Abstract
Anterograde viral tracers are powerful and essential tools for dissecting the output targets of a brain region of interest. They have been developed from herpes simplex virus 1 (HSV-1) strain H129 (H129), and have been successfully applied to map diverse neural circuits. Initially, the anterograde polysynaptic tracer H129-G4 was used by many groups. We then developed the first monosynaptic tracer, H129-dTK-tdT, which was highly successful, yet improvements are needed. Now, by inserting another tdTomato expression cassette into the H129-dTK-tdT genome, we have created H129-dTK-T2, an updated version of H129-dTK-tdT that has improved labeling intensity. To help scientists produce and apply our H129-derived viral tracers, here we provide the protocol describing our detailed and standardized procedures. Commonly-encountered technical problems and their solutions are also discussed in detail. Broadly, the dissemination of this protocol will greatly support scientists to apply these viral tracers on a large scale.
Keywords: HSV-1 strain H129 (H129), Anterograde transneuronal tracer, Polysynaptic tracer, Monosynaptic tracer, Production, Application, Neural circuit, Neural circuit tracing
Introduction
Viral tracers have been widely employed in neuroscience for mapping neural circuit structure and/or function [1]. Though adeno-associated viruses (AAVs) have low toxicity and well-characterized cell tropism, they are deficient in replication, which makes them unsuitable for transneuronal circuit tracing [2]. Rabies virus is the most well-developed neural circuit tracer to date, but it is a retrograde tracer and labels input networks [3, 4]. Meanwhile, the development of an anterograde neural circuit tracer for mapping output networks has fallen far behind.
Our group has pioneered the use of the herpes simplex virus type 1 (HSV-1) strain H129 as the foremost anterograde transneuronal tracer [5–15]. In recent years, H129 anterograde tracing technology has been widely applied by neuroscientists worldwide to map projection circuits and output networks in a variety of animal models [5–15]. But these studies have used the early versions of either wild-type H129 or recombinant H129 expressing a fluorescent protein with low labeling intensity, which requires immunostaining for visualization [8, 15]. Our laboratory developed the polysynaptic anterograde tracer H129-G4 and the very first monosynaptic anterograde tracer H129-dTK-tdT [16] (Fig. 1A). Recently, we generated an improved version of H129-dTK, named H129-dTK-T2, by inserting another tdTomato expression cassette between US7 and US8 in the H129-dTK-tdT genome (Fig. 1A).
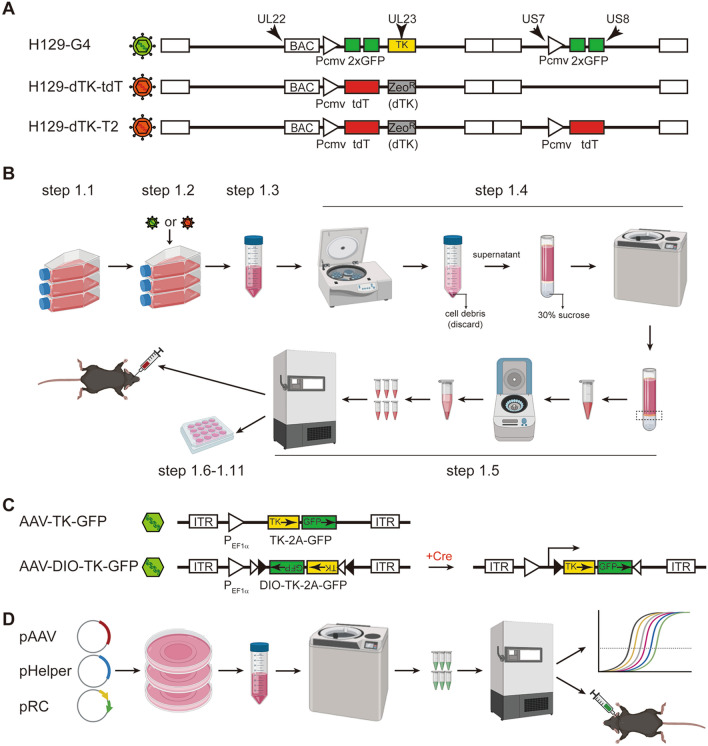
Fig. 1.
Schematic of the genome structure and flowchart for the preparation of viral tracers. A Schema of the genome structure of the H129-derived tracers. PCMV, CMV promoter; 2×GFP, GFP-2A-GFP expression cassette; tdT, tdTomato coding gene; ZeoR, zeocin-resistant gene; dTK, deleted viral TK gene (replaced by ZeoR). B Flowchart of the preparation of H129-derived tracers. C Schema of the genome structure of AAVs as helpers for H129-derived monosynaptic tracers. AAV-TK-GFP and AAV-DIO-TK-GFP (Cre-dependent) are helper viruses for anterograde monosynaptic tracing from starter cells. D Brief flowchart of the preparation of helper AAVs.
Here, we describe our detailed protocol for the production of all three H129 viruses (H129-G4, H129-dTK-tdT, and H129-dTK-T2). They can be produced by following a generic HSV-1 propagation protocol (Fig. 1B). However, similar to the well-developed retrograde G-protein-deleted monosynaptic rabies virus tracer [17], our TK-deficient H129 tracers (H129-dTK), including H129-dTK-tdT and H129-dTK-T2, also require appropriate helper AAVs to assist conditioned replication. For example, we use rAAV2/9 (AAV2/9-TK-GFP or AAV2/9-DIO-TK-GFP) as helpers (Fig. 1C), which express thymidine kinase (TK) and GFP to assist H129-dTK replication and label neurons in the injection site. AAV2/9 helpers are produced following the standard protocol described previously [18] (Fig. 1D).
The polysynaptic tracer H129-G4 transmits anterogradely through multiple synapses and labels all the infected neurons with high fluorescence intensity (Fig. 2A), which has been confirmed in our previous work [16]. So far, H129-G4 is an ideal tool to map output networks, with high transmission efficiency and labeling intensity, and is compatible with fMOST (fluorescence micro-optical sectioning tomography) for whole-brain high-resolution mapping [16]. After intracranial administration of an optimized dose of H129-G4 by stereotaxic injection, the tracing results can normally be obtained within 8 days (Fig. 2B). The progression of H129-G4 polysynaptic transmission can be estimated by the time post-injection according to the features of virus replication and spread [19–21].
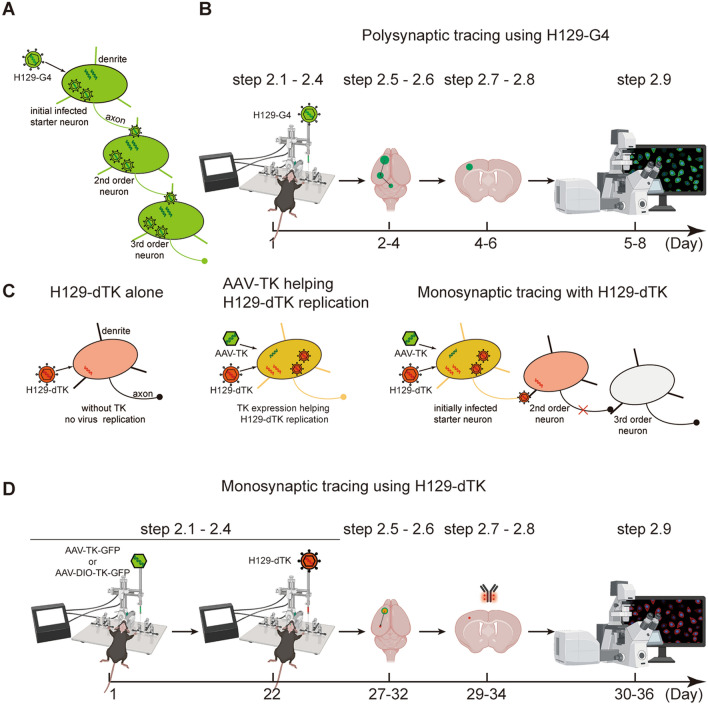
Fig. 2.
Overview of polysynaptic versus monosynaptic tracing with H129-derived tracers. A Schematic of the mechanism of polysynaptic tracing using H129-G4. B Flowchart and timeline for polysynaptic tracing using H129-G4. C Schematic of the mechanism of monosynaptic tracing using H129-dTK. H129-dTK virus can infect neurons normally by itself, but cannot replicate in neurons due to TK deficiency. TK expression by helper AAV-TK assists with H129-dTK replication, and offspring viruses transmit to the 2nd-order neurons and label them with the fluorescent protein. D Flowchart and timeline for monosynaptic tracing using H129-dTK tracers.
To prevent polysynaptic progression, our H129-dTK tracers are replication-deficient in neurons due to the deletion of the thymidine kinase (TK) gene (also known as UL23), but replication is rescued by the complementary TK expressed by the AAV helper AAV2/9-TK-GFP or AAV2/9-DIO-TK-GFP as described above. After conditioned replication, offspring viruses are transmitted to the post-synaptic neurons which are in turn labeled by the fluorescent protein tdTomato (tdT) expressed by H129-dTK (Fig. 2C), resulting in guaranteed monosynaptic labeling. Tracing with H129-dTK tracers requires the helper AAV, which is normally injected 3 weeks ahead, and the tracing results can be obtained at about 5 to 10 days after injecting H129-dTK (Fig. 2D). Recently, by inserting an additional tdT expression cassette into the H129-dTK-tdT genome, we created H129-dTK-T2, an improved version of H129-dTK-tdT (Fig. 1A). The double tdTomato expression cassettes result in a higher level of tdT protein and increased labeling intensity.
All of the H129-derived tracers (H129-G4, H129-dTK-tdT, and H129-dTK-T2) have been shared with neuroscience research groups worldwide. Here, we disseminate a standardized protocol to introduce the details to the field, and show how to produce and apply these tracers to neural circuit tracing in animal models.
Protocol
Caution
H129-derived tracers and helper AAVs are biohazardous materials with potential risk of infection. Therefore, all operations and experiments involving H129 tracers must be performed in a BSL-2 facility and handled according to governmental and institutional regulations.
Overview
This protocol has two parts: production of the viral tracers (Fig. 1) and application of the viral tracers (Fig. 2). The recommended parameters for production (Table 1) and application (Tables 2, 3, 4, 5, 6) are also shown based on our verification and optimization.
Table 1.
