Abstract
Introduction
Gastric cancer (GC) has a high incidence and mortality rate, especially in East Asians, and about 90% of GCs are adenocarcinomas. Histological and etiological heterogeneity and ethnic diversity make molecular subtyping of GC complicated, thus making it difficult to determine molecular division systems and standard treatment modalities. Limited cohorts from South Korea, Singapore, Australia, and Japan have been studied; however, the mutational landscape of gastric adenocarcinomas in Chinese patients is still unknown.
Methods
We performed a targeted sequencing panel focusing on cancer‐related genes and tumor‐associated microorganisms of 529 gastric adenocarcinoma samples with matched blood controls. We identified 449 clinically relevant gene mutations.
Results
Approximately 47.1% of Chinese patients with GC harbored at least one actionable mutation. The top somatic mutations were TP53, ARID1A, LRP1B, PIK3CA, ERBB2, CDH1, KRAS, FAT4, CCNE1, and KMT2D. Truncation mutations of ARID1A, KMT2D, RNF43, TGFBR2, and CIC occurred in patients with high tumor mutational burden. Gene amplifications of ERBB2, CCNE1, CDK12, and CCND1 were detected in patients with low tumor mutational burden. Pathway analysis revealed common gene alterations in the Wnt and PI3K/Akt signaling pathways. The ratio of patients with high microsatellite instability was significantly lower than other cohorts, and high microsatellite instability and Epstein‐Barr virus (EBV)–positive features seemed mutually inclusive in Chinese patients with GC. In 44 (8.3%) patients, 45 germline mutations were identified, among which SPINK1 mutations, all SPINK1 c.194 + 2T > C, were present in 15.9% (7/44) of patients. Microorganisms found in Chinese patients with GC included Helicobacter pylori, EBV, hepatitis B virus, and human papillomavirus types 16 and 18.
Conclusion
Identification of varied molecular features by targeted next‐generation sequencing provides more insight into patient stratification and offers more possibilities for both targeted therapies and immunotherapies of Chinese patients with GC.
Implications for Practice
This study investigated the genomic alteration profile of 529 Chinese patients with gastric adenocarcinoma by deep targeting sequencing, which might be the largest Chinese cohort on the genomic research of gastric adenocarcinoma up to now.
Keywords: Gastric adenocarcinoma, Next‐generation sequencing, Tumor mutational burden, Microsatellite instability, Microorganisms
Short abstract
Novel molecular targeted therapies for gastric cancer are urgently needed. This article explores the mutational landscape covering somatic, germline, and microorganisms among a cohort of Chinese patients with gastric cancer. Findings provide insight on patient stratification and potential molecular targets, and for developing new treatment strategies for patients in China and worldwide.
Introduction
Gastric cancer (GC) is the second leading cause of cancer‐related deaths globally, and more than half of patients with GC come from East Asia [1, 2]. The vast majority of GCs are adenocarcinomas [3]. GC is both genetically and phenotypically heterogeneous; however, current approaches for both first‐line and second‐line treatments of patients with GC are limited, benefitting only a small portion of patients [4]. A one‐size‐fits‐all approach is not appropriate for treating such a heterogeneous disease, and novel molecular targeted therapies are urgently needed [5].
To better understand the heterogeneity and develop new treatment strategies for GC, several classification systems have been proposed. The Lauren classification, both diffuse and intestinal, and the World Health Organization classification, tubular, papillary, mucinous, poorly cohesive, and mixed carcinomas, are the most widely adopted classification systems at the histological level, but both have limited clinical utility [6, 7]. At the molecular level, three major molecular division systems of GC were recently proposed by distinct study groups where different standard treatment modalities for advanced GC were recommended [6]. They analyzed tumor samples with different cohort sizes that were collected from South Korea, Singapore, and Australia, and the subtypes were described by considering the differences of microsatellite stability and microsatellite instability (MSI), infectious pathogens, DNA methylation, genomic alterations, Lauren classification, and TP53 mutation status [5, 8, 9]. However, the subtypes divided by each study group are largely different with respect to the differences of population and method; thus more studies on other populations located in different geographical regions are still needed.
Here, we performed a targeted sequencing panel focusing on both cancer‐related genes and tumor‐associated microorganisms of 529 gastric adenocarcinoma samples. We explored the mutational landscape covering somatic, germline, and microorganisms among these patients and provided a comprehensive mutational profile of the Chinese GC cohort. In addition, we analyzed several signaling pathways that are frequently altered in cancers. We divided these patients into five tiers according to targetable genomic alterations. These findings provide more insight into patient stratification and potential molecular targets and for developing new treatment strategies of patients with GC in China and worldwide.
Materials and Methods
Subject Inclusion and Sample Collection
This study was approved by the institutional review board according to the Declaration of Helsinki. A total of 529 patients who were diagnosed with phase I–IV GC were enrolled in this study. Tumor samples with peripheral blood controls were collected from December 2016 to January 2019 and transferred to OrigiMed (Shanghai, China) for genetic sequencing. Informed consent was obtained from all enrolled patients. Tumor and germline genomic DNA were extracted from the formalin‐fixed paraffin‐embedded (FFPE) and matched blood samples using QIAamp DNA FFPE Tissue Kit and QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The concentration of DNA was measured by Qubit and normalized to 20–50 ng/μL.
Detection and Curation of Genomic Alterations
Deep coverage next‐generation sequencing (NGS) targeting 7,029 exons of 450 cancer‐related genes, 64 selected introns of 39 genes, and 111 regions of 26 tumor‐associated microorganisms was performed on both tumor and germline DNA samples using the YuanSuTM450 gene panel (OrigiMed). All regions were captured and sequenced with a mean coverage of 800× using an Illumina NextSeq500 instrument (Illumina Inc., San Diego, CA). All classes of genomic alterations, including single‐nucleotide variations (SNVs), short and long insertions and deletions, copy number variations (CNVs), and gene rearrangements, were analyzed. Tumor mutational burden (TMB) and microsatellite instability (MSI) status were acquired by an NGS algorithm [10]. The true presence of a microorganism was determined by a cutoff that was validated by polymerase chain reaction methods. For germline mutations, common single‐nucleotide polymorphisms, defined as those from the dbSNP database (version 147), a frequency over 1.5% from the Exome Sequencing Project 6500, or over 1.5% from the 1000 Genomes Project, were excluded. Finally, all detected mutations were classified as clinically relevant mutations or variants of unknown significance based on the American College of Medical Genetics (ACMG) standard.
Statistical Analysis
Fisher's exact test or the chi‐square test was used to examine the association between clinical characteristics and genetic mutations. All probability values were adjusted by Bonferroni correction. Values of p < .05 were considered statistically significant. All the statistical analyses were performed using R version 3.5.1.
