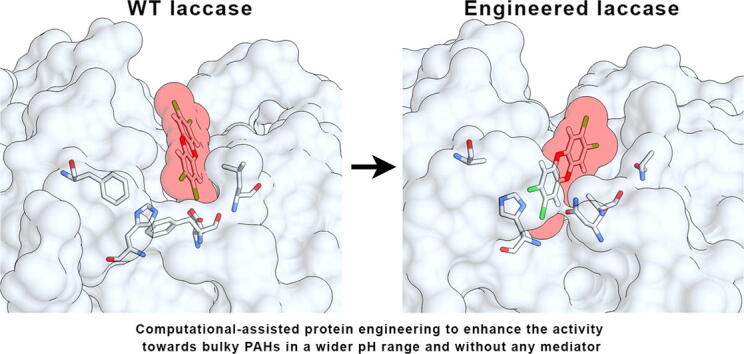
Graphical abstract
Keywords: Trametes versicolor laccase, Site directed mutagenesis, Rational design, Computational docking, Polycyclic aromatic hydrocarbon, Organic dye degradation
Highlights
-
•
Computational-assisted protein engineering of the binding pocket of laccases.
-
•
Mutants have activity increased up to ~ 300% in a broader pH range compared to the WT.
-
•
Enhanced activity towards bulky PAHs in comparison to the WT enzyme.
-
•
Ability to oxidize harmful PAH model compounds (dyes) that the WT enzyme cannot modify.
-
•
Higher oxidation levels without mediators compared to the WT laccase with mediators.
Abstract
Laccases are among the most sought-after biocatalyst for many green applications, from biosensors to pollution remedial, because they simply need oxygen from the air to oxidize and degrade a broad range of substrates. However, natural laccases cannot process large and toxic polycyclic aromatic hydrocarbons (PAHs) except in the presence of small molecules, called mediators, which facilitate the reaction but are inconvenient for practical on-field applications. Here we exploited structure-based protein engineering to generate rationally modified fungal laccases with increased ability to process bulky PAHs even in a mediator-less reaction. Computational simulations were used to estimate the impact of mutations in the enzymatic binding pocket on the ability to bind and oxidize a selected set of organic compounds. The most promising mutants were produced and their activity was evaluated by biochemical assays with phenolic and non-phenolic substrates. Mutant laccases engineered with a larger binding pocket showed enhanced activity (up to ~ 300% at pH 3.0) in a wider range of pH values (3.0–8.0) in comparison to the wild type enzyme. In contrast to the natural laccase, these mutants efficiently degraded bulky and harmful triphenylmethane dyes such as Ethyl Green (up to 91.64% after 24 h), even in the absence of mediators, with positive implications for the use of such modified laccases in many green chemistry processes (e.g. wastewater treatment).
1. Introduction
Laccases are glycosylated multicopper oxidase enzymes, biologically well characterized and constitutively expressed during primary metabolism by fungi, plants, prokaryotes and insects [1]. Depending on the source, these enzymes exhibit various catalytic functions roughly divided into three categories: cross-linking of small molecules, degradation of polymers and ring cleavage of aromatic compounds [2]. The broad substrate specificity of the ligand-binding pocket allows laccases to react with a large number of natural and anthropogenic compounds, mostly phenols or arylamines [1], [3]. The ability to metabolize various phenolic and non-phenolic molecules, as well as many xenobiotics, depends on the redox potential of the enzyme: the higher the potential, the greater the oxidation capabilities [4]. This redox potential is directly connected to the enzyme source and structure of the copper centers: laccases from white-rot fungi show higher redox potential than those from bacteria and plants [5]. Furthermore, in contrast to other oxidases that use H2O2 to oxidize substrates, the presence of copper allows laccases to exploit O2, a much more readily available electron acceptor [6]. These features make laccases the ideal biocatalysts for a wide range of biotechnological applications [4], [7]. For instance, naturally occurring fungal laccases have been already utilized in different technological sectors such as the food, paper, textile and paint industry, as well as for biofuels, bioremediation and biosensors [5], [7].
Polycyclic aromatic hydrocarbons (PAHs) and chlorinated aromatic compounds are the main pollutants present in water and soil, in which they accumulate due to their low solubility [8]. Moreover, they are toxic and most of them carcinogenic [9], [10], [11]. Laccases have been shown to be useful for the bioremediation of PAHs through oxidative degradation [9], [12], [13]. In particular, laccases from Trametes versicolor can aid the removal of a wide range of xenobiotics, including emerging contaminants and endocrine disrupting chemicals (EDCs) [8], [9], [10], [12], [13], [14]. Several authors demonstrated this activity by means of decolorization assays [15], [16], [17], [18]: a clear relationship between the oxidation of pollutants and the decolorization of specific dyes has been established both for peroxidases and laccases.
Laccases can directly degrade only small PAHs due to their low redox potential (450–800 mV, compared to > 1 V for peroxidases) or to steric hindrance [15], [19]. The oxidation of large, recalcitrant and toxic aromatic compounds, including bulky PAHs that require higher redox potentials, is not usually achievable [12], [20]. Minor laccase activity was observed only against hydroxylated pollutants such as 2-hydroxydibenzofuran [21], hydroxyl-PCBs [22], [23] and chlorinated hydroxybiphenyls [24], [25]. Unfortunately, the enzymatic reactivity towards these xenobiotics is negligible for practical applications [12], [15].
Redox mediators, small molecules such as N-hydroxybenzotriazole (HBT), can increase the laccase oxidation rate by mediating its radical-catalyzed activity towards large compounds with high redox potentials [7], [26], [27]. However, these compounds are usually expensive, toxic or otherwise unsuitable for practical applications [28]. Indeed, the need for mediators increases the operational costs of the enzymatic biotransformations (e.g. for biofuels production), as well as the complexity of in field applications such as environmental bioremediation or monitoring through biosensing.
In the last decade, to increase laccase performance and satisfy process requirements this biocatalyst was subjected to protein engineering, whose best results were recently reviewed [5], [6], [27], [29]. Among these, even the most successful examples rely on the presence of mediator to increase the reaction yields [6], [30], [31], [32]. As an attractive alternative to the use of mediators, we set forth to rationally modify a natural laccase to optimize its catalytic activity for the degradation of bulky and toxic PAHs. Fungal laccases are best suited to this aim because i) they have redox potential higher than laccases from other species; ii) they have high stability in extracellular environments due to their glycosylation pattern [33]; iii) they can be adapted to non-natural environments including organic solvents, human blood and ionic liquids [5]. On the downside, their production usually requires eukaryotic heterologous hosts (e.g. yeasts).
Fungal laccases from T. versicolor are the most active against xenobiotics and emerging pollutants [34], [35]. In particular, the T. versicolor lccβ is the most active isoform toward PAHs and has been successfully expressed in yeast (e.g. Pichia pastoris), showing high stability in different conditions [36]. Therefore, we chose this isoform as starting point for the rational design of a mutated enzyme that is either more efficient in catalysis or capable of oxidizing a wider range of toxic PAHs in a mediator-less reaction. We used a combination of computational simulations and molecular biology to generate rationally engineered T. versicolor laccases with increased ability to process large and persistent xenobiotics in the absence of any mediator. These mutated enzymes were expressed in P. pastoris, purified and characterized by means of biophysical and biochemical assays. Finally, the ability of the engineered laccases to metabolize PAHs was assessed by decolorization assays of selected dyes, showing increased reactivity toward big aromatic compounds even in a mediator-less configuration.
