Summary
Pancreatic ductal adenocarcinoma (PDAC) tumors have a nutrient poor, desmoplastic, and highly innervated tumor microenvironment. Although, neurons can release stimulatory factors to accelerate PDAC tumorigenesis, the metabolic contribution of peripheral axons has not been explored. We found that peripheral axons release serine (Ser) to support the growth of exogenous Ser (exSer)-dependent PDAC cells during Ser/Gly (glycine)-deprivation. Ser-deprivation resulted in ribosomal stalling on two of the six Ser codons, TCC and TCT, and allowed the selective translation and secretion of nerve growth factor (NGF) by PDAC cells to promote tumor innervation. Consistent with this, exSer-dependent PDAC tumors grew slower and displayed enhanced innervation in mice on a Ser/Gly-free diet. Blockade of compensatory neuronal innervation using LOXO-101, a Trk-NGF inhibitor, further decreased PDAC tumor growth. Our data indicate that axonal-cancer metabolic crosstalk is a critical adaptation to support PDAC growth in nutrient poor environments.
In Brief
The high level of innervation seen in pancreatic ductal adenocarcinoma tumors supplies serine and serine-deprived conditions promote tumor innervation to support growth in nutrient poor environments.
Graphical Abstract
Introduction
A common symptom of PDAC patients is neuropathic pain that can necessitate invasive procedures for palliation. Indeed, a majority of PDAC tumors display perineural invasion, which is associated with poor patient outcomes (Ceyhan et al., 2009b; Liebl et al., 2014). Along these lines, genetic analyses of PDAC has revealed frequent mutations in regulators of axon guidance, suggesting a role for neuronal support in tumorigenesis (Biankin et al., 2012). Furthermore, surgical denervation or ablation of nerves with capsaicin increases survival of PDAC-bearing mice (Renz et al., 2018a; Saloman et al., 2016), thus supporting a pro-oncogenic role for nerves in PDAC. Sensory neurons (Sinha et al., 2017) from the dorsal root ganglion (DRG) and sympathetic nerves have been reported to accelerate PDAC tumorigenesis, whereas cholinergic nerves (Renz et al., 2018b) suppressed growth. To date, the mechanisms have focused on the release of neurotrophic factors or neurotransmitters into the tumor.
Another defining feature of PDAC is an extraordinary metabolic plasticity, allowing them to adapt to nutrient poor environments by upregulating scavenging pathways, such as autophagy (Perera et al., 2015) and macropinocytosis (Commisso et al., 2013), and utilizing metabolites released from stromal fibroblasts and macrophages (Auciello et al., 2019; Halbrook et al., 2019; Sousa et al., 2016). Neurons also are known to release amino acid-derived neurotransmitters, such as glycine (Gly) and D-serine (Monaghan et al., 1989; Seal and Amara, 1999), which raises the possibility that neurons can support the metabolic needs of PDAC cells in nutrient poor environments, via their peripheral axons. However, the metabolic contribution of nerves in PDAC tumors has not been explored.
Serine (Ser) is a conditionally essential amino acid (AA) that can be obtained from the microenvironment or produced via the Ser biosynthesis pathway (SBP) from glucose and is the second most abundant AA found in human proteins (Mauro and Chappell, 2014). Ser is also used to generate Gly, which can be used to synthesize proteins or glutathione. It also participates in the production of certain lipids, and the folate cycle to generate one-carbon units required for the synthesis of nucleotides and methylation units (Gantner et al., 2019; Gao et al., 2018; Labuschagne et al., 2014; Yang and Vousden, 2016). Because Ser is necessary for multiple metabolic pathways, it can become rate limiting for growth and survival for several types of cancers (Sullivan et al., 2019). However, mouse PDAC cells derived from the genetically engineered KPC (KrasG12D/+; Tp53flox/+; Pdx-Cre) model can upregulate SBP enzymes, and are insensitive to Ser limitation (Maddocks et al., 2017). Whether human PDAC cells also upregulate the SBP or whether there are other mechanisms of obtaining extracellular Ser in not clear.
In this study, we investigated the role of peripheral axons in providing metabolic support to PDAC cells in nutrient deprived environments. We found a subset of human PDAC cell lines lacking expression of the SBP enzymes and were dependent on exogenous Ser (exSer) for growth. We demonstrate that axons release AA, such as Ser, and this was able to rescue growth of exSer-dependent PDAC cells in Ser/Gly-deprived conditions. Surprisingly, Ser-deprivation specifically decreased the mRNA translation efficiency of two out of six Ser codons, TCC and TCT, and allowed the selective translation and secretion of nerve growth factor (NGF). Consistent with these observations, exSer-dependent PDAC tumors from mice on Ser/Gly-free diet were smaller and exhibited increased tumor innervation when Ser is limiting. These data establish a metabolic axis between PDAC and neurons to promote tumor growth under nutrient-poor conditions.
Results
Axons release AA, such as Ser, into nutrient-deprived environments
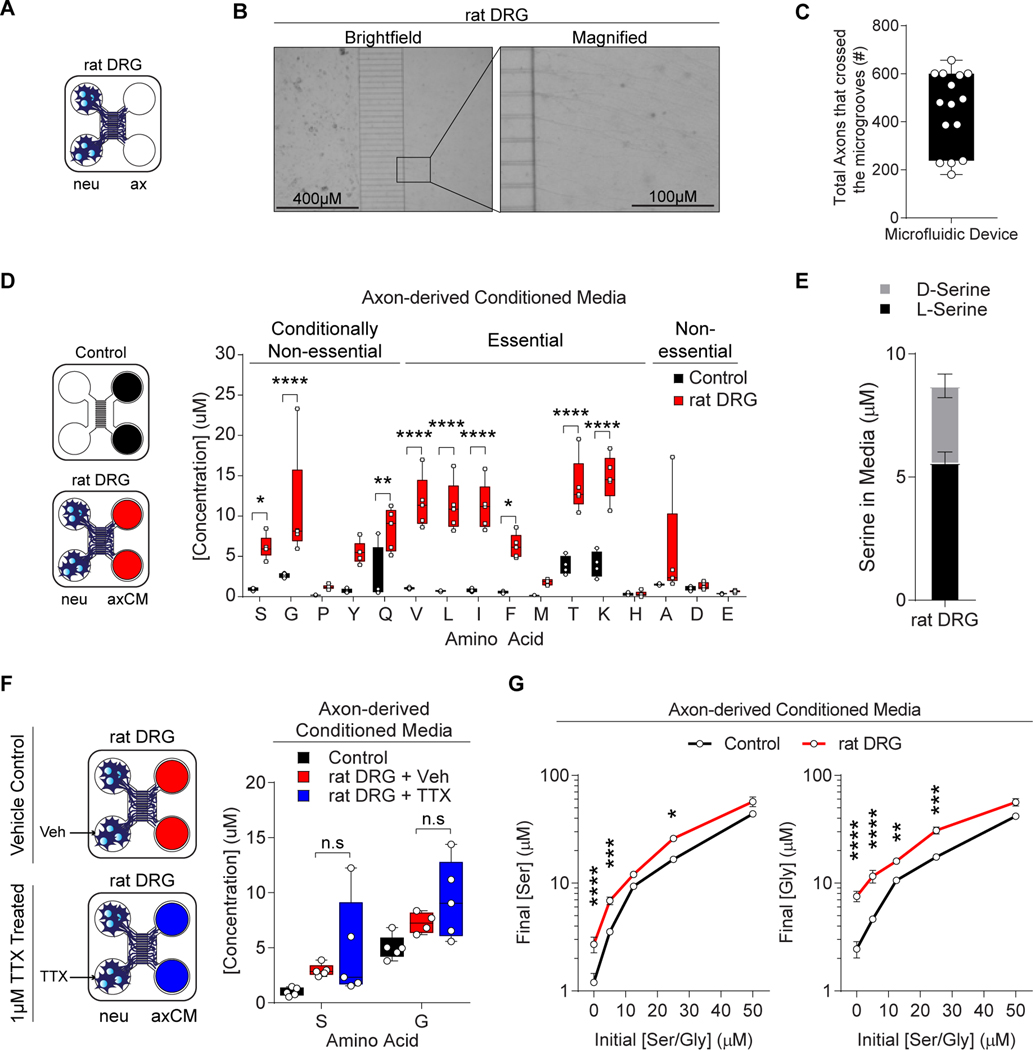
Neurons that innervate tumors are conceptually ideal to supply nutrients to poorly perfused areas in the tumor microenvironment. While the axons may be located in the nutrient poor tumor, the neuronal cell body has access to high levels of nutrients in the circulation. We assessed whether the axons of sensory nerves could supply nutrients through secretion of metabolites. Rat dorsal root ganglion cells (DRGs) were grown in microfluidic devices that isolated axons from the neuronal body (Figure 1A–C) (Taylor et al., 2005), simulating the in vivo situation whereby the tumors are only exposed to the axons. Hydrostatic pressure was established to isolate the nutrient-deprived axonal media from the nutrient-rich neuronal media. Conditioned media collected from the axonal side had increased levels of several AA, including Ser (35% D-Ser and 65% L-Ser), and Gly (Figure 1D–E). We focused on Ser because, unlike other AA that can be obtained by macropinocytosis, serum albumin does not contribute to the intracellular Ser pools in PDAC (Davidson et al., 2017). Ser has gained recent attention as critical in supporting growth of several cancer types. Interestingly, the release of Ser and Gly from axons was not action potential-dependent (Figure 1F and S1A) and axons were able to secrete Ser and Gly when cultured in a range of Ser/Gly concentrations (Figure 1G). Together these data indicate that axons can release AA into nutrient-poor environments.
Figure 1.
Axons secrete serine to support exSer-dependent PDAC growth.
A, Schematic of microfluidic device with rat DRGs on the neuronal side (neu) and axons on the axonal side (ax).
B, Representative images of axons from rat DRGs in microfluidic devices from A.
C, Quantification of the number of axons that crossed the microgrooves in the microfluidic device (n=15).
D, AA concentrations were measured from conditioned-media from the axonal side without (n=4) and with (n=5) rat DRGs. neu: neuronal side; axCM: axonal conditioned media.
E, L- and D-Ser concentrations were measured from conditioned media collected from the axCM of the microfluidic device with rat DRGs after 24 hours (n=5).
F, Ser and Gly concentrations were measured from axCM of rat DRGs treated with vehicle (Veh) or 1μM TTX as shown (n=4–5).
G, Ser and Gly concentrations were measured after 24 hours from axCM containing axons from rat DRGs. The axon side was cultured at the indicated initial Ser/Gly concentrations at the beginning of the assay (n=5).
“n” are displayed as individual points and represent the number of biologically independent devices. Graphs represent (median ± max/min (C, D, F) and mean ± s.e.m. (E, G)) were compared by two-way ANOVA (D, F-G), followed by Bonferroni post-hoc test (*p<0.05, **p<0.01, ****p<0.0001).
Axons support growth of exSer-dependent PDAC cells in Ser/Gly-deprived conditions
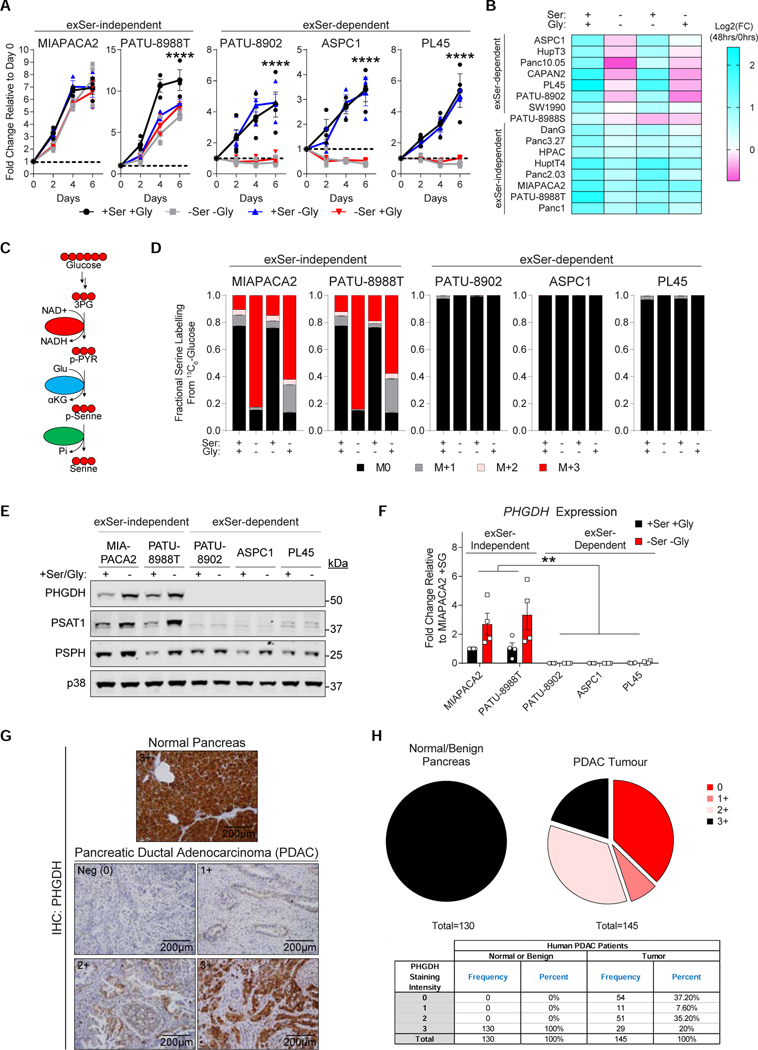
Mouse KPC PDAC cells can upregulate Ser synthesis and are insensitive to Ser/Gly starvation (Figure S1B). By contrast, we find that human PDAC cells show heterogeneity in their dependency on exSer with ~40% of human PDAC cell lines being completely dependent on exSer for proliferation under various nutrient conditions (Figure 2A–B and S1C). Furthermore, exSer-dependent PDAC cells exhibit a dose-dependent growth over a range of Ser concentrations (Figure S1D), and are incapable of adapting even after long periods of Ser/Gly-deprivation (Figure S1E). Unlike exSer-independent PDAC cells, exSer-dependent PDAC cells lack the expression and ability to upregulate PHGDH and PSAT1 to convert glucose into Ser (Figure 2C–F and S1F–G). Ectopic expression of PHGDH and/or PSAT1 were not sufficient to restore growth of exSer-dependent PDAC cells in Ser/Gly-deprived conditions. However after long-term culture, only exSer-dependent PDAC clones expressing both PHGDH and PSAT1 were able to adapt to the absence of Ser/Gly (Figure S1H–I). Adapted PHGDH and PSAT1-expressing PDAC cells expressed higher levels of the SBP enzymes and were able to grow in the absence of Ser/Gly (Figure S1I–J). Together, these data indicate that high expression of SBP enzymes are required to grow in the absence of Ser/Gly. We also found that ~40% of human PDAC tumors lacked PHGDH (Figure 2G–H). However, expression of SBP genes were not associated with molecular subtypes in PDAC cell lines or clinical samples in the TCGA (The Cancer Genome Atlas) (Figure S1K–L and Table S1). The inability to express and upregulate sufficient levels of SBP genes was observed to be negatively associated with methylation levels (Figure S1M–O), suggestive of epigenetic or transcriptional issues in exSer-dependent PDAC cells.
Figure 2.
A subset of human PDAC cells require exogenous Ser for growth.
A, Human PDAC cells were grown in the presence or absence of Ser and/or Gly (n=3 biologically independent experiments).
B, Heatmap of PDAC cell growth with or without Ser and/or Gly after 48 hours (n=2). FC reflects the number of cells at 48 hours normalized to 0 hours.
C, Schematic of SBP from 13C6-glucose. Each red and black circle represents one 13C and 12C, respectively.
D, PDAC cells were labelled with 13C6-glucose for 24 hours and fractional labelling of Ser was measured using GC-MS. (n=5).
E, Immunoblots of SBP enzymes (PHGDH, PSAT1 and PSPH) in PDAC cells grown with and without Ser/Gly. p38 serves as a loading control.
F, PHGDH expression in exSer-independent and -dependent human PDAC cells that were grown in the presence or absence of Ser/Gly after 24 hours (n=4).
G-H, Representative image (G), and scoring and frequency (H) of immunohistochemical (IHC) staining of PHGDH intensity scores in normal pancreas and PDAC tissue.
Where “n” represents the number of biologically independent replicates. Graphs (mean ± s.e.m.) were compared using two-way ANOVA (A, D, F), followed by Bonferroni post-hoc test (**p<0.01, ***p<0.005).
exSer-dependent PDAC growth in Ser/Gly-deprived media was rescued when co-cultured with axons (Figure 3A–B). Because axons were found to release both L- and D-Ser, and reported to form synaptic gap junctions with gliomas (Venkataramani et al., 2019; Venkatesh et al., 2019), we wanted to assess these effects on exSer-dependent PDAC cell growth. We found that the D-Ser was not able to restore growth in exSer-dependent PDAC cells under Ser/Gly-deprived conditions (Figure S2A). Furthermore, we did not observe gap junctions between axons and PDAC cells (Figure S2B–C) (Abbaci et al., 2008). Although axons were found to release ~5uM of Ser after 24 hours, which is not sufficient to support proliferation in monoculture assays, axons likely restore growth by replenishing steady-state levels of Ser that is consumed by exSer-dependent PDAC cells in co-culture assays. Indeed, repeated supplementation of low dose of Ser was able to stimulate growth of exSer-dependent PDAC cells (Figure S2D). Together, these observations indicate that neurons are able to release sufficient levels of L-Ser via their axons to metabolically support PDAC cells.
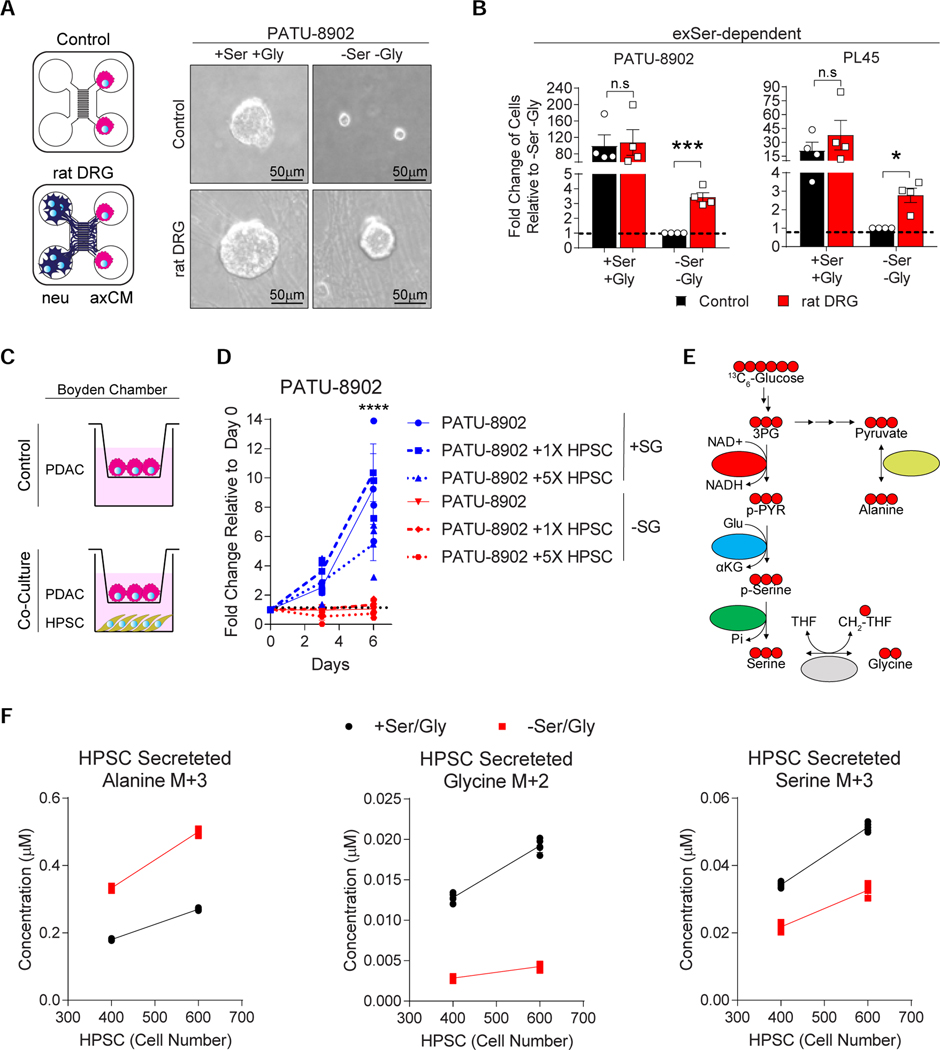
Figure 3.
Axons can metabolically support exSer-dependent PDAC cells in Ser/Gly-deprived conditions.
A, Representative images of axons supporting exSer-dependent PATU-8902 growth under Ser/Gly-deprived conditions in the channels of microfluidic device.