Results
Basic Characteristics of the Patients
A total of 529 patients with GC, including 356 men and 173 women, were included in this study. Patients ranged in age from 25 to 86 years, and the median age at diagnosis was 60 years. When diagnosed, 20% of patients were found to have a relatively early stage disease (stage I or II), whereas over 60% of patients were in late stage (stage III and IV). Of all patients with GC, 60 (11.3%) had microsatellite instability‐high (MSI‐H) tumors, whereas the third quartile of TMB value in patients with GC was 10.8 mutations per megabase (Mut/Mb), and a total of 70 patients (13.2%) had GCs with TMB ≥20 Mut/Mb. Here, we defined TMB‐high (TMB‐H) patients as those with high TMB (≥10.8 Mut/Mb). The summarized characteristics of all patients with GC are shown in supplemental online Table 1.
Somatic Mutation Profiling
To identify somatic mutations in GC, we performed a targeted sequencing panel focusing on exons of 450 cancer‐related genes (supplemental online Table 2) and selected introns of 39 genes that are frequently rearranged in solid tumors among 529 GC tumor‐normal pairs. We identified clinically relevant mutations in 449 genes. Alteration frequencies of 38 genes were over 5%, with the most frequently altered genes in GC being TP53 (59.7%), ARID1A (21.9%), LRP1B (14.7%), PIK3CA (13.8%), ERBB2 (13.4%), CDH1 (13.0%), KRAS (11.7%), FAT4 (11.5%), CCNE1 (10.6%), KMT2D (10.4%), and RNF43 (10.4%) (Fig. 1A). In examining the SNV spectrum, we observed that C > T was the most predominant pattern, accounting for 62.6% of the total SNVs, and this trend was more prevalent in TMB‐H patients. We also profiled the CNV landscape in the same cohort and found that gene amplifications and gene deletions showed an uneven distribution along the Chinese GC cohort. We identified 171 clinically related CNV events, including 152 gene amplifications and 19 gene deletions. Amplifications were more frequent than deletions (88.4% vs. 11.6%, respectively) among all CNV changes. The most frequently amplified genes and regions were CCNE1 (n = 55, 10.4%), ERBB2 (n = 44, 8.3%), 11q13 (including CCND1, FGF19, FGF4, and FGF3; n = 30, 5.7%), GATA4 (n = 26, 4.9%), and FGFR2 (n = 25, 4.7%), whereas the most commonly deleted region was CDKN2A/B (n = 7, 1.3%) (Fig. 1B). Interestingly, we also observed that the truncation mutations of several genes, including ARID1A, KMT2D, RNF43, TGFBR2, and CIC, occurred in TMB‐H patients, whereas gene amplifications, such as ERBB2, CCNE1, CDK12, and CCND1, were usually detected in TMB‐L patients (Fig. 1A). There were 358 gene rearrangements involving 175 different genes that were detected in 37.2% (197/529) of patients. Nearly half of these rearrangements (n = 140), which included 82 different genes and 105 patients, belonged to the clinically relevant genomic alterations (CRGAs) based on the ACMG standard. We found that CRGA fusion/rearrangements mostly occurred in BRCA1, CDKN2A, ETV6, ARID1B, and NOTCH1, accounting for 5%, 3.6%, 3.6%, 2.9%, and 2.9%, respectively (Fig. 1C).
Figure 1.
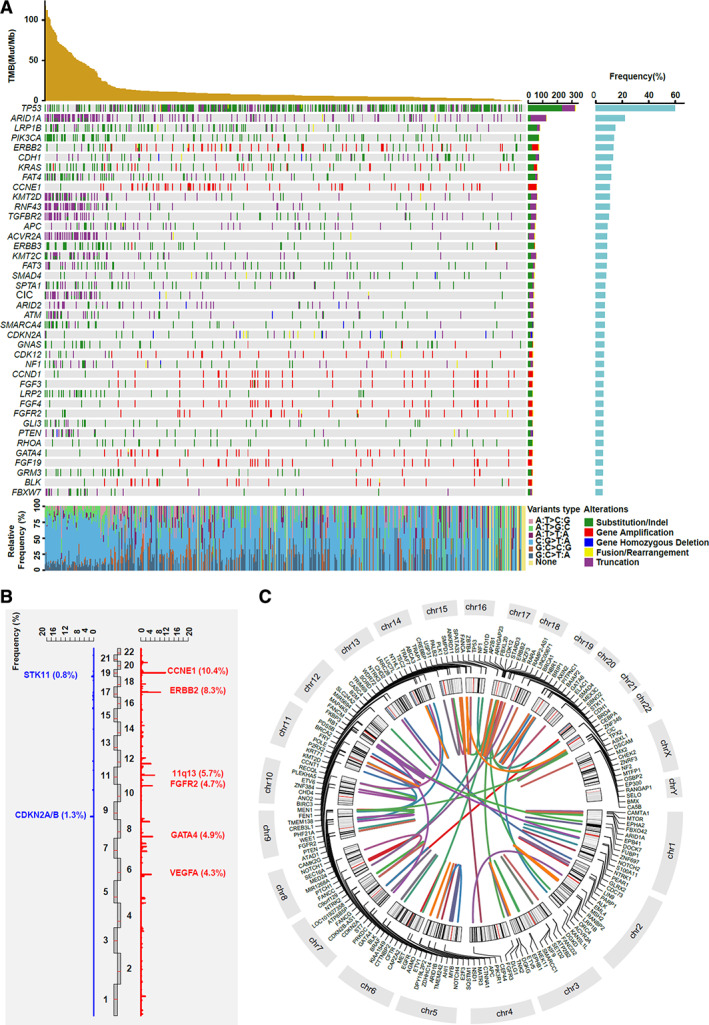
Summary of clinically relevant genetic alterations in 529 patients with gastric cancer. (A): The mutational oncoprint of recurrent genetic alterations. Rows and columns represent genes and patients, respectively. The top panel shows the TMB value for each patient. The bottom panel reflects single nucleotide variations of each patient. (B): The copy number variation (CNV) landscape. Red indicates copy number gain and blue indicates copy number loss. The significant oncogenes and tumor suppressor genes were labeled and the CNV frequency of each gene was marked. (C): Circus plot of clinically relevant gene fusion/arrangements. Both the 5’ and 3’ partner genes of each fusion/arrangement are shown. Abbreviations: Mut/Mb, mutations per megabase; TMB, tumor mutational burden.
