Abstract
Non-steroidal anti-inflammatory drugs (NSAIDs) showed promising clinical efficacy toward COVID-19 (Coronavirus disease 2019) patients as potent painkillers and anti-inflammatory agents. However, the prospective anti-COVID-19 mechanisms of NSAIDs are not evidently exposed. Therefore, we intended to decipher the most influential NSAIDs candidate(s) and its novel mechanism(s) against COVID-19 by network pharmacology. FDA (U.S. Food & Drug Administration) approved NSAIDs (19 active drugs and one prodrug) were used for this study. Target proteins related to selected NSAIDs and COVID-19 related target proteins were identified by the Similarity Ensemble Approach, Swiss Target Prediction, and PubChem databases, respectively. Venn diagram identified overlapping target proteins between NSAIDs and COVID-19 related target proteins. The interactive networking between NSAIDs and overlapping target proteins was analyzed by STRING. RStudio plotted the bubble chart of the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis of overlapping target proteins. Finally, the binding affinity of NSAIDs against target proteins was determined through molecular docking test (MDT). Geneset enrichment analysis exhibited 26 signaling pathways against COVID-19. Inhibition of proinflammatory stimuli of tissues and/or cells by inactivating the RAS signaling pathway was identified as the key anti-COVID-19 mechanism of NSAIDs. Besides, MAPK8, MAPK10, and BAD target proteins were explored as the associated target proteins of the RAS. Among twenty NSAIDs, 6MNA, Rofecoxib, and Indomethacin revealed promising binding affinity with the highest docking score against three identified target proteins, respectively. Overall, our proposed three NSAIDs (6MNA, Rofecoxib, and Indomethacin) might block the RAS by inactivating its associated target proteins, thus may alleviate excessive inflammation induced by SARS-CoV-2.
Subject terms: Biotechnology, Computational biology and bioinformatics, Genetics, Molecular biology, Health care, Medical research
Introduction
An initial outbreak of pneumonia caused by unknown etiology was first reported at Wuhan in Hubei Province, China, and alerted to the World Health Organization (WHO) by the Wuhan Municipal Health Commission on 31 December 20191. Later, the infectious disease experts detected severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), can rapidly transmit from person to person through interaction or respiratory droplets2. As a consequence of its tremendous spread globally, WHO announced a changing level from epidemic to pandemic disease (COVID-19) on March 11, 20203. Although the symptoms were identical to pneumonia, however, many COVID-19 patients showed no physical sign, thus transmitting the virus to others, as silently spread4.
Due to a reliable vaccine's unavailability, clinicians utilize antiviral drugs and NSAIDs as a significant viable option for COVID-19 patients5. A recent study has reported that the use of NSAIDs is safe for COVID-19 treatment without exposing specific adverse effects6. Though there is a lack of evidence whether combined NSAIDs treatment could worsen COVID-19 symptoms7, but researchers suggested that anti-inflammatory therapies might suppress the fatal cytokine storm of COVID-19 patients8. Additionally, WHO announced that no evidence of unwanted side effects was reported, particularly the risk of death with NSAIDs' administration in COVID-19 patients9.
NSAIDs are commonly used to treat diverse anti-inflammatory symptoms due to their excellent therapeutic efficacy10. For example, some evidence suggests that NSAIDs are associated with mitigating depression, bladder function recovery, reduction of psychiatric events, and a decrease in cancer risk, all of which connected directly to anti-inflammatory effects11. Most NSAIDs are known as inhibitors of COX-1 (Cyclooxygenase-1) or/and COX-2 (Cyclooxygenase-2) involved in the synthesis of prostaglandin and thromboxanes12. Recently, both COX-1 and COX-2 are expressed in inflamed tissues constitutively, and Indomethacin inhibited the two forms of COX effectively13. Also, Indomethacin is a potent NSAIDs against rheumatoid arthritis, used as an immune response enhancer against HIV (Human Immunodeficiency Virus), inhibiting harmful immune response induced by COX-213,14. Generally, COX-1 is expressed in most normal cells, and COX-2 is induced by an abundance of physiological stimulus15. An animal test demonstrated that the inhibition of COX-1 up-regulates COX-2 expression level, results in the prevention of aggravated effects against inflammatory response16. It was reported that the administration of dual COX-1/2 inhibitors slightly diminished viral DNA replication but could not induce viral DNA cleavage17. Hence, COX-1/2 inhibitors are involved in host immune responses while cannot take effect on pathogens. However, one potential drug is Indomethacin, which possesses both anti-inflammatory and antiviral properties. Its antiviral potentiality was first identified in 2006 during the outbreak of SARS-CoV18, and subsequent attribution was also observed against SARS-CoV-219. A study on canine coronavirus (in vitro) revealed that Indomethacin could significantly suppress virus replication, thus protecting host cell from virus-induced damage. A similar antiviral effect was also observed during in vivo assessment where an average anti-inflammatory dose was found very effective19,20. Although many NSAIDs may have possible therapeutic interventions against COVID-19, but lack of scientific evidence has limited their broad application to COVID-19 patients. Hence, we aimed to identify the most influential NSAIDs and their mechanism(s) against COVID-19 through network pharmacology.
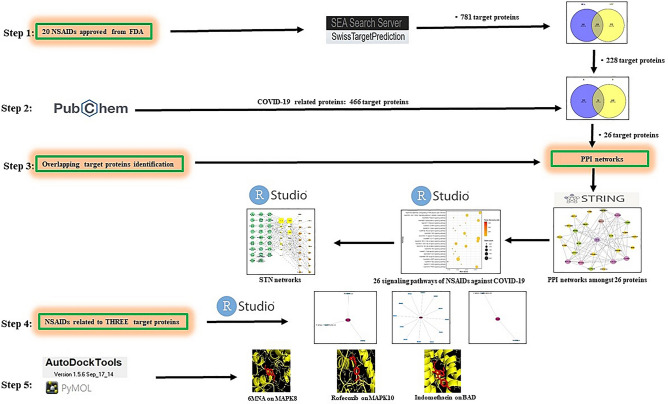
Network pharmacology can decode the mechanism(s) of drug action with an overall viewpoint21, which focuses on pattern changing from "single protein target, single drug" to "multiple protein targets, multiple drugs"22,23. Currently, network pharmacology has been extensively utilized to explore multiple targets and unknown additional mechanism(s) against diverse diseases24. In this research, network pharmacology was applied to investigate the most potent NSAIDs and their novel mechanisms of action against COVID-19. Firstly, FDA approved NSAIDs (nineteen active drugs and one prodrug) were selected via using public websites. The selected NSAIDs and COVID-19 related target proteins were also identified using public databases. Next, the extracted overlapping target proteins were discovered as target proteins for analyzing anti-COVID-19 properties. Finally, pathway enrichment analysis was performed to reveal the mechanism(s) of the most potent NSAIDs against COVID-19. Figure 1 shows the overall workflow.
Figure 1.
Workflow of network pharmacology analysis of NSAIDs against COVID-19.
Results
Information of NSAIDs
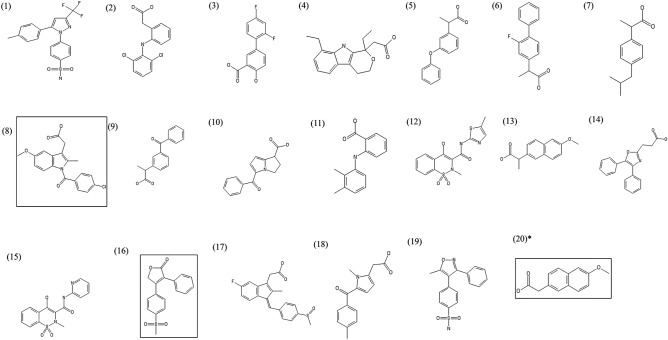
A total of nineteen NSAIDs and one prodrug (FDA approved) were selected. Table 1 displayed the NSAIDs' chemical information and TPSA (Topological Polar Surface Area). Among the selected NSAIDs, nineteen NSAIDs were found as an active drug, and one "nabumetone" was a prodrug, and its metabolite form is 6-methoxy-2-naphthylacetic acid (6MNA). Figure 2 exhibited the chemical structure of these NSAIDs.
Table 1.
