Coronavirus disease 2019 (COVID-19) is an acute pneumonia caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). According to the latest statistics for COVID-19 released by Johns Hopkins University on April 6, 2021, there were 132.45 million confirmed cases and 2.87 million deaths globally. Because COVID-19 has spread as a global pandemic, the development of therapeutics for this disease, such as neutralizing antibodies that can efficiently block SARS-CoV-2 infection, is urgently needed.
During the infection of host cells by SARS-CoV-2, the spike (S) glycoprotein of SARS-CoV-2 plays the most crucial roles in viral entry and cell fusion. The S protein includes two components, the S1 and S2 subunits. The S1 subunit is vital in determining tissue tropism and host ranges and consists of the N-terminal domain and C-terminal receptor-binding domain (RBD).1,2 Of note, the RBD of the S protein (S-RBD) supports the binding of the S protein to angiotensin-converting enzyme 2 (ACE2) on host cells, contributing to cell entry by SARS-CoV-2.3 Therefore, the S-RBD is widely used for the development of neutralizing antibodies, small-molecule inhibitors and vaccines.4 Previous studies have reported several neutralizing antibodies against the S-RBD that were identified from convalescent COVID-19 patients, transgenic mice or llamas immunized with the SARS-CoV-2 S protein and found to inhibit the interaction between the S protein and ACE2.5–7 In addition to isolating antibodies from patients and immunized animals, an engineered monoclonal antibody (mAb) library provides a powerful tool for screening therapeutic mAbs, with the advantages of safety, ease of performance and high efficiency.8
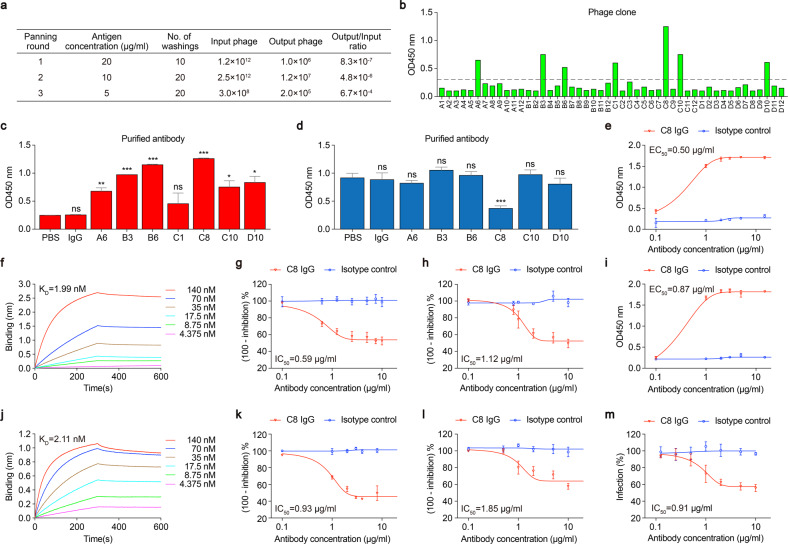
To identify specific antibodies against the S-RBD, we performed human scFv phage library screening using an S-RBD protein as the target. The Tomlinson I + J scFv phage libraries from Geneservice contain 1.47 × 108 and 1.37 × 108 human scFv fragments, respectively. During the screen, binders specific for the S-RBD were enriched after each round of selection. The enrichment rate was calculated as the ratio of the number of output phages to that of input phages (Fig. 1a). After three rounds of selection, 48 phage clones were selected and validated by monoclonal phage ELISA using the S-RBD as the target. Among these isolates, seven phage clones showed relatively high S-RBD binding signals (Fig. 1b). The seven scFv clones were reformatted to full-length human IgG1 by cloning the human variable heavy and light chain regions into a human IgG1 isotype backbone, followed by transient expression in 293T cells and purification. We first examined the capacity of these mAbs to bind the S-RBD protein. The results showed that six of them significantly bound to the S-RBD protein (Fig. 1c). Next, we investigated the inhibitory effects of these mAbs on the binding of the S-RBD to ACE2. Using an ELISA-based inhibition assay, we found that C8 IgG showed the strongest inhibition of S-RBD–ACE2 binding among the mAbs against the S-RBD (Fig. 1d). Overall, we identified the mAb C8 targeting the S-RBD (Fig. S1), which inhibited the S-RBD–ACE2 interaction.
