Abstract
Purpose:
High resolution visualization of the ocular surface and intact eyeball is critical and essential for our understanding and treatment of eye diseases. This study is to achieve this goal using advanced tissue clearing and three-dimensional (3D) imaging technologies.
Methods:
Wild type and fluorescently labelled transgenic mice of Prox-1-GFP (green fluorescent protein) or Thy1-YFP (yellow fluorescent protein) were used in the study. Eyeballs were harvested from normal or a disease model of corneal inflammation. Samples were infused with hydrogel monomers and heated for polymerization. Lipids were removed by electrophoresis. The transparent tissue-hydrogel hybrids of the anterior segments or intact eyeballs with immunolabeling or endogenous fluorescence were imaged by an advanced light sheet fluorescent microscope. High resolution 3D images and videos were captured for a wide array of structures and cell types.
Results:
Optical transparency was achieved from intact eyeballs of both normal and diseased conditions. A variety of important structures and cell types, such as blood and lymphatic vessels, Schlemm’s canal, nerves and endothelial cells, were detected with their natural morphology, location and organizational network.
Conclusions:
This study provides the first comprehensive and 3D high resolution imaging of the intact eyeball using tissue clearing and advanced light sheet microscopy. Given that the eye is the window of the body, we anticipate this advanced technology will facilitate diverse applications in biomedical research inside and outside the eye.
Keywords: cornea, conjunctiva, lymphatic vessel, blood vessel, nerve, tissue clearing, light sheet microscopy
1. Introduction
Optical tissue clearing has been used to study a variety of tissues and organs in recent years1–8. However, to date, there are still a lack of reports on high resolution three-dimensional (3D) imaging of the pigmented eyeball. As a specialized organ for visual function, the eye is highly heterogenic in structural, tissue and cellular compositions. It consists of a series of transparent structures in the visual pathway, such as the collagen packed cornea and lens, and the gel-like vitreous body. It also hosts a spectrum of opaque or semitransparent structures for other important functions, such as light conversion into electro-chemical impulses, intraocular pressure regulation, nutrient delivery and waste removal. These multifaceted functions are carried out by assorted tissues, such as the retina and the uveal tract (iris, ciliary body and choroids). The uveal tract is densely pigmented to prevent uncontrolled reflection and confusing images. Given such high complexity and heterogeneity, it is very challenging to work with this organ for optical tissue clearing.
In this study, we report a successful effort of tissue clearing and high-resolution 3D imaging of the intact pigmented eyeballs in mice, the most commonly employed mammalian research model. We have demonstrated the major features and advantages of this technology using a newly developed protocol that combines pigment removal, a tissue clearing system derived from CLARITY (Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/Immunostaining/In situ hybridization-compatible Tissue-hYdrogel), advanced light sheet microscopy, and a panel of wild type and fluorescently labeled transgenic mice for specific structure or cell type labeling1, 9–12. Several representative examples are presented for applying this new technology to study various structures and cellular components of the eyeball with an emphasis on the anterior segment where a corneal inflammation model is also employed. We show that the imaging processes can be performed with perspective, orthogonal, serial sectional, and rotational view and the results can be presented in 3D images, videos, and surface rendered models. Given the broad applications of eye research, we anticipate this new and powerful technology would have widespread applications in biomedical research.
2. Materials and methods
2.1. Animals
Eight- to 12-week-old C57BL/6 wild type and Thy1-YFP (yellow fluorescent protein)12 transgenic mice were purchased from Jackson Laboratory (Sacramento, CA). Prox-1 (prospero homeobox protein-1)-GFP (green fluorescent protein) transgenic mice were generated as reported previously11. All mice were treated according to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research, and the protocols were approved by the Animal Care and Use Committee, University of California, Berkeley. Mice were anesthetized using a mixture of ketamine, xylazine, and acepromazine (50 mg, 10 mg, and 1 mg/kg body weight, respectively) for each surgical procedure.
2.2. Corneal inflammatory neovascularization
The suture placement model was used to induce corneal inflammatory neovascularization as previously reported11, 13, 14. Briefly, four sutures (11–0 nylon, AROSurgical, Newport Beach, CA) were placed into corneal stroma without penetrating into the anterior chamber, followed by the application of antibiotic ointment. Eyeballs were harvested at 14 days post procedure for further analysis.
2.3. Tissue clearing
Intact eyeballs with optic nerves were harvested and fixed in 4% paraformaldehyde at 4°C for 24 hours. For pigment removal of C57BL/6 mice, samples were heated in 15% hydrogen peroxide at 55°C for 2 hours. Samples were washed in 1× phosphate-buffered saline (PBS) for several times and incubated in X-CLARITY™ hydrogel-initiator mixture solution at 4°C for 24 hours. Samples were polymerized in a vacuum of −70kPa at 37°C for 3 hours, gently shaken for 1 minute, and washed in 1 × PBS for 5 minutes and 3 times. The tissue-hydrogel hybrids were subsequently cleared in an SDS (sodium dodecyl sulfate)-based solution by the X-CLARITY™ electrophoretic tissue clearing system (Logos Biosystems, Annandale, VA, USA) at 37°C for 20 hours with 1.0-A current and 50 rpm pump speed. Cleared samples were washed in 1 × PBS for several times and incubated in 1 × PBS overnight at room temperature for further analysis.