B, Quantification of PATU-8902 and PL45 growth in the presence or absence of Ser/Gly and rat DRGs after 6 and 12 days, respectively (n=4).
C, Schematic of co-culture of exSer-dependent PDAC cells and human pancreatic stellate cells (HPSCs) in a Boyden Chamber.
D, exSer-dependent PDAC cells were co-cultured with HPSC cells with (+SG) and without (-SG) Ser/Gly as shown in C (n=3).
E, Schematic of 13C6-glucose labelling to produce 13C3-Ser (M+3), 13C2-Gly (M+2) and 13C3-alanine (M+3).
F, Concentrations of 13C6-glucose-derived alanine, Gly and Ser produced and secreted by 400 and 600 HPSCs (n=5).
Where “n” represents the number of biologically independent replicates. Graphs (mean ± s.e.m.) were compared by one-sample t-test (B) or two-way ANOVA (D, F), followed by Bonferroni post-hoc test (*p<0.05, ***p<0.005, ****p<0.0001).
We previously found that human pancreatic stellate cells (HPSCs) can support PDAC growth and metabolism under nutrient limiting conditions by secreting alanine (Sousa et al., 2016). Because PSCs reside in the same microenvironment as PDAC cells, we used Boyden chambers to assess whether HPSCs can support exSer-dependent PDAC cells in Ser/Gly-deprived conditions. Using a PDAC to HPSC ratio of 1:5 (an overestimation that assumes the whole stroma is HPSCs (Seymour et al., 1994)), HPSCs were not able release sufficient levels of Ser to support exSer-dependent PDAC cell growth in Ser/Gly-deprived conditions (Figure 3C–F). This indicates that axons are supplying significantly more Ser than HPSCs to exSer-dependent PDAC cells.
PDAC cells require Ser for efficient mRNA translation
Next, we assessed the role of Ser in PDAC cells. We found that exSer-dependent PDAC growth in Ser/Gly-deprived media was not restored by the addition of exogenous formate or nucleosides (Figure S2E–F). Only in the presence of high Gly did exogenous formate partially restore growth in some, but not all, exSer-dependent PDAC cells grown in Ser-deprived media (Figure S2F–G). Partial rescue by Gly and formate was likely due to SHMT1/2-dependent Gly-to-Ser conversion (Figure S2H–I). In addition, S-adenosyl-methionine:S-adenosylhomocysteine (SAM:SAH) and reduced:oxidized glutathione (GSH:GSSG) ratios were not decreased under Ser/Gly-depleted conditions in exSer-dependent PDAC cells (Figure S2J). Although PDAC cells incorporated Ser into GSH/GSSG, SAM/SAH, and purines, the changes of these metabolites upon Ser/Gly-deprivation were not significantly decreased between exSer-dependent and - independent PDAC cells (Figure S2K–M). This is consistent with the observation that exogenous nucleosides and formate (one-carbon units for methylation and nucleotides), and Gly (glutathione synthesis) could not rescue exSer-dependent PDAC cell growth in Ser-deprived conditions. Together these results indicate that Ser was not limiting for one-carbon metabolism, glutathione or SAM pathway activity in exSer-dependent PDAC cells.
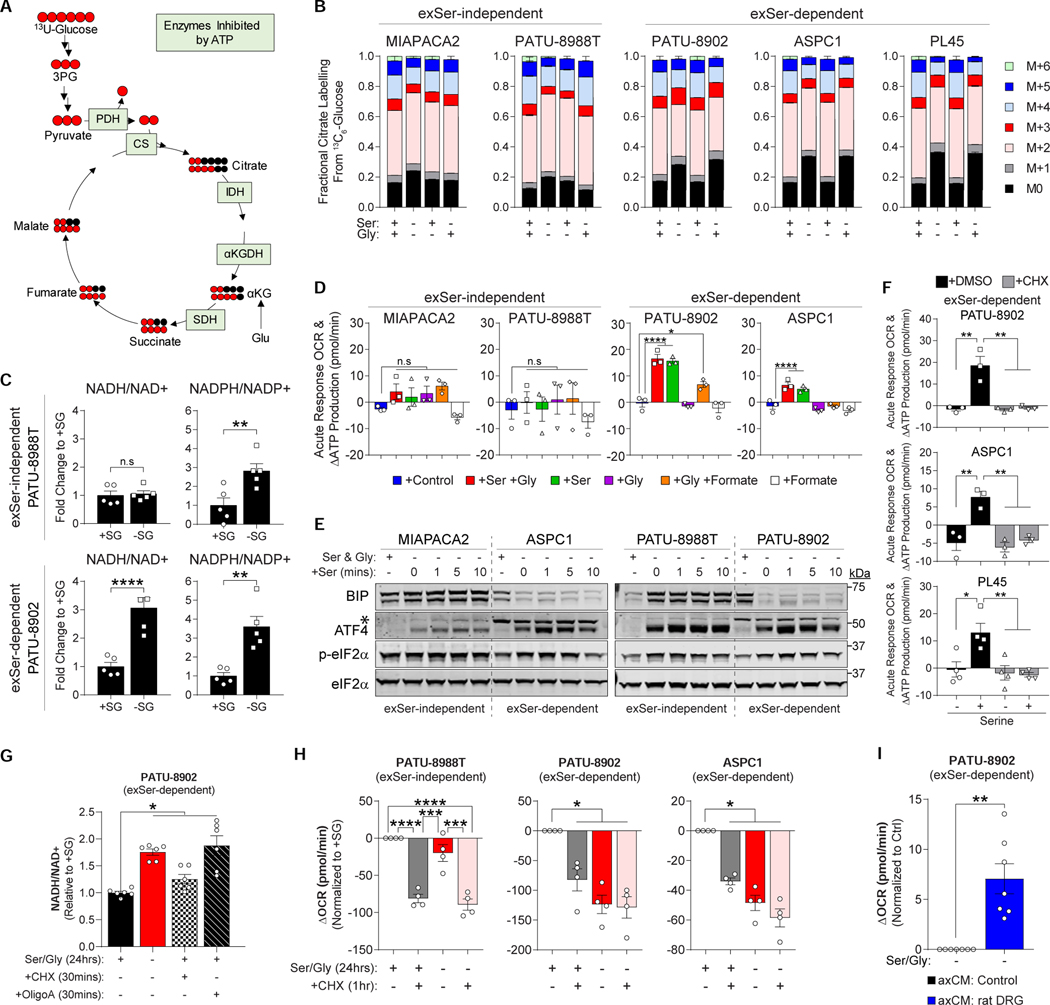
We noticed that glucose incorporation into citrate was decreased in exSer-dependent, but not exSer-independent, PDAC cells deprived of Ser (Figure 4A–B and S3A). exSer-dependent, but not exSer-independent, PDAC cells also displayed higher NADH/NAD+ ratios with Ser/Gly starvation (Figure 4C). These data suggested that exSer-dependent PDAC cells decrease mitochondrial activity during Ser limitation. To assess mitochondria activity in PDAC cells, we measured the oxygen consumption rate (OCR) under Ser/Gly-deprived conditions and stimulated with different combinations of Ser, Gly and formate. Injection of Ser/Gly and Ser alone increased OCR within minutes in exSer-dependent PDAC cells. By contrast, injection of Gly, Gly and formate, and formate alone had minimal effect on OCR (Figure 4D and S3B). Interestingly, the higher NADH/NAD+ ratios in exSer-dependent PDAC cells upon Ser/Gly-deprivation was positively associated with the acute response of OCR upon Ser stimulation (Figure S3C–D). Furthermore, Ser did not stimulate OCR in permeabilized cells, which directly measures mitochondrial OCR (Divakaruni et al., 2014), suggesting that Ser-induced increase in OCR was triggering a response in the cytosol in a rapid fashion (Figure S3E–F).
Figure 4.
Serine indirectly regulates mitochondrial activity in PDAC cells, via its importance for protein synthesis.
A, Schematic of 13C6-glucose labelling into the TCA cycle. Each red and black circle represent one 13C and 12C, respectively. Enzymes negatively regulated by ATP are indicated in green.
B, Fractional labelling of citrate from 13C6-glucose in PDAC cells measured with or without Ser and/or Gly after 24 hours (n=5).
C, Fold change of NADH/NAD+ and NADPH/NADP+ ratios in PDAC cells grown with and without Ser/Gly after 24 hours (n=5).
D, Acute response in oxygen consumption rate (OCR) after metabolite(s) stimulation (n=3).
E, Immunoblots of BIP, ATF4 and p-eIF2α in response to 400μM Ser stimulation in PDAC cells starved of Ser/Gly after 24 hours. eIF2α serves as a loading control. Asterisk (*) represents non-specific band.
F, Acute response in OCR to Ser in exSer-dependent PDAC cells pretreated with or without 50μM cyclohexamide (CHX) for 1 hour (n=3-4).
G, Fold change in NADH/NAD+ was assessed in exSer-dependent PDAC cells grown in Ser/Gly-rich and –poor conditions for 24 hours, and treated with or without 50μM CHX and 2μM oligomycin A (OligoA) for 30 mins (n=6).
H, Change in OCR in exSer-independent and –dependent in response to Ser-deprivation, with or without 50μM CHX treatment (n=4).
I, Change in OCR in exSer-dependent PDAC cells grown in 50% Ser/Gly-deprived media with 50% axon-derived conditioned media (axCM) from microfluidic devices without (Control) and with axons from rat DRGs for 24 hours (n=7).
Where “n” represents the number of biologically independent experiments for each group and condition. Graphs (mean ± s.e.m.) were compared by one-sample (I), two-tailed Student t-test (C), two (B) and one (D, F, G-H) -way ANOVA, followed by Bonferroni post-hoc test (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
The rapid stimulation of OCR was suggestive of a signaling mechanism, as transcriptional changes would be too slow. There are two major AA sensing signaling pathways, GCN2-ATF4 and the mTOR pathway (Chantranupong et al., 2015; Gonzalez and Hall, 2017). Under AA-deprived conditions, mTOR is inactivated and cell proliferation is suppressed. Whereas, AA-deprivation increases uncharged tRNAs and activates GCN2, which phosphorylates eIF2α to allow efficient translation of ATF4. ATF4 is a critical transcription factor that upregulates stress-response genes, such as CHOP, HSPA5 (a.k.a. BIP), as well as itself (Cho et al., 2013; Dai et al., 2017). There was no difference in mTOR signaling when Ser/Gly-deprived PDAC cells were stimulated with Ser (Figure S3G). Similarly, pre-treatment with mTOR inhibitors did not block the Ser-dependent increase in OCR (Figure S3H–I). Both exSer-independent and -dependent PDAC cells were able to activate the GCN2-eIF2α-ATF4 response under Ser/Gly-deprived conditions (Figure 4E). However, BIP levels were rapidly decreased under Ser/Gly-deprived conditions in exSer-dependent PDAC cells despite maintaining, or in some cases even increasing, its transcript levels. (Figure 4E and S3J). In addition, while both ATF4 transcript and protein increased upon Ser deprivation, its protein levels demonstrated a further increase within minutes of Ser re-stimulation. The timeframe indicates that the increase upon Ser stimulation is unlikely at the transcript level (Figure 4E) and suggested that Ser stimulation was selectively regulating mRNA translation rates.
Because tRNA charging and protein synthesis are one of the major consumers of ATP (Buttgereit and Brand, 1995), we hypothesized that the decreased mitochondrial activity we observed under Ser/Gly-deprived conditions was due to a decrease in ATP demand resulting from decreased protein synthesis rates (Figure S3K). Consistent with this, the Ser-induced increase in OCR was completely blocked when exSer-dependent PDAC cells were pre-treated with cycloheximide (CHX), a protein synthesis inhibitor (Figure 4F). In addition, acute CHX treatment increased the NADH/NAD+ ratios and decreased OCR in PDAC cells (Figure 4G–H). Interestingly, axon-derived conditioned media was able to increase OCR in the exSer-dependent PDAC cells grown in the absence of Ser/Gly, albeit at a lower magnitude than full Ser concentrations found in media (Figure 4I). Therefore, Ser-deprivation selectively reduces mRNA translation rates, which lowers intracellular ATP demand, resulting in decreased mitochondrial activity.
Ser-deprivation decreases mRNA translation efficiencies (TE) of TCC and TCT Ser codons
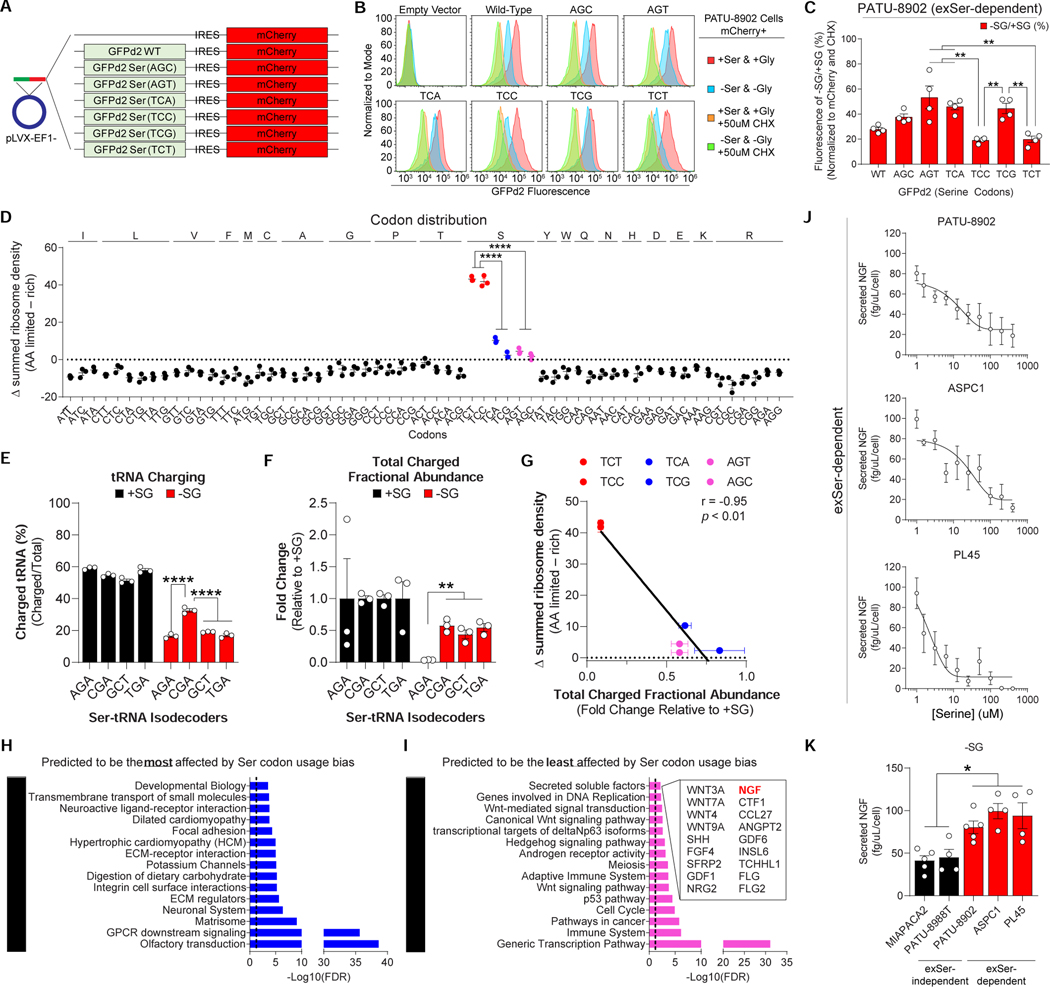
Next, we wanted to elucidate the effects of selective mRNA translation during Ser-deprivation. Because there are no obvious changes to the mRNA, protein half-life, mTOR signaling and GCN2-ATF4 signaling within minutes of Ser stimulation (Figure 4E and S3), we reasoned that the effects on mRNA translation rates were dependent on ribosomes and/or the coding sequence of genes. Ribosome stalling on codons and rare codon usage have been well-studied as mechanisms that regulate mRNA translation rates during nutrient deprived conditions in bacteria, but have only recently been identified in eukaryotes (Darnell et al., 2018; Loayza-Puch et al., 2016). This raised the possibility that differences in Ser codon usage could explain selective protein synthesis rates during Ser/Gly-deprivation. To address this question, we constructed several destabilized green fluorescent protein (GFPd2, ~2 hour half-life) (Li et al., 1998)-reporter constructs where all Ser codons were replaced with one of six Ser codons and the signal was normalized to IRES-mCherry (24 hour half-life) as an internal control (Figure 5A). Treatment with CHX decreased GFPd2, but not mCherry, fluorescence in Ser/Gly-rich and -deprived conditions (Figure 5B and S4A). Ser/Gly-deprivation also decreased GFPd2 fluorescence, with constructs containing TCC and TCT Ser codons displaying the largest drop in fluorescence (Figure 5B–C), and no changes in expression of the reporters at the RNA level (Figure S4B). This was specific to Ser-deprivation and not due to a general AA starvation response because depletion of other AA, such as tyrosine or phenylalanine, did not result in the same differences in mRNA TE of Ser codons (Figure S4C). These experiments suggest that there is decreased mRNA TE against coding sequences (CDS) containing Ser codons, TCT and TCC, during Ser-deprivation.
Figure 5.
Ser/Gly starvation regulates protein synthesis via differences in mRNA TE of Ser codons.
A, PDAC cells were infected with pLVX-EF1-GFPd2-IRES-mCherry constructs, where all Ser codons were replaced with indicated Ser codons. mCherry serves as an internal control.
B-C, Representative flow plots (B) and quantification (C) of GFPd2 fluorescence in PATU-8902 cells expressing Ser codon constructs from A, grown with and without Ser/Gly or 50μM CHX (n=4).
D, Quantification of ribosomal density at specific codons in PATU-8902 after being grown in Ser/Gly-depleted media (n=3).
E-F, Percent tRNA charging (E), and total fractional abundance of charged (F) Ser-tRNA isodecoders in PATU-8902 cells grown with and without Ser/Gly (n=3).
G, Ribosome density was plotted against change in total fractional abundance of charged Ser-tRNA isodcoders in exSer-dependent PDAC cells grown with or without Ser/Gly (n=3). Pearson correlation value is represented by r.
H-I, Top 20 canonical pathways that are predicted to be most (H) or least (I) affected by TCT and TCC Ser codon usage bias. Secreted factors predicted to be least affected by Ser codon usage bias are listed.
J, Measurements of NGF in the media of PDAC cells grown in different concentrations of Ser after 24 hours (n=4–5).
K, Secreted NGF from PDAC cells grown in Ser/Gly-free conditions after 24 hours from J and Figure S5O (n=4–5).
“n” are displayed as individual points and represent the number of biologically independent devices. Graphs (mean ± s.e.m.) were compared by one-way ANOVA (C-F, K), followed by Bonferroni post-hoc test (**p<0.01, ****p<0.0001).
We performed ribosome profiling (Ingolia et al., 2012) to assess the effects of Ser/Gly-deprivation on translation at a global scale in exSer-dependent PDAC cells. Consistent with the GFPd2 reporters, the codon-specific ribosome density during Ser/Gly-deprivation was higher on Ser codons in the ribosome protected fragments, but not total mRNA, with TCC and TCT showing the highest density of ribosomes (Figure 5D and Figure S4D). This confirmed that under Ser/Gly-deprivation, ribosomal stalling is occurring on TCC and TCT Ser codons. Furthermore, we found that exSer-independent PDAC cells did not exhibit decreased mRNA TE of TCC and TCT Ser codons, under Ser/Gly-deprived conditions (Figure S4E–F). We postulated that this was because exSer-independent PDAC cells can generate their own Ser when Ser/Gly is limiting because they express PHGDH. Therefore, we treated exSer-independent PDAC cells with a PHGDH inhibitor, which blocks SBP (Ngo et al., 2020; Rodriguez et al., 2019). exSer-independent PDAC cells treated with the PHGDH inhibitor exhibited the same decreased mRNA TE against TCC and TCT Ser codons under Ser/Gly-depleted conditions (Figure S4E–F). Similar effects were observed in breast cancer (MDA-MB-231, PHGDH low) and melanoma (SKMEL5, PHGDH high) cells, indicating that changes in mRNA TE of Ser codons occurs in multiple tumor types and is not specific to PDAC (Figure S4E, G–H).
To understand when differences in Ser codon usage was occurring, we monitored GFPd2 fluorescence, BIP levels and eIF2α phosphorylation (p-eIF2α), as an indirect measure of uncharged tRNAs under a wide-range of Ser concentrations (Figure S5A–D). We observed that GFPd2 and BIP levels exhibit the same initial decay in signal as Ser concentrations decreases, whereas p-eIF2α sharply increases at ~25–50µM. Interestingly, we noticed that GFPd2 fluorescence diverges among the Ser codon optimized variants at ~50μM. This indicates that the initial decrease in BIP and GFPd2 fluorescence is due to inhibition of general protein synthesis, and that differences in mRNA TE of Ser codons occurs when there are uncharged tRNAs. Furthermore, there was no association between the Ser codons and cognate tRNA gene copy number or expected usage frequency (Figure S5E–G), suggesting that differences in mRNA TE of Ser codons is not regulated by these factors. Together, these data suggest that there may be differences in the abundance or charging state of Ser tRNA that drives differences in mRNA TE of Ser codons when Ser/Gly becomes limiting.
