Abstract
Long non-coding RNA homeobox A11-antisense RNA (HOXA11-AS) has been implicated in cisplatin (DDP) resistance in multiple types of cancer. The purpose of the present study was to investigate the role of HOXA11-AS in DDP-resistant nasopharyngeal carcinoma (NPC) cells. The expression levels of HOXA11-AS were examined using reverse transcription-quantitative PCR. Cell viability was measured using a Cell Counting Kit-8 assay, and a TUNEL assay was utilized to assess cell apoptosis. The expression levels of apoptosis-related factors (Bax and Bcl-2) were detected by western blot analysis. The interaction between microRNA-98 (miR-98) and HOXA11-AS or pre-B-cell leukemia homeobox 3 (PBX3) was demonstrated using bioinformatics analysis, dual-luciferase reporter assays and RNA immunoprecipitation assays. HOXA11-AS and PBX3 expressions levels were upregulated, whereas miR-98 levels were downregulated in DDP-resistant NPC tissues. Patients with NPC with high HOXA11-AS expression had a low survival rate. Knockdown of HOXA11-AS enhanced the DDP sensitivity of DDP-resistant NPC (5-8F/DDP and SUNE1/DDP) cells, which was demonstrated by the accelerated apoptosis. In addition, HOXA11-AS inhibited the expression levels of miR-98 through direct interaction. Furthermore, miR-98 inhibition counteracted the inductive effect of HOXA11-AS-knockdown on the DDP sensitivity of NPC cells. PBX3 was a target of miR-98 and was positively modulated by HOXA11-AS. Overexpression of PBX3 reversed the suppressive effect of HOXA11-AS silencing on the DDP resistance of NPC cells. The data demonstrated that HOXA11-AS enhanced DDP resistance in NPC via the miR-98/PBX3 axis, providing a potential therapeutic target for patients with DDP-resistant NPC.
Keywords: homeobox A11-antisense RNA, microRNA-98, pre-B-cell leukemia homeobox 3, cisplatin resistance, nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is a multifaceted malignant tumor that originates from the nasopharynx epithelium (1,2). Currently, radiotherapy combined with chemotherapy is the main effective method for the treatment of NPC (3). Despite significant progress being made in the development of novel treatment methods for NPC, its 5-year survival rate remains <40%, which is mainly due to drug resistance (4,5). Therefore, it is vital to understand the mechanism of cisplatin (DDP) resistance in NPC.
Long non-coding RNAs (lncRNAs) are a type of non-coding RNA >200 nucleotides in length (6). Previous reports have revealed the regulatory role of lncRNAs in multiple types of cancer (7–9). In addition, lncRNAs have been demonstrated to exert crucial roles in the chemoresistance of human cancer types, including NPC (10). For instance, lncRNA MAGI2-antisense RNA 3 contributes to the DDP resistance of NPC by regulating the microRNA (miRNA/miR)-218-5p/glycerophosphodiester phosphodiesterase domain containing 5/SEC61 translocon subunit α1 axis (11). The lncRNA long intergenic non-protein coding RNA 346 promotes DDP resistance in NPC cells by sponging miR-342 (12). The knockdown of deleted in lymphocytic leukemia 1 sensitizes NPC to DDP by upregulating baculoviral IAP repeat containing 6 expression via miR-381 inhibition (13). lncRNA colon cancer associated transcript 1 regulates cytoplasmic polyadenylation element binding protein 2 expression by competitively binding to miR-181a, which results in the suppression of NPC cell paclitaxel sensitivity (14). However, the role of homeobox A11-antisense RNA (HOXA11-AS) in the DDP resistance of NPC remains uncertain.
miRNAs are a class of small non-coding RNA 20–22 nucleotides in length that can regulate multiple biological processes, including cell proliferation, metastasis, differentiation, proliferation and apoptosis (15,16). Previous studies have indicated that dysregulated miRNAs are frequently involved in the progression of tumors via various different mechanisms, including drug resistance (17,18). In addition, several miRNAs have been reported to modulate the DDP resistance of NPC. For example, miR-1278 suppresses the DDP resistance of NPC cells by targeting autophagy-related 2B (19). The inhibition of miR-125b expression facilitates DDP resistance and decreases DDP-induced apoptosis in NPC (20). Nevertheless, to the best of our knowledge, the involvement of miR-98 in DDP-resistant NPC remains elusive.
The present study aimed to investigate the biological role and molecular mechanism of HOXA11-AS in the chemoresistance of NPC, which might provide therapeutic strategies against the resistance of NPC to DDP.
Materials and methods
Patient samples
A total of 40 NPC tissues and adjacent normal tissues (2 cm away from the tumor tissues) were collected from patients (age range, 32–67 years; mean age, 56 years) who underwent surgical resection surgery at The Shanghai Ninth People's Hospital (Shanghai, China) between February 2017 and October 2019. Patients who had recurrent disease within 6 months of completing primary chemotherapy were classified as DDP-resistant (n=25). Patients with recurrence beyond 6 months or without recurrence were classified as DDP-sensitive (n=15). All patients who only had primary NPC were included in the present study. Patients with NPC who had received chemotherapy or radiotherapy prior to undergoing surgery were excluded from the present study. All specimens were quickly frozen in liquid nitrogen and stored at −80°C for subsequent use. The present study was approved by the Ethics Committee of Shanghai Ninth People's Hospital (Shanghai, China). All participants signed the relevant informed consent forms. The classification of clinical stage of patients with NPC was performed according to the American Joint Committee on Cancer (AJCC) guidelines 6th Edition (21).
