Abstract
CINSARC, a multigene expression signature originally developed in sarcomas, was shown to have prognostic impact in various cancers. We tested the prognostic value for disease-free survival (DFS) of CINSARC in a series of 6035 early-stage invasive primary breast cancers. CINSARC had independent prognostic value in the Luminal B subtype and not in the other subtypes. In Luminal B patients receiving adjuvant endocrine therapy but no chemotherapy, CINSARC identified patients with different 5-year DFS (90% [95%CI 86–95] in low-risk vs. 79% [95%CI 75–84] in high-risk, p = 1.04E−02). Luminal B CINSARC high-risk tumors were predicted to be less sensitive to endocrine therapy and CDK4/6 inhibitors, but more vulnerable to homologous recombination targeting and immunotherapy. We concluded that CINSARC adds prognostic information to that of clinicopathological features in Luminal B breast cancers, which might improve patients’ stratification and better orient adjuvant treatment. Moreover, it identifies potential therapeutic avenues in this aggressive molecular subtype.
Subject terms: Prognostic markers, Breast cancer
Introduction
During the last decades, significant progresses have been achieved in early breast cancer management, most notably through the routine use of post-operative systemic treatment including adjuvant cytotoxic chemotherapy and endocrine therapy1,2. Yet, the benefits conferred by these treatments are not uniformly distributed across the various molecular subtypes of disease described from gene expression profiling3. Thus, only endocrine receptor (ER)-positive breast cancers benefit from endocrine therapy, whereas cytotoxic chemotherapy, without and with anti-HER2 agents, has maximum efficacy in triple-negative and HER2-positive subtypes, respectively. In ER-positive/HER2-negative breast cancer, the so-called luminal-like breast cancer, only a minor subset of patients, with either a large tumor burden or a highly proliferative and aggressive biology, derive an actual benefit from chemotherapy. Accordingly, various prognostic signatures have been established and made commercially available to help identify these patients, and are now increasingly used in the clinic4,5. These signatures distinguish patients with low-, intermediate-, and high-risk of unfavorable outcome, the latter being recommended for adjuvant chemotherapy. Nevertheless, in those patients with molecularly-defined high-risk disease, the level of therapeutic discrepancy remains significant, because some patients receiving adjuvant chemotherapy will relapse and die, while a relatively high number of those high-risk patients could still achieve cure with endocrine therapy alone. Thus, alternative or additional molecular predictors are needed in this population.
The CINSARC (Complexity INdex in SARComas) signature was originally elaborated as a predictor of clinical outcome in soft tissue sarcomas with complex genetics and was subsequently demonstrated to have prognostic impact in different tumor types, including breast cancer6,7. CINSARC classifies the tumor samples into high-risk or low-risk of relapse. It includes genes implicated in mitosis and maintenance of chromosomes integrity, the deregulation of which may result in elevated genomic instability. Moreover, aberrant expression of CINSARC proteins was also suggested to favor higher migration and invasion abilities8,9. All of these features are associated with increased tumor aggressiveness and may explain the potential of this signature to prognosticate the recurrence of cancer across multiple malignancies.
Regarding the prognostic value of CINSARC in breast cancer, it is necessary to examine how it compares with the classical clinicopathological prognostic features in multivariate analysis. Thus, to further examine the potential prognostic value of CINSARC in breast cancer, we examined a set of 6035 early-stage, invasive primary breast cancers with publicly available gene expression and clinicopathological annotations including survival. We found that CINSARC had independent prognostic value in the Luminal B subtype but not in the other subtypes, notably in patients treated with adjuvant endocrine therapy without chemotherapy, thus identifying a subset of luminal B breast cancer in which therapeutic de-escalation might be possible. In addition, we identified in CINSARC high-risk patients an enrichment in gene signatures associated with response to PARP inhibitors and immunotherapy, thus providing potential clues to treat these poor-prognosis patients.
Results
The prognostic value of CINSARC in breast cancer is not independent
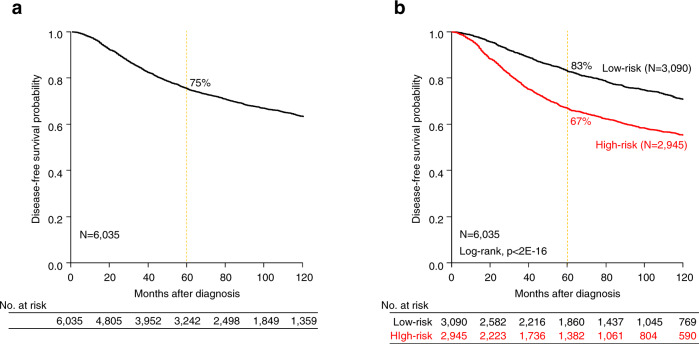
We analyzed our database of 8930 patients with early breast cancer, including 6,035 treated with primary surgery and with available DFS data (Supplementary Table 1). With a median follow-up of 77 months (range 1–382), 1759 experienced a DFS event, while 4276 remained disease-free for a 5-year DFS of 75% (95%CI, 74–76) (Fig. 1a). Applying CINSARC to this population identified 2945 CINSARC-high risk (49%) and 3090 CINSARC-low risk (51%) breast cancer patients (Fig. 1b), with significantly different 5-year DFS (67% vs. 83%, respectively; p < 2E−16, log-rank test). In univariate analysis (Table 1), CINSARC-high risk patients had an 80% increase in risk of DFS event as compared to the CINSARC-low risk (Hazard Ratio HR = 1.80, 95%CI 1.64–1.98, p = 9.44E−34, Wald test). Other variables associated with shorter DFS included younger age, pathological lymph node involvement, type, grade and tumor size, PAM50 molecular subtypes, delivery of adjuvant chemotherapy, and absence of adjuvant endocrine therapy. However, in multivariate analysis, the pathological lymph node involvement, tumor size and type, molecular subtypes and lack of endocrine therapy remained independently associated with survival outcome, whereas CINSARC lost its significance (HR = 1.19, 95%CI 0.97–1.46, p = 0.095, Wald test).
Fig. 1. Disease-free survival in early breast cancer patients.
a Kaplan–Meier disease-free survival (DFS) curve in 6035 informative early breast cancer patients. b Similar to a, but according to the two CINSARC classes (high-risk vs. low-risk).
Table 1.
