Abstract
Duchenne muscular dystrophy is an X-linked recessive hereditary monogenic disorder caused by inability to produce dystrophin protein. In most patients, the expression of dystrophin lost due to disrupting mutations in open reading frame. Despite the efforts in a large number of different therapeutic approaches to date, the treatments available for DMD remain mitigative and supportive to improve the symptoms of the disease, rather than to be curative. The advent of CRISPR/Cas9 technology has revolutionized genome editing scope and considered as pioneer in effective genomic engineering. Deletions or excisions of intragenic DNA by CRISPR as well as a similar strategy with exon skipping at the DNA level induced by antisense oligonucleotides, are new and promising approaches in correcting DMD gene, which restore the expression of a truncated but functional dystrophin protein. Also, CRISPR/Cas9 technology can be used to treat DMD by removing duplicated exons, precise correction of causative mutation by HDR-based pathway and inducing the expression of compensatory proteins such as utrophin. In this study, we briefly explained the molecular genetics of DMD and a historical overview of DMD gene therapy. We in particular focused on CRISPR/Cas9-mediated therapeutic approaches that used to treat DMD.
Keywords: CRISPR/Cas9, Duchenne muscular dystrophy, Genome editing, Therapeutic approaches, X-linked recessive
Introduction
Duchenne muscular dystrophy (DMD), a deadly disease with X-linked inheritance formerly reported with average worldwide incidence about 1:3500, but now it is demonstrated that the average global DMD incidence is near to 1:5000 males born.1,2 DMD is the result of dystrophin loss which leads to progressive muscle degeneration, wasting, and finally death in early adulthood due to heart or respiratory failure.3 The disease often occurs due to the destructive open reading frame (ORF) mutations in DMD gene, which is one of the largest genes in the human genome with 79 exons. This gene is also one of genes with the highest mutation rate.4 Different types of mutations, including deletions, duplications, point mutations, can produce an out of frame transcript, which cause inaccurate translation and lack of dystrophin expression.4 Dystrophin is a bar-shaped protein that connects actin fibers and intracellular contractile machinery of the cytoskeleton to the extracellular matrix and maintains the integrity of plasma membrane. In the absence of dystrophin, muscle cells degenerate.5 Despite numerous researches to date, the available treatments for DMD are mitigative and supportive to improve the symptoms of the disease such as pharmaceutical therapies with corticosteroids. In addition to pharmaceutical treatments with corticosteroids, other therapeutic approaches are under investigation, including induction of exon skipping by antisense oligonucleotides (ASOs) (like eteplirsen, a Morpholino-based anti-sense oligonucleotide that causes jumps from exon 51), increasing the expression of utrophin by drugs (SMT C1100), and the replacement with mini/micro-dystrophin.6 Among the vast range of DMD mutations, the majority of mutations (60% <) are large deletions, which removes one or more exons and destroys ORF. Those deletions of the DMD gene that maintain ORF will lead to in frame transcripts which produce truncated dystrophin protein, but otherwise functional. This cause a milder phenotype called Becker muscular dystrophy (BMD). These observations provide a logical and strong justification for the development of ASOs approaches which accomplish the expression of a truncated form of dystrophin protein resulted from intragenic exon deletion. Induction of exon skipping by ASOs as an encouraging approach in the treatment of DMD requires permanent delivery of therapeutic molecules as this approach works at the mRNA level. This repeated delivery is associated with toxicity and absorption by different tissue. Also, if necessary to skip from several exons to retrieve ORF, for each desired exon a separate ASOs must be designed which leads to increased costs and reduced efficiency.7 An alternative way to exon skipping is to simulate this approach at the DNA level by eliminating the intended exons, nowadays easily accessible by using genome editing technologies and in particular CRISPR/Cas9, but before the advent of this technology genome surgery was a big challenge. CRISPR/Cas9 technology is derived from the acquired immune system of bacteria, archea, against bacteriophages, and invasive plasmids which is driven by a sole gRNA directs Cas9 nucleases into a predefined location, creating a double-strand break (DSB) in the target DNA, after that, it activates the inherent mechanisms of DNA breakage repairing, including HDR and NHEJ (Fig. 1).8, 9, 10 The deletions or excisions of DNA induced by CRISPR in order to restore ORF is a new and encouraging approach in treating patients caused by deletion and duplication mutations that disrupt ORF. Also, the precise correction of the mutation through CRISPR-induced homologous directed repair (HDR) is another hallmark that brings by CRISPR/Cas9 technology for the treatment of DMD. In addition to the ability of manipulating and engineering the genome, recent changes in CRISPR/Cas9 technology have made it possible to regulate gene expression. Accordingly, this technology can be used to induce gene expression of compensatory proteins that ameliorate the phenotype of DMD, such as utrophin. In this review, the mechanism of CRISPR/Cas9 technology in genome editing very briefly described, then an overview of molecular genetics of DMD and a history of DMD gene therapy provided, finally therapeutic approaches based on CRISPR/Cas9 technology that has been used to treat Duchenne muscular dystrophy described (Table 1).
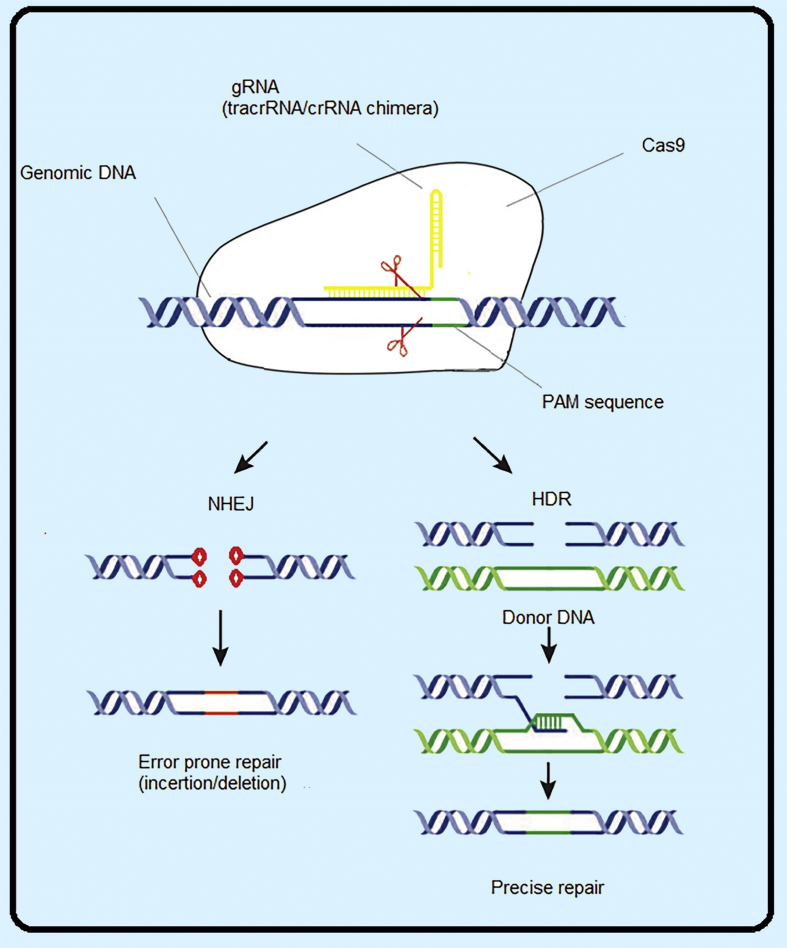
Figure 1.
