Abstract
Molecular subtypes-based therapies offer new potential framework for desired and precise outcome in clinical settings. Current treatment strategies in colorectal cancer are largely ‘one drug fit all’ model for patients that display same pathological conditions. However, CRC is a very heterogenous set of malignancy that does not support for above criteria. Each subtype displays different pathological and genetic signatures. Based on these features, therapeutic stratification for individual patients may be designed, which may ultimately lead to improved therapeutic outcomes. In this comprehensive review, we have attempted to briefly outline major CRC pathways. A detailed overview of molecular subtypes and their clinical significance has been discussed. Present and future methods, governing CRC subtyping in the era of personalized therapy with a special emphasis on CMS subtypes of CRC has been reviewed. Together, discovery and validation of new CRC patient stratification methods, screening for novel therapeutic targets, and enhanced diagnosis of CRC may improve the treatment outcome.
Keywords: Biomarkers, CMS subtypes, Immunotherapy, Molecular screening, Personalized medicine
Introduction
Colorectal cancer (CRC) is an important public health issue in developed as well as developing countries. It has emerged as an alarming health threat in terms of cancer related deaths throughout the world. Statistically, CRC is the third most common type of cancer in terms of incidence and second most dangerous cancer in terms of cancer related deaths. It was estimated that there will be over 1.8 million new cases of colorectal cancer and 881,000 deaths in 2018, representing about 1 in 10 cancer cases and deaths. Colorectal cancer incidence rates are about 3-fold higher in transitioned versus transitioning countries.1 In 2019, new cancer cases are projected to be 1,762,450 and cancer deaths are estimated to be 606,880 in the USA alone.2
The key driver events in CRC progression, RAS and RAF mutations along with TNM staging may help in the clinical management of CRC.3 Moreover, pathological staging and MSI status helps the clinicians in selection of adjuvant therapy. Further to this, mutational status of KRAS (exon2), BRAF (V600E) and PIK3CA shows the path for anti-EGFR therapy of mCRCs.4,5 Since molecular events play a crucial role in prognosis and deciding the therapy regimen it becomes essential to identify and characterize a molecular subtype of an individual tumor. Technical advancement in the various high throughput techniques now provide a large-scale data profile of diverse cancers, which enables the researchers to comprehensively characterize tumors in a more organized manner. With the advent of such technical advancements for molecular subtyping, there is a paradigm shift toward more specific and reliable approach based on omics data instead of mutation centric molecular subtyping. In this comprehensive review, we have attempted to briefly outline major CRC pathways. A detailed overview of molecular subtypes and their clinical significance has been discussed. Present and future methods, governing CRC subtyping in the era of personalized therapy with a special emphasis on CMS subtypes of CRC has been reviewed.
Molecular pathways in CRC
Colorectal cancer arises through multiple genetic events including mutations and epigenetic modifications in gene(s) that transform normal glandular epithelium cell into a benign neoplasm.6 In CRC, the progression of adenoma to carcinoma is a multistep process. Apparently, genomic instability is currently identified as a central molecular feature that leads to the accretion of other potential genetic aberrations responsible for the CRC transformation. It is a very decisive process for carcinogenesis and regulates the extent of the neoplastic evolutionary process.7 The cumulative findings of about past 35 odd years demonstrate the existence of at least three pathways involved in CRC origin and progression: Chromosomal Instability (CIN); Microsatellite Instability (MSI); and CpG Island Methylator Phenotype (CIMP).8
CIN pathway
It represents 85% of total CRCs and has a hallmark feature of APC loss.9 Karyotyping studies of CIN tumors reveal presence of chromosomal abnormalities that are specific to a set of tumor suppressor genes and tumor promoter (oncogene) gene loci. These mutations and the role of each abnormal gene is well established in tumor progression pathways.
MSI pathway
MSI comprises recurrent alterations in the microsatellite zone, without altering apparent structural and numerical changes in the genome. It is prominently reported in various studies that approximately 15% of all CRCs have high frequency of MSI due to germline mutations in mismatch repair (MMR) system or somatic inactivation by promoter hypermethylation of MLH1 gene.10 HNPCC (Hereditary Non polyposis Colorectal cancer) or Lynch syndrome (~3%) is an immensely invasive autosomal dominant disorder having ~80% lifetime risk for cancer relapse. It is induced by a germline mutation in MMR genes (MLH1, MSH2 in 70% cases and MSH6, PMS2 in 30% cases).11 Out of these genetic mutations, germline deletion of EPCAM gene at 3’ end in last exon (gene upstream to MSH2) also eventually leads to epigenetic inactivation of MSH2. Is described as one of the cause of Lynch syndrome.12
CIMP pathway
Another major pathway responsible for CRC is CIMP. Overall CIMP+ cancers represent approximately 20–30% of total CRCs predominantly in women, 30–40% of CIMP CRC are in the proximal colon and 3–12% in distal colon.13,14 CIMP status of CRC has been approved by several scientists from Toyota et al (2000), Wiesenberger et al (2006), Ogino et al (2007) to Curtin et al (2011). Various gene panels have been proposed to characterize CIMP+ CRCs and each criterion has its own merits and demerits but the outcome of these analyses revealed that CIMP tumors have distinct etiology, molecular features, and epigenetic landscape.15, 16, 17, 18
Toyota et al (1999) first proposed CIMP phenomenon in colorectal cancer. They found hypermethylation in 7 loci of MINT genes. At least 3 out of 7 were strongly associated with CDKN2A and MLH1 hypermethylation, representing a classic panel of CIMP in allocating CIMP (+/−) status.19 In 2006, Wiesenberger et al, classified CRCs into CIMP (+) and CIMP (−) based on 5 gene panel (CACNA1G, IGF2, NEUROG1, RUNX3, SOCS1) using real-time PCR.18 Shen et al, more precisely subtyped CRCs into three different subgroups; CIMP1, CIMP2, and CIMP negative and having a different genetic correlation with BRAF, KRAS, and TP53. CIMP1 tumors have signature BRAF (54%) mutations and often are MSI-H while CIMP2 have KRAS (92%) mutation without any TP53 and BRAF mutations.20 Similarly, Ogino et al, proposed a 5-marker (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1) based panel and later on updated to 8 marker panel (RUNX3, CACNA1G, IGF2, MLH1, NEUROG1, CRABP1, SOCS1and CDKN2A) to stratify CIMP status of tumor.16 More recently a combination of MALDI-TOF MS and ChIP on Chip analysis of CRC cell line and 149 CRC samples identified about 44 methylation biomarkers that efficiently classify colon cancer into CIMP high, CIMP intermediate and CIMP low in a twostep marker panel.21 Wiesenberger et al, (2015) in a recent study, correlated that CIMP + tumors are most frequently located in the right-sided colon and associated with older age and female gender with a signature mutation in BRAF (V600E), hypermethylated MLH1promoter and TP53 loss.14
Molecular classification of colorectal cancer
Currently, CRC pathogenesis is much more affected by the clinical and molecular characteristics of the tumor: MSI (+/−), RAS & RAF mutations including PIK3CA have been frequently used as biomarkers in clinical settings for CRC treatment. Although, based on these classification criteria, the patient's outcome in response to therapy differs significantly. Sjoblom (2006), gave the first insight towards molecular classification of colon and breast cancer based on their CAN (candidate cancer gene) genes concept.22 Several other classification systems have been established by scientists, however, these classifications are mainly based on genetic, epigenetic, gene expression parameters and single cell profiling system. These classification systems present remarkable differences between them. Number of subtypes suggested, varied from 3 subtypes to 6 subtypes with overlapping and mixed subtypes and none were able to classify the CRCs explicitly. This ambiguity may be attributed to methodological differences, different platform used to generate data and most importantly statistical and algorithm bias.22, 23, 24, 25
CRC is highly heterogeneous disease and to a great extent, its behavior depends on its molecular subtype. Molecular classification provides an insight toward the etiology and characteristics of cancer, which, eventually leads to enhanced understanding and better therapies. A vast majority of groups are trying to classify colon cancer according to tumor fate, genetic features, cellular specifications, tumor microenvironment, and more recently immunological characters. Each and every classification system acquires its own unique importance and clinical outcome. In this review, we have attempted to comprehensively review all the recent reported molecular subtypes and their clinical significance.
