Abstract
Several chemotherapeutic options are available for patients with metastatic colorectal cancer (mCRC), making it important to individualize treatment regimens. Individualization requires the clinical application of biomarkers for regimen selection, which is presently insufficient. miRNAs serve an important role in the control of biological processes in several types of cancer, acting as plasma biomarkers. The current study aimed to evaluate novel plasma microRNAs for predicting chemo-resistance in chemotherapy for patients with colorectal cancer (CRC) by employing a Toray 3D-Gene microRNA array-based approach, which compared plasma content before and during treatment. Specific miRNAs that acted as biomarkers of the fluoropyrimidine (FP) + oxaliplatin (OX) + bevacizumab (BEV) regime, a common first-line treatment for mCRC, were searched. The plasma samples of 110 patients with mCRC who had received the FP+OX+BEV regimen were subjected to microarray analyses using the 3D-Gene miRNA microarray platform, after which miRNAs levels were quantified via reverse transcription- quantitative PCR. Patients exhibiting complete response, partial response (PR) and reduced stable disease (SD) were defined as responders. Patients with extended SD and progression disease (PD) were defined as non-responders. Following microarray analysis, miR-33a-5p was selected as the candidate miRNA as it was upregulated in non-responder plasma samples. The expression of miR-33a-5p was upregulated in the non-responders (n=15) compared with the responders (n=95) (P=0.032). The high expression group demonstrated significantly poor progression-free survival (P<0.01). To evaluate whether miR-33a-5p can serve as a marker of chemo-resistance, miR-33a-5p expression levels were assessed at the following three time-points: Pre-point (before chemotherapy); PR-point (3-months after chemotherapy began); and PD-point (the time at which recurrence or progression was recorded). The results revealed that expression levels were significantly increased at the PD-point when compared with that at the pre-point (P=0.024). The current study determined that the miR-33a-5p expression level in the plasma may serve as a predictive marker of efficacy and as a biomarker of chemo-resistance.
Keywords: colorectal cancer, biomarker, chemo-resistance, microRNA, microRNA-33a-5p
Introduction
Colorectal cancer (CRC) is the fourth-most common cause of cancer deaths across the world and the second-most common cause of cancer deaths in Japan (1). The median overall survival (OS) of metastatic CRC (mCRC) in the past 20 years has remarkably improved from 6–12 months to >30 months (2) as a result of the development of multidisciplinary therapies (3,4). In earlier-line chemotherapy, cytotoxic agents such as fluoropyrimidine (FP), oxaliplatin (OX), irinotecan, anti-angiogenesis inhibitors, and anti-epidermal growth factor (EGFR) antibodies are used (5). Presently, RAS mutations and tumor locations are employed as predictive markers for the effect of anti-EGFR antibody drugs, and BRAF mutations are clinically applied as a poor prognosis marker, as recommended by the European Society for Medical Oncology (ESMO) guidelines (6). It is therefore important to select a suitable regimen for each patient; however, the clinical application of biomarkers for regimen selection is presently insufficient. Therefore, it is believed that the identification of predictive biomarkers can guide treatment decisions for the better management of CRC patients (2). The discovery of biomarkers for the prediction of treatment responses as well as resistance to such therapies can help in the selection of the most favorable, first-line chemotherapy regimen for mCRC. Biomarkers that can determine the degree of chemo-resistance are also considered useful for designing appropriate treatment strategies, including modifications in the regimen. The identification of such biomarkers would facilitate the elucidation of the mechanism of acquired resistance and in developing methods to overcome it. Clarification of the mechanism of the acquired resistance would provide a background for developing drugs that can overcome resistance.
According to recent evidences, miRNAs, which are small nucleotide sequences of noncoding RNA, play an important role in the control of biological processes, such as cellular development, differentiation, proliferation, apoptosis, and metabolism (7). It is already known that miRNA secreted or deviated from the cancer cells exist in the blood in a stable condition. Recent studies have demonstrated that miRNA is associated with the pathogenesis of numerous diseases, including cancer. MiRNA can act as transacting factors by suppressing translation or inducing messenger RNA (mRNA) degradation of the target genes (8). MiRNAs suppress important cancer-related genes and are differentially regulated in different human cancer types, which suggest their role as tumor suppressors or oncogenes (9). Aberrant expression of miRNAs has been reported in cancers of various tissues, such as the lung, breast, liver, colon, rectum, and prostate (10). Moreover, recent studies have suggested that the aberrant miRNA expression can play an important role in the chemotherapy response (11). Some past studies have reported that some miRNAs are involved in CRC chemo-resistance (12–15). Furthermore, another study has reported that miR-375 mediates the acquired chemo-resistance in cervical cancer (16).