Schematic mechanism of CRISPR/cas9 in genomic editing. Cas9 nuclease guided by a gRNA to a target DNA site adjacent to PAM. The Cas9-gRNA complex generates a double-strand break, which is repaired by non-homologous end joining or homology-directed repair.
Table 1.
Therapeutic approaches used to treatment of DMD using CRISPR/Cas9 technology.
| Target site | delivery method | nuclease | in-vivo/in-vitro | study situation | reference |
|---|---|---|---|---|---|
| HDR | |||||
| Precise correction | |||||
| Exon 23 | dual Adv vector | SpCas9 | in-vitro | pre-clinical | 34 |
| Exon 23 | 1-cell embryo injection | SpCas9 | in-vivo | pre-clinical | 35 |
| Exon 23 | Nanoparticle | SpCas9 | in-vivo | pre-clinical | 36 |
| Exon 53 | dual AAV-6 vector | SpCas9 | in-vivo | pre-clinical | 33 |
| NHEJ | |||||
| Direct exon skipping | |||||
| Exon 23 | dual AAV-9 vector | SaCas9 | in-vivo | pre-clinical | 40 |
| Exon 23 | dual AAV-8 vector | SaCas9 | in-vivo | pre-clinical | 39 |
| Exon 23 | dual AAV-9 vector | SpCas9 | iv-vivo | pre-clinical | 41 |
| Exon 21–23 | single Adv vector | SpCas9 | in-vivo | pre-clinical | 42 |
| Exon 23 | dual AAV-8 and 9 | SaCas9 | in-vivo | pre-clinical | 68 |
| Exon 20–23 | single AAV rh74 vector | SaCas9 | in-vivo | pre-clinical | 43 |
| Exon 3–9 | Nucleofection | SpCas9 | in-vitro | pre-clinical | 45 |
| Exon 6–9 | Nucleofection | SpCas9 | in-vitro | pre-clinical | 45 |
| Exon 7-11 | Nucleofection | SpCas9 | in-vitro | pre-clinical | 45 |
| Exon 51 | single Adv vector | SpCas9 | in-vitro | pre-clinical | 47 |
| Exon 51 | electroporation | SpCas9 | in-vitro | pre-clinical | 46 |
| Exon 53 | single Adv vector | SpCas9 | in-vitro | pre-clinical | 48 |
| Exon 53 | single Adv vector | SpCas9 | in-vitro | pre-clinical | 49 |
| Exon 52–53 | single, dual AAV6 vector | SpCas9, SaCas9 | in-vivo | pre-clinical | 33 |
| Exon 45–55 | electroporation | SpCas9 | in-vitro | pre-clinical | 46 |
| Exon 45–55 | nucleofection | SpCas9 | in-vitro/in-vivo | pre-clinical | 51 |
| Exon 45–55 | electroporation | SpCas9 | in-vivo | pre-clinical | 50 |
| Exon 44–54 | single Adv vector | SpCas9 | in-vitro | pre-clinical | 48 |
| Exon 44–54 | single Adv vector | SpCas9 | in-vitro | pre-clinical | 49 |
| Classical exon skipping | |||||
| Exon 51, 53 | single Adv vector | SpCas9 | in-vitro | pre-clinical | 49 |
| Exon 51 | dual AAV9 vector | SpCas9 | in-vivo | pre-clinical | 53 |
| Exon 43, 45 | dual AAV9 vector | SpCas9 | in-vivo/in-vitro | pre-clinical | 54 |
| Exon (6–8, 50–53, 55, 43–46) | Nucleofection | SpCas9 | in-vitro | pre-clinical | 52 |
| Indel derived reframing | |||||
| Exon 53 | dual AAV6 vector | SpCas9 | in-vivo | pre-clinical | 33 |
| Exon 23 | micro-injection | SpCas9 | in-vivo | pre-clinical | 35 |
| Exon 44–55 | electroporation | SpCas9 | in-vitro | pre-clinical | 46 |
| Exon 51, 53 | single Adv vector | SpCas9 | in-vitro | pre-clinical | 49 |
| Exon 23 | single AAV2/9 | CjCas9 | in-vivo | pre-clinical | 55 |
| Exon 43, 45 | dual AAV9 vector | SpCas9 | in-vivo/in-vitro | pre-clinical | 54 |
| Exon 51 | dual AAV9 vector | SpCas9 | in-vivo | pre-clinical | 56 |
| Reframing duplication mutations | |||||
| Exon 2 | single lentiviral vector | SpCas9 | in-vitro | pre-clinical | 58 |
| Exon 18–30 | single lentiviral vector | SpCas9 | in-vitro | pre-clinical | 59 |
| CRISPRa | |||||
| Klotho and Utrophin | dual AAV9 vector | (dead) SpCas9 | in-vivo | pre-clinical | 66 |
| Utrophin | electroporation | (dead) SpCas9 | in-vitro | pre-clinical | 59 |
| LAMA-1 | electroporation | (dead) SpCas9 | in-vivo/in-vitro | pre-clinical | 67 |
Adenoviral vector (Adv); Adeno-associated virus (AAV); Streptococcus pyogenes Cas9 (SpCas9); Campylobacter jejuni Cas9 (CjCas9); Staphylococcus aureus Cas9 (SaCas9).