Germline Mutation Number and Spectrum
To identify genetic variants that may affect risk for gastric cancer, we analyzed the germline mutations in 529 patients with gastric cancer. There were 45 mutations identified in 44 (8.3%) patients, 38.6% (17/44) being splicing site mutations and 52.3% (23/44) being truncation mutations. A total of 21 genes with germline mutations were detected. The most frequent gene mutations were SPINK1 (n = 7, 15.9%), FANCA (n = 5, 11.4%), ATM (n = 4, 9.1%), BARD1 (n = 3, 6.8%), BRCA2 (n = 3, 6.8%), PALB2 (n = 3, 6.8%), BRCA1 (n = 2, 4.5%), MSH2 (n = 2, 4.5%), RAD51D (n = 2, 4.5%), and SDHA (n = 2, 4.5%). As the most frequently mutated gene, all SPINK1 germline mutations in patients with GC were SPINK1 c.194 + 2T > C. Among all the patients who harbored germline mutations, 20.5% (9/44) of patients had a family history, 56.8% (25/44) harbored “BRCAness” and homologous recombination deficiency signaling changes, and 9.1% (4/44) had DNA mismatch repair (MMR) signaling changes (Fig. 2). Most of these mutations had been previously associated with other types of tumors.
Figure 2.
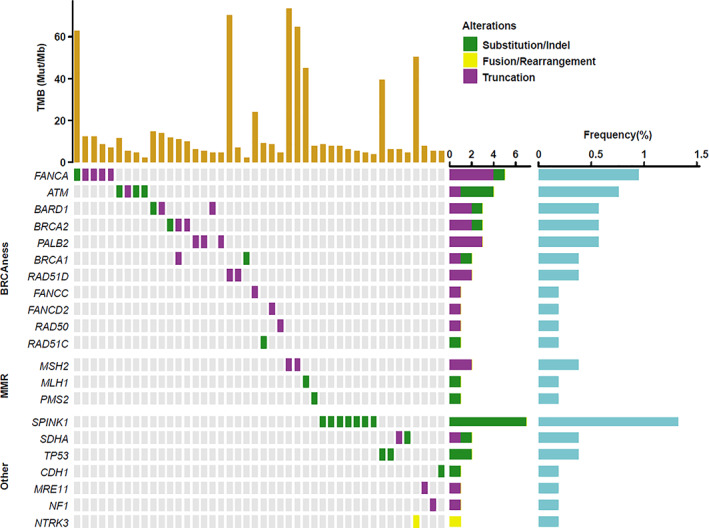
Profiling of germline mutations in 44 patients with gastric cancer. All germline mutations were classified into “BRCAness,” DNA mismatch repair, and other signaling pathways. Abbreviations: indel, insertion or deletion; MMR, mismatch repair; Mut/Mb, mutations per megabase; TMB, tumor mutational burden.
Microbial Community Profiling
In order to determine the characteristics of microbial pathogens in the Chinese GC population, we designed probes targeting 26 cancer‐related microorganisms and performed targeted sequencing in the 528 patients who were available for microbe analysis. In 104 patients (19.7%), we detected five different types of microorganisms, including Helicobacter pylori (49.0%, n = 51), Epstein‐Barr virus (EBV; 51.9%, n = 54), hepatitis B virus (HBV; 7.7%, n = 8), human papillomavirus type 16 (HPV16; 3.8%, n = 4), and human papillomavirus type 18 (HPV18; 1.0%, n = 1) (Fig. 3A). Although most coinfections are rare events, a coinfection of EBV and H. pylori is relatively common and was detected in 8.7% (n = 9) of microbe‐positive patients with GC (Fig. 3B). Furthermore, we analyzed the genomic alterations of patients with GC and the top four ranking infection subtypes, which were EBV infection alone (Fig. 3C), H. pylori infection alone (Fig. 3E), a coinfection of EBV and H. pylori (Fig. 3D), and HBV infection alone (Fig. 3F). A high frequency of TP53 and ARID1A mutations was observed in each group. In the EBV‐positive group, mutation frequencies of several genes such as PIK3CA (33% vs. 11.7%, respectively; p = .0156) and SMARCA4 (20.4% vs. 5.1%, respectively; p = .0326) were much higher than EBV‐negative groups.
Figure 3.
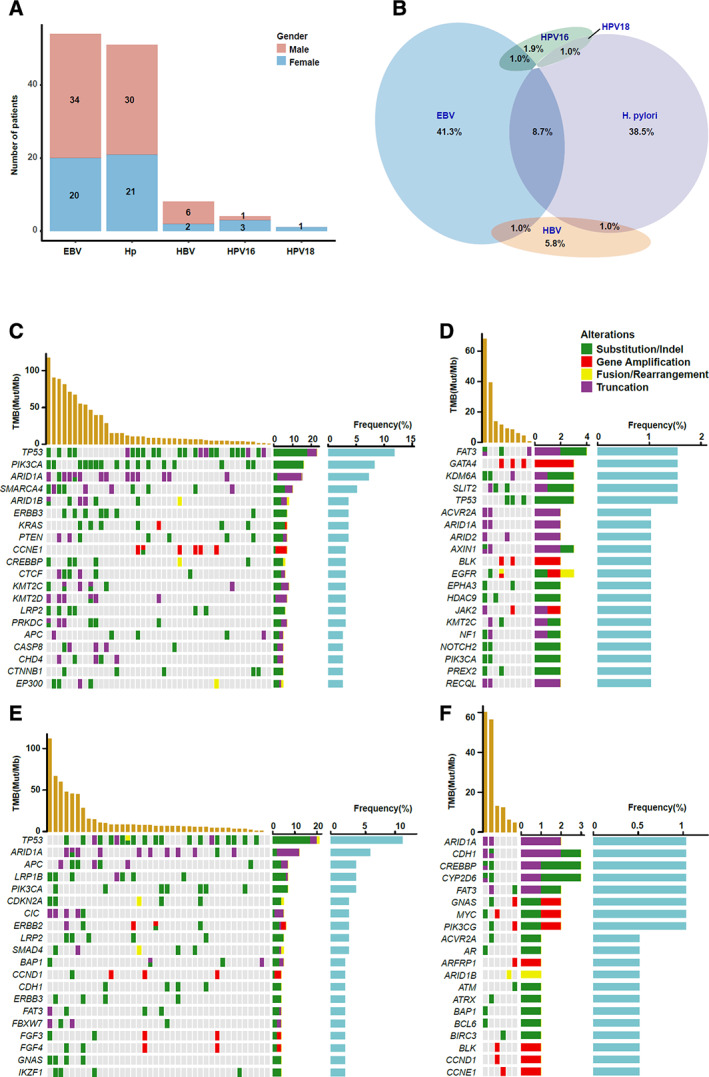
Microbial community analysis and profiling of microbe‐associated molecular features. (A): The microbial infection status was shown across genders among 104 patients with gastric cancer (GC). (B): The coinfection frequency of different GC‐associated microbes. (C): The genomic alterations of patients with GC with EBV infection alone. (D): The genomic alterations of patients with GC and H. pylori infection alone. (E): The genomic alterations of patients with GC and coinfection of EBV and H. pylori. (F): The genomic alterations of patients with GC and HBV infection alone. Abbreviations: EBV, Epstein‐Barr virus; HBV, hepatitis B virus; Hp, H. pylori; HPV16, human papillomavirus type 16; HPV18, human papillomavirus type 18; indel, insertion or deletion; Mut/Mb, mutations per megabase; TMB, tumor mutational burden.
