from squamous cell carcinoma. Methods Mol Biol. 2019;1879:407–414. doi: 10.1007/7651_2018_162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowry W.E., Flores A., White A.C. Exploiting mouse models to study ras-induced cutaneous squamous cell carcinoma. J Investig Dermatol. 2016;136(8):1543–1548. doi: 10.1016/j.jid.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 63.Malkoski S.P., Haeger S.M., Cleaver T.G. Loss of transforming growth factor beta type II receptor increases aggressive tumor behavior and reduces survival in lung adenocarcinoma and squamous cell carcinoma. Clin Cancer Res. 2012;18(8):2173–2183. doi: 10.1158/1078-0432.CCR-11-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwarz M., Munzel P.A., Braeuning A. Non-melanoma skin cancer in mouse and man. Arch Toxicol. 2013;87(5):783–798. doi: 10.1007/s00204-012-0998-9. [DOI] [PubMed] [Google Scholar]
- 65.Huang P.Y., Balmain A. Modeling cutaneous squamous carcinoma development in the mouse. Cold Spring Harb Perspect Med. 2014;4(9):a013623. doi: 10.1101/cshperspect.a013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schober M., Fuchs E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-beta and integrin/focal adhesion kinase (FAK) signaling. Proc Natl Acad Sci USA. 2011;108(26):10544–10549. doi: 10.1073/pnas.1107807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Owens D.M., Watt F.M. Contribution of stem cells and differentiated cells to epidermal tumours. Nat Rev Cancer. 2003;3(6):444–451. doi: 10.1038/nrc1096. [DOI] [PubMed] [Google Scholar]
- 68.Genander M., Cook P.J., Ramskold D. BMP signaling and its pSMAD1/5 target genes differentially regulate hair follicle stem cell lineages. Cell Stem Cell. 2014;15(5):619–633. doi: 10.1016/j.stem.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jian Z., Strait A., Jimeno A., Wang X.J. Cancer stem cells in squamous cell carcinoma. J Investig Dermatol. 2017;137(1):31–37. doi: 10.1016/j.jid.2016.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erfani E., Roudi R., Rakhshan A., Sabet M.N., Shariftabrizi A., Madjd Z. Comparative expression analysis of putative cancer stem cell markers CD44 and ALDH1A1 in various skin cancer subtypes. Int J Biol Mark. 2016;31(1):e53–e61. doi: 10.5301/jbm.5000165. [DOI] [PubMed] [Google Scholar]
- 71.Tijink B.M., Buter J., de Bree R. A phase I dose escalation study with anti-CD44v6 bivatuzumab mertansine in patients with incurable squamous cell carcinoma of the head and neck or esophagus. Clin Cancer Res. 2006;12(20 Pt 1):6064–6072. doi: 10.1158/1078-0432.CCR-06-0910. [DOI] [PubMed] [Google Scholar]
- 72.Huang P.Y., Kandyba E., Jabouille A. Lgr6 is a stem cell marker in mouse skin squamous cell carcinoma. Nat Genet. 2017;49(11):1624–1632. doi: 10.1038/ng.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzuki K., Yamaguchi Y., Villacorte M. Embryonic hair follicle fate change by augmented beta-catenin through Shh and Bmp signaling. Development. 2009;136(3):367–372. doi: 10.1242/dev.021295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. H&E images of typical skin cancer specimens and their adjacent normal skin tissues. Bar=100μm. Supplemental Figure 2. Correlations of fold change (FC) of the tumor and matched normal among all 35 samples. Supplemental Figure 3. (A) Numbers of DEGs and C2H2-type ZF proteins in different groups of DEGs. (B) Percentage of C2H2-type ZF proteins in each set of genes of interest. The dashed line represents the percentage of C2H2-type ZF proteins in all expressed genes in our study. Supplemental Figure 4. Heatmap of DEGs recognized in both BCC and SCC groups.
RNA-seq statistics on 70 paired normal and tumor specimens from 25 BCC and 10 SCC patients. The table lists essential information on each sample, including raw reads, reads aligned in pairs, and mapping ratio.
List of DEGs in either BCC or SCC or both, and their corresponding gene annotation for mouse SCC-SC, Ep-SC, HF-SC, and TAC, which were selected from previous publications. The genes encoding Zinc fingers C2H2-type binding proteins are indicated in the table.
Data Availability Statement
Both raw and analyzed data of RNA-seq have been deposited in the GEO database with the accession number GSE125285. The list of DEGs is available as Supplementary Table 2 in the paper.