The molecular genetics of DMD
DMD disease that inherited in an X-linked recessive pattern is caused by mutations in DMD gene with one-third of all occurrences derived from novel mutations. DMD gene spanning 2.2 Mb (10 times bigger than any other gene) in chromosomal region Xp21. Multiple isoforms are expressed in muscular and non-muscular tissues from seven unique promoters by alternative splicing. Among these isoforms, full-length dystrophin or the canonical Dp427 expressed in muscle, brain, and lymphocytes, which is driven by promoters upstream of exon 1. The huge size of this locus is probably a major cause of such high spontaneous mutation rate (the highest known spontaneous mutation frequency). This gene contains 79 exons coding for a 14-kb transcript (that had an 11.2-kb ORF) to produce a 427-kDa protein named dystrophin. This protein includes an actin-binding domain at the N-terminus, 24 spectrin-like repeat units that interspersed with 4 hinge domains, a cysteine-rich dystroglycan binding site, and a C-terminal domain (Fig. 2A).11 Based on a comprehensive analysis of the TREAT-NMD DMD Global database (http://umd.be/TREAT_DMD/) that analyzed genetic data for 7149 DMD mutations held within the database, the frequency of different types of dystrophin mutations are as follows: Large mutations (≥1exon) are accounting for ~79% of total mutations, including deletions (~68%) and duplications (~11%). Small mutations (<1exon) are accounting for ~20% of total mutations, comprising of small deletions (~5%), insertions (~2%), splice sites (<10 bp from exon, ~3%), and point mutations (nonsense ~10% and Missense ~0.4%). The rest are due to mid-intronic mutations (~0.3%).12 More than 90% of mutations in DMD disrupt ORF and cause the loss of dystrophin expression.13 According to the ‘reading frame rule’, the severe phenotype associated with DMD is most often resulted from out-of-frame mutations that lead to complete loss of dystrophin. In-frame mutations that allow for the synthesis of a shorter lower molecular weight but otherwise partially functional dystrophin are associated with a milder phenotype known as Becker muscular dystrophy or X-linked dilated cardiomyopathy. However, exceptions to this rule occur in <10% of all DMD mutations, in other words this rule has failed to explain approximately <10% of cases.14, 15, 16 Fig. 2B briefly summarized various types of DMD gene mutations and its consequences.
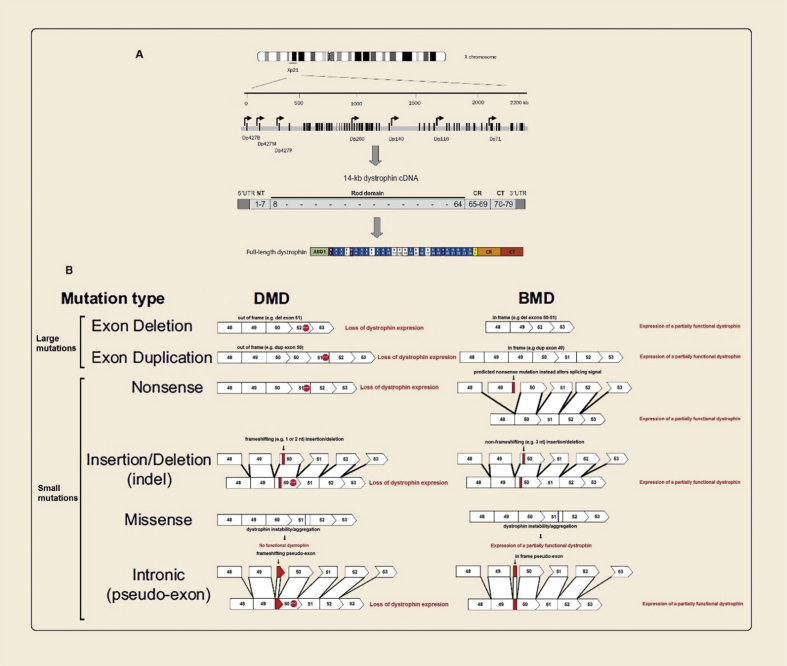
Figure 2.
(A) On the top; schematic outline of the dystrophin gene, located in Xp21. The black vertical lines represent the 79 exons of the dystrophin gene. The arrows indicate different promoters used in different tissues. In the middle; the 14-kb dystrophin cDNA, the numbers in the cDNA refers to exon number. In the bottom; full-length dystrophin contains an actin binding domain 1 (ABD1), 24 spectrin-like repeats (R1 to R24), four hinges (H1 toH4), a cysteine-rich domain (CR), and a C-terminal domain (CT). (B) The consequence of various types of DMD mutations on dystrophin expression and the subsequent phenotype. ABD1, actin-binding domain 1; CR, cysteine-rich domain; CT, C-terminal domain; NT, N-terminal domain; UTR, untranslated region.
A historical overview of DMD gene therapy
Gene therapy approaches for DMD had been under investigation for nearly 30 years, some methods developed in these years are; gene addition therapy, exon skipping therapy, induction of surrogate genes to substitute or compensate for the absence of dystrophin, and recently genome editing approaches. Since mid-ninetieth century, DMD-like manifestation have been characterized that finally after the publication of a clinical monograph in 1968 by French physician, DMD was emerged.17 Immediately after DMD gene identification by Kunkel et al. in 1987, gene addition therapy was proposed as a promising therapy.18 Because of the sheer size of the full-length dystrophin, early proof of concept efforts used simple intramuscular injection of DNA constructs by naked plasmid, which is an inefficient system for in vivo muscle gene therapy due to some limitations, i.e. this approach can only be applied locally at the injection site and provides only transient dystrophin expression.19, 20, 21 In 1990, the discovery of a truncated dystrophin but highly functional D17–48 or mini-dystrophin protein, by Davies et al., gene therapy approaches changed.22 Lentiviruses and adenoviruses vectors were used for delivery of both full length and mini-dystrophin Although encouraging results were achieved in animal models, but these vectors have not been translated to human DMD gene therapy, because each of them had its own limitation: adenoviral residual induce a strong immune response and retroviral vectors are difficult to use for in vivo gene therapy, also both of them aren't effective for systemic delivery.23 So researchers started studies to identify more suitable vector systems. Some studies displayed that intramuscular injection of adeno-associated viruses (AAVs) could lead to an efficient long-term gene transfer into muscle tissue of mice.11 But, one problem still exists which is related to the loading capacity of AAV virus. The maximum capacity of this vector is only 5 kb (1.2 kb smaller than the 6.2 kb mini-dystrophin gene). In 1997, the presentation of the first synthetic micro-dystrophin (3.7 kb DDysM3 gene by Yuasa et al.) surmounted this problem.24 The next breakthrough occurred in 2004 by systemic AAV delivery in rodents that can notably modify the clinical course of the DMD.25 In 2009 Duan et al, improved synthetic micro-dystrophin by the inclusion of dystrophin spectrin-like repeats 16 and 17 (R16/17) of neuronal nitric oxide synthase (nNOS)-binding domain, which increased therapeutic potency of these synthetic gene.26 Today, three distinct clinical trials are in progress for DMD patients by AAVs for delivery of micro-dystrophin.27 During advancement of gene addition therapy, other gene therapy approaches also developed. For example, the proof of concept that the antisense exon-skipping approach indeed could restore dystrophin protein expression was achieved by the early 2000s in patient-derived cell cultures and mdx mouse model.28,29 These studies followed by Systemic delivery treatment experiments in mouse and dog models.30 Around 2005, two open-label phase 1 and 2 clinical trials were initiated using local and systemic injection of drisapersen, which is based on a 2ʹ-O- methyl phosphorothioate (2OMePS) modification, and eteplirsen, which is based on a phosphorodiamidate morpholino oligomer (PMO) modification that target the skipping of exon 51.3 Recently, Eteplirsen has marketed (FDA) with confirmatory phase III trial.27 Exon skipping is likely to be the first gene therapy for DMD to reach the clinic. Another therapeutic option is surrogate gene therapy, a phase 1/2 clinical trial gene transfer study for DMD using rAAVrh74.MCK.GALGT2 is ongoing. GALGT2 gene therapy reinforces the structural integrity of muscles in processes that compensate for the loss of dystrophin protein, by increasing expression of proteins not lost or mutated in the DMD.27,31 Genome editing approaches are recently developed and showed a great hope for the treatment of a wide range of human disorders,32 below we provide a comprehensive review for CRISPR/Cas9 mediated DMD therapy.
