Abstract
Background
Our previous work shows Autophagy enhanced resistance to cisplatin in seminoma. The expression of the N6-methyladenosine (m6A) methyltransferases METTL3 was significantly increased in the cisplatin-resistant TCam-2 cell line of seminoma. We aimed to investigate the role of m6A methylation in autophagy and the chemosensitivity of seminoma cells.
Methods
Plasmid and siRNA were used to overexpress and knockdown METTL3. Autophagy was detected by western blot and immunofluorescence, respectively. The expression of downstream targets of METTL3 was detected by quantitative real-time PCR (qRT-PCR) and western blot, and the m6A level of them was detected by MeRIP-qPCR. Chemosensitivity of the TCam-2 cell line was identified through MTT assay.
Results
Upon METTL3 overexpression, autophagy of TCam-2 cell line was enhanced and its sensitivity to cisplatin was decreased. The use of autophagy inhibitors 3-methyladenine (3-MA) could reverse the protective effect of METTL3 on TCam-2 cells. We found that the up-regulation of METTL3 could increase the m6A modification level of ATG5 transcript, thus increased expression of ATG5. Moreover, knockdown of ATG5 reduced METTL3-induced autophagy, suggesting that ATG5 was a potential target for METTL3 to promote autophagy.
Conclusions
In summary, our research unveiled the unique mechanism by which m6A methylation regulates autophagy and chemosensitivity of the TCam-2 cell line and METTL3 was a potential target to overcome the cisplatin resistance of seminoma.
Keywords: Seminoma, m6A methylation, autophagy, chemosensitivity, METTL3
Introduction
Testicular germ cell tumor (TGCT) is one of the most common malignant solid tumors among adolescents and young men between 15 to 40 years of age, and it is the main cause of cancer-related mortality and morbidity in this age group (1,2). Epidemiological investigation shows that the incidence of TGCT has been increasing in recent years, especially in men of European origin (3). Seminoma accounts for about 55% of TGCTs. Because of its unique sensitivity to cisplatin [cis-diamminedichloroplatinum II, (CDDP)], seminoma is curable even at the stage of advanced metastatic disease (1,2,4,5). Cisplatin-based chemotherapy makes up an important part of adjuvant therapy for seminoma. Although most seminomas are sensitive to cisplatin therapy, the recurrence of a small number of refractory patients and the emergence of chemotherapy resistance can still become obstacles to the treatment of seminoma (4,6). In the present study, the concrete mechanism of cisplatin resistance in seminoma remains largely undefined and needs to be explored.
N6-methyladenosine (m6A) modification is usually enriched in the 5' end of the terminal exon, and it is the most common post-transcriptional modification of mRNA in mammals and plays an important role in a variety of physiological processes and development of diseases (7-9). m6A modification is a reversible process, which is dynamically regulated by three kinds of m6A regulators: the RNA methyltransferases (writers), the demethylases (erasers), and the m6A-binding proteins (readers). The m6A ‘writer’ is responsible for the installation of m6A, the m6A ‘reader’ can specifically bind to m6A and perform different biological functions, and the m6A ‘eraser’ is responsible for the removal of m6A (7,9). Studies have shown that m6A methylation can regulate the expression of cancer-related genes, and then promotes or inhibits the tumorigenesis of cancer (10,11). As a writer of m6A, METTL3 functions as a promoter in certain types of cancers, such as ovarian cancer, colorectal cancer, gastric cancer, and liver cancer (12-15).
Similar to apoptosis, autophagy is a significant cellular physiological process. Autophagy can capture and degrade intracellular proteins and organelles, and recycle the degradation products in biosynthesis, which plays an important role in maintaining intracellular metabolic homeostasis (16). Recent research shows that autophagy can promote resistance to chemotherapeutic drugs in a variety of tumors, and inhibition of autophagy may be a new potential target for tumor therapy (17,18). Our previous work has shown that autophagy can reduce the chemosensitivity to cisplatin therapy in seminoma, which is a crucial mechanism of drug resistance in seminoma (19). However, no research has reported the mutual regulatory network of m6A methylation and autophagy in seminoma, and the underlying relationship between them is worthy of further investigation.
In this study, we found that METTL3 was highly expressed in the cisplatin-resistant TCam-2 cell line (TCam-2/CDDP) which has been constructed in our previous study. Overexpression of METTL3 could desensitize the TCam-2 cell line, and this phenomenon was caused by autophagy induced by METTL3. We further explored the mechanism of autophagy induced by METTL3 in TCam-2 cells. Several autophagy-associated genes were detected and ATG5 was found overexpressed in TCam-2/CDDP cell line. Overexpression of METTL3 up-regulated ATG5’s expression and m6A modification level. ATG5 knockdown alleviated autophagy induced by METTL3. Therefore, we identify ATG5 as a downstream target of METTL3. Taken together, our data highlight a functional role for METTL3 in seminoma and uncover METTL3 as a therapeutic target for seminoma. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1411).
