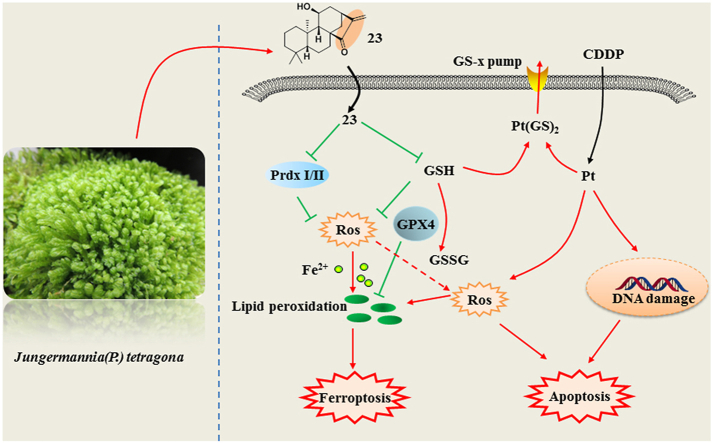
Abstract
Reactive oxygen species (ROS) induction is an effective mechanism to kill cancer cells for many chemotherapeutics, while resettled redox homeostasis induced by the anticancer drugs will promote cancer chemoresistance. Natural ent-kaurane diterpenoids have been found to bind glutathione (GSH) and sulfhydryl group in antioxidant enzymes covalently, which leads to the destruction of intracellular redox homeostasis. Therefore, redox resetting destruction by ent-kaurane diterpenoids may emerge as a viable strategy for cancer therapy. In this study, we isolated 30 ent-kaurane diterpenoids including 20 new samples from Chinese liverworts Jungermannia tetragona Lindenb and studied their specific targets and possible application in cancer drug resistance through redox resetting destruction. 11β-hydroxy-ent-16-kaurene-15-one (23) possessed strong inhibitory activity against several cancer cell lines. Moreover, compound 23 induced both apoptosis and ferroptosis through increasing cellular ROS levels in HepG2 cells. ROS accumulation induced by compound 23 was caused by inhibition of antioxidant systems through targeting peroxiredoxin I/II (Prdx I/II) and depletion of GSH. Furthermore, compound 23 sensitized cisplatin (CDDP)-resistant A549/CDDP cancer cells in vitro and in vivo by inducing apoptosis and ferroptosis. Thus, the ent-kaurane derivative showed potential application for sensitizing CDDP resistance by redox resetting destruction through dual inhibition of Prdx I/II and GSH in cancer chemotherapy.
Keywords: ent-kaurane diterpenoids, Cancer resistance, Apoptosis, Ferroptosis, Prdx I/II, GSH
Abbreviations: ROS, reactive oxygen species; SOD, superoxide dismutase; GSH, glutathione; CAT, catalase; GPx, glutathione peroxidase; Trx, thioredoxin; Prdx, peroxiredoxin; CDDP, Cisplatin; MMP, mitochondrial membrane potential; NSCLC, non-small cell lung cancer; EGFR, epidermal growth factor receptor
Graphical abstract
Highlights
-
•
Thirty ent-kaurane diterpenoids were isolated from the Chinese liverworts, Jungermannia tetragona.
-
•
11β-hydroxy-ent-16-kaurene-15-one (23) was identified to induce apoptosis and ferroptosis for the first time.
-
•
Compound 23 could disorder the intracellular redox system by directly targeting Prdx I/II and GSH.
-
•
Compound 23 could sensitize A549/CDDP cancer cells in vitro and in vivo through redox resetting destruction.
1. Introduction
ent-Kauranes are complex tetracyclic diterpenoids, mainly distributed in Jungermannia species of liverworts [[1], [2], [3]] and Isodon species of higher plants [4]. Numerous studies reported the α,β-unsaturated ketone was the active center for cytotoxic activity. The currently accepted mechanism for ent-kauranes is the Michael addition of thiols and protein sulfhydryl groups onto the α,β-unsaturated ketone moiety, which leads to induce ROS (reactive oxygen species) accumulation by the deactivation of SH enzymes or glutathione (GSH) [[5], [6], [7]].
ROS are generated during cell metabolism and possess multifaceted functions depending on concentrations [[8], [9], [10]]. The moderate levels of ROS aid tumor development and aggressiveness by activating various signaling pathways, while high levels of ROS are toxic to tumor cells by causing oxidative damage [[11], [12], [13], [14]]. Therefore, a variety of chemotherapeutic agents (e.g., adriamycin, imatinib and cisplatin) with direct or indirect effects on ROS induction have been used for effective cancer therapies [15,16].
Excessive free radicals will adversely modify cellular components and increase various pathogenesis, such as lipid, protein and DNA damage [17]. ROS may lead to the activation of intrinsic apoptosis by mitochondria-specific endometrium phospholipids and subsequent products of lipid peroxidation [18]. Recently, ferroptosis, a new form of programmed cell death, also occurs due to elevated ROS levels [19]. It has been found that ferroptosis is caused by lipid peroxidation, which is due to increased intracellular iron concentration and a depletion of antioxidant GSH [20]. Ferroptosis, as a cell death mode completely independent from apoptosis, may provide a new therapeutic method for cancer treatment [21]. Given that ent-kaurane diterpenoids can induce ROS level by targeting antioxidant proteins and depletion of GSH, it is still unclear whether ferroptosis exists in the cell death mechanism of ent-kaurane diterpenoids.
To restrict ROS levels below a threshold in adaption to the oxidative stress, cancer cells develop drug resistance for “redox resetting” to acquire a new redox balance through generating more GSH and overexpressing many anti-oxidative genes such as superoxide dismutase (SOD), glutathione peroxidase (Gpx), peroxiredoxin (Prdx), and thioredoxin (Trx) [[22], [23], [24]]. Resistance to Cisplatin (CDDP) is often accompanied by a significant increased expression of Prdx systems, as well as the elevated levels of GSH [[25], [26], [27]]. Among many ways used to overcome cancer resistance, redox resetting disruption through targeting redox proteins and GSH provides an alternative way to overcome drug-induced resistance [13,28,29]. Considering ent-kauranes targeted antioxidant system, we hypothesized that ent-kauranes with α,β-unsaturated ketone would contribute to overcoming CDDP drug resistance by disrupting the resettled redox homeostasis.
Our previous research on ent-kaurane diterpenoids revealed that their anticancer activities were caused by ROS formation through Michael modification of the protein thiols and depletion of GSH unselectively [30]. However, the underlined mechanisms and potential applications of ent-kaurane diterpenoids in reversing drug resistance and sensitizing chemotherapeutics are rarely studied.
In this research, thirty ent-kaurane derivatives including twenty new (1–20) were isolated and identified from the Chinese liverworts Jungermannia tetragona Lindenb. Among these compounds, the representative compound 23 which possessed strong inhibitory activity against several cancer cell lines was chosen for further studies. More importantly, compound 23 induced apoptosis and ferroptosis through accumulation of ROS in HepG2 cells. Further research found that 23 induced cell apoptosis and ferroptosis by inhibiting the activity of Prdx I/II and depleting reduced GSH. Besides, combination therapy of 23 and CDDP were executed on A549/CDDP cells in vitro and in vivo respectively. And the results demonstrated a significant sensitizing effect of 23 on CDDP anti-tumor activity by targetting redox resetting destruction through dual inhibition of Prdx I/II and GSH. Collectively, compound 23 may serve as a promising lead candidate for sensitizing redox resetting-induced drug resistance through inducing apoptosis and ferroptosis via targeting Prdx I/II and GSH in cancer chemotherapy.
2. Materials and methods
2.1. General experimental procedures and materials
Melting points were determined on an X-6 melting point apparatus (Beijing TECH Instrument Co. Ltd., Beijing, China) and are uncorrected. Optical rotations were acquired by a Perkin-Elmer 241 MC polarimeter at 20°C. HRESIMS data were determined on a Finnigan LC-QDECA mass spectrometer. ECD spectra were performed on a Chirascan spectropolarimeter. UV data were recorded using a UV-2550 spectrophotometer (Shimadzu, Japan). IR spectra were obtained on a Nicolet iN 10 Micro FTIR spectrometer. NMR spectra were recorded on a Bruker Avance spectrometer operating at 600 (1H) and 150 (13C) MHz with TMS as an internal standard. HPLC were performed on a Shimadzu LC-20AT pump equipped with a DGU-20A5R degasser, a SPD-M20A detector, and a Shim-pack GIST-C18 5 μm column (10 × 250 mm). The compounds were visualized under UV (254 nm) light and by spraying with H2SO4- ethanol (EtOH) (1:9, v/v) followed by heating. DFO, Fer-1 and MitoQ were obtained from MCE (New jersey, USA). Z-VAD-FMK, DCFHDA, JC-1, Hoechst 33342, DAPI, and BCA protein assay kit were purchased from Beyotime (Shanghai, China). Ac-DEVD-CHO and Z-LEHD-FMK TFA were obtained from Selleck Chemicals (Houston, USA). Amplex Red and C11-BODIPY were obtained from Thermo Fisher Scientific (Waltham, USA). Annexin V-FITC apoptosis detection kit was purchased from BD Biosciences (CA, USA).
2.2. Plant material
Whole plants of J. tetragona were collected at the Wenzhou, Zhejiang Province, China, in May 2017, and identified by Dr. Jinchuan Zhou (Linyi University, Linyi, China). A voucher specimen (No. 20170905) has been deposited at the Department of Natural Products Chemistry (Shandong University, Jinan, China).