2. Material and methods
2.1. Computational docking and rational design
Computational docking simulations were conducted as previously reported [37]. Mutants model structures were predicted by I-TASSER server [38] using as template the x-ray structure of T. versicolor laccase complexed with 2,5-xylidine (1KYA) [3], hereby referred to as wild type (WT) enzyme. PDB files of WT and mutant model structures were prepared for docking using the “dockprep” tool in UCSF Chimera package [39]. A selection of known substrates (2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS), 2,6-dimethoxyphenol (2,6-DMP) and N-hydroxybenzotriazole (HBT) and pollutants (anthracene (ANT), fluoranthene (FLUO), benzo-a-pyrene (BP), dibenzofuran (DF), 2,3,7,8-tetrachloro-p-dibenzodioxin (TCDD), pentachlorophenol (PCP), triclosan (TRI), 17β-estradiol (BE) and bisphenol A (BPA)) was computationally docked to WT and engineered laccases. These molecules were prepared for docking using MarvinSketch [Marvin 6.1.7, 2014 (http://www.chemaxon.com/)] and OpenBabel [40]. Computational docking was performed with the SwissDock server (www.swissdock.ch) based on the docking software EADock DSS [41] with user-defined parameters such as a simulation box (15x15x15 Å) centered on the known catalytic binding site. Molecular analysis of docking results was performed with the UCSF Chimera package [39] and Pymol [42].
2.2. P. pastoris culture conditions for laccase expression
The WT and mutated laccases were produced and purified from P. pastoris to measure their activity experimentally. To this aim, P. pastoris transformants (carrying WT and mutated laccase gene, see Supplementary data for details on mutagenesis and yeast transformation) were grown in 50 mL BMGY at 30 °C and 150 rpm to an OD600 of 10–15 (preculture phase); 0.2 mL from −80 °C stocks were used as inoculum. Overnight cultures were harvested by centrifugation (3000 rpm, 10 min, 4 °C) and washed twice with PBS. The pellets were resuspended in 500 mL Buffered Minimal Methanol (BMM, 13.4 g YNB, 0.1 M NaP pH 6.0, 0.5% (v/v) methanol, 0.4 mg biotin) supplemented with 0.3 mM CuSO4 to 1.0 OD600 (methanol induction phase). Then, they were cultured at 20 °C, 150 rpm and methanol (final concentration 0.5% (v/v)) was added daily to maintain the induction. Samples were taken daily for spectrophotometric determination of cell growth (OD600), protein concentration and laccase activity (see 2.4). The optimization of this protocol is reported in the Supplementary data (see Fig. S1, S2, Table S1 and their discussion).
2.3. Laccase purification
Yeast cells were harvested by centrifugation (3000 rpm, 10 min, 4 °C) 12–96 h after the methanol induction. The supernatants were concentrated with concentrator (VivaSpin 15R, cut-off 5 kDa, Sartorius Stedim Biotech GmbH) or by means of a Cogent (Millipore, cut-off 50 kDa) and then dialyzed overnight (cut-off 5 kDa) against 20 mM NaP pH 7.5, 1 M NaCl. Dialyzed supernatants were loaded on an affinity column (Hi-Trap Chelating HP, 5 mL, GE Healthcare) previously equilibrated with 0.1 M NiSO4 and then with 20 mM NaP pH 7.5, 1 M NaCl. When the sample was loaded, the column was washed with 20 mM NaP, pH 7.5, 200 mM NaCl, and then eluted with 20 mM NaP, pH 7.5, 150 mM NaCl, 200 mM imidazole. The laccase activity of the eluted fractions was assayed as stated below and the one with the highest activity was concentrated and loaded on a size exclusion column (Superdex 200 10/300 GL, 25 mL, GE Healthcare), previously equilibrated with 20 mM NaP, pH 7.5, 150 mM NaCl. The laccase activity of eluted fractions was evaluated (see 2.4), and the ones with the highest activity were blended and concentrated. The protein concentration was determined by measuring the OD at 280 nm using the laccase parameters (MW = 54.5 kDa; ε280 = 60,975 L/mol*cm) calculated by Expasy [43].
2.4. Enzyme activity assays
The enzymatic activity of laccase (50 μL of culture supernatant or diluted purified enzyme) was evaluated in 96 well plates (Corning Incorporated, 3799) at 22 °C with a phenolic (1 mM 2,6-DMP) or non-phenolic substrate (0.1 mM ABTS), using a Biotek Powerwave 340 plate-reader. The reaction mixture was prepared by diluting the enzyme sample in 20 mM sodium acetate pH 3.0–6.0 or 20 mM NaP buffer pH 6.0–8.0. The oxidation of ABTS led to an increase of absorbance at 420 nm (ε420 = 36,000 L/mol*cm), while the oxidation of 2,6-DMP led to an increase at 468 nm (ε468 = 49,600 L/mol*cm) [44]. One activity unit (UI) is defined as the amount of enzyme that oxidizes 1 μmol of substrate per minute. The specific activity values (UI/mg) were computed considering the total protein concentration (mg/mL) of the enzyme solution. All the measurements were performed at least twice.
2.5. Decolorization assays
Purified laccases (WT and mutants) were tested for enzymatic decolorization of dyes listed in Table S2. The test was performed with or without 1 mM redox mediator (Glycine, Cysteine, Imidazole, HBT) in 20 mM sodium acetate buffer pH 3.0–6.0, containing 8 μg/mL of purified enzyme. The reaction mixtures were incubated in 96-well plates, in the dark, at 22 °C and the decolorization activity of WT and mutated laccases was determined spectrophotometrically during time, as a relative decrease of OD (D % = (OD0 – ODt)/OD0 × 100, where OD0 is the initial OD of the reaction mixture and ODt is the residual value after the treatment) at the maximum wavelength of absorption of the dyes (Table S2). Controls without enzyme and in the presence of different mediators were included. All the tests were performed at least in duplicate.
