Abstract
Lung adenocarcinoma, a type of non-small cell lung cancer, is the leading cause of cancer death worldwide. Great efforts have been made to identify the underlying mechanism of adenocarcinoma, especially in relation to oncogenes. The present study by integrating computational analysis with western blotting, aimed to understand the role of the upregulation of glucosamine-phosphate N-acetyltransferase 1 (GNPNAT1) in carcinogenesis. In the present study, publicly available gene expression profiles and clinical data were downloaded from The Cancer Genome Atlas to determine the role of GNPNAT1 in lung adenocarcinoma (LUAD). In addition, the association between LUAD susceptibility and GNPNAT1 upregulation were analyzed using Wilcoxon signed-rank test and logistic regression analysis. In LUAD, GNPNAT1 upregulation was significantly associated with disease stage [odds ratio (OR)=2.92, stage III vs. stage I], vital status (dead vs. alive, OR=1.89), cancer status (tumor status vs. tumor-free status, OR=1.85) and N classification (yes vs. no, OR=1.75). Cox regression analysis and the Kaplan-Meier method were utilized to evaluate the association between GNPNAT1 expression and overall survival (OS) time in patients with LUAD. The results demonstrated that patients with increased GNPNAT1 expression levels exhibited a reduced survival rate compared with those with decreased expression levels (P=8.9×10−5). In addition, Cox regression analysis revealed that GNPNAT1 upregulation was significantly associated with poor OS time [hazard ratio (HR): 1.07; 95% confidence interval (CI): 1.04–1.10; P<0.001]. The gene set enrichment analysis revealed that ‘cell cycle’, ‘oocyte meiosis’, ‘pyrimidine mediated metabolism’, ‘ubiquitin mediated proteolysis’, ‘one carbon pool by folate’, ‘mismatch repair progesterone-mediated oocyte maturation’ and ‘basal transcription factors purine metabolism’ were differentially enriched in the GNPNAT1 high-expression samples compared with GNPNAT1 low-expression samples. The aforementioned pathways are involved in the pathogenesis of LUAD. The findings of the present study suggested that GNPNAT1 upregulation may be considered as a promising diagnostic and prognostic biomarker in patients with LUAD. In addition, the aforementioned pathways may be pivotal pathways perturbed by the abnormal expression of GNPNAT1 in LUAD. The findings of the present study demonstrated the therapeutic value of the regulation of GNPNAT1 in lung adenocarcinoma.
Keywords: glucosamine-phosphate N-acetyltransferase 1, upregulation, lung adenocarcinoma, poor survival, independent predictor
Introduction
Lung cancer is a deadly disease with an incidence rate of 11.4% worldwide in 2020 (1–4). The pathogenesis of lung cancer is associated with genetic and epigenetic factors, such as MYC amplification, deregulated expression and epigenetic inactivation of Ras Association Domain Family 1 (5–8). In the USA, lung cancer is the leading cause of cancer-related deaths in both men and women, with a mortality rate of 12.7%. This disease is an aggressive type of cancer, with a 5-year overall survival rate of 14% for advanced stage disease (3). There are two major types of lung cancer, namely small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) (9). The former one is responsible for ~15% of all lung cancer cases (10). SCLC tumors tend to be more aggressive and may be not diagnosed until they have already metastasized (11). NSCLC is the most common subtype of lung cancer, being responsible for ~85% of lung cancer cases (12). Among the different subtypes of NSCLC, lung adenocarcinoma (LUAD) is the most common type, accounting for >50% of NSCLC cases with an increasing incidence rate (13).
The 5-year overall survival rate for stage I SCLC and NSCLC is 50 and 60–70%, respectively (14,15). However, the survival rate in patients with advanced lung cancer is almost 15% (12). In total, ~70% of patients with lung cancer present with advanced stage of the disease at the time of diagnosis (12), supporting the lack of efficient methods for early diagnosis. The early signs of lung cancer are usually subtle or non-specific, such as cough, irritating dry cough or choking cough (16). In clinical practice, spiral computed tomography and fluorescence bronchoscopy are commonly used to detect tumors with a size of >1 mm (17,18). However, this resolution is commonly insufficient to diagnose stage I lung cancer (19). Hence, the diagnosis of patients at a very early stage of the disease and the massive screening of individuals at increased risk of developing lung cancer warrants the need to investigate the genetic basis of carcinogenesis. In terms of DNA mutations, point mutations in the KRAS gene (20), frame shift deletions or insertions in TP53 (21), and microsatellite alterations may trigger the occurrence of lung cancer (22). In addition, at the transcriptional level, hypermethylated gene promoters may serve as biomarkers for the early detection of lung cancer (23). A study demonstrated that cyclin-dependent kinase inhibitor 2A (CDKN2A), which is involved in the cell cycle, and O6-methylguanine DNA methyltransferase, which is involved in DNA repair, were both downregulated in lung cancer samples compared with paracarcinoma tissue (24).
Glucosamine-phosphate N-acetyltransferase 1 (GNPNAT1) encodes an enzyme involved in the pathway mediating the biosynthesis of uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), an important donor substrate for N-linked glycosylation, which is in turn involved in metabolism in eukaryotic cells (25). For example, silencing of GNPNAT1 in pancreatic β-cells modulated insulin secretion, while its increased methylation status was associated with reduced risk of developing type 2 diabetes mellitus (26). It has been reported that metabolism-related genes serve crucial roles in tumor progression (27). The levels of UDP-GlcNAc are known to affect hyaluronan synthesis and protein O-GlcNAcylation (28). Growing evidence has suggested that O-GlcNAcylation promotes cell survival via the aberrant metabolic state of malignant tumors (29).