Seed virus and cell parameters for inoculation
| Viral tracer | Seed virus for inoculation | Host cell (Vero-E6) | ||||
|---|---|---|---|---|---|---|
| MOI | Virion # (pfu) | Tite(pfu/mL) | Vol. (mL) | Cell / flask | Flask # | |
| H129-G4 | 0.1 | 2.0 × 106 | 5.0 × 107 | 0.48 | 2.0 × 107 | 12 |
| H129-dTK-tdT | 0.1 | 2.0 × 106 | 5.0 × 106 | 4.80 | 2.0 × 107 | 12 |
| H129-dTK-T2 | 0.1 | 2.0 × 106 | 5.0 × 106 | 4.80 | 2.0 × 107 | 12 |
Vol. volume
Table 2.
Recommended parameters for H129-G4 tracer
| Animal | Brain region | Coordinates (mm)a | H129-G4 tracer | Perfusion time (dpi)c | |||
|---|---|---|---|---|---|---|---|
| AP | ML | DV | Dose (pfu) | Vol. (μL)b | |||
| C57BL/6 | Retina [16] | NA | NA | NA | 5.0 × 106 | 1.00 | 4 |
| C57BL/6 | OB [16] | + 4.28 | + 0.50 | − 2.50 | 2.0 × 106 | 0.40 | 3 |
| C57BL/6 | M1 [16] | + 1.54 | − 1.70 | − 1.75 | 1.0 × 106 | 0.20 | 3 |
| C57BL/6 | AD | − 0.82 | − 0.75 | − 2.75 | 4.0 × 105 | 0.08 | 3 |
| C57BL/6 | CeA [23] | − 1.00 | − 2.64 | − 4.30 | 3.0 × 105 | 0.06 | 1.5 |
| C57BL/6 | αBLA [22] | − 1.34 | + 3.40 | − 4.80 | 5.0 × 105 | 0.10 | 1–2 |
| C57BL/6 | VPM [16] | − 1.82 | + 1.40 | − 3.62 | 1.0 × 106 | 0.20 | 3 |
| C57BL/6 | CA1 [34] | − 2.00 | + 1.50 | − 1.60 | 2.9 × 105 | 0.12 | 1.5 |
| C57BL/6 | LGN [16] | − 2.30 | + 2.13 | − 2.75 | 5.0 × 105 | 0.10 | 3 |
| C57BL/6 | pBLA [22] | − 2.30 | + 3.40 | − 4.85 | 5.0 × 105 | 0.10 | 1–2 |
| C57BL/6 | AuD | − 2.80 | − 4.13 | − 1.68 | 2.0 × 105 | 0.04 | 3 |
| C57BL/6 | V2L | − 2.80 | − 3.15 | − 1.25 | 2.0 × 105 | 0.04 | 3 |
| C57BL/6 | V1 [16] | − 3.52 | + 2.30 | − 1.15 | 5.0 × 105 | 0.10 | 3 |
| C57BL/6 | IC | − 5.20 | + 1.40 | − 2.00 | 2.0 × 105 | 0.04 | 3 |
| SD rat | M1 [24] | + 3.00 | − 2.60 | − 2.20 | 5.0 × 105 / 2.5 × 106 | 0.10 / 0.50 | 3 |
| Tree shrew | M1 [16] | + 1.85 | − 2.60 | − 2.10 | 1.5 × 106 | 0.30 | 6 |
aCoordinates according to the mouse brain in stereotaxic coordinates [31].
bInjection volume should not exceed 0.5 μL for brain regions.
cExtending the waiting time before perfusion normally results in higher labeling intensity, wider tracer spread, and more orders of anterograde transmission. However, the incidence of animal deaths is also increased due to H129-G4 toxicity [16].
Table 3.
Tested brain regions and coordinates for H129-dTK-tdT monosynaptic tracer
| Animal | Brain region | Coordinates (mm)a | ||
|---|---|---|---|---|
| AP | ML | DV | ||
| C57BL/6 | MOB [16]] | + 4.28 | − 0.48 | − 2.30 |
| C57BL/6 | αBLA [22] | − 1.34 | + 3.40 | − 4.80 |
| C57BL/6 | VPM [16] | − 1.82 | − 1.42 | − 3.55 |
| C57BL/6 | LGN [16] | − 2.30 | + 2.13 | − 2.75 |
| C57BL/6 | pBLA [22] | − 2.30 | + 3.40 | − 4.85 |
| PV-Cre | nRT [16] | − 0.82 | + 1.50 | − 3.75 |
| DAT-Cre | VTA [16] | − 3.28 | − 0.40 | − 4.30 |
aCoordinates according to the mouse brain in stereotaxic coordinates [31].
Table 4.
Injection parameters of H129-dTK-tdT monosynaptic tracer
| Animal | Brain region | Helper AAV | H129-dTK-tdT | Perfusion time (day)b | |||||
|---|---|---|---|---|---|---|---|---|---|
| AAVs (1 × 1012 vg/mL) | Vol.a (μL) | Inject. time | Dose (pfu) | Vol.a (μL) | Inject. time | For starter neuron | For postsynaptic neuron | ||
| C57BL/6 | MOB [16] | AAV2/9-TK-GFP | 0.45 | Day 1 | 2.00 × 105 | 0.40 | Day 22 | Day 25 | Day 29 |
| C57BL/6 | αBLA [22] | AAV2/9-TK-GFP | 0.20 | Day 1 | 5.00 × 104 | 0.10 | Day 22 | Day 25 | Day 27 |
| C57BL/6 | VPM [16] | AAV2/9-TK-GFP | 0.20 | Day 1 | 1.00 × 105 | 0.20 | Day 22 | Day 25 | Day 32 |
| C57BL/6 | LGN [16] | AAV2/9-TK-GFP | 0.20 | Day 1 | 1.00 × 105 | 0.20 | Day 22 | Day 25 | Day 32 |
| C57BL/6 | pBLA [22] | AAV2/9-TK-GFP | 0.20 | Day 1 | 5.00 × 104 | 0.10 | Day 22 | Day 25 | Day 27 |
| PV-Cre | nRT [16] | AAV2/9-DIO-TK-GFP | 0.35 | Day 1 | 1.25 × 105 | 0.25 | Day 22 | Day 25 | Day 32 |
| DAT-Cre | VTA [16] | AAV2/9-DIO-TK-GFP | 0.35 | Day 1 | 1.25 × 105 | 0.25 | Day 22 | Day 25 | Day 32 |
aInjected volume should not exceed 0.5 μL for brain regions.
bExtending the waiting time before perfusion normally results in higher labeling intensity and more labeled postsynaptic neurons. However, the starter neurons are also more severely damaged due to H129-dTK-tdT toxicity when conditional replication occurs.
Table 5.
Tested brain regions and coordinates for H129-dTK-T2 monosynaptic tracer
| Animal | Brain region | Coordinates (mm)a | ||
|---|---|---|---|---|
| AP | ML | DV | ||
| C57BL/6 | MOB | + 4.28 | − 0.48 | − 2.30 |
| C57BL/6 | DG | − 1.70 | − 0.70 | − 1.88 |
| C57BL/6 | AuD | − 2.80 | − 4.13 | − 2.38 |
| C57BL/6 | IC | − 5.20 | + 1.40 | − 2.00 |
aCoordinates according to the mouse brain in stereotaxic coordinates [31].
Table 6.
Injection parameters of H129-dTK-T2 monosynaptic tracer
| Animal | Brain region | Helper AAV | H129-dTK-T2 | Perfusion time (Day)b | ||||
|---|---|---|---|---|---|---|---|---|
| AAV (1 × 1012 vg/mL) | Vol.a (μL) | Inject. time | Dose (pfu) | Vol.a (μL) | Inject. time | |||
| C57BL/6 | MOB | AAV2/9-TK-GFP | 0.45 | Day 1 | 2.00 × 105 | 0.40 | Day 22 | Day 27 |
| C57BL/6 | DG | AAV2/9-TK-GFP | 0.20 | Day 1 | 1.00 × 105 | 0.20 | Day 22 | Day 27 |
| C57BL/6 | AuD | AAV2/9-TK-GFP | 0.20 | Day 1 | 5.00 × 104 | 0.10 | Day 22 | Day 28 |
| C57BL/6 | IC | AAV2/9-TK-GFP | 0.20 | Day 1 | 5.00 × 104 | 0.10 | Day 22 | Day 27 |
aInjected volume should not exceed 0.5 μL for brain regions.
bExtending the waiting time before perfusion normally results in brighter neural labeling and more labeled post-synaptic neurons. However, the starter neurons are also more severely damaged due to H129-dTK-T2 toxicity when replication occurs.
To produce high-titer H129-derived tracers, H129-G4 or H129-dTK are inoculated as seed virus to pre-cultured Vero cells at a multiplicity of infection (MOI) of 0.1 (Step 1.1–1.2). At 3 days–4 days post-inoculation, the supernatant containing the indicated tracer virus is collected, and H129-derived tracers are concentrated by centrifugation after removing cell debris, then aliquoted and stored at − 80°C (Steps 1.3–1.5). The tracer stocks are titrated by plaque-forming assays (Steps 1.6–1.12). The whole process of tracer production takes 6 to 7 days from virus seed inoculation to tracer titration.
H129-derived tracers are applied to map the innervation pathway of a nucleus of interest in vivo, either alone (for H129-G4) or along with the appropriate helper AAV (for H129-dTK). The helper AAV (when necessary) and H129-derived tracers are sequentially injected into the brain region by stereotaxic injection (Steps 2.1–2.4). At appropriate time points, animals are perfused, then the brains are collected and processed by cryo-sectioning, immunostaining, and imaging by microscopy (Steps 2.5–2.9). The process from viral tracer injection to data collection takes 5 days–36 days, which varies with the different H129-derived tracers.
To obtain reliable and accurate tracing results, the parameters for viral tracer application need to be optimized according to different brain regions, transgenic mouse lines, and animal models. We and collaborators have tested these H129-derived tracers in the retina [16], main olfactory bulb (MOB) [16], primary motor cortex (M1) [16], ventral posteromedial thalamic nucleus (VPM) [16], lateral geniculate nucleus (LGN) [16], primary visual cortex (V1) [16], α-basolateral amygdala (BLA) [22], posterior basolateral amygdala (pBLA) [22], central amygdala (CeA) [23], anterodorsal thalamic nucleus (AD), dorsal auditory cortex (AuD), secondary visual cortex lateral area (V2L), and inferior colliculus in wild-type C57BL/6 mice; in the nucleus of the reticular thalamus (nRT) in PV-Cre transgenic mice [16]; in the ventral tegmental area (VTA) in DAT-Cre transgenic mice [16]; in M1 in Sprague-Dawley rats [24], and in tree shrews [16]. In the present protocol, the optimized parameters are listed in Tables 2, 3, 4, 5 and 6.