Methods
Cell culture
The human TCam-2 cell line was kindly gifted by Dr. Riko Kitazawa, Department of Diagnostic Pathology, Ehime University Hospital, Matsuyama, Japan. Cisplatin-resistant TCam-2 cell line (TCam-2/CDDP) was obtained from TCam-2 cells by long-term intermittent exposure to low-dose CDDP (20). The two cell lines were cultured in RPMI-1640 medium (Roswell Park Memorial Institute Medium-1640) containing 10% fetal bovine serum and 1% penicillin/streptomycin. TCam-2/CDDP cells were routinely treated with low-dose cisplatin to maintain their drug resistance. All cells were cultured in a 37 °C incubator containing 5% CO2. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Real-time quantitative PCR (RT-qPCR)
Total RNA was extracted from cell samples by Trizol (Invitrogen). cDNA was synthesized by RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas). SYBR Green PCR Master Mix (ABI) was used for Realtime-PCR. The expression level of mRNA was calculated by 2–ΔΔCt, and the results were standardized as the expression of GAPDH. The primers used for PCR are shown in Table 1.
Table 1. Primers used for PCR.
| Gene | Primers sequence (5' to 3') |
|---|---|
| GAPDH | |
| Forward | GGAGCGAGATCCCTCCAAAAT |
| Reverse | GGCTGTTGTCATACTTCTCATGG |
| ATG12 | |
| Forward | TAGAGCGAACACGAACCATCC |
| Reverse | CACTGCCAAAACACTCATAGAGA |
| ATG5 | |
| Forward | AAAGATGTGCTTCGAGATGTGT |
| Reverse | CACTTTGTCAGTTACCAACGTCA |
| NRBF2 | |
| Forward | CAGACGAGCAGACCGTTTATT |
| Reverse | TGCTGGGCTTTCAATCTTTCTT |
| VMP1 | |
| Forward | GACCAGAGACGTGTAGCAATG |
| Reverse | ACAATGCTTTGACGATGCCATAA |
| SH3GLB1 | |
| Forward | ATTACCAGACTTCTGCTAGAGGG |
| Reverse | GGATGGAAAACTTCCCAGTTGTT |
| MAP3K7 | |
| Forward | ATTGTAGAGCTTCGGCAGTTATC |
| Reverse | CTGTAAACACCAACTCATTGCG |
| FADD | |
| Forward | GCTGGCTCGTCAGCTCAAA |
| Reverse | ACTGTTGCGTTCTCCTTCTCT |
Western blot analysis
RIPA lysis buffer containing protease inhibitor was used to extract total proteins from cell samples. After adding a loading buffer, the protein samples were heated at 100 °C for 3 minutes for degeneration. The equal quantities of protein (20 µg) were separated by 10% SDS-PAGE and transferred to the PVDF membranes. Blocking the membranes with 5% non-fat milk (PBS configuration) at room temperature for 2 hours, then incubating the membranes with the indicated primary antibody for 2 hours and washing them with PBST for 4 times. The membranes were added with HRP-conjugated secondary antibody and incubated overnight at 4 °C, then washed with PBST for 4 times. Finally, an appropriate dose of enhanced chemiluminescence reagent was added to visualize the protein bands, and the positive bands were analyzed by Gelpro4.0 gel optical density analysis software. Antibodies used in this study were ATG12 (Abcam, ab109491), ATG5 (Abcam, ab109490), FADD (Ptgcn, 14906-1-AP), METTL3 (Omnimabs, OM284614), LC3B (CST, #3868S), Beclin1 (Abcam, ab210498), GAPDH (Abcam, ab125247).
Cell viability assay
MTT method was used to observe the chemosensitivity of TCam-2 cell line and TCam-2/CDDP cell line to cisplatin under different conditions. The cells were inoculated in a 96-well plate (1×104/well) and cultured in a 37 °C incubator containing 5% CO2 for 24 hours. The cells were treated with CDDP for 0, 12, 24, 48, and 72 hours, respectively. According to the experimental instructions, MTT was added to incubate for 4 hours and DMSO was added to dissolve formazan. The absorbance of each well was measured at 570 nm by enzyme-labeling instrument.
RNA m6A quantification and MeRIP-qPCR
The m6A modification level of total RNA was measured by EpiQuik m6A RNA Methylation Quantification Kit. The total RNA was bound in the detection hole with a solution with a high binding degree of RNA. m6A was detected by a specific capture m6A antibody. The absorbance was read at 450 nm, then the OD value was measured by colorimetry and the total m6A content was calculated. EZ-Magna RIP kit (Millipore) was used to evaluate the modification level of m6A in the specific gene according to the manufacturer’s instructions. In short, the RNA containing m6A methylated was coated with m6A antibody and immunoprecipitated on magnetic beads A/G. The enriched RNA-antibody complex was digested with protease to obtain m6A modified RNA. Then qRT-PCR was used to quantify the enriched RNA.
Cell transfection
The plasmid overexpressing METTL3 was constructed and provided by Nkbiotechnology company (Changsha, China). siRNAs for METTL3 and ATG5 were designed. The siRNA sequences were as follows: siRNA-METTL3, sense: 5'-GCAAGUAUGUUCACUAUGATT-3', antisense: 5'-UCAUAGUGAACAUACUUGCAG-3'; siRNA-ATG5, sense: 5'-GUCCAUCUAAGGAUGCAAUTT-3', antisense: 5'-AUUGCAUCCUUAGAUGGACTT-3'. Both plasmids and siRNA were transfected with LipofectamineTM 3000 reagent and transfected when the cells met at 70% confluent.
