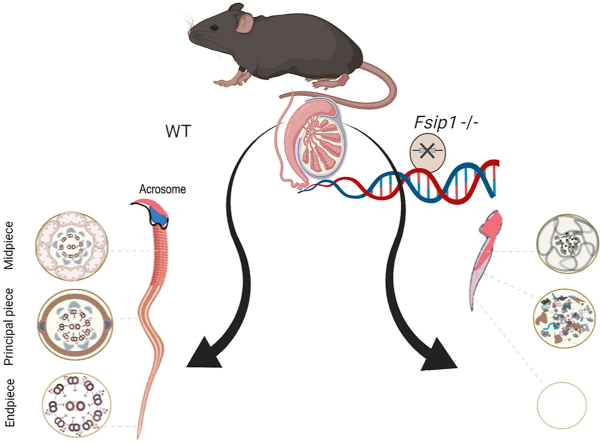
Abstract
Fibrous sheath interacting protein 1 (Fsip1) is a cytoskeletal structural protein of the sperm flagellar proteome. A few studies have reported that it plays a vital role in the tumorigenesis and cancer progression. However, little is known about the role of Fsip1 in spermatogenesis and mammalian sperm flagellogenesis. Fsip1 protein showed the highest expression in round spermatids, and was translocated from nucleus to the anterior region of the elongating spermatid head. To investigate its role we constructed homozygous Fsip1 null (Fsip1−/−) mice. We found that the homozygous Fsip1−/− mutant mice were infertile, with a low sperm count and impaired motility. Interestingly, a subtle phenotype characterized by abnormal head shape, and flagella deformities was observed in the sperm of Fsip1−/− mutant mice similar to the partial globozoospermia phenotype. Electron microscopy analysis of Fsip1−/− sperm revealed abnormal accumulation of mitochondria, disrupted axoneme and retained cytoplasm. Testicular sections showed increased cytoplasmic vacuoles in the elongated spermatid of Fsip1–/–mice, which indicated an intraflagellar transport (IFT) defect. Using proteomic approaches, we characterized the cellular components and the mechanism underlying this subtle phenotype. Our result indicated that Fsip1–/–downregulates the formation of acrosomal membrane and vesicles proteins, intraflagellar transport particles B, and sperm flagellum components. Our results suggest that Fsip1 is essential for normal spermiogenesis, and plays an essential role in the acrosome biogenesis and flagellogenesis by attenuating intraflagellar transport proteins.
Keywords: Fsip1, Ift20, Spermiogenesis, Intraflagellar transport particles B, Globozoospermia
Graphical abstract
Highlights
-
•
Disruption of Fsip1 leads to infertility with partial globozoospermia phenotype.
-
•
Homozygous deletion of Fsip1 alters spermiogenesis.
-
•
Fsip1 Knockout disrupts acrosome vesicle formation.
-
•
Fsip1 motif analysis involves in internal fertilization.
1. Introduction
Spermatogenesis is a highly organized differentiation process, that begins with the differentiation of diploid spermatogonium and ends with the generation of haploid spermatozoa [1]. Spermatogenesis involved three main cellular stages: spermatogonial mitosis, spermatocyte meiosis, and spermatid spermiogenesis [2]. Spermiogenesis is a complex metamorphosis process, during which round spermatids undergo structural modification and become mature spermatozoa [3]. In this process, spermatocytes undergo unique and orchestrated morphological changes, which cannot be seen in any other cell types [4]. Spermiogenesis include five major processes: chromatin-remodeling, acrosome biogenesis, mitochondrial rearrangement, flagellum assembly and cytoplasmic removal [5]. Morphologically, spermiogenesis can be divided into 16 steps in the mouse, which can be distinguished by the shape of the acrosome in developing round spermatids [6]. The acrosome membrane is a unique organelle located at the anterior part of the sperm head, that plays a crucial role in fertilization through sperm capacitation and acrosome reaction [7].
Globozoospermia is a severe form of monomorphic teratozoospermia, characterized by abnormally headed spermatozoa, lack of acrosome, and flagellum defects [8]. It is responsible for less than 0.1% of male infertility [9]. Until now, few genes defects have been identified that cause globozoospermia including, DPY19L2 [10], DNAH17 [11], SPATA16 [12], PICK1, CCIN, C7orf61, ZPBP, and CCDC62 and C2CD6 [13]. However, due to the lack of in vitro models, the molecular mechanisms of globozoospermia remain largely unknown.
Fibrous sheath interacting protein 1 (FSIP1) has been recently recognized as a tumor/testis antigen [14]. A few studies reported that FSIP1 may play a vital role in tumorigenesis and cancer progression [[14], [15], [16], [17], [18], [19], [20]]. Downregulation of FSIP1 promotes autophagy, and attenuates mitochondrial biogenesis by enhancing the activation of AMP-activated protein kinase in breast cancer [17]. In addition, knockdown of FSIP1 decreases cell proliferation and induces apoptosis in bladder urothelial carcinoma via inhibition of the PI3K/AKT pathway [21]. However, little is known about the role of this gene in spermatogenesis and mammalian sperm flagellogenesis. In order to fill this gap, we disrupted the Fsip1 gene and explored the role of Fsip1 in spermatogenesis and spermiogenesis.
The intraflagellar transport (IFT) complex is a unique bi-directional transport system, that plays an important role for appropriate assembly and maintenance of ciliated/flagellar cells and intracellular vesicular trafficking [22]. IFT protein complexes can be devided into A-complex and B-complex, which are functionally involved in the movement of large protein complexes, called “IFT particles” from the cell body compartment to the flagellum tip [23,24]. Among the IFT proteins, IFT20 is the most dynamic. It is responsible for the transfer of ciliary membrane proteins from the Golgi complex to the cilia [25].
In the current study, we constructed homozygous Fsip1 deficient (Fsip1−/−) mice, which were infertile, and displayed a remarkable decrease in sperm counts and motility. In addition, Fsip1−/− mutant mice showed abnormal morphology including a misshapen sperm head, and a dysmorphic/absent midpiece and principal piece. In order to clarify these alterations at the molecular level, we used tandem mass tag-based (TMT) proteomics to identify differentially expressed proteins and potential underlying mechanisms. Our results suggest that Fsip1 knockout affects the expression of spermiogenesis core proteins and attenuates intraflagellar transporters associated with acrosome and flagellogenesis. Our study shows for first time that Fsip1 plays an essential role in spermiogenesis, and it plays a crucial role in the acrosome biogenesis and flagella assembly.
2. Materials and methods
2.1. Ethical statement
All the animal experiments in this study were conducted according to the ethical guidelines for the use of experimental animals issued by the Animals Care and Use Committee of Guangzhou Medical University. All protocols were conducted in accordance with the national and institution guidelines.
