Abstract
Background
The Glycine N-acyltransferase (GLYAT) gene encodes a protein that catalyzes the transfer of acyl groups from acyl CoA to glycine, resulting in acyl glycine and coenzyme A. Aberrant GLYAT expression is associated with several malignant tumors, but its clinical importance in human breast cancer (BC), has yet to be fully addressed. This study aims to evaluate the clinical function of GLYAT in BC patients.
Methods
GLYAT expression was determined by immune blot and immunohistochemistry in three BC cell lines and primary cancer tissues. The MDA-MB 231 cell line was used for GLYAT gene knockdown experiments while the MCF7 cell line for overexpression experiments. Colony formation experiments, soft agar experiments, and transwell assays were utilized for further inspection of cell proliferation and migration capabilities. Immunofluorescence and western blot were used to detect markers of the epithelial-mesenchymal transition (EMT) and changes in the PI3K/AKT/Snail pathway. The role of GLYAT in tumor growth and metastasis was also assessed in nude mice in vivo. Also, a correlation analysis was performed between clinicopathological features and GLYAT expression in BC patients.
Results
GLYAT was decreased in human BC tissues and cell lines. Functional analysis showed that knockdown of GLYAT augmented BC cell proliferation in vitro and in vivo. However, this phenomenon was reversed when GLYAT was overexpressed in the transfected cells. Moreover, downregulation of GLYAT promoted the migratory properties of BC cells, likely through the activation of PI3K/AKT/Snail signaling, which subsequently induced the EMT. IHC analysis indicated that GLYAT was decreased in human BC tissues and lower GLYAT expression was correlated with histological grade, tumor TNM stage, Ki-67 status, and poorer survival in BC patients. Furthermore, lower GLYAT expression seemed as an independent risk factor related to poor prognosis in BC patients based on Cox regression analyses.
Conclusion
Our findings demonstrate that downregulation of GLYAT expression in human breast cancer is correlated with EMT via the PI3K/AKT/Snail pathway and is also associated with histological grade, tumor TNM stage, Ki-67 status, and poor survival in breast cancer patients.
Keywords: GLYAT, breast cancer, EMT, PI3K/AKT, clinicopathological features, prognosis
Introduction
Breast cancer (BC) amongst women has been on the rise and it is fast becoming the most common malignancy in this population (1). Leading cancer statistics show that the incidence and mortality of breast cancer remain high worldwide (2, 3). Although surgery, chemoradiotherapy, endocrine therapy, and molecular targeted therapy have greatly improved the survival of BC patients, those with advanced BC are often left with very limited therapeutic options (4, 5). Many factors influence the prognosis and probability of responding to systemic therapies (e.g., TNM stage, ER, PR, and HER2 status), but clinically, we have observed that even patients with the same TNM stage, molecular typing, and other diagnostic markers still experience varied prognosis despite being given the same treatment modalities (4). While early diagnosis and initiation of treatment are crucial in improving survival rates of this debilitating disease, there is an urgent need for a deeper understanding of its molecular biology in order to aid in the development of more personalized and targeted treatment.
The EMT involves the dedifferentiation of epithelial cells into mesenchymal cells (6, 7). Cells which have undergone EMT demonstrate more malignant features that promote metastasis, treatment resistance, as well as relapse (8–10). The EMT participates in tumor invasion and metastasis through multiple pathways. Many studies have indicated the EMT is strongly linked to breast cancer development (11–13). The EMT is regulated by multiple factors, among which the primary signaling pathway is the PI3K/AKT pathway (14).
The glycine N-acyltransferase (GLYAT) gene was first discovered in the mitochondria of the bovine liver in 1953 and isolated in the human liver and kidneys in 1976 (15). It contains more than 23,000 base pairs over six exons and is situated chromosome 11 at position 11q12 (16). GLYAT catalyzes acyl group transfer from acyl CoA to glycine, resulting in acyl glycine and coenzyme A (acyl-CoA) (17). Several catabolic and anabolic reactions are catalyzed by acyl-CoA esters (18). Almost all catabolic reactions produce acyl-CoA, the product of which is an important source of oxidative phosphorylation and lipogenesis. Studies have shown that GLYAT expression is suppressed in human hepatocellular carcinomas and may be a critical molecule in the transition between the differentiation and carcinogenesis of liver cells (19). Nevertheless, literature is scarce surrounding GLYAT expression and its impact on human breast cancer.
The current investigation uncovered markedly suppressed GLYAT expression in breast cancer cells and tissues, which correlated with poorer prognosis and highly malignant clinicopathologic features in individuals with breast cancer. Interestingly, the EMT pathway appeared to be inhibited in the presence of GLYAT, working to decrease in vivo and in vitro breast cancer cell migration through alteration of the PI3K/ATK/Snail signaling pathway. In conclusion, our study is the first of its kind to implicate GLYAT to be a breast cancer anti-oncogene. These findings lay the foundation for future research on the biological behavior and targeted therapy of BC.
Materials and Methods
Datasets and GLYAT Expression Analysis
The public data used in this study were obtained from UALCAN dataset (http://ualcan.path.uab.edu/index.html) (20), GEPIA (http://gepia.cancer-pku.cn/index.html) (21), and the Human Protein Atlas dataset (http://www.proteinatlas.org) (22). For UALCAN data, we analyzed the heat map of GLYAT expression between normal and breast cancer samples; For GEPIA data, we scrutinized differently GLYAT expression level and survival data in breast cancer. The relationship between patient survival and GLYAT levels were also analyzed using information available from the Human Protein Atlas data.
Cell Lines
Human BC cell lines MDA-MB-231, MCF-7, and SKRB-3 were supplied by The Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China). Cells were maintained at 37°C in a humidified cubicle containing 5% CO2 and 10% fetal bovine serum (FBS) in DMEM (all from Gibco, Carlsbad, CA, USA).
Plasmid Formation of GLYAT Knockdown and Overexpression in Breast Cancer Cells
The pGPU6/mCherry/Puro-shRNA-GLYAT plasmid was purchased from GenePharma Company (Shanghai, China). The pOGP-T2A-CKNeo-GLYAT overexpression plasmid was purchased from JTS Scientific Company (Wuhan, China). The target sequences of GLYAT were indicated in Supplementary Table 1 . Lipofectamine 3000 (Thermo Fisher Scientific, US) was used to transfect plasmids into cells as reported by the manufacturer’s instructions. Cells transfected with GPU6/mCherry/Puro-shNC or pOGP-T2A-CKNeo-NC were used as controls. Transfected cells were screened using 3 ug/mL puromycin or 400 ug/mL G418 for 2 weeks. Stable cells were cultured in complete medium with 0.25 ug/mL puromycin or 100 ug/mL G418 (Beyotime, Nanjing, China). Positive clones were then selected and amplified for further analyses.