Univariate and multivariate Cox regression analyses for DFS in breast cancer and per molecular subtype.
| DFS | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| N | HR [95% CI] | p-value | N | HR [95% CI] | p-value | ||
| All breast cancers | |||||||
| Patients’ age | >50 vs. ≤50 | 4707 | 0.81 [0.72–0.91] | 3.20E−04 | 2265 | 0.92 [0.76–1.11] | 0.384 |
| Pathological tumor type | Lobular vs. ductal | 3602 | 0.95 [0.75–1.21] | 2.69E−04 | 2265 | 1.45 [1.08–1.95] | 1.28E−02 |
| Other vs. ductal | 0.61 [0.47–0.77] | 2265 | 0.65 [0.48–0.88] | 5.57E−03 | |||
| Pathological grade | 2 vs. 1 | 4018 | 1.68 [1.35–2.09] | 2.46E−22 | 2265 | 1.12 [0.79–1.59] | 0.512 |
| 3 vs. 1 | 2.56 [2.06–3.17] | 2265 | 1.06 [0.74–1.53] | 0.738 | |||
| Pathological axillary lymph node status | Positive vs. negative | 5165 | 1.63 [1.47–1.82] | 3.16E−19 | 2265 | 1.83 [1.49–2.23] | 4.80E−09 |
| Pathological tumor size | pT2 vs. pT1 | 4719 | 1.59 [1.40–1.80] | 1.68E−22 | 2265 | 1.46 [1.23–1.73] | 1.92E−05 |
| pT3 vs. pT1 | 2.58 [2.10–3.17] | 2265 | 2.11 [1.56–2.87] | 1.64E−06 | |||
| Adjuvant chemotherapy | yes vs. no | 4442 | 1.52 [1.33–1.73] | 6.02E−10 | 2265 | 1.14 [0.90–1.43] | 0.272 |
| Adjuvant hormone therapy | yes vs. no | 4382 | 0.77 [0.68–0.87] | 2.95E−05 | 2265 | 0.71 [0.58–0.86] | 4.89E−04 |
| PAM50-derived molecular subtype | ERBB2 vs. Basal | 6035 | 1.11 [0.96–1.27] | 1.40E−45 | 2265 | 1.23 [0.97–1.55] | 0.088 |
| Luminal A vs. Basal | 0.45 [0.39–0.52] | 2265 | 0.6 [0.44–0.82] | 1.43E−03 | |||
| Luminal B vs. Basal | 0.85 [0.75–0.97] | 2265 | 1.08 [0.84–1.40] | 0.536 | |||
| Normal vs. Basal | 0.47 [0.39–0.57] | 2265 | 0.67 [0.46–0.98] | 4.03E−02 | |||
| CINSARC classes | High vs. Low-risk | 6035 | 1.80 [1.64–1.98] | 9.44E−34 | 2265 | 1.19 [0.97–1.46] | 0.095 |
| Per molecular subtype | |||||||
| CINSARC classes in Luminal A | High vs. Low-risk | 1753 | 1.40 [1.03–1.91] | 3.40E−02 | |||
| CINSARC classes in Luminal B | High vs. Low-risk | 1438 | 1.43 [1.18–1.73] | 2.18E−04 | |||
| CINSARC classes in Basal | High vs. Low-risk | 1241 | 1.03 [0.76–1.40] | 0.841 | |||
| CINSARC classes in ERBB2-enriched | High vs. Low-risk | 911 | 1.01 [0.79–1.30] | 0.925 | |||
| CINSARC classes in Normal-like | High vs. Low-risk | 692 | 1.27 [0.83–1.92] | 0.269 | |||
Because prognostic signatures may have clinical interest restricted to specific molecular subtypes, we repeated the same univariate analysis in each PAM50 subtype (Table 1). Interestingly, CINSARC had a significant prognostic impact only in the Luminal subtypes (HR = 1.40 [95%CI 1.03–1.91], p = 3.40E−02, and 1.43 [95%CI 1.18–1.73], p = 2.18E−04 in Luminal A and Luminal B, respectively). Accordingly, subsequent analyses were focused on these subtypes.
CINSARC has an independent prognostic value in the Luminal B subtype
Prognostic analyses were done in each Luminal subtype separately. In Luminal A breast cancer (n = 1592), the CINSARC high-risk class was associated with higher pathological grade (p = 7.60E-05) and tumor size (p = 4.00E−02), and with lower 5-year DFS (78%, 95%CI 72–86) as compared to the CINSARC low-risk class (88% [95%CI 86–89], p = 3.31E−02, log-rank test; Supplementary Table 2). In univariate analysis, CINSARC high-risk (HR = 1.40 [95%CI 1.03–1.91], p = 3.40E−02, Wald test) together with younger age, pathological lymph node involvement, higher grade and tumor size were associated with higher risk of DFS event (Supplementary Table 3). However, in multivariate analysis, CINSARC did not maintain its prognostic value (HR = 1.21 [95%CI 0.77–1.91], p = 0,398, Wald test). In addition, no prognostic value of CINSARC was found when focusing on the patients with Luminal A subtype treated with adjuvant endocrine treatment but without adjuvant chemotherapy (Supplementary Table 3).
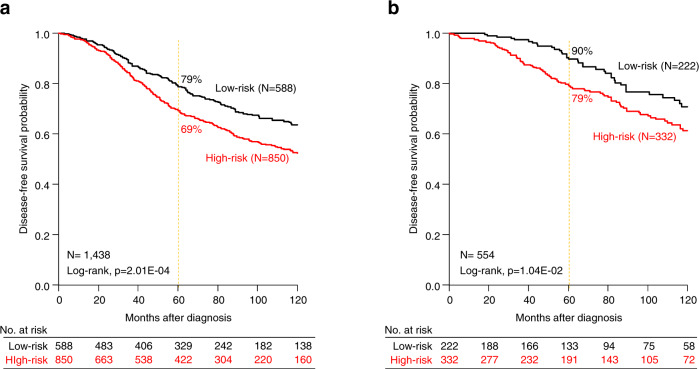
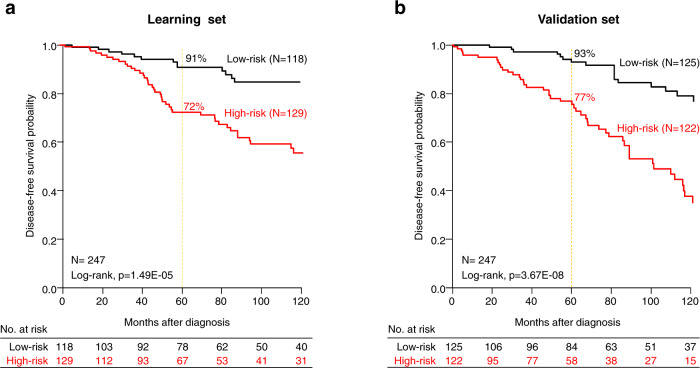
The results were different in the Luminal B subtype (n = 1438). The CINSARC high-risk class was associated with higher pathological grade (p = 2.18E−04) and lymph node involvement (p = 1.18E−02) (Table 2), and shorter DFS with 69% 5-year DFS (95%CI 66–73) as compared to the CINSARC low-risk class (79% [95%CI 75–83], p = 2.01E−04, log-rank test; Fig. 2a). Importantly and by contrast with what was observed in the Luminal A subtype, CINSARC demonstrated significant prognostic value in both univariate (HR = 1.43 [95%CI 1.18–1.73], p = 2.18E−04, Wald test) and multivariate analyses (HR = 1.46 [95%CI, 1.09–1.96], p = 1.20E−02, Wald test) (Table 3). Other clinicopathological features independently associated with shorter DFS included pathological lymph node involvement, tumor size, and type. Such prognostic complementarity between the clinicopathological variables and CINSARC was tested using the likelihood ratio (LR) test: CINSARC added prognostic information to that provided by the combination of clinicopathological variables (ΔLR-X2 = 6.53, p = 1.06E−02). Because a major aim of using a prognostic signature in Luminal breast cancer is therapeutic de-escalation, we assessed the prognostic value of CINSARC in the 554 Luminal B patients treated with adjuvant endocrine therapy only, without adjuvant chemotherapy. As shown in Fig. 2b, CINSARC identified 222 low-risk Luminal B patients with 90% 5-year DFS (95%CI 86–95), significantly better than the CINSARC high-risk patients (79% 5-year DFS [95%CI, 75–84]; p = 1.04E−02, log-rank test). Of note in this population also, CINSARC had independent prognostic value in multivariate analysis (HR = 1.62 [95%CI 1.11–2.37], p = 1.16E−02, Wald test), together with pathological lymph node involvement and tumor size (Table 3), and added independent prognostic information to these clinicopathological features (ΔLR-X2 = 6.71, p = 9.58E−03). We built a prognostic clinico-genomic model based on these three variables in a randomly defined learning set of 247 samples and tested its prognostic value in the validation set of 247 remaining samples: as shown in Fig. 3, the model was robust and identified an even lower risk subgroup with 5-year DFS of 93% (95%CI [89–97]).
