Abstract
Purpose
MicroRNAs (miRNAs) are small non-coding RNAs and are involved in the development, proliferation, and pathogenesis of prostate cancer (PCa). Urinary miRNAs are promising non-invasive biomarkers for PCa diagnosis because of their stability in urine. Here, we evaluated the diagnostic value of urinary miR-1913 to miR-3659 ratio in PCa patients and benign prostate hyperplasia (BPH) controls.
Materials and Methods
Candidate miRNAs were identified from urinary microarray data and tested by real-time PCR. The urinary miR-1913 to miR-3659 expression ratio was selected and tested in 83 urine samples (44 PCa and 39 BPH) to confirm its validity as a non-invasive diagnostic biomarker for PCa.
Results
The expression ratio of urinary miR-1913 to miR-3659 was significantly higher in PCa than in BPH (p=0.002) and showed a higher area under the receiver operating characteristic curve than prostate-specific antigen (PSA; 0.821 vs. 0.518) in patients within the PSA gray zone (tPSA: 3–10 ng/mL), with sensitivity of 75.0% and specificity of 78.6% (p=0.003).
Conclusions
The urinary miR-1913 to miR-3659 expression ratio was increased in PCa and may serve as a useful supplemental biomarker to PSA for the diagnosis of PCa, particularly in patients within the PSA gray zone.
Keywords: Biomarkers, Diagnosis, Gene expression, MicroRNA, Prostate neoplasms
INTRODUCTION
Prostate cancer (PCa) is a common cancer with high mortality among men worldwide, and its incidence is rising rapidly [1]. Serum prostate-specific antigen (PSA) is currently used as the primary marker in preliminary screening for PCa. However, this serum marker lacks specificity and is limited in its ability to distinguish tumor from non-cancerous tissue, which can lead to over-diagnosis and a high risk of false-positive results [2]. Numerous studies have aimed to identify PCa biomarkers to overcome the limitations of PSA [3]; however, most tests show poor reproducibility, or their protocols are too complex for clinical application. Thus, identifying a novel PCa biomarker that can be tested in the clinic with accurate reproducibility is important.
MicroRNAs (miRNAs) are small (18–24 nt) non-protein-coding RNAs that post-transcriptionally regulate gene expression. miRNAs are involved in numerous biological functions including tumor cell proliferation and apoptosis [4]. Studies assessing the value of miRNAs as biomarkers were conducted using surgery tissue samples [5,6]. However, biomarker identification from tissues is not ideal because it involves an invasive procedure [7]. Moreover, solid tumor tissues are extremely heterogeneous, which limits their reliability for biomarker studies [8].
Recent studies suggest that analysis of miRNAs in biofluids such as urine is an attractive method with potential clinical application. Urine is more likely to represent the genomic landscape of the whole tumor than local tumor samples, and it is an easily approachable source that does not require invasive procedures [9]. Furthermore, the exosome protects miRNAs from damage in the microenvironment, such as RNA degradation, which increases the potential of urinary miRNAs as tumor biomarkers [10]. An increasing number of urinary cell-free miRNA studies suggest that analysis of this liquid biospecimen is a promising non-invasive approach to PCa diagnosis [11]. Particularly considering the limitations of solid tumor tissues, urinary miRNAs could serve as reliable biomarkers in cancer.
The aim of this study was to identify novel non-invasive diagnostic biomarkers specific to PCa. Urine miRNA microarray profiling was performed to identify candidate biomarker genes in the urine of PCa patients compared with that of benign prostate hyperplasia (BPH). The clinical performance of candidate miRNAs was evaluated using ratio analysis (ratio of up- to down- or similarly expressed gene), which improves the accuracy of real-time PCR (RT-PCR) data.
