Abstract
Developing robust methods to detect the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), a causative agent for the current global health pandemic, is an exciting area of research. Nevertheless, the currently used conventional reverse transcription-polymerase chain reaction (RT-PCR) technique in COVID-19 detection endures with some inevitable limitations. Consequently, the establishment of rapid diagnostic tools and quick isolation of infected patients is highly essential. Furthermore, the requirement of point-of-care testing is the need of the hour. Considering this, we have provided a brief review of the use of very recently reported robust spectral tools for rapid COVID-19 detection. The spectral tools include, colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) and matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), with the admittance of principal component analysis (PCA) and machine learning (ML) for meeting the high-throughput and fool-proof platforms for the detection of SARS-CoV-2, are reviewed. Recently, these techniques have been readily applied to screen a large number of suspected patients within a short period and they demonstrated higher sensitivity for the detection of COVID-19 patients from unaffected human subjects.
Keywords: COVID-19, SARS-CoV-2, RT-PCR, RT-LAMP, MALDI-MS, Machine learning
1. Introduction
The current pandemic COVID-19 is caused by an etiological agent, a coronavirus named SARS-CoV-2 [1]. The striking feature of the COVID-19 outbreak is, it has symptoms similar to that of other common respiratory diseases and even some infected individuals can be asymptomatic [2]. This unusual characteristic has made tracing this novel coronavirus a laborious task. A real-time reverse transcription-polymerase chain reaction (RT-PCR) is the gold standard technique for identifying the viral nucleic acids and thus finds its applications in the diagnosis of SARS-CoV-2 viral infections [3]. However, low viral load containing swab specimens, improper sampling, and lack of high purity RNA sample accounts for the poor sensitivity and false-negative results of RT-PCR-based detection of COVID-19 [4], [5]. The impact of false negatives could be severe and also lead to grave consequences of transmission of viral infections. In the current scenario, China has recorded false positives rates for RT-PCR-based viral detection kits as high as 20–40% [4]. One of the crucial parameters for arriving at false-negative results is attributed to the quality of the RT-PCR test kit, mainly the detection limit of viral RNA. Among the 6 RT-PCR kits - Liferiver, Huada, GeneoDx, DAAN, Sansure, and BioGerm approved by China National Medical Products Administration (NMPA), GeneoDx and BioGerm showed a poor limit of detection (LOD) with 7744 copies/mL and 968 copies/mL, respectively [6]. Poor LOD and low sensitivity of these RT-PCR kits led to improper diagnosis of viral infections in the pandemic situation, which eventually result in false-negative results. Given this milieu, new screening techniques are required to correctly identify infected patients from probable cases and control the spread of these viral infections.
In this review, we aim at introducing matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) based and reverse transcription loop-mediated isothermal amplification (RT-LAMP)-based diagnostic tools with the intervention of artificial intelligence (AI) and machine learning (ML) techniques for the rapid and accurate detection of SARS-COV-2 virus. The application of AI and ML automation in health maintenance starts with the development of the first ML system called MYCIN [7], which was programmed to suggest antibiotics for the treatment of bacterial infected patients by using a data of 450 rules collected from a team of medical experts. Subsequently, several ML techniques have been employed as promising technology against various contagious (SARS [8], [9], [10], EBOLA [11], HIV [12], [13]) and non-contagious (Cancer [14], Diabetic [15], Heart disease [16], and Stroke [17]) epidemic outbreaks. These aforementioned pieces of shreds of evidence encourage researchers to face down the current epidemic using effective approaches of ML and AI technologies.
We believe that this review could provide essential information to researchers who have interests in developing MALDI-MS and RT-LAMP-based modern rapid tools for detection of the COVID-19, as well as pose a potential picture on the extensive use of these useful diagnostic methods for battling the current pandemic.