Cell culture
The NP-69 human normal nasopharyngeal epithelial cell line (cat. no. BNCC338439), and the 5–8F (cat. no. BNCC353307) and SUNE1 (cat. no. BNCC342441) human NPC cell lines were purchased from BeNa Culture Collection (Beijing Beina Chunglian Biotechnology Research Institute). 5–8F and SUNE1 cells were cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) containing streptomycin (100 U/ml) and 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified 5% CO2 environment. DDP-resistant NPC (5-8F/DDP and SUNE1/DDP) cell lines were established by continuous exposure to progressively increasing DDP (Sigma Aldrich; Merck KGaA) concentrations (0.5, 2, 4, 6 and 8 µg/ml) every 4 weeks. DDP-resistant NPC cells were cultured in RPMI-1640 medium supplemented with 2 µg/ml DDP and 10% FBS at 37°C in a humidified incubator with 5% CO2.
Cell transfection
Small interfering RNA (si) sequences specific for HOXA11-AS (si-HOXA11-AS) or PBX3 (si-PBX3), scrambled control sequences [si-negative control (NC)], miR-98 inhibitor, scrambled inhibitor control (NC inhibitor), miR-98 mimics and scrambled mimic negative control (NC mimics) were obtained from Sangon Biotech Co., Ltd. The sequences used were as follows: si-HOXA11-AS-1, 5′-CUACCAUCCCUGAGCCUUA-3′; si-PBX3, 5′-GCGAGCAUGUUUGTUGUUUU-3′; si-NC, 5′-UCAAGUCCACGACGACUUUG-3′; miR-98 mimics, 5′-UGAGGUAGUAAGUUGUAUUGUU-3′; NC mimics, 5′-GUGUAACACGUCUAUACGCCCA-3′; miR-98 inhibitor, 5′-AACAAUACAACUUACUACCUC-3′; and NC inhibitor, 5′-ACUAUUGGAGAUUAGGUAUGUA-3′. To establish the role of HOXA11-AS or PBX3, their corresponding overexpression plasmids were constructed by cloning the full-length HOXA11-AS or PBX3 sequences into the pcDNA3.1 plasmid vector (Invitrogen; Thermo Fisher Scientific, Inc.). The final plasmids were referred to as pcDNA3.1-HOXA11-AS (HOXA11-AS) or pcDNA3.1-PBX3 (PBX3). pcDNA3.1 was used as a control. Cell were transfected with 50 nM miRNA mimic, 50 nM miRNA inhibitor, 50 nM si-HOXA11-AS, 50 nM si-PBX3, 50 nM si-NC or 4.0 µg pcDNA3.1-HOXA11-AS, 4.0 µg pcDNA3.1-PBX3 and 4.0 µg pcDNA3.1 using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 15 min according to the manufacturer's protocol. After 48 h of transfection, the transfected NPC cells were used for subsequent experiments.
Drug sensitivity assay
A Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.) was utilized to evaluate the sensitivity of NPC cells to DDP. 5–8F/DDP and SUNE1/DDP cells (1×104 cells/well) were plated into 96-well plates and treated with DDP (10 µg) for 48 h. Subsequently, 10 µl CCK-8 solution was added to each well and incubated with the cells for 1 h. The absorbance was recorded using a microplate reader (BioTek China) at 450 nm. DDP sensitivity was assessed using the IC50.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from NPC tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was obtained following the reverse transcription of RNA using a PrimeScript RT Reagent kit (Takara Bio, Inc.) according to the manufacturer's protocols. RT-qPCR was conducted using the SYBR Green RT-qPCR Master mix (Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Pre-denaturation at 95°C for 1 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec. The data were analyzed using the 2−ΔΔCq method (22). GAPDH or U6 was used as the internal control for HOXA11-AS and PBX3 or miR-98, respectively. The primer sequences were as follows: HOXA11-AS forward, 5′-ACGCTAGGCACCACTTTGTT-3′ and reverse, 5′-CCGGCTACTAGTCAGTGTGA-3′; miR-98-5p forward, 5′-ACACTCCCUAUACAACUUAC-3′ and reverse, 5′-GGGAAAGUAGUGAGGCCTCAGA-3′; PBX3 forward, 5′-CGAGGCGCAAGCAAAGAAAC-3′ and reverse, 5′-TGCCAAAAGCATATTGTCCAGT-3′; GAPDH forward, 5′-CCCACTCCTCCACCTTTGAC-3′ and reverse, 5′-GGATCTCGCTCCTGGAAGATG-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
TUNEL staining
5-8F/DDP and SUNE1/DDP cells were washed with PBS, and fixed in 4% paraformaldehyde solution for 1 h at 4°C. The cells were permeabilized in a solution containing 0.1% Triton X-100 for 2 min, and incubated in TUNEL reaction mixture for 1 h at 37°C. Subsequently, the TUNEL-stained cells were washed with PBS 3 times and counterstained with DAPI (2 µg/ml; Beyotime Institute of Biotechnology) under antifade mounting medium (cat. no. P0126; Beyotime Institute of Biotechnology) for 15 min at room temperature. The number of apoptotic cells was counted in ≥5 fields of view under a fluorescence microscope.
Bioinformatics prediction and dual luciferase reporter assay
The downstream target genes of HOXA11-As and miR-98 were screened using the Starbase version 2.0 (http://starbase.sysu.edu.cn) and TargetScan version 7.2 (http://www.targetscan.org/vert_72/) websites, respectively. The wild-type (WT) and mutant (Mut) sequences of HOXA11-AS and PBX3 were cloned into the pmirGLO vector (Promega Corporation). Subsequently, these constructed vectors were co-transfected with NC mimics (5′-GUGUAACACGUCUAUACGCCCA-3′) and miR-98 mimics (5′-UGAGGUAGUAAGUUGUAUUGUU-3′) from Sangon Biotech Co., Ltd into 5–8F/DDP and SUNE1/DDP cells for 48 h using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the relative luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP)
The RIP assay was conducted to assess the association between HOXA11-AS and miR-98 using the EZMagna RIP kit (EMD Millipore). The cells were lysed with 100 µl RIP Lysis Buffer (EMD Millipore) for 5 min at 4°C, the obtained cell lysate (100 µl) was centrifuged at 40,000 × g at 4°C for 10 min and incubated with 50 µl A/G magnetic beads conjugated with argonaute 2 (Ago2; 5 µg; cat. no. 2897; Cell Signaling Technology, Inc.) antibody and IgG (5 µg; cat. no. PP64B; EMD Millipore) antibody. Subsequently, the beads were washed 3 times using RIP Wash Buffer. RNA was extracted from the immunoprecipitation complex using 50 µg/ml Proteinase K (cat. no. P2308; Sigma-Aldrich; Merck KGaA) at 55°C for 30 min to digest the protein and the enrichment levels of HOXA11-AS and miR-98 were measured using RT-qPCR as mentioned previously.