2.3. Extraction and isolation
The fresh and milled plant material of J. tetragona (1500 g) was extracted with 95% EtOH (3 × 1.5 L, each for one week) at room temperature and filtered. The filtrate was evaporated under reduced pressure at 40 °C to afford the crude extract (20.0 g), which was separated by MCI gel column chromatography [methanol (MeOH) –H2O, 3:7 to 9:1] and gave fractions 1–3. Fraction 2 (2.7 g) were chromatographed using a silica gel column [petroleum ether (60–90 °C)-acetone, 200:1 to 0:1] to give seven subfractions (2.1–2.7). Further separation of fraction 2.3 (33.4 mg) was subjected to a Sephadex LH-20 column (MeOH) and an RP-18 silica gel column (MeOH–H2O, 4:6 to 9:1) and then was purified using HPLC (acetonitrile/H2O, 70:30, 3.0 mL/min) to afford 19 (7.1 mg) and 28 (2.9 mg). Fraction 2.4 (99.0 mg) was chromatographed on Sephadex LH-20 (MeOH) and then separated by a RP-18 silica gel column (MeOH/H2O, 5:5 to 10:0) to obtain two fractions (2.4.1–2.4.2). Fraction 2.4.1 was purified by HPLC (MeOH/H2O, 83:17) to give 15 (5.9 mg), 26 (11.9 mg) and 27 (2.0 mg). Fraction 2.4.2 was purified by HPLC (MeOH/H2O, 73:27) to give 13 (5.9 mg). Fraction 2.5 (206.7 mg) was chromatographed on a Sephadex LH-20 column (MeOH–CHCl3, 1:1) and a RP-18 silica gel column (MeOH/H2O, 5:5 to 10:0) to obtain three fractions (2.5.1, 2.5.2 and 2.5.3). Fraction 2.5.1 was purified by HPLC (MeOH/H2O, 70:30, 3.0 mL/min) to give 20 (1.9 mg) and 25 (6.4 mg). Fraction 2.5.2 was purified by HPLC (MeOH/H2O, 78:22, 3.0 mL/min) to give 4 (7.3 mg), 7 (5 mg), 8 (8.5 mg), 16 (4.5 mg), 29 (4.8 mg) and 30 (4.1 mg). Fraction 2.5.3 was purified by HPLC (MeOH/H2O, 85:15, 3.0 mL/min) to give 23 (80.0 mg), and 24 (4.0 mg). Fraction 2.6 (281.7 mg) was subjected to a Sephadex LH-20 column (MeOH) and an RP-18 silica gel column (MeOH–H2O, 4:6 to 9:1) and then was purified using HPLC (MeOH–H2O, 68:32, 3.2 mL/min) to afford 1 (9.3 mg), 6 (5.6 mg), 9 (46.7 mg), 10 (13.5 mg), 22 (20.0 mg), and four fractions (2.6.1, 2.6.2, 2.6.3 and 2.6.4). These four fractions were further purified by HPLC using different proportions of acetonitrile/H2O as mobile phase to give 3 (17.6 mg), 11 (1.9 mg), 14 (2.1 mg), 17 (2.5 mg), 18 (4.5 mg) and 21 (4.7 mg). Fraction 2.7 (209.6 mg) was applied to a Sephadex LH-20 column (MeOH–CHCl3, 1:1) and a RP-18 silica gel column (MeOH/H2O, 5:5 to 10:0), followed by purification with HPLC using different proportions of MeOH/H2O to afford 2 (13.5 mg), 5 (18.5 mg) and 12 (34.7 mg). The purity of all isolated compounds is more than 95% as determined either by HPLC or NMR. Detailed procedure on the isolation of chemical constituents from J. tetragona and the X-ray crystallographic analysis of compounds 4, 5, 13 and 25 has been summarized in Supplementary data.
2.4. Cell culture
The A549, HepG2, 7860, A2870, HBE, HL-7702, HUVEC and A549/CDDP cell lines were obtained from the Chinese Academy of Sciences, Shanghai Institute for Biological Sciences (Shanghai, China). The cell lines were cultured in RPMI 1640 medium, which was supplemented with penicillin G (100 U/mL), streptomycin (100 μg/mL), and 10% fetal bovine serum, and incubated at 37°C in a humidified 5% CO2 incubator.
2.5. Cell viability and morphological changes assay
Cells (5 × 103 cells/well) were seeded in 96-well plates and cultured overnight. After incubation with several concentrations of the compounds for indicated time, MTT (5 mg/mL) was added to the cells and incubated for 4 h. The produced crystals were dissolved in DMSO and the cell viability was detected using a microplate reader at 570 nm (BioTek Winooski, USA). All cell viability data were normalized to the DMSO vehicle condition. Experiments were performed three independent times. IC50 values and error values were computed using GraphPad Prism 7.0. For examination of morphological changes, the cells were treated as above and the images were photographed using an inverted microscope (Olympus, Tokyo, Japan).
2.6. Time-lapse imaging
HepG2 cells were plated in a 20 mm glass bottom culture dish and incubated overnight. Then cells were treated with 23 at 2.5 μM. The dish was placed in a live cell chamber with 5% CO2 and 37 °C. Live phase-contrast images were acquired every 5 min for 24 h with a Lionheart FX microscope (BioTek, Winooski, USA).
2.7. Annexin V-FITC/PI double staining
Cells were seeded in 6-well plates (1.6 × 105 cells/well) and exposed to reagent for the indicated duration. After treatment, cells were trypsinized, washed and suspended in 100 μL 1X binding buffer, and stained with Annexin-V and PI according to the manufacturer's instructions. The fluorescence was measured with excitation at 488 nm and emission at 520 nm by a FACSC alibur flow cytometer (BD Biosciences, CA, USA).
2.8. ROS detection assay
Cells were seeded in 6-well plates (1.6 × 105 cells/well) and exposed to the indicated reagents. Cells were trypsinized, washed, suspended in PBS containing 10 μM DCFH-DA for additional 30 min according to manufacturer's protocols. Green fluorescence intensity, which was proportionate to the level of intracellular ROS, was measured on the FL1 channel using FACSC alibur flow cytometer (BD Biosciences, CA, USA).
2.9. Mitochondrial membrane potential (ΔΨm) assay
Cells were seeded in 6-well plates (1.6 × 105 cells/well) and exposed to the indicated reagents. Briefly, after treatment, the cells were incubated at 37 °C for 1 h with 5 mg/mL JC-1, then washed twice with PBS and placed in fresh medium without serum. The ratios of red/green fluorescent densities were measured and calculated by a FACSC alibur flow cytometer (BD Biosciences, CA, USA).
2.10. Detection of ferroptosis by C11-BODIPY lipid peroxidation assay
Cells plated in 6-well plates (1.6 × 105 cells/well) were exposed to reagent for the indicated duration. Cells were trypsinized, washed, suspended in PBS containing 1.5 μM C11-BODIPY and incubated at 37 °C for 15 min. Cells were pelleted and resuspended in PBS. Fluorescence intensity was measured on the FL1 channel using FACSC alibur flow cytometer (BD Biosciences, CA, USA).
2.11. Western blotting
Cells were collected and lysed with RIPA buffer containing a fresh protease inhibitor mixture (50 μg/mL aprotinin, 0.5 mM phenylmethanesulfonyl fluoride (PMSF), 1 mM sodium orthovanadate, 10 mM NaF, and 10 mM glycerol phosphate). Protein concentrations were quantified using BCA assay kit. Equal amounts of proteins were separated by SDS-PAGE (12%) and electrotransferred onto nitrocellulose membranes. The membranes were blocked with 5% nonfat milk in TBST buffer (20 mM Trisbuffered saline and 0.5% Tween 20) for 1 h at room temperature followed by incubation with the corresponding primary antibodies overnight at 4 °C. Primary antibodies are listed in Table S5. After washing with TBST buffer, secondary antibodies were used at 1:2000 dilutions for 1 h at room temperature. Protein detection was performed based on an enhanced chemiluminescence (ECL) method and photographed by using a BioSpectrum Gel Imaging System (Bio-Rad, Hercules, USA).
2.12. Pull-down and LC-MS/MS analysis
Biotin-tagged 23 and its intermediates were synthesized for the identification of 23-bound proteins (Supplementary data). The positive probe (biotin-23) and DMSO were each preincubated with streptavidin agarose (Thermo Fisher Scientific, Waltham, USA) for 4 h at RT. Then, the beads were washed three times in lysis buffer to remove the free probes, followed by incubation with HepG2 cell lysates overnight at 4 °C. The beads were washed six times with PBS on the following day, and the bead-bound proteins in SDS loading buffer were boiled for 10 min. The beads were centrifuged and the supernatant was collected, with the eluted proteins separated by SDS-PAGE and visualized by silver staining. The protein-containing bands in the gel were excised, followed by in-gel digestion. The samples were analyzed by LC-MS/MS analysis (Applied Protein Technology, Shanghai, China).
2.13. Molecular docking analysis
The dimeric Prdx I (PDB: 1QQ2) was used for the computational study in Schrodinger 2018 suite (San Diego, USA). After the four rat-specific residues Ser14, Ile88, Ile144, and Ile156 were mutated to human-specific residues Asn14, Val88, Val144, and Thr156, and the Ser83 were mutated back to cysteine, the protein was added hydrogens and minimized with OPLS_2005 force field. The docking grid was generated by selecting critical amino acid residues in dimerization pocket. The ligand (23) was docked into the dimerization pocket by Glide with extra precious and keeping other setting default. Then the complex was minimized in MacroModel using OPLS_2005 force field with water as solvent, and the Polak-Ribiere Conjugate Gradient (PRCG) method was employed to search the low energy conformation with energy convergence gradient threshold of 0.03 kJ/mol. Residues within 5 Å were set to be free, other atoms were constrained by a force of 200 kcal/(mol*A2).
2.14. Preparation of recombinant Prdx I/II and Prdx I/II activity assay
Human Prdx I and Prdx II were cloned into pET28a vector and the primers for Prdx constructs were described in Table S6. These proteins were expressed in the Escherichia coli strain BL21 and purified [5]. Two-cys Prdx activity was studied by Amplex Red Enzyme assay. Briefly, in the 96-well plate detection system, the required reagents were diluted with reaction buffer (50 mM sodium phosphate buffer, pH 7.4) to the corresponding concentration. 10 μL Prdxs (30 μM) and 40 μL compound were added to the 96-well plate and incubated for 30 min at room temperature. Then 30 μL hydrogen peroxide (10 μM) was added and incubated for 10 min, and finally 10 μL Amplex Red (50 μM) and 10 μL HRP (1 U/mL) were added and incubated for 10 min. The fluorescence (excitation 535 nm, emission 595 nm) was measured on a microplate reader (BioTek, Winooski, USA).
