Highlights
-
•
Cancer-associated cachexia is mainly characterized by skeletal muscle and adipose loss, release of pro-cachectic factors by cancer and immune cells and energy imbalance.
-
•
Cancer cachexia is prominent in many types of cancer and it is associated with poor prognosis.
-
•
MiRNAs and lncRNAs implicated in muscle atrophy, lipolysis, adipose browning and inflammation are deregulated in muscle or adipose tissue in cancer cachexia patients or animal models of cancer cachexia.
-
•
Deregulated ncRNAs are found in circulation or in circulating exosomes in cancer cachexia patients.
Keywords: Cachexia, Cancer, miRNAs, ncRNAs, lncRNAs, Circulating RNAs
Abstract
Cachexia is a multifactorial syndrome characterized by skeletal muscle loss, with or without adipose atrophy, irreversible through nutritional support, in the context of systemic inflammation and metabolic disorders. It is mediated by inflammatory reaction and affects almost 50% of all cancer patients, due to prominent systemic inflammation associated with the disease. The comprehension of the molecular mechanisms that are implicated in cancer cachexia sheds light on its pathogenesis and lays the foundations for the discovery of new therapeutic targets and biomarkers. Recently, ncRNAs, like microRNAs as well as lncRNAs and circRNAs seem to regulate pathways that are implicated in cancer cachexia pathogenesis, as it has been observed in animal models and in cancer cachexia patients, highlighting their therapeutic potential. Moreover, increasing evidence highlights the involvement of circulating and exosomal ncRNAs in the activation and maintenance of systemic inflammation in cancer and cancer-associated cachexia. In that context, the present review focuses on the clinical significance of ncRNAs in cancer-associated cachexia.
Graphical abstract
1. Introduction
Cachexia is a devastating multifactorial syndrome defined as skeletal muscle loss with or without adipose tissue loss, in the context of chronic systemic inflammation and metabolic disorders, and it is not reversed by nutritional support [1]. Cachexia results from the activity of cytokines, such as tumor necrosis factor alpha (TNFα), interleukin 1 (IL-1), IL-6 and interferon gamma (IFN-γ), which modify the metabolism of macronutrients, reducing protein synthesis, increasing protein degradation, enhancing lipolysis, intensifying the production of acute phase proteins in liver, increasing gluconeogenesis and inducing insulin resistance [2,3]. Sequentially, these alterations result in anorexia accompanied by muscle and fat loss [1].
According to 2011 Delphi International Consensus “cancer cachexia is defined as a multifactorial syndrome characterized by an ongoing loss of skeletal muscle mass (with or without loss of fat mass) that cannot be fully reversed by conventional nutritional support and leads to progressive functional impairment” [4]. Cancer cachexia is classified to three stages, depending on the severity of the disease: precachexia, which is characterized by weight loss ≤5%, anorexia and metabolic change, cachexia, in which weight loss >5% or body mass index (BMI) <20 and weight loss >2% or sarcopenia and weight loss >2% is observed, and refractory cachexia, which is characterized by low performance score and variable degrees of cachexia, procatabolic and not responsive cancer disease to treatment and very short (<3 months) expected survival [4]. The impact of cachexia on quality of life and survival becomes worse in cachexia and refractory cachexia, while less is the impact of precachexia on prognosis [5].
Cachexia can sometimes present together with sarcopenia and it can be hard to distinguish due to the lack of consensus regarding clinical definition, diagnostic criteria and classification of the syndrome, although in cancer patients cachexia and sarcopenia are two distinct muscle-wasting conditions [6]. Sarcopenia is defined as a progressive and generalized skeletal muscle disorder, which is characterized by muscle loss and function and results in adverse effects such as falls, impaired functions, weakening and mortality [7]. Sarcopenia is usually correlated with thinness, but can sometimes co-exist with obesity (BMI 30.0 kg/m2), which makes muscle mass quantification difficult and can lead to underestimation of the disease, leading to disability or death [8]. Moreover, skeletal muscle depletion is prognostic for cancer patients’ survival, independently of BMI [9]. Cachexia should be distinguished from sarcopenia or simple muscle wasting or atrophy, as it is a multifactorial syndrome and different mechanisms underlie the etiology of muscle atrophy in sarcopenia and cachexia. The age-related reduction in endocrine signaling and low calories intake affect muscle loss in sarcopenia, while muscle atrophy in cachexia is caused mainly by inflammation and oxidative stress and leads to muscle and fat loss [6].
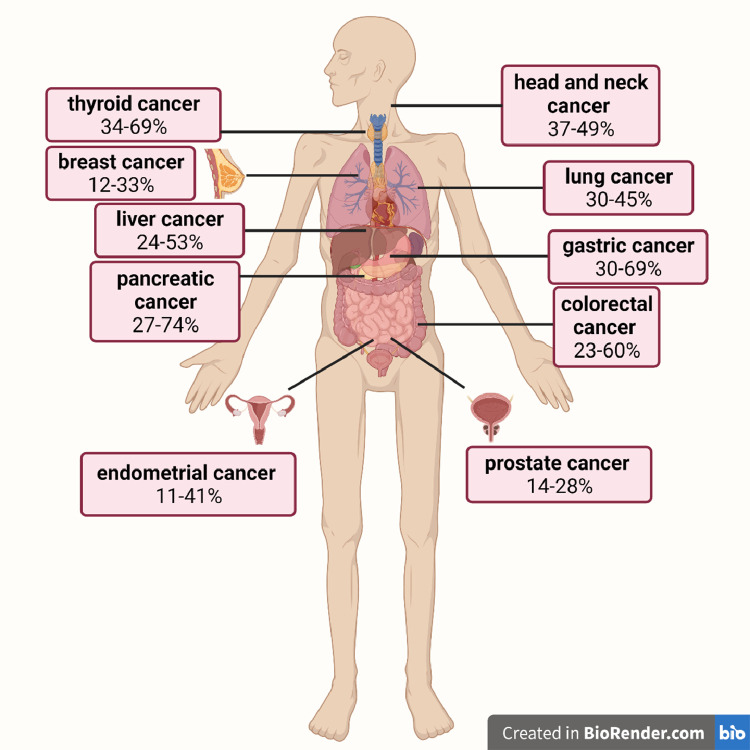
It has been documented that loss of 30% of the body weight is significantly associated with mortality of cancer patients [10]. As cachexia is mediated by inflammatory response, it affects almost 50% of all cancer patients, due to the prominent cancer-associated systemic inflammation. However, its incidence differs depending on cancer localization, with patients with solid tumors and especially gastric, pancreatic, lung, colorectal and head and neck cancer to present higher rates [5,11] (Fig. 1). Recently, Freire et al. revealed a tumor-specific cachexia-inducing factors profile, which shows that different solid tumors have different expression profile of secreted genes-inducers of cachexia, explaining why specific cancer types are more likely to develop cachexia [12]. Almost one third of cancer patients will lose >5% of their body weight after cancer diagnosis, while up to 80% of them with advanced disease will be diagnosed with cachexia [10,13,14]. Moreover, pretreatment weight loss in cancer patients significantly reduces overall survival, even in early disease stage [15].
Fig. 1.
Prevalence of cancer cachexia in different cancer types. Based on data from the systematic review by Anker et al. [11] (Created with BioRender.com).
2. Cachexia pathophysiology
Most data on cachexia pathophysiology have been derived from animal models. Clinical data from patients are limited, since cachexia presents in advance stage, when patients’ vulnerability does not permit conducting invasive tests. Moreover, cancer cachexia is usually diagnosed in advanced stages, thus the number of patients that can be under observation over time is limited [16].
Reduced food intake, which leads to weight loss is associated with cancer cachexia altering thus the energy balance and normal control of homeostasis. Energy intake is typically lower than energy expended for maintenance of homeostasis [17,18]. Moreover, reduced protein synthesis in muscle of cancer patients that lose weight can be re-activated through nutritional intake. Additionally, tumors compete with organs and tissues for the energy reserves, increasing the energy imbalance even more [19].
Cytokines as well as pro-cachectic factors are mainly implicated in cancer cachexia. Tumor cells, tumor microenvironment and immune cells secrete factors that promote catabolism in tissue-targets, such as pro-inflammatory cytokines [20]. Increased inflammation, which is enhanced by tumors, also participates in catabolic pro-inflammatory factors production. These factors mediate the homeostatic control in Central Nervous System leading to catabolic neuronal effects, as well as in neuroendocrine effects, such as adrenal corticosteroids release and weakness (anorexia, fatigue). These inflammatory, neuronal and behavioral effects directly activate proteolysis and lipolysis in organs-targets and particularly in skeletal muscle, adipose tissue and cardiac muscle [21]. Prostaglandins and particularly prostaglandin E2 are known mediators of tumor-induced bone absorption and paraneoplastic hypercalcemia and have been also documented as mediators of excessive catabolism in skeletal muscle [22]. Among peptidic inflammatory cachexia regulators are included IL-6, IL-1, TNF and IFNγ. These factors bind to their cell receptors leading to activation of specific transcription factors, such as STA3 and NFκ-B, which can induce autophagy and degradation of muscle proteins [23].
In cancer cachexia the interaction of various organs is prominent. It has been documented that Central Nervous System exercises primary control in cachexia pathogenesis through cytokines identification as molecular signals of the disease [24]. Pro-inflammatory cytokines produced by tumor and immune cells interaction can act in the Central Nervous System enhancing the inflammation in hypothalamus, leading to anorexia and skeletal muscle atrophy, while lipolytic factors induce lipolysis of adipose tissue [25], [26], [27].
In addition to muscle loss, adipose tissue loss is also responsible for weight loss in cancer patients [28]. Studies have shown that fat loss occurs mostly due to lipolysis, rather than non-reversible adipose cells degeneration due to apoptosis, and that the overall increase of lipolysis in cancer patients is about 50% [29]. Although many explanations have been suggested for tumor-induced lipolysis, as presence of inflammatory cytokines released from macrophages that infiltrate the tumor, induction of triglyceride lipase in fat and loss of activated protein kinase of 5’ AMP, the mechanism through which fat loss contributes to skeletal muscle atrophy remains unclear [30], [31], [32], [33]. It has also been suggested that white adipose tissue browning as well as vicious biochemical cycles that take place in brown adipose tissue and during which oxidative phosphorylation is not linked to ATP synthesis and only heat is produced, lead to increase and ineffective energy consumption, contributing to cachexia [34], [35], [36].
Regarding cardiac muscle atrophy, data derived from animal models with induced cancer cachexia have confirmed it along with cardiac dysfunction [37]. Moreover, data on cardiac atrophy mechanisms derived from animal models are typical of the skeletal muscles’ atrophy as well, including inflammation, proteolysis, apoptosis and autophagy [21].
3. Prognostic and predictive role of cachexia
Cachexia, sarcopenia and obesity have been associated with prognosis and effectiveness of therapy in many types of cancer. In non-small cell lung cancer underweight patients bearing EGFR mutations had poor prognosis (time to disease progression and survival) after treatment with tyrosine kinase inhibitors (ΤΚΙs) [38]. In a recent meta-analysis, cachexia was related to shorter overall survival of lung cancer patients, both small cell and non-small cell lung cancer [39]. In bladder cancer, cachexia has been associated with increased rate of perioperative complications during radical cystectomy, as well as poor prognosis of patients that had, or not, undergone surgery, while patients with urothelial cancer who had received platinum-based chemotherapy and regained skeletal muscle mass had good prognosis [40,41]. In colorectal cancer, developing cachexia after cancer diagnosis has been associated with poor prognosis (disease-free survival and overall survival) [42]. Similarly, in pancreatic cancer, where cachexia accounts for almost 80% of mortality, cachectic patients that had surgery or palliative therapy had shorter survival compared to non-cachectic patients [43]. In another study in advanced gastric cancer, patients who had palliative chemotherapy and cachexia, as confirmed with computed tomography, had poor prognosis [44].
Furthermore, cachexia has also been associated with cancer prognosis and prediction of response or therapy toxicities as well [45]. Metabolic changes that take place during cachexia result in reduced immunity against cancer, which negatively affects immunotherapy, that is implemented today in many types of cancer. Recent studies in non-small cell lung cancer patients, with cachexia presented before therapy or cachexia developed during therapy with immune checkpoint inhibitors, reported lower response rate, shorter disease-free survival and overall survival [46,47]. Besides, high Body Mass Index has been associated with higher response rates to immune therapy and better prognosis of patients with lung or other types of cancer [48], [49], [50]. Additionally, in a very recent systematic review published by Guzman-Prado et al. sarcopenia was linked to increased risk for adverse events in cancer patients treated with immune checkpoint inhibitors [51]. In metastatic renal cancer, treatment with sorafenib can intensify muscle wasting, even in patients with normal or high BMI and has been associated with weakness, fatigue and physical disability [52].