Immunofluorescence
Immunofluorescence staining: the cells in groups of different treatments were fixed with 4% paraformaldehyde for 20 minutes. After PBS washing, 0.5% TritonX-100 was used to permeate TCam-2 cells at room temperature for 15 minutes. The cells were blocked with non-immune goat serum (Maixin, Fujian) at room temperature for 20 minutes and then incubated with the primary antibody (Anti-LC3B, Abcam) and the secondary antibody (Goat Anti-Rabbit IgG H&L, Alexa Fluor® 555). Then the nucleus was stained with DAPI. The morphology of the cells was observed under a laser confocal microscope.
Statistical analysis
The data were analyzed by GraphPad Prism 8 software. All the experiments were repeated 3 times, and the results were expressed as mean ± standard deviation (S.D.). The results were analyzed by student’s t-test or one-way ANOVA. P value less than 0.05 is considered to be statistically significant.
Results
METTL3 confers resistance to cisplatin in TCam-2 cell line
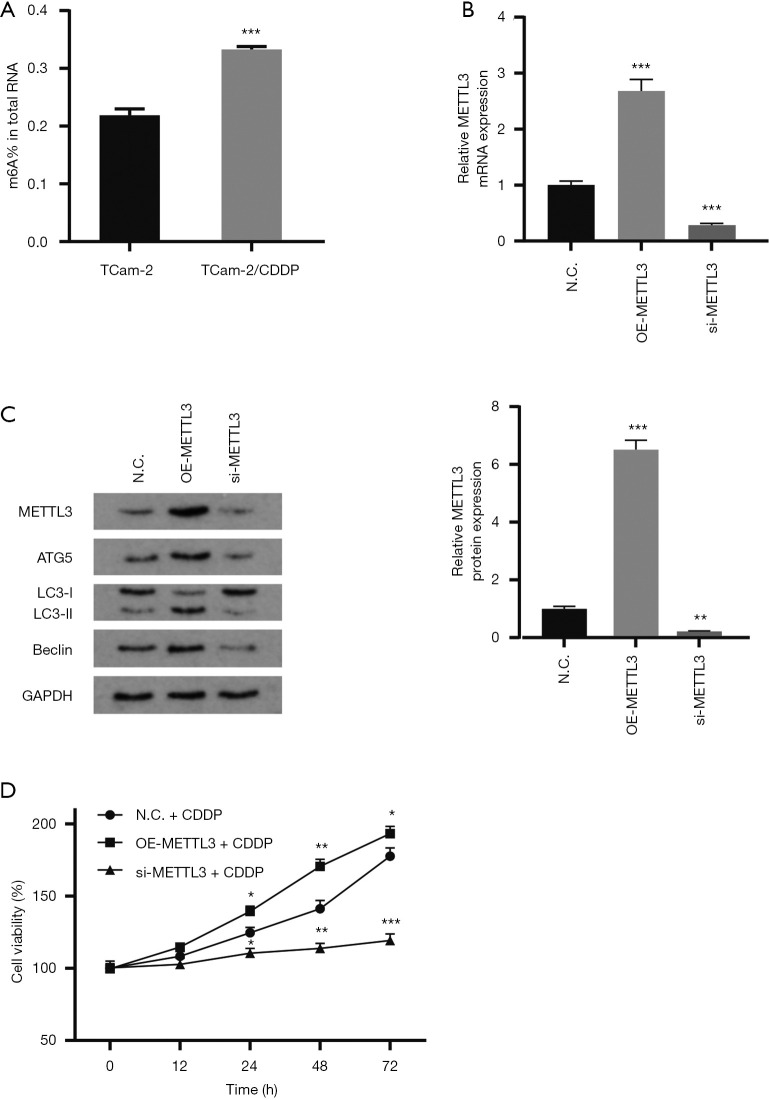
M6A modification is involved in the occurrence and development of tumor drug resistance (21,22). The expression of methyltransferase METTL3 in cisplatin-resistant TCam-2/CDDP cell line was significantly higher than that in cisplatin-sensitive TCam-2 cell line in our previous work, which suggested the potential function of m6A modification in the occurrence of drug resistance in seminoma (23). To elucidate the relationship between m6A methylation and chemosensitivity of seminoma cells, we detected m6A modification of total RNA in TCam-2 cell line and TCam-2/CDDP cell line. It was found that the m6A modification in total RNA of TCam-2/CDDP cells was significantly higher than that of TCam-2 cells (Figure 1A). To further verify our conjecture, siRNA or plasmid was used for knockdown (si-METTL3) or overexpression (OE-METTL3) of METTL3 in TCam-2 cell lines, respectively. The transfection efficiency was validated by qRT-PCR and western blot (Figure 1B,C). Based on previous studies, we have determined the IC25 concentration of cisplatin-treated TCam-2 cell line (2.55 µmol/L) (24). KO-METTL3 group and OE-METTL3 group were treated with cisplatin at IC25 concentration. The toxicity of cisplatin to these two groups of cells was detected by MTT assay. The result showed that the cell proliferation rate in METTL3 overexpression group was significantly higher than that in METTL3 knockdown group (Figure 1D). In summary, these results indicate that the overexpression of METTL3 can enhance the resistance to cisplatin in TCam-2 cells.
Figure 1.