GNPNAT1 serves as a biomarker for predicting prostate and colorectal cancer biochemical recurrence (30). A study demonstrated that the levels of lactate dehydrogenase A, lysophosphatidylglycerol acyltransferase 1, GNPNAT1, prostaglandin E synthase and thymidylate synthase were increased in lung cancer tissues compared with para-carcinoma tissues (31). However, the association between GNPNAT1 expression and the early diagnosis and prognosis of LUAD has not been fully investigated.
Hence, the present study aimed to comprehensively evaluate the diagnostic and prognostic potential of GNPNAT1 expression in human LUAD based on publicly available data from The Cancer Genome Atlas (TCGA). In addition, gene set enrichment analysis (GSEA) was carried out to identify the biological pathways involved in LUAD, perturbated by the GNPNAT1 regulatory network. The changes in the protein expression levels of GNPNAT1 in patients with LUAD were validated by western blotting. The findings of the present study demonstrated the therapeutic value of the regulation of GNPNAT1 in lung adenocarcinoma.
Materials and methods
Gene expression data and bioinformatics analysis
Gene expression data and corresponding clinical information of 585 individuals were downloaded from the TCGA official website (https://portal.gdc.cancer.gov; Project ID: TCGA-LUAD (32,33). Hence, the clinical data of 522 patients are shown in Table I. In addition, 63 healthy subjects were also included in this study. Also, the differential expression of GNPNAT1 between tumor and paracarcinoma tissues was conducted. The majority of the patients (81.0%) were >55 years and 53.6% were female. Boxplots and dot plots were used to visualize the differences in gene expression among different groups analyzed using the downloaded data (34). The classification systems was based on the Tumor Node Metastasis (TNM) classification of malignant tumors, 5th edition, 1997 (35).
Table I.
Characteristics of patients with LUAD (n=522) according to datasets from The Cancer Genome Atlas.
| Clinical characteristics | No. of patientsa | %b |
|---|---|---|
| Age at diagnosis, years | ||
| ≤55 | 80 | 15.3 |
| >55 | 423 | 81.0 |
| Stage | ||
| I | 279 | 53.4 |
| II | 124 | 23.8 |
| III | 85 | 16.3 |
| IV | 26 | 5.0 |
| Sex | ||
| Male | 242 | 46.4 |
| Female | 280 | 53.6 |
| T classification | ||
| T1 | 172 | 33.0 |
| T2 | 281 | 53.8 |
| T3 | 47 | 9.0 |
| T4 | 19 | 3.6 |
| N classification | ||
| N0 | 335 | 64.2 |
| N1 | 98 | 18.8 |
| N2 | 75 | 14.4 |
| N3 | 2 | 0.4 |
| M classification | ||
| M0 | 353 | 67.6 |
| M1 | 25 | 4.8 |
| Recurrence | ||
| No | 279 | 53.4 |
| Yes | 146 | 28.0 |
| Survival status | ||
| Death | 188 | 36.0 |
| Survival | 334 | 64.0 |
| Neoplasm cancer status | ||
| With tumor | 141 | 27.0 |
| Tumor free | 248 | 47.6 |
| Histological type | ||
| Acinar cell neoplasms | 22 | 4.2 |
| Adenomas and adenocarcinomas | 486 | 93.1 |
| Cystic, mucinous and serous neoplasms | 14 | 2.7 |
| Radiation therapy | ||
| No | 377 | 72.2 |
| Yes | 58 | 11.1 |
| Cigarette history | ||
| No | 166 | 31.8 |
| Yes | 356 | 68.2 |
The sum of all the numbers in each category is not always 522, because not everyone has every piece of diagnostic information.
The percentage is calculated according to the proportion of the actual number of the category in the total number of 522 people. T, tumor; N, node; M, metastasis; LUAD, lung adenocarcinoma.
GSEA
GSEA (v.4.1.0; http://www.gsea-msigdb.org/gsea/index.jsp) as performed as previously described (36). GSEA was performed to reveal the significant survival differences between the high [FPKM (fragments per kilo- base of transcript per million mapped reads) ≥ 10] and low GNPNAT1 (FPKM<10) expression groups. All genes were then ranked according to their association with the GNPNAT1 high-expression phenotype. The nominal (NOM) P-value and normalized enrichment score (NES) were utilized to sort the pathways enriched in each phenotype (36,37). In GSEA, pathways showing NOM P-value ≤0.05 or false discovery rate (FDR) q-value ≤0.05 were considered as significant (38).
Western blotting
Frozen tissues of 35 patients, those without any other diagnosed type of cancers, including 9 males and 26 females, age range, 32–77 years (median age, 60 years) were used to perform western blotting. All patients were recruited to the Department of Thoracic and Cardiovascular Surgery, The Second Affiliated Hospital of Nantong University (Nantong, China) during April of 2020. The present study was approved by the Ethics Committee of The Second Affiliated Hospital of Nantong University and in compliance with the Declaration of Helsinki. Written patient consent for use of their tissues in research was obtained. After surgical resection of the tissue, the central non-necrotic area was taken as the cancer tissue sample with sterile scissors, and then adjacent tissue 2–3 cm away from the edge of the cancer was obtained as the paracarcinoma tissue with another new sterile scissors and the remaining part was sent for biopsy. The frozen tissue samples were pulverized under −80°C using a mortar and pestle to extract proteins. Briefly, the cultured cells were rinsed thrice with precooled PBS. Subsequently, the cells were lysed with RIPA buffer (cat. no. P0013K) supplemented with phenylmethanesulfonylfluoride (PMSF; cat. no. ST506; both from Beyotime Institute of Biotechnology) at 4°C for 30 min and centrifuged at 14,000 × g at 4°C for 30 min. The supernatant containing the protein extracts was collected and proteins were quantified using the BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). Proteins (50 µg/lane) were separated by 8% SDS-PAGE (cat. no. ab139604; Abcam), and then transferred onto polyvinylidene difluoride membranes (EMD Millipore). Following blocking with 5% skimmed milk dissolved in Tris-buffered saline Tween-20 (0.1% TBST) for 1 h at room temperature, the membranes were rinsed with 0.1% TBST thrice. Subsequently, the membranes were incubated with primary antibodies at 4°C overnight. The following primary antibodies were used: Anti-GNPNAT1 polyclonal antibody (1:1,0000; cat. no. K107882P) and anti-GAPDH polyclonal antibody (1:1,0000, cat. no. K106389P; both from Beijing Solarbio Science & Technology Co., Ltd.). The next day, the membranes were incubated with the corresponding HRP-conjugated mouse anti-Human IgG1 FC secondary antibody (1:1,0000 dilution cat. no. AS16-3223; Agrisera AB). The immunoreactive bands were visualized using a developing and fixing kit (P0020-1; Beyotime Institute of Biotechnology). GAPDH was used as the loading control.