4. Epigenetics and cancer cachexia
Epigenetic mechanisms typically include DNA methylation, histone modifications and non-coding RNAs (ncRNAs) [53]. The role of DNA methylation in cancer has been established in every step of cancer progression, from initial carcinogenesis to metastasis [54,55]. However, few studies have reported the association of DNA methylation and cachexia. In a recent study, He et al. analyzed the blood methylation pattern of sarcopenic and non-sarcopenic women, 65–80 years old, and found that sarcopenic women had higher methylation levels in genomic regions before the transcription initiation site [56]. The methylated regions concerned genes related to muscle function, energy metabolism and cytoskeleton regulation, such as HSPB1, PBX4, CNKSR3, ORMDL3, MIR10A, ZNF619 and CRADD [56]: HSPB1 promotes the correct folding of other proteins, PBX4 play critical roles in embryonic development, CNKSR3 coordinates assembly of a multiprotein epithelial sodium channel (ENaC)-regulatory complex, ORMDL3 is highly expressed in fat and regulates cytokine production, ZNF619 is associated with muscular dystrophies and CRADD encodes a protein containing a death domain (DD) motif.
DNA methylation seems to play significant role in memory contribution or reprogramming of skeletal muscles in muscle loss. In vitro cultured muscle cells seem to remember the initial environment from which they were isolated [57]. Particularly, myoblasts that were treated initially with TNFα continued exhibiting high levels of myoD methylation even after 30 cell divisions and cells showed decreased morphological and biochemical differentiation. Since TNFα is chronically increased in cancer cachexia and high levels are associated with muscle loss, its lasting effect on myoD seems to play a critical role in muscle loss [57].
Regarding histone modifications, sirtuin 1 (Sirt1) is a NAD-dependent deacetylase of proteins which are key-regulators of cell defense and survival as response to stress, thus contributing to cell regulation. In myoblasts cultures Sirt1 was found increased in stress conditions and contributed to myoblasts death decrease, while after treatment with the antioxidant reservatrol, it regulated skeletal muscle survival and enhanced differentiation [58]. In another study, Byun et al. showed that HDAC11 can suppress myoblasts differentiation through MyoD-dependent regulation of transcription [59]. More specifically, in myoblasts cell line C2C12, HDAC11 expression pattern increased during myoblasts differentiation, while HDAC11 ectopic expression inhibited myoblasts differentiation, while mutated and inactive HDAC11 (H142/143A) did not. Moreover, wild type HDAC11 suppressed MyoD-induced activity of MEF2C and MYOG (Myogenin) and reduced histone acetylation close to E-boxes, MyoD and MEF2C and MYOG promoters, whereas mutated HDAC11 did not [59].
Trichostatin A (TSA), a histone deacetylase inhibitor has been shown to abolish muscle atrophy in a mouse model, through MuRF1 regulation [60]. Particularly, TSA reduced muscle atrophy, prevented type I and IIα fibers loss and reversed slow fibers transition to fast fibers. Autophagy-lysosomes pathway remained non activated, while MurF1 expression was suppressed, without transcription factor Foxo3 being affected [60].
Among epigenetic mechanisms, non-coding RNAs have emerged as the most prominent ones involved in cancer cachexia.
4.1. Non-coding RNAs and cancer cachexia
The term non-coding RNAs (ncRNAs) describes a large group of transcripts with low or no potential of translation to proteins, considering that only 2% of total transcribed RNAs is translated to proteins [61], [62], [63]. The majority of the genomes of mammals is transcribed into ncRNAs, many of which are further processed to alternative splicing and generate smaller molecules, which nevertheless are not translated to proteins [64]. NcRNAs can differ in length, and can be short, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), small nuclear RNAs (snRNAs), piwi-interacting RNAs (piRNAs) and other, or long, such as long ncRNAs (lncRNAs). NcRNAs play a significant role in phenotypic variation, chromatin architecture, transcription, RNA editing, translation, and other functions, thus are implicated in many physiological and pathophysiological processes, mainly via binding to multiple RNA molecules and regulating their expression [64]. Among the different ncRNAs, the most well studied ones are miRNAs, on which, along with lncRNAs, the present review will focus.
Recently, ncRNAs have been associated with cancer cachexia, modifying substantial functions, such as skeletal muscle and adipose tissue turnover. Moreover, it has been demonstrated that circulating ncRNAs can function as potential biomarkers for cancer cachexia risk [65,66].
4.1.1. miRNAs and animal studies in cancer cachexia
MiRNAs are short ncRNAs, 19-24 nt long, that regulate almost 60% of all genes coding proteins [67]. This regulation can occur through miRNAs binding to mRNA and sequential mRNA degradation or suppression of translation. Their role in cancer is well established, as they are implicated in every step of carcinogenesis, due to their involvement in proliferation, apoptosis, migration and invasion [61,68,69].
In an in vivo study using wild type and Parp-1−/− and Parp-2−/− (poly ADP-ribose polymerase) mouse models with lung cancer and cachexia an association was demonstrated between Parp-1/2 expression levels and muscle-specific miRNAs [70]. It was shown that miR-1 was downregulated in skeletal muscles, while in cachectic wild type and Parp-2−/− mice miR-133a expression was reduced in diaphragm and gastrocnemius compared to control mice, whereas in Parp-1−/− mice it was reduced only in diaphragm. In cachectic Parp-1−/− and Parp-2−/− mice miR-206 expression was also reduced in diaphragm, as well as in gastrocnemius and diaphragm of wild type cachectic mice. MiR-486 expression was lower in diaphragm and gastrocnemius of wild type and Parp-2−/− cachectic mice, compared to non-cachectic mice, whereas in Parp-1−/− cachectic mice no such difference was observed. Moreover, inhibition of Parp-1 via miR-133a, miR-206 and miR-486 promoted proliferation and differentiation of muscle cells, while inhibition of Parp-2 through miR-206 promoted differentiation of muscle cells in gastrocnemius of cachectic mice with lung cancer. Overall the above results in Parp-1−/− and Parp-2−/− mouse models highlight the association between Parp expression and signaling pathways regulated by miRNAs implicated in cancer cachexia [70].
A study by Lee et al. showed that miRNAs analysis of anterior tibialis muscle from 8 week old C57BL/6J mice with Lewis lung carcinoma that developed cachexia revealed 9 miRNAs with differential expression [71]. More specifically, miR-147-3p, miR-299a-3p, miR-1933-3p, miR-511-3p, miR-3473d, miR-223-3p, miR-431-5p, miR-665-3p and miR-205-3p were differentially expressed. These miRNAs are implicated in various functions, including cell signaling and growth, and inflammatory responses and were suggested to be involved in promoting a catabolic muscle state. However, the validation of the functional role of the above miRNAs needs to be established in mature skeletal muscle [71].
Integrated microRNAs-mRNAs omics analyses have become an additional tool in comprehension of transcriptional and post-transcriptional regulatory networks implicated in muscle loss in cancer cachexia. In a recent study by Fernandez et al. mRNA and miRNAs expression profiles of anterior tibialis muscle from mice with Lewis lung carcinoma and cachexia were analyzed [72]. The analysis revealed 1,008 mRNAs and 18 microRNAs with differential expression between cachectic and control mice. Despite the heterogeneity in mRNA expression profile, genes that were uniformly regulated in cachectic mice and were implicated in extracellular matrix, proteolysis, and inflammatory response were identified. Further analysis of transcription factors’ binding sites revealed activation of transcription factors associated with atrophy, such as NF-κB, Stat3, AP-1, and FoxO. The mRNA-miRNA expression profile integration showed a post-transcriptional regulation by miRNAs suggesting a key role of extracellular matrix remodeling in muscle atrophy in cachexia [72].
In Table 1 are presented deregulated muscle or adipose ncRNAs in animal models of cancer cachexia.
Table 1.
Deregulated muscle or adipose tissue ncRNAs in animal models of cancer cachexia and the pathway that are possibly implicated in (strong experimental evidence or predicted), according to miRPathDB v2.0 [75].
| ncRNA | Pathway possibly implicated in | Expression | Tissue | Reference |
|---|---|---|---|---|
| hsa-miR-1 | Genes targeted by miRNAs in adipocytes | Decreased | Diaphragm and gastrocnemius from cachectic mice with lung cancer | [70] |
| hsa-miR-133a | Potassium channel complex | |||
| hsa-miR-206 | Generic Transcription Pathway | |||
| hsa-miR-486 | Somatroph axis (GH) and its relationship to dietary restriction and aging | |||
| Decreased | Skeletal muscles from mice with breast cancer | [73] | ||
| mmu-miR-147-3p | Hippo signaling pathway | Increased | Tibialis anterior from cachectic mice with Lewis lung carcinoma | [71] |
| mmu-miR-205-5p | Axon guidance | |||
| mmu-miR-511-3p | phospholipase C-activating G protein-coupled receptor signaling pathway | |||
| mmu-miR-223-3p | Focal adhesion | |||
| mmu-miR-299a-3p | Axon guidance | Decreased | ||
| mmu-miR-1933-3p | Peptide GPCRs | |||
| mmu-miR-3473d | FoxO signaling pathway | |||
| mmu-miR-431-5p | Ascorbate and aldarate metabolism | |||
| mmu-miR-665-3p | Insulin signaling pathway | |||
| CAAlnc1 | Regulates the expression of C/EBP‐α and PPAR‐γ | Increased | Liver and brown adipose tissue from cachectic mice with cancer | [74] |
4.1.2. MiRNAs in cancer cachexia patients
MyomiRs represent a set of miRNAs, with high expression levels in skeletal muscles. Among them are included miR-1, miR-133a, miR-133b, miR-206, miR-208, miR-208b, miR-486, and miR-499. MyomiRs are transcriptionally controlled by myogenic regulatory factors and regulate skeletal muscle growth and muscle development and maintenance [76]. Since myomiRs are strongly regulated during resistance exercise, it has been suggested that they could be used as biomarkers for cachectic patients follow-up, in order to avoid detrimental exercise, or as companion biomarkers for drugs that mimic exercise, such as trimetazidine [77].
Narasimhan et al. in an interesting study analyzed miRNAs from rectus abdominis muscle biopsies from cachectic and non-cachectic cancer patients using next generation sequencing [78]. They revealed 8 miRNAs which were upregulated in cancer cachexia patients and were implicated in muscle metabolism, myogenesis and inflammation, namely, hsa-let-7d-3p, hsa-miR-345-5p, hsa-miR-423-5p, hsa-miR-532-5p, hsa-miR-1296-5p, hsa-miR-3184-3p, hsa-miR-423-3p and hsa-miR-199a-3p [78]. The authors demonstrated that let-7d-3p promotes downregulation of transferrin receptor pathway affecting muscle cells proliferation and myogenic differentiation, while miR-345-5p downregulates NOV and COL1A1 and consequently upregulates CYR61; NOV and CYR61 are implicated in insulin-like growth factor 1, Akt and mTOR pathways, reducing the ability for protein synthesis. In the same study, it was shown that miR-423-5p and miR-3184-3p downregulate SQLE and FADS2, which are involved in lipids biosynthesis, while miR-423-5p can also regulate leptin and other genes involved in energy balance and downregulates DLK1, which is implicated in muscle hypertrophy. Furthermore, it was shown that miR-423-3p promotes calcium signaling impairment, affecting CAMK2A and that miR-3184-3p is implicated in Wnt/β-catenin signaling, attenuating myogenic differentiation and that it can regulate BMPR1B and GREM1, affecting TGF-β and BMP signaling. In addition, it was demonstrated that miR-532-5p targets SULF1 which is related to BMP signaling affecting somits development, RPS6KA6, which contributes to CNTF (ciliary neurotrophic factor) activities and NPY1R, which is implicated in appetite regulation. Overall Narasimhan et al. suggested that different miRNAs play a critical role in modulation of the pathways involved in muscle atrophy in cachectic cancer patients.
A recent meta-analysis regarding miRNA–mRNA interaction networks associated with muscle loss in cancer cachexia revealed 52 differentially expressed genes in muscle tissue from patients and rodent models of cancer cachexia [79]. Protein-protein interaction network in muscle wasting in cancer cachexia highlighted most interaction components being transcriptional factors and proteins associated with the apoptotic process, metabolism and inflammatory response. Moreover, the study predicted new miRNA–mRNA interactions, which can contribute to muscle loss during cancer cachexia, like miR-27a/Foxo1, miR-27a/Mef2c, miR-27b/Cxcl12, miR-27b/Mef2c, miR-140/Cxcl12, miR-199a/Cav1 and miR-199a/Junb [79].
The post-transcriptional pathway which includes HuR-RNA binding protein seems also to play a critical role for cancer cachexia initiation and especially in muscle loss [80]. HuR can change its function from muscle fibers formation inducer to muscle loss inducer by binding initially to the 3’UTR of STAT3 mRNA, in a region close to a complementary to miR-330. HuR binding inhibits miR-330 binding to STAT3 mRNA and consequently its translation inhibition. The negative interaction of HuR with miR-330 implies a mechanism through which muscle fibers can modulate STAT3 expression in order to define their future in response to stress [80].