Upregulation of METTL3 expression in TCam-2 cells is associated with cisplatin resistance. (A) m6A modification level in total RNA of TCam-2 cells and TCam-2/CDDP cells. (B,C) Relative mRNA and protein expression of METTL3 in N.C. cells, OE-METTL3 cells and si-METTL3 cells to show the transfection efficiency. GAPDH served as the internal control. The results are normalized to the expression of N.C. cells. (D) Cell viability analysis of TCam-2 cells treated with IC25 concentration of CDDP. The data are shown as the means ± S.D. *, P<0.05; **, P<0.01; ***, P<0.001. CDDP, cis-diamminedichloroplatinum II; m6A, N6-methyladenosine; S.D., standard deviation.
METTL3 potentiates resistance to cisplatin by enhancing autophagy
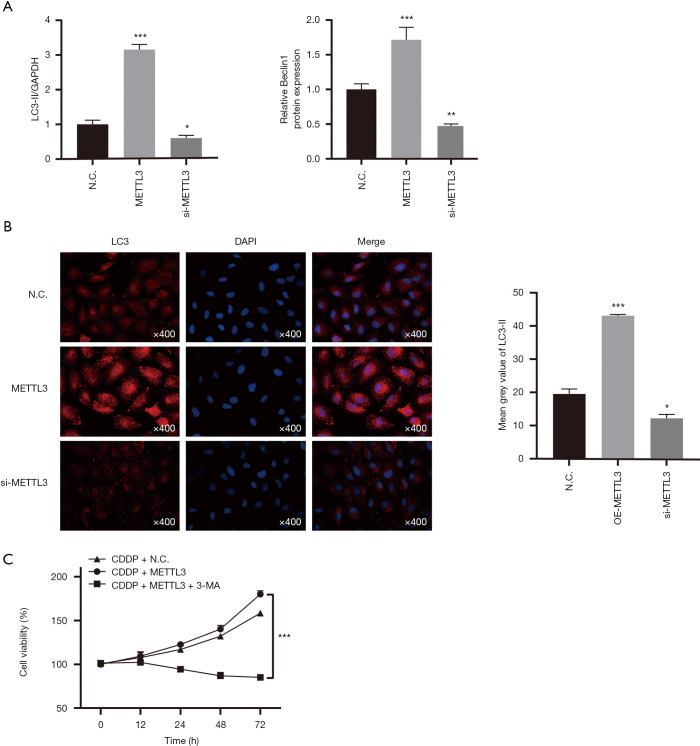
Autophagy has been well documented to help support the survival of cancer cells in radiation and chemotherapy reactions (17). In particular, our previous studies have shown that promoting autophagy can induce resistance to cisplatin in TCam-2 cells. Here, we want to figure out whether autophagy is involved in the process of METTL3 regulating the chemosensitivity of the TCam-2 cells to cisplatin. Gain- and loss-function experiments were conducted in TCam-2 cells to demonstrate the role of METTL3 in autophagy. To measure the autophagy level in TCam-2 cells, we analyzed the protein expression of autophagy-associated proteins LC3 (microscopic-associated protein 1 light chain 3) and Beclin1 by western blot (Figure 1C and Figure 2A). Compared with the control group, the silencing of METTL3 significantly decreased the expression of LC3-II and Beclin1. By contrast, the overexpression of METTL3 significantly increased the expression of LC3-II and Beclin1. Similarly, in immunofluorescence staining, TCam-2 cells with METTL3 overexpression had more LC3-II fluorescence spots (Figure 2B). These results indicate a positive correlation between METTL3 and autophagy. In addition, in MTT assay, autophagy inhibitor 3-methyladenine (3-MA) could reverse the enhancement of cisplatin resistance induced by METTL3 overexpression (Figure 2C). These data support that METTL3 up-regulates the cisplatin resistance of TCam-2 cells by enhancing autophagy.
Figure 2.
METTL3 promotes resistance to cisplatin by enhancing autophagy. (A) LC3-II/GAPDH ratio and Beclin1 levels were determined by western blot analysis after overexpression or knockdown of METTL3 in TCam-2 cells. GAPDH served as the internal control. (B) Immunofluorescence of LC3-II was performed in TCam-2 cell line with different METTL3 expression. The red signal represents the LC3-II levels. (magnification, 400×). (C) Cell viability after 72 h treatment with IC25 concentration of CDDP. The METTL3-overexpressing TCam-2 cell were treated with or without autophagy inhibitor 3-MA. The data are shown as the means ± S.D. *, P<0.05; **, P<0.01; ***, P<0.001. 3-MA, 3-methyladenine; S.D., standard deviation; CDDP, cis-diamminedichloroplatinum II.