MATERIALS AND METHODS
1. Overview
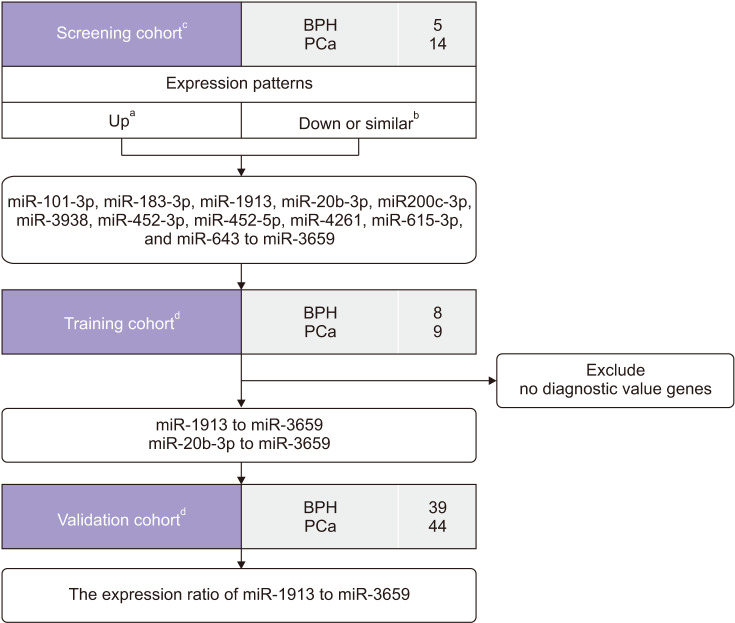
A workflow and overall study design are shown in Fig. 1. Urine samples were provided by Chungbuk National University, a member of the National Biobank of Korea, which is supported by the Ministry of Health, Welfare and Family Affairs. All urine samples used in the study were obtained from the National Biobank of Korea between January 2016 and August 2018 (approval number: 2010-12-010-006). miRNA microarray analysis was performed to identify PCa-specific miRNAs. A flexible and adaptive test based on p-value comparison, fold-change, and expression stability was used. Twelve candidate miRNAs (11 miRNAs up-regulated in PCa urine: miR-1913, miR-20b-3p, miR-3938, miR-101-3p, miR-615-3p, miR-452-5p, miR-452-3p, miR-200c-3p, miR-643, miR-4261, and miR-183-3p; and one miRNA which showed similar expression in PCa urine: miR-3659) were identified by RT-PCR. For ratio analysis (up- to down-regulated ratio), the expression levels of up-regulated genes were compared with that of miR-3659 as; the down-regulated miRNA in PCa, instead of using internal controls. The diagnostic power of 11 miRNA ratios were examined in the training cohort (data not shown), and two ratios, miR-1913:miR-20b-3p and miR-1913:miR-3659, were selected as potential PCa-specific biomarkers. The two miRNA ratios selected were evaluated in the validation cohort.
Fig. 1. Overview of the study. Twelve candidate miRNAs were identified in the screening cohort (11 up-regulated miRNAs and one down-regulated miRNA in PCa). For ratio analysis, the expression levels of 11 up-regulated miRNAs were compared with that of miR-3659 as the down-regulated miRNA. The diagnostic validity of 11 miRNA ratios was evaluated in the training cohort, and two miRNA ratios (miR-1913 to miR-3659 and miR-20b-3p to miR-3659) were selected for further analysis. In the validation cohort, only the expression ratio of urinary miR-1913 to miR-3659 showed a significant difference between PCa patients and BPH controls. BPH, benign prostate hyperplasia; PCa, prostate cancer; RT-PCR, real-time polymerase chain reaction. a:Candidate showed higher expression in PCa. b:Candidate showed lower expression in PCa or similar expression in PCa and BPH. c:miRNA microarray-based cohort. d:RT-PCRbased cohorts.
2. Cases
Table 1 and Fig. 1 summarizes the characteristics of the current study subjects. The microarray data were based on urine samples from 14 PCa patients and five BPH patients. The training set included 17 urine samples from nine PCa patients and eight BPH patients. The validation cohort consisted of 83 urine samples from 44 PCa patients and 39 BPH patients. All urine samples used in the study were collected in the morning and centrifuged at 2,500 rpm for 15 minutes; aliquoted supernatants were stored at −20℃ until use. PCa samples were obtained from patients with histologically confirmed primary adenocarcinoma who underwent radical prostatectomy or palliative transurethral resection of the prostate (TURP). Patients with BPH who underwent TURP were selected as controls. Gleason scores were assigned to specimens obtained from 12-core transrectal biopsies, TURP, or radical prostatectomy. Tumor stage was estimated from specimens obtained by radical prostatectomy, or from computed tomography, magnetic resonance imaging, or bone scans. The collection and analysis of all biospecimens were approved by the Institutional Review Board of Chungbuk National University Hospital, and written informed consent was obtained from each subject (approval number: 2010-12-010-006). The study methodologies conformed with the standards set by the Declaration of Helsinki.