Production of H129 Tracers (H129-G4, H129-dTK-tdT, or H129-dTK-T2)
The production of H129 tracers is modified from the previously-described protocol for HSV-1 propagation [25] (Fig. 1B, Table 1). The entire procedure takes 6 to 7 days to produce ready-to-use H129 tracers. During the process, the virus titration step can be done before using the tracer, so that a pause between virus propagation (step 1.5) and titration (step 1.6) is acceptable. For one batch of H129 tracer, 12× T175 flasks of Vero-E6 cells yield ~400 μL of ready-to-use tracers with titers of 5.0 × 109 plaque-forming units (pfu)/mL (H129-G4) or 5.0 × 108 pfu/mL (H129-dTK).
Required Expertise and Materials
Expertise and resources: Basic virology and cell culture techniques are critical for H129-derived tracer preparation. A BSL-2 laboratory is required, and investigators should be trained and certificated to work in such a laboratory, following the standard operating protocols for all work with H129-derived tracers [26].
Biological materials: H129-derived tracers (H129-G4, H129-dTK-tdT, and H129-dTK-T2) are recombinant viruses generated as previously described [16]. African green monkey kidney Vero-E6 cells were purchased from the American Type Culture Collection (# CRL-1586).
Reagents: Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Technologies, Cat. #12100046) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Technologies, Cat. #12483020) and 1% penicillin–streptomycin (pen–strep, Thermo Fisher Technologies, Cat. #15140122), 1 × phosphate-buffered saline (PBS, Thermo Fisher Technologies, Cat. #70013-016), 0.25% trypsin (Thermo Fisher Technologies, Cat. #15400054), sterile 2% agar (Biowest, Cat.#182195)/H2O solution.
Equipment: Class II Biosafety cabinet (NuAire, Type A2), 37°C and 5% CO2 incubator (Thermo Fisher Technologies, Heracell 150i), centrifuge (Eppendorf, 5424R, with rotor F-34-6-38, and adaptor for 15-mL and 50-mL Falcon tubes), ultracentrifuge (Beckman, Optima XE-100, with rotor SW32), fluorescence microscope for observation of GFP and tdTomato (Nikon, eclipse Ti), 37°C water bath, microwave oven, − 80°C freezer (Haier, DW-86L626A).
Propagation of H129-Derived Tracers (3 days–4 days, Fig. 1B)
-
1.1
Cell preparation. Prepare 12 × T175 flasks of Vero-E6 cells for virus inoculation. There are ~2.0 × 107 Vero-E6 cells in each T175 flask at 90% confluence. Change to fresh medium prior to virus infection.
-
1.2
Virus infection. Quickly transfer the frozen virus seed vials of H129-G4, H129-dTK-tdT, or H129-dTK-T2 from a − 80°C freezer to a 37°C water bath for rapid thawing. Mix and add a specific amount of the thawed virus seed (Table 1) to fresh medium, make up to a total volume of 12.5 mL, add 1 mL to the prepared cells at an MOI of 0.1 (0.1 virions per cell), and gently agitate the flask to achieve an even infection.
Note 1: Thawing should be rapid and thorough to maintain the maximum virus activity. The virus seed is ready for inoculation when the ice crystals of the stock have melted. Try to avoid thawing for too long, which may decrease virus activity and eventually lead to low propagation efficiency and a low titer of the viral tracer.
It is necessary to calculate the amount of virus seed stock according to the virus titer. Thaw a proper amount of the virus seed stock; prepare the infection solution by adding fresh medium to the virus seed to a total volume up to 13 mL (for 12 flasks); add 1 mL to each flask. The average titers of H129-G4, H129-dTK-tdT, and H129-dTK-T2 yielded from virus propagation without concentration (from Step 1.4), and the corresponding required amount of each virus seed are listed in Table 1.
-
1.3
Virus propagation and harvesting. Keep the inoculated cells in a 37°C incubator with 5% CO2 without changing the medium, and allow the infection to progress until all cells show a cytopathic effect (CPE). When only <50% of cells remain attached to the bottom surface of the flask, collect the supernatant, which contains released viruses, from all 12 flasks in multiple 50-mL conical tubes.
Note 2: CPE appearance and progress should be constantly monitored under a microscope. Typically, H129-derived tracers show CPEs characterized by morphological change (cell rounded up and detached from the surface). Progression can be monitored based on the fluorescence of the tracers [25]. It takes 3 days–4 days from inoculation to harvest.
Note 3: About 20 mL of supernatant is normally collected from each flask, and five 50-mL conical tubes are needed for 12 flasks. Depending on the laboratory equipment, centrifuge tubes with larger volumes can also be used.
-
1.4
Virus purification and concentration. After centrifuging the supernatant sample at 2000×g at 4°C for 10 min to remove cell debris, collect the supernatant and treat with a 0.45-μm Millex PVDF filter (Merck Millipore, Cat. #SLHV033R13) to further get rid of cell debris. Prepare 40 mL of 30% sucrose in PBS, filter with a 0.22-μm Millex PVDF filter (Merck Millipore, Cat. #SLGV033RB), and add 5 mL to the bottom of each ultracentrifuge tube (Beckman Coulter, Cat.#355631). Layer the filtered virus supernatant (33 mL) onto the sucrose-PBS cushion (5 mL), and then centrifuge at 25,000×g at 4°C for 2.5 h.
Note 4: Before ultracentrifugation, save 5 mL of virus supernatant and store in 1-mL aliquots (in 1.5-mL microtubes) at − 80°C as a virus seed stock for future virus production. We recommend making a big batch of virus seed stock at a low passage which keeps the quality of the original viral tracer for future virus production. Long-term passaging may result in unexpected mutations, recombinations, degeneration, low infection, low transmission efficiency, and low labeling efficiency and intensity.
Note 5: The propagated H129-derived tracers can also be collected and purified by density gradient ultracentrifugation using sucrose or Iodixanol, followed by dialysis, which may increase the purity of the H129-derived tracers.
-
1.5
Storage of viral tracer. After carefully removing the supernatant and sucrose-PBS cushion, re-suspend the pelleted virus in 500 μL PBS, and centrifuge at 5,000×g at 4°C for 3 min to remove cell debris. Aliquot the virus suspension into a 4 μL/vial–10 μL/vial in 0.2 mL microtubes for application in neural circuitry mapping. Label the aliquots of H129 tracers, and store them at − 80°C until use.
Note 6: The H129 tracer aliquots can be stably maintained at − 80°C for up to 2 years, despite a slight drop in virus titer. However, it is crucial to avoid repeated freeze-thaw cycles. The aliquot is very sensitive to temperature change due to the small volume. Therefore, it is recommended to keep the tracer aliquots in the back of the − 80°C freezer rather than close to the door where the temperature increases rapidly whenever the freezer is opened.
Titration of H129-Derived Tracers (3 days, Fig. 1B)
-
1.6
Prepare titer plates. Re-suspend 3 × 106 Vero-E6 cells in 12.5 mL medium (DMEM +10% FBS +1% Pen–strep), and plate 0.5 mL cell suspension into a 24-well plate where each well contains 1.25 × 105 cells. Incubate the 24-well plate in a 37°C incubator with 5% CO2 for 4 h to let the cells attach to the surface.
-
1.7
Serial dilution of H129-derived tracer. Rapidly thaw in your hand one 10-μL aliquot of the H129 tracer aliquots stored at − 80°C and then add 90 μL medium to make a 101-times dilution. Then transfer all 100 μL to a 1.5 mL microtube with 900 μL medium, to make a 102-times dilution. Similarly prepare 10-fold serial dilutions from 102-times to 109-times with medium. We recommend preparing the dilutions in 1.5 mL–2.0 mL microtubes with a final volume of 1 mL.
Note 7: Mix each dilution well by repeatedly pipetting before taking it to the next dilution. Improper serial dilution results in incorrect titration, and in turn leads non-optimal tracing results.
Note 8: The virus seed obtained from Step 1.4 can also be titrated following the same method.
-
1.8
Inoculate diluted virus. Remove 250 μL medium from each well of the 24-well plate from Step 1.6, and add 200 μL of each dilution from 102- to 109-times to corresponding wells. Triplicate wells of each dilution are required. Incubate the plates in a 37°C incubator with 5% CO2 for 3 h.
-
1.9
Agar overlay. Pre-warm 21 mL of medium in a 37°C water bath, and melt 2% soft agar/H2O solution in a microwave oven. When the temperature of the agar/H2O solution drops to ~60°C, mix 5 mL soft agar with the pre-warmed DMEM in a 50-mL conical tube by gentle rotation. Do not vortex or shake the tube to avoid bubbles. Immediately add 1 mL of the soft agar/medium mixture to each well of the 24-well plate in Step 1.8 before the mixture solidifies.
Note 9: When holding the bottle of melted 2% soft agar solution with the bare hand, a hot but bearable feeling indicates the temperature is roughly 60°C, or measure the temperature with a sterile thermometer.
Note 10: Vortexing or shaking the soft agar/medium mixture results in bubbles, which cause difficulties in counting the plaques in Step 1.11.
-
1.10
Plaque formation. When the mixture solidifies, which normally takes about 15 min, transfer the plate into a 37°C incubator with 5% CO2, and keep for 3 days.
-
1.11
Count plaques and calculate virus titer. Count plaque numbers (N) in each well under a fluorescence microscope, and calculate the virus titer according to the following formula:Note 11: N1, N2, N3 indicate the plaque numbers in the triplicate wells; 10A indicates the dilution times (102 to 109).
-
1.12
Titer of yields. When being propagated from 12 T175 flasks and re-suspended in a 400 µL volume, H129-G4, H129-dTK-tdT, and H129-dTK-T2 normally yield a final titer of 5.0 × 109, 5.0 × 108, and 5.0 × 108 pfu/mL, respectively. The titrated H129 tracers that were aliquoted and stored at − 80°C in Step 1.5, are now ready to use for further application in mapping neural circuitry.
Application of H129-Derived Tracers: H129-G4, H129-dTK-tdT, or H129-dTK-T2
The period for neural circuitry tracing varies from 1 day to 1 month, depending on the different H129 tracers and purposes of the experiment. The main procedures include tracer administration by stereotaxic injection, brain sample processing, and imaging for tracing results (Fig. 2B, D, Tables 2, 3, 4, 5, 6).