Fig. 1.
Identification and characterization of a monoclonal antibody targeting the S-RBD and blocking SARS-CoV-2 pseudovirus infection. a The phage library enrichment efficiency of each round of panning. b Phage ELISA of clones screened for binding to the S-RBD after the third round of selection. The dotted line shows an OD450 value of 0.3. Selected phage clones were converted into full-length human IgGs. Binding to the S-RBD (c) and inhibition of the S-RBD–ACE2 interaction (d) were examined by ELISA. e Binding of C8 IgG to the immobilized S-RBD (5 μg/ml, corresponding 166 μM) determined by ELISA. f Binding kinetics of C8 IgG to the S-RBD by BLI in OctetRED96. g Inhibitory effect of C8 IgG on the interaction between ACE2 and the S-RBD determined by ELISA. h Inhibitory effect of C8 IgG on the binding of the S-RBD to ACE2-expressing 293 T cells determined by flow cytometry. i Binding of C8 IgG to the immobilized S protein (30 μg/ml, corresponding 166 μM) determined by ELISA. j Binding kinetics of C8 IgG to the S protein by BLI in OctetRED96. k Inhibitory effect of C8 IgG on the interaction between ACE2 and the S protein determined by ELISA. l Inhibitory effect of C8 IgG on the binding of the S protein to ACE2-expressing 293T cells determined by flow cytometry. m Inhibition of luciferase-encoding SARS-CoV-2-type pseudovirus entry into ACE2-expressing 293T cells by C8 IgG. Data are shown as the mean ± SD (unpaired t-test; ***p < 0.001, **p < 0.01, *p < 0.05, ns not significant)
To further evaluate the binding capacity of C8 IgG toward the immobilized S-RBD, the half-maximal effective concentration (EC50) value of C8 IgG was calculated by ELISA and found to be 0.50 μg/ml (Fig. 1e). Next, a biolayer interferometry (BLI) assay was used to kinetically characterize the binding affinity of C8 IgG for the S-RBD. The biotinylated S-RBD protein was immobilized on a streptavidin-conjugated sensor, and the concentration of C8 IgG was ranged from 140 to 4.375 nM with half-fold serial dilution. The results showed the binding of C8 IgG to the S-RBD occurred with fast-on (kon = 8.11 × 104 Ms−1) and slow-off (koff = 1.62 × 10−4 s−1) rates. The KD value was calculated to be 1.99 nM (Fig. 1f). Next, we investigated the inhibitory effect of C8 IgG on the interaction between the S-RBD and ACE2. The results from an ELISA-based inhibition assay showed that C8 IgG significantly inhibited S-RBD–ACE2 binding with a half-maximal inhibitory concentration (IC50) of 0.59 μg/ml (Fig. 1g). Furthermore, flow cytometric analysis was performed to examine the inhibition of soluble S-RBD binding to ACE2-expressing 293T cells (293T-ACE2) by C8 IgG. The results showed that C8 IgG strongly inhibited S-RBD binding to 293T-ACE2 cells with an IC50 of 1.12 μg/ml (Fig. 1h).
To examine the binding capacity of C8 IgG to the full-length S protein and inhibition of the S protein-ACE2 interaction by C8, the soluble full-length S protein was expressed and purified as previously described.1 Compared with an isotype control, C8 IgG showed strong binding to the immobilized S protein with an EC50 value of 0.87 μg/ml (Fig. 1i). BLI results showed the binding of C8 IgG to the full-length S protein occurred with fast-on (kon = 1.38 × 105 Ms−1) and slow-off (koff = 2.92 × 10−4 s−1) rates. The KD value was 2.11 nM (Fig. 1j), which is similar to that determined with the S-RBD. Next, an ELISA-based inhibition assay showed that C8 IgG efficiently blocked the interaction between the S protein and ACE2 with an IC50 of 0.93 μg/ml (Fig. 1k). Moreover, flow cytometric analysis was performed to examine the inhibitory effect of C8 IgG on the binding of the soluble full-length S protein to 293T-ACE2 cells. Consistent with the results of the ELISA-based inhibition assay, C8 IgG significantly inhibited S protein binding to 293T-ACE2 cells with an IC50 of 1.85 μg/ml (Fig. 1l).