We assessed the levels of Ser-tRNA and Ser-tRNA charging by charged DM-tRNA-seq in exSer-dependent PDAC cells grown with and without Ser/Gly (Evans et al., 2017). There are four types of Ser-tRNA isodecoders (i.e. tRNA genes that have the same anti-codon), which contain AGA, CGA, GCT, or TGA anti-codon sequences and recognize the six Ser codons during mRNA translation. Because Ser-tRNA(AGA) is modified at position A34 to I34, it can recognize both TCC and TCT Ser codons. As expected, Ser-deprivation decreased tRNA charging of all Ser-tRNA isodecoders (Figure 5E). However, the percentage of Ser-tRNA(AGA) charging was not significantly different to the other Ser-tRNAs. Instead, we found that relative to the other Ser-tRNA isodecoders, the levels of total (uncharged + charged) Ser-tRNA(AGA) was significantly decreased upon Ser/Gly-deprivation (Figure S5H). The difference was still apparent even when factoring the percentage of charged tRNAs to calculate the total levels of charged Ser-tRNAs (Figure 5F). We found a strong negative association between the change in levels of total (uncharged + charged) or charged Ser-tRNA abundance and the ribosome density in the absence of Ser/Gly (Figure 5G and S5I–J). Together these data indicate that the decreased levels of Ser-tRNA(AGA) could explain the decreased mRNA TE of TCC and TCT Ser codons in exSer-dependent PDAC under Ser-deprived conditions.
High and low TCC and TCT containing genes are enriched in specific pathways
Next, we identified genes that would be most and least affected by differences in mRNA TE of Ser codons. We calculated the Ser usage bias (as indicated by Z-Score) of Ser codons TCT and TCC for 18,660 CDS (Figure S5K and Table S2). CDS containing high TCT and TCC Ser codons were enriched in canonical pathways related to developmental biology, and GPCR signaling (Figure 5H). Consistent with these results, BIP was among the top 5% of CDS predicted to have decreased mRNA TE due to Ser codon usage bias. In addition, a majority of the genes predicted to be most affected by slower TCC and TCT mRNA translation rates have decreased protein levels in the absence of Ser/Gly (Figure S5L and Table S3). In contrast, proteins such as RAS, which are not predicted to be affected by Ser codon usage bias, were not changed during Ser/Gly-deprivation (Figure S5L–N). CDS that are least likely to be affected by Ser codon usage bias were enriched in canonical pathways related to transcription, and secreted soluble factors (Figure 5I). Among the list of secreted factors, NGF was predicted to be the least affected by Ser codon usage bias during Ser/Gly starvation. Indeed, Ser/Gly-deprivation increased secreted NGF levels by exSer-dependent PDAC cells (Figure 5J). exSer-independent PDAC also exhibited NGF secretion in the absence of Ser/Gly, but at significantly lower levels than exSer-dependent PDAC cells (Figure 5K and S5O). Because NGF is a reported ATF4 target gene, its transcript also was increased upon Ser/Gly-deprivation (Figure S5P). To decouple transcription from translation, we ectopically expressed wild-type (84% non-stalled Ser codons), TCC (stalled)-, and TCA (non-stalled)-codon optimized NGF. We found that TCC-optimized NGF secretion rate was decreased compared to wild-type and TCA-optimized NGF in Ser/Gly-depleted media, but not Ser/Gly-rich media (Figure S5Q). This is consistent with the notion that NGF translation is minimally affected by differences in mRNA TE of Ser codons during Ser-deprived conditions and also shows how PDAC cells can stimulate tumor innervation during Ser/Gly-starvation by ensuring the effective translation of the upregulated transcript.
Ser/Gly-deprived PDAC tumors exhibit enhanced nerve infiltration
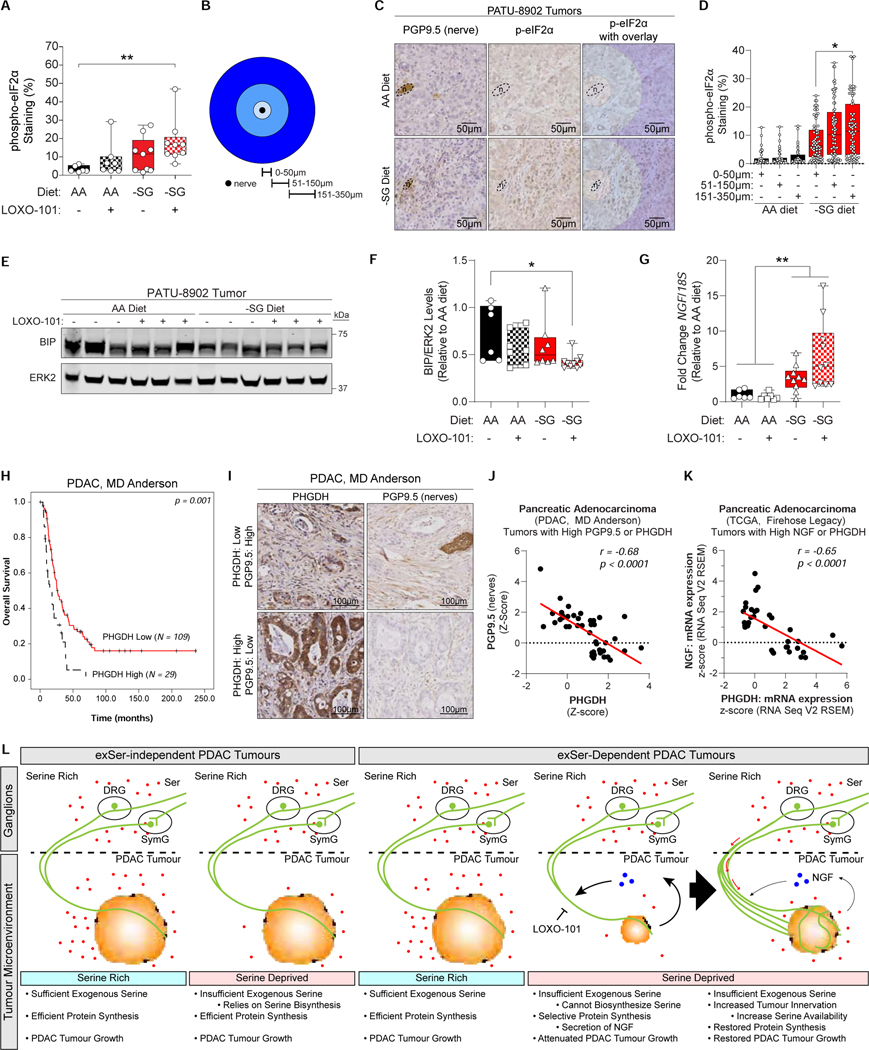
To determine whether Ser/Gly-deprivation could affect exSer-dependent PDAC cells in vivo, exSer-dependent PDAC cells were orthotopically injected into the pancreata of nude mice that were placed on a complete AA and Ser/Gly-free (-SG) diet (Figure 6A and Table S4). Plasma Ser and Gly in mice on the -SG diet were lower than that of mice on AA diet for the majority of the experiment (Figure S6A). However, at the end of 5 weeks, plasma Ser and multiple other AA dropped in the tumor-bearing mice fed the AA diet compared to non-tumor bearing mice, suggesting that tumors can exert systemic metabolic changes on AA plasma levels (Figure S6B). This may be reflective of cachexia that is often observed in PDAC patients. Importantly, Ser/Gly deprived tumors were significantly smaller compared to controls, indicating that Ser/Gly availability affects tumor growth (Figure 6B). Tumors from mice on -SG diet had lower p-histone 3 and no change in cleaved caspase 3, indicating that tumor weight difference was due to decreased proliferation (Figure 6C and S6C). We also observed increased tumor innervation by sympathetic (tyrosine hydroxylase (Th)+) and sensory (transient receptor potential cation channel subfamily V member 1 (Trpv1)+), but not parasympathetic (vesicular acetylcholine transporter (VAchT)+), nerves in mice on -SG diet compared to controls by IHC and 3D-imaging (Figure 4D–E and S6D–H). These differences in innervation could be explained by changes in the levels of axon guidance cues that repel (e.g. SEMA3A) or attract (e.g. SEMA7A) nerves upon Ser/Gly-deprivation (Figure S6I) (Koncina et al., 2007; Pasterkamp et al., 2003). Nonetheless, the observation is consistent with the fact that ~90% of human PDAC display neural alterations, and that NGF can recruit sensory and sympathetic nerves (Aloe et al., 2012; Ceyhan et al., 2009a). Because axons were found to secrete Ser, and PDAC cells can secrete NGF during Ser starvation, we hypothesized that nerves were recruited to metabolically support tumors when Ser levels are low.
Figure 6.
Ser/Gly-free diet and LOXO-101 affects tumor growth and nerve infiltration of exSer-dependent PDAC (PATU-8902) tumors.
A, Athymic nude mice were placed on AA or -SG diet for two weeks before orthotopic injection of PATU-8902 cells into the pancreas. Plasma and tumors were harvested at the indicated times.
B, PATU-8902 tumor weight from mice on AA (n=10) or -SG (n=9) diet after five weeks post-injection.
C, Quantification of IHC of p-Histone 3 and cleaved caspase 3 staining of PATU-8902 tumors collected from mice on AA (n=10) and –SG (n=9) diet.
D, Representative 3D fluorescent images of nerves (PGP9.5) in PATU-8902 tumors from mice on AA and -SG diets.
E, Quantification of nerves (PGP9.5) in PATU-8902 tumors from mice on AA (n=10) and -SG diets (n=9).
F, LOXO-101 treatment was started one week post-injection and given by oral gavage daily.
G-H, PATU-8902 tumor weight (G), and quantification of PGP9.5 and p-Histone 3 IHC staining (H) from mice on AA or -SG diet treated with or without LOXO-101 (n≥6).
I, Representative images and quantification of PGP9.5 from H (n≥6).
Where “n” and each point represents the number of biologically independent experiments for each group and condition. Graphs (median ± max/min) were compared using two-tailed Student t-test (B, C, E), or one-way ANOVA, followed by Holm-Sidak post-hoc test (G-I). (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Blocking nerve innervation with LOXO-101, decreases PDAC tumor burden in mice on Ser/Gly-diet
To assess whether nerves can support tumor survival or growth during Ser-deprivation, we treated mice on control and -SG diet with an FDA-approved TRK (the receptor for NGF) inhibitor, LOXO-101, after orthotopic injection of PDAC cells to reduce tumor innervation (Ghilardi et al., 2010) (Figure 6F). LOXO-101 had no effect on PDAC growth or on the ability of axons to release Ser from rat DRG neurons in vitro (Figure S7A–B). Furthermore, TRK levels were not detectable in exSer-dependent PDAC cells or tumors under Ser/Gly-rich and -deprived conditions with or without LOXO-101, indicating that any observable effects would be non-cell autonomous (Figure S7C–D). Plasma Ser and Gly were not affected by LOXO-101 treatment (Figure S7E) and there was no effect on tumor burden by LOXO-101 treatment alone. However, tumor weight, proliferation and neuronal innervation were further decreased when mice on -SG diet were treated with LOXO-101 (Figure 6G–I and S7F–G). In addition to the significant decrease in proliferation, LOXO-101 caused a small, but significant, increase in cell death compared to control tumors (Figure S7F–G). Tumors from mice on -SG diet and LOXO-101 treatment exhibited higher stress levels than control tumors as determined by p-eIF2α staining, which is likely related to decreased tRNA charging caused by Ser deprivation experienced in the tumor (Figure 7A and S7G). Interestingly, p-eIF2α staining was higher at distances furthest from nerves in mice on -SG diets (Figure 7B–D), indicating that there is less nutrient stress surrounding the nerves. In addition, tumors from mice on -SG diet and LOXO-101 treated had significantly lower levels of BIP and higher NGF expression (Figure 7E–G). Together, these results indicate that Trk inhibition is acting in a cell non-autonomous manner by blocking tumor innervation that metabolically supports PDAC cells under Ser/Gly-deprived conditions.
Figure 7.
Axons metabolically support regional limitation of Ser in PDAC.
A, Quantification of phospho-eIF2α staining of PATU-8902 tumors from mice on AA or - SG diet treated with (+) or without (−) LOXO-101, five weeks after orthotopic injection (n≥6). Representative images in Figure S7G.
B, Concentric circles of distances used to measure phospho-eIF2α staining from nerves.
C, Representative images of phospho-eIF2α staining around nerves (n) in PATU-8902 tumors from mice on AA or -SG diet. Overlay of concentric circles are shown.
D, Quantification of phospho-eIF2α staining at several distances around nerves from C (n = 40 (AA) or 73 (-SG) independent nerves).
E, Immunoblots of BIP levels from PATU-8902 tumors from mice on AA or –SG diet treated with and without LOXO-101. ERK2 serves as a loading control.
F-G, Quantification of BIP levels (F) and NGF expression (G) normalized to ERK2 or 18S, respectively, of tumors from mice on AA or -SG diet, treated with vehicle or LOXO-101 (n≥6).
H, Overall survival of PHGDH high and low expressing PDAC tumors from a TMA dataset.
I, Representative images of high and low PHGDH and PGP9.5 (nerve) staining from PDAC tumors from human primary samples.
J, PHGDH and PGP9.5 staining from PDAC tumors containing high levels of PHGDH or PGP9.5 from I.
K, Expression of NGF and PHGDH from PDAC tumors expressing high levels of PHGDH and NGF from the TCGA dataset.
“n” are displayed as individual points and represent the number of biologically independent replicates. Graphs represents median ± max/min, and were compared using one-way ANOVA, followed by Holm-Sidak post-hoc test (A, D, F-G). Pearson correlation values are represented by r (J-K). Scale bars are as indicated. *p<0.05, **p<0.01, ****p<0.0001.
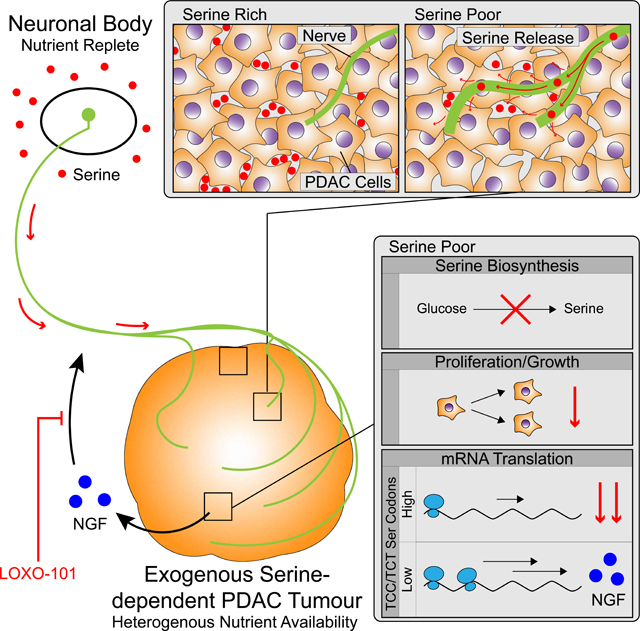
L, Proposed model depicting the interaction of exSer-independent and –dependent PDAC tumors and nerves in response to Ser-rich and –deprived regions in the tumor microenvironment. The main body of sensory and sympathetic neurons are located in nutrient replete environments of the DRG and sympathetic ganglion (SymG), respectively. Blocking nerve recruitment by TRK inhibitors, such as LOXO-101, can further decrease exSer-dependent PDAC tumor growth during Ser-limitation.
Next, we assessed whether patients with PHGDH high and low PDAC tumors exhibit similar effects on innervation and signaling. PDAC patients with high expression or levels of PHGDH have a significantly lower overall survival (Figure 7H and S7H). This suggests that the PDAC microenvironment does not contain sufficient levels of extracellular Ser to allow maximal tumor growth in patients. In addition, patients with high PHGDH expressing tumors are more poorly differentiated, and have larger tumors (Figure S7I–J). Furthermore, human PDAC tumors with high PHGDH levels had low levels of nerves, as determined by PGP9.5 staining. Whereas, tumors with high PGP9.5 staining had low levels of PHGDH staining (Figure 7I–J). Similarly, PDAC tumors with high PHGDH expression had low expression of NGF, and vice versa (Figure 7K). These observations are consistent with Ser being limiting in the PDAC microenvironment, and that Ser-deprivation is associated with increased tumor innervation.
Discussion
Our results identify a previously undescribed role for neurons, via their peripheral axons, to metabolically support PDAC cell growth under nutrient limiting conditions (Figure 7L). Multiple studies have found that neurons lack Phgdh (Furuya et al., 2000; Yamasaki et al., 2001) and therefore have to obtain Ser from external sources, such as supporting glial cells (Ehmsen et al., 2013; Yamasaki et al., 2001; Yamashita et al., 2003) and/or the circulation. Because sensory and sympathetic neuronal bodies reside in the DRG, spinal cord and sympathetic ganglia, and that axons travel alongside blood vessels (Mukouyama et al., 2002), peripheral neurons are exposed to plasma concentrations of AA (Jimenez-Andrade et al., 2008). Our data demonstrates that innervation may provide another mechanism to supply AA from nutrient rich plasma to nutrient poor environments in PDAC tumors (Figure 7L). Although cells can adapt to nutrient deprivation by upregulating transcription and cap-independent translation of stress response genes (Gonzalez and Hall, 2017), ribosomal stalling can still occur when essential AA are severely limiting. Under Ser-deprived conditions, exSer-dependent PDAC cells appear to allow the translation of TCC- and TCT-poor transcripts, such as NGF, to adapt to stress and allow the recruitment of nerves. In turn, this would establish a neuro-cancer crosstalk to metabolically support PDAC cells in nutrient-deprived environments (Figure 7L).
We found that a large fraction of human PDAC cells lack SBP enzymes and cannot generate Ser. Although PDAC cells exhibit enhanced autophagy and macropinocytosis, these pathways do not appear to generate sufficient levels of Ser to promote growth in PDAC cells under Ser/Gly-deprivation. Therefore, PDAC cells must import exSer or increase SBP. Upregulation of SBP enzymes in mouse KPC cells is driven by Kras (Maddocks et al., 2017). However, the overwhelming majority of the human exSer-dependent PDAC cells used in this study also harbor KRAS mutations, which indicates that there are additional genetic and epigenetic aberrations that inactivate the SBP in many human PDAC tumors. It also is unclear as to why PDAC tumors would lose their ability to synthesize Ser. It is possible that 1) another driver mutation is suppressing the transcription of PHGDH and/or PSAT1, 2) that Ser biosynthesis is energy expensive, or 3) the pathway may generate toxic metabolic intermediates. Elucidating the answer to these questions will be important for understanding tumorigenesis of PDAC and other malignancies.
PDAC cells often adapt to the nutrients in their environment by rewiring their metabolism. exSer-dependent PDAC cells do not have observable defects in Ser-dependent metabolites, such as one carbon units, glutathione and nucleotide levels. Instead, we found that Ser is required for efficient mRNA translation of TCC and TCT codons and remains a bottleneck for PDAC tumor growth. Ser codon TE in bacteria and yeast relies on tRNA abundance and rare codon usage, such as AGT and AGC, in Ser-deprived media (Subramaniam et al., 2013a; Subramaniam et al., 2013b). Similar to work with arginine limitation in eukaryotes, we did not find an association of tRNA copy number or the calculated codon usage frequency with ribosome stalling upon Ser/Gly-limitation in PDAC.
Ribosomal stalling has been observed to occur in many different scenarios. For example, proline limitation was identified in kidney and breast tumors using ribosomal profiling (Loayza-Puch et al., 2016). Arginine limitation also resulted in ribosomal stalling in mammalian cells (Darnell et al., 2018). Interestingly, ribosome stalling on Ser codons has not been observed in eukaryotic cells, most likely because many cells in these studies can biosynthesize their own Ser. Consistent with this, we found that SBP can prevent differences in Ser codon TE from occurring during Ser/Gly-limiting conditions (Figure S4F–H). Ribosomal stalling caused by arginine, and proline deprivation is associated with a respective decrease in the percentage of charged tRNAs. By contrast, we found decreased levels of Ser-tRNA(AGA) in the absence of Ser, which can result in lower mRNA TE of TCC and TCT Ser codons. The cause of the selective decrease in Ser-tRNA(AGA) under Ser/Gly-deprivation remains to be elucidated.