Comparison of Partial Reported Molecular Features Between Different Cohorts
Previous studies by The Cancer Genome Atlas (TCGA) and Asian Cancer Research Group (ACRG) have reported some common features in GC, such as EBV positivity, high MSI, and recurrent mutations of TP53, ARID1A, and PIK3CA [4, 8]. Here, we compared the similarities and differences of some representative features of our data with the TCGA and ACRG GC tumor cohorts. All three data sets showed similarities in the mutation frequencies of PIK3CA and RHOA (Fig. 4A) and the predilection of PIK3CA mutations in EBV‐positive GC (Fig. 4B). However, we saw several differences between cohorts. Our results revealed that only 11.3% of patients with GC were MSI‐H, which was significantly lower than that in both the TCGA (21.7%) and ACRG (22.7%) cohorts; however, the percentage of EBV‐positive patients (10.2%) was slightly higher than in the other two cohorts (Fig. 4A). In our cohort, MSI‐H patients and EBV‐positive patients could not be separated independently according to the classification method by TCGA. We found that 23.3% (14/60) of MSI‐H patients were EBV‐positive and that 25.9% (14/54) of EBV‐positive patients were MSI‐H, whereas RHOA mutations seemed to define a particular subtype, which was basically exclusive with both high MSI and EBV positivity (Fig. 4B).
Figure 4.
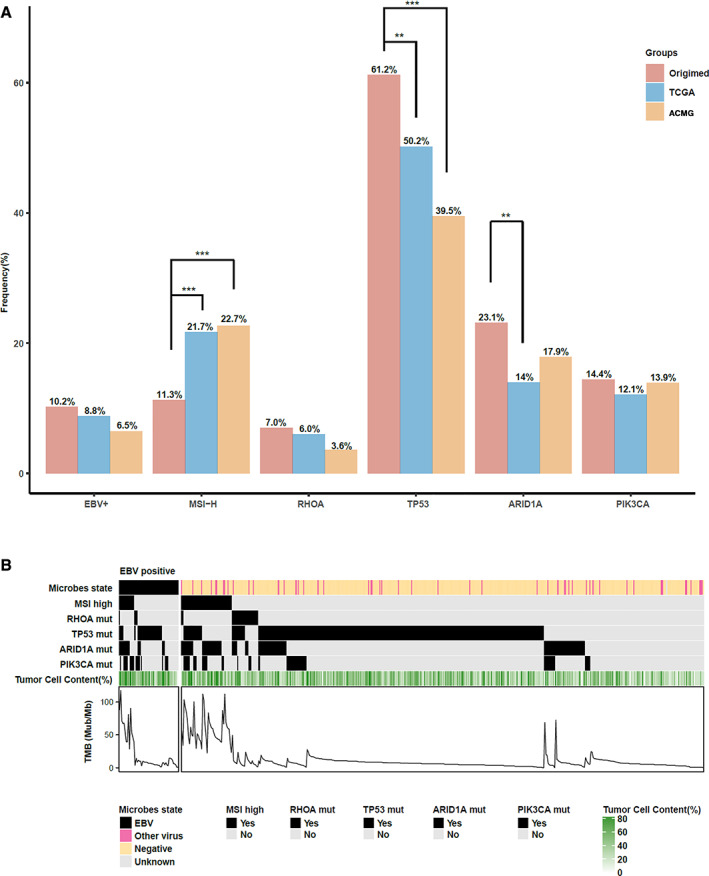
The comparison of partial molecular features of patients with gastric cancer (GC) between our Chinese cohort and TCGA and ACRG cohorts. (A): The frequency of patients infected with EBV, identified with high microsatellite instability and mutations in RHOA, TP53, ARID1A, or PIK3CA in three GC cohorts are shown here. The chi‐square test was used to examine the differences of three cohorts among each group. **, p < .01; ***, p < .001. (B): The overlap of six characteristics in 529 Chinese patients with GC. Unknown of microbe state means the microbe status in this patient is unknown. Abbreviations: ACRG, Asian Cancer Research Group; EBV, Epstein‐Barr virus; MSI, microsatellite instability; MSI‐H, MSI‐high; mut, mutation; Mut/Mb, mutations per megabase; TCGA, The Human Genome Atlas; TMB, tumor mutational burden.
Alterations of Cancer‐Related Signaling Pathways in GC
Inactivation or activation of various signaling pathways has been described in several cancers, and certain types of cancers seem to favor certain signaling changes. To describe the recurrent signaling alterations in GC, we analyzed six signaling pathways that are commonly altered in cancers among the 525 patients with GC who had genetic alterations. The most frequently mutated signaling pathways were Wnt, PI3K/Akt, and cell cycle signaling. Among the patients with GC, 42.3% (n = 222) carried 314 genetic alterations in the Wnt signaling pathway, and the most frequently mutated genes belonging to the Wnt signaling pathway were LRP1B and APC, accounting for 24.8% and 15%, respectively (Fig. 5A). The Food and Drug Administration (FDA) has approved several inhibitors targeting the P13K/Akt and ERBB signaling pathways. We found that 39% and 29.7% of patients with GC harbored mutations in the PI3K/Akt and ERBB signaling pathways, respectively, and that the comutation rate was 13.1% (Fig. 5B).
Figure 5.