2.15. Glutathione (GSH) assay
HepG2 cells exposed to reagent for the indicated duration. The cells were collected and resuspended in a protein removal agent to collect supernatant by centrifugation after repeated freezing and thawing. The determination of GSH and GSSG was performed by GSH and GSSG Assay Kit according to the manufacturer's instructions. Colorimetric determination was conducted using a microplate reader (BioTek, Winooski, USA).
2.16. Measuring the direct reaction of compound 23 with GSH in HepG2 cells
Compound 23-GSH was synthesized as positive molecule for the identification of 23-GSH in 23 treated cells (Supplementary data). HepG2 cells were seeded in a 6-well plate at a density of 3 × 105 cells and cultured overnight. After treatment with 2.5 μM 23 for 8 h, the cells were collected and lysed using RIPA lysis buffer. The lysate was extracted with dichloromethane and dried with nitrogen. And the compound 23-GSH was detected by UPLC-MS (Supplementary data).
2.17. Measuring the reactivity of compound 23 with GSH in vitro
Compound 23 (1.2 mg) were incubated with an excess of GSH (2.4 mg) in deuterated phosphate buffer (10 mM KD2PO4, 10 mM K2DPO4 pH 7.4, 0.02 mL) and DMSO‑d6 (0.50 mL) at 37 °C. The reaction was monitored by the change of representative hydrogen signals with 1H NMR analysis.
2.18. Cell transfection
Prdx I and Prdx II siRNA (Table S7) were synthesized by Sangon Biotech (Shanghai, China). Human Prdx I or Prdx II genes were inserted in pcDNA3.1 (+) vector and the empty vector was used as the negative control. A549/CDDP cells were plated in 6-well plates (1.6 × 105 cells/well) and cultured overnight. Then cells were transfected with 100 pM siRNA or 2 μg vector DNA, using 5 μL HighGene Transfection reagents following the manufacturer's instruction (Abclonal, Wuhan, China). After treatment, the cells were harvested and processed for further analysis.
2.19. Tumor xenograft
Thirty male athymic (Balb/c-nu) mice (4-week-old) were purchased from the SPF (Beijing) biotechnology (Beijing, China) and allowed to acclimatize for 1 week. A549/CDDP cells (5 × 106 cells) were injected subcutaneously into the right anterior flanks of the mice. Two weeks after the injection of cells, when the tumors became palpable (around 100 mm3), mice were randomly divided into four groups (n = 6 per group). Tumor-bearing mice were received equal amount of solvent, CDDP (4 mg/kg) or compound 23 (10 mg/kg) or combination CDDP and 23 were injected via intraperitoneal injection. Administration was performed every 3 days for 30 days.
Tumor growth was measured through calipers, and tumor volumes and body weight were in record. The calculation of tumor volumes was according to formula 0.5 × length × width2. All animal treatments were performed strictly in accordance with the institutional guidelines of Animal Care and Use Committee at Shandong University.
2.20. Histology, immunohistochemical, TUNEL, C-11 BODIPY and DCFH-DA staining
One day after the last measurement of tumor size, tumor tissues and other main organs of mice were excised. The tissues were fixed in 4% formaldehyde and embedded in paraffin. Paraffin-embedded tumor and organ sections (5 mm) were deparaffinized for H&E staining and immunohistochemical analysis. H&E staining was performed using routine methods. The tumor slides were boiled for 10 min in 0.01 M sodium citrate buffer (pH 6.0) for antigen retrieval. Subsequently, sections were allowed to cool down and incubated with γ-H2AX antibody (1:200, Rabbit) at 4 °C overnight. After staining with Anti-mouse/rabbit immunohistochemical detection kit (Proteintech Group, Wuhan, China), the samples were counterstained with hematoxylin, and images were captured by a Fluorescence Microscope (Olympus, Tokyo, Japan).
Analysis of apoptotic cells was performed by TUNEL apoptosis assay kit following the manufacturer's direction (Beyotime, Shanghai China). Apoptotic index was calculated by dividing the number of TUNEL-positive cells by the total number of cells in the field.
For C-11 BODIPY and DCFH-DA staining, the frozen tissue sections were incubated with 1 μM C11-BODIPY (581/591) or 1 μM DCFH-DA for a final 30 min. Fluorescence of C11-BODIPY and DCFH-DA were measured using a Fluorescence Microscope (Olympus, Tokyo, Japan).
2.21. Statistical analysis
Data are presented as the mean ± SD for triplicate experiments. Statistically significant differences were analyzed using one-way ANOVA followed by the Tukey post-hoc test with GraphPad Prism 7.0 (San Diego, CA), and p < 0.05 was considered statistically significant.
3. Results and discussion
3.1. Isolation and structure elucidation of natural ent-kaurane diterpenoids and their activities against cancer cells
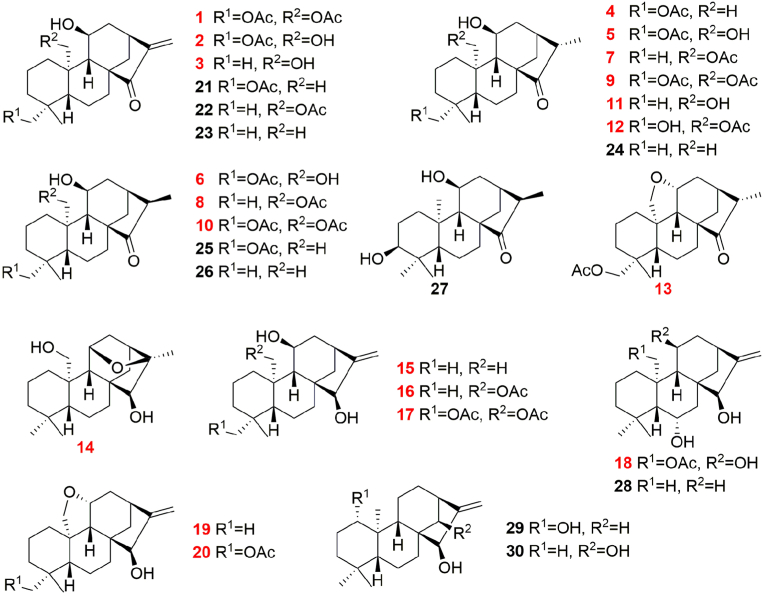
The EtOH extract of J. tetragona was repeatedly chromatographed over MCI gel, silica gel, Sephadex LH-20, and HPLC to yield 20 new (1–20) and 10 known (21–30) ent-kaurane diterpenoids (Fig. 1). These structures of new compounds were determined by extensive nuclear magnetic resonance (NMR) and high-resolution mass spectrometry (HRMS) analyses, including their configurations as defined by nuclear overhauser effect spectroscopy (NOESY) data, coupling constant analysis, electronic circular dichroism (ECD) comparisons, and single crystal X-ray diffraction measurements. The other reported compounds were identified by combined analyses of their spectroscopic data (Supplementary data).
Fig. 1.
Structures of compounds 1–30. The new compounds are indicated with red color. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
All of the kaurane diterpenoids were screened for their cytotoxicities against HepG2, A2780, 7860 and A549 cancer cells, as well as against a normal lung cell line, HBE (Table 1). The results confirmed that compounds 1–3 and 21–23, which contain an exo-methylene cyclopentanone system, possess strong activity against most of the tested cell lines. Among the tested compounds in the cytotoxicity assay, 23 showed the most significant activity (Table 1), and enough material was also available to complete further experiments. Meanwhile, The MTT assay results showed that the toxicity of 23 to several normal cells (HBE, HUVEC, HL-7702 cells) was significantly lower than that of A549 cancer cells (Fig. S5A).
Table 1.
Cytotoxicities of compounds against various human cell lines.
| Compounds | IC50 (μM) |
||||
|---|---|---|---|---|---|
| HBE | HepG2 | A549 | A2780 | 7860 | |
| 1 | 2.4 ± 0.1 | 5.5 ± 0.2 | >20 | 18.8 ± 0.1 | >20 |
| 2 | 15.0 ± 0.5 | 6.0 ± 0.3 | >20 | 19.5 ± 2.5 | >20 |
| 3 | >20 | 4.2 ± 0.1 | 13.0 ± 1.8 | >20 | >20 |
| 4 | 20.0 ± 0.8 | >20 | >20 | >20 | >20 |
| 5 | >20 | >20 | 9.8 ± 0.4 | >20 | 3.8 ± 0.5 |
| 10 | >20 | >20 | >20 | >20 | 4.3 ± 0.2 |
| 12 | >20 | >20 | >20 | >20 | 1.5 ± 0.1 |
| 21 | 8.1 ± 0.1 | 15.5 ± 1.5 | 19.0 ± 1.1 | 2.4 ± 0.5 | 8.0 ± 0.3 |
| 22 | 9.6 ± 0.3 | 8.0 ± 0.3 | 4.0 ± 0.1 | 2.6 ± 0.1 | 8.8 ± 0.9 |
| 23 | 5.1 ± 0.8 | 4.2 ± 0.1 | 3.8 ± 0.2 | 0.9 ± 0.1 | 4.9 ± 0.4 |
| Adenanthin | 7.7 ± 1.1 | 7.3 ± 0.8 | 8.5 ± 1.0 | 5.4 ± 0.9 | 5.5 ± 0.6 |
| Cisplatin | 9.2 ± 1.4 | 8.5 ± 0.4 | 9.1 ± 0.6 | 9.3 ± 0.8 | 13.5 ± 1.3 |
Results are expressed as the mean ± standard deviation (SD). The other compounds not shown in the table were inactive (IC50 > 20 μM) toward all of the selected cell lines. The experiments were performed three times.