The unique and complex microenvironment and metabolic plasticity of PDAC tumors have contributed to the poor outcomes of patients. By limiting Ser/Gly availability, through dietary manipulation, we can impair PDAC growth in exSer-dependent tumors in mice. However, PDAC cancer cells in patients may experience more severe nutrient-deprived than in mouse PDAC tumor models. While orthotopic PDAC models can display desmoplasia, the levels in human PDAC tumors are often greater, leading to the collapse of blood vessels and limiting the nutrient supply (Chauhan et al., 2013). Indeed, Ser concentrations in human PDAC tumors have been shown to be ~60% less than in benign adjacent tissues (Kamphorst et al., 2015). As these differences were measured in bulk tumors, it would not be surprising if there were local regions of even more severe nutrient deprivation in PDAC tumors. Taken together, Ser limitation, either by dietary restriction and/or SBP or uptake inhibitors, may be able to slow PDAC tumor growth in patients. However, in response, PDAC cells may alter the tumor microenvironment to obtain Ser, such as recruiting nerves.
While it has been shown that nerves can affect PDAC growth by releasing neurotrophic factors and neurotransmitters (Renz et al., 2018a; Sinha et al., 2017), our data reveals that nerves also can supply AA, such as Ser, to nutrient-deprived areas of PDAC tumors to promote growth. PDAC patients that have undergone denervation (Lillemoe et al., 1993) or received β-blockers (Renz et al., 2018a), which can impair nerve innervation, have been documented to have better overall survival. Similar observations were found in animal studies (Saloman et al., 2016), and anti-NGF therapy also decreases neurogenic inflammation, perineural invasion, and metastasis in the immune-competent KPC PDAC mouse model (Saloman et al., 2018). We found that PDAC growth can be further impaired by targeting the recruitment of neuronal axons in response to low Ser conditions with TRK inhibition. Further studies will be needed to elucidate whether other cell types in the PDAC microenvironment, such as macrophages and stellate cells, which have been reported to secrete NGF, also can contribute to neuronal innervation of the tumor under nutrient deprived conditions (Caroleo et al., 2001; Jiang et al., 2020). Although, we did not observe TRK expression in exSer-dependent PDAC cells, NGF was reported to stimulate proliferation in mouse PDAC cells that express Trk (Renz et al., 2018a). This is likely due to heterogeneity across PDAC and that in some circumstances it may act in a cell autonomous fashion and in the situation of exSer-dependent PDAC, acts non-autonomously. Together, these observations suggest that targeting nerves and Trk signaling in PDAC patients could decrease tumor recurrence by limiting both the supply of neurotrophic factors and nutrients to tumors. As TRK inhibitors are approved in the treatment of cancers with TRK alterations, it is tempting to speculate that they could be used in combination with or without Ser restriction following surgery in certain PDAC patients. Our findings highlight the importance of Ser availability, axon-cancer metabolic crosstalk and selective mRNA translation in PDAC cells and offers potential therapeutic options in the treatment of this disease.
STAR★ Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Alec C. Kimmelman (alec.kimmelman@nyulangone.org).
Material availability
The unique reagents generated in this study are available from the Lead Contact Alec C. Kimmelman (alec.kimmelman@nyulangone.org).
Data and code availability
The accession number for the raw high throughput sequencing files for ribosome profiling and charge DM-tRNA-seq experiments is GEO: GSE139910. All codes used in this study were publicly available from published studies as indicated.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Pancreatic cancer, MDA-MB-231, and SKMEL5 cell lines were purchased from the American Type Culture Collections and Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). No lines used in this study are found in the ICLAC or NCBI Biosample databases of commonly misidentified cell lines. Cultures were routinely verified to be negative for mycoplasma by PCR. Cell lines were authenticated by periodic fingerprinting and visual inspection, and low passage cultures were carefully maintained in a central lab cell bank (Uphoff and Drexler, 2011). PATU-8902 cells were infected with lentivirus-expressing PHGDH and/or PSAT1, and selected with puromycin (2 μg/mL) and/or blasticidin (10 μg/mL), respectively. PATU-8902 ectopically expressing PHGDH and PSAT1 were adapted in the absence of Ser/Gly for 21 days. All pancreas cancer cells were maintained in DMEM+10%FBS. All cells were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2. See Key Resource Table for complete list of cell lines
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-PHGDH | Sigma: Protein Atlas | HPA021241 |
| Rabbit anti-PSAT1 | Abnova | H00029968-A01 |
| Rabbit anti-PSPH | Sigma: Protein Atlas | HPA020376 |
| Mouse anti-p38 | Cell Signaling | #8690 |
| Rabbit anti-BIP | Cell Signaling | #3177 |
| Rabbit anti-ATF4 | Cell Signaling | #11815 |
| Rabbit anti-phospho-eIF2α (S51) | Cell Signaling | #3398 |
| Mouse anti-eIF2α | Cell Signaling | #2103 |
| Rabbit anti-phospho-4-EBP1 (T37/46) | Cell Signaling | #2855 |
| Rabbit anti-4-EBP1 | Cell Signaling | #9644 |
| Rabbit anti-phospho-mTOR (S2448) | Cell Signaling | #5536 |
| Rabbit anti-phospho-ULK1 (S757) | Cell Signaling | #6888 |
| Rabbit anti-phospho-AKT1 (S473) | Cell Signaling | #4060 |
| Rabbit anti-phospho-ERK (T202/Y204) | Cell Signaling | #4376 |
| Rabbit anti-phospho-S6 (S240/244) | Cell Signaling | #5364 |
| Rabbit anti-pan-Ras | Cell Signaling | #3339 |
| Mouse anti-Kras | Millipore Sigma | OP24 |
| Rabbit anti-Tyrosine hydroxylase | Millipore Sigma | AB152MI |
| Rabbit anti-Trpv1 | Alomone labs | ACC-030 |
| Rabbit anti-VAChT | Synaptic Systems | 139103 |
| Rabbit anti-pan-Trk | Abcam | ab181560 |
| Rabbit anti-TrkA | Cell Signaling | #2510 |
| Rabbit Phospho-TrkA (Tyr674/675)/TrkB (Tyr706/707) | Cell Signaling | #4621 |
| Rabbit anti-phospho-H3 (S10) | Cell Signaling | #9701 |
| Rabbit anti-Cleaved Caspase 3 | Cell Signaling | #9664 |
| Rabbit anti-PGP9.5 | Abcam | ab108986 |
| IRDye® 680RD Goat anti-Mouse IgG (H + L) | LiCOR | 926-68070 |
| IRDye® 800CW Goat anti-Rabbit IgG (H + L) | LiCOR | 926-32211 |
| Bacterial and Virus Strains | ||
| N/A | ||
| Biological Samples | ||
| Human PDAC tissue microarray | MD Anderson | IRB: LAB05-0854 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Oligomycin A | Cayman Chemicals | 11342 |
| FCCP (trifluorocarbonylcyanide phenylhydrazone) | Cayman Chemicals | 370-86-5 |
| Rotenone | Cayman Chemicals | 13995 |
| Antimycin A | Sigma Millipore | A8674 |
| Cycloheximide | Sigma Millipore | 239765 |
| Torin1 | Sigma Millipore | 475991 |
| Rapamycin | Sigma Millipore | R8781 |
| Formate | Sigma Millipore | 71539 |
| EmbryoMax® Nucleosides (100X) | Sigma Millipore | ES-008-D |
| 13C6-Glucose | Cambridge Isotope | CLM-1396 |
| 13C3-Serine | Cambridge Isotope | CLM-1574-H |
| NGF 2.5S Native Mouse Protein | ThermoFisher Scientific | 13257019 |
| B-27™ Supplement (50X), serum free | ThermoFisher Scientific | 17504044 |
| Neurobasal™-A Medium (1X) liquid | ThermoFisher Scientific | 10888022 |
| Uridine | Sigma | U3003 |
| 5-Fluoro-2′-deoxyuridine | Sigma | F0503 |
| Teterodoxin | Abcam | ab120054 |
| Poly-D-Lysine solution, 1.0 mg/mL | Sigma | A-003-E |
| Laminin from Engelbreth-Holm-Swarm murine sarcoma basement membrane | Sigma | L2020 |
| 150μm AXIS axon isolation device | Millipore | AX15010 |
| LOXO-101 sulfate (Larotrectinib) | InVivoChem | V2599 |
| Labrafac PG | GATTEFOSSÉ | |
| growth factor-reduced Matrigel | Fisher Scientific | CB-40230C |
| THINCERT cell culture insert for24 well plates (1μm) | Greiner Bio-One | 662610 |
| 40% Acrylamide Solution | Bio-rad | 1610140 |
| Paraformaldehyde, 16% (Aqueous) | Tedpella | 18505 |
| Histodenz | Sigma | D2158 |
| SYBR Green Supermix | Bio-Rad | 1725275 |
| Turbo DNase | ThermoFisher Scientific | AM2238 |
| RNAse I | ThermoFisher Scientific | AM2294 |
| SuperScript III | ThermoFisher Scientific | 18080-093 |
| GlycoBlue | ThermoFisher Scientific | AM9515 |
| Universal miRNA Cloning Linker | NEB | S1315S |
| T4 RNA ligase 2, truncated | NEB | M0242S |
| T4 polynucleotide kinase | NEB | M0201S |
| CircLigase™ ssDNA Ligase | Lucigen | CL4115K |
| CircLigase™ II ssDNA Ligase | Lucigen | CL9021K |
| 3K Amicon Ultra centrifugal filter | Sigma Millipore | UFC800324 |
| Seahorse XF Plasma Membrane Permeabilizer | Agilent | 102504-100 |
| Pyridine | Fisher | AC364425000 |
| N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-Butyldimethylchlorosilane | Sigma | 375934 |
| Sodium periodate | Sigma | 311448 |
| Critical Commercial Assays | ||
| CyQuant Proliferation Assay | ThermoFisher Scientific | C7026 |
| PureLink RNA mini kit | ThermoFisher Scientific | 12183025 |
| Superscript Vilo IV | ThermoFisher Scientific | 11766050 |
| RNA Clean & Concentrator-5 | Zymo Research | R1015 |
| RNA Clean & Concentrator™−25 | Zymo Research | R1017 |
| DL-Serine Assay kit | Abcam | ab241027 |
| Human beta-NGF DuoSet ELISA | R&D Systems | DY256 |
| NAD/NADH-Glo™ Assay | Promega | G9071 |
| rtStar™ tRNA-optimized First-Strand cDNA Synthesis Kit | Array Star | AS-FS-004 |
| Deposited Data | ||
| Ribosomal Profiling | This study | GSE139910 |
| tRNA-seq | This study | GSE139910 |
| Experimental Models: Cell Lines | ||
| MIAPACA2 | ATCC | CRL-1420 |
| PATU-8988T | DSMZ | ACC 162 |
| PATU-8902 | DSMZ | ACC 179 |
| ASPC1 | ATCC | CRL-1682 |
| PL45 | ATCC | CRL-2558 |
| HupT3 | DSMZ | ACC 259 |
| Panc10.05 | ATCC | CRL-2547 |
| CAPAN2 | ATCC | HTB-80 |
| SW1990 | ATCC | CRL-2172 |
| PATU-8988S | DSMZ | ACC 204 |
| DanG | DSMZ | ACC 249 |
| Panc03.27 | ATCC | CRL-2549 |
| HPAC | ATCC | CRL-2119 |
| HupT4 | DSMZ | ACC 223 |
| Panc02.03 | ATCC | CRL-2553 |
| Panc1 | ATCC | CRL-1469 |
| MDA-MB-231 | ATCC | HTB-26 |
| SKMEL5 | ATCC | HTB-70 |
| 293T | ATCC | CRL-3216 |
| Primary rat Dorsal Root Ganglion cells | Lonza | R-DRG-505 |
| Experimental Models: Organisms/Strains | ||
| Athymic nude (CrTac:NCr-Foxn1nu) | Taconic | NCRNU-F |
| Oligonucleotides | ||
| See Table S5 | This study | N/A |
| Recombinant DNA | ||
| pLVX-EF1a- IRES-mCherry | Clontech | 631987 |
| pLVX-EF1a- GFPd2 (WT)- IRES-mCherry | This study | N/A |
| pLVX-EF1a- GFPd2 (AGT)- IRES-mCherry | This study | N/A |
| pLVX-EF1a- GFPd2 (AGC)- IRES-mCherry | This study | N/A |
| pLVX-EF1a- GFPd2 (TCA)- IRES-mCherry | This study | N/A |
| pLVX-EF1a- GFPd2 (TCC)- IRES-mCherry | This study | N/A |
| pLVX-EF1a- GFPd2 (TCG)- IRES-mCherry | This study | N/A |
| pLVX-EF1a- GFPd2 (TCT)- IRES-mCherry | This study | N/A |
| psPAX2 | (Roca et al., 2008) | Didier Trono (Addgene #12260) |
| pMD2.G | (Roca et al., 2008) | Didier Trono (Addgene #12259) |
| Software and Algorithms | ||
| Xcalibur | ThermoFisher | ThermoFisher |
| GraphPad Prism 8 | Version 8.2.0 | GraphPad |
| MATLAB | Version 9.2 | MathWorks |
| OpenChrom | (Wenig and Odermatt, 2010) | https://lablicate.com/platform/openchrom |
| Imagescope | Version 12.4.3 | Leica |
| Cutadapt | Version 1.18 | http://gensoft.pasteur.fr/docs/cutadapt/1.18/installation.html |
| Bowtie2 | Version 2.3.4.1 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Bowtie | Version 1.2.3 | http://bowtie-bio.sourceforge.net/index.shtml |
| Tophat | Version 2.1.1 | https://ccb.jhu.edu/software/tophat/index.shtml |
| R | Version 3.5.0 | https://www.r-project.org |
| Other | ||
| Amino acid control diet | ENVIGO | TD.01084 |
| Ser/Gly-free diet | ENVIGO | TD.180296 |
Primary cell culture
Primary rat dorsal root ganglia (rat DRGs, Lonza) cells were grown in 150μm AXIS axon isolation device (Millipore, AX15010) as described (Taylor et al., 2005). Briefly, AXIS devices were coated with 0.5mg/mL poly-D-lysine and 0.02mg/mL laminin. Approximately 50,000 rat DRGs were seeded into one of the channels in the AXIS device (neuron side) with Neurobasal A media supplemented with B27, glutamine, uridine, 5-fluoro-2-deoxyuridine (5-FU), and NGF. Media on the neuronal and axon side was changed after 24 hours with media without or with NGF, respectively to stimulate axon growth through the grooves and into the axon side. Hydrostatic pressure was established to prevent NGF from the axon side from moving to the neuronal side. Approximately 6–10 days after initial seeding, ~400 peripheral nerves should be established on the axonal side (Figure 1A–C). Cells were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Human samples
Human pancreatic ductal adenocarcinoma (PDAC) tissue microarrays (TMAs) were obtained from M.D. Anderson (IRB protocol: LAB05–0854). The human PDAC TMA contained 138 normal and PDAC matched samples, from 77 male and 61 female patients, and belonged to the age groups of <60 (n=42), 60–70 (n=55), and >70 (n=41) years old. Predetermined sample size calculations were not made because TMAs were generated previously as part of the human PDAC tissue array.
Mouse pancreas orthotopic xenografts
All animal studies were approved by the NYULMC IACUC (Institutional Animal Care and Use Committee, Protocol #IA16–00507 and IA16–01862). Mice used in this study were 7–8 weeks old, and not involved in previous procedures.
Female immunocompromised athymic nude mice (CrTac:NCr-Foxn1nu) were randomly placed on complete amino acid (TD.01084, ENVIGO) or Ser/Gly-free (TD.180296, ENVIGO) diets for two weeks before orthotopic injection of tumor cells into the pancreas. Composition of diets can be found in Table S4. Mice were anaesthetized with ketamine (120mg/kg) and xylazine (10mg/kg) before surgery. PATU-8902 (1 × 104) cells were suspended in 20 uL of 50% growth factor-reduced Matrigel (BD Science), and injected into the pancreas. Mice were treated with buprenex every 12 hours after surgery for 48 hours. For LOXO-101 treatment, mice were given 100mg/kg/day of LOXO-101 sulfate (InVivoChem) by oral gavage for four weeks after one week post-injection. LOXO-101 was resuspended in Labrafac PG (GATTEFOSSÉ). Based on our laboratory’s prior experience, 10 mice per condition in each experiment was chosen to ensure adequate power to detect a two-fold change in tumor size. All mice were housed in a temperature controlled ABSL-2 facility with 12 hour day-night light cycles. Mice were cared for by the husbandry staff at NYULH DCM (division of comparative medicine), and diet and water were provided ad libitum.
METHOD DETAILS
Reagents
Oligomycin A, FCCP (trifluorocarbonylcyanide phenylhydrazone), rotenone and antimycin A were purchased from Cayman Chemicals. Cyclohexamide, Torin1, rapamycin, formate and nucleosides were purchased from Sigma-Millipore. 13C6-glucose, 13C3-serine and labelled amino acid internal standards were purchased from Cambridge isotopes. Nerve growth factor (NGF) was purchased from ThermoFisher Scientific. LOXO-101 sulfate (Larotrectinib) was purchased from InVivoChem (V2599) and Labrafac PG was obtained from GATTEFOSSÉ. Teterodoxin (ab120054) was purchased from abcam. Antibodies were purchased and used for immunoblotting at manufacturer-recommended concentrations (See Key Resource Table for complete list of reagents).
Growth Assays
For growth assays, PDAC cells were washed with PBS before changing to the indicated media condition. Because glucose concentrations are at least 60% decreased in pancreatic tumors versus benign tissue (Kamphorst et al., 2015), unless otherwise stated, growth curves and experiments were performed with DMEM (2.78mM (0.5g/L) glucose, 4mM glutamine, and 1mM pyruvate) + 10% dialyzed FBS, to mimic the glucose availability for serine biosynthesis. For 2X daily spike-in, Ser/Gly was added to PATU-8902 cells twice a day for the duration of the assay. Growth curves were measured by CyQUANT Cell Proliferation Assay kit (ThermoFisher) following manufacturer’s instructions.
Co-culture Assays
For PDAC co-culture experiments with axons, 5,000 PDAC cells were evenly seeded into the wells on the axonal side after nerves have been established. The media was changed to Neurobasal A + B27, glutamine, and NGF on the neuronal side and DMEM (2.78mM glucose. +4mM glutamine and +1mM pyruvate) with or without serine and glycine on the axonal side. Hydrostatic pressure was established every day to prevent the flow of media from the neuronal side into the axonal side. For assessing gap junctions between axons and PDAC cells, neurons were completely labelled with calcein-AM on the neuronal side after six days. Afterwards, exSer-dependent PDAC cells were monitored for calcein fluorescence. At the end of the experiment, images were taken, and cells were trypsinized and counted.
For PDAC co-culture experiments with human pancreatic stellate cells (HPSCs), 2,500 PDAC cells were seeded into a 1.0μm Boyden chamber insert with or without 2,500 or 12,500 HPSCs in the bottom on the well, and allowed to attach overnight. The media was changed to DMEM (2.78mM glucose. +4mM glutamine and +1mM pyruvate) + 10% dialyzed FBS with or without serine and glycine. exSer-dependent PDAC cells were counted at the indicated times.
Conditioned Media
For axon conditioned media (axCM), the wells and channels were washed with PBS twice and the media was changed to Neurobasal A + B27, glutamine, and NGF, without uridine and 5-FU, on the neuronal side and DMEM (2.78mM glucose and +1mM pyruvate) without amino acids on the axonal side or only with different concentrations of Ser/Gly as indicated. For teterodoxin and LOXO-101 experiments, the neuronal side was treated with or without 1uM TTX, 10nM or 1000nM LOXO-101. As a positive control, the NYU Langone Ion Channel and Immunology Core performed whole-cell patch clamp to assess TTX activity, as described below. LOXO-101 activity was assessed by monitoring Trk phosphorylation in rat DRGs treated with LOXO-101 for 24 hours, and stimulated with or without 50ng/mL NGF for 30mins. Hydrostatic pressure was established to prevent the flow of neuronal media into the axonal side. Conditioned media from the axonal side was collected after 24 hours and prepared for analysis by gas chromatograph mass spectrometry (GC-MS).
To measure the amount of alanine, glycine and Ser produced and released by HPSCs, cells were seeded at different densities and allowed to adhere overnight. Wells were washed in PBS before changing media to DMEM (2.78mM 13C6-glucose. +4mM glutamine and +1mM pyruvate) + 10% dialyzed FBS with or without serine and glycine. After 24 hours, cells were counted and media was collected to measure the amount of 13C6-glucose-derived alanine, glycine, and serine released by HPSCs. Samples were prepared for GC-MS analysis as described in the Metabolomics section.
Rat DRG Patch clamp
Whole-cell recordings were made using patch clamp techniques at 22–24°C (Axopatch 200B; Molecular Devices, Sunnyvale, CA). Currents were low-pass filtered with 8-pole Bessel filter (−3dB @ 1 Hz) and digitized at 3 kHz (DigiData 1550A, Molecular Devices) using pClamp v10.5 software. Patch electrodes were manufactured (Zeitz, Germany) using borosilicate glass (1.5 mm OD; World Precision Instruments, Sarasota, FL). Rat DRG neurons were grown on 12-mm coverslips in 24-well culture plate, which were placed on the stage of an inverted microscope (DIAPHOT 300, Nikon). Standard patch clamp techniques were employed.