Table 2.
Correlations of CINSARC classes with clinicopathological characteristics in Luminal B breast cancer patients.
| Characteristics | CINSARC classes | ||||
|---|---|---|---|---|---|
| N | Low-risk | High-risk | p-value | ||
| Patients’ age | 0.233 | ||||
| ≤50 | 283 | 104 (23%) | 179 (26%) | ||
| >50 | 843 | 345 (77%) | 498 (74%) | ||
| Pathological tumor type | 0.178 | ||||
| Ductal | 723 | 289 (80%) | 434 (81%) | ||
| Lobular | 76 | 26 (7%) | 50 (9%) | ||
| Other | 96 | 46 (13%) | 50 (9%) | ||
| Pathological grade | 3.13E−04 | ||||
| 1 | 65 | 35 (9%) | 30 (5%) | ||
| 2 | 427 | 198 (51%) | 229 (42%) | ||
| 3 | 446 | 156 (40%) | 290 (53%) | ||
| Pathological axillary lymph node status | 1.18E−02 | ||||
| Negative | 676 | 295 (60%) | 381 (52%) | ||
| Positive | 545 | 199 (40%) | 346 (48%) | ||
| Pathological tumor size | 0.322 | ||||
| pT1 | 398 | 169 (37%) | 229 (33%) | ||
| pT2 | 664 | 252 (55%) | 412 (59%) | ||
| pT3 | 91 | 38 (8%) | 53 (8%) | ||
| Adjuvant chemotherapy | 0.012 | ||||
| No | 880 | 375 (83%) | 505 (77%) | ||
| Yes | 230 | 77 (17%) | 153 (23%) | ||
| Adjuvant hormone therapy | 0.709 | ||||
| No | 457 | 188 (42%) | 269 (41%) | ||
| Yes | 647 | 258 (58%) | 389 (59%) | ||
| Follow-up median, months (min–max) | 1438 | 71 (0–243) | 63 (0–294) | 0.677 | |
| DFS event (%) | 1438 | 164 (28%) | 309 (36%) | 9.19E−04 | |
| 5-year DFS | 1438 | 79% [75–83] | 69% [66–73] | 2.01E−04 | |
Fig. 2. Disease-free survival in Luminal B early breast cancer patients according to CINSARC signature.
a Kaplan–Meier disease-free survival (DFS) in Luminal B breast cancer patients according to the two CINSARC classes (high-risk versus low-risk) in the overall population. b Similar to a, but only in patients receiving adjuvant endocrine therapy but no adjuvant chemotherapy.
Table 3.
Univariate and multivariate Cox regression analyses for DFS in Luminal B breast cancer patients.
| All patients | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| N | HR [95% CI] | p-value | N | HR [95% CI] | p-value | ||
| Patients’ age | >50 vs. ≤50 | 1126 | 0.66 [0.52–0.84] | 5.73E−04 | 759 | 0.69 [0.47–1.03] | 0.070 |
| Pathological tumor type | lobular vs. ductal | 895 | 1.86 [1.21–2.84] | 1.47E−02 | 759 | 2.17 [1.38–3.39] | 7.18E-04 |
| other vs. ductal | 0.94 [0.60–1.47] | 759 | 1.19 [0.73–1.92] | 0.485 | |||
| Pathological grade | 2 vs. 1 | 938 | 1.23 [0.75–2.00] | 0.077 | |||
| 3 vs. 1 | 1.51 [0.93–2.46] | ||||||
| Pathological axillary lymph node status | positive vs. negative | 1221 | 1.69 [1.38–2.07] | 4.75E−07 | 759 | 2.55 [1.79–3.63] | 1.83E−07 |
| Pathological tumor size | pT2 vs. pT1 | 1153 | 1.48 [1.17–1.88] | 2.24E−07 | 759 | 1.52 [1.10–2.10] | 1.14E−02 |
| pT3 vs. pT1 | 2.96 [1.99–4.38] | 759 | 3.25 [1.89–5.58] | 2.09E−05 | |||
| Adjuvant chemotherapy | yes vs. no | 1110 | 1.51 [1.13–2.01] | 5.41E−03 | 759 | 1.08 [0.72–1.64] | 0.702 |
| Adjuvant hormone therapy | yes vs. no | 1104 | 0.74 [0.60–0.92] | 6.93E−03 | 759 | 0.69 [0.47–0.99] | 4.48E−02 |
| CINSARC classes | High vs. Low-risk | 1438 | 1.43 [1.18–1.73] | 2.18E−04 | 759 | 1.46 [1.09–1.96] | 1.20E−02 |
| Patients with adjuvant HT and without adjuvant CT | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| N | HR [95% CI] | p-value | N | HR [95% CI] | p-value | ||
| Patients’ age | >50 vs. ≤50 | 517 | 0.99 [0.50–1.96] | 0.985 | |||
| Pathological tumor type | lobular vs. ductal | 482 | 1.71 [0.94–3.12] | 0.195 | |||
| other vs. ductal | 0.92 [0.49–1.71] | ||||||
| Pathological grade | 2 vs. 1 | 407 | 0.84 [0.38–1.85] | 0.871 | |||
| 3 vs. 1 | 0.81 [0.37–1.78] | ||||||
| Pathological axillary lymph node status | positive vs. negative | 535 | 2.88 [1.98–4.19] | 3.13E−08 | 494 | 2.9 [1.91–4.40] | 6.23E−07 |
| Pathological tumor size | pT2 vs. pT1 | 513 | 1.87 [1.24–2.82] | 1.76E−06 | 494 | 1.76 [1.16–2.66] | 7.68E−03 |
| pT3 vs. pT1 | 5.21 [2.76–9.83] | 494 | 4.71 [2.47–8.98] | 2.41E−06 | |||
| CINSARC classes | High vs. Low-risk | 554 | 1.60 [1.11–2.30] | 1.11E−02 | 494 | 1.62 [1.11–2.37] | 1.16E−02 |
Fig. 3. Disease-free survival in Luminal B early breast cancer patients only treated with adjuvant endocrine therapy without adjuvant chemotherapy according to a clinico-genomic model combining CINSARC, pathological tumor size and lymph node status.
a Kaplan–Meier disease-free survival (DFS) in Luminal B breast cancer patients according to the two classes defined by the model (high-risk versus low-risk) in the learning set. b Similar to a, but in the validation set.