Western blot analysis
Total proteins were extracted from the treated cells using RIPA lysis buffer (Beyotime Institute of Biotechnology). The protein concentration was detected using a BCA protein quantitation kit. The proteins (40 µg/lane) were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Following blocking for 2 h with 5% skimmed dry milk at room temperature, the membranes were incubated with antibodies against Bcl-2 (1:1,000; cat. no. ab32124; Abcam), Bax (1:1,000; cat. no. ab32503; Abcam) and GAPDH (1:1,000; cat. no. ab9485; Abcam) at 4°C overnight. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (1:1,000; cat. no. ab205718; Abcam) for 1 h at 37°C. Finally, the protein expression levels were examined using Pierce™ ECL A (Pierce; Thermo Fisher Scientific, Inc.) and analyzed using Quantity One software version 4.6 (Bio-Rad Laboratories, Inc.).
Statistical analysis
All experiments were repeated in triplicate. Statistical analysis was performed using SPSS version 16.0 (SPSS, Inc.) and the results are presented as the mean ± standard deviation. An unpaired Student's t-test was performed to determine differences between two groups, and a paired Student's t-test was used to determine differences between NPC tissues and adjacent normal tissues. Differences among multiple groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. Kaplan-Meier analysis and a log-rank test were used to assess the association between HOXA11-AS expression and patient survival, and patients were divided into high and low expression groups based on the mean value of HOXA11-AS expression in patients with NPC. In addition, the association between PBX3 expression and clinicopathological characteristics of patients with NPC was analyzed using χ2 tests, and the cut-off value (1.92) was the mean expression of PBX3 in patients with NPC. P<0.05 was considered to indicate a statistically significant difference.
Results
HOXA11-AS levels are increased in DDP-resistant NPC tissues and cells
The expression levels of HOXA11-AS were increased in NPC tissues compared with the levels in normal tissues (Fig. 1A). Furthermore, HOXA11-AS expression was higher in DDP-resistant NPC tissues compared with that in DDP-sensitive NPC tissues (Fig. 1B). To determine whether DDP-resistant NPC cell lines (5-8F/DDP and SUNE1/DDP) were successfully established, the IC50 of DDP was measured using a CCK-8 assay. The results indicated that 5–8F/DDP and SUNE1/DDP cells displayed low sensitivity to DDP, as demonstrated by the increased IC50 values (Fig. 1C and D). Furthermore, compared with NP-69 cells, 5–8F and SUNE1 cells exhibited elevated expression levels of HOXA11-AS, which were further upregulated in DDP-resistant NPC cells (5-8F/DDP and SUNE1/DDP cells; Fig. 1E and F). In addition, patients with high HOXA11-AS expression exhibited shorter survival times compared with patients with low expression levels of HOXA11-AS (Fig. 1G). Overall, the data suggested that HOXA11-AS upregulation may be involved in NPC DDP resistance.
Figure 1.
HOXA11-AS expression is enhanced in DDP-resistant NPC tissues and cells. (A) Expression levels of long non-coding RNA HOXA11-AS were determined in normal tissues (n=40) or NPC tissues (n=40) using RT-qPCR. (B) HOXA11-AS expression in DDP-sensitive or -resistant NPC tissues was detected by RT-qPCR. (C) IC50 values of DDP in 5–8F/DDP and 5–8F were measured using a Cell Counting Kit-8 assay. (D) IC50 values of DDP in SUNE1/DDP and SUNE1 cells were measured using a Cell Counting Kit-8 assay. (E) RT-qPCR assays were used to assess the expression levels of HOXA11-AS in normal nasopharyngeal epithelial NP-69, 5–8F/DDP and 5–8F cells. (F) RT-qPCR assays were used to assess the expression levels of HOXA11-AS in NP-69, SUNE1/DDP and SUNE1 cells. (G) Overall survival was evaluated by Kaplan-Meier curve analysis for high and low HOXA11-AS expression groups. Data are presented as the mean ± SD. *P<0.05. DDP, cisplatin; HOXA11-AS, homeobox A11-antisense RNA; HR, hazard ratio; NPC, nasopharyngeal carcinoma; RT-qPCR, reverse transcription-quantitative PCR.
Knockdown of HOXA11-AS attenuates DDP resistance of NPC cells
To further estimate the effect of HOXA11-AS on DDP-resistant NPC cells, si-HOXA11-AS was transfected into 5–8F/DDP and SUNE1/DDP cells to knockdown HOXA11-AS expression (Fig. 2A). Notably, HOXA11-AS knockdown increased the DDP sensitivity of 5–8F/DDP and SUNE1/DDP cells (Fig. 2B and C). Furthermore, silencing of HOXA11-AS promoted the apoptosis of 5–8F/DDP and SUNE1/DDP cells, which indicated that the downregulation of HOXA11-AS increased the chemosensitivity of DDP-resistant NPC cells (Fig. 2D). The expression levels of apoptosis-associated proteins (Bax and Bcl-2) were assessed by western blotting. HOXA11-AS depletion increased the protein expression levels of Bax and inhibited Bcl-2 expression in DDP-resistant NPC cells (Fig. 2E). Collectively, the data indicated that HOXA11-AS-knockdown suppressed DDP resistance in NPC cells.
Figure 2.