In a very recent study van de Worp et al. analyzed the expression levels of 754 miRNAs in vastus lateralis biopsies from 8 cachectic patients with non-small cell lung cancer and 8 healthy individuals and validated the results in 15 cachectic patients with non-small cell lung cancer, 11 non- cachectic patients with non-small cell lung cancer and 22 healthy individuals [81]. They demonstrated that 28 miRNAs exhibited differential expression, 5 were upregulated and 23 downregulated. Further analysis of the miRNAs revealed 158 potential target genes, which were involved in 22 pathways associated with muscle regenerative or degenerative processes, such as IL-6, TGF-β, TNFα, insulin and Akt. More specifically, miR-424-5p, miR-424-3p and miR-450a-5p had higher expression levels in the muscles of cachectic patients, whereas miR-144-5p and miR-451a had lower expression levels, compared to healthy ones. In non-cachectic patients only miR-424-3p was significantly upregulated compared to controls. Moreover, although the number of the patients was small, the combination of miR-450-5p and miR-451a expression had prognostic value for patients’ survival [81].
In addition to muscle atrophy, lipolysis and fat browning are also prominent features of cancer cachexia, in which miRNAs are implicated (Table 2, Fig. 2) [82]. In gastrointestinal cancer, when abdominal subcutaneous tissue from cachectic and non-cachectic patients was compared, downregulation of miR-483-5p, miR-23a, miR-744 and miR-99b and upregulation of miR-378 was observed in cachectic patients, which was positively related to catecholamines-induced lipolysis [83]. In in vitro experiments, where human adipocytes from healthy donors were cultured, inhibition of miR-378 led to impaired lipolysis induced from norepinephrine and downregulation of genes with key role in lipolysis, such as LIPE, PNPLA2 and PLIN1 [83].
Table 2.
Deregulated muscle or adipose tissue ncRNAs in cancer cachexia patients and the pathway that are possibly implicated in (strong experimental evidence), according to miRPathDB v2.0 [75].
| ncRNA | Pathway possibly implicated in | Expression | Tissue | Reference |
|---|---|---|---|---|
| hsa-miR-3184-3p | Photodynamic therapy-induced HIF-1 survival signaling | Increased | Muscles from cachectic patients with colorectal or pancreatic cancer | [78] |
| hsa-miR- 423-5p | Carbohydrate metabolic process | |||
| hsa-let-7d-3p | mRNA Processing | |||
| hsa-miR-1296-5p | Protein-containing complex | |||
| hsa-miR-345-5p | Cancer, Intestinal immune network for IgA production | |||
| hsa-miR-532-5p | Lung fibrosis | |||
| hsa-miR-423-3p | Chromosomal and microsatellite instability in colorectal cancer | |||
| hsa-miR-199a-3p | Focal Adhesion | |||
| hsa-miR-424-5p | Cell cycle | Increased | Vastus lateralis from cachectic patients with non-small cell lung cancer | [81] |
| hsa-miR-424-3p | Transcriptional regulation by RUNX1 | |||
| hsa-miR-450a | Herpes simplex infection | |||
| hsa-miR-451a | Hepatitis B | Decreased | ||
| hsa-miR-144-5p | Androgen receptor signaling pathway | |||
| hsa-miR-483-5p | Envelope proteins and their potential roles in EDMD physiopathology | Decreased | Abdominal subcutaneous tissue from cachectic patients with gastrointestinal cancer | [83] |
| hsa-miR-23a | Muscle myosin complex | |||
| hsa-miR-744 | LncRNA involvement in canonical Wnt signaling and colorectal cancer | |||
| hsa-miR-99b | Somatroph axis (GH) and its relationship to dietary restriction and aging | |||
| hsa-miR-378 | Determination of heart left/right asymmetry | Increased | ||
| VLDLR-AS1 | Response to chemotherapy | Increased | Adipose tissue from cachectic patients with gastrointestinal cancer | [84] |
| MALAT1 | Transcriptional regulator for genes genes involved in cancer metastasis and cell migration | Decreased | Subcutaneous white adipose tissue from cachectic patients with cancer | [85] |
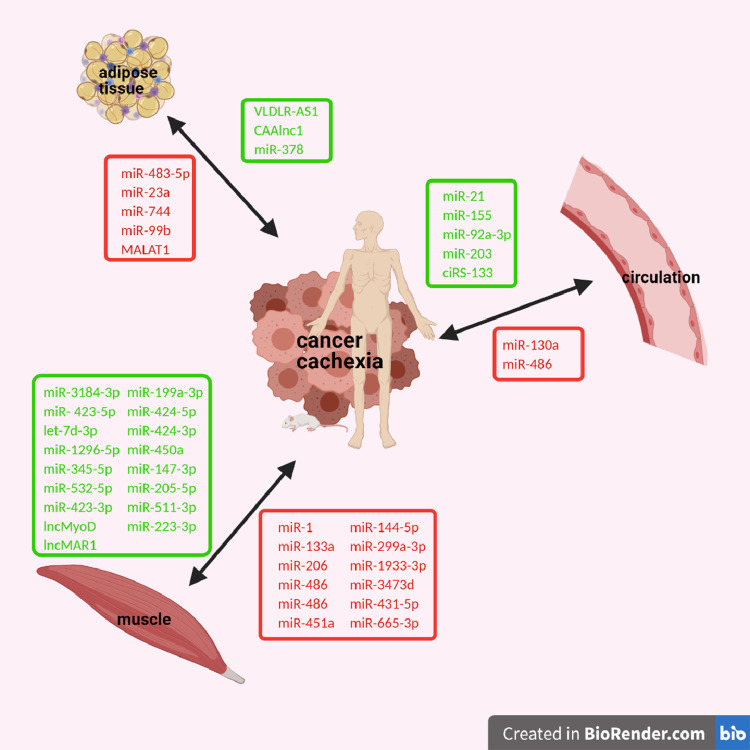
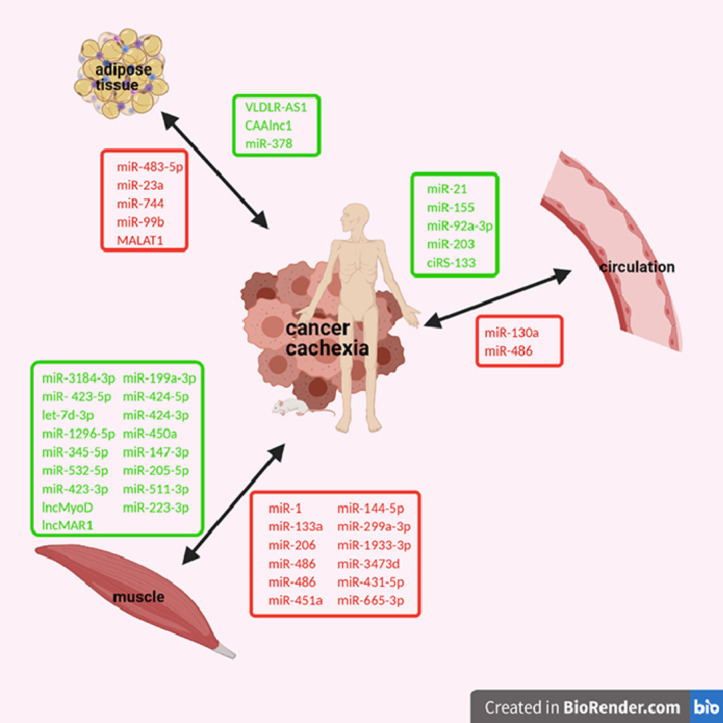
Fig. 2.
Deregulated ncRNAs in muscle or adipose tissue or in circulation of patients or animal models with cancer cachexia. Downregulated ncRNAs are colored in red and upregulated ncRNAs are colored in green. (Created with BioRender.com).
In Table 2 are presented deregulated muscle or adipose ncRNAs in cancer cachexia patients.
4.1.3. Circulating miRNAs in cancer cachexia patients
Additionally to their intracellular localization, miRNAs can be secreted actively by the cell, or released through cell membrane as a reaction to various stimuli, leading in their circulation in blood or other biological fluids [86]. This gets particular value for muscle loss, as very few blood biomarkers have been associated with muscle mass, such as myostatin and arginine [87], [88], [89].
Circulating miRNAs can serve as potential biomarkers, not only for muscle loss, but also muscle regeneration, effectiveness of therapy and early detection of cachexia as well (Table 3, Fig. 2). It has been reported that patients with head and neck cancer and low miR-130a plasma levels have high TNF-α plasma levels and increased risk for cachexia [90]. Additionally, miR-130a expression can separate cachectic from mildly malnourished patients, with sensitivity and specificity values of 76.4% and 80.8%, respectively. When low miR-130a levels were combined high TNF-α levels, sensitivity and specificity values increased to 83.3% and 91.7%, respectively. In patients with weight loss >5% nutritional evaluation was improved when miR-130a levels were used, resulting in sensitivity and specificity 88.6% and 94.3%, respectively, suggesting that miR-130a is a potential biomarker for cachexia prediction, as well as improvement of cachexia diagnosis in patients with head and neck cancer [90].
Table 3.
Deregulated circulating ncRNAs in cancer cachexia and the pathway that are possibly implicated in (strong experimental evidence), according to miRPathDB v2.0 [75].
| ncRNA | Pathway possibly implicated in | Expression | Circulation | Reference |
|---|---|---|---|---|
| hsa-miR-21 | Cancer, regulation of catalytic activity | Increased | Lung or pancreatic cancer-derived exosomes | [113] |
| Serum from cachectic patients with colorectal cancer | [91] | |||
| hsa-miR-155 | Chromosome, nuclear lumen | Increased | Breast cancer-derived exosomes | [109] |
| hsa-miR-92a-3p | Activation of BIM and translocation to mitochondria | Increased | Chronic myeloid leukemia-derived exosomes | [111] |
| hsa-miR-130a | Negative regulation of blood vessel endothelial cell migration | Decreased | Plasma from cachectic patients with head and neck cancer | [90] |
| hsa-miR-203 | Hepatitis B, Pancreatic cancer, Androgen receptor signaling pathway | Increased | Serum from patients with colorectal cancer | [92] |
| hsa-miR-486 | Role of phospholipids in phagocytosis | Decreased | Serum from patients with breast cancer | [73] |
| ciRS-133 | Circular RNA sponge for miR-133 | Increased | Gastric cancer-derived exosomes | [65] |
Okugawa et al showed that colorectal cancer patients with low psoas muscle index had high serum miR-21 levels making miR-21 a useful parameter for the clinician to decide the nutritional strategy for these patients [91]. Later, the same group showed that patients with low psoas muscle index had high miR-203 serum levels, whereas intramuscular fat was not associated with serum or tissue miR-203 levels [92]. Moreover, high miR-203 levels were shown to be an independent prognostic factor for myopenia for colorectal cancer patients [92]. In in vitro experiments, human skeletal muscle cells after transfection with miR-203 mimics became less proliferative and more apoptotic, indicating a possible prognostic value of serum miR-203 in myopenia, and a novel therapeutic target for inhibition of myopenia in colorectal cancer [92].
ΜiR-486, a muscle-enriched miRNA, has been found downregulated in the circulation of breast cancer patients and animal models compared to healthy controls [73]. In breast cancer mice, miR-486 levels were low in muscles, whereas miR-486 target genes, such as PTEN and FOXO1A were increased and the signaling was attenuated through the PI3K/AKT pathway. MyoD expression, which is a transcription factor that regulates miR-486 expression, was shown to be low in skeletal muscles as well. Moreover, when supernatant medium from breast cancer cells cultures was added to C2C12 myoblasts cultures, miR-486 levels were decreased, while PTEN and FOXO1A levels increased. Further analysis of the medium revealed TNFα and 4 more cytokines as mediators of miR-486 expression in myoblasts, implicating TNFα-dependent miRNA circuitry in muscle differentiation and survival pathways in cancer. [73].
MyomiRs have also been found to be released in circulation by rhabdomyosarcoma, a malignant tumor arising from skeletal muscles, leading to high serum levels of miR-1, miR-133a, miR-133b, and miR-206 [93]. Changes in circulating myomiRs have been demonstrated in different types of cancer. For instance, in gastric cancer patients, an increase in miR-1 in circulation has been reported, while in non-small cell lung cancer, melanoma, astrocytoma and osteosarcoma, lower levels of circulating myomiRs have been observed [94], [95], [96], [97]. MiR-1, miR-133a, miR-133b and miR-206 which are tumor-suppressive miRNAs are mostly downregulated in several types of cancers, while, expression of serum miR-1, miR-20a, miR-27a, miR-34 and miR-423- 5p could be used to detect gastric cancer [94,98]. In lung cancer, a risk-score signature consisted of miR-486, miR-30d, miR-1 and miR-499 serum levels could differentiate patients in two groups, in long and short survival groups [96]. Additionally, low serum miR-133b miR-206 expression had negative prognostic value for both overall survival and disease-free survival in osteosarcoma patients [99].
4.1.4. Exosomal miRNAs in cancer cachexia
Paracrine and endocrine effects of miRNAs have been associated with expansion of systemic inflammation, development of metastasis and activation of muscle loss-promoting pathways [100]. Alternatively, miRNAs can be transferred in circulation by enclosure into exosomes [101]. Exosomes are small membrane vesicles of 30–100 nm which can effectively transfer proteins, RNA, cytokines and other molecules, with great stability avoiding their degradation in circulation [102], [103], [104].