3. Results and discussion
3.1. Rational engineering of fungal laccase
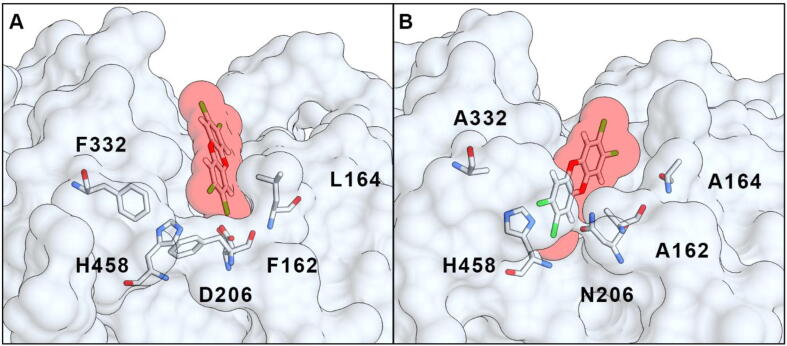
In order to change the substrate specificity and improve the enzymatic activity of the T. versicolor lccβ laccase, sequence and three-dimensional structure analysis were employed to mutate specific residues in its active site. As a first step, computational docking simulations were used to obtain 3D atomic models of various known laccase substrates (ABTS, 2,6-DMP, HBT) and pollutants (ANT, FLUO, BP, DF, TCDD, PCP, TRI, BE, BPA) bound to lccβ, since no experimental structure of laccase complexed with a PAH is available. The x-ray structure of lccβ [3] was used as initial template. Docking of the small ligands to lccβ was restricted to the known binding site in order to improve local sampling and accuracy. This analysis showed how some xenobiotics can interact with the catalytic residues in the binding pocket (e.g. His458, suggested to be a primary electron acceptor [3]). It is known, for example, that anthracene can be faintly oxidized by laccase, a reaction enhanced by the presence of mediators [15], [19]; indeed, this compound can be fully accommodated in the enzyme binding pocket in close proximity to His458. On the other hand, larger PAHs such as TCDD could not fit in the binding pocket of the WT enzyme in a pose suitable to efficiently interact with the catalytic His458 (Fig. 1A). This likely explains the lack of enzymatic activity for such compounds [20]. Indeed, steric hindrance negatively affects the laccase oxidation capabilities and rates [12]. Redox mediators can allow degradation of some larger PAHs but are expensive and practically inconvenient for industrial and environmental applications [28]. Therefore, we aimed to improve the catalysis of large PAHs by designing laccase mutants (Table 1) that would enlarge the catalytic binding pocket to make it more accessible to large PAHs. A second set of mutants aimed to alter the distribution of charged residues within the binding pocket to foster more favorable electrostatic interactions between enzyme and chlorinated or other negatively charged compounds. A strategy similar to the former was effectively applied to enable the degradation of TCDD by a rat cytochrome P450 by replacing a phenylalanine with an alanine (F240A), resulting in a more accessible active site [45].
Fig. 1.
Computational docking model of the TCDD molecule binding to WT laccase (A) or its M6 (F162A/L164A/D206N/F332A) mutant (B).
Table 1.
Free energy of binding (ΔG, kcal/mol) values of laccase-ligand complexes estimated by docking simulations. For each substrate, values more negative than the WT are green-shaded (theoretically resulting in better affinity) and values more positive red-shaded. The most favorable value for each compound is in bold. The column ‘size’ indicates mutations that enlarge the binding pocket; ‘charge’ indicates mutations that alter the charge distribution. The leftmost column indicates the name used in this manuscript for each mutant, M1–M6.
 |
In order to enlarge the lccβ active site the bulky Phe162, Leu164 and Phe332 were mutated to smaller but still hydrophobic alanine residues. Phe265 and Phe337 were not considered for mutagenesis due to the known detrimental effect of their substitution [36], [46]. Although Leu164 does not appear to occupy the binding cavity in the available x-ray structure of laccases, our computational simulations showed that its long side-chain can occlude the binding pocket, reducing substrate accessibility. It is plausible that some of the plasticity typical of protein structure might be escaping crystallography investigation. As illustrated later in the manuscript, the L164A substitution yielded an enzyme with higher activity than the WT. Sole reliance on crystallographic data without the aid of computational simulations would not have considered such mutation.
In the second set of mutants Asp206 was mutated to Asn, which has similar steric properties but lacks a negative charge. Sequence alignment of the binding site of fungal, bacterial and plants laccases showed that this aspartate is conserved in laccases from Basidiomycetes and Ascomycetes but is mutated to asparagine in plants and alanine in bacteria. Asp206 is supposed to stabilize the cation intermediate formed during the oxidation of phenolic substrates [3]; we mutated it to evaluate the influence of its negative charge on the binding and degradation of chlorinated PAHs.
Overall, we designed and tested six lccβ laccase mutants, hereby called M1 to M6, resulting from the combination of the above substitutions as summarized in Table 1 and Fig. S3. We then used computational simulations to investigate their effect on the binding and interaction with known substrates and PAHs. First, the 3D structure of each mutated laccase was predicted according to standard homology modelling techniques. Then, each substrate/pollutant was docked with each mutant, evaluating their ability to bind in the proximity of the catalytic residues both by visual structural analysis and by simulated binding energy.
The computational docking process generates several thousand models for each enzyme/substrate pair, which are evaluated by a “scoring function” that is supposed to recognize favorable binding interaction between enzyme and substrate. However, although surprisingly accurate and precise models are typically found among the thousands, scoring functions often fail to discriminate accurate from inaccurate solutions. The model with the best score, in other words, is not always the most accurate. Therefore, in order to limit the impact of modelling errors, instead of relying on the single model with the best computational score, for each substrate-mutant combination we first clustered all models by structural similarity; the best-scoring representative models within each cluster were kept and further analyzed based on structural and visual considerations, all others discarded. For instance, a model that would put the reactive moiety far away from the active enzymatic residues would be discarded. For substrates with known degradation pathway, we also analyzed the models in view of the known intermediates. All models equally satisfying the above computational and structural analysis (up to ~ 100) were taken into consideration for mutant design.
Docking can provide also a computational estimate of the free energy of binding (ΔG) but these values are notoriously error-prone. In order to evaluate the impact of mutations on protein–ligand interaction we, therefore, also relied on visual inspection and structural considerations, taking into consideration that proteins are flexible and able to accommodate ligands in different configurations. In other words, we exploit computational models to visualize what it could be rather than believing that a single model can provide a reliable picture of what it is. We have employed this approach for the rational design of protein mutants with considerable success in several cases ranging from small molecules to antibody binding [37], [47], [48].
The estimated free energy of binding (ΔG) obtained by the best models after the application of the visual refinement was reported in Table 1.
Mutations aimed at enlarging the laccase binding pocket to increase its accessibility had the bigger impact on substrate binding, not surprisingly. Computational simulations showed that the mutants with the largest pocket were able to accommodate bulky PAHs that cannot fit in the natural, WT active site. For instance, TCDD cannot enter the WT binding pocket (Fig. 1A) whereas it binds close to the catalytic residues in the M6 (F162A/L164A/D206N/F332A) engineered laccase (Fig. 1B), as reflected in more favorable calculated binding energy (Table 1, M6). By contrast, when small ligands (such as 2,6-DMP, HBT or ANT) are docked to the mutants with the largest binding pocket, they can assume several different conformations; however, not all of them lead to favorable contact between substrate and catalytic residues.
