Abstract
Thousands of long non-coding RNAs (lncRNAs) have been discovered in human genomes by gene chip, next-generation sequencing, and/or other methods in recent years, which represent a significant subset of the universal genes involved in a wide range of biological functions. An abnormal expression of lncRNAs is associated with the growth, invasion, and metastasis of various types of human cancers, including hepatocellular carcinoma (HCC), which is an aggressive, highly malignant, and invasive tumor, and a poor prognosis in China. With a more in-depth understanding of lncRNA research for HCC and the emergence of new molecular-targeted therapies, the diagnosis, treatment, and prognosis of HCC will be considerably improved. Therefore, this review is expected to provide recommendations and directions for future lncRNA research for HCC.
Keywords: ceRNAs, HCC, lncRNAs, miRNAs, ncRNAs, snoRNAs
Abbreviations: HCC, Hepatocellular carcinoma; RNA, Ribonucleic acid; mRNA, Messenger RNA; ncRNAs, Non-coding RNAs; lncRNAs, Long non-coding RNAs; miRNAs, microRNAs; ceRNAs, Competing endogenous RNAs; snoRNAs, Small nucleolar RNAs; AASLD, American Association for the Study of Liver Diseases
Introduction
Depending on the differences in their structure and function, RNAs have been mainly divided into three categories, namely, messenger RNAs (mRNAs), transport RNAs (tRNAs), and ribosomal RNA (rRNAs). The mRNAs are transcribed according to the DNA in the nucleus, which is a template for the synthesis of proteins. The tRNAs are used to identify the genetic codons on the mRNAs and transport the amino acids. The rRNA is a component of ribosomes, which is a workplace for protein synthesis. In most eukaryotic cells, an abundance of different types of RNAs is different, and the rRNAs account for 80%–85% of the cellular RNA mass roughly, followed by tRNAs and mRNAs.1, 2, 3, 4 At some stage of germ cell development, these RNA mass ratios might change because of the PIWI-interacting RNA (piRNA) expression.5
With the rapid development of molecular biology and molecular diagnostic technology, an increasing number of non-coding RNAs (ncRNAs)6 in the human genome have been discovered by gene chip, next-generation sequencing, and/or other methods in recent years, such as long non-coding RNAs (lncRNAs), microRNAs (miRNAs), competing endogenous RNAs (ceRNAs), and small nucleolar RNAs (snoRNAs).7 However, the specific molecular mechanisms of lncRNAs, miRNAs, and ceRNAs are unclear. The ncRNAs that have occupied a vast majority of chromosomes are involved in many biological processes and have been increasingly seen as important transcriptional regulators. Initially, they were considered “junk genes” that did not encode proteins; then, they were gradually discovered to play an important regulatory role in various biological processes8, 9, 10, 11, 12, 13, 14 and perhaps as a regulator of the innate immune cell development and inflammatory gene expressions, biomarkers, and therapeutic targets for liver fibrosis or tumors, particularly in the process of tumor occurrence and development. This could be attributed to the intricate molecular mechanism between the lncRNAs and the other genes, which have rapidly become star molecules in the field of genetic research, particularly in the study of the correlation between lncRNAs and tumors.15, 16, 17, 18, 19 This article will focus on the latest advances in lncRNA research for HCC.
Characteristics of lncRNAs
LncRNAs have been identified as non-protein coding RNA transcripts with a length of approximately more than 200 nt, which have been shown to be involved in diverse molecular functions and pathological processes in epigenetic regulation.20 LncRNAs often have a poly(A) tail structure, which accounts for approximately 80% of the ncRNAs and is regulated by a variety of transcription factors.21 At the beginning, researchers thought that lncRNAs were a by-product of RNA polymerase II transcription. With an in-depth understanding of lncRNAs, a number of studies have shown that lncRNAs may have multiple origins: (1) chromatin recombination, (2) replicon concatenation, (3) transposon insertion into the gene, (4) reverse translocation during ncRNA replication, and (5) structural disruption of coding genes.22
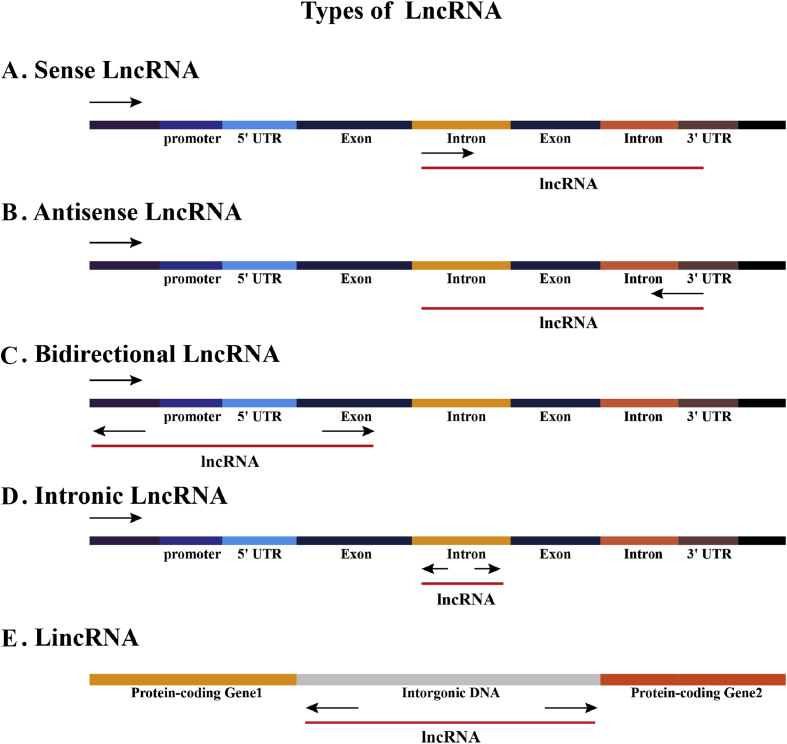
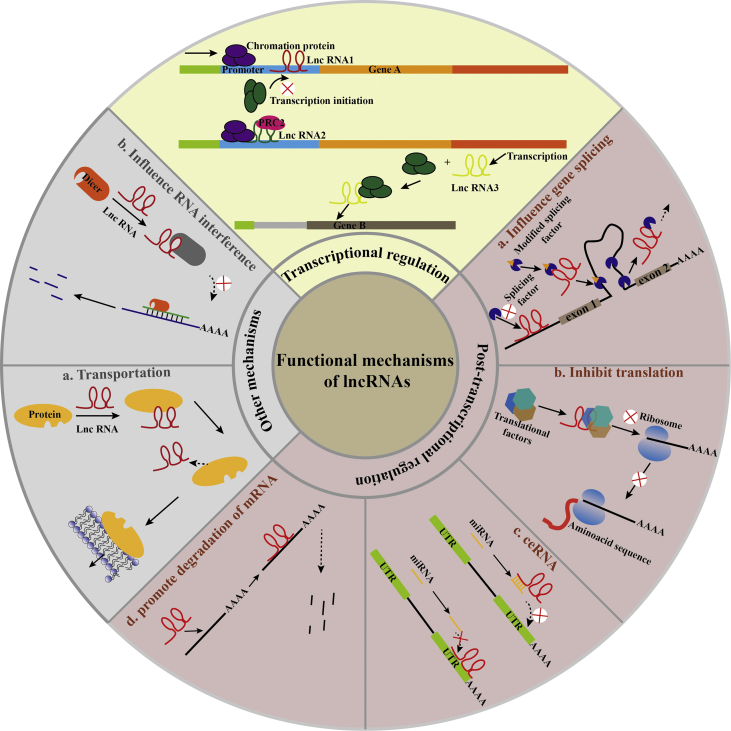
However, currently, there is no unified standard for the classification of lncRNAs. For the classification method, some researchers23,24 have suggested that it can be carried out according to the following characteristics: (1) According to the position of the genome, lncRNAs can be divided into five types: sense lncRNAs, antisense lncRNAs, bidirectional lncRNAs, intronic lncRNAs, and intergenic lncRNAs (lincRNAs) (Fig. 1). (2) According to the mode of action based on genes, they can be divided into cis-acting, trans-acting, and so on. (3) According to the functional mechanisms, they can be divided into the transcription level, post-transcription level, translation level, and so on (Fig. 2). (4) According to the mechanism of searching for target genes, they can be divided into the signal mode, molecular decoy mode, guidance mode, molecular framework mode, and so on.
