Abstract
Serpinb1a, a serine protease inhibitor family protein, has been implicated in immunoregulation and several metabolic disorders, such as diabetes and obesity; however, its roles in bone remain unknown. Therefore, we herein investigated the physiological functions of Serpinb1a in osteoclastic and osteoblastic differentiation using mouse cell lines. Serpinb1a overexpression markedly reduced the number of tartrate-resistant acid phosphatase (TRAP)- and calcitonin receptor-positive multinucleated cells increased by receptor activator nuclear factor κB ligand (RANKL) in mouse preosteoclastic RAW 264.7 cells. Moreover, it significantly decreased the mRNA levels of nuclear factor of activated T-cells, cytoplasmic 1 (NFATc1), TRAP and cathepsin K in these cells. Regarding osteoblasts, Serpinb1a overexpression significantly reduced the mRNA levels of alkaline phosphatase (ALP) and osteocalcin as well as ALP activity induced by bone morphogenetic protein-2 (BMP-2) in mouse mesenchymal ST2 cells, although it did not alter osteoblast differentiation in mouse osteoblastic MC3T3-E1 cells. Concerning the pathophysiological relevance of Serpinb1a, Serpinb1a mRNA levels were decreased in the soleus and gastrocnemius muscles of mice 4 weeks after bilateral sciatic nerve resection. In conclusion, we herein revealed for the first time that Serpinb1a inhibited osteoclast formation induced by RANKL in RAW 264.7 cells and suppressed BMP-2-induced ALP activity in ST2 cells.
Keywords: Serpinb1, Serine protease inhibitor, Osteoclast, Osteoblast, Mechanical stress
Highlights
-
•
Serpinb1a overexpression markedly suppressed TRAP- and calcitonin receptor-positive osteoclast-like cell formation.
-
•
Serpinb1a overexpression significantly suppressed the mRNA levels of bone resorption-related genes in these cells.
-
•
Serpinb1a overexpression did not affect osteoblast differentiation in mouse osteoblastic cells.
-
•
Serpinb1a mRNA levels were reduced in the soleus and gastrocnemius muscles of mice after bilateral sciatic nerve resection.
-
•
Serpinb1a may be one of the crucial protease inhibitors related to unloading-induced muscle atrophy and osteoporosis.
1. Introduction
The serine protease inhibitor family, which includes 16 types of serpins, are important protease inhibitors [1,2]. Serpins are ubiquitously expressed in various tissues and have been implicated in pathophysiological states. Serpinb1a, a member of this family, was initially shown to play a role in innate immunity, and its involvement in the regulation of host innate immunity through the inhibition of neutrophil elastases has since been extensively examined [[3], [4], [5]]. A previous study reported that lung defenses were preserved by enhancements in neutrophil elastases in Serpinb1-deficient mice infected with Pseudomonas aeruginosa [6]. Moreover, proteinase 3-dependent caspase cleavage modulated neutrophil death and inflammation through the inhibition of caspase activation by Serpinb1 [7,8]. On the other hand, Takebayashi et al. showed that plasma Serpinb1 levels were correlated with insulin resistance and fat mass in patients with type 2 diabetes and obesity [9]. Although the roles of Serpinb1 in the diabetic state currently remain unclear, Serpinb1a was shown to directly promote the proliferation of pancreatic islet β cells [10,11]. These findings suggest that Serpinb1a exhibits various pathophysiological functions through the inhibition of serine protease or mechanisms other than its function as an endogenous serine protease inhibitor.
Some serpins have been shown to function in bone. Plasminogen activator inhibitor-1 (PAI-1) (SerpinE1), an inhibitor of plasminogen activators, is involved in the pathophysiology of various bone metabolic disorders through its negative effects on bone tissues as an adipokine [12]. We previously reported that PAI-1 contributed to glucose intolerance, osteopenia, delayed bone repair after bone injury and muscle wasting induced by diabetes or excess glucocorticoid levels in mice [[13], [14], [15], [16], [17]]. Moreover, a PAI-1 deficiency was shown to inhibit the differentiation of mesenchymal stem cells into osteoblasts in mice [18]. We recently revealed that Serpina3n, which is expressed in mouse osteoblasts in female-dominant manner, suppresses the osteoblast phenotypes, such as type 1 collagen expression and alkaline phosphatase (ALP) activity in mouse differentiated osteoblasts [19]. These findings suggest that serpins play significant physiological and pathophysiological roles in bone; however, the function of Serpinb1a currently remains unknown.
In the present study, we investigated the effects of Serpinb1a on mouse osteoblasts and osteoclasts using a cell line and Serpinb1a expression vector. Moreover, we tried to explore some physiological significance of Serpinb1a actions on bone related to the muscle/bone interactions.
2. Materials and methods
2.1. Cell culture
Mouse osteoblastic MC3T3-E1 cells (provided by Dr. Kodama, Ohu Dental Collage, Koriyama, Japan) were cultured in α-minimum essential medium (MEM) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. ST2 cells (RIKEN, Tsukuba, Japan), a mouse mesenchymal cell line, were maintained in RPMI1640 with 10% FBS and 1% penicillin/streptomycin. RAW 264.7 cells, a mouse monocyte-macrophage cell line (ATCC, Manassas, VA), were cultured in high glucose Dulbecco's Modified Eagle's Medium (DMEM) containing 10% FBS and 1% penicillin/streptomycin.