Jass classification of CRC26
Jass et al (2007), proposed that accretion of malignancy phenotypes more reliably depends upon perturbation of key growth-promoting signaling pathways. Based on genetic and epigenetic landscape of given tumors, 5 basic subtypes were proposed.
-
i)
CIMP high (12%) methylated MLH1, BRAF mutation, chromosomally stable, MSI-H is hallmark of sporadic polyps.
-
ii)
CIMP high, partial methylation of MLH1 gene, BRAF mutation, Chromosomally stable MSS and MSS-L origin in serrated polyps (8%).
-
iii)
CIMP low, KRAS mutated, MGMT methylation, chromosomal instability, MSS or MSS low, origin in adenoma and serrated polyps (20%).
-
iv)
CIMP negative, chromosomal instability, MSS, origin in adenomas (may be sporadic), FAP-associated or MUTYH, associated polyposis (57%).
-
v)
Lynch syndrome, CIMP negative, BRAF negative, chromosomally stable, MSI-H, origin in adenomas (3%).
Clinical significance of Jass classification
Bee et al, analyzed the CIMP and MSI status of 734 tumors and classified them according to Jass classification. The frequency of CIMP 0/non-MSI-H subtypes covers 63% of total surgically resected tumors and CIMP-H/MSI-H had least occurrence frequency. CIMP high/MSI (−) subtypes were found to be associated with right-sided colon, luminal serration, distant nodal metastasis, poor differentiation, and BRAF mutation. MSI-H/CIMP-0/L subtypes were associated with early onset, moderate differentiation and BRAF negative compared with CIMP 0/non-MSI-H. MSI-H tumors share some common clinical features regardless of right-sidedness, Crohn like lymphoid reaction, mucinous histology, dirty necrosis and low KRAS mutations than MSI low subtypes. According to Ward et al, 2004, adjuvant therapy treated patients (stage I–IV) having CIMP-H/non-MSI- H subtypes of cancer had the shortest overall survival rate.27, 28, 29 Ogino et al, (2008), showed that CIMP 0, L and non-MSI-H subtype in different stages of CRC patients have worst Disease Free Survival (DFS).30 In a clinical trial (C89803) the authors tried to establish an association between molecular subtypes and treatment responses in various stages; stage III cancer patients were treated with adjuvant (FU/LV Fluorouracil/Leucovorin Calcium) in combination with (FU/LV + irinotecan) or alone irinotecan. The study suggested that CIMP-H/non-MSI-H subtype was significantly correlated with worst DFS in patients treated without irinotecan.31,32 Han et al (2013) studied the effect of FOLFOX (5-Flourouracil, leucovorin, oxaliplatin) therapy over stage III and high-risk stage II patients and found no significant difference in overall survival rate between the 4 subtypes.33 Sinicrope et al, reported the significance of KRAS mutation in non MSI-H colon cancer and reported that mutated KRAS and BRAF were associated with poorer DFS in comparison to wild-type KRAS/BARF and non-MSI-H.34
Ogino classification system30
Ogino and Goel (2008), established a correlation between epigenetic and genetic variations and subtypes.31 Molecular correlation helps in avoiding selection of nonrandom alterations in tumors succeeded by multiple genetic processes. The ultimate goal resides in the credentials of clinically useful biomarkers which may provide a lead for treatment and genetic counseling.35 Following are given subtypes for Ogino classification:
-
i)
Group 1 (MSI-H, CIMP-H)
It represents 10% of total CRC with the characteristic feature of CIN (−), TP53 (wild-type) BRAF mutated (V600E), MLH1 methylation, intact p21 expression,35 mucinous histology, and poor differentiation.
-
ii)
Group 2 (MSI-H, CIMP low/0)
This type occurs at 5% of total CRC. It includes lynch syndrome (1–3% of total population). Most of the tumors are histologically moderately differentiated, mucinous and tends toward proximal colon. Genetically; KRAS mutated, TP53 (wild-type) and CIN negative with fatty acid synthase overexpression.36
-
iii)
Group 3 (MSS-L/MSS, CIMP high)
These represent class of BRAF mutated, TP53 wildtype, CIN negative and histologically poor differentiated signet ring cells. Frequently present in older female and right-sided colon.37,38
-
iv)
Group 4 (MSI-L & CIMP low)
Characteristic features are mutated KRAS and MGMT methylation. MGMT methylation is primarily responsible for MSI low and CIMP low tumors.39
-
v)
Group 5 (MSS, CIMP low)
30–35% of total CRCs, genetically KRAS mutated, CIN (−) and occurs more frequently in male.35
-
vi)
Group 6 (MSI-L/MSS, CIMP 0)
About 40% of total CRCs. Remarkable features are CIN positive with wild-type KRAS and BRAF frequently located at distal colon without any gender bias.40
Colon cancer subtypes (CCS)23
De souse and coauthors (2013), initially performed a study on 90 patients of stage II colon cancer in an unsupervised manner and subsequently validated in over 1100 patients. They derived a 146 gene classifier that reliably grouped the patients into three major cancer subtypes (colon cancer subtypes). Transcriptomic and epigenomic data were taken into account to robustly classify these given subgroups.
-
i)
CCS1 (49%)
Gene mapping expression data reveals the CIN positive tumors having a frequent mutation in KRAS and TP53. WNT pathway in this type of tumors is remarkably overexpressed.
-
ii)
CCS2 (24%)
These are highly associated with MSI and CIMP+ subset of tumors. The inflammatory cell infiltration is more frequently reported in this subtype.
-
iii)
CCS3 (27%)
These are a highly diverse group of tumors that may have MSI and CIN like characteristics but overexpresses genes related to EMT transition, matrix remodeling, and cell migration. It also represents a large no. of tumors that have acquired a mutation in KRAS and BRAF genes. TGFβ signaling pathway becomes more activated in this subtype and mainly responsible for aggressiveness of serrated tumors41
Clinical significance of CCSs
Based on previous studies this classification system supported the fact that CCS1– CIN tumors are mainly left sided and CCS2-MSI tumors are positioned in the right of the colon whereas CCS3 tumors did not have such correlation. CCS tumors are poorly differentiated with worst overall survival rate and DFS. More than 50% of the patients with CCS3 subtype have DFS up to two years. CMS1 and CMS2 subtypes are well documented for their genetic makeup. CMS3 largely having some features of CIMP/MSS tumors but mutationally not defined and may represent more aggressive sessile serrated adenomas. In vitro studies, supported the fact that anti-tumor drugs and antibodies have significantly different treatment response. A seminal work on metastatic CCS3 patients show the poor response to anti-EGFR therapy independent of KRAS mutation. The study proposed the importance of CCS3 subtypes for patient management.42
Sadanandam CRC assigner system43
Sadanandam et al, (2013). Revolutionized the approach of molecular classification of CRC by associating the gene expression pattern to corresponding therapeutic outcome of patients treated with cetuximab. They analyze two gene expression dataset (GSE 13294 & GSE 14333)44,45 using several statistical methods. A total of 1290 CRC tumors were profiled for genetic clustering. Expression of these genetic cluster shared some common expression patterns in different cell type present in the normal colon crypt. These 5 subtypes are classified as:
-
i)
Goblet type
Having well-differentiated gene expression pattern and under-expressed WNT and stem cell markers. High mRNA expression of MUC2 and TFF3 (goblet like).