Mining ATG5 as a downstream target of METTL3-mediated m6A modification
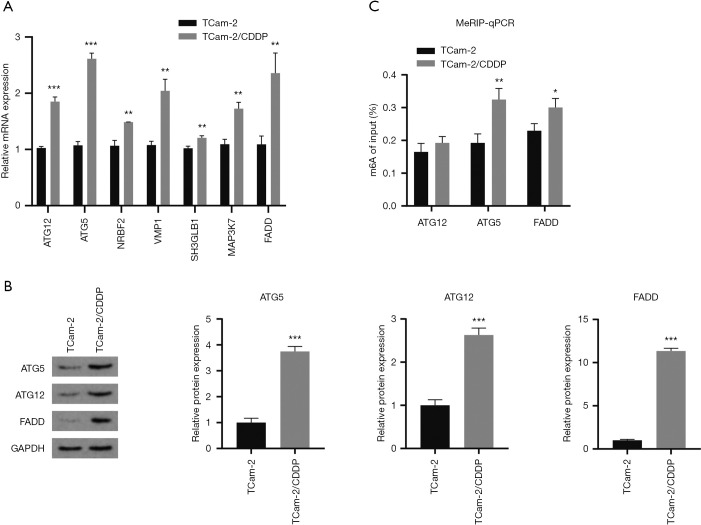
To date, researchers have suggested that m6A modification can induce the occurrence of autophagy by regulating the expression of autophagy-associated genes (25,26). We speculated that METTL3 can promote the expression of autophagy-associated genes through m6A modification and thus induce autophagy in TCam-2 cells. According to our previous gene microarray results, we found autophagy-associated gene ATG12, ATG5, NRBF2, VMP1, SH3GLB1, MAP3K7, and FADD were increased in TCam-2/CDDP cell line (27). Moreover, predicted by online tools (RMBase V2.0), these genes have potential m6A modification sites. We detected the transcriptional expression of these genes in TCam-2 cells and TCam-2/CDDP cells by qRT-PCR. The expression of mRNA in TCam-2/CDDP cells group was standardized in TCam-2 cells group. Compared with TCam-2 cells, the expression levels of these genes in TCam-2/CDDP cells increased in varying degrees (Figure 3A). The molecules with significant difference in mRNA expression were selected for western blot detection. It was found that the protein levels of ATG5, ATG12, and FADD in TCam-2/CDDP cells were higher than those in TCam-2 cells (Figure 3B). We further detected the m6A modification level of these three genes. MeRIP-qPCR showed that ATG5 and FADD had higher m6A abundance in TCam-2/CDDP group (Figure 3C). Among them, ATG5 had the most obvious difference, which indicated ATG5 as the key target for m6A modification to induce autophagy. Therefore, we chose ATG5 for our further research.
Figure 3.
Mining potential downstream targets of METTL3-mediated m6A modification. (A) Relative mRNA expression of ATG12, ATG5, NRBF2, VMP1, SH3GLB1, MAP3K7 and FADD in TCam-2 and TCam-2/CDDP cell line by qRT-PCR. (B) Western blot analysis of ATG5, ATG12 and FADD in TCam-2 and TCam-2/CDDP cell line. GAPDH served as the internal control. (C) MeRIP-qPCR analysis to show the m6A modification of the specific gene. m6A level of ATG5, ATG12 and FADD was detected in TCam-2 and TCam-2/CDDP cell line. All data were measurement data and expressed as mean ± S.D. *, P<0.05; **, P<0.01; ***, P<0.001. m6A, N6-methyladenosine; CDDP, cis-diamminedichloroplatinum II; qRT-PCR, quantitative real-time PCR; S.D., standard deviation.
METTL3 promotes autophagy and resistance to cisplatin by targeting ATG5
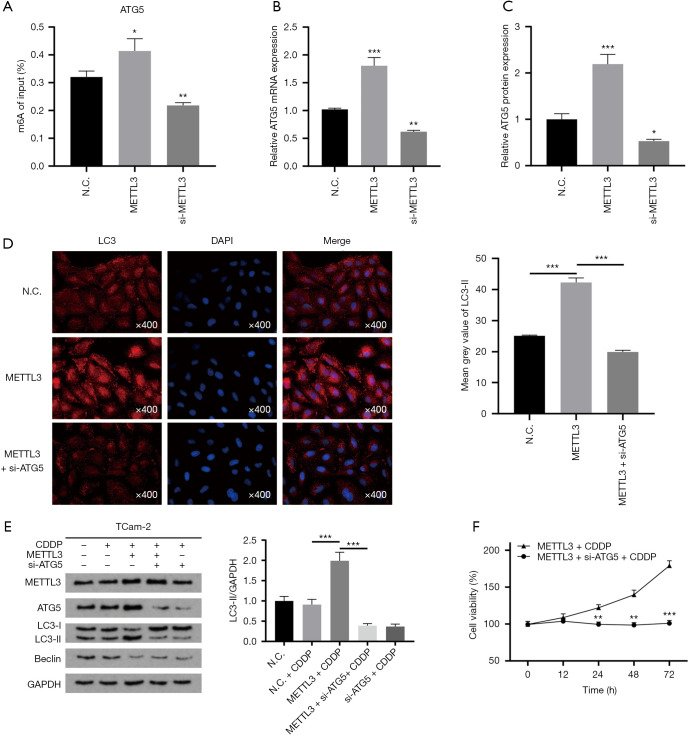
ATG5 functions as an autophagy elongation protein in autophagosome formation and plays a crucial role in cancer (28-30). To identify the role of ATG5 in METTL3-mediated autophagy, we first explored the relationship between METTL3 and ATG5. We examined the change of m6A modification level of ATG5 after overexpression or knockdown of METTL3 in TCam-2 cells. The results show that the level of m6A modification in ATG5 was positively correlated with the expression of METTL3 (Figure 4A). The impact of METTL3 on ATG5 expression was confirmed by qRT-PCR and western blot. qRT-PCR showed that the mRNA level of ATG5 in METTL3 overexpression group was significantly higher than that in the control group, while the mRNA level of ATG5 in METTL3 knockdown group was lower than that in the control group (Figure 4B). Similarly, the increase or decrease of METTL3 expression can up-regulate or down-regulate the protein level of ATG5, respectively (Figure 1C and Figure 4C). These results demonstrate that METTL3 can directly target ATG5 transcripts and promote the expression of ATG5 in an m6A-dependent manner.
