Abstract
Background and Aims
Bowel function requires coordinated activity of diverse enteric neuron subtypes. Our aim was to define gene expression in these neuron subtypes to facilitate development of novel therapeutic approaches to treat devastating enteric neuropathies, and to learn more about enteric nervous system function.
Methods
To identify subtype–specific genes, we performed single-nucleus RNA-seq on adult mouse and human colon myenteric plexus, and single-cell RNA-seq on E17.5 mouse ENS cells from whole bowel. We used immunohistochemistry, select mutant mice, and calcium imaging to validate and extend results.
Results
RNA-seq on 635 adult mouse colon myenteric neurons and 707 E17.5 neurons from whole bowel defined seven adult neuron subtypes, eight E17.5 neuron subtypes and hundreds of differentially expressed genes. Manually dissected human colon myenteric plexus yielded RNA-seq data from 48 neurons, 3798 glia, 5568 smooth muscle, 377 interstitial cells of Cajal, and 2153 macrophages. Immunohistochemistry demonstrated differential expression for BNC2, PBX3, SATB1, RBFOX1, TBX2, and TBX3 in enteric neuron subtypes. Conditional Tbx3 loss reduced NOS1-expressing myenteric neurons. Differential Gfra1 and Gfra2 expression coupled with calcium imaging revealed that GDNF and neurturin acutely and differentially regulate activity of ∼50% of myenteric neurons with distinct effects on smooth muscle contractions.
Conclusion
Single cell analyses defined genes differentially expressed in myenteric neuron subtypes and new roles for TBX3, GDNF and NRTN. These data facilitate molecular diagnostic studies and novel therapeutics for bowel motility disorders.
Keywords: Calcium Imaging, Pou3f3 (Brn1), Transcription Factors, Human and Mouse Colon
Abbreviations used in this paper: ACSF, artificial cerebrospinal fluid; BSA, bovine serum albumin; DBPBS, Dulbecco-modified phosphate-buffered saline; DMEM, Dulbecco’s modified Eagle medium; EFS, electrical field stimulation; EGFP, enhanced green fluorescent protein; ENK, enkephalin; ENS, enteric nervous system; FACS, fluorescence-activated cell sorter; FBS, fetal bovine serum; GDNF, glial cell line–derived neurotrophic factor; GO, Gene Ontology; HBSS, Hank’s balanced salt solution; HSCR, Hirschsprung disease; IACUC, Institutional Animal Care and Use Committee; ICC, interstitial cells of Cajal; IRB, institutional review board; NADPH, nicotinamide adenine dinucleotide phosphate; NDS, normal donkey serum; NRTN, neurturin; NOS1, neuronal nitric oxide synthase; OCT, optimal cutting temperature; PBS, phosphate-buffered saline; PFA, paraformaldehyde; RIN, RNA integrity number; RNA-seq, RNA sequencing; RT, room temperature; RT-PCR, reverse-transcription polymerase chain reaction; SMC, smooth muscle cell; SP, substance P; t-SNE, t-distributed stochastic neighbor embedding; TTX, tetrodotoxin; UMI, unique molecular identifier
Summary.
Using single-cell RNA analysis and advanced imaging methods on the enteric nervous system, we define many differentially expressed genes in myenteric neuron subtypes, validate numerous findings, and describe new functions for TBX3, glial cell line–derived neurotrophic factor, and neurturin in the bowel.
The enteric nervous system (ENS) is a network of neurons and glia within the bowel wall that is crucial for normal bowel function.1 ENS defects include developmental anomalies (Hirschsprung disease [HSCR] and primary chronic intestinal pseudo-obstruction) and acquired defects (achalasia, gastroparesis, Parkinson’s disease, and Chagas disease). Unfortunately, we have limited ability to identify missing or defective ENS cell populations in people with bowel dysmotility, and therapies are inadequate. Promising new approaches like regenerative medicine2 or glial cell line–derived neurotrophic factor (GDNF)–induced regeneration of the ENS3 would benefit from a more thorough characterization of the transcription factors, receptors, and signaling pathways that define enteric neuron subclasses.
Several laboratories, ours included, previously used bulk RNA sequencing (RNA-seq) or microarray analyses to delineate ENS gene expression patterns. Memic et al,4 in particular, combined bulk RNA-seq with immunohistochemistry to identify transcription factors in enteric neurons with distinct neurochemical signatures at different embryonic ages. While this was a significant advance, frozen sections provided limited information regarding co-localization of transcription factors with other markers and there was little quantitative data.
We hypothesized that ENS subpopulations could be more definitively characterized using single-cell RNA sequencing. As our studies proceeded, other groups pursued similar work, each with a unique focus.5, 6, 7, 8, 9 Zeisel et al6 used single-cell sequencing to define enteric neuron subtypes in the small intestine myenteric plexus of young adult mice. Morarach et al extended their analysis with greatly increased numbers of young adult mouse myenteric neurons as well as developing (E15.5 and E18.5) mouse small intestine.5 They identified 12 neuron subtypes and showed depletion of CALB+ enteric neurons in Pbx3–/– mice.5,6 May-Zhang et al7 identified 14 distinct neuron clusters in adult mouse ENS and highlighted differences between bowel regions. They then compared murine data to bulk human ENS RNA-seq analysis and discovered distinct mouse and human intrinsic primary afferent neuron subtypes.7 Drokhlyansky et al8 tackled the difficult problem of sequencing rare human enteric neurons, circumventing the issue of neuron scarcity by overloading nuclei into droplets and retroactively filtering out doublets, a technique they dubbed MIRACL-seq. Using this strategy, they sequenced 1445 human enteric neurons and identified 14 neuron subsets in the colon. Other papers provided additional data about single cells in developing mouse ENS on a smaller scale.9 Collectively, these manuscripts, each with a unique focus, define 8–14 myenteric neuron types and >20 total enteric neuron types, plus 4–7 types of enteric glia, consistent with anatomic and functional analyses over decades.10, 11, 12 Our study provides additional RNA-seq data from the ENS of whole bowel at E17.5 and from adult mouse colon, new data from human colon cells that control bowel motility, extensive validation using state-of-the-art confocal imaging, and unique analysis of ENS structure in 4 mutant mouse lines deficient in transcription factors preferentially expressed in cholinergic or nitrergic neurons. We also leveraged our data to identify novel roles for GDNF and neurturin (NRTN) in the ENS.
We focused on distinctions between myenteric nitrergic (nitric oxide–producing) and cholinergic (acetylcholine-producing) neurons because they comprise >89% of human and >95% of mouse myenteric neuron types.13,14 We found many cell type–specific ion channels, neurotransmitters, adhesion proteins, and signaling pathways supporting functional connectivity, and identified >40 differentially expressed transcriptional regulators, including Tbx3. Tbx3 is preferentially expressed in neuronal nitric oxide synthase (NOS1)–producing myenteric neurons, and mice lacking Tbx3 in ENS had a 30% reduction in NOS1 neurons. Another intriguing finding was that nitrergic neurons express Gfra1, but cholinergic neurons express Gfra2 in adult and E17.5 mouse myenteric plexus. GFRA1 and GFRA2 are cell surface receptors that bind preferentially to GDNF and NRTN, respectively. Using calcium imaging, we tested the hypothesis that GDNF and NRTN acutely affect ENS function and bowel motility in adulthood and discovered strikingly different effects of these trophic factors on GCaMP6s activity in enteric neurons, with procontractility effects of GDNF. Collectively, our single-cell RNA-seq data provide highly validated information about gene expression in mouse and human ENS, supporting a new foundation for ENS cell classification.
Results
Single Nucleus RNA-Seq Defines Mouse Distal Colon Enteric Neuron Subpopulations
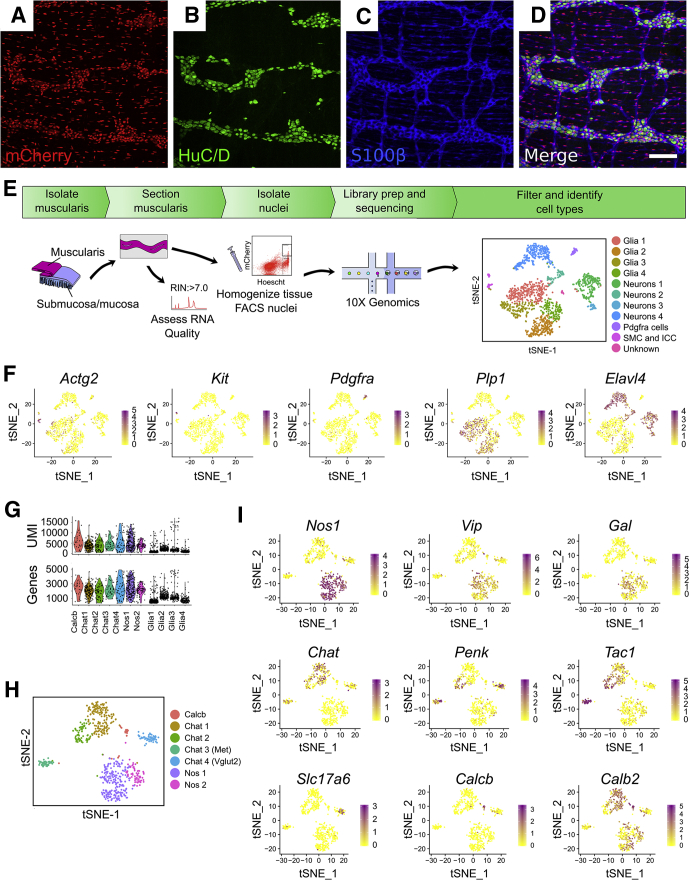
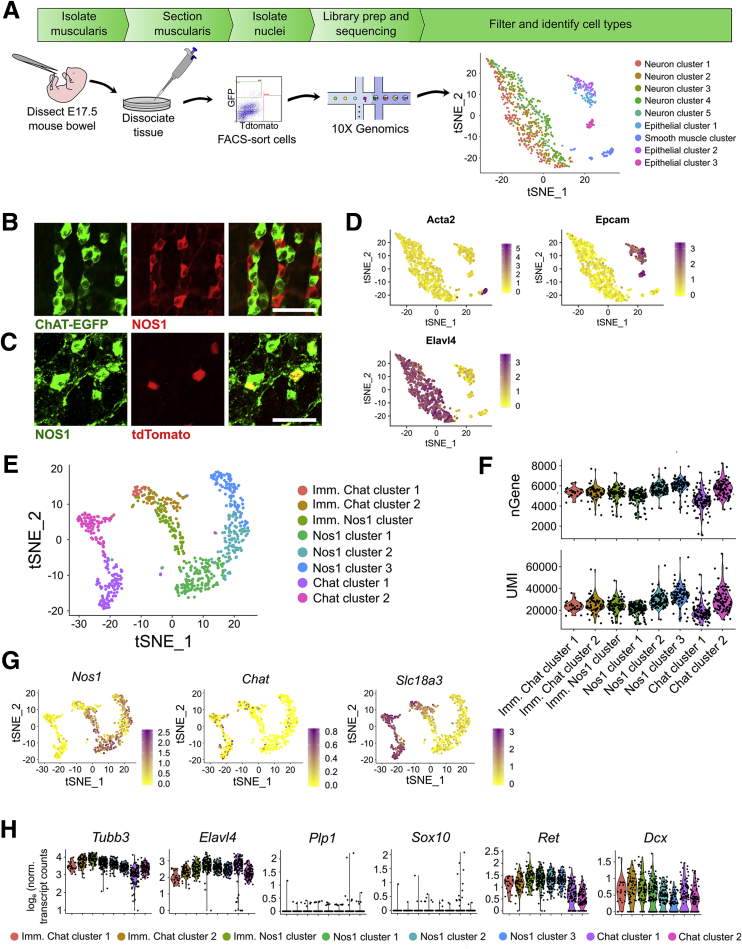
We tried many approaches for isolating the ENS from the adult mouse distal colon for sequencing (Table 1). Ultimately, myenteric plexus nuclei were isolated from 47- to 52-day-old Wnt1-CreCre/wt; R26R-LSL-H2B-mCherry mice that express the fluorescent nuclear protein Histone 2B-mCherry after CRE-induced DNA recombination (Figure 1A–D). We used Wnt1-Cre to induce H2B-mCherry expression because prior studies show that this Wnt1-Cre model induces recombination in almost all enteric neurons and glia at E13.5 and almost all colonic neurons in E18.5 and adult mice, with no ectopic expression in other bowel cells.15,16 We observed mCherry in ∼73% of adult colon myenteric neurons in our colony, a smaller percentage than previously reported, but decided to proceed with analyses. Tissue was frozen and cryosectioned to disrupt smooth muscle cells, improving yield. Dounce homogenization released nuclei (Figure 1E). The 10x Genomics platform (10x Genomics, Pleasanton, CA) generated data from 1520 mCherry+ neuronal and glial nuclei. Because of low read depth, we included intronic reads, yielding a mean of 2970 unique molecular identifiers (UMIs) and 1549 genes per cell.
Table 1.
Methods Used to Isolate Cells or Nuclei for RNA-Seq
| Genotype | Whole Cells, or Nuclei? | Description |
|---|---|---|
| Wnt1-Cre;R26R-EGFP | Cells | The EGFP signal in Wnt1-Cre;R26R-EGFP animals was too weak to flow sort effectively given the high degree of background in the 488 channel. |
| Wnt1-Cre;R26R-tdTomato | Cells | The localization of tdTomato to neurites in our Wnt1-Cre;R26R-Tdtomato line was problematic, since we desired clean separation of single cells. Sorting myenteric plexus from this mouse line often resulted in preps with neurites attached to tdTomato- cells. We tried dissociating with different proteases (cold active protease, dispase, and collagenase), different incubation times (15 min, 30 min), multiple methods of trituration (pipette-based, needle-based), and different bowel layers, with little improvement in outcome |
| Wnt1-Cre;ROSANT-NG | Nuclei | Wnt1-Cre;RosaNT-NG mice had tdTomato in their nuclei at baseline; with CRE-induced recombination, they accumulated EGFP in their nuclei instead of tdTomato. Unfortunately, these mice lost fluorescent signal during the Dounce homogenization procedure. We hypothesize that membrane damage associated with homogenization led to diffusion of GFP and loss of signal. |
| Wnt1-Cre;Rosa26 LSL H2B mCherry | Nuclei | Successful and used to generate data in Figure 1, Figure 2, Figure 3, Figure 4. |
| Wild-type | Nuclei | We attempted to use directly conjugated NeuN and PHOX2B antibodies to isolate mouse ENS nuclei with flow sorting, since some neuronal nuclei in mouse stain with this NeuN antibody by immunohistochemistry. We were unsuccessful. |
EGFP, enhanced green fluorescent protein; ENS, enteric nervous system; GFP, green fluorescent protein; RNA-seq, RNA sequencing.
Figure 1.
Adult mouse distal colon myenteric plexus RNA-seq defines neuron and glia subtypes. (A–D) H2B-mCherry fluorescence (red) in 50-day-old Wnt1-crecre/wt; R26R-H2B-mCherrych/wt mice colocalizes with HuC/D+ neurons (green) and S100β+ glia (blue) in the ENS. ∼73% of HuC/D+ neurons were H2B-mCherry+. Scale bar = 100 μm. (E) RNA-seq workflow with t-SNE containing all cells. (F) Feature plots for Actg2, Kit, Pdgfrα, Plp1, and Elavl4 indicate the locations of SMC, PDGFRα+ cells, ICC, neurons, and glia, respectively. Color key represents loge(normalized gene expression). (G) Violin plots of neuronal and glial clusters. (H) t-SNE of neuron clusters (I) Feature plots for selected markers highlight neuron subtypes. Color shows loge(normalized gene expression).
Using CellRanger and Seurat,17 we identified 12 clusters: 4 glial (Plp1+, Sox10+), 4 neuronal (Elavl4+), and several non-ENS (6.5% of total), including Pdgfra+ (PDGFRα+ cells), Kit+ (interstitial cells of Cajal [ICC]), and Actg2+ (smooth muscle cells [SMCs]) (Figure 1F). Mean UMI and gene counts were higher for neurons than glia (neurons: 4629 UMIs, 2217 genes; glia: 1780 UMIs, 1069 genes) (Figure 1G). To refine analyses, we reclustered neuronal groups (Figure 1H) and identified 7 clusters (635 neurons total).
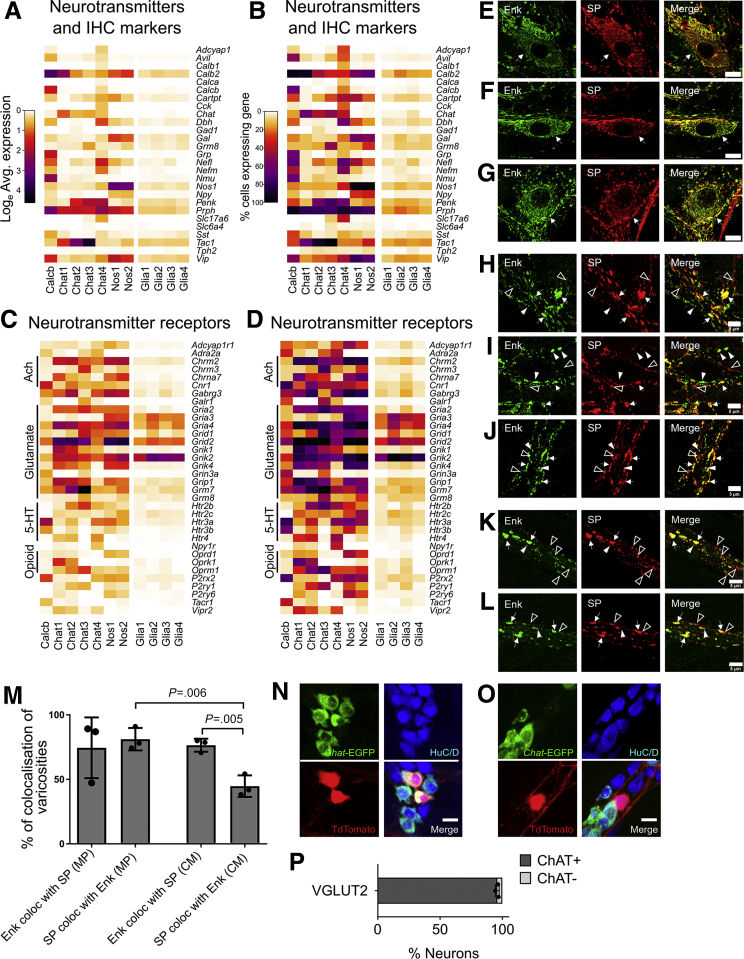
Two neuron groups (Nos1 and Nos2) express Nos1, Vip, and Gal, consistent with known coexpression in NOS1+/VIP+ inhibitory motor neurons and interneurons.12,18 Chat1 cells express choline acetyl-transferase (Chat) and preprotachykinin (substance P [SP] precursor, Tac1), but not proenkephalin (Penk), consistent with excitatory motor or intrinsic sensory neurons.19,20 Chat2 cells express Chat, Tac1, and Penk, which marks guinea pig ileum excitatory motor neurons and ascending interneurons. Chat3 cells express Chat, Tac1, Penk, and hepatocyte growth factor receptor Met (Figures 1I, 2A and B, and 3). We tested if Tac1 and Penk expression could differentiate between motor neurons and interneurons. Immunohistochemistry showed that in circular smooth muscle, 55.3 ± 8.4% of SP-containing varicosities do not contain enkephalin (ENK) (Figure 2K–M), but in myenteric ganglia, ∼25% of SP terminals lack ENK and vice versa (Figure 2E–J and M). An additional clustered neuron group appeared heterogeneous. We manually divided these neurons into Chat4, which express Chat, Penk, and Slc17a6 (encoding VGLUT2), and Calcb, which express Calcb, Grp, and Nmu. We hypothesize that Chat4 are interneurons since VAChT+/VGLUT2+ terminals were reported in mouse distal colon myenteric ganglia but not in muscle.21 Chat/Penk/Vglut2 neurons also express Calb1, a calcium-binding protein in mouse interneurons.12,22 To further validate Vglut2/Chat coexpression, we bred Vglut2-IRES-Cre;TdTomato × ChAT-L10A-EGFP and found 95.9 ± 0.9% of TDTOMATO+ neurons in myenteric plexus coexpressed enhanced green fluorescent protein (EGFP) (Figure 2N–P).
Figure 2.
Expression patterns of neurotransmitters, neurotransmitter receptors, and common immunohistochemistry (IHC) markers, and validation of ENK and SP coexpression in adult mouse colon. (A) Average expression for neurotransmitters and IHC markers that were differentially expressed between distinct neuron and glial subclasses. (B) Proportion of cells per cluster with expression values >0 for differentially expressed neurotransmitters and IHC markers. (C) Average expression for neurotransmitter receptors and subunits that were differentially expressed between distinct neuron and glial subclasses. (D) Proportion of cells per cluster with expression values >0 for differentially expressed neurotransmitter receptors and subunits. (A, C) Color key represents loge(normalized average gene expression within each cluster). (E–G) ENK (green) colocalizes with SP (red) in myenteric neuron cell bodies in (E) proximal, (F) mid, and (H) distal colon. (H–J) Most myenteric intraganglionic neuron varicosities in (H) proximal, (I) mid, and (J) distal colon express both ENK (green) and SP (red). (K, L) Most enkephalin+ (green) neuron varicosities in circular smooth muscle in (K) mid colon and (L) distal colon also express SP (red), but only a subset of substance P-expressing neuron varicosities in circular smooth muscle express ENK. (E–L) Images representative of n = 3 preparations per colon region and n = 3 images per preparation. (M) Quantification of colocalization of ENK and SP in varicosities within mid colon myenteric ganglia and within circular smooth muscle. (N) EGFP (green) fluorescence signal colocalizes with TDTOMATO+ (red) neurons (blue) in Vglut2-IRES-Cre; R26R-TdTomato; ChAT-EGFP-L10A mice. (O) In a small subset of HuC/D+ neurons (blue), TDTOMATO+ (red) does not colocalize with EGFP fluorescence signal in Vglut2-IRES-Cre; R26R-TdTomato; ChAT-EGFP-L10A mice. (P) Quantification of the colocalization of EGFP fluorescence with TDTOMATO fluorescence in Vglut2-IRES-Cre; R26R-TdTomato mice. (E–G) Scale bar = 10 μm. (H–L) Scale bar = 5 μm. (E–L) White arrows point toward cells and varicosities that express both ENK and SP. (H–L) White arrowheads point toward varicosities that express enkephalin only. Empty arrowheads point toward varicosities that express SP only. (M, P) Mean ± SD.
Figure 3.
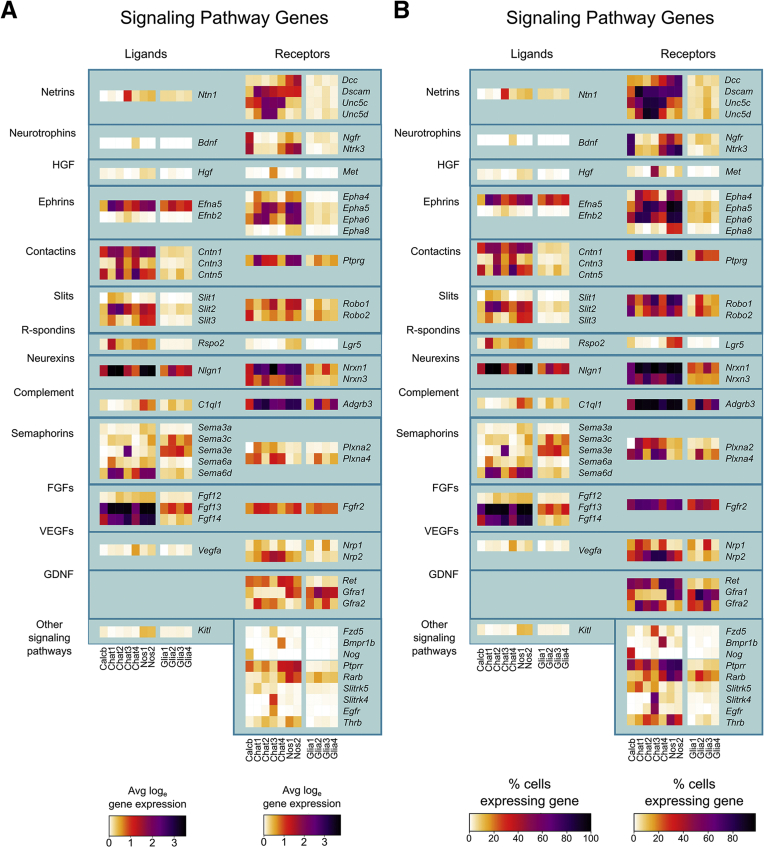
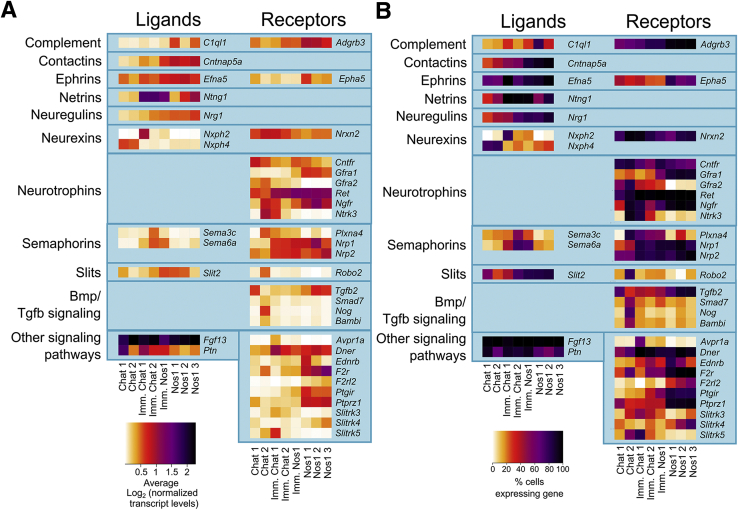
Average expression and percent cells per cluster with detectable levels of differentially expressed signaling pathway molecules in adult distal mouse colon. (A) Average expression for selected ligands (left) and receptors (right) that were differentially expressed between distinct neuron and glial subclasses. Color key represents loge(normalized average gene expression within each cluster). (B) Proportion of cells per cluster with expression values >0 for differential expressed ligands (left) and receptors (right).