2.2. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells using TriSure reagent (Nippon Genetics, Tokyo, Japan) in accordance with the manufacturer's instructions. The ReverTra Ace qPCR RT Master Mix with a gDNA Remover (Toyobo, Osaka, Japan) was used for the reverse transcription reaction. SYBR Green-based real-time PCR was performed using a StepOnePlus Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) with the THUNDERBIRD qPCR Mix (Toyobo) and specific primers. Each PCR primer set used in the present study is shown in Supplemental Table 1. mRNA expression levels were normalized to the relative amount of the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
2.3. DNA construction
The coding region of murine Serpinb1a was amplified by PCR with the KAPA HiFi DNA Polymerase enzyme (KAPA Biosystems, Wilmington, MA, USA) using cDNA from the mouse neonatal liver as a template and primers (5′-ATCAAGCTAGCATGGAGCAGCTGAGTTCAGC-3′ as the forward primer and 5′-ATCAACTCGAGCTATGGGGAACAAACCCTGC-3′ as the reverse primer). Serpinb1a cDNA was inserted into the mammalian expression vector, pcDNA3.1(+) (Invitrogen, Thermo Fisher Scientific) at the NheI/XhoI sites and the sequence was verified.
2.4. Gene transfection
Regarding transient transfection, the Serpinb1a expression vector was transfected into MC3T3-E1 cells, ST2 cells and RAW 264.7 cells using the jetPRIME reagent (Polyplus-transfection SA, Illkirch, France) according to the manufacturer's protocol. Transfection efficiency was confirmed with qRT-PCR for Serpinb1a for each experiment.
2.5. Osteoclast formation
RAW 264.7 cells transiently expressing Serpinb1a or the empty vector were seeded onto 96-well plates at 1000 cells/well. To achieve osteoclast differentiation, cells were cultured with αMEM, 10% FBS and 50 ng/ml of receptor activator nuclear factor κB ligand (RANKL) (FUJIFILM Wako Pure Chemicals, Osaka, Japan) for 6 days. Cells were then fixed with 4% formaldehyde, and tartrate-resistant acid phosphatase (TRAP) staining was performed with the TRAP Stain kit (FUJIFILM Wako Pure Chemicals) according to the manufacturer's protocol. TRAP-positive multinucleated cells (MNCs) with three and more nuclei were counted as osteoclasts. Immunocytochemistry for the calcitonin receptor was performed as previously described [20]. Briefly, cells were fixed in 10% formaldehyde at room temperature for 20 min. Cells were depleted of endogenous peroxidase activity using 3% H2O2 for 15 min, blocked with 5% FBS for 60 min, and then incubated with a mouse monoclonal anti-calcitonin receptor (CTR) antibody (Clone 2F7, Cat# sc-293299, Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4 °C overnight. Cells were incubated with a horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody (Cat#58802S, Cell Signaling Technology, Beverly, MA, USA) at room temperature for 60 min followed by 3, 3′ diaminobenzidine tetrahydrochloride (DAB) substrate (TaKaRa Bio, Otsu, Japan) according to the manufacturers' protocols. Images of samples were captured using a KEYENCE photomicroscope (original magnification, × 100). CTR-positive MNCs with three and more nuclei were counted as osteoclasts.
2.6. ALP activity
Samples were centrifuged at 15000 rpm at 4 °C for 10 min and ALP activity in supernatants was analyzed using the Lab assay ALP kit (FUJIFILM Wako Pure Chemicals). Total protein concentrations were measured using the Protein Assay BCA Kit (FUJIFILM Wako Pure Chemicals), as described previously [19]. Absorbance was measured at 405 nm (for ALP activity) and 562 nm (for the BCA assay) by the Multiskan Go microplate spectrophotometer (Thermo Fisher Scientific). ALP activity was defined as [units/total protein (μg)].
2.7. Sciatic-neurectomized (SNX) mice
Animal experiments were performed according to the guidelines of the National Institutes of Health and the institutional rules for the use and care of laboratory animals at Kindai University. All animal experiments were approved by the Experimental Animal Welfare Committee of Kindai University (Permit number: KAME-27-029). All efforts were made to minimize suffering. Mice were euthanized with excess isoflurane. SNX or sham surgery was performed on 7-week-old male C57BL/6J mice, as previously described [21].
2.8. Statistical analysis
Data were expressed as the mean ± the standard error of the mean (SEM). Results represent experiments performed independently three times. The significance of differences was evaluated using the Mann–Whitney U test for comparisons of 2 groups. Two-way analyses of variance followed by the Tukey–Kramer test were conducted for multiple comparisons. The significance level was set at P < 0.05. All statistical analyses were performed using GraphPad PRISM 7.00 software.