-
ii)
Enterocyte type
It is characterized by differentially upregulated genes of enterocytes of intestinal epithelium.
-
iii)
Stem cell type
Characterize by higher expression of WNT signaling pathway targets and demonstrating myoepithelial, mesenchymal and stem cell feature. These subtypes represent a stem or progenitor cell phenotype with least expression of differentiation markers.
-
iv)
Inflammatory type
This subtype is characterized by marked increase in expression of chemokines and interferon associated genes.
-
v)
Transit-amplifying type
In this subgroup, a mixed/heterogeneous population of cells is reported with altered WNT targets and stemness-related genes.
Clinical significance of Sadanandam classification
This classification system reveals different clinical outcome in different treatment groups. Since each subtype have a characteristic genetic and histological feature, clinical significance also varies accordingly. A follow up of 197 patients after surgery receiving adjuvant chemotherapy did not show any significant association with DFS and stage of the tumor. However, in the untreated group, DFS varied significantly. Stem cell-like subtypes represented the shortest DFS, enterocyte subtype, and inflammatory subtype demonstrated moderate DFS while goblet like and transit amplifying subtypes showed good DFS. The data also supports that adjuvant chemotherapy and radio-chemotherapy patients have an improved DFS in stem-like subtypes whereas, for goblet like and transit amplifying type, it exerts detrimental effect on patients. Out of above three subtypes, Transit-amplifying subtype does not potentially respond to cetuximab but may be inhibited by cMET receptor tyrosine kinase inhibitor in filamin A overexpressed subtypes. Rest two subtypes may respond better for FOLFIRI treatment in metastatic CRC.43
Colon cancer molecular subtype (2013)46
This study was primarily based on mRNA expression profile of a huge multicenter cohort of 750 patients of all stages (I–IV) of CRC patients. Genomic alteration in major driver genes (KRAS, BRAF & TP53), CIMP (+/−) status and MSI were studied along with a transcriptomic study. Unsupervised hierarchical clustering analysis of gene expression dataset was performed to establish a significant association between clinicopathological characteristics and following molecular subtypes were proposed.
-
i)
C1 subtype
This contributes to 21% of total CRC samples. This has a characteristic mutation in KRAS, TP53 gene and displays CIN phenotype. Pathway related to immune system and EMT/motility are down-regulated.
-
ii)
C2 Subtype
Represents 19% of studied CRC samples. Marked by high expression of MMR genes, strongly CIMP+ (59%), BRAF mutated (40%), suppression of WNT pathway but upregulation of immune system and proliferation pathway.
-
iii)
C3 Subtypes
Show remarkable features of CIMP low (18%), KRAS mutated, MSS and downregulation of pathway involved in EMT transition and immune system. A total of 13% samples were classified into this subtype.
-
iv)
C4 subtypes
Display both CIN and CIMP (moderate 30%) phenotype along with a frequent mutation in KRAS, BRAF, and TP53 gene. Proliferation-related pathways are down-regulated while EMT transition is upregulated. It was found in 10% of total CRC samples.
-
v)
C5 subtype
Strongly CIN associated, frequent KRAS and TP53 mutated and differentially upregulated WNT related pathway genes. Most predominant form of CRC subtype with 27% total occurrence.
-
vi)
C6 subtype
These are normal cell-like tumors having no mutation in KRAS and BRAF gene (doubtful) but acquire CIN characteristics. C6 subtype has associated with down-regulation of death and cell growth pathway and overexpression of EMT pathway. This type occurs at 10% frequency among studied cohort.
Clinical significance of colon cancer molecular subtype
In this study, survival analysis was confined to stage II & III tumors. The prognosis in each subtype differs slightly but not significantly. C4 and C6 subtype display poorer RFS (relapse-free survival) compared to other four subtypes. C1, C5, and C6 subtypes, share shares some common genetic features such as CIN (high), CIMP (−), TP53 (mutated) but the rest (C2, C4 & C3) display clear discrimination between each other. Broadly these subgroups may be grouped into two well-known molecular pathways i.e. serrated pathway and conventional CRC like phenotype. The biological significance of all six subtypes is well documented through their prognostic behavior, which enhances the possibility to improve the prognostic model and targeted therapy.46
Colorectal cancer intrinsic subtypes (2014)47
A large patient's dataset (n = 188) were analyzed and validated in a cohort of stage I–IV CRC using whole genome analysis in an unsupervised manner to classify the CRC into 3 intrinsic molecular subtypes. The system is primarily rely on following hallmarks; epithelial to mesenchymal transition, higher frequency of mutation rate and cellular proliferation.
-
i)
Type A; MMR deficient epithelial subtype
Having strong MSI phenotype (mainly dMMR), epithelial-like cells, a higher mutation in BRAF gene and represent good prognosis. They represent about 20–30% of all CRC subtypes identified in this study.
-
ii)
Type B; proliferative epithelial subtype
Most frequent (50–60%) and strong association with epithelial phenotype, remarkably BRAF (wt) and MSS. Patients receiving chemotherapy show positive response but prognosis is likely to intermediate to poor.
-
iii)
Type C; Mesenchymal subtype
Rest of the patients (~10–30%) share type C featuring EMT and MSI high signatures. This subtype displays discrete features of poor prognosis and resistance against adjuvant chemotherapy. It also displays lower proliferative activity48
Clinical significance of colorectal cancer intrinsic subtypes
Clinical significance of this study primarily revolves between three hallmark features such as MSI/dMMR and strong association with CIMP+ phenotype, hypermutable characteristics with right sidedness.49 MSI status may vary with different stages and have good prognosis and DFS in comparison to MSS tumors.50 Along with molecular and immunohistochemical based techniques, improvement in this classification system may expand the treatment strategy apart for CRC patients.
Consensus molecular subtypes (2015)51
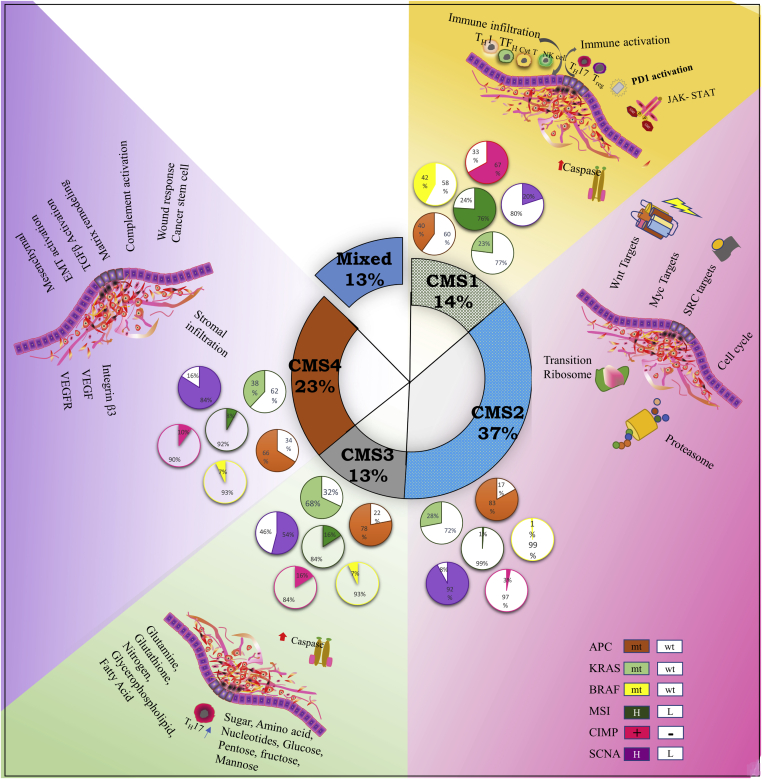
Previously described molecular subtyping methods hold some interesting and exclusive features that may be true for a particular set of patient groups. However, they may be quite different in terms of their methodology, study's inclusion and exclusion criteria etc. This perhaps made the classification of tumor types and their clinical significance invariably overlapping and ambiguous. So there was a need to establish a gold standard that may be able to normalize the differences in the selection of cohort which are generated by virtue of data processing, algorithm bias, and sample preparation methods and basis of classification. The group analyzed a large data set (n = 4151 patients (stage II & III)) generated from different gene expression platform and from different tumor types (FFPE & fresh frozen samples). The outcome of this organized method categorized subtypes of colon cancer into four subtypes. These may differ significantly in genetic epigenetic and signaling pathway they follow. The study also reported a fifth class of subtype that represents a mixed phenotype without any clear designation (Fig. 1).