For recording the Na+ currents in the whole-cell patch clamp configuration, pipettes had tip resistances of 1.8 – 2.5 MΩ when filled with (in mM): 50 CsCl, 60 CsF, 10 TEA-Cl, 20 EGTA, 5 Na2ATP, 10 HEPES, and pH 7.2 adjusted with CsOH. Cells were initially kept in Tyrode’s solution, consisting of (in mM): 140 NaCl, 4 KCl, 1 MgCl2, 10 HEPES, 1 CaCl2, 5 glucose, 0.1 CdCl2 and adjusted to pH 7.37 with NaOH. Following Giga-Ohm seal formation and successfully establishing the whole-cell mode, the extracellular solution was changed to the ‘test’ solution consisting of (in mM): 30 NaCl, 110 CsCl, 4 KCl, 1 CaCl2, 1 MgCl2, 0.1 CdCl2, 10 HEPES, 5 glucose and pH 7.35 with CsOH. During this time, the whole-cell capacitance and series resistance were compensated to levels greater than 80%. Current-voltage relationships were constructed with a voltage clamp protocol that consisted of 20 ms voltage steps between −100 mV to +80 mV were applied in 10 mV increments at a frequency of 2 Hz. TTX was applied using a rapid solution exchange system (RSC-160, Bio-Logic Science Instruments, Seyssinet-Pariset, France). Data were not corrected for the liquid junction potential, calculated to be 3.1 mV. Current density (pA/pF) was calculated by dividing the measured current by the calculated cell capacitance.
For action potential recordings, the pipette resistances were 2.0 – 2.8 MΩ when filled with pipette solution consisting of (in mM): 140 KCl, 1 MgCl2, 5 Na2ATP, 10 HEPES, and pH 7.2 with KOH. The bath solution consisted of (in mM): 140 NaCl, 4 KCl, 1 CaCl2, 1 MgCl2, 10 HEPES, 5 Glucose and pH 7.35 with NaOH. Spontaneous action potentials were first recorded, if none, action potentials were evoked by depolarization pulse. Membrane potentials were adjusted by calculated junction potential (4.9 mV).
Mouse plasma and tumor studies
Blood samples were collected by retro-orbital bleeding and plasma was used for mass spectrometry (MS) analysis at the indicated times. Tumors were harvested and weighed at the end of the experiment, 5 weeks post-injection. Tumors were fixed in 10% buffered formalin and embedded in paraffin for histology or fixed in paraformaldehyde (PFA) for 3D imaging. Sectioning, H&E staining, Trichrome staining, and slide scanning were performed by the NYULMC Experimental Pathology Research Laboratory (NYULMC, New York, NY, USA).
3D-fluorescent imaging
Samples were prepared for Lightsheet imaging as described (Butterworth et al., 2018). PFA-fixed tissue were embedded in a hydrogel (4% acrylamide, 1% PFA in 1X PBS) for three days at 4°C and protected from light. The hydrogel was then polymerized by degassing the oxygen from the solution using gaseous N2 and placed in a 37°C incubator for three hours. The tissue was then cleared in a solution containing 4% SDS in 1X PBS at pH 8.5 and placed in a 37°C incubator for four weeks. The clearing solution was frequently changed. After clearing, the tissues were stained with primary and secondary antibodies in PACT staining buffer (1X PBS, 0.01% sodium azide and 0.1% triton-x-100) + 2% goat serum for four days each. Finally tissues are equilibrated in the refractive index matched solution (RIMS) buffer (50% histodenz in 1X PBS) for four days and imaged using the Zeiss Z1 Lightsheet microscope in the NYULMC Microscopy Laboratory (NYULMC, New York, NY, USA). Images were processed using Zeiss Zen software and Imaris (bitplane).
Immunohistochemistry
Paraffin sections were deparaffinized and antigens were unmasked with citrate (pH 6) and heat. Sections were treated with 3% hydrogen peroxide in Methanol before being blocked with 1% goat serum in PBS. Samples that were unmasked with citrate buffer were stained with phospho-H3, cleaved caspase 3, PGP9.5, Th, Trpv1, VAchT,pan-Trk, and phospho-eIF2α. Sections were then stained using Vectastain anti-rabbit HRP kit (Vector Laboratories) and DAB (3,3’-diaminobenzidine) peroxidase substrate kit (Vector Laboratories). Briefly, sections were then incubated with biotinylated anti-rabbit antibodies, then by streptavidin-HRP and developed with DAB. Slides were scanned at the NYULMC Experimental Pathology Research Laboratory (NYULMC, New York, NY, USA) and whole tissue staining was quantified by Aperio ImageScope software (Leica Biosystems) in non-necrotic areas.
Human PDAC TMAs (IRB protocol: LAB05–0854) were stained for PHGDH at the NYULMC Center for Biospecimen Research and Development laboratory. TMAs also were stained with PGP9.5 as described above. Tissues were scored from 0 to 3 for PHGDH intensity by a GI pathologist or quantified by Aperio ImageScope software.
Quantitative Real-Time PCR
Cellular and tumor RNA was extracted using the PureLink RNA mini kit (ThermoFisher Scientific) or TRIzol, respectively. Purified RNA was reversed transcribed using Superscript Vilo IV (ThermoFisher Scientific). The resultant cDNA was mixed with SYBR Green Supermix (Bio-rad), along with primers specific to PHGDH, ATF4, HSPA5 (BIP), DDIT3 (CHOP), NGF, GFPd2, and mCherry and 18S (See Table S5 for complete list of primers). Reactions were performed on the CFX96 real-time PCR machine (Bio-Rad). ΔCT was calculated using 18S as a loading control. Fold-change was calculated relative to the indicated control.
Immunoblotting
For immunoblotting, whole cell lysates were generated in modified radioimmunoprecipitation (RIPA) buffer (50mM Tris-HCl pH 8.0, 150mM NaCl, 2mM EDTA, 1% NP-40, and 0.1% SDS, without sodium deoxycholate supplemented with protease (ThermoFisher Scientific) and phosphatase (10mM NaF, 1mM Na3VO4, 10mM β-glycerophosphate, and 10mM sodium pyrophosphate) inhibitors. Bulk tumors were washed in PBS and homogenized in modified RIPA buffer containing phosphatase and protease inhibitors, as described above. Protein lysates were loaded into 4–12% gradient Boltz gels (Life Technologies) and transferred with 1X transfer buffer (Tris-glycine) and 10% methanol. Membranes were incubated with their respective primary antibodies, and visualized with IRDye infrared secondary antibodies, using an Odyssey Infrared imaging system (Li-Cor Biosciences). Band intensities were quantified for BIP, phosho-eIF2α, eIF2α, and ERK2 using Image Studio Lite (Li-Cor Biosciences).
Metabolomics
Briefly, cells were washed in 0.9% saline and metabolites were extracted with 46% methanol:23% H2O:31% chloroform that contained labelled amino acid standards (MSK-A2–1.2, Cambridge Isotope Laboratories, Inc). Plasma samples were extracted using 75% acetonitrile: 25% methanol: 0.1% formic acid that contained labelled amino acid standards. Extracts were placed into 1.5mL Eppendorf tubes, vortexed at 4°C for 10mins and centrifuged at 4°C for 10mins at 17,200 × g in a microcentrifuge. The aqueous phase was transferred to vials for GC-MS or Eppendorf tubes for LC-MS. Samples were evaporated in a SpeedVac and samples were resolved by GC-MS or LC-MS.
For GC-MS, the metabolite extracts were derivatized with a solution of methoxamine (Sigma) dissolved in pyridine (Sigma) for 30mins at 37°C. An equal volume of tert-butyldimethylsilyl (TBDMS) was added to the mixture and incubated at 37°C for 30mins. Samples were resolved by GC (Agilent, 122–3832UI), peaks were picked using OpenChrom(Wenig and Odermatt, 2010) and analyzed using MATLAB.
For LC-MS, metabolite extracts were submitted to the NYULMC Metabolomics Laboratory. Briefly, metabolite extracts were resolubilized and resolved by liquid chromatography using a p-HILIC column, peak heights were analyzed using Xcalibur (ThermoFisher Scientific).
All metabolites were normalized to the closest labelled amino acid standard and also the respective cell number of the sample.
NADH/NAD+ Measurements
NADH/NAD+ ratios were measured using a NADH and NAD+ Assay kit (Promega), performed per manufacturer’s instructions with slight differences. Briefly, 200,000 PDAC cells were grown in the presence and absence of Ser/Gly for 24 hours. When indicated, PDAC cells were treated with 50μM CHX or 2μM oligomycin A for 15 minutes prior to collection. Cells were washed in 0.9% saline, and metabolites were collected with 46% methanol:23% H2O:31% chloroform, as described above. Dried down samples were resuspended in PBS solution to measure NADH and NAD+ according to the manufacturer’s instructions.
DL-Serine Assay
D- and L-Ser was measured using a DL-Serine Assay kit (abcam), performed as per manufacturer’s instructions. Briefly, conditioned media was pretreated with sample cleanup mix, deproteinized and transferred to 3K Amicon Ultra centrifugal filter (Millipore-Sigma). Samples and D-Ser standards were measured and quantified following manufacturer’s protocol.
Oxygen consumption measurements
Oxygen consumption rate (OCR) was measured with an XF96 analyzer (Seahorse Bioscience). Briefly, cells (18,000–20,000/well) were seeded in DMEM+10%FBS and allowed to adhere overnight. Media was changed to DMEM (2.78mM glucose, 4mM glutamine, 1mM pyruvate, with or without serine and glycine)+10% dialyzed FBS for 24 hours before measurements. Before placement into the analyzer, media was changed to DMEM to DMEM (2.78mM glucose, 4mM glutamine, 1mM pyruvate, with or without serine, glycine, phenol red and bicarbonate) + 10% dialyzed FBS ± 50μM CHX, and allowed to equilibrate for 1hr at 37°C, no CO2. Injection ports were loaded to achieve final working concentrations: serine (400μM), glycine (400μM), formate (1mM), oligomycin A (2μM), FCCP (0.5μM), rotenone (1μM) and antimycin A (1μM). For effects of axon-derived conditioned media (axCM) on OCR, cells were cultured in 50% DMEM (2.78mM glucose, 4mM glutamine, 1mM pyruvate, without serine and glycine)+10% dialyzed FBS and 50% axCM from control or rat DRG containing devices for 24 hours. The media was refreshed 1 hour prior to measuring OCR. The OCR measurements were normalized to changes in CyQUANT values.
Mitochondrial respiration measurements on isolated mitochondria was performed as described (Divakaruni et al., 2014). Briefly, cells were permeabilized with Seahorse XF Plasma Membrane Permeabilizer (PMP,1nM) and the assay was immediately performed in 1X MAS buffer (220mM Mannitol, 70mM Sucrose, 10mM KH2PO4, 5mM MgCl2, 2mM HEPES, 1mM EGTA, 5mM pyruvate, 10mM malate, 10mM glutamate, 4mM ADP and pH 7.2). Injection ports were loaded to achieve final working concentrations: serine (400μM), oligomycin A (2μM), FCCP (0.5μM), rotenone (1μM) and antimycin A (1μM) in 1X MAS buffer.
Flow cytometry
GFPd2 and mCherry fluorescence was measured by flow cytometry. Briefly, PDAC MDA-MB-231, and SKMEL cells were grown with and without Ser/Gly, PH719 (Raze Therapeutics), and/or cyclohexamide (CHX) for 24hrs. As a control, PDAC cells also were grown with or without tyrosine or phenylalanine, and/or CHX for 24hrs. For Ser gradient studies, exSer-dependent PDAC cells were grown in several concentrations of Ser for 24hrs. Cells were trypsinized and recovered by centrifugation at 300 x g in a table top centrifuge (Beckman Coulter). The samples were collected using the CytoFLEX flow cytometer (Beckman Coulter) and the data was analyzed using FlowJo. The geometric means were used to normalize GFPd2 to mCherry fluorescence to control for expression, and the value of CHX treated cells were subtracted from all the samples to calculate the maximum effects of tyrosine, phenylalanine, and Ser/Gly deprivation on GFPd2 fluorescence. Gating strategy is shown in Methods S1.
ELISA
ELISA assays to measure NGF (R&D, DY256) in conditioned media was assayed per manufacturer’s instructions. Briefly, PDAC cells were grown with different concentrations of Ser without Gly for 24hrs. For NGF secretion rate of wild-type and codon-optimized NGF, exSer-dependent PDAC cells expressing pLVX-EF1α-NGF-IRES-mCherry were grown with or without Ser/Gly for 24hrs. Media was refreshed and collected at the indicated time points. Cells were centrifuged at 300 x g in a table top centrifuge and media was collected for NGF measurements. NGF secretion was normalized to cell number or mCherry signal.
Quantitative Proteomics
Materials
The following reagents were employed: Isobaric TMT reagents (Thermo Fisher Scientific), BCA protein concentration assay kit (Thermo Fisher Scientific), Empore-C18 material for in-house made StageTips (3 M), Sep-Pak cartridges (100 mg, Waters), solvents for Liquid chromatography (LC) (J.T. Baker), mass spectrometry (MS)-grade trypsin (Thermo Fisher Scientific), Lys-C protease (Wako), and cOmplete protease inhibitors (Millipore Sigma). Unless otherwise noted, all other chemicals were purchased from Thermo Fisher Scientific.
MS sample processing
Cell pellets from PATU-8902 cells were lysed using 8 M urea, 200 mM 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid (EPPS) at pH 8.5 with protease inhibitors. Samples were further homogenized, and DNA was sheered via sonication using a probe sonicator (20 × 0.5sec pulses; level 3). Total protein was determined using a BCA assay and stored at −80°C until future use. A total of 100 μg of protein was aliquoted for each condition and TMT channel for further downstream processing. Protein extracts were reduced using 5 mM dithiothreitol (DTT) for 30 min at room temperature. Next samples were alkylated with 20 mM iodoacetamide for 45 min in the dark at room temperature. To facilitate the removal of incompatible reagents, proteins were precipitated using chloroform methanol. Briefly, to 100 μL of each sample, 400 μL of methanol was added, followed by 100 μL of chloroform with thorough vortexing. Next, 300 μL of HPLC grade water was added and samples were vortexed thoroughly. Each sample was centrifuged at 14,000 x g for 5 min at room temperature. The upper aqueous layer was removed, and the protein pellet was washed twice with methanol and centrifuged at 14 000 × g for 5 min at room temperature. Protein pellets were resolubilized in 200 mM EPPS buffer and digested overnight with Lys-C (1:100, enzyme: protein ratio) at room temperature. The next day, trypsin (1:100 ratio) was added and incubated at 37°C for an additional 6 hrs in a ThermoMixer set to 1,200 RPM. Digested samples were labeled immediately or stored at −80°C until future use.
TMT labeling
To each digested sample, 30% anhydrous acetonitrile was added and 100 μg of peptides were labeled using 200 μg of TMT reagent (TMT1-TMT11). To equalized protein loading, a ratio check was performed by pooling 2 μg of each TMT-labeled sample. Samples were combined and desalted using in-house packed C18 StageTips and analyzed by LC-MS/MS. Normalization factors were calculated from this label check, samples were mixed 1:1 across all TMT channels and desalted using a 100 mg Sep-Pak solid phase extraction cartridge. Eluted pooled peptides were fractionated immediately or stored at −80°C until future use.
Basic Ph Reversed-phase (bRP) fractionation
Pooled TMT labeled peptide samples were resuspended in 10 mM ammonium bicarbonate, 5% acetonitrile, pH 8.0 buffer and were fractionated with basic-pH reverse-phase (bRP) HPLC using an Agilent 300 extend C18 column and collected into a 96 deep-well plate. Peptides were subjected to a 50 min linear gradient from 13 to 43% buffer B (10 mM ammonium bicarbonate, 90% acetonitrile, pH 8.0) at a flow rate of 0.6 mL/min. Samples were consolidated into 24 fractions as previously described, and 24 fractions were desalted using StageTips prior to analyses using LC-MS/MS (Navarrete-Perea et al., 2018; Paulo, 2014; Paulo and Gygi, 2017).
MS data acquisition
All mass spectrometry data was acquired using an Orbitrap Fusion Lumos mass spectrometer in-line with a Proxeon NanoLC-1000 UHPLC system. Peptides were separated using an in-house 100 um capillary column packed with 40 cm of Accucore 150 resin (2.6 um, 150 Å) (ThermoFisher Scientific) using a 120 min LC gradient from 4 to 24% acetonitrile in 0.125% formic acid per fraction. Eluted peptides were acquired using a synchronous precursor selection (SPS-MS3) method for TMT quantification as previously described (McAlister et al., 2014). Briefly, MS1 spectra were acquired at 120K resolving power with a maximum of 50 ms in the Orbitrap. MS2 spectra were acquired by selecting the top 10 most abundant features via collisional induced dissociation (CID) in the ion trap using an automatic gain control (AGC) of 15K, quadrupole isolation width of 0.5 m/z and a maximum ion time of 50 ms. For MS3 acquisition, a synchronous precursor selection of 10 fragment ions was acquired with an AGC of 150K for 150 ms and a normalized collision energy of 65.
MS data analysis
All acquired data were processed using Comet (Eng et al., 2015; Eng et al., 2013) and a previously described in-house informatics pipeline (Elias and Gygi, 2007; Eng et al., 1994; Huttlin et al., 2010). Briefly, peptide spectral libraries were first filtered to a peptide false discovery rate (FDR) of less than 1% using linear discriminant analysis employing a target decoy strategy. Spectral searches were done using a custom fasta formatted database which included common contaminants, reversed sequences (Uniprot Human, 2014) and the following parameters: 50 ppm precursor tolerance, fully tryptic peptides, fragment ion tolerance of 0.9 Da and a static modification of TMT (+229.163 Da) on lysine and peptide N-termini, carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification. Resulting peptides were further filtered to obtain a 1% protein FDR and proteins were collapsed into groups. Reporter ion intensities were adjusted to correct for impurities during synthesis of different TMT reagents according to the manufacturer’s specifications. For quantitation, a total sum signal-to-noise of all report ion ions of 100 was required for analysis. Lastly, protein quantitative values were normalized so that the sum of the signal for all protein in each channel was equal to account for sample loading.
Ribosomal profiling
Ribosomal profiling was performed as described (Ingolia et al., 2012). Briefly, cells were washed in cold PBS containing 100μg/mL CHX and RNA was extracted from cells using 1X polysome lysis buffer (20mM Tris-HCl pH 7.4, 150mM NaCl, 5mM MgCl2, 1% Triton-X-100, 1mM DTT, 10U Turbo DNase, 100μg/mL CHX, and 0.1% NP-40. Lysates were clarified by centrifugation at 4°C at 20,000 × g for 10mins and separated for total RNA and ribosome protected fragments (RPFs).
For RPFs samples, RNA was digested with RNase I for 45mins at room temperature and purified using MicroSpin S-400 columns and Zymo Research RNA clean & Concentrator kit. The RPF footprints were run on a TBE-Urea gel and the bands corresponding to the 26–34 nucleotide (nt) region was excised and the RNA was extracted for further preparation. Total RNA was purified using Zymo Research RNA clean & Concentrator kit and heat fragmented for further preparation. RNA end-repair were performed using T4 PNK for both the total RNA fragments and RPFs, and purified using the Zymo Research RNA clean & Concentrator kit. A 3’ adaptor was ligated to the total RNA fragments and RPFs using a universal preadenylated linker (NEB), ran on a TBE-Urea gel and purified by gel excision and extraction. Samples were reversed transcribed and cDNAs were excised and extracted from a TBE-Urea gel. Next, cDNAs were circularized using circLigase and rRNA was depleted using a set of biotinylated rRNA depletion oligomers. Sequencing library was amplified from rRNA-depleted cDNAs using Phusion HF and the products were excised and extracted from a TBE gel. Sequencing library was submitted to NYULMC genome technology core for analysis on the HiSeq4000.
Ribosome profiling data analysis
Ribosome profiling analysis was performed as described by Darnell A.M, et al (Darnell et al., 2018). The 50 nucleotide (nt) single-end raw sequencing reads were trimmed for the adapter sequence 5’-CTGTAGGCACCATCAAT-3’ using cutadapt (Martin, 2011) with a minimum length of 13 nt. Ribosomal RNA were removed using bowtie2(Langmead and Salzberg, 2012) and the remaining sequences were aligned to the transcriptome using rsem (Li and Dewey, 2011) and bowtie2. To calculate the ribosome density at codons, and because the precise frame information was not required, 12nt were trimmed from both sides to smooth the ribosome density profiles as described by multiple studies (Darnell et al., 2018; Oh et al., 2011; Subramaniam et al., 2014). The average ribosome density around each codon was calculated by considering only transcripts with a minimum average read density of 1 read per codon, which were then normalized to the mean read count for that transcript. For each codon, the average read coverage was found for each position in a 150nt window on either side of all occurrences of that codon. The change in ribosome density around each codon upon Ser/Gly-deprivation was calculated by subtracting the average ribosome density at each position in the 150nt window around the codon in the Ser/Gly-rich conditions from the Ser/Gly-deprived conditions. This determined the changes in ribosome density on all codons upon Ser/Gly-limitation.