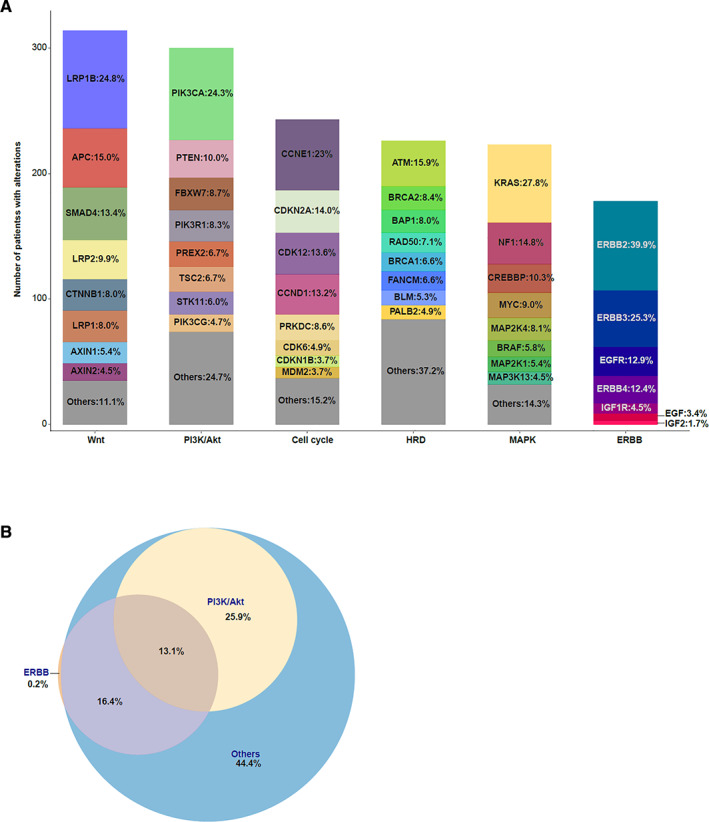
Analysis of signaling changes in patients with gastric cancer (GC). (A): Genetic alterations of 529 patients with GC were classified by cell signaling pathways, including Wnt, PI3K/Akt, cell cycle, HRD, MAPK, and ERBB. The y‐axis shows the number of GC samples with alterations. (B): Coexisting mutations analysis of PI3K/Akt and ERBB signaling in GC. The Venn diagram show the percentage of samples with each signaling change and coalterations of both pathways. Abbreviation: HRD, homologous recombination deficiency.
Targetable Genomic Alterations
To assess the clinical utility of prospective molecular profiling in GC and to determine how many targetable genomic alterations were identified, we systematically evaluated all genetic mutations according to Memorial Sloan Kettering Cancer Center (MSKCC) criteria [1, 11] based on the Precision Oncology Knowledge Base (OncoKB; data version: v1.21; https://oncokb.org/) and classified them into six tiers (1, 2A, 2B, 3A, 3B, and 4) for therapeutic implications. The evidence level of each patient was defined as the highest level across all actionable mutations. Altogether, 47.1% (249/529) of patients harbored at least one actionable mutation, and four types of evidence levels, including level 1 (9.5%), level 2B (19.1%), level 3B (7.8%), and level 4 (10.8%), were identified (Fig. 6). We also showed the summary of genes and alteration types in each kind of evidence level. Among 50 patients who had biomarkers predictive of response to an FDA‐approved drug in GC, 88% of patients harbored ERBB2 gene amplification, whereas the remaining 12% had NTRK mutations. In 101 patients who harbored biomarkers predictive of response to an FDA‐approved drug in other indications besides GC, 13 genes involving 17 kinds of mutation types were identified, and more than half of the patients harbored PIK3CA substitution or insertion or deletion (indel) mutations. In 41 patients whose biomarkers were predictive of response to drugs in other indications, but neither biomarkers nor drugs were standard care, KRAS substitution or indel mutations accounted for 26.3% (Fig. 6).
Figure 6.
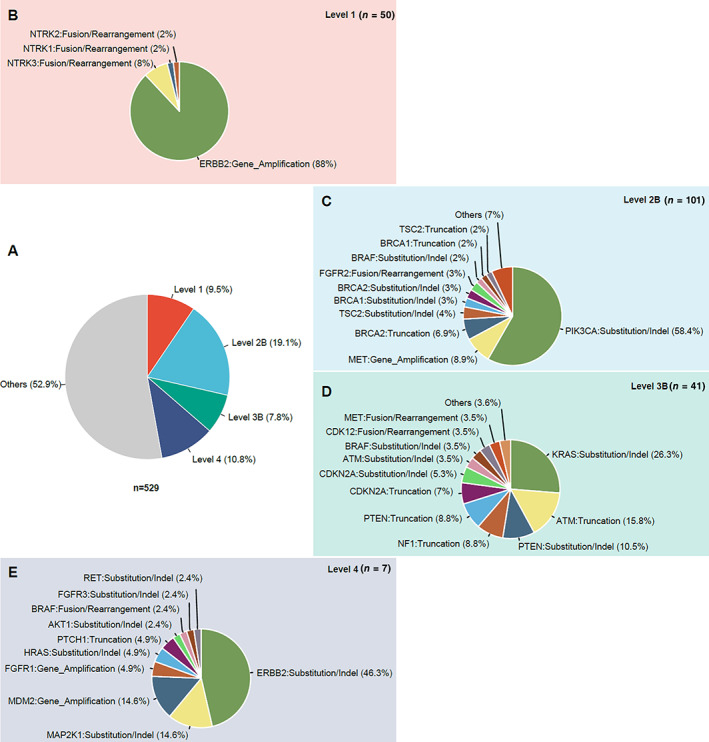
Clinical actionable genomic alterations defined by Memorial Sloan Kettering Cancer Center (MSKCC) levels of evidence in 529 patients with gastric cancer (GC). (A): The larger pie chart shows the percentage of the highest level of actionable mutations in patients with GC. (B–E): The smaller pie charts show the distribution of different genes and mutation types in each level: level 1 (B), level 2B (C), level 3B (D), and level 4 (E). Abbreviation: indel, insertion or deletion.
Immunotherapy Implications
In addition to targetable therapies, checkpoint inhibitor–based immunotherapies have achieved impressive success in the treatment of diverse cancer types including GC. Previous studies have mentioned that anti–PD‐1/PD‐L1 therapy was more effective to a subset of patients with PD‐L1 positivity, high MSI, EBV positivity, or high TMB [12, 13]. To identify precise predictive biomarkers to better select patients with GC who may benefit most from immune inhibitors, here we analyzed the patients who may benefit from immunotherapies and further explored the mutation signatures of two immunotherapy‐associated features: high MSI and high TMB in GC. We found that all MSI‐H patients were TMB‐H, but only a very small portion of TMB‐H patients were microsatellite stable (MSS). PD‐L1–positive or EBV‐positive patients were randomly distributed in TMB‐L and TMB‐H groups or MSI‐H and MSS groups (Fig. 7A). In other words, both PD‐L1 positivity and EBV positivity had no relevance with TMB or MSS status. Furthermore, we revealed that certain gene mutations were significantly associated with both high TMB and high MSI. In patients with GC with high TMB and high MSI, the mutation rates of several genes, including ACVR2A, ARID1A, B2M, CIC, ERBB3, and KMT2C, were much higher than in TMB‐L and MSS patients (Fig. 7B).