Note 12: For monosynaptic tracing, H129-dTK-tdT and H129-dTK-T2 require the assistance of TK expression by helper AAVs to replicate and transmit to downstream neurons (Fig. 2C). The present protocol uses AAV2/9-TK-GFP (1.0 × 1012 viral genomes (vg)/mL) and AAV2/9-DIO-TK-GFP (1.0 × 1012 vg/mL) (Fig. 1C), whose packaging follows the standard protocol introduced previously [18] (Fig. 1D).
Required Expertise and Materials
Expertise and resources: Applying H129 tracers requires experience with animal work and a basic knowledge of histology. The animal facility must meet ABSL-2 criteria, and all investigators should be approved for animal work and certificated to work in such a laboratory. The animal experiments must be approved by the Ethics Committee, and performed strictly following the standard operation protocols [26].
Biological materials: H129 tracers (H129-G4, H129-dTK-tdT, and H129-dTK-T2) and helper AAVs (AAV2/9-TK-GFP and AAV2/9-DIO-TK-GFP as representatives in the present protocol), and experimental animals. Animals tested for tracing with H129-derived tracers include C57BL/6 mice [16, 22–24, 27–29], PV-Cre transgenic mice [16], DAT-Cre transgenic mice [16], Sprague-Dawley rats [24], and tree shrews [16]. Regular-sized adult mice (6–8 weeks old) and rats (8–10 weeks old) are recommended. Results shown in the present protocol are from wild-type C57BL/6 mice (Figs. 3, 4, 5, 6).
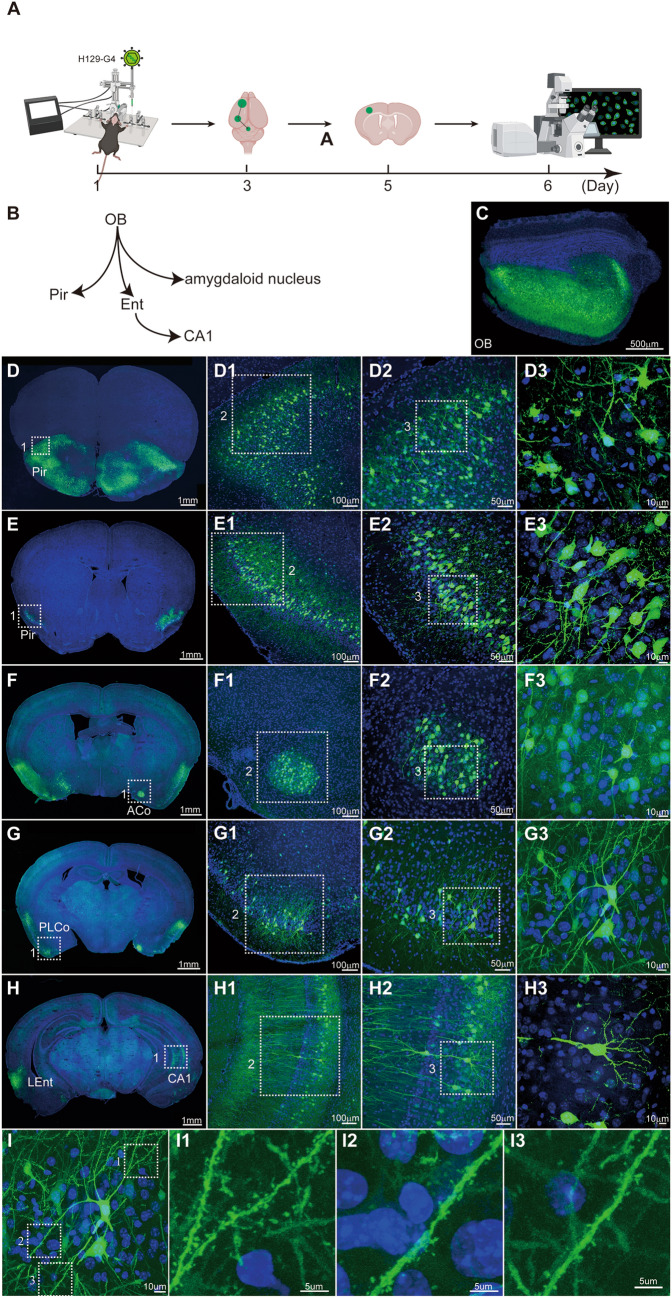
Fig. 3.
Mapping the output pathways of the olfactory bulb (OB) with the polysynaptic anterograde tracer H129-G4. A Timeline for tracing the OB output pathways with H129-G4. B Schema of the simplified OB projection pathways. OB, olfactory bulb; Pir, piriform cortex; Ent, entorhinal cortex; CA1, field CA1 of the hippocampus. C Representative image of a H129-G4 injection site in the OB. H129-G4, 5.0 × 109 pfu/mL, 400 nL, perfusion time: day 3. D–H Representative images of H129-G4-labeled downstream regions. The boxed areas are presented in the right panels at a higher magnification. Coronal brain slices were counterstained with DAPI. ACo, anterior cortical amygdaloid nucleus; PLCo, posterolateral cortical amygdaloid nucleus; LEnt, lateral entorhinal. I Reconstruction of a H129-G4-labeled neuron with fine structural details. A representative GFP-labeled neuron in PLCo is shown, and the magnified images of the dendritic segments are presented in the right panels (I1–I3).
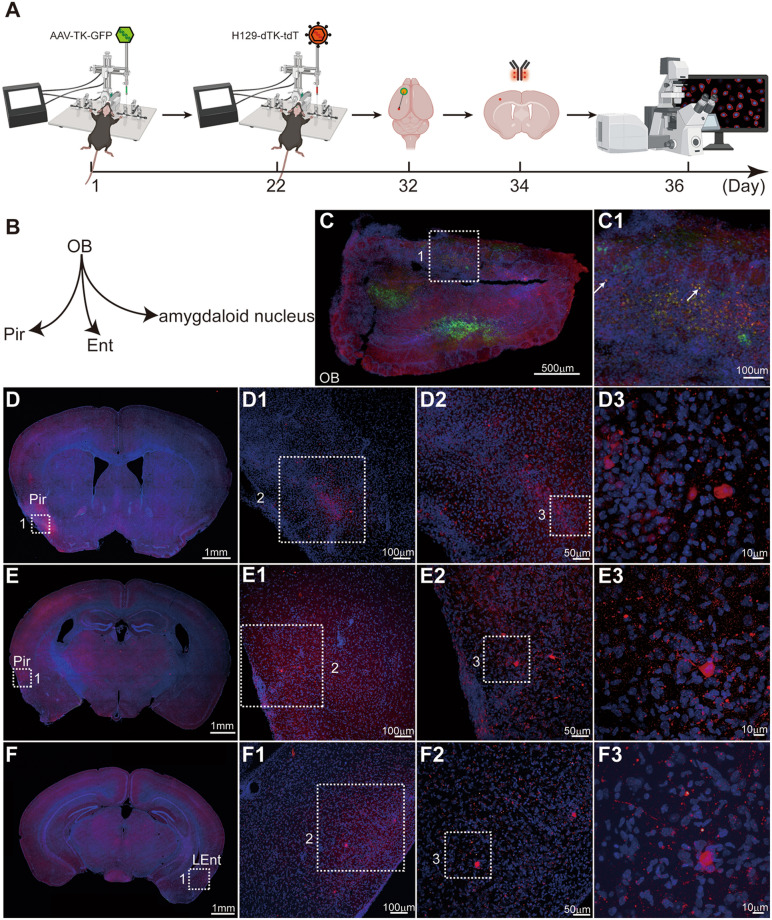
Fig. 4.
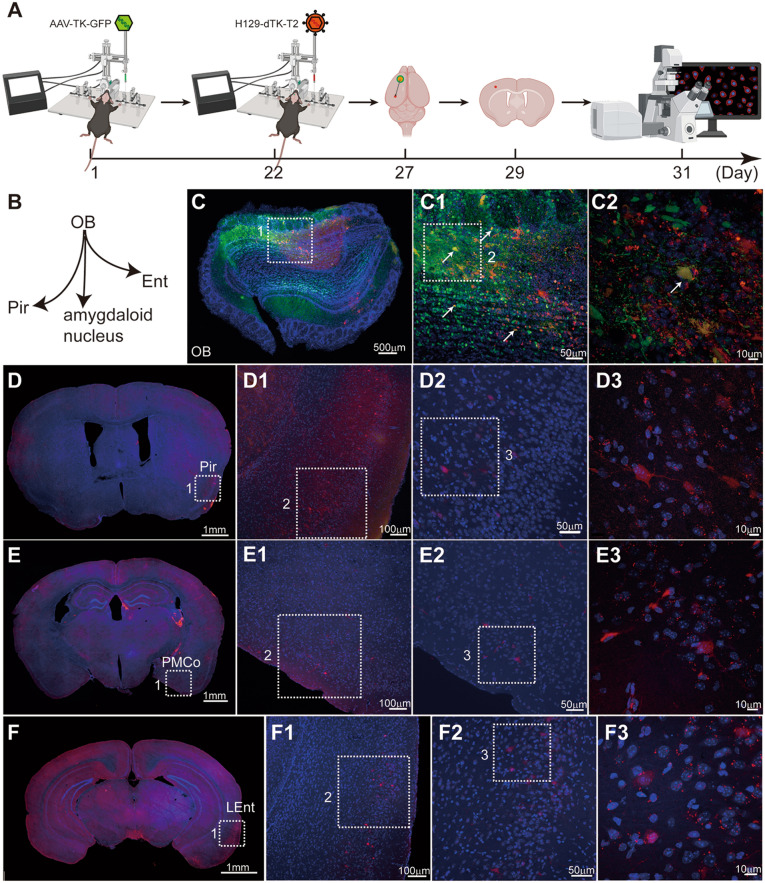
Mapping the direct projection target regions of the OB with our monosynaptic anterograde tracing system (H129-dTK-tdT + helper AAV2/9-TK-GFP). A Timeline for mapping the direct OB output pathways using H129-dTK-tdT along with AAV2/9-TK-GFP helper. Immunostaining against tdTomato is required for robust detection of H129-labeled postsynaptic neurons. B Schema of the simplified OB projection pathways. AAV2/9-TK-GFP, 1.0 × 1012 vg/mL, 450 nL, H129-dTK-tdT, 5.0 × 108 pfu/mL, 400 nL, perfusion time: day 32. C Representative image of the virus injection site in the OB. The boxed areas are presented in the right panels at a higher magnification. The starter neurons labeled by both GFP and tdTomato are indicated with white arrows. D–F Representative tracing images of the regions innervated by the OB. To visualize the labeled post-synaptic neuron, a brain slice was immunostained with tdTomato antibody. The boxed areas are presented in the right panels at a higher magnification. Coronal brain slices were counterstained with DAPI, and tdTomato signals were amplified by immunostaining with the anti-DsRed polyclonal antibody and Alexa Fluor 594-conjugated goat anti-rabbit IgG (H+L). LEnt, lateral entorhinal cortex.