3. Results
3.1. Effects of Serpinb1a on osteoclast formation
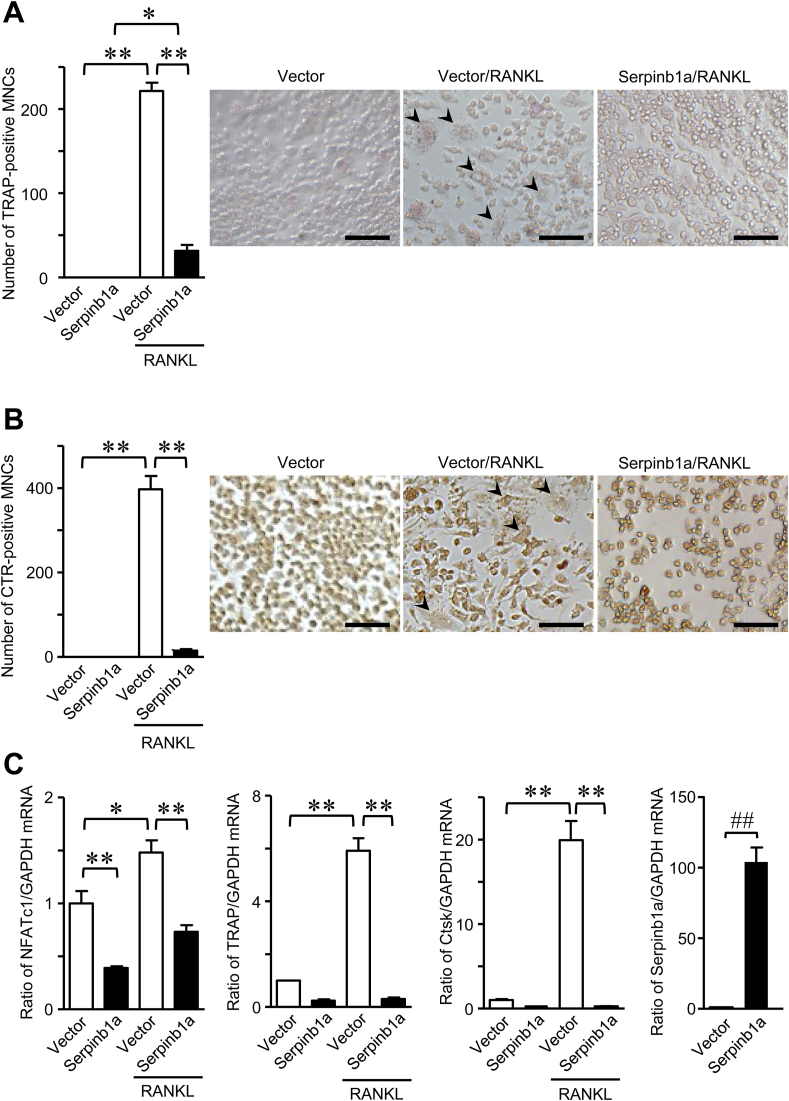
We examined the effects of Serpinb1a on osteoclast formation using mouse preosteoclastic RAW 264.7 cells. The number of TRAP-positive MNCs enhanced by RANKL was significantly lower in RAW 264.7 cells transiently overexpressing Serpinb1a than in empty vector-transfected cells (Fig. 1A). CTR is well known as one of osteoclast-specific marker [22]. Since Serpinb1a may influence TRAP activity in MNCs, we examined CTR-positive MNCs to assess osteoclast numbers. As shown in Fig. 1B, the number of CTR-positive MNCs induced by RANKL was markedly lower in RAW 264.7 cells transiently overexpressing Serpinb1a than in empty vector-transfected cells. Moreover, the mRNA levels of NFATc1, TRAP and cathepsin K induced by RANKL were lower in RAW 264.7 cells transiently overexpressing Serpinb1a than in empty vector-transfected cells (Fig. 1C).
Fig. 1.
Effects of Serpinb1a on osteoclast formation. RAW 264.7 cells transiently expressing Serpinb1a or an empty vector were cultured with αMEM, 10% FBS and 50 ng/ml RANKL for 6 days. (A) The number of TRAP-positive MNCs was counted. (B) The number of CTR-positive MNCs was counted. Data shows the representative images (A, B). Scale bar indicates 50 μm (A, B). Arrowheads indicates TRAP- or CTR-positive MNCs (A, B). (C) Total RNA was extracted, and qRT-PCR for NFATc1, TRAP, cathepsin K (Ctsk), Serpinb1a or GAPDH was performed. Data are expressed relative to GAPDH mRNA. Data represent the mean ± SEM (n = 4 for the empty vector or Serpinb1a without RANKL group and 8 for the empty vector or Serpinb1a with RANKL group). **p < 0.01, *p < 0.05 by the Tukey-Kramer test. ##p < 0.01 by the Mann-Whitney U test.