Table 1. Clinical characteristics of the study subjects.
| Variable | Training cohort | Validation cohort | ||||
|---|---|---|---|---|---|---|
| BPH | PCa | p-value* | BPH | PCa | p-value* | |
| No. of patients | 8 | 9 | 39 | 44 | ||
| Age (y) | 69.63±2.08 | 61.00±3.16 | 0.066 | 71.41±8.44 | 68.57±5.31 | 0.051 |
| PSA (ng/mL) | 2.72±1.60 | 10.79±7.59 | 0.054 | 1.92±3.53 | 10.76±95.33 | <0.001 |
| Operation | ||||||
| TURP | 8 (100.0) | 39 (100.0) | ||||
| Radical prostatectomy | 9 (100.0) | 44 (100.0) | ||||
| Gleason score | ||||||
| 6 | 1 (2.0) | |||||
| 7 (3+4) | 6 (67.0) | 29 (66.0) | ||||
| 7 (4+3) | 3 (33.0) | 10 (23.0) | ||||
| ≥8 | 4 (9.0) | |||||
| TNM stage | ||||||
| T2 | 2 (22.0) | 24 (55.0) | ||||
| T3 | 6 (67.0) | 15 (34.0) | ||||
| T4 or metastasis | 1 (11.0) | 5 (11.0) | ||||
Values are presented as number only, mean±standard deviation, or number (%).
BPH, benign prostate hyperplasia; PCa, prostate cancer; PSA, prostate-specific antigen; TURP, transurethral resection of the prostate.
*All p-values were obtained by the Mann–Whitney U-test for comparisons between urine from BPH patients and PCa patients in the study.
3. Microarrays
Total RNA was extracted from each sample using the miRNA Microarray System labeling kit (Illumina, San Diego, CA, USA), and RNA quantity and integrity were assessed using the RNA 6000 Pico Chip Kit (Agilent Technologies, Santa Clara, CA, USA) and the Agilent 2100 Bioanalyzer. miRNA profiling was performed using the Agilent Human miRNA Microarray Release 16.0 platform, which contains 1,205 human and 144 viral miRNAs [12]. The protocol used to generate microarray gene expression datasets was reported previously [13].
4. miRNA purification from urine
A Genolution urine miRNA purification kit (Genolution Pharmaceuticals Inc., Seoul, Korea) was used to purify the urine samples. A volume of 500 µL supernatant from each urine sample was added to a tube containing the Genolution proprietary miRNA separation solution and vortexed for 20 seconds. Next, 200 µL chloroform was added and vortexed for 10 seconds, followed by centrifugation at 13,000 rpm for 10 minutes at 4℃. A sample of 650 µL of the top aqueous phase was removed without disturbing the white precipitate and was transferred into a new 1.5 mL tube, to which 0.8 mL isopropanol was added, followed by centrifugation for 20 minutes at 15,000 rpm at 4℃. The solution was decanted by tilting the tube in the opposite direction of the expected RNA pellet, and 500 µL of 70% EtOH was added, followed by centrifugation for 20 minutes at 15,000 rpm at 4℃. After removing the leftover ethanol, the pellet was dissolved in 40 µL RNase-free water and stored at −80℃ until use.
5. cDNA synthesis from urinary miRNAs
The concentration of isolated RNA was measured using the Quant-IT RiboGreen RNA Reagent and Kit (Invitrogen, Grand Island, NY, USA). The cDNA was synthesized using the Mir-X™ miRNA First Strand cDNA Synthesis Kit (TAKARA BIO, Otsu, Japan) according to the manufacturer's protocol.
6. Real-time PCR
The expression of miRNAs derived from the miRNA microarray (GSE54010) was quantified by RT-PCR using the Rotor-Gene Q instrument (Qiagen, Valencia, CA, USA) and SYBR Premix EX Taq (TAKARA BIO) in micro-reaction tubes (Corbett Research, Mortlake, Australia) in a final volume of 10 µL. Chemically synthesized RNA oligonucleotides (Integrated DNA Technologies [IDT], Seoul, Korea) corresponding to the target miRNAs were used to generate standard curves. The standard curves ranged from 2.25×105 to 2.25×108 copies. All samples were run in triplicate, and RT-PCR conditions were those described in the manufacturer's protocol. Rotor-Gene Q software 2.3.1.49 was used for capturing and analyzing spectral data.