-
i)
CMS1
Figure 1.
CMS classification of colorectal cancer: Major characteristics of consensus molecular subtypes in CRC. Each group is elaborated with its special features and clinical significance. Currently, CRC is subdivided in to four primary major classes and one mixed subtype. CMS 1 tumor display a higher percentage of MSI, hypermethylation, and hypermutation, and associated with improved survival. CMS2; These contributes to larger subset of subtypes approximately 37% of all tumor subtypes. These tumors have SCNA high, microsatellite stable, activated WNT & Myc pathway and elevated EGFR with mutated TP53 gene. CMS 3 tumors harbors high mutation in KRAS gene and KRAS mutation is highly heterogenous at the gene expression level and having epithelial characteristics. CMS 4 has a high CpG methylator phenotype with strong stromal infiltration, upregulated angiogenic features and hyperactivated TGF-beta.
Represent hypermutable characteristics, MSI high along with CIMP+ phenotype with BRAF frequently mutated, low Somatic Copy Number Alteration (SCNA), immune infiltration, and worse prognosis. They represented 14% of total CRC tumors.
-
ii)
CMS2 Canonical
These contributes to 37% of all tumor subtypes. These tumors have SCNA high, microsatellite stable, activated WNT & Myc pathway and elevated EGFR with the mutated TP53 gene.
-
iii)
CMS3 Metabolic
These had global genomic and epigenomic pattern with mixed characteristics. About 30% of tumors were hypermutated, moderate/low MSI and intermediate CIMP status. Another characteristic feature is elevated multiple metabolic signatures, moderately activated WNT/Myc signaling pathway along with mutated KRAS, PIK3CA, and overexpressed IGBP3. A total of 13% of all tumors were classified into this subtype.
-
iv)
CMS4 mesenchymal
This subtype indicates remarked upregulation of gene involved in EMT transition, matrix remodeling pathway, angiogenesis, TGFβ signaling, and inflammatory-related system.
Clinical significance of CMS subtypes
The ultimate goal of given molecular subtype is to improve prognosis and diagnosis of colorectal cancer at defined stages and to define a targeted therapy for improved DFS. Vast amount of data generated from various types of omics studies has been successfully correlated to demographic and phenotypic characteristics and associated with various molecular subtypes.52 Recently, Sveen A et al (2018) established a preclinical model to explore targetable cancer cell dependencies in an in-vitro model. First, they stratified cell lines and PDX (patient derived xenograft) model system by optimizing cellular intrinsic signal and precisely integrating high throughput drug screening (n = 459) and then classified subtypes sensitive drug regimens; CMS2 subtype was the best respondent to anti-EGFR and HER2 inhibitors while CMS1 & CMS4 display high against HSP like inhibitors. Interestingly CMS4 subtype displays a distinct chemoresistance against a combination of 5-FU and luminespib drugs.53
Molecular subtype alone does not appear to be very helpful in targeted therapy due to lack of integration of transcriptomic, genomic and proteomic data into a reproducible and reliable cluster that may help in preclinical testing. However, phenotypic signatures displayed by each subgroup may help in disease stratification in routine pathology. Therefore, subtypes-based signature analysis is required to simplify the subtyping methods for patient's clinical outcome. CMS subtype categorically addresses this limitation of single method. We have discussed the clinical significance of CMS in detail in the following section.
Treatment strategies based on CMS subtypes of colorectal cancer
Treatment based on CMS1 (MSI, CIMP+ and BRAF mt)
CMS 1 subtype is remarkably MSI high and total 15–20% of CMS1 tumors represent MSI (+) features. MSI high tumors display worse prognosis in sporadic tumors while lynch syndrome appears to have better prognosis.54 It was reported that dMMR tumors have better prognosis than MSS. dMMR tumors having germline mutations treated with 5FU based therapy shows significant DFS, while sporadic CRCs does not comply with therapy.34 Prognostic effect of adjuvant chemotherapy (FOLFOX ± cetuximab) on dMMR tumors provides location-specific treatment response. dMMR with proximal colon presents better therapeutic outcome while distal tumors display poor outcome. KRAS and BRAF mutations are independently associated with worse outcome.55 CMS1 tumors are also characterized by promoter hypermethylation (CIMP high) and are strongly associated with improved prognosis and DFS than CIMP low tumors. However, CIMP status (high/low) invariably does not correlate with tumor stages and mortality rate.56 MSI-H may be accounted for one of the significant factor for CMS1 subtype, however, other subtypes also harbor moderate to low MSI status. These features in isolation were not complete to characterize the CMS subtype, therefore, there was a need for other phenotypic components which can specifically characterize the CMS subtypes.
It is suggested in several findings that infiltration of immune cells is significantly associated with better prognosis in MSI tumors.57 It is reported that local infiltration is highly enriched with tumor-infiltrating CTLs (cytotoxic T lymphocytes) in core tumoral area and surrounding peritumoral area. MSI tumors are enriched with CD3+, CD4+, CD20+, and CD68+ in the intraepithelial region compared with MSS tumors. MSI tumors have higher angiogenic potential as evident with higher microvessel density (MVD) than MSS counterpart.58 The local inflammatory response is widely reported in tumor progression in most of the cancers and presence of TILs (tumor infiltrating lymphocytes:CD4+ &CD8+) in the tumor microenvironment are most important in the suppression of tumor progression and invasion. Presence of CD8+ TILs at the margins of the tumor periphery may also be significantly associated with better prognosis. Therefore TILs are more important prognostic components in CMS1 than MSI.59 Inhibitors, such as checkpoint-based inhibitors that stimulate TILs have been proposed to regulate CRC progression in CRCs. PD1 blocker exert a significant clinical outcome in MSI tumors. PD1 blockers + pembrolizumab given in combination or as monotherapy is still debatable and needs more intensive clinical research.60, 61, 62
Tumor immunogenicity is potentially regulated by signature genes which play a significant role in escaping immune surveillance. The molecular mediators of this immune escape follow three major mechanisms; i) reduced cancer antigen presentation ii) survival and resistance promoted by oncogenic mutation iii) building of tumor suppressive microenvironment.63 Interestingly patients receiving immune checkpoint blockade therapy attain a good clinical outcome in MSI and hypermutated MSS tumors and display various convenient features such as infiltration of CTLs and elevated expression of neoantigens. Le et al (2017), proposed that mutant neoantigen in dMMR cancers increases the sensitivity against the immune checkpoint blockade. The efficiency of immune checkpoint blockade in advanced dMMR tumors was achieved as expected in 53% and complete response in (21%) of the patients.61