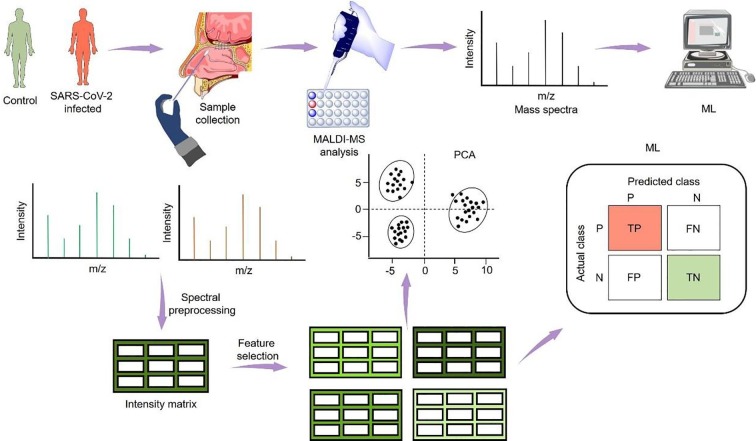
2. MALDI-MS-based SARS-CoV-2 detection
The research group from the University of Talca and Autonomous University of Chile reported a holistic approach for the detection of SARS-CoV-2 in nasal swabs using MALDI-MS and ML (Fig. 1 ) [18]. In this study, a total of 362 nasal mucous secretion swab samples collected from three different laboratories located in different countries were tested by the standard RT-PCR technique. Results showed that 211 nasal swab samples were COVID-19 positive and the remaining were negative. The same sets of samples were subjected to MALDI-MS and spectra generated were preprocessed to obtain an intensity matrix with identified peak. The most important aspect of this study was the identification of the correct peak which delineates the information whether samples being handled were COVID-19 positive or negative. Further, feature selection (FS)–information gain (Ig)-based and correlation-based (Cfs) tools were applied to obtained intensity matrices to establish the signature peaks with significant differences between COVID-19 positive and negative mean spectra. All the obtained data were individually processed for principal component analysis (PCA) and no distinct separation between the control and the deceased group was observed. Owing to this, six ML algorithms such as decision tree (DT), naive Bayes (NB), k-nearest neighbors (KNN), random forest (RF), support vector machine with a radial kernel (SVM-R), support vector machine with a linear kernel (SVM-L), were tested by the research group, for no FS, Ig FS, and Cfs to precisely distinguish COVID-19 positive samples from negative. Among the ML models tested, SVM-R ML Model exhibited an area under the curve of 0.99 (no FS), 0.98 (Ig FS), and (Cfs). Moreover, SVM-R with no FS demonstrated better accuracy with sensitivity and specificity of 0.947 and 0.926, respectively. This method could serve as a robust diagnostic tool for the identification of SARS-CoV-2 infected patients whose RT-PCR detection with cyclic threshold (Ct) value ranges from 17 to 37. Moreover, the nasal swab samples can be used directly for MALDI-MS without any specific purification step. Overall, the concordance rate between MALDI-MS and RT-PCR for the clinical diagnosis of SARS-CoV-2 was >80%.
Fig. 1.
Nasal swabs were used to acquire MALDI-MS spectra. The obtained data were subjected to PCA and ML techniques. Figure adapted and redrawn from Ref. [18].
In a separate study, a mass spectrometry (MS)-based approach was employed to detect the presence of viral proteins in a gargle solution of COVID-19 patients [19]. The key constituents present in the gargle solution are acetone and trypsin which were used for precipitation and digestion of proteins, respectively. MS analysis identified the presence of unique peptide fragments originated from SARS-CoV-2 nucleoprotein based on their mass/charge (m/z) value, which forms the assay principle for the clinical diagnosis of COVID-19.
Another study has been reported by Rocca et al. wherein they have demonstrated a potential tool for the detection of SARS-CoV-2 by combining, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) with multivariate ML algorithms [20]. They collected nasopharyngeal swab samples in 2 ml of saline solution and stored them at −20 °C. They recorded the MALDI spectra within a range of 2 kDa-20 kDa, and thereafter differentiated the SARS-COV-2 positive and negative sample peaks by using two software such as Flex analysis v3.4 and ClinPro Tools software v3.0. The performance of the combined tool has been estimated by evaluating, accuracy (67.6%), sensitivity (71.7%), specificity (61.7%), positive prediction (60%), and negative prediction (73%) by using the ClinPro Tools.
Very recently, Yan et al. [21] have reported a serum peptidome profiling method based on MALDI-TOF MS for efficient and rapid detection of SARS-CoV-2. After processing the MS data, they have implemented 8 ML methods to construct the classification models based on which a very high accuracy rate of 99%, with a specificity of 100% and sensitivity of 98% have been achieved. They focused on the analysis within a mass range of 5000 to 30,000 m/z, which corresponds to the serum peptidome and small proteins. A very minute volume of sample (5 μL) has been analyzed within 1 min with a cost of < 1 USD per sample.