7. Statistical analysis
Receiver operating characteristic (ROC) curves were used to estimate the optimal cutoff point yielding high sensitivity and specificity for the miR-1913 to miR-3659 ratio in both training and validation cohorts. Differences in the hsv2-miR-H9 to hsa-miR-3659 ratio were identified using the Mann–Whitney U-test in each cohort. Statistical analysis was performed using IBM SPSS 21.0 (IBM, Armonk, NY, USA). Results with p-values <0.05 were considered significant.
RESULTS
1. Screening cohort: miRNA microarray
PCa-specific miRNAs in urine specimens were identified using a miRNA microarray assay as described in the Materials and Methods section. In the microarray-based screening cohort, we identified 12 miRNAs with potential to distinguish PCa from controls, including 11 up-regulated miRNAs (miR-101-3p, miR-183-3p, miR-1913, miR-20b-3p, miR-200c-3p, miR-3938, miR-452-3p, miR-452-5p, miR-4261, miR-615-3p, miR-643) and one miRNA (miR-3659) which showed similar expression in PCa and BPH controls (Table 2).
Table 2. The microarray data of 12 miRNA candidates.
| Expression | Gene | FC of log | BPH/Raw | PCa/Raw | p-value* |
|---|---|---|---|---|---|
| Down or similar | hsa-miR-3659 | 1.61 | 788.399 | 6,893.83 | 0.737 |
| Up | hsa-miR-101-3p | 7.98 | -0.83044 | 8.379992 | 0.0545 |
| hsa-miR-183-3p | 9.04 | -1.66955 | 7.628885 | 0.0000395 | |
| hsa-miR-1913 | 8.70 | -2.5046 | 9.96242 | 0.00104 | |
| hsa-miR-20b-3p | 5.95 | -2.46165 | 1.318423 | 0.0458 | |
| hsa-miR-200c-3p | 8.19 | -1.86396 | 11.526 | 0.0065 | |
| hsa-miR-3938 | 7.93 | -1.42385 | 10.66832 | 0.058 | |
| hsa-miR-452-3p | 8.10 | -2.29163 | 17.37931 | 0.00867 | |
| hsa-miR-452-5p | 8.01 | -2.01941 | 5.322599 | 0.000595 | |
| hsa-miR-4261 | 9.02 | -1.85743 | 10.23327 | 0.000595 | |
| hsa-miR-615-3p | 10.88 | -1.67958 | 48.16512 | 0.000595 | |
| hsa-miR-643 | 6.09 | -2.45995 | 2.827512 | 0.252 |
BPH, benign prostate hyperplasia; PCa, prostate cancer.
*All p-values were obtained by the Mann–Whitney U-test for comparisons between urine from BPH patients and PCa patients.
2. Training cohort: Diagnostic ability of selected miRNAs
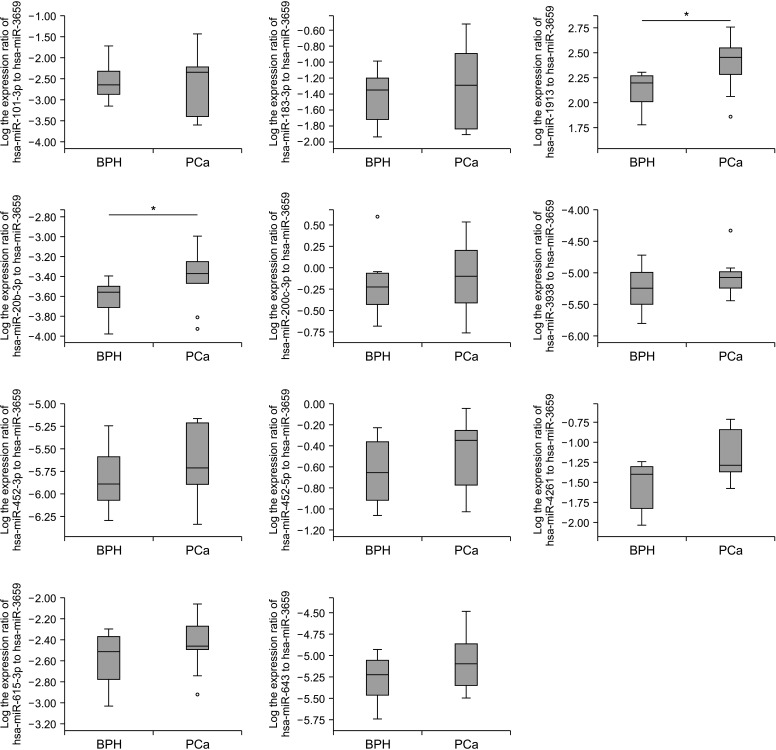
Seventeen urine samples (eight BPH and nine PCa) in the training cohort were used to determine the diagnostic ability of the candidate miRNAs. We used ratio analysis (measurement of the expression ratio of up- to down-regulated genes) to improve the accuracy of the data and avoid the instability of internal controls. The ratios of the expression of 11 up-regulated miRNAs to that of miR-3659 were evaluated in PCa and BPH urine. The results showed that the miR-1913 to miR-3659 ratio and the miR-20b-3p to miR-3659 ratio differed significantly between PCa patients and controls (both p=0.043; Fig. 2).