3.2. Compound 23 induces apoptosis in HepG2 cells
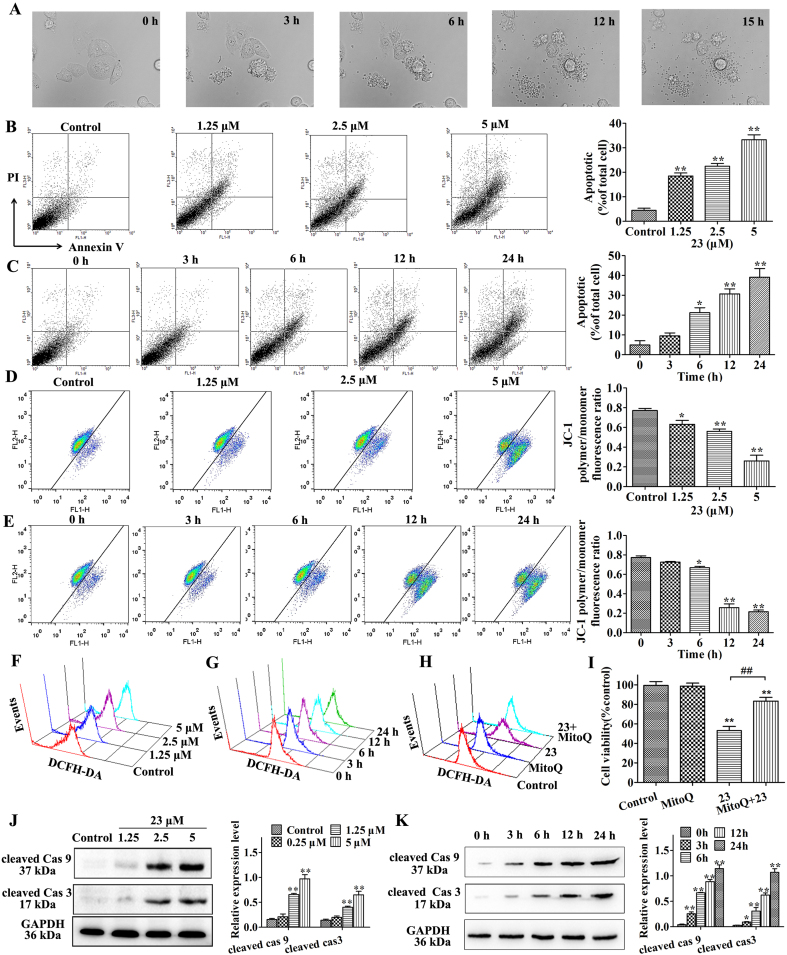
Morphological changes were detected by time-lapse phase-contrast microscopy. After exposure to 23 at 2.5 μM, the morphologies of HepG2 cells were altered, including cell membrane shrinkage and granular apoptotic body formation (Fig. 2A and Supplementary Video 1). Subsequently, cells were stained with AV/PI and analyzed by flow cytometry. And a significant dose- and time-dependent increase in the percentages of AV-positive cells was observed after treatment with 23 when compared with the control cells (Fig. 2B and C). JC-1 dye was used to further investigate the effect of 23 on the mitochondrial membrane potential (MMP, ΔΨm) in HepG2 cells. Compound 23 caused a loss of ΔΨm in HepG2 cells, and a gradual decrease in MMP was observed in response to different dose and treatment time of compound 23 (Fig. 2D and E). Next, we measured the cellular ROS levels after treatment with compound 23. The results demonstrated 23 elevated the ROS levels in a dose- and time-dependent manner (Fig. 2F and G, Fig. S5B and C). Further research showed that 23 also increased ROS and decreased ΔΨm at concentration of 10 μM in HUVEC and HL-7702 normal cell lines (Fig. S5D and E). We used the mitochondria-targeted antioxidant MitoQ to verify the role of ROS in 23-induced cell death. The appropriate concentration of MitoQ in HepG2 cells was 1 μM through MTT and DCFH-DA staining assay (Fig. S5F and G). The effects of MitoQ on the 23-induced ROS accumulation and cell viability were detected and the results showed that MitoQ reversed the 23-induced ROS accumulation and cell viability inhibition (Fig. 2H and I, Fig. S5H). Meanwhile, the results demonstrated that 23 significantly increased the expression of cleaved of caspase-9 and caspase-3 after treatment with compound 23 (Fig. 2J and K).
Fig. 2.
Compound 23 induces apoptosis in HepG2 cells. (A) Morphological changes were detected by time-lapse phase-contrast microscopy. Cells were treated with 2.5 μM 23 and bright field images were captured every 5 min by a time-lapse microscope. (B, C) Cells were exposed to 1.25, 2.5 and 5 μM 23 for 24 h (B) or 2.5 μM 23 for 0, 3, 6, 12, 24 h (C). Cells were collected and stained with Annexin V and PI, *P < 0.05, **P < 0.01, relative to control or 0 h. (D, E) ΔΨm was measured by flow cytometry. Cells were treated with 1.25, 2.5 and 5 μM 23 for 24 h (D) or 2.5 μM 23 for 0, 3, 6, 12, 24 h (E), *P < 0.05, **P < 0.01, relative to control or 0 h. (F, G) The intracellular ROS in HepG2 cells was detected by DCFH-DA staining assay. Cells were exposed to 1.25, 2.5 and 5 μM 23 for 24 h (F) or 2.5 μM 23 for 0, 3, 6, 12, 24 h (G). (H) The intracellular ROS was detected by DCFH-DA staining assay. HepG2 cells were pretreated with Mito-Q (1 μM) for 1 h and incubated with 2.5 μM of 23 for 24 h. (I) Cell viability was measured using the MTT assay. HepG2 cells were pretreated with MitoQ (1 μM) for 1 h and incubated with 2.5 μM of 23 for 24 h **P < 0.01, relative to control, ##P < 0.01 relative to 23. (J, K) The expression of cleaved caspase-3 and cleaved caspase-9 were detected by Western blot. HepG2 cells were treated with 1.25, 2.5 and 5 μM 23 for 24 h (J) or 2.5 μM 23 for 0, 3, 6, 12, 24 h (K), *P < 0.05, **P < 0.01, relative to control or 0 h. Results are expressed as mean ± SD, (n = 3).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.redox.2021.101977
The following is the supplementary data related to this article:
Video 1
3.3. Compound 23 induces ferroptosis in HepG2 cells
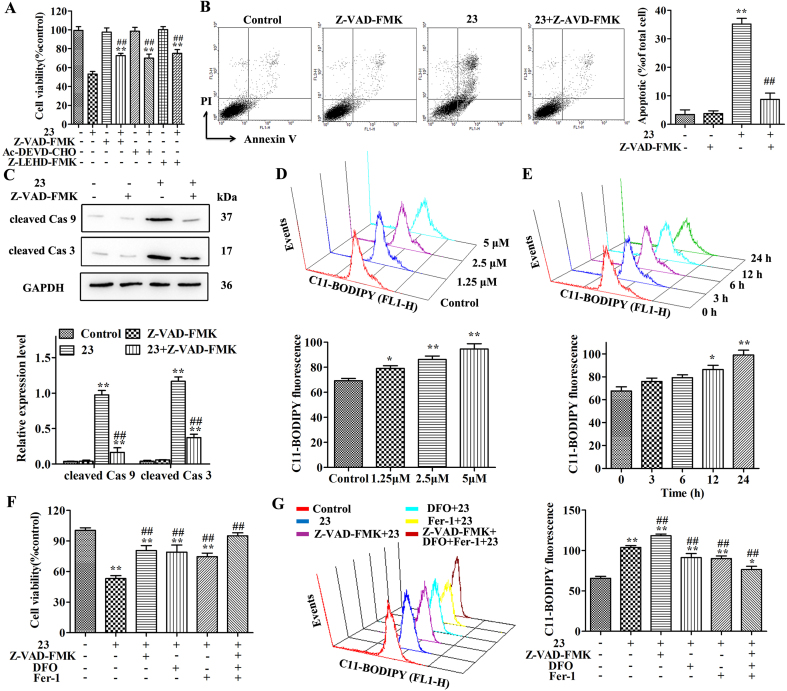
Then, after pretreating the cells with the Z-VAD-FMK (pan-caspase inhibitor), Ac-DEVD-CHO (caspase-3 inhibitor), or Z-LEHD-FMK TFA (caspase-9 inhibitor) for 2 h, the effects of 23 on cell viability were partially reversed in HepG2 cells by caspase inhibitors (Fig. 3A). In addition, the effects of Z-VAD-FMK on cell apoptosis, cell morphological changes, caspase activation, ROS and MMP were detected respectively. The results showed that the effects of 23 on cell morphological changes, cell apoptosis and caspase activation were markedly blocked by Z-VAD-FMK (Fig. 3B and C, Fig. S5I). However, Z-VAD-FMK has no effect on the changes in ROS and MMP caused by 23 (Fig. S5J and K).
Fig. 3.
Compound 23 induces ferroptosis in HepG2 cells. (A) Cell viability was measured using the MTT assay. HepG2 cells were pretreated with Z-VAD-FMK (10 μM), Ac-DEVD-CHO (50 μM), or Z-LEHD-FMK TFA (20 μM) for 2 h and incubated with 2.5 μM of 23 for 24 h **P < 0.01 relative to control, ##P < 0.01 relative to 23. (B) Cells were pretreated with Z-VAD-FMK (10 μM) for 2 h and incubated with 2.5 μM of 23 for 24 h. And the cells were collected and stained with Annexin V and PI. **P < 0.01 relative to control, ##P < 0.01 relative to 23. (C) The expression of cleaved caspase-3 and cleaved caspase-9 were detected by Western blot. HepG2 cells were treated as above. *P < 0.01 relative to control, ##P < 0.01 relative to 23. (D, E) Lipid peroxidation was measured by flow cytometry. Cells were exposed to 1.25, 2.5 and 5 μM 23 for 24 h (D) or 2.5 μM 23 for 0, 3, 6, 12, 24 h (E), *P < 0.05, **P < 0.01, relative to control or 0 h. (F) Cell viability was measured using the MTT assay. HepG2 cells were pretreated with Fer-1 (10 μM) or DFO (10 μM) or Z-VAD-FMK (10 μM) for 1 h and incubated with 2.5 μM of 23 for 24 h **P < 0.01 relative to control, ##P < 0.01 relative to 23. (G) Lipid peroxidation was measured by flow cytometry. Cells were treated as above. **P < 0.01 relative to control, ##P < 0.01 relative to 23. Results are expressed as mean ± SD, (n = 3).