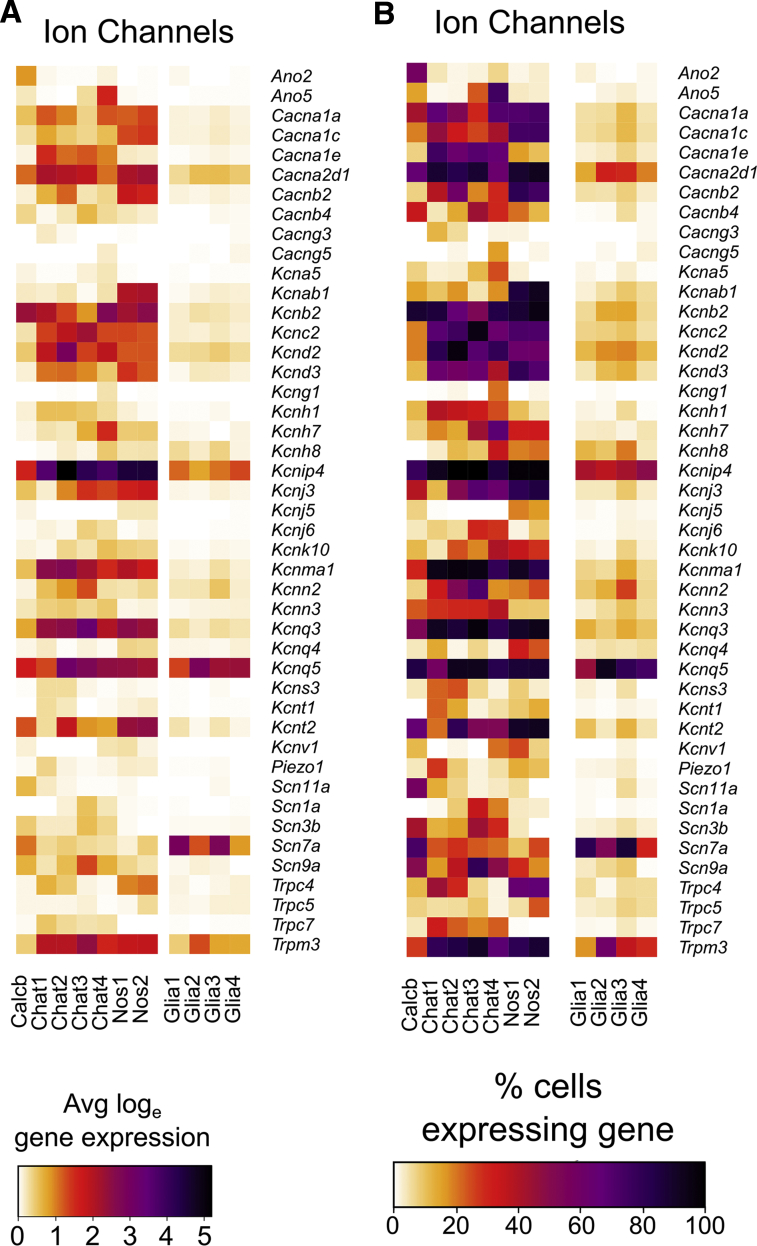
Many neurotransmitters (Figure 2A and B) and neurotransmitter receptors (Figure 2C and D) were differentially expressed between neuron subgroups, including receptors for acetylcholine, glutamate, serotonin, opioids, and purines. Neuron groups also differentially expressed signaling molecules (Figure 3), ion channels (Figure 4), axon guidance molecules, adhesion proteins, survival factors, and other receptors.
Figure 4.
Average expression and percent cells per cluster with detectable levels of differentially expressed ion channels in adult distal mouse colon. (A) Average expression of differentially expressed ion channel (subunit) genes for different neuron and glial groups. Color key represents loge(normalized average gene expression within each cluster). (B) Proportion of cells per cluster with expression values >0 for differentially expressed ion channels.
Single-Nucleus Sequencing Reveals Distinct Roles for GDNF and NRTN in Mouse Distal Colon
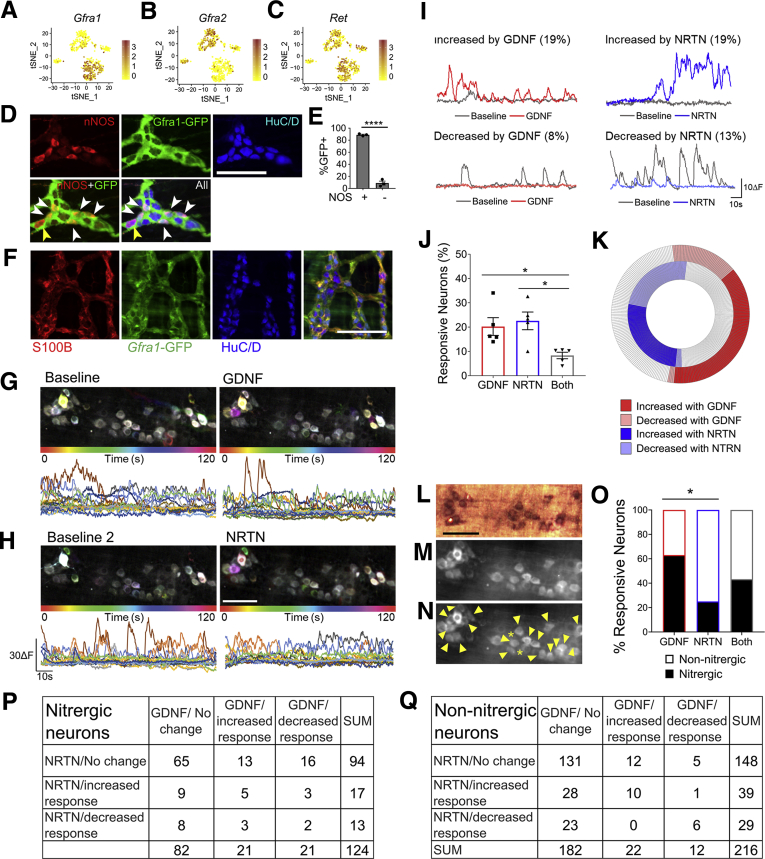
We were particularly intrigued that Gfra1 and Gfra2, preferred receptors for GDNF and NRTN, respectively, were abundant in glia but differentially expressed in neuron subsets. Gfra2 was primarily in Chat+ neurons (Figure 5A–C), consistent with prior studies.23 Gfra1 was primarily in Nos/Vip/Gal+ neurons. We could not find reports of Gfra1-restricted expression in adult mouse ENS, so we tested if NOS1 colocalized with GFP in the distal colon of Gfrα1Gfp/wt mice that express GFP from a Gfra1 locus.24 GFP was in 88.9 ± 1.8% of NOS1+ neurons, 8.8 ± 5.7% of NOS1-negative neurons (Figure 5D and E), and in S100β+ glia (Figure 5F) consistent with RNA-seq. Coupled with the observation that GDNF acts acutely in the ascending peristaltic response,25 our data suggest that GDNF and NRTN have opposing acute effects on bowel contractility, although both activate RET.
Figure 5.
GDNF and NRTN acutely influence GCaMP activity of largely nonoverlapping adult distal colon myenteric neuron populations. (A–C) Feature plots show Gfra1 primarily in Nos1/Vip/Gal neurons (A), Gfra2 in Chat neurons (B), and Ret in almost all neurons except Chat3 (C). (D) Whole mount immunohistochemistry shows GFP (green) in most NOS1+ (red) neurons of Gfra1Gfp/wt distal colon. White arrowheads show GFP+NOS1+ neurons. Yellow arrowhead shows GFP-NOS1+ neuron. (E) Most NOS1+ neurons are GFP+. Most NOS1– neurons are GFP-, consistent with RNA-seq (P < .0001, Student’s t test, n = 3 mice (Gfra1Gfp/wt)). (F) Whole mount immunohistochemistry using Gfra1Gfp/wt distal colon shows colocalization of GFP (green) with S100B+ (red) glia and HuC/D (blue) neurons. Scale bar = 100 μm. (G, H) Time-lapse images (top) (pixels were assigned color based on transients timing; color = activity) and traces (bottom) of GCaMP6s activity from regions of interest on myenteric neurons during baseline (left) and after adding 10 nM GDNF (G) or 10 nM NRTN (H) (right). (I) Sample traces from neurons with activity increased (top) or decreased (bottom) by GDNF (red) or NRTN (blue). Baseline (gray) and percent neurons (in parentheses) with increased or decreased activity (>2 SD change). (J) Percent neurons responding to only GDNF (red), only NRTN (blue), or both (gray) (P < .05, 1-way analysis of variance, Tukey’s multiple comparisons test). (K) Iris plot of GDNF and NRTN responsive myenteric neurons (n = 260 of 523 [49.7%] of total). GDNF is shown in the outer circle (red), NRTN is shown in the inner circle (blue) (n = 5 mice), and gray indicates no ligand-induced activity change. Light shades of red and blue indicate decreased activity after ligand addition. Most responsive neurons are affected by either GDNF or NRTN, not both. (L) NADPH diaphorase stained colon identifies nitric oxide–producing neurons. (M) Corresponding GCaMP6s imaging field. (N) GCaMP6s imaging field shown in M, where yellow arrowheads identify putative nitrergic myenteric neurons and asterisks indicate putative NADPH diaphorase positive neurons with low GCaMP6s signal. (O) Quantification of GDNF- and NRTN-responsive nitrergic neurons (n = 3 fields from separate experiments, P = .0298, Fisher exact test, 2 × 2 contingency table [NOS+/NOS– and GDNF response/NRTN response]). (P, Q) Quantitative data for GCaMP6s imaging fields stained post hoc for NADPH diaphorase, indicating response to GDNF and NRTN. (P) Responsive nitrergic neurons. (Q) Responsive non-nitrergic neurons. (E, J) Mean ± SD. (D, F, G, H) Scale bar = 100 μm. (L–Q) n = 3 fields of view from separate mice. ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
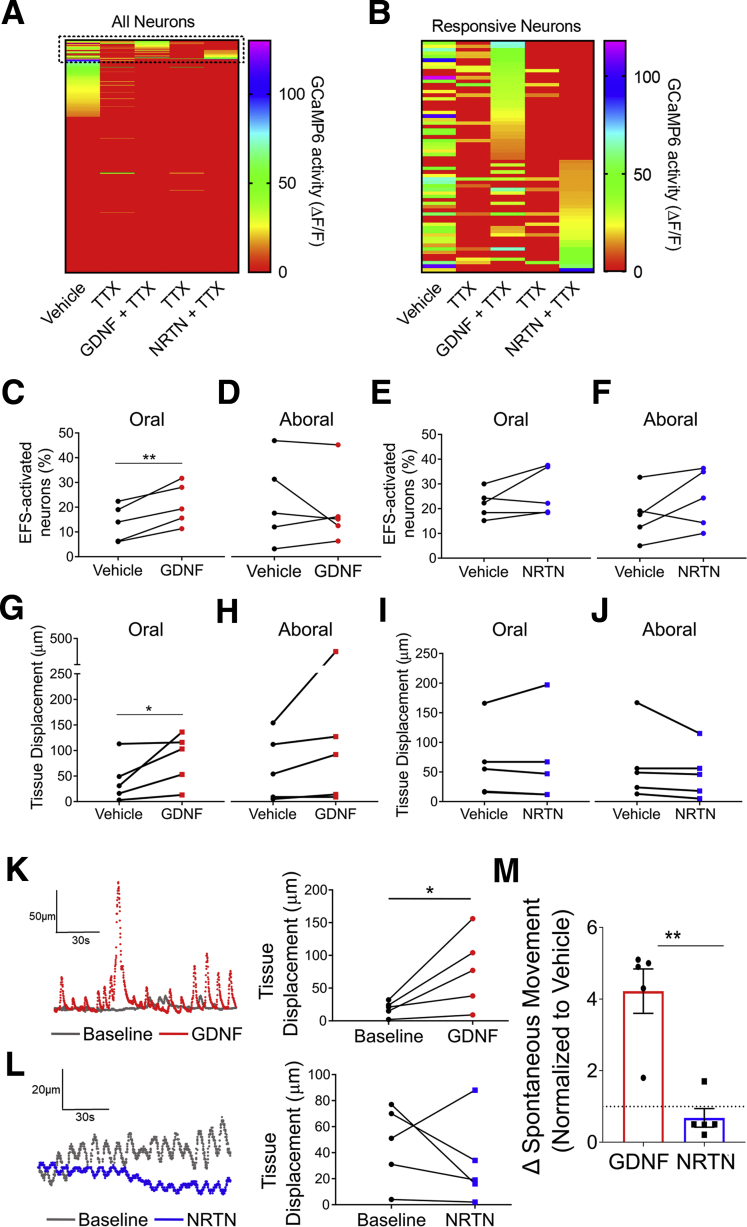
To test this, we recorded myenteric neuron calcium transients in GDNF or NRTN-treated full-thickness colon of E2a-Cre;R26R-GCaMP6s mice that express the calcium indicator GCaMP6s in all cells (Figure 5G and H). We also simultaneously monitored colon movement, an indirect measure of muscle tension.26 GDNF and NRTN increased or decreased spontaneous GCaMP6s activity in 50% (260/523) of myenteric neurons (Figure 5I–K). Only 8.3% responded to GDNF and NRTN (Figure 5J), suggesting that most effects are via their preferred receptors, GFRA1 and GFRA2, respectively. To test this, we identified nitrergic neurons post hoc in 3 fields. A total of 63% (n = 29 of 48) of neurons responding only to GDNF were nitrergic and 25% (n = 17 of 68) NRTN-only responders were nitrergic (Figure 5L–Q). The high percentage of GDNF-responsive non-nitrergic neurons suggested some GCaMP6s activity changes were due to synaptic connectivity. Remarkably, when we added the voltage-gated sodium channel blocker tetrodotoxin (TTX), only 8%–9% of myenteric neurons had GDNF- or NRTN-induced changes in GCaMP6s signal with minimal overlap between GDNF- and NRTN-influenced cells (Figure 6A and B). This suggests GDNF and NRTN have direct and indirect effects on GCaMP6s activity in distinct myenteric neurons.
Figure 6.
GDNF modulates activity in some myenteric neurons and enhances colon muscularis movement. (A, B) Heatmaps of GCaMP6s activity before and after adding TTX and during sequential GDNF and NRTN addition. Colors indicate GCaMP6s amplitude (red, no activity). (A) Heatmap of all neurons (n = 803 from 7 fields of view in 3 mice). Dotted box shows GDNF- and NRTN-responsive neurons. (B) Expanded heatmap of 34 neurons that were only GDNF-responsive, 20 neurons only NRTN-responsive, and 12 neurons that responded to both. (C–J) Percent of total observed neurons per GCaMP6s imaging field with increased activity after EFS applied 5 mm (C, E) oral or (D, F) aboral relative to the imaging field at baseline (vehicle) and in the presence of (C, D) GDNF and (E, F) NRTN. (C,) GDNF increases the percent of activated neurons after orally applied EFS (P = .0068, ratio paired t test). (G–J) Tissue displacement (micrometers) after EFS applied 5 mm (G, I) oral or (H, J) aboral relative to the imaging field at baseline (vehicle) and in the presence of (G, H) GDNF and (I, J) NRTN. (G) Tissue displacement is increased in the presence of GDNF after orally applied EFS (P = .0230, ratio paired t test). Sample traces (left) and graph (right) of tissue displacement at baseline and after (K) GDNF (P = .0022, ratio paired t test) or (L) NRTN (P = .1583, ratio paired t test). (M) GDNF, but not NRTN, enhanced tissue movement (P < .01, unpaired t test). (M) Mean ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
GDNF also increased myenteric neurons responding to oral electrical field stimulation (EFS) applied 5 mm from the imaged field by 71.3 ± 17.5% (P = .0068) but did not affect responses to EFS applied 5 mm aboral to recorded ganglia (P = .49) (Figure 6C and D). In contrast, NRTN did not affect the percentage of EFS-responsive myenteric neurons, regardless of stimulus location (P = .116 aboral, P = .188 oral) (Figure 6E and F). GDNF and NRTN both altered which neurons responded to oral and/or aboral EFS (data not shown). GDNF also robustly increased spontaneous and oral EFS-induced movement in the imaging field (P = .0230) (Figure 6G, H, K, and M). NRTN decreased movement in 3 of 5 preparations, but this was not statistically significant (Figure 6I, J, L, and M). These observations suggest that GDNF and NRTN influence distinct myenteric neurons and GDNF acutely enhances colon contractility.
Sequencing Suggests Combinatorial Transcription Factor Codes Define Neuron Subtypes
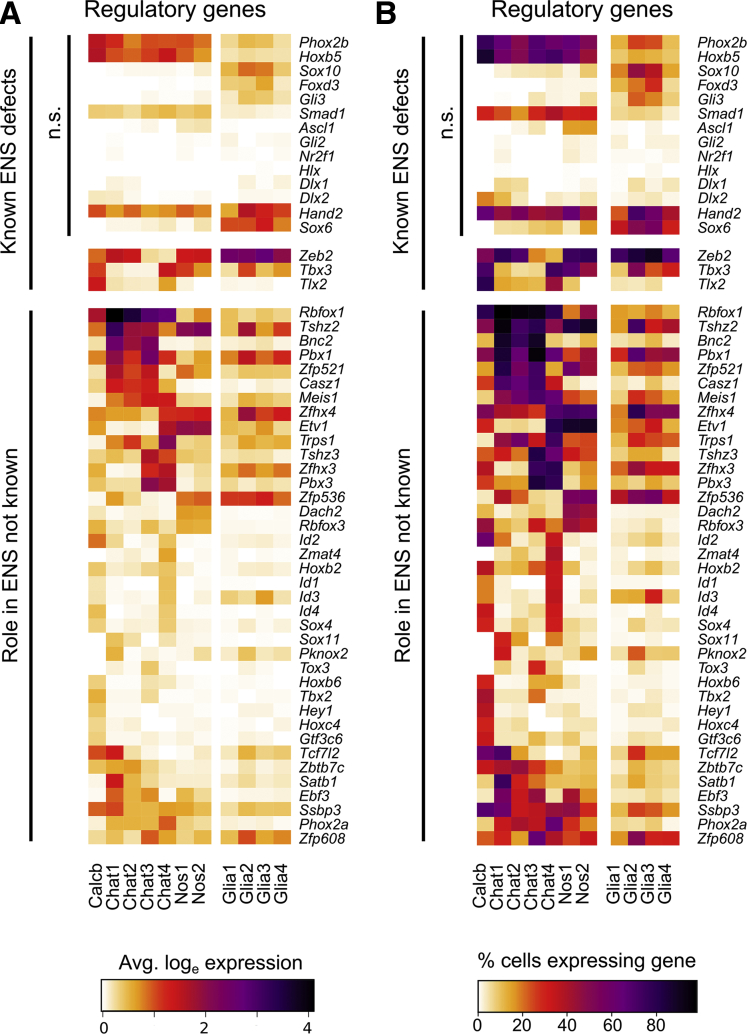
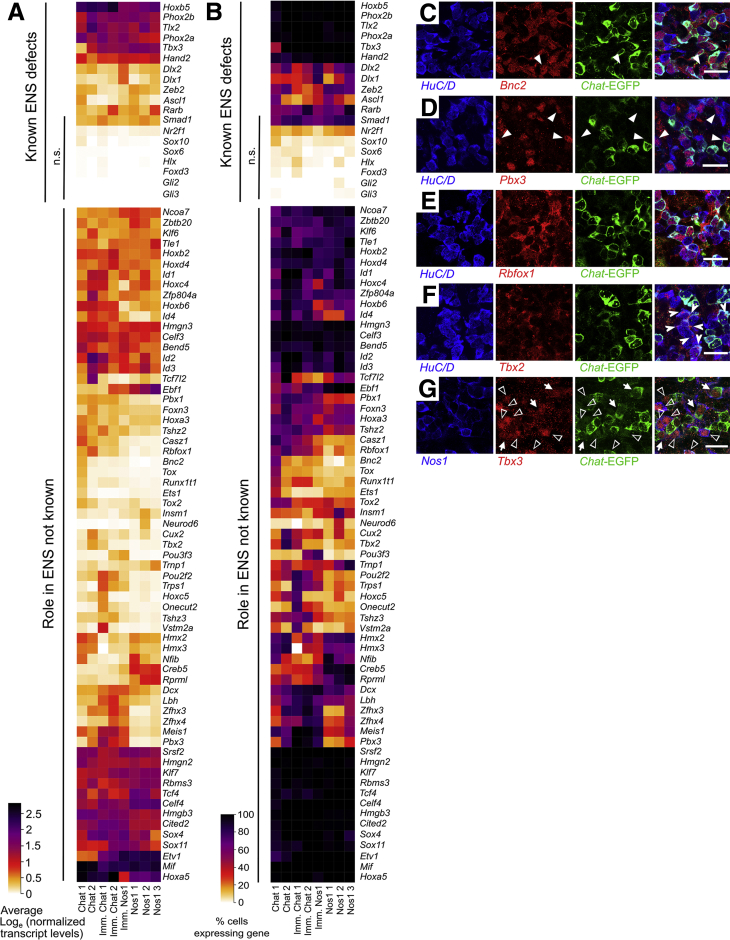
To gain insight into neuronal subtype identity, we examined transcription and splicing factors (regulatory genes) in adult colon myenteric neurons. Some genes that affect ENS subtype ratios (eg, Ascl1, Hand2, Sox6) were not differentially expressed in adult neuron subtypes, but others not known to affect subtype specification (eg, Tbx3, Tlx2, Zeb2)27,28 were differentially expressed (Figure 7). Many other transcription factors had distinct expression patterns in neuron subgroups including Pbx3, Etv1, Casz1, Bnc2, and Zfhx3 (Figure 7).
Figure 7.
Average expression and percent cells per cluster with detectable levels of differentially expressed transcription and splicing factors in adult distal mouse colon. (A) Transcription and splicing factors with known roles in ENS development (top), and newly identified differentially expressed factors (bottom). (B) Proportion of cells per cluster with expression values >0 for transcription and splicing factors with known roles in ENS development (top), and regulatory genes (transcription factors and splicing factors) newly identified in the ENS in this study (bottom). n.s., not significantly differentially expressed between neuron clusters.