Statistical analysis
R statistical software v.3.5.3 (39) was used to perform the statistical analyses. The association between cancer susceptibility and the GNPNAT1 high-expression phenotype was evaluated using both the Wilcoxon signed-rank test and logistic regression. Cox regression analysis and the Kaplan-Meier method were performed to determine the association between GNPNAT1 expression and overall survival (OS) time in patients with LUAD. Multivariate Cox analysis was conducted to evaluate the association between the expression of GNPNAT1 and other clinical features, such as clinical stage, sex, tumor (T) classification, node (N) classification, metastasis (M) classification, recurrence, survival status, cancer status, histological type, radiation therapy and smoking status. Tests among multiple groups of samples, such as advanced clinical stage (stage I vs. stage II vs. stage III vs. stage IV), were assayed by Kruskal Wallis test and the post hoc Dunn's test. In the survival analysis, the cut-off value of GNPNAT1 expression was set to 10. P<0.05 was considered to indicate a statistically significant difference and P<0.01 was considered to indicate a highly statistically significant difference.
Results
Patient characteristics
As shown in Table I, the clinical and gene expression data of 522 patients were obtained from TCGA. The majority of the patients (81.0%) were >55 years. A total of 279 patients (53.4%) were of stage I, 124 (23.8%) of stage II, 85 (16.3%) of stage III and 26 (5.0%) of stage IV. The majority of tumor samples (93.1%; n=486) were classified as adenomas and adenocarcinomas, which were significant standard clinical indexes. Among the 522 patients, 146 (28.0%) experienced cancer recurrence. Regarding cancer status, 248 patients (47.6%) were of tumor-free status and 141 of tumor status (27.0%), while 11.1% (n=58) underwent intensity-modulated radiation therapy. Finally, 68.2% of all participants had history of cigarette smoking, supporting the strong association between smoking and LUAD.
GNPNAT1 expression increases in LUAD
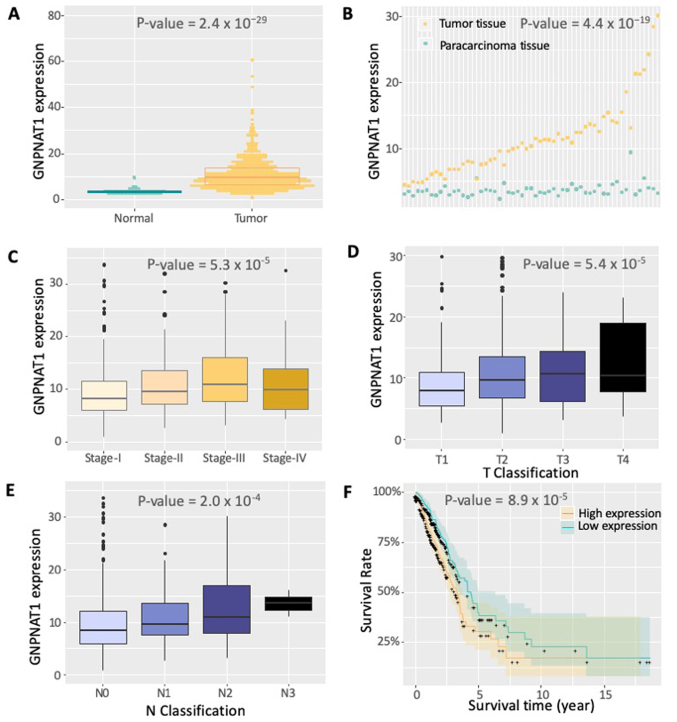
The expression data of GNPNAT1 from 522 samples were obtained from TCGA. As shown in Fig. 1, GNPNAT1 was differentially expressed between normal and tumor samples (Fig. 1A; normal vs. tumor samples; Wilcoxon test, P=2.4×10−29). The significant differential expression of GNPNAT1 was also observed between tumor and paracarcinoma tissues (Fig. 1B). The expression levels of GNPNAT1 were also associated with clinical stages (Fig. 1C), that is, higher GNPNAT1 expression was associated with advanced clinical stage (stage I vs. stage II vs. stage III vs. stage IV; Kruskal Wallis test, P=5.3×10−5). The slight decrease in GNPNAT1 expression in stage IV compared with stage III could be due to the sole effective radiation therapy in this advanced stage (Fig. 1C). The significant differential expression of GNPNAT1 could be also observed under diverse classification standards. As the tumor proliferated from lung (T1), principle bronchus (T2), chest walls (T3) to heart and great vessels (T4), the expression of GNPNAT1 increased gradually (Fig. 1D). Under a smaller proliferation region, from no proliferation (N0), ipsilateral trachea (N1) to ipsilateral mediastinum (N2), the expression of GNPNAT1 increased gradually, except a slightly decrease at N3 stage (proliferated to contralateral mediastinum) (Fig. 1F). The aforementioned results supported the reliability of GNPNAT1 expression on patient demographics and histories, diagnostic criteria and staging, pathology and even initial treatment. In addition, the results obtained from the western blotting of 35 patients revealed that the protein expression levels of GNPNAT1 in tumor tissues is higher compared with paracarcinoma tissues (Fig. 2). This experimental result was consistent with those observed at the transcriptional level from bioinformatics analysis that both the protein and mRNA expression of GNPNAT1 were higher in the tumor tissues compared with paracarcinoma tissues.