Treatment based on CMS2 and CMS 3 subtypes
CMS2 and CMS 3 subtypes relatively similar share some significant features. So, these may follow several common therapeutic targets to increase OS and PFS. Somatic Copy Number Alterations (SCNA) that is detected by whole genome sequencing occurs during meiosis is the main feature of CMS 2 subtype and characterized by the loss or gain in various chromosome (20q, 13q, 8q, 7 gain and 4, 8p, 18q, and 17p loss). These SCNA affects the WNT and MAPK pathway which drives the CRC progression. β catenin is another gene which is related to worse prognosis and is crucial for CRC progression and may serve an as a potential genomic marker for this subtype.64
Patients grouped in CMS 3 subtype display frequent mutation in KRAS gene (mainly exon 2) resulting in constitutive activation MAPK pathway which is associated with poorer clinical outcome.65 It has been noted that KRAS mutated tumors tend to be associated with lung metastasis that may possibly be the effect of MAPK invasion.66 NRAS (~5%) mutation alter the clinical outcome with poor prognosis. Hagland et al (2013) established an association between KRAS mutation and altered metabolic pathway in cancer cells. This metabolic reprogramming is refered as “Warburg effect” that shifts the cells to the glycolytic pathway to maintain the survival and cellular proliferation.67 Therefore an understanding of glucose metabolic pathway in cancer may also be seen as novel therapeutic targets.68 Chen G et al (2015) studied the effect of halofugine on Warburg effect and reported that it can regulate the glucose metabolism via suppression of Akt/mTOR pathway. It diminishes the glycolytic and TCA flux in mice and HCT116 cell lines.69 Similarly, Propofol reduced the glycolysis in tumor cells through suppressed NMDAR-CAMKII- ERK pathway in tumor cells. In future, these molecules may be proposed as an antitumor metabolic agent in clinical settings.70 In a multi-omic study, Myc gene expression was found to be strongly associated with metabolic reprogramming in adenoma to carcinoma sequence. It regulates the genes involved in mitochondrial biogenesis hence molecules targeting Myc expression could be a potential target for cancer therapy.71
Treatment based on CMS 4 subtypes
This subtype display SCNA in tumors cells as CMS2 subtype with increased expression of TFGβ signaling pathway. It is reported that stromal cells are highly rich with TGF beta cytokine and therefore could be targeted for study in cancer progression and metastasis in CRC. Increased tumor stromal cells percentage is associated with poor prognosis for CRC (HR 2.46 95% CI 1.56–3.89).72 TSP (tumor stromal percentage) status may provide an idea to stratify CMS 4 into TSP low or high. High TSP is supposed to inhibit the immune infiltration into the tumor area and associated with worst prognosis.62 EMT is supposed to be promoted by aberrant activation of TGF beta signaling pathway in elevated stromal cells tumor. Therefore, targeting TGF-β, actin and stromal cell proliferation can potentially suppress the distant metastasis in patients.73 Currently, there are many drugs, ranging from small molecules, monoclonal antibody, vaccines, and antisense oligonucleotides that are being used in clinical trials to inhibit the TGF β activity. Small molecule inhibitors that target dihydropyrrolo pyrazole scaffold on TGF β are developed by ELI Lilly company. One of them galunisertib, is the most preferred, tested drug which displayed interesting and promising outcome in phase II trials. Similarly, fresolimumab (mab), lucanix, vigil (vaccine) and trabedersen (antisense oligonucleotides) limited the progression of metastatic CRC through compromising the TGF β targets.74
Stem cells like CMS4 tumors are significantly associated with positive clinical outcome to FOLFIRI treatment.75 On the other hand, there are several other molecules (thioridazine, tranilast, and metformin) discovered that target the cancer stem cell's intrinsic pathways. These are classical drugs targetting specific cell surface molecules (CD44, CD47, Lgr 5, EPCAM).76
Anti VEGF therapies targeting VEGFR JAK3, CLDN5 and FLT4 may also serve as a good option in therapeutic settings. It was reported that anti-angiogenic (VEGFR) treatment after surgery is the first-line of treatment and provides a better clinical outcome and significantly improves OS in metastatic CRCs.77 In the therapeutic setting inhibitors for FLT4 (sorafenib, telatinib, sunitinib, and pazopanib) also have demonstrated their potential. Similarly, Src inhibitors may also provide better results in EMT subtypes through regulation of multiple signaling pathways involved in invasion and cytoskeleton formation.78
Current strategies in personalized medicine for CRC treatment
Personalized medicine is primarily aimed for customized treatment of an individual. It entirely helps the clinicians in disease management, accurate therapy, and better prevention (Fig. 2). This aspect of therapy increases the treatment response by many folds such as, it may reduce unwanted intervention, moreover, target based therapy improves the clinical response. Prognostic biomarkers help the clinicians for personalizing therapy, while predictive biomarkers provide an important idea about therapeutic outcome for an individual patient. At present clinicians routinely decide the therapy on the basis of stage of tumor and their location. Stage II and stage III patients primarily receives adjuvant chemotherapy (FOLFOX & FOLFIRI) while advanced stage CRC (mCRC) patients receive various systematic therapies that include monoclonal antibody (anti EGFR, anti VEGFR) along with the chemotherapies.79
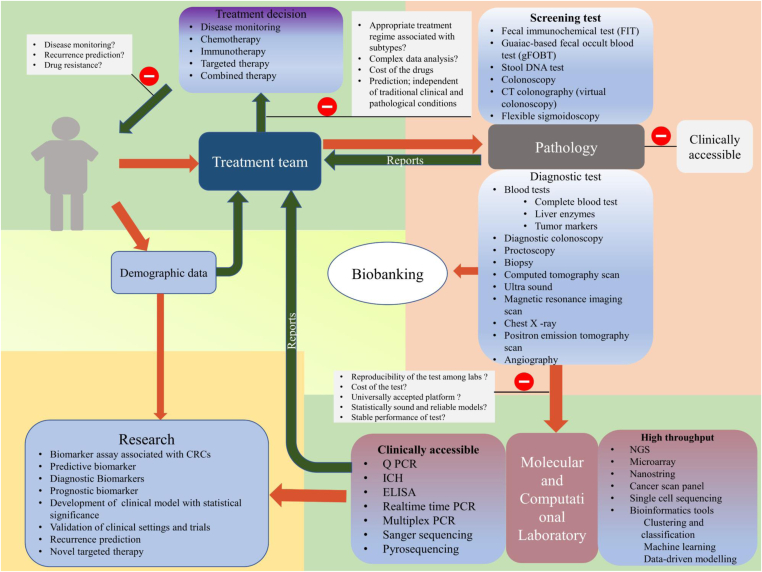
Figure 2.
A proposed flow chart of therapeutic settings and major bottleneck factors in CRC personalized therapy: Personalized therapy for CRC patients follows a route from primary screening to molecular testing that ultimately provide a precise information to clinical team in therapy selection. The first step is the patient approches to the clinician and checked for disease symptoms in pathology. The sample then screened for molecular testing to check molecular profile of specific tumors. Then, pathology and molecular test reports are analyzed by the team of clinician and therapy for a particular patient have to be decided on the basis of pervious data available from research and outcome given by the various drugs of interest (Red arrow indicates up line of screening and diagnosis while green line represents reports of the tests that leads toward the treatment team). This personalized treatment flow faces several hurdles that have to be overcome in future treatment management to improve the therapeutic outcome.