2.4. Immunostaining
Cleared samples were labeled with staining antibodies using the DeepLabel antibody labeling kit (Logos Biosystems, Annandale, VA, USA). Samples were immersed in solution A at 37°C for 24 hours, incubated with the primary antibodies (1:100 diluted in solution B) at 37°C for 3 days in sealed plate, washed in 1 × PBS for 6 hours with the replacement of fresh PBS every 2 hours. Samples were then incubated with the secondary antibodies (1:100 diluted in solution B) at 37°C for 3 days in sealed plate, washed and stored in 1 × PBS. The primary antibodies include rabbit anti-mouse glial fibrillary acidic protein (GFAP) (Dako-Agilent, Santa Clara, CA, USA) and goat anti-mouse CD31 (pan-endothelial marker) (R&D, Minneapolis, MN, USA). Rabbit isotype control (Abcam, Cambridge MA, USA) and normal goat IgG (R&D) were tested for the immunostaining protocols. The secondary antibodies are Cy3 conjugated donkey anti-rabbit IgG, Cy3 conjugated donkey anti-goat IgG, and FITC conjugated donkey anti-goat IgG (Jackson ImmunoResearch, West Grove, PA, USA).
2.5. Imaging and post-processing
Samples were immersed in X-CLARITY™ mounting solution for refractive index (RI) matching before imaging by a light sheet fluorescent microscopy (Zeiss Lightsheet Z.1, Carl Zeiss AG, Germany)15. Samples were mounted to a customized adapter and immersed in the chamber filled with the mounting solution. For illumination, 488-nm and 561-nm laser lines, and Illumination Optics Lightsheet Z.1 5×/0.1 were used. For detection, Lightsheet Z.1 detection optics 5×/0.16 were used. Images and videos were processed with Bitplane Imaris x64 software, version 9.2.0 (Bitplane AG, Zurich, Switzerland). For displaying purposes, images were 3D cropped to show certain regions, snapshot at 15° perspective view or orthogonal view, and gamma corrected. Surface rendered 3D models were generated using the Surface function, and pseudo-colors were aligned by automatically computed surface object statistics, value - area. Videos were generated under 3D view in animation mode.
3. Results
3.1. Stepwise images showing major clearing steps of an intact pigmented eyeball
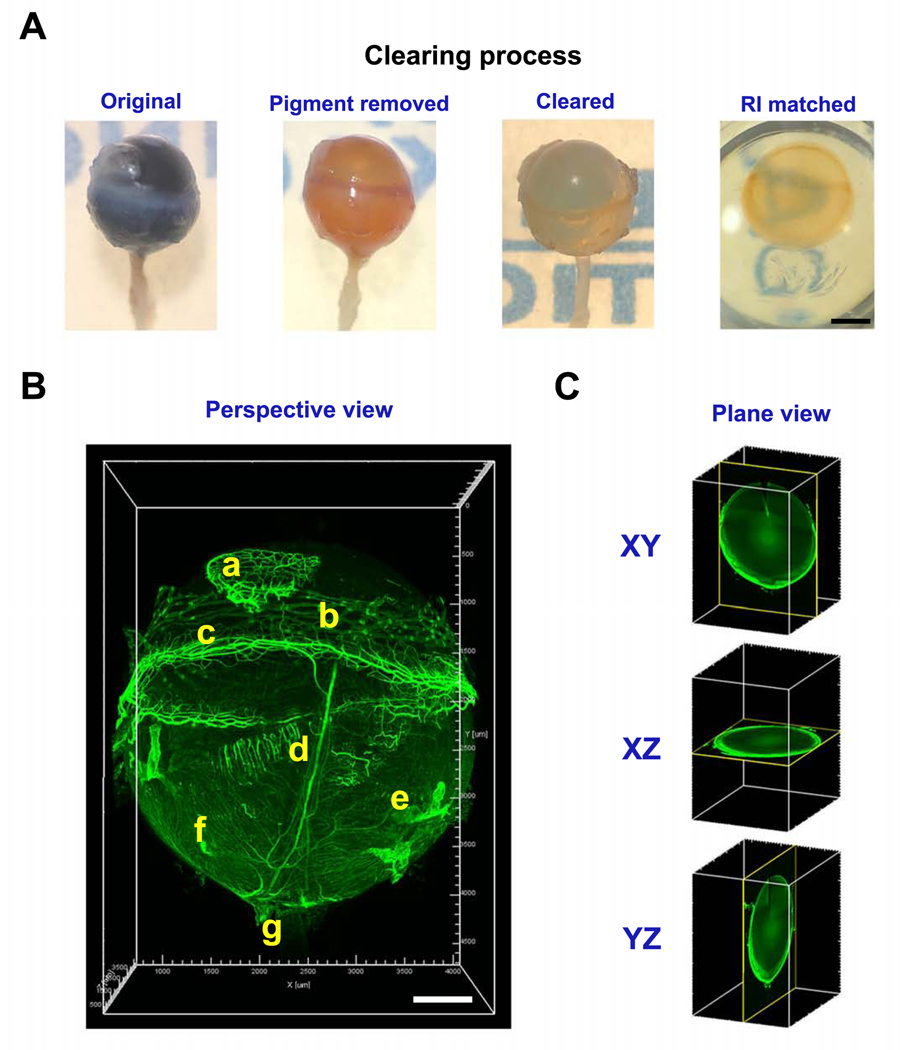