Knockdown of HOXA11-AS attenuates DDP resistance of NPC cells. (A) Reverse transcription-quantitative PCR was used to detect HOXA11-AS expression in 5–8F/DDP and SUNE1/DDP cells transfected with si-NC or si-HOXA11-AS. IC50 values of DDP in (B) 5–8F/DDP and (C) SUNE1/DDP cells transfected with si-NC or si-HOXA11-AS were measured using a Cell Counting Kit-8 assay. (D) Cell apoptosis of 5–8F/DDP and SUNE1/DDP cells transfected with si-HOXA11-AS or si-NC was detected by TUNEL staining. Magnification, ×200. (E) Protein expression levels of Bax and Bcl-2 in 5–8F/DDP and SUNE1/DDP cells transfected with si-HOXA11-AS or si-NC were detected by western blot analysis. Data are presented as the mean ± SD. *P<0.05. DDP, cisplatin; HOXA11-AS, homeobox A11-antisense RNA; NC, negative control; NPC, nasopharyngeal carcinoma; si, small interfering RNA.
HOXA11-AS acts as a molecular sponge for miR-98
At present, multiple studies have suggested that lncRNAs exert their biological function by interacting with specific miRNAs (23,24). Therefore, StarBase version 2.0 was used to uncover potential miRNAs, which could interact with HOXA11-AS. The data demonstrated that miR-98 contained complementary bases pairing with HOXA11-AS (Fig. 3A). RT-qPCR indicated that miR-98 expression was upregulated following miR-98 mimic transfection in 5–8F/DDP and SUNE1/DDP cells compared with in cells transfected with NC mimics (Fig. 3B). Luciferase reporter assays revealed that miR-98 mimics decreased the luciferase activity of HOXA11-AS-WT, whereas this effect was not noted for HOXA11-AS-Mut (Fig. 3C and D). In addition, HOXA11-AS and miR-98 were highly enriched by Ago2, while IgG was not significantly enriched (Fig. 3E and F). These results demonstrated that HOXA11-AS could directly bind to miR-98. The HOXA11-AS overexpression plasmid increased the expression levels of HOXA11-AS in both 5–8F/DDP and SUNE1/DDP cells compared with cells transfected with si-NC (Fig. 3G). To investigate whether HOXA11-AS could regulate the expression levels of miR-98, si-HOXA11-AS and HOXA11-AS overexpression plasmids were transfected into 5–8F/DDP and SUNE1/DDP cells. The results of RT-qPCR indicated that knockdown of HOXA11-AS increased miR-98 expression, whereas the transfection with HOXA11-AS plasmid decreased miR-98 expression (Fig. 3H and I). Furthermore, miR-98 expression was markedly decreased in NPC tissues and cell lines, particularly in DDP-resistant NPC tissues and cell lines (Fig. 3J-N). In summary, the results demonstrated that HOXA11-AS acted as a miR-98 sponge in NPC cells.
Figure 3.
HOXA11-AS serves as a molecular sponge for miR-98. (A) Putative binding regions of HOXA11-AS and miR-98 predicted using biological software. (B) RT-qPCR was used to detect the expression levels of miR-98 in 5–8F/DDP and SUNE1/DDP cells transfected with NC mimics or miR-98 mimics. Luciferase activity of WT or Mut HOXA11-AS in (C) 5–8F/DDP and (D) SUNE1/DDP cells following transfection with NC mimics or miR-98 mimics as determined by a luciferase reporter assay. The interaction between HOXA11-AS and miR-98 was further confirmed using an RNA immunoprecipitation assay in (E) 5–8F/DDP and (F) SUNE1/DDP cells. (G) Expression levels of HOXA11-AS in 5–8F/DDP and SUNE1/DDP cells transfected with NC or HOXA11-AS were detected by RT-qPCR. miR-98 expression was detected in (H) 5–8F/DDP and (I) SUNE1/DDP cells transfected with si-HOXA11-AS or HOXA11-AS overexpression plasmid. (J) miR-98 expression was measured in NPC tissues (n=40) or normal tissues (n=40). (K) miR-98 expression in DDP-sensitive or DDP-resistant NPC tissues was measured by RT-qPCR. (L) miR-98 expression in NP-69, 5–8F and 5–8F/DDP cells was measured by RT-qPCR. (M) miR-98 expression in NP-69, SUNE1 and SUNE1/DDP cells was measured by RT-qPCR. Data are presented as the mean ± SD. *P<0.05. Ago2, argonaute RISC catalytic component 2; DDP, cisplatin; HOXA11-AS, homeobox A11-antisense RNA; miR-98, microRNA-98; Mut, mutant; NC, negative control; NPC, nasopharyngeal carcinoma; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; WT, wild-type.
miR-98 inhibition abolishes the effects of si-HOXA11-AS on DDP resistance in NPC cells
The levels of miR-98 were markedly decreased in 5–8F/DDP and SUNE1/DDP cells transfected with miR-98 inhibitor (Fig. 4A). To further explore whether HOXA11-AS regulated the DDP resistance of NPC via miR-98, 5–8F/DDP and SUNE1/DDP cells were transfected with si-NC, si-HOXA11-AS and si-HOXA11-AS + miR-98 inhibitor. RT-qPCR analysis indicated that HOXA11-AS interference upregulated the expression levels of miR-98, whereas the treatment of the cells with the miR-98 inhibitor partially counteracted the induced expression of miR-98 (Fig. 4B). Furthermore, knockdown of HOXA11-AS increased the DDP sensitivity of 5–8F/DDP and SUNE1/DDP cells, which was reversed following the transfection of the cells with the miR-98 inhibitor (Fig. 4C). Furthermore, the miR-98 inhibitor neutralized the promoting effects of si-HOXA11-AS on the induction of cell apoptosis (Fig. 4D). The depletion of HOXA11-AS increased the protein expression levels of Bax and inhibited Bcl-2 expression, which was reversed following the transfection of cells with the miR-98 inhibitor (Fig. 4E). The data indicated that HOXA11-AS silencing promoted the DDP sensitivity of NPC cells by sponging miR-98.