Grouping patients on basis of molecular subtypes helps in decision making to treat CRC. It relies upon the mutational, transcriptomic and proteomics data analysis for prospective personalized therapy.80 MSI plays a significant role as a predictive and prognostic biomarker in analyzing therapeutic response and clinical outcome. MSI-H patients treated with adjuvant therapy does not respond well while some studies reported that if a large deletion in the T intronic repeats of HSP 110 is present then approximately 20–25% stage II & stage III CRC patients responded well with better OS against adjuvant therapy.81 In another retrospective study on 59,475 stage II CRC patients, MSS patients were benefitted more with chemotherapy and they exhibit improved survival (HR 0.54, 95% CI 0.43–0.70; P < 0.001). On the contrary, MSI patients were less benefitted with adjuvant chemotherapy (HR 0.81, 95% CI 0.38–1.78; P < 0.595).82
These seminal studies have created a platform for personalized medicine in CRC treatment. Advancement in the sequencing techniques has opened a new door to identify specific biomarkers, making clinicians to understand the tumors to a great extent. This in turn has made the researchers to identify global molecular pathways for progression and resistance against therapy in CRC. For example, EGFR resistance now a days become major concern for clinicians, which directly correlates to multiple genetic alteration in RAS, RAF, EGFR, and ERBB2 & MET amplification.5 This data is helping clinicians to deliver the right therapy to patients. Evidence suggests that mutated RAS (KRAS, HRAS & NRAS) are resistant against anti EGFR (cetuximab and panitumumab) therapy83, 84, 85 BRAF is also reported as a biomarker for resistance to anti-EGFR therapy. It is a downstream gene to RAS and frequently co-exist with MSI and poor prognosis. BRAF mutated and KRAS wild type tumors have been reported to display poor OS and PFS even if these patients receive anti- EGFR therapy with standard chemotherapy.86 FOLFOX plus bevacizumab may offer a good therapy regime for BRAF mutated metastatic CRC patients. In TRIBE, a study on 28 BRAF mutated tumors reported that FOLFOX plus bevacizumab treated patients were conclusively associated with improved overall survival (HR 0.54, 95% CI 0.24–1.20) and PFS (HR 0.57, 95% CI 0.27–1.23) in comparison to FOLFIRI + bevacizumab therapy.87 For the BRAF mutant with MSI-H patients, there is still an absence of defined criteria whether immunotherapy or BRAF inhibitor along with other drugs may improve the therapeutic outcome and propose a better therapeutic approach. As evident from the two big studies CHECKMATE 120 (25% with nivolumab alone Vs 33.3% nivolumab + ipilimumab) and SWAG S1406 (16% with vemurafenib, cetuximab, and irinotecan) combined immunotherapy have been associated with better treatment response as compared to monotherapy for BRAF mutated and MSI-H patients.88,89 PD1 blockers are unique in its category that they serve as a multiple target biomarker in different cancers such as non-cell lung cancer, metastatic CRC, renal cell carcinoma, melanoma, and Merkel cell carcinoma.90, 91, 92, 93 Current studies suggest that MMR status of individual tumor may serve as potent biomarker to monitor and screen the response of immune check point blocker (ICB) on solid tumors. In the KEYNOTE trail with 149 patients having 15 different MSI status (MSI H) enrolled in five trials receiving pembrolizumab at 200 mg dose for defined time intervals in designated trail groups were studied. The final outcome of study reported that overall response rate (ORR) was 39.6% (95%, CI 31.7–47.9). Pembrolizumab displays a sustained drug response along with improved survival in MSI H chemotherapy resistant metastatic solid tumors. On the basis of above positive therapeutic response in 2017 FDA initialize the approval of pembrolizumab as a second or higher line drug choice for wider coverage of patients with un resectable, MSI H solid tumors.60,94,95 Another key immune check point blocker nivolumab was analyzed with dMMR/MSI H in an open label multicenter phase II study CheckMate142. The efficiency of nivolumab was evaluated on 74 mCRC patients having received first line conventional chemotherapy. At the last, it displayed a remarkable improvement, and about 31.1% patients attain an objective response, 69% patients had achieved disease free survival for more than two weeks, PFS and OS at 12 months was 50% and 70% respectively.89 Immune check point blockers have increased their therapeutic efficiency in combined therapy. In CheckMate142 trial 119 mCRC (MSI H) patients having received nivolumab plus ipilimumab showed a better ORR (55%), DCR for more than 12 weeks (80%), 12 months PFS (71%) and corresponding OS was 85%.93,96 The immune check pint blockers have the potential to prevent the disease progression, but all above and available studies/trials needs to be validated among diverse clinical groups.
Future prospects in CRC management and personalized therapy
Cell free DNA offers a real time monitoring of frequent genotypic subpopulation in a diagnosed CRC patient. This technique utilizes only few millions of cells to perform Next generation sequencing and NGS biomarker panels assists in decision making for an individual. These panels authorize the clinicians to trace the clonal dynamics and cellular response against therapy during the course of treatment.97 Immune therapies including CTLA4, PDCD1 (PD-1), and CD274 (PD-L1) check point blockers have been used to improve and monitoring the treatment response in MSI H CRCs. Together, the tumoral and immune factors may offer a new cancer classification system TIME (Tumor Immunity in the MicroEnvironment). However, variable immune response limited the use of immunoscore classification system in routine clinical practices.98
ERBB2 amplification and mutation is reported in approximately 3–4% and 1–2% mCRC patients and is associated with resistance to anti EGFR therapy and poor PFS.99 According to PETACC8 trial, stage III CRC patients receiving adjuvant therapy having ERBB2 amplification or mutations were associated with shorter relapse time (HR 1.55, 95% CI 1.02–2.36; P = 0.04) and reduced OS (HR 1.57, 95% CI 0.99–2.5; P = 0.05)100 Fusion transcripts/protein, a novel category of structural alteration in chromosomal assembly, currently does not have any molecular signature to fall in any of CMS subtypes of CRC. But they covers at least 10% of total CRC with R spondin fusion that are mutually reported with aberrant APC.101 Similarly, ALK and RET fusion proteins are also important in development of novel therapeutic agent. R-spondin inhibitor Porcupine that block the Wnt secretion results in improved clinical outcome in some patients.102
Authors contribution
Conceptualization SS and MPS, writing original draft MPS, AP and SR, review and editing NKS and SS.
Conflict of Interests
The authors declare that the study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
The study is supported by Department of Biotechnology grant no. 6242-P103/RGCB/PMD/DBT/SMSV/2015. Authors are thankful to the Ministry of Human Resource and Development, and CSIR Govt. of India, New Delhi, India for providing scholarship during this tenure. The authors acknowledge Dr. Tonima Kamat for critically reviewing the manuscript.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Manish Pratap Singh, Email: manish.biophd@mnnit.ac.in.
Nand K. Singh, Email: singhnand@gmail.com.