Figure 1.
GNPNAT1 expression in LUAD from The Cancer Genome Atlas. (A) Expression levels of GNPNAT1 in the same anatomical sites of the lung tissue between normal subjects and patients with LUAD are shown. (B) Expression levels of GNPNAT1 in tumor and para-carcinoma tissues isolated from the same patient are shown. The data in each column are paired from the same individual. (C) Expression levels of GNPNAT1 in the same anatomical sites of the lung tissue in patients of different LUAD stages are shown. (D) Expression levels of GNPNAT1 in different T stages are shown. (E) Expression levels of GNPNAT1 in different N stages are shown. (F) Survival rate fitting curve of the patients based on the expression of GNPNAT1. The cut-off value for the expression of GNPNAT1 was set to 10. GNPNAT1, glucosamine-phosphate N-acetyltransferase 1; LUAD, lung adenocarcinoma; T, tumor; N, node; M, metastasis.
Figure 2.
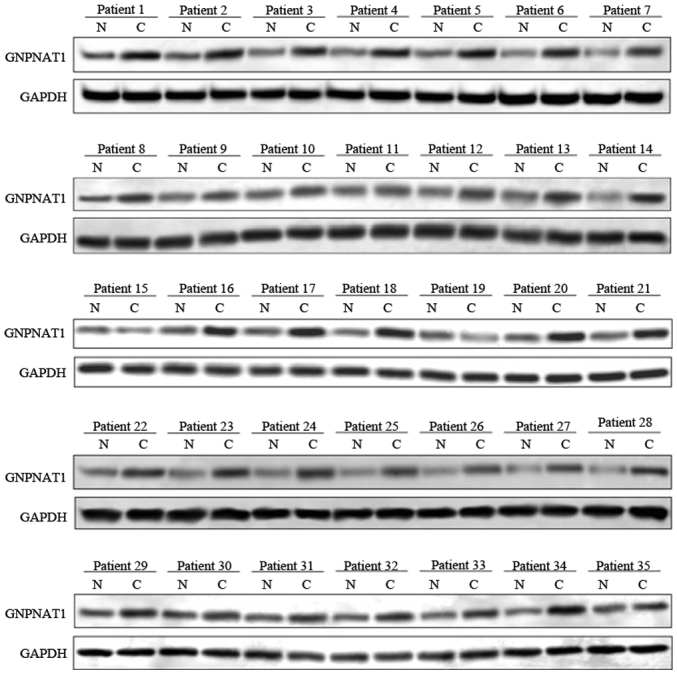
Protein expression levels of GNPNAT1 were determined in tumor and para-carcinoma tissues isolated from 35 patients with LUAD using western blotting. N, para-carcinoma normal tissues; C, tumor tissues; GNPNAT1, glucosamine-phosphate N-acetyltransferase 1; LUAD, lung adenocarcinoma.
Logistic regression analysis revealed that the expression of GNPNAT1 was associated with the clinical stage of LUAD [odds ratio (OR)=2.92; 95% confidence interval (CI), 1.76–4.96, P-value=4.88×10−5; stage III vs. stage I; Table II]. In addition, the GNPNAT1 high-expression phenotype in tumors was notably associated with vital status (dead vs. alive, OR=1.89), tumor status (tumor status vs. tumor-free status, OR=1.85), N classification (yes vs. no, OR=1.75) and clinical stage (stage II vs. stage I, OR=1.66; all P<0.05; Table II). The aforementioned findings indicated that subjects with higher GNPNAT1 expression levels may be more susceptible to LUAD. Additionally, patients with GNPNAT1 high-expression phenotype may be more likely to develop advanced stage LUAD compared with those with lower GNPNAT1 expression levels.
Table II.
Logistic regression analysis for the association between the expression of GNPNAT1 and clinicopathological characteristics of patients with LUAD.
| Clinical characteristics | Total (N) | Odds ratio in GNPNAT1 expression | P-value |
|---|---|---|---|
| Stage (II vs. I) | 395 | 1.66 (1.08–2.56) | 0.021 |
| Stage (III vs. I) | 358 | 2.92 (1.76–4.96) | 4.88×10−5 |
| Status (with tumor vs. tumor-free) | 389 | 1.85 (1.22–2.84) | 0.004 |
| Age (≥55 vs. <55 years) | 503 | 0.81 (0.50–1.31) | 0.391 |
| Radiation therapy (yes vs. no) | 435 | 1.77 (1.01–3.16) | 0.050 |
| Vital status (dead vs. alive) | 522 | 1.89 (1.31–2.73) | 0.001 |
| Cigarettes history (yes vs. no) | 522 | 1.28 (0.88–1.86) | 0.194 |
| M classification (M1 vs. M0) | 378 | 1.10 (0.48–2.50) | 0.825 |
| N classification (yes vs. no) | 510 | 1.75 (1.21–2.55) | 0.003 |
| New tumor event after initial treatment (yes vs. no) | 425 | 1.50 (1.00–2.26) | 0.051 |
| Disease type (adenomas and adenocarcinomas vs. acinar cell neoplasms adenocarcinomas vs. acinar cell neoplasms | 508 | 1.49 (0.63–3.67) | 0.370 |
| Disease type (cystic, mucinous and serous neoplasms vs. acinar cell neoplasms) | 36 | 0.754 | 0.755 |
N, node; M, metastasis, GNPNAT1, Glucosamine-phosphate N-acetyltransferase 1; LUAD, lung adenocarcinoma.