A list of NSAIDs (19 active drugs and one prodrug) approved by FDA and TPSA (Å2) values.
| No | Drug name | PubChem CID | Mechanism of action | TPSA (< 140 Å2) |
|---|---|---|---|---|
| 1 | Flubiprofen | 3394 | Nonselective COX inhibitor | 37.30 |
| 2 | Ibuprofen | 3672 | Nonselective COX inhibitor | 37.30 |
| 3 | Indomethacin | 3715 | Nonselective COX inhibitor | 68.53 |
| 4 | Ketorolac | 3826 | Nonselective COX inhibitor | 59.30 |
| 5 | Mefenamic acid | 4044 | Nonselective COX inhibitor | 49.33 |
| 6 | Piroxicam | 54,676,228 | Nonselective COX inhibitor | 107.98 |
| 7 | Diflunisal | 3059 | Prostaglandin synthesis inhibitor | 57.53 |
| 8 | Fenoprofen | 3342 | Prostaglandin synthesis inhibitor | 46.53 |
| 9 | Naproxen | 156,391 | Prostaglandin synthesis inhibitor | 46.53 |
| 10 | Sulindac | 1,548,887 | Prostaglandin synthesis inhibitor | 73.58 |
| 11 | Tolmetin | 5509 | Prostaglandin synthesis inhibitor | 59.30 |
| 12 | Ketoprofen | 3825 | Selective COX-1 inhibitor | 54.37 |
| 13 | Oxaprozin | 4614 | Selective COX-1 inhibitor | 63.33 |
| 14 | Celecoxib | 2662 | Selective COX-2 inhibitor | 86.36 |
| 15 | Rofecoxib | 5090 | Selective COX-2 inhibitor | 68.82 |
| 16 | Valdecoxib | 119,607 | Selective COX-2 inhibitor | 94.57 |
| 17 | Diclofenac | 3033 | Selective COX-2 inhibitor | 49.33 |
| 18 | Etodolac | 3308 | Selective COX-2 inhibitor | 62.32 |
| 19 | Meloxicam | 54,677,470 | Selective COX-2 inhibitor | 136.22 |
| 20a | 6MNA | 32,176 | Selective COX-2 inhibitor | 46.53 |
a6MNA (Active form) of Nabumetone (Prodrug); TPSA (Topological Polar Surface Area).
Figure 2.
Structure of 19 NSAIDs and 1 prodrug. (1) Celecoxib (2) Diclofenac (3) Diflunisal (4) Etodolac (5) Fenoprofen (6) Flubiprofen (7) Ibuprofen (8) Indomethacin (9) Ketoprofen (10) Ketorolac (11) Mefenamic acid (12) Meloxicam (13) Naproxen (14) Oxaprozin (15) Piroxicam (16) Rofecoxib (17) Sulindac (18) Tolmetin (19) Valdecoxib *(20): Nabumetone (prodrug of 6MNA). The three NSAIDs in box line are the most potent NSAIDs candidates against COVID-19.
NSAIDs connected to the 781 target proteins or COVID-19 targeted proteins
A total of 781 NSAIDs related proteins were identified (Supplementary Table S1) through two public databases (SEA and STP). Figure 3A showed the overlapping target proteins (228 target proteins) selected from the two databases (Supplementary Table S2). The number of 466 COVID-19 targeted proteins was identified from the PubChem database (Supplementary Table S3). Figure 3B illustrated that the final 26 overlapping target proteins were selected between 228 overlapped target proteins and 466 COVID-19 targeted proteins (Supplementary Table S4).
Figure 3.

(A) Overlapping target proteins (228 target proteins) of NSAIDs related target proteins identified from SEA (529 target proteins) and STP (480 target proteins). (B) Overlapping target proteins (26 target proteins) between NSAIDs related 228 overlapped target proteins and COVID-19 related 466 target proteins.
Protein–protein interaction (PPI) networks and KEGG pathway enrichment analysis
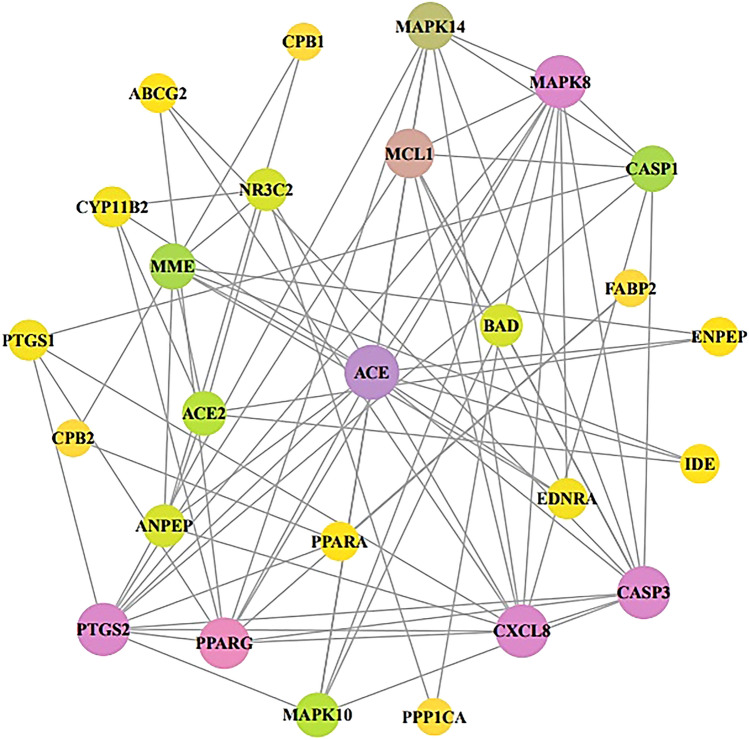
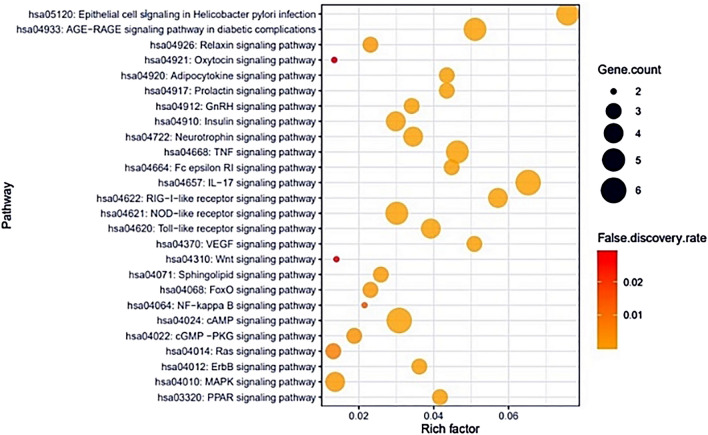
Figure 4 demonstrated that the final 26 overlapping target proteins were selected by STRING, which represents 26 nodes and 78 edges. According to the KEGG pathway enrichment analysis, 13 target proteins were connected to 26 signaling pathways (False Discovery Rate < 0.05). Table 2 showed the description of 26 signaling pathways. Figure 5 displayed that 13 out of the final 26 overlapping target proteins were strongly associated with 26 signaling pathways against COVID-19; moreover, the RAS signaling pathway with the lowest Rich factor was identified as a hub signaling pathway.
Figure 4.
Protein protein interaction (PPI) networks with 26 nodes and 78 edges in NSAIDs against COVID-19 via STRING analysis. Node: the number of units; Edge: the number of interactions between the units.
Table 2.
Target proteins in 26 signaling pathways enrichment related to COVID-19.
| KEGG ID & description | Target proteins | Rich factor | False discovery rate |
|---|---|---|---|
| hsa04014: Ras signaling pathway | MAPK8, MAPK10, BAD | 0.013157895 | 0.0071 |
| hsa04921: Oxytocin signaling pathway | PTGS2, PPP1CA | 0.013422819 | 0.0294 |
| hsa04010: MAPK signaling pathway | MAPK8, MAPK10, MAPK14, CASP3 | 0.013651877 | 0.0016 |
| hsa04310: WNT signaling pathway | MAPK8, MAPK10 | 0.013986014 | 0.0276 |
| hsa04022: cGMP -PKG signaling pathway | ENDRA, BAD, PPP1CA | 0.01875 | 0.003 |
| hsa04064: NF-kappa B signaling pathway | CXCL8, PTGS2 | 0.021505376 | 0.0136 |
| hsa04926: Relaxin signaling pathway | MAPK8, MAPK10, MAPK14 | 0.023076923 | 0.0018 |
| hsa04068: FoxO signaling pathway | MAPK8, MAPK10, MAPK14 | 0.023076923 | 0.0018 |
| hsa04071: Sphingolipid signaling pathway | MAPK8, MAPK10, MAPK14 | 0.025862069 | 0.0014 |
| hsa04910: Insulin signaling pathway | MAPK8, MAPK10, BAD, PPP1CA | 0.029850746 | 0.00013 |
| hsa04621: NOD-like receptor signaling pathway | MAPK8, MAPK10, MAPK14, CXCL8, CASP1 | 0.030120482 | 0.0000197 |
| hsa04024: cAMP signaling pathway | ENDRA, MAPK8, BAD, MAPK10, PPP1CA, PPARA | 0.030769231 | 0.00000373 |
| hsa04912: GnRH signaling pathway | MAPK8, MAPK10, MAPK14 | 0.034090909 | 0.00072 |
| hsa04722: Neurotrophin signaling pathway | MAPK8, MAPK10, MAPK14, BAD | 0.034482759 | 0.00000853 |
| hsa04012: ErbB signaling pathway | MAPK8, MAPK10, BAD | 0.036144578 | 0.00062 |
| hsa04620: Toll-like receptor signaling pathway | MAPK8, MAPK10, MAPK14, CXCL8 | 0.039215686 | 0.0000581 |
| hsa03320: PPAR signaling pathway | PPARA, PPARG, FABP2 | 0.041666667 | 0.00044 |
| hsa04920: Adipocytokine signaling pathway | MAPK8, MAPK10, PPARA | 0.043478261 | 0.00041 |
| hsa04917: Prolactin signaling pathway | MAPK8, MAPK10, MAPK14 | 0.043478261 | 0.00041 |
| hsa04664: Fc epsilon RI signaling pathway | MAPK8, MAPK10, MAPK14 | 0.044776119 | 0.00039 |
| hsa04668: TNF signaling pathway | MAPK8, MAPK10, MAPK14, CASP3, PTGS2 | 0.046296296 | 0.00000413 |
| hsa04370: VEGF signaling pathway | MAPK8, MAPK10, BAD | 0.050847458 | 0.00029 |
| hsa04933: AGE-RAGE signaling pathway in diabetic complications | MAPK8, MAPK10, MAPK14, CXCL8, CASP1 | 0.051020408 | 0.00000373 |
| hsa04622: RIG-I-like receptor signaling pathway | MAPK8, MAPK10, MAPK14, CXCL8 | 0.057142857 | 0.0000197 |
| hsa04657: IL-17 signaling pathway | MAPK8, MAPK10, MAPK14, CXCL8, CASP3, PTGS2 | 0.065217391 | 0.000000135 |
| hsa05120: Epithelial cell signaling in Helicobacter pylori infection | MAPK8, MAPK10, MAPK14, CXCL8, CASP3 | 0.075757576 | 0.000000954 |
Figure 5.