The specifically altered miRNA expression patterns in the plasma may serve as diagnostic or prognostic markers (10). Because liquid biopsy is convenient to perform and minimally invasive in nature, plasma miRNAs can be measured repeatedly and monitored during chemotherapy over a period of time. The miRNAs level in the plasma may be related to the efficacy of chemotherapy and chemo-resistance. It has also been reported that the miRNA level in the plasma can act as a predictive biomarker of adjuvant chemotherapy for gastric cancer. In the past, miR-1229-3p was detected in the plasma of gastric cancer patients by using the Toray 3D-Gene microRNA array, and the clinical usefulness and the mechanism of chemo-resistance were studied in gastric cancer patients (17). The combination chemotherapy with FP, OX, and bevacizumab (BEV) is one of the most common regimens used in the first-line of chemotherapy for mCRC patients.
In this study, we identified miR-33a-5p as the predictive biomarker for FP+OX+BEV therapy by using the Toray 3D-Gene microRNA array. The clinical usefulness of miR-33a-5p and the association between miR-33a-5p and chemo-resistance were analyzed by quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR) using the plasma samples of mCRC patients who received the FP+OX+BEV therapy.
Materials and methods
Patients and samples
The plasma samples examined in this study were obtained from a total of 110 mCRC patients who received the FP+OX+BEV regimen as the first-line of chemotherapy in Tokyo Medical and Dental University Hospital between June 2011 and August 2018. All patients received CT examination every 3 months after chemotherapy, and the chemo-efficacy was evaluated with reference to the Response Evaluation Criteria in Solid Tumors (18). The complete response, partial response (PR), and reduced stable disease (SD) were defined as responders, and the extended SD and progression disease (PD) were defined as non-responders in this study.
Microarray analysis
The microarray analyses of the plasma samples were performed using the 3D-Gene miRNA Microarray Platform (Toray Industries) (19). We collected 300 µl of the pretreatment plasma samples from the mCRC patients. RNA extraction and microarray analysis were performed as per the manufacturer's instructions (20). In brief, the amount of total RNAs in the plasma was very little; therefore, 2–4 µl of the extracted total RNAs from 300 µl of the plasma samples were used in the microarray experiments. The extracted RNAs were labeled using the 3D-Gene miRNA Labeling Kit (Toray) and hybridized at 32°C for 16 h on the 3D-Gene Chip. The 3D-Gene miRNA Microarray (Human_miRNA_17v1.0.0; Toray Industries) can mount 41500 miRNAs based on the Human miRNA Version17 of MirBase (http://microrna.-sanger.ac.uk/) (21). The microarray was scanned, and the images obtained were enumerated by using the 3D-GeneH Scanner 3000 (Toray Industries). The expression level of each miRNA was globally normalized using the background-subtracted signal intensity of the entire set of miRNAs in each microarray. The obtained microarray images were analyzed using the GenePix Pro™ (Molecular Devices).
miRNA expression assay
Total RNA was extracted from 200 µl of plasma by using the miRNeasy Serum/Plasma Kit (Qiagen), as per the manufacturer's instructions. Then, 5.6×108 copies cel-miR-39 synthetic RNA/sample were used as the spike-in control (Qiagen). The RNA was eluted in 15-µl nuclease-free water supplied with the kit. The reverse transcription reaction was performed with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). We used cel-miR-39 (miRNeasy Serum/Plasma Spike-In Control: Qiagen) as the internal control. We then mixed 15 µl solution containing 1 µl of the extracted RNAs, 0.15 µl of 100 mM dNTPs, 1.00 µl of Multiscribe Reverse Transcriptase (50 U/µl), 1.50 µl of 10X Reverse Transcription Buffer, 0.19 ml of RNase inhibitor (20 U/µl), 3 µl of gene-specific primers (hsa-miR-33a-5p; assay ID: 465396, cel-miR-39, assay ID: 000200), and 8.16 µl of nuclease-free water. To synthesize cDNA, the reaction mixtures were incubated at 16°C for 30 min, at 42°C for 30 min, and then at 85°C for 5 min, followed by a final holding at 4°C.