One of the main mechanisms involved in cancer cachexia initiation is white adipose tissue induction of circulating inflammatory cytokines [31]. It has been documented that white adipose tissue can secrete exosomes containing miRNAs, which regulate inflammatory processes in tissues and immune cells [105,106]. Moreover, adipose tissue exosomes can trigger and regulate tumor growth, progression and aggressiveness [105]. Similarly, various studies have shown than tumor-derived exosomes contain miRNAs which play a role in inflammation activation in cancer [107,108]. These cancer exosomes induce production and release of proinflammatory cytokines, promoting cancer cells’ survival through their interaction with mesenchymal stem cells [107]. Moreover, tumor-derived exosomes can induce tumor progression, modifying the tumor microenvironment and promoting metastasis [107,108].
Tumor-derived exosomes can induce cancer cachexia as Wu et al. showed recently [109]. More specifically, when adipocytes were cultured in presence of 4T1 breast cancer cells, they exhibited upregulation of UCP1 (uncoupling protein 1) and downregulation of PPARγ (peroxisome proliferator activated receptor gamma) and phosphorylated (P)-PPARγ, which are implicated in fat accumulation, as well as downregulation of phosphorylated ERK1/2 (extracellular signal-regulated kinase) and upregulation of phosphorylated p38 [109]. Furthermore, after co-culturing muscle cells with breast cancer cells, muscle cells lost myosin heavy chain and had increased death rate, myotubes atrophied, UCP3 levels increased, P-p38 was upregulated, while P-ERK1/2, PPARγ and P-PPARγ were downregulated, suggesting that tumor-derived exosomes contain miRNAs which promote catabolism in adipocytes and muscle fibers [109]. Moreover, miR-155 was upregulated in tumor-derived exosomes and PPARγ targeting in adipocytes promoted remodeling of adipocytes metabolism and browning. It is noteworthy that cancer cells that were co-cultured with adipocytes or myocytes displayed increased invasiveness via epithelial-mesenchymal transition, while tumor-derived exosomes increased catabolism and metabolites release in adipocytes and myocytes [110].
In agreement with the above study, injection of leukemia exosomes to mice led to weight and fat loss [111]. Moreover, mesenchymal stem cells of adipose tissue could receive chronic myeloid leukemia cells (Κ562) exosomes and subsequently suppress adipogenicity [111]. In Κ562 cells as well as in K562-derived exosomes upregulation of miR-92a-3p was observed, which could suppress adipogenicity through downregulation of Cebpa (CCAAT enhancer binding protein alpha), suggesting a key role of exosomal microRNAs, in cancer cachexia mediation.
Furthermore, exosomal miRNAs have been documented to contribute to initiation, as well as maintenance of cancer cachexia-associated inflammation [100,112]. Overexpression of miR-21 in lung or pancreatic cells-derived microvesicles induced apoptosis in skeletal muscle cells [113]. Myoblasts isolated from TLR7−/− (toll-like receptor 7) mice and cultured with supernatant medium from Lewis carcinoma cells or serum from cachectic patients with pancreatic cancer exhibited reduced cell death rate [113]. Moreover, exogenous addition of miR-21 in myoblasts from TLR7+/+ increased cell death rate, while in proliferating myoblasts after transfection with miR-21, JNK and c-Jun were upregulated, suggesting that miR-21 promotes cell death through activation of TLR7 signaling downstream of JNK [113].
In summary, these findings highlight the role of exosomal miRNAs in initiation and maintenance of inflammation in cancer cachexia. Systemic inflammation affects negatively muscle metabolism, regulating signaling pathways involved in muscle proteins synthesis and degradation, contributing to muscle loss, which characterizes cancer cachexia [100,114,115].
In Table 3 are presented deregulated circulating ncRNAs in cancer cachexia.
4.1.5. Long non-coding RNAs in animal models of cancer cachexia
LncRNAs (long non-coding RNAs) are transcripts longer than 200 RNAs with no potential of translation [116]. It is well established that lncRNAs are implicated in many cell functions; they can regulate and remodel chromatin, they can regulate transcription and nuclear bodies, they are implicated in mRNA cycle, translation and post-translational modifications [116]. LncRNAs exert their regulatory effects mainly through interacting with miRNAs [116,117]. They are implicated in various diseases including cancer and cachexia, while few studies implicate lncRNAs in cancer cachexia [74,84,118,119]. LncRNAs play a significant role in myoblast differentiation and muscle development, mainly through regulating cell cycle inhibitors and myogenic factors, thus it is not surprising that lncRNAs are dysregulated in muscle regeneration and various muscular diseases, such as atrophy and hypertrophy, muscular dystrophy etc [120].
LncIRS1 (insulin receptor substrate 1) is a lncRNA that was recently revealed to act as an endogenous competitive RNA in mouse skeletal muscles where it is upregulated [121]. LncIRS1 was shown to regulate myoblasts proliferation and differentiation in vitro and in vivo and muscle mass and sectional area of fibers in vivo [121]. LncIRS1 can act as miR-15 family sponge regulating IRS1 expression. When overexpressed it activates the IGF1-PI3K/Akt signaling pathway increasing skeletal muscle proliferation and muscle mass and modulates the expression of important atrophy genes, while its suppression results to decreased IGF1 levels and weight loss in mice. All above point to a potential therapeutic role of lncIRS1 for treating muscle atrophy [121], [122], [123].
LncRNA MAR1 (muscle anabolic regulator 1), another lncRNA associated with cachexia, is upregulated in mice skeletal muscles and has been positively associated with muscular differentiation and growth, in vitro and in vivo, through interacting with miR-487b, which regulates Wnt5a (Wnt family member 5A) and promotes muscular differentiation and regeneration [124]. When overexpressed, it attenuated muscle atrophy, while muscle strength and mass was retained, making it a possible candidate as therapeutic target for muscle atrophy associated with cancer cachexia, for example being overexpressed and delivered using adeno-associated virus particles (AAV) [124].
LncRNAs have also been involved in adipose tissue loss of cachectic mice. In particular, Shen et al. analyzed the transcriptome of white adipose tissue from cachectic vs non-cachectic mice that had been injected with colon adenocarcinoma (C26) cells and identified a functional CAAlnc1 (cachexia-related anti-adipogenesis lncRNA 1) which suppressed adipogenesis through interaction with HuR (necessary protein for adipogenesis) leading to fat loss [74].
4.1.6. LncRNAs in cancer cachexia patients
In a study conducted by Liu et al. adipose tissue from cachectic and non-cachectic patients with gastrointestinal cancer was analyzed and VLDLR-AS1 (VLDLR Antisense RNA 1) was related to cachexia and fat loss [84]. Further analysis and prediction models suggested that VLDLR-AS1 regulated GOLGA3 (golgin A3), DUSP14 (dual specificity phosphatase 14) and UCHL1 (ubiquitin C-terminal hydrolase L1) through interaction with miR-600 and ZNF219 (zinc finger protein 219), as well as RNF141 (ring finger protein 141) and CALU (calumenin) through binding to miR-1224-3p. However, the mechanism through which VLDLR-AS regulates fat loss in cancer cachexia has not been established [84].
Since deregulated myogenesis is associated with cachexia, Gong et al. suggested that lncMyoD, a lncRNA which is activated during myoblasts differentiation directly by MyoD (myogenic differentiation 1), is also possibly associated to cachexia [125]. When it is active lncMyoD binds to IMP2 (IGF2-mRNA-binding protein 2) and negatively regulates the IMP2-mediated translation of critical for proliferation genes, such as N-Ras (neuroblastoma ras oncogene) and c-Myc (myelocytomatosis oncogene) [125]. LncMyoD suppression has been reported to inhibit muscular differentiation and cell cycle exit, pointing to its potential association with cachexia [125].
Very recently, Han et al. suggested that lncRNA MALAT1 (metastasis associated lung adenocarcinoma transcript 1) can regulate PPAR-γ expression at the transcriptional level and it is associated with adipose loss in cancer-associated cachexia. MALAT1 expression in the subcutaneous white adipose tissue of cancer-associated cachexia patients was low and was prognostic of low fat mass index and poor clinical outcome of the patients [85].
4.1.7. circRNAs in cancer cachexia
Circular RNAs (circRNAs) are generated when specific exons are circularized through covalent bonds [126]. It has been documented that circRNAs can act as miRNAs sponge, since they contain many binding sites for miRNAs and seem to play significant role in various diseases [127,128].
Regarding the role of circRNAs in cancer cachexia, there is only one study in gastric cancer, which reports that ciRS-133 (circRNA Hsa_circ_0010522) expression was positively associated with brown fat mass and body adipose percentage [65]. In vivo studies revealed that ciRS-133 interacts with miR-133. Exosomes derived from gastric cancer cell line (SGC7091) had high expression of ciRS-133, and when added to 3T3L1 culture medium resulted to PRDM16 (PR-domain containing 16) and UCP1 upregulation. On the other hand, miR-133 overexpression downregulated PRDM16 and UCP1, whereas suppression of miR-133 reversed this effect [65]. Injection of SGC7901 cells with overexpression of ciRS-133 in mice led to increase of ciRS-133 expression in cancer tissue, of serum exosomes and inguinal adipose tissue. Moreover, the mice presented with reduced inguinal adipose tissue mass and browning, suggesting that ciRS-133 worsens cancer cachexia, probably through fat browning [65].
5. NcRNAs in cancer-associated cachexia: clinical implications and future perspectives
Detection of cachexia in early stages (precachexia or cachexia) is vital for prevention of refractory cachexia, relief of symptoms and improvement of quality of life and prognosis. Circulating miRNAs, such as miR-21 can aid to this direction as an indication to the clinicians for the nutritional strategy they should suggest to high risk colorectal cancer patients [91]. MyomiRs and other miRNAs that are released in circulation by various tumors (ie miR-450-5p and miR-451a) and have prognostic value for patients’ survival can also be used for identification of high risk patients with urgent need for treatment [81,93]. Other circulating miRNAs, such as miR-130a, can separate cachectic from mildly malnourished head and neck cancer patients, which is also crucial for effective treatment and delay of refractory cachexia [90]. Moreover, miRNAs such as myomiRs that are regulated during resistance exercise, have been suggested to be used as companion biomarkers for drugs that mimic exercise, such as trimetazidine [77].
As long as several pathways are implicated in cancer cachexia, therapeutic approaches modifying multiple targets would be ideal. Such a therapeutic approach for cancer cachexia is provided via epigenetic therapy, which can cause reversible changes in gene regulation, without affecting DNA sequence [129].
When considering treatment of multifactorial background, miRNAs have an advantage over gene therapy, as they can regulate the expression of multiple genes at the same time. In the context of a new therapeutic strategy, miRNAs can be used either as knockdown therapy or as replacement therapy, as they bind with high specificity on their complementary mRNA target leading to translational suppression. In the case of knockdown therapy, antagomiRs consist of specific oligomers which are complementary to specific miRNAs and they compete with mRNA targets in order to bind to the miRNAs. In the replacement therapy, miRNA mimics interfere with the mRNA target, functioning as endogenous miRNA [130].
Currently, siRNAs which have similar mechanism of action to that of miRNAs, are used for the treatment of spinal muscular atrophy and hereditary transthyretin -mediated amyloidosis (Nusinersen and Patisiran, respectively) [131,132]. In cancer therapy, miRNAs may be used as knockdown or replacement therapy, when targeting miRNAs which function as oncogenes or tumor suppressor genes, respectively.
Two types of carriers are usually used for miRNAs transfer, viral and non-viral vectors [133]. However, the efforts for the development of viral vectors for miRNAs transfer have failed, mainly due to the immune response trigger, leading the researchers towards non-viral alternatives [134]. Although results from preclinical studies were promising, translational clinical studies results using miRNAs replacement therapy have proved rather disappointing. A phase I clinical study which implied miR-34 replacement therapy in cancer patients failed to pass to phase II, due to its toxic side effects (NCT01829971, NCT02862145) [135]. On the other hand, a phase I clinical study using miR‐16 mimics in mesothelioma or non-small cell lung cancer patients has been successfully completed and despite some complications, it is expected to continue to phase II (NCT02369198) [136].
One of the main challenges in miRNA replacement therapy is the effectiveness and accuracy of miRNAs in target cells. An ideal transfer system should protect miRNAs from degradation, transfer them and facilitate their intake form the target cells, while they should not trigger immune response [137], [138], [139]. Extracellular vesicles satisfy those criteria and preclinical studies with extracellular vesicles transferring nucleic acids have shown promising results in tumors [140,141]. MiRNAs or antagomiRs can be integrated in extracellular vesicles and be used as therapeutic agents in the clinical setting [142]. A phase I clinical study is ongoing which evaluates the effectiveness and side effects of exosomes carrying IGFR1 antisense oligodeoxynucleotides (NCT02507583). In another ongoing clinical study, mesenchymal exosomes carrying a small siRNA which targets mutant KRAS is used in pancreatic cancer patients (NCT03608631). These clinical studies, as well as many more, highlight the possible use of extracellular vesicles as short-RNAs carriers which can modulate gene expression and proliferation of different tumors. Although the results from preclinical studies are promising, many obstacles need to be overcome before miRNAs are applied for therapeutic purposes in the clinical setting.