2.2. Generation of homozygous Fsip1 null mice
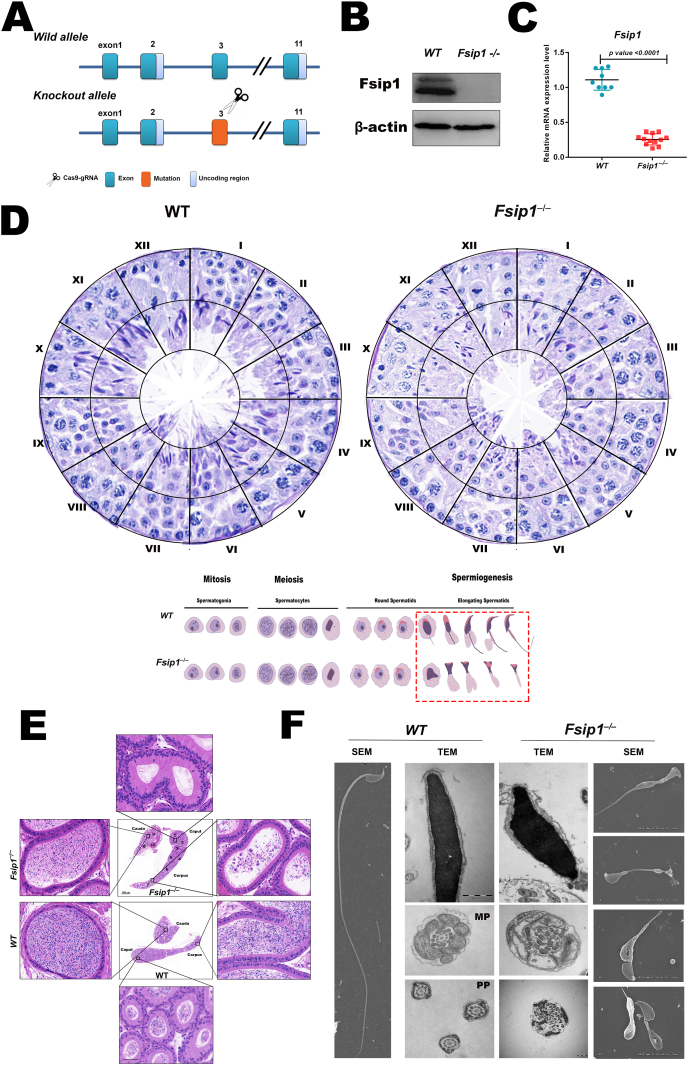
The Fsip1 null mice line (Fsip1−/−) was generated on C57BL/6J genetic background by non-homologous end joining (NHEJ) using CRISPR/Cas9 technology. The Fsip1 gene (ENSMUSG00000027344) is located on the chromosome-2 (Chr2:118130424-118256966), contains 11 exons. Cas9 mRNA and small guide RNAs (sgRNA) were performed as previously described elsewhere [26]. Four sgRNAs targeting the 3rd exon of Fsip1 gene were designed (Fig. 1 A). The mixture of Cas9 mRNA and sgRNA was injected into fertilized embryos of super-ovulated C57BL/6J female mouse. The injected zygotes were cultured in vitro at 37 °C and 5% CO2, and the obtained blastocysts were then transferred into the uterus of pseudo pregnant C57BL/6J female mouse. In order to determine the genotype of the offspring, PCR was carried out with genomic DNA. The primers used were as follows: forward: 5′ CCCTGGGGTGTATCTGCTTCTA 3′ and reverse: 5′ GGCTGGCTTACTCACCATTACCT 3'. F0 mice were selected to mate with wild type C57BL/6J mice, and F1 mice were obtained and genotyped. Homozygous Fsip1 null (Fsip1 −/−) mice were produced by mating between Fsip1 +/− heterozygous mice.
Fig. 1.
Effect of Fsip1 knockout on spermatogenesis (A) Schematic diagram showing the targeted mutation site used to construct the Fsip1 knockout (Fsip1−/−) mice model. (B) Western blot analysis showing the testicular protein expression level of Fsip1 in the WT and Fsip1–/–mice. (C) Real time qPCR results presented as dot plot showing the testicular expression level of Fsip1 mRNA in WT and Fsip1–/–mice (D) Wheel graph representing bright field images of testicular tissue sections stained with Periodic Acid-Schiff (PAS), showing the gross morphology, and spermatogenesis alterations between in Fsip1−/− compared with WT mice at the 12 stages of spermatogenesis. The schematic-diagram represents the morphological changes between WT and Fsip1−/− mice. (E) H&E staining of WT and Fsip1−/− mice epididymal sections. (F) Scanning and transmission electron microscope assessment of epididymal cauda spermatozoa.
2.3. Fertility assessment
To examine the Fsip1−/− mice fertility. We cohabited a sexually mature Fsip1−/− male mouse (8–12 weeks old) with two virgin wild-type (WT) female mice for more than one month, and checked the vaginal plug every morning. Normal mating behavior was indicated if vaginal plug was observed, then the female mouse was placed in a separate cage, and another female mouse was placed in the cage for another round of mating. The male was deemed as infertile if it didn't show evidence of mating (presence of copulatory plug) or induced pregnancy after two months of co habitation. The female mice were euthanized to confirm the lack of pregnancy at the end of the two months. Fertility test was performed on at least 10 mice of each genotype.
2.4. Evaluation of sperm motility and morphology
Sperm quality parameters including sperm concentration, and progressive sperm motility were evaluated according to WHO standard. Cauda epididymis were collected immediately from adult male mice after euthanization, and washed with PBS, then several incisions were made and placed in 1 ml of Sperm Rinse™ medium (Vitrolife). The Cauda epididymal spermatozoa were released for evaluation by the swim-up method for 30 min at 37 °C, and then the supernatant was collected in a clean tube. For sperm motility assessment, 10 μl of the supernatant was placed in Makler chamber, and examined under phase contrast microscope using Computer-assisted semen Analyzer (CASA) system (Microptic S.L.).
2.5. Scanning and transmission electron microscope
Sperm morphology was evaluated by scanning electron microscope (SEM). Briefly, sperm samples were isolated as described above, washed in PBS, and fixed overnight at 4 °C in 2.5% glutaraldehyde and 2% paraformaldehyde in 0.15 M sodium phosphate buffer. Then the sperm was washed and collected on nucleopore filters or glass cover slip, dried at critical point, and coated with gold/palladium. Samples were examined at 20 kV using HITACHI S-3000 N scanning electron microscope.
2.6. Transmission electron microscope
Testis and sperm were fixed with 2.5% glutaraldehyde in 0.2 M cacodylate buffer overnight, washed with 0.2 M PBS, then the tissues were cut into about 1 mm3, and soaked in 1% OsO4 in 0.2 M cacodylate buffer for 2 h at 4 °C. The samples were dehydrated in a series of graded ethanol and embedded in resin. Ultrathin sections were cut using an ultramicrotome, stained with uranyl acetate and lead citrate, and observed using HITACHI H-7500 transmission electron microscope at 80 kV.
2.7. Sperm immunostaining
The cauda epididymal spermatozoa were isolated as described early, and smears were made on clean glass slides, allowed to dry in air, and fixed with 4% PFA or 95% ethanol, permeabilized with freshly prepared 0.3% Triton X-100 in PBS, blocked with 10% goat serum for 1 h at room temperature (RT), and then incubated overnight at 4 °C with optimal dilution of primary antibody diluted in blocking buffer (Table S1). On the second day, the slides were allowed to stand for 1 h at RT, washed with PBS, and then incubated with Secondary Antibody Alexa Fluor 488 Goat anti-Rabbit IgG (H + L) for 1 h at RT. Finally, the slide was washed and mounted using anti-fade mounting media with DAPI. In order to observe the sperm midpiece (mitochondria) the sperms were stained with Mito-tracker Deep Red (Invitrogen, USA). For the co-distribution of two different proteins, we used two antibodies from different species (e.g., Rabbit anti mouse target 1 and mouse anti mouse target 2, if the target is mouse protein), and a mixture of two secondary antibodies raised in different species (with two different fluorochromes). The images were captured with a high magnification fluorescence microscope or confocal microscope (Leica Microsystems).
2.8. Duolink proximity ligation assay
The proximity ligation assay (PLA) was conducted using Duolink In Situ reagents (Sigma, USA). Briefly, sperm cells were fixed and incubated with rabbit anti-Fsip1 and mouse anti-Acrv1 antibodies. Then a pair of oligonucleotide-labeled anti-rabbit and anti-mouse PLA probes used to label the primary antibodies, if the two proteins in close proximity a PLA red fluorescent signal will be generated. Finally, the slide washed and mounted using anti-fade mounting media with DAPI. The images were taken using confocal microscope (Leica Microsystems).
2.9. Testicular tissue processing and morphological evaluation
Testis/epididymis tissues were excised and fixed in modified Davidson's Fluid (mDF) (ServiceBio, Wuhan, China). The tissues were dehydrated in ascending grades of ethanol, and embedded in paraffin. Then 5 μm thick sections were prepared. For histological evaluation, sections were stained with hematoxylin-eosin according to the manufacturer's instructions. PAS staining was used to stain the testicular sections with periodic acid solution and Schiff's reagent (Cat# 395B, Sigma-Aldrich). Histology and morphology were observed and captured using PANNORAMIC slide scanners (3DHITECH, Hungary), and images were processed and exported using CaseViewer™ (version 2.4).