Fig. 5.
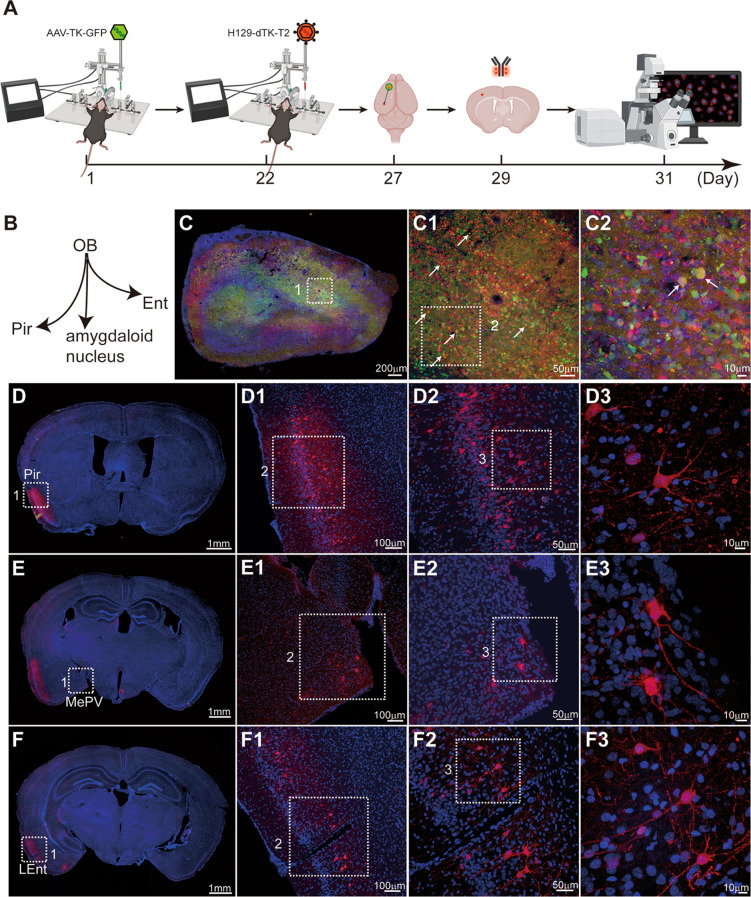
Mapping the direct projection areas of the OB using our improved monosynaptic anterograde tracing system (H129-dTK-T2 + helper AAV2/9-TK-GFP). A Timeline for mapping the direct OB output pathways using H129-dTK-T2 along with AAV2/9-TK-GFP helper. This improved monosynaptic anterograde tracing system allows for detection of postsynaptic labeled neurons with native tdTomato signals (no immunostaining enhancement). B Schema of the simplified OB projection pathways. AAV2/9-TK-GFP, 1.0 × 1012 vg/mL, 450 nL, H129-dTK-T2, 5.0 × 108 pfu/mL, 400 nL, perfusion time: day 27. C Representative image of the virus injection site in the OB. The boxed areas are presented in the right panels at a higher magnification. The starter neurons labeled by both GFP and tdTomato are indicated by white arrows. D–F Representative labeling images of the regions innervated by the OB. The boxed areas are presented in the right panels at a higher magnification. Coronal brain slices were counterstained with DAPI. LEnt, lateral entorhinal cortex; PMCo, posteromedial cortical amygdaloid nucleus.
Fig. 6.
Our H129-dTK-T2-based tracing system enables visualization of detailed structures in postsynaptic neurons, aided by immunostaining enhancement of tdTomato signals. A Timeline for mapping the direct OB output pathways using H129-dTK-T2 along with AAV2/9-TK-GFP helper. Immunostaining against tdTomato is applied. B Schema of the simplified OB projection pathways. AAV2/9-TK-GFP, 1.0 × 1012 vg/mL, 450 nL, H129-dTK-T2, 5.0 × 108 pfu/mL, 400 nL, perfusion time: day 27. C Representative image of the virus injection site in the OB. The boxed areas are presented in the right panels at a higher magnification. The starter neurons labeled by both GFP and tdTomato are indicated by white arrows. D–F Representative labeling images of the regions innervated by the OB. To improve observation of the labeled post-synaptic neuron, the brain slice was immunostained with tdTomato antibody. The boxed areas are presented in the right panels at a higher magnification. Coronal brain slices were counterstained with DAPI, and tdTomato signals were amplified by immunostaining with the anti-DsRed polyclonal antibody and Alexa Fluor 594-conjugated goat anti-rabbit IgG (H+L). LEnt, lateral entorhinal cortex; MePV, medial amygdaloid.
Reagents: 0.7% pentobarbital, 70% ethanol, eye ointment (Vidisic, Carbomer 980), liquid paraffin (Aladdin, CAS: B042-47-5), 1 × PBS, sterile normal saline, 4% paraformaldehyde (PFA, Sinopharm Chemical Reagent, Cat. #80096618), 30% sucrose in PBS, cryoprotectant (glycerin: isopropanol: 1 × PBS = 2:3:5), DAPI (Roche, Cat. #10236276001), mounting media (National Diagnostics, Cat. #HS-106), embedding matrix for frozen sections (Cell Path, Cat. #80202-0001), and antibodies. The antibodies used in the present protocol include anti-DsRed polyclonal antibody (Takara, Cat. #632496) and Alexa Fluor 594-conjugated goat anti-rabbit IgG (H+L) (Invitrogen, Cat. #A-11037), but they may be changed according to the requirements of experiments and resources.
Equipment: Stereotaxic injection system (RWD, CAX01-011), micropipette puller (Sutter Instruments, P-1000), microliter syringes (Hamilton, Cat. #86259), heating pad, microtome (Thermo Fisher Scientific, Microm HM550), confocal fluorescence microscope (Nikon A1R MP+), and surgical equipment (microdrill, electric clipper, fine-tip forceps, suture scissors, needles, thread).
Administration of H129 Tracers to Mouse Intracranially (~40 min per Animal)
-
2.1
Anesthesia. The experimental animal is anesthetized by intraperitoneal (i.p.) injection of 0.7% pentobarbital (0.1 mL/g body weight).
Note 13: Virus injection and the management of infected animals must be performed in a BSL-2 animal facility.
Note 14: In the present protocol, we use 0.7% pentobarbital by i.p. injection for easy access. Other anesthetics, such as isoflurane (recommended) and avertin can also be used as alternatives.
-
2.2
Injection of helper AAV (for H129-dTK-tdT and H129-dTK-T2). Inject the appropriate amount of helper AAV into the region of interest (Tables 3, 4, 5, 6) by stereotaxic injection 21 days prior to H129-dTK tracer injection (Tables 4, 6).
Note 15: This step is only necessary for monosynaptic tracing using H129-dTK-tdT or H129-dTK-T2. The recommended amount for injection of AAV helpers is shown in Tables 4 and 6. To trace the direct output pathway of a specific neuron type in certain brain regions, transgenic mice expressing Cre recombinase in this specific neuron type are required, and the AAV-DIO-TK-GFP should be chosen accordingly.
Note 16: Helper AAVs, either commercially available or home-made, normally have a titer between 1.0 × 1012 vg/mL and 1.0 × 1013 vg/mL. To standardize the experimental parameters, the AAVs are diluted to the working titer of 1.0 × 1012 vg/mL in 1 × PBS, aliquoted, and stored at − 80°C for further use.
Note 17: The stereotaxic injection was performed following a well-established standard protocol [30]. The injection coordinates were obtained from a mouse brain atlas [31].
-
2.3
Injection of H129 tracers. Inject an appropriate amount of H129-derived tracers. The multisynaptic tracer H129-G4 alone is sufficient to map output networks by injecting into the region of interest. To map direct monosynaptic output, H129-dTK-tdT or H129-dTK-T2 should be injected into the location where the helper AAV has been injected as described in Step 2.2 at 21 days after AAV administration.
Note 18: To standardize the experimental parameters, the H129 tracers are adjusted to the same titer with 1 × PBS prior to injection: 1.0 × 109 pfu/mL for H129-G4 and 5 × 108 pfu/mL for H129-dTK.
Note 19: The amount of tracer should be carefully optimized for different species, lines, brain regions, and experimental purposes. Over-dosage of H129 tracers leads to stronger toxicity and may cause the death of the animal, while insufficient tracer leads to insufficient infection, low transneuronal transmission, and poor neuronal labeling. We and collaborators have tested H129-G4 and H129-dTK in multiple brain regions. The detailed experimental parameters, including the regions tested, coordination of injections, optimal tracer dose, and recommended time points for observation, are listed in Tables 2, 3, 4, 5 and 6.
Note 20: The stereotaxic injection in the present protocol, for both helper AAVs and H129 tracers, was performed following the well-established standard protocol described previously [30]. The injection coordinates were from a mouse brain atlas [31].
-
2.4
Management of the animal. Remove the injected animal from the stereotaxic frame, and place it on a 37°C heating pad after suturing the wound. Transfer the awake animal into a cage and keep it in the ABSL-2 facility until perfusion.
Note 21: Monitor the animals constantly. Terminate the experiment immediately when the animal meets the euthanasia criteria.
Note 22: H129-G4 may anterogradely transmit multiple orders through neural circuits, and this can be roughly controlled by carefully choosing the perfusion time. According to previous reports, H129-G4 transmits to the 2nd, 3rd, and 4th order neurons at 36–44, 52–60, and 68–76 h after virus injection [20, 21]. However, the transmission time may vary according to the connection type and projection distance. Therefore, circuit order cannot precisely be determined by H129-G4.
Brain sample Processing and Imaging (4 days)
-
2.5
Perfusion. Anesthetize the animal by i.p. injection of 0.7% pentobarbital (0.1 mL/g body weight), perform cardiac perfusion with 1 × PBS followed by 4% PFA following the protocol described previously [32].
Note 23: For one mouse brain, normal perfusion with 20 mL 1 × PBS is sufficient to remove the blood, and 20 mL 4% PFA is sufficient for brain tissue fixation. For one rat brain, 60 mL of each is recommended.
-
2.6
Brain collection, fixation, and dehydration. Carefully dissect and obtain the intact brain, immerse in 4% PFA for 12 h for post-fixation, then dehydrate in 30% PBS-buffered sucrose for 36 h at 4°C. The well-fixed and sufficiently dehydrated brain should sink to the bottom of the sucrose solution.