Figure 1.
Types of lncRNAs.
Figure 2.
Types of functional mechanisms of lncRNAs.
Biological function of lncRNAs
With an in-depth study,25, 26, 27, 28, 29, 30, 31 it was found that lncRNAs can participate in not only the normal physiological activities of cells but also in the pathophysiological activity of various diseases, particularly tumors, through the modification of chromosomes, splicing, transcriptional activation, mRNA degradation, translation regulation, and so on. The molecular mechanisms of action of lncRNAs can be explained as follows: (1) acting on the promoter regions of the encoding gene, which can interfere with the transcriptional process; (2) forming a double strand with a transcript that can encode the gene by base complementation and then interfere with the cleavage of the mRNA or generate endogenous interfering RNA by using the Dicer enzyme; (3) influencing gene expression by interfering with the activity of RNA polymerase II, mediating chromatin remodeling; (4) playing a biological function by binding to specific proteins; (5) altering the cytoplasmic localization of proteins; and (6) playing a biological function as a structural component of protein or as a precursor molecule of small RNA.
Biological function of lncRNAs in carcinomas
In the initial stage of lncRNA research, some scholars suggested that there is a difference in the expression of lncRNAs between tumor tissues and adjacent normal tissues. An increasing number of studies32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65 have shown that lncRNAs may play an important biological role in the regulation of cancer development, as the biological function of lncRNAs in carcinomas is to promote cancer cell proliferation and invasion, suppress cancer cell proliferation and invasion, estimate the prognosis and efficacy, and act as potential biomarkers, as summarized in Table 1.
Table 1.
Biological function of lncRNAs in carcinomas.
| Biological Function | LncRNA | Target Gene/Pathway | Cancer Type | Reference |
|---|---|---|---|---|
| Promote proliferation, migration, and invasion | LINC00052 | miR-608/EGFR | Head and neck cancer | 32 |
| AC009022.1 | miR-497-5p | Colorectal cancer | 33 | |
| DLGAP1-AS1 | miR-486-5p | Hepatocellular cancer | 34 | |
| GHSROS | – | Breast cancer | 35 | |
| LINC00337 | TIMP2/DNMT1 | Non-small-cell lung cancer | 36 | |
| AK001058 | ADAMTS12 | Colorectal cancer | 37 | |
| FOXD2-AS1 | miR-185-5p | Thyroid cancer | 38 | |
| LINC00460 | – | Colorectal cancer | 39 | |
| LINC00908 | Sox-4 | Hepatocellular cancer | 40 | |
| PVT1 | Smad3/miR-140-5p | Cervical cancer | 41 | |
| RAIN | RUNX2 | Breast and thyroid cancer | 42 | |
| LINC00673 | miR-515-5p/MARK4/Hippo | Breast cancer | 43 | |
| TTN-AS1 | KLF15 | Colorectal cancer | 44 | |
| SNHG4 | ZIC5 | Prostate cancer | 45 | |
| SOX2-OT | miR-369-3p/CFL2 | Prostate cancer | 46 | |
| LINC01559 | YAP | Pancreatic cancer | 47 | |
| VCAN-AS1 | p53 | Gastric cancer | 48 | |
| Suppress proliferation and invasion | OSER1-AS1 | miR-372-3p/Rab23 | Hepatocellular cancer | 49 |
| ZEB1-AS1 | ZEB1 | Esophageal cancer | 50 | |
| NBAT-1 | PKM2 | Esophageal cancer | 51 | |
| ENST00000489676 | MiR-922 | Thyroid cancer | 52 | |
| CASC2c | ERK1/2, Wnt/β-catenin | Hepatocellular cancer | 53 | |
| GAS5 | YAP | Colorectal cancer | 54 | |
| ADAMTS9-AS2 | CDH3 | Esophageal cancer | 55 | |
| TCONS_00020456 | Smad2/PKCα | Glioblastoma | 56 | |
| Estimate prognosis and efficacy | UCA1, H19 | 5-fluorouracil | Rectal cancer | 57 |
| ADAMTS9-AS2 | FUS/MDM2 | Glioblastoma | 58 | |
| INCAC112721.1, AL356479.1, LINC00466 | hsa-miR-204 | Breast cancer | 59 | |
| GAS5, HOTAIR, H19, MALAT | – | Colorectal cancer | 60 | |
| HOXA-AS3 | HOXA3 | Non-small-cell lung cancer | 61 | |
| Act as potential biomarkers | MALAT1 | – | Breast cancer | 62 |
| HOTAIR | – | Breast cancer | 63 | |
| PURPL, NONHSAT062994 | – | Gastric cancer | 47 | |
| SNHG11 | – | Colorectal cancer | 64 | |
| SNHG12 | – | Pan-cancer | 65 |
Note: References 32–48 discuss the promotion of cancer cell proliferation and invasion, references 49–56 discuss the suppression of cancer cell proliferation and invasion, references 57–61 discuss the estimation of the prognosis, and references 62–66 discuss efficacy or potential biomarkers.