Although computational docking cannot predict the enzymatic activity, the simulated free energy of binding (Table 1) suggests that different mutants might be better suited to different substrates. For instance, M1 (F162A/L164A) has a more favorable binding energy for HBT than the WT. The binding of larger PAHs, instead, is favored by the introduction of more substitutions: e.g., the M6 binding pocket (F162A/L164A/D206N/F332A) has a higher calculated affinity to FLUO, BP, DF, TCDD and BPA, if compared with other mutants. The predicted affinity of the M3 mutant (F332A) for BE, a large compound that carry a hydroxyl moiety, is worse than the WT, possibly due to the loss of aromatic and Van der Vaals interactions between Phe332 and the organic ligand, in line with existing literature [46]. At the same time, this mutation does not affect BPA, whose computed affinity is comparable in both WT and F332A, in agreement with the reported degradation of this xenobiotic (72% and 79%, respectively), previously measured by Galli et al. [46].
Structural analysis of the substrates docked to mutants carrying the D206N substitution did not reveal evident advantages for this mutation; if anything, the simulations tended to find multiple conformations for each enzyme-bound substrate, suggesting that D206N could destabilize the interaction. In particular, the D206N mutation leads to an impaired interaction of the enzyme with all the molecules that carry an –OH group (2,6-DMP, HBT, BE, BPA) and with ABTS, but shows better interaction with chlorinated molecules (TCDD, PCP and TRI), as expected by the rational design.
Overall, the models obtained from docking simulations showed promising results in agreement with our proposed goals. We thus proceeded to produce WT and mutated laccase and test their catalytic activity with biochemical assays.
3.2. Mutants preliminary characterization: evaluation of the specific activity toward phenolic and non-phenolic compounds
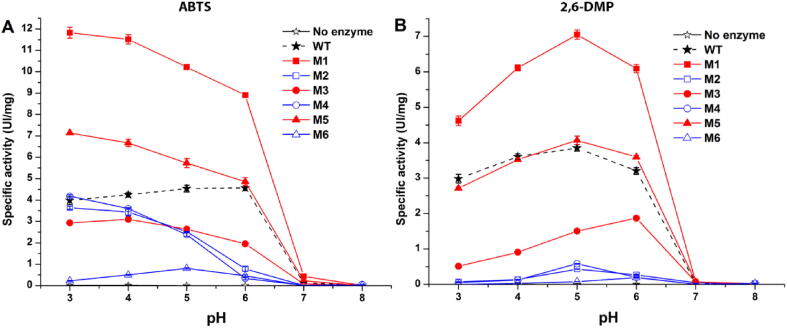
Laccases are able to process both phenolic and non-phenolic compounds. The optimal specific activity is achieved at different pH for different compounds [44]. By mutating the binding pocket the substrate specificity can be changed that is one of the main goals of our work. Indeed, we aimed to change the binding pocket to accommodate larger molecules such as PAHs, but any variation in the binding site could have affected the activity towards both phenolic and non-phenolic substrates, too. Therefore, as a first assessment of the result of the protein engineering, the enzymatic activity of WT and mutants was evaluated in the 3 to 8 pH range by a colorimetric assay with either a non-phenolic (ABTS, Fig. 2A) or a phenolic compound (2,6-DMP, Fig. 2B) that are often used to test the activity of laccases.
Fig. 2.
Enzymatic activity of WT and mutant laccases against non-phenolic ABTS (A) and phenolic 2,6-DMP (B) at different pH (x axis). The average of at least 3 repetitions is shown; the error bars are the standard deviation. WT laccase is in black; mutants carrying the D206N substitution are in blue, those with a larger binding pocket are in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Two mutants (M1 and M5) showed higher enzymatic activity (up to ~ 300% at pH 3.0) than the WT towards ABTS (Fig. 2A). M1 also had increased activity (183% more active than WT at pH 5.0) for 2,6-DMP (Fig. 2B). Instead, all mutants with the D206N charge-altering substitution (M2, M4 and M6, blue in Fig. 2) had lower activity than the WT with both substrates. The D206N mutation, therefore, influences not only the oxidation of phenolic compounds like 2,6-DMP, as previously reported [49], but also that of non-phenolic ABTS.
Enlargement of the active site had positive but not easily predictable effects on the enzymatic activity. M1 (F162A, L164A) had increased activity for both ABTS and 2,6-DMP: if the former molecule is large and only partially buried in the WT binding pocket [50], the latter is small and should have no problem fitting in the WT pocket. So, no benefit would be expected by pocket enlargement. Similarly, if accessibility was the only criteria it would be hard to explain why M1 (F162A, L164A) resulted in higher activity whereas the equally accessible M5 (F162A, L164A, F332A) caused diminished activity. Evidently, hydrophobic and aromatic interactions lost with the F332A substitution as well as shape complementarity are also important. Incidentally, the M3 (F332A) mutant also has reduced activity for both substrates, as an evidence of the importance of this residue, as previously speculated from the docking results in paragraph 3.1.
Another partially unexpected outcome is that altering the size and hydrophobicity of the active pocket (M1, M3 and M5 mutants) results in altered pH dependency for ABTS but not for 2,6-DMP. WT lccβ has peak ABTS activity between pH 5.0 and 6.0. By contrast, the activity of the M1, M3 and M5 mutants decreases with pH. It is plausible that the larger binding pocket allows the interaction of ABTS in a different orientation, resulting in altered atomic interactions and pH dependence. All WT and mutated laccases showed no activity at pH 7.0 or above.
In contrast to ABTS, the pH-dependent activity profiles for 2,6-DMP are very similar for WT and mutants (Fig. 2B): a typical bell-shaped trend with a maximum at pH 5.0 is observed (except for M3 that has a maximum shifted at pH 6.0). It was suggested that this bell-profile is due to the balance of two opposite mechanisms: the redox potential difference between the reducing substrate and the T1 copper, favored by higher pH values for a phenolic substrate, and the binding of a hydroxide anion to the copper in the T2/T3 center, which inhibits the activity of laccase at higher pH [51]. These effects are negligible for non-phenolic substrates such as ABTS and the pH-activity profile decreases monotonically as the pH increases (Fig. 2A). It is noteworthy that the pH value corresponding to the maximal activity obtained with 2,6-DMP is 1.5 pH unit higher than that reported by other authors for the same enzyme expressed in the yeast Yarrowia lipolytica [52], [53]. Maybe this derives from a specific glycosylation pattern by different yeast strains, since it is known that the extent and pattern of glycosylations are crucial for laccase activity [54].
The change in substrate specificity of the mutants is highlighted also by calculating the activity ratio of the two different substrates (ABTS/2,6-DMP), as reported in Fig. S3. The D206N mutation generated the greatest variation of substrate specificity. At the same time, the results of the simulations were compared to the experimentally measured enzymatic activity, showing that it qualitatively agrees with the estimated free energy of binding calculated during computational docking (Fig. S4).
3.3. Evaluation of the reactivity against PAH by means of decolorization assay
As previously reported, the decolorization of dyes is a reliable indicator of the enzymatic biodegradation of xenobiotics by laccases [16], [28]. Indeed, one of the simplest, fastest and safest methods to evaluate their activity toward PAHs involves the enzymatic breakdown of dyes with chemical structure resembling that of aromatic organic pollutants, whose oxidation is easily and cheaply measured by the loss of color. Although degradation of the chromophore does not strictly imply degradation of the entire molecule, there is ample literature supporting the use of decolorization assays as a proxy for overall degradation [15], [18], [55]. Furthermore, many dyes used in industrial processes are toxic, mutagenic or carcinogenic [54], [56]; it is conceivable to use laccases to degrade and safely remove their presence in the environment, especially if mediators can be avoided. Thus, we developed a simple decolorization assay to assess the ability of our engineered laccases to degrade dyes and, by relation, toxic organic pollutants.