An important mechanism in ferroptosis-mediated cell death is ROS production and oxidative stress [31,32]. The increased ROS in 23 treated cells raises the possibility that 23 may contribute to cell death through ferroptosis. To test this possibility, we detected the lipid peroxidation in HepG2 cells and the dose- and time-dependent increase in fluorescence intensity of the C11-BODIPY was monitored after 23 treatment by flow cytometry (p < 0.01) (Fig. 3D and E). Further research results showed that the effects of 23 on cell morphological changes and cell viability were markedly blocked by pretreatment with DFO (iron chelator), Fer-1 (ferroptosis inhibitor) or Z-VAD-FMK (Fig. 3F, Fig. S5L). Meanwhile, the lipid peroxidation was attenuated in the presence of DFO or Fer-1 (Fig. 3G). We also measured the expression of Glutathione peroxidase 4 (GPX4) in cells exposed to 23. The Western blot results showed that 23 has no effect on the expression of GPX4 in HepG2 cells (Fig. S5M).
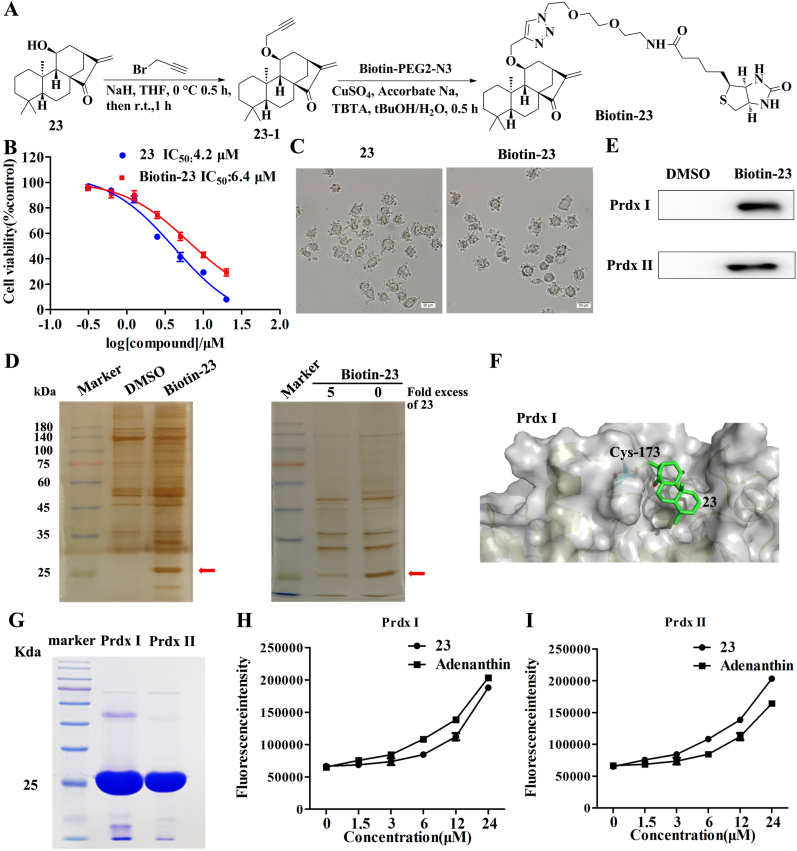
3.4. Compound 23 directly targets Prdx I and Prdx II
To further explore the functional target(s) of compound 23, a biotin-tagged probe biotin-23 was synthesized (Fig. 4A and Supplementary data). Compared with 23, biotin-23 showed similar cytotoxic activity and the same changes in cell morphology (Fig. 4B and C). To identify the potential cellular targets of 23, pull-down experiments using biotin-23 were performed. The streptavidin agarose beads were incubated with DMSO or biotin-23 and then incubated with HepG2 cell lysates. The proteins precipitated by streptavidin agarose beads were resolved on SDS-PAGE and stained with silver. A band of approximately 23 kDa was detected only in the cell lysates incubated with biotin-23 (Fig. 4D). The band was competed away by high concentrations of unlabeled 23, indicating that the protein that bound biotin-23 also bound 23 (Fig. 4D). MS analysis revealed that the 23-bound protein is Prdx I/II (Figure S6). Thus, we further blotted the precipitates with antibodies against Prdx I and Prdx II, which were effectively pulled down by biotin-23 (Fig. 4E). To further explore how 23 inactivates Prdx I, molecular dynamics (MD) simulations using the dimeric structure of Prdx I (PDB: 1QQ2) and 23 were conducted. It was found that when binding to Prdx I, 23 docks into the protein pocket formed by the dimerization of Prdx I and disrupts the Cys173-Cys52 disulfide bond by modifying Cys173 (Fig. 4F).
Fig. 4.
Compound 23 directly targets Prdx I and Prdx II. (A) Scheme for synthesis of probe Biotin-23. (B) Cell viability was measured using the MTT assay. HepG2 cells were treated with various concentrations of biotin-23 and 23 for 24 h. (C) Cell morphological changes were detected by a phase-contrast microscopy after treatment with 5 μM biotin-23 or 23 for 12 h. (D, E) HepG2 cell lysates were incubated with biotin-23 in the absence or presence of a 5-fold excess of unlabeled 23, followed by pull-down with streptavidin-agarose. The precipitates were resolved by SDS-PAGE, and the gel was stained with silver or detected by western blotting for Prdx I/II as indicated. (F) Model for binding of 23 to the pocket formed by Prdx I dimerization (PDB: 1QQ2). 23 docks into the protein pocket formed by the dimerization of Prdx I and disrupts the Cys173-Cys52 disulfide bond by modifying Cys173. (G) The recombinant Prdx I and Prdx II proteins on the gel were stained with Commassie Blue. (H, I) The peroxidase activity of recombinant Prdx I (H) and Prdx II (I) proteins was detected by Amplex Red probe. Results are expressed as mean ± SD, (n = 3). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To determine whether 23 directly binds to Prdx I and Prdx II, recombinant Prdx I and Prdx II were expressed and purified in a prokaryotic expression system (Fig. 4G, Supplementary data). We measured the peroxidase activities of recombinant Prdx I and Prdx II proteins in the presence of 23 or adenanthin (Prdx I/II inhibitor). The results demonstrated that 23 and adenanthin effectively inhibited the peroxidase activity of recombinant Prdx I and Prdx II (Fig. 4H and I).
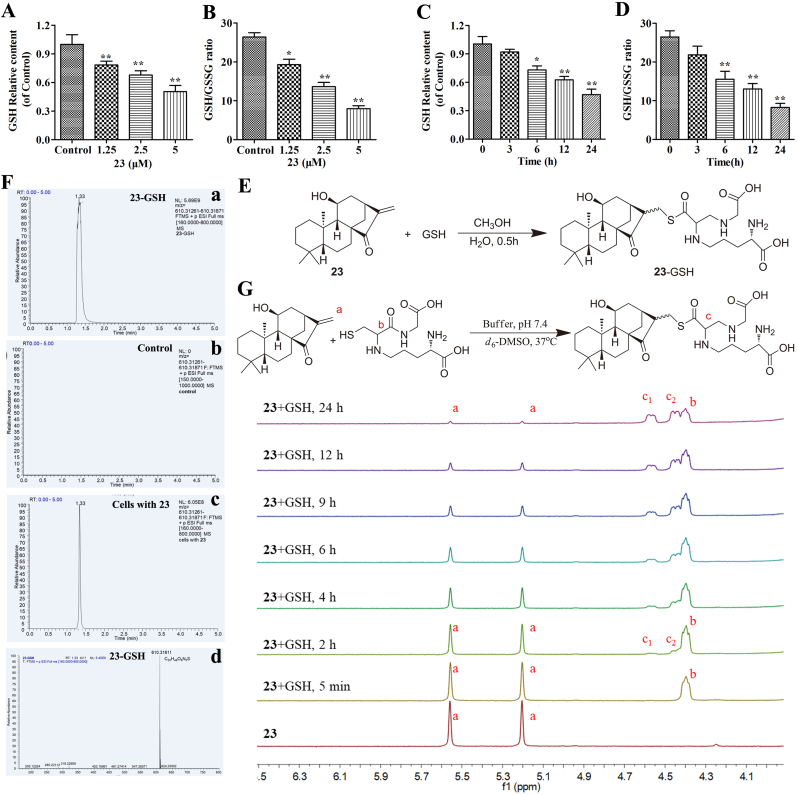
3.5. Compound 23 directly reacts with GSH
GSH, a thiol-based intracellular antioxidant, is also a major ROS scavenging agent in cells [33]. Depletion of GSH results in ROS accumulation and cell death [34]. We next detected the effects of 23 on the levels of cellular GSH and the ratio of GSH/GSSG. And the results showed that the GSH content and the ratio of GSH/GSSG in HepG2 cells were reduced upon treatment with 23 in a dose- and time-dependent manner (Fig. 5A, B, C and D). GSH possesses a mercapto moiety, which has the potential to react with the α,β-unsaturated carbonyl moiety of diterpenoids. We first obtained the conjugated compound 23-GSH by incubating 23 with GSH (Fig. 5E and Supplementary data). As expected, conjugated 23-GSH was detected in the treated cells by HPLC-MS/MS (Fig. 5F). Collectively, these results clearly demonstrated that 23 depleted intracellular GSH through a conjugation reaction.
Fig. 5.