We had previously shown the prognostic complementarity and independence for DFS of commercial prognostic proliferation-based signatures (70_gene, Recurrence Score, ROR-P) and the ICR immune signature10. Thus we tested such independence between CINSARC and immune signatures including the Palmer’s metagenes (B-cells, T-cells, and CD8 T-cells)11, the Rooney’ cytolytic activity score12, and the three signatures predictive for response to immune therapy (ICR, TIS, and TLS). Multivariate analysis (Table 4) showed that in each case, CINSARC remained significant as well as each immune signature, suggesting independent prognostic value.
Table 4.
Multivariate Cox regression analyses for DFS in Luminal B breast cancer including immune signatures.
| DFS | Multivariate | ||
|---|---|---|---|
| N | HR [95%CI] | p-value | |
| ICR score | 1438 | 0.84 [0.73–0.97] | 1.95E−02 |
| CINSARC classes | 1438 | 1.47 [1.22–1.78] | 7.29E−05 |
| TIS score | 1438 | 0.89 [0.79–1.01] | 0.074 |
| CINSARC classes | 1438 | 1.46 [1.20–1.76] | 1.09E−04 |
| TLS score | 1438 | 0.83 [0.72–0.95] | 8.46E−03 |
| CINSARC classes | 1438 | 1.47 [1.22–1.78] | 6.79E−05 |
| Palmer, B-cells module | 1438 | 0.32 [0.23–0.45] | 4.62E−11 |
| CINSARC classes | 1438 | 1.44 [1.19–1.74] | 1.50E−04 |
| Palmer, CD8 T-cells module | 1438 | 0.60 [0.53–0.68] | 3.58E−15 |
| CINSARC classes | 1438 | 1.43 [1.19–1.73] | 1.90E−04 |
| Palmer, T-cells module | 1438 | 0.35 [0.25–0.48] | 2.32E−10 |
| CINSARC classes | 1438 | 1.45 [1.20–1.75] | 1.19E−04 |
| Rooney, Cytolytic activity | 1438 | 0.86 [0.77–0.95] | 3.09E−03 |
| CINSARC classes | 1438 | 1.45 [1.20–1.75] | 1.37E−04 |
CINSARC classes and therapeutic vulnerability in the Luminal B subtype
Beyond prognostication, multigene signatures may also help identify therapeutic targets that might improve survival of patients with high risk of recurrence. Thus, in the whole population of 2028 Luminal B patients of our database, we wondered whether the two CINSARC classes displayed different probabilities of response to specific systemic therapies routinely used or under development in breast cancer (Table 5).
Table 5.
Correlations of CINSARC classes with therapeutic response/vulnerability in Luminal B breast cancers.
| Therapies | Characteristics | N | CINSARC classes | p-value | ||
|---|---|---|---|---|---|---|
| Low-risk | High-risk | |||||
| Chemotherapy | 107-gene signature | No pCR-like | 1587 | 663 (83%) | 924 (75%) | 1.43E−04 |
| pCR-like | 441 | 140 (17%) | 301 (25%) | |||
| pCR | No pCR | 124 | 49 (88%) | 75 (80%) | 0.270 | |
| pCR | 26 | 7 (12%) | 19 (20%) | |||
| Hormone therapy | E2F4-activation signature | Low | 496 | 348 (43%) | 148 (12%) | 3.73E−57 |
| High | 1532 | 455 (57%) | 1077 (88%) | |||
| CDK4/6 inhibitors | RBsig signature | Score | 2028 | 0.01 (−0.9–1.6) | 0.44 (−0.6–2.6) | 1.57E−100 |
| E2F regulon signature | Score | 2028 | 0.11 (−0.6–0.6) | 0.3 (−0.5–0.73) | 5.36E−56 | |
| PARP inhibitors | HRD signature | Low | 248 | 94 (94%) | 154 (81%) | 2.59E−03 |
| High | 42 | 6 (6%) | 36 (19%) | |||
| Immune checkpoint inhibitorss | ICR signature | Score | 2028 | −0.33 (−2.31–2.6) | −0.21 (−1.8–3.1) | 8.07E−04 |
| TIS signature | Score | 2028 | −0.35 (−2.3–2.2) | −0.24 (−2.1–2.8) | 2.68E−03 | |
| TLS signature | Score | 2028 | −0.32 (−2.2–1.8) | −0.17 (−3.2–2.3) | 1.69E−05 | |
| IA ESCAT alterations | ERBB2 amplification1 | No | 764 | 305 (100%) | 459 (98%) | 0.156 |
| Yes | 8 | 1 (0%) | 7 (2%) | |||
| PIK3CA mutation | No | 575 | 219 (72%) | 356 (76%) | 0.151 | |
| (E542K, E545K/A, H1047R/L) | Yes | 197 | 87 (28%) | 110 (24%) | ||
| IIA ESCAT alterations | ESR1 mutation | No | 772 | 306 (100%) | 466 (100%) | — |
| (E380Q, Y537S/C/N, D538G) | Yes | 0 | 0 (0%) | 0 (0%) | ||
| PTEN loss2 | No | 766 | 305 (100%) | 461 (99%) | 0.411 | |
| Yes | 6 | 1 (0%) | 5 (1%) | |||
| IIB ESCAT alterations | AKT1 mutation | No | 752 | 296 (97%) | 456 (98%) | 0.361 |
| (E17K) | Yes | 20 | 10 (3%) | 10 (2%) | ||
| ERBB2 mutation3 | No | 761 | 302 (99%) | 459 (98%) | 1 | |
| Yes | 11 | 4 (1%) | 7 (2%) | |||
pCR: pathological complete response; 1: >= 6 copies; 2homozigous deletion, truncated mutations and kown inactivating missense mutations (e.g., R130Q/G); 3hotspot activating missense mutations (e.g., S310F/Y, L755S, V777L), inframe insertion exon 2 O (e.g., Y772_A775dup).
Regarding chemotherapy, 94 CINSARC high-risk and 56 CINSARC low-risk cases were informative about achievement or not of a pathological complete response (pCR) after anthracycline/taxane-based neoadjuvant chemotherapy. High-risk patients had a numerical but non-statistically significant increase in pCR rate (20%) as compared to low-risk patients (12%, p = 0.270, Fisher exact test). Close percentages were observed when considering the probability of pCR as defined using an expression signature of pathological response to neoadjuvant chemotherapy in breast cancer13: 25% of high-risk patients were predicted with pCR versus 17% of low-risk patients, and the difference was significant (p = 1.43E−04). By contrast, CINSARC high-risk patients were associated with a lower probability of sensitivity to hormone therapy (88%) according to the E2F4-activation signature14, as compared to CINSARC low-risk patients (57%; p = 3.73E−57). Altogether, these results suggested that CINSARC high-risk patients might be more sensitive to chemotherapy and less sensitive to hormone therapy than low-risk patients.
We also examined the potential vulnerabilities of CINSARC classes to targeted therapies, using predictive gene signatures. We observed higher RBsig15 and E2F regulon16 scores in high-risk patients (p = 1.57E−100 and p = 3.36E−56, respectively), suggesting higher probability of RB1-pathway inactivation and of resistance to CDK4/6 inhibitors. Conversely, high-risk patients displayed a signature predictive of Homologous Recombination Deficiency17 more frequently than low-risk patients (19% versus 6%: p = 2.59E−03), which may indicate higher sensitivity to PARP inhibitors. We also compared the proportion of patients in each class displaying an actionable genomic alteration with high evidence level according to ESCAT scale18. There was no significant difference between CINSARC high-risk and low-risk patients regarding the proportion of ESCAT I level alterations (ERBB2 amplification, and PIK3CA mutation), and ESCAT II level alterations (ESR1 mutation, PTEN loss, AKT1 mutations, ERBB2 mutation).