Bubble chart of 26 signaling pathways related to the occurrence and progression of COVID-19.
A signaling pathway-target protein—NSAID (STN) network analysis
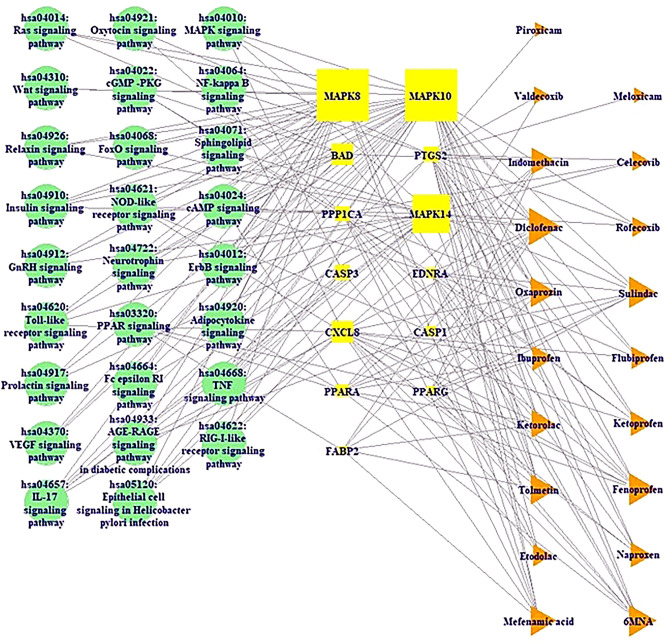
Figure 6 revealed 26 signaling pathways—13 target proteins—19 NSAIDs networks (58 nodes and 194 edges). Among 20 NSAIDs, Diflunisal has no association with signaling pathways against COVID-19.
Figure 6.
Signaling pathway-Target protein-NSAID (STN) networks. Green circle: signaling pathway; Yellow square: target protein; Orange triangle: NSAID.
The nodes indicated a total number of signaling pathways, target proteins, and NSAIDs. The edges represented the relationships of the three components. The STN relationship suggested that the network might be potential therapeutic efficacy against COVID-19. The STN network displayed that 13 target proteins associated with 26 signaling pathways built by a size map. Among the 13 target proteins, both MAPK8 and MAPK10 have the highest degree (22), followed by MAPK14 (15), BAD (7), and CXCL8 (7). Additionally, among the 19 NSAIDs, Diclofenac has the greatest degree (10), followed by 6MNA (9), Fenoprofen (8), and Sulindac (8).
MDT of 3 target proteins and 15 NSAIDs associated with RAS signaling pathway
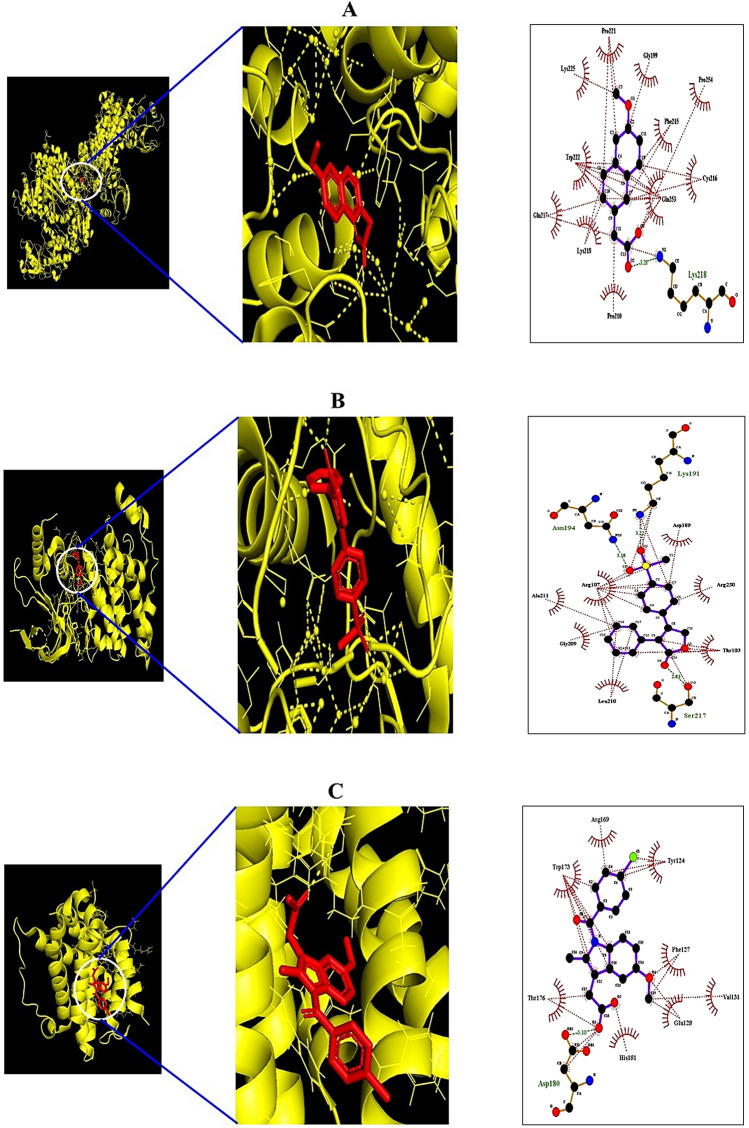
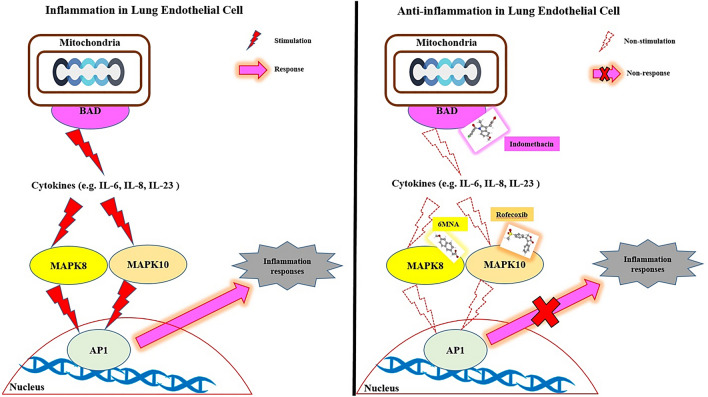
From the SEA and STP databases, it was revealed that MAPK8 was associated with three NSAIDs (6MNA, Mefenamic acid, and Etodolac), MAPK10 was related to twelve NSAIDs (Mefenamic acid, Naproxen, Tolmetin, Fenoprofen, Ketorolac, Ketoprofen, Ibuprofen, Flurbiprofen, Oxaprozin, Sulindac, Diclofenac, and Rofecoxib), BAD was involved with two NSAIDs (6MNA and Indomethacin). The MDT was performed to evaluate NSAIDs' binding affinity on target three proteins (MAPK8, MAPK10, and BAD) connected directly to the RAS signaling pathway. The MDT score of three NSAIDs on MAPK8 (PDB ID: 4YR8) was analyzed in the "Homo Sapiens" mode. Table 3 indicated that 6MNA (− 7.1 kcal/mol) revealed the highest binding energy, followed by Mefenamic acid (− 6.4 kcal/mol), and Etodolac (− 6.3 kcal/mol) on MAPK8 (PDB ID: 4YR8). Figure 7A exhibited the MDT of the 6MNA-MAPK8 (PDB ID: 4YR8) complex with the highest binding affinity. The MDT score of twelve NSAIDs on MAPK10 (PDB ID: 3TTJ) was analyzed in the "Homo Sapiens" mode. Table 4 indicated that Rofecoxib (− 7.5 kcal/mol) exposed the highest binding energy, followed by Sulindac (− 7.4 kcal/mol), Oxaprozin (− 7.1 kcal/mol), Ketorolac (− 7.1 kcal/mol), Flubiprofen (− 6.9 kcal/mol), Tolmetin (− 6.7 kcal/mol), Diclofenac (− 6.7 kcal/mol), Fenoprofen (− 6.5 kcal/mol), Ketoprofen (− 6.5 kcal/mol), Mefenamic acid (− 6.4 kcal/mol), Naproxen (− 6.1 kcal/mol), Ibuprofen (− 5.6 kcal/mol) on MAPK10 (PDB ID: 3TTJ). Figure 7B presented the MDT of the Rofecoxib—MAPK10 (PDB ID: 3TTJ) complex with the highest binding affinity. The MDT score of two NSAIDs on BAD (PDB ID: 1G5J) was analyzed in the "Homo Sapiens" mode. Figure 7C exhibited the MDT of the Indomethacin—BAD (PDB ID: 1G5J) complex with the highest binding affinity. Table 5 showed that Indomethacin (− 7.1 kcal/mol) was the highest binding energy, followed by 6MNA (− 6.8 kcal/mol) on BAD (PDB ID: 1G5J). Figure 8 depicted that MAPK-6MNA complex and MAPK10-Rofecoxib complex inhibit AP1 induced inflammatory responses; simultaneously, blocked AP1 inhibits cytokine productions. As an upstream region, the BAD-Indomethacin complex also interrupts cytokine production against COVID-19.