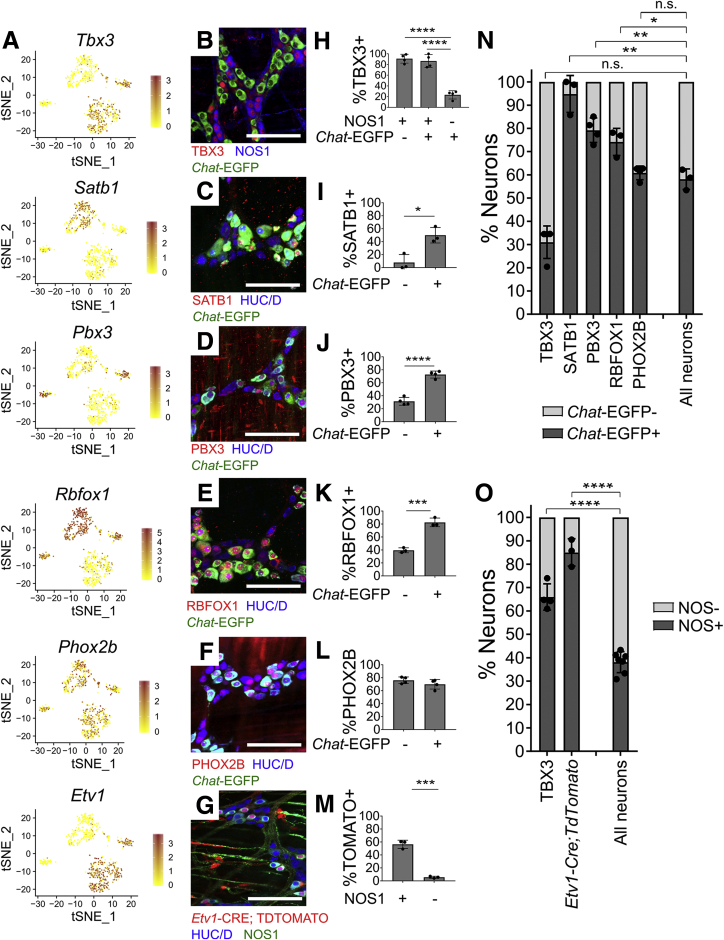
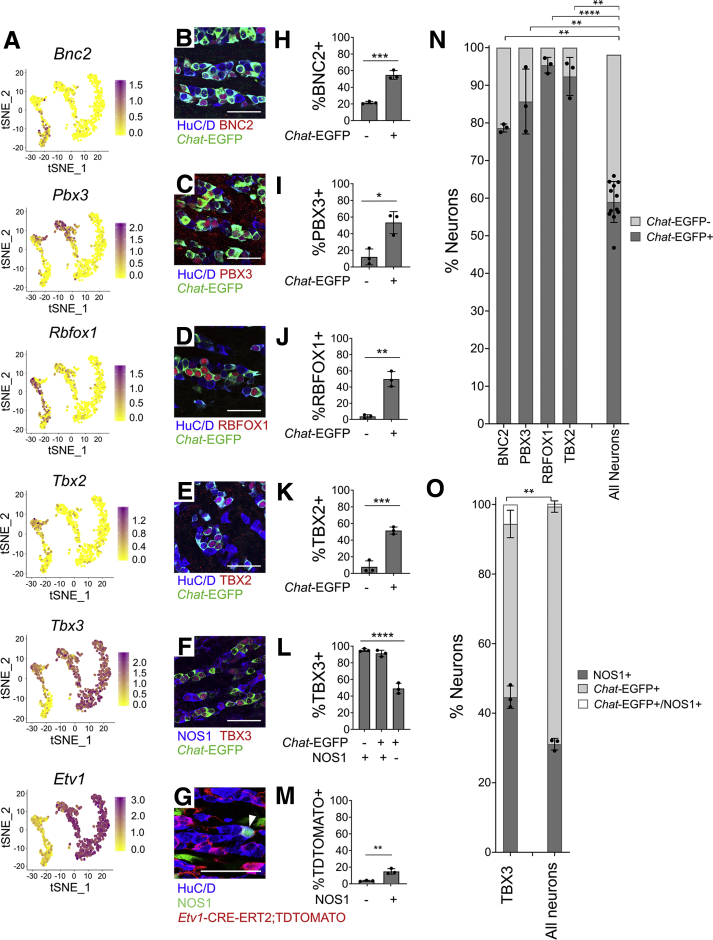
We pursued immunohistochemistry on regulatory genes differentially expressed in cholinergic or nitrergic neurons, as most myenteric neurons express either Chat or Nos1. We used ChAT-EGFP-L10A mice that reliably express EGFP at high levels in CHAT+ cells, as CHAT antibody staining is often weak.29 Consistent with RNA-seq, SATB1, RBFOX1, and PBX3 are preferentially in Chat-EGFP+ neurons (Figure 8A, C–E, I–K). TBX3 was primarily in NOS1+ neurons (Figure 8A, F, and L). RNA-seq also suggested Etv1 enrichment in NOS1+ neurons, which we validated by treating Etv1-CreERT2Cre/wt;R26R-TdTomato mice with high-dose tamoxifen to induce TDTOMATO from Etv1+ regulatory elements (Figure 8A, G, and M). Also consistent with RNA-seq, PHOX2B protein had similar abundance in Chat-EGFP+ and Chat-EGFP–myenteric neurons (Figure 8A, F, and L). Distribution of these transcription factors between Chat-EGFP+ and NOS1+ neurons (summarized in Figure 8N and O) shows substantial correlation between protein and RNA-seq data (Figure 7).
Figure 8.
Regulatory genes are expressed in distinct adult ENS subsets. (A) Feature plots of select regulatory genes. Color key shows loge(normalized gene expression). (B–F) Myenteric plexus wholemount immunohistochemistry in young adult ChAT-EGFP-L10A distal colon. (G) Whole mount myenteric plexus immunohistochemistry in distal colon from tamoxifen-treated Etv1-CreErt2;R26R-TdTomato mice. TDTOMATO is in many NOS1+ neurons and some non-neuronal cells with ICC morphology. (H–L) Immunohistochemistry quantification. TBX3 is found mostly in (H) nitrergic (NOS1+) neurons (P < .0001, n = 3). (I) SATB1 (P = .013, n = 3), (J) PBX3 (P < .0001, n = 3), and (K) RBFOX1 (P = .0006, n = 3) are mostly in cholinergic (EGFP+) neurons, consistent with single-cell RNA data. (L) PHOX2B is equally abundant in Chat-EGFP+ and Chat-EGFP–cells (P = .2193, n = 4). (M) Quantification of G shows that 56.4 ± 3.6% of NOS1+ neurons are TDTOMATO+. Only 5.6 ± 0.9% of NOS1– neurons are TDTOMATO+ (P = .0002, n = 3). (N) Cholinergic (Chat-EGFP+) and (O) nitrergic (NOS1+) identity for cells expressing select factors. SATB1 (P = .0022), PBX3 (P = .0026), and RBFOX1 (P = .019) are largely restricted to cholinergic (Chat-GFP+) neurons. PHOX2B is present in both cholinergic and noncholinergic neurons (P = .370) (P values compare Transcription factor+EGFP+/Transcription factor+ vs EGFP+/Total neuron ratios). TBX3 (P < .0001) and ETV1 (P < .0001) are primarily in nitrergic (NOS1+) neurons (P values compare Transcription factor+NOS1+/Transcription factor+ vs NOS1+/Total neuron ratios). (I–O) Student’s t test. (H) Analysis of variance with Tukey’s post hoc test. Scale bar = 100 μm. ChAT-EGFP-L10A = Chat-EGFP. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
Adult Neuron Subtype Transcription Factor Patterns Are Largely Established by E17.5
In mice, CHAT+ and NOS1+ enteric neurons are identifiable by late gestation and appear lineage committed.30,31 To identify lineage specific transcription factors at E17.5, we performed RNA-seq using whole bowel from ChAT-L10a-EGFP wt/wt; Nos1-CreERT2Cre/wt;TdTomato+ and ChAT-L10a-EGFP+; Nos1-CreERT2wt/wt;TdTomato+ littermates after E15.5 tamoxifen treatment. In these mice, EGFP marks CHAT-lineage and TDTOMATO identifies NOS1-lineage neurons (Figure 9A–C; see also Supplementary Figure 2).29 These lineages were chosen to minimize the presence of less-differentiated precursors in the sample and were sequenced together. A total of 1005 cells were sequenced at 83% saturation. We normalized after removing outliers, leaving 707 neurons (Elavl4+), 179 epithelial (Epcam+), and 67 smooth muscle cells (Actg2+) (Figure 9A and D). Reclustering without epithelia and muscle yielded 8 neuron clusters (Figure 9E and F). All had high Ret expression and expressed Dcx, an immature neuron marker.32 Enteric glia (Plp1+, Sox10+) were rare (Figure 9H). Chat transcripts were also rare as others reported,5,8 but vesicular acetylcholine transporter Slc18a3 was abundant in neuron clusters with low Nos1 (Figure 9G). Three clusters appeared less mature, prominently expressing neurite projection and cytoskeleton reorganization genes, based on Gene Ontology (GO) analysis. Other clusters expressed synaptogenesis and maintenance genes suggesting mature neurons (Figure 10A and B). Many neurotransmitters, receptors, ion channel subunits, and signaling molecules were differentially expressed between E17.5 subgroups (Figures 10C–E and 11), including Gfra1 and Gfra2 (Figure 11), and there was substantial overlap between E17.5 and adult subgroup expression of regulatory genes including Casz1, Bnc2, Etv1, Pbx3, Tbx2, Tbx3, and Rbfox1, a subset of which were validated using immunohistochemistry (Figure 7, Figure 8, Figure 12, and 13). Similar to our adult data, we also validated Etv1 enrichment in NOS1+ neurons by treating Etv1-CreERT2Cre/wt;R26R-TdTomato mice with high-dose tamoxifen to induce TDTOMATO from Etv1+ regulatory elements (Figure 13A, G, and M).
Figure 9.
Single-cell RNA-seq of E17.5 ChAT-EGFP-L10A+ and Nos1-CreERT2Cre/wt;R26R-TdTomato+ neurons show distinct nitrergic and cholinergic clusters. (A) RNA-seq workflow with t-SNE plot of all cell groups isolated from E17.5 bowel. (B) ChAT-EGP (green) and NOS1 (red) do not overlap at E17.5. Scale bar = 50 μm. (C) NOS1 (green) and nNOS-Cre-ERT2;R26R-tdTomato (red) overlap. (D) Feature plots of Acta2 as smooth muscle marker, Epcam as an intestinal epithelial marker, and Elavl4 as a pan-neuronal marker. Color key represents loge(normalized gene expression). (E) After removing the epithelial and smooth muscle cells from the dataset and reclustering, t-SNE plot reveals multiple neuron subpopulations. (F) Violin plots show subgroups had 25,319.7 ± 5562.2 unique RNA transcripts (UMIs) and 5291.6 ± 558.8 detected genes (nGenes) per cell. (G) Expression of Nos1 and the cholinergic marker Slc18a3/VachT verifies the presence of the expected cholinergic and nitergic neuronal subpopulations. Chat expression was low throughout, but overlapped with Slc18a3 expression. Color key represents loge(normalized gene expression). (H) All neurons have high expression of pan-neuronal markers Tubb3 and Elavl4. There is minimal contamination with glial cells based on the expression of enteric glial marker Plp1. Sox10, which marks enteric glia and enteric neural crest precursors was also low. Ret, which is expressed in ENS precursors and many neurons, was present in all clusters, and all neurons still express the immature pan-neuronal marker Dcx. This suggests that these cells are lineage-restricted immature neurons.
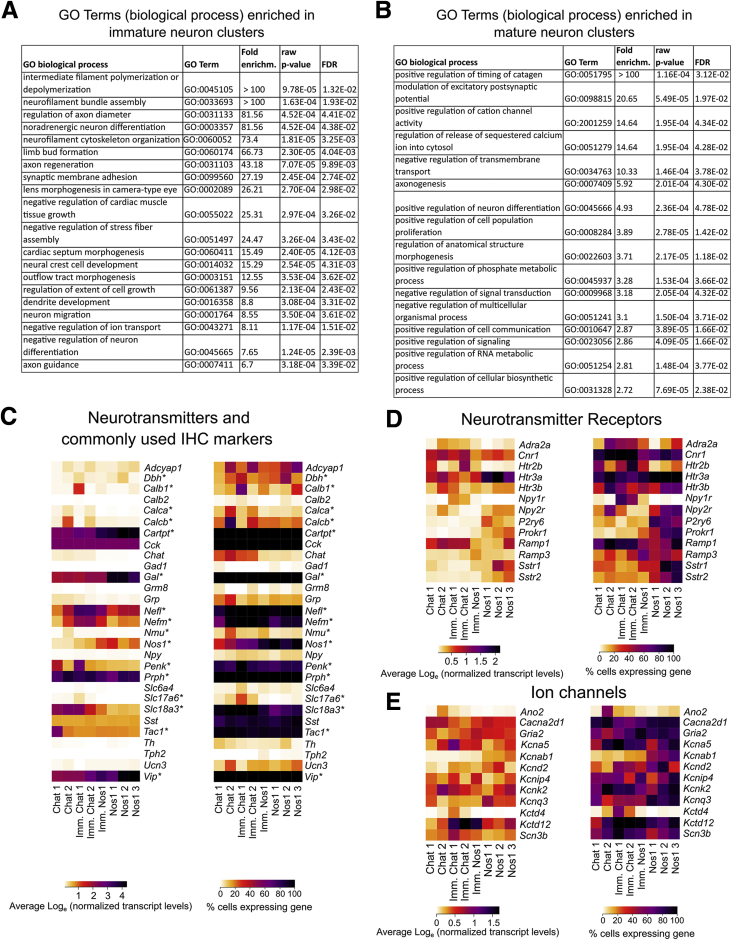
Figure 10.
GO term analysis of differentially expressed genes in E17.5 bowel shows more immature and mature neuron clusters, with unique combinations of neurotransmitters, receptors, and ion channels. (A) GO term analysis of differentially expressed genes in the 3 more immature neuron clusters (immature Chat cluster 1, immature Chat cluster 2, immature Nos1 cluster) compared with all other clusters indicates that these neurons are actively involved in cytoskeletal reorganization and neurite extension. (B) GO term analysis of differentially expressed genes in the 5 more mature neuron clusters (Chat cluster 1, Chat cluster 2, Nos1 cluster 1, Nos1 cluster 2, and Nos1 cluster 3) compared with the immature clusters (immature Chat cluster 1, immature Chat cluster 2, and immature Nos1 cluster) indicates that these neurons are actively involved in synapse formation. (C–E) Average expression (left) and proportion of cells per cluster (right) with expression values >0 (right) for neurotransmitters and commonly used (C) immunohistochemistry markers, (D) neurotransmitter receptors, and (E) ion channels for different 17.5 groups. Color key for left panels represents loge(normalized average gene expression within each cluster). (C) Asterisk indicates differential expression across neuron subtypes. (D, E) Genes listed are differentially expressed across subtypes. imm, immature.
Figure 11.
Average expression and percent cells per cluster with detectable levels of differentially expressed signaling pathway molecules in E17.5 mouse bowel. (A) Average expression of differentially expressed signaling pathway genes for distinct neuron clusters. Color key represents loge(normalized average gene expression within each cluster). (B) Proportion of cells per cluster with expression values >0 for differentially expressed signaling pathway molecules.
Figure 12.
Differentially expressed transcription and splicing factors in E17.5 mouse bowel. (A) Transcription and splicing factors with known roles in ENS development (top), and newly identified differentially expressed factors (bottom). (B) Proportion of cells per cluster with expression values >0 for transcription and splicing factors with known roles in ENS development (top), and regulatory genes (transcription factors and splicing factors) newly identified in the ENS in this study (bottom). (C–G) Whole mount immunohistochemistry of select regulatory genes in E17.5 ChAT-EGFP-L10A reporter mouse mid colon shows gene localization in neuron subsets. (C) BNC2, (D) PBX3, and (E) RBFOX1 are predominantly expressed in cholinergic (EGFP+) neurons. (C, D) White arrowheads indicate neurons that express the regulatory gene in question but are not cholinergic. (F) TBX2 does not have nuclear or diffuse cytoplasmic staining in the E17.5 colon. Instead, TBX2 immunoreactive cytoplasmic aggregates were detected in a subset of cholinergic (EGFP+) neurons (notched white arrowheads). (G) TBX3 is expressed in some cholinergic (EGFP+, white arrows) and most nitrergic (NOS1+, empty arrowheads) neurons. (C–G) ChAT-EGFP-L10A reporter = Chat-EGFP. Scale bar = 20 μm (C–G). n.s., not significantly differentially expressed between neuron clusters.
Figure 13.
E17.5 data show many regulatory genes are differentially expressed in patterns resembling adult colon myenteric plexus. (A) Feature plots. Colors show loge (normalized gene expression). (B–F) Whole mount immunohistochemistry of E17.5 ChAT-EGFP-L10A mid small intestine. (G) Whole mount myenteric plexus immunohistochemistry of E17.5 mid small intestine from tamoxifen-treated Etv1-CreErt2;R26R-TdTomato mice. TDTOMATO is expressed in many NOS1– neurons and some NOS1+ neurons. White arrowhead points to a NOS1+TDTOMATO+ neuron. Scale bar = 50 μm. (H–L) Immunohistochemistry quantification. (H) BNC2 (P = .0005, n = 3), (I) PBX3 (P = .0119, n = 3), (J) RBFOX1 (P = .0012, n = 3), and (K) TBX2 (P < .001, n = 3) are primarily in cholinergic (Chat-GFP+) neurons. (L) TBX3 (P < .0001, n = 3, analysis of variance with Tukey’s post hoc test) is primarily in NOS1+ neurons. (M) Quantification of G shows preferential TDTOMATO expression in NOS1+ neurons (P = .0042, n = 3). Quantification of (N) cholinergic (Chat-EGFP+) and (O) nitrergic (NOS1+) identity. Neurons expressing BNC2 (P = .0018), PBX3 (P = .0165), RBFOX1 (P < .0001), and TBX2 (P = .0016) are primarily cholinergic (Chat-GFP+) (P values compare Transcription factor+EGFP+/Transcription factor+ vs EGFP+/Total neuron ratios). TBX3+ neurons (P = .003) are primarily nitrergic (P values compare Transcription factor+NOS1+/Transcription factor+ vs NOS1+/Total neuron ratios). (H–L) Mean ± SD. (B–O) ChAT-EGFP-L10A reporter=Chat-EGFP. (H–K, M–O) Student’s t test. (L) Analysis of variance with Tukey’s post hoc test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
Conditional Tbx3 Loss Reduced NOS1+ Myenteric Neuron Density, But Casz1, Tbx2, and Rbfox1 Loss Did Not Alter Chat-EGFP+/Total Neuron Subtype Ratios
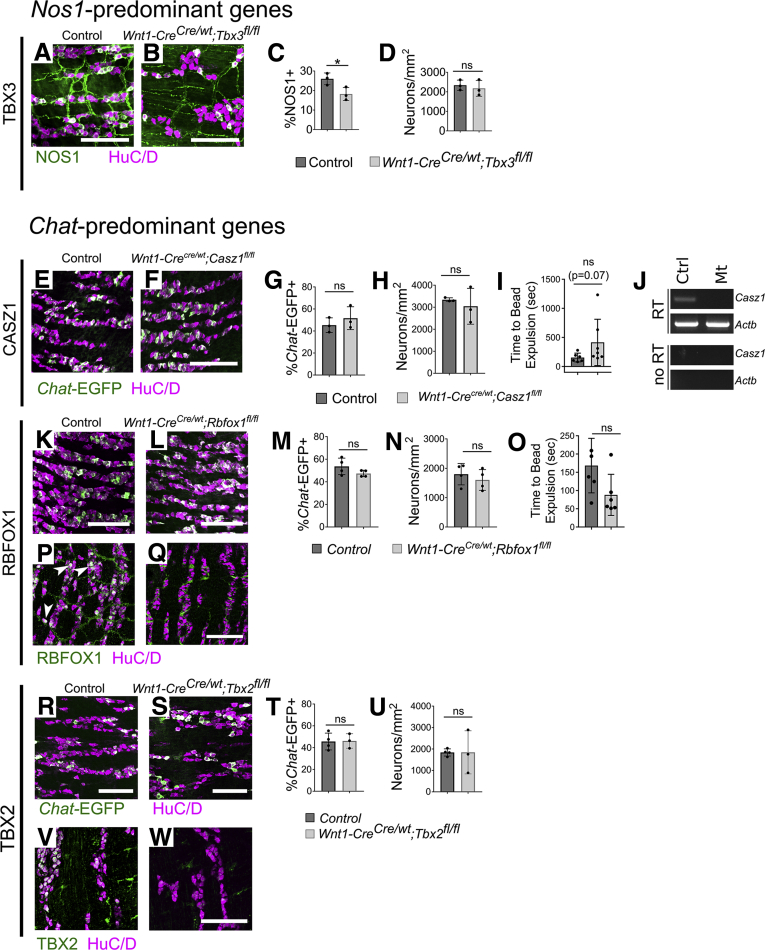
For several differentially expressed regulatory genes, we determined if mutations altered nitrergic or cholinergic enteric neuron abundance. We found that loss of TBX3 in ENS led to ∼30% fewer NOS1+ neurons in small bowel of P0 Wnt1-Cre;Tbx3fl/fl;R26R-Tdtomato mice, despite normal total neuron density (Figure 14A–D). In contrast, Chat-EGFP+ neuron density was normal in Casz1 mutants (Wnt1-Cre;Casz1fl/fl;Chat-EGFP-L10A), in Rbfox1 mutants (Wnt1-Cre;Rbfox1fl/fl;Chat-EGFP-L10A), and in Tbx2 mutants (Wnt1-Cre;Tbx2fl/fl;Chat-EGFP-L10A) despite enrichment of these genes in cholinergic neurons (Figure 14E–H, K–N, and R–U). Colonic bead expulsion latency tests were also normal in adult Casz1 (Figure 14I) and Rbfox1 (Figure 14O) mutant mice, suggesting intact ENS-mediated distal colon motility. We were unable to perform similar motility experiments in Tbx2 mutant mice, which died shortly after birth. Collectively, these findings suggest TBX3 impacts NOS1+ neurons, but conditional loss of Tbx2, Casz1, and Rbfox1 does not alter CHAT/NOS1 neuron numbers.
Figure 14.
Tbx3 loss reduces NOS1+ myenteric neurons. Casz1, Rbfox1, and Tbx2 mutants had normal abundance of Chat-EGFP+ neurons and Casz1 and Rbfox1 had normal colon bead expulsion. (A, B) Confocal maximum intensity projections of P0 Tbx3 mutant and control small bowel myenteric plexus revealed a (C, D) 30% reduction in NOS1+/Total neuron ratio in (C) Wnt1-CreCre/wt;Tbx3fl/fl P0 small bowel (P = .041, n = 3 per group) despite (D) normal total neuron density (P = .601, n = 3 per group). (E) P0 small bowel myenteric plexus of control and (F) Wnt1-CreCre/wt;Casz1fl/fl mice also carrying the Chat-EGFP-L10A reporter. (G) Quantification shows normal Chat-EGFP+/Total neuron ratios (P = .424, n = 3 in each group) and normal neuron density (H; P = .700, n = 3 in each group) in Casz1 mutant P0 small bowel. (I) Colonic bead expulsion latency is normal in Wnt1-CreCre/wt;Casz1fl/fl mice (P = .073, n = 6 [control, male = 2/female = 4, P44–70 at start of assay], n = 7 [Wnt1-CreCre/wt;Casz1fl/fl, male = 2/female = 5, P42–50 at start of assay]). (J) RT-PCR of TDTOMATO+ cells from Wnt1-CreCre/wt;Casz1fl/fl;TdTomato+ mice lacks the Casz1 band, consistent with loss of Casz1 expression. Single confocal planes from P0 small bowel of (K) control and (L) Wnt1-CreCre/wt;Rbfox1fl/fl bred to Chat-EGFP-L10A. (M, N) Quantification shows that proportion of EGFP+ neurons (P = .162, n = 4 per group) and total neuron density (P = .470, n = 4 per group) are normal in Rbfox1 mutants. (O) Colon bead expulsion latency is normal in Wnt1-CreCre/wt;Rbfox1fl/fl mice (P = .2677, n = 5 [control, male = 2/female = 3, P44–46 at start of assay], n = 6 [Wnt1-CreCre/wt;Rbfox1fl/fl, male = 2/female = 4, P46–58 at start of assay]). (P, Q) Control mice at P0 showed robust RBFOX1 immunofluorescence in neuronal nuclei (white/green, examples shown with white arrowheads), whereas Wnt1-CreCre/wt;Rbfox1fl/fl had no neuronal nuclear RBFOX1 immunofluorescence. (R, S) Single confocal planes from P0 small bowel myenteric plexus from control and Wnt1-CreCre/wt;Tbx2fl/fl bred to Chat-EGFP-L10A. (T, U) Quantification shows that proportion of EGFP+ neurons (P = .926, n = 4 [control] and n = 3 [Wnt1-CreCre/wt;Tbx2fl/fl]) and total neuron density (P = .857, n = 4 [control] and n = 3 [Wnt1-CreCre/wt;Tbx2fl/fl]) are normal in Tbx2 mutants. (V, W) Control mice at P0 showed robust TBX2 immunofluorescence in neuronal nuclei (white/green), whereas Wnt1-CreCre/wt;Tbx2fl/fl had no neuronal nuclear TBX2 immunofluorescence. (C, D, G–I, M, O, T, U) Mean ± SD. (C, D, G, H, M, N, T) Student’s t test. (I, O, U) Mann-Whitney. (A, B, E, F, K, L, P–S, V, W) Scale bar = 100 μm. ChAT-EGFP-L10A=Chat-EGFP. ∗P < .05. Ctrl, control; Mt, Wnt1-CreCre/wt;Casz1fl/fl;TdTomato+.
Pou3f3 Is in Colon, But Not Small Bowel ENS
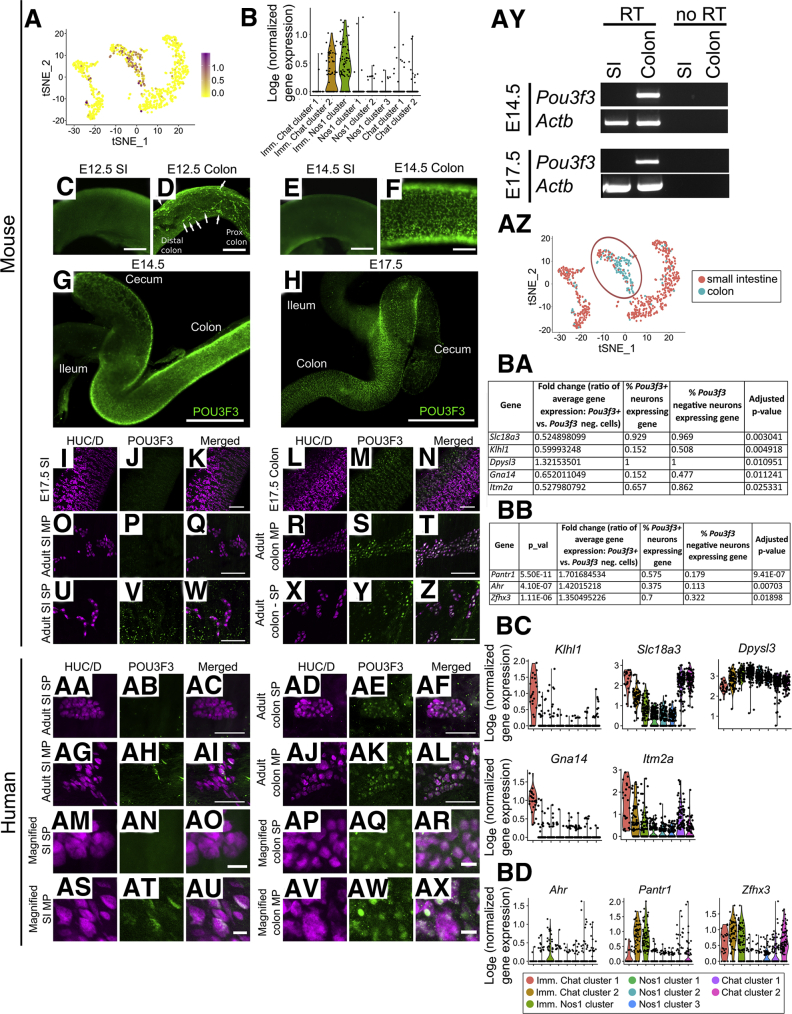
In addition to subtype identity, regional differences in neurons could promote distinct motility patterns. We were intrigued by the observation that Pou3f3 (aka Brn1), a transcription factor important for CNS development, was mostly restricted to E17.5 immature Chat2 and immature Nos1 clusters (Figure 15A and B). Pou3f3 enrichment in colon ENS could explain these results, a hypthesis confirmed by RT-PCR (Figure 15AY). Immunohistochemistry using 2 antibodies confirmed POU3F3 marks all immature and mature colon neurons, but no small bowel neurons (Figure 15C–Z), as recently reported.33 Human colon also had POU3F3 in the nucleus of all submucosal and myenteric neurons. Human small bowel submucosal neurons lacked POU3F3, but some small bowel myenteric neurons had cytoplasmic POU3F3 immunoreactivity of uncertain significance (Figure 15AA–AX). These data significantly extend recent reports that colon, but not small bowel, enteric neurons express POU3F3.