Charge DM-tRNA-seq and analysis
tRNA charging and fractional abundance measurements were performed as described with some modifications (Evans et al., 2017). Briefly, RNA was extracted using TRIzol, treated with 50mM of sodium periodate to determine charge and uncharged tRNAs, and repaired with 60mM sodium borate (pH 9.5). tRNAs were gel purified, and demethylated (ArrayStar tRNA RT-PCR Kit). RNA end-repair were performed using T4 PNK on demethylated tRNAs, and purified. A 3’ adaptor was ligated to the demethylated tRNAs using a universal preadenylated linker (NEB), ran on a TBE-Urea gel and purified by gel excision and extraction. Samples were reversed transcribed and cDNAs were excised and extracted from a TBE-Urea gel. Next, cDNAs were circularized using circLigase II, and Sequencing library were amplified using Phusion HF. The products were excised and extracted from a TBE gel. Sequencing library was submitted to NYULMC genome technology core for analysis on the HiSeq4000. Analysis of tRNA charging and fractional abundance were performed (Evans et al., 2017). Codes and scripts were obtained from Dr. Tao Pan’s lab, and are available at https://github.com/Jessica-Pan/DM-tRNA-seq-ref-genomes.
Estimation of usage bias for pause-site serine codons
Analysis was performed as described (Darnell et al., 2018). Briefly, a binomial probability distribution for each gene was used to estimate the probability of the observed number of TCC and TCT codons to the genome-wide average of Ser codon frequencies. To avoid skew due to differences in TCN vs AGN (where N, can be A, C, T, and G) bias/frequencies in our analyses, we only considered pause-site (TCT/TCC) and non-pause-site (TCG/TCA) serine codons. The average frequency of stall sites relative to other TCN codons (p = 0.66). The Z-score was then calculated using the calculated theoretical binomial probability distribution, the standard deviation of that theoretical binomial probability distribution, and the observed number of stalled Ser codons for each gene. Low and high Z-scores predict genes that would least and most likely affected by Ser codon usage bias, respectively. The resulting Z-score ranged from −12.3 to 9.79 and represents bias against (low scores) or toward (high scores) the use of pause-site serine codons to encode serine in each gene. The 95th (1.89) and 5th (−2.04) percentile Z-scores were used as cutoff values for further analyses (Table S2). Using R and dlpyr, a list of genes that overlapped with the proteins identify by quantitative proteomics was generated to determine the association with the calculated Z-Score (Table S3). The protein abundance of the top 50 genes predicted to be most or least affected according to changes calculated Z-Score was plotted as a waterfall plot. Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) was used to identify enrichment for Canonical Pathways in the MSigDB collections (Liberzon et al., 2015) in genes with biased usage of pause-site and non-pause-site serine codons. Canonical pathways with a false-discovery rate adjusted p-value of <0.05 were considered significant.
Survival, Molecular Subtyping and Correlation Analyses
For the MD Anderson TMA dataset, samples were grouped into PHGDH low (grade 0, 1, and 2), and high (grade 3+) based on the PHGDH staining score. The overall survival of PHGDH low and high tumors was plotted, and the frequency differentiation status and tumor size were compared. The correlation of PHGDH and PGP9.5 staining from TMA sections were performed by comparing tumors with high PHGDH or PGP9.5, by setting a greater than one standard deviation cutoff. The TCGA PDAC dataset (RNA-seq, methylation, and survival data) was obtained using cBioportal (TCGA, Firehose Legacy) (Cerami et al., 2012; Gao et al., 2013). For overall survival, samples were grouped into PHGDH high and low expressing tumors, using an expression cutoff of less than −0.5 standard deviation. The Basal and Classical molecular subtypes of PDAC tumors from the TCGA were obtained from a published study to assess differences in PHGDH expression (Cancer Genome Atlas Research Network. Electronic address and Cancer Genome Atlas Research, 2017). The correlation of PHGDH and NGF expression from TCGA dataset was performed by comparing tumors with high PHGDH and NGF expression, by setting a cutoff of a greater than one standard deviation. The RNA-seq and methylation for PDAC cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE) (Ghandi et al., 2019). Analyses were performed on exSer-dependent and –independent PDAC cells. The molecular subtypes of the PDAC cell lines were obtained from a published study (Yu et al., 2019). All plots were analyzed using Pearson’s correlation.
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample sizes (n), what “n” represents, definition of center, dispersion and precision measures, and statistical tests for each experiment are denoted in the figure legends. Each immunoblot was performed at least three times. All orthotopic xenografts represent one independent experiment with the indicated biological replicates. All metabolomics data represent n=5 biological replicates for each group. All immunohistochemical images show samples from a single mouse of each group, and quantification was performed on all biological replicates in the experiment, with the number of data points indicating the number of independent biological replicates. For datasets that were skewed or contained outliers as determined by the ROUT test, dataset was transformed, followed by the indicated statistical analysis. The between-group variances were similar, and the data were normally distributed. All analyses and graphs were generated with GraphPad Prism 8. A p-value of <0.05 was considered significant, precise p-values can be found in the figures.
Supplementary Material
Methods S1. Related to STAR Methods. Gating strategy for flow cytometry analysis. Gating strategy used for GFPd2-mCherry flow cytometry analysis. Single cells were gated on FSC:H and SSC:H followed by FSC:W and SSC:H. Live (DAPI-) cells were used to set up gates for mCherry expression. mCherry+ cells were selected to determine the geometric mean of GFP fluorescence.
Supplementary Table 1. Related to Figure 2. Information of PDAC cell lines used in this study.
Supplementary Table 2. Related to Figure 5. Calculated Z-Scores of stalled Ser codons across 18,660 human coding sequences.
Supplementary Table 3. Related to Figure 5. Overlapping list of proteins measured by quantitative proteomics and calculated Z-Scores from Supplementary Table 2.
Supplementary Table 4. Related to Figure 6. Composition of amino acid (AA) and Ser/Gly-free (-SG) mouse diets.
Supplementary Table 5. Related to STAR Methods. List of oligonucleotides used in study.
Supplementary Figure 1. A subset of human, but not mouse, PDAC cells require exSer for growth. Related to Figure 1 and 2.
A, Representative traces (left) of spontaneous action potentials, depolarization-evoked action potentials and Na+ currents in the absence (black), presence (red), and washout (blue) of 1μM TTX in rat dorsal root ganglion (DRG) neurons. Quantification of spontaneous action potential (AP) frequency (n=4 cells), evoked AP amplitudes (n=6 cells), and Na+ current densities (n=10 cells) is presented on the right.
B, Mouse KPC PDAC cells grown in the presence or absence of Ser and/or Gly (n=3 technical replicates for three independently derived cell lines).
C, exSer-dependent PDAC cells were grown in media containing high (25mM) or low (2.78mM) glucose with or without 1mM pyruvate and Ser/Gly for the indicated times (n=3 for each group and condition).
D, exSer-dependent PDAC cell growth was measured after 24 hours in several different media concentrations of Ser (n=4 for each condition).
E, exSer-dependent PDAC cells were cultured in Ser/Gly-rich and –deprived conditions for 18 days (n=3).
F, Immunoblot of SBP enzymes, PHGDH, PSAT1, and PSPH, in exSer-independent and –dependent PDAC cells grown in Ser/Gly-deprived conditions for the indicated times. p38 serves as a loading control.
G, PDAC cells were labelled with 13C6-glucose for 24 hours and total levels of unlabeled (black) and labelled (red) intracellular Ser was measured using GC-MS. (n=5 biologically independent wells).
H, exSer-dependent PDAC cells expressing empty vector, Flag-PSAT1, PHGDH, or both were adapted in Ser/Gly-deprived conditions for 21 days. Growth was observed in a subset of adapted exSer-dependent PDAC cells expressing Flag-PSAT1 and PHGDH. (n=3).
I, exSer-dependent PDAC cells expressing empty vector, Flag-PSAT1, PHGDH, or both in Ser/Gly-rich and –depleted conditions for the indicated times (n=3). exSer-dependent PDAC cells expressing both PHGDH and Flag-PSAT1 were adapted in Ser/Gly-deprived conditions for 21 days before assessing growth.
J, Immunoblot of SBP enzymes, PHGDH, PSAT1, and PSPH, in exSer-independent and -dependent PDAC cells expressing empty vector, Flag-PSAT1, PHGDH, or both grown in Ser/Gly-rich and -deprived conditions for 24 hours. exSer-dependent PDAC cells expressing both PHGDH and Flag-PSAT1 were adapted in Ser/Gly-deprived conditions for 21 days before assessing levels of protein. p38 serves as a loading control.
K-L, PHGDH expression in basal and classical subtypes of the PDAC cell lines used in this study (K) and the TCGA database (L).
M-N, PHGDH (M) and PSAT (N) expression and methylation levels in exSer-independent and –dependent PDAC cell lines measured from the CCLE database. The highest methylation score was plotted against expression.
O, PHGDH expression and methylation levels in PDAC tumors from the TCGA database.
“n” represents the number of biologically independent replicates. Graphs (mean ± s.e.m.) were compared using one (A) or two (B, C, G)-way ANOVA, followed by Dunnett’s (A) or Bonferroni (B, C, G) post-hoc test. Pearson correlation values are represented by r (M-O). (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Supplementary Figure 3. Acute increase in the oxygen consumption rate (OCR) by exSer is not directly acting on mitochondria or the mTOR signaling pathway. Related to Figure 4.
A, Total levels of unlabeled (black) and labelled (red and blue) citrate from 13C6-glucose in PDAC cells measured in the presence or absence of Ser and/or Gly after 24 hours (n=5 biologically independent wells).
B, OCR of PDAC cells that were Ser/Gly-deprived for 24 hours. PDAC cells were stimulated with the respective metabolite(s), oligomycin A, FCCP, and rotenone and antimycin A at the indicated times. Related to Figure 4D.
C, Fold change in NADH/NAD+ was assessed in exSer-independent and -dependent PDAC cells grown in Ser/Gly-rich and –poor conditions for 24 hours (n=6).
D, Positive association of the acute oxygen consumption rate (OCR) response upon Ser stimulation, and the fold change of NADH/NAD+ ratio in exSer-independent and – dependent PDAC cells grown in the absence and presence of Ser/Gly from C.
E, Mitochondrial OCR from permeabilized exSer-dependent PDAC cells (PATU-8902) that were deprived of Ser and Gly for 24 hours and stimulated with control or Ser as indicated.
F, Acute change in mitochondrial OCR from permeabilized exSer-dependent PDAC cells after stimulation with control or Ser from E (n=3 technical replicates).
G, Immunoblot of mTOR signaling in PATU-8902 cells that were grown in the presence or absence of Ser and Gly for 24 hours and stimulated with 400μM Ser for 1, 5, and 10 mins.
H, OCR measurements of exSer-dependent PDAC cells pretreated with mTOR inhibitors for 1 hour and stimulated with 400μM Ser after 24 hours of Ser and Gly deprivation.
I, Acute response to OCR after pre-treatments with or without mTOR inhibitors as described in H (n=3–5 technical replicates).
J, Gene expression of ATF4, DDIT3 (CHOP), and HSPA5 (BIP) in exSer-independent and –dependent PDAC cells that were grown with or without Ser and Gly for 24 hours (n=3 biologically independent experiments).
K, In Ser-rich conditions, cells have high rates of seryl-tRNA charging and protein synthesis, which results in a high ATP demand. Mitochondria will have higher activity and OCR to keep up with the ATP demand of the cell. By contrast, Ser becomes limiting for seryl-tRNA charging and protein synthesis during Ser-deprived conditions, which would decrease the intracellular ATP demand. In response to lower ATP demand, the mitochondria will be less active and have a lower OCR.
Graphs (mean ± s.e.m.) were compared using one (C) or two-way (J) ANOVA, followed by Bonferroni post-hoc test. Pearson correlation values are represented by r (D). (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Supplementary Figure 2. exSer-dependent PDAC cells require L-Ser from nerves to grow in Ser/Gly-deprived media. Related to Figure 3.
A, Growth of PATU-8902 (exSer-dependent) cells in the absence of L-Ser and Gly or L-Ser alone in presence of several concentrations of exogenous D-Serine after 48 hours (n=3 technical replicates and repeated once).
B, Schematic of outcomes if synaptic gap junctions form between nerves and exSer-dependent PDAC cells 6 days after co-culture in the microfluidics device. Although cell-impermeable, calcein is small enough to pass through gap junctions. If gap junctions are formed between nerves and PDAC cells, then PDAC cells would be labelled with calcein as strongly as the nerves. Control devices were used as a negative control for autofluorescence.
C, Representative image of exSer-dependent PDAC cells cultured in control and rat DRG microfluidic devices after six days, with the neuronal side stained with calcein-AM.
D, exSer-dependent PDAC cells were grown in medium containing different concentrations of serine for the indicated amount of time (n=3 biologically independent experiments). Control samples only had one dose of exSer at the beginning of the experiment. In comparison in the “2X Daily” samples, the same initial concentration of serine was added twice a day.
E, Schematic of the multiple role of serine for proteins, for the generation of glycine and glutathione (GSH), one-carbon cycle and transmethylation cycle. γGC: γ-glutamylcysteine, THF: tetrahydrofolate, CH2-THF: 5,10-methylenetetrahydrofolate, CH-THF: 5,10-methenyltetrahydrofolate, CHO-THF: 10-formyltetrahydrofolate, dTMP: deoxythymidine monophosphate, Met: methionine, SAM: S-adenosylmethionine, SAH: S-adenosylhomocysteine, HCys: homocysteine.
F, Growth of exSer-dependent PDAC cells (PATU-8902) in the presence or absence of Ser and/or Gly and treated with or without 1mM formate or 1X nucleosides after two days (n=3 technical replicates and repeated once).
G, Growth of exSer-dependent PDAC cells in the presence or absence of Ser and/or Gly and treated with or without 1mM formate after two days (n=4).
H, exSer-dependent PDAC cells were labelled with 13C2-Gly for 24 hours with or without Ser, Gly, and/or 1mM formate. Total levels of unlabeled (black) and labelled (red) Ser and Gly were measured using GC-MS (n=5).
I, Schematic of Gly-to-Ser conversion in exSer-dependent PDAC cells in the presence of Ser, Gly, and/or formate.
J, Reduced to oxidized glutathione (GSH:GSSG) ratios and S-adenosylmethionine (SAM) to S-adenosylhomocysteine (SAH) ratios in exSer-independent (PATU-8988T) and -dependent (PATU-8902) cells after 24 hours in the presence or absence of Ser and Gly (n=5 biologically independent wells).
K, exSer-independent and -dependent PDAC cells were labelled with 13C3-Ser for 24 hours in Ser/Gly-rich or –deprived conditions. Total levels of unlabeled (black) and labelled (blue) ATP, GTP, GSH, GSSG, SAM, and SAH were measured using LC-MS (n=5).
L, Fold change of Ser-derived metabolites in exSer-independent and –dependent PDAC cells grown in the absence and presence of Ser/Gly from K, M (n=5).
M, Total levels of NADH and NAD+ from exSer-independent and –dependent PDAC cells labelled with 13C3-Ser for 24 hours in Ser/Gly-rich or –deprived conditions (n=5).
Graphs (mean ± s.e.m.) were compared using two-tailed t-test (J) and two-way ANOVA (F-H), followed by Bonferroni post-hoc test (***p<0.005).
Supplementary Figure 4. Analysis of Ser codon usage bias in exSer-dependent PDAC (PATU-8902) and other cancer cells. Related to Figure 5.
A, Representative flow plots of mCherry levels, an internal control, in cells expressing the GFPd2 Ser codon reporter constructs from Figure 5A. PATU-8902 cells were grown in the presence and absence of Ser and Gly and treated with or without 50μM cycloheximide (CHX).
B, Fold change and ratio of GFPd2 and mCherry expression relative to Ser/Gly-rich conditions in exSer-dependent PDAC cells grown with or without Ser/Gly for 24 hours (n=3 technical replicates and repeated once).
C, Quantification of GFPd2 fluorescence in exSer-dependent PDAC cells (n=3 technical replicates and repeated once for both conditions) grown with or without tyrosine or phenylalanine for 24 hours.
D, Ribosome density analysis of total mRNA as a negative control from PATU-8902 that were grown in the presence or absence of Ser and Gly for 24 hours (n=3 biologically independent replicates).
E, Immunoblots of SBP enzymes, PHGDH, PSAT1, and PSPH in PDAC, breast cancer and melanoma cells grown with or without Ser/Gly for 24 hours. ERK2 serves as a loading control.
F-H, Quantification of GFPd2 fluorescence in exSer-independent PDAC (F, n=3), breast cancer (G, n=5), and melanoma (H, n=3) cells, and grown with or without Ser/Gly and 1μM PH719, a PHGDH inhibitor. Dotted-line represents mean fluorescence of AGC (F) and AGT (G, H) GFPd2 fluorescence.
Where “n” represents the number of biologically independent experiments for each group and condition. Graphs represent mean ± s.e.m. and were compared using one-way ANOVA (D, F-H), followed by Bonferroni post-hoc test (*p<0.05, **p<0.01, ****p<0.0001).
Supplementary Figure 5. Ser codon usage bias occurs when GCN2 is activated, and is not correlated with tRNA gene copy number. Related to Figure 5.
A, GFPd2 fluorescence of the Ser codon-optimized reporters were measured across a range of Ser concentrations in exSer-dependent PDAC cells (n=3 technical replicates, performed at least twice).
B, Immunoblot of BIP and phospho-eIF2α levels in exSer-dependent PDAC cells grown across a range of Ser concentrations for 24 hours. eIF2α serves as a loading control.
C-D, Quantification of phospho-eIF2α (C) and BIP (D) levels normalized to eIF2α from B (n=4 biologically independent replicates). Graphs represent mean ± s.e.m. and the EC50 was determined.
E-G, Plots of summed ribosome densities of the ribosome protected mRNA against the expected codon frequency (E), cognate Ser tRNA gene copy number (F) or the calculated codon usage frequency (G) of the Ser codons. Line of best fit is shown in black and the data was analyzed using Pearson’s correlation.
H, Total fractional abundance of charged Ser-tRNA isodecoders (AGA, CGA, GCT, and TGA) in exSer-dependent PDAC cells grown in Ser/Gly-rich and –depleted conditions (n=3).
I-J, Ribosome density was plotted against percent tRNA charging (I), and the change in total fractional abundance (J) of charged Ser-tRNA isodecoders (AGA, CGA, GCT, and TGA) in exSer-dependent PDAC cells grown in Ser/Gly-rich and –depleted conditions (n=3).
K, Distribution of Z-scores reflecting pause-site Ser codon usage bias in ~18,000 CDS (see methods for details). TCY: TCT and TCC, TCR: TCA and TCG.
L, Waterfall plots of the change in protein abundance for the top 50 genes predicted to be most and least affected by differential mRNA translation of Ser codons, TCC and TCT. Protein levels were measured exSer-dependent PDAC cells grown in Ser/Gly-rich or –deprived conditions after 24 and 48 hours (n=3). A Chi square test was used to assess the significance of the observed (red line) to expected (black line) frequencies. p-values are indicated in each graph.
M, Immunoblot of RAS levels and ERK activation in exSer-dependent PDAC cells expressing shGFP or shKRAS grown with or without Ser/Gly for 24 hours. ERK2 serves as a loading control.
N, Quantitative proteomics of RAS isoforms in exSer-dependent PDAC cells grown in Ser/Gly-rich and –deprived conditions for 24 (n=3) and 48 (n=2 for +Ser/Gly and n=3 for –Ser/Gly) hours by mass spectrometry. Graphs represent mean ± s.e.m.
O, NGF levels in the media of exSer-independent PDAC cells grown in different concentrations of Ser after 24 hours (n=4–5).
P, NGF mRNA levels were measured in exSer-dependent PDAC cells after being grown in Ser/Gly-rich and –poor media for 24 hours (n=3).
Q, exSer-dependent PDAC cells were grown in Ser/Gly-rich and –deprived media for 24 hours. Secreted NGF levels were measured at the indicated time points from exSer-dependent PDAC cells expressing empty vector, wild-type (WT containing 3 stalled and 16 non-stalled Ser codons), TCC-, and TCA- codon optimized NGF followed by IRES-mCherry. Levels of secreted NGF was normalized to mCherry signal (n=4).
Where “n” presents the number of biologically independent replicates. Graph represent (mean ± s.e.m.) and was compared using a one-sample t-test (P), one- (H), or two-way ANOVA (Q), followed by Bonferroni post-hoc test. Pearson correlation values are represented by r (I-J). (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Supplementary Figure 6. The effects of –SG (Ser/Gly-free)-diet on exSer-dependent PDAC tumors. Related to Figure 6.