Cell Proliferation Assay
For colony forming experiments, six-well plates were used to house stable GLYAT-shRNA and NC cells. The cells were maintained at a concentration of 500 cells/well for 12 days in DMEM supplemented with 10% FBS, with media changed once every three days. Cells were then methanol-fixed and treated with crystal violet (Sigma, St Louis, MO, USA) prior to manual counting and photographing of the visible colonies. For soft agar experiments, Cells are harvested and pipetted well to become single-cell suspension in complete culture media in 1x 106/ml. A mixture of 0.9 ml 4% soft-agar (Sigma) with 4.1 ml pre-warmed 10% FBS DMEM was added into a 60-mm culture dish to make the bottom layer. The top layer contained 3 x 104 cells in 3 ml of 10% FBS DMEM and 0.36% agar. The soft-agar colony dish was marked and placed at a 37°C incubator for 3 weeks. cells were harvested and fully mixed to form a single-cell suspension in media of 1 x 106/ml. The upper layer included 3 x 104 cells, 3 ml of 10% fetal bovine serum, DMEM, combined with 0.36% agar (Sigma), while the bottom layer contained 0.9 ml 4% soft-agar, 4.1 ml pre-heated 10% fetal bovine serum, and DMEM in a 60-mm culture dish. Labeled colony dishes were placed in an incubator at 37°C for 3 weeks. Ten fields of each group were selected and the colony size was measured using Image J software (vision 1.53 National Institutes of Health, USA). All experiments were performed in triplicate and repeated at least three times.
Transwell Migration Assay
Costar chambers containing 8 μm pore inserts were used (Millipore-Sigma, Danvers, MA, USA). The top chamber was used to house suspended cells transfected with GLYAT KD, GLYAT OE, or NC in 200 μl of serum-free medium, while media with 20% FBS was applied to the bottom chamber. The upper chamber was removed after 36 hours of incubation, with the bottom cartridge fixed with methanol and DAPI-stained to determine the migrated number of cells using a microscope to visualize the cells across five randomly selected fields (Olympus, Tokyo, Japan). Cell numbers were quantified manually.
Western Blot Analysis
The proteins in BC cells were first extracted and separated. Skimmed milk (5%) was used to block endogenous reactions. Membranes were left to incubate with antibodies of GLYAT (1:1000, Abcam, Cambridge, MA, USA), E-cadherin, vimentin, N-cadherin, fibronectin (1:1000 dilution respectively, Cell Signaling Technology, Boston, MA, USA), p-AKT Ser473, AKT (1:1000 and 1:5000 dilution, Signalway Antibody, Baltimore, MD, USA), PI3K p58 and SNAI1 (1:10000 and 1:1000 dilution, Proteintech, Rosemont, IL, USA) in agreement with the protocols set by the manufacturer. Anti-β-actin (1:10000 dilution, Proteintech, Rosemont, IL, USA) was used as the internal control. After three rinses with TBS-T for 5 minutes, the membranes were incubated with the corresponding secondary antibody and observed with an ECL Plus kit. The relative levels of individual target proteins to the control were determined by the densitometric analysis using the ImageJ software.
Immunofluorescence Staining
Cells were subjected into 8 well chamber slides (Millipore-Sigma, Danvers, MA, USA) and for 20 hours incubation then rinsed with PBS containing 10% FBS. Pre-cooled methanol was used to fix the cells prior to further incubation with 0.2% Triton. BSA (5%) was used to block cells. The cells were next incubated overnight with primary antibodies E-cadherin, vimentin (1:200 dilution respectively) before being washed and re-incubated with fluorescein-conjugated secondary antibodies (1:500 ZSGB-Bio, Beijing China) for one hour. Then, DAPI and a fluorescence microscope (Olympus, Tokyo, Japan) were used for visualization and intensity analysis. For dewaxed sections, the process was similar like above, and primary antibodies dilution of E-cadherin, vimentin and p-AKT Ser473 were 1:100 respectively.
In Vivo Metastasis Assay
The HFK BIOSCIENCE CO., LTD (Beijing, China) provided 5 weeks old BALB/c (nu/nu) female nude mice (18–20 g). These animals were divided into four cohorts with five mice each. Food and drinking water were provided every day. All mice received subcutaneous injections into their left breast fat pads with 100 microliters of PBS containing 2.0 × 106 MCF7 cells with or without GLYAT OE and MDA-MB-231 cells with or without GLYAT KD. At time of MCF7 cells injection, E2 pellets (60-day release, 1.5 mg/pellet; Innovative Research of America, Sarasota, Florida USA) was implanted subcutaneously at the mammary fat pad. Four days measurements and volume calculation ((mm3) =length × width2/2) of the tumors were performed. The mice were sacrificed and underwent tumor removal after 24 days. The tumors were processed with 4% paraformaldehyde solution for immunofluorescence staining analysis.
Patients and Samples
Two independent breast cancer patient cohorts from the Shengjing Hospital of China Medical University were collected for this investigation. The first cohort consisted of 21 breast cancer patients (seven for Luminal A/B, seven for HER2 positive, and seven for TNBC) and 20 normal controls. Samples from the cancer patients and normal controls were analyzed for GLYAT expression. The second cohort comprised of 310 chemoradiotherapy-naïve breast cancer patients who had distinctive pathological diagnoses and possessed complete follow-up data from 2006 to 2008. Additional follow-up of these patients was carried out until September 2012.
Immunohistochemistry
The dewaxed sections were retrieved with Tris/EDTA (pH9.0) and quenched with 0.3% H2O2. Sections then underwent an overnight incubation with GLYAT primary antibody (1:100, Abcam, Cambridge, MA, USA). PBS was used to rinse the samples prior to repeat incubation with secondary biotinylated antibodies at room temperature for 45 minutes. Coloration was visualized with 3,3 ‘-diaminobenzidine tetrachoric acid (Sigma-Aldrich), Vector hematoxylin QS (Vector Laboratories) was reverse-stained, Zeiss Mirax MidiSlide scanner was analyzed, and image collection was performed with 3 CCD color cameras and Panoramic Audience (3DHISTECH, Budapest Hungary). H-score was calculated as the percentage of positive tumor cells multiplied by the intensity of staining (0, no staining; 1, weak; 2, mild to moderate; 3, strong staining) and the overall H-score ranged from 0 to 300. An H-score of more than 100 was set as cut-off value to define high/low level for GLYAT expression.