In short, according to the function of lncRNAs in tumors, lncRNAs can be divided into carcinogenic lncRNAs and anticancer lncRNAs. For example, the lncRNA DANCR can promote the proliferation and invasion of gastric cancer cells, but in the case of knockdown lncRNA DANCR, the migration and invasion of GC cells via the suppression of epithelial–mesenchymal transition (EMT) can be inhibited.66 It has been reported that67 the lncRNA NEAT1 has emerged as an important participant of the complex transcription network in cancer development. Recently, further studies have shown that68 the lncRNA-meditated gene expression can also be at the post-transcriptional level, which includes gene splicing, mRNA stability, protein stability, and nuclear trafficking. Some studies have shown that lncRNAs can act as tumor-promoting factors sometimes but at other times, they can act as tumor suppressors, such as H19. It has been reported that69 the level of lncRNA H19 is highly correlated with a higher tumor burden of papillary thyroid carcinoma, which also contributes to the EMT process and then promotes the migration and invasion of papillary thyroid carcinoma cells. However, in the another research,70 even in the case of the same type of tumor (papillary thyroid carcinoma), the lncRNA H19 has an opposite effect that can inhibit cell migration and invasion in the thyroid carcinoma cells. Professor Wu71 reported that the H19-mTOR-4E- BP1 axis can regulate pituitary tumor growth, which may be a potential therapeutic target and be more effective than cabergoline treatment in the suppression of human pituitary carcinomas. The lncRNA H19 can also be used as an important biomarker for prognosis and efficacy evaluation.72,73 To date, more than 8000 lncRNAs have been discovered in cancer cells. Their increased number and high expression specificity make these molecules a valuable source of biomarkers and potential therapeutic targets.74 As the research progresses, the regulatory network in the tumor by lncRNAs will become more complicated.
Biological function of lncRNAs in HCC
HCC is one of the most common malignancies in the world, and its mortality rate ranks the third among cancer-related deaths.75 However, thus far, no effective systematic research has been conducted for the early diagnosis and treatment of HCC. Moreover, the discovery and diagnosis of HCC is often in the late stage of the disease, which may cause a very poor prognosis. The American Association for the Study of Liver Diseases (AASLD) recommends that high-risk patients be regularly screened and monitored.76 The past treatments for this disease include liver transplantation, surgical liver resection, chemotherapy, radiotherapy, vascular embolization, and ablation.77 The early screening and diagnosis of HCC, particularly with the rapid development of the liquid biopsy technology, will help the prognosis of patients with HCC. Therefore, some researchers have paid more attention to the molecular targets of the HCC treatment and hope to find specific molecular markers for the diagnosis and treatment of HCC. With the deepening of research, many studies have shown that some lncRNAs are directly related to the occurrence and metastasis of HCC.78, 79, 80, 81, 82, 83, 84 HULC (lncRNA highly upregulated in the cases of liver cancer), the first discovered lncRNA particularly upregulated in HCC, was found to be more highly expressed in HCC tissues than in normal liver tissues, and the expression level of HULC correlated with the prognosis of patients.85, 86, 87 LncRNAs have complex regulatory effects on the development of HCC; as early as 2010, some researchers showed that CREB could upregulate HULC with miR-372, which was regarded as a ceRNA.88 A number of studies have shown that lncRNAs can play an important biological role in the regulation of HCC development; the biological function of lncRNAs in HCC is to promote cancer cell proliferation and invasion, suppress cancer cell proliferation and invasion, estimate prognosis and efficacy, and act as potential biomarkers, as summarized in Table 2.
Table 2.
Biological function of lncRNAs in HCC.
| Biological Function | LncRNA | Target Gene/Pathway | Reference(s) |
|---|---|---|---|
| Promote proliferation, migration, and invasion | HOTAIR | OGFr, miR-122, SETD2 | 90, 91, 92, 93, 94 |
| IHS | ERK- and AKT/GSK-3β | 96 | |
| HOXD-AS1 | SOX4 | 97 | |
| PDPK2P | PDK1/AKT/Caspase 3 | 98 | |
| ATB | autophagy-related protein 5 | 99 | |
| Ftx | PPARγ | 100 | |
| lncCAMTA1 | CAMTA1 | 101 | |
| SNHG15 | miR-490-3p/histone deacetylase 2 axis | 102 | |
| RNA LINC00908 | Sox-4 | 103 | |
| MCM3AP-AS1 | miR-194-5p/FOXA1 axis | 104 | |
| Suppress proliferation and invasion | GAS8-AS1 | GAS8 | 105 |
| SVUGP2 | MMP2 and 9 | 106 | |
| uc.134 | LATS1 | 107 | |
| TCONS_00006195 | ENO1 | 108 | |
| EPB41L4A-AS2 | miR-301a-5p/FOXL1 | 109 | |
| FENDRR | miR-423-5p | 110 | |
| SchLAH | – | 111 | |
| GAS5 | miR-182/ANGPTL1 | 112 | |
| MIR31HG | microRNA-575 | 113 | |
| MIR22HG | miR-10a-5p/NCOR2 | 114 | |
| Estimate prognosis and efficacy | HOTAIR | – | 92 |
| MIR22HG, CTC-297N7.9, CTD-2139B15.2, RP11-589N15.2, RP11-343N15.5, and RP11-479G22.8 | – | 115 | |
| MALAT1, HOTAIR, MDIG | Caspase-8/LSD1/H3K9me3/ | 116 | |
| miR503HG | HNRNPA2B1/NF-κB | 117 | |
| RP11-466I1.1 | – | 118 | |
| Act as potential biomarkers | PVT1, uc002mbe.2 | – | 119 |
| UCA1 | – | 120 | |
| LRB1 | – | 121 | |
| RP11-486O12.2, RP11-863K10.7, LINC01093 and RP11-273G15.2 | – | 122 | |
| ELMO1-AS1 | – | 123 |
Note: References 91–105 discuss the promotion of cancer cell proliferation and invasion, References 106–115 discuss the suppression of cancer cell proliferation and invasion, References 93 and 116–119 discuss the estimation of prognosis, and References 120–124 discuss the efficacy or potential biomarkers.