Figure 4.
miR-98 inhibition abolishes the effects of si-HOXA11-AS on DDP resistance of nasopharyngeal carcinoma cells. (A) RT-qPCR was performed to examine the expression levels of miR-98 in 5–8F/DDP and SUNE1/DDP cells transfected with NC inhibitor or miR-98 inhibitor. (B) RT-qPCR analysis was performed to determine miR-98 expression in 5–8F/DDP and SUNE1/DDP cells transfected with si-NC, si-HOXA11-AS or si-HOXA11-AS + miR-98 inhibitor. (C) IC50 of DDP was evaluated in 5–8F/DDP and SUNE1/DDP cells transfected with si-NC, si-HOXA11-AS or si-HOXA11-AS + miR-98 inhibitor using a Cell Counting Kit-8 assay. (D) Cell apoptosis was detected by TUNEL staining (magnification, ×200) in 5–8F/DDP and SUNE1/DDP cells transfected with si-NC, si-HOXA11-AS or si-HOXA11-AS + miR-98 inhibitor. (E) Protein expression levels of Bax and Bcl-2 in 5–8F/DDP and SUNE1/DDP cells transfected with si-NC, si-HOXA11-AS or si-HOXA11-AS + miR-98 inhibitor were detected by western blot analysis. Data are presented as the mean ± SD. *P<0.05. DDP, cisplatin; HOXA11-AS, homeobox A11-antisense RNA; miR-98, microRNA-98; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA.
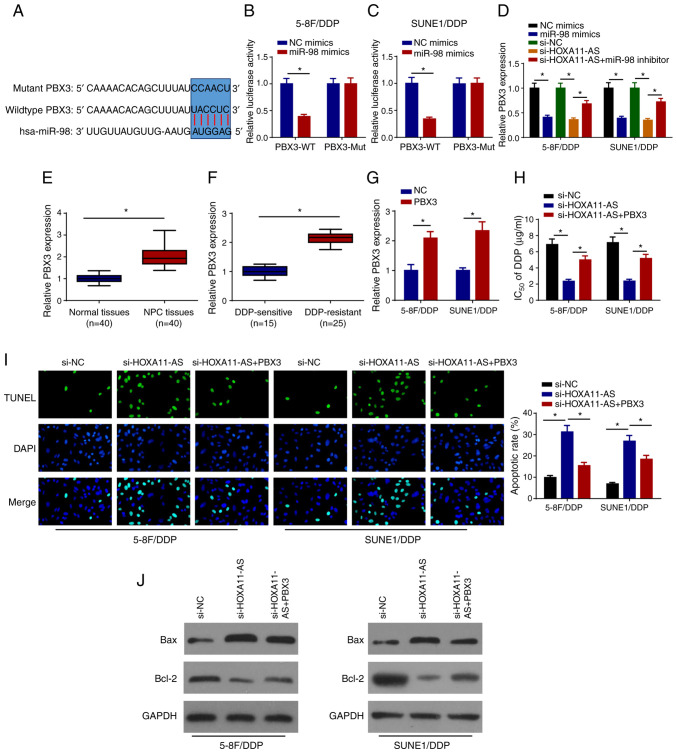
HOXA11-AS confers DDP resistance by upregulating PBX3 expression in NPC cells via miR-98 sponging
TargetScan version 7.2 was used to predict the candidate targets of miR-98. As shown in Fig. 5A, PBX3 contained a miR-98 binding sequence in the 3′-untranslated region. Furthermore, it was revealed that miR-98 mimics markedly decreased the relative luciferase activity of PBX3-WT, while the activity of PBX3-Mut remained unchanged, suggesting that PBX3 was a direct target of miR-98 (Fig. 5B and C). The effects of miR-98 and HOXA11-AS on the regulation of PBX3 expression were investigated in 5–8F/DDP and SUNE1/DDP cells following transfection with NC mimics, miR-98 mimics, si-NC, si-HOXA11-AS and si-HOXA11-AS + miR-98 inhibitor. RT-qPCR analysis revealed that PBX3 expression was downregulated in DDP-resistant NPC cells transfected with miR-98 mimics and si-HOXA11-AS, whereas the inhibitory effects of HOXA11-AS-knockdown on PBX3 expression were abrogated by miR-98 inhibition (Fig. 5D). Upregulated levels of PBX3 were directly positively associated with lymph node metastasis, distant metastasis and clinical stage (Table I). In addition, PBX3 levels were upregulated in NPC tissues and DDP-resistant NPC tissues (Fig. 5E and F). To further explore whether HOXA11-AS was associated with NPC DDP resistance by regulating PBX3 expression, 5–8F/DDP and SUNE1/DDP cells were transfected with si-NC, si-HOXA11-AS or si-HOXA11-AS + PBX3. RT-qPCR revealed that PBX3 overexpression plasmid transfection increased PBX3 expression in 5–8F/DDP and SUNE1/DDP cells compared with the cells transfected with NC (pcDNA3.1) (Fig. 5G), and the overexpression of PBX3 abolished the effects of HOXA11-AS silencing on the DDP sensitivity of 5–8F/DDP and SUNE1/DDP cells (Fig. 5H). Furthermore, the overexpression of PBX3 reversed the inductive effects of si-HOXA11-AS on cell apoptosis (Fig. 5I). The interference of HOXA11-AS increased the protein expression levels of Bax and inhibited Bcl-2 expression, which was partially rescued by the overexpression of PBX3 (Fig. 5J). In summary, these findings indicated that HOXA11-AS promoted DDP resistance in NPC cells by regulating the miR-98/PBX3 axis.
Figure 5.
