Abstract
The article introduces two complementary datasets intended for the development of data-driven solutions for cranial implant design, which remains to be a time-consuming and laborious task in current clinical routine of cranioplasty. The two datasets, referred to as the SkullBreak and SkullFix in this article, are both adapted from a public head CT collection CQ500 (http://headctstudy.qure.ai/dataset) with CC BY-NC-SA 4.0 license. The SkullBreak contains 114 and 20 complete skulls, each accompanied by five defective skulls and the corresponding cranial implants, for training and evaluation respectively. The SkullFix contains 100 triplets (complete skull, defective skull and the implant) for training and 110 triplets for evaluation. The SkullFix dataset was first used in the MICCAI 2020 AutoImplant Challenge (https://autoimplant.grand-challenge.org/) and the ground truth, i.e., the complete skulls and the implants in the evaluation set are held private by the organizers. The two datasets are not overlapping and differ regarding data selection and synthetic defect creation and each serves as a complement to the other. Besides cranial implant design, the datasets can be used for the evaluation of volumetric shape learning algorithms, such as volumetric shape completion. This article gives a description of the two datasets in detail.
Keywords: cranial implant design, cranioplasty, deep learning, volumetric shape learning, skull, autoimplant
Specification Table
| Subject | Information |
| Specific subject area | Computer Vision and Pattern Recognition |
| Type of data | Image |
| How data were acquired | The two datasets were adapted from a public head CT collection CQ500 with CC BY-NC-SA 4.0 license |
| Data format | Raw |
| Parameters for data collection | The selection of DICOM files from the CQ500 head CT collection was based on the image quality (e.g., slice thickness, fracture, scanning protocol) |
| Description of data collection | The datasets were adapted from the CQ500 CT data. The adaptation process involves pre-processing (data format conversion, selection, transformation, skull segmentation, post-processing (e.g., noise removal) artificial defect injection). |
| Data source location | The dataset was adapted from the public head CT collection CQ500 with CC BY-NC-SA 4.0 license. The SkullFix dataset was first released to participants of the AutoImplant (https://autoimplant.grand-challenge.org/) challenge. |
| Data accessibility | The SkullFix dataset can be downloaded from the AutoImplant challenge website at https://autoimplant.grand-challenge.org/. Besides, we also provided the direct download links for the two datasets: SkullBreak (https://www.fit.vutbr.cz/~ikodym/skullbreak_training.zip and https://www.fit.vutbr.cz/~ikodym/skullbreak_evaluation.zip). SkullFix (https://files.icg.tugraz.at/f/2c5f458e781a42c6a916/?dl=1). |
| Related research articles | Jianning Li, Antonio Pepe, Christina Gsaxner, Gord von Campe, and Jan Egger. title: A baseline approach for autoimplant: the miccai 2020 cranial implant design challenge, MICCAI CLIP 2020. DOI: https://doi.org/10.1007/978-3-030-60946-7_8. reference: [1] |
| Oldřich Kodym, Michal Španěl, and Adam Herout. title: Skull shape reconstruction using cascaded convolutional networks. DOI: https://doi.org/10.1016/j.compbiomed.2020.103886. reference: [2] |
Value of the Data
-
•
Researchers from academia and industry can use the dataset to develop automatic cranial implant design solutions, which can improve the current clinical routines for cranioplasty.
-
•
The dataset can also be used as a benchmark for volumetric shape learning tasks such as volumetric shape completion. Compared to other 3D shape datasets, the dataset contains high-resolution () shapes.
-
•
The dataset can be used to create 3D surface models of human skulls for 3D printing.
1. Data Description
The dataset described in this article was adapted from a public head CT collection from CQ500 (http://headctstudy.qure.ai/dataset), which was originally intended for the detection of critical findings in head CTs. The head CT collection is originally provided by the Centre for Advanced Research in Imaging, Neurosciences and Genomics(CARING), New Delhi, IN. In total, the adapted dataset consists of 880 triplets of a defective skull, a corresponding complete skull and an implant, which is given by the difference between the defective and the complete skull. All data samples are represented as binary volumes and saved in NRRD format. The dataset is split into two tracks: the SkullFix track and the SkullBreak track.