As shown in Figure 1A in the stepwise images from an intact pigmented eyeball of a C57BL/6 mouse, the pigment was first removed before tissue clearing and after refractive index (RI) matching, the entire eyeball was optically transparent. In Figure 1B and Supplementary Video 1, the entire vascular system of the eyeball was revealed by immunolabeling with CD31, a pan-endothelial marker. Both 3D images and videos can be captured and rotated in 360°, and the analysis can be performed at various planes to locate a specific region or spot. This capacity allows accurate tracking of the vascular tree or a particular branch, such as the ciliary artery, from the anterior to the posterior segment, in their natural location, and from different respective views (Figure 1C and Supplementary Videos 2 and 3).
Figure 1. Representative images showing major clearing steps of a pigmented eyeball.
A. Stepwise images of a C57BL/6 mouse eyeball during tissue clearing process. RI: refractive index. B. Left panel, a 15° perspective view (all images unless otherwise indicated) of a cleared intact eyeball showing vasculature structures labeled by CD31 staining. a, nictitating membrane; b, conjunctiva; c, limbus; d, ciliary artery; e, vorticose vein; f, choroidal blood vessels; g, optic nerve. C. 2D views at different axial planes. Scale bar: 1 mm (A). 500 μm (B).
3.2. Limbal structures
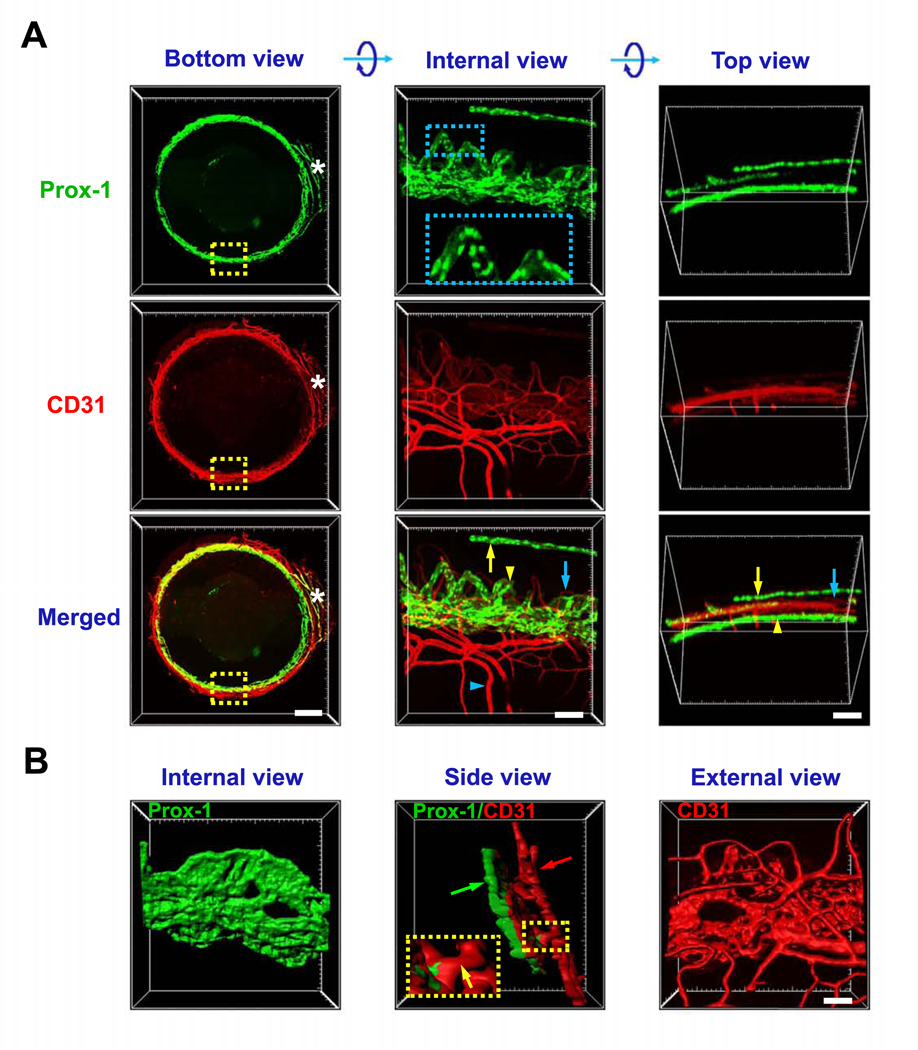
We next assessed the limbal region where three important vascular structures are closely located: limbal blood vessels, limbal lymphatic vessels, and the Schlemm’s canal of the anterior chamber angle. In this case, a fluorescently labeled transgenic mouse of Prox-1, the master control gene for lymphatic determination11, 16, was used in the study. The eyeballs were optically cleared and immunolabeled with CD31. Because Prox-1 is also expressed on the lens, the anterior segments were dissected for imaging purpose. As shown in Figure 2, the endogenous fluorescence of Prox-1-GFP was successfully maintained after the tissue clearing. In combination with CD31 immunolabeling, all three types of vascular structures can be captured and clearly demonstrated. These include limbal blood vessels (CD31strongProx-1−), limbal lymphatics (Prox-1+CD31weak), and the Schlemm’s canal (Prox-1+CD31weak). As shown in Figure 2A and Supplementary Video 4, 3D images and videos were obtained from the bottom, top and internal views, reflecting their original spatial locations. The bottom view showed the complete