The miRNA levels were quantified by RT-qPCR using the human TaqMan MicroRNA Assay Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). cDNA (1.33 µl) was amplified using 10 µl of the TaqMan Universal Master Mix II (Applied Biosystems; Thermo Fisher Scientific, Inc.), 1.0 µl of gene-specific primers/probe, and 7.67 µl of nuclease-free water in a final volume of 20 µl. Then, RT-qPCR was run on the ABI Prism 7300 Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.), and the reaction mixtures were incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The cycle threshold (Ct) values were calculated with the SDS v1.4 with RQv1.0 software (Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative quantities (RQ) of target miRNAs were calculated using the ΔΔCq method (22).
The results were normalized using the spike-in control cel-miR-39 as a reference target and were expressed in a relative quantity to a single reference sample (19). The relative miRNA levels were normalized to those of cel-miR-39 and calculated by using the equation 2−ΔΔCq. The RQ values for miRNA-33a-5p were then normalized to the average expressions of each of these miRNAs in healthy individuals (15,20).
Statistical analyses
Data are presented as the mean ± standard deviation. Experiments were repeated in triplicate. To estimate the differences between the groups, Chi-square test were used. To estimate the relationship between the candidate miRNA expression and the response rate, Mann-Whitney U test was applied. The PFS was defined as the time from the initiation of chemotherapy to its progression or recurrence. The OS was calculated from the start of chemotherapy to the death from any cause. The survival curves were estimated using the Kaplan-Meier method, and the curves were compared by using the log-rank test. The factors affecting the PFS and OS were examined by univariate analyses using the Cox proportional hazards model. The paired t-test was used for the chemo-resistance biomarker analysis. P<0.05 was considered to be statistically significant.
All statistical analyses were performed with the EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), which is a graphical user-interface for R (The R Foundation for Statistical Computing, Vienna, Austria). More precisely, it is a modified version of R commander that has been designed to add statistical functions frequently used in biostatistics.
Results
Characteristics of the patients
Table I displayed the characteristics of patients who were enrolled in this study. We noted that the non-responders were significantly younger than the responders. The patients' age range was 44–86 years, and the median age was 66 years. We enrolled 110 patients, which included 65 men and 45 women. We compared each item between the two groups and found no significant differences. Moreover, we obtained similar result for tumor markers.
Table I.
Investigation of the relationship between responder and non-responder characteristics.
| Parameter | All patients (n=110) | Responders (n=95) | Non-responders (n=15) | P-value |
|---|---|---|---|---|
| Age (years) | 0.033 | |||
| Average | 66.4 | 67.2 | 61.9 | |
| Median (range) | 67 (44–86) | 68 (46–86) | 63 (44–75) | |
| Sex (n) | 0.057 | |||
| Male | 65 | 60 | 5 | |
| Female | 45 | 35 | 10 | |
| Stage (n) | 0.810 | |||
| Metachronous | 36 | 32 | 4 | |
| Synchronous | 74 | 63 | 11 | |
| Primary tumor location (n) | 0.742 | |||
| Right-sided colon | 30 | 26 | 4 | |
| Left-sided colon | 43 | 39 | 4 | |
| Rectum | 40 | 33 | 7 | |
| Histological type (n) | 0.611 | |||
| Well/not well | 63 | 53 | 10 | |
| Other | 47 | 42 | 5 | |
| Lymph node metastasis (n) | 0.056 | |||
| Absent | 43 | 41 | 2 | |
| Present | 67 | 54 | 13 | |
| Number of metastatic sites (n) | 0.530 | |||
| 1 | 56 | 50 | 6 | |
| >1 | 54 | 45 | 9 | |
| RAS (n) | 0.911 | |||
| Mutation | 56 | 47 | 9 | |
| All wild | 6 | 5 | 1 | |
| KRAS wild | 45 | 40 | 5 | |
| Median CEA (ng/ml) | 21.3 | 29.2 | 10.4 | 0.214 |
| Median CA19-9 (U/ml) | 20.3 | 19.1 | 38.8 | 0.711 |
| Median PFS (months) | 10 | 11 | 3 | <0.010 |
| Median OS (months) | 21 | 23 | 15 | 0.310 |
Responder, cases with complete response, partial response and reduced stable disease; non-responder, cases with extended stable disease and progression disease; metachronous, cases in which distant metastases were not observed at the time of diagnosis of colorectal cancer and chemotherapy was started when distant metastasis recurrence was later observed; synchronous, cases in which distant metastases were observed at the time of colorectal cancer diagnosis, after which chemotherapy was started; right-sided colon, cecum, ascending and transverse colon; left-sided colon, descending and sigmoid colon; well, tubular adenocarcinoma; not well, poorly differentiated adenocarcinoma; other, papillary adenocarcinoma, mucinous adenocarcinoma and signet-ring cell carcinoma; PFS, recurrence free survival; OS, overall survival; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9.