HOXA11-AS confers DDP resistance via upregulation of PBX3 expression in NPC cells by sponging miR-98. (A) Binding sites between PBX3 and miR-98 were predicted. Luciferase activities of WT or Mut PBX3 in (B) 5–8F/DDP and (C) SUNE1/DDP cells following transfection of NC mimics or miR-98 mimics were detected using a luciferase reporter assay. (D) RT-qPCR was used to detect PBX3 expression in 5–8F/DDP and SUNE1/DDP cells transfected with NC mimics, miR-98 mimics, si-NC, si-HOXA11-AS, si-HOXA11-AS + miR-98 inhibitor. (E) RT-qPCR was used to detect the expression levels of PBX3 in NPC and normal tissues. (F) PBX3 expression in DDP-sensitive or DDP-resistant NPC tissues was detected using RT-qPCR. (G) Expression levels of PBX3 were detected by RT-qPCR in 5–8F/DDP and SUNE1/DDP cells transfected with NC or PBX3. (H) IC50 of DDP was measured in 5–8F/DDP and SUNE1/DDP cells transfected with si-NC, si-HOXA11-AS or si-HOXA11-AS + PBX3 using a Cell Counting Kit-8 assay. (I) Cell apoptosis was measured by TUNEL staining (magnification, ×200) in 5–8F/DDP and SUNE1/DDP cells transfected with si-NC, si-HOXA11-AS or si-HOXA11-AS + PBX3. (J) Protein expression levels of Bax and Bcl-2 in 5–8F/DDP and SUNE1/DDP cells transfected with si-NC, si-HOXA11-AS or si-HOXA11-AS + PBX3 were assessed by western blot analysis. Data are presented as the mean ± SD. *P<0.05. DDP, cisplatin; HOXA11-AS, homeobox A11-antisense RNA; miR-98, microRNA-98; Mut, mutant; NC, negative control; NPC, nasopharyngeal carcinoma; PBX3, pre-B-cell leukemia homeobox 3; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; WT, wild-type.
Table I.
Association between clinicopathological characteristics and PBX3 expression in nasopharyngeal carcinoma.
| Variable | No. of patients | Low PBX3 expression, n | High PBX3 expression, n | P-value |
|---|---|---|---|---|
| Age, years | 0.934 | |||
| <55 | 19 | 10 | 9 | |
| ≥55 | 21 | 11 | 10 | |
| Sex | 0.973 | |||
| Male | 22 | 12 | 10 | |
| Female | 18 | 9 | 9 | |
| Lymph node metastasis | 0.001 | |||
| No | 23 | 16 | 7 | |
| Yes | 17 | 5 | 12 | |
| Distant metastasis | 0.012 | |||
| No | 22 | 14 | 8 | |
| Yes | 18 | 7 | 11 | |
| Clinical stage | 0.001 | |||
| I/II | 21 | 15 | 6 | |
| III/IV | 19 | 6 | 13 |
PBX3, pre-B-cell leukemia homeobox 3.
Discussion
Chemoresistance has been considered to be a barrier to the effective treatment of NPC (25,26). Therefore, it is necessary to clarify the underlying mechanism of chemoresistance in NPC. In the present study, knockdown of HOXA11-AS enhanced the DDP sensitivity of NPC cells by sponging miR-98 and suppressing PBX3. Preliminary evidence suggested that the HOXA11-AS/miR-98/PBX3 axis was involved in the regulation of DDP resistance in NPC.
The expression levels of HOXA11-AS have been investigated in multiple studies (27–29). According to previous studies, HOXA11-AS has been identified as an oncogene in several types of cancer, including gastric cancer (30), glioma (31) and hepatocellular carcinoma (32). In addition, HOXA11-AS promotes the DDP resistance of lung cancer via regulation of the miR-454/STAT3 axis (33). Similarly, HOXA11-AS contributes to the DDP resistance and proliferation of oral squamous cell carcinoma by inhibiting miR-214 expression (34). In the present study, HOXA11-AS expression was increased in DDP-resistant NPC tissues and cells. Furthermore, knockdown of HOXA11-AS promoted the sensitivity and apoptosis of DDP-resistant NPC cells. These data revealed that HOXA11-AS conferred DDP resistance in NPC cells.
Accumulating evidence has demonstrated that lncRNAs act as competing endogenous RNAs by sponging miRNAs to participate in tumor progression and drug resistance (35). In the present study, HOXA11-AS served as a molecular sponge for miR-98. miR-98 has been demonstrated to be a cancer-associated miRNA, with abnormal expression in multiple cancer types (36–38). For instance, miR-98 suppresses the migration of breast cancer by targeting E2F transcription factor 5 (39). Upregulation of miR-98 expression suppresses the development of non-small cell lung cancer by targeting MAPK6 (40). In addition, miR-98 sensitizes DDP-resistant lung adenocarcinoma cells by regulating high mobility group AT-hook 2 (41). miR-98 overexpression impedes proliferation and enhances the sensitivity of leukemia cells to Adriamycin (42). The present study demonstrated that miR-98 was a direct target of HOXA11-AS. Overexpression of HOXA11-AS suppressed miR-98 expression, whereas knockdown enhanced miR-98 expression. Furthermore, HOXA11-AS-knockdown enhanced the DDP sensitivity of NPC cells by sponging miR-98.
PBX3 is a member of the PBX family, which is implicated in the development of various cancer types (43). For example, the overexpression of PBX3 accelerates the development of gastric cancer (44). Knockdown of PBX3 inhibits the cervical cancer cell epithelial-mesenchymal transition process (45). To the best of our knowledge, the present study provided the first preliminary evidence regarding the interaction between PBX3 (target gene) and miR-98. PBX3 expression was markedly increased in NPC tissues, notably in DDP-resistant NPC tissues. miR-98 overexpression and HOXA11-AS-knockdown markedly decreased the expression levels of PBX3, whereas miR-98 inhibition partly abolished the repressive effects of HOXA11-AS-knockdown on PBX3 expression. In addition, overexpression of PBX3 weakened the increased DDP sensitivity of 5–8F/DDP and SUNE1/DDP cells caused by HOXA11-AS silencing.