Precise gene correction mediate by HDR
DMD disease-causing gene mutations can be accurately corrected using HDR approach. With the delivery of nucleases Cas9, gRNA and a single-stranded oligodeoxynucleotide (ssODN) as a DNA template, HDR approach can be used. So far, this approach has been used to correct mutations in exon 23 of mdx mice, DMD mouse models that have a nonsense mutation in this exon, and exon 53 in mdx4CV mice which have a nonsense mutation in this exon.33, 34, 35, 36 HDR based therapies have the potential to back the wild type sequence, so unlike other DMD gene therapy approaches which add a partially functional dystrophin to a patient's cells, but leave the original dysfunctional copy of the gene intact, this therapy can cut out the error, therefore the development of HDR based treatments is logically very interesting. Although the achievement of precise correction of genes is very valuable, but the current HDR-based gene editing approaches are limited by these facts: first, HDR occurs at much lower frequency than NHEJ in cell or in other words NHEJ is dominant in cells, second, HDR in post-mitotic cells, such as muscles has a very low efficiency, third gene editing via HDR in vivo is challenging because HDR requires the delivery of Cas9, gRNA, and donor DNA. AAVs are currently the most advanced methodology for delivering Cas9 in vivo and AAVs have a small packing size thus more than one viruses are needed to deliver these components in vivo. This issue declines the HDR efficiency of AAV based Cas9 delivery methods.36,37 Non-Homologous End Joining (NHEJ)-based therapeutic approaches due to high efficacy in all cell types specially post mitotic cells, with no DNA template delivery, and the ability to be used in a wider range of patients with different mutations compared to HDRs that is limited to the correction of unique patient mutations, is the predominant therapeutic approach in studies, and most studies for the treatment of DMD have been done by this method.
NHEJ-based therapeutic approaches
DMD mutations are often frameshift deletions of one or several exons. In-frame deletions, do not disrupt ORF and produce a truncated but yet functional protein. These mutations are associated with much milder symptoms of disease which is termed BMD.22 By providing a rational justification, these observations led to the development of approaches of creating a dystrophin protein similar to the dystrophin expressed in BMD patient which has deletions but still retain ORF. One of these approaches is dystrophin replacement therapy by delivering a mini/micro-dystrophin coding sequence to patients, as a gene therapy method. The other is ASOs-induced exon skipping in order to process pre-mRNA into a BDD-like mature transcript. ASOs to skip exon 51 can be used to target 13% of DMD patients. By targeting other exons, up to 83% of patients can be covered.38 The advent of CRISPR/Cas9 technology revolutionized hopes for efficient restoration of dystrophin expression through simulating exon skipping by removing one or more exons at the genomic DNA level. Due to the operation in the genome level, this technology may bring about long-term efficacy, no need for repeated treatment, and the proper expression of dystrophin under control of natural promoter. So, with regard to the ability of CRISPR/Cas9 technology to induce NHEJ, eliminate internal DNA, and induce small deletions and inserts (indel), several NHEJ-based therapeutic approaches have been used to treat DMD.
Direct exon skipping by deletions of one or more exons
By the simultaneous generation of DSBs via targeting the two sides of the desired region by a pair of gRNAs and removing the internal genomic sequence, the desired exons can be removed to restore ORF and produce an internally truncated, but partially functional, dystrophin protein. In 2016, several proofs of concept studies were conducted to evaluate the feasibility of this idea. In these studies, exon 23 and exons 21–23 in mdx mice was targeted and removed by two gRNA which target both sides of this exon. The results of these studies represented a successful ORF restoration and subsequent expression of dystrophin by a significant improvement of skeletal and cardiac muscle function, and increased survival of mdx mice.39, 40, 41, 42 In summary, these studies provided enough proof to patronage the efficiency of in-vivo genome editing to restore the expression of dystrophin in dystrophic mice. In a study in 2017, exons 20–23 were targeted to examine the efficacy of this approach in restoring the expression of dystrophin and its function in cardiac muscle in mdx/Utr+/- mice.43 To date, the mutation hot spots in the DMD gene, which is between the exons of 2–10 adjacent the 5′ end of the gene and exons 45–55 that clustered in the middle region of the gene were targeted as well.12,14 Mutations that disrupt the N-terminal actin-binding domain 1 (ABD-1) of dystrophin are the second prevalent cause of DMD. This domain that is encoded by exons 2–8, contains 3 actin binding sites (ABS1-3) and is vital for the fixation of muscle membrane by dystrophin. Studies that reported some cases of DMD with deletions in exons 3–9 with no obvious phenotype of DMD, led to a theory that the use of CRISPR/Cas9 technology for targeted deletion of these exons may help the correction of mutations in the ABD-1.44 Kyrychenko et al. used 3 strategies for the removal of exons 3–9, including, the removal of exons 3–9, 6–9, and 7–11, which among them the removal of exons 3–9 was the most effective approach to retrieve the expression and function of dystrophin.45 The major hotspot region of deletion mutations is located between exons 45–55, mutations in this region account for 60% of DMD cases. So far, several studies have been conducted to remove exon 51 and, as noted earlier, this elimination caused ORF restoration in 13% of patients.46,47 Other studies performed to remove exon 53 and exons 52–53 in mdx4CV mice have shown the retrieval of dystrophin expression and improved muscle function. These targeted exon deletions can be used in 8–10% of patients.33,48,49 Another approach is the removal of exons 45–55, regarding the reports that showed in-frame deletions of exons 45–55 leads to the generation of an internally deleted dystrophin with a very mild BMD phenotype, so that some patients remain asymptomatic even in their sixth decade of life.46,50,51 The advantage of this approach is, as the occurrence of 60% of DMD mutations are in this area, with a single deletion, up to 60% of the patients can be treated irrespective of which exon was deleted in this region. In two other studies, the removal of exons 44–54 was used to reframe dystrophin.48,49
Classical exon skipping at DNA level
Another NHEJ-based approach that used to restore the expression of dystrophin and treatment of DMD, was the knockout of conserved RNA splice acceptor sites of mutation or out of frame exons for skipping from these exons at the DNA level.49,52, 53, 54 This approach is similar to ASOs-based exon skipping, but acts at the DNA level and therefore does not require permanent treatment. The Cas9 nuclease derived from Streptococcus pyogenes (SpCas9) is the most used nuclease for genome editing which uses NGG or NAG sequences as PAM sequences. Due to the accidental similarity of SpCas9 PAM sequence and the conserved splice acceptor and splice donor sequences (NAG or NGG, as SpCas9 PAM sequence, are in accord with the universal splice acceptor sequence [AG] and most of the donor sequences [GG]), by targeting Cas9 to these conserved sequences and disruption or knockout of these sequences one can induce small indel, for effective exon skipping. This approach only requires a sole gRNA for inducing DSB.