3.2. Effects of Serpinb1a on osteoblast differentiation
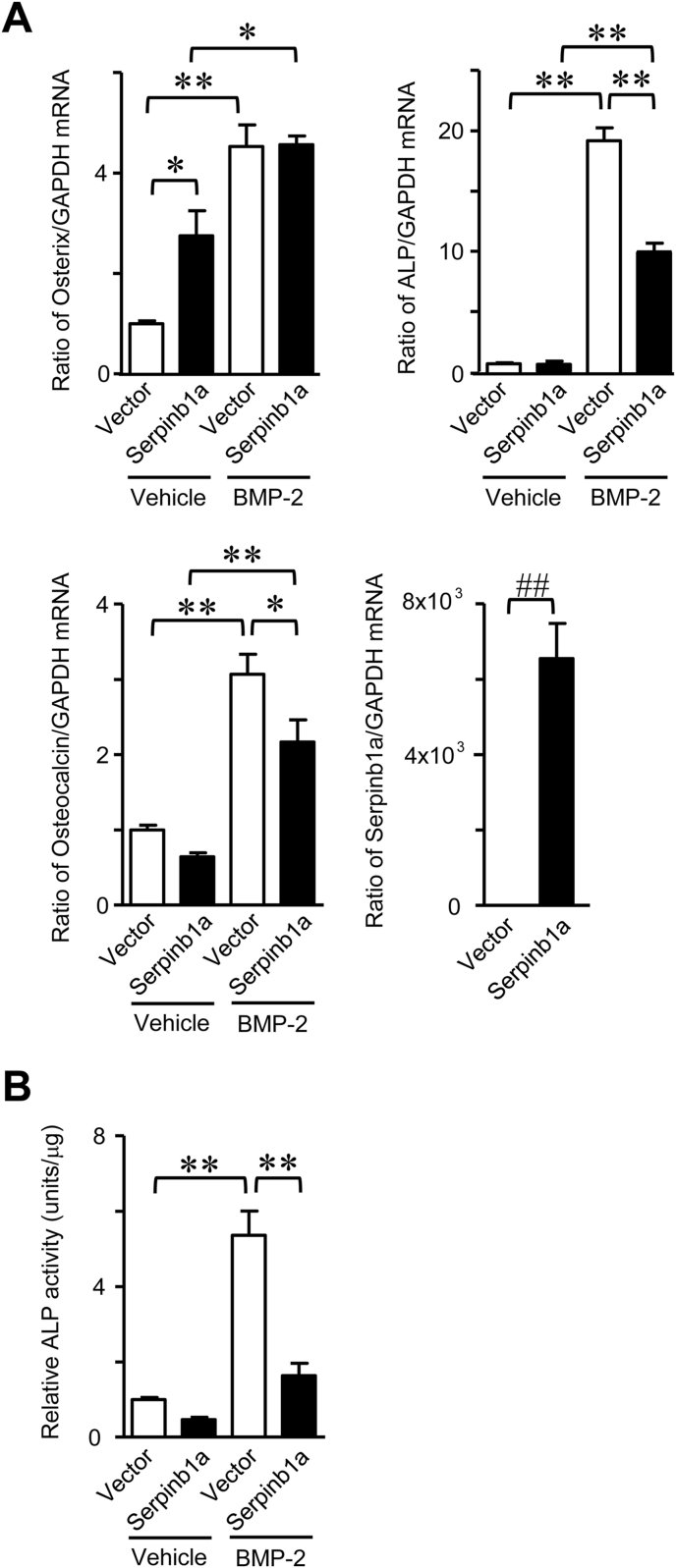
We examined the effects of Serpinb1a overexpression on osteoblastic differentiation. We utilized ST2 cells, the mouse mesenchymal cell line, to examine the effects of Serpinb1a overexpression on the differentiation of mesenchymal cells into osteoblastic cells. Transient Serpinb1a overexpression significantly suppressed the mRNA levels of ALP and osteocalcin as well as ALP activity induced by BMP-2 in ST2 cells (Fig. 2A and B), but did not affect Osterix mRNA levels induced by the BMP-2 stimulation (Fig. 2A).
Fig. 2.
Effects of Serpinb1a on osteoblastic differentiation. (A) Total RNA was extracted from transiently empty vector- or Serpinb1a-transfected ST2 cells with vehicle (0.5% sucrose, 2.5% glycine, 5 mM glutamic acid, 5 mM sodium chloride and 0.01% Tween 80) or 200 ng/ml BMP-2 treatment for 72 h. Real-time PCR for Osterix, ALP, osteocalcin or GAPDH was performed. Data represent the mean ± SEM (n = 6 in each group). **p < 0.01, *p < 0.05 by the Tukey-Kramer test. ##p < 0.01 by the Mann-Whitney U test. (B) ALP activity was measured in Serpinb1a- or empty vector-transfected ST2 cells with vehicle (0.5% sucrose, 2.5% glycine, 5 mM glutamic acid, 5 mM sodium chloride and 0.01% Tween 80) or 200 ng/ml BMP-2 treatment for 72 h, as described in the Methods. Data represent the mean ± SEM (n = 6 in each group). **p < 0.01 by the Tukey-Kramer test.