Figure 7.
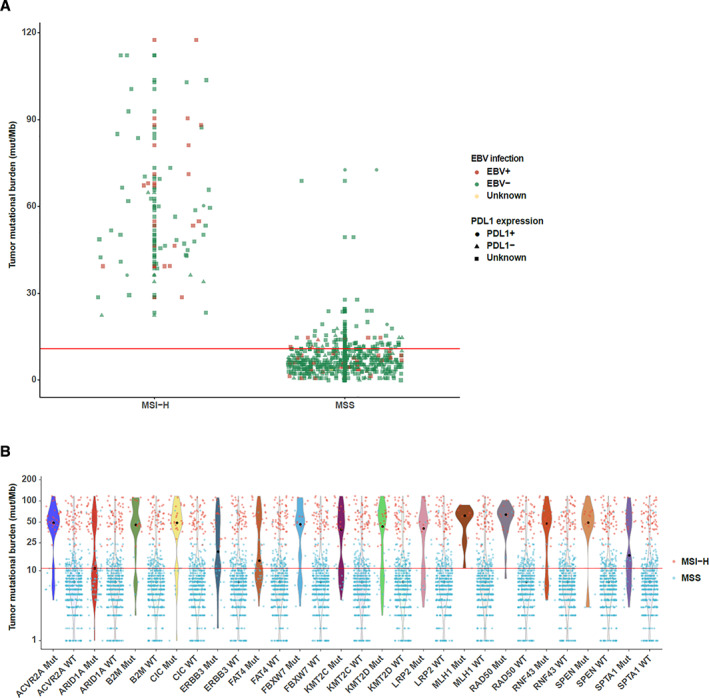
Immunotherapy‐associated features in 529 patients with gastric cancer (GC). (A): The distribution of patients with different status in microsatellite instability, tumor mutational burden, Epstein‐Barr virus infection, and PD‐L1. (B): Genetic alterations associated with both high microsatellite instability and high tumor mutational burden in patients with GC. Abbreviations: EBV, Epstein‐Barr virus; MSI‐H, high microsatellite instability; MSS, microsatellite stability; mut/Mb, mutations per megabase.
Discussion
The widely used hybrid capture–based NGS technology makes it possible to comprehensively understand the molecular features of each tumor and helps to lay out the foundation of patient stratification. More precise stratification strategies may provide more opportunities for precision therapy. Here, we comprehensively profiled the genomic features of Chinese patients with GC and compared some of these features with profiles from other cohorts, and we provided some new implications for both targeted therapies and immunotherapies.
In this study, besides previously known recurrent mutations, we identified several unique molecular characteristics in Chinese patients with GC. For somatic mutations, gene amplifications such as ERBB2, CCND1, FGF3/4/19, and FGFR2 were more frequent than in other cohorts [5, 14]. In our cohort, ERBB2 alteration was found in 71 (13.4%) patients. Of these, 44 (8.3%) patients had ERBB2 amplification, and 31 (5.9%) patients had mutations in ERBB2. The most frequent mutation site in ERBB2 was R678Q (11 patients), followed by S310F, D769Y, and V842I (three patients, respectively). These substitutions have all been considered as activating and oncogenic [11, 15]. The reported rate of ERBB2 amplification in patients with gastric cancer ranged widely from 7% to 34% because of the different proportions of tumor location, grade, and histologic subtype among the cohorts [16, 17, 18]. Likewise, we consider that those differences may cause the lower rate of the ERBB2 amplification in our cohort than that in most studies.
For germline mutations, we identified SPINK1 c.194 + 2T > C as the most frequently appearing variant, which is associated with idiopathic chronic pancreatitis in East Asians and has been widely reported in pancreatic cancer but, to our knowledge, has not been reported in GC [19, 20, 21]. SPINK1 c.194 + 2T > C has been reported to abolish SPINK1 mRNA expression [22]. The loss of SPINK1 expression in gastric cancer tissue has been reported by other cohorts and may be associated with adverse prognosis [23, 24, 25]. On the other hand, high SPINK1 expression in gastric cancer was also been reported and associated with favorable outcome [25, 26]. Thus SPINK1 may be conferred a biologic function responsible for gastric cancer progression, although the mechanism remains to be elucidated. FANCA is the most prevalent mutated gene in Fanconi anemia, which is an autosomal recessive DNA disorder and has a high risk of developing hematological and solid malignancies [27, 28]. In our study, FANCA germline mutations found in five patients included truncations (four of five) and mutation in splicing site (one of five). FANCA is recognized as a tumor suppressor, and its truncated or inactivated mutations may be involved in the promotion of genetic instability, which would promote oncogenesis [27, 29]. Based on these observations, there are reasons to suspect that SPINK1 and FANCA germline mutations may increase the risk of gastric cancer.
In addition, we noticed that the most widely adopted TCGA subtyping system may not be suitable for Chinese patients. By analyzing 295 patients from across the world, the TCGA divided GC into four subtypes: EBV‐positive tumors, microsatellite unstable tumors (MSI‐H), genomically stable tumors, and tumors with chromosomal instability. EBV positivity and high MSI seemed to be mutually exclusive events in the TCGA GC cohort, but in Chinese patients with GC, EBV positivity and high MSI were mutually inclusive. However, the overlap of MSI‐H and EBV‐positive tumors is not an exception. Martinez‐Ciarpaglini et al. reported that 3 of 13 (23%) EBV‐positive cases were also MSI‐H [30]. These findings may result from the different detection methods and cutoffs for defining positivity. Here, when we used 10 mapping reads on the EBV genome as a cutoff for EBV, 14 out of 54 (26%) EBV‐positive cases were MSI‐H. When the cutoff for EBV was 100, only two (14%) of the 14 EBV‐positive cases were MSI‐H. However, when 1,000 was the cutoff, EBV positivity and high MSI became mutually exclusive. In order to know the correspondence of EBV DNA level with the EBV encoding region in situ hybridization (EBER ISH) detection result, we used targeted sequencing in parallel with EBER ISH for EBV evaluation; the results showed that EBV DNA more than 1,000 may have a positive detection in EBER ISH (unpublished data). Interestingly, we found that in these EBV‐low‐load (EBV DNA levels 10 ~ 1,000) MSI‐H tumors, ARID1A mutations were found in 10 cases (71%), and no MMR genes had germline mutations. ARID1A loss was reported to have significant association with EBV positivity, loss of MMR protein expression, and MSI‐H status [31]. EBV‐positive tumors reported by TCGA had a higher prevalence of DNA hypermethylation, but they lacked the MLH1 hypermethylation characteristic of MSI‐H tumors [9]. Our results showed that the higher EBV DNA level detected in patients, the lower frequency of high MSI occurred, indicating a regulation between virus‐induced hypermethylation and MLH1 hypermethylation–induced high MSI, which needs confirmation and further studies. RHOA mutations were almost incompatible with EBV positivity or high MSI, consistent with RHOA mutating almost exclusively in genomically stable gastric tumors [9].