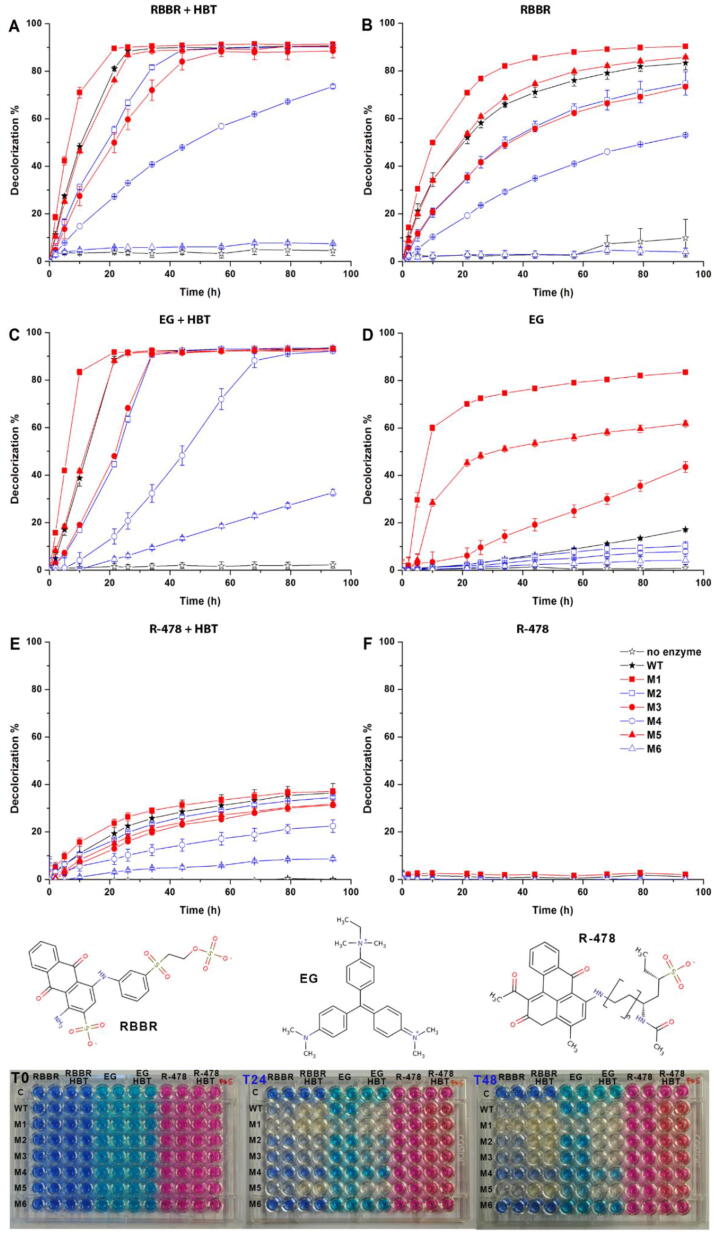
Initially, we tested the ability of a commercial T. versicolor laccase (WT enzyme) to decolorize a selection of dyes responsible for important environmental pollution [56] with or without a mediator in the reaction mixture. Only three of the tested dyes were effectively processed by the WT enzyme in the absence of a mediator (Table S2). We then evaluated the ability of our WT and engineered laccases to decolorize three dyes belonging to different structural classes: the anthraquinone Remazol Brilliant Blue R (RBBR); the triphenylmethane Ethyl Green (EG); the polymeric R-478 (Fig. 3). Among them, only RBBR was effectively degraded by the natural protein in the absence of any mediator. The HBT molecule, the most effective mediator during preliminary tests, was selected for the following decolorization assays.
Fig. 3.
Decolorization % of selected dyes performed at pH 5.0 with ((A) RBBR, (C) EG, (E) R-478) or without HBT ((B) RBBR, (D) EG, (F) R-478). Representative pictures of the 96-well plates decolorization assay are shown at the bottom.
RBBR is particularly relevant for the textile industry because it is commonly used as a raw material for the production of polymeric dyes. It belongs to a widespread class of toxic and recalcitrant organopollutants whose degradation is achieved only by high-redox-potential enzymes or by using redox mediators [17], [56].
The highest oxidation of RBBR at 24 h was obtained by M1 with (90.08 ± 0.53%) or without HBT (76.75 ± 0.01%). The mutant performs better than the WT both in term of percentage of oxidized molecules and in reaction rate (Fig. 3A and B). Notably, the M1 mutant without mediator reaches the same level of oxidation as the WT with mediator, strengthening the idea that our mutants may find practical use in mediator-free reactions.
The oxidation rate and efficiency followed the same order (M1 > WT > M5 > M2 > M3 > M4 > M6) with or without HBT, but it was faster in its presence (steeper slope of the curves). The presence of the mediator had a larger impact on the activity of the enzymes with smaller binding pocket, suggesting that a larger pocket allows favorable interaction between substrate (RBBR) and catalytic residues. Finally, mutants carrying the D206N mutation had lower activity, similarly to the previously illustrated cases with ABTS and 2,6-DMP.
The decolorization of EG in the presence of HBT was similar to that of RBBR (M1 > WT > M5 > M3 > M2 > M4 > M6), with M1 achieving the highest oxidation at 24 h (91.64 ± 0.79%). Remarkably, whereas the WT enzyme was not able to oxidize EG in the absence of mediators, the dye was efficiently degraded by the mutants with larger binding pocket (M1, M3 and M5), with M1 achieving the highest oxidation (72.50 ± 0.72%) at 24 h (Fig. 3C and D).