Candidate miRNAs
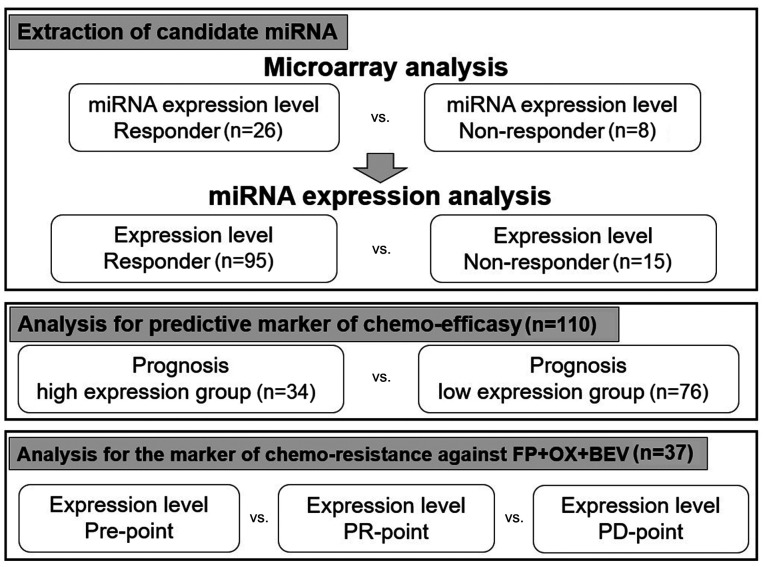
Our study design was depicted in Fig. 1. To extract the candidate miRNAs associated with chemo-efficacy, the miRNA expression levels before chemotherapy were compared between the responders and non-responders in 34 mCRC patients by microarray analysis. We selected nine candidate miRNAs whose expression levels were significantly upregulated in eight non-responders when compared with those in 26 responders (Table II).
Figure 1.
Schematic representation of the study design. miRNA, microRNA; FP, fluoropyrimidine; OX, oxaliplatin; BEV, bevacizumab; PR, partial response; PD, progression disease.
Table II.
Identification of the candidate miRNA that are highly expressed in the pretreatment plasma collected from non-responders.
| miRNA | Non-responder/responder | P-value |
|---|---|---|
| Hsa-miR-19b-1-5p | 1.580 | 0.008 |
| Hsa-miR-144-3p | 2.122 | 0.011 |
| Hsa-miR-33a-5p | 1.901 | 0.032 |
| Hsa-miR-301b | 1.565 | 0.033 |
| Hsa-miR-142-3p | 1.738 | 0.044 |
| Hsa-miR-101-3p | 1.631 | 0.046 |
| Hsa-miR-99b-5p | 1.560 | 0.047 |
| Hsa-miR-484 | 1.343 | 0.048 |
| Hsa-miR-340-5p | 1.625 | 0.049 |
Responder, case with complete response, partial response and reduced stable disease; non-responder, case with extended stable disease and progression disease. miRNA/miR, microRNA.
The nine miRNAs are reportedly associated with various malignant tumors, including CRC. However, the high expression of eight miRNA, except miR-33a-5p, has been associated with decreased malignancy of cancer and better prognosis in various cancer tissues (23–30). Furthermore, some past studied have demonstrated that miR-33a-5p can be an oncomir. For example, it has been reported that miR-33a-5p plays an important role as an oncomir in hepatocellular carcinoma (HCC) and has been associated with chemo-resistance against cisplatin in osteosarcoma (31,32). Therefore, we focused on miR-33a-5p for further detailed analyses as an oncomir in this study.