Figure 4.
ATG5 is a downstream target of METTL3 for autophagy regulation. (A) m6A modification level of ATG5 in TCam-2 cells upregulating or downregulating METTL3. (B,C) mRNA and protein expression of ATG5 in TCam-2 cells transfected with METTL3 plasmid or si-METTL3. GAPDH served as the internal control. (D) Immunofluorescence image of LC3-II (red) in TCam-2 cells. TCam-2 cells were treated with METTL3 transfection or si-ATG5. (E) Western blot analysis of LC3II/GAPDH ratio and Beclin1. GAPDH served as the internal control. (F) Cell viability of METTL3 overexpressing TCam-2 cells treated with or without si-ATG5 after incubated with CDDP (IC25) for 72 h. All data are shown as the means ± S.D. *, P<0.05; **, P<0.01; ***, P<0.001. m6A, N6-methyladenosine; CDDP, cis-diamminedichloroplatinum II; S.D., standard deviation.
Next, we investigated whether ATG5 is the downstream target of METTL3 by which it regulates autophagy. We used siRNA for ATG5 to transfect TCam-2 cells and detected LC3-II by immunofluorescence. The fluorescence intensity of LC3-II in TCam-2 cells overexpressing METTL3 was increased, while siRNA for ATG5 decreased the fluorescence intensity of LC3-II in TCam-2 cells overexpressing METTL3 (Figure 4D). Western blot analysis further confirmed these findings. The ratio of LC3-II/GAPDH and the expression of Beclin1 were increased in TCam-2 cells overexpressing METTL3, while these changes could be reversed by knockdown of ATG5 (Figure 4E). TCam-2 cells were treated with IC25 concentration of CDDP, and the viability of TCam-2 cells was tested by MTT assay. After ATG5 knockdown, the proliferation rate of TCam-2 cells overexpressing METTL3 decreased significantly (Figure 4F). To sum up, ATG5 functions as the downstream target of METTL3. METTL3 induces autophagy by regulating ATG5’s expression and then enhances the cisplatin resistance of TCam-2 cells.
Discussion
Seminoma has an exquisite sensitivity to cisplatin, and cisplatin-based chemotherapy plays an important role in the treatment of seminoma (4). Unfortunately, not all patients with seminoma have a good result because of cisplatin resistance (6,31). The studies of the mechanism of cisplatin resistance in seminoma will help to provide a new and reasonable combination therapy for refractory seminoma, and even stimulate the development of new treatment strategies for other tumors. In our work, we found that the expression of m6A methyltransferase METTL3 was increased in drug-resistant TCam-2/CDDP cells. This suggests that m6A modification may be involved in the process of chemotherapy resistance of seminoma. Our previous studies have already shown that autophagy is involved in the development of cisplatin resistance in seminoma, whereas it is not clear whether the autophagy in TCam-2/CDDP cells is related to the change of METTL3 expression level (19). In this study, we aimed to investigate whether METTL3 can regulate autophagy and cisplatin chemosensitivity in seminoma.
m6A modification can regulate not only the expression of oncogenes but also the expression of tumor suppressor genes. It functions as a double-edged sword in cancer (10). Although the current research on m6A modification in TGCTs is relatively rare, the same cytogenetic background and heterogeneity between different subtypes suggest that epigenetic modifications may play an important role in the pathogenesis of TGCTs (32). Lobo et al.’s analysis of the TCGA database indicated that the expression of m6A-related markers METTL4, VIRMA, and YTHDF3 are related to the monitoring and identification of TGCTs, and subsequent detection in tissue found that VIRMA and YTHDF3 have a co-occurrent effect in the maintenance of seminoma phenotype (33,34). Nettersheim et al. found that the biomarkers METTL3, ALKB3H5, YTHDC1/F1/F2 and HNRNPC increased at the level of TGCTs’ cell lines and tissues, and this increase seems to be related to the differentiation induction of TGCTs (35). A more complete understanding of mechanisms underlying the function of m6A biomarkers in oncogenesis and chemoresistance could potentially allow us to identify new m6A-targeted approaches for cancer therapy.
As a m6A ‘writer’, METTL3 is considered to be a promoter of malignant tumors. In ovarian cancer, METTL3 promotes tumor growth and invasion by regulating AXL translation and EMT (12). In gastric cancer, METTL3 promotes tumor progression by targeting MYC pathway (14). In colorectal cancer, METTL3 promotes tumor metastasis through miR-1246/SPRED2/MAPK pathway (36). As for TGCTs, METTL3 can promote the proliferation, invasion, migration and EMT of TGCTs cell line (37). In this study, we found that the expression of METTL3 was increased in drug-resistant TCam-2 cells. Knockdown of METTL3 can increase the chemosensitivity to cisplatin in TCam-2 cells, while overexpression of METTL3 had the opposite effect. These results suggest that METTL3 can promote cisplatin resistance in seminoma.