In conclusion, the present study demonstrated that HOXA11-AS conferred DDP resistance of NPC via the miR-98/PBX3 axis. These findings suggested that HOXA11-AS may be a valuable target for overcoming DDP resistance in patients with NPC. In future studies, the mechanism by which the HOXA11-AS/miR-98/PBX3 axis increases DDP resistance of NPC should be explored.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HNL, YZ and QW designed the present study. JH and SY performed the experiments. HBL and HNL analyzed the data and prepared the figures. HNL and YZ drafted the initial manuscript. QW reviewed and revised the manuscript. The authenticity of all the raw data must have been assessed by HNL and QW. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Shanghai Ninth People's Hospital (Shanghai, China). All participants provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhang W, Zhang Y, Xi S. Upregulation of lncRNA HAGLROS enhances the development of nasopharyngeal carcinoma via modulating miR-100/ATG14 axis-mediated PI3K/AKT/mTOR signals. Artif Cells Nanomed Biotechnol. 2019;47:3043–3052. doi: 10.1080/21691401.2019.1640233. [DOI] [PubMed] [Google Scholar]
- 2.Fan C, Tang Y, Wang J, Wang Y, Xiong F, Zhang S, Li X, Xiang B, Wu X, Guo C, et al. Long non-coding RNA LOC284454 promotes migration and invasion of nasopharyngeal carcinoma via modulating the Rho/Rac signaling pathway. Carcinogenesis. 2019;40:380–391. doi: 10.1093/carcin/bgy143. [DOI] [PubMed] [Google Scholar]
- 3.Kang M, Zhou P, Li G, Yan H, Feng G, Liu M, Zhu J, Wang R. Validation of the 8th edition of the UICC/AJCC staging system for nasopharyngeal carcinoma treated with intensity-modulated radiotherapy. Oncotarget. 2017;8:70586–70594. doi: 10.18632/oncotarget.19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei F, Wu Y, Tang L, He Y, Shi L, Xiong F, Gong Z, Guo C, Li X, Liao Q, et al. BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM. Br J Cancer. 2018;118:233–247. doi: 10.1038/bjc.2017.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamran SC, Riaz N, Lee N. Nasopharyngeal carcinoma. Surg Oncol Clin North Am. 2015;24:547–561. doi: 10.1016/j.soc.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Kim ED, Sung S. Long noncoding RNA: Unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 2012;17:16–21. doi: 10.1016/j.tplants.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y, Liu Y, Zhang N, Yang Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234:9105–9117. doi: 10.1002/jcp.27587. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X, Tai S. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233. doi: 10.1016/j.cca.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Feng L, Liu P, Duan W. ANRIL promotes chemoresistance via disturbing expression of ABCC1 by regulating the expression of Let-7a in colorectal cancer. Biosci Rep. 2018;38:BSR20180620. doi: 10.1042/BSR20180620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao C, Zhou S, Hu J. Long noncoding RNA MAGI2-AS3/miR-218-5p/GDPD5/SEC61A1 axis drives cellular proliferation and migration and confers cisplatin resistance in nasopharyngeal carcinoma. Int Forum Allergy Rhinol. 2020;10:1012–1023. doi: 10.1002/alr.22562. [DOI] [PubMed] [Google Scholar]
- 12.Cui Z, Pu T, Zhang Y, Wang J, Zhao Y. Long non-coding RNA LINC00346 contributes to cisplatin resistance in nasopharyngeal carcinoma by repressing miR-342-5p. Open Biol. 2020;10:190286. doi: 10.1098/rsob.190286. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Li H, Huang J, Yu S, Lou Z. Long non-coding RNA DLEU1 Up-regulates BIRC6 expression by competitively sponging miR-381-3p to promote cisplatin resistance in nasopharyngeal carcinoma. Onco Targets Ther. 2020;13:2037–2045. doi: 10.2147/OTT.S237456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Zhang W, Hao S. LncRNA CCAT1 modulates the sensitivity of paclitaxel in nasopharynx cancers cells via miR-181a/CPEB2 axis. Cell Cycle. 2017;16:795–801. doi: 10.1080/15384101.2017.1301334. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Lenkala D, LaCroix B, Gamazon ER, Geeleher P, Im HK, Huang RS. The impact of microRNA expression on cellular proliferation. Hum Genet. 2014;133:931–938. doi: 10.1007/s00439-014-1434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tufekci KU, Meuwissen RL, Genc S. The role of microRNAs in biological processes. Methods Mol Biol. 2014;1107:15–31. doi: 10.1007/978-1-62703-748-8_2. [DOI] [PubMed] [Google Scholar]
- 17.Mihanfar A, Fattahi A, Nejabati HR. MicroRNA-mediated drug resistance in ovarian cancer. J Cell Physiol. 2019;234:3180–3191. doi: 10.1002/jcp.26060. [DOI] [PubMed] [Google Scholar]
- 18.van Jaarsveld MT, Helleman J, Berns EM, Wiemer EA. MicroRNAs in ovarian cancer biology and therapy resistance. Int J Biochem Cell Biol. 2010;42:1282–1290. doi: 10.1016/j.biocel.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Wang P, Wu Q. miR-1278 sensitizes nasopharyngeal carcinoma cells to cisplatin and suppresses autophagy via targeting ATG2B. Mol Cell Probes. 2020;53:101597. doi: 10.1016/j.mcp.2020.101597. [DOI] [PubMed] [Google Scholar]
- 20.Yuan TZ, Zhang HH, Lin XL, Yu JX, Yang QX, Liang Y, Deng J, Huang LJ, Zhang XP. microRNA-125b reverses the multidrug resistance of nasopharyngeal carcinoma cells via targeting of Bcl-2. Mol Med Rep. 2017;15:2223–2228. doi: 10.3892/mmr.2017.6233. [DOI] [PubMed] [Google Scholar]