MiRNAs-based approaches are not used in cancer cachexia treatment yet, although the implication of miRNAs in cancer-associated cachexia pathogenesis makes them an attractive therapeutic target. In a preclinical mouse model, a miRNA was used to target serum amyloid 1 and 2 (SAA), which are typical apolipoproteins induced in response to inflammatory cytokines and promote muscular atrophy. In C26 colon adenocarcinoma mice injected with AAV–miRNA targeting SAA1/2, serum levels of SAA1/2 were 4 times reduced, although no difference was observed in cachexia progression [143]. Further liver-specific RNA and proteomic analysis revealed a high level of overlapping and their exploration can pave the way to novel treatment strategies [143]. However, from bench to bedside a translational failure is often observed, with most of the successful preclinical studies turning sour in clinical studies with human patients, thus impeding drug discovery. The main reason for this translational failure lies in species differences as well as other external and internal validity issues [144]. Nevertheless, animal models and preclinical studies still remain the cornerstone for understanding the molecular mechanisms underlying cancer cachexia and have to be interpreted critically [145].
MiRNAs-based therapeutic approaches for cancer cachexia targeting specific pathways could potentially restore homeostasis of chronically deregulated networks and predispose muscles to positive response to therapy including exercise and diet. Therefore, miRNAs could be used additionally to increase the effectiveness of already existing therapies [146]. Animal models are essential for validation of candidate drugs for cancer cachexia treatment before being applied in clinical trials and more studies should be focused to this direction.
6. Conclusion
The comprehension of the molecular mechanisms that are implicated in cancer cachexia is significant not only for understanding the pathophysiology of the disease, but for novel therapeutic targets and biomarkers identification as well. NcRNAs, such as microRNAs, regulate the signaling pathways implicating in cancer cachexia pathophysiology making them attractive therapeutic targets and promising biomarkers for cancer cachexia diagnosis, prognosis and follow up.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
Author contributions
Conceptualization, AT; Investigation, AK; Writing - Original Draft, AK; Writing - Review & Editing; AK, F-ID, AT; Supervision, AT
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.Evans WJ, Morley JE, Argilés J, Bales C, Baracos V, Guttridge D. Cachexia: a new definition. Clin. Nutr. 2008;27:793–799. doi: 10.1016/j.clnu.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Jeejeebhoy KN. Malnutrition, fatigue, frailty, vulnerability, sarcopenia and cachexia: overlap of clinical features. Curr. Opin. Clin. Nutr. Metab. Care. 2012:213–219. doi: 10.1097/MCO.0b013e328352694f. [DOI] [PubMed] [Google Scholar]
- 3.Kotler DP. Cachexia. Ann. Intern. Med. 2000:622–634. doi: 10.7326/0003-4819-133-8-200010170-00015. American College of Physicians. [DOI] [PubMed] [Google Scholar]
- 4.Fearon K, Strasser F, Anker SD, Bosaeus I, Bruera E, Fainsinger RL. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 2011:489–495. doi: 10.1016/S1470-2045(10)70218-7. Elsevier. [DOI] [PubMed] [Google Scholar]
- 5.Tan BHL, Fearon KCH. Cachexia: prevalence and impact in medicine. Curr. Opin. Clin. Nutr. Metab. Care. 2008:400–407. doi: 10.1097/MCO.0b013e328300ecc1. [DOI] [PubMed] [Google Scholar]
- 6.Peterson SJ, Mozer M. Differentiating sarcopenia and cachexia among patients with cancer [internet] Nutr. Clin. Pract. 2017:30–39. doi: 10.1177/0884533616680354. http://www.ncbi.nlm.nih.gov/pubmed/28124947 SAGE Publications Inc.[cited 2020 Apr 9]Available from: [DOI] [PubMed] [Google Scholar]
- 7.Cruz-Jentoft AJ, Bahat G, Bauer J, Boirie Y, Bruyère O, Cederholm T. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing [Internet] 2019;48:16–31. doi: 10.1093/ageing/afy169. http://www.ncbi.nlm.nih.gov/pubmed/30312372 [cited 2020 Apr 9]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barazzoni R, Bischoff S, Boirie Y, Busetto L, Cederholm T, Dicker D. Sarcopenic obesity: time to meet the challenge [Internet] Obes. Facts. 2018:294–305. doi: 10.1159/000490361. http://www.ncbi.nlm.nih.gov/pubmed/30016792 S. Karger AG[cited 2020 May 14]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin L, Birdsell L, MacDonald N, Reiman T, Clandinin MT, McCargar LJ. Cancer cachexia in the age of obesity: skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. J Clin Oncol. 2013;31:1539–1547. doi: 10.1200/JCO.2012.45.2722. [DOI] [PubMed] [Google Scholar]
- 10.Fox KM, Brooks JM, Gandra SR, Markus R, Chiou CF. Estimation of cachexia among cancer patients based on four definitions. J Oncol [Internet] 2009 doi: 10.1155/2009/693458. https://www.ncbi.nlm.nih.gov/pubmed/?term=Estimation+of+cachexia+among+cancer+patients+based+on+four+definitions [cited 2020 May 19]; Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anker MS, Holcomb R, Muscaritoli M, Von Haehling S, Haverkamp W, Jatoi A. 2019. Orphan Disease Status of Cancer Cachexia in the USA And in the European Union: A Systematic Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freire PP, Fernandez GJ, de Moraes D, Cury SS, Dal Pai-Silva M, dos Reis PP. The expression landscape of cachexia-inducing factors in human cancers. J Cachexia Sarcopenia Muscle [Internet] 2020;11:947–961. doi: 10.1002/jcsm.12565. https://pubmed.ncbi.nlm.nih.gov/32125790/ Wiley Blackwell[cited 2021 Mar 29]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewys WD, Begg C, Lavin PT, Band PR, Bennett JM, Bertino JR. Prognostic effect of weight loss prior to chemotherapy in cancer patients. Eastern Cooperative oncology group. Am J Med [Internet] 1980;69:491–497. doi: 10.1016/s0149-2918(05)80001-3. http://www.ncbi.nlm.nih.gov/pubmed/7424938 [cited 2020 May 19]Available from: [DOI] [PubMed] [Google Scholar]
- 14.Argilés JM, Busquets S, Stemmler B, López-Soriano FJ. Cachexia and sarcopenia: Mechanisms and potential targets for intervention. Curr. Opin. Pharmacol. 2015:100–106. doi: 10.1016/j.coph.2015.04.003. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 15.Gannavarapu BS, Lau SKM, Carter K, Cannon NA, Gao A, Ahn C. Prevalence and survival impact of pretreatment cancer-associated weight loss: a tool for guiding early palliative care. J Oncol Pract [Internet] 2018;14:e238–e250. doi: 10.1200/JOP.2017.025221. https://ascopubs.org/doi/10.1200/JOP.2017.025221 American Society of Clinical Oncology[cited 2021 Mar 24]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fearon K, Arends J, Baracos V. Understanding the mechanisms and treatment options in cancer cachexia. Nat. Rev. Clin. Oncol. 2013:90–99. doi: 10.1038/nrclinonc.2012.209. [DOI] [PubMed] [Google Scholar]
- 17.Kubrak C, Olson K, Jha N, Scrimger R, Parliament M, McCargar L. Clinical determinants of weight loss in patients receiving radiation and chemoirradiation for head and neck cancer: a prospective longitudinal view. Head Neck. Head Neck; 2013;35:695–703. doi: 10.1002/hed.23023. [DOI] [PubMed] [Google Scholar]
- 18.Silver HJ, Dietrich MS, Murphy BA. Changes in body mass, energy balance, physical function, and inflammatory state in patients with locally advanced head and neck cancer treated with concurrent chemoradiation after low-dose induction chemotherapy. Head Neck. Head Neck; 2007;29:893–900. doi: 10.1002/hed.20607. [DOI] [PubMed] [Google Scholar]
- 19.Friesen DE, Baracos VE, Tuszynski JA. Modeling the energetic cost of cancer as a result of altered energy metabolism: Implications for cachexia. Theor Biol Med Model [Internet] 2015;12 doi: 10.1186/s12976-015-0015-0. https://pubmed.ncbi.nlm.nih.gov/26370269/ BioMed Central Ltd.[cited 2020 Jul 12]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, Liu Z, Ding H, Zhou Y, Doan HA, Sin KWT. Tumor induces muscle wasting in mice through releasing extracellular Hsp70 and Hsp90. Nat Commun [Internet] 2017;8 doi: 10.1038/s41467-017-00726-x. https://pubmed.ncbi.nlm.nih.gov/28928431/ Nature Publishing Group[cited 2020 Jul 12]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy KT. The pathogenesis and treatment of cardiac atrophy in cancer cachexia [Internet] Am. J. Physiol. - Hear. Circ. Physiol. 2016:H466–H477. doi: 10.1152/ajpheart.00720.2015. https://pubmed.ncbi.nlm.nih.gov/26718971/ American Physiological Society[cited 2020 Jul 12]Available from: [DOI] [PubMed] [Google Scholar]
- 22.Tashjian AH. Role of prostaglandins in the production of hypercalcemia by tumors. Cancer Res. [Internet] 1978;38:4138–4141. https://pubmed.ncbi.nlm.nih.gov/698957/ [cited 2020 Jul 12]Available from: [PubMed] [Google Scholar]
- 23.Fearon KCH, Glass DJ, Guttridge DC. Cancer cachexia: mediators, signaling, and metabolic pathways [Internet] Cell Metab. 2012:153–166. doi: 10.1016/j.cmet.2012.06.011. https://pubmed.ncbi.nlm.nih.gov/22795476/ [cited 2020 Jul 12]Available from: [DOI] [PubMed] [Google Scholar]
- 24.Burfeind KG, Michaelis KA, Marks DL. The central role of hypothalamic inflammation in the acute illness response and cachexia [Internet] Semin. Cell Dev. Biol. 2016:42–52. doi: 10.1016/j.semcdb.2015.10.038. https://pubmed.ncbi.nlm.nih.gov/26541482/ Academic Press[cited 2020 Jul 13]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grossberg AJ, Scarlett JM, Zhu X, Bowe DD, Batra AK, Braun TP. Arcuate nucleus proopiomelanocortin neurons mediate the acute anorectic actions of leukemia inhibitory factor via gp130. Endocrinology [Internet] 2010;151:606–616. doi: 10.1210/en.2009-1135. https://pubmed.ncbi.nlm.nih.gov/20016025/ Endocrine Society[cited 2020 Jul 13]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodnar RJ, Pasternak GW, Paul D, Warren R, Donner DB, Mann PE. Mediation of anorexia by human recombinant tumor necrosis factor through a peripheral action in the rat. Cancer Res [Internet] 1989;49:6280–6284. https://pubmed.ncbi.nlm.nih.gov/2804974/ [cited 2020 Jul 13]Available from: [PubMed] [Google Scholar]
- 27.Braun TP, Zhu X, Szumowski M, Scott GD, Grossberg AJ, Levasseur PR. Central nervous system inflammation induces muscle atrophy via activation of the hypothalamic-pituitary-adrenal axis. J Exp Med [Internet] 2011;208:2449–2463. doi: 10.1084/jem.20111020. https://pubmed.ncbi.nlm.nih.gov/22084407/ J Exp Med[cited 2020 Jul 13]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouladiun M, Körner U, Bosaeus I, Daneryd P, Hyltander A, Lundholm KG. Body composition and time course changes in regional distribution of fat and lean tissue in unselected cancer patients on palliative care - correlations with food intake, metabolism, exercise capacity, and hormones. Cancer [Internet] 2005;103:2189–2198. doi: 10.1002/cncr.21013. https://pubmed.ncbi.nlm.nih.gov/15822132/ Cancer[cited 2020 Jul 13]Available from: [DOI] [PubMed] [Google Scholar]
- 29.Hall KD, Baracos VE. Computational modeling of cancer cachexia [Internet] Curr. Opin. Clin. Nutr. Metab. Care. 2008:214–221. doi: 10.1097/MCO.0b013e3282f9ae4d. https://pubmed.ncbi.nlm.nih.gov/18403915/ [cited 2020 Jul 12]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camargo RG, dos Reis Riccardi DM, Ribeiro HQT, Carnevali LC, de Matos-Neto EM, Enjiu L. Nf-κbp65 and expression of its pro-inflammatory target genes are upregulated in the subcutaneous adipose tissue of cachectic cancer patients. Nutrients [Internet] 2015;7:4465–4479. doi: 10.3390/nu7064465. https://pubmed.ncbi.nlm.nih.gov/26053616/ MDPI AG[cited 2020 Jul 13]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neves RX, Rosa-Neto JC, Yamashita AS, Matos-Neto EM, Riccardi DMR, Lira FS. White adipose tissue cells and the progression of cachexia: Inflammatory pathways. J Cachexia Sarcopenia Muscle [Internet] 2016;7:193–203. doi: 10.1002/jcsm.12041. https://pubmed.ncbi.nlm.nih.gov/27493872/ Wiley Blackwell[cited 2020 Jul 13]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohm M, Schäfer M, Laurent V, Üstünel BE, Niopek K, Algire C. An AMP-activated protein kinase-stabilizing peptide ameliorates adipose tissue wasting in cancer cachexia in mice. Nat Med [Internet]. 2016;22:1120–1130. doi: 10.1038/nm.4171. https://pubmed.ncbi.nlm.nih.gov/27571348/ Nature Publishing Group[cited 2020 Jul 13]Available from: [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science (80-) [Internet] 2004;306:1383–1386. doi: 10.1126/science.1100747. https://pubmed.ncbi.nlm.nih.gov/15550674/ Science[cited 2020 Jul 13]Available from: [DOI] [PubMed] [Google Scholar]