Statistical Analysis
Data were analyzed using SPSS version 25.0 (SPSS, Inc., Chicago, IL, USA) and expressed as means ± SD. The Kaplan-Meier method was used to estimate survival curves. Cox regression model was applied to carry out univariate and multivariate statistical analysis. Comparison between two groups was assessed by Chi-square. P value of less than 0.05 was considered statistically significant.
Results
Characterization of GLYAT Expression Profiling in Human BC
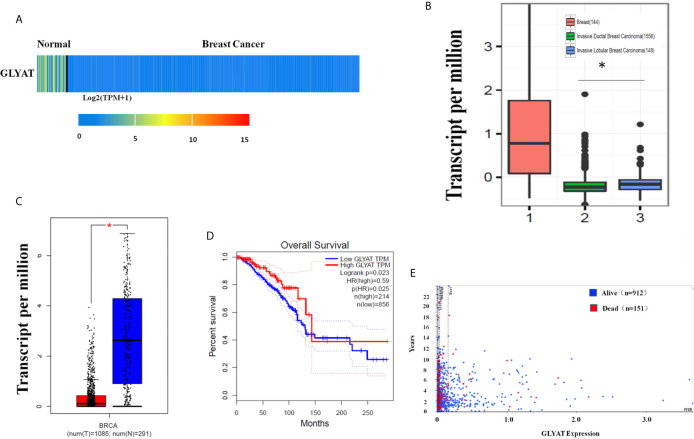
The heat map and box plot analysis of the UALCAN and GEPIA dataset showed lower expression of GLYAT protein and mRNA in BC tissues in contrast to healthy samples ( Figures 1A, C ). However, the expression of GLYAT was not statistically significant change in other cancer types, besides BRCA, LIHC and CHOL ( Supplementary Figure 1 ), which indicated that GLYAT may have specific function in breast cancer. Additionally, analysis of data published in another study in Nature (23) which compared mRNA expression levels of GLYAT in normal breast tissues with invasive ductal and invasive lobular carcinoma revealed that two breast carcinoma subtypes had lower GLYAT mRNA levels ( Figure 1B ). A Kaplan-Meier analysis from GEPIA dataset and a scatter plot from the Human Protein Atlas dataset suggested that patients with low GLYAT expression in breast cancer tissues had worse overall survival (OS) ( Figures 1D, E ), further highlighting the suppressive function of GLYAT in BC progression.
Figure 1.
Characterization of GLYAT expression profiling in human breast cancer. (A) Heatmaps showing the clustering patterns of differentially expressed GLYAT between normal and breast cancer specimens. (B) Box plot analysis of the GLYAT mRNA levels in IDBC, ILBC and normal breast tissue samples. (C) Box plot analysis of the GLYAT mRNA levels in BC and NC tissues. T, tumor; N, normal. (D, E) Kaplan-Meier plots of OS in BC patients with different levels of GLYAT. *p < 0.05.
GLYAT Suppresses BC Cell Proliferation and Metastasis
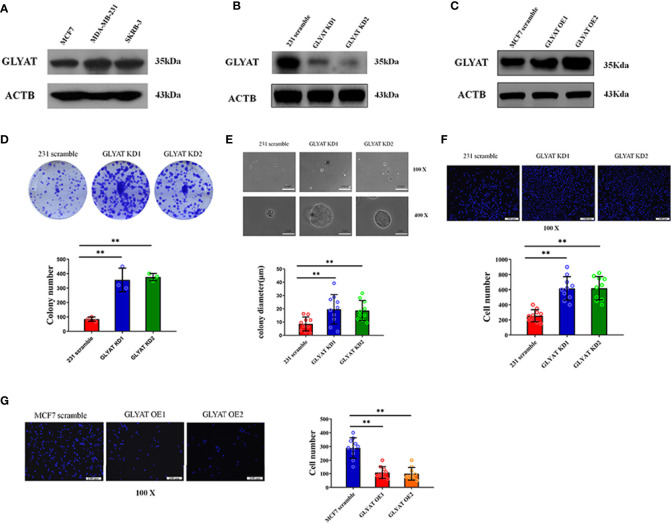
Three typical breast cancer cell lines were scrutinized for GLYAT expression. These cell lines included SK-BR3 (Her-2 positive subtype), MDA-MB-231 (triple negative subtype) and MCF-7 (luminal A subtype). Western blotting showed that the MDA-MB-231 cell line had the highest GLYAT expression, while the MCF-7 cell line had the lowest amount ( Figure 2A ). Therefore, these two cell lines above were selected for further experiments.
Figure 2.
GLYAT suppresses BC cell proliferation and metastasis. (A) GLYAT protein level was assessed in different BC cell lines. (B) MDA-MB-231 cells were transfected with GLYAT KD plasmid and a scrambled plasmid as control. The suppression of GLYAT in MDA-MB-231 cells was confirmed at the protein level. (C) MCF-7 cells were transfected with GLYAT OE plasmid and a scrambled plasmid as control. The over expression of GLYAT in MCF-7 cells was confirmed at the protein level. (D, E) GLYAT KD in MDA-MB-231 cells significantly increased colony numbers and size compared with the scramble cells. (F) Transwell assay revealed that the migration ability of MDA-MB-231 cells was significantly increased following transfection with GLYAT KD1 and KD2 compared with the scramble control. (G) Transwell assay revealed that the migratory ability was significantly decreased in MCF-7 cells GLYAT OE compared with scramble control. **p < 0.01. Scale bars for (E) are 100μm and 25μm, and for (F, G) are 100μm.
In vitro assays were carried out in MDA-MB-231 cells with GLYAT knockdown (KD) and overexpression (OE) in MCF-7 cells to determine the impact of GLYAT expression on breast cancer behavior ( Figures 2B, C ). The tablet colony formation and soft agar colony formation experiments revealed that GLYAT KD in MDA-MB-231 cells significantly increased colony numbers and size in contrast to control cell lines ( Figures 2D, E ). Transwell migration assay revealed artificially altered MDA-MB-231 cells with GLYAT KD also had stronger migratory abilities in contrast to the control cells ( Figure 2F ). In line with these results, the converse was seen in MCF-7 cells with OE GLYAT ( Figure 2G ). These results indicate that GLYAT may hinder the proliferative and metastatic abilities of breast cancer cells.