In addition, we profiled the comprehensive infectious landscape of 26 cancer‐related microbes in GC and detected five different types of microorganisms, including EBV, H. pylori, HBV, HPV16, and HPV18 in GC, which is partially similar to previous studies, but the frequency of each infection type was a bit different [32, 33, 34]. Consistent with prior reports, we observed high mutation frequencies of PIK3CA and ARID1A in EBV‐positive tumors [9]. In contrast to the rare TP53 mutations in TCGA EBV‐positive subtype, we observed a high mutation rate of TP53 because of our lower threshold for EBV positivity. In gastric cancer with H. pylori infection, a high frequency of TP53 mutations (three of nine) were observed, as suggested by prior studies [35, 36]. To our knowledge, this is the first mutational landscape of GCs infected with EBV infection alone, H. pylori infection alone, coinfection of EBV and H. pylori, and HBV infection alone. Recent studies suggest that survival may be better in patients with GC tumors that harbor EBV, and negative H. pylori status is associated with poor prognosis in patients with GC [37, 38]. However, the underlying molecular mechanisms were not well studied, and the clinical outcomes of other kinds of infections in GC are largely unknown. Our microbial profiling data combined molecular changes with certain types of infection, which may provide new perspectives on prognosis prediction and help to develop new prognostic tumor markers to improve clinical outcomes for patients with GC.
We found that many patients with GC may be able to benefit from either targeted therapy or immunotherapy. There were 47.1% (249/529) of patients who harbored at least one actionable mutation according to MSK levels of evidence. For the remaining 52.9% of patients, there were still 25% (70/280) of patients with at least one feature of PD‐L1 positivity, high MSI, EBV positivity, or high TMB. We also observed a relatively high rate of EBV‐positive patients in Chinese GC, which may be due to methodology differences, but this information is still valuable and, at the very least, offers complementary information on identifying potential patients who may benefit from immunotherapy. Simultaneous inhibition of the PI3K/ERBB pathway has been tested in clinical trials for the treatment of various cancers, and, given that the co‐occurring mutation of PI3K and ERBB signaling is relatively common in Chinese GC, combination therapy of both PI3K and ERBB inhibitors may be a feasible option [39]. Furthermore, we identified several genes such as ACVR2A, ARID1A, CIC, ERBB3, FAT4, and KMT2C with significantly higher mutation rates in both TMB‐H and MSI‐H patients with GC. Even though the mechanisms are not very clear, this phenomenon has been observed in certain genes in other types of cancer, such as ARID1A in ovarian cancer [40]. Hence, recurrent mutation of these genes may add predictive value for immunotherapy response in GC.
Conclusion
In this study, we provided comprehensive mutational landscapes and identified unique molecular features covering somatic mutations, germline mutations, and microorganisms in 529 Chinese patients with gastric adenocarcinoma. The mutation frequency of PIK3CA and RHOA and the predilection of PIK3CA mutations in EBV‐positive GC are consistent with previous reports, but the ratio of MSI‐H and EBV‐positive patients in Chinese patients are completely different from both the TCGA and ACRG cohorts, and the MSI‐H and EBV‐positive features seem to be mutually inclusive in Chinese patients with GC. Based on the unique molecular alterations, we further analyzed their potential implications on targeted, combined, and immunotherapies. We hope these results will facilitate the development of clinical trials to explore new biomarkers and more precision therapy, ultimately deepening our understanding and improving patient survival of GC.
Author Contributions
Conception/design: Yi‐an Du, Xiangdong Cheng
Provision of study material or patients: Pengfei Yu, Yusheng Wang, Ling Huang
Collection and/or assembly of data: Pengfei Yu, Yanfei Yu, Aodi Wang, Yuan Zhang
Data analysis and interpretation: Pengfei Yu, Yusheng Wang, Wenjing Liu, Haiyan Wu
Manuscript writing: Pengfei Yu, Ming Yao, Xiangdong Cheng, Yi‐an Du
Final approval of manuscript: Pengfei Yu, Yusheng Wang, Yanfei Yu, Aodi Wang, Ling Huang, Yuan Zhang, Wenjing Liu, Haiyan Wu, Ming Yao, Yi‐An Du, Xiangdong Cheng
Disclosures
Yanfei Yu: OrigiMed (E); Aodi Wang: OrigiMed (E); Yuan Zhang: OrigiMed (E); Wenjing Liu: OrigiMed (E); Haiyan Wu: OrigiMed (E); Ming Yao: OrigiMed (E). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Table S1 Characteristics of the patients
Table S2. Gene lists for OM 450‐gene panel.
Acknowledgments
The study team is grateful to all investigators and patients who contributed to this study and also to the study coordinators for their contributions. This study was supported by the Natural Science Foundation of Zhejiang Province of China (LY18H160032 to Pengfei Yu) and Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (2020KY471).
No part of this article may be reproduced, stored, or transmitted in any form or for any means without the prior permission in writing from the copyright holder. For information on purchasing reprints contact commercialreprints@wiley.com. For permission information contact permissions@wiley.com.
Disclosures of potential conflicts of interest may be found at the end of this article.
Contributor Information
Ming Yao, Email: yaom@origimed.com.
Yi‐An Du, Email: duyajim@126.com.
Xiangdong Cheng, Email: abdsurg@163.com.