-
2.7
Cryo-sectioning. Cut brain sections 40 μm thick on a freezing microtome, then mount the sections on slides.
-
2.8
Staining or immunostaining. Perform appropriate immunostaining or histological staining according to different experimental purposes. Seal the slides with mounting medium after staining.
Note 24: In the present protocol, all slides shown were counterstained with DAPI for nuclear staining. For H129-dTK-tdT tracing, we used rabbit anti-DsRed polyclonal antibody and Alexa Fluor 594-conjugated goat anti-rabbit IgG to amplify the tdTomato signal.
Note 25: The procedure can be paused here, and the slides can be stored at 4°C for future examination. The slides are stable at 4°C for one month.
-
2.9
Imaging and data collection. Obtain the tracing results using an appropriate imaging system. It is recommended to first examine all slides with a fluorescence slide scanner, and then capture higher-resolution images of the areas of interest under a confocal microscope.
Note 26: Cryo-section and regular fluorescence microscopy imaging were introduced in the present protocol. But H129 tracers are also compatible with other brain processing protocols for different imaging methods, such as the transparent brain, fMOST [16], and volumetric imaging with synchronized on-the-fly-scan and readout (VISoR) [33], etc.
Representative Tracing Results
H129 transmits transneuronally in an anterograde manner [5–14], and the H129-derived tracers maintain this property. Therefore, the tracers are mainly used to map the output pathways of brain regions [16, 22–24, 27–29].
Representative Tracing Results Using H129-G4
H129-G4 (Figs 1A, 2A, B) is the anterograde tracer with the most intense fluorescence labeling so far [8, 15, 16]. As shown in Fig. 3, H129-G4 is capable of spreading transneuronally through the olfactory bulb (OB) output pathways (Fig. 3A–C) and efficiently labeling the neurons of downstream brain regions (Fig. 3D–H) within 3 days. The labeling intensity is high enough to visualize the neuronal morphology and fine structure details, such as dendritic spines (Fig. 3I). Based on the labeling efficiency and intensity, H129-G4 is suitable for exploring the novel innervation targets of a specific brain region or to verify a novel projection pathway identified by other methods. Our collaborators have successfully used these H129-derived tracers (H129-G3 and H129-G4) in different brain regions for different purposes. For instance, Li et al. applied H129-G3, a previous version of H129-G4 with dimmer labeling intensity, to verify the projection from entorhinal cortex to dorsal CA1 pyramidal cell synapses [28]; Yu et. al. applied H129-G4 to verify the projection from the lateral CeA to the lateral amygdala (LA) [27]; Pi et al. mapped the innervation between aBLA or pBLA and ventral CA1 using H129-G4 and H129-R4 (H129-R4 is similar to H129-G4 but with 4 × m Cherry instead of 4 × GFP) [22]; and Zhu et al. used H129-G4 to verify the projection from CeA to glutamatergic neurons in the parafascicular nucleus [23]. H129-G4 can be also used to compare the projection strength between specific brain regions under different conditions. Using H129-G4, Wang et al. revealed that the connectivity from M1 to the subthalamic nucleus is impaired in unilateral 6-hydroxydopamine-lesioned parkinsonian rats [24], and Ma et al. identified a conditioning-strengthened indirect circuit from CA1 to BLA through the postrhinal cortex participates in morphine withdrawal memory retrieval [34].
Representative tracing results using H129-dTK-tdT
H129-dTK tracers are currently the best choice to directly map monosynaptic anterograde projections, especially from a genetically-defined neuronal population (such as Cre+), in which the recently-reported transneuronal AAV tracer fails [35, 36]. H129-dTK is a useful tool to confirm the direct projection from a known brain region, as well as to verify the innervation targets of a defined neuronal type from a region of interest. Monosynaptic anterograde tracing using H129-dTK requires the assistance of helpers, mostly recombinant AAVs expressing the TK gene (Figs. 1C, 2C). Briefly, the H129-dTK tracers H129-dTK-tdT or H129-dTK-T2 and the helpers AAV-TK-GFP or AAV-DIO-TK-GFP (for starter neuron type-specific monosynaptic anterograde tracing in Cre-transgenic mouse lines) are intracranially injected into the same site, and the brains are obtained at the appropriate time points and processed for imaging (Fig. 2D).
Representative tracing results from the OB using H129-dTK monosynaptic tracers are shown in Figs. 4, 5 and 6, where the differences in labeling efficiency and intensity of H129-dTK-tdT and H129-dTK-T2 are clear. First, AAV-TK-GFP and H129-dTK-tdT were sequentially injected into the OB of wild-type C57BL/6 mice on days 1 and 22 as shown in the timeline schematic. Then, brains were harvested on day 32 (Fig. 4A). Coronal brain sections were counterstained with DAPI (Roche, Cat. #10236276001), and the tdTomato signals were amplified by immunostaining with the anti-DsRed polyclonal antibody (Cat. #632496, Takara) and Alexa Fluor 594-conjugated goat anti-rabbit IgG (H+L) (Cat. #A-11037, Invitrogen) (Fig. 4). Since 10 days are required between H129-dTK-tdT injection and perfusion, the co-infected neurons at the injection site were dead due to cytotoxicity caused by H129-dTK replication with AAV assistance (Fig. 4C1). tdTomato-labeled neurons were seen in the downstream regions, including the piriform cortex (Pir), and lateral entorhinal cortex (LEnt) (Fig. 4D–F). The labeling intensity of H129-dTK-tdT was too low to directly observe the post-synaptic neuron (data not shown), so signal amplification of the fluorescent protein tdTomato by immunostaining was required. Even so, the labeling signal remained weak in the post-synaptic neurons. Thus, the application of H129-dTK-tdT is limited by poor labeling intensity.
Representative Tracing Results Using H129-dTK-T2
To improve the labeling efficiency of H129-dTK-tdT, we generated H129-dTK-T2 by inserting an additional tdTomato expression cassette between US7 and US8 of the H129-dTK-tdT genome (Fig. 1A). AAV-TK-GFP and H129-dT-T2 were injected into the same site in the OB of C57BL/6 mice with the same parameters as for H129-dTK-tdT. The time between H129-dTK-T2 injection and perfusion was shortened to 5 days (Figs. 5A, 6A), so neurons labeled with tdTomato and GFP were observed in the injected OB (Figs. 5C, 6C). The labeling signals of the post-synaptic neurons in the Pir, posteromedial cortical amygdaloid nucleus, and LEnt were visible without immunostaining, although dim (Fig. 5D–F). Immunostaining with the antibody against tdTomato clearly boosted the labeling intensity of H129-dTK-T2 (Fig. 6), and the morphological details of post-synaptic neurons were better visualized than the results without immunostaining and results from labeling by H129-dTK-tdT. Notably, the observation time with H129-dTK-T2 was brought forward to 27 (Figs. 5, 6) rather than day 32 for H129-dTK-tdT (Fig. 4). These results indicate that the additional fluorescent protein expression cassette improved the labeling efficiency and intensity, highlighting that H129-dTK-T2 is a better version than H129-dTK-tdT.
Troubleshooting and Discussion
The H129 tracers’ production, storage, application, and result analysis are influenced by various factors. According to previous experience from our and other laboratories, we have summarized them in the following troubleshooting list. Investigators are encouraged to further optimize the protocols to achieve the most optimal tracing result in their brain regions of interest.
Problem 1
Viral yield is much lower than expected (Table 1).
Possible reason: Host cell condition, mycoplasma contamination, inoculation amount, harvesting timing, incorrect titration, and/or repeated freezing/thawing of the virus.
Potential solution: Use freshly recovered mycoplasma-free cells (Step 1.1). Carefully keep the cells in the best condition without overgrowth, and precisely count the cell number (Step 1.1). Inoculate the cells with virus seed stock without repeated freezing/thawing, which should be pre-titrated and maintained to guarantee the inoculation dose (Steps 1.2 and 1.3). Redo the titration properly using a newly-thawed viral tracer aliquot (Steps 1.6–1.11). Keep the virus on ice and avoid repeated freezing/thawing.
Problem 2
Alarge volume (>5 μL) of tracer needs to be injected intracranially.
Possible reason: Incorrect calculation, incorrect titration, the titer of H129 tracer may be too low.
Potential solution: Redo the calculation and/or titration. Re-concentrate the virus by centrifugation if the original volume is large. Choose a different batch of virus tracer with a higher titer.
Problem 3
Rapid death of injected animals.
Possible reason: Improper injection, overdosed viral tracer (especially for H129-G4), originally unhealthy animal, brain region being traced is essential to animal survival.
Potential solution: Carefully perform the stereotactic injection without causing unnecessary damage. Check the titer and recalculate to verify the viral dose. Check the health status of animals before experiments, and let transported animals rest in the new environment for a few days before tracer injection. Try alternative brain regions if possible, or use an alternative tracer other than those derived from H129.
Problem 4
Severe inflammation at the injection site.
Possible reason: Immune response and inflammation induced by viral infection; tracer is not pure and contains cell debris. Helper AAV may induce strong immune responses, but the H129 tracer is more likely to be the major trigger.
Potential solution: Reproduce the virus properly following the present protocol. Optimize the virus concentration and purification protocol, such as density gradient centrifugation dialysis, and ultrafiltration.
Problem 5
Too many cells labeled with H129-G4.
Possible reason: overdose of the injected H129-G4; waited too long to perfuse.
Potential solution: Reduce the amount of tracer injected, or the waiting time, by careful titration and optimization of the dose or perfusion timing.
Problem 6
No or few neurons labeled with H129-G4 at the injection site.
Possible reason: Improper injection, improper perfusion timing, incorrect tracer titration, improper dose of injected tracer, degraded or inactive H129-G4, low susceptibility in animal species/strain.
Potential solution: Perform the stereotactic injection carefully, and keep the needle in the injection site for 5 to 15 min after tracer injection to avoid spill-over, so that the injected tracers stay in the site. Adjust the perfusion timing to be neither too soon nor too late post-injection. Harvest 3 mice each day from 1 days to 7 days after H129-G4 injection, and compare the tracing results to choose the best timepoints for the target tracing experiment. Titration needs to be done following the protocol described above. The dose can be optimized by injecting serial amounts of H129-G4 (from 1 × 104 pfu to 1 × 107 pfu) and comparing the results. Check the calculation to make sure the dose is correct, as described above. Store the tracers deep inside the − 80°C freezer without frequent and dramatic temperature fluctuations, and avoid using tracers left over from the previous injection vial. Switch the experimental animal to a susceptible species/strain if possible.