Indel derived reframing
NHEJ repair is an error-prone repairing approach that directly ligates two DNA termini created by chromosomal DSBs and often leads to the introduction of small insertions and deletions (indels) at the junction. Regarding this issue, another NHEJ-based method for the treatment of DMD is the induction of small insertion and deletions (indel) in the out of frame mutated exons or sequences to convert an out of frame Dmd to in frame Dmd. This approach is also performed by a single gRNA, which removes or inserts a small number of nucleotides upstream of the mutation in the exon or the corresponding sequence and causes a targeted frame-shift which restores ORF at the downstream of this small indel.33,35,46,49,54, 55, 56 For example, in Min et al. study a single adenosine (A) insertion in 42% of the sequenced patient-derived induced pluripotent stem cells (iPSCs) clones led to reframing of exon 45 and restored the dystrophin protein ORF.54
CRISPR/Cas9 technology has been applied in large-animal model such as a deltaE50-MD canine model of DMD. This model was created by a splice site missense mutation in 5′ intron 50 that causes the out-of-frame skipping of exon 50, which causes a lack of dystrophin. Amoasii et al. designed a gRNA that targets a sequence on exon 51 located beside the intron 50 splice acceptor site, which is intended to either induce indels that would reframe exon 51or would interrupt the acceptor site leading to exon 51 skipping.56 The results of targeting efficiency by PCR amplification indicated that an insertion of adenosine (A) immediately 3′ to the Cas9 genomic cutting site is the most commonly identified mutation that results in reframing of exon 51 and restores dystrophin expression. Also, the deletions resulted from this cutting site contain a highly predicted exonic splicing enhancer (ESE) site for exon 51 that causes skipping from exon 51 and restores dystrophin expression. Their study showed a restoration range from 3 to 90% of normal dystrophin expression, depending on muscle type.
Reframing of duplication mutations by removal of duplicated exon(s)
Therapeutic approaches that target exon duplications are less studied than those targeting deletions, however these mutations account for about 10–15% of the DMD cases.57 Theoretically, by removing a copy of the duplicated exon, ORF can be restored. In fact by the presence of the complete sequence of the gene, a wild type transcript of the gene can be restored. So far, two studies have used an alternative approach with a single gRNA to remove duplicated exon 2 and exons 18–30.58,59 Due to the nature of a tandem or head-to-tail duplication, designed gRNA target site will be detected twice, so leads to the creation of two DSBs and the removal of the intervening sequence, a segment of the genome is removed which is equal to the total size of the duplication. As we need just a single gRNA, several advantages are derived from this approach, including, using the minimum constituents of CRISPR which is a very advantageous feature in Adeno-associated viruses (AAVs) vector with a limited loading capacity, increasing targeting efficiency, and eliminates the limitation of gRNA design for the specific sequences near the breakpoints by providing a wide region for designing gRNA in any portion of the duplicated sequence. Restoration of dystrophin expression in patients with deletions leads to a truncated, but still functional, version of dystrophin. It has been estimated that the restoration of a truncated dystrophin in patients up to 20% of the normal levels is sufficient to reduce the severity of the disease, while studies in mice suggest approximately 5% of full-length dystrophin can improve disease pathology.60,61 This approach demonstrates high efficiency and safety which can be considered as an invaluable approach for future therapeutic developments. Fig. 3 represent a schematic view of CRISPR/Cas9 based DMD gene correction.
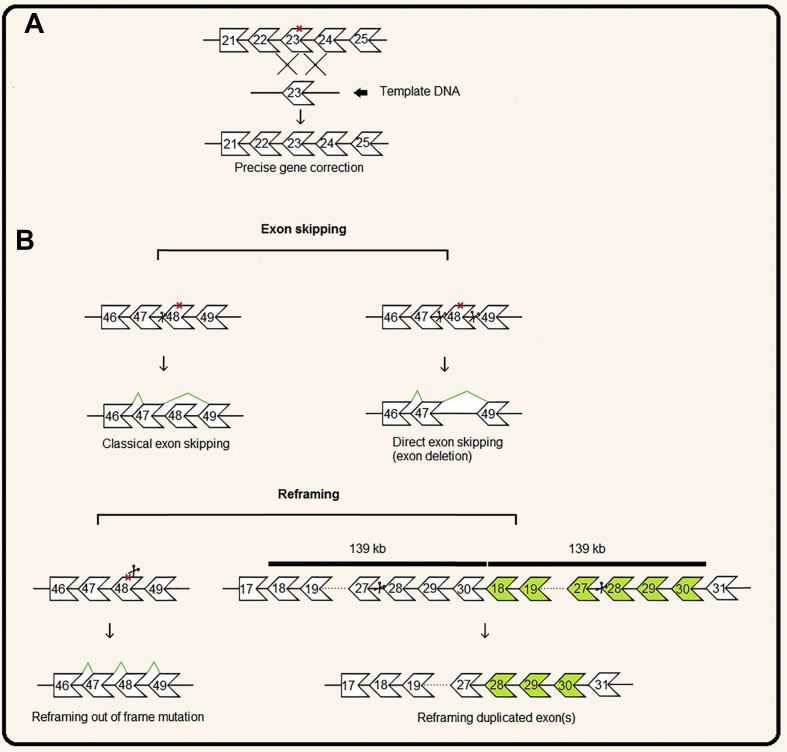
Figure 3.