The lncRNA HOTAIR (which stands for HOX transcript antisense intergenic RNA)89 is considered to be a carcinogenic lncRNA involved in the regulation of most human cancers; it is a 2158-nt lncRNA discovered by Howard Chang's group (lncRNA Database, http://www.lncrnadb.org/) and is located on human chromosome 12q13.13. Some studies have reported that as a human tumorigenesis regulatory gene, HOTAIR is dysregulated in HCC frequently. Ying90 reported that after the inhibition of HOTAIR, there are, in all, 673 transcripts and 293 proteins the can be dysregulated in HCC and promote HCC cell proliferation by regulating the opioid growth factor receptor (OGFr). In the case of knockdown HOTAIR, the HCC cell proliferation can be inhibited dramatically and tumorigenicity can be suppressed by upregulating the miR-122 expression.91 The expression of HOTAIR92 is strongly associated with the prognosis in HCC, which is an indicator of poor prognosis, may be a therapeutic target in HCC, and can promote the HCC cell proliferation by the suppressed RNA binding motif protein 38.93 HOTAIR can promote the malignant growth of HCC stem cells by the downregulation of SETD2.94 HOTAIR can also promote the release of exosomes by inducing MVB transport to the plasma membrane, which is regulated by RAB35 and SNAP23.95
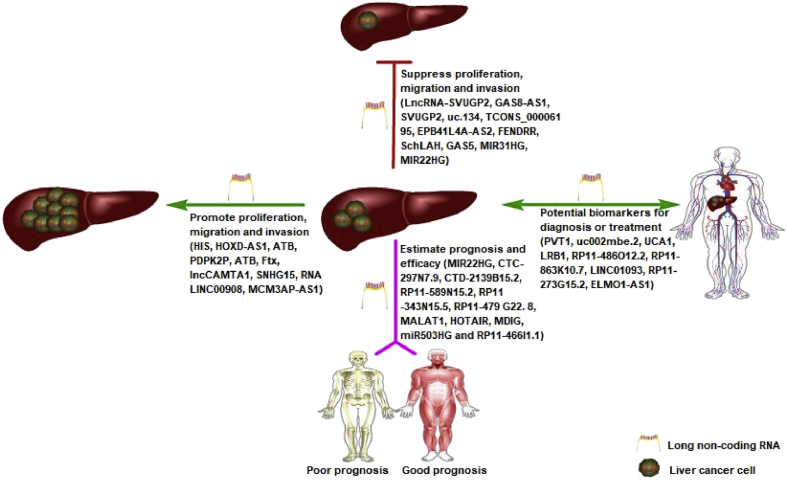
A number of lncRNAs96, 97, 98, 99, 100, 101, 102, 103, 104 can promote the progress (such as proliferation, migration, and invasion) of HCC. The lncRNAs HIS, HOXD-AS1, ATB, PDPK2P, ATB, Ftx, lncCAMTA1, SNHG15, RNA LINC00908, and MCM3AP-AS1 can accelerate the proliferation, migration, and invasion of hepatocellular carcinoma. However, some researchers have shown that the lncRNAs105, 106, 107, 108, 109, 110, 111, 112, 113, 114 can also inhibit the development of HCC. The lncRNA GAS8-AS1 can activate GAS8 epigenetically and then inhibit the malignant transformation of hepatocytes. The progression (such as proliferation, migration, and invasion) of hepatocellular carcinoma can be suppressed by a mass of lncRNAs, such as LncRNA-SVUGP2, GAS8-AS1, SVUGP2, uc.134, TCONS_000061 95, EPB41L4A-AS2, FENDRR, SchLAH, GAS5, MIR31HG, and MIR22HG.
In contrast, the lncRNAs MIR22HG, CTC-297N7.9, CTD-2139B1 5.2, RP11-589N15.2, RP11-343N15.5, RP11-479 G22. 8, MALAT1, HOTAIR, MDIG, miR503HG, RP11-466I1.1 PVT1, uc002mbe.2, UCA1, LRB1, RP11-486O12.2, RP11-863K10.7, LINC01093, RP11-273G15.2, and ELMO1-AS1 can estimate the prognosis and efficacy or maybe act as potential biomarkers for the diagnosis or treatment in HCC.92,115, 116, 117, 118, 119, 120, 121, 122, 123 In the meantime, some researchers106,124,125 have also reported that the HCC with different viral causes was regulated by different lncRNAs, and the lncRNAs can regulate the progression (such as proliferation, migration, and invasion) of hepatocellular carcinoma by regulating various pathways.
Conclusion and perspective
As is known to all, HCC is a type of malignant tumor with high morbidity and mortality in the world. However, there are still some bottlenecks in the early screening and postoperative efficacy of HCC, and the high metastaticity and the lack of the specific molecular-targeted drugs lead to the poor prognosis of liver cancer. At present, the research on lncRNA in HCC is mostly at the level of cell or mouse experiments, the clinical research is relatively little, and the study on the role of lncRNAs in the regulation of gene expression is just the tip of the iceberg, which may seriously restrict the application of lncRNA in HCC. Although some progress and achievements have been made in the research on lncRNAs in HCC, and in fact, the biological function of lncRNAs in HCC is extremely complicated, this article is only a review of the recent research on lncRNAs in HCC; further research on the role of lncRNAs in the regulation of the development of HCC should be conducted more extensively.
This review provides an overall view of lncRNAs in HCC. The lncRNAs have been divided into four categories according to the regulatory mechanisms in HCC (Fig. 3). The biological functions of lncRNAs in HCC are as follows: The first aspect is to promote the proliferation, migration, and invasion of HCC; the second aspect is to suppress the proliferation and invasion of HCC; the third aspect is to estimate the prognosis and efficacy of HCC; the fourth aspect is to act as potential biomarkers that can be used for the diagnosis and treatment of HCC. In fact, the biological function of lncRNAs in HCC is intertwined; most of the lncRNAs that have been clearly studied belong to the tumor gene activation category, which mainly promotes the occurrence and development of HCC by activating the Wnt, STAT-3, EMT, and other signaling pathways, but these lncRNAs may have the potential to act as tumor markers for clinical diagnosis or treatment.
Figure 3.
Types of lncRNAs in HCC.
Although many studies126, 127, 128 have confirmed that the biological function of lncRNA can be mutually regulated with miRNA, and the regulatory axis of lncRNA-miRNA-mRNA has been proved to exist in other tumors, no complete regulatory network has been established in HCC. In order to fully grasp the regulatory role of lncRNA in the development of HCC, it is important to explore the regulatory network of miRNA–lncRNA–mRNA in HCC and the other undiscovered signaling pathways. The multidimensional regulatory mechanisms of lncRNA in HCC should be further explored and analyzed, and the comprehensive and detailed lncRNA-related databases should be established as soon as possible. There is increasing evidence that an abnormal expression of lncRNAs is associated with diseases, particularly tumors. We need to find specific lncRNAs that are closely related to HCC and study their specific mechanism in depth to carry out more related work and find new directions for HCC diagnosis and treatment. The multidimensional methods of molecular biology, bioinformatics, and other fields should be combined to study the lncRNAs in HCC in order to establish a sophisticated regulatory network successfully. Therefore, this review is expected to be used to provide recommendations and directions for future lncRNA research for HCC.