GLYAT Suppresses the EMT Phenotype in BC Cells
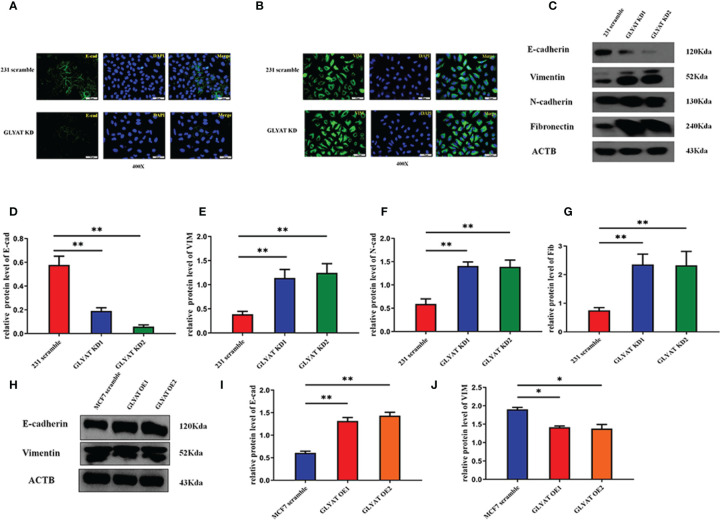
Given the prominent role of EMT in tumor spread, we postulated that GLYAT may be involved in the EMT process. In vitro assays were performed in MDA-MB-231 with GLYAT KD and in MCF with GLYAT OE. Both vimentin and E-cadherin were analyzed for their nucleic location via immunofluorescence assays. In GLYAT KD cells, we found an increase in vimentin but a decrease in E-cadherin expression ( Figures 3A, B ). Western blotting of EMT-related proteins was consistent with the immunofluorescence results, showing a reduction in E-cadherin, but increase in levels of vimentin, N-cadherin, and fibronectin in MDA-MB-231 GLYAT KD cells ( Figures 3C–G ). On the contrary, MCF-7 GLYAT OE cells demonstrated raised E-cadherin protein expression and lowered vimentin levels ( Figures 3H–J ). These results suggest that EMT formation could be suppressed by GLYAT.
Figure 3.
GLYAT suppress EMT phenotype in BC cells. (A, B) Immunofluorescence assay showed that E-cadherin expression was decreased and the vimentin expression was increased after the treatment of GLYAT KD. (C–G) Protein levels of E-cadherin was reduced, whereas the levels of Vimentin, N-cadherin, and Fibronectin were increased in MDA-MB-231 cells following GLYAT inhibition. (H–J) Protein levels of E-cadherin was increased whereas the expression of Vimentin was reduced in GLYAT OE MCF-7 cells. *p < 0.05, **p < 0.01. Scale bars are 25μm.
GLYAT Suppresses the EMT of BC Cells via the PI3K/Akt/Snail Pathway
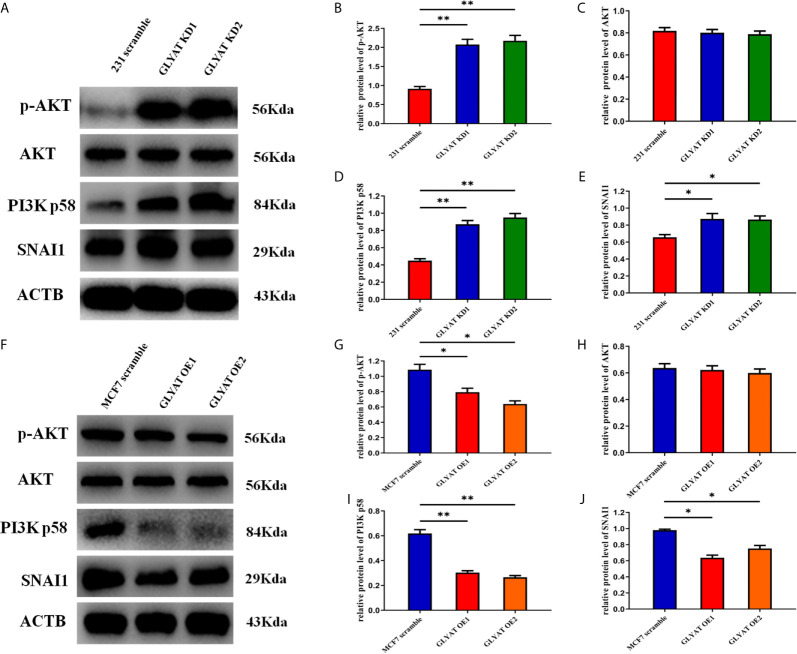
EMT process is highly regulated by the PI3k/Akt/Snail pathway. We therefore hypothesized that GLYAT might suppress breast cancer metastasis by modulation of this pathway. Western blot assays indicated that p-AKT, AKT, PI3K p58, and SNAI1 were significantly activated in cells with GLYAT KD ( Figures 4A–E ). OE cells on the other hand, demonstrated markedly suppressed levels of p-AKT, AKT, PI3K p58, and SNAI1 in contrast to control cells ( Figures 4F–J ). Our results indicate the involvement of the PI3K/Akt/Snail signaling pathway in GLYAT-mediated EMT suppression in BC cells.
Figure 4.
GLYAT regulated breast cancer cells EMT progression through PI3K/Akt signaling Pathway. (A–E) Protein levels of p-AKT,AKT, PI3K and SNAI1 were significantly activated in GLYAT KD MDA-MB-231 cells. (F–J) Protein levels of p-AKT,AKT, p-PI3K and SNAI1 were significantly reduced in GLYAT over expressed OE MF-7 cells. *p < 0.05, **p < 0.01, and p < 0.05 was considered to be statistically significant.
GLYAT Suppresses In Vivo BC EMT, Proliferation and Metastasis
In vivo experiments were performed on nude mice that were received subcutaneous injection with either stable MDA-MB-231 GLYAT KD or MCF-7 GLYAT OE cells ( Figure 5A ). The mice injected with stable GLYAT KD cells developed bigger and heavier tumors ( Figures 5B–D ). In contrast, the mice injected with stable GLYAT OE MCF-7 cells had markedly smaller and lighter tumors ( Figures 5E–G ). Then immunofluorescence assays for E-cadherin, vimentin, and p-AKT were performed using serial sections of mouse tumor tissues. These assays revealed that GLYAT silencing in MDA-MB-231 cells significantly decreased E-cadherin levels, but increased vimentin and p-AKT levels ( Figure 5H ). In contrast, in GLYAT OE MCF-7 cells, E-cadherin was increased, whereas vimentin and p-AKT were decreased ( Figure 5I ).
Figure 5.