2.10. Tissue immunostaining
Paraffin fixed sections were hydrated in descending grades of ethanol, then antigen retrieval carried out using citrate buffer (pH = 6) in low heat microwave for 20 min. Then slides were permeabilized with 0.3% Triton X-100, blocked with 10% goat serum at RT for 1 h, and incubated overnight with primary antibodies in humidity chamber at 4 °C (Table S1). The slides were then allowed to stand for 1 h at RT, followed by two washes with PBS for 5 min, then incubated with stages marker fluorescence lectin, followed by the secondary antibody (Goat anti-rabbit Alexa Fluor 488 or 568), and mounted with fluoroshield medium with DAPI (Cat# ab104139, abcam). PANNORAMIC slide scanner was used to acquire images (3DHITECH, Hungary), and CaseViewer software was used to process and export images (version 2.4).
2.11. Western blotting
The total protein was extracted for western blot analysis. Briefly, about 50–100 mg of testes were homogenized on ice with pre-chilled RIPA lysis buffer containing a 10 mM cocktail protease inhibitor (Roche) and PMSF, the homogenate allowed to set for 30 min with occasional shaking, and centrifuged at 14000 rpm for 15 min. The protein concentration was measured by BCA method, and then denatured with 4× Laemmli Sample Buffer (Cat#1610747, BioRad) at 95 °C for 10 min. Denatured protein was separated in SDS-polyacrylamide gel, and transferred to PVDF membrane (Millipore®). After blocking with skimmed milk, the membrane was incubated with the optimal antibody dilution (Table S1). β-actin was used as the loading control.
2.12. Immunoprecipitation (IP) and Co-immunoprecipitation (CO-IP)
For investigating the protein-protein interaction, we used Pierce Classic Magnetic Immunoprecipitation (IP) and Co-immunoprecipitation (CO-IP) kit (Cat # 88804, Thermo Scientific, USA). Briefly, the testes were homogenized with lyses buffer containing PMSF and cocktail protease inhibitors. Then about 1500 μg protein was incubated with 5 μg of IP antibody (Table S1) or IgG (as a negative control) overnight at 4ᵒC on rotator, followed by incubation with A/G magnetic beads for 1 h at RT. Finally, the mixture was washed and eluted. The whole cell lysate (Input) and the immune-precipitant or IgG was subjected to western blotting to confirm the interaction.
2.13. Tandem mass tag (TMT) proteomic analysis
Total proteins were extracted from testis, and Tandem mass tags (TMT) proteomics were conducted as described elsewhere [27]. Gene set enrichment analysis was performed using GSEA software v3.1.0 (www.broadinstitute.org/gsea/index.jsp) separately against different protein sets: Acrosomal vesicle associated proteins, Midpiece associated proteins, principal piece associated proteins; Sperm motility associated proteins; and Intraflagellar transporter proteins.
2.14. Domain and conserved motifs analysis
Protein domains of Fsip1 were predicted by the NCBI's Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Conserved motifs were predicted by MEME (Multiple EM for Motif Elicitation) motif discovery tool [28] (http://meme-suite.org/tools/meme). Amino acid sequence of Fsip1 of eight species, Homo sapiens (NP_001311267.1), Mus musculus (NP_001361601.1), Gallus gallus (XP_015143418.1), Pelodiscus sinensis (XP_014429496.1), Salmo salar (XP_014065924.1), Acanthaster planci (XP_022107759.1), Mizuhopecten yessoensis (XP_021347285.1) and Stylophora pistillata (PFX17399.1) were included. The minimum and maximum width of each motif was set to be 30 aa and 100 aa, respectively. Each motif must be recognized in at least 2 sequences. The visual representations of motifs were generated by TBtools [29].
2.15. Homology modeling and protein-protein docking
Three-dimensional structural models of Fsip1 and Ift20 were predicted by the I-TASSER web tool [29] (http://zhang.bioinformatics.ku.edu/I-TASSER/). The models with the highest C-score were selected for docking. Flexible protein-protein docking for Fsip1 and Ift20 was performed using HADDOCK webserver (https://bianca.science.uu.nl/haddock2.4/submit/1) [30]. The active and passive residues of Fsip1 and Ift20 used to define the ambiguous interaction restraints (AIRs) in docking were predicted by the CPORT server (http://alcazar.science.uu.nl/services/CPORT/) [31]. The 1st complex model with the top cluster (the lowest HADDOCK score) was selected. Visual representation and analysis of models were conducted using software package visual molecular dynamics (VMD) [32].
2.16. RNA isolation and quantitative real time PCR
The total RNA was isolated from the frozen testicular tissues by homogenization in TRIzol Reagent (Cat# 15596026, Life technologies, USA). The synthesis of cDNA was performed on 5 μg of total RNA using ThermoScript™ RT-PCR system kit (Cat# 11146-024, Invitrogen, USA). The cDNA obtained was used as a qPCR template mixed with PowerUp SYBR Green Master Mix (Cat#A25742, Applied Biosystems) as recommended by the manufacturer, the reaction mixture processed on Quant 6 flexi Applied Biosystems platform. The primers used Fsip1-forward: 5' CAGCCAGGGTTTCATTAGGCA 3'; Reverse: 5' GCACTGGAGAGTCCTGAGTC 3'. The ddCt comparative method used to analyze the data. β-actin was used as an internal reference control.
3. Results
3.1. Fsip1 homozygous null mice reveal abnormal spermiogenesis
The mice model of Fsip1–/–was successfully generated using CRISPR/Cas9 genome editing technology. Homozygous loss of Fsip1 was genotyped and confirmed. The lack of Fsip1 expression in the mutant mice was examined and validated using western blotting (Fig. 1B) and qPCR (Fig. 1C). Histological examination of testicular sections showed that there were no gross anomalies in mitosis and meiosis in Fsip1–/–mice compared with wild-type (WT) mice. However, in the final developmental phase of spermiogenesis, when the round spermatid undergoes structural modulation to become an elongated spermatid,a subtle phenotype was observed in Fsip1 −/− mutant mice sperm that could be defined as an abnormal shape of the elongated spermatid head; this was first identified in step 9 (Fig. 1D). In normal conditions, from step 9 to 12 the structure of the mice elongating sperm head undergoes structural modification and becomes scimitar-shaped with a pointed tip. The alteration of the sperm head occurring in this transition phase in Fsip1−/− mice indicated an essential role for Fsip1 in the development of post-meiotic spermatids. In steps 10–16, compared with WT mice, the Fsip1−/− phenotype had a tendency to be accentuated, with decreased spermatid count. Most of the remaining spermatids had an abnormal head shape, and short or incompletely formed flagella. Moreover, we observed spermatide phagocytosis in the seminiferous epithelium of the Fsip1−/− mice. In addition, the sperm cells that had completed the process of spermatogenesis were further tracked by analyzing the epididymal sections. We found that the morphology of most Fsip1−/− sperm was abnormal, and in drastically reduced number compared with WT sperm (Fig. 1E).
To further characterize the Fsip1–/–phenotype, we isolated and examined the epididymal cauda spermatozoaby electron microscopy. Our results showed that the morphology of epididymal cauda spermatozoa in Fsip1–/–mice was abnormal. Some spermatozoa had abnormal head morphology, midpieces were missing, and the flagella were short and incomplete. Moreover, by using transmission electron microscopy (TEM), we confirmed the misassembly of the main ultrastructural components of sperm flagella, such as mitochondria, outer dense fiber and “9 + 2” axonemal structures (Fig. 1F). These characteristics indicated that the sperm cell development was impaired due to improper spermatid assembly. Based on this observation, we propose that the role Fsip1 is not only a fibrous sheath assembly protein, but also an essential protein involved in the basic organization of the elongated spermatid structure.