Figure 15.
Pou3f3 is expressed in mouse colon ENS but not small intestine ENS. In adult humans, nuclear POU3F3 immunoreactivity was also restricted to colon enteric neurons. (A) Feature plot shows scattered E17.5 enteric neurons expressing Pou3f3 throughout the neuron clusters. Color key represents loge(normalized gene expression). (B) Violin plot of Pou3f3 expression in E17.5 enteric neurons indicates that Pou3f3 is predominantly expressed in Immature Chat cluster 2 and Immature Nos1 cluster. (C–Z) Whole mount immunohistochemistry confirms that POU3F3 immunoreactivity (green) is not detected in small bowel enteric neurons at (C) E12.5, (E) at E14.5, (H–N) at E17.5, or (O–Z) in adulthood. POU3F3 immunoreactivity is easily detected in (D) proximal colon at E12.5 (arrows) and throughout the colon (F) at E14.5, (H, M) at E17.5, and (S, Y) in adulthood. (N, T, Z) At E17.5 and in adulthood, colonic POU3F3 co-localizes with the enteric neuron marker HuC/D. Confocal z-stack maximum intensity projections at (AA–AL) lower magnification and (AM–AX) high magnification of whole mount immunohistochemistry for POU3F3 in (AA–AC, AG–AI, AM–AO, AS–AU) adult human small intestine and (AD–AF, AJ–AL, AP–AR, AV–AX) adult human colon shows nuclear POU3F3 localization only in colonic neurons. (AA–AC, AM–AO) No POU3F3 staining was detectable in human small intestine submucosal plexus, whereas (AD–AF, AP–AR) clear nuclear POU3F3 staining could be seen in human colon submucosal neurons. (AG–AI, AS–AU) Cytoplasmic POU3F3 antibody staining was present in a subpopulation of human small intestine myenteric neurons. (AJ–AL, AV–AX) In the human colon myenteric plexus, all neurons showed clear nuclear POU3F3 localization. (AY) Representative RT-PCR for flow sorted TDTOMATO+ ENS cells from Wnt1-Cre;R26R-TdTomato mice confirms Pou3f3 in fetal colon but not small intestine (SI). (AZ) Feature plot showing scattered E17.5 enteric colonic neurons. Cells expressing Pou3f3 were assigned colonic identity. Red circle marks immature clusters: immature Chat cluster 1, immature Chat cluster 2, and immature Nos1 cluster. (BA) Genes differentially expressed by cells assigned colon identity compared with cells assigned small intestine identity within the 3 immature clusters (immature Chat cluster 1, immature Chat cluster 2, immature Nos1 cluster). (BB) Genes differentially expressed by cells assigned colon identity compared with cells assigned small intestine identity within the 3 mature clusters (Chat cluster 1 and 2 and Nos1 cluster 1, 2, and 3). (BC) Violin plots showing expression of the differentially expressed genes across all E17.5 neuron clusters indicate that the genes identified in BA are specific to immature Chat cluster 1 and not colon or small intestine (Pearson correlation between the expression of the identified gene and Pou3f3 supports this conclusion: P > .1 for all, except Dpysl3; correlation coefficient = 0.0819, P = .0294). (BD) Violin plots showing expression of the differentially expressed genes across all E17.5 neuron clusters suggest that the expression of genes Ahr and Pantr1 and possibly Zfhx3 are specific to colon myenteric neurons (Pearson correlation between the expression of the identified gene and Pou3f3 supports this conclusion: Ahr, correlation coefficient = 0.3581, P < 2.2 × 10-6; Pantr1, correlation coefficient = 0.5640, P < 2.2 × 10-6; Zfhx3, correlation coefficient = 0.4034, P < 2.2 × 10-6). Images are representative of 3 independent biological replicates. Scale bar = 100 μm (C, E–AL), 200 μm (D), 20 μm (AM–AX) 500 μm (G), 1 mm (H). ChAT-EGFP-L10A=Chat-EGFP. MP, myenteric plexus; SP, submucosal plexus,
Using Pou3f3 to mark E17.5 colon neurons (Figure 15AZ) and restricting comparisons to only mature or immature clusters, we identified 5 genes differentially expressed in Pou3f3+ vs Pou3f3- enteric neurons of immature clusters (Slc18a3, Klhl1, Dpysl3, Gna14, and Itm2a) (Figure 15BA and BC) and 3 genes differentially expressed in Pou3f3+ vs Pou3f3- neurons of mature clusters (Ahr, Pantr1, Zfhx3) (Figure 15BB and BD). Ahr, Pantr1, and Zfhx3 patterns resemble Pou3f3 (Pearson correlation) (compare Figure 15B–BD), and others recently reported Ahr enrichment in colon ENS.33
Adult Human Colon Myenteric Plexus
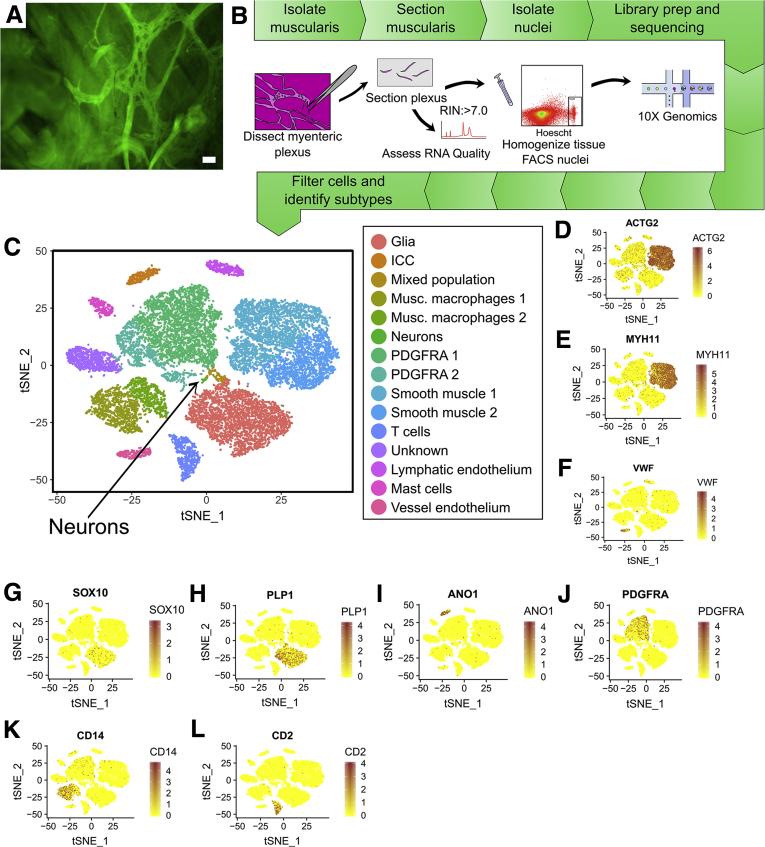
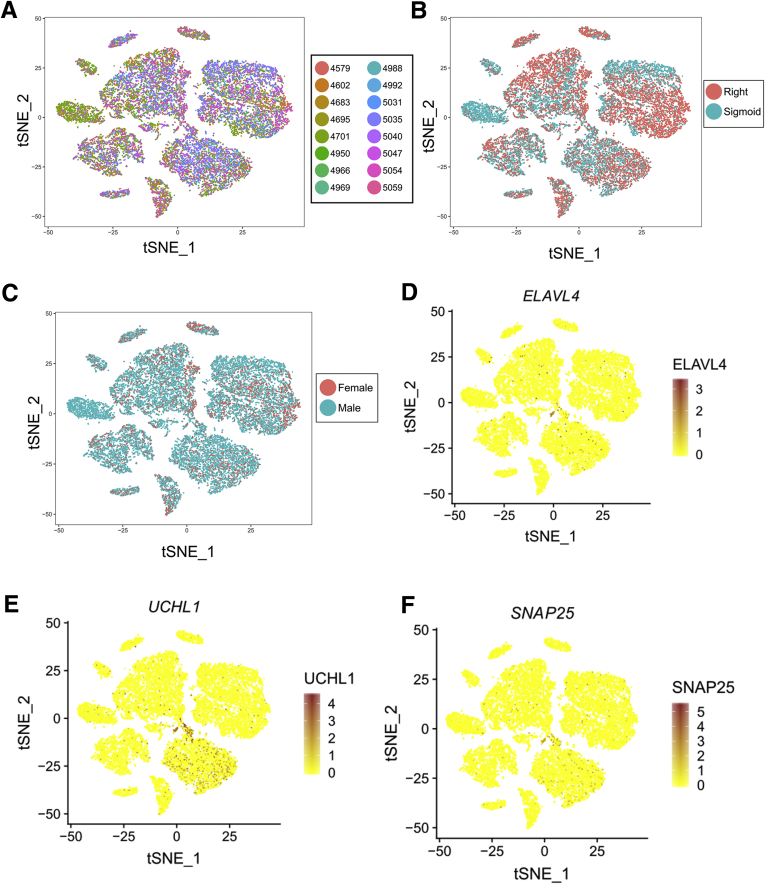
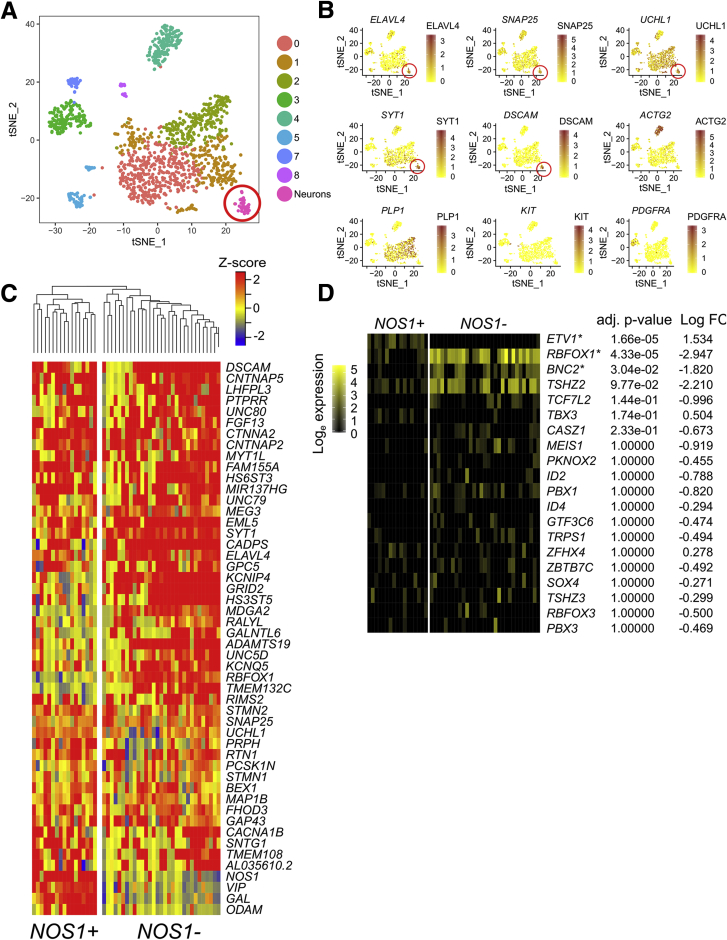
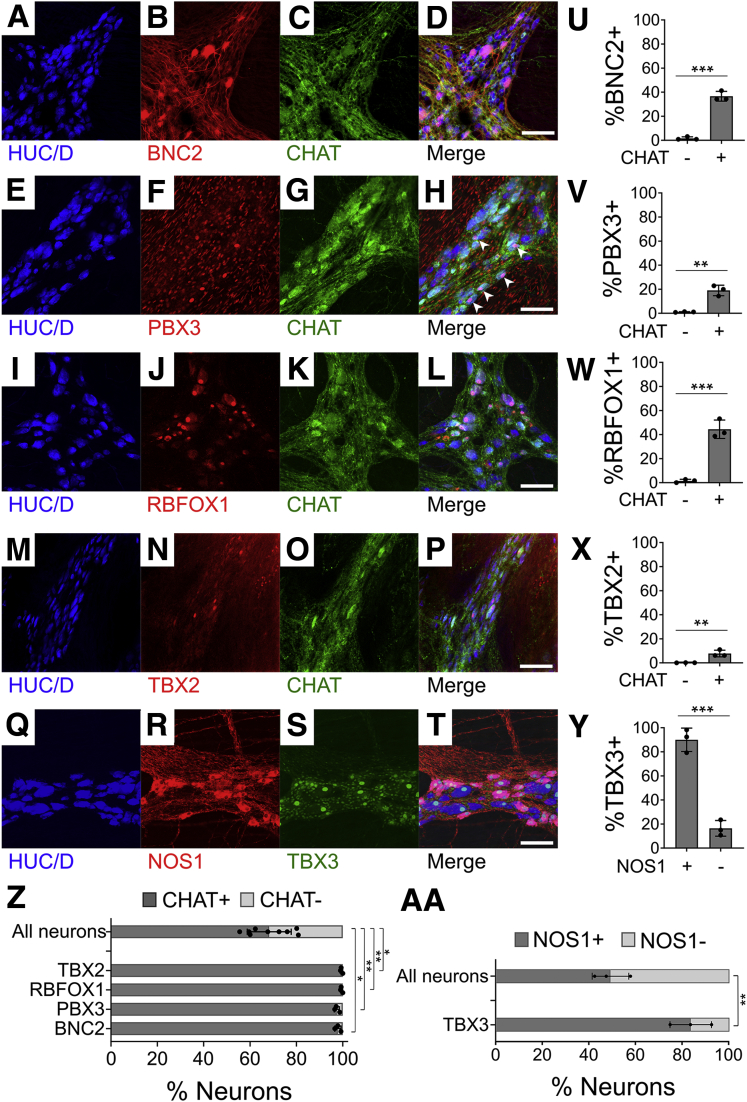
We next sequenced single nuclei from human colon myenteric plexus and surrounding cells. Isolating ENS cells was challenging. Neither whole cell sequencing nor immunoselection of neuronal nuclei using NeuN or PHOX2B antibodies was successful. Ultimately, we obtained data from a limited number of human colon myenteric neuron nuclei by microdissecting live plexus labeled with 4-Di-2-Asp (4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide) (a dye that accumulates in presynaptic nerve terminal mitochondria) (Figure 16A), freezing in optimal cutting temperature compound (OCT), sectioning, Dounce homogenizing, and flow sorting Hoechst+ single nuclei from cell debris. 10x Genomics (Figure 16B) yielded RNA-seq data from 20,167 nuclei of 16 adult colons (Table 2) with a mean of 1455 UMIs and 894 genes per nucleus (Table 3). Our data included many smooth muscle, ICC, PDGFRα+ cells, enteric glia, muscularis macrophage, and vascular endothelial and hematopoietic cells (Figure 16B–L) (ie, most cells that impact bowel motility). One cluster of 48 cells was strongly positive for neuronal genes like ELAVL4, SNAP25, and UCHL1 (Figure 17D–F). Reclustering all human nuclei expressing ELAVL4, UCHL1, or SNAP25 led to unbiased identification of this same group, plus other cells that are likely doublets, confirming neuron identity of this group (Figure 18A and B). Because of small cell numbers, we used hierarchical Euclidean clustering based on the top 50 markers distinguishing neurons from other cells. This gave 2 distinct clusters (Figure 18C) that differ in NOS1/VIP/GAL+ expression. CHAT and SLC18A3 (VAChT) transcripts were not detected in either subgroup, probably due to low read depth. Low CHAT was also noted by others.5,8 We next asked if any of the 41 transcription or splicing factors differentially expressed in mouse Nos1+ vs Chat+ myenteric neurons were also differentially expressed in human myenteric neuron subtypes. Consistent with mouse data, ETV1 was most abundant in NOS1+ neurons, while RBFOX1 and BNC2 were enriched in NOS1– neurons (Figure 18D) after multiple comparison testing. Immunohistochemistry confirmed BNC2 (Figure 19A–D and U), PBX3 (Figure 19E–H and V), RBFOX1 (Figure 19I–L and W), TBX2 (Figure 19M–P and X), and TBX3 (Figure 19Q–T and Y) were differentially expressed in human enteric neuron subtypes, with patterns similar to mouse (ie, most RBFOX1+, PBX3+, BNC2+, and TBX2+ cells are CHAT+; most TBX3+ cells are NOS+) (Figure 19Z and AA).
Figure 16.
Human single-nucleus RNA-seq analysis from 20,167 cells yielded data from many cells that impact bowel motility including SMC, ICC, PDGFRA+ cells, muscularis macrophage, and glia. (A) Human myenteric plexus after incubation with 4-Di-2-Asp (4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide), with muscle layers partially peeled away. Scale bar = 50 μm. (B) RNA-seq workflow for adult human colon myenteric plexus. (C) t-SNE plot of 20,167 nuclei shows glia, ICC, muscularis macrophages, PDGFRA+ cells, smooth muscle, T cells, endothelium, and unknown groups. Neurons comprise 1 small cluster (∼48 cells). (D–L) Feature plots showing genes expressed highly in (D, E) adult human smooth muscle (ACTG2, MYH11), (F) vessel endothelial cells (VWF), (G, H) glial cells (PLP1, SOX10), (I) ICC (ANO1), (J) PDGFRA+ cells (PDGFRA), (K) muscularis macrophages (CD14), and (L) T cells (CD2). Color key represents loge(normalized gene expression).
Table 2.
Characteristics of Colon Resection Samples
| Sample ID | Age (y) | Sex | History | Colon region | RNA Integrity number: plexus | RNA Integrity number: surrounding muscle |
|---|---|---|---|---|---|---|
| 4579 | 54 | M | Cecal polyp | Right | 7.6 | 7.9 |
| 4602 | 75 | M | Cecal lesion | Right | 7.5 | 6.9 |
| 4683 | 38 | M | Goblet cell carcinoma | Right | 7.9 | 8 |
| 4695 | 77 | F | Colonic mass | Right | 7.4 | 7.4 |
| 4701 | 78 | M | Rectal cancer | Sigmoid | 8.2 | 6.3 |
| 4950 | 78 | M | Bowel obstruction | Sigmoid colon | 6.50, 6.90, and 7.10 (3 pieces) | 7.6 |
| 4969 | 83 | M | Adenocarcinoma | Right colon | 6.5 | 7.90, 5.80 (2 pieces) |
| 4966 | 71 | F | Bowel obstruction | Right colon | 7.2 | 8.1, 7.9 (2 pieces) |
| 4988 | 65 | F | Colon polyp | Right colon | 5.20 and 2.90 (RNA concentration very low) | 7.5 and 7.4 (2 pieces) |
| 4992 | 47 | M | Rectal carcinoma | Sigmoid colon | 4.4 (RNA concentration very low)a | 7.1 |
| 5031 | 70 | M | Colon polyp | Right colon | 7.8 | 7.4 and 7.2 |
| 5035 | 24 | M | Volvulus | Sigmoid colon | 8 | 6.6 and 7.2 |
| 5040 | 44 | M | Colonic mass | Right colon | 7.7 | 7.5 and 7.8 |
| 5047 | 65 | M | Rectal adenocarcinoma | Sigmoid colon | 6.6 | 7.4 |
| 5054 | 36 | F | Bowel adhesions | Right colon | 7.6 | 7.20 and 7.70 |
| 5059 | 59 | F | Adenocarcinoma | Right | 7.2 | 7.50 and 6.40 |
F, female; M, male.
RNA concentrations which are too low can yield sub-par RIN.
Table 3.
UMI and Gene Counts From Colon Resection Samples
| Sample ID | Cells or nuclei loaded (predicted from FACS) | Cells with RNA-Seq data | Ratio of sequenced cells to loaded cells | Average UMI count with intronic reads mapped | Average gene count with intronic reads mapped |
|---|---|---|---|---|---|
| 4579 | 4000 | 680 | 0.17 | 2474 | 1559 |
| 4602 | 6900 | 2316 | 0.3356522 | 2004 | 1268 |
| 4683 | 6300 | 2081 | 0.3303175 | 1397 | 876 |
| 4695 | 4000 | 833 | 0.20825 | 1487 | 974 |
| 4701 | 12300 | 4414 | 0.3588618 | 1296 | 780 |
| 4950 | 5200 | 1237 | 0.2378846 | 1415 | 910 |
| 4969 | 5300 | 653 | 0.1232075 | 1262 | 1262 |
| 4966 | 2100 | 432 | 0.2057143 | 1856 | 1193 |
| 4988 | 1200 | 237 | 0.1975 | 2354 | 1554 |
| 4992 | 673 | 60 | 0.089153 | 3939 | 2336 |
| 5031 | 5200 | 1728 | 0.3323077 | 1538 | 967 |
| 5035 | 7100 | 2524 | 0.355493 | 1886 | 1119 |
| 5040 | 2300 | 755 | 0.3282609 | 1941 | 1212 |
| 5047 | 5000 | 957 | 0.1914 | 1701 | 1013 |
| 5054 | 3100 | 1219 | 0.3932258 | 2071 | 1216 |
| 5059 | 1200 | 338 | 0.2816667 | 2470 | 1474 |
FACS, fluorescence-activated cell sorter; RNA-seq, RNA sequencing; UMI, unique molecular identifier.
Figure 17.
Human single-nucleus RNA-seq analysis showed minimal batch effects and yielded data from 48 definitive neurons. (A–C) t-SNE plots of human nuclei colored by (A) sample number, (B) colon location (right vs sigmoid), and (C) sex. Cells from different colon regions and different sexes largely form the same t-SNE clusters. (D–F) Feature plots show location of (D) ELAVL4, (E) UCHL1, and (F) SNAP25 expression suggest that a small population of neurons expressing all 3 is present in this dataset, but most of the 20,000 nuclei are not ELAVL4, SNAP25, or UCHL1-positive. Color key represents loge(normalized gene expression).
Figure 18.
Human myenteric plexus NOS1/VIP/GAL+ and NOS1/VIP/GAL– neurons differentially express many regulatory genes also differentially expressed in mouse ENS. (A) t-SNE plot of all human nuclei expressing ELAVL4, UCHL1, or SNAP25 reveals many populations that may be doublets because they cluster with nuclei expressing non-neuronal cell markers. For this paper, we only describe in detail expression data for the tight cluster of cells we believe are single neurons based on high expression of ELAVL4, SNAP25, UCHL1, SYT1, and DSCAM (highlighted with red circle). (B) Feature plots of neuronal markers (ELAVL4, SNAP25, UCHL1, SYT1, and DSCAM), SMC markers (ACTG2), glial cell markers (PLP1), ICC markers (KIT), and PDGFRA+ cell markers (PDGFRA) suggest that other populations are not neurons. Color key represents loge(normalized gene expression). (C) Heatmap shows 50 genes with the highest fold difference between neurons and other cells. Hierarchical clustering suggests 2 subgroups: NOS1+/VIP+/GAL+ (17 neurons) and NOS1–/VIP-/GAL– (31 neurons). (D) Heatmap shows transcription and splicing factors differentially expressed in mouse colon that were in >10% of human myenteric neurons. RBFOX1, ETV1, and BNC2 were differentially expressed between NOS1/VIP/GAL+ and NOS1/VIP/GAL– human neurons (Wilcoxon rank sum test, Bonferroni correction).
Figure 19.
Immunohistochemistry of human colon myenteric plexus shows preferential expression of select transcription factors in CHAT+ and NOS+ neuron subtypes. (A–T) Maximum intensity projections of adult human colonic ENS confocal Z-stacks: (A–D) BNC2, (E–H) PBX3 (white arrowheads indicate nuclear PBX3 in neurons), (I–L) RBFOX1, (M–P) TBX2, and (Q–T) TBX3 are present in subsets of human myenteric neurons. (U–Y) Immunohistochemistry quantification: (U) BNC2 (P = .0001, n = 3), (V) PBX3 (P = .0021, n = 3), (W) RBFOX1 (P = .0007, n = 3), and (X) TBX2 (P = .0089, n = 3) are primarily in CHAT+ neurons. (Y) TBX3 (P = .0004, n = 3) is primarily in NOS1+ neurons. Quantification of (Z) CHAT+ and (AA) NOS1+ reveals that BNC2 (P = .0102), PBX3 (P = .0021), RBFOX1 (P = .0016), and TBX2 (P = .0162) are largely restricted to CHAT+ neurons in adult human ENS (P values compare Transcription factor+EGFP+/Transcription factor+ vs EGFP+/Total neuron ratios). TBX3 (P = .0073) is primarily in NOS1+ neurons (P values compare Transcription factor+NOS1+/Transcription factor+ vs NOS1+/Total neuron ratios). (U–AA) Mean ± SD and Student’s t test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. Scale bar = 100 μm.