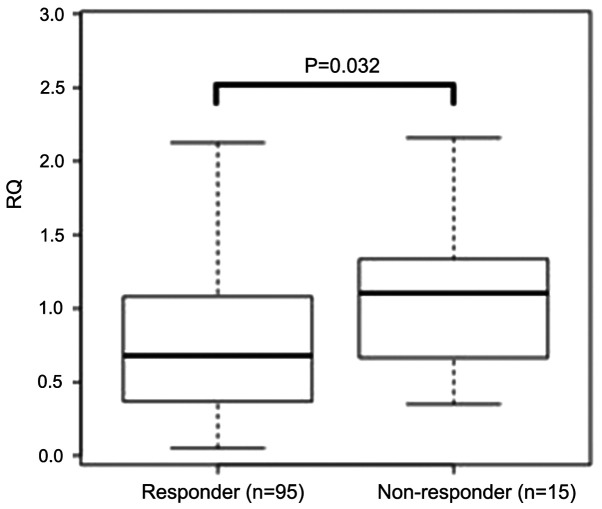
The result of miR-33a-5p was confirmed in whole patients by RT-qPCR method. We found that the expression levels of miR-33a-5p in 15 non-responders were significantly higher than those in 95 responders (P=0.032; Fig. 2).
Figure 2.
Expression of miR-33a-5p in the responders (n=95) and non-responders (n=15) included in the current study at the pre-treatment stage. The bar indicates the median relative quantification. Mann-Whitney U test was performed to compare expression levels.
Analyses for predictive marker of chemo-efficacy
To explore the predictive value of miR-33a-5p for the FP+OX+BEV chemotherapy, we compared the clinical outcomes (responders vs. non-responders) between patients with high and low miR-33a-5p expressions. The expression levels before chemotherapy were categorized into two groups based on the cut-off value calculated by receiver operator characteristic (ROC) analysis. Comparing each factor in these two groups yielded no significant difference (Table III).
Table III.
Characteristics of patients in the high and low expression groups.
| Parameter | All patients (n=110) | High expression group (n=34) | Low expression group (n=76) | P-value |
|---|---|---|---|---|
| Age (years) | 0.061 | |||
| Mean | 66.4 | 64.4 | 67.4 | |
| Median (range) | 67 (44–86) | 54 (44–86) | 68 (46–83) | |
| Sex (n) | 0.863 | |||
| Male | 65 | 21 | 44 | |
| Female | 45 | 13 | 32 | |
| Stage (n) | 0.780 | |||
| Metachronous | 36 | 10 | 26 | |
| Synchronous | 74 | 24 | 50 | |
| Primary tumor location (n) | 0.714 | |||
| Right-sided colon | 30 | 8 | 22 | |
| Left-sided colon | 43 | 13 | 30 | |
| Rectum | 40 | 14 | 26 | |
| Histopathological subtype (n) | 0.216 | |||
| Well/not well | 63 | 23 | 40 | |
| Other | 47 | 11 | 36 | |
| Lymph node (n) | 0.353 | |||
| Absent | 43 | 16 | 27 | |
| Present | 67 | 18 | 49 | |
| Number of metastatic sites (n) | 0.741 | |||
| 1 | 56 | 16 | 40 | |
| >1 | 54 | 18 | 36 | |
| KRAS (n) | 0.770 | |||
| Mutation | 56 | 19 | 37 | |
| Wild | 51 | 15 | 36 | |
| Median CEA (ng/ml) | 21.3 | 39.5 | 28.4 | 0.683 |
| Median CA19-9 (U/ml) | 20.3 | 39.4 | 19.6 | 0.312 |
Metachronous, cases in which distant metastases were observed at the time of diagnosis of colorectal cancer and chemotherapy was started; synchronous, cases in which distant metastases were not observed at the time of diagnosis of colorectal cancer, and chemotherapy was started when distant metastasis recurrence was later observed; right-sided colon, included cecum, ascending and transverse colon; left-sided colon, included descending and sigmoid colon; well, tubular adenocarcinoma; not well, poorly differentiated adenocarcinoma; other, papillary adenocarcinoma, mucinous adenocarcinoma and signet-ring cell carcinoma