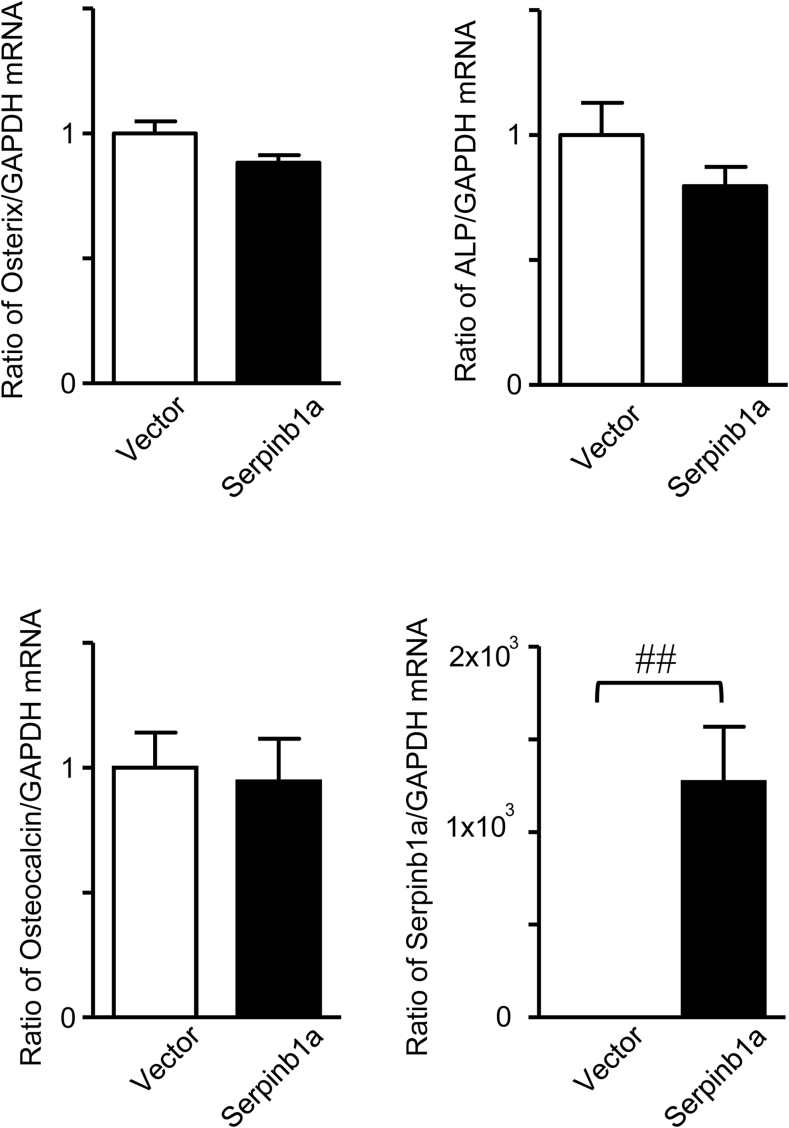
On the other hand, transient Serpinb1a overexpression did not affect the mRNA levels of Osterix, ALP or osteocalcin in the mouse osteoblastic cell line MC3T3-E1 (Fig. 3).
Fig. 3.
Effects of Serpinb1a on osteoblast phenotypes. Total RNA was extracted from transiently empty vector- or Serpinb1a-transfected MC3T3-E1 cells. Real-time PCR for Osterix, ALP, osteocalcin, Serpinb1a or GAPDH was performed. Data represent the mean ± SEM (n = 8 in each group). ##p < 0.01 by the Mann-Whitney U test.
3.3. Serpinb1a expression in skeletal muscles of SNX mice
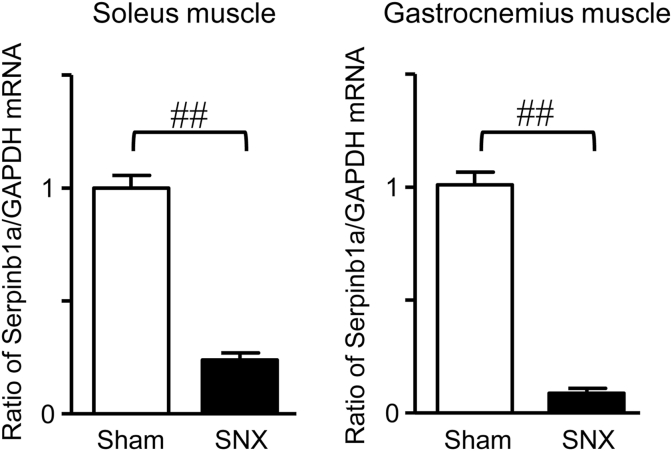
We previously used male SNX mice to examine the effects of mechanical unloading on muscle and bone [21]. The findings obtained revealed that mechanical unloading reduced the tissue weight of the soleus muscle and trabecular bone mineral density at the tibia 4 weeks after SNX. We herein examined the expression of Serpinb1a using the soleus and gastrocnemius muscles of the same mice. As shown in Fig. 4, Serpinb1a mRNA levels in the soleus and gastrocnemius muscles were significantly lower in SNX mice than in sham-operated mice.
Fig. 4.
Expression of Serpinb1a in SNX mice. Total RNA was extracted from the soleus and gastrocnemius muscles of mice 4 weeks after SNX or sham surgery. qRT-PCR was performed for Serpinb1a or GAPDH. Data represent the mean ± SEM (n = 6 mice in each group). ##p < 0.01 by the Mann-Whitney U test.
4. Discussion
Osteoclasts differentiated from monocyte-macrophage lineage cells when stimulated with RANKL, and endogenous proteases were associated with osteoclast function [23,24]. Serpinb1a has been shown to regulate the number and maturation of macrophages in mice partly through the inhibition of cathepsin and endogenous proteases [6,25,26]. In the present study, we showed that Serpinb1a overexpression decreased the numbers of TRAP- and CTR-positive MNCs as well as the expression levels of NFATc1, TRAP and cathepsin K, which are crucial osteoclast-related genes, in RAW 264.7 cells. These results indicated that Serpinb1a potently suppressed RANKL-induced osteoclast formation in RAW 264.7 cells. Therefore, we speculated that Serpinb1a may inhibit endogenous elastase in monocytes; however, the mechanisms by which Serpinb1a suppresses osteoclast formation remain unclear in our study.