Fig. 2. The expression ratio of 11 urinary miRNA ratios (start left top: miR-101-3p, miR-183-3p, miR-1913, miR-20b-3p, miR-200c-3p, miR-3938, miR-452-3p, miR-452-5p, miR-4261, miR-615-3p, miR-643 to miR-3659) in the training cohort. Two urinary miRNA ratios; miR-1913 to miR-3659 and miR-20b-3p to miR-3659, showed significant differences between PCa patients and BPH controls. BPH, benign prostate hyperplasia; PCa, prostate cancer. A p-value was determined by the Mann–Whitney U-test. *p<0.05.
3. Validation cohort: miR-1913 to miR-3659 ratio
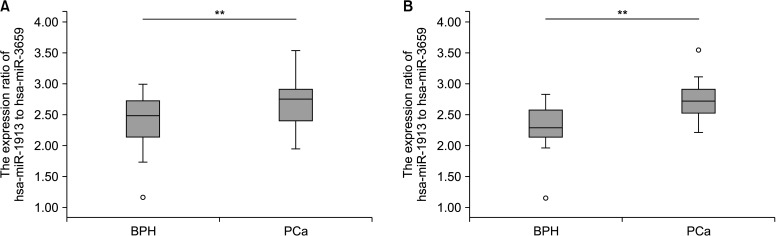
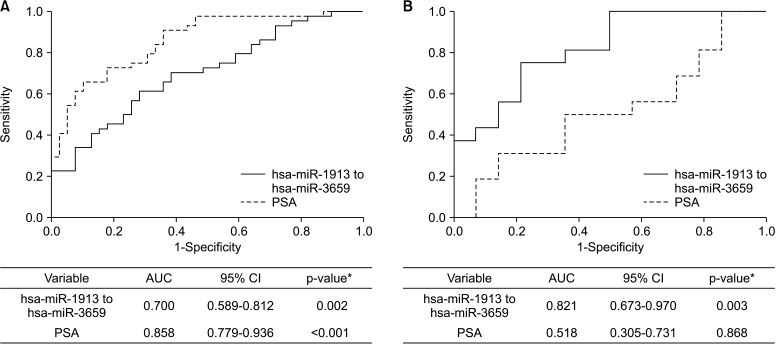
Two expression ratios (miR-1913 to miR-3659 and miR-20b-3p to miR-3659) were tested in the validation cohort. The results showed that urine miR-1913 to miR-3659 ratio differed significantly between PCa patients and BPH controls (p<0.01; Fig. 3A). In cases of PSA gray zone (tPSA, 3–10 ng/mL), the miR-1913 to miR-3659 expression ratio was remarkably higher in the urine of PCa patients than in that of controls (p<0.01; Fig. 3B). ROC curve analysis was performed to determine the sensitivity and specificity of urinary miR-1913 to miR-3659 ratio for detecting PCa. The area under the ROC curve (AUC) of urinary miR-1913 to miR-3659 ratio was 0.700 (95% confidence interval [CI]=0.589–0.812), with a sensitivity of 61.4% and a specificity of 71.8% (p=0.002), respectively. The sensitivity and specificity of PSA assay was 65.9% and 89.7% (AUC 0.858 [95% CI=0.779–0.936]), with a cutoff point 7.65 (p<0.001) (Fig. 4A). Nevertheless, when using the standard cutoff of PSA (>3 ng/mL) for PCa detection, the McNemar Test showed a negative correlation between PSA assay and pathological diagnosis (p=0.012), indicating the limitation of PSA in PCa diagnosis; although, no difference was explored between urinary miR-1913 to miR-3659 ratio and pathological diagnosis (p=0.345). Moreover, a statistical difference was identified when compared urinary miR-1913 to miR-3659 ratio to PSA (p=0.007) (Supplementary Table 1). For patients within the PSA gray zone, the AUC of the miR-1913 to miR-3659 ratio was considerably higher than that of the PSA assay (0.821, 95% CI=0.673–0.970), with sensitivity of 75.0% and specificity of 78.6% (p=0.003) (Fig. 4B). However, there was no significant relationship between the expression ratio of target miRNAs and pathological characteristics (Gleason score ≤7 vs. ≥8; and ≤T3 vs. T4 or metastasis; data not shown).