circle of the limbus with the nictitating membrane as an orientation marker. The internal and top views showed the well-maintained spatial organization of these vessels at a higher magnification. With enlarged internal view, the details of Prox-1 expression on the nuclei of Schlemm’s canal endothelial cells were clearly observed. With the surface rendering function of post image processing, a 3D model can be reconstructed as well, which not only revealed the external, internal and side surface of the structures but also offered an intuitive view of the delicate collector channels between the Schlemm’s canal and episcleral veins (Figure 2B and Supplementary Video 5).
Figure 2. Representative images showing limbal structures of blood vessels, lymphatic vessels and Schlemm’s canal.
A. Bottom view shows the intact circle of limbus area with nictitating membrane as the orientation mark (white asterisk). Internal and top views show the enlarged boxed area from bottom view after rotation along X axis. Limbal blood (blue arrow) and lymphatic (yellow arrow) vessels, Schlemm’s canal (yellow arrowhead) and episcleral vein (blue arrowhead) are shown in Prox-1-GFP (green, nuclear labeling) mice with CD31 staining (red). Boxed area in internal view of Prox-1 shows details of Prox-1 expression in Schlemm’s canal. B. Surface rendered 3D model showing structural orientation of Schlemm’s canal (green arrow) and limbal vessels (red arrow). Boxed area shows the collector channel between Schlemm’s canal and episcleral vein (yellow arrow). Scale bars: 500 μm (bottom), 100 μm (side and top), 50 μm (surface).
3.3. Corneal inflammatory neovascularization
We next assessed pathological neovascularization in the cornea using Prox-1-GFP mice and a well-established and widely used suture placement model11. Figure 3A shows a quarter of the well-maintained dome-shaped cornea occupied by the newly formed blood (CD31strongProx-1−) and lymphatic (Prox-1+CD31weak) vessels in their original organization. The spatial location of the neo-vessels and the adjacent Schlemm’s canal was assessed by the surface rendered 3D model. Pseudo-colors were assigned to distinguish between the Prox-1 positive limbal lymphatics and Schlemm’s canal (Figure 3B and Supplementary Video 6).
Figure 3. Representative images showing newly formed vessels in the inflamed cornea.