Table 3.
Binding energy and interactions of potential three NSAIDs on MAPK8 (PDB ID:4YR8).
| Protein | Ligand | PubChem ID | Symbol | Binding energy (kcal/mol) | Hydrogen bond interactions | Hydrophobic interactions |
|---|---|---|---|---|---|---|
| Amino acid residue | Amino acid residue | |||||
| 4YR8 | 6MNA | 32,176 | M1 | − 7.1 | Lys-218 | Pro-221, Gly-199 |
| Pro-254, Phe215 | ||||||
| Cys-216, Gln-253 | ||||||
| Pro-210, Lys-218 | ||||||
| Glu-217, Lys-225 | ||||||
| Mefenamic acid | 4044 | M2 | − 6.4 | Glu-217 | Trp-222, Val-211 | |
| Arg-208, Cys-216 | ||||||
| Asn-193, Lys-218 | ||||||
| Pro-211 | ||||||
| Etodolac | 3308 | M3 | − 6.3 | n/a | Tyr-202, Lys-203 | |
| Met-200, Gly-201 | ||||||
| Pro-221, Lys-218 | ||||||
| Lys-251, Ser-307 | ||||||
| Ala-306 |
Figure 7.
Molecular docking interaction between best docked NSAIDs and target proteins. (A) 6MNA on MAPK 8 (PDB ID:4YR8) (B) Rofecoxib on MAPK 10 (PDB ID:3TTJ). (C) Indomethacin on BAD (PDB ID: 1G5J).
Table 4.
Binding energy and interactions of potential twelve NSAIDs on MAPK10 (PDB ID: 3TTJ).
| Protein | Ligand | PubChem ID | Symbol | Binding energy (kcal/mol) | Hydrogen bond interactions | Hydrophobic Interactions |
|---|---|---|---|---|---|---|
| Amino acid residue | Amino acid residue | |||||
| 3TTJ | Mefenamic acid | 32,176 | R1 | − 6.4 | Arg-107 | Asp-207, Gln-75 |
| Leu-206, Lys-93 | ||||||
| Asn-194, Asp-207 | ||||||
| Naproxen | 4044 | R2 | − 6.1 | Asn-194, Lys-93 | Arg-107, Asp-189 | |
| Val-225, Lys-191 | ||||||
| Gln-75, Gly-73 | ||||||
| Tolmetin | 3308 | R3 | − 6.7 | Asn-194, Asp-189 | Lys-106, Leu-210 | |
| Ala-211, Arg-110 | ||||||
| Arg-230, Lys-191 | ||||||
| Arg-107, Thr-103 | ||||||
| Fenoprofen | 3342 | R4 | − 6.5 | Lys-93, Lys-191 | Ser-193, Ser-72 | |
| Asn-194 | Val-78, Gly-73 | |||||
| Gln-75, Ala-74 | ||||||
| Arg-107 | ||||||
| Ketorolac | 3826 | R5 | − 7.1 | Glu-111, Arg-107 | Asp-207, Leu-206 | |
| Asn-194, Lys-93 | Gln-75, Ser-193 | |||||
| Ser-72, Val-78 | ||||||
| Gly-73 | ||||||
| Ketoprofen | 3825 | R6 | − 6.5 | Lys-93, Asn-194 | Val-78, Leu-206 | |
| Ser-193 | Arg-107, Gln-75 | |||||
| Gly-73 | ||||||
| Ibuprofen | 3672 | R7 | − 5.6 | Lys-93, Ser-193 | Leu-206, Ala-74 | |
| Asn-194 | Gly-73, Gln-75 | |||||
| Val-78 | ||||||
| Flubiprofen | 3394 | R8 | − 6.9 | Lys-191, Asp-189 | Lys-93, Val-78 | |
| Asn-194 | Gly-73, Arg-107 | |||||
| Gln-75 | ||||||
| Oxaprozin | 4614 | R9 | − 7.1 | Asn-194, Arg-107 | Asp-189, Thr-103 | |
| Lys-191 | Ser-217, Val-225 | |||||
| Arg-230 | ||||||
| Sulindac | 1,548,887 | R10 | − 7.4 | Asn-152 | Arg-107, Asn-194 | |
| Lys-93, Ser-72 | ||||||
| Gly-73, Ser-193 | ||||||
| Ala-74 | ||||||
| Diclofenac | 3033 | R11 | − 6.7 | Asn-194 | Ser-72, Gly-73 | |
| Ser-193, Gln-75 | ||||||
| Arg-107, Lys-93 | ||||||
| Leu-206, Val-78 | ||||||
| Gly-71 | ||||||
| Rofecoxib | 5090 | R12 | − 7.5 | Asn-194, Lys-191 | Asp-189, Arg-230 | |
| Ser-217 | Thr-203, Leu-210 | |||||
| Gly-209, Ala-211 | ||||||
| Arg-107 |
Table 5.
Binding energy and interactions of potential two NSAIDs on BAD (PDB ID: 1G5J).
| Protein | Ligand | PubChem ID | Symbol | Binding energy (kcal/mol) | Hydrogen bond interactions | Hydrophobic interactions |
|---|---|---|---|---|---|---|
| Amino acid residue | Amino acid residue | |||||
| 1G5J | 6MNA | 32,176 | B1 | − 6.8 | Trp-173, His-181 | Arg-169, Tyr-124 |
| Phe-127, Tyr-177 | ||||||
| Thr-176 | ||||||
| Indomethacin | 3715 | B2 | − 7.1 | Asp-180 | Arg-169, Tyr-124 | |
| Phe-127, Val-131 | ||||||
| Glu-128, His-181 | ||||||
| Thr-176, Trp-173 |
Figure 8.
Anti-inflammation mechanisms of promising NSAIDs against COVID-19.