Fig. 3. Expression ratio of urinary miR-1913 to miR-3659 in the validation cohort. (A) All patients. (B) Patients within the PSA gray zone (3–10 ng/mL). BPH, benign prostate hyperplasia; PCa, prostate cancer; PSA, prostate-specific antigen. A p-value was determined by the Mann–Whitney U-test. **p<0.01.
Fig. 4. Receiver operating characteristic curve for discriminating PCa from BPH controls in the validation cohort. (A) All patients. (B) Patients within the PSA gray zone (3–10 ng/mL). AUC, area under the curve; BPH, benign prostate hyperplasia; CI, confidence interval; PCa, prostate cancer; PSA, prostate-specific antigen. *A p-value was determined by the Z-score.
DISCUSSION
The current study identified the miR-1913 to miR-3659 expression ratio as a promising non-invasive biomarker for PCa diagnosis. This biomarker may be useful as a complement to the serum PSA test, particularly in patients within the PSA gray zone (tPSA, 3–10 ng/mL).
PSA, the most commonly used serum marker for diagnosing PCa, lacks the required sensitivity and specificity to detect tumor tissues [14]. Because of the inherent limitations of the PSA test, a novel biomarker to replace or refine the diagnostic accuracy of the PSA test is urgently needed.
Prostate cancer antigen 3 (PCA3) mRNA, for which the encoding gene maps to chromosome 9q21-22, has been widely reported as a specific biomarker that is elevated in men with PCa compared with controls [15]. The ConfirmMDx, an epigenetic assay for determining the risk of cancerization at the DNA level was suggested as a useful method to identify high-risk patients for further testing or accurate clinical decision making [16]. The prostate health index test measures three markers using the formula [−2] proPSA/fPSA×PSA1/2 to determine the need for biopsy in cases within the PSA gray zone [17]. Decipher measures the expression levels of 22 RNAs involved in aggressive PCa [18]. These tests and others have been examined as potential biomarker assays for the detection of PCa [19]. However, few tests have been approved for clinical practice because most of the suggested markers show unsatisfactory reproducibility, and the optimal threshold cutoff values for these markers remain undefined [20,21]. Moreover, protocol to analyze most markers mentioned above, is too complex to utilize as clinical implementations and the invasive procedure is inevitably included as the origin sources of these markers are generally derived from tumor tissues or blood. The identification of a biomarker that can be analyzed using non-invasive and cost-effective methods has thus been challenging.
MiRNAs have been widely studied as potential biomarkers because of their roles as key regulators of many biological processes, including tumorigenesis, cell proliferation, and apoptosis [12,22]. miRNA expression profiling studies have been performed using solid tumor specimens [11]; however, the heterogeneity of tissues often leads to unreliable results [8]. Liquid biopsy (serum, plasma, or urine) specimens are considered ideal materials for analyzing the events associated with tumorigenesis, and tumors release miRNAs into biofluids directly or indirectly [23,24]. Among various sources of miRNAs, urine has become an attractive biospecimen for PCa biomarker studies [25,26]. Because the exosome encapsulates and protects miRNAs from most RNA-degrading agents, urinary cell-free miRNAs are often used in biomarker studies [10,27]. Furthermore, urine-based miRNA screening is non-invasive, and the ease of collection ensures the possibility of repeated analysis. These facts have increased the interest in urinary cell-free miRNAs as relevant potential molecules for biomarker studies. In the current study, we used RT-PCR to identify urinary miRNAs specific to PCa. This assay is the gold standard for the quantification of miRNA expression and is generally applicable to biomarker studies because of its simplicity and cost-effectiveness [28].