Compound 23 directly reacts with GSH. (A, B, C, D) HepG2 cells were treated with 1.25, 2.5 and 5 μM 23 for 24 h or 2.5 μM 23 for 0, 3, 6, 12, 24 h, and the levels of GSH (A, C) and the ratio of GSH/GSSG (B, D) were analyzed using GSH detection kits. *P < 0.05, **P < 0.01, relative to control or 0 h. (E) Scheme for synthesis of 23-GSH. (F) The 23-GSH was detected by UPLC-MS/MS. Standard substances (a); Control group (b); 23 treated group (c); Ion extraction of 23-GSH in the 23 treated group (d). After treatment with 0 or 2.5 μM 23 for 8 h, the cells were collected and lysed using RIPA lysis buffer. (G) The reaction was monitored by the change of representative hydrogen signals with 1H NMR analysis. Compound 23 (1.2 mg) were incubated with an excess of GSH (2.4 mg) in deuterated phosphate buffer at pH 7.4 (0.02 mL) and DMSO‑d6 (0.50 mL) at 37 °C. Results are expressed as mean ± SD, (n = 3).
In order to detect the reactivity window of 23, we utilized NMR spectroscopy to determine the reaction rates of 23 with GSH. Compound 23 was incubated at 37 °C with 1.5-2 equiv of GSH in 0.5 mL of DMSO‑d6 and 0.02 mL of sodium phosphate deuterium oxide buffer (pH 7.4). The samples were measured by 1H NMR spectrum at seven time points. Based on the 1H NMR peak c generated from the reaction, t1/2 was identified as approximately 4-6 h (Fig. 5G). Therefore, these results indicated that the D-ring enone of 23 is a drug like pharmacophore.
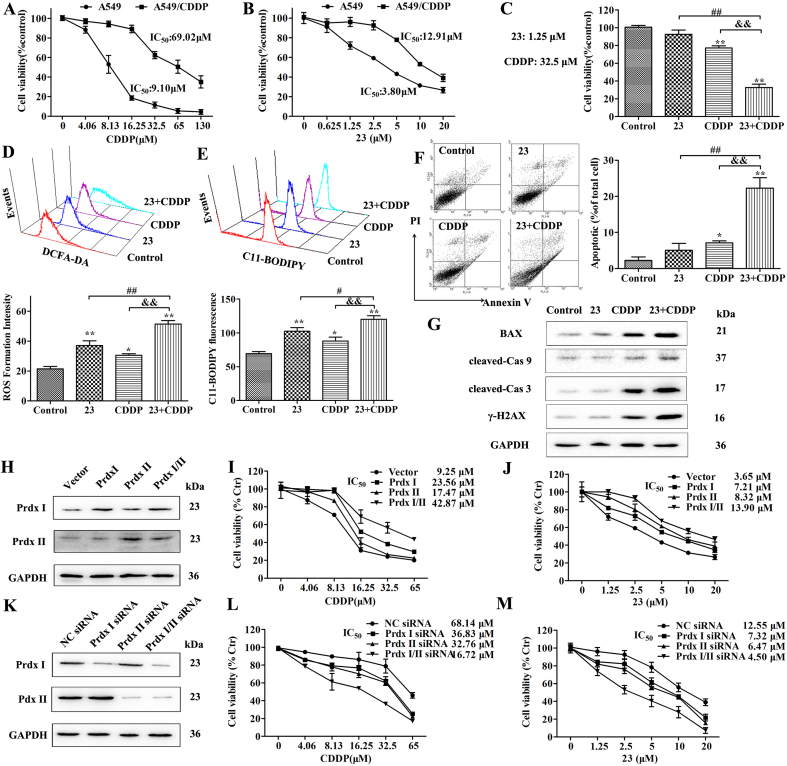
3.6. Compound 23 sensitizes CDDP-induced cell death against A549/CDDP cells
Since compound 23 could disrupt redox resetting by restraining Prdx I/II and GSH, we explored the possibility whether inhibition of Prdx I/II and GSH by 23 could sensitize CDDP-resistant cancer cells. To substantiate the above concept, we first proved that CDDP exhibited weak cytotoxicity in A549/CDDP cells than that in A549 cells (Fig. 6A). Then, we assayed the expression of Prdx I/II and the GSH level in A549 and A549/CDDP cells. Compared with A549 cells, the levels of Prdx I/II and GSH were remarkably increased in A549/CDDP cells (Fig. S7A, B and C). The MTT assay showed that compound 23 displayed less toxicity in A549/CDDP cells than that in A549 cells (Fig. 6B). Next, the A549/CDDP cells were treated with 23 (1.25 μM) and CDDP (32.5 μM). The drug combination of 23 and CDDP showed significant inhibitory activity against A549/CDDP cells than the groups of 23 or CDDP (Fig. 6C). Moreover, the ROS formation intensity and C11-BODIPY fluorescence intensity of A549/CDDP cells induced by the combination treatment group was higher than the groups of 23 or CDDP (Fig. 6D and E). Next, cells were stained with annexin V/PI, and a significant increase in the percentages of apoptotic cells was observed after combination treatment with 23 (1.25 μM) and CDDP (32.5 μM) (Fig. 6F). The expression of apoptosis-related proteins in A549/CDDP cells was also investigated. The expression levels of cleaved caspase-3, caspase-9, γ-H2AX and Bax were upregulated more apparently in the combination group than that in the groups of 23 or CDDP (Fig. 6G, Fig. S7D).
Fig. 6.
Compound 23 sensitizes CDDP-induced cell death against A549/CDDP cells. (A) Cytotoxicity of CDDP against the A549 and A549/CDDP cells was determined by MTT assay. Cells were treated with various concentrations CDDP for 24 h. (B) Cytotoxicity of 23 against the A549 and A549/CDDP cells were determined by MTT assay. Cells were treated with various concentrations 23 for 24 h. (C) Cells were treated with indicated dose of 23 (1.25 μM) or CDDP (32.5 μM) for 24 h. Cell viability was detected by MTT assay. (D) Effect of combined treatment on the ROS in A549/CDDP cells was detected by flow cytometry. Cells were treated as above and stained with DCFH-DA. (E) Effect of combined treatment on lipid peroxidation was detected by flow cytometry. Cells were treated as above and stained with C11-BODIPY. (F) Effect of combined treatment on the cell apoptosis was detected by flow cytometry. Cells were treated as above and stained with AV and PI. **P < 0.01, *P < 0.05, relative to control group, ##P < 0.01 relative to 23 group, &&P < 0.01 relative to CDDP group. (G) Western blot was performed to examine the effects of combined treatment on the expression of BAX, cleaved caspase-3, cleaved caspase-9 and γ–H2AX. (H) Western blot was performed to examine the overexpression of Prdx I/II in A549 cells. Infected cells were harvested at the indicated times and cell lysates were blotted for proteins as indicated. (I, J) MTT assay was performed to examine the effect of Prdx I/II on cell viability induced by 23 and CDDP. Infected cells were treated with different concentration of 23 or CDDP as indicated for 24 h. (K) Western blot was performed to examine the effect of Prdx I siRNA and Prdx II siRNA on expression of Prdx I/II in A549/CDDP cells. Infected cells were harvested at the indicated times and cell lysates were blotted for proteins as indicated. (L, M) MTT assay was performed to examine the effect of Prdx I/II on cell viability induced by 23 and CDDP. Infected cells were treated with different concentration of 23 or CDDP as indicated for 24 h.
To validate that Prdx I/II are the endogenous targets of 23 and play an important role in CDDP-resistant A549 cells, we knocked down and overexpressed Prdx I/II in A549 or A549/CDDP cells. We confirmed the transfection efficiency using western blot analysis (Fig. 6H, Fig. S7E). Cell viability assay showed that Prdx I/II overexpression significant decreased the effect of 23 and CDDP on cell viability in A549 cells (Fig. 6I and J). Prdx I siRNA inhibited Prdx I but not Prdx II expression, whereas Prdx II siRNA almost completely eliminated Prdx II but not Prdx I expression, indicating the specificity of these siRNAs (Fig. 6K, Fig. S7F). Silencing of Prx I or Prx II expression also increased the effect of 23 and CDDP on cell viability in A549/CDDP cells (Fig. 6L and M).
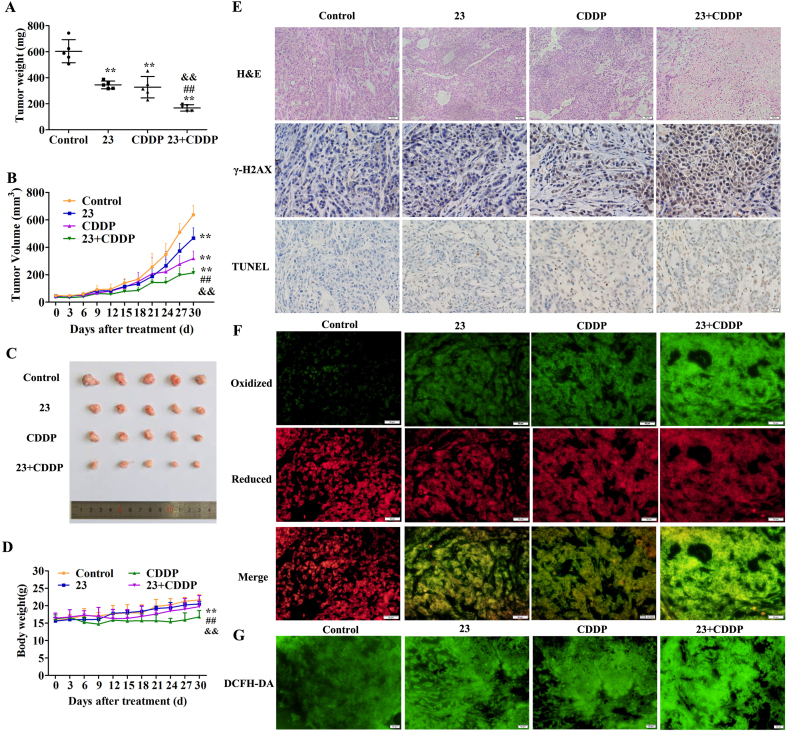
3.7. Compound 23 increases the antitumor efficacy of CDDP in nude mice bearing A549/CDDP cells
The drug combination of 23 and CDDP showed significant inhibitory activity against A549/CDDP cells in vitro. Therefore, the antitumor therapeutic potential in vivo was further investigated. The combination of 23 and CDDP significantly reduced the final tumor weight in the mice compared to the other three groups, basically matching the tumor volume curves (p < 0.01) (Fig. 7A, B, C). The curves indicated that the body weight in combination treatment group is higher than that of the CDDP treatment group (p < 0.01) (Fig. 7D). The mice treated with CDDP showed the myocardial fiber break, hepatocyte necrosis and apoptosis, alveolar damage, and vacuolar degeneration of renal cells (black arrow). In comparison, there was no obvious tissue damage in the combined treatment group, control group, and 23 treatment group (Fig. S8A). The AST and ALT test results also confirmed that the combination group had lower liver toxicity (Fig. S8B and C).