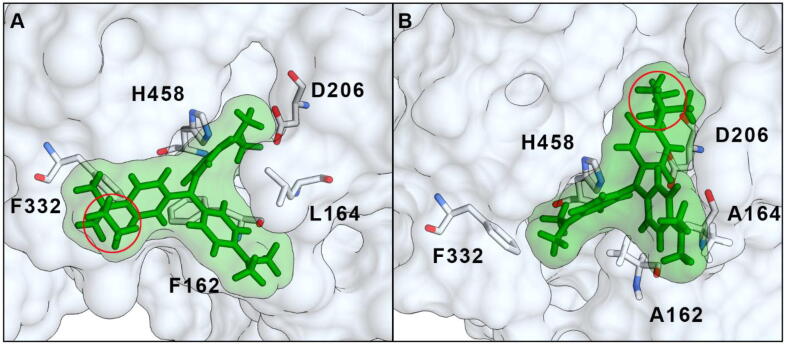
These results highlight the effectiveness of the rational engineering. Indeed, triphenylmethane dyes such as EG are non-planar and, apart from the smaller molecules belonging to this structural class such as Malachite Green or Fuchsin, it is suggested that these compounds are not oxidized by laccases alone because they are larger than the binding pocket. An extreme example is reported by Chhabra et al.: a laccase from the white-rot Cyathus bulleri was able to fully degrade Malachite Green even without mediators, whereas the decolorization of the bulky Acid Violet 17 was possible only in the presence of a redox mediator [57]. Similarly, the results obtained with Malachite Green and Methyl Violet by Salami et al. highlighted that only one N-methyl moiety can drastically reduce the reactivity of laccases towards similar triphenylmethane dyes and only the presence of HBT produced a comparable decolorization of these two compounds [58]. The presence of mediators is thus necessary for the reaction to take place. Enlarging the binding pocket, as in our mutants, allows these bulky substrates to get in contact with the catalytic residues without the need for mediators. Indeed, the reactivity of the M1 laccase towards EG did not change significantly with or without HBT because of the broader substrate specificity of this mutant. Actually, computational docking shows that although the EG molecule can enter the binding pocket of the WT laccase, the enlarged pocked of M1 allows reorientation of the molecule with the ethyl moiety (situated outside the enzymatic site in the WT laccase) positioned in close proximity to His458 and Asp206. The central quaternary C atom of EG is also closer to His458 in the M1 mutant than in the WT (Fig. 4). Probably, the closest position of the central quaternary carbon atom to the catalytic residues of M1 enhanced the degradation of the dye compared to the WT, in accordance with the proposed degradation pathway of triphenylmethane dyes that can be demethylated, with no decolorization, or hydroxylated (on the quaternary C) to the carbinol form with the destruction of the chromophore group [55], [57], [59], [60]. It is worth noticing that factors other than steric hindrance (e.g. higher redox potential) maybe contributed to the better activity of M1 towards EG, but further studies must be carried out to disclose these aspects. Anyway, the attempts to modify the redox potential towards higher values by engineering the enzyme by random or site-specific mutagenesis resulted effective only in case of low redox potential laccases [26]. Moreover, Monza et al. recently verified that no significant changes of the redox potential can be registered after the site-directed mutagenesis of the active site [61]. Then, we do not think that the redox potential of the mutants we obtained by rational design was changed, compared to the starting WT enzyme.
Fig. 4.
Comparison between the docking simulation of EG with WT (A) and M1 mutant (B). The highest ranking selected models are shown. The red circles indicate the ethyl moiety of EG. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Finally, the R-478 dye, usually exploited to screen for xenobiotics degraders among white rot fungi isolates [16], was not oxidized by either natural or mutated laccases in the absence of mediator and only limited oxidation (26.36 ± 1.73% at 24 h by M1, Fig. 3E) was obtained in the presence of HBT, with ranking of activity analogous to the previous dyes (M1 marginally better than WT). Polymeric R-478 is significantly larger than the other two dyes and cannot fit in the laccases binding pocket even when enlarged as in the engineered mutants. For this reason it can be only degraded by high-redox potential enzymes in the presence of mediators, as broadly reported [8], [12], [15], [16], [30].
Overall, all of these results highlighted the effectiveness of the rational design of the T. versicolor laccase lccβ, to boost, even in the absence of mediators, its ability to bind and metabolize aromatic dyes resembling large PAHs that are not efficiently degraded by the WT enzyme.
4. Conclusions
Structure-based protein engineering guided by computational simulations increased the laccase enzymatic activity up to ~ 300% towards known phenolic and non-phenolic substrates, in a broader pH range, compared to the WT enzyme. This is expected to facilitate on-field applications by relaxing the need for strict pH control. Mutants with an enlarged binding pocket were able to degrade organic dyes often used as models for the degradation of bulky PAHs, including those for which the WT has little to no enzymatic activity such as. EG. Remarkably, degradation to a level comparable to that of the WT with mediators was observed for the mutants even in the absence of mediators. These results highlight the potentiality of our engineered laccases for the efficient degradation of recalcitrant PAHs and, most likely, of other organic pollutants of similar structure. This may prove valuable in practical field applications. In particular, the best mutant (M1, F162A/L164A) could find use in the mediator-less degradation of PAHs in bioremediation, wastewater treatment (e.g. textile wastewater) or industrial processes (Kraft pulp bleaching). The ability to process pollutants in the absence of mediators is expected to reduce cost and simplify the design and usage of apparatus and protocols for on-field applications.
Further work may very well allow the design of other mutants capable of degrading other small organic molecules without the use of expensive and inconvenient mediators, for a future application of our engineered laccase in green processes.
Author contributions
AC, FB, LM and LV conceived the presented idea; All authors discussed and decided the methods to be used; FB, MP, LS, LM, and LV supervised the project; AC and MB carried out the experiments; AC and LS analysed the data and prepared figures, tables and artworks; AC prepared the first draft of the manuscript; AC, LM and LV obtained the funding. All authors discussed the results and contributed to the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to acknowledge the Erasmus LLP program exchange for the studentship used by A.C. for his visiting period at the IRB of Bellinzona and the Swiss National Science Foundation (SNF) for the grant 31003A_182270.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.03.017.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Giardina P., Faraco V., Pezzella C., Piscitelli A., Vanhulle S., Sannia G. Laccases: a never-ending story. Cell Mol Life Sci. 2010;67:369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dwivedi U.N., Singh P., Pandey V.P., Kumar A. Structure – function relationship among bacterial, fungal and plant laccases. J Mol Catal B Enzym. 2011;68:117–128. doi: 10.1016/j.molcatb.2010.11.002. [DOI] [Google Scholar]
- 3.Bertrand T., Jolivalt C., Briozzo P., Caminade E., Joly N., Madzak C. Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics. Biochemistry. 2002;41:7325–7333. doi: 10.1021/bi0201318. [DOI] [PubMed] [Google Scholar]
- 4.Alcalde M. Engineering the ligninolytic enzyme consortium. Trends Biotechnol. 2015;33:155–162. doi: 10.1016/j.tibtech.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Mate D.M., Alcalde M. Laccase engineering: from rational design to directed evolution. Biotechnol Adv. 2015;33:25–40. doi: 10.1016/j.biotechadv.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Stanzione I., Pezzella C., Giardina P., Sannia G., Piscitelli A. Beyond natural laccases: extension of their potential applications by protein engineering. Appl Microbiol Biotechnol. 2020;104:915–924. doi: 10.1007/s00253-019-10147-z. [DOI] [PubMed] [Google Scholar]
- 7.Moreno A.D., Ibarra D., Eugenio M.E., Tomás-Pejó E. Laccases as versatile enzymes: from industrial uses to novel applications. J Chem Technol Biotechnol. 2020;95:481–494. doi: 10.1002/jctb.6224. [DOI] [Google Scholar]