Discussion
Regenerative medicine and trophic factor modulation hold promise for treating life-threatening enteric neuropathies.2,3 To facilitate reprogramming and improve bowel function, better understanding of the molecular identity and characteristics of ENS subtypes is needed.2 We used single-cell transcriptomics to identify hundreds of neurotransmitters, receptors, ion channels, signaling molecules, and messenger RNA regulatory genes differentially expressed in 7 mouse myenteric neuron subclasses in adult distal colon and 8 neuron groups at E17.5. Immunohistochemistry validated RNA-seq, and functional studies identified new ENS roles for TBX3, GDNF, and NRTN. Our analyses complement recently published manuscripts,5, 6, 7, 8,33 providing a foundation for novel diagnostics and disease management.
Several transcription and splicing factors were differentially expressed in nitrergic vs cholinergic neurons, with similar patterns in mouse and human RNA-seq (ETV1, RBFOX1, BNC2) and immunohistochemistry (BNC2, PBX3, RBFOX1, TBX3, and TBX2) data. Some of these genes were previously reported in Memic et al,4 who used bulk RNA analysis via microarray and immunohistochemistry to study transcription factor expression during fetal development. However, unlike their study, which lacked the resolution of single-cell sequencing and had no quantitative assessment of transcription factor expression,4 we provide rigorous quantitative data by combining immunohistochemistry with 3-dimensional confocal imaging. For a subset of genes, we tested if loss altered Chat or Nos1 subtype abundance and discovered that Tbx3 loss reduced NOS1+ myenteric neuron density. Total neuron density was normal in Tbx3 mutant mice, confirming our prior analysis.27 In that prior study, we also found loss of Tbx3 reduced enteric glia but had no discernible effects on gastric and small intestine transit time at P0.27 Unfortunately, Tbx3 mutant mice die at P0, which precludes analysis of colonic or total gastrointestinal transit. Intriguingly, Tbx3 expression in limb bud is induced by the transcription factor HAND2,34 and Hand2 mutations cause complete loss of NOS1+/VIP+ enteric neurons,35 suggesting TBX3 may act downstream of HAND2 in the ENS.
In contrast, Chat-EGFP+ neuron density and total neuron density were normal in Wnt1-CreCre/wt;Casz1fl/fl, Wnt1-CreCre/wt;Rbfox1fl/fl, and Wnt1-CreCre/wt;Tbx2fl/fl mice. Casz1, Rbfox1, and Tbx2 were chosen for in vivo study because most adult ChAT+ myenteric neurons express Casz1 and Rbfox1, and because Tbx2 was expressed in many ChAT+ neurons at E17.5. Furthermore, RBFOX1 regulates CNS neuron subtype differentiation, and CASZ1 and TBX2 regulate subtype specification in many cells including photoreceptors.36, 37, 38, 39, 40 Normal colonic bead expulsion latency in Casz1 and Rbfox1 mutant mice suggests redundant ENS mechanisms prevent obvious anatomic or functional defects.
Another striking observation was that the transcription factor POU3F3 is expressed in colon ENS, but not small bowel ENS, as others recently confirmed.7,33 POU3F3 regulates cortical neuron migration and neurogenesis, but ENS function is unknown.41 Human mutations cause intellectual disability, speech delay, autism, epilepsy, craniofacial anomalies, and cryptorchidism,42 but effects of POU3F3 mutation on bowel function were not reported. Restricted colon expression is interesting because motility and ENS structure differ between colon and small bowel,22 and we know little about underlying mechanisms.
Human RNA-seq yielded data from 17 NOS1+ and 31 NOS1– myenteric neurons as well as from many other cells that impact bowel motility. Guided by RNA-seq, we found that ETV1 was more abundant in human nitrergic neurons, while RBFOX1 and BNC2 were enriched in non-nitrergic neurons, suggesting that subtype-specific genes may be conserved between humans and mice. Immunohistochemistry confirmed that TBX3, PBX3, and TBX2 are also present in subsets of human enteric neurons as predicted by mouse data.
Many other genes differentially expressed in enteric neuron subclasses may merit investigation. For example, ZEB2 mutations cause Mowat-Wilson syndrome, characterized by HSCR, intellectual disability, and distinctive facial features. Postsurgical outcomes in Mowat-Wilson syndrome are worse than for isolated HSCR, and ZEB2 mutations cause chronic constipation without HSCR,43,44 suggesting that Zeb2 might have roles beyond bowel colonization by ENS precursors. Zfhx3, another HSCR-associated gene,45 drives neuronal differentiation in other regions, but roles in ENS subtype specification are unexplored.
Integrating New and Old Data About ENS Subtypes
Using neurotransmitter-associated markers in adults (Table 4) and at E17.5 (Table 5), we correlated our RNA-seq data with known ENS subtype markers. Subtype designations at E17.5 include small bowel and colon neurons (distinguished by Pou3f3) and immature clusters defined by GO term modules (Figure 10A and B). The similarities between the E17.5 small bowel and adult colon suggest that some aspects of neuron differentiation are similar across bowel regions and completed by E17.5. Many newly identified differentially expressed genes are highlighted in Tables 4 and 5. Some genes known to be expressed in enteric neuron subtypes were not detected (see Table 4), but lack of expression is difficult to interpret because of limited read depth in adult colon nuclei.
Table 4.
Predicted Adult Enteric Neuron Subtype Classification Based on RNA-Seq
| Cluster name (adult colon) | Chat 1 | Chat 2 | Chat 3 (Met) | Chat 4 (Vglut2) | Calcb | Nos 1 | Nos 2 |
|---|---|---|---|---|---|---|---|
| Suggested neuron type | Intrinsic sensory neurons, interneurons (with mechanosensitive properties) | Interneurons or excitatory motor neurons | Interneurons or excitatory motor neurons | Interneuron | Intrinsic Sensory Neuron | Inhibitory motor neurons ± interneurons? | Inhibitory motor neurons ± interneurons? |
| Expressed markers supporting conclusion | Chat,aSlc18a3, Tac1, (Piezo1)a | Chat, Tac1,aPenka | Chat, Met,aPenk,aTac1a | Slc18a3,aChat, Nos1, Vip, Calb1,aPenk,aNefma | Calcb,aNefm, Scn11a,aCalb2,aTacr1,aHtr3a,aHtr3b,aP2rx2a | Nos1,aVip,aGal,aNpya | Nos1,aVip, Gal, Npy |
| Other genes of note differentially expressed | Calb2a | Slc17a6a | Nmu,aGrp,aAvila | Htr3a,aP2rx2a | |||
| Corresponding neuron subtype designations from Drokhlyansky et al8(Adult Mouse Colon) | PEMN 1, 3, 4, 6 (based on Chat, Tac1, Piezo1) | PEMN 2, or PIN1, PIN2 (based on Chat, Penk, Tac1, and absent Piezo1) | PIN1, PIN2 or PEMN 2 (based on Chat, Penk, Tac1, Calcrl, Ramp1) | PIN3 or PSN3 (based on Chat, Slc17a6, Nxph2) | PSN_1 (based on Nmu, Grp, Calcb) | PIMN_1-7 (based on Nos1, Vip, Gal) | PIMN_1-7 (based on Nos1, Vip, Gal) |
| Corresponding neuron subtype designations in Morarach et al5(P21 mouse small intestine)—very limited data available | ENC1,7 (Chat, Tac1, absent Penk) | ENC2-4 (Chat, Tac1, Penk) | ENC4 (Chat, Tac1, Penk, Fut9, Nfatc1), | ENC12 > ENC7 (Slc17a6, Penk, Nxph2, Nefm) | ENC6 > ENC5 (Calcb, Nmu, Nog, Sst) | ENC8-10 (Nos, Vip, Gal) | ENC8-10 (Nos, Vip, Gal) |
| Corresponding neuron subtype designations from Zeisel et al6(P21 mouse small intestine):http://mousebrain.org/genesearch.html? | ENT5 (Chat, Tac1, Dmkn, Hoxb5, absent Penk) | No equivalent cluster in this small intestine dataset. Penk is only expressed in ENT6 and ENT7, but neither of these clusters are a good match due to Slc17a6 expression and lack of Fut9 and Nfatc1 expression in this cluster. |
No equivalent cluster in this small intestine dataset. ENT6 is closest (Chat, Tac1, Penk, Fut9, Nfatc1, but ENT6 is missing Met expression) |
ENT7 (based on Chat, Penk, Slc17a6, Calb1, Nxph2, Phox2a) | ENT9 (Nmu, Myl1, P2rx2) | ENT2 > ENT1, EN3 (Cartpt, Nos1, Ass1, Bglap) | ENT1,3 > ENT2 (Nos1, Ass1, Cox8b, Gal, Moxd1) |
| Markers missing | Kcnn4, Piezo1 | ||||||
| Citations | 19,20,48,63 | 19,20 | 49 | 5,47,64 | 47 | 10,18 | 10,18 |
RNA-seq, RNA sequencing.
Differential expression.
Table 5.
Predicted E17.5 Enteric Neuron Subtype Classification Based on RNA-Seq
| Cluster name (E17.5 Whole Bowel) | Immature chat cluster 1 | Chat cluster 1 | Chat cluster 2 | Nos1 cluster 1 | Nos1 cluster 2 | Nos1 cluster 3 | Immature chat cluster 2 | Immature nos1 cluster |
|---|---|---|---|---|---|---|---|---|
| Suggested neuron type | Small intestine interneurons | Small intestine excitatory motor neurons | Small intestine intrinsic sensory neurons | Small intestine inhibitory motor neurons or interneurons | Small intestine interneurons | Small intestine inhibitory motor neurons or interneurons | Colon (± some small intestine) cholinergic neurons | Colon (± some small intestine) nitrergic neurons |
| Differentially expressed markers supporting conclusion | Slc18a3, Chat, Penk, Calb1 | Slc18a3, Chat, Penk, Tac1 | Calca, Calcb, Htr3b, Nmu | Nos1, Vip, Gal | Nos1, Vip, Gal, Neurod6 | Nos1, Vip, Gal | Slc18a3 | Nos1, Pou3f3 |
| Other genes of note differentially expressed | Slc17a6, Npy1r, Avpr1a | Tgfb2, Htr2b | Nmu, Nog, Bambi, Smad7, Adra2a, Ntrk3, Npy2r | Ednrb, C1ql1, P2ry6, Htr3a, Cartpt, Npy2r | Dbh, Sstr1, Sstr2, Cartpt, Htr3a, Npy2r | Sstr1, Sstr2, Cartpt, Prokr1, Npy2r | Htr2b, Npy1r, Ramp1 | Npy1r, Ramp3 |
| Markers missing | ± Npy | ± Npy | ± Npy | |||||
| Corresponding P21 small intestine neuron subtype designations from Morarach et al5 | ENC12 (based on Chat, Slc18a3, Slc17a6, Calb1, Penk) | ENC2 > ENC3 (based on Chat, Slc18a3, Penk, Tac1, Gda, Ndufa4l2) | ENC6 (based on Calcb, Nmu) | ENC8 or ENC9 (based on Nos1, Vip, Gal, C1ql1) | ENC10 (based on Nos1, Vip, Gal, Neurod6) | ENC8 or ENC9 (based on Nos1, Vip, Gal) | N/A | N/A |
| Corresponding P21 small intestine neuron subtype designations from Zeisel et al6 | ENT7 (based on Chat, Slc18a3, Slc17a6, Calb1, Penk) | ENT6 > ENT6 and ENT4 (based on Chat, Slc18a3, Penk, Tac1, Gda, Ndufa4l2) | ENT9 (based on Calcb, Nmu, Nog) | ENT3 > ENT2 (based on Nos1, Vip, Gal, Cartpt) | ENT1 (based on Nos1, Vip, Gal, Cartpt, Neurod6) | ENT2 > . ENT3 (based on Nos1, Vip, Gal, Cartpt, Ltk) | N/A | N/A |
| Citations | 5,12,22,47,64 |
12,18,22 Our own data |
5,18,47 | 12,63,65 | 5,65 | 12,64,65 | Our own data | Our own data |
N/A, not available.
In adults, Calcb neurons appear to be AH/Dogiel type II intrinsic sensory neurons that express neurofilament M (Nefm), Nav1.9 (Scn11.1a), calretinin (Calb2), NK1 tachykinin receptor (Tacr1), serotonin receptors Htr3A and Htr3b, and purine receptor P2rx2.18,46,47 Chat1 neurons express the mechanosensitive channel Piezo1, suggesting that these cells are stretch sensitive intrinsic sensory neurons, but other sensory neuron markers are absent. There are reports of mechanosensitive interneurons and motor neurons,48 so Chat1 neurons may correspond to these. Tac1 and Penk coexpression make Chat2 and Chat3 clusters excitatory motor neurons or interneurons (Figure 1I).18 Unexpectedly, however, Chat3 neurons also express hepatocyte growth factor receptor Met and do not express calcitonin gene-related peptide (Calcb). This is an interesting finding, which suggests that Met+ colonic neurons differ from small bowel, where MET and calcitonin gene-related peptide were found in the same neurons by immunohistochemistry and single-cell sequencing.49,50 Similar to small bowel, however, Met and Ret appear to be mutually exclusive in colon, with all groups except Chat3 expressing Ret. Finally, Chat4 cells appear to be interneurons based on coexpression of Slc18a3, Chat, Nos1, Vip, and Calb1. Many findings fit well with known neuron subtypes, and immunohistochemistry correlates well with RNA-seq, providing high confidence in our results. We point out unexpected findings to suggest that there is more to learn and that future studies based on our RNA-seq are needed to fully define adult enteric neuron subtypes.
Defining neuron subtypes at E17.5 presents an added challenge because differentiation is incomplete. Table 5 highlights some markers differentially expressed in neuron subtypes. Because almost all cholinergic and nitrergic neurons at E17.5 remain cholinergic or nitrergic, respectively, in adults,30,31 we extrapolate from adult data. Assessment of maturity is based on GO term analysis. Colon neurons were identified by Pou3f3 expression. Chat cluster 1 and Chat cluster 2 markers suggest small bowel excitatory motor neurons and intrinsic sensory neurons, respectively. Chat cluster 2 also expresses bone morphogenetic protein (BMP) and transforming growth factor β signaling antagonists (Nog, Bambi, Smad7), which may provide new insights into ENS development. Immature Chat cluster 1 appears to be developing myenteric interneurons that express Slc17a6, similar to the adult Chat4 cluster. Nos1 clusters 1 and 3 are most likely inhibitory motor neurons but could also be interneurons. Nos1 cluster 2 represents possible inhibitory interneurons based on the expression of the transcription factor Neurod6.5 Interestingly, Nos1 cluster 1 differentially expresses Ednrb, but the role of Ednrb/EDN3 signaling at this time is not known. We highlight these observations to emphasize how mining our data might provide new insight into development.
GDNF and NRTN Acutely Affect Calcium Signaling in Distinct Adult Enteric Neuron Populations
GDNF/GFRA1/RET signaling is essential for ENS precursor survival, proliferation, and differentiation during gestation and early ENS maturation.28 In adult ENS, GDNF increases the strength of bowel contraction in the ascending limb of the peristaltic response.25 Here, we confirm and extend prior observations by showing GDNF alters calcium signaling in some colon neurons and acutely enhances basal and EFS-evoked motor patterns. In contrast, although NRTN/GFRA2/RET are known to influence bowel motility and support acetylcholinesterase+ and SP+ neurites,23,51 acute effects of NRTN on the ENS or bowel motility had not been demonstrated. We found that Gfra1 and Gfra2, the preferred GDNF and NRTN co-receptors, respectively, are expressed in largely nonoverlapping Ret+ E17.5 and adult myenteric neuron groups. GFRA1 is in ∼90% of NOS1+ myenteric neurons, but very few NOS1– neurons, so we anticipated GDNF would affect primarily NOS1+ neurons. However, 44% (n = 33 of 75) of GDNF-influenced neurons were non-nitrergic. This might occur because GDNF activates RET via GFRA2 (NRTN’s preferred co-receptor) at high (pharmacologic) concentrations,52, 53, 54 but this seems unlikely because GCaMP6s signaling was altered by both GDNF and NRTN in only 8% of myenteric neurons. Instead, the observation that some neurons had increased and others decreased GCaMP activity in response to each trophic factor suggests that both direct and indirect effects were measured and that responses depend on synaptic inputs, as confirmed by TTX data. The hypothesis that GDNF and NRTN sensitize responsive neurons to incoming stimuli is also supported by our prior studies of dorsal root ganglia, where these factors acutely potentiate TRPV1-induced calcium signals and induce hyperalgesia when injected into hindpaw.55 Although we do not know the source of calcium (eg, intra- or extracellular) or whether evoked GCaMP signals were activity dependent, our data collectively show that NRTN and GDNF acutely influence nonoverlapping sets of myenteric neurons and differentially regulate bowel motility. These new data may in part explain our prior studies showing that Nrtn–/–, Gfra1±, and Ret± mice have normal numbers of enteric neurons, but a >90% reduction in colon contractility and a 50%–90% (depending on mouse line) reduction in VIP and SP release in response to electric field stimulation.56
Summary
Enteric neuropathies remain challenging to treat, and we are hampered by inadequate data about enteric neuron subtypes. Here, we used single-cell RNA-seq to identify many subtype–specific regulatory molecules, and demonstrated new roles for GDNF, NRTN, and Tbx3.
Materials and Methods
All authors had access to the study data and reviewed and approved the final manuscript.
Study Approval
Studies adhere to ARRIVE (Animals in Research: Reporting In Vivo Experiments) guidelines.57 Mouse experiments were performed in accordance with Institutional Animal Care and Use Committee (IACUC) approval from Children’s Hospital of Philadelphia (IACUC#19-001041) and University of Pittsburgh (IACUC#IS00013297) or University of Melbourne Animal Experimentation Ethics Committee (project number 1714308.2). Human colon was acquired with institutional review board (IRB) approval from Perelman School of Medicine at University of Pennsylvania (IRB#804376) and from Children’s Hospital of Philadelphia (IRB#13-010357).
Animals
B6.FVB(Cg)-Tg(Chat-EGFP/Rpl10a,Slc18a3)DW167Htz/J mice, referred to as ChAT-EGFP-L10a (RRID:IMSR_JAX:030250; C57BL/6J), were a kind gift from Dr Joseph Dougherty at Washington University School of Medicine in St Louis. Tbx3tm3.1Moon mice (referred to as Tbx3; RRID:MGI:5538564; described previously; mixed background)22 were a kind gift from Dr. Anne Moon at the Geisinger Commonwealth School of Medicine. B6.129S2-Rbfox1tm1.1Dblk/J mice (referred to as Rbfox1; RRID:IMSR_JAX:014089; C57BL6/J) were a kind gift from Dr. Douglas Black at the University of California, Los Angeles. Gfra1tm2.1Jmi mice (referred to as Gfra1-Gfp; RRID:MGI:3715157, C57BL/6) were a kind gift of Dr. Sanjay Jain at Washington University School of Medicine in St Louis. C57BL/6J mice (RRID:IMSR_JAX:000664), H2az2(Tg(Wnt1cre)11Rth mice (referred to as Wnt1-Cre; RRID:IMSR_JAX:003829), B6;129S6-Gt(ROSA)26Sortm1(CAG-tdTomato∗,-EGFP∗)Ees/J mice (referred to as ROSAnT-nG; RRID:IMSR_JAX:023035), B6;129S-Gt(ROSA)26Sortm1.1Ksvo/J mice (referred to as R26R-H2b-mCherry; RRID:IMSR_JAX:023139), B6(Cg)-Etv1tm1.1(cre/ERT2)Zjh/J mice (referred to as Etv1-CreERT2 mice; RRID:IMSR_JAX:013048), Slc17a6tm2(cre)Lowl/J mice (referred to as Vglut2-IRES-Cre mice; RRID:IMSR_JAX:016963), B6;129S-Nos1tm1.1(cre/ERT2)Zjh/J mice (referred to as Nos1-CreERT2; RRID:IMSR_JAX:014541), Gt(ROSA)26Sortm9(CAGtdTomato)Hze mice (referred to as R26R-TdTomato; RRID:IMSR_JAX:007909), B6.FVB-Tg(EIIa-cre)C5379Lmgd/J (referred to as E2a-Cre RRID:IMSR_JAX:003724; C57BL/6), and B6J.Cg-Gt(ROSA)26Sortm96(CAG-GCaMP6s)Hze/MwarJ (referred to R26R-Gcamp6s; RRID:IMSR_JAX:028866; C57BL/6) were obtained from the Jackson Laboratory (Bar Harbor, ME). Wnt1-Cre mice on a mixed C57BL/6J × CBA/J)F1 background were bred into R26R-H2b-mCherry mice on a mixed 129S4/SvJaeSor × C57BL/6J background and maintained on a mixed C57BL/6J × 129S4/SvJaeSor × CBA/J)F1 background. Etv1-CreERT2 mice were bred to R26R-TdTomato mice and ChAT-EGFP-L10a mice to generate Etv1-CreERT2;R26R-TdTomato mice on a C57BL/6J background. Vglut2-IRES-Cre mice on a C57BL/6;FVB;129S6 background were bred to R26R-TdTomato on a C57BL/6J background and maintained on a mixed C57BL/6;FVB;129S6 background. Nos1-CreERT2 mice were crossed with R26R-TdTomato mice and ChAT-EGFP-L10a mice to generate ChAT-EGFP-L10a; Nos1-CreERT2; R26R-TdTomato mice and maintained on a C57BL/6 background. Rbfox1;Wnt1-Cre;R26R-TdTomato;ChAT-EGFP-L10a mice were generated by crossing Rbfox1 mice to Wnt1-Cre mice, ChAT-EGFP-L10a mice, and R26R-TdTomato mice and were maintained on a mixed background. Wnt1-Cre;Tbx3 mice were generated by crossing Wnt1-Cre mice to Tbx3 mice and were maintained on a mixed background. E2a-Cre mice were crossed with R26R-Gcamp6s mice to generate mice that express GCaMP6s (a genetically encoded calcium indicator) in all cells. E2a-Cre;R26R-GCaMP6s mice were maintained on a pure C57BL/6 background. Genotyping was performed using previously published and novel primers (Table 6) and by Transnetyx (Cordova, TN). Vaginal plug day was considered day E0.5. Experimental animals of the correct strain, genotype, and age were randomly selected from the total pool of mice available Animal husbandry information can be found in Table 7.
Table 6.
List of PCR Primers for Mouse Genotyping
| Gene (strain) | Primer sequence | Band size | Genotyping solution | Reference |
|---|---|---|---|---|
| Tbx3 | F: 5′-GTG TGA GAC AGA GAA ATC AGT GG-3′ R: 5′-CCA ACT GGT ATC TTG ATA AAC CTC-3′ |
Mut: 480 bp WT: 320 bp |
KAPA (KAPA Biosystems; Cat. #KK7352), Taq (NEB; Cat. #M0271L) | 27 |
| Gfp (ChAT-EGFP-L10a) | F: 5′-TCA TAG AGG CGC AGA GTT CC-3′ R: 5′-CTG AAC TTG TGG CCG TTT AC-3′ |
Mut: 250 bp | KAPA (KAPA Biosystems; Cat. #KK7352) | JAX genotyping protocol (Stock No: 030250) |
| Rbfox1 | F: 5′-ATGCCCATGCAGTGAAAAAT-3′ R: 5′-TGCAGCACATTGAAACCTTC-3′ |
Mut: 397 bp WT: 294 bp |
KAPA (KAPA Biosystems; Cat. #KK7352) | 69 |
| Gfra1-Gfp | Common F: 5′-CTTCCAGGTTGGGTCGGAACTGAACCC-3′ Mut R: 5′-GCCGTTTACGTCGCCGTCCAGCTCGACCAG-3′ WT R: 5′-AGAGAGCTCAGCGTGCAGAGATC-3′ |
WT: ∼200 bp Mut: ∼300 bp |
KAPA (KAPA Biosystems; Cat. #KK7352) | 24 and personal correspondence with Dr Sanjay Jain’s laboratory |
| Cre (Wnt1-Cre) | F: 5′-GCA TTA CCG GTC GAT GCA ACG AGT GAT GAG-3′ R: 5′-GAG TGA ACG AAC CTG GTC GAA ATC AGT GCG-3′ |
408 bp | KAPA (KAPA Biosystems; Cat. #KK7352), Taq (NEB) | https://mgc.wustl.edu/protocols/pcr_genotyping_primer_pairs |
| R26R-mCherry (R26R-H2b-mCherry) | Common F: 5′-AAA GTC GCT CTG AGT TGT TAT-3′ Mutant R: 5′-TTA TGT AAC GCG GAA CTC CA-3′ WT R: 5′GGA GCG GGA GAA ATG GAT ATG-3′ |
Mut: 309 bp WT: 603 bp |
KAPA (KAPA Biosystems; Cat. #KK7352) | JAX genotyping protocol (Stock No: 023139) |
| Cre (Vglut2-IRES-Cre) | F: 5′-GCA TTA CCG GTC GAT GCA ACG AGT GAT GAG-3′ R: 5′-GAG TGA ACG AAC CTG GTC GAA ATC AGT GCG-3′ |
408 bp | KAPA (KAPA Biosystems; Cat. #KK7352), Taq (NEB; Cat. #M0271L) | https://mgc.wustl.edu/protocols/pcr_genotyping_primer_pairs |
| Cre (Nos1-CreERT2) | F: 5′-GCA TTA CCG GTC GAT GCA ACG AGT GAT GAG-3′ R: 5′-GAG TGA ACG AAC CTG GTC GAA ATC AGT GCG-3′ |
408 bp | KAPA (KAPA Biosystems; Cat. #KK7352), Taq (NEB; Cat. #M0271L) | https://mgc.wustl.edu/protocols/pcr_genotyping_primer_pairs |
| R26R-TdTomato | Common F: 5′-AAAGTCGCTCTGAGTTGTTAT-3′ Mut R: 5′-GCGAAGAGTTTGTCCTCAACC-3′ WT R: 5′-GGAGCGGGAGAAATGGATATG-3′ |
Mut: ∼350 bp WT: ∼600 bp |
KAPA (KAPA Biosystems; Cat. #KK7352), Taq (NEB; Cat. #M0271L) | JAX genotyping protocol (Stock No: 007900) |
| Etv1-CreERT2 | WT F: 5′-CCC TCC CCT CTC ATT TTC TC-3′ Mut F: 5′-TGG TTT GTC CAA ACT CAT. CAA-3′ Common R: 5′-ACA GTT TCT CCC ACG CTG AC-3′ |
Mut: 170 bp WT: 459 bp |
KAPA ((KAPA Biosystems; Cat. #KK7352) | JAX genotyping protocol (Stock No: 013048) |
| Cre (E2a-Cre) | F: 5′-GCG GTC TGG CAG TAA AAA CTA TC-3′ R: 5′-GTG AAA CAG CAT. TGC TGT CAC TT-3′ |
100 bp | HotStar Taq Kit (Qiagen; Cat. #203443) | JAX genotyping protocol (Stock No: 003724) |
| R26R-GCaMP6s | Mut F: 5′-ACG AGT CGG ATC TCC CTT TG-3′ Mut R: 5′-AGA CTG CCT TGG GAA AAG CG-3′ WT F: 5′-AAG GGA GCT GCA GTG GAG TA-3′ WT R: 5′-CCG AAA ATC TGT GGG AAG TC-3′ |
Mut: 356 bp WT: 297 bp |
KAPA (KAPA Biosystems; Cat. #KK7352) | JAX genotyping protocol (Stock No: 028866) |
Mut, mutant; PCR, polymerase chain reaction; WT, wild-type.