Discussion
STN networking analysis demonstrated that the therapeutic effect of NSAIDs against COVID-19 was directly related to 26 signaling pathways—13 target proteins—19 NSAIDs. The results of the KEGG pathway enrichment analysis of 13 target proteins suggested that 26 signaling pathways were associated with the occurrence and development of the COVID-19 symptoms. The relationships of 26 signaling pathways with COVID-19 symptoms were succinctly discussed as follows. PPAR (Peroxisome Proliferator-Activated Receptor) signaling pathway: a report shows that PPARγ (Peroxisome Proliferator-Activated Receptor-gamma), PPARα (Peroxisome Proliferator-Activated Receptor-alpha), and PPARβ/δ (Peroxisome Proliferator-Activated Receptor-beta/delta) agonists have anti-inflammatory and immunomodulatory functions25. MAPK (Mitogen-Activated Protein Kinase) signaling pathway: The mechanisms of p38 MAPK inactivation might be an effective therapy against the SARS infected cells26. Additionally, MAPK stimulates cytokine production such as IL-10 (Interleukin 10), TNF-α (Tumor Necrosis Factor-Alpha), IL-4 (Interleukin 4), and IFN-γ (Interferon-gamma)27. It is evident that MAPK inhibitor can alleviate inflammatory responses against COVID-19. ErbB (Erythroblastic Leukemia Viral Oncogene Homolog) signaling pathway: ErbB signaling reduces the proinflammatory activation in cardiac cells28. RAS (Renin-Angiotensin System) signaling pathway: The inhibition of ACE (Angiotensin Converting Enzyme) connected to RAS signaling pathway could reduce tissue damage in COVID-19 patients29. Thus, the study indicates that blocking of the RAS signaling pathway can reduce inflammatory response level. cGMP-PKG (Cyclic GMP-Protein Kinase G) signaling pathway: the activation of cGMP-PKG signaling inhibits inflammatory response in the prostate, and also decreases CCL5 (C–C Motif Chemokine Ligand 5) released in CD8 + T cells (Cluster of Differentiation 8 T cells)30. cAMP (Cyclic Adenosine Monophosphate) signaling pathway: the elevation of cAMP leads to diverse cellular effects, such as airway smooth muscle relaxation, repressed effects on cellular inflammation, and immune responses31. NF-κB (Nuclear Factor kappa-light-chain-enhancer of activated B cells) signaling pathway: activation of the NF-κB signaling pathway gives rise to the inflammation induced by the SARS-CoV infection. In contrast, NF-κB inhibitors are the potential antivirals, even against SARS-CoV and can also contribute to other pathogenic human coronaviruses32. FOXO (Forkhead box protein O1) signaling pathway: decrease of FOXO3 (Forkhead box protein O3) in T cells inhibits apoptosis, enhances multifunction of CD8 cells, and elevates viral control33. Sphingolipid signaling pathway: Sphingolipids play a vital role to protect the lung from damages, and its control may give a good therapeutic efficacy34. WNT (Wingless/Integrated) signaling pathway: WNT signaling involves with the prime inflammatory pathways like intestinal inflammation. Also, the understanding mechanism of WNT ligands and cytokines manifest new treatment strategies for chronic colitis and other inflammatory diseases35. VEGF (Vascular Endothelial Growth Factor) signaling pathway: a report suggested that the activation of ACE2 (Angiotensin-Converting Enzyme 2) inhibits VEGFA (Vascular Endothelial Growth Factor A), which elevates vascular permeability and severity of endothelial damage36. TLR (Toll-like receptor) signaling pathway: Toll-like receptors (TLRs) play a pivotal role in the innate immune system and contribute to defending host cells by recognizing PAMPs (Pathogen-Associated Molecular Patterns) induced by various microbes37. TLRs' activation triggers an array of responses resulting in the expression of different cytokines and chemokines, phagocytosis, and even apoptotic case activation to induce programmed cell death38. NOD-like receptor (NLR) signaling pathway: Nod-like receptors (NLRs) have been revealed as the major microbial signals that take part in the universal immune responses to infection and contribute to the prevention of infections39. RIG-I-like receptor (RLR) signaling pathway: RIG-I-like receptors (RLRs) play a vital role in the pathogen sensor of RNA virus infection, enhancing the antiviral immunity by sensing foreign RNA40. IL-17 (Interleukin-17) signaling pathway: IL-17 receptor inhibitors are widely used to ameliorate the inflammatory acuteness to date. Furthermore, it is a potential target to suppress severe inflammation induced by COVID-1941. Fc epsilon RI signaling pathway: Fc epsilon RI interconnecting causes mast cell degranulation and synthesis of proinflammatory mediators42. TNF (Tumor Necrosis Factor) signaling pathway: TNF deficit is associated with dysfunctional secretion of inflammatory cytokine, leading to lung pathology and death during respiratory poxvirus infection, and thus TNF is a very significant element for regulating inflammation43. Neurotrophin signaling pathway: COVID-19 causes severe brain damage and destruction of the central nervous system derived from neurotrophin44,45. Insulin signaling pathway: Obesity-oriented insulin resistance is associated with the induction of proiifnnflammatory macrophage, leads to inflammation of adipose tissue46. GnRH (Gonadotropin-Releasing Hormone) signaling pathway: BBB (Blood Brain Barrier) disrupted by a viral infection, lymphocytes (B and T cells), monocytes, and granulocytes can penetrate in the brain parenchyma, which induces inflammation, resulting in dysregulation of GnRH neurons. Additionally, the inflammation of GnRH neurons inhibits GnRH transport through proinflammatory cytokines by impairing the cytoskeleton47. Prolactin signaling pathway: HIV patients have greater prolactin quantity compared to others. Besides, prolactin is regarded as a cytokine to stimulate the immune system48,49. Adipocytokine signaling pathway: Adipocytokines stimulate inflammation and disrupt immune response, which induces proinflammation in RA (Rheumatoid Arthritis) patients, leading to the development of bone damage50. Oxytocin signaling pathway: oxytocin interrupts proinflammatory cytokines' production by inactivating the eIF-2α–ATF4 (Eukaryotic Initiation Factor-2 alpha-Activating Transcription Factor 4) pathway51. Relaxin signaling pathway: relaxin inhibitors are good therapeutic targets to suppress inflammation caused by airway dysfunction52. AGE-RAGE (Advanced Glycation End product-Receptor of Advanced Glycation End product) signaling pathway in diabetic complications: The binding of AGE to its receptor RAGE can trigger cytokine production, thus, can cause tissue damages, while the blockage of AGE-RAGE can effectively ameliorate the inflammation53. Epithelial cell signaling in Helicobacter pylori infection: Helicobacter pylori interrupts T and B cell signaling to work the immune system. It is apparent that COVID-19 patients with Helicobacter pylori might be vulnerable to inflammatory responses54.
Generally, SARS-CoV-2 invades the lungs and throat, induces excessive inflammation, which causes cytokines' secretion, resulting in severe complications like acute respiratory failure, pneumonia, and acute liver injury55. The leading cause is that the downregulation of ACE2 results in an angiotensin-II (Ang II) increase, which might spur the progression of COVID-19 through activated RAS56.
It was discovered that ACE2 is the functional receptor for the SARS-CoV-2 to trigger an infection in the lung alveolar epithelial cells. The internalization of the virus leads to downregulating the ACE2 on the host cell surface that could cause the elevation and demotion of Ang II and angiotensin 1–7 (Ang 1–7), respectively. Such an imbalance between these angiotensins may induce deleterious effects in the lung and heart. Thus, the SARS-CoV-2 affects humans through this mechanism57–60. Therefore, RAS blockade may restore the RAS balance by reducing the deleterious effects associated with Ang II61. Recent evidence showed that RAS inhibitors might be a promising target for relieving acute-severe pneumonia caused by the COVID-1962.
Interestingly, our study identified that the three target proteins (MAPK8, MAPK10, and BAD) are mainly associated with the RAS signaling pathway. MAPK8 and MAPK10 are members of the MAPK family which are the key mediators of inflammation, vasoconstriction, and thrombosis. Besides, overwhelming heart and lung injury in COVID-19 infection might be due to the overactivation of MAPK63. Therefore, these proteins' inactivation can also be a viable strategy for relieving COVID-19 induced organ injury. In addition, disposal of inflammatory cells by promoting cell death can be an innovative approach to control excessive inflammation. In this regard, the anti-apoptotic Bcl-2 gene's inhibition can also be a potential target to lessen inflammation64,65. Our findings also explored that MAPK8 MAPK10 and BAD proteins are related to three, 12, and two NSAIDs, respectively. During the molecular docking analysis, 6MNA, Rofecoxib, and Indomethacin revealed promising binding affinity along with the highest docking score against MAPK8, MAPK10 and BAD proteins, respectively. The result suggested that the three NSAIDs' key mechanism against COVID-19 might be to inhibit inflammation of lung cells by inactivating the RAS signaling pathway, and blockers of MAPK8, MAPK10 and BAD might suppress cytokine storm. Among various NSAIDs, Indomethacin is a current drug of interest to clinicians. Primary care physicians (New York) reported that Indomethacin had been prescribed to a large number of COVID-19 patients and observed quick recovery from cough, pain, and other symptoms. Such improvements and well-being benefits were not evident in the case of ibuprofen and hydroxychloroquine implementation ( Little66). Notably, many researchers previously reported varying degrees of Indomethacin antiviral activity against herpesvirus67, pseudorabies virus68, cytomegalovirus69, hepatitis B virus70, vesicular stomatitis virus71, rotavirus 72, and canine coronavirus18. In contrast, 6MNA (an active metabolite of Nabumetone) and Rofecoxib are also potential anti-inflammatory drugs, but studies disclosed that they are less potent than Indomethacin73,74. A clinical study recently demonstrated that Indomethacin has potent anti-inflammatory (decrease in IL-6) and antiviral efficacy; taking SRF (Sustained Release Formulation) with 75 mg twice a day achieved full effects in 3 days for patients infected by COVID-1975.