- 8.Husain M., Husain Q. Applications of redox mediators in the treatment of organic pollutants by using oxidoreductive enzymes: a review. Crit Rev Environ Sci Technol. 2013:1–42. doi: 10.1080/10643380701501213. [DOI] [Google Scholar]
- 9.Xu P., Du H., Peng X., Tang Y., Zhou Y., Chen X. Degradation of several polycyclic aromatic hydrocarbons by laccase in reverse micelle system. Sci Total Environ. 2020;708 doi: 10.1016/j.scitotenv.2019.134970. [DOI] [PubMed] [Google Scholar]
- 10.Shakerian F., Zhao J., Li S. Recent development in the application of immobilized oxidative enzymes for bioremediation of hazardous micropollutants – A review. Chemosphere. 2020;239 doi: 10.1016/j.chemosphere.2019.124716. [DOI] [PubMed] [Google Scholar]
- 11.Li L., Lai C., Huang F., Cheng M., Zeng G., Huang D. Degradation of naphthalene with magnetic bio-char activate hydrogen peroxide: Synergism of bio-char and Fe–Mn binary oxides. Water Res. 2019;160:238–248. doi: 10.1016/j.watres.2019.05.081. [DOI] [PubMed] [Google Scholar]
- 12.Majeau J., Brar S.K., Tyagi R.D. Laccases for removal of recalcitrant and emerging pollutants. Bioresour Technol. 2010;101:2331–2350. doi: 10.1016/j.biortech.2009.10.087. [DOI] [PubMed] [Google Scholar]
- 13.Bilal M., Iqbal H.M.N., Barceló D. Persistence of pesticides-based contaminants in the environment and their effective degradation using laccase-assisted biocatalytic systems. Sci Total Environ. 2019;695 doi: 10.1016/j.scitotenv.2019.133896. [DOI] [PubMed] [Google Scholar]
- 14.Gasser C.A., Ammann E.M., Shahgaldian P., Corvini P.F.X. Laccases to take on the challenge of emerging organic contaminants in wastewater. Appl Microbiol Biotechnol. 2014;98:9931–9952. doi: 10.1007/s00253-014-6177-6. [DOI] [PubMed] [Google Scholar]
- 15.Alcalde M., Bulter T., Arnold F.H. Colorimetric assays for biodegradation of polycyclic aromatic hydrocarbons by fungal laccases. J Biomol Screen. 2002;7:547–553. doi: 10.1177/1087057102238629. [DOI] [PubMed] [Google Scholar]
- 16.Field J.A., de Jong E., Feijoo-Costa G., de Bont J.A.M. Screening for ligninolytic fungi applicable to the biodegradation of xenobiotics. Trends Biotechnol. 1993;11:44–49. doi: 10.1016/0167-7799(93)90121-O. [DOI] [Google Scholar]
- 17.Palmieri G., Cennamo G., Sannia G. Remazol Brilliant Blue R decolourisation by the fungus Pleurotus ostreatus and its oxidative enzymatic system. Enzyme Microb Technol. 2005;36:17–24. doi: 10.1016/j.enzmictec.2004.03.026. [DOI] [Google Scholar]
- 18.Chroma L., Macek T., Demnerova K., Mackova M. Decolorization of RBBR by plant cells and correlation with the transformation of PCBs. Chemosphere. 2002;49:739–748. doi: 10.1016/s0045-6535(02)00397-1. [DOI] [PubMed] [Google Scholar]
- 19.Canas A.I., Alcalde M., Plou F., Jesus Martinez M., Martinez A.T., Camarero S. Transformation of polycyclic aromatic hydrocarbons by laccase is strongly enhanced by phenolic compounds present in soil. Environ Sci Technol. 2007;41:2964–2971. doi: 10.1021/es062328j. [DOI] [PubMed] [Google Scholar]
- 20.Camoni I., Di Muccio A., Pontecorvo D., Rubbiani M., Silano V., Vergori L. Lack of in vitro oxidation of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in the presence of laccase from Polyporus versicolor fungus. Chemosphere. 1983;12:945–949. [Google Scholar]
- 21.Jonas U., Hammer E., Schauer F., Bollag J. Transformation of 2-hydroxydibenzofuran by laccases of the white rot fungi Trametes versicolor and Pycnoporus cinnabarinus and characterization of oligomerization products. Biodegradation. 1998;8:321–328. doi: 10.1023/a:1008220120431. [DOI] [PubMed] [Google Scholar]
- 22.Fujihiro S., Higuchi R., Hisamatsu S., Sonoki S. Metabolism of hydroxylated PCB congeners by cloned laccase isoforms. Appl Microbiol Biotechnol. 2009;82:853–860. doi: 10.1007/s00253-008-1798-2. [DOI] [PubMed] [Google Scholar]
- 23.Schultz A., Jonas U., Hammer E., Schauer F. Dehalogenation of chlorinated hydroxybiphenyls by fungal laccase. Appl Environ Microbiol. 2001;67:4377–4381. doi: 10.1128/AEM.67.9.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullah M.A., Bedford C.T., Evans C.S. Reactions of pentachlorophenol with laccase from Coriolus versicolor. Appl Microbiol Biotechnol. 2000;53:230–234. doi: 10.1007/s002530050013. [DOI] [PubMed] [Google Scholar]
- 25.Kordon K., Mikolasch A., Schauer F. Oxidative dehalogenation of chlorinated hydroxybiphenyls by laccases of white-rot fungi. Int Biodeterior Biodegradation. 2010;64:203–209. doi: 10.1016/j.ibiod.2009.10.010. [DOI] [Google Scholar]
- 26.Rodgers C.J., Blanford C.F., Giddens S.R., Skamnioti P., Armstrong F.A., Gurr S.J. Designer laccases: a vogue for high-potential fungal enzymes? Trends Biotechnol. 2009;28:63–72. doi: 10.1016/j.tibtech.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Guan Z.B., Luo Q., Wang H.R., Chen Y., Liao X.R. Bacterial laccases: promising biological green tools for industrial applications. Cell Mol Life Sci. 2018;75:3569–3592. doi: 10.1007/s00018-018-2883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zucca P., Cocco G., Sollai F., Sanjust E. Fungal laccases as tools for biodegradation of industrial dyes. Biocatalysis. 2016;1:82–108. doi: 10.1515/boca-2015-0007. [DOI] [Google Scholar]
- 29.Pardo I., Camarero S. Laccase engineering by rational and evolutionary design. Cell Mol Life Sci. 2015;72:897–910. doi: 10.1007/s00018-014-1824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mateljak I., Monza E., Lucas M.F., Guallar V., Aleksejeva O., Ludwig R. Increasing redox potential, redox mediator activity, and stability in a fungal laccase by computer-guided mutagenesis and directed evolution. ACS Catal. 2019;9:4561–4572. doi: 10.1021/acscatal.9b00531. [DOI] [Google Scholar]
- 31.Novoa C., Dhoke G.V., Mate D.M., Martínez R., Haarmann T., Schreiter M. KnowVolution of a fungal laccase toward alkaline pH. ChemBioChem. 2019;20:1458–1466. doi: 10.1002/cbic.201800807. [DOI] [PubMed] [Google Scholar]
- 32.Yin Q., Zhou G., Peng C., Zhang Y., Kües U., Liu J. The first fungal laccase with an alkaline pH optimum obtained by directed evolution and its application in indigo dye decolorization. AMB Express. 2019;9:1–13. doi: 10.1186/s13568-019-0878-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh Arora D., Kumar Sharma R., Arora D.S., Sharma R.K. Ligninolytic Fungal Laccases and Their Biotechnological Applications. Appl Biochem Biotechnol. 2010;160:1760–1788. doi: 10.1007/s12010-009-8676-y. [DOI] [PubMed] [Google Scholar]
- 34.Ba S., Arsenault A., Hassani T., Jones J.P., Cabana H. Laccase immobilization and insolubilization: from fundamentals to applications for the elimination of emerging contaminants in wastewater treatment. Crit Rev Biotechnol. 2012;33:1–15. doi: 10.3109/07388551.2012.725390. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen L.N., Hai F.I., Yang S., Kang J., Leusch F.D.L., Roddick F. Removal of pharmaceuticals, steroid hormones, phytoestrogens, UV-filters, industrial chemicals and pesticides by Trametes versicolor: Role of biosorption and biodegradation. Int Biodeterior Biodegrad. 2014;88:169–175. doi: 10.1016/j.ibiod.2013.12.017. [DOI] [Google Scholar]
- 36.Koschorreck K., Richter S.M., Swierczek A., Beifuss U., Schmid R.D., Urlacher V.B. Comparative characterization of four laccases from Trametes versicolor concerning phenolic C-C coupling and oxidation of PAHs. Arch Biochem Biophys. 2008;474:213–219. doi: 10.1016/j.abb.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Ferrero V.E.V., Pedotti M., Chiadò A., Simonelli L., Calzolai L., Varani L. Rational modification of estrogen receptor by combination of computational and experimental analysis. PLoS ONE. 2014;9:1–10. doi: 10.1371/journal.pone.0102658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF Chimera – A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delano WL. The PyMOL Molecular Graphics System 2002.