Conflict of Interests
The authors declare no conflict of interests.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant number 81871653), the Program for Innovation Team of Higher Education in Chongqing (grant number CXTDX201601015) and the Natural Science Foundation of Chongqing (grant number cstc2016jcyjA0269). The authors are grateful to the study participants.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Dominissini D., Moshitch-Moshkovitz S., Schwartz S. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 2.Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci. 2013;38(4):204–209. doi: 10.1016/j.tibs.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu N., Parisien M., Dai Q. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19(12):1848–1856. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlile T.M., Rojas-Duran M.F., Zinshteyn B. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weick E.M., Miska E.A. piRNAs: from biogenesis to function. Development. 2014;141(18):3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- 6.Guttman M., Amit I., Garber M. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jankowsky E., Harris M.E. Specificity and non-specificity in RNA-protein interactions. Nat Rev Mol Cell Biol. 2015;16(9):533–544. doi: 10.1038/nrm4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teng K.Y., Ghoshal K. Role of noncoding RNAs as biomarker and therapeutic targets for liver fibrosis. Gene Expr. 2015;16(4):155–162. doi: 10.3727/105221615X14399878166078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meller V.H., Joshi S.S., Deshpande N. Modulation of chromatin by noncoding RNA. Annu Rev Genet. 2015;49:673–695. doi: 10.1146/annurev-genet-112414-055205. [DOI] [PubMed] [Google Scholar]
- 10.Gromesová B., Kubaczková V., Bollová B. Potential of long Non- coding RNA molecules in diagnosis of tumors. Klin Onkol. 2016;29(1):20–28. [PubMed] [Google Scholar]
- 11.Elling R., Chan J., Fitzgerald K.A. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur J Immunol. 2016;46(3):504–512. doi: 10.1002/eji.201444558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng Hua, Tan Xiao-Di. Functional diversity of long non-coding RNAs in immune regulation. Genes Dis. 2016;3(1):72–81. doi: 10.1016/j.gendis.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y., Wang X. Role of long noncoding RNAs in malignant disease (Review) Mol Med Rep. 2016;13(2):1463–1469. doi: 10.3892/mmr.2015.4711. [DOI] [PubMed] [Google Scholar]
- 14.Liu Yamin, Han Ting-Li, Luo Xiaofang. The metabolic role of LncZBTB39-1:2 in the trophoblast mobility of preeclampsia. Genes Dis. 2018;5(3):235–244. doi: 10.1016/j.gendis.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briggs J.A., Wolvetang E.J., Mattick J.S. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88(5):861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 16.Yu X., Li Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol Lett. 2015;10(4):1953–1958. doi: 10.3892/ol.2015.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugihara H., Ishimoto T., Miyake K. Noncoding RNA expression aberration is associated with cancer progression and is a potential biomarker in esophageal squamous cell carcinoma. Int J Mol Sci. 2015;16(11):27824–27834. doi: 10.3390/ijms161126060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao H., Yang J.J., Zhou X. Emerging role of long noncoding RNAs in lung cancer: current status and future prospects. Respir Med. 2016;110:12–19. doi: 10.1016/j.rmed.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Ouyang J., Hu J., Chen J.L. lncRNAs regulate the innate immune response to viral infection. Wiley Interdiscip Rev RNA. 2016;7(1):129–143. doi: 10.1002/wrna.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Z., Hu Y., Lai S. Long Noncoding RNA: its partners and their roles in cancer. Neoplasma. 2015;62(6):846–854. doi: 10.4149/neo_2015_103. [DOI] [PubMed] [Google Scholar]
- 21.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Kumar Mohan M., Goyal Ravi. LncRNAs as a therapeutic target for angiogenesis. Curr Top Med Chem. 2017;17(15):1750–1757. doi: 10.2174/156802661766616111644744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P., Zhang G., Li J. Long noncoding RNA RGMB-AS1 indicates a poor prognosis and modulates cell proliferation, migration and invasion in lung adenocarcinoma. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0150790. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Wilk R., Hu J., Blotsky D. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016;30(5):594–609. doi: 10.1101/gad.276931.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K., Zhao B.S., He C. Nucleic acid modifications in regulation of gene expression. Cell Chem Biol. 2016;23(1):74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Sun J., Wang J. Long noncoding RNAs in gastric cancer: functions and clinical applications. OncoTargets Ther. 2016;9:681–697. doi: 10.2147/OTT.S95412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Xu A.M., Zhang J.Y. Prognostic significance of long non-coding RNA MALAT-1 in various human carcinomas: a meta-analysis. Genet Mol Res. 2016;29(1):e15. doi: 10.4238/gmr.15017433. [DOI] [PubMed] [Google Scholar]
- 30.Li C., Chen J., Zhang K. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36(2):423–434. doi: 10.1159/000430109. [DOI] [PubMed] [Google Scholar]
- 31.Sun Lin, Guo Yujie, Peng He. Genome-wide profiling of long noncoding RNA expression patterns and CeRNA analysis in mouse cortical neurons infected with different strains of borna disease virus. Genes Dis. 2019;6(2):147–158. doi: 10.1016/j.gendis.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouyang T., Zhang Y., Tang S. Long non-coding RNA LINC00052 regulates miR-608/EGFR axis to promote progression of head and neck squamous cell carcinoma. Exp Mol Pathol. 2019;111:e104321. doi: 10.1016/j.yexmp.2019.104321. [DOI] [PubMed] [Google Scholar]
- 33.Yu C., Zhang F. LncRNA AC009022.1 enhances colorectal cancer cells proliferation, migration, and invasion by promoting ACTR3B expression via suppressing miR-497-5p. J Cell Biochem. 2020;121(2):1934–1944. doi: 10.1002/jcb.29428. [DOI] [PubMed] [Google Scholar]
- 34.Peng X., Wei F., Hu X. Long noncoding RNA DLGAP1-AS1 promotes cell proliferation in hepatocellular carcinoma via sequestering miR-486-5p. J Cell Biochem. 2020;121(2):1953–1962. doi: 10.1002/jcb.29430. [DOI] [PubMed] [Google Scholar]