Hence, such compelling outcomes indicate that Indomethacin can be considered alone or in combination for antiviral therapy, which may assist in combating human coronavirus (SARS-CoV-2). However, there are also some limitations to our analysis. This study has provided a predictive viewpoint of NSAIDs' mechanism against COVID-19 through public databases. Thus, further experimental results should be validated to achieve the reliability of predicted outputs through in vitro and in vivo followed by NGS (Next Generation Sequencing) technique. Finally, our analysis did not consider the target gene expression level practically after treating the selected NSAIDs (6MNA, Rofecoxib, and Indomethacin), which should be considered and implemented in the future. The proposed (three) NSAIDs against COVID-19 might be significant for clinical application, mainly depending on the genetic, ethnic, and underlying diseases associated with the therapeutic method.
Conclusion
This study suggests that 6MNA, Rofecoxib, and Indomethacin are the most potent NSAIDs against COVID-19. The basis of this research is an understanding of how these NSAIDs (which stimulates anti-inflammatory processes against COVID-19) work against COVID-19 patients. That scientific evidence informs the selection of NSAIDs, in turn, provides for clinical design against COVID-19. Our research suggests that BAD-Indomethacin's inhibition with the other two hub proteins, MAPK8-6MNA, MAPK10-Rofecoxib might play cumulative actions by inactivating the RAS signaling pathway against COVID-19. Most recently, the efficacy of Indomethacin against COVID-19 has been approved clinically. Our study presents that Indomethacin is a potent therapeutic candidate to relieve COVID-19 symptoms, which is in line with the many previous studies. However, further clinical trial on Indomethacin should be warranted in COVID-19 patients to slow down the progression of SARS-CoV-2 and mitigate the severity.
Materials and methods
NSAIDs linked to selected proteins or COVID-19 related proteins
FDA (U.S. Food & Drug Administration) approved NSAIDs (nineteen active drugs and one prodrug) were used in this study. Based on SMILES, targeted gene(s) of NSAIDs were identified through Similarity Ensemble Approach (SEA) (http://sea.bkslab.org/)76 and Swiss Target Prediction (STP) (http://www.swisstargetprediction.ch/)77 with the "Homo sapience" mode78. Additionally, COVID-19 targeted proteins were identified by retrieving COVID-19 in PubChem (https://pubchem.ncbi.nlm.nih.gov/). Also, the topological polar surface area (TPSA) value identified by SwissADME is included to verify the NSAIDS' cell permeability; particularly, its permeability is typically limited when the TPSA value is more than 140 Å279. The final overlapping proteins between NSAIDs and COVID-19—targeted proteins were identified and visualized by Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/).
PPI network between NSAIDs and COVID-19 targeted proteins
The protein–protein interaction network (PPI) between NSAIDs and COVID-19 targeted proteins were selected by STRING (https://string-db.org/)80 and finally plotted by RStudio.
Signaling pathway enrichment analysis of overlapping proteins via KEGG database
KEGG database provides correlation of target proteins and signaling pathways through functional annotation81. A bubble chart of signaling pathways associated with COVID-19 infection plotted by RStudio. The bubble chart demonstrates a hub signaling pathway (Lowest rich factor) between NSAIDs and COVID-19 related proteins.
The construction of STN network
The Signaling pathway(s)—Target protein(s)—NSAIDs were used to construct a signaling pathway—target protein—NSAID (STN) network. In this STN network, different colors and shapes (nodes) stand for the signaling pathways (green circle), target proteins (yellow square), and NSAIDs (orange triangles). Gray lines (edges) indicated the interaction of signaling pathways—target proteins—NSAIDs.
The STN networks were utilized to construct a size map, based on degree of values. In this network, green circles (nodes) represented signaling pathways; yellow squares (nodes) represented target proteins, and orange triangles (nodes) represented NSAIDs; its size represented degree value. The size of yellow squares stands for the number of connectivity with signaling pathways; the size of orange triangles stands for the number of connectivity with target proteins. The merged networks were constructed by using RStudio.
Preparation for MDT of NSAIDs
The ligand molecules were converted .sdf from PubChem into .pdb format using Pymol, and the ligand molecules were converted into .pdbqt format through Autodock.
Preparation for MDT of target proteins
Three target proteins of a hub signaling pathway, i.e. MAPK8 (PDB ID: 4YR8), MAPK10 (PDB ID: 3TTJ), and BAD (PDB ID: 1G5J), were identified by RCSB PDB (https://www.rcsb.org/). The proteins selected as .pdb format converted into .pdbqt format via Autodock (http://autodock.scripps.edu/).
NSAIDs- target protein(s) docking
The ligand molecules were docked with target proteins utilizing autodock4 by setting-up 4 energy range and 8 exhaustiveness as default to obtain 10 different poses of ligand molecules82. The active site's grid box size was x = 20.973 Å, y = 25.96 Å and z = 41.239 Å. The 2D binding simulation was identified via LigPlot + v.2.2 (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/). After docking, NSAIDs of the lowest binding energy (highest affinity) were selected to visualize the 3D docking simulation in Pymol.
Supplementary Information
Acknowledgements
This research was acknowledged by the Department of Bio-Health Convergence, College of Biomedical Science, Kangwon National University, Chuncheon, 24341, Korea.
Abbreviations
- ACE
Angiotensin-Converting Enzyme
- ACE2
Angiotensin-Converting Enzyme 2
- AGE-RAGE
Advanced Glycation End product /Receptor of Advanced Glycation End product
- Angiotensin 1–7
Ang 1–7
- Angiotensin-II
Ang II
- BAD
Bcl-2-Associated Death promoter
- BBB
Blood Brain Barrier
- cAMP
Cyclic Adenosine MonoPhosphate
- CCL5
C–C Motif Chemokine Ligand 5
- CD8+ T cells
Cluster of Differentiation 8T cells
- cGMP-PKG
Cyclic Guanosine MonoPhosphate-Protein Kinase G
- COX-1
Cyclooxygenase-1
- COX-2
Cyclooxygenase-2
- COVID-19
Coronavirus disease 2019
- eIF-2α-ATF4
Eukaryotic Initiation Factor-2 alpha-Activating Transcription Factor 4
- ErbB
Erythroblastic Leukemia Viral Oncogene Homolog
- FDA
US Food & Drug Administration
- FOXO
FOrkhead boX protein O
- GnRH
Gonadotropin-Releasing Hormone
- HIV
Human Immunodeficiency Virus
- IFN-γ
Interferon gamma
- IL-4
Interleukin 4
- IL-10
Interleukin 10
- IL-17
Interleukin 17
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
Mitogen-Activated Protein Kinase
- MAPK8
Mitogen-Activated Protein Kinase 8
- MAPK10
Mitogen-Activated Protein Kinase 10
- MDT
Molecular Docking Test
- NF-κB
Nuclear Factor Kappa-light-chain-enhancer of activated B cells
- NLR
Nod-Like Receptor
- NLRs
Nod-Like Receptors
- NSAIDs
Non-Steroidal Anti-Inflammatory Drugs
- PPAR
Peroxisome Proliferator-Activated Receptors
- PPARα
Peroxisome Proliferator-Activated Receptor-alpha
- PPARγ
Peroxisome Proliferator-Activated Receptor-gamma
- PPARβ/δ
Peroxisome Proliferator-Activated Receptor-beta/delta
- PAMPs
Pathogen-Associated Molecular Patterns
- PPI
Protein–protein interaction
- RA
Rheumatoid Arthritis
- RAS
Renin Angiotensin System
- RLR
RIG-I-Like Receptor
- RLRs
RIG-I-Like Receptors
- SARS-CoV-2
Severe Acute Respiratory Syndrome CoronaVirus 2
- SEA
Similarity Ensemble Approach
- STN
Signaling pathway-Target protein-NSAID
- STP
Swiss Target Prediction
- TLR
Toll-like receptor
- TLRs
Toll-like receptors
- TNF
Tumor Necrosis Factor
- TNF-α
Tumor Necrosis Factor-Alpha
- TPSA
Topological Polar Surface Area
- VEGF
Vascular Endothelial Growth Factor
- VEGFA
Vascular Endothelial Growth Factor A
- WHO
World Health Organization
- WNT
Wingless/Integrated
Author contributions
K.K.O.: Conceptualization, Methodology, Formal analysis, Investigation, Data Curation, Writing—Original Draft. K.K.O. and M.A.: Software, Investigation, Data Curation. M.A.: Validation, Writing—Review & Editing. D.H.C.: Supervision, Project administration.