A schematic view of DMD gene correction by CRISPR/Cas9. (A) Precise correction of DMD gene mediated by HDR approach in a hypothetical patient with a point mutation in exon 23 (marked by an X) that creates a premature stop codon. (B) On the top; Direct and Classical exon skipping mediated by NHEJ approach in a hypothetical patient with a point mutation in exon 48 (marked by an X) that creates a premature stop codon. In the bottom; reframing an out of frame mutation in a hypothetical patient with a point mutation in exon 48 mediated by NHEJ and reframing of a duplication mutation in a hypothetical patient with duplication of exon 18 to 30 (duplicated exons mark with green). Boxes represent exons; lines between boxes represent introns.
Each of these NHEJ based therapeutic approaches has their own sets of pros and cons that is discussed below. One of the advantages of direct exon skipping by CRISPR/Cas9 is that by a single cut with a pair of gRNA we can treat the majority of DMD patients, because approximately 60% of mutations causing Duchenne occur between DMD exons 45–55, so by deletion of DMD exons 45–55 we can reach this goal. Second, it offers a higher gRNA target-site selection within normally extensive introns. Also, this approach eliminates the required steps for designing, evaluation of efficiency, accuracy and safety for each unique deletion which reduces time and cost for the development of a CRISPR platform.51 On the other hand, this approach uses two guide RNA that decrease the efficiency of DMD correction and also increases the potential off target effects, which raise as a potential concern for gene editing in vivo. Classical exon skipping and reframing of indels, which are repaired by NHEJ, use only one gRNA, so benefit from its advantages. These single gRNA gene editing approaches need minimal modification of the genome to induce ORF reframing, thereby increases the efficiency of dystrophin restoration. Also, the use of a single gRNA decreases the likelihood of off target events that might be incurred with multiple gRNAs and bypasses the problems of limited loading capacity.53 However, the process of indel reframing is accidental in nature and only part of indels appropriately ORF reframed the rest results in products harboring possibly immunogenic epitopes. The disadvantage of classical exon skipping may be a high context dependency, for example, difficult accessibility to proper Cas9 target sites adjacent to conservative splicing elements.49 Finally, reframing of duplication mutations, not only benefits from advantages of using a single gRNA, but also provides some additional advantages, such as, a higher gRNA target-site selection within any portion of the duplicated sequence than classical exon skipping that leads to greatly reduced possible off-target effects. The removal of duplication restores the full-length dystrophin not a partially functional shorter dystrophin that only ameliorates the disease phenotype to the extent of making it alike to BMD phenotype.58,59
CRISPRa, transcriptional activation of compensatory protein
Latest alterations to CRISPR/Cas9 technology have been repurposed this technology and provides new capabilities to regulate gene expression. This new capability obtained by the use of a catalytically inactive or “dead” Cas9 (dCas9) to suppress gene expression by sterically impeding RNA polymerase machinery probably through hindering transcriptional elongation, CRISPRi. One can also activates transcription of a desired gene by fusing dCas9 into a transcription activation domain (VP160 or VP16 or VP64) and convert Cas9 into a synthetic transcriptional activator, CRISPRa. The transcription activator domain operates as a recruiter for the multiple parts of the pre-initiation complex.62,63 CRISPRa platform provides a novel opportunity for disease treatment by modulating the expression of genes that contribute to the pathogenesis of the disease. Utrophin, a very similar protein with dystrophin (80% similarity), is one of the genes that meliorates the progression of DMD. Therefore, activation of endogenous utrophin gene by the use of CRISPRa system partially compensates for the loss of dystrophin in patients.64 Some studies have estimated that increased expression of utrophin to over 2 fold more than basal levels is enough to modify the progression of the disease.65 Accordingly a drug called SMT C1100 is currently at the clinical trials to increase the expression of utrophin.
CRISPRa activates the expression of the desired gene without the induction of DSBs, thus, is free from any unwanted mutations and potential off-target effects originated from DSBs. It also would be suitable for all DMD patients regardless of their mutations, which is not the case for treatments derived from exon skipping or gene editing with CRISPR, because some specific exons may be required for protein function and therefore cannot be removed. CRISPRa platform has so far been used to activate utrophin, Klotho, and laminin subunit alpha 1 (LAMA1) to modifying the progression of DMD.59,66,67 However, further studies on the safety of this approach, the development of its specificity by designing tissue-specific promoters, and the use of AAVs vectors with specific tissue tropism are required for its use in the clinic.
Discussion
The ease of use, flexibility, and efficiency of CRISPR/Cas9 technology in targeted genome editing has revolutionized biology and revived great ambition for using genome editing to treat a wide range of diseases, including infectious diseases, cancer and monogenic disorders. Therefore, the potential capability of CRISPR/Cas9 in treating a variety of human diseases has attracted much attention and increased its use in therapeutic approaches.
Recently, a study on long-term evaluation of AAV vector-based CRISPR genome editing has been done for the treatment of Duchenne muscular dystrophy.68 In this study, the long-term persistence and safety of CRISPR gene editing to excise exon 23 of the DMD gene in mdx mice at least 1 year after a single administration assessed. The results of this study indicated that the restoration of dystrophin expression was sustained for 1 year. The AAV-CRISPR was well-tolerated over this period with no sign of toxicity, however a host response to Cas9 and persistent unintended genome modifications was shown. On the other hand, the study of Amoasii et al56 in a canine model of DMD in spite of small sample size and short study term provided important proof of concept for a shift from rodents to large mammals. In fact, this study in the canine model may fill the gap between mice and humans that lacks equivalent results in human patients in comparison with mice. However, moving these therapeutic approaches from research to clinic requires further studies to assess safety and efficiency in larger animal models of DMD. Also CRISPR-associated issues still to be solved. Reducing off target effects to the minimum possible level or even pushing them to zero is a key priority for the clinical application of this technology. The other challenges include improving efficiency, safety, and the improvement of tissue-specific delivery methods.