- 35.Thomas P.B., Seim I., Jeffery P.L. The long non-coding RNA GHSROS facilitates breast cancer cell migration and orthotopic xenograft tumour growth. Int J Oncol. 2019;55(6):1223–1236. doi: 10.3892/ijo.2019.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Gong J., Lu J. Long noncoding RNA LINC00337 accelerates the non-small-cell lung cancer progression through inhibiting TIMP2 by recruiting DNMT1. Am J Transl Res. 2019;11(9):6075–6083. [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng S., Lin F., Zhang M. Long non-coding RNA AK001058 regulates tumor growth and angiogenesis in colorectal cancer via methylation of ADAMTS12. Am J Transl Res. 2019;11(9):6117–6123. [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Han Q., Chen Y. Upregulation of the long non-coding RNA FOXD2-AS1 is correlated with tumor progression and metastasis in papillary thyroid cancer. Am J Transl Res. 2019;11(9):5457–5471. [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H., Lu Y., Wu J. LINC00460 hypomethylation promotes metastasis in colorectal carcinoma. Front Genet. 2019;30(10):e880. doi: 10.3389/fgene.2019.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu X., Li Q., Zhang J. The long noncoding RNA LINC00908 facilitates hepatocellular carcinoma progression via interaction with Sox-4. Cancer Manag Res. 2019;11:8789–8797. doi: 10.2147/CMAR.S216774. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Chang Q.Q., Chen C.Y., Chen Z. LncRNA PVT1 promotes proliferation and invasion through enhancing Smad3 expression by sponging miR-140-5p in cervical cancer. Radiol Oncol. 2019;53(4):443–452. doi: 10.2478/raon-2019-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi T., Pistoni M., Sancisi V. RAIN is a novel Enhancer-associated lncRNA that controls RUNX2 expression and promotes breast and thyroid cancer. Mol Cancer Res. 2020;18(1):140–152. doi: 10.1158/1541-7786.MCR-19-0564. [DOI] [PubMed] [Google Scholar]
- 43.Qiao K., Ning S., Wan L. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38(1):e418. doi: 10.1186/s13046-019-1421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Jiang F., Xiong Y. LncRNA TTN-AS1 sponges miR-376a-3p to promote colorectal cancer progression via upregulating KLF15. Life Sci. 2019;11:e116936. doi: 10.1016/j.lfs.2019.116936. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z.Y., Duan Y., Wang P. SP1-mediated upregulation of lncRNA SNHG4 functions as a ceRNA for miR-377 to facilitate prostate cancer progression through regulation of ZIC5. J Cell Physiol. 2020;235(4):3916–3927. doi: 10.1002/jcp.29285. [DOI] [PubMed] [Google Scholar]
- 46.Wo Q., Zhang D., Hu L. Long noncoding RNA SOX2-OT facilitates prostate cancer cell proliferation and migration via miR-369-3p/CFL2 axis. Biochem Biophys Res Commun. 2019;520(3):586–593. doi: 10.1016/j.bbrc.2019.09.108. [DOI] [PubMed] [Google Scholar]
- 47.Lou C., Zhao J., Gu Y. LINC01559 accelerates pancreatic cancer cell proliferation and migration through YAP-mediated pathway. J Cell Physiol. 2020;235(4):3928–3938. doi: 10.1002/jcp.29288. [DOI] [PubMed] [Google Scholar]
- 48.Feng L., Li J., Li F. Long noncoding RNA VCAN-AS1 contributes to the progression of gastric cancer via regulating p53 expression. J Cell Physiol. 2020;1235(5):4388–4398. doi: 10.1002/jcp.29315. [DOI] [PubMed] [Google Scholar]
- 49.Fan J., Zhang J., Huang S., Li P. lncRNA OSER1-AS1 acts as a ceRNA to promote tumorigenesis in hepatocellular carcinoma by regulating miR-372-3p/Rab23 axis. Biochem Biophys Res Commun. 2020;521(1):196–203. doi: 10.1016/j.bbrc.2019.10.105. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y., Wang N., Zhang X. LncRNA ZEB1-AS1 down-regulation suppresses the proliferation and invasion by inhibiting ZEB1 expression in oesophageal squamous cell carcinoma. J Cell Mol Med. 2019;23(12):8206–8218. doi: 10.1111/jcmm.14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao B., Cao P., Hu S. LncRNA-NBAT-1 modulates esophageal cancer proliferation via PKM2. Am J Transl Res. 2019;11(9):5978–5987. [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W., Xia S., Zhan W. The long non-coding RNA ENST00000489676 influences papillary thyroid cancer cell proliferation and invasion through regulating MiR-922. J Cancer. 2019;10(22):5434–5446. doi: 10.7150/jca.31751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q.Y., Yang K., Liu F.G. Long noncoding RNA CASC2c inhibited cell proliferation in hepatocellular carcinoma by inactivated ERK1/2 and Wnt/β-catenin signaling pathway. Clin Transl Oncol. 2020;22(3):302–310. doi: 10.1007/s12094-019-02223-7. [DOI] [PubMed] [Google Scholar]
- 54.Ni W., Yao S., Zhou Y. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. 2019;18(1):e143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu D., Wu K., Yang Y. Long noncoding RNA ADAMTS9-AS2 suppresses the progression of esophageal cancer by mediating CDH3 promoter methylation. Mol Carcinog. 2020;59(1):32–44. doi: 10.1002/mc.23126. [DOI] [PubMed] [Google Scholar]
- 56.Tang C., Wang Y., Zhang L. Identification of novel LncRNA targeting Smad2/PKCα signal pathway to negatively regulate malignant progression of glioblastoma. J Cell Physiol. 2020;235(4):3835–3848. doi: 10.1002/jcp.29278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yokoyama Y., Sakatani T., Wada R. In vitro and in vivo studies on the association of long non-coding RNAs H19 and urothelial cancer associated 1 with the susceptibility to 5-fluorouracil in rectal cancer. Int J Oncol. 2019;55(6):1361–1371. doi: 10.3892/ijo.2019.4895. [DOI] [PubMed] [Google Scholar]
- 58.Yan Y., Xu Z., Chen X. Novel function of lncRNA ADAMTS9-AS2 in promoting temozolomide resistance in glioblastoma via upregulating the FUS/MDM2 ubiquitination Axis. Front Cell Dev Biol. 2019;7:e217. doi: 10.3389/fcell.2019.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J.J., Huang Y.Q., Song W. Comprehensive analysis of the lncRNA-associated competing endogenous RNA network in breast cancer. Oncol Rep. 2019;42(6):2572–2582. doi: 10.3892/or.2019.7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bermúdez M., Aguilar-Medina M., Lizárraga-Verdugo E. LncRNAs as regulators of autophagy and drug resistance in colorectal cancer. Front Oncol. 2019;9:e1008. doi: 10.3389/fonc.2019.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin S., Zhang R., An X. LncRNA HOXA-AS3 confers cisplatin resistance by interacting with HOXA3 in non-small-cell lung carcinoma cells. Oncogenesis. 2019;8(11):e60. doi: 10.1038/s41389-019-0170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miao Y., Fan R., Chen L. Clinical significance of long non- coding RNA MALAT1 expression in tissue and serum of breast cancer. Ann Clin Lab Sci. 2016;46(4):418–424. [PubMed] [Google Scholar]
- 63.Avazpour N., Hajjari M., Tahmasebi Birgani M. HOTAIR: a promising long non-coding RNA with potential role in breast invasive carcinoma. Front Genet. 2017;8:e170. doi: 10.3389/fgene.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu W., Zhou G., Wang H. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer. 2020;146(10):2901–2912. doi: 10.1002/ijc.32747. [DOI] [PubMed] [Google Scholar]