Pseudoviruses have similar infectivity to authentic viruses and thus have been widely applied to carry out research on the intrusion mechanism of viruses with high infectivity and pathogenicity. To evaluate the inhibitory effect of C8 IgG on the entry of SARS-CoV-2 pseudoviruses into 293T-ACE2 cells, we added C8 IgG to a SARS-CoV-2 pseudovirus infection assay system at a series of concentrations. The results showed that C8 IgG efficiently inhibited SARS-CoV-2 pseudovirus cell entry in vitro with an IC50 of 0.91 μg/ml (Fig. 1m). Thus, C8 IgG may serve as a neutralizing antibody against SARS-CoV-2 by blocking the binding of the S protein to ACE2.
COVID-19 is a serious threat to the worldwide health system and is devastating to elderly people and people with comorbidities. Given the continuous spread of COVID-19 around the world, the development of effective blocking antibodies or inhibitors to prevent SARS-CoV-2 infection has become a hot spot in current research. Recently, the Chinese Antibody Society developed the “COVID-19 Antibody Therapeutics Tracker” to provide free and open access to a global database for tracking the ongoing clinical and preclinical development of antibody-based therapeutics for the prevention and treatment of COVID-19. These antibodies mainly recognize the SARS-CoV-2 S protein, thus inhibiting viral cell entry by blocking the S protein–ACE2 interaction.9
Phage display technology has been used for antibody screening for a long time and plays an important role in the discovery of therapeutic antibodies against infectious diseases.8 The greatest advantage of this approach is that it has no safety or ethical concerns, particularly as it avoids the direct use of patient samples in a pandemic situation.10 In this study, we used phage display technology to screen monoclonal antibodies blocking the S-RBD–ACE2 interaction. Although C8 IgG efficiently inhibited SARS-CoV-2 pseudovirus infection in ACE2-expressing 293T cells in vitro, the inhibition of authentic SARS-CoV-2 infection in vitro and in vivo requires further investigation. C8 IgG has potential in the prevention and treatment of COVID-19.
Supplementary information
Acknowledgements
This research was supported by the National Key Research and Development Program of China (2020YFA0509000 to J.F.C.); the National Natural Science Foundation of China (32030024, 31830112, and 31525016 to J.F.C. and 31970702 to C.D.L.); the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA12010101 to J.F.C.); the Program of Shanghai Academic Research Leaders (19XD1404200); National Ten Thousand Talents, the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2020266); and the Young Elite Scientist Sponsorship Program by CAST (2019QNRC001). The authors gratefully acknowledge the support of the SA-SIBS scholarship program and Bio-Research innovation Center Suzhou, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences.
Author contributions
C.D.L. and J.F.C. conceptualized the project and designed the experiments. M.Y.Y., Y.Y.W., and C.L. performed the experiments and data analysis. Y.L. and Z.Y.L. helped with protein purification and proofreading the manuscript. C.D.L. and J.F.C. interpreted the results. The manuscript was drafted by Y.Y.W. and C.L. and edited by C.D.L. and J.F.C.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: MengYa Yuan, YaoYing Wan, Cui Liu
Contributor Information
ChangDong Lin, Email: linchangdong@sibcb.ac.cn.
JianFeng Chen, Email: jfchen@sibcb.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00684-x.
References
- 1.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walls AC, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shang J, et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl Acad. Sci. USA. 2020;117:11727–34.. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Premkumar, L. et al. The receptor-binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. 5, 48 (2020). [DOI] [PMC free article] [PubMed]
- 5.Wang C, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D, et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1436–1441. doi: 10.1016/j.cell.2020.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roncolato EC, et al. Phage display as a novel promising antivenom therapy: a review. Toxicon. 2015;93:79–84. doi: 10.1016/j.toxicon.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Shi R, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 10.Kumar R, Parray HA, Shrivastava T, Sinha S, Luthra K. Phage display antibody libraries: a robust approach for generation of recombinant human monoclonal antibodies. Int. J. Biol. Macromol. 2019;135:907–918. doi: 10.1016/j.ijbiomac.2019.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.