A, Plasma concentrations of Ser and Gly from mice on AA (n=5–10) or –SG (n=5–9) diet at the indicated times. As a control, plasma concentrations of Ser and Gly also were measured from non-tumor bearing mice on AA and –SG diet (n=3) at the indicated times.
B, Changes in AA plasma concentrations at week 5 from non-tumor bearing (n=3–5), and exSer-dependent tumor-bearing mice on AA (n=5–10) or –SG (n=5–9) diet from A.
C, Representative images of H&E, trichrome, phospho-Histone 3 and cleaved caspase 3 staining of PATU-8902 tumors from mice on AA and -SG diet after five weeks post-injection (from Figure 6C).
D, Representative images of nerves (PGP9.5) of PATU-8902 tumors from mice on AA and -SG diet after five weeks post-injection (from Figure 6E).
E-F, Representative images of immunohistochemistry staining for sympathetic (E, tyrosine hydroxylase (Th+)) and sensory (F, transient receptor potential cation channel subfamily V member 1 (Trpv1+)) nerves in exSer-dependent PDAC tumors from mice on control and Ser/Gly-free diet.
G, Two representative images of lack of parasympathetic (vesicular acetylcholine transporter (VAchT+)) nerve staining in exSer-dependent tumors grown in mice on Ser/Gly-free diet. Mouse brain serves as a positive control and PGP9.5 staining serves as a nerve staining control.
H, 3D-images of clarified exSer-dependent PDAC tumors stained with PGP9.5 to detect innervation of tumors from mice on AA and –SG diet after five-week post-injection. The magnified image of tumor #4 (AA diet) and tumor #1 (-SG diet) is also shown in Figure 6D.
I, Protein abundance fold change of axon guidance cues in exSer-dependent PDAC cells grown in Ser/Gly-deprived conditions for 24 and 48 hours (n=3).
“n” presents the number of biologically independent replicates. Graph represent (mean ± s.e.m.) and was compared using permutation-based FDR (I) or two-way ANOVA (A), followed by Bonferroni post-hoc test (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Supplementary Figure 7. The effects of –SG-diet and LOXO-101 on PDAC cells in vitro and in vivo. Related to Figure 6 and 7.
A, In vitro growth of exSer-independent and -dependent PDAC cells treated with different concentrations of LOXO-101 in the presence or absence of Ser and Gly (n=3 technical replicates and repeated once for five independent cell lines).
B, Ser (S) and Gly (G) concentrations were measured from axCM (axon-derived conditioned media) of rat DRGs treated with Veh or LOXO-101 as shown (n=4–5). As a positive control for LOXO-101, rat DRG neurons were treated with LOXO-101, followed by 30min NGF stimulation as indicated. Phosphorylation of TrkA at Y684/685 was used to assess Trk activity and the effect of LOXO-101. ERK2 serves as a loading control.
C, Immunoblot of TRK levels in exSer-dependent PDAC cells grown with or without Ser/Gly, and tumors from mice on AA or –SG diet treated with or without LOXO-101. ERK2 serves as a loading control. Mouse brain was used a positive control.
D, Representative TRK staining of exSer-dependent PDAC tumors from mice on AA or –SG diet. Mouse brain was used a positive control.
E, Plasma concentrations of Ser and Gly from mice on AA or -SG diet and treated with vehicle or LOXO-101 at the indicated times (n≥6 for each group and timepoint).
F, Quantification of cleaved caspase 3 staining in tumors from mice fed amino acid (AA) or Ser/Gly-free (-SG) diet, treated with vehicle or LOXO-101 from G (n≥6).
G, Representative images of p-Histone 3, cleaved caspase 3, and phospho-eIF2α staining of PATU-8902 tumors from Figure 6.
H, Overall survival of PHGDH high and low expressing PDAC tumors from TCGA dataset.
I-J, Observed frequencies of the differentiation state (I) and tumor size (J) of PHGDH low and high PDAC tumors from patients at MD Anderson. A Chi square test was used to assess the significance of the observed to expected frequencies.
Where “n” represents the number of biologically independent experiments for each group and condition. Graphs (mean ± s.e.m. for A, D) or (median ± max/min for B, F) were compared using one-way (B, F) or two-way ANOVA (E), followed by Bonferroni or Holm-Sidak post-hoc test (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Highlights.
Nerves secrete amino acids, such as serine, to support PDAC growth and survival.
PDAC cells exhibit metabolic plasticity, but protein synthesis is a bottleneck.
Serine deprivation decreases mitochondria activity by affecting mRNA translation.
Serine deprivation decreases mRNA translation of TCC and TCT serine codons.
Acknowledgements
We acknowledge NYULH Metabolomics Core, Applied Bioinformatics Laboratories, Genome Technology Center, Experimental Pathology Research Laboratory, Microscopy Laboratory, and Ion Channel and Immunology Core for their assistance. These shared resources are partially supported by P30CA016087. We thank Dr. Steven Gygi for use of CORE for mass spectrometry data analysis software and Dr. Tao Pan for providing scripts to analyze the tRNA sequencing data. We thank Raze therapeutics for PH719. R.S.B. is a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2348-18). D.E.B. is supported by a Ruth L. Kirschstein Institutional National Research Service Award, T32 CA009161 (Levy), and the NCI F99/K00 award (F99 CA245822). K.Y. was supported by an Uehara Memorial Foundation Research Fellowship. M.E.P. is supported by a Mary Kay Foundation Cancer Research Grant (017-32), the Shifrin-Myers Breast Cancer Discovery Fund at NYU, a V Foundation V Scholar Grant funded by the Hearst Foundation (V2017-004), and an NCI K22 Career Transition Award (1K22CA212059). This work was supported by NCI Grants R01CA157490, R01CA188048, P01CA117969, R35CA232124; ACS RSG-13-298-01-TBG; NIH grant R01GM095567; and the Lustgarten Foundation, and SU2C to A.C.K.; by NIH T32 CA9161-41 to B.W.; by NCI grant 1RO1CA178509 and Breast Cancer Research Foundation grant BCRF-16-143 to R.J.S.; by Burroughs Wellcome Fund Career Award for Medical Scientists, Brigham and Women’s Hospital MFCD Award, Dana-Farber Cancer Institute Claudia Adams Barr Program for Innovative Cancer Research Award, and the Hale Family Center for Pancreatic Cancer Research to J.D.M. We thank the members of the Kimmelman and Pacold labs for their helpful suggestions and comments.
Footnotes
Declaration of Interests.
M.E.P. has options in Raze Therapeutics and received travel funds from Thermo Fisher Scientific. J.D.M is an inventor on a patent pertaining to the autophagic control of iron metabolism. A.C.K. has financial interests in Vescor Therapeutics, LLC. A.C.K. is an inventor on patents pertaining to KRAS regulated metabolic pathways, redox control pathways in pancreatic cancer, targeting GOT1 as a therapeutic approach, and the autophagic control of iron metabolism. A.C.K is on the SAB of Rafael/Cornerstone Pharma. A.C.K has been a consultant for Deciphera Pharma. The other authors declare no competing interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbaci M, Barberi-Heyob M, Blondel W, Guillemin F, and Didelon J. (2008). Advantages and limitations of commonly used methods to assay the molecular permeability of gap junctional intercellular communication. BioTechniques 45, 33–62. [DOI] [PubMed] [Google Scholar]
- Aloe L, Rocco ML, Bianchi P, and Manni L. (2012). Nerve growth factor: from the early discoveries to the potential clinical use. J Transl Med 10, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auciello FR, Bulusu V, Oon C, Tait-Mulder J, Berry M, Bhattacharyya S, Tumanov S, Allen-Petersen BL, Link J, Kendsersky ND, et al. (2019). A Stromal Lysolipid-Autotaxin Signaling Axis Promotes Pancreatic Tumor Progression. Cancer Discov 9, 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. (2012). Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth E, Dickerson W, Vijay V, Weitzel K, Cooper J, Atkinson EW, Coleman JE, Otto KJ, and Campbell-Thompson M. (2018). High Resolution 3D Imaging of the Human Pancreas Neuro-insular Network. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, and Brand MD (1995). A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 312 ( Pt 1), 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Electronic address, a.a.d.h.e., and Cancer Genome Atlas Research, N. (2017). Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 32, 185–203 e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroleo MC, Costa N, Bracci-Laudiero L, and Aloe L. (2001). Human monocyte/macrophages activate by exposure to LPS overexpress NGF and NGF receptors. J Neuroimmunol 113, 193–201. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceyhan GO, Bergmann F, Kadihasanoglu M, Altintas B, Demir IE, Hinz U, Muller MW, Giese T, Buchler MW, Giese NA, et al. (2009a). Pancreatic neuropathy and neuropathic pain--a comprehensive pathomorphological study of 546 cases. Gastroenterology 136, 177–186 e171. [DOI] [PubMed] [Google Scholar]
- Ceyhan GO, Demir IE, Rauch U, Bergmann F, Muller MW, Buchler MW, Friess H, and Schafer KH (2009b). Pancreatic neuropathy results in “neural remodeling” and altered pancreatic innervation in chronic pancreatitis and pancreatic cancer. Am J Gastroenterol 104, 2555–2565. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, and Sabatini DM (2015). Nutrient-sensing mechanisms across evolution. Cell 161, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan VP, Martin JD, Liu H, Lacorre DA, Jain SR, Kozin SV, Stylianopoulos T, Mousa AS, Han X, Adstamongkonkul P, et al. (2013). Angiotensin inhibition enhances drug delivery and potentiates chemotherapy by decompressing tumour blood vessels. Nat Commun 4, 2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Wu M, Zhang L, Thompson R, Nath A, and Chan C. (2013). Signaling dynamics of palmitate-induced ER stress responses mediated by ATF4 in HepG2 cells. BMC Syst Biol 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. (2013). Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F, Lee H, Zhang Y, Zhuang L, Yao H, Xi Y, Xiao ZD, You MJ, Li W, Su X, et al. (2017). BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc Natl Acad Sci U S A 114, 3192–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell AM, Subramaniam AR, and O’Shea EK (2018). Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells. Mol Cell 71, 229–243 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Jonas O, Keibler MA, Hou HW, Luengo A, Mayers JR, Wyckoff J, Del Rosario AM, Whitman M, Chin CR, et al. (2017). Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat Med 23, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AS, Rogers GW, and Murphy AN (2014). Measuring Mitochondrial Function in Permeabilized Cells Using the Seahorse XF Analyzer or a Clark-Type Oxygen Electrode. Curr Protoc Toxicol 60, 25 22 21–16. [DOI] [PubMed] [Google Scholar]
- Ehmsen JT, Ma TM, Sason H, Rosenberg D, Ogo T, Furuya S, Snyder SH, and Wolosker H. (2013). D-Serine in Glia and Neurons Derives from 3-Phosphoglycerate Dehydrogenase. The Journal of Neuroscience 33, 12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, and Gygi SP (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4, 207–214. [DOI] [PubMed] [Google Scholar]
- Eng JK, Hoopmann MR, Jahan TA, Egertson JD, Noble WS, and MacCoss MJ (2015). A deeper look into Comet--implementation and features. J Am Soc Mass Spectrom 26, 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, Jahan TA, and Hoopmann MR (2013). Comet: An open-source MS/MS sequence database search tool. PROTEOMICS 13, 22–24. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, and Yates JR (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Evans ME, Clark WC, Zheng G, and Pan T. (2017). Determination of tRNA aminoacylation levels by high-throughput sequencing. Nucleic Acids Res 45, e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya S, Tabata T, Mitoma J, Yamada K, Yamasaki M, Makino A, Yamamoto T, Watanabe M, Kano M, and Hirabayashi Y. (2000). L-serine and glycine serve as major astroglia-derived trophic factors for cerebellar Purkinje neurons. Proc Natl Acad Sci U S A 97, 11528–11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner ML, Eade K, Wallace M, Handzlik MK, Fallon R, Trombley J, Bonelli R, Giles S, Harkins-Perry S, Heeren TFC, et al. (2019). Serine and Lipid Metabolism in Macular Disease and Peripheral Neuropathy. N Engl J Med 381, 1422–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Lee K, Reid MA, Sanderson SM, Qiu C, Li S, Liu J, and Locasale JW (2018). Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Rep 22, 3507–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, Barretina J, Gelfand ET, Bielski CM, Li H, et al. (2019). Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilardi JR, Freeman KT, Jimenez-Andrade JM, Mantyh WG, Bloom AP, Kuskowski MA, and Mantyh PW (2010). Administration of a tropomyosin receptor kinase inhibitor attenuates sarcoma-induced nerve sprouting, neuroma formation and bone cancer pain. Mol Pain 6, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, and Hall MN (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J 36, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbrook CJ, Pontious C, Kovalenko I, Lapienyte L, Dreyer S, Lee HJ, Thurston G, Zhang Y, Lazarus J, Sajjakulnukit P, et al. (2019). Macrophage-Released Pyrimidines Inhibit Gemcitabine Therapy in Pancreatic Cancer. Cell Metab 29, 1390–1399 e1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, and Gygi SP (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Bai J, Qin T, Wang Z, and Han L. (2020). NGF from pancreatic stellate cells induces pancreatic cancer proliferation and invasion by PI3K/AKT/GSK signal pathway. J Cell Mol Med 24, 5901–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Andrade JM, Herrera MB, Ghilardi JR, Vardanyan M, Melemedjian OK, and Mantyh PW (2008). Vascularization of the dorsal root ganglia and peripheral nerve of the mouse: implications for chemical-induced peripheral sensory neuropathies. Mol Pain 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, et al. (2015). Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res 75, 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncina E, Roth L, Gonthier B, and Bagnard D. (2007). Role of semaphorins during axon growth and guidance. Adv Exp Med Biol 621, 50–64. [DOI] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, and Maddocks OD (2014). Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep 7, 1248–1258. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, and Kain SR (1998). Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem 273, 34970–34975. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, and Tamayo P. (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl F, Demir IE, Mayer K, Schuster T, D’Haese JG, Becker K, Langer R, Bergmann F, Wang K, Rosenberg R, et al. (2014). The impact of neural invasion severity in gastrointestinal malignancies: a clinicopathological study. Ann Surg 260, 900–907; discussion 907–908. [DOI] [PubMed] [Google Scholar]
- Lillemoe KD, Cameron JL, Kaufman HS, Yeo CJ, Pitt HA, and Sauter PK (1993). Chemical splanchnicectomy in patients with unresectable pancreatic cancer. A prospective randomized trial. Ann Surg 217, 447–455; discussion 456–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza-Puch F, Rooijers K, Buil LC, Zijlstra J, Oude Vrielink JF, Lopes R, Ugalde AP, van Breugel P, Hofland I, Wesseling J, et al. (2016). Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature 530, 490–494. [DOI] [PubMed] [Google Scholar]
- Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, Mackay GM, Labuschagne CF, Gay D, Kruiswijk F, et al. (2017). Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 544, 372–376. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, 3. [Google Scholar]
- Mauro VP, and Chappell SA (2014). A critical analysis of codon optimization in human therapeutics. Trends Mol Med 20, 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, and Gygi SP (2014). MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 86, 7150–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan DT, Bridges RJ, and Cotman CW (1989). The excitatory amino acid receptors: their classes, pharmacology, and distinct properties in the function of the central nervous system. Annu Rev Pharmacol Toxicol 29, 365–402. [DOI] [PubMed] [Google Scholar]
- Mukouyama YS, Shin D, Britsch S, Taniguchi M, and Anderson DJ (2002). Sensory nerves determine the pattern of arterial differentiation and blood vessel branching in the skin. Cell 109, 693–705. [DOI] [PubMed] [Google Scholar]
- Navarrete-Perea J, Yu Q, Gygi SP, and Paulo JA (2018). Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho)proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. J Proteome Res 17, 2226–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo B, Kim E, Osorio-Vasquez V, Doll S, Bustraan S, Liang RJ, Luengo A, Davidson SM, Ali A, Ferraro GB, et al. (2020). Limited Environmental Serine and Glycine Confer Brain Metastasis Sensitivity to PHGDH Inhibition. Cancer Discov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. (2011). Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasterkamp RJ, Peschon JJ, Spriggs MK, and Kolodkin AL (2003). Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature 424, 398–405. [DOI] [PubMed] [Google Scholar]
- Paulo JA (2014). Nicotine alters the proteome of two human pancreatic duct cell lines. JOP 15, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo JA, and Gygi SP (2017). Nicotine-induced protein expression profiling reveals mutually altered proteins across four human cell lines. Proteomics 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. (2015). Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 524, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz BW, Takahashi R, Tanaka T, Macchini M, Hayakawa Y, Dantes Z, Maurer HC, Chen X, Jiang Z, Westphalen CB, et al. (2018a). beta2 Adrenergic-Neurotrophin Feedforward Loop Promotes Pancreatic Cancer. Cancer Cell 33, 75–90 e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz BW, Tanaka T, Sunagawa M, Takahashi R, Jiang Z, Macchini M, Dantes Z, Valenti G, White RA, Middelhoff MA, et al. (2018b). Cholinergic Signaling via Muscarinic Receptors Directly and Indirectly Suppresses Pancreatic Tumorigenesis and Cancer Stemness. Cancer Discov 8, 1458–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca H, Varsos Z, and Pienta KJ (2008). CCL2 protects prostate cancer PC3 cells from autophagic death via phosphatidylinositol 3-kinase/AKT-dependent survivin up-regulation. J Biol Chem 283, 25057–25073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AE, Ducker GS, Billingham LK, Martinez CA, Mainolfi N, Suri V, Friedman A, Manfredi MG, Weinberg SE, Rabinowitz JD, et al. (2019). Serine Metabolism Supports Macrophage IL-1beta Production. Cell Metab 29, 1003–1011 e1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saloman JL, Albers KM, Li D, Hartman DJ, Crawford HC, Muha EA, Rhim AD, and Davis BM (2016). Ablation of sensory neurons in a genetic model of pancreatic ductal adenocarcinoma slows initiation and progression of cancer. Proc Natl Acad Sci U S A 113, 3078–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saloman JL, Singhi AD, Hartman DJ, Normolle DP, Albers KM, and Davis BM (2018). Systemic Depletion of Nerve Growth Factor Inhibits Disease Progression in a Genetically Engineered Model of Pancreatic Ductal Adenocarcinoma. Pancreas 47, 856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal RP, and Amara SG (1999). Excitatory amino acid transporters: a family in flux. Annu Rev Pharmacol Toxicol 39, 431–456. [DOI] [PubMed] [Google Scholar]
- Seymour AB, Hruban RH, Redston M, Caldas C, Powell SM, Kinzler KW, Yeo CJ, and Kern SE (1994). Allelotype of pancreatic adenocarcinoma. Cancer Res 54, 2761–2764. [PubMed] [Google Scholar]
- Sinha S, Fu YY, Grimont A, Ketcham M, Lafaro K, Saglimbeni JA, Askan G, Bailey JM, Melchor JP, Zhong Y, et al. (2017). PanIN Neuroendocrine Cells Promote Tumorigenesis via Neuronal Cross-talk. Cancer Res 77, 1868–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. (2016). Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Deloughery A, Bradshaw N, Chen Y, O’Shea E, Losick R, and Chai Y. (2013a). A serine sensor for multicellularity in a bacterium. Elife 2, e01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Pan T, and Cluzel P. (2013b). Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc Natl Acad Sci U S A 110, 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Zid BM, and O’Shea EK (2014). An integrated approach reveals regulatory controls on bacterial translation elongation. Cell 159, 1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MR, Mattaini KR, Dennstedt EA, Nguyen AA, Sivanand S, Reilly MF, Meeth K, Muir A, Darnell AM, Bosenberg MW, et al. (2019). Increased Serine Synthesis Provides an Advantage for Tumors Arising in Tissues Where Serine Levels Are Limiting. Cell Metab 29, 1410–1421 e1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, and Jeon NL (2005). A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods 2, 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphoff CC, and Drexler HG (2011). Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol Biol 731, 93–103. [DOI] [PubMed] [Google Scholar]
- Venkataramani V, Tanev DI, Strahle C, Studier-Fischer A, Fankhauser L, Kessler T, Körber C, Kardorff M, Ratliff M, Xie R, et al. (2019). Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573, 532–538. [DOI] [PubMed] [Google Scholar]
- Venkatesh HS, Morishita W, Geraghty AC, Silverbush D, Gillespie SM, Arzt M, Tam LT, Espenel C, Ponnuswami A, Ni L, et al. (2019). Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenig P, and Odermatt J. (2010). OpenChrom: a cross-platform open source software for the mass spectrometric analysis of chromatographic data. BMC Bioinformatics 11, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Yamada K, Furuya S, Mitoma J, Hirabayashi Y, and Watanabe M. (2001). 3-Phosphoglycerate dehydrogenase, a key enzyme for l-serine biosynthesis, is preferentially expressed in the radial glia/astrocyte lineage and olfactory ensheathing glia in the mouse brain. J Neurosci 21, 7691–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Sakai K, Furuya S, and Watanabe M. (2003). Selective expression of L-serine synthetic enzyme 3PGDH in Schwann cells, perineuronal glia, and endoneurial fibroblasts along rat sciatic nerves and its upregulation after crush injury. Archives of Histology and Cytology 66, 429–436. [DOI] [PubMed] [Google Scholar]
- Yang M, and Vousden KH (2016). Serine and one-carbon metabolism in cancer. Nat Rev Cancer 16, 650–662. [DOI] [PubMed] [Google Scholar]
- Yu K, Chen B, Aran D, Charalel J, Yau C, Wolf DM, van ‘t Veer LJ, Butte AJ, Goldstein T, and Sirota M. (2019). Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat Commun 10, 3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods S1. Related to STAR Methods. Gating strategy for flow cytometry analysis. Gating strategy used for GFPd2-mCherry flow cytometry analysis. Single cells were gated on FSC:H and SSC:H followed by FSC:W and SSC:H. Live (DAPI-) cells were used to set up gates for mCherry expression. mCherry+ cells were selected to determine the geometric mean of GFP fluorescence.