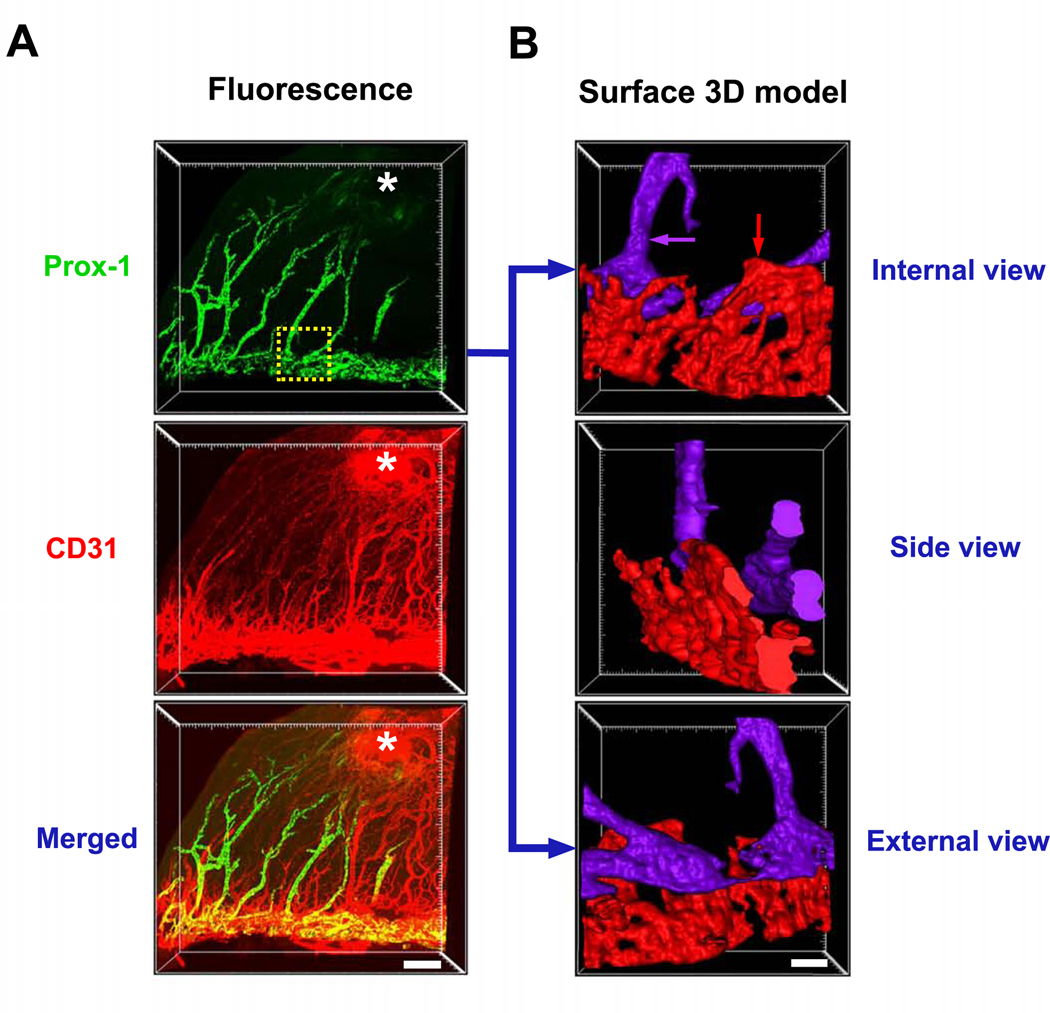
A. Inflammatory blood (CD31+Prox-1−) and lymphatic (CD31−Prox-1+) vessels induced by suturing (indicated by white asterisk) in Prox-1- GFP (green) mice. B. Surface rendered 3D model of the boxed area showing structural orientation of Schlemm’s canal (red arrow) and corneal neo-lymphatics (purple arrow) in pseudo-colors. Scale bar: 200 μm (A), 50 μm (B).
3.4. Corneal nerves and conjunctival vessels
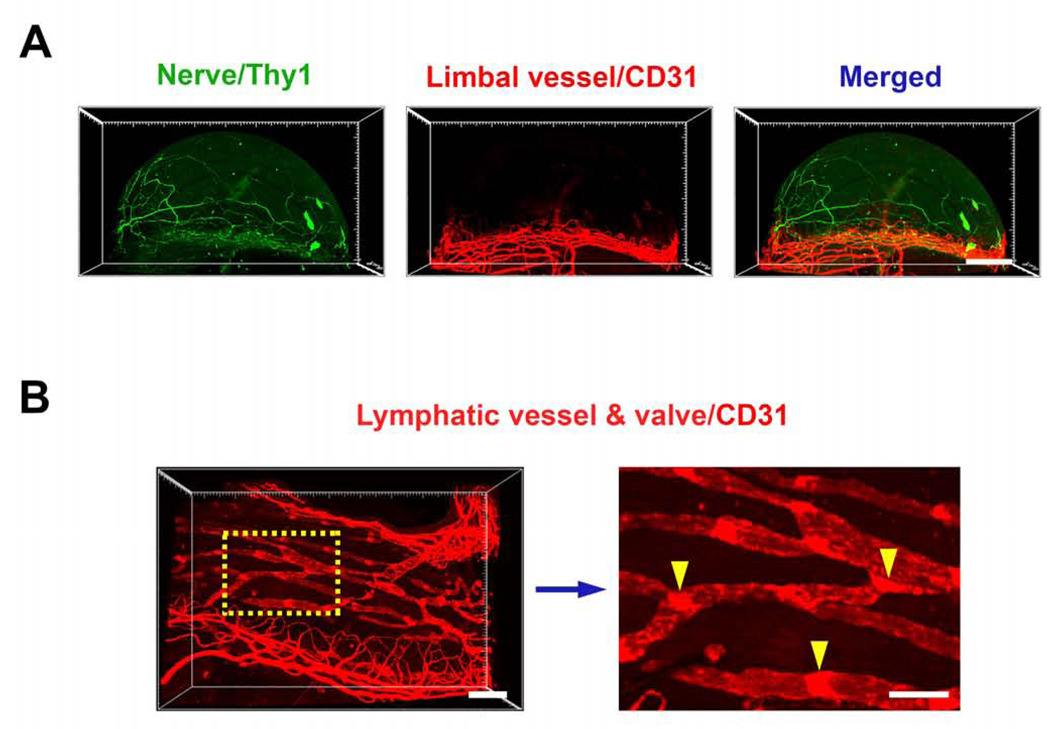
We also studied another important structure of the cornea, the nerves, together with the limbal and conjunctival vessels using the Thy1-YFP mice for neuronal recognition12 and CD31 immunolabeling, as shown in Figure 4A. Supplementary video 7 shows the interwoven network of these two structures near normal limbal area. The conjunctival lymphatics with their luminal valves were detected as well (Figure 4B).