- 65.Tamang S., Acharya V., Roy D. SNHG12: an LncRNA as a potential therapeutic target and biomarker for human cancer. Front Oncol. 2019;9:e901. doi: 10.3389/fonc.2019.00901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pan L., Liang W., Gu J. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget. 2017;9(2):1915–1930. doi: 10.18632/oncotarget.23019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li S., Li J., Chen C., Zhang R., Wang K. Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Genes Dis. 2018;5(1):27–35. doi: 10.1016/j.gendis.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He R.Z., Luo D.X., Mo Y.Y. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6(1):6–15. doi: 10.1016/j.gendis.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang W.Q., Zeng D., Chen C.F. Long noncoding RNA H19 is a critical oncogenic driver and contributes to epithelial-mesenchymal transition in papillary thyroid carcinoma. Cancer Manag Res. 2019;11:2059–2072. doi: 10.2147/CMAR.S195906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang P., Liu G., Xu W., Liu H., Bu Q., Sun D. Long noncoding RNA H19 inhibits cell viability, migration, and invasion via downregulation of IRS-1 in thyroid cancer cells. Technol Cancer Res Treat. 2017;16(6):1102–1112. doi: 10.1177/1533034617733904. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Wu Z.R., Yan L., Liu Y.T. Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat Commun. 2018;9(1):e4624. doi: 10.1038/s41467-018-06853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao H., Hao G., Sun Y., Li L., Wang Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Onco Targets Ther. 2018;11:8001–8012. doi: 10.2147/OTT.S172379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao L.M., Xu S.F., Zheng Y. Long non-coding RNA H19 is responsible for the progression of lung adenocarcinoma by mediating methylation-dependent repression of CDH1 promoter. J Cell Mol Med. 2019;23(9):6411–6428. doi: 10.1111/jcmm.14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gugnoni M., Ciarrocchi A. Long noncoding RNA and epithelial mesenchymal transition in cancer. Int J Mol Sci. 2019;20(8):e1924. doi: 10.3390/ijms20081924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng G.S. Conflicting roles of molecules in epatocarcino- genesis: paradigm or paradox. Cancer Cell. 2012;21(2):150–154. doi: 10.1016/j.ccr.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang N., Ekanem N.R., Sakyi C.A. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev. 2015;81:62–74. doi: 10.1016/j.addr.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 77.Maluccio M., Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62(6):e394. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 78.Mehra M., Chauhan R. Long noncoding RNAs as a key player in hepatocellular carcinoma. Biomark Cancer. 2017;9 doi: 10.1177/1179299X17737301. e1179299X1 7737301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lanzafame M., Bianco G., Terracciano L.M. The role of long non-coding RNAs in hepatocarcinogenesis. Int J Mol Sci. 2018;19(3):e682. doi: 10.3390/ijms19030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qiu L., Tang Q., Li G. Long non-coding RNAs as biomarkers and therapeutic targets: recent insights into hepatocellular carcinoma. Life Sci. 2017;191:273–282. doi: 10.1016/j.lfs.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 81.He Y., Meng X.M., Huang C. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344(1):20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 82.Cao M.R., Han Z.P., Liu J.M. Bioinformatic analysis and prediction of the function and regulatory network of long non-coding RNAs in hepatocellular carcinoma. Oncol Lett. 2018;15(5):7783–7793. doi: 10.3892/ol.2018.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gramantieri L., Baglioni M., Fornari F. LncRNAs as novel players in hepatocellular carcinoma recurrence. Oncotarget. 2018;9(80):35085–35099. doi: 10.18632/oncotarget.26202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guerrieri F. Long non-coding RNAs era in liver cancer. World J Hepatol. 2015;7(16):1971–1973. doi: 10.4254/wjh.v7.i16.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang B.G., Lv Z., Ding H.X. The association of lncRNA-HULC polymorphisms with hepatocellular cancer risk and prognosis. Gene. 2018;670:148–154. doi: 10.1016/j.gene.2018.05.096. [DOI] [PubMed] [Google Scholar]
- 86.Xiong H., Li B., He J. lncRNA HULC promotes the growth of hepatocellular carcinoma cells via stabilizing COX-2 protein. Biochem Biophys Res Commun. 2017;490(3):693–699. doi: 10.1016/j.bbrc.2017.06.103. [DOI] [PubMed] [Google Scholar]
- 87.Sonohara F., Sugimoto H., Hayashi M. Prognostic value of long non-coding RNA HULC and MALAT1 following the curative resection of hepatocellular carcinoma. Sci Rep. 2017;7(1):e16142. doi: 10.1038/s41598-017-16260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J., Liu X., Wu H. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chi Y., Wang D., Wang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):e1015. doi: 10.3390/cells8091015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu Y., Xiong Q., Li S., Yang X., Ge F. Integrated proteomic and transcriptomic analysis reveals long noncoding RNA HOX transcript antisense intergenic RNA (HOTAIR) promotes hepatocellular carcinoma cell proliferation by regulating opioid growth factor receptor (OGFr) Mol Cell Proteom. 2018;17(1):146–159. doi: 10.1074/mcp.RA117.000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng D., Deng J., Zhang B. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine. 2018;36:159–170. doi: 10.1016/j.ebiom.2018.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yao Y., Li J., Wang L. Large intervening non-coding RNA HOTAIR is an indicator of poor prognosis and a therapeutic target in human cancers. Int J Mol Sci. 2014;15(10):18985–18999. doi: 10.3390/ijms151018985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu L., Zhang L., Zheng S. Role of the long non-coding RNA HOTAIR in hepatocellular carcinoma. Oncol Lett. 2017;14(2):1233–1239. doi: 10.3892/ol.2017.6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li H., An J., Wu M., Zheng Q. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget. 2015;6(29):27847–27864. doi: 10.18632/oncotarget.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang L., Peng X., Li Y. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol Cancer. 2019;18(1):e78. doi: 10.1186/s12943-019-0990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Z., Yu W., Zhou Q. A novel lncRNA IHS promotes tumor proliferation and metastasis in HCC by regulating the ERK- and AKT/GSK-3β-Signaling pathways. Mol Ther Nucleic Acids. 2019;16:707–720. doi: 10.1016/j.omtn.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang H., Huo X., Yang X.R. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:e136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pan W., Li W., Zhao J. lncRNA-PDPK2P promotes hepatocellular carcinoma progression through the PDK1/AKT/Caspase 3 pathway. Mol Oncol. 2019;13(10):2246–2258. doi: 10.1002/1878-0261.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang C.Z., Yan G.X., Dong D.S. LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J Gastroenterol. 2019;25(35):5310–5322. doi: 10.3748/wjg.v25.i35.