References
- 1. Zehir A, Benayed R, Shah RH et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 3. Karimi P, Islami F, Anandasabapathy S et al. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao JP, Xu W, Liu WT et al. Tumor heterogeneity of gastric cancer: From the perspective of tumor‐initiating cell. World J Gastroenterol 2018;24:2567–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cristescu R, Lee J, Nebozhyn M et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–456. [DOI] [PubMed] [Google Scholar]
- 6. Cislo M, Filip AA, Arnold Offerhaus GJ et al. Distinct molecular subtypes of gastric cancer: From Laurén to molecular pathology. Oncotarget 2018;9:19427–19442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lauren P. The two histological main types of gastric carcinoma: Diffuse and so‐called intestinal‐type carcinoma. An attempt at a histo‐clinical classification. Acta Pathol Microbiol Scand 1965;64:31–49. [DOI] [PubMed] [Google Scholar]
- 8. Lei Z, Tan IB, Das K et al. Identification of molecular subtypes of gastric cancer with different responses to PI3‐kinase inhibitors and 5‐fluorouracil. Gastroenterology 2013;145:554–565. [DOI] [PubMed] [Google Scholar]
- 9. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao J, Chen L, Li H et al. An accurate and comprehensive clinical sequencing assay for cancer targeted and immunotherapies. Oncologist 2019;24:e1294–e1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chakravarty D, Gao J, Phillips SM et al. OncoKB: A precision oncology knowledge base. JCO Precis Oncol 2017;2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen C, Zhang F, Zhou N et al. Efficacy and safety of immune checkpoint inhibitors in advanced gastric or gastroesophageal junction cancer: A systematic review and meta‐analysis. Oncoimmunology 2019;8:e1581547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim ST, Cristescu R, Bass AJ et al. Comprehensive molecular characterization of clinical responses to PD‐1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–1458. [DOI] [PubMed] [Google Scholar]
- 14. Wang K, Yuen ST, Xu J et al. Whole‐genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573–582. [DOI] [PubMed] [Google Scholar]
- 15. Subramanian J, Katta A, Masood A et al. Emergence of ERBB2 mutation as a biomarker and an actionable target in solid cancers. The Oncologist 2019;24:e1303–e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanayama K, Imai H, Usugi E et al. Association of HER2 gene amplification and tumor progression in early gastric cancer. Virchows Arch 2018;473:559–565. [DOI] [PubMed] [Google Scholar]
- 17. Jiang W, Jin Z, Zhou F et al. High co‐expression of SP1 and HER‐2 is correlated with poor prognosis of gastric cancer patients. Surg Oncol 2015;24:220–225. [DOI] [PubMed] [Google Scholar]
- 18. Zhou F, Li N, Jiang W et al. Prognosis significance of HER‐2/Neu overexpression/amplification in Chinese patients with curatively resected gastric cancer after the TOGA clinical trial. World J Surg Oncol 2012;10:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun C, Liao Z, Jiang L et al. The contribution of the SPINK1 c.194+2T>C mutation to the clinical course of idiopathic chronic pancreatitis in Chinese patients. Dig Liver Dis 2013;45:38–42. [DOI] [PubMed] [Google Scholar]
- 20. Boortalary T, Jalaly NY, Moran RA et al. Metastatic pancreatic adenocarcinoma in a patient with chronic calcific pancreatitis and a heterozygous SPINK1 c.194+2T>C mutation. Pancreas 2018;47:e24–e25. [DOI] [PubMed] [Google Scholar]
- 21. Hegyi E, Sahin‐Toth M. Genetic risk in chronic pancreatitis: The trypsin‐dependent pathway. Dig Dis Sci 2017;62:1692–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kereszturi E, Kiraly O, Sahin‐Toth M. Minigene analysis of intronic variants in common SPINK1 haplotypes associated with chronic pancreatitis. Gut 2009;58:545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lei KF, Liu BY, Zhang XQ et al. Development of a survival prediction model for gastric cancer using serine proteases and their inhibitors. Exp Ther Med 2012;3:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiksten JP, Lundin J, Nordling S et al. Comparison of the prognostic value of a panel of tissue tumor markers and established clinicopathological factors in patients with gastric cancer. Anticancer Res 2008;28:2279–2287. [PubMed] [Google Scholar]
- 25. Rasanen K, Itkonen O, Koistinen H et al. Emerging roles of SPINK1 in cancer. Clin Chem 2016;62:449–457. [DOI] [PubMed] [Google Scholar]
- 26. Wiksten JP, Lundin J, Nordling S et al. High tissue expression of tumour‐associated trypsin inhibitor (TATI) associates with a more favourable prognosis in gastric cancer. Histopathology 2005;46:380–388. [DOI] [PubMed] [Google Scholar]
- 27. Tischkowitz MD, Morgan NV, Grimwade D et al. Deletion and reduced expression of the Fanconi anemia fanca gene in sporadic acute myeloid leukemia. Leukemia 2004;18:420–425. [DOI] [PubMed] [Google Scholar]
- 28. Alter BP. Fanconi's anemia and malignancies. Am J Hematol 1996;53:99–110. [DOI] [PubMed] [Google Scholar]
- 29. Castella M, Pujol R, Callen E et al. Origin, functional role, and clinical impact of Fanconi anemia fanca mutations. Blood 2011;117:3759–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez‐Ciarpaglini C, Fleitas‐Kanonnikoff T, Gambardella V et al. Assessing molecular subtypes of gastric cancer: Microsatellite unstable and Epstein‐Barr virus subtypes. Methods for detection and clinical and pathological implications. ESMO Open 2019;4:e000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Han N, Kim MA, Lee HS et al. Loss of ARID1A expression is related to gastric cancer progression, Epstein‐Barr virus infection, and mismatch repair deficiency. Appl Immunohistochem Mol Morphol 2016;24:320–325. [DOI] [PubMed] [Google Scholar]
- 32. Zeng ZM, Luo FF, Zou LX et al. Human papillomavirus as a potential risk factor for gastric cancer: A meta‐analysis of 1,917 cases. Onco Targets Ther 2016;9:7105–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wei XL, Qiu MZ, Jin Y et al. Hepatitis B virus infection is associated with gastric cancer in China: An endemic area of both diseases. Br J Cancer 2015;112:1283–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh S, Jha HC. Status of Epstein‐Barr virus coinfection with Helicobacter pylori in gastric cancer. J Oncol 2017;2017:3456264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shimizu T, Marusawa H, Matsumoto Y et al. Accumulation of somatic mutations in TP53 in gastric epithelium with Helicobacter pylori infection. Gastroenterology 2014;147:407–417.e403. [DOI] [PubMed] [Google Scholar]
- 36. Li N, Xie C, Lu NH. P53, a potential predictor of Helicobacter pylori infection‐associated gastric carcinogenesis? Oncotarget 2016;7:66276–66286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marrelli D, Pedrazzani C, Berardi A et al. Negative Helicobacter pylori status is associated with poor prognosis in patients with gastric cancer. Cancer 2009;115:2071–2080. [DOI] [PubMed] [Google Scholar]
- 38. Camargo MC, Kim WH, Chiaravalli AM et al. Improved survival of gastric cancer with tumour Epstein‐Barr virus positivity: An international pooled analysis. Gut 2014;63:236–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Crafter C, Vincent JP, Tang E et al. Combining AZD8931, a novel EGFR/HER2/HER3 signalling inhibitor, with AZD5363 limits AKT inhibitor induced feedback and enhances antitumour efficacy in HER2‐amplified breast cancer models. Int J Oncol 2015;47:446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen J, Ju Z, Zhao W et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med 2018;24:556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Table S1 Characteristics of the patients
Table S2. Gene lists for OM 450‐gene panel.