Figure 4. Representative images showing corneal nerves and conjunctival lymphatic vessels.
A. Corneal nerves and limbal vessels in Thy1-YFP (green) mice with CD31 immunolabeling (red), respectively and merged. B. Left panel: conjunctival lymphatic vessels and valves labeled by CD31. Right panel: boxed area from left panel in a higher magnification, showing lymphatic vessel valves (yellow arrowheads). Scale bar: 500 μm (A), 200 μm (B left), 100 μm (B right).
3.5. Others
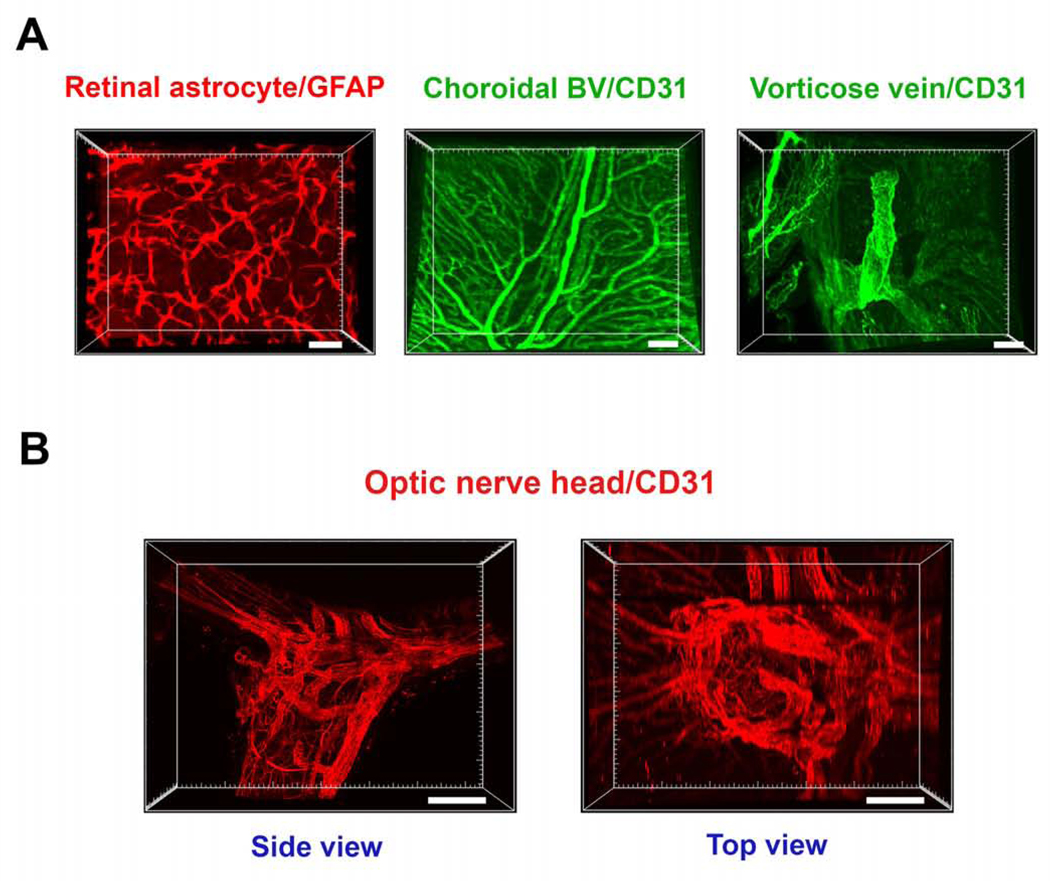
While the posterior segment of the eyeball is not the focus of the current report, we herein presented several examples for demonstration. With the macromolecule-permeable feature after clearing, Figure 5A shows three representative cell types and structures, respectively, including the GFAP+ astrocytes in the retina, CD31+ blood vessels in the choroids, and the CD31+ vorticose vein draining out of the sclera. At the optic nerve head area with complex vascular supply, the CD31+ blood vessels were distinctly displayed at both side and top views. Detailed vascular distribution and branching can be tracked by image rotation as well (Figure 5B and Supplementary Video 8). All these structures were immunolabeled and imaged with intact pigmented C57BL/6 eyeballs.