Sameer Srivastava, Email: sameers@mnnit.ac.in.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA: A Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama G., Tanaka C., Kodera Y. Current options for the diagnosis, staging and therapeutic management of colorectal cancer. Gastrointest Tumours. 2013;1(1):25–32. doi: 10.1159/000354995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao B., Wang L., Qiu H. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. OncoTarget. 2016;8(3):3980–4000. doi: 10.18632/oncotarget.14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misale S., Di Nicolantonio F., Sartore-Bianchi A., Siena S., Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4(11):1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 6.Mundade R., Imperiale T.F., Prabhu L., Loehrer P.J., Lu T. Genetic pathways, prevention, and treatment of sporadic colorectal cancer. Oncoscience. 2014;1(6):400–406. doi: 10.18632/oncoscience.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worthley D.L. Colorectal carcinogenesis: road maps to cancer. World J Gastroenterol. 2007;13(28):e3784. doi: 10.3748/wjg.v13.i28.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pino M.S., Chung D.C. The chromosomal instability pathway IN colon cancer. Gastroenterology. 2010;138(6):2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteller M., Corn P.G., Baylin S.B., Herman J.G. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- 10.Gatalica Z., Vranic S., Xiu J., Swensen J., Reddy S. High microsatellite instability (MSI-H) colorectal carcinoma: a brief review of predictive biomarkers in the era of personalized medicine. Fam Cancer. 2016;15(3):405–412. doi: 10.1007/s10689-016-9884-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hewish M., Lord C.J., Martin S.A., Cunningham D., Ashworth A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat Rev Clin Oncol. 2010;7(4):197–208. doi: 10.1038/nrclinonc.2010.18. [DOI] [PubMed] [Google Scholar]
- 12.Ligtenberg M.J.L., Kuiper R.P., Chan T.L. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3’ exons of TACSTD1. Nat Genet. 2009;41(1):112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 13.Migliore L., Migheli F., Spisni R., Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:e792362. doi: 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisenberger D.J., Levine A.J., Long T.I. Association of the colorectal CpG island methylator phenotype with molecular features, risk factors, and family history. Cancer Epidemiol Biomark Prev. 2015;24(3):512–519. doi: 10.1158/1055-9965.EPI-14-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtin K., Slattery M.L., Samowitz W.S. CpG island methylation in colorectal cancer: past, present and future. Pathol Res Int. 2011;2011 doi: 10.4061/2011/902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogino S., Kawasaki T., Kirkner G.J., Kraft P., Loda M., Fuchs C.S. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9(3):305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyota M., Ohe-Toyota M., Ahuja N., Issa J.P. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97(2):710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisenberger D.J., Siegmund K.D., Campan M. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 19.Toyota M., Ahuja N., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.-P.J. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L., Waterland R.A. Methods of DNA methylation analysis. Curr Opin Clin Nutr Metab Care. 2007;10(5):576–581. doi: 10.1097/MCO.0b013e3282bf6f43. [DOI] [PubMed] [Google Scholar]
- 21.Yagi K., Akagi K., Hayashi H. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16(1):21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 22.Sjöblom T., Jones S., Wood L.D. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 23.De Sousa E Melo F., Wang X., Jansen M. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 24.Leary R.J., Lin J.C., Cummins J. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci USA. 2008;105(42):16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Kinzler K.W. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jass J.R. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50(1):113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee S., Cho N.-Y., Choi M., Yoo E.J., Kim J.-H., Kang G.H. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int. 2008;58(2):104–113. doi: 10.1111/j.1440-1827.2007.02197.x. [DOI] [PubMed] [Google Scholar]
- 28.Nosho K., Kawasaki T., Ohnishi M. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10(6):534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward R.L., Cheong K., Ku S.-L., Meagher A., O'Connor T., Hawkins N.J. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21(20):3729–3736. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- 30.Ogino S., Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10(1):13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saltz L.B., Niedzwiecki D., Hollis D. Irinotecan plus fluorouracil/leucovorin (IFL) versus fluorouracil/leucovorin alone (FL) in stage III colon cancer (intergroup trial CALGB C89803) J Clin Orthod. 2004;22(14_suppl):e3500. [Google Scholar]
- 32.Shiovitz S., Bertagnolli M.M., Renfro L.A. CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage 3 colon cancer. Gastroenterology. 2014;147(3):637–645. doi: 10.1053/j.gastro.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han S.-W., Lee H.-J., Bae J.M. Methylation and microsatellite status and recurrence following adjuvant FOLFOX in colorectal cancer. Int J Cancer. 2013;132(9):2209–2216. doi: 10.1002/ijc.27888. [DOI] [PubMed] [Google Scholar]
- 34.Sinicrope F.A., Okamoto K., Kasi P.M., Kawakami H. Molecular biomarkers in the personalized treatment of colorectal cancer. Clin Gastroenterol Hepatol. 2016;14(5):651–658. doi: 10.1016/j.cgh.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino S., Kawasaki T., Kirkner G.J., Loda M., Fuchs C.S. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8(5):582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogino S., Kawasaki T., Ogawa A., Kirkner G.J., Loda M., Fuchs C.S. Fatty acid synthase overexpression in colorectal cancer is associated with microsatellite instability, independent of CpG island methylator phenotype. Hum Pathol. 2007;38(6):842–849. doi: 10.1016/j.humpath.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 37.Goel A., Nagasaka T., Arnold C.N. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132(1):127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Samowitz W.S., Albertsen H., Herrick J. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129(3):837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 39.Ogino S., Kawasaki T., Kirkner G.J., Suemoto Y., Meyerhardt J.A., Fuchs C.S. Molecular correlates with MGMT promoter methylation and silencing support CpG island methylator phenotype-low (CIMP-low) in colorectal cancer. Gut. 2007;56(11):1564–1571. doi: 10.1136/gut.2007.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogino S., Kawasaki T., Kirkner G.J., Ohnishi M., Fuchs C.S. 18q loss of heterozygosity in microsatellite stable colorectal cancer is correlated with CpG island methylator phenotype-negative (CIMP-0) and inversely with CIMP-low and CIMP-high. BMC Canc. 2007;7(1):e72. doi: 10.1186/1471-2407-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van der Flier L.G., Sabates-Bellver J., Oving I. The intestinal Wnt/TCF signature. Gastroenterology. 2007;132(2):628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 42.Khambata-Ford S., Garrett C.R., Meropol N.J. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25(22):3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 43.Sadanandam A., Lyssiotis C.A., Homicsko K. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorissen R.N., Lipton L., Gibbs P. DNA copy-number alterations underlie gene expression differences between microsatellite stable and unstable colorectal cancers. Clin Cancer Res. 2008;14(24):8061–8069. doi: 10.1158/1078-0432.CCR-08-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jorissen R.N., Gibbs P., Christie M. Metastasis-associated gene expression changes predict poor outcomes in patients with dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15(24):7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marisa L., Reyniès A de, Duval A. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roepman P., Schlicker A., Tabernero J. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134(3):552–562. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh A., Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roth A.D., Tejpar S., Delorenzi M. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28(3):466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 51.Guinney J., Dienstmann R., Wang X. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garnett M.J., Edelman E.J., Heidorn S.J. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sveen A., Bruun J., Eide P.W. Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin Cancer Res. 2018;24(4):794–806. doi: 10.1158/1078-0432.CCR-17-1234. [DOI] [PubMed] [Google Scholar]