Table 7.
ARRIVE Guidelines: General Animal Husbandry Information
| Location of animal facility (types of experiments) | Children’s Hospital of Philadelphia (all experiments, except listed elsewhere) | University of Pittsburgh (live calcium imaging experiments) | University of Melbourne (High-magnification images of myenteric plexus stained with substance P and Enkephalin) |
|---|---|---|---|
| Facility type | Conventional: cages are opened at room air, face masks are not required when handling mice | Barrier housing and maintained with caging opened under Animal Transfer Station/Biosafety Cabinets | IVC (Tecniplast): cages opened under Animal Transfer Station |
| Facility type | Specific pathogen free, pathogens detected in room within the past 2 y: MNV, Helicobacter not tested but likely present | Specific pathogen free; sentinel testing for excluded pathogens and MNV | Specific pathogen free; sentinel testing for excluded pathogens and MNV |
| Animal welfare assessment | Daily | Daily | Daily (food and water) Weekly (health check) |
| Bedding | ¼ inch corn cob (The Andersons, Product 4B) | P.J. Murphy Coarse Certified Aspen Sani-chip | Compressed paper-based bedding |
| Cage type | Lab Products (Seaford, DE) 75 inches2 ventilated. | Allentown mouse IVC | Tecniplast IVC (green line) |
| Cage cleaning/sterilization | Standard tunnel washer | Standard tunnel washer, boxes autoclaved before use | Rack washer (Tecniplast), steam sterilisation |
| Mouse diet | Mouse Diet 5015 (Lab Diet), direct from manufacturer: not autoclaved, not irradiated | Prolab Isopro RMH 3000 (5P75) irradiated | Jackson labs: LabDiet 5K52 formulation (6% fat). Animal Facility: Barastoc WEHI Mice Cubes, irradiated |
| Light/dark cycle, h | 12/12 | 12/12 | 12/12 |
| Temperature | 72 ± 2°F | 72 ± 4°F | 18–24 ± 1°C |
| Humidity | 30%–70% depending on the day/season | 30%–70% depending on the day/season | 30%–70% depending on the day/season |
| Water pH and quality | Reverse osmosis, pH ∼7, Edstrom automatic watering system | Reverse osmosis, pH ∼7, Edstrom automatic watering system | Filtered water bottles (down to 0.2 microns), pH ∼7 |
| Access to food and water | Continuous | Continuous | Continuous |
| Number of mice per cage | 1–5 (20–30 g) | 1–4 males, 1–5 females | 1–5 |
| Cage enrichment | house/dome (Bio-Serv, S3174) and nestlet (Ancare) | house/dome (Bio-Serv, S3174) and nestlet (Ancare) | House/dome, tissues, chewing wooden block for single-housed mice |
| Mating strategy | Continuous | Continuous | Not applicable in this study |
| Age at weaning | 19–20 d | 19–28 d | 19–21 d (not applicable for this study) |
IVC, individually ventilated cage; MNV, mouse norovirus.
Human Colon
Colon tissue was acquired using the Abramson Cancer Center Tumor Bank (Philadelphia, PA). Colon was de-identified, providing limited clinical data. Resected colons were maintained at ambient temperature until arrival in pathology, where they were transferred to ice-cold Dulbecco-modified phosphate-buffered saline (DPBS) 1–4 hours after resection. Pathologists selected regions of colon without gross abnormalities, and colon specimens were transferred to the laboratory in DPBS on ice.
Tamoxifen Treatment
Tamoxifen (10 mg/mL or 20 mg/mL; Sigma-Aldrich, St Louis, MO; Cat. #T5648) was prepared by first adding 200 μL ethanol and then 1800 μL sunflower oil (Sigma-Aldrich; Cat. #S5007). Tamoxifen was dissolved by alternately vortexing and incubating in a 37°C water bath. Nos1-creERT2;Chat-EGFP-L10A;R26-TdTomato E15.5 dams were injected once with 200-mg/kg tamoxifen at 10-mg/mL concentration. Etv1-CreERT2;R26-TdTomato E15.5 dams were gavaged once with tamoxifen at 200-mg/kg concentration at 20-mg/mL concentration, and pups were dissected at E17.5. Adult Etv1-CreERT2;R26-TdTomato adult mice were gavaged twice (2 days apart) with 233-mg/kg tamoxifen at 20-mg/mL concentration 3 weeks prior to dissection.
Preparing Young Adult Mouse Colon for Nuclei Isolation
Wnt1-CreCre/wt;R26R-H2b-mCherry+ 47–52-day-old mice (8 females, 1 male) were euthanized with CO2. Colon was removed, flushed with cold, sterile DPBS using a 20-mL syringe to remove luminal contents, and placed in fresh cold, sterile DPBS. Colon was carefully cut along the mesentery, pinned mucosal side down on dishes treated with Sylgard Elastomer 184 Silicone Encapsulant Clear (Dow Corning, Midland, MI). Muscularis was carefully peeled from the mucosa and placed in cold OCT compound (Fisher Healthcare Tissue-Plus OCT Compound; Thermo-Fisher Scientific, Hampton, NH; Cat. #23-730-571) in a biopsy specimen cryomold (VWR, Radnor, PA; Cat. #4565), frozen in methylbutane on dry ice, and stored at –80°C.
Preparing Adult Human Colon for Nuclei Isolation
Using insect pins (Fine Science Tools, Foster City, CA; Cat. #26002-20), colons were maximally stretched and pinned serosa side up on 30-mm dishes treated with Sylgard Elastomer 184. Tissue was incubated in 9 parts carboxygenated (95% oxygen, 5% CO2) Krebs-Ringer solution (118-mM NaCl [Sigma-Aldrich; Cat. #S6191], 4.6-mM KCl [Thermo-Fisher Scientific; Cat. #BP366-500], 2.5-mM CaCl2 [Cat. #C7902], 1.2-mM MgSO4 [Sigma-Aldrich; Cat. #M-7506], 1-mM NaH2PO4 [Sigma-Aldrich; Cat. #S0751], 11-mM D-(+)-Glucose [Sigma-Aldrich; Cat. #G-7021], 25-mM NaHCO3 [Thermo-Fisher Scientific; Cat. #BP328-500], pH 7.4) and 1 part 4-Dia-2-Asp (4-(4-(dimethylamino)styryl)-N-methylpyridinium iodide) (Abcam, Cambridge, United Kingdom; Cat. #ab145266) at room temperature in sterile DPBS. After 10 minutes, the tissue was transferred to ice-cold carboxygenated Krebs-Ringers solution under a SteREO Discovery.V20 fluorescence dissecting scope (ZEISS, Oberkochen, Germany; 488-nm filter). Fluorescent ENS was carefully dissected from muscularis by peeling away longitudinal muscle strips with Dumont #5 forceps (Fine Science Tools; Cat. #11251-30), placed in cold OCT compound (Fisher Healthcare Tissue-Plus OCT Compound; Cat. #23-730-571) in a biopsy specimen cryomold (VWR; Cat. #4565), frozen in methylbutane (Thermo-Fisher Scientific; Cat. #O3551-4) on dry ice, and stored at –80°C.
RNA Extraction for RNA Integrity Number Assessment Prior to Sequencing
One to 3 days before sequencing, frozen mouse or human tissues were sectioned on a Microm HM 505 E Cryostat (GMI, Ramsey, MN; 100-μM sections, –20°C). RNA was extracted from several (n = 1–5) 100-μM sections using the Qiagen RNeasy Plus Micro kit (Qiagen, Hilden, Germany; Cat. #74034) and using Qiagen's RNase Free DNase Set (Qiagen; Cat. #79254). Samples were run on an 2100 Agilent Bioanalyzer using an RNA 6000 Pico Kit (Agilent, Santa Clara, CA; Cat. #5067-1513), and tissue was used if RNA integrity number (RIN) was >7.0.
Nuclei Isolation and Sorting
To isolate nuclei, frozen colon sections (100 μm) in ice-cold lysis buffer (10-mM Tris HCl pH 7.5 (Cat. #15567-027), 10-mM NaCl (Sigma-Aldrich; Cat. #S6191), 3-mM MgCl2 (Cat. #BP214-500), and 0.005% Nonidet P40 Substitute (Thermo-Fisher Scientific; Cat. #AM9010; Sigma-Aldrich; Cat. #74385) were chopped rapidly with iridectomy scissors (1 minute), transferred to a precooled Dounce homogenizer (VWR; Cat. #357538) on ice, homogenized 15 strokes with the loose pestle and 40 strokes with the tight pestle, filtered through MACS SmartStrainer (Miltenyi Biotec, Bergisch Gladbach, Germany; Cat. #130-098-458), centrifuged (590 g, 8 minutes, 4°C), and resuspended in staining buffer (1× PBS, 1% w/v Ultrapure bovine serum albumin [BSA] [Life Technologies, Carlsbad, CA; Cat. #AM2618], 0.2U/mL Protector RNase inhibitor, [Sigma-Aldrich; Cat. #3335399001]); 2.5μg/mL Hoechst 33342 Trihydrochloride Trihydrate (Thermo-Fisher Scientific; Cat. #H3570) was added to a final concentration of 2.5 μg/mL before filtering (FlowMi strainer; VWR; Cat. #H13680-0040) and flow sorting (MoFlo Astrios; Beckman Coulter, Indianapolis, IN) into 5-μL staining buffer using a 70-μm nozzle to isolate Hoechst+/mCherry+ nuclei (mouse) or Hoechst+ nuclei (human).
Whole Cell Isolation From E17.5 Mice
An E17.5 Nos1-CreERT2;ChAT-L10A-EGFP;R26R-TdTomato dams, tamoxifen treated (200 mg/kg) at E15.5 was euthanized, and pups were rapidly dissected on ice (2 TdTomato+, 3 GFP+). For RNA-seq, full-length small intestines and colon dissociated together were used for RNA-seq. For RT-PCR, TdTomato+ E14.5 and E17.5 small intestine and colon from Wnt1-Cre+;R26-TdTomato+ mice were analyzed separately. Tissue in carboxygenated 1× Hank’s balanced salt solution (HBSS; Thermo-Fisher Scientific; Cat. #14025092) was cut into small pieces with insulin needles, and dissociated with Liberase (Sigma-Aldrich; Cat. #5401135001) plus DNase I (0.02 U/μL; Roche, Basel, Switzerland; Cat. #04716728001), MgCl2 (6 mM; Cat. #BP214-500), and CaCl2 (1 mM; Cat. #C7902) in HBSS (37°C, 40 minutes) with repeated trituration (P1000). Dissociated cells were passed 2–3 times through Falcon 35-μm filters (Corning, Corning, NY; Cat. #352235), pelleted (170 g, 3 minutes, 4°C) in 10% fetal bovine serum (FBS)/Iscove's Dulbecco’s modified Eagle medium (DMEM) (Corning; Cat. #10-016-CM), resuspended in fluorescence-activated cell sorter (FACS) buffer (0.04% w/v BSA [Thermo-Fisher Scientific; Cat. #AM2618] in HBSS [Thermo-Fisher Scientific; Cat. #14025092]), filtered again, and sorted (BD FACSJazz, 100μm nozzle) into 300-μL FACS buffer for sequencing. For RT-PCR, tdTomato+ cells were collected in 1-mL Iscove's DMEM/10% FBS, centrifuged (600 g, 5 minutes, 4°C) and resuspended in Buffer RLT plus (Qiagen; Cat. #74034).
Reverse-Transcription Polymerase Chain Reaction
RNA prepared using RNeasy Plus Micro Kit (Qiagen, Hilden, Germany; Cat. #74034 and Cat. #79254) or PicoPure RNA Isolation Kit (Thermo-Fisher Scientific, Waltham, MA; Cat. #KIT0204) with RIN >7.0 was reverse-transcribed using Superscript II RNase H (Thermo-Fisher Scientific; Cat. #18064022). RT-PCR used KAPA mixture (KAPA Biosystems, Wilmington, MA; Cat. #KK7352; Brn1) or GoTaq Green (Promega, Madison, WI; Cat. #M7122; Casz1) and previously described primers (Table 8).
Table 8.
List of RT-PCR Primers
| Gene | Primer sequence | Band size | Genotyping solution | Reference |
|---|---|---|---|---|
| Pou3f3 (mouse) | 5′-CAACAGCCACGACCCTCACT-3′ 5′-CAGAACCAGACCCGCACGAC-3′ |
450 bp | KAPA (KAPA Biosystems; Cat. #KK7352) | 66 |
| Actb (mouse) | 5′-GAGAGGGAAATCGTGCGTGAC-3′ 5′-AGCTCAGTAACAGTCCGCCTA-3′ |
534 bp | KAPA (KAPA Biosystems; Cat. #KK7352) or GoTaq Green (Promega; Cat. #M7122) | 67 |
| Casz1 (mouse) | 5′-GCAGAAGAGCCCTCAAAAGATAA-3′ 5′-GAAGCAGCGTAGTCCCTCAGA-3′ |
113 bp | GoTaq Green (Promega; Cat. #M7122) | 68 |
RT-PCR, reverse-transcription polymerase chain reaction.
Library Generation, Sequencing, and Data Processing
Libraries prepared with Chromium Single Cell 3′ Reagent Kits v2 (10x Genomics; Cat. #120237) were sequenced on an Illumina HiSeq 2500 (Illumina, San Diego, CA). Cell Ranger pipeline (10x Genomics) was used to convert BCL files into FASTQ files, perform STAR alignment,58 filter, count UMIs, and generate gene-barcode matrices. Cell Ranger Aggr pipeline (10x Genomics; v. 3.0.0) was used to aggregate multiple samples, normalize outputs, and recompute gene-barcode matrices on combined data.
Data Availability
Raw and processed single cell and single nucleus RNA sequencing data are available at GEO Accession: GSE156905.
Analysis of Murine Single-Nucleus and Single-Cell Sequencing Data
Using Seurat,17,59 gene-barcode matrices were imported into R, version 3.5.1 (R Foundation for Statistical Computing, Vienna, Austria), filtered to remove low expressors or doublets (nGenes = 200–5000 for adult distal colon; nGenes = 1000–9000 for E17.5 whole bowel) and mitochondrial contaminants (percent mitochondria <10% for adult distal colon, percent mitochondria <5% for E17.5 whole bowel), normalized, and scaled to regress out UMIs and percent mitochondria. Cells were clustered using the most statistically significant principal components. To cluster the E17.5 mouse data, we excluded genes located on the X and Y chromosomes from the principal components and included principal components up to the point where either any additional principal component contributed <5% of SD and the principal components cumulatively contributed to 90% of the SD or when the variation changes by <0.1% between consecutive principal components.60 This turned out to be 15 principal components. For adult mouse distal colon data, we found that taking all of the recommended statistically significant principal components resulted in difficult-to-interpret clustering, possibly owing to batch effects in our dataset, and that the first ∼11 components were sufficient to cluster our data in a biologically meaningful way. When reclustering was performed on subsets of data (ie, to filter out contaminants), subsetted data were rescaled and reclustered using the same method described previously, minus the normalization step. To identify genes that were differentially expressed between clusters, clusters were compared 1 at a time to all other cells in the dataset using a Wilcoxon rank sum test with Bonferroni multiple testing correction (Seurat's FindAllMarkers command). A cutoff of at least 10% of cells in a group expressing a gene of interest was required for genes to be tested, and only genes with loge(fold change) >0.25 were included in the analysis. Heatmaps (Figure 2, Figure 3, Figure 4, 7, Figure 10, Figure 11, Figure 12) were generated by taking the mean of the log-normalized data scaled by Seurat for cells within each cluster. Except where indicated, only genes that were differentially expressed between neuronal clusters are shown. Hierarchical clustering (Figure 18C) was performed using the complete linkage method. For the GO term analysis, we combined the 3 putative immature clusters into 1 and compared gene expression for these 3 clusters with the gene expression of all other clusters using a Wilcoxon rank sum test with Bonferroni multiple testing correction (Seurat's FindMarkers command). We performed GO enrichment analysis for GO biological processes (PANTHER Overrepresentation Test, GO database released 2020-02-21) using Fisher's exact test with false discovery rate correction. We submitted differentially expressed genes for the immature clusters (immature Chat cluster1, immature Chat cluster 2, immature Nos1 cluster) and compared these to the reference gene list for mus musculus (22265 genes). We also submitted differentially expressed genes for the more mature cluster (Chat cluster 1, Chat cluster 2, Nos1 cluster 1, Nos1 cluster 2, Nos1 cluster 3) and compared these with the reference gene list for Mus musculus (22265 genes). We reported the 20 most enriched GO terms. To identify genes differentially expressed between colon and small intestine, we defined colonic neurons as all cells with detectable Pou3f3 expression. We compared gene expression using a Wilcoxon rank sum test with Bonferroni multiple testing correction (Seurat's FindMarkers command) for neurons within the immature clusters (immature Chat cluster 1, immature Chat cluster 2, immature Nos1 cluster) or within the mature clusters (Chat cluster 1, Chat cluster 2, Nos1 cluster 1, Nos1 cluster 2, and Nos1 cluster 3) to normalize for the relatively more immature state of development of the majority of Pou3f3-expressing cells (40 mature Pou3f3-expressing neurons vs 503 other mature neurons and 94 immature Pou3f3-expressing neurons vs 62 other immature neurons). We followed up the identification of potentially colon-specific genes by correlating the gene expression of the gene in question with Pou3f3 expression in all neurons (pairwise Pearson correlation).
Analysis of Human Single-Nucleus Sequencing Data
Using Seurat,17,59 gene-barcode matrices were imported into R, filtered to remove low-expressors or doublets (nGenes = 200–5000) and mitochondrial contaminants (percent mitochondria <10%), normalized, and scaled to regress out UMIs and percent mitochondria. Nuclei were clustered using the most statistically significant principal components identified by elbow plot (11 principle components). Because unbiased clustering initially did not detect the small group of neurons as a distinct group, we identified 48 neurons by isolating all nuclei expressing any copies of ELAVL4, UCHL1, or SNAP25 (Figure 18A). These nuclei were reclustered. We saw 7 subgroups that we believe are doublets containing neuronal and non-neuronal nuclei based on coexpression of genes like ACTG2, PLP1, KIT 9 (Figure 18B). One subgroup appeared to be pure single-neuron data based on high expression of other neuronal markers like SYT1 and DSCAM and the absence of markers for other well-defined cell populations (Figure 18B). The strongly positive cluster (48 neurons) was mapped back onto the t-distributed stochastic neighbor embedding (t-SNE) plot. To identify neuron-specific genes, we compared this cluster of neurons against all other cell populations using a Wilcoxon rank sum test with Bonferroni multiple testing correction (Seurat’s FindMarkers command). A cutoff of at least 25% of cells in a group with expression >0 was required for genes to be tested. Neuron gene expression for the top 50 neuron-specific genes was graphed and neurons were clustered using the Euclidean distance method (Figure 18C). This revealed 2 groups of neurons, 1 which was NOS1+ and another that was NOS1–. We imported a list of 36 transcription and splice factors identified as significantly different in mouse adult colon Nos1 and Chat expressing neurons. Using this list and Seurat’s FindMarkers command, we compared expression for these genes between NOS1+ and NOS1– neurons and found differential expression for a subset of transcription factors (Figure 18D).
Preparing Bowel for Whole Mount Immunochemistry
E17.5 bowel was washed in PBS, straightened with stainless steel insect pins (Fine Science Tools; Cat. #26002-20) on Sylgard Elastomer 184 and fixed (4% paraformaldehyde [PFA], 20 minutes, room temperature [RT]). P0 small intestine in ice-cold PBS was opened along mesenteric border, pinned with insect pins serosal side up without stretching onto Sylgard Elastomer 184, before fixing (4% PFA, 20–30 minutes, RT) and peeling muscularis from mucosa/submucosa. Adult distal colon flushed with ice-cold PBS was opened along the mesenteric border. Muscle layers were peeled from mucosa/submucosa using Dumont #5 forceps (Fine Science Tools; Cat. #11251-30), maximally stretched, pinned (insect pins) to Sylgard Elastomer 184, fixed (4% PFA, 20–30 minutes, RT), briefly washed (1× PBS), equilibrated in 50% glycerol/50% PBS (30 minutes, RT; or overnight, 4°C) before storage (–20°C). For high magnification anti-enkephalin and anti-SP staining, colon opened along mesenteric border was stretched, pinned, and fixed (4% PFA, overnight, 4°C), rinsed in 1× PBS (3 × 10 minutes), and then dissected with fine forceps to remove mucosa, submucosa, and circular muscle from myenteric plexus attached to longitudinal muscle.
Anti-enkephalin and anti-SP antibody studies used C57BL6 wild-type mice. Proximal, mid, and distal colon tissue was removed, opened along the mesenteric border, cleaned of fecal matter, stretched and pinned to Sylgard Elastomer 184, and postfixed (4% PFA, overnight, 4°C). After rinsing 3 times (10 min/rinse) in 1× PBS, mucosa, submucous plexus and circular muscle were removed with fine forceps, leaving preparations of myenteric plexus attached to longitudinal muscle.
Whole Mount Immunohistochemistry
Bowel stored at –20°C in 50% glycerol/50% PBS was rinsed in PBS, blocked (2 hours, PBS + 0.5% Triton X-100 (Sigma-Aldrich; Cat. #T8787) with 5% normal donkey serum (NDS) (Jackson ImmunoResearch Laboratory, West Grove, PA; RRID: AB_2337258), incubated in primary antibody (Table 9) with gentle rocking (4°C, 2 days), except for ANNA-1, which was incubated overnight (4°C). After washing (3 × 5 minutes, 0.5% PBS + 0.5% Triton X-100) and secondary antibody (Table 9) incubation in blocking solution with gentle rocking (1 hour, RT), tissue was washed (3 × 5 minutes, PBS) and mounted in 50% glycerol/50% PBS.
Table 9.