Survival outcomes and multivariate analysis
As shown in Fig. 1F, Kaplan-Meier survival analysis revealed that patients with higher expression of GNPNAT1 exhibited a lower survival rate compared with those with a low expression of GNPNAT1 (Wilcox test, P=8.9×10−5). In addition, multivariate analysis demonstrated that the GNPNAT1 high-expression phenotype was significantly associated with cancer status (HR=68.65; 95% confidence interval=11.77–400.24; P<0.001; Fig. 3]. In addition, follow up treatment success was also associated with poor survival in 198 patients who had detailed clinical information (HR=0.49; 95% CI: 0.24–0.97; P-value=0.04; Fig. 3). The above findings further demonstrated that the expression of GNPNAT1 could predict the survival time and regard as a treatment index.
Figure 3.
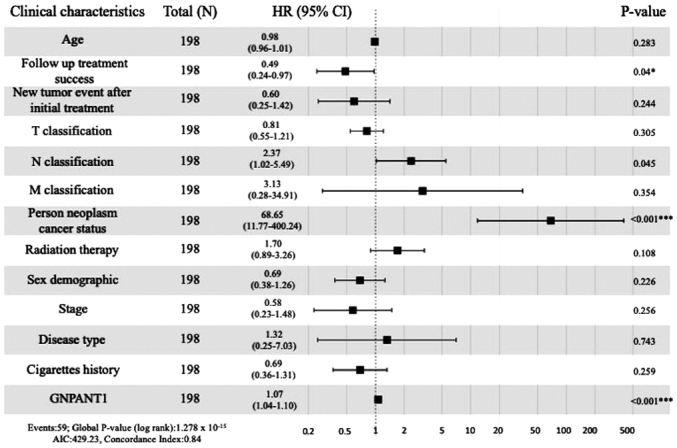
Association between OS and the clinicopathological characteristics of patients with LUAD from TCGA by Cox regression analysis. *0.01<P<0.05, ***P<0.001. OS, overall survival; TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma; T, tumor; N, node; M, metastasis.
Biological pathways associated with GNPNAT1 expression according to GSEA
GSEA between normal and high GNPNAT1 expression datasets was performed to identify biological pathways differentially enriched in LUAD. A total of 16 significant pathways were found, while only 9 pathways were the most significantly enriched biological pathways, with cut-off values of NOM P-value=0 and FDR q-value ≤0.05 (Table III) in the present study. ‘Cell cycle’, ‘oocyte meiosis’, ‘pyrimidine mediated metabolism’, ‘ubiquitin mediated proteolysis’, ‘one carbon pool by folate’, ‘mismatch repair’, ‘progesterone mediated oocyte maturation’, ‘basal transcription factors’, and ‘purine metabolism’ were the most differentially enriched pathways in the GNPNAT1 high-expression phenotype (Table III). According to the types of these pathways, some of them are prone to somatic mutation, and some of them affect metabolism. This is consistent with the previous observation that somatic cell mutation is high in lung cancer (40).
Table III.
Glucosamine-phosphate N-acetyltransferase 1-related biological pathways according to Gene Set Enrichment Analysis.
| Gene set name | NES | NOM P-value | FDR q-value |
|---|---|---|---|
| Cell cycle | 2.33 | 0 | 3.24×10−4 |
| Oocyte meiosis | 2.34 | 0 | 4.86×10−4 |
| Pyrimidine metabolism | 2.23 | 0 | 6.55×10−4 |
| Ubiquitin mediated proteolysis | 2.39 | 0 | 9.72×10−4 |
| One carbon pool by folate | 2.06 | 0 | 0.009 |
| Mismatch repair | 2.02 | 0 | 0.013 |
| Progesterone mediated oocyte maturation | 1.95 | 0 | 0.021 |
| Basal transcription factors | 1.95 | 0 | 0.022 |
| Purine metabolism | 1.96 | 0 | 0.023 |
| p53 signaling pathway | 1.88 | 0.002 | 0.031 |
| DNA replication | 1.94 | 0.002 | 0.021 |
| Cysteine and methionine metabolism | 1.92 | 0.002 | 0.022 |
| Aminoacyl tRNA biosynthesis | 2.11 | 0.002 | 0.005 |
| RNA degradation | 2.25 | 0.002 | 0.001 |
| Homologous recombination | 1.87 | 0.004 | 0.031 |
| Nucleotide excision repair | 1.86 | 0.008 | 0.034 |
Gene sets with NOM P-value ≤0.05 and FDR q-values ≤0.05 were considered as significant. NES, normalized enrichment score; NOM, nominal; FDR, false discovery rate.
Discussion
Nothing was previously known about GNPNAT1 in lung carcinomas. Recently, the expression and function of GNPNAT1 in cancer have been extensively reported (31). GNPNAT1 is an essential enzyme involved in the biosynthesis of UDP-GlcNAc and metabolism in eukaryotic cells (28). GNPNAT1 upregulation may affect the occurrence and development of LUAD by disturbing cell metabolism (27). Until now, the expression of GNPNAT1 and its potential prognostic value in LUAD has not been fully investigated. Hence, the present study aimed to evaluate the potential role of GNPNAT1 in LUAD.