In bone remodeling, osteoclastic bone resorption is followed by osteoblastic bone formation [27]. Osteoblasts are generated by the differentiation of mesenchymal stem cells or mesenchymal cells. Therefore, Serpinb1a may affect osteoblastic differentiation and osteoblast function as well as osteoclast formation. We showed that Serpinb1a overexpression significantly suppressed the expression of osteocalcin and ALP, but not Osterix, as well as ALP activity induced by BMP-2 in mouse mesenchymal ST2 cells, but did not affect the expression of Osterix, ALP or osteocalcin in mouse osteoblastic MC3T3-E1 cells. These results suggest that Serpinb1a suppressed the differentiation of mesenchymal cells into osteoblasts, but did not affect well-differentiated osteoblasts. However, we speculate that Serpinb1a may not affect the differentiation of mesenchymal cells into preosteoblasts at the early stage because Osterix is a crucial transcription factor for the commitment of preosteoblasts [28] and Serpinb1a overexpression did not affect Osterix expression induced by BMP-2 in ST2 cells.
Mullin et al. recently reported that Serpinb1 is one of 32 genes that possess a human osteoclast-specific expression quantitative trait locus, related to bone mineral density by genome-wide association studies [29]. In that study, Serpinb1 variants in osteoclasts were derived from patients with postmenopausal osteoporosis [29]. The present results suggested that Serpinb1a markedly suppressed osteoclast formation, whereas its effects on osteoblastic cells seemed to be less potent in mouse cell lines, which may lead to the increase in bone mineral density in mice. Therefore, Serpinb1a may exert positive and protective effects for bone remodeling and osteoporosis, respectively. Further in vivo studies are needed to clarify the roles of Serpinb1a in bone metabolism and the pathophysiology of osteoporosis.
Sarcopenia has recently been recognized as a public health issue, and the prevention and treatment of sarcopenia due to disuse or mechanical unloading in the elderly is a clinically important task. Mice with bilateral sciatic nerve resection are a useful mouse model for examining mechanical unloading-induced muscle wasting and osteopenia, as previously reported [21]. Regarding serpins, we previously showed that the expression levels of Serpina3n were higher in female than in male mouse osteoblasts, which exerted negative effects on osteoblastic function in mouse cells [19]. Moreover, Serpina3n, identified by mass spectroscopic analyses using excess glucocorticoid-treated mouse muscles, appeared to protect against muscle atrophy in Serpina3n transgenic mice [30,31]. Although the significance of Serpinb1a in the pathological states of musculoskeletal disorders and muscle/bone interactions remain unclear, these findings raised us the possibility that Serpinb1a may be related to unloading-induced muscle wasting and osteopenia. Therefore, we herein examined the expression of Serpinb1a in the soleus and gastrocnemius muscles of SNX mice, a mechanical unloading mouse model. The soleus and gastrocnemius muscles are representative muscles with type I and type IIb myofiber-dominant muscles, respectively. In the present study, mechanical unloading by SNX reduced the expression levels of Serpinb1a in both the soleus and gastrocnemius muscles of mice. This result suggested that mechanical unloading suppressed Serpinb1a expression in skeletal muscles. Taken together with the Serpinb1a-induced suppression of osteoclast formation, mechanical unloading-suppressed Serpinb1a secretion from muscles may enhance bone resorption and subsequent increases in bone mineral density in mice. Interestingly, in proteomics analyses of dystrophic muscles conducted by Arecco et al. Serpinb1a accumulated in these muscles over several weeks, and elastase levels and activity were enhanced in the dystrophic muscles of older mice [32]. Moreover, the addition of recombinant elastase markedly repressed myoblast proliferation, survival, myotube formation as well as MyoD expression. These findings suggest that Serpinb1a inhibits endogenous elastase to disturb muscle function. Xu et al. also identified Serpinb1a as one of the genes expressed in injured muscles and during muscle regeneration by cumulative cDNA microarray analyses [33].
The vitamin D receptor (VDR), a member of the nuclear receptor superfamily, plays a crucial role in the physiological effects of vitamin D [34]. The expression of many of the genes involved in not only calcium/phosphate homeostasis, but also cellular proliferation and differentiation as well as immune responses is controlled by VDR responses [34,35]. He et al. revealed that the up-regulation of Serpinb1a expression by active vitamin D inhibited elastase activity to protect against chlamydial infection in mice [36]. Furthermore, the expression of Serpinb1a was shown to be induced by active vitamin D in both human transformed and normal mammary cells using cumulative gene expression analyses with RNA-seq [37]. Moreover, Serpinb1 was identified as a vitamin D-responsive gene in both human breast malignant and normal tissues [38]. Hui et al. showed that active vitamin D inhibited human keratinocyte HaCaT cell proliferation through Serpinb1 [39]. However, we previously observed that a vitamin D deficiency did not affect Serpinb1 mRNA levels in the tibia and gastrocnemius muscles of VD-deficient diet-fed mice (data not shown) [40,41]. Therefore, VDR signaling may not be important for the roles of Serpinb1a expression in muscle/bone interactions in mice, although further studies using active vitamin D-treated mice are needed to elucidate the relationships between vitamin D and Serpinb1a in muscle and bone.
In conclusion, we herein demonstrated for the first time that Serpinb1a suppressed osteoclast formation enhanced by RANKL in RAW 264.7 cells. Moreover, it decreased BMP-2-induced ALP activity in ST2 cells. Therefore, Serpinb1a would be one of the crucial protease inhibitor in mechanical unloading-induced muscle atrophy and osteoporosis. However, the mechanisms by which Serpinb1a suppresses osteoclast formation and osteoblast differentiation have remained unclear in the present study. Further studies are necessary to clarify this issue.