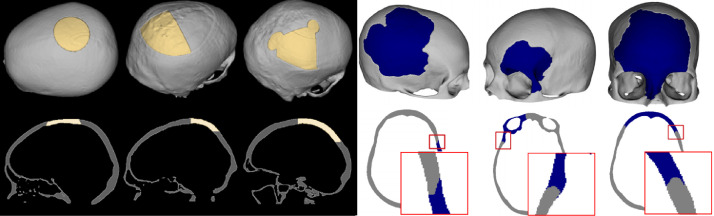
The SkullFix track consists of the data used in the MICCAI 2020 AutoImplant challenge. This track is split into a training set with 100 triplets, a test set with 100 triplets and a test set with 10 triplets. The defects in this track have rectangular shape with craniotome drill holes in the corners, as often encountered in the craniotomy procedures. The defects are mostly located in the back of the skulls, depending on patient position during the CT data acquisition, with the exception of the test set with 10 triplets, where the shapes, sizes and positions of the defect are different as seen Fig. 1. These 10 defects were used in the AutoImplant challenge to test robustness of the proposed methods. The dimension of the data in SkullFix is and is the number of axial slices.
Fig. 1.
Examples of the 3D model renders (top) and slices (bottom) through the skull defect data. 3 defects from the 10 test data of the SkullFix track and 3 defects from the training data of the SkullBreak track (taken from [2]), respectively.
The SkullBreak track consists of the data originally used in Kodym et al. [2]. The dataset was adapted so that there is no overlap between training and test sets of both tracks presented in this article. 114 training and 20 test skulls were used to create this track. On each skull in this track, five different synthetic defects were created:
-
•
unilateral defect in the parieto-temporal area
-
•
unilateral defect in the fronto-orbital area
-
•
bilateral defect
-
•
two random defects
This resulted in a training set with 570 triplets and a test set with 100 triplets. The defects in this track were created with random shapes. Several examples from this track can be seen in Fig. 1. The dimension of the data in SkullBreak is .
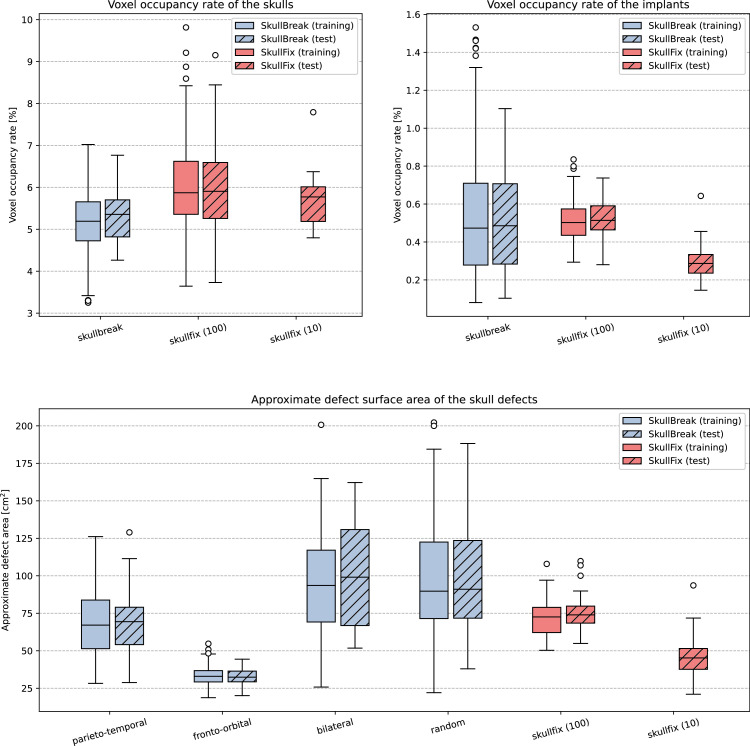
In addition to position and shape of the skull defects, the defect size is another factor that has effect on difficulty of implant design. Therefore, we report voxel occupancy rate (VOR) and approximate defect surface area (DSA) to illustrate properties of the described dataset. Figure 2 shows the statistics of the dataset, including the VOR of the complete skulls and the implants and the DSA. The VOR is defined as the percentage of occupied voxels in the whole image volume: where is the number of occupied voxels in the volume and is the total voxel number (occupied and unoccupied). The DSA was approximated by isolating only the implant voxels located on the outer surface of the original complete skull and using the voxel size information to approximate the total area of the isolated surface. The differences in DSA between individual defect groups in the dataset are most apparent in the SkullBreak track, owing to the fact that the groups are constrained to different areas. The unilateral defects (parieto-temporal and fronto-orbital) are smaller because they are restricted only to one side of the skull, while the others are unconstrained. The fronto-orbital defects are further constrained to the space around orbits while the parieto-temporal can span the whole braincase area. The SkullFix defects have less variability, mostly being average in their size with the exception of the 10 test cases which are smaller.