- 34.Beck SA, Tisdale MJ. Effect of cancer cachexia on triacylglycerol/fatty acid substrate cycling in white adipose tissue. Lipids [Internet] 2004:1187–1189. doi: 10.1007/s11745-004-1346-8. https://pubmed.ncbi.nlm.nih.gov/15736914/ Lipids[cited 2020 Jul 12]Available from: [DOI] [PubMed] [Google Scholar]
- 35.Kir S, Spiegelman BM. Cachexia and brown fat: a burning issue in cancer [Internet] Trends in Cancer. 2016:461–463. doi: 10.1016/j.trecan.2016.07.005. https://pubmed.ncbi.nlm.nih.gov/28459108/ Cell Press[cited 2020 Jul 12]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biswas AK, Acharyya S. Understanding cachexia in the context of metastatic progression. Nat. Rev. Cancer. 2020 doi: 10.1038/s41568-020-0251-4. Nature Research. [DOI] [PubMed] [Google Scholar]
- 37.Kazemi-Bajestani SMR, Becher H, Fassbender K, Chu Q, Baracos VE. Concurrent evolution of cancer cachexia and heart failure: bilateral effects exist. J. Cachexia. Sarcopenia Muscle. 2014:95–104. doi: 10.1007/s13539-014-0137-y. Wiley Online Library. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minami S, Ihara S, Nishimatsu K, Komuta K. Low body mass index is an independent prognostic factor in patients with non-small cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitor. World J Oncol. 2019;10:187–198. doi: 10.14740/wjon1244. Elmer Press, Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang M, Shen Y, Tan L, Li W. Prognostic value of sarcopenia in lung cancer: a systematic review and meta-analysis. Chest. 2019;156:101–111. doi: 10.1016/j.chest.2019.04.115. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- 40.Fukushima H, Takemura K, Suzuki H, Koga F. Impact of sarcopenia as a prognostic biomarker of bladder cancer. Int. J. Mol. Sci. 2018 doi: 10.3390/ijms19102999. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukushima H, Kataoka M, Nakanishi Y, Sakamoto K, Takemura K, Suzuki H. Posttherapeutic skeletal muscle mass recovery predicts favorable prognosis in patients with advanced urothelial carcinoma receiving first-line platinum-based chemotherapy. Urol Oncol Semin Orig Investig [Internet] 2018;36:156.e9–156.e16. doi: 10.1016/j.urolonc.2017.09.016. https://pubmed.ncbi.nlm.nih.gov/29051030/ Elsevier Inc.[cited 2020 Jul 10]Available from: [DOI] [PubMed] [Google Scholar]
- 42.Deng CY, Lin YC, Wu JS, Cheung YC, Fan CW, Yeh KY. Progressive sarcopenia in patients with colorectal cancer predicts survival. Am J Roentgenol. 2018;210:526–532. doi: 10.2214/AJR.17.18020. American Roentgen Ray Society. [DOI] [PubMed] [Google Scholar]
- 43.Bachmann J, Heiligensetzer M, Krakowski-Roosen H, Büchler MW, Friess H, Martignoni ME. Cachexia worsens prognosis in patients with resectable pancreatic cancer. J Gastrointest Surg. 2008;12:1193–1201. doi: 10.1007/s11605-008-0505-z. [DOI] [PubMed] [Google Scholar]
- 44.Lee JS, Kim YS, Kim EY, Jin W. Prognostic significance of CT-determined sarcopenia in patients with advanced gastric cancer. PLoS One. 2018:13. doi: 10.1371/journal.pone.0202700. Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis MP, Panikkar R. Sarcopenia associated with chemotherapy and targeted agents for cancer therapy. Ann. Palliat. Med. 2019:86–101. doi: 10.21037/apm.2018.08.02. AME Publishing Company. [DOI] [PubMed] [Google Scholar]
- 46.Roch B, Coffy A, Jean-Baptiste S, Palaysi E, Daures JP, Pujol JL. Cachexia - sarcopenia as a determinant of disease control rate and survival in non-small lung cancer patients receiving immune-checkpoint inhibitors. Lung Cancer. 2020;143:19–26. doi: 10.1016/j.lungcan.2020.03.003. Elsevier Ireland Ltd. [DOI] [PubMed] [Google Scholar]
- 47.Nishioka N, Uchino J, Hirai S, Katayama Y, Yoshimura A, Okura N. Association of sarcopenia with and efficacy of anti-PD-1/PD-L1 therapy in non-small-cell lung cancer. J Clin Med. 2019;8:450. doi: 10.3390/jcm8040450. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortellini A, Bersanelli M, Buti S, Cannita K, Santini D, Perrone F. A multicenter study of body mass index in cancer patients treated with anti-PD-1/PD-L1 immune checkpoint inhibitors: When overweight becomes favorable. J Immunother Cancer [Internet] 2019;7 doi: 10.1186/s40425-019-0527-y. https://pubmed.ncbi.nlm.nih.gov/30813970/ BioMed Central Ltd[cited 2020 Oct 1]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimitrakopoulos F-ID, Nikolakopoulos A, Kottorou AE, Liolis E, Frantzi T, Pyrousis I. PIOS ratio: predicting the best response of non-small-cell lung cancer (NSCLC) patients treated with immune checkpoint inhibitors. J Clin Oncol. Am. Soc. Clin. Oncol. (ASCO); 2020;38:e21507. –e21507. [Google Scholar]
- 50.Dimitrakopoulos FI, Nikolakopoulos A, Kottorou A, Kalofonou F, Liolis E, Frantzi T. Pios (Patras immunotherapy score) score is associated with best overall response, progression-free survival, and post-immunotherapy overall survival in patients with advanced non-small-cell lung cancer (NSCLC) treated with anti-program cell death-1 (PD-1) Cancers (Basel) [Internet] 2020;12 doi: 10.3390/cancers12051257. https://pubmed.ncbi.nlm.nih.gov/32429368/ MDPI AG[cited 2020 Oct 1]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guzman-Prado Y, Ben Shimol J, Samson O. Sarcopenia and the risk of adverse events in patients treated with immune checkpoint inhibitors: a systematic review. Cancer Immunol Immunother [Internet] 2021 doi: 10.1007/s00262-021-02888-6. https://pubmed.ncbi.nlm.nih.gov/33625531/ Springer Science and Business Media Deutschland GmbH[cited 2021 Mar 26]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antoun S, Birdsell L, Sawyer MB, Venner P, Escudier B, Baracos VE. Association of skeletal muscle wasting with treatment with sorafenib in patients with advanced renal cell carcinoma: Results from a placebo-controlled study. J Clin Oncol. 2010;28:1054–1060. doi: 10.1200/JCO.2009.24.9730. J Clin Oncol. [DOI] [PubMed] [Google Scholar]
- 53.Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals [Internet] Cell. Mol. Life Sci. 2009:596–612. doi: 10.1007/s00018-008-8432-4. https://pubmed.ncbi.nlm.nih.gov/18985277/ [cited 2020 Jul 16]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cock-Rada A, Weitzman JB. The methylation landscape of tumour metastasis. Biol Cell [Internet] 2013;105:73–90. doi: 10.1111/boc.201200029. http://www.ncbi.nlm.nih.gov/pubmed/23198959 2012/12/04. Unidad de Genetica Medica, Facultad de Medicina, Universidad de Antioquia, Medellin, Colombia.Available from: [DOI] [PubMed] [Google Scholar]
- 55.Carmona FJ, Esteller M. DNA methylation in early neoplasia. Cancer Biomark [Internet] 2010;9:101–111. doi: 10.3233/CBM-2011-0184. http://www.ncbi.nlm.nih.gov/pubmed/22112471 2011/11/25. Cancer Epigenetics and Biology Program (PEBC), Bellvitge Institute for Biomedical Research (IDIBELL), 08907 L'Hospitalet de Llobregat, Barcelona, Catalonia, Spain.Available from: [DOI] [PubMed] [Google Scholar]
- 56.He L, Khanal P, Morse CI, Williams A, Thomis M. Differentially methylated gene patterns between age-matched sarcopenic and non-sarcopenic women. J Cachexia Sarcopenia Muscle. 2019;10:1295–1306. doi: 10.1002/jcsm.12478. Wiley Blackwell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharples AP, Polydorou I, Hughes DC, Owens DJ, Hughes TM, Stewart CE. Skeletal muscle cells possess a ‘memory’ of acute early life TNF-α exposure: role of epigenetic adaptation. Biogerontol. Springer Netherlands; 2016;17:603–617. doi: 10.1007/s10522-015-9604-x. [DOI] [PubMed] [Google Scholar]
- 58.Saini A, Al-Shanti N, Sharples AP, Stewart CE. Sirtuin 1 regulates skeletal myoblast survival and enhances differentiation in the presence of resveratrol. Exp Physiol [Internet] 2012;97:400–418. doi: 10.1113/expphysiol.2011.061028. https://pubmed.ncbi.nlm.nih.gov/22125309/ Blackwell Publishing Ltd[cited 2020 Jul 17]Available from: [DOI] [PubMed] [Google Scholar]
- 59.Byun SK, An TH, Son MJ, Lee DS, Kang HS, Lee EW. HDAC11 inhibits myoblast differentiation through repression of myod-dependent transcription. Mol Cells. Korean Soc. Mole. Cell. Biol.; 2017;40:667–676. doi: 10.14348/molcells.2017.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dupré-Aucouturier S, Castells J, Freyssenet D, Desplanches D. Trichostatin A, a histone deacetylase inhibitor, modulates unloaded-induced skeletal muscle atrophy. J Appl Physiol. 2015;119:342–351. doi: 10.1152/japplphysiol.01031.2014. American Physiological Society. [DOI] [PubMed] [Google Scholar]
- 61.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell [Internet] 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. http://www.ncbi.nlm.nih.gov/pubmed/19167326 2009/01/27. Howard Hughes Medical Institute, Massachusetts Institute of Technology, Cambridge, MA 02139, USA. dbartel@wi.mit.eduAvailable from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costa FF. Non-coding RNAs: lost in translation? Gene [Internet] 2007;386:1–10. doi: 10.1016/j.gene.2006.09.028. http://www.ncbi.nlm.nih.gov/pubmed/17113247 2006/11/23. Cancer Biology and Epigenomics Program, Children's Memorial Research Center and Northwestern University's Feinberg School of Medicine, 2300 Children's Plaza, Box 220, Chicago, IL 60614, USA.Available from: [DOI] [PubMed] [Google Scholar]
- 63.Dermitzakis ET, Reymond A, Antonarakis SE. Conserved non-genic sequences - an unexpected feature of mammalian genomes. Nat. Rev. Genet [Internet] 2005;6:151–157. doi: 10.1038/nrg1527. http://www.ncbi.nlm.nih.gov/pubmed/15716910 2005/02/18. Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SA, UK. md4@sanger.ac.uk;Available from: [DOI] [PubMed] [Google Scholar]
- 64.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet [Internet] 2006;15 doi: 10.1093/hmg/ddl046. https://academic.oup.com/hmg/article/15/suppl_1/R17/632705 Oxford Academic[cited 2021 Mar 26]Spec No:R17–29. Available from: [DOI] [PubMed] [Google Scholar]
- 65.Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int. J. Cancer. 2019;144:2501–2515. doi: 10.1002/ijc.31977. Wiley-Liss Inc. [DOI] [PubMed] [Google Scholar]
- 66.Santos JMO, Peixoto da Silva S, Gil da Costa RM, Medeiros R. The emerging role of micrornas and other non-coding RNAs in cancer cachexia. Cancers (Basel) [Internet] 2020;12:1004. doi: 10.3390/cancers12041004. https://www.mdpi.com/2072-6694/12/4/1004 Multidisciplinary Digital Publishing Institute[cited 2020 Apr 29]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ha M, Kim VN. Regulation of microRNA biogenesis [Internet] Nat. Rev. Mol. Cell Biol. 2014:509–524. doi: 10.1038/nrm3838. https://pubmed.ncbi.nlm.nih.gov/25027649/ Nature Publishing Group[cited 2020 Jul 30]Available from: [DOI] [PubMed] [Google Scholar]
- 68.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer [Internet] 2010;126:1283–1290. doi: 10.1002/ijc.25014. https://pubmed.ncbi.nlm.nih.gov/19877123/ Int J Cancer[cited 2020 Aug 4]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chacon-Cabrera A, Fermoselle C, Salmela I, Yelamos J, Barreiro E. MicroRNA expression and protein acetylation pattern in respiratory and limb muscles of Parp-1-/- and Parp-2-/- mice with lung cancer cachexia. Biochim Biophys Acta - Gen Subj. 2015;1850:2530–2543. doi: 10.1016/j.bbagen.2015.09.020. Elsevier. [DOI] [PubMed] [Google Scholar]