Future directions and perspectives
Future research should be addressed some limitations, including the outcomes of long-term expression of Cas9 nuclease in vivo, the longevity of dystrophin expression, likely immunological responses to nuclease as well as dystrophin protein. Future studies have to evaluate this technology in larger animals and, ultimately, patients with DMD. So far only one long-term study has been done, and also only one large animal model (DeltaE50-MD dogs) study described treatment with CRISPR/Cas9 platform, so, additional studies needed before translation to clinic. Also, there is a need for more in vivo studies monitoring the off-target effects resulted from the therapy and the potential immune responses activated by viral delivery vectors by more sensitive assays to mitigate the potential immunogenicity. Finally, based on the results of the first long term study future preclinical studies should be focused on increasing efficiency and increasing the proportion of the intended gene modifications by optimizing delivery and gene editing strategy.
Conflict of Interests
The authors declare no conflict of interests.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Sussman M. Duchenne muscular dystrophy. J Am Acad Orthop Surg. 2002;10(2):138–151. doi: 10.5435/00124635-200203000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Zhang K., Yang X., Lin G., Han Y., Li J. Molecular genetic testing and diagnosis strategies for dystrophinopathies in the era of next generation sequencing. Clin Chim Acta. 2019;491:66–73. doi: 10.1016/j.cca.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Guiraud S., Aartsma-Rus A., Vieira N.M., Davies K.E., van Ommen G.J., Kunkel L.M. The pathogenesis and therapy of muscular dystrophies. Annu Rev Genom Hum Genet. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 4.Nallamilli B.R., Ankala A., Hegde M. Molecular diagnosis of Duchenne muscular dystrophy. Curr Protoc Hum Genet. 2014;83 doi: 10.1002/0471142905.hg0925s83. 9.25.1-9.25.29. [DOI] [PubMed] [Google Scholar]
- 5.Lapidos K.A., Kakkar R., McNally E.M. The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ Res. 2004;94(8):1023–1031. doi: 10.1161/01.RES.0000126574.61061.25. [DOI] [PubMed] [Google Scholar]
- 6.Mendell J.R., Rodino-Klapac L.R. Duchenne muscular dystrophy: CRISPR/Cas9 treatment. Cell Res. 2016;26(5):513–514. doi: 10.1038/cr.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salmaninejad A., Valilou S.F., Bayat H. Duchenne muscular dystrophy: an updated review of common available therapies. Int J Neurosci. 2018;128(9):854–864. doi: 10.1080/00207454.2018.1430694. [DOI] [PubMed] [Google Scholar]
- 8.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):e1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 9.Mollanoori H., Shahraki H., Rahmati Y., Teimourian S. CRISPR/Cas9 and CAR-T cell, collaboration of two revolutionary technologies in cancer immunotherapy, an instruction for successful cancer treatment. Hum Immunol. 2018;79(12):876–882. doi: 10.1016/j.humimm.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Teimourian S., Abdollahzadeh R. Technology developments in biological tools for targeted genome surgery. Biotechnol Lett. 2015;37(1):29–39. doi: 10.1007/s10529-014-1656-5. [DOI] [PubMed] [Google Scholar]
- 11.Chamberlain J.R., Chamberlain J.S. Progress toward gene therapy for Duchenne muscular dystrophy. Mol Ther: J Am Soc Gene Ther. 2017;25(5):1125–1131. doi: 10.1016/j.ymthe.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bladen C.L., Salgado D., Monges S. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat. 2015;36(4):395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nance M.E., Hakim C.H., Yang N.N., Duan D. Nanotherapy for Duchenne muscular dystrophy. Wiley interdisciplinary Rev Nanomedicine Nanobiotechnol. 2018;10(2):1–30. doi: 10.1002/wnan.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aartsma-Rus A., Van Deutekom J.C., Fokkema I.F., Van Ommen G.J., Den Dunnen J.T. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34(2):135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 15.Koenig M., Beggs A.H., Moyer M. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989;45(4):498–506. [PMC free article] [PubMed] [Google Scholar]
- 16.Bladen C.L., Rafferty K., Straub V. The TREAT-NMD Duchenne muscular dystrophy registries: conception, design, and utilization by industry and academia. Hum Mutat. 2013;34(11):1449–1457. doi: 10.1002/humu.22390. [DOI] [PubMed] [Google Scholar]
- 17.Rondot P. 2005. G.B.A. Duchenne de Boulogne (1806–1875) 252(7):866–867. [DOI] [PubMed] [Google Scholar]
- 18.Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50(3):509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 19.Duan D. Myodys, a full-length dystrophin plasmid vector for Duchenne and Becker muscular dystrophy gene therapy. Curr Opin Mol Ther. 2008;10(1):86–94. [PubMed] [Google Scholar]
- 20.Wolff J.A., Malone R.W., Williams P. Direct gene transfer into mouse muscle in vivo. Science. 1990;247(4949 Pt 1):1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 21.Acsadi G., Dickson G., Love D.R. Human dystrophin expression in mdx mice after intramuscular injection of DNA constructs. Nature. 1991;352(6338):815–818. doi: 10.1038/352815a0. [DOI] [PubMed] [Google Scholar]
- 22.England S.B., Nicholson L.V., Johnson M.A. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343(6254):180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 23.Chamberlain J.S. Gene therapy of muscular dystrophy. Hum Mol Genet. 2002;11(20):2355–2362. doi: 10.1093/hmg/11.20.2355. [DOI] [PubMed] [Google Scholar]
- 24.Yuasa K., Ishii A., Miyagoe Y., Takeda S. Introduction of rod-deleted dystrophin cDNA, delta DysM3, into mdx skeletal muscle using adenovirus vector. Jpn J Clin Med. 1997;55(12):3148–3153. Nihon rinsho. [PubMed] [Google Scholar]
- 25.Gregorevic P., Blankinship M.J., Allen J.M. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat Med. 2004;10(8):828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai Y., Thomas G.D., Yue Y. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J Clin Investig. 2009;119(3):624–635. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhaart I.E.C., Aartsma-Rus A. Therapeutic developments for Duchenne muscular dystrophy. Nat Rev Neurol. 2019;15(7):373–386. doi: 10.1038/s41582-019-0203-3. [DOI] [PubMed] [Google Scholar]
- 28.Mann C.J., Honeyman K., Cheng A.J. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc Natl Acad Sci USA. 2001;98(1):42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Deutekom J.C., Bremmer-Bout M., Janson A.A. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum Mol Genet. 2001;10(15):1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- 30.Lu Q.L., Rabinowitz A., Chen Y.C. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc Natl Acad Sci USA. 2005;102(1):198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chicoine L.G., Rodino-Klapac L.R., Shao G. Vascular delivery of rAAVrh74.MCK.GALGT2 to the gastrocnemius muscle of the rhesus macaque stimulates the expression of dystrophin and laminin alpha 2 surrogates. Mol Ther: J Am Soc Gene Ther. 2014;22(4):713–724. doi: 10.1038/mt.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mollanoori H., Teimourian S. Therapeutic applications of CRISPR/Cas9 system in gene therapy. Biotechnol Lett. 2018;40(6):907–914. doi: 10.1007/s10529-018-2555-y. [DOI] [PubMed] [Google Scholar]