- 43.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein Identification and Analysis Tools on the ExPASy Server. Proteomics Protoc. Handb., Totowa, NJ: Humana Press; 2005, p. 571–607. doi:10.1385/1-59259-890-0:571.
- 44.Johannes C., Majcherczyk A. Laccase activity tests and laccase inhibitors. J Biotechnol. 2000;78:193–199. doi: 10.1016/s0168-1656(00)00208-x. [DOI] [PubMed] [Google Scholar]
- 45.Shinkyo R., Sakaki T., Takita T., Ohta M., Inouye K. Generation of 2,3,7,8-TCDD-metabolizing enzyme by modifying rat CYP1A1 through site-directed mutagenesis. Biochem Biophys Res Commun. 2003;308:511–517. doi: 10.1016/S0006-291X(03)01439-6. [DOI] [PubMed] [Google Scholar]
- 46.Galli C., Gentili P., Jolivalt C., Madzak C., Vadalà R. How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Appl Microbiol Biotechnol. 2011;91:123–131. doi: 10.1007/s00253-011-3240-4. [DOI] [PubMed] [Google Scholar]
- 47.Simonelli L., Pedotti M., Beltramello M., Livoti E., Calzolai L., Sallusto F. Rational engineering of a human anti-dengue antibody through experimentally validated computational docking. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0055561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.La Spina R., Ferrero V.E.V., Aiello V., Pedotti M., Varani L., Lettieri T. Label-free biosensor detection of endocrine disrupting compounds using engineered estrogen receptors. Biosensors. 2018;8:1–15. doi: 10.3390/bios8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kallio J.P., Auer S., Jänis J., Andberg M., Kruus K., Rouvinen J. Structure-Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds. J Mol Biol. 2009;392:895–909. doi: 10.1016/j.jmb.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 50.Enguita F.J., Marcal D., Martins L.O., Grenha R., Henriques A.O., Lindley P.F. Substrate and dioxygen binding to the endospore coat laccase from bacillus subtilis. J Biol Chem. 2004;279:23472–23476. doi: 10.1074/jbc.M314000200. [DOI] [PubMed] [Google Scholar]
- 51.Xu F., Berka R.M., Wahleithner J.A., Nelson B.A., Shuster J.R., Brown S.H. Site-directed mutations in fungal laccase: effect on redox potential, activity and pH profile. Biochem J. 1998;334:63–70. doi: 10.1042/bj3340063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madzak C., Mimmi M.C., Caminade E., Brault A., Baumberger S., Briozzo P. Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Eng Des Sel. 2006;19:77–84. doi: 10.1093/protein/gzj004. [DOI] [PubMed] [Google Scholar]
- 53.Jolivalt C., Madzak C., Brault A., Caminade E., Malosse C., Mougin C. Expression of laccase IIIb from the white-rot fungus Trametes versicolor in the yeast Yarrowia lipolytica for environmental applications. Appl Microbiol Biotechnol. 2005;66:450–456. doi: 10.1007/s00253-004-1717-0. [DOI] [PubMed] [Google Scholar]
- 54.Herkommerová K., Dostál J., Pichová I. Decolorization and detoxification of textile wastewaters by recombinant Myceliophthora thermophila and Trametes trogii laccases. 3 Biotech. 2018;8:1–13. doi: 10.1007/s13205-018-1525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shanmugam S., Ulaganathan P., Swaminathan K., Sadhasivam S., Wu Y.R. Enhanced biodegradation and detoxification of malachite green by Trichoderma asperellum laccase: Degradation pathway and product analysis. Int Biodeterior Biodegrad. 2017;125:258–268. doi: 10.1016/j.ibiod.2017.08.001. [DOI] [Google Scholar]
- 56.Mechichi T., Mhiri N., Sayadi S. Remazol Brilliant Blue R decolourization by the laccase from Trametes trogii. Chemosphere. 2006;64:998–1005. doi: 10.1016/j.chemosphere.2005.12.061. [DOI] [PubMed] [Google Scholar]
- 57.Chhabra M., Mishra S., Sreekrishnan T.R. Laccase/mediator assisted degradation of triarylmethane dyes in a continuous membrane reactor. J Biotechnol. 2009;143:69–78. doi: 10.1016/j.jbiotec.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 58.Salami F., Habibi Z., Yousefi M., Mohammadi M. Covalent immobilization of laccase by one pot three component reaction and its application in the decolorization of textile dyes. Int J Biol Macromol. 2018;120:144–151. doi: 10.1016/j.ijbiomac.2018.08.077. [DOI] [PubMed] [Google Scholar]
- 59.Yang J., Yang X., Lin Y., Ng T.B., Lin J., Ye X. Laccase-catalyzed decolorization of malachite green: Performance optimization and degradation mechanism. PLoS ONE. 2015;10:1–14. doi: 10.1371/journal.pone.0127714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Casas N., Parella T., Vicent T., Caminal G., Sarrà M. Metabolites from the biodegradation of triphenylmethane dyes by Trametes versicolor or laccase. Chemosphere. 2009;75:1344–1349. doi: 10.1016/j.chemosphere.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 61.Monza E., Lucas M.F., Camarero S., Alejaldre L.C., Martínez A.T., Guallar V. Insights into laccase engineering from molecular simulations: Toward a binding-focused strategy. J Phys Chem Lett. 2015;6:1447–1453. doi: 10.1021/acs.jpclett.5b00225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.