Fig. 7.
Compound 23 increases the antitumor efficacy of CDDP in nude mice bearing A549/CDDP cells. (A) Differences in tumor weight in mice treated with 0.9% saline, 23 (10 mg/kg), CDDP (4 mg/kg) and combination of 23 (10 mg/kg) and CDDP (4 mg/kg). (B) Progression of tumor volumes of the four groups in mice during 30 days treatment. (C) Images of tumor of the four groups in mice after 30 days treatment. (D) Changes in body weight of mice in the four groups during 30 days treatment. *P < 0.05, **P < 0.01, relative to control group, ##P < 0.01 relative to 23 group, &&P < 0.01 relative to CDDP group. (E) H&E staining, immunohistochemical staining of γ-H2AX and TUNEL staining in tumor tissues. Scale bar, 20 μm. (F) Effect of combination of 23 and CDDP on cell lipid peroxidation. Scale bar, 50 μm. (G) Effect of combination of 23 and CDDP on intracellular ROS level. Scale bar, 50 μm.
In addition, the tumor sections were assessed by histological and immunohistochemical analysis. The H&E staining results showed that administration of 23 and CDDP markedly induced tumor cell damage compared to the CDDP group (Fig. 7E). The number of γ-H2AX-positive and TUNEL-positive tumor cells was increased, confirming 23 could enhance CDDP-induced DNA damage and apoptotic index (Fig. 7E, Fig. S9A and B). Next, we evaluated the effects of the combined treatment with 23 and CDDP on lipid peroxidation and cellular ROS. The results showed that the combination group significantly increased lipid peroxidation and ROS in the mice compared with other three groups (Fig. 7F and G, Fig. S9C and D).
4. Discussion
New therapeutic drugs that specifically and effectively target cancer cells are essential for the development of promising cancer treatment modalities [35]. ent-Kauranes have shown a variety of encouraging biological activities, including anti-inflammatory, antimicrobial, antioxidant, and antitumor properties [36,37].
Herein, we identified thirty ent-kaurane derivatives from the Chinese liverworts J. tetragona. Among them, compound 23 showed strong inhibitory activity against several cancer cell lines. Further research displayed that compound 23 induced apoptosis through a loss of the ΔΨm, activation of caspases and accumulation of cellular ROS. Adenanthin, a natural diterpenoid isolated from the leaves of Isodon adenanthus, induced leukemia cell differentiation by targeting Prdx I/II [5]. JDA-202, another diterpenoid, was proved to inhibit the activity of Prdx I and possess strong anti-proliferative activities on esophageal cancer cells [38]. Through chemical biology studies, we found that compound 23 targeted Prdx I/II and selectively bound to conserved cysteine residues of Prdx I/II. Moreover, compound 23 effectively inhibited the peroxidase activity of recombinant Prdx I/II. We also found that compound 23 reacted directly with GSH in HepG2 cells and in vitro.
Cancer cells have higher levels of ROS than normal cells as a result of hypermetabolism, but the redox balance is maintained in cancer cells due to their marked antioxidant capacity [39]. Destruction of redox balance by small molecules can selectively kill tumor cells, which will provide an alternative way to fight cancer [40]. Herein, the data discovered that compound 23 induced cancer cell death by redox resetting destruction through targeting Prdx I/II and depletion of GSH. GPX4 has been shown to be an important regulator of ferroptosis, which was mediated by GSH [41]. The biosynthesis of GSH is essential to protect cells from oxidative damage, and the cysteine-glutathione pathway is one of the most critical upstream mechanisms for ferroptosis [[42], [43], [44]]. Peroxiredoxins (Prdxs), a ubiquitous family of redox regulatory proteins, are reported of potential to eliminate various ROS [45]. Prdx I/II belong to the typical 2-Cys subfamily and were initially identified as natural enhancing factors [46]. The Prdxs, especially Prdx I and Prdx II, are overexpressed in several human cancer types [47,48]. Loss of Prdx I contributed to ferroptosis induced by ferrostatin-1 through upregulation of lipid ROS [49]. The current study is the first to report the ent-Kauranes induce ferroptosis through targeting Prdx I/II and depletion of GSH.
Compounds modulating cellular ROS levels can enhance multidrug resistance cancer cell death and sensitize multidrug resistance cancer cells to chemotherapeutic drugs [50]. CDDP-based chemotherapy is still the most effective strategy for lung cancer treatment [51]. Sustaining the CDDP efficacy for lung cancer is quintessential due to prevailing drug resistance [26,52]. The mechanisms leading to CDDP resistance are highly complex and include: altered cellular accumulation of drug, altered DNA repair and cytosolic inactivation of drug [53,54]. Increasing data from in vitro and in vivo research has identified that CDDP resistance is closely related to redox resetting, which is produced mainly through the high expression of antioxidant substances such as GSH, Prdx, and Trx [29]. Our study also showed that Prdx I/II were related to CDDP resistance and compound 23 enhanced the sensitivity of A549/CDDP to CDDP. Therefore, the current data and others’ studies implies that impairing antioxidant capacity of CDDP-resistant cancers emerges as a good strategy to overcome CDDP resistance.
Many ROS modulators which regulate the antioxidant system, including natural products and approved drugs, are undergoing clinical trials to treat certain drug-resistant cancer patients [55]. The current study provides more options for pharmacologists to develop formulations composed of certain drug combinations. Despite having broad antitumor spectrum, the application of compound 23 was hindered due to its moderate efficacy and poor selectivity. A series of enmein-type diterpenoid amino acid ester derivatives have been designed and synthesized, which displayed high efficiency in cancer cells and almost no cytotoxicity in normal cells [56]. So we can also synthesize some new amino acid ester derivatives of 23 to obtain high efficiency and selectivity in the follow-up research.
In conclusion, the present study determined that compound 23 induced both ferroptosis and apoptosis through GSH depletion and inactivation of Prdx I/II. The synergistic antitumor effect of compound 23 combined with CDDP suggested that ent-kaurane diterpenoids modulate ROS to overcome CDDP resistance in cancer. Therefore, ent-kaurane diterpenoids provide a novel sensitized approach for the clinical application of CDDP by redox resetting destruction, and a bright future for overcoming tumor resistance.
Author contributions
Yong Sun, Yanan Qiao, and Hongxiang Lou conceived and designed the experiments. Yong Sun, Yanan Qiao, Yue Liu, Jinchuan Zhou, and Xue Wang performed the experiments. Yong Sun, Yanan Qiao, and Chunyang Zhang analyzed the data. Yong Sun, Yanan Qiao, Hongbo Zheng, Zejun Xu, Jiaozhen Zhang, Yi Zhou, and Lilin Qian contributed reagents/materials/analysis tools. Yong Sun, Yanan Qiao, and Hong-xiang Lou wrote the draft, and checked and revised. All authors approved to submit this version to this publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81630093, 81874293) and Foundation of Ministry of Science and Technology of the People's Republic of China (2019YFA0905701), and Major Basic Research Program of Shandong Provincial Natural Science Foundation (ZR2019ZD26). We thank Dr. Shuqi Wang for NMR measurements.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101977.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Detailed procedure on the extraction and isolation of chemical constituents from J. tetragona, the chemical structures of purified constituents, and supplementary figures have been summarized in supplementary data.