We next investigated the mechanism of METTL3 regulating the sensitivity of TCam-2 cells. It was found that METTL3 could up-regulate the expression of autophagy-associated markers LC3-II and Beclin1. In addition, autophagy inhibitor 3-MA can inhibit METTL3-mediated cisplatin resistance. These results verify the hypothesis that METTL3 promotes drug resistance of seminoma through enhancing autophagy.
To further explore the mechanism of METTL3 regulating autophagy, we selected candidate targets through literature retrieval and online tool prediction. Through qRT-PCR, western blot, and MeRIP-qPCR, ATG5 was finally selected as our research object. After overexpression of METTL3 in TCam-2 cells, the expression of ATG5 was increased, as well as the m6A abundance of ATG5. This indicates that METTL3 can promote the expression of ATG5 through m6A modification. Knockdown of ATG5 could reverse the potentiation effect of METTL3 on autophagy. Furthermore, knockdown of ATG5 could sensitize TCam-2 cells overexpressing METTL3, and then reduced the viability of TCam-2 cells treated with CDDP. These results strongly validate that ATG5 is a downstream target for METTL3 to regulate autophagy.
In general, we revealed the mechanism of METTL3 promoting drug resistance of seminoma through autophagy in vitro, suggesting the potential role of m6A modification in the progression of seminoma. However, some limitations should be acknowledged in this study. It remains unclear how METTL3 promotes the expression of ATG5. Our data only show the relationship between METTL3 and drug resistance in seminoma, while drug resistance in TGCTs occurs mostly in non-seminoma. The relationship between METTL3 and non-seminoma deserves further exploration.
Conclusions
In summary, our study emphasizes the important role of METTL3 in mediating acquired cisplatin resistance in seminoma. Mechanically, METTL3 can enhance autophagy by targeting ATG5, and then induce resistance to cisplatin in seminoma (Figure 5). Therefore, targeting METTL3 may be a potential therapeutic strategy for chemoresistance of seminoma.
Figure 5.
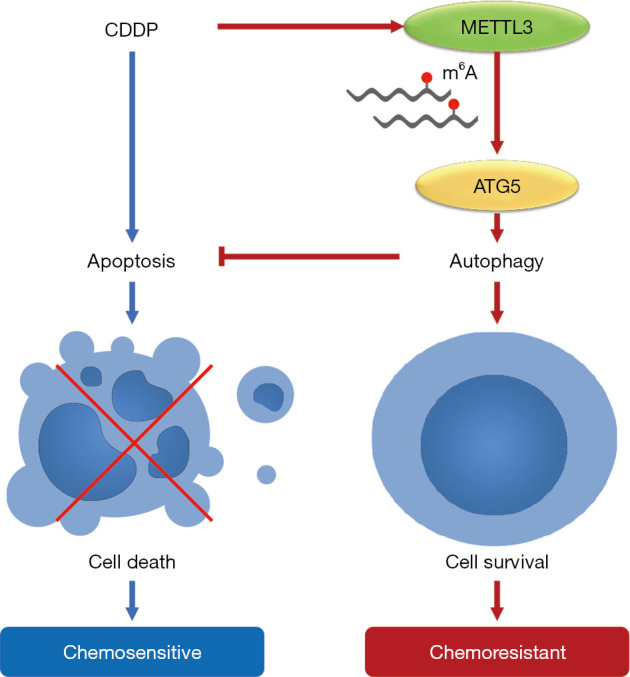
Schematic diagram of the role of METTL3 in regulating autophagy and chemosensitivity of TCam-2 cells during cisplatin treatment. CDDP, cis-diamminedichloroplatinum II.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study is supported by National Natural Science Foundation of China, grant/award number: #81571432, YT; #81372181, JY; New Xiangya Talent Project of the Third Xiangya Hospital of Central South University, grant/award number: #20180305, JY; Natural Science Foundation of Hunan Province, grant/award number: #2019JJ80097, YT.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1411
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-1411
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1411). The authors have no conflicts of interest to declare.