Most miRNA studies normalize RT-PCR results using U6; however, the instability of this internal control limits its use [29]. To overcome issues linked to data normalization and to improve the accuracy of the results, a new strategy is needed. Here, we used ratio analysis, which consists of measuring and comparing the expression ratios of up-regulated to down-regulated miRNAs in PCa and control samples. This method has been suggested as an attractive strategy for RT-PCR analysis as an alternative to the use of internal controls [30].
The present results showed that urinary miR-1913 to miR-3659 expression ratio was significantly higher in patients with PCa than in controls. ROC curve analysis showed that urinary miR-1913 to miR-3659 ratio could discriminate PCa patients from controls with appropriate sensitivity and specificity. Furthermore, the diagnostic performance of this expression ratio was superior to that of PSA assays, showing good sensitivity and specificity in cases within the PSA gray zone. There was no significant association between urinary miR-1913 to miR-3659 ratio and clinicopathological parameters of PCa (data not shown), which suggests that the urinary miRNA biomarker is limited for predicting the prognosis of PCa. However, to our knowledge, the roles of these two miRNAs; hsa-miR-1913 and hsa-miR-3659 in prostate have not been studied yet.
The present study had several limitations. First, the single center retrospective design of the study may have resulted in selection bias. Second, the small sample size could have reduced the statistical accuracy. Further collaborative studies are needed to overcome these limitations. Despite these shortcomings, the present results may be clinically important because the simplicity and cost-effectiveness of the method proposed overcome several challenges associated with existing techniques. Moreover, this urinary miRNA biomarker can be assessed using easy and non-invasive methods, and it is not influenced by unstable internal controls.
Collectively, we showed that urinary miR-1913 to miR-3659 ratio has high potential as a diagnostic biomarker for PCa, especially for patients within the PSA gray zone. This candidate biomarker may serve as a relevant non-invasive supplemental tool to the serum PSA assay for the diagnosis of PCa.
CONCLUSIONS
The present study shows that the expression ratio of urinary mir-1913 to mir-3659 could discriminate between PCa and BPH, especially for patients within the PSA gray zone of 3–10 ng/mL total PSA as valuable supplemental biomarker for diagnosing PCa.
ACKNOWLEDGMENTS
This research was supported by Osong Medical Innovation foundation funded by Chungcheongbuk-do (No.: AG200904005, AG200902001).
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
- Research conception and design: Young Joon Byun and Wun-Jae Kim.
- Data acquisition: Young Joon Byun, Xuan-Mei Piao, Pildu Jeong, Ho Won Kang, Sung Phil Seo, Hee Youn Lee, and Won Tae Kim.
- Statistical analysis: Young Joon Byun and Jong-Young Lee.
- Data analysis and interpretation: Young Joon Byun.
- Drafting of the manuscript: Young Joon Byun and Xuan-Mei Piao.
- Critical revision of the manuscript: Pildu Jeong, Ho Won Kang, Sang-Cheol Lee, Yung Hyun Choi, Sung-Kwon Moon, Won Tae Kim, Eun-Jong Cha, Seok Joong Yun, and Wun-Jae Kim.
- Administrative, technical, or material support: Wun-Jae Kim.
- Supervision: Seok Joong Yun and Wun-Jae Kim.
- Approval of the final manuscript: all authors.
SUPPLEMENTARY MATERIAL
Supplementary material can be found via https://doi.org/10.4111/icu.20200488.