Finally, high-risk patients displayed higher score for three signatures associated with response to immune checkpoint inhibitors: ICR19 (p = 8.07E−04), TIS20 (p = 2.68E−03), and TLS signature21 (p = 1.69E−05), suggesting a potential higher response to immunotherapy in Luminal B CINSARC high-risk patients than low-risk patients.
Biological correlates of CINSARC classes in the Luminal B subtype
To further elucidate the biological differences between the two CINSARC classes and identify potential therapeutic targets, we compared their whole-exome mutational, whole-genome copy number and transcriptional, and proteomic (RPPA) profiles by using the TCGA dataset, which included 297 Luminal B cases. No genomic alteration was differentially mutated, deleted or amplified between the two classes (Supplementary Tables 4–6). A total of 510 genes were differentially expressed between the two classes (Supplementary Fig. 1, Supplementary Table 7). The robustness of this gene list was confirmed in the METABRIC independent validation set, and ontology analysis revealed a large preponderance of mitotic processes, including mitotic spindle assembly and chromosomal segregation, and DNA repair among the genes upregulated in the high-risk class (Supplementary Table 8). Proteomic analysis using RPPA results identified 16 proteins with differential expression between the two CINSARC classes (Table 6, Supplementary Table 9), including proteins involved in the cell cycle (cyclin B1, p27kip1, cyclin E2), cell proliferation (FOXM1 and its 14-3-3_zeta regulator22, ASNS23), DNA repair (KU80, RAD50, ERCC5, MSH6), AKT/mTOR pathway (4E-BP1, p70S6K), and epigenetic regulator (GCN5L2).
Table 6.
List of 16 proteins/phosphoproteins differentially expressed between the two CINSARC classes in Luminal B TCGA breast cancers.
| Gene#Protein | CINSARC, high- vs. low-risk | Expression status | |||
|---|---|---|---|---|---|
| N | Odds ratio [95%CI] | p-value | q-value | ||
| CCNB1#Cyclin_B1 | 240 | 1.66 [1.41–1.96] | 6.53E−07 | 1.46E−04 | up CINSARC high-risk |
| MSH6#MSH6 | 240 | 1.32 [1.19–1.47] | 1.38E−05 | 1.03E−03 | up CINSARC high-risk |
| FOXM1#FoxM1 | 240 | 1.31 [1.20–1.44] | 2.31E−06 | 2.58E−04 | up CINSARC high-risk |
| RPS6KB1#p70S6K | 240 | 1.28 [1.11–1.47] | 3.86E−03 | 0.086 | up CINSARC high-risk |
| SYK#Syk | 240 | 1.26 [1.10–1.44] | 4.65E−03 | 0.093 | up CINSARC high-risk |
| ENY2#ENY2 | 218 | 1.23 [1.10–1.37] | 2.46E−03 | 0.063 | up CINSARC high-risk |
| YWHAZ#14-3-3_zeta | 240 | 1.20 [1.09–1.33] | 2.21E−03 | 0.063 | up CINSARC high-risk |
| ASNS#ASNS | 240 | 1.20 [1.08–1.33] | 5.48E−03 | 0.093 | up CINSARC high-risk |
| KAT2A#GCN5L2 | 218 | 1.19 [1.07–1.32] | 6.22E−03 | 0.093 | up CINSARC high-risk |
| XRCC5#Ku80 | 240 | 1.15 [1.06–1.25] | 6.65E−03 | 0.093 | up CINSARC high-risk |
| CCNE2#Cyclin_E2 | 240 | 1.13 [1.06–1.20] | 1.10E−03 | 0.061 | up CINSARC high-risk |
| EIF4EBP1#4E-BP1_pT70 | 240 | 1.12 [1.05–1.20] | 6.61E−03 | 0.093 | up CINSARC high-risk |
| CDKN1B#p27_pT198 | 240 | 1.09 [1.03–1.14] | 6.07E−03 | 0.093 | up CINSARC high-risk |
| RAD50#Rad50 | 240 | 0.88 [0.83–0.94] | 1.71E−03 | 0.063 | down CINSARC high-risk |
| ERCC5#ERCC5 | 240 | 0.87 [0.81–0.94] | 2.17E−03 | 0.063 | down CINSARC high-risk |
| MAPK8#JNK_pT183_pY185 | 240 | 0.86 [0.79–0.93] | 2.56E−03 | 0.063 | down CINSARC high-risk |
Discussion
By examining the prognostic value of CINSARC signature in a large population of early breast cancers, we found that CINSARC was independently associated with survival outcome in the Luminal B subtype. In this subtype, CINSARC also identified potential vulnerabilities to specific therapeutics, including innovative classes of compounds that have been recently approved in HER2-negative breast cancer, as well as biological features that could be exploited as future therapeutic targets. These results may provide insights in the clinical development and use of prognostic signatures, and open perspectives for a further stratified management of breast cancer.
First, our study reinforces the need for integrating any new prognostic multigene signature together with other important clinical and biological features which are specifically related to a given tumor type. CINSARC signature was recently demonstrated to outperform histological grade in predicting metastatic outcome in soft tissue sarcomas6,24, and is currently prospectively tested to guide treatment in these tumors. CINSARC was also demonstrated to have prognostic value in various other tumor types and was proposed as a universal prognostic biomarker7. Based on a multivariate analysis involving several hundreds of clinically and biologically annotated breast cancers, our results demonstrate that CINSARC is not independently associated with survival in this disease and that its prognostic importance is dependent on the molecular subtypes. Thus, it is likely that in ERBB2-positive and basal-like breast cancers, other drivers than CINSARC genes are prominently leading the metastatic process, while in Luminal A breast cancers estrogen receptor signaling plays a major role. In Luminal B, the main biological processes that are captured by CINSARC, such as mitosis and chromosomal instability, may be of particular interest to predict clinical outcomes. And multivariate analyses showed that such prognostic value was also independent from that of immune signatures, clearly suggesting that mitosis and chromosomal instability and immune response provide complementary prognostic information. Of course because of a few limitations inherent to retrospective studies and associated biases), further validation in larger and prospective studies is warranted.
Second, while Luminal B breast cancer is thought to be an aggressive subtype and thus is almost always candidate to adjuvant chemotherapy, CINSARC also allowed identifying a population of patients with favorable outcome while only receiving adjuvant endocrine treatment without any adjuvant chemotherapy. In addition, combining CINSARC with clinical features, such as tumor size and lymph node status, identified a low-risk class of patients with a 93% probability of being disease-free at 5 years. All current prognostic signatures in breast cancer aim to separate low-risk patients, in which adjuvant chemotherapy may be safely spared and endocrine therapy alone may guarantee a high level of cure, from high-risk patients, in which endocrine treatment is not enough and adjuvant chemotherapy should be added. Yet, in the latter subgroup, 60–70% of patients would still be cured by endocrine treatment alone, which represents a high level of residual therapeutic inadequacy. Thus, CINSARC could be helpful in detecting those patients with “low-risk” Luminal B subtype in which the benefit of adjuvant chemotherapy remains questionable and might be replaced by alternative less toxic approaches.