Supplementary Table 1. Related to Figure 2. Information of PDAC cell lines used in this study.
Supplementary Table 2. Related to Figure 5. Calculated Z-Scores of stalled Ser codons across 18,660 human coding sequences.
Supplementary Table 3. Related to Figure 5. Overlapping list of proteins measured by quantitative proteomics and calculated Z-Scores from Supplementary Table 2.
Supplementary Table 4. Related to Figure 6. Composition of amino acid (AA) and Ser/Gly-free (-SG) mouse diets.
Supplementary Table 5. Related to STAR Methods. List of oligonucleotides used in study.
Supplementary Figure 1. A subset of human, but not mouse, PDAC cells require exSer for growth. Related to Figure 1 and 2.
A, Representative traces (left) of spontaneous action potentials, depolarization-evoked action potentials and Na+ currents in the absence (black), presence (red), and washout (blue) of 1μM TTX in rat dorsal root ganglion (DRG) neurons. Quantification of spontaneous action potential (AP) frequency (n=4 cells), evoked AP amplitudes (n=6 cells), and Na+ current densities (n=10 cells) is presented on the right.
B, Mouse KPC PDAC cells grown in the presence or absence of Ser and/or Gly (n=3 technical replicates for three independently derived cell lines).
C, exSer-dependent PDAC cells were grown in media containing high (25mM) or low (2.78mM) glucose with or without 1mM pyruvate and Ser/Gly for the indicated times (n=3 for each group and condition).
D, exSer-dependent PDAC cell growth was measured after 24 hours in several different media concentrations of Ser (n=4 for each condition).
E, exSer-dependent PDAC cells were cultured in Ser/Gly-rich and –deprived conditions for 18 days (n=3).
F, Immunoblot of SBP enzymes, PHGDH, PSAT1, and PSPH, in exSer-independent and –dependent PDAC cells grown in Ser/Gly-deprived conditions for the indicated times. p38 serves as a loading control.
G, PDAC cells were labelled with 13C6-glucose for 24 hours and total levels of unlabeled (black) and labelled (red) intracellular Ser was measured using GC-MS. (n=5 biologically independent wells).
H, exSer-dependent PDAC cells expressing empty vector, Flag-PSAT1, PHGDH, or both were adapted in Ser/Gly-deprived conditions for 21 days. Growth was observed in a subset of adapted exSer-dependent PDAC cells expressing Flag-PSAT1 and PHGDH. (n=3).
I, exSer-dependent PDAC cells expressing empty vector, Flag-PSAT1, PHGDH, or both in Ser/Gly-rich and –depleted conditions for the indicated times (n=3). exSer-dependent PDAC cells expressing both PHGDH and Flag-PSAT1 were adapted in Ser/Gly-deprived conditions for 21 days before assessing growth.
J, Immunoblot of SBP enzymes, PHGDH, PSAT1, and PSPH, in exSer-independent and -dependent PDAC cells expressing empty vector, Flag-PSAT1, PHGDH, or both grown in Ser/Gly-rich and -deprived conditions for 24 hours. exSer-dependent PDAC cells expressing both PHGDH and Flag-PSAT1 were adapted in Ser/Gly-deprived conditions for 21 days before assessing levels of protein. p38 serves as a loading control.
K-L, PHGDH expression in basal and classical subtypes of the PDAC cell lines used in this study (K) and the TCGA database (L).
M-N, PHGDH (M) and PSAT (N) expression and methylation levels in exSer-independent and –dependent PDAC cell lines measured from the CCLE database. The highest methylation score was plotted against expression.
O, PHGDH expression and methylation levels in PDAC tumors from the TCGA database.
“n” represents the number of biologically independent replicates. Graphs (mean ± s.e.m.) were compared using one (A) or two (B, C, G)-way ANOVA, followed by Dunnett’s (A) or Bonferroni (B, C, G) post-hoc test. Pearson correlation values are represented by r (M-O). (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Supplementary Figure 3. Acute increase in the oxygen consumption rate (OCR) by exSer is not directly acting on mitochondria or the mTOR signaling pathway. Related to Figure 4.
A, Total levels of unlabeled (black) and labelled (red and blue) citrate from 13C6-glucose in PDAC cells measured in the presence or absence of Ser and/or Gly after 24 hours (n=5 biologically independent wells).
B, OCR of PDAC cells that were Ser/Gly-deprived for 24 hours. PDAC cells were stimulated with the respective metabolite(s), oligomycin A, FCCP, and rotenone and antimycin A at the indicated times. Related to Figure 4D.
C, Fold change in NADH/NAD+ was assessed in exSer-independent and -dependent PDAC cells grown in Ser/Gly-rich and –poor conditions for 24 hours (n=6).
D, Positive association of the acute oxygen consumption rate (OCR) response upon Ser stimulation, and the fold change of NADH/NAD+ ratio in exSer-independent and – dependent PDAC cells grown in the absence and presence of Ser/Gly from C.
E, Mitochondrial OCR from permeabilized exSer-dependent PDAC cells (PATU-8902) that were deprived of Ser and Gly for 24 hours and stimulated with control or Ser as indicated.
F, Acute change in mitochondrial OCR from permeabilized exSer-dependent PDAC cells after stimulation with control or Ser from E (n=3 technical replicates).
G, Immunoblot of mTOR signaling in PATU-8902 cells that were grown in the presence or absence of Ser and Gly for 24 hours and stimulated with 400μM Ser for 1, 5, and 10 mins.
H, OCR measurements of exSer-dependent PDAC cells pretreated with mTOR inhibitors for 1 hour and stimulated with 400μM Ser after 24 hours of Ser and Gly deprivation.
I, Acute response to OCR after pre-treatments with or without mTOR inhibitors as described in H (n=3–5 technical replicates).
J, Gene expression of ATF4, DDIT3 (CHOP), and HSPA5 (BIP) in exSer-independent and –dependent PDAC cells that were grown with or without Ser and Gly for 24 hours (n=3 biologically independent experiments).
K, In Ser-rich conditions, cells have high rates of seryl-tRNA charging and protein synthesis, which results in a high ATP demand. Mitochondria will have higher activity and OCR to keep up with the ATP demand of the cell. By contrast, Ser becomes limiting for seryl-tRNA charging and protein synthesis during Ser-deprived conditions, which would decrease the intracellular ATP demand. In response to lower ATP demand, the mitochondria will be less active and have a lower OCR.
Graphs (mean ± s.e.m.) were compared using one (C) or two-way (J) ANOVA, followed by Bonferroni post-hoc test. Pearson correlation values are represented by r (D). (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Supplementary Figure 2. exSer-dependent PDAC cells require L-Ser from nerves to grow in Ser/Gly-deprived media. Related to Figure 3.
A, Growth of PATU-8902 (exSer-dependent) cells in the absence of L-Ser and Gly or L-Ser alone in presence of several concentrations of exogenous D-Serine after 48 hours (n=3 technical replicates and repeated once).
B, Schematic of outcomes if synaptic gap junctions form between nerves and exSer-dependent PDAC cells 6 days after co-culture in the microfluidics device. Although cell-impermeable, calcein is small enough to pass through gap junctions. If gap junctions are formed between nerves and PDAC cells, then PDAC cells would be labelled with calcein as strongly as the nerves. Control devices were used as a negative control for autofluorescence.
C, Representative image of exSer-dependent PDAC cells cultured in control and rat DRG microfluidic devices after six days, with the neuronal side stained with calcein-AM.
D, exSer-dependent PDAC cells were grown in medium containing different concentrations of serine for the indicated amount of time (n=3 biologically independent experiments). Control samples only had one dose of exSer at the beginning of the experiment. In comparison in the “2X Daily” samples, the same initial concentration of serine was added twice a day.
E, Schematic of the multiple role of serine for proteins, for the generation of glycine and glutathione (GSH), one-carbon cycle and transmethylation cycle. γGC: γ-glutamylcysteine, THF: tetrahydrofolate, CH2-THF: 5,10-methylenetetrahydrofolate, CH-THF: 5,10-methenyltetrahydrofolate, CHO-THF: 10-formyltetrahydrofolate, dTMP: deoxythymidine monophosphate, Met: methionine, SAM: S-adenosylmethionine, SAH: S-adenosylhomocysteine, HCys: homocysteine.
F, Growth of exSer-dependent PDAC cells (PATU-8902) in the presence or absence of Ser and/or Gly and treated with or without 1mM formate or 1X nucleosides after two days (n=3 technical replicates and repeated once).
G, Growth of exSer-dependent PDAC cells in the presence or absence of Ser and/or Gly and treated with or without 1mM formate after two days (n=4).
H, exSer-dependent PDAC cells were labelled with 13C2-Gly for 24 hours with or without Ser, Gly, and/or 1mM formate. Total levels of unlabeled (black) and labelled (red) Ser and Gly were measured using GC-MS (n=5).
I, Schematic of Gly-to-Ser conversion in exSer-dependent PDAC cells in the presence of Ser, Gly, and/or formate.
J, Reduced to oxidized glutathione (GSH:GSSG) ratios and S-adenosylmethionine (SAM) to S-adenosylhomocysteine (SAH) ratios in exSer-independent (PATU-8988T) and -dependent (PATU-8902) cells after 24 hours in the presence or absence of Ser and Gly (n=5 biologically independent wells).
K, exSer-independent and -dependent PDAC cells were labelled with 13C3-Ser for 24 hours in Ser/Gly-rich or –deprived conditions. Total levels of unlabeled (black) and labelled (blue) ATP, GTP, GSH, GSSG, SAM, and SAH were measured using LC-MS (n=5).
L, Fold change of Ser-derived metabolites in exSer-independent and –dependent PDAC cells grown in the absence and presence of Ser/Gly from K, M (n=5).
M, Total levels of NADH and NAD+ from exSer-independent and –dependent PDAC cells labelled with 13C3-Ser for 24 hours in Ser/Gly-rich or –deprived conditions (n=5).
Graphs (mean ± s.e.m.) were compared using two-tailed t-test (J) and two-way ANOVA (F-H), followed by Bonferroni post-hoc test (***p<0.005).
Supplementary Figure 4. Analysis of Ser codon usage bias in exSer-dependent PDAC (PATU-8902) and other cancer cells. Related to Figure 5.
A, Representative flow plots of mCherry levels, an internal control, in cells expressing the GFPd2 Ser codon reporter constructs from Figure 5A. PATU-8902 cells were grown in the presence and absence of Ser and Gly and treated with or without 50μM cycloheximide (CHX).
B, Fold change and ratio of GFPd2 and mCherry expression relative to Ser/Gly-rich conditions in exSer-dependent PDAC cells grown with or without Ser/Gly for 24 hours (n=3 technical replicates and repeated once).
C, Quantification of GFPd2 fluorescence in exSer-dependent PDAC cells (n=3 technical replicates and repeated once for both conditions) grown with or without tyrosine or phenylalanine for 24 hours.
D, Ribosome density analysis of total mRNA as a negative control from PATU-8902 that were grown in the presence or absence of Ser and Gly for 24 hours (n=3 biologically independent replicates).
E, Immunoblots of SBP enzymes, PHGDH, PSAT1, and PSPH in PDAC, breast cancer and melanoma cells grown with or without Ser/Gly for 24 hours. ERK2 serves as a loading control.
F-H, Quantification of GFPd2 fluorescence in exSer-independent PDAC (F, n=3), breast cancer (G, n=5), and melanoma (H, n=3) cells, and grown with or without Ser/Gly and 1μM PH719, a PHGDH inhibitor. Dotted-line represents mean fluorescence of AGC (F) and AGT (G, H) GFPd2 fluorescence.
Where “n” represents the number of biologically independent experiments for each group and condition. Graphs represent mean ± s.e.m. and were compared using one-way ANOVA (D, F-H), followed by Bonferroni post-hoc test (*p<0.05, **p<0.01, ****p<0.0001).
Supplementary Figure 5. Ser codon usage bias occurs when GCN2 is activated, and is not correlated with tRNA gene copy number. Related to Figure 5.
A, GFPd2 fluorescence of the Ser codon-optimized reporters were measured across a range of Ser concentrations in exSer-dependent PDAC cells (n=3 technical replicates, performed at least twice).
B, Immunoblot of BIP and phospho-eIF2α levels in exSer-dependent PDAC cells grown across a range of Ser concentrations for 24 hours. eIF2α serves as a loading control.
C-D, Quantification of phospho-eIF2α (C) and BIP (D) levels normalized to eIF2α from B (n=4 biologically independent replicates). Graphs represent mean ± s.e.m. and the EC50 was determined.
E-G, Plots of summed ribosome densities of the ribosome protected mRNA against the expected codon frequency (E), cognate Ser tRNA gene copy number (F) or the calculated codon usage frequency (G) of the Ser codons. Line of best fit is shown in black and the data was analyzed using Pearson’s correlation.
H, Total fractional abundance of charged Ser-tRNA isodecoders (AGA, CGA, GCT, and TGA) in exSer-dependent PDAC cells grown in Ser/Gly-rich and –depleted conditions (n=3).
I-J, Ribosome density was plotted against percent tRNA charging (I), and the change in total fractional abundance (J) of charged Ser-tRNA isodecoders (AGA, CGA, GCT, and TGA) in exSer-dependent PDAC cells grown in Ser/Gly-rich and –depleted conditions (n=3).
K, Distribution of Z-scores reflecting pause-site Ser codon usage bias in ~18,000 CDS (see methods for details). TCY: TCT and TCC, TCR: TCA and TCG.
L, Waterfall plots of the change in protein abundance for the top 50 genes predicted to be most and least affected by differential mRNA translation of Ser codons, TCC and TCT. Protein levels were measured exSer-dependent PDAC cells grown in Ser/Gly-rich or –deprived conditions after 24 and 48 hours (n=3). A Chi square test was used to assess the significance of the observed (red line) to expected (black line) frequencies. p-values are indicated in each graph.
M, Immunoblot of RAS levels and ERK activation in exSer-dependent PDAC cells expressing shGFP or shKRAS grown with or without Ser/Gly for 24 hours. ERK2 serves as a loading control.
N, Quantitative proteomics of RAS isoforms in exSer-dependent PDAC cells grown in Ser/Gly-rich and –deprived conditions for 24 (n=3) and 48 (n=2 for +Ser/Gly and n=3 for –Ser/Gly) hours by mass spectrometry. Graphs represent mean ± s.e.m.
O, NGF levels in the media of exSer-independent PDAC cells grown in different concentrations of Ser after 24 hours (n=4–5).
P, NGF mRNA levels were measured in exSer-dependent PDAC cells after being grown in Ser/Gly-rich and –poor media for 24 hours (n=3).
Q, exSer-dependent PDAC cells were grown in Ser/Gly-rich and –deprived media for 24 hours. Secreted NGF levels were measured at the indicated time points from exSer-dependent PDAC cells expressing empty vector, wild-type (WT containing 3 stalled and 16 non-stalled Ser codons), TCC-, and TCA- codon optimized NGF followed by IRES-mCherry. Levels of secreted NGF was normalized to mCherry signal (n=4).
Where “n” presents the number of biologically independent replicates. Graph represent (mean ± s.e.m.) and was compared using a one-sample t-test (P), one- (H), or two-way ANOVA (Q), followed by Bonferroni post-hoc test. Pearson correlation values are represented by r (I-J). (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Supplementary Figure 6. The effects of –SG (Ser/Gly-free)-diet on exSer-dependent PDAC tumors. Related to Figure 6.
A, Plasma concentrations of Ser and Gly from mice on AA (n=5–10) or –SG (n=5–9) diet at the indicated times. As a control, plasma concentrations of Ser and Gly also were measured from non-tumor bearing mice on AA and –SG diet (n=3) at the indicated times.
B, Changes in AA plasma concentrations at week 5 from non-tumor bearing (n=3–5), and exSer-dependent tumor-bearing mice on AA (n=5–10) or –SG (n=5–9) diet from A.
C, Representative images of H&E, trichrome, phospho-Histone 3 and cleaved caspase 3 staining of PATU-8902 tumors from mice on AA and -SG diet after five weeks post-injection (from Figure 6C).
D, Representative images of nerves (PGP9.5) of PATU-8902 tumors from mice on AA and -SG diet after five weeks post-injection (from Figure 6E).
E-F, Representative images of immunohistochemistry staining for sympathetic (E, tyrosine hydroxylase (Th+)) and sensory (F, transient receptor potential cation channel subfamily V member 1 (Trpv1+)) nerves in exSer-dependent PDAC tumors from mice on control and Ser/Gly-free diet.
G, Two representative images of lack of parasympathetic (vesicular acetylcholine transporter (VAchT+)) nerve staining in exSer-dependent tumors grown in mice on Ser/Gly-free diet. Mouse brain serves as a positive control and PGP9.5 staining serves as a nerve staining control.
H, 3D-images of clarified exSer-dependent PDAC tumors stained with PGP9.5 to detect innervation of tumors from mice on AA and –SG diet after five-week post-injection. The magnified image of tumor #4 (AA diet) and tumor #1 (-SG diet) is also shown in Figure 6D.
I, Protein abundance fold change of axon guidance cues in exSer-dependent PDAC cells grown in Ser/Gly-deprived conditions for 24 and 48 hours (n=3).
“n” presents the number of biologically independent replicates. Graph represent (mean ± s.e.m.) and was compared using permutation-based FDR (I) or two-way ANOVA (A), followed by Bonferroni post-hoc test (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Supplementary Figure 7. The effects of –SG-diet and LOXO-101 on PDAC cells in vitro and in vivo. Related to Figure 6 and 7.
A, In vitro growth of exSer-independent and -dependent PDAC cells treated with different concentrations of LOXO-101 in the presence or absence of Ser and Gly (n=3 technical replicates and repeated once for five independent cell lines).
B, Ser (S) and Gly (G) concentrations were measured from axCM (axon-derived conditioned media) of rat DRGs treated with Veh or LOXO-101 as shown (n=4–5). As a positive control for LOXO-101, rat DRG neurons were treated with LOXO-101, followed by 30min NGF stimulation as indicated. Phosphorylation of TrkA at Y684/685 was used to assess Trk activity and the effect of LOXO-101. ERK2 serves as a loading control.
C, Immunoblot of TRK levels in exSer-dependent PDAC cells grown with or without Ser/Gly, and tumors from mice on AA or –SG diet treated with or without LOXO-101. ERK2 serves as a loading control. Mouse brain was used a positive control.
D, Representative TRK staining of exSer-dependent PDAC tumors from mice on AA or –SG diet. Mouse brain was used a positive control.
E, Plasma concentrations of Ser and Gly from mice on AA or -SG diet and treated with vehicle or LOXO-101 at the indicated times (n≥6 for each group and timepoint).
F, Quantification of cleaved caspase 3 staining in tumors from mice fed amino acid (AA) or Ser/Gly-free (-SG) diet, treated with vehicle or LOXO-101 from G (n≥6).
G, Representative images of p-Histone 3, cleaved caspase 3, and phospho-eIF2α staining of PATU-8902 tumors from Figure 6.
H, Overall survival of PHGDH high and low expressing PDAC tumors from TCGA dataset.
I-J, Observed frequencies of the differentiation state (I) and tumor size (J) of PHGDH low and high PDAC tumors from patients at MD Anderson. A Chi square test was used to assess the significance of the observed to expected frequencies.
Where “n” represents the number of biologically independent experiments for each group and condition. Graphs (mean ± s.e.m. for A, D) or (median ± max/min for B, F) were compared using one-way (B, F) or two-way ANOVA (E), followed by Bonferroni or Holm-Sidak post-hoc test (*p<0.05, **p<0.01, ***p<0.005, ****p<0.0001).
Data Availability Statement
The accession number for the raw high throughput sequencing files for ribosome profiling and charge DM-tRNA-seq experiments is GEO: GSE139910. All codes used in this study were publicly available from published studies as indicated.