Problem 7
For H129-G4 tracing, no or few labeled downstream neurons with normal labeling at the injection site.
Possible reason: Too short a time prior to perfusion, poor virus replication efficiency or transmission efficiency, poor projection from the injection site.
Potential solution: Wait longer from injection to perfusion. Check the H129-G4 tracer titer and use the growth curve to confirm it replicates normally. To map new brain regions, set up positive controls by injecting H129-G4 into a region known to trace well. For a susceptible animal species/strain, use animals confirmed to work well.
Problem 8
For H129-dTK tracers, no or few AAV-labeled neurons at the injection site.
Possible reason: Insufficient time since AAV injection, improper AAV, incorrect animal strain or mouse line, not enough Cre transgenic neurons at the injection site.
Potential solution: Wait longer to allow the helper AAV to express the target genes, fluorescent protein, and TK expression, before H129-dTK-tdT or H129-T2 injection and perfusion. Try different serotypes of helper AAVs with a more suitable promoter driving the fluorescence marker and TK expression, and determine the one with best expression efficiency. Make sure the Cre-transgenic line is used for the AAV-DIO-TK-GFP injection. Examine the numbers of Cre-expressing neurons by histological or immunostaining assays.
Problem 9
No or few neurons labeled by H129-dTK tracers at the injection site, with normal AAV labeling.
Possible reason: Insufficient fluorescent protein and TK expression from the AAV helper, non-overlapped injection sites of AAV and H129-dTK tracer, injection of an insufficient amount of H129-dTK tracer, degraded or inactive H129-dTK tracers.
Potential solution: Confirm sufficient expression of the fluorescent protein and TK expression by immunostaining, which is essential to support H129-dTK replication. When necessary, extend the intervals between the AAV and H129-dTK injections, or use an AAV with a stronger or more suitable promoter for fluorescent protein and TK expression. Check the titer and carefully calculate it for the H129-dTK tracer right before injection, and increase the dose of H129-dTK tracers if necessary. Properly store the virus and avoid repeated freezing/thawing of H129-dTK tracer as described above. H129 (enveloped virus) is much more sensitive than AAV (non-enveloped virus), and the H129 titer drops greatly with dramatic temperature fluctuations and freezing/thawing.
Problem 10
Weak labeling intensity of the neurons labeled by H129-dTK tracers.
Possible reason: Insufficient time for the florescent protein to accumulate, low fluorescent protein level due to poor replication of H129-dTK, non-optimal immunostaining, photobleaching of the fluorescent protein.
Potential solution: Wait longer after H129-dTK injection prior to perfusion to allow H129-dTK to express the fluorescent protein. Amplify the fluorescence signal by immunostaining with antibody against tdTomato expressed by H129-dTK. Adjust the concentration and amount of primary and secondary antibodies for better tdTomato signal amplification. Put the slides in a box to avoid exposure to strong light, optimize the exposure time during imaging
Problem 11
No or few post-synaptic neurons are labeled, while the injection site is well labeled by AAV and H129-dTK.
Possible reason: Insufficient time for H129-dTK transmission to the post-synaptic (2nd order) neurons, fluorescent signal hardly visible, low transmission efficiency, poor projection from the injection site.
Potential solution: Wait longer after H129-dTK injection prior to perfusion to allow H129-dTK transmission to the post-synaptic neurons. Amplify the tdTomato signal by immunostaining as described above. A positive control helps to judge the results, as described above.
Problem 12
Nearby regions are labeled by H129-dTK, but not the known downstream regions far from the injection site.
Possible reason: Distance from the injection site.
Potential solution: Virus transmission is associated with the transmission speed and distance. It takes less time for tracers to transmit to close regions than distant ones. Wait longer to let the H129-dTK tracer transmit further and express abundant fluorescent proteins.
Problem 13
No or few AAV and H129-dTK double-labeled neurons at the injection site, but well-labeled downstream neurons.
Possible reason: Co-infected neurons at the injection site are dead due to cytotoxicity from H129-dTK replication upon AAV support.
Potential solution: Shorten the time between H129-dTK injection and perfusion. However, shortening the time may lead to less transmission of H129-dTK to the post-synaptic regions and lower labeling efficiency of post-synaptic neurons. Thus, optimization of the time point for examination helps to solve this problem. Alternatively, examine the injection site and innervation target at two different time points from two different animal groups, especially if there is no perfect timing at which to observe both sites.
Problem 14
Unexpected labeling at regions upstream of the injection site.
Possible reason: Nonspecific retrograde labeling caused by the potential axonal terminal invasion of H129, unknown projection to the regions considered to be “upstream”.
Potential solution: Always include a proper control, such as Cholera Toxin B (CTB) or RetroAAV, to rule out potential nonspecific retrograde labeling by H129 terminal invasion. Apply other methods, such as electrophysiology or retrograde tracers, to confirm the potential unknown projection.
Problem 15
For H129-dTK tracers, how can it be confirmed that the labeled neurons or regions after transmission are really direct projection targets?
Potential solution: Always include negative controls by injecting H129-dTK alone without helper AAVs, which rules out the potential leaked transmission of H129-dTK. Though there is no evidence showing H129 is transmitted exclusively through synapses, new evidence of axonal transport of H129 to axonal terminals strongly supports trans-synaptic transmission [19].
Limitation of Current H129-Derived Tracers
Several polysynaptic and monosynaptic tracers derived from various viruses have been developed in the past decades (Table 7) [8, 15, 16, 35]. They all have unique properties and have contributed to mapping output neuronal circuits. Compared to other polysynaptic tracers, H129-G4 displays the highest intensity so far, and it has been used in rats and tree shrews as well as mice [16]. AAV2/1 and AAV2/9 have been used as monosynaptic tracers [35], but their transneuronal transmission efficiency and labeling intensity are so low that they have to be used together with a reporter system. Besides, AAV monosynaptic tracers cannot initiate tracing from a defined neuron population, such as Cre-expressing neurons, limited by their transmission mechanism [36]. The failure of starter cell specificity greatly limits their application. Compared to AAV, H129-dTK represents more promising monosynaptic tracers so far. However, despite these advantages and applications, H129-G4 and H129-dTK have certain drawbacks to be improved or optimized.
Table 7.
Currently-available anterograde trans-neuronal viral tracers
| Anterograde tracers | Virus | Properties | Limitations |
|---|---|---|---|
| Polysynaptic | |||
| H129-G4[16] | HSV-1 (H129) |
Brightest anterograde tracer so far Compatible with fMOST Alternative tracer with red fluorescence Can be used in tree shrew and rat |
Potential retrograde labeling caused by axonal terminal pickup High toxicity No starter cell-specificity |
| H129dTK-TT [15] | HSV-1 |
The first H129-derived anterograde tracer with fluorescence labeling Polysynaptic tracing from Cre+ neuron population |
High toxicity Relatively low labeling intensity Cannot trace from naïve neuron population without Cre |
| H129-EGFP [8] | HSV-1 (H129) |
First H129-derived anterograde tracer with EGFP labeling First H129-derived anterograde tracer initiating polysynaptic tracing from naïve neuron populations |
High toxicity Relatively low labeling intensity |
| Monosynaptic | |||
|
AAV2/1 [35] AAV2/9 [35] |
AAV |
Easy access Very low toxicity Compatible with functional assays |
No starter cell specificity Very low transneuronal transmission efficiency Very low labeling intensity, requiring a reporter system |
| H129-dTK [16] | HSV-1 (H129) |
First anterograde monosynaptic viral tracer Suitable for both starter neuron specific and nonspecific tracing Having an alternative tracer with green fluorescence |
Relatively low labeling intensity in post-synaptic neurons Relatively high cytotoxicity in starter neurons |
As described above, strong toxicity is the main intrinsic disadvantage of all current H129-derived transneuronal tracers [8, 15, 16, 37]. H129-G4 causes cell death within 24 h post-infection (MOI = 1) on primary neurons in vitro, and the normal tracing dose of H129-G4 kills the injected mouse or rat within 5–7 days, depending on different doses and the injected brain regions. TK deficiency of H129-dTK-tdT and H129-dTK-T2 impairs virus replication, which results in much weaker cytotoxicity in the post-synaptic neurons. But severe cell damage and neuronal death still occur at the injection site upon the support of AAV-TK-GFP. Therefore, current H129 tracers are not suitable for in vivo electrophysiology, optogenetics assays, and Ca2+ imaging for functional circuit mapping. This highlights that cytotoxicity limits the application of the tracers.
Potential retrograde labeling is another concern in the application of H129-derived tracers. Although H129 mainly enters neurons from the soma and transmits anterogradely [19], it is known to invade neurons from axon terminals at low incidence, which can potentially lead to retrograde labeling [5, 9, 10, 15, 38]. Therefore, when using H129-G4 or H129-dTK to identify novel innervation targets, it is strongly recommended to perform appropriate controls to label the upstream regions, using CTB or AAV-Retro.
The major limitation of H129-dTK monosynaptic tracers is the weak labeling intensity of the downstream post-synaptic neurons, which is a result of TK-deficiency associated with the impairment of H129 replication. Although H129-dTK-T2 improves the labeling intensity, it is still not strong enough to easily visualize long-distance innervated regions. Some other factors, such as non-optimal experimental parameters or few Cre-expressing neurons in the injection site, may dampen the labeling effect of the H129-dTK tracer.
Acknowledgements
We appreciate Prof. Bo Li (Cold Spring Harbor Laboratory, USA), Xiaohui Zhang (Beijing Normal University, China), Xiao-Min Wang (Capital Medical University, China), Jian-Zhi Wang (Huazhong University of Science and Technology, China), Zhi Zhang (University of Science and Technology of China, China), and Ping Zheng (Fudan University, China) for providing the tracing parameters and results. We would like to thank the Animal Experiment Center and Core Facilities and Analytical Center of the Wuhan Institute of Virology for their support in animal experiments and imaging. This work was supported by the National Key Basic Research Program of China (2015CB755601), the National Natural Science Foundation of China (81427801, 81571355, and 81601206), and the National Institutes of Health RF1 Funding of China (RF1MH120020-01).