GLYAT suppress breast cancer proliferation, metastasis and EMT in vivo. (A) Schematic diagram of the metastasis model in mice. (B–D) The mice injected with stable GLYAT KD MDA-MB-231 cells had markedly bigger and heavier tumors. (E–G) The mice injected with stable GLYAT OE MCF-7 cells had markedly smaller and lighter tumors. (H) Immunofluorescence assay of serial sections mouse tumor tissues revealed that GLYAT KD MDA-MB-231 cells significantly decreased the expression of E-cadherin whereas increased the expression of Vimentin and p-AKT. (I) Immunofluorescence assay of serial sections mouse tumor tissues revealed that, in GLYAT OE MCF-7 cells, the expression of E-cadherin was increased whereas Vimentin and p-AKT were decreased. (J) The schematic diagram of the role of GLYAT in BC. *p < 0.05, **p < 0.01. Scale bars are 50μm.
Lower GLYAT Expression Is Correlated With Poorer Prognosis and Malignant Clinicopathological Features in Human Breast Cancer Tissues
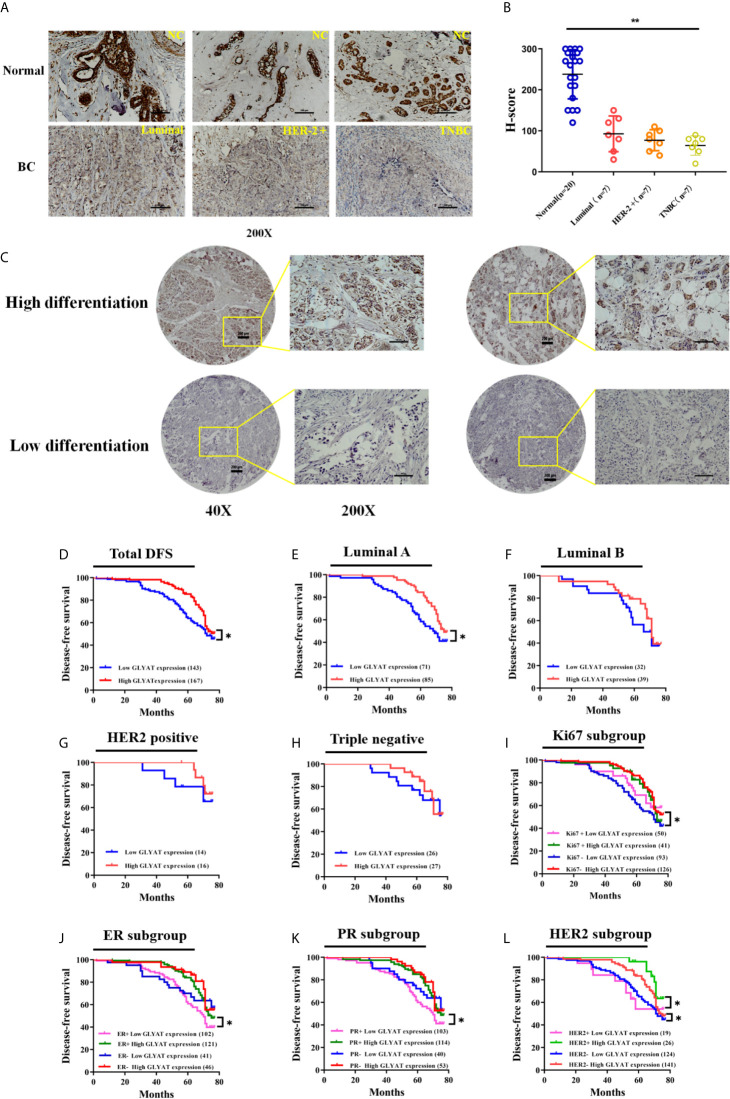
Immunohistochemical experiments found that breast cancer tissues possessed markedly decreased GLYAT expression in contrast to healthy breast tissue ( Figures 6A, B ). Additionally, GLYAT expression in BC tissues correlated positively with the degree of pathological differentiation. ( Figure 6C ). Based on these results, we conclude GLYAT may act as an anti-oncogene in BC.
Figure 6.
GLYAT is less expressed in BC tissues and correlated with worse survival. (A, B) Immunohistochemical staining showed that GLYAT was downregulated in BC tissues compared with NC. (C) Immunohistochemical staining showed that the lower differentiation of the breast cancer, the less expressed of GLYAT in breast cancer tissues. (D) Kaplan-Meier analysis of DFS in BC patients with different levels of GLYAT. (E–H) Kaplan-Meier analysis of DFS in luminal A, luminal B, HER2 positive and triple negative BC patients with different levels of GLYAT. (I–L) Kaplan-Meier analysis for DFS by Ki67, ER, PR, HER2 status in BC patients with different levels of GLYAT. *p < 0.05, **p < 0.01. Scale bars for (A) are 200μm, and for (B) are 50μm and 200μm.
We then studied the correlation between clinicopathological features and GLYAT expression in 310 patients who were recruited for this investigation. Table 1 demonstrates their characteristics. All individuals were women between 29 to 78 years old. No one had a previous history of malignancy and were chemoradiotherapy-naïve. The patients were grouped based on their GLYAT levels-low (n=143) or high (n=167) expression groups. We found GLYAT expression correlated with TNM stage, histological grade, and Ki-67 status ( Table 1 ; all P<0.05). Additionally, we did not uncover any relationship between GLYAT level and menopausal status, age, receptor status (HER2, ER, PR), tumor size, node status, pathologic or molecular subtype ( Table 1 ; all P>0.05). These findings suggest GLYAT is associated with malignant clinicopathological characters of BC.
Table 1.
Correlation between GLYAT expression level and clinicopathological features in patients with breast cancer.
| Clinical characteristics | No. (n = 310) | GLYAT | P value | |
|---|---|---|---|---|
| Low (143) | High (167) | |||
| Age (years) | ||||
| <50 | 139 | 59 | 80 | 0.241 |
| ≥50 | 171 | 84 | 87 | |
| Menopausal status | ||||
| Premenopausal | 165 | 73 | 92 | 0.477 |
| Postmenopausal | 145 | 70 | 75 | |
| Tumor Size (cm) | ||||
| ≤2 | 193 | 93 | 100 | 0.351 |
| >2 | 117 | 50 | 67 | |
| Nodes status | ||||
| Positive | 102 | 41 | 61 | 0.142 |
| Negative | 208 | 102 | 106 | |
| TNM stage | ||||
| I | 128 | 40 | 68 | 0.033 |
| II | 133 | 64 | 69 | |
| III | 49 | 39 | 30 | |
| Pathologic type | ||||
| Invasive ductal carcinoma | 202 | 97 | 105 | 0.361 |
| Others | 108 | 46 | 62 | |
| Histological grade | ||||
| I | 51 | 16 | 35 | 0.040 |
| II | 201 | 95 | 106 | |
| III | 58 | 32 | 26 | |
| ER status | ||||
| Positive | 223 | 102 | 121 | 0.826 |
| Negative | 87 | 41 | 46 | |
| PR status | ||||
| Positive | 217 | 103 | 114 | 0.471 |
| Negative | 93 | 40 | 53 | |
| Her-2 status | ||||
| Positive | 45 | 19 | 26 | 0.570 |
| Negative | 265 | 124 | 141 | |
| Ki-67 status | ||||
| High | 91 | 50 | 41 | 0.045 |
| Low | 219 | 93 | 126 | |
| Molecular subtype | ||||
| Luminal A | 156 | 71 | 85 | 0.971 |
| Luminal B | 71 | 32 | 39 | |
| HER2 Positive | 30 | 14 | 16 | |
| Triple Negative | 53 | 26 | 27 | |
GLYAT, Glycine N-acyltransferase; TNM, tumor-node-metastasis.