Data availability
All data generated or analyzed during this study are included in this published article (and its “Supplementary Information S1” files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88313-5.
References
- 1.Harapan, H. et al. Coronavirus disease 2019 (COVID-19): A literature review. J. Infect. Public Health. 13, 667–673 (2020). [DOI] [PMC free article] [PubMed]
- 2.Pericàs JM, et al. COVID-19: From epidemiology to treatment. Eur. Heart J. 2020 doi: 10.1093/eurheartj/ehaa462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astuti I, Ysrafil Y. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab. Syndrome Clin. Res. Rev. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 2020 doi: 10.7326/m20-3012. [DOI] [PubMed] [Google Scholar]
- 5.Wu R, et al. An update on current therapeutic drugs treating COVID-19. Curr. Pharmacol. Rep. 2020;6:56–70. doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozili P. COVID-19 in Africa: Socio-economic impact, policy response and opportunities. Int. J. Sociol. Soc. Policy. 2020 doi: 10.1108/IJSSP-05-2020-0171. [DOI] [Google Scholar]
- 7.Capuano A, Scavone C, Racagni G, Scaglione F. NSAIDs in patients with viral infections, including Covid-19: Victims or perpetrators? Pharmacol. Res. 2020;157:104849. doi: 10.1016/j.phrs.2020.104849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, W. et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The experience of clinical immunologists from China. Clin. Immunol. 214, 1–5 (2020). [DOI] [PMC free article] [PubMed]
- 9.Sodhi M, Etminan M. Safety of ibuprofen in patients with COVID-19: Causal or confounded? Chest. 2020;158:55–56. doi: 10.1016/j.chest.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meek IL, van de Laar MAFJ, Vonkeman HE. Non-steroidal anti-inflammatory drugs: An overview of cardiovascular risks. Pharmaceuticals. 2010;3:2146–2162. doi: 10.3390/ph3072146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wongrakpanich S, Wongrakpanich A, Melhado K, Rangaswami J. A comprehensive review of non-steroidal anti-inflammatory drug use in the elderly. Aging Dis. 2018;9:143–150. doi: 10.14336/AD.2017.0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fokunang, C. Overview of non-steroidal anti-inflammatory drugs (nsaids) in resource limited countries. MOJ Toxicol.4, 5–13 (2018).
- 13.Bourinbaiar AS, Lee-Huang S. The non-steroidal anti-inflammatory drug, indomethacin, as an inhibitor of HIV replication. FEBS Lett. 1995;360:85–88. doi: 10.1016/0014-5793(95)00057-G. [DOI] [PubMed] [Google Scholar]
- 14.Prebensen C, et al. Immune activation and HIV-specific T cell responses are modulated by a cyclooxygenase-2 inhibitor in untreated HIV-infected individuals: An exploratory clinical trial. PLoS ONE. 2017;12:e0176527. doi: 10.1371/journal.pone.0176527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams CS, Mann M, DuBois RN. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene. 1999;18:7908–7916. doi: 10.1038/sj.onc.1203286. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka A, Araki H, Hase S, Komoike Y, Takeuchi K. Up-regulation of COX-2 by inhibition of COX-1 in the rat: A key to NSAID-induced gastric injury. Aliment. Pharmacol. Therap. Suppl. 2002;16:90–101. doi: 10.1046/j.1365-2036.16.s2.22.x. [DOI] [PubMed] [Google Scholar]
- 17.Ray N, Bisher ME, Enquist LW. Cyclooxygenase-1 and -2 are required for production of infectious pseudorabies virus. J. Virol. 2004;78:12964–12974. doi: 10.1128/JVI.78.23.12964-12974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amici C, et al. Indomethacin has a potent antiviral activity against SARS coronavirus. Antivir. Ther. 2006;11:1021. [PubMed] [Google Scholar]
- 19.Xu, T., Gao, X., Wu, Z., Selinger, D. W. & Zhou, Z. Indomethacin has a potent antiviral activity against SARS CoV-2 in vitro and canine coronavirus in vivo. bioRxiv. 10.1101/2020.04.01.017624 (2020).
- 20.Russell, B., Moss, C., Rigg, A. & Van Hemelrijck, M. COVID-19 and treatment with NSAIDs and corticosteroids: Should we be limiting their use in the clinical setting? ecancermedicalscience. 10.3332/ecancer.2020.1023 (2020). [DOI] [PMC free article] [PubMed]
- 21.Oh KK, Adnan M, Ju I, Cho DH. A network pharmacology study on main chemical compounds from Hibiscus cannabinus L. leaves. RSC Adv. 2021;11:11062–11082. doi: 10.1039/D0RA10932K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh KK, Adnan M, Cho DH. Network pharmacology of bioactives from Sorghum bicolor with targets related to diabetes mellitus. PLoS ONE. 2020;15:e0240873. doi: 10.1371/journal.pone.0240873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh KK, Adnan M, Cho DH. Active ingredients and mechanisms of Phellinus linteus (grown on Rosa multiflora) for alleviation of Type 2 diabetes mellitus through network pharmacology. Gene. 2020 doi: 10.1016/j.gene.2020.145320. [DOI] [PubMed] [Google Scholar]
- 24.Oh, K. K., Adnan, M. & Cho, D. H. Network pharmacology approach to bioactive chemical compounds identified from Lespedeza bicolor lignum methanol extract by GC–MS for amelioration of hepatitis. Gene Rep. 10.1016/j.genrep.2020.100851 (2020).
- 25.Belvisi MG, Mitchell JA. Targeting PPAR receptors in the airway for the treatment of inflammatory lung disease. Br. J. Pharmacol. 2009;158:994–1003. doi: 10.1111/j.1476-5381.2009.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 2004;319:1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Souza, A. P. et al. MAPK involvement in cytokine production in response to Corynebacterium pseudotuberculosis infection. BMC Microbiol.14, 1–9 (2014). [DOI] [PMC free article] [PubMed]
- 28.Ryzhov S, et al. ERBB signaling attenuates proinflammatory activation of nonclassical monocytes. Am. J. Physiol. Heart Circ. Physiol. 2017;312:H907–H918. doi: 10.1152/ajpheart.00486.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bian J, Li Z. Angiotensin-converting enzyme 2 (ACE2): SARS-CoV-2 receptor and RAS modulator. Acta Pharmaceutica Sinica B. 2021;11:1–12. doi: 10.1016/j.apsb.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin S, et al. Activation of cGMP/PKG/p65 signaling associated with PDE5—Is downregulates CCL5 secretion by CD8+ T cells in benign prostatic hyperplasia. Prostate. 2019;79:909–919. doi: 10.1002/pros.23801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan K, Gao LN, Cui YL, Zhang Y, Zhou X. The cyclic AMP signaling pathway: Exploring targets for successful drug discovery (review) Mol. Med. Rep. 2016;13:3715–3723. doi: 10.3892/mmr.2016.5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeDiego ML, et al. Inhibition of NF- B-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014;88:913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan JA, Kim EH, Plisch EH, Suresh M. FOXO3 Regulates the CD8 T cell response to a chronic viral infection. J. Virol. 2012;86:9025–9034. doi: 10.1128/JVI.00942-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abu-Farha, M. et al. The role of lipid metabolism in COVID-19 virus infection and as a drug target. Int. J. Mol. Sci.21, 1–11 (2020). [DOI] [PMC free article] [PubMed]
- 35.Moparthi L, Koch S. Wnt signaling in intestinal inflammation. Differentiation. 2019;108:24–32. doi: 10.1016/j.diff.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Turkia M. COVID-19 as an endothelial disease and its relationship to vascular endothelial growth factor (VEGF) and iodide. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3604987. [DOI] [Google Scholar]
- 37.Kawasaki, T. & Kawai, T. Toll-like receptor signaling pathways. Front. Immunol.5, 1–8 (2014). [DOI] [PMC free article] [PubMed]
- 38.Ahmadabad RA, Ghadiri MK, Gorji A. The role of Toll-like receptor signaling pathways in cerebrovascular disorders: The impact of spreading depolarization. J. Neuroinflamm. 2020;17:1–13. doi: 10.1186/s12974-019-1655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carneiro LAM, Travassos LH, Girardin SE. Nod-like receptors in innate immunity and inflammatory diseases. Ann. Med. 2007;39:581–593. doi: 10.1080/07853890701576172. [DOI] [PubMed] [Google Scholar]