Abbreviations
- HSV-1
Herpes simplex virus 1
- H129
Herpes simplex virus 1 (HSV-1) strain H129
- tdT
tdTomato
- fMOST
Fluorescence Micro-Optical Sectioning Tomography
- TK
Thymidine kinase
- AAV
Adeno-associated virus
- CTB
Cholera Toxin B
- MOI
Multiplicity of infection
- CPE
Cytopathic effects
- FBS
Fetal bovine serum
- PBS
Phosphate-buffered saline
- Pen–strep
Penicillin–streptomycin
- PFA
Paraformaldehyde
- OB
Olfactory bulb
- M1
Primary motor cortex
- Pir
Piriform cortex
- LEnt
Lateral entorhinal cortex
- Ent
Entorhinal cortex
- CA1
Field CA1 of the hippocampus
- BLA
Basolateral amygdala
Conflict of interest
All authors claim that there are no conflicts of interest. The Wuhan Institute of Virology has filed a patent application for H129-G4 (US Patent No. 201615747742), and a patent application for H129-dTK-tdT and H129-dTK-T2 is still pending. The authors declare no other competing interests.
Ethics Approval and Consent to Participate
The standards of performance and animal studies were approved by the Institutional Review Board and Institutional Animal Welfare Committee (WIVA10201502), including intracerebral inoculation of mice and rats with viral tracers. All the experiments with viruses were performed in bio-safety level 2 laboratory and animal facilities.
Contributor Information
Wen-Bo Zeng, Email: zengwb@wh.iov.cn.
Fei Zhao, Email: zhaofei@cibr.ac.cn.
Min-Hua Luo, Email: luomh@wh.iov.cn.
References
- 1.Nassi JJ, Cepko CL, Born RT, Beier KT. Neuroanatomy goes viral! Front Neuroanat. 2015;9:80. doi: 10.3389/fnana.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schultz BR, Chamberlain JS. Recombinant adeno-associated virus transduction and integration. Mol Ther. 2008;16:1189–1199. doi: 10.1038/mt.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly RM, Strick PL. Rabies as a transneuronal tracer of circuits in the central nervous system. J Neurosci Methods. 2000;103:63–71. doi: 10.1016/S0165-0270(00)00296-X. [DOI] [PubMed] [Google Scholar]
- 4.Wickersham IR, Sullivan HA, Seung HS. Axonal and subcellular labelling using modified rabies viral vectors. Nat Commun. 2013;4:2332. doi: 10.1038/ncomms3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett EM, Evans GD, Sun N, Perlman S, Cassell MD. Anterograde tracing of trigeminal afferent pathways from the murine tooth pulp to cortex using herpes simplex virus type 1. J Neurosci. 1995;15:2972–2984. doi: 10.1523/JNEUROSCI.15-04-02972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garner JA, LaVail JH. Differential anterograde transport of HSV type 1 viral strains in the murine optic pathway. J Neurovirol. 1999;5:140–150. doi: 10.3109/13550289909021996. [DOI] [PubMed] [Google Scholar]
- 7.McGovern AE, Davis-Poynter N, Farrell MJ, Mazzone SB. Transneuronal tracing of airways-related sensory circuitry using herpes simplex virus 1, strain H129. Neuroscience. 2012;207:148–166. doi: 10.1016/j.neuroscience.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 8.McGovern AE, Davis-Poynter N, Rakoczy J, Phipps S, Simmons DG, Mazzone SB. Anterograde neuronal circuit tracing using a genetically modified herpes simplex virus expressing EGFP. J Neurosci Methods. 2012;209:158–167. doi: 10.1016/j.jneumeth.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 9.Rinaman L, Schwartz G. Anterograde transneuronal viral tracing of central viscerosensory pathways in rats. J Neurosci. 2004;24:2782–2786. doi: 10.1523/JNEUROSCI.5329-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song CK, Schwartz GJ, Bartness TJ. Anterograde transneuronal viral tract tracing reveals central sensory circuits from white adipose tissue. Am J Physiol Regul Integr Comp Physiol. 2009;296:R501–511. doi: 10.1152/ajpregu.90786.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaughan CH, Bartness TJ. Anterograde transneuronal viral tract tracing reveals central sensory circuits from brown fat and sensory denervation alters its thermogenic responses. Am J Physiol Regul Integr Comp Physiol. 2012;302:R1049–1058. doi: 10.1152/ajpregu.00640.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun N, Cassell MD, Perlman S. Anterograde, transneuronal transport of herpes simplex virus type 1 strain H129 in the murine visual system. J Virol. 1996;70:5405–5413. doi: 10.1128/JVI.70.8.5405-5413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dum RP, Levinthal DJ, Strick PL. The spinothalamic system targets motor and sensory areas in the cerebral cortex of monkeys. J Neurosci. 2009;29:14223–14235. doi: 10.1523/JNEUROSCI.3398-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly RM, Strick PL. Cerebellar loops with motor cortex and prefrontal cortex of a nonhuman primate. J Neurosci. 2003;23:8432–8444. doi: 10.1523/JNEUROSCI.23-23-08432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo L, Anderson DJ. A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng WB, Jiang HF, Gang YD, Song YG, Shen ZZ, Yang H, et al. Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Mol Neurodegener. 2017;12:38. doi: 10.1186/s13024-017-0179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etessami R, Conzelmann KK, Fadai-Ghotbi B, Natelson B, Tsiang H, Ceccaldi PE. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J Gen Virol. 2000;81:2147–2153. doi: 10.1099/0022-1317-81-9-2147. [DOI] [PubMed] [Google Scholar]
- 18.Chen YH, Keiser MS, Davidson BL. Adeno-associated virus production, purification, and titering. Curr Protoc Mouse Biol. 2018;8:e56. doi: 10.1002/cpmo.56. [DOI] [PubMed] [Google Scholar]
- 19.Dong X, Zhou J, Qin HB, Xin B, Huang ZL, Li YY, et al. Anterograde viral tracer herpes simplex virus 1 strain H129 transports primarily as capsids in cortical neuron axons. J Virol 2020, 94. [DOI] [PMC free article] [PubMed]
- 20.Tang H, Wu GS, Xie J, He X, Deng K, Wang H, et al. Brain-wide map of projections from mice ventral subiculum. Neurosci Lett. 2016;629:171–179. doi: 10.1016/j.neulet.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Deng K, Yang L, Xie J, Tang H, Wu GS, Luo HR. Whole-brain mapping of projection from mouse lateral septal nucleus. Biol Open 2019, 8. [DOI] [PMC free article] [PubMed]
- 22.Pi G, Gao D, Wu D, Wang Y, Lei H, Zeng W, et al. Posterior basolateral amygdala to ventral hippocampal CA1 drives approach behaviour to exert an anxiolytic effect. Nat Commun. 2020;11:183. doi: 10.1038/s41467-019-13919-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu X, Zhou W, Jin Y, Tang H, Cao P, Mao Y, et al. A central amygdala input to the parafascicular nucleus controls comorbid pain in depression. Cell Rep. 2019;29(3847–3858):e3845. doi: 10.1016/j.celrep.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang YY, Wang Y, Jiang HF, Liu JH, Jia J, Wang K, et al. Impaired glutamatergic projection from the motor cortex to the subthalamic nucleus in 6-hydroxydopamine-lesioned hemi-parkinsonian rats. Exp Neurol. 2018;300:135–148. doi: 10.1016/j.expneurol.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Blaho JA, Morton ER, Yedowitz JC. Herpes simplex virus: propagation, quantification, and storage. Curr Protoc Microbiol 2005, Chapter 14: Unit 14E 11. [DOI] [PubMed]
- 26.Services USDH . Control CfD, Prevention, Health Do, Services H. Createspace Independent Pub: Health NIo. Biosafety in Microbiological and Biomedical Laboratories; 2013. [Google Scholar]
- 27.Yu K, Ahrens S, Zhang X, Schiff H, Ramakrishnan C, Fenno L, et al. The central amygdala controls learning in the lateral amygdala. Nat Neurosci. 2017;20:1680–1685. doi: 10.1038/s41593-017-0009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Xu J, Liu Y, Zhu J, Liu N, Zeng W, et al. A distinct entorhinal cortex to hippocampal CA1 direct circuit for olfactory associative learning. Nat Neurosci. 2017;20:559–570. doi: 10.1038/nn.4517. [DOI] [PubMed] [Google Scholar]
- 29.Jin Y, Meng Q, Mei L, Zhou W, Zhu X, Mao Y, et al. A somatosensory cortex input to the caudal dorsolateral striatum controls comorbid anxiety in persistent pain. Pain. 2020;161:416–428. doi: 10.1097/j.pain.0000000000001724. [DOI] [PubMed] [Google Scholar]
- 30.Keiser MS, Chen YH, Davidson BL. Techniques for intracranial stereotaxic injections of adeno-associated viral vectors in adult mice. Curr Protoc Mouse Biol. 2018;8:e57. doi: 10.1002/cpmo.57. [DOI] [PubMed] [Google Scholar]
- 31.Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. Elsevier Academic Press, 2004.
- 32.Gage GJ, Kipke DR, Shain W. Whole animal perfusion fixation for rodents. J Vis Exp. 2012;65:3564. doi: 10.3791/3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Zhu Q, Ding L, Shen Y, Yang CY, Xu F, et al. Scalable volumetric imaging for ultrahigh-speed brain mapping at synaptic resolution. Nat Sci Rev. 2019;6:982–992. doi: 10.1093/nsr/nwz053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Q, Fu Y, Cao Z, Shao D, Song J, Sheng H, et al. A conditioning-strengthened circuit from CA1 of dorsal hippocampus to basolateral amygdala participates in morphine-withdrawal memory retrieval. Front Neurosci 2020, 14. [DOI] [PMC free article] [PubMed]
- 35.Zingg B, Chou XL, Zhang ZG, Mesik L, Liang F, Tao HW, et al. AAV-mediated anterograde transsynaptic tagging: mapping corticocollicular input-defined neural pathways for defense behaviors. Neuron. 2017;93:33–47. doi: 10.1016/j.neuron.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao F, Jiang HF, Zeng WB, Shu Y, Luo MH, Duan S. Anterograde trans-synaptic tagging mediated by adeno-associated virus. Neurosci Bull. 2017;33:348–350. doi: 10.1007/s12264-017-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su P, Wang H, Xia J, Zhong X, Hu L, Li Y, et al. Evaluation of retrograde labeling profiles of HSV1 H129 anterograde tracer. J Chem Neuroanat. 2019;100:101662. doi: 10.1016/j.jchemneu.2019.101662. [DOI] [PubMed] [Google Scholar]
- 38.Wojaczynski GJ, Engel EA, Steren KE, Enquist LW, Patrick Card J. The neuroinvasive profiles of H129 (herpes simplex virus type 1) recombinants with putative anterograde-only transneuronal spread properties. Brain Struct Funct. 2015;220:1395–1420. doi: 10.1007/s00429-014-0733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]