The percent progressive motility of Fsip1–/–sperm were significantly decreased (Table 1). Compared to the WT mice, most spermatozoa in Fsip1−/− mutant mice were obviously immotile. Remarkably, the parameters of sperm movement in forward direction were significantly decreased, including curvilinear velocity (VCL), straight‐line velocity (VSL), and angular path velocity (VAP) (Supplementary video).
Table 1.
Sperm motility assessment using Computer-assisted semen analyzer (CASA system).
| Parameter | WT |
Fsip1−/− |
P value |
|---|---|---|---|
| Mean ± SD | Mean ± SD | ||
| Concentration Mill/ml | 41.2 ± 14.93 | 23.18 ± 9.80 | 0.0538 |
| Motility | |||
| A(%) | 7.42 ± 0.97 | 0.00 ± 0.00 | 0.0000049 |
| B(%) | 24.37 ± 4.60 | 0.03 ± 0.04 | 0.00004 |
| C(%) | 22.96 ± 1.38 | 0.27 ± 0.18 | 0.00000005 |
| D(%) | 45.24 ± 6.87 | 99.70 ± 0.19 | 0.000004 |
| VCL | 124.7 ± 3.68 | 13.74 ± 9.78 | 0.000001 |
| VSL | 38.2 ± 3.79 | 3.62 ± 2.61 | 0.000007 |
| VAP | 56.6 ± 4.89 | 6.59 ± 3.92 | 0.000004 |
| LIN % | 30.6 ± 2.41 | 37.82 ± 33.26 | 0.4588 |
| STR % | 67.5 ± 1.21 | 61.05 ± 28.72 | 0.79 |
| WOB % | 45.4 ± 2.94 | 56.07 ± 22.68 | 0.40618 |
The motility of Fsip1–/–sperm is significantly reduced (n = 5000 sperm analyzed from each mouse, and (n = 5) from each genotype for each group; data are mean ± SEM; significance assessed by two-tailed t-test). Motility Parameters indicated according to WHO criteria.
3.2. Expression and distribution of Fsip1 during spermatogenesis
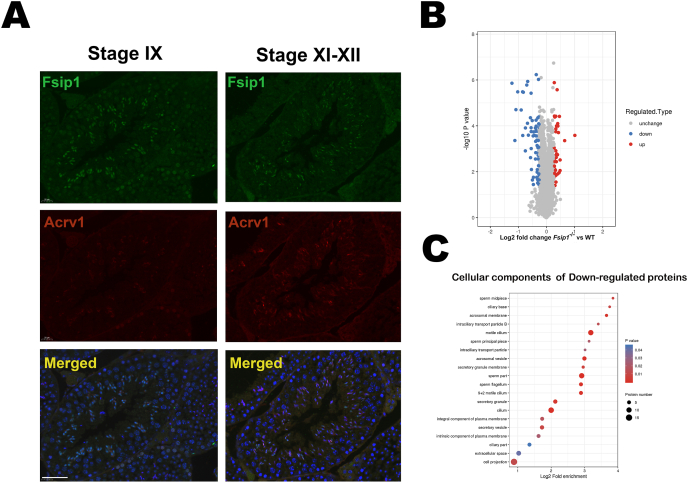
Before exploring the potential molecular function of Fsip1 in spermatogenesis and spermiogenesis, we first investigated the expression and distribution of the Fsip1 in the seminiferous tubules. The Fsip1 protein expression at different stages of spermatogenesis was examined by immunofluorescence. Interestingly, Fsip1 protein was expressed in all cellular components of the spermatogenesis. However, we observed that Fsip1 had differential expression and localization at the different stages of spermatogenesis. The intensity of Fsip1 staining was the highest and localized at the nucleus in in round spermatid steps 9–10 (Fig. 2A), and the signal was translocated from the nucleus to the anterior part of the elongating spermatid head at steps 11–12. These results suggest that Fsip1 may play a crucial role in head shaping of elongating spermatids.
Fig. 2.
Expression and localization of the Fsip1 protein during spermatogenesis. (A) The expression and localization of Fsip1 (green) in the testicular tissue sections at stages IX and XI-XII. The acrosome protein (Acrv1) was used as stage marker (Red). Scale bar 50 μm. (B) Volcano blot representing differentially expressed proteins from the proteomics data. (C) The top 20 downregulated cellular components at log2 fold enrichment. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Effect of Fsip1 deletion on the spermiogenesis proteome
To characterize the phenotype associated with the loss of Fsip1, and to address the possible underlying mechanism. Total testicular proteins were isolated and subjected to quantitative TMT based proteomics. A total of 109 differentially expressed proteins were identified with a 1.2-fold change in the testes of Fsip1−/− mutant mice compared to WT mice (Fig. 2B), of which 69 were downregulated and 40 were upregulated. The cellular components of down-regulated proteins in Fsip1−/− mice (Fig. 3C) indicated a significant decrease in acrosomal membrane and vesicles, intraciliary transport particles B, motile cilium, and sperm flagellum components (sperm midpiece, sperm flagellum, principal piece). In conclusion, homozygous deletion of the Fsip1 gene leads to abnormal sperm morphology and development.
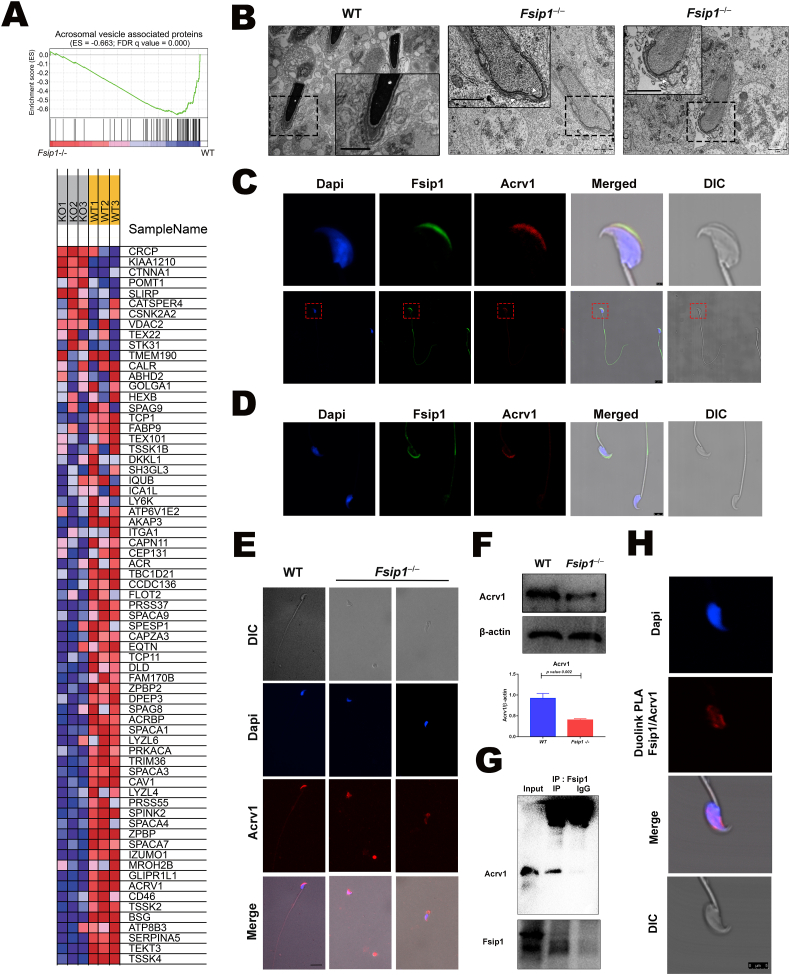
Fig. 3.