- 71.Lee DE, Brown JL, Rosa-Caldwell ME, Blackwell TA, Perry RA, Brown LA. Cancer cachexia-induced muscle atrophy: evidence for alterations in microRNAs important for muscle size. Physiol Genomics [Internet] 2017;49:253–260. doi: 10.1152/physiolgenomics.00006.2017. www.physiolgenomics.org [cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 72.Fernandez GJ, Ferreira JH, Vechetti IJ, de Moraes LN, Cury SS, Freire PP. MicroRNA-mRNA co-sequencing identifies transcriptional and post-transcriptional regulatory networks underlying muscle wasting in cancer cachexia. Front. Genet. [Internet] 2020;11 doi: 10.3389/fgene.2020.00541. https://pubmed.ncbi.nlm.nih.gov/32547603/ Frontiers Media S.A.[cited 2020 Aug 26]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen D, Goswami CP, Burnett RM, Anjanappa M, Bhat-Nakshatri P, Muller W. Cancer affects microRNA expression, release, and function in cardiac and skeletal muscle. Cancer Res. [Internet] 2014;74:4270–4281. doi: 10.1158/0008-5472.CAN-13-2817. http://cancerres.aacrjournals.org/ American Association for Cancer Research Inc.[cited 2020 Aug 31]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen L, Han J, Wang H, Meng Q, Chen L, Liu Y. Cachexia-related long noncoding RNA, CAAlnc1, suppresses adipogenesis by blocking the binding of HuR to adipogenic transcription factor mRNAs. Int. J. Cancer [Internet] 2019;145:1809–1821. doi: 10.1002/ijc.32236. https://pubmed.ncbi.nlm.nih.gov/30807648/ Wiley-Liss Inc.;[cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 75.Kehl T, Kern F, Backes C, Fehlmann T, Stöckel D, Meese E. MiRPathDB 2.0: a novel release of the miRNA pathway dictionary database. Nucleic Acids Res. [Internet] 2020;48:D142–D147. doi: 10.1093/nar/gkz1022. https://academic.oup.com/nar/article/48/D1/D142/5613673 Oxford University Press[cited 2021 Mar 24]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma M, Juvvuna PK, Kukreti H, McFarlane C. Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front. Physiol. [Internet] 2014 doi: 10.3389/fphys.2014.00239. https://pubmed.ncbi.nlm.nih.gov/25018733/ Frontiers Research Foundation[cited 2020 Jul 30];5 JUN. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Molinari F, Pin F, Gorini S, Chiandotto S, Pontecorvo L, Penna F. The mitochondrial metabolic reprogramming agent trimetazidine as an ‘exercise mimetic’ in cachectic C26-bearing mice. J. Cachexia Sarcopenia Muscle [Internet] 2017;8:954–973. doi: 10.1002/jcsm.12226. https://pubmed.ncbi.nlm.nih.gov/29130633/ Wiley Blackwell[cited 2020 Jul 30]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Narasimhan A, Ghosh S, Stretch C, Greiner R, Bathe OF, Baracos V. Small RNAome profiling from human skeletal muscle: novel miRNAs and their targets associated with cancer cachexia. J. Cachexia Sarcopenia Muscle. 2017;8:405–416. doi: 10.1002/jcsm.12168. Wiley Blackwell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Freire PP, Fernandez GJ, Cury SS, De Moraes D, Oliveira JS, De Oliveira G. The pathway to cancer Cachexia: Microrna-regulated networks in muscle wasting based on integrative meta-analysis. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20081962. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mubaid S, Ma JF, Omer A, Ashour K, Lian XJ, Sanchez BJ. HuR counteracts miR-330 to promote STAT3 translation during inflammation-induced muscle wasting. Proc Natl Acad Sci USA. Natl. Acad. Sci. 2019:116. doi: 10.1073/pnas.1905172116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van de Worp WRPH, Schols AMWJ, Dingemans AMC, Op den Kamp CMH, Degens JHRJ, Kelders MCJM. Identification of microRNAs in skeletal muscle associated with lung cancer cachexia. J Cachexia Sarcopenia Muscle. 2020;11:452–463. doi: 10.1002/jcsm.12512. Wiley Blackwell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Daas SI, Rizeq BR, Nasrallah GK. Adipose tissue dysfunction in cancer cachexia. J Cell Physiol [Internet] 2018;234:13–22. doi: 10.1002/jcp.26811. https://pubmed.ncbi.nlm.nih.gov/30078199/ Wiley-Liss Inc.[cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 83.Kulyté A, Lorente-Cebrián S, Gao H, Mejhert N, Agustsson T, Arner P. MicroRNA profiling links miR-378 to enhanced adipocyte lipolysis in human cancer cachexia. Am. J. Physiol. - Endocrinol Metab. 2014:306. doi: 10.1152/ajpendo.00249.2013. [DOI] [PubMed] [Google Scholar]
- 84.Liu H, Zhou T, Wang B, Li L, Ye D, Yu S. Identification and functional analysis of a potential key lncRNA involved in fat loss of cancer cachexia. J. Cell. Biochem. [Internet] 2018;119:1679–1688. doi: 10.1002/jcb.26328. https://pubmed.ncbi.nlm.nih.gov/28782835/ Wiley-Liss Inc.[cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 85.Han J, Shen L, Zhan Z, Liu Y, Zhang C, Guo R. The long noncoding RNA MALAT1 modulates adipose loss in cancer-associated cachexia by suppressing adipogenesis through PPAR-γ. Nutr. Metab. [Internet] 2021;18 doi: 10.1186/s12986-021-00557-0. BioMed Central Ltd[cited 2021 Mar 30]Available from: https://pubmed.ncbi.nlm.nih.gov/33691715/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Siracusa J, Koulmann N, Banzet S. Circulating myomiRs: a new class of biomarkers to monitor skeletal muscle in physiology and medicine [Internet] J. Cachexia. Sarcopenia Muscle. 2018:20–27. doi: 10.1002/jcsm.12227. http://www.ncbi.nlm.nih.gov/pubmed/29193905 Wiley Blackwell[cited 2020 Apr 29]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scherbakov N, Knops M, Ebner N, Valentova M, Sandek A, Grittner U. Evaluation of C-terminal Agrin Fragment as a marker of muscle wasting in patients after acute stroke during early rehabilitation. J. Cachexia Sarcopenia Muscle [Internet] 2016;7:60–67. doi: 10.1002/jcsm.12068. https://pubmed.ncbi.nlm.nih.gov/27066319/ Wiley Blackwell[cited 2020 Jul 30]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nishikawa H, Enomoto H, Ishii A, Iwata Y, Miyamoto Y, Ishii N. Elevated serum myostatin level is associated with worse survival in patients with liver cirrhosis. J. Cachexia Sarcopenia Muscle [Internet] 2017;8:915–925. doi: 10.1002/jcsm.12212. https://pubmed.ncbi.nlm.nih.gov/28627027/ Wiley Blackwell[cited 2020 Jul 30]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Calvani R, Marini F, Cesari M, Tosato M, Anker SD, Von Haehling S. Biomarkers for physical frailty and sarcopenia: state of the science and future developments [Internet] J. Cachexia. Sarcopenia Muscle. 2015:278–286. doi: 10.1002/jcsm.12051. https://pubmed.ncbi.nlm.nih.gov/26675566/ Wiley Blackwell[cited 2020 Jul 30]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Powrózek T, Mlak R, Brzozowska A, Mazurek M, Gołębiowski P, Małecka-Massalska T. MiRNA-130a significantly improves accuracy of SGA nutritional assessment tool in prediction of malnutrition and cachexia in radiotherapy-treated head and neck cancer patients. Cancers (Basel) [Internet] 2018;10 doi: 10.3390/cancers10090294. https://pubmed.ncbi.nlm.nih.gov/30200243/ MDPI AG[cited 2020 Jul 27]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okugawa Y, Yao L, Toiyama Y, Yamamoto A, Shigemori T, Yin C. Prognostic impact of sarcopenia and its correlation with circulating miR-21 in colorectal cancer patients. Oncol. Rep. [Internet] 2018;39:1555–1564. doi: 10.3892/or.2018.6270. http://www.ncbi.nlm.nih.gov/pubmed/29484416 Spandidos Publications[cited 2020 Apr 29]Available from: [DOI] [PubMed] [Google Scholar]
- 92.Okugawa Y, Toiyama Y, Hur K, Yamamoto A, Yin C, Ide S. Circulating miR-203 derived from metastatic tissues promotes myopenia in colorectal cancer patients. J. Cachexia Sarcopenia Muscle. 2019;10:536–548. doi: 10.1002/jcsm.12403. Wiley Blackwell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miyachi M, Tsuchiya K, Yoshida H, Yagyu S, Kikuchi K, Misawa A. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem. Biophys. Res. Commun. [Internet] 2010;400:89–93. doi: 10.1016/j.bbrc.2010.08.015. https://pubmed.ncbi.nlm.nih.gov/20696132/ Biochem Biophys Res Commun[cited 2020 Jul 30]Available from: [DOI] [PubMed] [Google Scholar]
- 94.Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur. J. Cancer [Internet] 2011;47:784–791. doi: 10.1016/j.ejca.2010.10.025. https://pubmed.ncbi.nlm.nih.gov/21112772/ Eur J Cancer[cited 2020 Jul 30]Available from: [DOI] [PubMed] [Google Scholar]
- 95.Li M, Zhang Q, Wu L, Jia C, Shi F, Li S. Serum miR-499 as a novel diagnostic and prognostic biomarker in non-small cell lung cancer. Oncol. Rep. [Internet] 2014;31:1961–1967. doi: 10.3892/or.2014.3029. https://pubmed.ncbi.nlm.nih.gov/24549225/ Spandidos Publications[cited 2020 Jul 30]Available from: [DOI] [PubMed] [Google Scholar]
- 96.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol [Internet] 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. https://pubmed.ncbi.nlm.nih.gov/20194856/ J Clin Oncol[cited 2020 Jul 30]Available from: [DOI] [PubMed] [Google Scholar]
- 97.Tian R, Liu T, Qiao L, Gao M, Li J. Decreased serum microRNA-206 level predicts unfavorable prognosis in patients with melanoma. Int. J. Clin. Exp. Pathol. [Internet] 2015;8:3097–3103. https://pubmed.ncbi.nlm.nih.gov/26045823/ [cited 2020 Jul 30]Available from: [PMC free article] [PubMed] [Google Scholar]
- 98.Nohata N, Hanazawa T, Enokida H, Seki N. MicroRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget [Internet] 2012;3:9–21. doi: 10.18632/oncotarget.424. https://pubmed.ncbi.nlm.nih.gov/22308266/ Impact Journals LLC[cited 2020 Jul 30]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang C, Yao C, Li H, Wang G, He X. Serum levels of microRNA-133b and microRNA-206 expression predict prognosis in patients with osteosarcoma. Int J Clin Exp Pathol [Internet] 2014;7:4194–4203. https://pubmed.ncbi.nlm.nih.gov/25120799/ [cited 2020 Jul 30]Available from: [PMC free article] [PubMed] [Google Scholar]
- 100.Camargo RG, Quintas Teixeira Ribeiro H, Geraldo MV, Matos-Neto E, Neves RX, Carlos Carnevali L. Cancer cachexia and microRNAs. Mediators Inflamm. [Internet] 2015;2015:1–5. doi: 10.1155/2015/367561. https://pubmed.ncbi.nlm.nih.gov/26504359/ [cited 2020 Aug 4]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nie M, Deng Z-L, Liu J, Wang D-Z. Noncoding RNAs, emerging regulators of skeletal muscle development and diseases. Biomed. Res. Int. [Internet] 2015;2015:1–17. doi: 10.1155/2015/676575. https://pubmed.ncbi.nlm.nih.gov/26258142/ [cited 2020 Aug 4]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang H, Wang B. Extracellular vesicle microRNAs mediate skeletal muscle myogenesis and disease (Review) Biomed Reports [Internet] 2016;5:296–300. doi: 10.3892/br.2016.725. https://pubmed.ncbi.nlm.nih.gov/27588172/ Spandidos Publications[cited 2020 Aug 4]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rashed MH, Bayraktar E, Helal GK, Abd-Ellah MF, Amero P, Chavez-Reyes A. Exosomes: From garbage bins to promising therapeutic targets. Int. J. Mol. Sci. [Internet] 2017;18 doi: 10.3390/ijms18030538. https://pubmed.ncbi.nlm.nih.gov/28257101/ MDPI AG[cited 2020 Aug 4]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Iraci N, Leonardi T, Gessler F, Vega B, Pluchino S. Focus on extracellular vesicles: Physiological role and signalling properties of extracellular membrane vesicles. Int. J. Mol. Sci. [Internet] 2016;17 doi: 10.3390/ijms17020171. https://pubmed.ncbi.nlm.nih.gov/26861302/ MDPI AG[cited 2020 Aug 4]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lazar I, Clement E, Dauvillier S, Milhas D, Ducoux-Petit M, LeGonidec S. Adipocyte exosomes promote melanoma aggressiveness through fatty acid oxidation: a novel mechanism linking obesity and cancer. Cancer Res. [Internet] 2016;76:4051–4057. doi: 10.1158/0008-5472.CAN-16-0651. https://pubmed.ncbi.nlm.nih.gov/27216185/ American Association for Cancer Research Inc.[cited 2020 Aug 4]Available from: [DOI] [PubMed] [Google Scholar]