BC patients were analyzed for their GLYAT status and prognosis via Kaplan-Meier survival analysis as well as log-rank tests after being followed up for an average of 61.75 months (range, 9–77 months). We found that those with decreased GLYAT levels experienced poorer disease-free survival (DFS) (Figure 6D; P=0.012). GLYAT expression was a significant indicator of DFS rates in breast cancer patients based on univariate Cox regression analysis, demonstrating a hazard ratio (HR) of 1.565 [P<0.05; 95% confidence interval (CI) 1.098-2.232] (Table 2). Further multivariate analysis revealed that GLYAT expression was significantly related to DFS (HR, 0.145; 95% CI, 0.031-0.690; P=0.015) (Table 2) and represented an independent risk factor of prognosis for BC.
Table 2.
Univariate and multivariate analyses of clinicopathological risk factors for disease-free survival among breast cancer patients.
| DFS | ||||
|---|---|---|---|---|
| Variables | Univariate analysis | Multivariate analysis | ||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age | 1.109 (0.778-1.580) | 0.567 | 1.779 (0.995-3.182) | 0.052 |
| Menopausal status | 0.835 (0.586-1.191) | 0.321 | 0.562 (0.313-1.009) | 0.053 |
| T stage | 1.139 (0.784-1.653) | 0.044 | 0.457 (0.245-0.851) | 0.014 |
| N stage | 0.951 (0.655-1.381) | 0.023 | 0.466 (0.235-0.927) | 0.029 |
| TNM stage | 0.630 (0.375-1.058) | 0.043 | 0.432 (0.225-0.741) | 0.004 |
| Pathological type | 0.914 (0.633-1.319) | 0.630 | 0.937 (0.635-1.383) | 0.744 |
| Histological grade | 1.035 (0.580-1.846) | 0.908 | 0.878 (0.403-1.914) | 0.743 |
| ER status | 0.726 (0.480-1.097) | 0.128 | 0.510 (0.215-1.210) | 0.127 |
| PR status | 0.737 (0.496-1.095) | 0.131 | 0.788 (0.422-1.469) | 0.453 |
| HER-2 status | 1.181(0.618-2.257) | 0.615 | 1.986 (0.689-5.726) | 0.204 |
| Ki-67 status | 0.864 (0.574-1.300) | 0.483 | 1.408 (0.621-3.192) | 0.412 |
| Molecular subtype | 1.677 (0.925-3.041) | 0.089 | 2.910 (0.696-4.177) | 0.143 |
| GLYAT status | 1.565 (1.098-2.232) | 0.013 | 1.475 (0.991-2.197) | 0.046 |
Patients were then divided into five subgroups based on their molecular subtype, Ki67, ER, PR and HER2 status in order to investigate the effect of GLYAT on prognosis.
In the subgroup analyses by molecular subtype, patients were grouped into triple negative or HER2 positive, Luminal A, or Luminal B subgroup. We found that GLYAT had a negative impact on DFS in all groups, especially in the Luminal A subgroup, where there was a statistically significant difference (P=0.014, Figures 6E–H ).
In the subgroup analyses based on Ki67, those with low GLYAT levels were found to have shorter DFS in comparison to those with high GLYAT, regardless of whether they had high or low Ki67. This difference was statistically significant in those with low Ki67 (P=0.014, Figure 6I ). Similarly, those with low GLYAT levels also experienced shorter DFS in contrast to those with high GLYAT, irrespective of ER+ or ER- status. There was a significantly statistical difference in those of ER+ status (P=0.011, Figure 6J ). The same pattern of findings also applied to subgroup analyses based on PR. We also observed a statistically significant difference in the subgroup of PR+ patients (P=0.011, Figure 6K ). Likewise, HER2 status did not change the impact of low GLYAT on shorter DFS, with statistically significant changes observed in both HER+ or HER- status groups (P=0.046 and P=0.045, Figure 6L ).
Discussion
Advanced breast cancer is a condition that carries high morbidity and mortality, despite the existence of several diagnostic markers and treatment strategies (24). Therefore, we sought to determine novel molecular components involved in the biology of BC that may be useful in the management of this condition.
A potential candidate is the GLYAT protein, which is involved in glycine conjugation of xenobiotics such as benzoic acid, and plays a role in many anabolic and catabolic reactions (25). In cancerous cells, GLYAT functions as a key metabolite that inhibits glycine uptake or biosynthesis, impairing growth of these cells likely through inhibition of nucleic acid synthesis (26). Previous reports have highlighted the role of this molecule in musculoskeletal growth and development as well as in hepatocellular carcinoma progression (19). Nevertheless, there has been no study on its oncological effect in breast cancer.
Our series of experiments found that human breast cancer cells and tissues contained remarkably suppressed levels of GLYAT. We further uncovered that lower GLYAT level was associated with more malignant clinicopathological characteristics, including higher TNM stage, histological grade, and Ki-67 status. DFS of those with lower GLYAT levels were also shorter. Our findings are consistent with information available in public databases. As breast cancer is related to hormone and has different molecular types, we further divided patients into five separate subgroups based on molecular subtype, Ki67 status, ER status, PR status, and HER2 status to analyze the correlation between GLYAT expression and prognosis of breast cancer. We found that breast cancer patients with low expression levels of GLYAT had poorer DFS regardless of the molecular subtype, Ki67 status, ER status, PR status, and HER2 status. Especially in the Luminal A subgroup, the Ki67 low status subgroup, the ER+ subgroup, the PR+ subgroup, and both the HER+ and HER- subgroups, there was a statistically significant difference (all P<0.05). We postulate that the lack of statistical significance for other subgroups, such as the Ki67 high status subgroup, the ER- subgroup, and the PR- subgroup, can be attributed to small sample sizes or an inadequate follow-up time. However, perhaps more importantly, ER and PR status are intricately linked to breast cancer initiation, development and prognosis. GLYAT may act in combination with ER or PR status to impact breast cancer prognosis. Additional experiments are needed to clarify this question in further research.