Effect of Fsip1 deletion on the expression and distribution of acrosome vesicle proteins (A) Gene set enrichment analysis (GSEA) and heatmap showing the differentially expressed acrosomal vesicle proteins. (B) Transmission electron microscope representing the elongated spermatid head. The white arrows indicate the morphological alterations of the acrosome vesicle and membrane during the development of elongated spermatids. Scale bars 2 μm. (C) and (D) Immunofluorescent staining of sperms from WT mice showing the expression and localization of Fsip1 and Acrv1. Scale bars 1 μm and 5 μm, respectively. (E) Immunofluorescent staining of Fsip1(green) and Acrv1(red) proteins in the sperms of WT and Fsip1–/–mice. (F) Western blotting assessment of Acrv1 protein expression in the testes of WT and Fsip1–/–mice. (G) Co-immunoprecipitation analysis of Fsip1 and Acrv1. Rabbit IgG was used as negative control. (H) Duolink in situ proximity ligation assay of Fsip1 and Acrv1 in the mice sperms. Scale bar 5 μm.
3.4. Disruption of Fsip1 alters the expression and distribution of the acrosome vesicle proteins
The acrosome membrane plays an essential role in fertilization process through sperm capacitation and acrosome reaction. Gene set enrichment analysis revealed that Fsip1 knockout resulted in a significantly downregulation of the acrosome vesicle associated proteins (Fig. 3A). Transmission electron microscopy evidenced the morphological alterations in the spermatid head and acrosome membrane in Fsip1–/–spermatids (Fig. 3B). Co-immunostaining of acrosome vesicle protein 1 (Acrv1) and Fsip1 showed their co-expression in the acrosome vesicle (Fig. 3C). Interestingly, the fluorescent signal of the acrosome vesicle protein Acrv1 was only observed in Fsip1 positive sperm (Fig. 3D). In addition, loss of Fsip1 resulted in translocation and diffusion of the Acrv1 protein (Fig. 3E). Western blot was used to further quantify the expression level of Acrv1 protein, which was significantly decreased in the testes of Fsip1−/− mice (Fig. 3F). Based on the above findings, we further examined and confirmed the protein-protein interactions between Fsip1 and Acrv1, usingCO-IP (Fig. 3G), and proximity ligation assay (PLA) (Fig. 3H). Therefore, our results indicated that Fsip1 may play an important role in the acrosome vesicle and membrane formation.
3.5. Disruption of Fsip1 impairs flagellogenesis
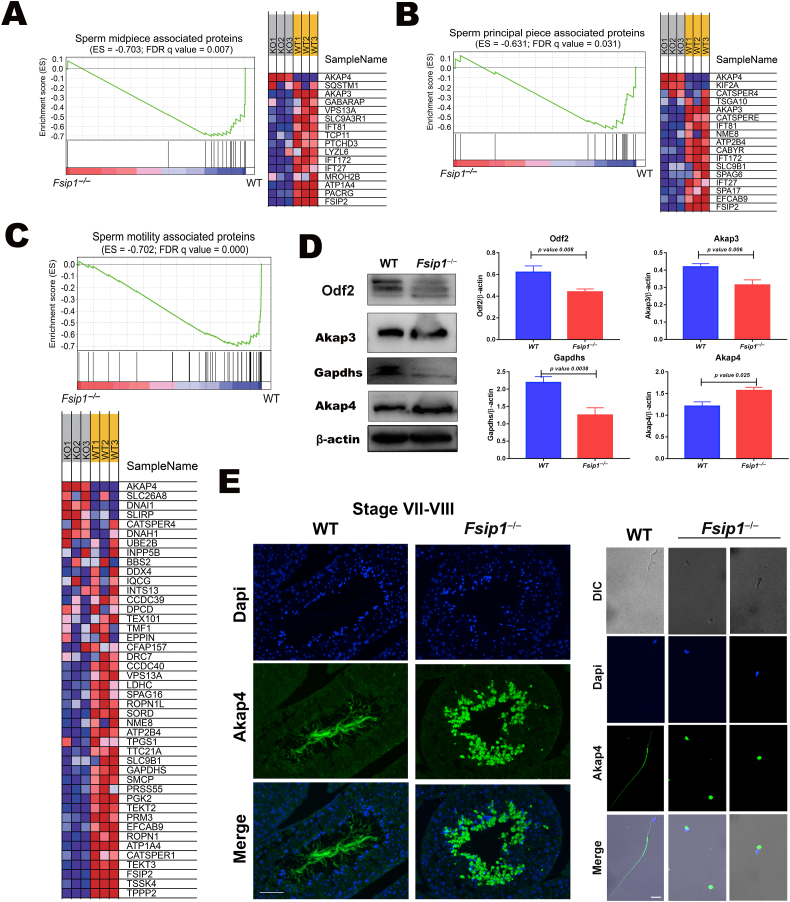
We observed that the homozygous knockout of Fsip1 provoked a subtle phenotype reminiscent to partial globozoospermia, characterized by abnormal sperm head and flagellum ultrastructure. TEM microscopy indicated that Fsip1 –/–testis displayed rare axonemes, and large vacuoles surrounded by mitochondria. GSEA enrichment analysis of proteomics data revealed that the differentially expressed proteins were related to midpiece (MP) and principal piece (PP) in sperm flagella (Fig. 4A and B, respectively), including significant downregulation of motility associated proteins (Fig. 4C). The protein expression levels of Odf2, Akap3, Gapdhs, and Akap4 were assessed using western blot (Fig. 4D). Odf2, Akap3, and Gapdhs were significantly downregulated in Fsip1–/–mice. However, unexpectedly, Akap4 was significantly upregulated. Immunostaining of testicular sections showed that Akap4 was retained in the cytoplasm of elongated spermatids in a condensate or compressed state, and the same was observed in the cauda epididymis spermatozoa (Fig. 4E).
Fig. 4.
Effect of Fsip1deficiency on flagellum formation proteins. (A) GSEA enrichment analysis and heatmap showed the expression level of the proteins associated with sperm midpiece associated proteins. (B) Principal piece associated proteins. (C) Sperm motility associated proteins. (D) Western blot analysis of Odf2, Akap3, Akap4 and Gapdhs. β-actin was used as internal control. The bar graph represents the statistical analysis of three independent samples. The p value was calculated using Student's two-tailed, unpaired t-test. (E) Immunofluorescent staining of Akap4 in Fsip1−/− and WT testicular sections and cauda epididymis spermatozoa. Scale bars 50 μm and 10 μm, respectively.
3.6. Fsip1 knockout attenuates intraflagellar transporters
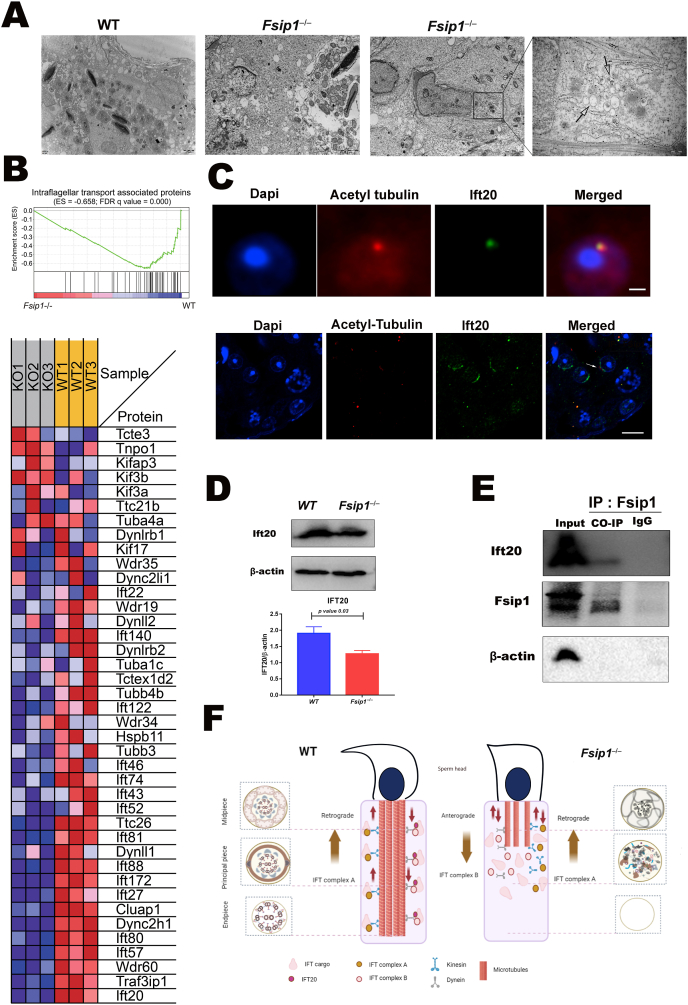
During spermiogenesis, the intraflagellar protein complex is essential for acrosome vesicle development and flagellogenesis. Compared with WT mice, the TEM results of testis sections from Fsip1 −/− mutant mice showed fewer or absent axonemal structures, and presence of cytoplasmic residuals with many large vacuoles in elongated spermatids (Fig. 5A). Some of the elongated spermatids were surrounded by mitochondria visible at high magnification (Fig. 5A right panel), indicating autophagosomes resulting from flagellum mis-assembly. In addition, our proteomics data showed that Fsip1 knockout significantly attenuated intraflagellar transporter proteins (Fig. 5B). Among the IFT proteins, the expression of Ift20 was downregulated most obviously. Immunofluorescent staining of Ift20 and acetyl-tubulin was used to detect the co-expression of Ift20 in early tail formation (Fig. 5C). Ift20 expression was significantly downregulated in the testis of Fsip1–/–mice using western blot (Fig. 5D). In addition, Ift20 co-immunoprecipitated with Fsip1 using (Fig. 5E). These findings suggest that Fsip1 is necessary for the expression of IFT proteins. Downregulation of IFT proteins in the Fsip1−/− may cause disordered flagellogenesis. Therefore, Fsip1 is necessary for sperm flagellum assembly.