- 54.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(6):2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinicrope F.A., Mahoney M.R., Smyrk T.C. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX-based adjuvant chemotherapy. J Clin Oncol. 2013;31(29):3664–3672. doi: 10.1200/JCO.2013.48.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogino S., Nosho K., Kirkner G.J. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deschoolmeester V., Baay M., Lardon F., Pauwels P., Peeters M. Immune cells in colorectal cancer: prognostic relevance and role of MSI. Canc Microenviron. 2011;4(3):377–392. doi: 10.1007/s12307-011-0068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Smedt L., Lemahieu J., Palmans S. Microsatellite instable vs stable colon carcinomas: analysis of tumour heterogeneity, inflammation and angiogenesis. Br J Canc. 2015;113(3):500–509. doi: 10.1038/bjc.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park J.H., Powell A.G., Roxburgh C.S.D., Horgan P.G., McMillan D.C., Edwards J. Mismatch repair status in patients with primary operable colorectal cancer: associations with the local and systemic tumour environment. Br J Canc. 2016;114(5):562–570. doi: 10.1038/bjc.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Le D.T., Uram J.N., Wang H. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le D.T., Durham J.N., Smith K.N. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh P.P., Sharma P.K., Krishnan G., Lockhart A.C. Immune checkpoints and immunotherapy for colorectal cancer. Gastroenterol Rep (Oxf) 2015;3(4):289–297. doi: 10.1093/gastro/gov053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teng M.W.L., Bowman E.P., McElwee J.J. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med. 2015;21(7):719–729. doi: 10.1038/nm.3895. [DOI] [PubMed] [Google Scholar]
- 64.Najdi R., Holcombe R.F., Waterman M.L. Wnt signaling and colon carcinogenesis: beyond APC. J Carcinog. 2011;10:e5. doi: 10.4103/1477-3163.78111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phipps A.I., Buchanan D.D., Makar K.W. KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. Br J Canc. 2013;108(8):1757–1764. doi: 10.1038/bjc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghidini M., Personeni N., Bozzarelli S. KRAS mutation in lung metastases from colorectal cancer: prognostic implications. Cancer Med. 2016;5(2):256–264. doi: 10.1002/cam4.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hagland H.R., Berg M., Jolma I.W., Carlsen A., Søreide K. Molecular pathways and cellular metabolism in colorectal cancer. Dig Surg. 2013;30(1):12–25. doi: 10.1159/000347166. [DOI] [PubMed] [Google Scholar]
- 68.Fang S., Fang X. Advances in glucose metabolism research in colorectal cancer. Biomed Rep. 2016;5(3):289–295. doi: 10.3892/br.2016.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen G.-Q., Tang C.-F., Shi X.-K. Halofuginone inhibits colorectal cancer growth through suppression of Akt/mTORC1 signaling and glucose metabolism. OncoTarget. 2015;6(27):24148–24162. doi: 10.18632/oncotarget.4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X., Li L.-Y., Jiang J.-L. Propofol elicits autophagy via endoplasmic reticulum stress and calcium exchange in C2C12 myoblast cell line. PLoS One. 2018;13(5):e0197934. doi: 10.1371/journal.pone.0197934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Satoh K., Yachida S., Sugimoto M. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci. August 2017;114(37):E7697–E7706. doi: 10.1073/pnas.1710366114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park J.H., Richards C.H., McMillan D.C., Horgan P.G., Roxburgh C.S.D. The relationship between tumour stroma percentage, the tumour microenvironment and survival in patients with primary operable colorectal cancer. Ann Oncol. 2014;25(3):644–651. doi: 10.1093/annonc/mdt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Staudacher J.J., Bauer J., Jana A. Activin signaling is an essential component of the TGF-β induced pro-metastatic phenotype in colorectal cancer. Sci Rep. 2017;7(1):e5569. doi: 10.1038/s41598-017-05907-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Gramont A., Faivre S., Raymond E. Novel TGF-β inhibitors ready for prime time in onco-immunology. OncoImmunology. 2016;6(1):e1257453. doi: 10.1080/2162402X.2016.1257453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sadanandam A., Wang X., de Sousa E Melo F. Reconciliation of classification systems defining molecular subtypes of colorectal cancer. Cell Cycle. 2014;13(3):353–357. doi: 10.4161/cc.27769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garza-Treviño E.N., Said-Fernández S.L., Martínez-Rodríguez H.G. Understanding the colon cancer stem cells and perspectives on treatment. Cancer Cell Int. 2015;15(1):e2. doi: 10.1186/s12935-015-0163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pogue-Geile K.L., Song N., Gavin P.G. Clinical outcome and benefit of oxaliplatin in colon cancer according to intrinsic subtypes: results from NRG Oncology/NSABP C-07. J Clin Orthod. 2016;34(15_suppl):e3510. [Google Scholar]
- 78.Qiu T., Chen W., Li P., Sun J., Gu Y., Chen X. Subsequent anti-VEGF therapy after first-line anti-EGFR therapy improved overall survival of patients with metastatic colorectal cancer. OncoTargets Ther. 2018;11:465–471. doi: 10.2147/OTT.S149110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Graham D.M., Coyle V.M., Kennedy R.D., Wilson R.H. Molecular subtypes and personalized therapy in metastatic colorectal cancer. Curr Colorectal Canc Rep. 2016;12:141–150. doi: 10.1007/s11888-016-0312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rodriguez-Salas N., Dominguez G., Barderas R. Clinical relevance of colorectal cancer molecular subtypes. Crit Rev Oncol Hematol. 2017;109:9–19. doi: 10.1016/j.critrevonc.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 81.Dorard C., de Thonel A., Collura A. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat Med. 2011;17(10):1283–1289. doi: 10.1038/nm.2457. [DOI] [PubMed] [Google Scholar]
- 82.Koenig J.L., Lin A.Y., Pollom E.L., Chang D.T. Microsatellite instability and adjuvant chemotherapy in stage II colon cancer. J Clin Orthod. 2018;36(4_suppl):e767. doi: 10.1097/COC.0000000000000554. [DOI] [PubMed] [Google Scholar]
- 83.Douillard J.-Y., Ostoros G., Cobo M. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Canc. 2014;110(1):55–62. doi: 10.1038/bjc.2013.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ciardiello F., Normanno N., Martinelli E. Cetuximab continuation after first progression in metastatic colorectal cancer (CAPRI-GOIM): a randomized phase II trial of FOLFOX plus cetuximab versus FOLFOX. Ann Oncol. 2016;27(6):1055–1061. doi: 10.1093/annonc/mdw136. [DOI] [PubMed] [Google Scholar]
- 85.Peeters M., Oliner K.S., Price T.J. Analysis of KRAS/NRAS mutations in a phase III study of panitumumab with FOLFIRI compared with FOLFIRI alone as second-line treatment for metastatic colorectal cancer. Clin Cancer Res. 2015;21(24):5469–5479. doi: 10.1158/1078-0432.CCR-15-0526. [DOI] [PubMed] [Google Scholar]
- 86.Rowland A., Dias M.M., Wiese M.D. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Canc. 2015;112(12):1888–1894. doi: 10.1038/bjc.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cremolini C., Loupakis F., Antoniotti C. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16(13):1306–1315. doi: 10.1016/S1470-2045(15)00122-9. [DOI] [PubMed] [Google Scholar]
- 88.Kopetz S., McDonough S.L., Morris V.K. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG 1406) J Clin Orthod. 2017;35(4_suppl):e520. doi: 10.1200/JCO.20.01994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Overman M.J., Kopetz S., McDermott R.S. Nivolumab ± ipilimumab in treatment (tx) of patients (pts) with metastatic colorectal cancer (mCRC) with and without high microsatellite instability (MSI-H): CheckMate-142 interim results. J Clin Orthod. 2016;34(15_suppl):e3501. [Google Scholar]
- 90.Brahmer J., Reckamp K.L., Baas P. Nivolumab versus docetaxel in advanced squamous-cell non–small-cell lung cancer. N Engl J Med. 2015;373(2):123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Apolo A.B., Infante J.R., Balmanoukian A. Avelumab, an anti–programmed death-ligand 1 antibody, in patients with refractory metastatic urothelial carcinoma: results from a multicenter, phase ib study. J Clin Oncol. 2017;35(19):2117–2124. doi: 10.1200/JCO.2016.71.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reck M., Rodríguez-Abreu D., Robinson A.G. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 93.Zhao P., Li L., Jiang X., Li Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol. 2019;12(1):e54. doi: 10.1186/s13045-019-0738-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marcus L., Lemery S.J., Keegan P., Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res. 2019;25(13):3753–3758. doi: 10.1158/1078-0432.CCR-18-4070. [DOI] [PubMed] [Google Scholar]
- 95.Lemery S., Keegan P., Pazdur R. First FDA approval agnostic of cancer site - when a biomarker defines the indication. N Engl J Med. 2017;377(15):1409–1412. doi: 10.1056/NEJMp1709968. [DOI] [PubMed] [Google Scholar]
- 96.Overman M.J., Lonardi S., Wong K.Y.M. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair–deficient/microsatellite instability–high metastatic colorectal cancer. J Clin Orthod. 2018;36(8):773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 97.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 98.Ogino S., Giannakis M. Immunoscore for (colorectal) cancer precision medicine. The Lancet. 2018;391(10135):2084–2086. doi: 10.1016/S0140-6736(18)30953-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martin V., Landi L., Molinari F. HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Canc. 2013;108(3):668–675. doi: 10.1038/bjc.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laurent-Puig P., Balogoun R., Cayre A. ERBB2 alterations a new prognostic biomarker in stage III colon cancer from a FOLFOX based adjuvant trial (PETACC8) Ann Oncol. 2016;27(6_suppl):vi149–vi206. [Google Scholar]
- 101.Seshagiri S., Stawiski E.W., Durinck S. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Madan B., Ke Z., Harmston N. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2016;35(17):2197–2207. doi: 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]