Third, CINSARC also revealed potential therapeutic vulnerabilities in Luminal B breast cancers that may impact the future management of this hard-to-treat subtype. We found that CINSARC high-risk tumors were predicted to be more sensitive to chemotherapy but more resistant to endocrine therapy. Importantly, these high-risk tumors were associated with RB1 inactivation, indicating a higher probability of resistance to CDK4/6 inhibitors25, a therapeutic class improving survival in ER/PR-positive/HER2-negative advanced breast cancers and currently under investigation in the adjuvant setting26–32. Therefore, a low-risk CINSARC signature could identify Luminal B breast cancers with both relatively favorable outcome and relative resistance to chemotherapy, but with sensitivity to endocrine therapy and CDK4/6 inhibitors, making this combination an attractive alternative to evaluate in this population. Moreover, in accordance with its tight biological relationship with chromosomal instability and rearrangements, we found that CINSARC signature predicted higher sensitivity to both DNA repair- and immune-targeting therapeutics. Thus, high-risk CINSARC tumors were found to display more frequently a high HRD score (in nearly 20% of patients). Although PARP inhibitors were only approved in HER2-negative advanced breast cancer with germline BRCA1/2 mutation (gBRCAm)33,34, including half of patients displaying ER-positive tumors, clinical trials are now underway to evaluate these compounds in other genetic contexts. Thus, CINSARC might contribute to better identify these tumors displaying gBRCA wild-type but HRD features that may also prove to be sensitive to PARP inhibitors and other DNA repair targeting therapeutics. High-risk patients were also predicted to be more sensitive to immunotherapy. Essentially developed in triple-negative breast cancer, with promising results in both advanced and early settings35,36, recent data indicate that immune checkpoint inhibitors might be also active in ER-positive breast cancer37, thus, CINSARC could be useful to identify those Luminal B patients who could be candidate to PD1/PD-L1 targeting agents.
Finally, our study also allowed describing biological features associated with CINSARC in Luminal B breast cancer and thus proposing new therapeutic avenues in the field. As expected, genes and proteins associated with high-risk signature were involved in mitotic processes, chromosomal segregation, cell cycle and proliferation, as well as DNA repair. Interestingly, cyclin E2 protein was found to be up-regulated in high-risk tumors. Both cyclin E2 and cyclin E1 are able to complex with CDK2 through G1-to-S-phases, allowing RB1 phosphorylation and thus cell cycle progression, and both were shown to promote resistance to endocrine treatment38 and CDK4/6 inhibitors13,39. Importantly, high cyclin E2 expression may predict activity of CDK2-targeted approaches that are in development, either as specific CDK2 inhibitors or pan-CDK inhibitors that include CDK2 in their spectrum of activity40. Other potentially actionable proteins upregulated in CINSARC high-risk tumors include 4E-BP1 and p70S6K, which are downstream effectors of mTOR and AKT pathways, respectively. While mTOR inhibitor everolimus has been registered in endocrine treatment-resistant advanced breast cancer and is under investigation in high-risk early breast cancer41, several AKT inhibitors are currently evaluated in advanced breast cancer, including endocrine treatment-resistant luminal disease42. Ultimately, CINSARC high-risk tumors may represent a favorable subpopulation to investigate those compounds in the early setting. Of note, histone acetyl transferase GCN5L2, which was shown to regulate TGFβ signaling pathway and induce expression of epithelial-mesenchymal transition43, was also upregulated in high-risk tumors and may indicate a potential for epigenetic treatment in this subtype.
In conclusion, we found that CINSARC, a multigene signature initially developed in sarcomas, has an independent prognostic value in breast cancer restricted to the Luminal B subtype. CINSARC may not only identify a subgroup of tumors with relatively favorable outcome, which may not require adjuvant chemotherapy, but also suggests clues to better select patients with a higher probability of benefit from therapeutics under investigation in early breast cancer, such as cell cycle inhibitors, DNA repair targeting agents, immune checkpoint inhibitors, AKT/mTOR inhibitors, and epigenetic regulating agents.
Methods
Breast cancer samples and molecular profiling
We analyzed our breast cancer gene expression database10 pooled from 36 public datasets (Supplementary Table 10), comprising 8982 invasive breast cancer samples. The details of Institutional Review Board and Ethical Committee approval and patients’ consent for all 36 studies are present in their corresponding publications listed in Supplementary Table 10. Our study is based upon public data from published studies in which ethics approval and informed consent to participate were already obtained by authors. This study was approved by our institutional review board (Comité d’Orientation Stratégique, COS). Gene expression profiles had been generated using DNA microarrays and RNA-Seq, and collected from the National Center for Biotechnology Information (NCBI)/Genbank GEO and ArrayExpress databases, and authors’ website. The final pooled data set contained 8930 non-redundant non-metastatic, non-inflammatory, primary, invasive breast cancers. Before analysis, data were processed as previously described10. Briefly, the pre-analytic processing first included normalization of each data set separately, and was done by Robust Multi-Array (RMA) with the oligo R package (version 1.46.0) for Affymetrix data and by quantile normalization with the limma R package (version 3.38.3) for other microarray platforms. When multiple probes mapped to the same GeneID, we retained the one with the highest variance in each data set. We log2-transformed the already normalized TCGA RNAseq data. We also collected DNA and proteomic processed data from TCGA (whole-exome sequencing (WES), array-CGH and HRD score, and RPPA) and METABRIC (targeted-NGS, array-CGH).
Analysis of molecular profiles
To avoid biases related to trans-institutional immunohistochemical analyses and thanks to the bimodal distribution of respective mRNA expression levels, the ER, progesterone receptor (PR), and HER2 statutes (negative/positive) were defined on transcriptional data of ESR1, PGR, and HER2 respectively, as previously described44. In addition to the CINSARC signature6, we applied to each dataset separately several multigene signatures: PAM505 allowing to define the Luminal A, Luminal B, ERBB2-enriched, Basal, and Normal subtypes, immune signatures including the Palmer’s B-cell, T-cell, and CD8+ T-cell signatures11, and the Rooney’ cytolytic activity score12, 107-gene signature predictive for pathological response to anthracycline-based neoadjuvant chemotherapy in breast cancer13, E2F4-activation signature predictive for response to hormone therapy in breast cancer14, Rbsig15 and E2F regulon16 signatures predictive for resistance to CDK4/6 inhibitors on breast cancer pre-clinical models14 and clinical samples of PALOMA-3 trial16, and immune signatures predictive for response to immune checkpoint inhibitors: ICR (Immune Constant of Rejection)19 and TIS (T cell-inflamed signature)20 signatures and a TLS (tertiary lymphoid structures) signature21.
We also compared the molecular profiles of CINSARC high-risk versus low-risk Luminal B samples by applying supervised analyses to TCGA and METABRIC data sets at different levels: WES mutational, copy number alterations (CNA), and RPPA data using logistic regression with significance thresholds of p ≤ 0.05 and q ≤ 0.10, and transcriptional data using moderated t-test with significance thresholds of fold-change |FC | > 1.5, p ≤ 0.05 and q ≤ 0.10. This later used the TCGA set as learning set and the METABRIC set as independent validation set. Ontology analysis of the resulting gene list was based on the GO biological processes of the Database for Annotation, Visualization and Integrated Discovery (DAVID; david.abcc.ncifcrf.gov/).