Fig. 5.
Fsip1 knockout attenuates intraflagellar transporters during flagellogenesis. (A) Transmission electron microscopic evaluation of testicular sections showing missing axoneme structures in Fsip1–/–mice compared with WT mice (left panel), and large vacuoles some with mitochondria in the cytoplasmic residuals of the Fsip1−/− mutant elongated spermatid. (B) GSEA enrichment analysis and related heatmap showing the intraflagellar transport associated proteins. (C) Immunofluorescent staining of Ift20 (green) and Acetyl-tubulin (red) in testicular sections. Scale bars 2 μm and 10 μm, respectively. (D) Western blotting analysis of Ift20. β-actin was used as internal loading control. The graph bar represents the statistical analysis of three independent samples. The p-value was calculated using Student's two-tailed, unpaired t-test. (E) Western blot analysis showed Immunoprecipitation (IP) of Fsip1, and it co-precipitation (Co-IP) with Ift20. IgG was used as IP negative control. β-actin was used as the Co-IP negative control. (F) Schematic diagram representing the sperm component alterations due to decreased intraflagellar transport proteins in the Fsip1 compared with WT mice.
3.7. Two feature motifs of Fsip1 are involved in its interaction with Ift20
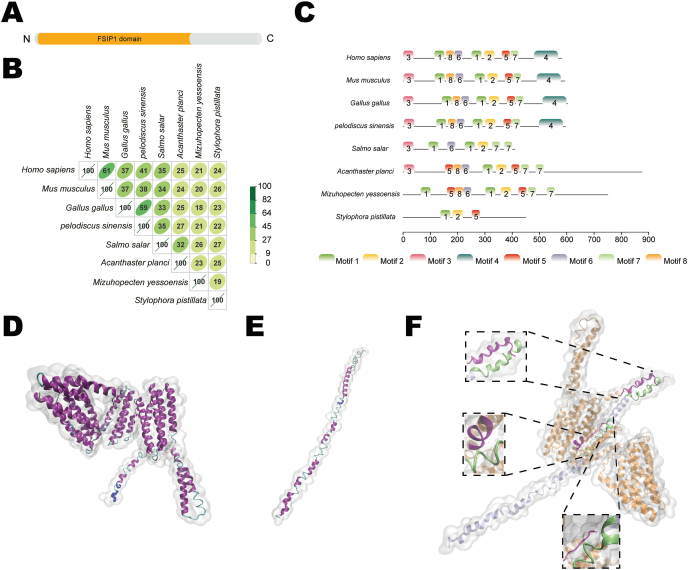
To map the binding site of Ift20 on Fsip1, we investigated the sequence and structural characteristics of Fsip1. First, the potential functional domains of Fsip1 were identified with a protein domain prediction tool. Only one Fsip1 domain was early identified (Asp4-Glu405, Fig. 6A), whereas the other parts of Fsip1 remained unknown. In light of this, we explored the conserved motifs of Fsip1 using the MEME online tool. Eight species, Homo sapiens, Mus musculus, Gallus gallus, Pelodiscus sinensis, Salmo salar, Acanthaster planci, Mizuhopecten yessoensis and Stylophora pistillata were investigated. In spite of the low pairwise similarities (≤60% except that between Homo sapiens and Mus musculus, Fig. 6B), a total of 8 motifs were identified; among them, the 1st motif (Glu264-Asp300), the 2nd motif (Gly310-Thr348), and the 5th motif (Thr374-Leu403) were conserved in all species (Fig. 6C). Notably, the 4th C-terminal motif (Asp491-Gln579) was only identified in Homo sapiens, Mus musculus, Gallus gallus and Pelodiscus sinensis, i.e., the internally fertilized species; the 3rd N-terminal motif (residual Met3-Gly41) was more conserved, appearing in Acanthaster planci and higher life forms.
Fig. 6.
Fsip1 interacts with Ift20 with two proteinic motifs. (A) Domain of Fsip1. (B) Sequence similarity between Fsip1 proteins across eight species. (C) Motifs identified in Mus musculus Fsip1 and seven other species. (D) and (E) Ribbon diagrams of three-dimensional structural models of Fsip1 and Ift20, colored according to their secondary structures. (F) Structural model of the Fsip1/Ift20 complex. Three regions on Fsip1 (in magenta helix: Met1-Ser20; helix: Ser249-Phe255; loop: Ser279-Val284) and corresponding binding sites on Ift20 (in lime color helix: Ile5-Glu25; helix: Lys43-Gly53; loop: Leu57-Lys63) are highlighted in opaque NewCartoon representations and indicated by close-up views. The other parts of Fsip1 and Ift20 are shown with transparent orange and ice-blue NewCartoon representations, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In parallel, structural models of Fsip1 and Ift20 were constructed by homology modeling. The overall structure of Fsip1 adopted a distorted “W” global scaffold, comprising α-helixes and random coils (Fig. 6D). In the meantime, the structure of Ift20 adopted a global scaffold of an extended helix, comprising α-helixes and random coils (Fig. 6E). On the basis of the structural models, we employed protein–protein flexible docking technology to assess intermolecular interactions between Fsip1 and Ift20. Three regions on Fsip1 (helix: Met1-Ser20; helix: Ser249-Phe255; loop: Ser279-Val284) were predicted to interact with Ift20 (helix: Ile5-Glu25; helix: Lys43-Gly53; loop: Leu57-Lys63) through intermolecular hydrogen bonds and hydrophobic interactions (Fig. 6F). Interestingly, two out of the three predicted regions on Fsip1 (helix: Met1-Ser20 and loop: Ser279-Val284) overlapped with the conserved motifs the 3rd motifMet3-Gly41 and 1st motif Glu264-Asp300.