Funding
This study was partly supported by a grant from The Nakatomi Foundation, Japan, to M.I. Grants-in-Aid for Scientific Research (C:20K09514) to H.K. and (C: 19K09659) to M.I. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and a Grant-in-Aid for Scientific Research on Innovative Areas (grant number 15H05935, “Living in Space”) to H.K. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author contributions
M.I. and H.K. contributed to the conception and design of the research. M.I., N.K., Y.M. and Y.T. performed experiments. M.I. analyzed data. M.I. and H.K. interpreted the results of the experiments. M.I. prepared figures. M.I. drafted the manuscript. M.I. and H.K. edited and revised the manuscript. All authors approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge the members of the Life Science Research Institute, Kindai University for their technical support and advice.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Silverman G.A., Bird P.I., Carrell R.W. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 2.Kellici T.F., Pilka E.S., Bodkin M.J. Small-molecule modulators of serine protease inhibitor proteins (serpins) Drug Discov. Today. 2021;26:442–454. doi: 10.1016/j.drudis.2020.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Lucas A., Yaron J.R., Zhang L. Overview of serpins and their roles in biological systems. Methods Mol. Biol. 2018;1826:1–7. doi: 10.1007/978-1-4939-8645-3_1. [DOI] [PubMed] [Google Scholar]
- 4.Lucas A., Yaron J.R., Zhang L. Serpins: development for therapeutic applications. Methods Mol. Biol. 2018;1826:255–265. doi: 10.1007/978-1-4939-8645-3_17. [DOI] [PubMed] [Google Scholar]
- 5.Carrell R.W., Read R.J. How serpins transport hormones and regulate their release. Semin. Cell Dev. Biol. 2017;62:133–141. doi: 10.1016/j.semcdb.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Benarafa C., Priebe G.P., Remold-O'Donnell E. The neutrophil serine protease inhibitor serpinb1 preserves lung defense functions in Pseudomonas aeruginosa infection. J. Exp. Med. 2007;204:1901–1909. doi: 10.1084/jem.20070494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loison F., Xu Y., Luo H.R. Proteinase 3 and Serpin B1: a novel pathway in the regulation of caspase-3 activation, neutrophil spontaneous apoptosis, and inflammation. Inflamm Cell Signal. 2014;1:e462. doi: 10.14800/ics.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loison F., Zhu H., Karatepe K. Proteinase 3-dependent caspase-3 cleavage modulates neutrophil death and inflammation. J. Clin. Invest. 2014;124:4445–4458. doi: 10.1172/JCI76246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takebayashi K., Hara K., Terasawa T. Circulating SerpinB1 levels and clinical features in patients with type 2 diabetes. BMJ Open Diabetes Res Care. 2016;4 doi: 10.1136/bmjdrc-2016-000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Ouaamari A., Dirice E., Gedeon N. SerpinB1 promotes pancreatic β cell proliferation. Cell Metabol. 2016;23:194–205. doi: 10.1016/j.cmet.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarasov A.I., Rorsman P. Dramatis personae in β cell mass regulation: enter SerpinB1. Cell Metabol. 2016;23:8–10. doi: 10.1016/j.cmet.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Kaji H. Adipose tissue-derived plasminogen activator inhibitor-1 function and regulation. Comp. Physiol. 2016;6:1873–1896. doi: 10.1002/cphy.c160004. [DOI] [PubMed] [Google Scholar]
- 13.Tamura Y., Kawao N., Okada K. Plasminogen activator inhibitor-1 is involved in streptozotocin-induced bone loss in female mice. Diabetes. 2013;62:3170–3179. doi: 10.2337/db12-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura Y., Kawao N., Yano M. Role of plasminogen activator inhibitor-1 in glucocorticoid-induced diabetes and osteopenia in mice. Diabetes. 2015;64:2194–2206. doi: 10.2337/db14-1192. [DOI] [PubMed] [Google Scholar]
- 15.Tamura Y., Kawao N., Shimoide T. Role of plasminogen activator inhibitor-1 in glucocorticoid-induced muscle change in mice. J. Bone Miner. Metabol. 2018;36:148–156. doi: 10.1007/s00774-017-0825-8. [DOI] [PubMed] [Google Scholar]
- 16.Shimoide T., Kawao N., Tamura Y. Role of macrophages and plasminogen activator inhibitor-1 in delayed bone repair in diabetic female mice. Endocrinology. 2018;159:1875–1885. doi: 10.1210/en.2018-00085. [DOI] [PubMed] [Google Scholar]
- 17.Okada K., Okamoto T., Okumoto K. PAI-1 is involved in delayed bone repair induced by glucocorticoids in mice. Bone. 2020;134:115310. doi: 10.1016/j.bone.2020.115310. [DOI] [PubMed] [Google Scholar]
- 18.Takafuji Y., Tatsumi K., Ishida M. Plasminogen activator inhibitor-1 deficiency suppresses osteoblastic differentiation of mesenchymal stem cells in mice. J. Cell. Physiol. 2019;234:9687–9697. doi: 10.1002/jcp.27655. [DOI] [PubMed] [Google Scholar]
- 19.Ishida M., Kawao N., Okada K. Serpina3n, dominantly expressed in female osteoblasts, suppresses the phenotypes of differentiated osteoblasts in mice. Endocrinology. 2018;159:3775–3790. doi: 10.1210/en.2018-00639. [DOI] [PubMed] [Google Scholar]