Relationshipa between miRNA ratio and PSA
References
- 1.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hafron JM, Yu H, Juang A, Vuong D, Kamer S, Carbonell L, et al. New developments in prostate cancer screening using a novel cancer-specific, non-PSA biomarker assay derived from autoantibody signatures. J Med Ther. 2017 Oct 13; doi: 10.15761/JMT.1000119. [Epub] [DOI] [Google Scholar]
- 3.Liu DF, Wu JT, Wang JM, Liu QZ, Gao ZL, Liu YX. MicroRNA expression profile analysis reveals diagnostic biomarker for human prostate cancer. Asian Pac J Cancer Prev. 2012;13:3313–3317. doi: 10.7314/apjcp.2012.13.7.3313. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem. 2010;56:998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- 7.Hessels D, Schalken JA. Urinary biomarkers for prostate cancer: a review. Asian J Androl. 2013;15:333–339. doi: 10.1038/aja.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mino-Kenudson M. Cons: can liquid biopsy replace tissue biopsy?-the US experience. Transl Lung Cancer Res. 2016;5:424–427. doi: 10.21037/tlcr.2016.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endzeliņš E, Melne V, Kalniņa Z, Lietuvietis V, Riekstiņa U, Llorente A, et al. Diagnostic, prognostic and predictive value of cell-free miRNAs in prostate cancer: a systematic review. Mol Cancer. 2016;15:41. doi: 10.1186/s12943-016-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mlcochova H, Hezova R, Stanik M, Slaby O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol Oncol. 2014;32:41.e1–41.e9. doi: 10.1016/j.urolonc.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–652. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao Z, Ji A, Yang K, He W, Hu Y, Zhang Q, et al. Diagnostic performance of PCA3 and hK2 in combination with serum PSA for prostate cancer. Medicine (Baltimore) 2018;97:e12806. doi: 10.1097/MD.0000000000012806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ploussard G, de la Taille A. The role of prostate cancer antigen 3 (PCA3) in prostate cancer detection. Expert Rev Anticancer Ther. 2018;18:1013–1020. doi: 10.1080/14737140.2018.1502086. [DOI] [PubMed] [Google Scholar]
- 16.Sartori DA, Chan DW. Biomarkers in prostate cancer: what's new? Curr Opin Oncol. 2014;26:259–264. doi: 10.1097/CCO.0000000000000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini S. PSA and beyond: alternative prostate cancer biomarkers. Cell Oncol (Dordr) 2016;39:97–106. doi: 10.1007/s13402-016-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohammed AA. Biomarkers in prostate cancer: new era and prospective. Med Oncol. 2014;31:140. doi: 10.1007/s12032-014-0140-3. [DOI] [PubMed] [Google Scholar]
- 19.Ziaran S, Varchulova Novakova Z, Bohmer D, Danisovic L. Biomarkers for determination prostate cancer: implication for diagnosis and prognosis. Neoplasma. 2015;62:683–691. doi: 10.4149/neo_2015_082. [DOI] [PubMed] [Google Scholar]
- 20.Anceschi U, Tuderti G, Lugnani F, Biava PM, Malossini G, Luciani L, et al. Novel diagnostic biomarkers of prostate cancer: an update. Curr Med Chem. 2019;26:1045–1058. doi: 10.2174/0929867325666180914115416. [DOI] [PubMed] [Google Scholar]
- 21.Yun SJ, Jeong P, Kang HW, Kim YH, Kim EA, Yan C, et al. Urinary microRNAs of prostate cancer: virus-encoded hsv1-miRH18 and hsv2-miR-H9-5p could be valuable diagnostic markers. Int Neurourol J. 2015;19:74–84. doi: 10.5213/inj.2015.19.2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 24.Stuopelyte K, Daniunaite K, Bakavicius A, Lazutka JR, Jankevicius F, Jarmalaite S. The utility of urine-circulating miRNAs for detection of prostate cancer. Br J Cancer. 2016;115:707–715. doi: 10.1038/bjc.2016.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuopelytė K, Daniūnaitė K, Jankevičius F, Jarmalaitė S. Detection of miRNAs in urine of prostate cancer patients. Medicina (Kaunas) 2016;52:116–124. doi: 10.1016/j.medici.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Kelly BD, Miller N, Healy NA, Walsh K, Kerin MJ. A review of expression profiling of circulating microRNAs in men with prostate cancer. BJU Int. 2013;111:17–21. doi: 10.1111/j.1464-410X.2012.11244.x. [DOI] [PubMed] [Google Scholar]
- 27.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Planell-Saguer M, Rodicio MC. Detection methods for microRNAs in clinic practice. Clin Biochem. 2013;46:869–878. doi: 10.1016/j.clinbiochem.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454:210–214. doi: 10.1016/j.bbrc.2014.10.064. [DOI] [PubMed] [Google Scholar]
- 30.Piao XM, Jeong P, Kim YH, Byun YJ, Xu Y, Kang HW, et al. Urinary cell-free microRNA biomarker could discriminate bladder cancer from benign hematuria. Int J Cancer. 2019;144:380–388. doi: 10.1002/ijc.31849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relationshipa between miRNA ratio and PSA