List of Antibodies
| Antibody | Concentration | Catalog number | Source |
|---|---|---|---|
| Rabbit anti-NOS1 | 1:200 (mouse), 1:100 (human) | AB5380 | Sigma-Aldrich; RRID:AB_91824 |
| Goat anti-CHAT | 1:100 (human) | AB144P | Sigma-Aldrich; RRID:AB_2079751 |
| ANNA-1 (HuC/D) | N/A (mouse) | N/A | Kind gift from Dr V. Lennon, Mayo Clinic |
| Mouse anti-HuC/D | 1:200 (human) | A21271 | Thermo-Fisher Scientific, RRID:AB_221448 |
| Goat anti-TBX3 | 1:100 (human and mouse) | AF4509-SP | R&D Systems, RRID:AB_2240328 |
| Goat anti-PHOX2B | 1:100 (mouse) | AF4940-SP | R&D Systems, RRID:AB_10889846 |
| Rabbit anti-SATB1 | 1:100 (mouse) | ab109122 | Abcam, RRID:AB_10862207 |
| Chicken anti-GFP | 1:500 (mouse) | #GFP-1020 | Aves Labs, RRID:AB_10000240 |
| Rabbit anti-S100B | 1:200 (mouse) | Ab52642 | Abcam; RRID:AB_882426 |
| Rabbit anti-PBX3 | 1:100 (human and mouse) | 12571-1-AP | Proteintech Group; RRID:AB_2160469 |
| Rabbit anti-TBX2 | 1:100 (human and mouse) | 22346-1-AP | Proteintech Group |
| Rabbit anti-POU3F3 | 1:100 (human and mouse) | HPA067151 | Sigma-Aldrich; RRID:AB_2685790 |
| Rabbit anti-RBFOX1 | 1:100 (human and mouse) | HPA040809 | Sigma-Aldrich; RRID:AB_10796228 |
| Goat anti-BRN1 (also called POU3F3 protein) | 1:100 (mouse) | Santa Cruz; no longer sold | |
| Rabbit anti-BNC2 | 1:100 (human and mouse) | HPA018525 | Sigma-Aldrich; RRID:AB_1845402 |
| Rabbit anti-Met Enkephalin | 1:500 (mouse) | AB5026 | Sigma-Aldrich; RRID:AB_91644 |
| Rat anti-Substance P (NC1/34) | 1:800 (mouse) | AB150349 | Abcam |
| Alexa Fluor goat anti-human 647 | 1:400 (mouse and human) | A21445 | Thermo-Fisher Scientific; RRID:AB_2535862 |
| Alexa Fluor donkey anti-rabbit 488 | 1:400 (mouse and human) | A21206 | Thermo-Fisher Scientific; RRID:AB_2535792 |
| Alexa Fluor donkey anti-rabbit 594 | 1:400 (mouse and human) | A21207 | Thermo-Fisher Scientific; RRID:AB_141637 |
| Alexa Fluor donkey anti-rabbit 647 | 1:400 (mouse and human) | A31573 | Thermo-Fisher Scientific; RRID:AB_2536183 |
| Alexa Fluor goat anti-rat 594 | 1:500 (mouse) | A11007 | Thermo-Fisher Scientific; RRID:AB_10561522 |
| Alexa Fluor donkey anti-goat 594 | 1:400 (mouse and human) | A11058 | Thermo-Fisher Scientific; RRID:AB_2534105 |
| Alexa Fluor goat anti-chicken 488 | 1:400 (mouse and human) | A11039 | Thermo-Fisher Scientific; RRID:AB_142924 |
| Alexa Fluor donkey anti-goat 647 | 1:400 (mouse and human) | A21447 | Thermo-Fisher Scientific; RRID:AB_141844 |
| Alexa Fluor donkey anti-goat 488 | 1:400 (mouse and human) | A11055 | Thermo-Fisher Scientific; RRID:AB_2534102 |
| Alexa Fluor goat anti-rabbit 488 | 1:400 (mouse and human) | A31556 | Thermo-Fisher Scientific; RRID:AB_221605 |
N/A, not available. (Note: We do not have a catalogue number or exact concentration for this antibody since it was generated in limited quantities from human serum and reused for each experiment).
For high-magnification imaging, longitudinal muscle with exposed myenteric plexus was placed in 10% Casblock (Thermo-Fisher Scientific; Cat. #008120) with 0.1% Triton X-100 (ProSciTech, Thuringowa, Queensland, Australia) in PBS (30 minutes, RT), then in primary antisera (24–48 hours, 4°C) (Table 9) before rinsing (PBS, 3 × 10 minutes), and secondary antisera staining (Table 9) (2 hours, RT). After rinsing (PBS, 3 × 10 minutes), preparations were mounted using fluorescence mounting medium (DAKO, Carpenteria, CA).
Nicotinamide Adenine Dinucleotide Phosphate Diaphorase Staining
Bowel was pinned flat and fixed (4% PFA, 20 minutes, RT), briefly washed (1× PBS), incubated in 0.1-M phosphate buffer, pH 7.4 with 1-mg/mL beta-nicotinamide adenine dinucleotide phosphate (beta-NADPH; Sigma-Aldrich; Cat. #N750), 0.1-mg/mL nitroblue tetrazolium (Sigma-Aldrich; Cat. #11383213001), 0.3% Triton-X 100 (1 hour, 37°C), washed in PBS, and imaged immediately in a custom-made holding chamber.
Preparing Human Colon for Whole Mount Immunochemistry
Human colon with fat trimmed was pinned serosa side up on Sylgard Elastomer 184 plates in ice-cold PBS using stainless steel insect pins. Tissue was maximally stretched during pinning to make the colon as thin as possible. Pinned tissue was fixed overnight (4°C, 4% PFA), washed in DPBS, and stored at 4°C in 50% PBS/50% glycerol/0.05% sodium azide until staining.
Immunofluorescence Staining of Human Colon Whole Mount
Following our new protocol,13 0.7–1 cm × 0.7–1 cm colon pieces were washed (1× PBS, RT), incubated in 100% methanol on ice (1 hour), treated with Dent's bleach (1 mL 30% hydrogen peroxide (Thermo-Fisher Scientific; Cat. #H1009), 1-mL dimethyl sulfoxide (Sigma-Aldrich; Cat. #D2650), 4 mL 100% methanol61 at room temperature (2 hours), washed again (1× PBS, 3 × 5 minutes, room temperature), blocked (0.5% Triton X-100, 5% NDS in PBS, 3 days), and then incubated 14 days in blocking solution with primary antibodies (37°C, 40–100 rpm) (Table 9). Following PBS washing (1 day, 3 washes, gentle rocker, RT), colon was incubated in secondary antibodies (3 days, 37°C, 40–100 rpm), washed in PBS over 1 day, dehydrated in a methanol series, and placed in Murray's clear (2:1 benzyl benzoate:benzyl alcohol, gentle rocking, RT) until translucent (30–60 minutes). Transparent colon mounted on glass slides in Murray's clear was imaged within 1 day (for details, see dx.doi.org/10.17504/protocols.io.wyeffte).
Microscopy
Images were acquired with a Zeiss LSM 710 confocal microscope with 20×/0.8 air or 63×/1.4 oil DIC M27 Plan-Apochromat objectives and Zeiss Zen software version 2.3 14.0.14.201 or upright fluorescent Olympus BX60 microscope (Shinjuku City, Tokyo, Japan) with Axiocam CCD camera and Axiovision software (Carl Zeiss Microscopy, North Ryde, New South Wales, Australia). ImageJ, version 2.35 (National Institutes of Health, Bethesda, MD) and Photoshop CS6 (Adobe, San Jose, CA) were used to crop and uniformly adjust color. High-magnification myenteric plexus images were acquired with a Zeiss LSM 880 Airyscan microscope with a Plan-Apochromat 63 × 1.40 Oil DIC M27 objective (Carl Zeiss Microscopy; Cat. #420782-990-799) at the Biological Optical Microscopy Platform, University of Melbourne.
Image Analysis
Counting for adult mouse distal colon and human colon was performed on >5 randomly selected ×20 fields per animal in each region using ImageJ's CellCounter module (Table 10). Because of high cell density at E17.5, only half of each ×20 field was counted. Investigators were blinded to genotype when comparing knockout to control mice. High-magnification myenteric plexus images were processed to quantify SP and enkephalin colocalization using Imaris 9.0.0 (Bitplane, Zürich, Switzerland).21
Table 10.
Numbers of Biological Replicates and Numbers of Cells/Terminals Counted
| Experiment | Biological replicate number | Total neurons or nerve terminals counted | Statistical tests used | Descriptive statistics |
|---|---|---|---|---|
| Figure 2: enkephalin and substance P staining quantification | 3 | 11,628 (myenteric ganglia) 2918 (circular muscle) |
Unpaired t test | Varicosities within myenteric ganglia: % of Enk+ varicosities that were SP+: 74.5 ± 23.6, 86.9 (47.3 - 89.2) % of SP+ varicosities that were Enk+: 81.2 ± 8.7, 78.0 (74.5 - 91.0) Varicosities in circular muscle: % of Enk+ varicosities that were SP+: 76.5 ± 5.0, 78.7 (70.7 - 80.0) % of SP+ varicosities that were Enk+: 44.7 ± 8.4, 41.9 (41.9 - 38.2 - 54.2) |
| Figure 2: Vglut-Ires-Cre;TdTomato × Chat-EGFP quantification (mouse adult distal colon) | 3 | 114 | N/A | % TDTOMATO+/EGFP+-/HUC+: 4.1 ± 1.5, 3.5 (3.0 - 5.8) % TDTOMATO+/EGFP+/HUC+: 95.9 ± 1.5, 96.5 (94.2 - 97.0) |
| Figure 5: Gfra1-GFP quantification (mouse adult distal colon) | 3 | 614 | Student’s t test | % of NOS1+ that were GFP+: 88.9 ± 1.8, 89.1 (87.0 - 90.6) % of NOS- neurons that were GFP+: 8.8 ± 5.7, 7.7 (3.8 - 15.0) |
| Figure 5: NADPH diaphorase staining | 5 | 340 | Fisher exact test (2 × 2 contingency table) | N/A |
| Figure 8: Etv1-CreERT2;TdTomato × NOS1 quantification (mouse adult distal colon) | 3 | 1101 | Student’s t test | % of NOS+ neurons that were TDT+: 56.4 ± 6.3, 59.7 (49.1 - 60.3) % of NOS- neurons that were TDT+: 5.6 ± 1.5, 5.6 (4.1 - 7.1) % of TDTOMATO+ neurons that were NOS+: 85.0 ± 5.8, 85.7 (78.9 - 90.5) % of TDTOMATO neurons that were NOS-: 15.0 ± 5.8, 14.3 (9.5 - 21.1) |
| Figure 8: PBX3 × Chat-EGFP quantification (mouse adult distal colon) | 4 | 2684 | Student’s t test | % of EGFP–neurons that were PBX3+: 31.4 ± 6.0, 29.5 (26.9 - 37.7) % of EGFP+ neurons that were PBX3+: 72.7 ± 5.4, 74.2 (67.1 - 76.9) % of PBX3+ neurons that were EGFP+: 79.2 ± 5.3, 79.4 (74.0 - 84.2) % of PBX3+ neurons that were EGFP-: 20.8 ± 5.3, 20.6 (15.8 - 26.0) |
| Figure 8: PHOX2B × Chat-EGFP quantification (mouse adult distal colon) | 4 | 2136 | Student’s t test | % of EGFP–neurons that were PHOX2B+: 69.9 ± 7.3, 72.0 (71.2 - 80.7) % of EGFP+ neurons that were PHOX2B+: 76.1 ± 5.1, 76.3 (62.1 - 75.6) % of PHOX2B+ neurons that were EGFP+: 60.8 ± 2.9, 62.1 (57.8 - 62.7) % of PHOX2B+ neurons that were EGFP-: 39.2 ± 2.9, 37.9 (37.3 - 42.2) |
| Figure 8: RBFOX1 × Chat-EGFP quantification (mouse adult distal colon) | 3 | 993 | Student’s t test | % of EGFP–neurons that were RBFOX1+: 39.4 ± 3.8, 38.7 (36.0 - 43.6) % of EGFP+ neurons that were RBFOX1+: 82.5 ± 6.7, 78.8 (78.4 - 90.2) % of RBFOX1+ neurons that were EGFP+: 74.2 ± 5.8, 76.9 (67.5 - 78.1) % of RBFOX1+ neurons that were EGFP-: 25.8 ± 5.8, 23.1 (21.9 - 32.5) |
| Figure 8: SATB1 × Chat-EGFP quantification (mouse adult distal colon) | 3 | 1028 | Student’s t test | % of EGFP–neurons that were SATB1+: 7.9 ± 12.3, 1.6 (0 - 22.1) % of EGFP+ neurons that were SATB1+: 50.0 ± 12.0, 45.3 (41.0 - 63.5) % of SATB1+ neurons that were EGFP+: 94.8 ± 7.9, 98.8 (85.7 - 100) % of SATB1+ neurons that were EGFP-: 5.2 ± 7.9, 1.2 (0 - 14.) |
| Figure 8: TBX3 × NOS1 quantification (mouse adult distal colon) | 4 | 1033 | Student’s t test | % of NOS+/EGFP–neurons that were TBX3+: 90.9 ± 8.2, 92.0 (82.5 - 98.1) % of NOS+/EGFP+ neurons that were TBX3+: 86.5 ± 12.1, 85.6 (75.7 - 98.3) % of NOS-/EGFP+ neurons that were TBX3+: 23.4 ± 7.7, 26.3 (15.4 - 28.5) % of TBX3+ neurons that were EGFP+: 31.0 ± 7.0, 34.5 (24.0 - 34.6) % of TBX3+ neurons that were EGFP-: 69.0 ± 7.0, 65.5 (65.4 - 76) % of TBX3+ neurons that were NOS+: 66.0 ± 5.6, 64.5 (61.7 - 71.7) % of TBX3+ neurons that were NOS-: 34.0 ± 5.6, 35.5 (28.3 - 38.3) |
| Figure 8: All neurons (mouse adult distal colon) | 3 (EGFP) 7 (NOS) |
N/A | % of total neurons that were EGFP+:58.1 ± 4.4, 58.8 (53.4 - 62.2) % of total neurons that were EGFP-: 41.9 ± 4.4, 41.2 (37.8 - 46.6) % of total neurons that were NOS+: 38.1 ± 4.5, 39.2 (33.3 - 41.2) % of total neurons that were NOS-: 61.9 ± 4.5, 60.8 (58.8 - 66.7) |
|
| Figure 13: BNC2 × Chat-EGFP quantification (mouse E17.5 mid small intestine) | 3 | 6186 | Student’s t test | BNC2+EGFP+/EGFP+: 54.8 ± 5.2, 52.3 (51.3 - 60.8) BNC2+EGFP–/EGFP-: 21.6 ± 1.5, 21.3 (20.3 - 23.2) BNC2+EGFP+/BNC2+: 78.7 ± 1.1, 78.7 (77.5 - 79.6) EGFP+HUC+/HUC+: 59.2 ± 4.4, 57.0 (56.3 - 64.3) |
| Figure 13: PBX3 × Chat-EGFP quantification (mouse E17.5 mid small intestine) | 3 | 10259 | Student’s t test | PBX3+EGFP+/EGFP+: 53.4 ± 13.3, 60.6 (38.1 - 61.5) PBX3+EGFP–/EGFP-: 12.1 ± 9.5, 9.7 (4.0 - 22.6) PBX3+EGFP+/PBX3+: 85.7 ± 8.6, 84.5 (77.7 - 94.9) EGFP+HUC+/HUC+: 56.2 ± 9.6, 55.8 (46.8 - 65.9) |
| Figure 13: RBFOX1 × Chat-EGFP quantification (mouse E17.5 mid small intestine) | 3 | 11522 | Student’s t test | RBFOX1+EGFP+/EGFP+: 49.7 ± 9.4, 48.3 (41.0 - 59.8) RBFOX1+EGFP–/EGFP-: 3.7 ± 2.1, 4.0 (1.5 - 5.7) RBFOX1+EGFP+/RBFOX1+: 95.3 ± 2.1, 95.7 (93.1 - 97.2) EGFP+HUC+/HUC+: 59.7 ± 2.8, 60.6 (56.6 - 61.9) |
| Figure 13: TBX2 × Chat-EGFP quantification (mouse E17.5 mid small intestine) | 3 | 8369 | Student’s t test | TBX2+EGFP+/EGFP+: 51.5 ± 4.3, 51.4 (47.3 - 55.8) TBX2+EGFP–/EGFP-: 7.8 ± 7.3, 3.7 (3.5 - 16.2) TBX2+EGFP+/TBX2+: 92.3 ± 5.1, 94.7 (86.5 - 95.8) EGFP+HUC+/HUC+: 60.9 ± 5.1, 63.3 (55.0 - 64.4) |
| Figure 13: TBX3 × Chat-EGFP × NOS1 quantification (mouse E17.5 mid small intestine) | 3 | 4235 | Analysis of variance with Tukey’s post hoc test | TBX3+EGFP+/EGFP+: 49.1 ± 6.3, 48.1 (43.3 - 55.8) TBX3+NOS1+/NOS1+: 95.2 ± 1.7, 94.6 (93.8 - 97.1) TBX3+NOS1+EGFP+/NOS1+EGFP+: 91.2 ± 3.9, 91.4 (87.2 - 95.0) TBX3+EGFP+/TBX3+: 0.50 ± 0.04, 0.5 (0.45 - 0.53) TBX3+NOS1+/TBX3+: 44.6 ± 3.2, 43.9 (41.8 - 48.1) TBX3+EGFP+NOS1+/TBX3+: 5.6 ± 0.9, 5.4 (4.9 - 6.5) EGFP+/(sum of EGFP+, NOS1+, NOS1+EGFP+): 68.3 ± 1.6, 67.5 (67.3 - 70.2) NOS1+/(sum of EGFP+, NOS1+, NOS1+EGFP+): 31.1 ± 1.7, 32.0 (29.2 - 32.1) EGFP+NOS1+/(sum of EGFP+, NOS1+, NOS1+EGFP+): 0.8 ± 0.0, 0.8 (0.8 - 0.9) |
| Figure 13: Etv1-CreERT2;TDTOMATO × NOS1 quantification (mouse E17.5 mid small intestine) | 3 | Student’s t test | TDTOMATO+NOS1+/NOS1+: 14.86 ± 3.59, 13.59 (12.41 - 18.57) TDTOMATO+/NOS1–/NOS1–: 3.59 ± 0.59, 3.65 (2.98 - 4.15) |
|
|
Figure 14: Wnt1-Cre;Casz1 flox neuron numbers (mouse P0 mid small intestine) |
3 (control), 3 (KO) | 4526 (control), 4126(KO) | Student’s t test | % Chat-EGFP+ neurons: KO: 51.6 ± 6.0, 48.3 (43.3 - 63.3) Control: 45.3 ± 3.9, 44.57 (39.0 - 52.3) Total neuron density: KO: 3044 ± 812.2, 2891 (2320 - 3922) Control: 3339 ± 89.0, 3294 (3283 - 3442) |
| Figure 14: Wnt1-Cre;Tbx3 flox neuron numbers (mouse P0 mid small intestine) | 3 (control), 3 (KO) | 8618 (control), 3983 (KO) | Student’s t test | % NOS1+ neurons: KO: 18.2 ± 3.3, 17.4 (15.3 - 21.9) Control: 26.0 ± 3.0, 26.4 (22.8 - 28.8) Total Neuron Density: KO: 2180 ± 403.2, 2075 (1839 - 2625) Control: 2337 ± 260.6, 2302 (2096 - 2613) |
| Figure 14: Wnt1-Cre;Rbfox1 flox neuron numbers (mouse P0 mid small intestine) | 4 (control), 4 (KO) | 6871 (control), 5348 (KO) | Student’s t test | % Chat-EGFP+ neurons: KO: 47.4 ± 2.9, 47.3 (44.8 - 50.0) Control: 53.6 ± 7.3, 53.6 (46.6 - 60.7) Total Neuron Density: KO: 1603 ± 352.2, 1589 (1273 - 1947) Control: 1789 ± 364.6, 1903 (1413 - 2078) |
| Figure 14: Wnt1-Cre;Tbx2 flox neuron numbers (mouse P0 mid small intestine) | 4 (control), 3 (KO) | 5911 (control), 2581 (KO) |
Student’s t test MWRST |
% Chat-EGFP+ neurons: KO: 46.15 ± 6.678, 46.3 (39.4 - 52.8) Control: 45.6 ± 7.741, 44.75 (38.6 - 53.5) Total Neuron Density: KO: 1844 ± 1005, 1754 (886.8 - 2891) Control: 1839 ± 166.8, 1846 (1680 - 1992) |
| Figure 19: BNC2 × CHAT quantification (human colon) | 3 | 2675 | Student’s t test | BNC2+CHAT+/CHAT+: 36.6 ± 4.1, 34.3 (34.2 - 41.3) BNC2+CHAT-/CHAT-: 1.6 ± 1.3, 0.8 (0.8 - 3.1) BNC2+CHAT+/BNC2+: 97.9 ± 1.3, 97.9 (96.6 - 99.2) CHAT+HUC+/HUC+: 66.6 ± 11.8, 60.0 (59.6 - 80.1) |
| Figure 19: PBX3 × CHAT quantification (human colon) | 3 | 2439 | Student’s t test | PBX3+CHAT+/CHAT+: 19.0 ± 4.3, 19.9 (14.3 - 22.8) PBX3+CHAT-/CHAT-: 1.1 ± 0.3, 1.1 (0.8 - 1.4) PBX3+CHAT+/PBX3+: 97.5 ± 1.1, 97.2 (96.5 - 98.7) CHAT+HUC+/HUC+: 68.7 ± 6.9, 67.7 (62.3 - 76.1) |
| Figure 19: RBFOX1 × CHAT quantification (human colon) | 3 | 2825 | Student’s t test | RBFOX1+CHAT+/CHAT+: 44.4 ± 7.7, 41.4 (38.8 - 53.1) RBFOX1+CHAT-/CHAT-: 1.5 ± 1.4, 1.6 (0 - 2.7) RBFOX1+CHAT+/RBFOX+: 99.4 ± 0.5, 99.3 (99.0 - 100.0) CHAT+HUC+/HUC+: 87.4 ± 2.7, 86.9 (85.0 - 90.3) |
| Figure 19: TBX2 × CHAT quantification (human colon) | 3 | 3427 | Student’s t test | TBX2+CHAT+/CHAT+: 7.8 ± 2.8, 6.2 (6.2 - 11.0) TBX2+CHAT-/CHAT-: 0.1 ± 0.1, 0.1 (0 - 0.2) TBX2+CHAT+/TBX2+: 99.4 ± 0.5, 99.3 (99.0 - 100.0) CHAT+HUC+/HUC+: 69.6 ± 12.9, 72.5 (55.5 - 80.9) |
| Figure 19: TBX3 × NOS1 quantification (human colon) | 3 | 972 | Student’s t test | TBX3+NOS1+/NOS1+: 90.0 ± 9.7, 92.4 (79.2 - 98.2) TBX3+NOS1–/NOS1–: 16.4 ± 6.5, 14.9 (10.7 - 23.5) TBX3+NOS1+/TBX3+: 83.5 ± 8.9, 83.3 (74.6 - 92.4) NOS1+HUC+/HUC+: 49.1 ± 7.8, 47.3 (42.4 - 57.7) |
Values are mean ± SD, median (range).
EGFP, enhanced green fluorescent protein; KO, knockout; N/A, not available; NADPH, nicotinamide adenine dinucleotide phosphate; NOS1, neuronal nitric oxide synthase.
RNA Extraction and RT-PCR for Pou3f3 Expression Analysis
Wnt1-Cre;R26-TdTomato E14.5 and E17.5 dams were euthanized with 5% CO2, and pups were removed from the mother. For each litter, all TdTomato+ small intestines and colons were combined to increase cell count. Small intestines and colons were dissected in ice-cold Dulbecco-modified PBS, transferred to HBSS (Thermo-Fisher Scientific; Cat No: 14025092), separated, divided into small pieces using insulin needles (Beckton Dickerson; Cat. #08290-3284-18), and dissociated for 30 minutes at 37°C in Liberase (Sigma-Aldrich; Cat. #5401135001) supplemented with DNase I (Roche; Cat. #04716728001), MgCl2 (6 mM) and CaCl2 (1 mM) in HBSS with P1000 trituration. Cells were filtered, washed with Iscove's DMEM (Cat. #10-016-CM), and resuspended in FACS buffer (10mM HEPES, 1mg/mL BSA, 1% penicillin and streptomycin (Thermo-Fisher Scientific; Cat. #15140122) in HBSS). Fluorescent TdTomato+ cells were sorted on a BD FACSJazz (BD Biosciences, San Jose, CA) and collected in Iscove's DMEM with 10% FBS. Cells were spun down at 600 g, resuspended in Buffer RLT plus and RNA was isolated using the Qiagen RNeasy Plus Micro Kit (Cat. #74034) with DNase treatment (Qiagen; Cat. #79254).
RNA integrity and concentrations were measured on a 2100 Agilent Bioanalyzer using an RNA 6000 Pico Kit (Agilent, Santa Clara, CA; Cat. #5067-1513). All samples used had RIN ≥7. Complementary DNA was generated using Superscript II RNase H (Thermo-Fisher Scientific; Cat. #18064022). RT-PCR was performed using KAPA mixture (Cat. #KK7352) and previously described primers (Table 8). Three replicates were run for all experiments.
Colon Bead Expulsion Studies
Young adult mice (P44–58 at begin of assay, P49–64 at end of assay) were anesthetized with 2 L/min carbogen with 2.5% (v/v) isoflurane (Piramal, Mumbai, India; Cat. #NDC 66794-017-10) for 1.5 minutes before the start of the experiment, anesthesia was continued until completed bead insertion. A glass bead (3 mm; Sigma-Aldrich; Cat. #Z143928) lubricated with sunflower seed oil (Sigma-Aldrich; Cat. #S5007) was inserted 2 cm into the colon using a custom-made 3-mm rounded glass rod. The glass rod was removed, isoflurane was stopped, mice were placed in empty cages, and time to bead expulsion was recorded. If a mouse regained consciousness before bead insertion, mice were anesthetized for an additional 2–4 minutes. The assay was repeated 3 times per mouse (48–96 hours between procedures). Mice had free access to food and water prior to the experiment.