We further discovered GLYAT to be an independent prognostic factor for BC. These findings indicated that GLYAT acts as antioncogene and is associated with malignant clinicopathological features that may enhance breast cancer metastasis and progression. Our findings based on DFS alone are suggestive of the potential of this molecule to prognosticate breast cancer. Our findings are consistent with bioinformatics analyses and the data reported by Matsuo M et al., who indicated that GLYAT was suppressed in human hepatocellular carcinomas (19). Given that our study is the first to analyze the correlation between GLYAT and breast cancer, future investigations and follow up investigations regarding OS information (not available in our study due to short follow-up time) are still required.
We further demonstrated that GLYAT regulates breast cancer migration and invasion via EMT modulation. EMT is reduced via alteration of the PI3K/ATK/Snail signaling pathway both in vitro and in vivo. The EMT process participate in cancer metastasis (27). Numerous studies reported that EMT is regulated by several signaling pathways, for example Notch-, Wnt-, PI3K/Akt- and NF-κB-dependent pathways (28–30). Previous studies have confirmed that the EMT enhances tumor cell mobility and invasiveness, thus, contributing to the development of chemotherapy resistance and metastasis (31, 32). Additionally, breast cancer recurrence and prognosis have been reported to be affected strongly by EMT (13). We believe there may be other mechanisms in addition to the PI3K/AKT/Snail pathway by which GLYAT promotes metastasis that need to be further explored. One example by Ren et al. using a Drosophila model demonstrated GLYAT to be a critical modulator of the c-Jun N-terminal kinase (JNK) signaling pathway (33). GLYAT downregulation suppresses JNK-dependent ROS activation, which is a key facilitator of several important biological functions.
Besides the impact on metastasis, the proliferation ability of BC cells was increased after inhibiting GLYAT both in vitro and in vivo, further highlighting the tumor suppressed role of GLYAT. Our research is the only study to illustrate the role of GLYAT in BC patients, revealing that down-regulated GLYAT induces EMT and tumor metastasis via PI3K/AKT/Snail signaling.
Conclusion
Collectively, our data revealed that GLYAT downregulated in human BC cells and tissues, and lower GLYAT expression was related to poor clinical outcomes. GLYAT suppresses BC cell proliferation and migration through EMT induction via the PI3K/AKT/Snail pathway in vitro and in vivo. Our study puts forth GLYAT as a novel biomarker in BC and may also represent a therapeutic target for BC treatment.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of Shengjing Hospital of China Medical University (NO.2018PS234K). The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the ethics committee of Shengjing Hospital of China Medical University (NO.2018PS234K). Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
SG conceived and designed the experiments as well as contributed to the writing of the manuscript. XT, NW, MJ, YZ, NM, and RW performed the experiments. GL provided the tumor tissue microarray and brief guidance about conception at the beginning of the study. XT performed clinical analysis and helped with drawing the figures. SG revised the paper. All authors contributed to the article and approved the submitted version.
Funding
The study was funded by the Doctoral start-up foundation of Liaoning Province (No. 20180540024), and 345 Talent Project, Shengjing Hospital of China Medical University (No. M0397).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential confliict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.641399/full#supplementary-material
Abbreviations
BC, Breast cancer; NC, Normal control; ER, Estrogen receptor; PR, Progesterone receptor; HER2, Human epidermal growth factor receptor 2; TNBC, Triple negative breast cancer; BRCA, Breast invasive carcinoma; IDBC, Invasive ductal breast carcinoma; ILBC, Invasive lobular breast carcinoma; LIHC, Liver hepatocellular carcinoma; CHOL, Cholangio carcinoma; ACC, Adrenocortical carcinoma; BLCA, Bladder Urothelial carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, Colon adenocarcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell carcinoma; KIRC, Kidney renal clear cell carcinoma; LAML, Acute Myeloid Leukemia; LGG, Brain Lower Grade Glioma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; STAD, Stomach adenocarcinoma; THCA, Thyroid carcinoma; THYM: Thymoma.
References
- 1. Yang H, Pawitan Y, He W, Eriksson L, Holowko N, Hall P, et al. Disease trajectories and mortality among women diagnosed with breast cancer. Breast Cancer Res (2019) 21:95. doi: 10.1186/s13058-019-1181-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3. Sancho-Garnier H, Colonna M. [Breast cancer epidemiology]. Presse Med (2019) 48:1076–84. doi: 10.1016/j.lpm.2019.09.022 [DOI] [PubMed] [Google Scholar]
- 4. Scimeca M, Urbano N, Bonfiglio R, Duggento A, Toschi N, Schillaci O, et al. Novel insights into breast cancer progression and metastasis: A multidisciplinary opportunity to transition from biology to clinical oncology. Biochim Biophys Acta Rev Cancer (2019) 1872:138–48. doi: 10.1016/j.bbcan.2019.07.002 [DOI] [PubMed] [Google Scholar]
- 5. Claessens A, Ibragimova K, Geurts S, Bos M, Erdkamp F, Tjan-Heijnen V. The role of chemotherapy in treatment of advanced breast cancer: an overview for clinical practice. Crit Rev Oncol Hematol (2020) 153:102988. doi: 10.1016/j.critrevonc.2020.102988 [DOI] [PubMed] [Google Scholar]
- 6. Boulding T, McCuaig RD, Tan A, Hardy K, Wu F, Dunn J, et al. LSD1 activation promotes inducible EMT programs and modulates the tumour microenvironment in breast cancer. Sci Rep (2018) 8:73. doi: 10.1038/s41598-017-17913-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lourenco AR, Ban Y, Crowley MJ, Lee SB, Ramchandani D, Du W, et al. Differential Contributions of Pre- and Post-EMT Tumor Cells in Breast Cancer Metastasis. Cancer Res (2020) 80:163–9. doi: 10.1158/0008-5472.CAN-19-1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krstic M, Kolendowski B, Cecchini MJ, Postenka CO, Hassan HM, Andrews J, et al. TBX3 promotes progression of pre-invasive breast cancer cells by inducing EMT and directly up-regulating Slug. J Pathol (2019) 248:191–203. doi: 10.1002/path.5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramesh V, Brabletz T, Ceppi P. Targeting EMT in Cancer with Repurposed Metabolic Inhibitors. Trends Cancer (2020) 6:942–50. doi: 10.1016/j.trecan.2020.06.005 [DOI] [PubMed] [Google Scholar]