In the present study, bioinformatics analysis of the expression data obtained from TCGA demonstrated that increased expression of GNPNAT1 in LUAD was associated with advanced clinical pathologic characteristics (stage, survival status and N classification). To further analyze and reveal the effects of GNPNAT1 expression in LUAD, GSEA was carried out. The analysis revealed that ‘cell cycle’, ‘oocyte meiosis’, ‘pyrimidine mediated metabolism’, ‘ubiquitin mediated proteolysis’, ‘one carbon pool by folate’, ‘mismatch repair’, ‘progesterone mediated oocyte maturation’, ‘basal transcription factors’ and ‘purine metabolism’ were enriched in the GNPNAT1 high-expression phenotype. This finding suggested that GNPNAT1 may be regarded as a potential prognostic biomarker and therapeutic target in LUAD. Cell cycle and DNA repair pathways are considered as the 2 most susceptible pathways in LUAD pathogenesis (21). In the cell cycle pathway, the inactivation of several cyclin genes, including CDKN2A, and cyclin-dependent kinases 4 and 6, promotes the escape of cells from the M0 checkpoint, eventually resulting in cellular immortalization, which is a characteristic of cancer cells (41). In the DNA repair pathway, breakdown of the repair system mediates the accumulation of mutations, especially those inactivating tumor suppressor genes and activating oncogenes (42), during the DNA replication process (43). For example, the mutations in AT rich interactive domain 2 results in truncated proteins through out-of-frame indels, nonsense mutations or splice site alterations in hepatocellular carcinoma (44). Somatic intronic mutations of oncogene Met led to an alternatively spliced transcript in lung cancer (45). GNPNAT1 may interfere with these pathways via regulating the activity of cyclin genes through post translational modifications (28). O-linked N-acetylglucosamine (GlcNAc) transferase (OGT) is necessary for the cell cycle since silencing of OGT prevents the synthesis of cyclin D1 (46).
The present study demonstrated that GNPNAT1 may be associated with LUAD carcinogenesis. A previous study demonstrated that silencing of GNPNAT1 attenuated cell proliferation, adhesion, and migration of cancer and fetal human colon cell lines (47). Hence, it was hypothesized that GNPNAT1 upregulation may promote cell migration during carcinogenesis (47). However, the molecular mechanisms underlying LUAD carcinogenesis are still poorly understood. Whether this phenotypic change was directly triggered by GNPNAT1 upregulation remains unknown (48). Hence, cytological evidence is required to further elucidate the biological function of GNPNAT1 in carcinogenesis.
In the future, knockdown of GNPNAT1 in a LUAD animal model could be performed to further evaluate the effects of GNPNAT1 in carcinogenesis. Abraxane®, a FDA approved drug is used to treat advanced breast, lung and pancreatic cancer, and it has been reported to be more effective compared with paclitaxel in the treatment of NSCLC (48). A comparative analysis in A549 lung cancer cells treated in parallel with abraxane and paclitaxel demonstrated that only GNPNAT1 was differentially expressed by 2-fold in A549 cells treated with different drugs (25). This finding indicated that the effects of abraxane may be mediated by GNPNAT1 downregulation, which may cause proliferative delay and cell adhesion defects. Once the role of GNPNAT1 upregulation in LUAD is determined, the screening of more effective and accessible antitumor drugs may be accelerated to benefit all patients suffering from LUAD. Therapeutic intervention based on the effects of GNPNAT1, possibly through mannose analogs, may also have a favorable effect on several diseases, including cancer, which could benefit from suppression of O-GlcNAc signaling and hyaluronan synthesis.
In conclusion, the expression of GNPNAT1 may be a potential and promising diagnostic and prognostic molecular marker of poor survival in patients with LUAD. In addition, the cell cycle and several metabolic pathways, such as pyrimidine metabolism and purine metabolism may be the key pathways regulated by GNPNAT1 in LUAD. However, further validation experiments are needed to verify the biologic effects of GNPNAT1.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the China postdoctoral science foundation (grant no. 2018M632352), Jiangsu Youth Medical Key Talents (grant no. QNRC2016405), Development of biodegradable anti-reflux anastomotic stent for reconstruction of digestive tract after esophagectomy (grant no. 18441902300), significance of imaging and immunohistochemical analysis in the differential diagnosis of solitary and multiple ground glass nodules of the lung (grant no. YLT1901) and Clinical study of vagus nerve preservation in minimally invasive surgery for early stage (IA1 and 2) lung cancer [grant no. ITJ(ZD)1906].
Funding
The present study was supported by the China postdoctoral science foundation (grant no. 2018M632352), Jiangsu Youth Medical Key Talents (grant no. QNRC2016405), Development of biodegradable anti-reflux anastomotic stent for reconstruction of digestive tract after esophagectomy (grant no. 18441902300), significance of imaging and immunohistochemical analysis in the differential diagnosis of solitary and multiple ground glass nodules of the lung (grant no. YLT1901) and Clinical study of vagus nerve preservation in minimally invasive surgery for early stage (IA1 and 2) lung cancer [grant no. ITJ(ZD)1906].