- 20.Xing R., Zhang Y., Li C. Interleukin-21 promotes osteoclastogenesis in RAW264.7 cells through the PI3K/AKT signaling pathway independently of RANKL. Int. J. Mol. Med. 2016;38:1125–1134. doi: 10.3892/ijmm.2016.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawao N., Moritake A., Tatsumi K. Roles of irisin in the linkage from muscle to bone during mechanical unloading in mice. Calcif. Tissue Int. 2018;103:24–34. doi: 10.1007/s00223-018-0387-3. [DOI] [PubMed] [Google Scholar]
- 22.Quinn J.M.W., Morfis M., Lam M.H.C. Calcitonin receptor antibodies in the identification of osteoclasts. Bone. 1999;25:1–8. doi: 10.1016/s8756-3282(99)00094-0. [DOI] [PubMed] [Google Scholar]
- 23.Boyle W.J., Simonet W.S., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 24.Suda T., Takahashi N., Martin T.J. Modulation of osteoclast differentiation. Endocr. Rev. 1992;13:66–80. doi: 10.1210/edrv-13-1-66. [DOI] [PubMed] [Google Scholar]
- 25.Burgener S.S., Leborgne N.G.F., Snipas S.J. Cathepsin G inhibition by serpinb1 and serpinb6 prevents programmed necrosis in neutrophils and monocytes and reduces GSDMD-driven inflammation. Cell Rep. 2019;27:3646–3656. doi: 10.1016/j.celrep.2019.05.065. e3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasumatsu R., Altiok O., Benarafa C. SERPINB1 upregulation is associated with in vivo complex formation with neutrophil elastase and cathepsin G in a baboon model of bronchopulmonary dysplasia. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;291:L619–L627. doi: 10.1152/ajplung.00507.2005. [DOI] [PubMed] [Google Scholar]
- 27.Sims N.A., Ng K.W. Implications of osteoblast-osteoclast interactions in the management of osteoporosis by antiresorptive agents denosumab and odanacatib. Curr. Osteoporos. Rep. 2014;12:98–106. doi: 10.1007/s11914-014-0196-1. [DOI] [PubMed] [Google Scholar]
- 28.Nakashima K., Zhou X., Kunkel G. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 29.Mullin B.H., Zhu K., Xu J. Expression quantitative trait locus study of bone mineral density GWAS variants in human osteoclasts. J. Bone Miner. Res. 2018;33:1044–1051. doi: 10.1002/jbmr.3412. [DOI] [PubMed] [Google Scholar]
- 30.Gueugneau M., d'Hose D., Barbe C. Increased Serpina3n release into circulation during glucocorticoid-mediated muscle atrophy. J Cachexia Sarcopenia Muscle. 2018;9:929–946. doi: 10.1002/jcsm.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tjondrokoesoemo A., Schips T., Kanisicak O. Genetic overexpression of Serpina3n attenuates muscular dystrophy in mice. Hum. Mol. Genet. 2016;25:1192–1202. doi: 10.1093/hmg/ddw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arecco N., Clarke C.J., Jones F.K. Elastase levels and activity are increased in dystrophic muscle and impair myoblast cell survival, proliferation and differentiation. Sci. Rep. 2016;6:24708. doi: 10.1038/srep24708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu P., Werner J.U., Milerski S. Diet-induced obesity affects muscle regeneration after murine blunt muscle trauma-a broad spectrum analysis. Front. Physiol. 2018;9:674. doi: 10.3389/fphys.2018.00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Zhu J., DeLuca H.F. Where is the vitamin D receptor? Arch. Biochem. Biophys. 2012;523:123–133. doi: 10.1016/j.abb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Dzik K.P., Kaczor J.J. Mechanisms of vitamin D on skeletal muscle function: oxidative stress, energy metabolism and anabolic state. Eur. J. Appl. Physiol. 2019;119:825–839. doi: 10.1007/s00421-019-04104-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Q., Ananaba G.A., Patrickson J. Chlamydial infection in vitamin D receptor knockout mice is more intense and prolonged than in wild-type mice. J. Steroid Biochem. Mol. Biol. 2013;135:7–14. doi: 10.1016/j.jsbmb.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons K.M., Beaudin S.G., Narvaez C.J. Gene signatures of 1,25-dihydroxyvitamin D3 exposure in normal and transformed mammary cells. J. Cell. Biochem. 2015;116:1693–1711. doi: 10.1002/jcb.25129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheng L., Anderson P.H., Turner A.G. Identification of vitamin D3 target genes in human breast cancer tissue. J. Steroid Biochem. Mol. Biol. 2016;164:90–97. doi: 10.1016/j.jsbmb.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Hui L., Yongxia Z., Zhili G. Calcitriol inhibits keratinocyte proliferation by upregulating leukocyte elastase inhibitor (serpin B1) J. Dermatol. 2014;41:393–398. doi: 10.1111/1346-8138.12434. [DOI] [PubMed] [Google Scholar]
- 40.Mao L., Tamura Y., Kawao N. Influence of diabetic state and vitamin D deficiency on bone repair in female mice. Bone. 2014;61:102–108. doi: 10.1016/j.bone.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 41.Tamura Y., Fujito H., Kawao N. Vitamin D deficiency aggravates diabetes-induced muscle wasting in female mice. Diabetol Int. 2017;8:52–58. doi: 10.1007/s13340-016-0278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.