3. RT-LAMP-based SARS-CoV-2 detection
Loop‐mediated isothermal amplification (LAMP) is a technology for nucleic acid amplification in 30 min, at one constant temperature (~65 °C). This method has been employed for the last few years to detect viral and bacterial pathogen [22], [23], [24]. Usually, the LAMP method is used for DNA amplification in presence of 4 to 6 specially designed primers to bind corresponding regions of the target DNA with very high specificity [25], [26]. However, to detect RNA sequence, the LAMP protocol has been incorporated with the reverse transcription method and collectively called RT-LAMP technology. In the RT-LAMP method, the viral RNA has been converted to complementary DNA that undergoes amplification at isothermal conditions [27]. The assay involved in the RT-LAMP method is colorimetric, wherein the presence of target viral RNA is detected by observing the change in color of the clinical samples, as shown in Fig. 2 .
Fig. 2.
Workflow of RT-LAMP process in the detection of SARS-CoV-2.
Recently, the colorimetric technique, RT-LAMP assay, for the detection of viral RNA of SARS-CoV-2 using N gene-specific primers has been reported by Dao et al. [28]. The reaction setup was demonstrated to capable of detecting 100 in vitro transcription (IVT) RNA molecules. They used 1 μL of isolated RNA solution (~100 RNA molecules) in 12.5 μL reaction volume, which displayed a change of color from red to yellow following an incubation period of 30 min at 65 °C. These results were plotted against the Ct readings of RT-qPCR performed using a primer set specific for the viral E gene. For Ct readings up to 30, a robust color change was noticed which determines the detection limit of this RT-LAMP assay. For the RNA samples isolated from a total of 768 pharyngeal swabs tested for COVID-19, this RT-LAMP assay detects viral RNA with the sensitivity and selectivity of 97.5% and 99.7%, respectively, for samples with Ct < 30. Further, the positive result of the LAMP assay was validated using the multiplexed LAMP-sequencing through the amplification of viral RNA sequences. Without RNA isolation step, direct swab (raw swab specimen) or Hot-swab (heat-treated swabs-95 °C for 5 min) to RT-LAMP assay displayed higher sensitivity for detecting samples with Ct values of up to 25 and <30, respectively (Table 1 ).
Table 1.
Testing of 592 clinical samples by RT-qPCR and RT-LAMP techniques classified into Ct value bins. The table is adapted from Ref. [28].
| Hot swab-to-RT-LAMP | RT-LAMP |
||||
|---|---|---|---|---|---|
| Ct | Pos | Neg | Sum | ||
| RT-qPCR | Pos | 0–25 | 38 | 4 | 42 |
| 25–30 | 17 | 5 | 22 | ||
| 30–35 | 5 | 23 | 28 | ||
| 35–40 | 0 | 36 | 36 | ||
| Neg | Neg | 1 | 214 | 215 | |
| Sum | 61 | 282 | 343 | ||
| Direct swab-to-RT-LAMP | RT-LAMP | ||||
| Ct | Pos | Neg | Sum | ||
| RT-qPCR | Pos | 0–25 | 15 | 1 | 16 |
| 25–30 | 6 | 11 | 17 | ||
| 30–35 | 2 | 21 | 23 | ||
| 35–40 | 3 | 23 | 26 | ||
| Neg | Neg | 9 | 144 | 153 | |
| Sum | 35 | 200 | 235 | ||
Similarly, Yu et al. described the isothermal LAMP-based method for COVID-19 (iLACO) for the rapid detection of SARS-CoV-2 [29]. The developed iLACO method that is based on ORF1ab gene-specific primers can detect a minimum of 10 copies of this gene. Further, they optimized the efficacy of the iLACO under incubation at 65 °C in a reaction volume of 1.5 ml and they observed that a significant color change appeared as the reaction time reached twenty minutes with a virus RNA concentration of 1000 copies per µL (i.e., using 1 µL sample in total 20 µL reaction). Among 248 COVID-19 samples tested, 89.9% samples showed positive signals which have detection threshold below 60 copies per μL and the Ct values lesser than 35.