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
PZ, SG and HH were responsible for conceiving the study, drafting and editing of the manuscript. CZ, ZL and XZ contributed to the acquisition of TCGA data and all the data analysis needed by R packages, and confirmed the authenticity of all the raw data. WW and SX were in charge of patient sample collection, storage and western blotting. KW and TL were responsible for the statistical analysis. YZ was in charge of the study design and manuscript review for important intellectual content. All authors have read and approved the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The Second Affiliated Hospital of Nantong University and in compliance with the Declaration of Helsinki. Written patient consent for use of their tissues in research was obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Dubey AK, Gupta U, Jain S. Epidemiology of lung cancer and approaches for its prediction: A systematic review and analysis. Chin J Cancer. 2016;35:71. doi: 10.1186/s40880-016-0135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathur P, Sathishkumar K, Chaturvedi M, Das P, Sudarshan KL, Santhappan S, Nallasamy V, John A, Narasimhan S, Roselind FS, ICMR-NCDIR-NCRP Investigator Group Cancer statistics, 2020: Report from national cancer registry programme, India. JCO Glob Oncol. 2020;6:1063–1075. doi: 10.1200/GO.20.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Didkowska J, Wojciechowska U, Mańczuk M, Łobaszewski J. Lung cancer epidemiology: Contemporary and future challenges worldwide. Ann Transl Med. 2016;4:150. doi: 10.21037/atm.2016.03.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Punturieri A, Szabo E, Croxton TL, Shapiro SD, Dubinett SM. Lung cancer and chronic obstructive pulmonary disease: Needs and opportunities for integrated research. J Natl Cancer Inst. 2009;101:554–559. doi: 10.1093/jnci/djp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobti RC, Sharma S, Janmeja AK, Jindal SK. Molecular Pathogenesis of Lung Cancer. In: Sobit RC, Obe G, Athwal RS, editors. Some Aspects of Chromosome Structure and Functions 2002. Springer; Dordrecht: 2002. pp. 193–205. [DOI] [Google Scholar]
- 7.Massó-Vallés D, Beaulieu ME, Soucek L. MYC, MYCL, and MYCN as therapeutic targets in lung cancer. Expert Opin Ther Targets. 2020;24:101–114. doi: 10.1080/14728222.2020.1723548. [DOI] [PubMed] [Google Scholar]
- 8.Burbee DG, Forgacs E, Zöchbauer-Müller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Virno F, Di Giorgio A, Di Lauro G, Bellezza F, Carrozzini AI. Small cell lung cancer (SCLC) and non small cell lung cancer (NSCLC): Comparative evaluation of survival after surgical treatment by computer. Lung Cancer. 1986;2:105–106. doi: 10.1016/S0169-5002(86)80420-2. [DOI] [Google Scholar]
- 10.Sabari JK, Lok BH, Laird JH, Poirier JT, Rudin CM. Unravelling the biology of SCLC: Implications for therapy. Nat Rev Clin Oncol. 2017;14:549–561. doi: 10.1038/nrclinonc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riihimäki M, Hemminki A, Fallah M, Thomsen H, Sundquist K, Sundquist J, Hemminki K. Metastatic sites and survival in lung cancer. Lung Cancer. 2014;86:78–84. doi: 10.1016/j.lungcan.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Brzezniak C, Satram-Hoang S, Goertz HP, Reyes C, Gunuganti A, Gallagher C, Carter CA. Survival and Racial Differences of Non-Small Cell Lung Cancer in the United States Military. J Gen Intern Med. 2015;30:1406–1412. doi: 10.1007/s11606-015-3280-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong S, Men W, Yang S, Xu S. Identification of lung adenocarcinoma biomarkers based on bioinformatic analysis and human samples. Oncol Rep. 2020;43:1437–1450. doi: 10.3892/or.2020.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerfolio RJ, Bryant AS, Scott E, Sharma M, Robert F, Spencer SA, Garver RI. Women with pathologic stage I, II, and III non-small cell lung cancer have better survival than men. Chest. 2006;130:1796–1802. doi: 10.1378/chest.130.6.1796. [DOI] [PubMed] [Google Scholar]
- 15.Moore W, Talati R, Bhattacharji P, Bilfinger T. Five-year survival after cryoablation of stage I non-small cell lung cancer in medically inoperable patients. J Vasc Interv Radiol. 2015;26:312–319. doi: 10.1016/j.jvir.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Molassiotis A, Smith JA, Bennett MI, Blackhall F, Taylor D, Zavery B, Harle A, Booton R, Rankin EM, Lloyd-Williams M, et al. Clinical expert guidelines for the management of cough in lung cancer: Report of a UK task group on cough. Cough. 2010;6:9. doi: 10.1186/1745-9974-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastarrika G, García-Velloso MJ, Lozano MD, Montes U, Torre W, Spiteri N, Campo A, Seijo L, Alcaide AB, Pueyo J, et al. Early lung cancer detection using spiral computed tomography and positron emission tomography. Am J Respir Crit Care Med. 2005;171:1378–1383. doi: 10.1164/rccm.200411-1479OC. [DOI] [PubMed] [Google Scholar]
- 18.Lam S, MacAulay C, leRiche JC, Palcic B. Detection and localization of early lung cancer by fluorescence bronchoscopy. Cancer. 2000;89(Suppl):2468–2473. doi: 10.1002/1097-0142(20001201)89:11+<2468::AID-CNCR25>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 19.Wisnivesky JP, Yankelevitz D, Henschke CI. Stage of lung cancer in relation to its size: part 2. Chest. 2005;127:1136–11369. doi: 10.1378/chest.127.4.1136. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez JL, Sarries C, de Castro PL, Roig B, Queralt C, Escuin D, de Aguirre I, Sanchez JM, Manzano JL, Margelí M, et al. Methylation patterns and K-ras mutations in tumor and paired serum of resected non-small-cell lung cancer patients. Cancer Lett. 2003;193:207–216. doi: 10.1016/S0304-3835(02)00740-1. [DOI] [PubMed] [Google Scholar]