4. Discussion
Sperm morphology plays an essential role in male fertility. Defective sperm morphology involves a wide range of phenotypes. The present study demonstrated that the Fsip1–/–mice provoke a partial globozoospermia phenotype characterized by impaired spermiogenesis [33]. Fsip1–/–mice sperm showed abnormal morphology, including a misshapen head, flagella assembly failure, and disrupted acrosome vesicle. Several genes have been reported to be associated with defective spermatozoa development. For example, subtle ultrastructure disorganizations were reported in Tektin4 knockout mice [34]. Axoneme defects were observed in Odf2 null mice [35]. Akap4, Fsip2 and Cabyr deficiency in mouse resulted in a lack of fibrous sheath [36]. The ablation of Ropn1 impaired the structure of sperm principal piece [37]. Until now, few genes defects were identified as direct causes of globozoospermia in human, including; DPY19L2, SPACA1, DNAH17, SPATA16, PICK1, CCIN, C7orf61, ZPBP, and CCDC62 and C2CD6. However, due to the lack of in vitro models, the molecular mechanisms causing globozoospermia remain largely unknown. This study demonstrates for the first time the potential effect of Fsip1 knockout on the spermatogenesis. Our findings indicated that Fsip1 is not only a fibrous sheath cytoskeletal protein, but also an essential protein involved in the organization of the basic structure of the elongated spermatids.
Scanning electron microscopy showed that there were obvious abnormalities in the Fsip1–/–sperm, including abnormal morphology of the spermatid head, and deformity or absence of midpiece and principal piece. The transmission electron microscopy analysis showed that the main ultrastructural components of sperm flagellum, such as mitochondria, outer dense fibers, and “9 + 2” axonemal structures, were mis-assembled, demonstrating that spermatid development was impaired due to improper assembly of sperm cell components. However, low motility was observed in Fsip1–/–sperm, possibly due to the abnormal morphology and structure of flagella.
Our results indicated that disruption of Fsip1−/− significantly downregulated most of the proteins involved in acrosome vesicle formation. The enrichment analysis of the cellular components of significantly downregulated proteins indicated a significant reduction in acrosomal membrane and acrosomal vesicle. Interestingly, genes such as ZBPB, SPACA1, PICK1 and DPYL19L2 which were recently identified as causes of globozoospermia significantly down regulated in our results. Deletion of developmental pluripotency-associated 19-like 2 (DPYL19L2) gene is the most well known cause of globozoospermia, with about 70% prevalence in patients [10]. DPY19L2 acts as a bridge between the nucleus and the acroplaxome and regulates the adhesion of the nuclear envelope to the acroplaxome [8]. DPY19L2 deficiency leads to a loss of the acrosome due to disruption of the nuclear/acroplaxome junction. Additionally, PICK1 facilitated formation of trafficking vesicles from the Golgi apparatus to the acrosome [38]. Zpbp1 deletion caused severe disorganization of the acrosome [39]. Nonetheless, truncating mutations in SPACA1 and SPINK2 leads to severe flagellum malformations in both humans and mice were also downregulated in our data [40,41]. In our study, we found Fsip1 to physically interact with Acrv1, an acrosome vesicle protein belonging to a family of proteins called bone morphogenetic protein (BMP) type I receptors [42]. BMP receptors span the cell membrane in such a way that one end protrudes from the outer surface of the cell and the other end of the protein remains inside the cell. Acrv1 has also been suggested as a seminiferous tubule stage marker [43]. Since the absence of Fsip1 impairs the acrosome vesicle localization and head shape, as well as being associated with downregulation of acrosome vesicle biogenesis proteins, it seems that the dynamics of Fsip1 translocation from the nucleus to the acrosome between steps 9–12 indicating an essential role of Fsip1 during the spermatid elongating stage.
Furthermore, gene set enrichment analysis of flagellum proteins revealed that the expression of several members of midpiece and principal piece proteins were downregulated. However, the role Fsip1 plays in the downregulation of these key proteins remains to be determined. Accumulated evidence showed that downregulation of these genes leads to defects in sperm flagellum development and resulted in abnormal morphology, which is consistent with our findings. Unexpectedly, Akap4, the most abundant protein in sperm flagellum was upregulated, and retained or accumulated in the cytoplasm of elongated spermatids. This may be due to Akap4 being unassembled in vivo until it is transported to the developing tail structure and integrated into the fibrous sheath [44]. It is believed that the retained cytoplasmic components mechanically hinder the elongation of sperm flagella and affect sperm development.
An increased number of cytoplasmic vesicles, fiber-like structures, and accumulation of mitochondria is representative of flagellum failure, which has been observed with absent/downregulated intraflagellar transporters [25]. During the development of elongated spermatids, the flagellum proteins proceed anterogradely and retrogradely by the IFT system along the tail of spermatozoa. IFT-B directs anterograde transport from the cell body to the distal axoneme, while IFT-A controls the retrograde return [22]. The loss of a single member of IFT-B or IFT-A usually leads to dysfunction of the whole subcomplex and loss of anterograde or retrograde function [45,46]. It is worth noting that the deletion of Fsip1 attenuates the anterograde IFT-B complex proteins and impairs their function, although they are structurally and functionally compromised, because flagella are not yet fully formed.
Among the IFT-B proteins, Ift20 is the most dynamic member, and co-localized in the Golgi apparatus during the acrosome formation of early round spermatocytes. Follit et al. reported that Ift20 Knockdown blocked ciliary assembly without affecting the Golgi structure [46], similarly to our results. The expression level of Ift20 in the testis of the Fsip1–/–mice was significantly reduced. This finding suggests the Fsip1 is not only a structural protein, but also a specific regulator of proteins delivery or flagellum assembly of anterograde IFT-B complex.
Fsip1 is an evolutionary conserved component of the flagellar proteome in diverse species. Although the interaction between Fsip1 and Ift20 was corroborated, we sought to identify the binding sites of Ift20 on Fsip1, which prompted us to investigate the sequence and structural characteristics of Fsip1. However, a conventional protein domain prediction tool could not provide sufficient information, since only one FSIP1 domain was identified and the other parts was still unknown. Using the motif discovery tool, we identified eight motifs in Fsip1 conserved in eight species. Coincidentally, previous findings predicted that three regions on the human FSIP1 (helix Ser479-Asn492; loop Pro541-Leu549 and loop Tyr310-Gln316) were predicted to interact with ULK1 (loop Ile22-Ala26; loop Ala179-Ser184) through hydrophobic interactions [29]. The involved regions overlapped with the 4th C-terminal motif (Gly481-Gln569) and the most conserved 2nd motif (Gly298-Thr336) of human FSIP1. This result indicated that the 2nd and 4th motifs of FSIP1 were involved in the interactions with ULK1. Here through homology modeling and flexible docking, we also generated a structural model for the Fsip1/Ift20 complex, from which the binding sites were also predicted. Interestingly, two of the four binding regions of Fsip1 the helix Met1-Ser20 and loop Ser279-Val284 overlapped with the 3rd N-terminal motif (residues Met3-Gly41) and the most conserved 1st motif (Glu264-Asp300) of mouse Fsip1, respectively, which suggests that the 1st and 3rd motifs of Fsip1 are involved in the interactions with Ift20.
Our findings suggest that Fsip1 is essential for normal spermiogenesis, and plays a crucial role in acrosome biogenesis and flagella formation by attenuating intraflagellar transport proteins.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgment
We acknowledge Linglong Huang for his kindly help in electron microscopy experiment. This work was supported by the National Natural Science Foundation of China (81701451), the Natural Science Foundation of Guangdong Province of China (2018A030313538), the China Postdoctoral Science Foundation (2019M662852), the Guangdong Science and Technology Department of Guangdong Province of China (2017A030223003) and the Key-Area Research and Development Program of Guangdong Province (2019B020227001).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101969.
Contributor Information
Hao Hu, Email: huh@cougarlab.org.