- 33.Bengtsson N.E., Hall J.K., Odom G.L. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8:e14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu P., Wu F., Mosenson J., Zhang H., He T.C., Wu W.S. CRISPR/Cas9-Mediated genome editing corrects dystrophin mutation in skeletal muscle stem cells in a mouse model of muscle dystrophy. Mol Ther Nucleic Acids. 2017;7:31–41. doi: 10.1016/j.omtn.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9–mediated editing of germline DNA. Science. 2014;345(6201):1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K., Conboy M., Park H.M. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng. 2017;1(11):889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jasin M., Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harbor Perspect Biol. 2013;5(11):a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aartsma-Rus A., Fokkema I., Verschuuren J. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30(3):293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 39.Nelson C.E., Hakim C.H., Ousterout D.G. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabebordbar M., Zhu K., Cheng J.K.W. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long C., Amoasii L., Mireault A.A. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351(6271):400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu L., Park K.H., Zhao L. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther: J Am Soc Gene Ther. 2016;24(3):564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El Refaey M., Xu L., Gao Y. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res. 2017;121(8):923–929. doi: 10.1161/CIRCRESAHA.117.310996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura A., Fueki N., Shiba N. Deletion of exons 3-9 encompassing a mutational hot spot in the DMD gene presents an asymptomatic phenotype, indicating a target region for multiexon skipping therapy. J Hum Genet. 2016;61(7):663–667. doi: 10.1038/jhg.2016.28. [DOI] [PubMed] [Google Scholar]
- 45.Kyrychenko V., Kyrychenko S., Tiburcy M. Functional correction of dystrophin actin binding domain mutations by genome editing. JCI insight. 2017;2(18):e95918. doi: 10.1172/jci.insight.95918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ousterout D.G., Kabadi A.M., Thakore P.I., Majoros W.H., Reddy T.E., Gersbach C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:e6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ehrke-Schulz E., Schiwon M., Leitner T. CRISPR/Cas9 delivery with one single adenoviral vector devoid of all viral genes. Sci Rep. 2017;7(1):e17113. doi: 10.1038/s41598-017-17180-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maggio I., Liu J., Janssen J.M., Chen X., Goncalves M.A. Adenoviral vectors encoding CRISPR/Cas9 multiplexes rescue dystrophin synthesis in unselected populations of DMD muscle cells. Sci Rep. 2016;6:e37051. doi: 10.1038/srep37051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maggio I., Stefanucci L., Janssen J.M. Selection-free gene repair after adenoviral vector transduction of designer nucleases: rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016;44(3):1449–1470. doi: 10.1093/nar/gkv1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young C.S., Mokhonova E., Quinonez M., Pyle A.D., Spencer M.J. Creation of a novel humanized dystrophic mouse model of Duchenne muscular dystrophy and application of a CRISPR/Cas9 gene editing therapy. J Neuromuscul Dis. 2017;4(2):139–145. doi: 10.3233/JND-170218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young C.S., Hicks M.R., Ermolova N.V. A single CRISPR-cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell stem cell. 2016;18(4):533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long C., Li H., Tiburcy M. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv. 2018;4(1) doi: 10.1126/sciadv.aap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amoasii L., Long C., Li H. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med. 2017;9(418) doi: 10.1126/scitranslmed.aan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Min Y.-L., Li H., Rodriguez-Caycedo C. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv. 2019;5(3):eaav4324. doi: 10.1126/sciadv.aav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koo T., Lu-Nguyen N.B., Malerba A. Functional rescue of dystrophin deficiency in mice caused by frameshift mutations using Campylobacter jejuni Cas9. Mol Ther: J Am Soc Gene Ther. 2018;26(6):1529–1538. doi: 10.1016/j.ymthe.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amoasii L., Hildyard J.C.W., Li H. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362(6410):86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuffery-Giraud S., Béroud C., Leturcq F. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD–DMD database: a model of nationwide knowledgebase. Hum Mutat. 2009;30(6):934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 58.Lattanzi A., Duguez S., Moiani A. Correction of the exon 2 duplication in DMD myoblasts by a single CRISPR/Cas9 system. Mol Ther Nucleic Acids. 2017;7:11–19. doi: 10.1016/j.omtn.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wojtal D., Kemaladewi D.U., Malam Z. Spell checking nature: versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am J Hum Genet. 2016;98(1):90–101. doi: 10.1016/j.ajhg.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicholson L.V., Johnson M.A., Bushby K.M. Integrated study of 100 patients with Xp21 linked muscular dystrophy using clinical, genetic, immunochemical, and histopathological data. Part 1. Trends across the clinical groups. J Med Genet. 1993;30(9):728–736. doi: 10.1136/jmg.30.9.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Putten M., Hulsker M., Young C. Low dystrophin levels increase survival and improve muscle pathology and function in dystrophin/utrophin double-knockout mice. FASEB (Fed Am Soc Exp Biol) J. 2013;27(6):2484–2495. doi: 10.1096/fj.12-224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi L.S., Larson M.H., Gilbert L.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mali P., Aach J., Stranges P.B. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:e833. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rafael J.A., Tinsley J.M., Potter A.C., Deconinck A.E., Davies K.E. Skeletal muscle-specific expression of a utrophin transgene rescues utrophin-dystrophin deficient mice. Nat Genet. 1998;19:e79. doi: 10.1038/ng0598-79. [DOI] [PubMed] [Google Scholar]
- 65.Fairclough R.J., Wood M.J., Davies K.E. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat Rev Genet. 2013;14:e373. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 66.Liao H.K., Hatanaka F., Araoka T. In vivo target gene activation via CRISPR/Cas9-Mediated trans-epigenetic modulation. Cell. 2017;171(7):1495–1507. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perrin A., Rousseau J., Tremblay J.P. Increased expression of laminin subunit alpha 1 chain by dCas9-VP160. Mol Ther Nucleic Acids. 2017;6:68–79. doi: 10.1016/j.omtn.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nelson C.E., Wu Y., Gemberling M.P. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med. 2019;25(3):427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]