- 21.Andriani F, Conte D, Mastrangelo T, Leon M, Ratcliffe C, Roz L, Pelosi G, Goldstraw P, Sozzi G, Pastorino U. Detecting lung cancer in plasma with the use of multiple genetic markers. Int J Cancer. 2004;108:91–96. doi: 10.1002/ijc.11510. [DOI] [PubMed] [Google Scholar]
- 22.Mao L, Lee DJ, Tockman MS, Erozan YS, Askin F, Sidransky D. Microsatellite alterations as clonal markers for the detection of human cancer. Proc Natl Acad Sci USA. 1994;91:9871–9875. doi: 10.1073/pnas.91.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 24.Kwon Y-J, Lee SJ, Koh JS, Kim SH, Lee HW, Kang MC, Bae JB, Kim YJ, Park JH. Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. J Thorac Oncol. 2012;7:20–33. doi: 10.1097/JTO.0b013e3182307f62. [DOI] [PubMed] [Google Scholar]
- 25.Zhao M, Li H, Ma Y, Gong H, Yang S, Fang Q, Hu Z. Nanoparticle abraxane possesses impaired proliferation in A549 cells due to the underexpression of glucosamine 6-phosphate N-acetyltransferase 1 (GNPNAT1/GNA1) Int J Nanomedicine. 2017;12:1685–1697. doi: 10.2147/IJN.S129976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bacos K, Gillberg L, Volkov P, Olsson AH, Hansen T, Pedersen O, Gjesing AP, Eiberg H, Tuomi T, Almgren P, et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat Commun. 2016;7:11089. doi: 10.1038/ncomms11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta E, Okuda H, Kobayashi A, Watabe K. Metabolic genes in cancer: Their roles in tumor progression and clinical implications. Biochim Biophys Acta. 2010;1805:141–152. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aquino-Gil M, Pierce A, Perez-Cervera Y, Zenteno E, Lefebvre T. OGT: A short overview of an enzyme standing out from usual glycosyltransferases. Biochem Soc Trans. 2017;45:365–370. doi: 10.1042/BST20160404. [DOI] [PubMed] [Google Scholar]
- 29.Oikari S, Makkonen K, Deen AJ, Tyni I, Kärnä R, Tammi RH, Tammi MI. Hexosamine biosynthesis in keratinocytes: Roles of GFAT and GNPDA enzymes in the maintenance of UDP-GlcNAc content and hyaluronan synthesis. Glycobiology. 2016;26:710–722. doi: 10.1093/glycob/cww019. [DOI] [PubMed] [Google Scholar]
- 30.Chu J, Li N, Gai W. Identification of genes that predict the biochemical recurrence of prostate cancer. Oncol Lett. 2018;16:3447–3452. doi: 10.3892/ol.2018.9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Lu Y, Liu Z, Li X, Wang Z, Cai Z. Identification Six Metabolic Genes as Potential Biomarkers for Lung Adenocarcinoma. J Comput Biol. 2020;27:1532–1543. doi: 10.1089/cmb.2019.0454. [DOI] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. Cancer Genome Atlas Research Network Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickham H. ggplot2. Wiley Interdiscip Rev Comput Stat. 2011;3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- 35.Sobin LH, Fleming ID. TNM Classification of Malignant Tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80:1803–1804. doi: 10.1002/(SICI)1097-0142(19971101)80:9<1803::AID-CNCR16>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 36.Wu H, Zhang J. Decreased expression of TFAP2B in endometrial cancer predicts poor prognosis: A study based on TCGA data. Gynecol Oncol. 2018;149:592–597. doi: 10.1016/j.ygyno.2018.03.057. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haley JA, Haughney E, Ullman E, Bean J, Haley JD, Fink MY. Altered Transcriptional Control Networks with Trans-Differentiation of Isogenic Mutant-KRas NSCLC Models. Front Oncol. 2014;4:344. doi: 10.3389/fonc.2014.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Core Team, corp-author. R: A language and environment for statistical computing. Vienna, Austria. http://www.R-project.org/ 2013 [Google Scholar]
- 40.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 41.Singhal S, Vachani A, Antin-Ozerkis D, Kaiser LR, Albelda SM. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: A review. Clin Cancer Res. 2005;11:3974–3986. doi: 10.1158/1078-0432.CCR-04-2661. [DOI] [PubMed] [Google Scholar]
- 42.Weinberg RA. Oncogenes and tumor suppressor genes. CA Cancer J Clin. 1994;44:160–170. doi: 10.3322/canjclin.44.3.160. [DOI] [PubMed] [Google Scholar]
- 43.Wei Q, Cheng L, Hong WK, Spitz MR. Reduced DNA repair capacity in lung cancer patients. Cancer Res. 1996;56:4103–4107. [PubMed] [Google Scholar]
- 44.Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, Holcomb T, Pujara K, Stinson J, Fu L, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 46.Olivier-Van Stichelen S, Drougat L, Dehennaut V, El Yazidi-Belkoura I, Guinez C, Mir AM, Michalski JC, Vercoutter-Edouart AS, Lefebvre T. Serum-stimulated cell cycle entry promotes ncOGT synthesis required for cyclin D expression. Oncogenesis. 2012;1:e36. doi: 10.1038/oncsis.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steenackers A, Olivier-Van Stichelen S, Baldini SF, Dehennaut V, Toillon RA, Le Bourhis X, El Yazidi-Belkoura I, Lefebvre T. Silencing the nucleocytoplasmic O-GlcNAc transferase reduces proliferation, adhesion, and migration of cancer and fetal human colon cell lines. Front Endocrinol (Lausanne) 2016;7:46. doi: 10.3389/fendo.2016.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sparreboom A, Scripture CD, Trieu V, Williams PJ, De T, Yang A, Beals B, Figg WD, Hawkins M, Desai N. Comparative preclinical and clinical pharmacokinetics of a cremophor-free, nanoparticle albumin-bound paclitaxel (ABI-007) and paclitaxel formulated in Cremophor (Taxol) Clin Cancer Res. 2005;11:4136–4143. doi: 10.1158/1078-0432.CCR-04-2291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.