References
- 1.Hanna NH, Einhorn LH. Testicular cancer--discoveries and updates. N Engl J Med 2014;371:2005-16. 10.1056/NEJMra1407550 [DOI] [PubMed] [Google Scholar]
- 2.Winter C, Albers P. Testicular germ cell tumors: pathogenesis, diagnosis and treatment. Nat Rev Endocrinol 2011;7:43-53. 10.1038/nrendo.2010.196 [DOI] [PubMed] [Google Scholar]
- 3.Rajpert-De Meyts E, McGlynn KA, Okamoto K, et al. Testicular germ cell tumors. Lancet 2016;387:1762-74. 10.1016/S0140-6736(15)00991-5 [DOI] [PubMed] [Google Scholar]
- 4.Voutsadakis IA. The chemosensitivity of testicular germ cell tumors. Cell Oncol (Dordr) 2014;37:79-94. 10.1007/s13402-014-0168-6 [DOI] [PubMed] [Google Scholar]
- 5.Marko J, Wolfman DJ, Aubin AL, et al. Testicular seminoma and its mimics: from the radiologic pathology archives. Radiographics 2017;37:1085-98. 10.1148/rg.2017160164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fankhauser CD, Curioni-Fontecedro A, Allmann V, et al. Frequent PD-L1 expression in testicular germ cell tumors. Br J Cancer 2015;113:411-3. 10.1038/bjc.2015.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer KD, Jaffrey SR. Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol 2017;33:319-42. 10.1146/annurev-cellbio-100616-060758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roignant JY, Soller M. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet 2017;33:380-90. 10.1016/j.tig.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 9.Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet 2014;15:293-306. 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 10.He L, Li H, Wu A, et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer 2019;18:176. 10.1186/s12943-019-1109-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother 2019;112:108613. 10.1016/j.biopha.2019.108613 [DOI] [PubMed] [Google Scholar]
- 12.Hua W, Zhao Y, Jin X, et al. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol 2018;151:356-65. 10.1016/j.ygyno.2018.09.015 [DOI] [PubMed] [Google Scholar]
- 13.Shen C, Xuan B, Yan T, et al. m6A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer 2020;19:72. 10.1186/s12943-020-01190-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang DD, Chen ZH, Yu K, et al. METTL3 promotes the progression of gastric cancer via targeting the MYC pathway. Front Oncol 2020;10:115. 10.3389/fonc.2020.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu GM, Zeng HD, Zhang CY, et al. Identification of METTL3 as an adverse prognostic biomarker in hepatocellular carcinoma. Dig Dis Sci 2021;66:1110-26. 10.1007/s10620-020-06260-z [DOI] [PubMed] [Google Scholar]
- 16.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004;6:463-77. 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- 17.Sui X, Chen R, Wang Z, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis 2013;4:e838. 10.1038/cddis.2013.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li YJ, Lei YH, Yao N, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer 2017;36:52. 10.1186/s40880-017-0219-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng D, Wei J, Gan Y, et al. Testis developmental related gene 1 regulates the chemosensitivity of seminoma TCam-2 cells to cisplatin via autophagy. J Cell Mol Med 2019;23:7773-84. 10.1111/jcmm.14654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi T, Fujii T, Jo Y, et al. Possible mechanism responsible for the acquisition of resistance to cis-diamminedichloroplatinum (II) by cultured human testicular seminoma cells. J Urol 2004;171:1929-33. 10.1097/01.ju.0000122901.70300.20 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Kang M, Zhang B, et al. m6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer 2019;18:185. 10.1186/s12943-019-1116-x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Jin D, Guo J, Wu Y, et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol 2019;12:135. Erratum in: J Hematol Oncol 2020;13:106. 10.1186/s13045-019-0830-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Wei J, Yin Y, Zhou J, et al. METTL3 potentiates resistance to cisplatin through m6A modification of TFAP2C in seminoma. J Cell Mol Med 2020;24:11366-80. 10.1111/jcmm.15738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Gan Y, Tan Z, et al. TDRG1 functions in testicular seminoma are dependent on the PI3K/Akt/mTOR signaling pathway. Onco Targets Ther 2016;9:409-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Wu R, Liu Y, et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 2020;16:1221-35. 10.1080/15548627.2019.1659617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin S, Zhang X, Miao Y, et al. m6A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res 2018;28:955-7. 10.1038/s41422-018-0069-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei J, Gan Y, Peng D, et al. Long non-coding RNA H19 promotes TDRG1 expression and cisplatin resistance by sequestering miRNA-106b-5p in seminoma. Cancer Med 2018;7:6247-57. 10.1002/cam4.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogov V, Dötsch V, Johansen T, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 2014;53:167-78. 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 29.Kong E, Kim HD, Kim J. Deleting key autophagy elongation proteins induces acquirement of tumor-associated phenotypes via ISG15. Cell Death Differ 2020;27:2517-30. 10.1038/s41418-020-0519-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H, Chitiprolu M, Roncevic L, et al. Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev Cell 2017;43:716-30.e7. 10.1016/j.devcel.2017.11.018 [DOI] [PubMed] [Google Scholar]
- 31.Koster R, van Vugt MATM, Timmer-Bosscha H, et al. Unravelling mechanisms of cisplatin sensitivity and resistance in testicular cancer. Expert Rev Mol Med 2013;15:e12. 10.1017/erm.2013.13 [DOI] [PubMed] [Google Scholar]
- 32.Van Der Zwan YG, Stoop H, Rossello F, et al. Role of epigenetics in the etiology of germ cell cancer. Int J Dev Biol 2013;57:299-308. 10.1387/ijdb.130017ll [DOI] [PubMed] [Google Scholar]
- 33.Lobo J, Barros-Silva D, Henrique R, et al. The emerging role of epitranscriptomics in cancer: focus on urological tumors. Genes (Basel) 2018;9:552. 10.3390/genes9110552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lobo J, Costa AL, Cantante M, et al. m6A RNA modification and its writer/reader VIRMA/YTHDF3 in testicular germ cell tumors: a role in seminoma phenotype maintenance. J Transl Med 2019;17:79. 10.1186/s12967-019-1837-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nettersheim D, Berger D, Jostes S, et al. N6-Methyladenosine detected in RNA of testicular germ cell tumors is controlled by METTL3, ALKBH5, YTHDC1/F1/F2, and HNRNPC as writers, erasers, and readers. Andrology 2019;7:498-506. [DOI] [PubMed] [Google Scholar]
- 36.Peng W, Li J, Chen R, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res 2019;38:393. 10.1186/s13046-019-1408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo Y, Sun Y, Li L, et al. METTL3 may regulate testicular germ cell tumors through EMT and immune pathways. Cell Transplant 2020;29:963689720946653. 10.1177/0963689720946653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as