Fig. 2.
Boxplot of the data information for the training set and test set of both dataset tracks, including VOR of the complete skulls (top left), VOR of the implants (top right) and approximate defect area (bottom).
2. Experimental Design, Materials and Methods
Cranial implant design is the main bottleneck for an optimized workflow for cranioplasty [4]. The development of automated cranial implant design solutions, especially deep learning based solutions, has been hindered by the lack of public datasets. The real defective skulls from brain tumor surgery or trauma are difficult to obtain in large quantities and there often lack the ground truth, i.e., the complete skulls or the implants. We hence devised a pipeline to convert publicly available head CT collections into datasets suitable for training deep learning models. Artificial skull defects are used to simulate the surgical and traumatic process. The original CQ500 data contain head CT scans and expert annotations from the Centre for Advanced Research in Imaging, Neurosciences and Genomics, New Delhi, IN. The scans that contained skull fractures according to the expert annotations were removed as well as those that were not acquired with thin plate scanning protocol. The skulls in the presented dataset were sampled from the remaining scans. In cases of multiple series, the series with the highest resolution were used for further processing of each scan. The chosen scans contained between 211 and 394 axial slices. Acquisition geometry regularization transform was applied to each scan to correct deformations caused by gantry tilt and the data were converted to the NRRD format.
Both dataset tracks were created from the original CQ500 data using three main steps; CT data preprocessing, skull segmentation and artificial defect injection. However, because they were originally created independently of each other, the individual processing steps differ. The different properties and the specific steps taken to create the two tracks are summarized in the Table 1.
Table 1.
Differences between the SkullFix and the SkullBreak dataset tracks.
| SkullFix | SkullBreak | |
|---|---|---|
| Training/test split | ||
| Volume size | ||
| Voxel size | various | 0.4 mm |
| Preprocessing | acquisition geometry regularization transform | acquisition geometry regularization transform and rigid alignment using the landmarks defining Frankfort-horizontal plane |
| Skull segmentation | thresholding at 150 HU, noise removal using connected components analysis | convolutional neural network and graph-cut [3] |
| Defect injection | binary defect shape subtraction from complete skull | binary defect shape subtraction from complete skull and defect border smoothing using morphological operations |
For preprocessing, the acquisition geometry regularization transform of SkullFix is performed using 3D Slicer to correct obvious deformation or tilting of the skull data. It involves several steps:
-
•
Go to Edit -> Application Settings -> DICOM and change “acquisition geometry regularization” to “apply regularization transform”.
-
•
While importing the DICOM, an item called “acquisition transform” is shown within the series.
-
•
In the transform hierarchy, right click on the deformed DICOM and press “harden transform”.
-
•
Save the transformed data as NRRD.