Ganguli et al. have reported a RT-LAMP isothermal assay with the combination of a simple point-of-care (POC) instrument for the detection of the novel coronavirus and demonstrated the assay on several clinical samples [30]. The incorporation of the POC device with the RT-LAMP technique has enhanced the simplicity and accessibility of the assay. The analysis of the swab samples has been performed in a viral transport medium (VTM) in absence of the RNA extraction kit. To perform a highly sensitive and specific RT-LAMP assay for the detection of SARS-CoV-2, the authors have used a basic local alignment search tool for nucleotides (BLASTn) analysis to identify ORF 1a, S, ORF 8, and N genes for primer design which code for the corresponding viral genomes. Finally, a portable POC device has been used to demonstrate clinical samples for the distinction of the positive and negative sample within 30 min.
In another study, Oliveria et al. have described a polystyrene-toner (PS-T) centrifugal microfluidic device for the diagnosis of the COVID-19 by RT-LAMP assay, with automated colorimetric detection [31]. The assay has been performed in a microchamber with a LOD of 5 μL, at 72 °C for 10 min. It has been demonstrated that even in reactions initiated with approximately 10−3 copies of SARS-CoV-2 RNA, the detection of SARS-CoV-2 is possible.
Additional techniques such as tandem MS [32], [33], lateral flow immunoassay [34], [35], [36], and few other enzymatic assays are established for the diagnosis of COVID-19 [37], [38], [39], [40], [41]. Tandem MS is a sophisticated technique operated by highly skilled experts and it requires relatively more time for sample preparation, data acquisition, and processing. Due to the lack of highly skilled personnel, it is inappropriate to implement Tandem MS for the rapid detection of viral infections during pandemic situations; however, in terms of analysis of samples for exploratory research, this method stands highly important.
As a natural response to viral infections, antibodies are secreted by the immune system through a regulated host defense mechanism. Antibodies profiling during the early phase of COVID-19 infection, as a part of serological testing, stands important [42]. In another study, Huang et al. reported a colloidal gold nanoparticle-based lateral flow immunoassay (AuNP-LF assay) as a qualitative approach for the detection of anti-human IgM antibodies against the SARS-CoV-2 virus [43]. The efficiency of the assay was optimized by them, in which a concentration of 1 mg/ml of SARS-CoV-2 NPs with 1.5 μg of anti-human IgM at pH 8 was found to be the best performing approach. The optimized volume of the serum sample was found to be 10–20 μL mixed with the corresponding amount of buffer. The sensitivity and specificity of the AuNP- LF assay were observed to be 100 and 93.9%, respectively. They also revealed several superiorities of this assay over the conventional RT-PCR method such as rapid test time, less sample consumption, simple operability, employment of inexpensive and non-sophisticated instruments and materials, and independence of high professional experience. However, as this technique involves SARS-CoV-2 antigen coating, the specificity of this assay is limited due to factors like the variation in antigen preparation, purity, and slow color development. Similarly, the enzyme-based detection of anti-SARS-CoV-2 antibodies using indirect ELISA or SARS-CoV-2 antigens using sandwich ELISA is also established [44]. However, major limitations with the ELISA technique [45] for COVID-19 diagnosis are (i) effortful and costly to produce a specific antibody owing to the employment of sophisticated technique, and expensive cell culture media, (ii) high probability of faulty test results because of inadequate blocking of the surface of microtiter plate impaired with antigen, (iii) vulnerability of the antibody due to the requirement of refrigerated transport and storage of the protein, (iv) precondition of a large amount of SARS-CoV-2 antigens, typically performed in 96-well format and requires 100–200 µL of sample for testing, and (v) long completion period of the assay, generally on the scale of 4–6 h, because of multiple wash steps and incubation times. A recent study showed that the time of detection of antibodies like IgA, IgG, IgM in the blood plasma of SARS-CoV-2 positive patients was found to vary with the days post-infection and symptom onset [46]. In this case, an integrated approach of IgM ELISA and RT-qPCR test is vital for the diagnosis of COVID-19 with reduced false positive rate and high detection efficiency.
In the context of rapid diagnosis of SARS-CoV-2, MALDI-MS and RT-LAMP assays are few sensitive techniques that are highly compatible with unprocessed specimen swabs and the presence of saliva. Although these diagnostic methods cannot handle complex matrices like blood specimens, the requirement of non-invasive samples like nasopharyngeal swab and saliva samples make them suitable for large-scale testing in this COVID-19 pandemic. The high initial cost of the instrument and the familiarity of ML algorithms are some of the limitations for MALDI-MS and the design of specific primers is a crucial factor for colorimetric RT-LAMP assay. Given these limitations, these techniques possess superior features such as high detection limit, less detection time, and the reduced false negative rate which warrants the rapid detection of SARS-CoV-2 in a large population to contain the spread of viral infections.