Na Li, Email: lina@cougarlab.org.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Table 1: List of antibodies
Multimedia component representing sperm motility by CASA: Fsip1-/- mutant spermFS
Multimedia component representing sperm motility by CASA: WT sperm
References
- 1.Cheng C.Y. Biology of spermatogenesis. Semin. Cell Dev. Biol. 2014;29:1. doi: 10.1016/j.semcdb.2014.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z. Sertoli cell-only phenotype and scRNA-seq define PRAMEF12 as a factor essential for spermatogenesis in mice. Nat. Commun. 2019;10(1):5196. doi: 10.1038/s41467-019-13193-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vries M. Chromatin remodelling initiation during human spermiogenesis. Biol Open. 2012;1(5):446–457. doi: 10.1242/bio.2012844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griswold M.D. Spermatogenesis: the commitment to meiosis. Physiol. Rev. 2016;96(1):1–17. doi: 10.1152/physrev.00013.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J. MNS1 is essential for spermiogenesis and motile ciliary functions in mice. PLoS Genet. 2012;8(3) doi: 10.1371/journal.pgen.1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.VanGompel M.J., Xu E.Y. A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum. Mol. Genet. 2010;19(12):2360–2369. doi: 10.1093/hmg/ddq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo J. Proteomic changes in human sperm during sequential in vitro capacitation and acrosome reaction. Front Cell Dev Biol. 2019;7:295. doi: 10.3389/fcell.2019.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celse T. Genetic analyses of a large cohort of infertile patients with globozoospermia, DPY19L2 still the main actor, GGN confirmed as a guest player. Hum. Genet. 2021;140(1):43–57. doi: 10.1007/s00439-020-02229-0. [DOI] [PubMed] [Google Scholar]
- 9.Chansel-Debordeaux L. Reproductive outcome in globozoospermic men: update and prospects. Andrology. 2015;3(6):1022–1034. doi: 10.1111/andr.12081. [DOI] [PubMed] [Google Scholar]
- 10.Noveski P. A homozygous deletion of the DPY19l2 gene is a cause of globozoospermia in men from the Republic of Macedonia. Balkan J. Med. Genet. 2013;16(1):73–76. doi: 10.2478/bjmg-2013-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nsota Mbango J.F. Genetic causes of male infertility: snapshot on morphological abnormalities of the sperm flagellum. Basic Clin Androl. 2019;29:2. doi: 10.1186/s12610-019-0083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujihara Y. Human globozoospermia-related gene Spata16 is required for sperm formation revealed by CRISPR/Cas9-Mediated mouse models. Int. J. Mol. Sci. 2017;18(10) doi: 10.3390/ijms18102208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oud M.S. Exome sequencing reveals novel causes as well as new candidate genes for human globozoospermia. Hum. Reprod. 2020;35(1):240–252. doi: 10.1093/humrep/dez246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman K.B. Elevated expression of cancer/testis antigen FSIP1 in ER-positive breast tumors. Biomarkers Med. 2013;7(4):601–611. doi: 10.2217/bmm.13.58. [DOI] [PubMed] [Google Scholar]
- 15.Chen M. Loss-of-function variants in FSIP1 identified by targeted sequencing are associated with one particular subtype of mucosal melanoma. Gene. 2020;759:144964. doi: 10.1016/j.gene.2020.144964. [DOI] [PubMed] [Google Scholar]
- 16.Li X. FSIP1 is correlated with estrogen receptor status and poor prognosis. Mol. Carcinog. 2020;59(1):126–135. doi: 10.1002/mc.23134. [DOI] [PubMed] [Google Scholar]
- 17.Liu C. FSIP1 regulates autophagy in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2018;115(51):13075–13080. doi: 10.1073/pnas.1809681115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu T. FSIP1 binds HER2 directly to regulate breast cancer growth and invasiveness. Proc. Natl. Acad. Sci. U. S. A. 2017;114(29):7683–7688. doi: 10.1073/pnas.1621486114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan M. Over-expression of FSIP1 promotes breast cancer progression and confers resistance to docetaxel via MRP1 stabilization. Cell Death Dis. 2019;10(3):204. doi: 10.1038/s41419-018-1248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H. Expression and clinicopathological significance of FSIP1 in breast cancer. Oncotarget. 2015;6(12):10658–10666. doi: 10.18632/oncotarget.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun M. Knockdown of fibrous sheath interacting protein 1 expression reduces bladder urothelial carcinoma cell proliferation and induces apoptosis via inhibition of the PI3K/AKT pathway. OncoTargets Ther. 2018;11:1961–1971. doi: 10.2147/OTT.S158275. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Ishikawa H., Marshall W.F. Intraflagellar transport and ciliary dynamics. Cold Spring Harb Perspect Biol. 2017;9(3) doi: 10.1101/cshperspect.a021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z.B., Wang H.Q., Zhang L. Intraflagellar transport is essential for spermiogenesis. Zhonghua Nan Ke Xue. 2019;25(3):195–201. [PubMed] [Google Scholar]
- 24.Zhang Z. Some thoughts about intraflagellar transport in reproduction. Mol. Reprod. Dev. 2021;88(2):115–118. doi: 10.1002/mrd.23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z. Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol. Biol. Cell. 2016;27(23):3705–3716. doi: 10.1091/mbc.E16-05-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen B. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23(5):720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang X. Proteomics and single-cell RNA analysis of Akap4-knockout mice model confirm indispensable role of Akap4 in spermatogenesis. Dev. Biol. 2019;454(2):118–127. doi: 10.1016/j.ydbio.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 28.Bailey T.L. Meme SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 30.van Zundert G.C.P. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 31.de Vries S.J., Bonvin A.M. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1) doi: 10.1016/0263-7855(96)00018-5. 33-38, 27-8. [DOI] [PubMed] [Google Scholar]
- 33.Dam A.H. Morphology of partial globozoospermia. J. Androl. 2011;32(2):199–206. doi: 10.2164/jandrol.109.009530. [DOI] [PubMed] [Google Scholar]
- 34.Roy A. Absence of tektin 4 causes asthenozoospermia and subfertility in male mice. FASEB J. 2007;21(4):1013–1025. doi: 10.1096/fj.06-7035com. [DOI] [PubMed] [Google Scholar]
- 35.Donkor F.F. Outer dense fibre protein 2 (ODF2) is a self-interacting centrosomal protein with affinity for microtubules. J. Cell Sci. 2004;117(Pt 20):4643–4651. doi: 10.1242/jcs.01303. [DOI] [PubMed] [Google Scholar]
- 36.Young S.A. CABYR is essential for fibrous sheath integrity and progressive motility in mouse spermatozoa. J. Cell Sci. 2016;129(23):4379–4387. doi: 10.1242/jcs.193151. [DOI] [PubMed] [Google Scholar]
- 37.Fiedler S.E. Loss of R2D2 proteins ROPN1 and ROPN1L causes defects in murine sperm motility, phosphorylation, and fibrous sheath integrity. Biol. Reprod. 2013;88(2):41. doi: 10.1095/biolreprod.112.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu G., Shi Q.W., Lu G.X. A newly discovered mutation in PICK1 in a human with globozoospermia. Asian J. Androl. 2010;12(4):556–560. doi: 10.1038/aja.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Y.N. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol. Cell Biol. 2007;27(19):6794–6805. doi: 10.1128/MCB.01029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujihara Y. SPACA1-deficient male mice are infertile with abnormally shaped sperm heads reminiscent of globozoospermia. Development. 2012;139(19):3583–3589. doi: 10.1242/dev.081778. [DOI] [PubMed] [Google Scholar]
- 41.Kherraf Z.E. SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol. Med. 2017;9(8):1132–1149. doi: 10.15252/emmm.201607461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang A. Developmental expression of ACRV1 in humans and mice. Andrologia. 2012;44(1):16–22. doi: 10.1111/j.1439-0272.2010.01095.x. [DOI] [PubMed] [Google Scholar]
- 43.Osuru H.P. The acrosomal protein SP-10 (Acrv1) is an ideal marker for staging of the cycle of seminiferous epithelium in the mouse. Mol. Reprod. Dev. 2014;81(10):896–907. doi: 10.1002/mrd.22358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blommaert D. Expression, localization, and concentration of A-kinase anchor protein 4 (AKAP4) and its precursor (proAKAP4) in equine semen: promising marker correlated to the total and progressive motility in thawed spermatozoa. Theriogenology. 2019;131:52–60. doi: 10.1016/j.theriogenology.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 45.Pazour G.J. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151(3):709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Follit J.A. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 2006;17(9):3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: List of antibodies
Multimedia component representing sperm motility by CASA: Fsip1-/- mutant spermFS
Multimedia component representing sperm motility by CASA: WT sperm