References
- 1.Liu N., Wu C., Wang P., Lou H.X. Diterpenoids from liverworts and their biological activities. Curr. Org. Chem. 2018;22:1847–1860. [Google Scholar]
- 2.Nagashima F., Kondoh M., Fujii M., Takaoka S., Watanabe Y., Asakawa Y. Novel cytotoxic kaurane-type diterpenoids from the New Zealand Liverwort Jungermannia species. Tetrahedron. 2005;61:4531–4544. [Google Scholar]
- 3.Nagashima F., Kondoh M., Uematsu T., Nishiyama A., Saito S., Sato M. Cytotoxic and apoptosis-inducing ent-kaurane-type diterpenoids from the Japanese liverwort Jungermannia truncata NEES. Chem. Pharm. Bull. (Tokyo) 2002;50:808–813. doi: 10.1248/cpb.50.808. [DOI] [PubMed] [Google Scholar]
- 4.Sun H.D., Huang S.X., Han Q.B. Diterpenoids from Isodon species and their biological activities. Nat. Prod. Rep. 2006;23:673–698. doi: 10.1039/b604174d. [DOI] [PubMed] [Google Scholar]
- 5.Liu C.X., Yin Q.Q., Zhou H.C., Wu Y.L., Pu J.X., Xia L. Adenanthin targets peroxiredoxin I and II to induce differentiation of leukemic cells. Nat. Chem. Biol. 2012;8:486–493. doi: 10.1038/nchembio.935. [DOI] [PubMed] [Google Scholar]
- 6.Santagata S., Xu Y.M., Wijeratne E.M., Kontnik R., Rooney C., Perley C.C. Using the heat-shock response to discover anticancer compounds that target protein homeostasis. ACS Chem. Biol. 2012;7:340–349. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wijeratne E.M., Bashyal B.P., Liu M.X., Rocha D.D., Gunaherath G.M., U'Ren J.M. Geopyxins A-E, ent-kaurane diterpenoids from endolichenic fungal strains Geopyxis aff. majalis and Geopyxis sp. AZ0066: structure-activity relationships of geopyxins and their analogues. J. Nat. Prod. 2012;75:361–369. doi: 10.1021/np200769q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickinson B.C., Chang C.J. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 2011;7:504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhee S.G. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 10.Schumacker P.T. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Canc. Cell. 2006;10:175–176. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Nathan C., Cunningham-Bussel A. Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nat. Rev. Immunol. 2013;13:349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacker P.T. Reactive oxygen species in cancer: a dance with the devil. Canc. Cell. 2015;27:156–157. doi: 10.1016/j.ccell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y., Li Q., Zhou L., Xie N., Nice E.C., Zhang H. Cancer drug resistance: redox resetting renders a way. Oncotarget. 2016;7:42740–42761. doi: 10.18632/oncotarget.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 15.Gorrini C., Harris I.S., Mak T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 16.Marullo R., Werner E., Degtyareva N., Moore B., Altavilla G., Ramalingam S.S. Cisplatin induces a mitochondrial-ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PloS One. 2013;8 doi: 10.1371/journal.pone.0081162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundgren C.A.K., Sjöstrand D., Biner O. Scavenging of superoxide by a membrane-bound superoxide oxidase. 2018;14:788–793. doi: 10.1038/s41589-018-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong H., Xiao M., Zarkovic K., Zhu M., Sa R., Lu J. Mitochondrial control of apoptosis through modulation of cardiolipin oxidation in hepatocellular carcinoma: a novel link between oxidative stress and cancer. Free Radic. Biol. Med. 2017;102:67–76. doi: 10.1016/j.freeradbiomed.2016.10.494. [DOI] [PubMed] [Google Scholar]
- 19.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J. Ferroptosis: a regulated cell death nexus linking metabolism. Redox Biology, and Disease, Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masaldan S., Clatworthy S.A.S., Gamell C., Meggyesy P.M., Rigopoulos A.T., Haupt S. Iron accumulation in senescent cells is coupled with impaired ferritinophagy and inhibition of ferroptosis. Redox Biol. 2018;14:100–115. doi: 10.1016/j.redox.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen T.H.P., Mahalakshmi B., Velmurugan B.K. Functional role of ferroptosis on cancers, activation and deactivation by various therapeutic candidates-an update. Chem. Biol. Interact. 2020;317:108930. doi: 10.1016/j.cbi.2019.108930. [DOI] [PubMed] [Google Scholar]
- 22.Ramanathan B., Jan K.Y., Chen C.H., Hour T.C., Yu H.J., Pu Y.S. Resistance to paclitaxel is proportional to cellular total antioxidant capacity. Canc. Res. 2005;65:8455–8460. doi: 10.1158/0008-5472.CAN-05-1162. [DOI] [PubMed] [Google Scholar]
- 23.Wang H., Jiang H., Corbet C., de Mey S., Law K., Gevaert T. Piperlongumine increases sensitivity of colorectal cancer cells to radiation: involvement of ROS production via dual inhibition of glutathione and thioredoxin systems. Canc. Lett. 2019;450:42–52. doi: 10.1016/j.canlet.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 24.Piska K., Koczurkiewicz P., Wnuk D., Karnas E., Bucki A., Wójcik-Pszczoła K. Synergistic anticancer activity of doxorubicin and piperlongumine on DU-145 prostate cancer cells - the involvement of carbonyl reductase 1 inhibition. Chem. Biol. Interact. 2019;300:40–48. doi: 10.1016/j.cbi.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Kalinina E.V., Berezov T.T., Shtil A.A., Chernov N.N., Glazunova V.A., Novichkova M.D. Expression of peroxiredoxin 1, 2, 3, and 6 genes in cancer cells during drug resistance formation. Bull. Exp. Biol. Med. 2012;153:878–881. doi: 10.1007/s10517-012-1849-7. [DOI] [PubMed] [Google Scholar]
- 26.Kwon T., Rho J.K., Lee J.C., Park Y.H., Shin H.J., Cho S. An important role for peroxiredoxin II in survival of A549 lung cancer cells resistant to gefitinib. Exp. Mol. Med. 2015;47:e165. doi: 10.1038/emm.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scarbrough P.M., Mapuskar K.A., Mattson D.M., Gius D., Watson W.H., Spitz D.R. Simultaneous inhibition of glutathione- and thioredoxin-dependent metabolism is necessary to potentiate 17AAG-induced cancer cell killing via oxidative stress. Free Radic. Biol. Med. 2012;52:436–443. doi: 10.1016/j.freeradbiomed.2011.10.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kachalaki S., Ebrahimi M., Mohamed Khosroshahi L., Mohammadinejad S., Baradaran B. Cancer chemoresistance; biochemical and molecular aspects: a brief overview. Eur. J. Pharmaceut. Sci. 2016;89:20–30. doi: 10.1016/j.ejps.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 29.Kalinina E.V., Chernov N.N. Why does chemotherapy stop affecting the cells of ovarian and breast tumors? Future Oncol. 2018;14:1137–1140. doi: 10.2217/fon-2018-0001. [DOI] [PubMed] [Google Scholar]
- 30.Lin Z.M., Guo Y.X., H Gao Y., Wang S.Q., Wang X.N., Xie Z.Y. Ent-kaurane diterpenoids from Chinese liverworts and their antitumor activities through Michael addition as detected in situ by a fluorescence probe. J. Med. Chem. 2015;58:3944–3956. doi: 10.1021/acs.jmedchem.5b00208. [DOI] [PubMed] [Google Scholar]
- 31.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao J.Y., Dixon S.J. Mechanisms of ferroptosis. Cell. Mol. Life Sci. 2016;73:2195–2209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu G., Fang Y.Z., Yang S., Lupton J.R., Turner N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 34.Lu S.C. Regulation of glutathione synthesis. Mol. Aspect. Med. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cognetti D.M., Weber R.S., Lai S.Y. Head and neck cancer: an evolving treatment paradigm. Cancer. 2008;113:1911–1932. doi: 10.1002/cncr.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M., Wang W.G., Sun H.D., Pu J.X. Diterpenoids from Isodon species: an update. Nat. Prod. Rep. 2017;34:1090–1140. doi: 10.1039/c7np00027h. [DOI] [PubMed] [Google Scholar]
- 37.Yao R., Chen Z., Zhou C., Luo M., Shi X., Li J. Xerophilusin B induces cell cycle arrest and apoptosis in esophageal squamous cell carcinoma cells and does not cause toxicity in nude mice. J. Nat. Prod. 2015;78:10–16. doi: 10.1021/np500429w. [DOI] [PubMed] [Google Scholar]
- 38.Shi X.J., Ding L., Zhou W., Ji Y., Wang J., Wang H. Pro-apoptotic effects of JDA-202, a novel natural diterpenoid, on esophageal cancer through targeting peroxiredoxin I. Antioxid. Redox Signal. 2017;27:73–92. doi: 10.1089/ars.2016.6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sosa V., Moliné T., Somoza R., Paciucci R., Kondoh H., Leonart Me L. Oxidative stress and cancer: an overview. Ageing Res. Rev. 2013;12:376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Hou G.X., Liu P.P., Zhang S., Yang M., Liao J., Yang J. Elimination of stem-like cancer cell side-population by auranofin through modulation of ROS and glycolysis. Cell Death Dis. 2018;9:89. doi: 10.1038/s41419-017-0159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skouta R., Dixon S.J., Wang J., Dunn D.E., Orman M., Shimada K. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J. Am. Chem. Soc. 2014;136:4551–4556. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Y., Hou W., Song X., Yu Y., Huang J., Sun X. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimada K., Stockwell B.R. tRNA synthase suppression activates de novo cysteine synthesis to compensate for cystine and glutathione deprivation during ferroptosis. Mol Cell Oncol. 2016;3 doi: 10.1080/23723556.2015.1091059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhee S.G., Kang S.W., Chang T.S., Jeong W., Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life. 2001;52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
- 46.Shau H., Kim A. Identification of natural killer enhancing factor as a major antioxidant in human red blood cells. Biochem. Biophys. Res. Commun. 1994;199:83–88. doi: 10.1006/bbrc.1994.1197. [DOI] [PubMed] [Google Scholar]
- 47.Suet-Hui Ow, Pei-Jou Chua, Boon-Huat Bay. Epigenetic regulation of peroxiredoxins: implications in the pathogenesis of cancer. Exp. Biol. Med. 2017;242:140–147. doi: 10.1177/1535370216669834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding C., Fan X., Wu G. Peroxiredoxin 1 - an antioxidant enzyme in cancer. J. Cell Mol. Med. 2017;21:193–202. doi: 10.1111/jcmm.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lovatt M., Adnan K., Kocaba V., Dirisamer M., Peh G.S.L., Mehta J.S. Peroxiredoxin-1 regulates lipid peroxidation in corneal endothelial cells. Redox Biol. 2020;30:101417. doi: 10.1016/j.redox.2019.101417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sznarkowska A., Kostecka A., Meller K., Bielawski K.P. Inhibition of cancer antioxidant defense by natural compounds. Oncotarget. 2017;8:15996–16016. doi: 10.18632/oncotarget.13723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D., Lippard S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 52.Amable L. Cisplatin resistance and opportunities for precision medicine. Pharmacol. Res. 2016;106:27–36. doi: 10.1016/j.phrs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Richon V.M., Schulte N., Eastman A. Multiple mechanisms of resistance to cis-diamminedichloroplatinum(II) in murine leukemia L1210 cells. Canc. Res. 1987;47:2056–2061. [PubMed] [Google Scholar]
- 54.Ferry K.V., Hamilton T.C., Johnson S.W. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells: role of ERCC1-XPF. Biochem. Pharmacol. 2000;60:1305–1313. doi: 10.1016/s0006-2952(00)00441-x. [DOI] [PubMed] [Google Scholar]
- 55.Cui Q., Wang J.Q., Assaraf Y.G., Ren L., Gupta P., Wei L. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist. Updates. 2018;41:1–25. doi: 10.1016/j.drup.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Hu X., Bai Z., Qiao J., Li H., Xu S., Wang X. Effective enmein-type mimics of clinical candidate HAO472: design, synthesis and biological evaluation. Eur. J. Med. Chem. 2019;171:169–179. doi: 10.1016/j.ejmech.2019.03.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1