- 40.Chan YK, Gack MU. RIG-I-like receptor regulation in virus infection and immunity. Curr. Opin. Virol. 2015;12:7–14. doi: 10.1016/j.coviro.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pacha O, Sallman MA, Evans SE. COVID-19: A case for inhibiting IL-17? Nat. Rev. Immunol. 2020;20:345–346. doi: 10.1038/s41577-020-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klemm S, Ruland J. Inflammatory signal transduction from the FcεRI to NF-κB. Immunobiology. 2006;211:815–820. doi: 10.1016/j.imbio.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Kels MJT, et al. TNF deficiency dysregulates inflammatory cytokine production, leading to lung pathology and death during respiratory poxvirus infection. Proc. Natl. Acad. Sci. USA. 2020;117:15935–15946. doi: 10.1073/pnas.2004615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jha NK, et al. Evidence of coronavirus (CoV) pathogenesis and emerging pathogen SARS-CoV-2 in the nervous system: A review on neurological impairments and manifestations. J. Mol. Neurosci. 2021;17:20. doi: 10.1007/s12031-020-01767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riley CP, Cope TC, Buck CR. CNS neurotrophins are biologically active and expressed by multiple cell types. J. Mol. Histol. 2004;35:771–783. doi: 10.1007/s10735-004-0778-9. [DOI] [PubMed] [Google Scholar]
- 46.Shimobayashi M, et al. Insulin resistance causes inflammation in adipose tissue. J. Clin. Investig. 2018;128:1538–1550. doi: 10.1172/JCI96139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barabás, K., Szabó-Meleg, E. & Ábrahám, I. M. Effect of inflammation on female gonadotropin-releasing hormone (GnRH) neurons: Mechanisms and consequences. Int. J. Mol. Sci.21, 1–17 (2020). [DOI] [PMC free article] [PubMed]
- 48.Acharya S, Fernando JJR, Gama R. Gynaecomastia, hyperprolactinaemia and HIV infection. Ann. Clin. Biochem. 2005;42:301–303. doi: 10.1258/0004563054255542. [DOI] [PubMed] [Google Scholar]
- 49.Borba VV, Zandman-Goddard G, Shoenfeld Y. Prolactin and autoimmunity: The hormone as an inflammatory cytokine. Best Pract. Res. Clin. Endocrinol. Metab. 2019;33:101324. doi: 10.1016/j.beem.2019.101324. [DOI] [PubMed] [Google Scholar]
- 50.Ruscitti, P. et al. Adipocytokines in rheumatoid arthritis: The hidden link between inflammation and cardiometabolic comorbidities. J. Immunol. Res.2018, 1–10 (2018). [DOI] [PMC free article] [PubMed]
- 51.Inoue T, et al. Oxytocin suppresses inflammatory responses induced by lipopolysaccharide through inhibition of the eIF-2α–ATF4 pathway in mouse microglia. Cells. 2019 doi: 10.3390/cells8060527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin B, et al. Relaxin reverses inflammatory and immune signals in aged hearts. PLoS ONE. 2018 doi: 10.1371/journal.pone.0190935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramasamy R, Yan SF, Schmidt AM. Receptor for AGE (RAGE): Signaling mechanisms in the pathogenesis of diabetes and its complications. Ann. N. Y. Acad. Sci. 2011;1243:88–102. doi: 10.1111/j.1749-6632.2011.06320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reyes, V. E. & Peniche, A. G. Helicobacter pylori deregulates T and B cell signaling to trigger immune evasion. in Current Topics in Microbiology and Immunology. 421, 229–265 (2019). [DOI] [PMC free article] [PubMed]
- 55.Gupta A, et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 56.Datta PK, Liu F, Fischer T, Rappaport J, Qin X. SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy. Theranostics. 2020;10:7448–7464. doi: 10.7150/thno.48076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaduganathan M, et al. Renin–angiotensin–aldosterone system inhibitors in patients with COVID-19. N. Engl. J. Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sama IE, et al. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin–angiotensin–aldosterone inhibitors. Eur. Heart J. 2020;41:1810–1817. doi: 10.1093/eurheartj/ehaa373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bloch MJ. Renin-angiotensin system blockade in COVID-19: Good, bad, or indifferent? J. Am. Coll. Cardiol. 2020;76:277. doi: 10.1016/j.jacc.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mascolo, A. et al. Renin-angiotensin system and Coronavirus disease 2019: A narrative review. Front. Cardiovasc. Med.7, 1–13 (2020). [DOI] [PMC free article] [PubMed]
- 61.Xu X, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. USA. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun ML, Yang JM, Sun YP, Su GH. Inhibitors of RAS might be a good choice for the therapy of COVID-19 pneumonia. Zhonghua jie he he hu xi za zhi = Zhonghua jiehe he huxi zazhi = Chin. J. Tuberculosis Respir. Diseases. 2020;43:219–222. doi: 10.3760/cma.j.issn.1001-0939.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 63.Grimes JM, Grimes KV. p38 MAPK inhibition: A promising therapeutic approach for COVID-19. J. Mol. Cell. Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bulanova D, et al. Antiviral properties of chemical inhibitors of cellular anti-apoptotic Bcl-2 proteins. Viruses. 2017;9:271. doi: 10.3390/v9100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian B, et al. Bcl-2 inhibitors reduce steroid-insensitive airway inflammation. J. Allergy Clin. Immunol. 2017;140:418–430. doi: 10.1016/j.jaci.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 66.Little, P. Non-steroidal anti-inflammatory drugs and COVID-19. BMJ 10.1136/bmj.m1185 (2020) [DOI] [PubMed]
- 67.Reynolds AE, Enquist LW. Biological interactions between herpesviruses and cyclooxygenase enzymes. Rev. Med. Virol. 2006;16:393–403. doi: 10.1002/rmv.519. [DOI] [PubMed] [Google Scholar]
- 68.Ray N, Bisher ME, Enquist LW. Cyclooxygenase-1 and-2 are required for production of infectious pseudorabies virus. J. Virol. 2004;78:12964–12974. doi: 10.1128/JVI.78.23.12964-12974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schröer J, Shenk T. Inhibition of cyclooxygenase activity blocks cell-to-cell spread of human cytomegalovirus. Proc. Natl. Acad. Sci. 2008;105:19468–19473. doi: 10.1073/pnas.0810740105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bahrami H, et al. Effects of indomethacin on viral replication markers in asymptomatic carriers of hepatitis B: A randomized, placebo-controlled trial. Am. J. Gastroenterol. 2005;100:856–861. doi: 10.1111/j.1572-0241.2005.41144.x. [DOI] [PubMed] [Google Scholar]
- 71.Chen N, Warner JL, Reiss CS. NSAID treatment suppresses VSV propagation in mouse CNS. Virology. 2000;276:44–51. doi: 10.1006/viro.2000.0562. [DOI] [PubMed] [Google Scholar]
- 72.Rossen JWA, Bouma J, Raatgeep RHC, Büller HA, Einerhand AWC. Inhibition of cyclooxygenase activity reduces rotavirus infection at a postbinding step. J. Virol. 2004;78:9721–9730. doi: 10.1128/JVI.78.18.9721-9730.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Melarange R, et al. Anti-inflammatory and gastrointestinal effects of nabumetone or its active metabolite, 6MNA (6-methoxy-2-naphthylacetic acid): Comparison with indomethacin. Agents Actions. 1992;36:C82–C83. doi: 10.1007/BF01996102. [DOI] [PubMed] [Google Scholar]
- 74.Van Der Heide HJL, Rijnberg WJ, Van Sorge A, Van Kampen A, Schreurs BW. Similar effects of rofecoxib and indomethacin on the incidence of heterotopic ossification after hip arthroplasty. Acta Orthop. 2007;78:90–94. doi: 10.1080/17453670610013475. [DOI] [PubMed] [Google Scholar]
- 75.Gomeni R, Xu T, Gao X, Bressolle-Gomeni F. Model based approach for estimating the dosage regimen of indomethacin a potential antiviral treatment of patients infected with SARS CoV-2. J. Pharmacokinet Pharmacodyn. 2020;47:189–198. doi: 10.1007/s10928-020-09690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keiser MJ, et al. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 77.Gfeller D, Michielin O, Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics. 2013;29:3073–3079. doi: 10.1093/bioinformatics/btt540. [DOI] [PubMed] [Google Scholar]
- 78.Chittepu, V. C. S. R., Kalhotra, P., Osorio-Gallardo, T., Gallardo-Velázquez, T. & Osorio-Revilla, G. Repurposing of FDA-approved NSAIDs for DPP-4 inhibition as an alternative for diabetes mellitus treatment: Computational and in vitro study. Pharmaceutics.11, 1–13 (2019). [DOI] [PMC free article] [PubMed]
- 79.Matsson P, Kihlberg J. How big is too big for cell permeability? J. Med. Chem. 2017;60:1662–1664. doi: 10.1021/acs.jmedchem.7b00237. [DOI] [PubMed] [Google Scholar]
- 80.Szklarczyk D, et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khanal P, Patil BM, Chand J, Naaz Y. Anthraquinone derivatives as an immune booster and their therapeutic option against COVID-19. Nat. Products Bioprospect. 2020;10:325–335. doi: 10.1007/s13659-020-00260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its “Supplementary Information S1” files).