For SkullBreak, the “dcm2niix” tool (https://github.com/rordenlab/dcm2niix) is used to convert DICOM series to 3D data. It corrects the deformation caused by gantry tilt. On each skull, four landmarks defining the Frankfort-horizontal plane, i.e., the left and right auditory meatus and the left and right orbital floor, were aligned onto the plane using rigid transformation. Finally, to be in conformity with the SkullFix dataset, the SkullBreak dataset is converted into NRRD format.
2.1. Artificial Defect Shapes
The skull defects in the presented dataset are created by subtracting a part of the healthy skull with the subtracted part serving as ground-truth for the implant shape. In real patients, the defects can have different causes, such as craniotomy due to brain or bone tumour, brain swelling or traumatic fractures. Consequently, the shape, position and size of the defects can also vary.
During craniotomy, the craniotome drill is often used to drill holes into the skull by the surgeon, leading to the defect having roundish holes in the corners. The defects in the SkullFix track emulate this property by injecting defects with such roundish corners into back of the complete skulls. The defects in the SkullBreak track, on the other hand, have completely random shape given by random combinations of elastically deformed spheres. The positions of the SkullBreak defects are also random, although they are structured into several categories, as discussed in the Data Description section. Furthermore, ongoing bone remodelling processes can deform the defect borders. To simulate this, the defect borders in the SkullBreak track are smoothed by morphological opening operation with structuring element in form of sphere with radius between 2 and 7 voxels. Examples of the defects in both dataset tracks can be seen in Figure 1.
Ethics Statement
The two datasets, SkullBreak and SkullFix described in the article are adapted from a public head CT collection CQ500 http://headctstudy.qure.ai/dataset), which is licensed under CC BY-NC-SA 4.0 and End User License Agreement (EULA). According to the usage notes provided in their website, we make the adapted datasets public under the same licenses as the original CQ500 collection.
CRediT Author Statement
Oldřich Kodym: Data curation, Writing - original draft; Jianning Li: Data curation, Writing - original draft; Antonio Pepe: Writing - original draft; Christina Gsaxner: Writing - original draft; Sasank Chilamkurthy: Resources; Jan Egger: Supervision; Michal Španěl: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the following funding bodies:
-
•
CAMed (COMET K-Project 871132, see also https://www.medunigraz.at/camed/), which is funded by the Austrian Federal Ministry of Transport, Innovation and Technology (BMVIT) and the Austrian Federal Ministry for Digital and Economic Affairs (BMDW), and the Styrian Business Promotion Agency (SFG)
-
•
The Austrian Science Fund (FWF) KLI 678-B31 (enFaced)
-
•
TU Graz Lead Project (Mechanics, Modeling and Simulation of Aortic Dissection)
-
•
BUT Department of Information Technology and Tescan 3Dim company joint research fund
Moreover, We thank the creators of the CQ500 dataset (http://headctstudy.qure.ai/dataset). Finally, we want to point out to our medical online framework Studierfenster (www.studierfenster.at), where an automatic cranial reconstruction and implant design system has been incorporated.
References
- 1.Li J., Pepe A., Gsaxner C., von Campe G., Egger J. Multimodal Learning for Clinical Decision Support and Clinical Image-Based Procedures. Springer; 2020. A baseline approach for autoimplant: the miccai 2020 cranial implant design challenge; pp. 75–84. [Google Scholar]
- 2.Kodym O., Španěl M., Herout A. Skull shape reconstruction using cascaded convolutional networks. Comput. Biol. Med. 2020;123:103886. doi: 10.1016/j.compbiomed.2020.103886. [DOI] [PubMed] [Google Scholar]
- 3.Kodym O., S̃panel M., Herout A. Segmentation of defective skulls from ct data for tissue modelling. arXiv preprint arXiv: 1911.08805. 2019 [Google Scholar]
- 4.Li J., Pepe A., Gsaxner C., Egger J. An online platform for automatic skull defect restoration and cranial implant design. Proc. SPIE 11598, Medical Imaging 2021: Image-Guided Procedures, Robotic Interventions, and Modeling, 115981Q. 2021 [Google Scholar]