- 10. Kar R, Jha NK, Jha SK, Sharma A, Dholpuria S, Asthana N, et al. A “NOTCH” Deeper into the Epithelial-To-Mesenchymal Transition (EMT) Program in Breast Cancer. Genes (Basel) (2019) 10:961. doi: 10.3390/genes10120961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim BG, Sung JS, Jang Y, Cha YJ, Kang S, Han HH, et al. Compression-induced expression of glycolysis genes in CAFs correlates with EMT and angiogenesis gene expression in breast cancer. Commun Biol (2019) 2:313. doi: 10.1038/s42003-019-0553-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun X, Chang X, Wang Y, Xu B, Cao X. Oroxylin A Suppresses the Cell Proliferation, Migration, and EMT via NF-kappaB Signaling Pathway in Human Breast Cancer Cells. BioMed Res Int (2019) 2019:9241769. doi: 10.1155/2019/9241769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ren Z, Yang T, Zhang P, Liu K, Liu W, Wang P. SKA2 mediates invasion and metastasis in human breast cancer via EMT. Mol Med Rep (2019) 19:515–23. doi: 10.3892/mmr.2018.9623 [DOI] [PubMed] [Google Scholar]
- 14. Jiang H, Zhou Z, Jin S, Xu K, Zhang H, Xu J, et al. PRMT9 promotes hepatocellular carcinoma invasion and metastasis via activating PI3K/Akt/GSK-3beta/Snail signaling. Cancer Sci (2018) 109:1414–27. doi: 10.1111/cas.13598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van der Sluis R, Badenhorst CP, Erasmus E, van Dyk E, van der Westhuizen FH, van Dijk AA. Conservation of the coding regions of the glycine N-acyltransferase gene further suggests that glycine conjugation is an essential detoxification pathway. GENE (2015) 571:126–34. doi: 10.1016/j.gene.2015.06.081 [DOI] [PubMed] [Google Scholar]
- 16. Guo YF, Zhang LS, Liu YJ, Hu HG, Li J, Tian Q, et al. Suggestion of GLYAT gene underlying variation of bone size and body lean mass as revealed by a bivariate genome-wide association study. Hum Genet (2013) 132:189–99. doi: 10.1007/s00439-012-1236-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dempsey DR, Bond JD, Carpenter AM, Rodriguez OS, Merkler DJ. Expression, purification, and characterization of mouse glycine N-acyltransferase in Escherichia coli. Protein Expr Purif (2014) 97:23–8. doi: 10.1016/j.pep.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Sluis R, Badenhorst CP, van der Westhuizen FH, van Dijk AA. Characterisation of the influence of genetic variations on the enzyme activity of a recombinant human glycine N-acyltransferase. GENE (2013) 515:447–53. doi: 10.1016/j.gene.2012.12.003 [DOI] [PubMed] [Google Scholar]
- 19. Matsuo M, Terai K, Kameda N, Matsumoto A, Kurokawa Y, Funase Y, et al. Designation of enzyme activity of glycine-N-acyltransferase family genes and depression of glycine-N-acyltransferase in human hepatocellular carcinoma. Biochem Biophys Res Commun (2012) 420:901–6. doi: 10.1016/j.bbrc.2012.03.099 [DOI] [PubMed] [Google Scholar]
- 20. Chandrashekar DS, Bashel B, Balasubramanya S, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. NEOPLASIA (2017) 19:649–58. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45:W98–W102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait BH, et al. A subcellular map of the human proteome. Science (2017) 356:eaal3321. doi: 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- 23. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature (2012) 486:346–52. doi: 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gu X, Xue J, Ai L, Sun L, Zhu X, Wang Y, et al. SND1 expression in breast cancer tumors is associated with poor prognosis. Ann N Y Acad Sci (2018) 1433:53–60. doi: 10.1111/nyas.13970 [DOI] [PubMed] [Google Scholar]
- 25. Badenhorst CP, van der Sluis R, Erasmus E, van Dijk AA. Glycine conjugation: importance in metabolism, the role of glycine N-acyltransferase, and factors that influence interindividual variation. Expert Opin Drug Metab Toxicol (2013) 9:1139–53. doi: 10.1517/17425255.2013.796929 [DOI] [PubMed] [Google Scholar]
- 26. Lino CC, Bourgine J, Cauffiez C, Allorge D, Lo-Guidice JM, Broly F, et al. Genetic polymorphisms of glycine N-acyltransferase (GLYAT) in a French Caucasian population. Xenobiotica (2010) 40:853–61. doi: 10.3109/00498254.2010.519407 [DOI] [PubMed] [Google Scholar]
- 27. Matysiak M, Kapka-Skrzypczak L, Jodlowska-Jedrych B, Kruszewski M. EMT promoting transcription factors as prognostic markers in human breast cancer. Arch Gynecol Obstet (2017) 295:817–25. doi: 10.1007/s00404-017-4304-1 [DOI] [PubMed] [Google Scholar]
- 28. De Francesco EM, Maggiolini M, Musti AM. Crosstalk between Notch, HIF-1alpha and GPER in Breast Cancer EMT. Int J Mol Sci (2018) 19:2011. doi: 10.3390/ijms19072011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu J, Luo Y, Wen Q. Nalbuphine suppresses breast cancer stem-like properties and epithelial-mesenchymal transition via the AKT-NFkappaB signaling pathway. J Exp Clin Cancer Res (2019) 38:197. doi: 10.1186/s13046-019-1184-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baek SH, Ko JH, Lee JH, Kim C, Lee H, Nam D, et al. Ginkgolic Acid Inhibits Invasion and Migration and TGF-beta-Induced EMT of Lung Cancer Cells Through PI3K/Akt/mTOR Inactivation. J Cell Physiol (2017) 232:346–54. doi: 10.1002/jcp.25426 [DOI] [PubMed] [Google Scholar]
- 31. Chaffer CL, San JB, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev (2016) 35:645–54. doi: 10.1007/s10555-016-9648-7 [DOI] [PubMed] [Google Scholar]
- 32. Cho ES, Kim NH, Yun JS, Cho SB, Kim HS, Yook JI. Breast Cancer Subtypes Underlying EMT-Mediated Catabolic Metabolism. Cells (2020) 9:2064. doi: 10.3390/cells9092064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ren P, Li W, Xue L. GLYAT regulates JNK-mediated cell death in Drosophila. Sci Rep (2017) 7:5183. doi: 10.1038/s41598-017-05482-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.