Calcium Imaging of Full Thickness Colon
Colons from E2a-Cre;R26R-Gcamp6s mice 12–16 weeks of age were cut longitudinally and pinned (mucosa facing down) in a Sylgard-lined dish superfused with carbogenated (95% O2, 5% CO2) artificial cerebrospinal fluid (ACSF),26 freshly prepared, maintained at 35°C. Nifedipine (1 μM, Sigma-Aldrich; Cat. #N7634) in ACSF improved stability for calcium imaging analyses. GCaMP6s in myenteric neurons was imaged with an upright DM6000FS Leica fluorescence microscope (Leica, Buffalo Grove, IL) and EMCCD camera (Photometrics; Roper Scientific, Tucson, AZ) using a ×20 objective. Images were collected with Metamorph, version 7.10.1.161 (Molecular Devices, San Jose, CA) at 10-Hz sampling rate. For each field, spontaneous activity was imaged 2 minutes, before two 30-second movies were collected in response to EFS (100-μs pulse, 20 Hz, 1 second) using concentric electrodes 5 mm oral or aboral to imaging fields (order randomized). Recombinant rat GDNF (Cat. #512-GF; R&D Systems, Minneapolis, MN) or NRTN (Cat. #450-11; PeproTech, Rocky Hill, NJ) were added at 10-nM final concentration (order of presentation randomized). We recorded spontaneous activity in 2-minute movies collected intermittently while ligands continuously superfused circulating ACSF for 10 minutes. Responses to oral and aboral stimulation were then reimaged. Colon was washed for 30 minutes, and baseline activity and responses were reimaged before the next receptor ligand was applied. In a subset of experiments, the addition of TTX (0.5 μM; Cat. #T8024; Sigma-Aldrich) blocked all neural transmission allowing for identification of myenteric neurons directly activated by GDNF or NRTN. Spontaneous activity was recorded (three 2-minute movies) at baseline, 10 minutes following TTX and during application of GDNF or NRTN (in the presence of TTX; order of presentation of GDNF/NRTN was randomized). Colon tissue was then “washed” with the TTX, activity was reimaged, and the next receptor ligand was applied while imaging responses.
Analysis of Live GCaMP6s Imaging
Metamorph (Molecular Devices, Downington, PA) image files exported to ImageJ were coded for blinded analysis. Circular regions of interest were drawn on all myenteric neurons in each field. Amplitude of GCaMP6s signals were analyzed and quantified as described.62 We calculated ΔF/F0 [% = ((F − F0)/F0) × 100], where F is peak fluorescence and F0 is baseline mean fluorescence; ΔF/F0 of 4SD > baseline was considered a response. Time control studies, in which GCaMP6s activity from individual neurons was measured over time, were used to determine whether changes in activity were significantly altered due to GDNF or NRTN application. Tissue movement along x- and y-axes was measured using ImageJ Template-Matching plugin. Time-lapse color-coded images created in ImageJ use pixel color to indicate when pixels reached maximum F, providing spatial and temporal information about activity.
Statistics
We used Prism 7.03 software (GraphPad Software, San Diego, CA). A cutoff of P < .05 was considered significant. Data represent mean ± SEM.
Acknowledgments
The authors thank Dr Emma Furth, Dr Federico Valdivieso, Dr Michael Feldman, Dr Ali Naji, Rachel H. Céron, Silvia Huerta López, Caitlin Feltcher, Andrew Kromer, Lauren Schmucker, and Jennifer Finan for help acquiring human tissue. The authors also thank Scott Gianino for POU3F3 initial studies.
CRediT Authorship Contributions
Christina M. Wright, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Sabine Schneider, BS (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Kristen M. Smith-Edwards, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting; Validation: Supporting; Visualization: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Fernanda Mafra, MS PhD (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Anita J.L. Leembruggen, MBiomedSci (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Michael V. Gonzalez, BS (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Deepika R. Kothakapa, BS Candidate (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Jessica B. Anderson, BA Candidate (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Beth A. Maguire, BS (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Tao Gao, PhD (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Tricia A. Missall, MD PhD (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Marthe J. Howard, PhD (Conceptualization: Supporting; Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Joel C. Bornstein, PhD (Conceptualization: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Brian M. Davis, PhD (Conceptualization: Supporting; Funding acquisition: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Robert O. Heuckeroth, MD PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of Interest This author discloses the following: Robert O. Heuckeroth is a consultant for BlueRock Therapeutics and has served on a scientific advisory board for Takeda. The remaining authors disclose no conflicts.
Funding This work was supported by the Irma and Norman Braman Endowment; the Children's Hospital of Philadelphia Research Institute; the Suzi and Scott Lustgarten Center Endowment; National Institutes of Health grants 5 F30 DK117546-02 (to Christina M. Wright), F32 DK120115 (to Kristen M. Smith-Edwards), and R01DK122798 (PI: Brian Davis; co-PIs: Robert Heuckeroth, Marthe Howard); and National Institutes of Health SPARC (Stimulating Peripheral Activity to Relieve Conditions) program OT2OD023859 (PI: Marthe Howard; co-PIs: Robert Heuckeroth, Brian Davis, Joel Bornstein).
Supplementary Material
References
- 1.Schneider S., Wright C.M., Heuckeroth R.O. Unexpected roles for the second brain: enteric nervous system as master regulator of bowel function. Annual Review of Physiology. 2019;81:235–259. doi: 10.1146/annurev-physiol-021317-121515. [DOI] [PubMed] [Google Scholar]
- 2.McCann C.J., Thapar N. Enteric neural stem cell therapies for enteric neuropathies. Neurogastroenterol Motil. 2018;30:e13369. doi: 10.1111/nmo.13369. [DOI] [PubMed] [Google Scholar]
- 3.Soret R., Schneider S., Bernas G., Christophers B., Souchkova O., Charrier B., Righini-Grunder F., Aspirot A., Landry M., Kembel S.W., Faure C., Heuckeroth R.O., Pilon N. Glial cell derived neurotrophic factor induces enteric neurogenesis and improves colon structure and function in mouse models of Hirschsprung disease. Gastroenterology. 2020;159:1824–1838.e7. doi: 10.1053/j.gastro.2020.07.018. [DOI] [PubMed] [Google Scholar]
- 4.Memic F., Knoflach V., Morarach K., Sadler R., Laranjeira C., Hjerling-Leffler J., Sundström E., Pachnis V., Marklund U. Transcription and signaling regulators in developing neuronal subtypes of mouse and human enteric nervous system. Gastroenterology. 2018;154:624–636. doi: 10.1053/j.gastro.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morarach K., Mikhailova A., Knoflach V., Memic F., Kumar R., Li W., Ernfors P., Marklund U. Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing. Nat Neurosci. 2021;24:34–46. doi: 10.1038/s41593-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L.E., La Manno G., Codeluppi S., Furlan A., Lee K., Skene N., Harris K.D., Hjerling-Leffler J., Arenas E., Ernfors P., Marklund U., Linnarsson S. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.May-Zhang A.A., Tycksen E., Southard-Smith A.N., Deal K.K., Benthal J.T., Buehler D.P., Adam M., Simmons A.J., Monaghan J.R., Matlock B.K., Flaherty D.K., Potter S.S., Lau K.S., Southard-Smith E.M. Combinatorial transcriptional profiling of mouse and human enteric neurons identifies shared and disparate subtypes in situ. Gastroenterology. 2020 Sep 30 doi: 10.1053/j.gastro.2020.09.032. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drokhlyansky E., Smillie C.S., Van Wittenberghe N., Ericsson M., Griffin G.K., Eraslan G., Dionne D., Cuoco M.S., Goder-Reiser M.N., Sharova T., Kuksenko O., Aguirre A.J., Boland G.M., Graham D., Rozenblatt-Rosen O., Xavier R.J., Regev A. The human and mouse enteric nervous system at single-cell resolution. Cell. 2020;182:1606–1622.e23. doi: 10.1016/j.cell.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lasrado R., Boesmans W., Kleinjung J., Pin C., Bell D., Bhaw L., McCallum S., Zong H., Luo L., Clevers H., Vanden Berghe P., Pachnis V. Lineage-dependent spatial and functional organization of the mammalian enteric nervous system. Science. 2017;356:722–726. doi: 10.1126/science.aam7511. [DOI] [PubMed] [Google Scholar]
- 10.Lomax A.E., Furness J.B. Neurochemical classification of enteric neurons in the guinea-pig distal colon. Cell Tissue Res. 2000;302:59–72. doi: 10.1007/s004410000260. [DOI] [PubMed] [Google Scholar]
- 11.Knowles C.H., Veress B., Kapur R.P., Wedel T., Farrugia G., Vanderwinden J.-M., Geboes K., Smith V.V., Martin J.E., Lindberg G., Milla P.J., De Giorgio R. Quantitation of cellular components of the enteric nervous system in the normal human gastrointestinal tract--report on behalf of the Gastro 2009 International Working Group. Neurogastroenterol Motil. 2011;23:115–124. doi: 10.1111/j.1365-2982.2010.01657.x. [DOI] [PubMed] [Google Scholar]
- 12.Sang Q., Young H.M. Chemical coding of neurons in the myenteric plexus and external muscle of the small and large intestine of the mouse. Cell Tissue Res. 1996;284:39–53. doi: 10.1007/s004410050565. [DOI] [PubMed] [Google Scholar]
- 13.Graham K.D., López S.H., Sengupta R., Shenoy A., Schneider S., Wright C.M., Feldman M., Furth E., Valdivieso F., Lemke A., Wilkins B.J., Naji A., Doolin E., Howard M.J., Heuckeroth R.O. Robust, 3-dimensional visualization of human colon enteric nervous system without tissue sectioning. Gastroenterology. 2020;158:2221–2235.e5. doi: 10.1053/j.gastro.2020.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y., Dong H., Eckmann L., Hanson E.M., Ihn K.C., Mittal R.K. Visualizing the enteric nervous system using genetically engineered double reporter mice: comparison with immunofluorescence. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake J.I., Avetisyan M., Zimmermann A.G., Heuckeroth R.O. Neural crest requires Impdh2 for development of the enteric nervous system, great vessels, and craniofacial skeleton. Dev Biol. 2016;409:152–165. doi: 10.1016/j.ydbio.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brokhman I., Xu J., Coles B.L.K., Razavi R., Engert S., Lickert H., Babona-Pilipos R., Morshead C.M., Sibley E., Chen C., van der Kooy D. Dual embryonic origin of the mammalian enteric nervous system. Dev Biol. 2019;445:256–270. doi: 10.1016/j.ydbio.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu Z.-D., Thacker M., Castelucci P., Bagyánszki M., Epstein M.L., Furness J.B. Immunohistochemical analysis of neuron types in the mouse small intestine. Cell Tissue Res. 2008;334:147–161. doi: 10.1007/s00441-008-0684-7. [DOI] [PubMed] [Google Scholar]
- 19.Furness J.B., Costa M., Miller R.J. Distribution and projections of nerves with enkephalin-like immunoreactivity in the guinea-pig small intestine. Neuroscience. 1983;8:653–664. doi: 10.1016/0306-4522(83)90001-5. [DOI] [PubMed] [Google Scholar]
- 20.Bornstein J.C., Costa M., Furness J.B., Lees G.M. Electrophysiology and enkephalin immunoreactivity of identified myenteric plexus neurones of guinea-pig small intestine. J Physiol. 1984;351:313–325. doi: 10.1113/jphysiol.1984.sp015247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swaminathan M., Hill-Yardin E.L., Bornstein J.C., Foong J.P.P. Endogenous glutamate excites myenteric calbindin neurons by activating group I metabotropic glutamate receptors in the mouse colon. Front Neurosci. 2019;13:426. doi: 10.3389/fnins.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sang Q., Young H.M. The identification and chemical coding of cholinergic neurons in the small and large intestine of the mouse. Anat Rec. 1998;251:185–199. doi: 10.1002/(SICI)1097-0185(199806)251:2<185::AID-AR6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 23.Heuckeroth R.O., Enomoto H., Grider J.R., Golden J.P., Hanke J.A., Jackman A., Molliver D.C., Bardgett M.E., Snider W.D., Johnson E.M., Jr., Milbrandt J. Gene targeting reveals a critical role for neurturin in the development and maintenance of enteric, sensory, and parasympathetic neurons. Neuron. 1999;22:253–263. doi: 10.1016/s0896-6273(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 24.Uesaka T., Jain S., Yonemura S., Uchiyama Y., Milbrandt J., Enomoto H. Conditional ablation of GFRα1 in postmigratory enteric neurons triggers unconventional neuronal death in the colon and causes a Hirschsprung’s disease phenotype. Development. 2007;134:2171–2181. doi: 10.1242/dev.001388. [DOI] [PubMed] [Google Scholar]
- 25.Grider J.R., Heuckeroth R.O., Kuemmerle J.F., Murthy K.S. Augmentation of the ascending component of the peristaltic reflex and substance P release by glial cell line-derived neurotrophic factor. Neurogastroenterol Motil. 2010;22:779–786. doi: 10.1111/j.1365-2982.2010.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith-Edwards K.M., Najjar S.A., Edwards B.S., Howard M.J., Albers K.M., Davis B.M. Extrinsic primary afferent neurons link visceral pain to colon motility through a spinal reflex in mice. Gastroenterology. 2019;157:522–536.e2. doi: 10.1053/j.gastro.2019.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López S.H., Avetisyan M., Wright C.M., Mesbah K., Kelly R.G., Moon A.M., Heuckeroth R.O. Loss of Tbx3 in murine neural crest reduces enteric glia and causes cleft palate, but does not influence heart development or bowel transit. Dev Biol. 2018;444(Suppl 1):S337–S351. doi: 10.1016/j.ydbio.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lake J.I., Heuckeroth R.O. Enteric nervous system development: migration, differentiation, and disease. Am J Physiol Gastrointest Liver Physiol. 2013;305:G1–G24. doi: 10.1152/ajpgi.00452.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright C.M., Garifallou J.P., Schneider S., Mentch H.L., Kothakapa D.R., Maguire B.A., Heuckeroth R.O. Dlx1/2 mice have abnormal enteric nervous system function. JCI Insight. 2020;5 doi: 10.1172/jci.insight.131494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao M.M., Bornstein J.C., Young H.M. Development of myenteric cholinergic neurons in ChAT-Cre;R26R-YFP mice. J Comp Neurol. 2013;521:3358–3370. doi: 10.1002/cne.23354. [DOI] [PubMed] [Google Scholar]
- 31.Bergner A.J., Stamp L.A., Gonsalvez D.G., Allison M.B., Olson D.P., Myers M.G., Jr., Anderson C.R., Young H.M. Birthdating of myenteric neuron subtypes in the small intestine of the mouse. J Comp Neurol. 2014;522:514–527. doi: 10.1002/cne.23423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M.-T., Kuan Y.-H., Wang J., Hen R., Gershon M.D. 5-HT4 receptor-mediated neuroprotection and neurogenesis in the enteric nervous system of adult mice. J Neurosci. 2009;29:9683–9699. doi: 10.1523/JNEUROSCI.1145-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obata Y., Castaño Á., Boeing S., Bon-Frauches A.C., Fung C., Fallesen T., de Agüero M.G., Yilmaz B., Lopes R., Huseynova A., Horswell S., Maradana M.R., Boesmans W., Vanden Berghe P., Murray A.J., Stockinger B., Macpherson A.J., Pachnis V. Neuronal programming by microbiota regulates intestinal physiology. Nature. 2020;578:284–289. doi: 10.1038/s41586-020-1975-8. [DOI] [PubMed] [Google Scholar]
- 34.Osterwalder M., Speziale D., Shoukry M., Mohan R., Ivanek R., Kohler M., Beisel C., Wen X., Scales S.J., Christoffels V.M., Visel A., Lopez-Rios J., Zeller R. HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev Cell. 2014;31:345–357. doi: 10.1016/j.devcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei J., Howard M.J. Targeted deletion of Hand2 in enteric neural precursor cells affects its functions in neurogenesis, neurotransmitter specification and gangliogenesis, causing functional aganglionosis. Development. 2011;138:4789–4800. doi: 10.1242/dev.060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monteiro C.B., Midão L., Rebelo S., Reguenga C., Lima D., Monteiro F.A. Zinc finger transcription factor Casz1 expression is regulated by homeodomain transcription factor Prrxl1 in embryonic spinal dorsal horn late-born excitatory interneurons. Eur J Neurosci. 2016;43:1449–1459. doi: 10.1111/ejn.13214. [DOI] [PubMed] [Google Scholar]
- 37.Mattar P., Stevanovic M., Nad I., Cayouette M. Casz1 controls higher-order nuclear organization in rod photoreceptors. Proc Natl Acad Sci U S A. 2018;115:E7987–E7996. doi: 10.1073/pnas.1803069115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Delfin K., Morris A.C., Snelson C.D., Gamse J.T., Gupta T., Marlow F.L., Mullins M.C., Burgess H.A., Granato M., Fadool J.M. Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proc Natl Acad Sci U S A. 2009;106:2023–2028. doi: 10.1073/pnas.0809439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho G.-S., Park D.-S., Choi S.-C., Han J.-K. Tbx2 regulates anterior neural specification by repressing FGF signaling pathway. Dev Biol. 2017;421:183–193. doi: 10.1016/j.ydbio.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X., Chen M.H., Wu X., Kodani A., Fan J., Doan R., Ozawa M., Ma J., Yoshida N., Reiter J.F., Black D.L., Kharchenko P.V., Sharp P.A., Walsh C.A. Cell-type–specific alternative splicing governs cell fate in the developing cerebral cortex. Cell. 2016;166:1147–1162.e15. doi: 10.1016/j.cell.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugitani Y., Nakai S., Minowa O., Nishi M., Jishage K.-I., Kawano H., Mori K., Ogawa M., Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snijders Blok L., Kleefstra T., Venselaar H., Maas S., Kroes H.Y., Lachmeijer A.M.A., van Gassen K.L.I., Firth H.V., Tomkins S., Bodek S., Õunap K., Wojcik M.H., Cunniff C., Bergstrom K., Powis Z., Tang S., Shinde D.N., Au C., Iglesias A.D., Izumi K., Leonard J., Abou Tayoun A., Baker S.W., Tartaglia M., Niceta M., Dentici M.L., Okamoto N., Miyake N., Matsumoto N., Vitobello A., Faivre L., Philippe C., Gilissen C., Wiel L., Pfundt R., Deriziotis P., Brunner H.G., Fisher S.E. De novo variants disturbing the transactivation capacity of POU3F3 cause a characteristic neurodevelopmental disorder. Am J Hum Genet. 2019;105:403–412. doi: 10.1016/j.ajhg.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dastot-Le Moal F., Wilson M., Mowat D., Collot N., Niel F., Goossens M. ZFHX1B mutations in patients with Mowat-Wilson syndrome. Hum Mutat. 2007;28:313–321. doi: 10.1002/humu.20452. [DOI] [PubMed] [Google Scholar]
- 44.Coyle D., Puri P. Hirschsprung’s disease in children with Mowat-Wilson syndrome. Pediatr Surg Int. 2015;31:711–717. doi: 10.1007/s00383-015-3732-x. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z., Li Q., Diao M., Liu N., Cheng W., Xiao P., Zou J., Su L., Yu K., Wu J., Li L., Jiang Q. Sporadic Hirschsprung disease: mutational spectrum and novel candidate genes revealed by next-generation sequencing. Sci Rep. 2017;7:14796. doi: 10.1038/s41598-017-14835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nurgali K., Stebbing M.J., Furness J.B. Correlation of electrophysiological and morphological characteristics of enteric neurons in the mouse colon. J Comp Neurol. 2004;468:112–124. doi: 10.1002/cne.10948. [DOI] [PubMed] [Google Scholar]
- 47.Furness J.B., Robbins H.L., Xiao J., Stebbing M.J., Nurgali K. Projections and chemistry of Dogiel type II neurons in the mouse colon. Cell Tissue Res. 2004;317:1–12. doi: 10.1007/s00441-004-0895-5. [DOI] [PubMed] [Google Scholar]
- 48.Mazzuoli G., Schemann M. Multifunctional rapidly adapting mechanosensitive enteric neurons (RAMEN) in the myenteric plexus of the guinea pig ileum. J Physiol. 2009;587:4681–4694. doi: 10.1113/jphysiol.2009.177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Avetisyan M., Wang H., Schill E.M., Bery S., Grider J.R., Hassell J.A., Stappenbeck T., Heuckeroth R.O. Hepatocyte growth factor and MET support mouse enteric nervous system development, the peristaltic response, and intestinal epithelial proliferation in response to injury. Journal of Neuroscience. 2015;35:11543–11558. doi: 10.1523/JNEUROSCI.5267-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morarach K., Memic F., Zeisel A., Hochgerner H., Marklund U., Linnarsson S. Single-cell molecular interrogation of enteric nervous system development. Mech Dev. 2017;145:S67. [Google Scholar]
- 51.Rossi J., Herzig K.-H., Võikar V., Hiltunen P.H., Segerstråle M., Airaksinen M.S. Alimentary tract innervation deficits and dysfunction in mice lacking GDNF family receptor alpha2. J Clin Invest. 2003;112:707–716. doi: 10.1172/JCI17995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Creedon D.J., Tansey M.G., Baloh R.H., Osborne P.A., Lampe P.A., Fahrner T.J., Heuckeroth R.O., Milbrandt J., Johnson E.M., Jr. Neurturin shares receptors and signal transduction pathways with glial cell line-derived neurotrophic factor in sympathetic neurons. Proc Natl Acad Sci U S A. 1997;94:7018–7023. doi: 10.1073/pnas.94.13.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baloh R.H., Tansey M.G., Golden J.P., Creedon D.J., Heuckeroth R.O., Keck C.L., Zimonjic D.B., Popescu N.C., Johnson E.M., Jr., Milbrandt J. TrnR2, a novel receptor that mediates neurturin and GDNF signaling through Ret. Neuron. 1997;18:793–802. doi: 10.1016/s0896-6273(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 54.Klein R.D., Sherman D., Ho W.H., Stone D., Bennett G.L., Moffat B., Vandlen R., Simmons L., Gu Q., Hongo J.A., Devaux B., Poulsen K., Armanini M., Nozaki C., Asai N., Goddard A., Phillips H., Henderson C.E., Takahashi M., Rosenthal A. A GPI-linked protein that interacts with Ret to form a candidate neurturin receptor. Nature. 1997;387:717–721. doi: 10.1038/42722. [DOI] [PubMed] [Google Scholar]
- 55.Malin S.A., Molliver D.C., Koerber H.R., Cornuet P., Frye R., Albers K.M., Davis B.M. Glial cell line-derived neurotrophic factor family members sensitize nociceptors in vitro and produce thermal hyperalgesia in vivo. J Neurosci. 2006;26:8588–8599. doi: 10.1523/JNEUROSCI.1726-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gianino S., Grider J.R., Cresswell J. GDNF availability determines enteric neuron number by controlling precursor proliferation. Development. 2003;130:2187–2198. doi: 10.1242/dev.00433. [DOI] [PubMed] [Google Scholar]
- 57.Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. Osteoarthritis Cartilage. 2012;20:256–260. doi: 10.1016/j.joca.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piper M., Pantano L., Mistry M., Khetani R. Single-cell RNA-seq: clustering analysis. in-depth-NGS-data-analysis-course. https://hbctraining.github.io/In-depth-NGS-Data-Analysis-Course/sessionIV/lessons/SC_clustering_analysis.html Available at:
- 61.Dent J.A., Polson A.G., Klymkowsky M.W. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- 62.Smith-Edwards K.M., DeBerry J.J., Saloman J.L., Davis B.M., Woodbury C.J. Profound alteration in cutaneous primary afferent activity produced by inflammatory mediators. Elife. 2016;5 doi: 10.7554/eLife.20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spencer N.J., Smith T.K. Mechanosensory S-neurons rather than AH-neurons appear to generate a rhythmic motor pattern in guinea-pig distal colon. J Physiol. 2004;558:577–596. doi: 10.1113/jphysiol.2004.063586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sang Q., Williamson S., Young H.M. Projections of chemically identified myenteric neurons of the small and large intestine of the mouse. J Anat. 1997;190:209–222. doi: 10.1046/j.1469-7580.1997.19020209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Branchek T.A., Gershon M.D. Time course of expression of neuropeptide Y, calcitonin gene-related peptide, and NADPH diaphorase activity in neurons of the developing murine bowel and the appearance of 5-hydroxytryptamine in mucosal enterochromaffin cells. J Comp Neurol. 1989;285:262–273. doi: 10.1002/cne.902850208. [DOI] [PubMed] [Google Scholar]
- 66.Jin Z., Liu L., Bian W., Chen Y., Xu G., Cheng L., Jing N. Different transcription factors regulate nestin gene expression during P19 cell neural differentiation and central nervous system development. J Biol Chem. 2009;284:8160–8173. doi: 10.1074/jbc.M805632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ngan E.S.-W., Garcia-Barceló M.-M., Yip B.H.-K., Poon H.-C., Lau S.-T., Kwok C.K.-M., Sat E., Sham M.-H., Wong K.K.-Y., Wainwright B.J., Cherny S.S., Hui C.-C., Sham P.C., Lui V.C.-H., Tam P.K.-H. Hedgehog/Notch-induced premature gliogenesis represents a new disease mechanism for Hirschsprung disease in mice and humans. J Clin Invest. 2011;121:3467–3478. doi: 10.1172/JCI43737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Z., Zhang X., Lei H., Lam N., Carter S., Yockey O., Xu M., Mendoza A., Hernandez E.R., Wei J.S., Khan J., Yohe M.E., Shern J.F., Thiele C.J. CASZ1 induces skeletal muscle and rhabdomyosarcoma differentiation through a feed-forward loop with MYOD and MYOG. Nat Commun. 2020;11:911. doi: 10.1038/s41467-020-14684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gehman L.T., Stoilov P., Maguire J., Damianov A., Lin C.-H., Shiue L., Ares M., Jr., Mody I., Black D.L. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed single cell and single nucleus RNA sequencing data are available at GEO Accession: GSE156905.