- 21.Carey LA, Metzger R, Dees EC, Collichio F, Sartor CI, Ollila DW, Klauber-DeMore N, Halle J, Sawyer L, Moore DT, Graham ML. American Joint Committee on Cancer tumor-node-metastasis stage after neoadjuvant chemotherapy and breast cancer outcome. J Natl Cancer Inst. 2005;97:1137–1142. doi: 10.1093/jnci/dji206. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Lou W, Ding B, Yang B, Lu H, Kong Q, Fan W. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging. 2019;11:2610–2627. doi: 10.18632/aging.101933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo H, Xu C, Le W, Ge B, Wang T. lncRNA CASC11 promotes cancer cell proliferation in bladder cancer through miRNA-150. J Cell Biochem. 2019;120:13487–13493. doi: 10.1002/jcb.28622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Xie T, Zhong X, Jiang HL, Li R, Wang BY, Huang XT, Cen BH, Yuan YW. Melatonin reverses nasopharyngeal carcinoma cisplatin chemoresistance by inhibiting the Wnt/beta-catenin signaling pathway. Aging (Albany NY) 2020;12:5423–5438. doi: 10.18632/aging.102968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R, Li SW, Liu L, Yang J, Huang G, Sang Y. TRIM11 facilitates chemoresistance in nasopharyngeal carcinoma by activating the beta-catenin/ABCC9 axis via p62-selective autophagic degradation of Daple. Oncogenesis. 2020;9:45. doi: 10.1038/s41389-020-0229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue JY, Huang C, Wang W, Li HB, Sun M, Xie M. HOXA11-AS: A novel regulator in human cancer proliferation and metastasis. Onco Targets Ther. 2018;11:4387–4393. doi: 10.2147/OTT.S166961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Zhang Y, You Q, Fu H, Zhao X, Lu K, Yan R, Yang D. LncRNA PTCSC3 alleviates the postoperative distant recurrence of gastric cancer by suppression of lncRNA HOXA11-AS. Cancer Manag Res. 2020;12:2623–2629. doi: 10.2147/CMAR.S229269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu X, Yang B, Liu F, Fang Q. LncRNA HOXA11-AS promotes OSCC progression by sponging miR-98-5p to upregulate YBX2 expression. Biomed Pharmacother. 2020;121:109623. doi: 10.1016/j.biopha.2019.109623. [DOI] [PubMed] [Google Scholar]
- 30.Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 31.Xu CH, Xiao LM, Liu Y, Chen LK, Zheng SY, Zeng EM, Li DH. The lncRNA HOXA11-AS promotes glioma cell growth and metastasis by targeting miR-130a-5p/HMGB2. Eur Rev Med Pharmacol Sci. 2019;23:241–252. doi: 10.26355/eurrev_201901_16770. [DOI] [PubMed] [Google Scholar]
- 32.Zhan M, He K, Xiao J, Liu F, Wang H, Xia Z, Duan X, Huang R, Li Y, He X, et al. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J Cell Mol Med. 2018;22:3758–3767. doi: 10.1111/jcmm.13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao X, Li X, Zhou L, Ni J, Yan W, Ma R, Wu J, Feng J, Chen P. LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via modulating miR-454-3p/Stat3. Cancer Sci. 2018;109:3068–3079. doi: 10.1111/cas.13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Li H, Shi J. LncRNA HOXA11-AS promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by suppression of miR-214-3p expression. Biomed Res Int. 2019;2019:8645153. doi: 10.1155/2019/8645153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie C, Chen B, Wu B, Guo J, Cao Y. LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in osteosarcoma by regulating miR-212-3p/FOXA1 axis. Biomed Pharmacother. 2018;97:1645–1653. doi: 10.1016/j.biopha.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Luo H, Yang L, Liu C, Wang X, Dong Q, Liu L, Wei Q. TMPO-AS1/miR-98-5p/EBF1 feedback loop contributes to the progression of bladder cancer. Int J Biochem Cell Biol. 2020;122:105702. doi: 10.1016/j.biocel.2020.105702. [DOI] [PubMed] [Google Scholar]
- 37.Yahya SM, Yahya SM. The effect of miR-98 and miR-214 on apoptotic and angiogenic pathways in hepatocellular carcinoma HepG2 cells. Indian J Clin Biochem. 2020;35:353–358. doi: 10.1007/s12291-019-00824-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu K, Xie Q, Jiang S, Lin T. miR-98-5p promotes apoptosis and inhibits migration and cell growth in papillary thyroid carcinoma through Bax/Caspase-3 by HMGA2. J Clin Lab Anal. 2020;34:e23044. doi: 10.1002/jcla.23044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485:272–278. doi: 10.1016/j.bbrc.2017.02.094. [DOI] [PubMed] [Google Scholar]
- 40.Wu F, Mo Q, Wan X, Dan J, Hu H. NEAT1/hsa-mir-98-5p/MAPK6 axis is involved in non-small-cell lung cancer development. J Cell Biochem. 2019;120:2836–2846. doi: 10.1002/jcb.26442. [DOI] [PubMed] [Google Scholar]
- 41.Xiang Q, Tang H, Yu J, Yin J, Yang X, Lei X. MicroRNA-98 sensitizes cisplatin-resistant human lung adenocarcinoma cells by up-regulation of HMGA2. Pharmazie. 2013;68:274–281. [PubMed] [Google Scholar]
- 42.Huang Y, Hong X, Hu J, Lu Q. Targeted regulation of MiR-98 on E2F1 increases chemosensitivity of leukemia cells K562/A02. Onco Targets Ther. 2017;10:3233–3239. doi: 10.2147/OTT.S126819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo H, Chu Y, Wang L, Chen X, Chen Y, Cheng H, Zhang L, Zhou Y, Yang FC, Cheng T, et al. PBX3 is essential for leukemia stem cell maintenance in MLL-rearranged leukemia. Int J Cancer. 2017;141:324–335. doi: 10.1002/ijc.30739. [DOI] [PubMed] [Google Scholar]
- 44.Wang S, Li C, Wang W, Xing C. PBX3 promotes gastric cancer invasion and metastasis by inducing epithelial-mesenchymal transition. Oncol Lett. 2016;12:3485–3491. doi: 10.3892/ol.2016.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Wang J, Xu F, Wang L, Sun G, Wang J, Yang Y. By downregulating PBX3, miR-526b suppresses the epithelial-mesenchymal transition process in cervical cancer cells. Future Oncol. 2019;15:1577–1591. doi: 10.2217/fon-2018-0575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.