Statistical analysis
Correlations between tumor classes and clinicopathological variables were analyzed using the one-way analysis of variance (ANOVA) or the Fisher’s exact test when appropriate. Disease-free survival (DFS) was calculated from the date of diagnosis until the date of disease recurrence or death from any cause. Follow-up was measured from the date of diagnosis to the date of last news for event-free patients. Survivals were calculated using the Kaplan–Meier method and curves were compared with the log-rank test. Uni- and multivariate prognostic analyses were done using Cox regression analysis (Wald test). The variables submitted to univariate analyses included patients’ age at diagnosis (≤50 years vs > 50), pathological type (lobular vs ductal vs other), pathological axillary lymph node status (pN: negative vs positive), pathological tumor size (pT1 vs pT2 vs pT3), pathological grade (1 vs 2 vs 3), PAM50-derived molecular subtypes (Luminal A vs Luminal B vs Normal vs Basal vs ERBB2-enriched), delivery of adjuvant chemotherapy (CT), delivery of adjuvant hormone therapy (HT), and CINSARC-based classifications. The likelihood ratio (LR) tests were used to assess the prognostic information provided beyond that of a clinical model, assuming a X2 distribution. Changes in the LR values (LR-ΔX2) measured quantitatively the relative amount of information of one model compared with another. All statistical tests were two-sided at the 5% level of significance. In the case of multiple testing, the p-values were replaced by the corrected q-values. Statistical analysis was done using the survival package (version 2.30) in the R software (version 2.9.1; http://www.cran.r-project.org/). We followed the reporting REcommendations for tumor MARKer prognostic studies (REMARK criteria)45.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Our work was supported by the the Ligue Nationale Contre le Cancer (label F.B.), SIRIC (INCa-DGOS-Inserm 6038 grant), Ruban Rose (F.B.), Association La Marie-Do (F.B.), and Fondation Groupe EDF (D.B.). The authors wish to thank the computing facilities DISC (Datacenter IT and Scientific Computing, CRCM) and DSIO (Institut Paoli Calmettes) for their technical support. We acknowledge the support of the Institut Paoli-Calmettes biobank (authorization number AC-2018-1905).
Author contributions
A.G. and F.B. were involved in the conception and design of study, analysis and interpretation of data, and draft of the manuscript. P.F. was involved in the acquisition, analysis and interpretation of data. All authors read critically and approved the final manuscript.
Data availability
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1435087146. All data sets of primary breast cancer were downloaded from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), ArrayExpress (https://www.ebi.ac.uk/arrayexpress/), Genomic Data Commons (GDC, https://portal.gdc.cancer.gov/) and cBioPortal (https://www.cbioportal.org/) databases. All accession IDs are provided in Supplementary Table 10 (Table S10 revised.xlsx), which is included with the data record. The data underlying the figures and tables are contained in the files ‘Goncalves_supporting_data.xlsx’ and ‘Table S8.xlsx’, which are included with the data record. A detailed list of the data underlying each figure and table is also available in the file ‘Goncalves_2021_underlying_data_list.xlsx’, which is included with the data record.
Code availability
Normalization of public data sets were done by Robust Multi-Array (RMA) with the oligo R package (version 1.46.0) for Affymetrix data and by quantile normalization with the limma R package (version 3.38.3) for other microarray platforms. Supervised analysis was done using a moderated t-test with empirical Bayes statistic included in the limma R package (version 3.38.3). For correction of the multiple-testing hypothesis, False Discovery Rate (FDR) was assessed using qvalue R package (version 2.14.1) (Storey et al., Annals of Statistics, 2003). Several multigene signatures were applied to each dataset separately: CINSARC6, PAM505, and 107-gene predictive signatures13, who were based on nearest-centroid classification using genes, data and distance method described in each respective study. Also were applied Rbsig15, E2F regulon16, ICR19, TIS20, TLS21, Palmer’s immune modules (B-cells, T-cells, and CD8 T-cells)11, and the Rooney’ cytolytic activity score12 signatures who were based on a Z-score metagene using gene list described in each respective study. Statistics analysis was done with the stats R package (version 3.5.2) and the survival R package (version 3.1-12) for survival analysis.
Competing interests
A.G. reports non-financial support (travel expenses, accommodation and meeting registration) from Astra Zeneca, Pfizer, Novartis, Roche. The remaining authors declare no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41523-021-00256-2.
References
- 1.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379:432–444. doi: 10.1016/S0140-6736(11)61625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet. 2015;386:1341–1352. doi: 10.1016/S0140-6736(15)61074-1. [DOI] [PubMed] [Google Scholar]
- 3.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 4.Sabatier R, Gonçalves A, Bertucci F. Personalized medicine: present and future of breast cancer management. Crit. Rev. Oncol. Hematol. 2014;91:223–33. doi: 10.1016/j.critrevonc.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chibon F, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat. Med. 2010;16:781–787. doi: 10.1038/nm.2174. [DOI] [PubMed] [Google Scholar]
- 7.Lesluyes T, Delespaul L, Coindre J-M, Chibon F. The CINSARC signature as a prognostic marker for clinical outcome in multiple neoplasms. Sci. Rep. 2017;7:5480. doi: 10.1038/s41598-017-05726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jemaà M, et al. Heterogeneity in sarcoma cell lines reveals enhanced motility of tetraploid versus diploid cells. Oncotarget. 2016;8:16669–16689. doi: 10.18632/oncotarget.14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee M, et al. MMTV-Espl1 transgenic mice develop aneuploid, estrogen receptor alpha (ERα)-positive mammary adenocarcinomas. Oncogene. 2014;33:5511–5522. doi: 10.1038/onc.2013.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertucci F, et al. The immunologic constant of rejection classification refines the prognostic value of conventional prognostic signatures in breast cancer. Br. J. Cancer. 2018;119:1383–1391. doi: 10.1038/s41416-018-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer C, et al. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rooney MS, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertucci, F. et al. Gene expression profiles of inflammatory breast cancer: correlation with response to neoadjuvant chemotherapy and metastasis-free survival. Ann. Oncol. 10.1093/annonc/mdt496 (2013). [DOI] [PMC free article] [PubMed]
- 14.Guerrero-Zotano AL, et al. ER+ breast cancers resistant to prolonged neoadjuvant letrozole exhibit an E2F4 transcriptional program sensitive to CDK4/6 inhibitors. Clin. Cancer Res. 2018;24:2517–2529. doi: 10.1158/1078-0432.CCR-17-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malorni L, et al. A gene expression signature of retinoblastoma loss-of-function is a predictive biomarker of resistance to palbociclib in breast cancer cell lines and is prognostic in patients with ER positive early breast cancer. Oncotarget. 2016;7:68012–68022. doi: 10.18632/oncotarget.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner NC, et al. Cyclin E1 expression and palbociclib efficacy in previously treated hormone receptor–positive metastatic breast cancer. J. Clin. Oncol. 2019;37:1169–1178. doi: 10.1200/JCO.18.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature490, 61–70 (2012). [DOI] [PMC free article] [PubMed]