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiao Li, Qi Zhao, Qi Jianni. lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARγ pathway in hepatocellular carcinoma. Int J Oncol. 2018;53(2):551–566. doi: 10.3892/ijo.2018.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ding L.J., Li Y., Wang S.D. Long noncoding RNA lncCAMTA1 promotes proliferation and cancer stem cell-like properties of liver cancer by inhibiting CAMTA1. Int J Mol Sci. 2016;17(10):e1617. doi: 10.3390/ijms17101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dai W., Dai J.L., Tang M.H. lncRNA-SNHG15 accelerates the development of hepatocellular carcinoma by targeting miR-490-3p/histone deacetylase 2 axis. World J Gastroenterol. 2019;25(38):5789–5799. doi: 10.3748/wjg.v25.i38.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu X., Li Q., Zhang J. The long noncoding RNA LINC00908 facilitates hepatocellular carcinoma progression via interaction with Sox-4. Cancer Manag Res. 2019;11:8789–8797. doi: 10.2147/CMAR.S216774. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Wang Y., Yang L., Chen T. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18:e28. doi: 10.1186/s12943-019-0957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pan W., Zhang N., Liu W. The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J Biol Chem. 2018;293(44):17154–17165. doi: 10.1074/jbc.RA118.003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hu J., Song C., Duan B. LncRNA-SVUGP2 suppresses progression of hepatocellular carcinoma. Oncotarget. 2017;8(58):97835–97850. doi: 10.18632/oncotarget.18279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wen N., Zhang Y., Zhan Z. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol. 2017;10(1):e91. doi: 10.1186/s13045-017-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu S., Li N., Huang Z. A novel lncRNA, TCONS_00006195, represses hepatocellular carcinoma progression by inhibiting enzymatic activity of ENO1. Cell Death Dis. 2018;9(12):e1184. doi: 10.1038/s41419-018-1231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y.G., Wang T., Shi M. Long noncoding RNA EPB41L4A-AS2 inhibits hepatocellular carcinoma development by sponging miR-301a-5p and targeting FOXL1. J Exp Clin Cancer Res. 2019;38(1):e153. doi: 10.1186/s13046-019-1128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu Z., Zhao H., Feng X. Long non-coding RNA FENDRR acts as a miR-423-5p sponge to suppress the treg-mediated immune escape of hepatocellular carcinoma cells. Mol Ther Nucleic Acids. 2019;17:516–529. doi: 10.1016/j.omtn.2019.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ge Z., Cheng Z., Yang X. Long noncoding RNA SchLAH suppresses metastasis of hepatocellular carcinoma through interacting with fused in sarcoma. Cancer Sci. 2017;108(4):653–662. doi: 10.1111/cas.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen F., Li Y., Li M., Wang L. Long noncoding RNA GAS5 inhibits metastasis by targeting miR-182/ANGPTL1 in hepatocellular carcinoma. Am J Cancer Res. 2019;9(1):108–121. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 113.Yan S., Tang Z., Chen K. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J Exp Clin Cancer Res. 2018;37(1):e214. doi: 10.1186/s13046-018-0853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu Y., Zhou Y., Huan L. LncRNA MIR22HG inhibits growth, migration and invasion through regulating the miR-10a-5p/NCOR2 axis in hepatocellular carcinoma cells. Cancer Sci. 2019;110(3):973–984. doi: 10.1111/cas.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu Y., Wang P.S., Wang B.G. Genomewide identification of a novel six-LncRNA signature to improve prognosis prediction in resectable hepatocellular carcinoma. Cancer Med. 2018;7(12):6219–6233. doi: 10.1002/cam4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li G., Zhang H., Wan X. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. BioMed Res Int. 2014;2014:e780521. doi: 10.1155/2014/780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang H., Liang L., Dong Q. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. 2018;8(10):2814–2829. doi: 10.7150/thno.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang J., Zhang D., Zhao Q., Qi J., Li X., Qin C. A distinctively expressed long noncoding RNA, RP11-466I1.1, may serve as a prognostic biomarker in hepatocellular carcinoma. Cancer Med. 2018;7(7):2960–2968. doi: 10.1002/cam4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu J., Han J., Zhang J. The long noncoding RNAs PVT1 and uc002mbe.2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Medicine (Baltim) 2016;95(31):e4436. doi: 10.1097/MD.0000000000004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zheng Z.K., Pang C., Yang Y., Duan Q., Zhang J., Liu W.C. Serum long noncoding RNA urothelial carcinoma-associated 1: a novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. J Int Med Res. 2018;46(1):348–356. doi: 10.1177/0300060517726441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Z.F., Hu R., Pang J.M. Serum long noncoding RNA LRB1 as a potential biomarker for predicting the diagnosis and prognosis of human hepatocellular carcinoma. Oncol Lett. 2018;16(2):1593–1601. doi: 10.3892/ol.2018.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li G., Shi H., Wang X. Identification of diagnostic long non-coding RNA biomarkers in patients with hepatocellular carcinoma. Mol Med Rep. 2019;20(2):1121–1130. doi: 10.3892/mmr.2019.10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Luo T., Chen M., Zhao Y. Macrophage-associated lncRNA ELMO1-AS1: a novel therapeutic target and prognostic biomarker for hepatocellular carcinoma. OncoTargets Ther. 2019;12:6203–6216. doi: 10.2147/OTT.S213833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Q., Matsuura K., Kleiner D.E., Zamboni F., Alter H.J., Farci P. Analysis of long noncoding RNA expression in hepatocellular carcinoma of different viral etiology. J Transl Med. 2016;14:e328. doi: 10.1186/s12967-016-1085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y., Huang J.C., Cai K.T. Long non-coding RNA HOTTIP promotes hepatocellular carcinoma tumorigenesis and development: a comprehensive investigation based on bioinformatics, qRT-PCR and meta-analysis of 393 cases. Int J Oncol. 2017;51(6):1705–1721. doi: 10.3892/ijo.2017.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zeng T., Wang D., Chen J. AF119895 regulates NXF3 expression to promote migration and invasion of hepatocellular carcinoma through an interaction with miR-6508-3p. Exp Cell Res. 2018;363(1):129–139. doi: 10.1016/j.yexcr.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 127.Wang J.Y., Yang Y., Ma Y. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother. 2020;121:e109627. doi: 10.1016/j.biopha.2019.109627. [DOI] [PubMed] [Google Scholar]
- 128.Yan S., Tang Z., Chen K. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J Exp Clin Cancer Res. 2018;37(1):e214. doi: 10.1186/s13046-018-0853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]