A brief comparison among the RT-PCR, RT-LAMP, and MALDI-MS methods for the SARS-CoV-2 detection is provided below with corresponding highlight features (Table 2 ).
Table 2.
| Parameters | RT-PCR | RT-LAMP | MALDI-MS |
|---|---|---|---|
| Target | N, ORF, E genes | N, ORF, E, S genes | M, S, N proteins |
| Assay reaction time (min) | ~120 | ~30 | ~1 |
| Detection basis | Ct values (qualitative) | Colorimetric (qualitative) | m/z signals |
| Cost (USD) | 15 | ~3–4 | <1 |
| Instrumentation | bulky, sophisticated and expensive | simple, portable, economical | Sophisticated and expensive |
| Sensitivity (%) | 30–60 | ~90 | ~98 |
| Specificity (%) | ~100 | ~97 | ~100 |
4. Conclusion and outlook
In conclusion, an emphasis on the use of well-established spectral techniques for the swift disclosure of SARS-CoV-2 has been presented. The use of MS and colorimetric detection system, RT-LAMP assay using UV/Vis spectroscopy have been recently developed to meet the limitations of RT-PCR used for the detection of SARS-CoV-2. From a diagnostic point of view, the advantages of these methods were the overall reduced false-negative rate, lesser time of detection, low sample requirement, and improved LOD of viral RNA. Importantly, these methods can detect the positive infected cases of COVID-19 directly using nasal swab specimens without the requirement of the RNA isolation step. Collectively, these features would be useful to diagnose the COVID-19 at a faster rate, and eventually, the spread of viral infections during the current pandemic can be controlled to a great extent. Soon, these techniques may be extended to study other infectious diseases. Indeed! the MALDI-TOF technology has been already demonstrated to recognize an ample mix of viruses such as human herpesviruses (HHV) [48], influenza viruses [49], and for the diseases that are ascribed to intense enterovirus infections such as echovirus, coxsackievirus A and B, and poliovirus [50], [51]. Further studies on the improvement of LAMP primers design could enhance the sensitivity and specificity of the RT-LAMP assays. Additionally, the development of a cost-effective, single-step, and closed-tube isothermal PCR assay may allow the assay to conduct a largescale diagnosis of the COVID-19 virus with a very high detection capacity. Future progress in the construction of more handy POC devices consists of portable power sources and their inclusion with the RT-LAMP tools could further simplify the assay operability. Finally, all these aforementioned aspects of both the MALDI-MS and RT-LAMP assay may lead to building a potential strategy for rapidly defending the ongoing global health pandemic.
CRediT authorship contribution statement
Abhijeet Mohanty: Writing-original draft, writing-review and editing. Adarsh P. Fatrekar: Writing original draft. Saravanan Krishnan: Writing original draft. Amit A. Vernekar: Conceptualization, supervision, writing-review and editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Department of Science and Technology (DST), India, for the DST-INSPIRE faculty award to AAV, and Council of Scientific and Research (CSIR) and Science and Engineering Research Board (SERB), New Delhi, India, for COVID-19 research funding support. AM thanks University Grants Commission (UGC), New Delhi, for a research fellowship. CSIR-CLRI communication number 1442.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., Engl N. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao H., Lu X., Deng Y., Tang Y., Lu J. Epidemiol. Infect. 2020;148 doi: 10.1017/S0950268820001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Euro Surveill. 2020;25:2000045. [Google Scholar]
- 4.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Radiology. 2020;296:E32–e40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winichakoon P., Chaiwarith R., Liwsrisakun C., Salee P., Goonna A., Limsukon A., Kaewpoowat Q. J. Clin. Microbiol. 2020;58:e00297–00220. doi: 10.1128/JCM.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X., Yao H., Xu X., Zhang P., Zhang M., Shao J., Xiao Y., Wang H. Clin. Chem. 2020;66:977–979. doi: 10.1093/clinchem/hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shortliffe E.H. Computer-Based Medical Consultations: Mycin. Elsevier. 1976:63–78. [Google Scholar]
- 8.Barbat M.M., Wesche C., Werhli A.V., Mata M.M. ISPRS J. Photogramm. Remote Sens. 2019;156:247–259. [Google Scholar]
- 9.Li H.-C., Yang G., Yang W., Du Q., Emery W.J. ISPRS J. Photogramm. Remote Sens. 2020;160:167–179. [Google Scholar]
- 10.Shang R., Qi L., Jiao L., Stolkin R., Li Y. Eng. Appl. Artif. Intell. 2014;31:53–67. [Google Scholar]
- 11.Colubri A., Hartley M.-A., Siakor M., Wolfman V., Felix A., Sesay T., Shaffer J.G., Garry R.F., Grant D.S., Levine A.C., Sabeti P.C. EClinicalMedicine. 2019;11:54–64. doi: 10.1016/j.eclinm.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chockanathan U., Dsouza A.M., Abidin A.Z., Schifitto G., Wismüller A. Comput. Biol. Med. 2019;106:24–30. doi: 10.1016/j.compbiomed.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nápoles G., Grau I., Bello R., Grau R. Expert. Syst. Appl. 2014;41:821–830. [Google Scholar]
- 14.A.R. Vaka, B. Soni, S.R. K, ICT Express 6 (2020) 320-324.