Figure 5. Representative images showing various vessels and cells at the posterior segment and optic nerve.
A. From left to right: retinal astrocytes labeled by GFAP, choroidal blood vessels (BV) and vorticose vein labeled by CD31. Scale bars: 50 μm (left), 100 μm (middle and right). B. Side and top view of CD31 labeled vascular networks around the optic nerve head area. Scale bar: 200 μm.
Discussion
In this study, we have shown that the newly developed technique of tissue clearing with CLARITY combined with advanced light sheet microscopy is highly capable and accurate to detect various components of the eyeball from the anterior to the posterior segment, to distinguish adjacent structures with complex connections, and to evaluate pathological changes in a diseased condition. Undoubtedly, this advanced technology has great advantages over conventional approaches with flat-mount or sectioned tissues, which is not only time and labor demanding but also fraught with inaccurate information due to the loss of sample integrity. This new technique allows us to study ocular components with their natural morphology, location and organizational networks.
To our knowledge, this technique provides the first chance for directly viewing the entire ring as well as various surfaces of the Schlemm’s canal, a critical structure for aqueous humor drainage and intraocular pressure regulation. This is an embedded angular structure that is not accessible by conventional approaches. It can now be directly and clearly examined from the inner to the outer surface and in association with the connecting channels and the adjacent limbal vessels at high resolution. The Schlemm’s canal has recently been redefined to possess lymphatic features14. This new technology that enables a holistic view of this important structure should greatly enhance our research capacity for glaucoma, a major eye disease leading to blindness.
This study provides the first comprehensive, high resolution, and 3D information of the eyeball. We have shown that the technology is applicable to various mouse strains (i.e. wild type or fluorescently labeled), and a spectrum of ocular components and cell types (i.e. nerves, blood vessels, lymphatic vessels and valves, Schlemm’s canal, astrocytes). In a few previous attempts using different clearing protocols, only non-intact eyeballs or dissected tissues, such as the retina17–19, or non-pigmented albino eyeball18 at low resolution were reported. A more recent study using the EyeCi protocol showed some vascular structures at the posterior segment20, however, no other structures were reported. Also, none of these studies showed high resolution cellular components inside the intact eyeball, or the maintenance of fluorescent signals of the transgenic mice. During the review process of this work, Prahst et al. reported a study on mouse retina using the PACT clearing protocol21. Most of the images were collected from dissected tissues, and the study focused on postnatal stages when the eyeballs were smaller, immature, and easier to handle and clear. All the images presented in the current work were achieved from adult mature eyes. As with all new techniques, it takes time and efforts to develop and optimize the protocols. We have taken steps to modify the parameters. For example, for the polymerization procedure, we tested different values of vacuum and found −70kPa yielded better results than −90kPa, which may compress the eyeballs.
Not yet demonstrated, it is possible to apply this new technique to investigate other structures and cell types inside the eyeball, and to assess pathological changes in other diseases. Given its high efficiency and accuracy and the broad implications of eye research, we expect this new technique will become a powerful tool for future investigation both inside and outside the eye.
Supplementary Material
3D fluorescent image of a CD31 labeled intact eyeball, rotated 360° horizontally and vertically.
3D fluorescent image of a CD31 labeled intact eyeball, tracking the ciliary artery (yellow arrow) from the anterior to the posterior segment.
Continuous cross-sectional view of the CD31 labeled vessels from the posterior segment to the optic nerve.
3D fluorescent image of CD31 (red) and Prox-1 (green) labeled limbal vessels and the Schlemm’s canal, rotated from the internal to the top view.
Surface rendered 3D image of CD31 (red) and Prox-1 (green) labeled limbal vessels and the Schlemm’s canal, rotated 180° horizontally, showing the location of the collector channel (yellow arrow).
Surface rendered 3D image of Prox-1 labeled Schlemm’s canal (red) and newly formed lymphatic vessels (purple), rotate 180° horizontally.
3D fluorescent image of Thy1 labeled corneal nerves (green) and CD31 labeled limbal vessels (red).
3D fluorescent image of CD31 labeled vessels in the optic nerve head area, rotated 180° horizontally and vertically.