- 106.Huang-Doran I, Zhang CY, Vidal-Puig A. Extracellular vesicles: novel mediators of cell communication in metabolic disease [Internet] Trends Endocrinol. Metab. 2017:3–18. doi: 10.1016/j.tem.2016.10.003. https://pubmed.ncbi.nlm.nih.gov/27810172/ Elsevier Inc.[cited 2020 Aug 4]Available from: [DOI] [PubMed] [Google Scholar]
- 107.Lobb RJ, Lima LG, Möller A. Exosomes: key mediators of metastasis and pre-metastatic niche formation. Semin Cell Dev Biol [Internet] 2017;67:3–10. doi: 10.1016/j.semcdb.2017.01.004. https://pubmed.ncbi.nlm.nih.gov/28077297/ Elsevier Ltd[cited 2020 Aug 4]Available from: [DOI] [PubMed] [Google Scholar]
- 108.Tomasetti M, Lee W, Santarelli L, Neuzil J. Exosome-derived microRNAs in cancer metabolism: Possible implications in cancer diagnostics and therapy. Exp Mol Med [Internet] 2017;49 doi: 10.1038/emm.2016.153. https://pubmed.ncbi.nlm.nih.gov/28104913/ Nature Publishing Group[cited 2020 Aug 4]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu Q, Sun S, Li Z, Yang Q, Li B, Zhu S. Tumour-originated exosomal miR-155 triggers cancer-associated cachexia to promote tumour progression. Mol Cancer [Internet] 2018;17:155. doi: 10.1186/s12943-018-0899-5. https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-018-0899-5 BioMed Central Ltd.[cited 2020 Apr 30]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Wu Q, Sun S, Li Z, Yang Q, Li B, Zhu S. Breast cancer-released exosomes trigger cancer-associated cachexia to promote tumor progression. Adipocyte [Internet] 2019;8:31–45. doi: 10.1080/21623945.2018.1551688. https://pubmed.ncbi.nlm.nih.gov/30474469/ Taylor and Francis Inc.[cited 2020 Aug 26]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 111.Wan Z, Chen X, Gao X, Dong Y, Zhao Y, Wei M. Chronic myeloid leukemia-derived exosomes attenuate adipogenesis of adipose derived mesenchymal stem cells via transporting miR-92a-3p. J. Cell. Physiol. [Internet] 2019;234:21274–21283. doi: 10.1002/jcp.28732. https://pubmed.ncbi.nlm.nih.gov/31062357/ Wiley-Liss Inc.[cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 112.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer - a brief overview. Adv. Biol. Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 113.He WA, Calore F, Londhe P, Canella A, Guttridge DC, Croce CM. Microvesicles containing miRNAs promote muscle cell death in cancer cachexia via TLR7. Proc. Natl. Acad. Sci. USA. 2014;111:4525–4529. doi: 10.1073/pnas.1402714111. National Academy of Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad. Sci. USA [Internet] 2012;109 doi: 10.1073/pnas.1209414109. https://pubmed.ncbi.nlm.nih.gov/22753494/ Proc Natl Acad Sci U S A[cited 2020 Aug 4]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fabbri M, Paone A, Calore F, Galli R, Croce CM. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol [Internet] 2013;10:169–174. doi: 10.4161/rna.23144. https://pubmed.ncbi.nlm.nih.gov/23296026/ Taylor and Francis Inc.[cited 2020 Aug 4]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol [Internet] 2019;21:542–551. doi: 10.1038/s41556-019-0311-8. https://pubmed.ncbi.nlm.nih.gov/31048766/ Nature Publishing Group[cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 117.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA–LncRNA interactions. Methods Mol Biol [Internet] 2016:271–286. doi: 10.1007/978-1-4939-3378-5_21. https://pubmed.ncbi.nlm.nih.gov/26721498/ Humana Press Inc.[cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 118.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. [Internet] 2009;10:155–159. doi: 10.1038/nrg2521. https://pubmed.ncbi.nlm.nih.gov/19188922/ Nat Rev Genet[cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 119.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 120.Sweta S, Dudnakova T, Sudheer S, Baker AH, Bhushan R. Importance of long non-coding RNAs in the development and disease of skeletal muscle and cardiovascular lineages. Front. Cell. Dev. Biol. [Internet] 2019;7:1–19. doi: 10.3389/fcell.2019.00228. Frontiers Media S.A.[cited 2021 Mar 29]Available from: /pmc/articles/PMC6813187/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Z, Cai B, Abdalla BA, Zhu X, Zheng M, Han P. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J. Cachexia Sarcopenia Muscle [Internet] 2019;10:391–410. doi: 10.1002/jcsm.12374. https://pubmed.ncbi.nlm.nih.gov/30701698/ Wiley Blackwell[cited 2020 Jul 27]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schiaffino S, Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: Insights from genetic models. Skelet Muscle [Internet] 2011;1 doi: 10.1186/2044-5040-1-4. https://pubmed.ncbi.nlm.nih.gov/21798082/ Skelet Muscle[cited 2020 Jul 27]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dong X, Park S, Lin X, Copps K, Yi X, White MF. Irs1 and Irs2 signaling is essential for hepatic glucose homeostasis and systemic growth. J Clin Invest [Internet] 2006;116:101–114. doi: 10.1172/JCI25735. https://pubmed.ncbi.nlm.nih.gov/16374520/ The American Society for Clinical Investigation[cited 2020 Jul 27]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang ZK, Li J, Guan D, Liang C, Zhuo Z, Liu J. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle. 2018;9:613–626. doi: 10.1002/jcsm.12281. Wiley Blackwell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell. [Internet] 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. https://pubmed.ncbi.nlm.nih.gov/26143994/ Cell Press[cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 126.Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer [Internet] 2019;18 doi: 10.1186/s12943-018-0934-6. https://pubmed.ncbi.nlm.nih.gov/30626395/ BioMed Central Ltd.[cited 2020 Jul 27]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee ECS, Elhassan SAM, Lim GPL, Kok WH, Tan SW, Leong EN. The roles of circular RNAs in human development and diseases. Biomed Pharmacother [Internet] 2019;111:198–208. doi: 10.1016/j.biopha.2018.12.052. https://pubmed.ncbi.nlm.nih.gov/30583227/ Elsevier Masson SAS[cited 2020 Jul 27]Available from: [DOI] [PubMed] [Google Scholar]
- 128.Patop IL, Wüst S, Kadener S. Past, present, and future of circ RNA s. EMBO J [Internet] 2019;38 doi: 10.15252/embj.2018100836. https://pubmed.ncbi.nlm.nih.gov/31343080/ EMBO[cited 2020 Jul 27]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Carr RM, Enriquez-Hesles E, Olson RLO, Jatoi A, Doles J, Fernandez-Zapico ME. Epigenetics of cancer-associated muscle catabolism. Epigenomics. 2017:1259–1265. doi: 10.2217/epi-2017-0058. Future Medicine Ltd. [DOI] [PubMed] [Google Scholar]
- 130.Terasawa K, Shimizu K, Tsujimoto G. Synthetic pre-miRNA-based shRNA as potent RNAi triggers. J. Nucleic Acids [Internet] 2011 doi: 10.4061/2011/131579. https://pubmed.ncbi.nlm.nih.gov/21776374/ J Nucleic Acids[cited 2020 Nov 3];2011. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang J. Patisiran for the treatment of hereditary transthyretin-mediated amyloidosis. Expert Rev. Clin. Pharmacol. [Internet] 2019;12:95–99. doi: 10.1080/17512433.2019.1567326. https://pubmed.ncbi.nlm.nih.gov/30644768/ Taylor and Francis Ltd[cited 2020 Jul 29]Available from: [DOI] [PubMed] [Google Scholar]
- 132.Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet [Internet] 2019;10:478. doi: 10.3389/fgene.2019.00478. https://www.frontiersin.org/article/10.3389/fgene.2019.00478/full Frontiers Media S.A.[cited 2020 Jul 29]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bakhshandeh B, Soleimani M, Hafizi M, Ghaemi N. A comparative study on nonviral genetic modifications in cord blood and bone marrow mesenchymal stem cells. Cytotechnology [Internet] 2012;64:523–540. doi: 10.1007/s10616-012-9430-9. https://pubmed.ncbi.nlm.nih.gov/22328133/ Cytotechnology[cited 2020 Nov 3]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet [Internet] 2014;15:541–555. doi: 10.1038/nrg3763. https://pubmed.ncbi.nlm.nih.gov/25022906/ Nature Publishing Group[cited 2020 Nov 3]Available from: [DOI] [PubMed] [Google Scholar]
- 135.Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer [Internet] 2020;122:1630–1637. doi: 10.1038/s41416-020-0802-1. https://pubmed.ncbi.nlm.nih.gov/32238921/ Springer Nature[cited 2020 Nov 3]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol [Internet] 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. https://pubmed.ncbi.nlm.nih.gov/28870611/ Lancet Publishing Group[cited 2020 Nov 3]Available from: [DOI] [PubMed] [Google Scholar]
- 137.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. https://pubmed.ncbi.nlm.nih.gov/17449137/ Adv Drug Deliv Rev [Internet] [cited 2020 Nov 3]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang X, Yu B, Ren W, Mo X, Zhou C, He H. Enhanced hepatic delivery of siRNA and microRNA using oleic acid based lipid nanoparticle formulations. J Control Release [Internet] 2013;172:690–698. doi: 10.1016/j.jconrel.2013.09.027. https://pubmed.ncbi.nlm.nih.gov/24121065/ J Control Release[cited 2020 Nov 3]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang T, Thakur A, Chen T, Yang L, Lei G, Liang Y. MicroRNA-15a induces cell apoptosis and inhibits metastasis by targeting BCL2L2 in non-small cell lung cancer. Tumor Biol. [Internet] 2015;36:4357–4365. doi: 10.1007/s13277-015-3075-1. https://pubmed.ncbi.nlm.nih.gov/25874488/ Kluwer Academic Publishers[cited 2020 Nov 3]Available from: [DOI] [PubMed] [Google Scholar]
- 140.György B, Hung ME, Breakefield XO, Leonard JN. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu Rev Pharmacol Toxicol [Internet] 2015;55:439–464. doi: 10.1146/annurev-pharmtox-010814-124630. http://www.annualreviews.org/doi/10.1146/annurev-pharmtox-010814-124630 Annual Reviews Inc.[cited 2020 Nov 3]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kalra H, Drummen GPC, Mathivanan S. Focus on extracellular vesicles: Introducing the next small big thing. Int. J. Mol. Sci. [Internet] 2016;17 doi: 10.3390/ijms17020170. https://pubmed.ncbi.nlm.nih.gov/26861301/ MDPI AG[cited 2020 Nov 3]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Keller A, Leidinger P, Bauer A, Elsharawy A, Haas J, Backes C. Toward the blood-borne miRNome of human diseases. Nat. Methods [Internet] 2011;8:841–843. doi: 10.1038/nmeth.1682. https://pubmed.ncbi.nlm.nih.gov/21892151/ Nat Methods[cited 2020 Nov 3]Available from: [DOI] [PubMed] [Google Scholar]
- 143.Ebner N, Anker SD, von Haehling S. Recent developments in the field of cachexia, sarcopenia, and muscle wasting: highlights from the 12th Cachexia Conference. J Cachexia Sarcopenia Muscle [Internet] 2020:274–285. doi: 10.1002/jcsm.12552. https://pubmed.ncbi.nlm.nih.gov/32049447/ Wiley Blackwell[cited 2020 Jul 29]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pound P, Ritskes-Hoitinga M. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail [Internet] J. Transl. Med. BioMed Central Ltd. 2018:304. doi: 10.1186/s12967-018-1678-1. https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-018-1678-1 [cited 2021 Mar 26]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mueller TC, Bachmann J, Prokopchuk O, Friess H, Martignoni ME. Molecular pathways leading to loss of skeletal muscle mass in cancer cachexia - can findings from animal models be translated to humans? BMC Cancer [Internet] 2016;16:1–14. doi: 10.1186/s12885-016-2121-8. https://bmccancer.biomedcentral.com/articles/10.1186/s12885-016-2121-8 BioMed Central Ltd.[cited 2021 Mar 30]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sannicandro AJ, McDonagh B, Goljanek-Whysall K. MicroRNAs as potential therapeutic targets for muscle wasting during cancer cachexia. Curr Opin Clin Nutr Metab Care. NLM (Medline); 2020;23:157–163. doi: 10.1097/MCO.0000000000000645. [DOI] [PubMed] [Google Scholar]