- 15.Nazir T., Irtaza A., Shabbir Z., Javed A., Akram U., Mahmood M.T. Artif. Intell. Med. 2019;99 doi: 10.1016/j.artmed.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Sharma P., Choudhary K., Gupta K., Chawla R., Gupta D., Sharma A. Artif. Intell. Med. 2020;102 doi: 10.1016/j.artmed.2019.101752. [DOI] [PubMed] [Google Scholar]
- 17.Liu T., Fan W., Wu C. Artif. Intell. Med. 2019;101 doi: 10.1016/j.artmed.2019.101723. [DOI] [PubMed] [Google Scholar]
- 18.Nachtigall F.M., Pereira A., Trofymchuk O.S., Santos L.S. Nat. Biotechnol. 2020;38:1168–1173. doi: 10.1038/s41587-020-0644-7. [DOI] [PubMed] [Google Scholar]
- 19.Ihling C., Tänzler D., Hagemann S., Kehlen A., Hüttelmaier S., Arlt C., Sinz A. J. Proteome Res. 2020;19:4389–4392. doi: 10.1021/acs.jproteome.0c00280. [DOI] [PubMed] [Google Scholar]
- 20.Rocca M.F., Zintgraff J.C., Dattero M.E., Santos L.S., Ledesma M., Vay C., Prieto M., Benedetti E., Avaro M., Russo M., Nachtigall F.M., Baumeister E. J. Virol. Methods. 2020;286 doi: 10.1016/j.jviromet.2020.113991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan L., Yi J., Huang C., Zhang J., Fu S., Li Z., Lyu Q., Xu Y., Wang K., Yang H., Ma Q., Cui X., Qiao L., Sun W., Liao P. Anal. Chem. 2021;93:4782–4787. doi: 10.1021/acs.analchem.0c04590. [DOI] [PubMed] [Google Scholar]
- 22.Hong T.C.T., Mai Q.L., Cuong D.V., Parida M., Minekawa H., Notomi T. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boehme C.C., Nabeta P., Henostroza G., Raqib R., Rahim Z., Gerhardt M., Sanga E., Hoelscher M., Notomi T., Hase T., Perkins M.D. J. Clin. Microbiol. 1936;2007:45. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori Y., Notomi T. J. Infect. Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, T. Hase, Nucleic Acids Res. 2000, 28, E63-E63. [DOI] [PMC free article] [PubMed]
- 26.Tomita N., Mori Y., Kanda H., Notomi T. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 27.Thompson D., Lei Y. Sensors Actuat., Rep. 2020;2 doi: 10.1016/j.snr.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Barreto Miranda I., Schnitzler P., Kräusslich H.-G., Knop M., Anders S. Sci. Transl. Med. 2020;12:eabc7075. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.H., Yin X. Clin. Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., Ramirez S.A.S.d., Valera E., Cunningham B.T., King W.P., Bashir R. Proc. Natl. Acad. Sci. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Oliveira K.G., Estrela P.F.N., Mendes G.d.M., dos Santos C.A., Silveira-Lacerda E.d.P., Duarte G.R.M. Analyst. 2021;146:1178–1187. doi: 10.1039/d0an02066d. [DOI] [PubMed] [Google Scholar]