- 18.Condorelli R, et al. Genomic alterations in breast cancer: level of evidence for actionability according to ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann. Oncol. 2019;30:365–373. doi: 10.1093/annonc/mdz036. [DOI] [PubMed] [Google Scholar]
- 19.Roelands J, et al. Oncogenic states dictate the prognostic and predictive connotations of intratumoral immune response. J. Immunother. Cancer. 2020;8:e000617. doi: 10.1136/jitc-2020-000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayers M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coppola D, et al. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am. J. Pathol. 2011;179:37–45. doi: 10.1016/j.ajpath.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergamaschi A, Christensen BL, Katzenellenbogen BS. Reversal of endocrine resistance in breast cancer: interrelationships among 14-3-3ζ, FOXM1, and a gene signature associated with mitosis. Breast Cancer Res. 2011;13:R70. doi: 10.1186/bcr2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin, C., Yang, X. & Zhan, Z. High expression of asparagine synthetase is associated with poor prognosis of breast cancer in Chinese population. Cancer Biother. Radiopharm. 10.1089/cbr.2019.3295 (2020). [DOI] [PubMed]
- 24.Le Guellec S, et al. Validation of the complexity INdex in SARComas prognostic signature on formalin-fixed, paraffin-embedded, soft-tissue sarcomas. Ann. Oncol. 2018;29:1828–1835. doi: 10.1093/annonc/mdy194. [DOI] [PubMed] [Google Scholar]
- 25.Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 26.Finn RS, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 27.Cristofanilli M, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016;17:425–439. doi: 10.1016/S1470-2045(15)00613-0. [DOI] [PubMed] [Google Scholar]
- 28.Hortobagyi, G. N. et al. Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N. Engl. J. Med. 10.1056/NEJMoa1609709 (2016). [DOI] [PubMed]
- 29.Im S-A, et al. Overall survival with ribociclib plus endocrine therapy in breast cancer. N. Engl. J. Med. 2019;381:307–316. doi: 10.1056/NEJMoa1903765. [DOI] [PubMed] [Google Scholar]
- 30.Slamon DJ, et al. Overall survival with ribociclib plus fulvestrant in advanced breast cancer. N. Engl. J. Med. 2020;382:514–524. doi: 10.1056/NEJMoa1911149. [DOI] [PubMed] [Google Scholar]
- 31.Sledge, G. W. et al. The effect of abemaciclib plus fulvestrant on overall survival in hormone receptor-positive, ERBB2-negative breast cancer that progressed on endocrine therapy-MONARCH 2: a randomized clinical trial. JAMA Oncol. 10.1001/jamaoncol.2019.4782 (2019). [DOI] [PMC free article] [PubMed]
- 32.Goetz MP, et al. MONARCH 3: abemaciclib as initial therapy for advanced breast cancer. J. Clin. Oncol. 2017;35:3638–3646. doi: 10.1200/JCO.2017.75.6155. [DOI] [PubMed] [Google Scholar]
- 33.Robson M, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 34.Litton JK, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid, P. et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 10.1056/NEJMoa1809615 (2018). [DOI] [PubMed]
- 36.Schmid P, et al. Pembrolizumab for early triple-negative. Breast Cancer N. Engl. J. Med. 2020;382:810–821. doi: 10.1056/NEJMoa1910549. [DOI] [PubMed] [Google Scholar]
- 37.Nanda, R. et al. Effect of pembrolizumab plus neoadjuvant chemotherapy on pathologic complete response in women with early-stage breast cancer: an analysis of the ongoing phase 2 adaptively randomized I-SPY2 trial. JAMA Oncol. 10.1001/jamaoncol.2019.6650 (2020). [DOI] [PMC free article] [PubMed]
- 38.Desmedt C, et al. Impact of cyclins E, neutrophil elastase and proteinase 3 expression levels on clinical outcome in primary breast cancer patients. Int. J. Cancer. 2006;119:2539–2545. doi: 10.1002/ijc.22149. [DOI] [PubMed] [Google Scholar]
- 39.Arnedos M, et al. Modulation of Rb phosphorylation and antiproliferative response to palbociclib: the preoperative-palbociclib (POP) randomized clinical trial. Ann. Oncol. 2018;29:1755–1762. doi: 10.1093/annonc/mdy202. [DOI] [PubMed] [Google Scholar]
- 40.Milioli, H., Alexandrou, S., Lim, E. & Caldon, C. E. Cyclins E1 and E2 in ER+ breast cancer: prospects as biomarkers and therapeutic targets. Endocr. Relat. Cancer10.1530/ERC-19-0501 (2020). [DOI] [PubMed]
- 41.Baselga J, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellis H, Ma CX. PI3K inhibitors in breast cancer therapy. Curr. Oncol. Rep. 2019;21:110. doi: 10.1007/s11912-019-0846-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhao L, Pang A, Li Y. Function of GCN5 in the TGF-β1-induced epithelial-to-mesenchymal transition in breast cancer. Oncol. Lett. 2018;16:3955–3963. doi: 10.3892/ol.2018.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McShane LM. Reporting recommendations for tumor marker prognostic studies (REMARK) Nat. Clin. Pract. Oncol. 2005;2:416–422. [PubMed] [Google Scholar]
- 46.Goncalves, A., Finetti, P., Birnbaum, D. & Bertucci, F. Metadata record for the manuscript: the CINSARC signature predicts the clinical outcome in patients with Luminal B breast cancer. 10.6084/m9.figshare.14350871 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1435087146. All data sets of primary breast cancer were downloaded from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), ArrayExpress (https://www.ebi.ac.uk/arrayexpress/), Genomic Data Commons (GDC, https://portal.gdc.cancer.gov/) and cBioPortal (https://www.cbioportal.org/) databases. All accession IDs are provided in Supplementary Table 10 (Table S10 revised.xlsx), which is included with the data record. The data underlying the figures and tables are contained in the files ‘Goncalves_supporting_data.xlsx’ and ‘Table S8.xlsx’, which are included with the data record. A detailed list of the data underlying each figure and table is also available in the file ‘Goncalves_2021_underlying_data_list.xlsx’, which is included with the data record.
Normalization of public data sets were done by Robust Multi-Array (RMA) with the oligo R package (version 1.46.0) for Affymetrix data and by quantile normalization with the limma R package (version 3.38.3) for other microarray platforms. Supervised analysis was done using a moderated t-test with empirical Bayes statistic included in the limma R package (version 3.38.3). For correction of the multiple-testing hypothesis, False Discovery Rate (FDR) was assessed using qvalue R package (version 2.14.1) (Storey et al., Annals of Statistics, 2003). Several multigene signatures were applied to each dataset separately: CINSARC6, PAM505, and 107-gene predictive signatures13, who were based on nearest-centroid classification using genes, data and distance method described in each respective study. Also were applied Rbsig15, E2F regulon16, ICR19, TIS20, TLS21, Palmer’s immune modules (B-cells, T-cells, and CD8 T-cells)11, and the Rooney’ cytolytic activity score12 signatures who were based on a Z-score metagene using gene list described in each respective study. Statistics analysis was done with the stats R package (version 3.5.2) and the survival R package (version 3.1-12) for survival analysis.