Acknowledgements
This work is supported in part by research grants from the National Institutes of Health and the University of California at Berkeley (L.C.). The funding sources had no role in study design, the collection, analysis and interpretation of data, the writing of the report, or the decision to submit the article for publication. We thank Young K. Hong at University of Southern California and the Mutant Mouse Regional Resource Centers (MMRRC) for providing the founder Prox-1 transgenic mice. Lightsheet microscope imaging was conducted at the CRL Molecular Imaging Center, Helen Wills Neuroscience Institute at the University of California at Berkeley.
Footnotes
Declarations of Interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chung K, Wallace J, Kim SY, et al. Structural and molecular interrogation of intact biological systems. Nature 2013;497:332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson DS, Lichtman JW. Clarifying Tissue Clearing. Cell 2015;162:246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erturk A, Mauch CP, Hellal F, et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat Med 2011;18:166–171. [DOI] [PubMed] [Google Scholar]
- 4.Yang B, Treweek JB, Kulkarni RP, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 2014;158:945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 2013;16:1154–1161. [DOI] [PubMed] [Google Scholar]
- 6.Susaki EA, Tainaka K, Perrin D, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 2014;157:726–739. [DOI] [PubMed] [Google Scholar]
- 7.Hama H, Kurokawa H, Kawano H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 2011;14:1481–1488. [DOI] [PubMed] [Google Scholar]
- 8.Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 2013;140:1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SY, Chung K, Deisseroth K. Light microscopy mapping of connections in the intact brain. Trends Cogn Sci 2013;17:596–599. [DOI] [PubMed] [Google Scholar]
- 10.Dodt HU, Leischner U, Schierloh A, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 2007;4:331–336. [DOI] [PubMed] [Google Scholar]
- 11.Kang GJ, Ecoiffier T, Truong T, et al. Intravital Imaging Reveals Dynamics of Lymphangiogenesis and Valvulogenesis. Sci Rep 2016;6:19459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan YA, Misgeld T, Lichtman JW, Sanes JR. Effects of neurotoxic and neuroprotective agents on peripheral nerve regeneration assayed by time-lapse imaging in vivo. J Neurosci 2003;23:11479–11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sessa R, Yuen D, Wan S, et al. Monocyte-derived Wnt5a regulates inflammatory lymphangiogenesis. Cell Res 2016;26:262–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Hamrah P, Cursiefen C, et al. Vascular endothelial growth factor receptor-3 mediates induction of corneal alloimmunity. Nat Med 2004;10:813–815. [DOI] [PubMed] [Google Scholar]
- 15.Lyons CE, Razzoli M, Larson E, et al. Optogenetic-induced sympathetic neuromodulation of brown adipose tissue thermogenesis. FASEB J 2020;34:2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truong TN, Li H, Hong YK, Chen L. Novel characterization and live imaging of Schlemm’s canal expressing Prox-1. PLoS One 2014;9:e98245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohberger B, Baumgart C, Bergua A. Optical clearing of the eye using the See Deep Brain technique. Eye (Lond) 2017;31:1496–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Y, Ma J, Langenbacher AD, et al. Multiscale light-sheet for rapid imaging of cardiopulmonary system. JCI Insight 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh JN, Nowlin TM, Seedorf GJ, Abman SH, Shepherd DP. Quantifying three-dimensional rodent retina vascular development using optical tissue clearing and light-sheet microscopy. J Biomed Opt 2017;22:76011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henning Y, Osadnik C, Malkemper EP. EyeCi: Optical clearing and imaging of immunolabeled mouse eyes using light-sheet fluorescence microscopy. Exp Eye Res 2019;180:137–145. [DOI] [PubMed] [Google Scholar]
- 21.Prahst C, Ashrafzadeh P, Mead T, et al. Mouse retinal cell behaviour in space and time using light sheet fluorescence microscopy. Elife 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D fluorescent image of a CD31 labeled intact eyeball, rotated 360° horizontally and vertically.
3D fluorescent image of a CD31 labeled intact eyeball, tracking the ciliary artery (yellow arrow) from the anterior to the posterior segment.
Continuous cross-sectional view of the CD31 labeled vessels from the posterior segment to the optic nerve.
3D fluorescent image of CD31 (red) and Prox-1 (green) labeled limbal vessels and the Schlemm’s canal, rotated from the internal to the top view.
Surface rendered 3D image of CD31 (red) and Prox-1 (green) labeled limbal vessels and the Schlemm’s canal, rotated 180° horizontally, showing the location of the collector channel (yellow arrow).
Surface rendered 3D image of Prox-1 labeled Schlemm’s canal (red) and newly formed lymphatic vessels (purple), rotate 180° horizontally.
3D fluorescent image of Thy1 labeled corneal nerves (green) and CD31 labeled limbal vessels (red).
3D fluorescent image of CD31 labeled vessels in the optic nerve head area, rotated 180° horizontally and vertically.