- 32.Gouveia D., Grenga L., Gaillard J.C., Gallais F., Bellanger L., Pible O., Armengaud J. Proteomics. 2020;20 doi: 10.1002/pmic.202000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardozo K.H.M., Lebkuchen A., Okai G.G., Schuch R.A., Viana L.G., Olive A.N., Lazari C.d.S., Fraga A.M., Granato C.F.H., Pintão M.C.T., Carvalho V.M. Nat. Commun. 2020;11:6201. doi: 10.1038/s41467-020-19925-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 35.Montesinos I., Gruson D., Kabamba B., Dahma H., Van den Wijngaert S., Reza S., Carbone V., Vandenberg O., Gulbis B., Wolff F., Rodriguez-Villalobos H. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen T., Huang C., Shi F.-J., Zeng X.-Y., Lu T., Jiao Y. Analyst. 2020;145:5345–5352. doi: 10.1039/d0an00629g. [DOI] [PubMed] [Google Scholar]
- 37.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo X., Geng P., Wang Q., Cao B., Liu B. J. Microbiol. Biotechnol. 2014;24:1445–1454. doi: 10.4014/jmb.1404.04024. [DOI] [PubMed] [Google Scholar]
- 39.Chen Q., Li J., Deng Z., Xiong W., Wang Q., Hu Y.Q. Intervirology. 2010;53:95–104. doi: 10.1159/000264199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.S.C. Moore, R. Penrice-Randal, M. Alruwaili, X. Dong, S.T. Pullan, D.P. Carter, K. Bewley, Q. Zhao, Y. Sun, C. Hartley, E.-m. Zhou, T. Solomon, M.B.J. Beadsworth, J. Cruise, D. Bogaert, D.W. Crook, D.A. Matthews, A.D. Davidson, Z. Mahmood, W. Aljabr, J. Druce, R.T. Vipond, L.F.P. Ng, L. Renia, P.J.M. Openshaw, J.K. Baillie, M.W. Carroll, M.G. Semple, L. Turtle, J.A. Hiscox, medRxiv (2020) 2020.2003.2005.20032011.
- 41.Park T.J., Hyun M.S., Lee H.J., Lee S.Y., Ko S. Talanta. 2009;79:295–301. doi: 10.1016/j.talanta.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J., Long X., Guo S., Zhao Z., Liu Y., Hu H., Xue H., Li Y. J. Infect. 2020;81:e28–e32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang C., Wen T., Shi F.-J., Zeng X.-Y., Jiao Y.-J. ACS Omega. 2020;5:12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakamoto S., Putalun W., Vimolmangkang S., Phoolcharoen W., Shoyama Y., Tanaka H., Morimoto S. J. Nat. Med. 2018;72:32–42. doi: 10.1007/s11418-017-1144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q.W., Xu S.Y., Zhu H.D., Xu Y.C., Jin Q., Sharma L., Wang L., Wang J. Clin. Infect. Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.X. Hu, Q. Deng, J. Li, J. Chen, Z. Wang, X. Zhang, Z. Fang, H. Li, Y. Zhao, P. Yu, W. Li, X. Wang, S. Li, L. Zhang, T. Hou, mSphere 5 (2020) e00808-00820. [DOI] [PMC free article] [PubMed]
- 48.Sjöholm M.I.L., Dillner J., Carlson J. J. Clin. Microbiol. 2008;46:540–545. doi: 10.1128/JCM.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chou T.-C., Hsu W., Wang C.-H., Chen Y.-J., Fang J.-M. J. Nanobiotechnol. 2011;9:52. doi: 10.1186/1477-3155-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piao J., Jiang J., Xu B., Wang X., Guan Y., Wu W., Liu L., Zhang Y., Huang X., Wang P., Zhao J., Kang X., Jiang H., Cao Y., Zheng Y., Jiang Y., Li Y., Yang Y., Chen W. PLOS ONE. 2012;7 doi: 10.1371/journal.pone.0042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng J., Yang F., Xiong Z., Guo J., Du J., Hu Y., Jin Q. J. Clin. Virol. 2013;56:170–174. doi: 10.1016/j.jcv.2012.10.020. [DOI] [PubMed] [Google Scholar]