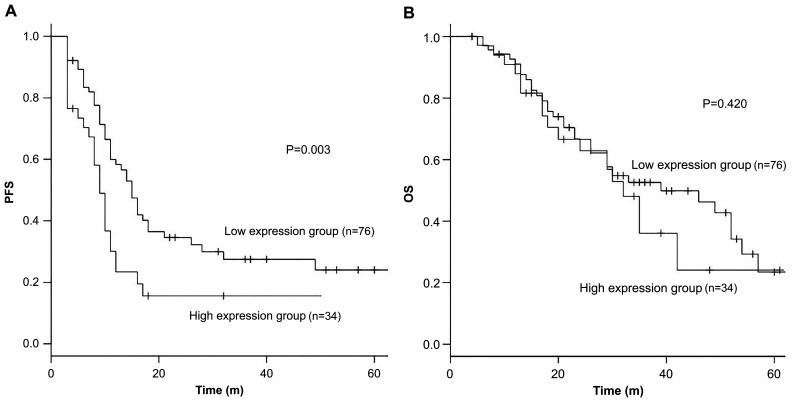
Patients with high miR-33a-5p expression showed a significantly shorter PFS when compared with those with low miR-33a-5p expression. The median PFS was 9 months for patients with high expression levels compared with 15 months for those with low expression levels [HR 1.51, 95% confidence interval (CI): 1.04–2.20, P<0.01] (Fig. 3). However, no significant difference was noted in the OS (median 39 vs. 32 months, HR 0.98, 95% CI: 0.60–1.59, P=0.42) (Fig. 3).
Figure 3.
PFS and OS according to microRNA-33a-5p expression. Kaplan-Meier survival curves presenting the association between (A) PFS, (B) OS and miR-33a-5p expression levels in the pretreatment plasma samples of patients with mCRC. The low expression group is indicated by a black line, and the high expression group by a gray line. The cut-off value was defined as the expression level resulting in the highest number of correctly classified samples in receiver operator characteristic analysis. The Kaplan-Meier survival curves were compared by performing log-rank test. PFS, progression-free survival; OS, overall survival; m, months.
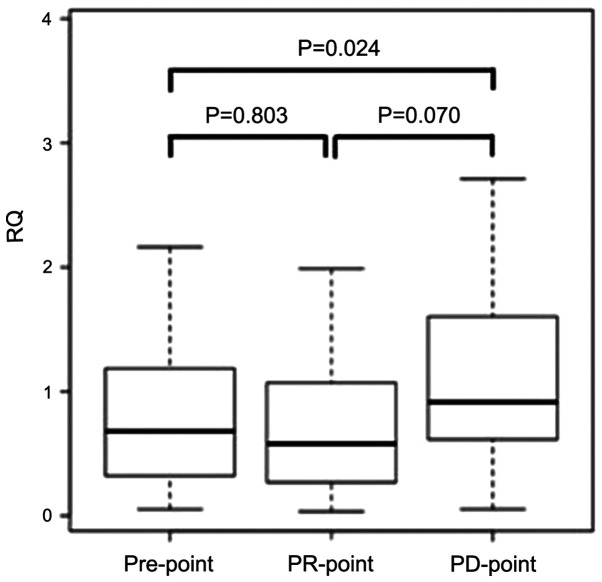
Analyses for the marker of chemo-resistance against FP+OX+BEV regime. To evaluate whether miR-33a-5p can act as a marker of chemo-resistance, we examined the expression levels of miR-33a-5p at three different time-points among 37 responders. We defined each time-point as follows: the time before chemotherapy as the ‘Pre-point’ the time 3 months after initiating the chemotherapy as the ‘PR-point’ and the time of recording recurrence or progression as the ‘PD-point’. The expression levels at each point were compared.
Although no significant differences were noted in the expression levels between the Pre-point and PR-point, the expression level was significantly increased at the PD-point when compared with that at the Pre-point (P=0.024). In addition, the expression level tended to increase at the PD-point when compared with that at the PR-point (P=0.070) (Fig. 4).
Figure 4.
Expression of micorRNA-33a-5p in 37 responders at the pre-point, PR-point and PD-point. The bar indicates the median relative quantification. To compare the expression levels, a paired t-test was used. PR, partial response; PD, progression disease.
Discussion
In this study, the plasma miR-33a-5p expression levels before chemotherapy were significantly associated with the response rate and the PFS from FP+OX+BEV chemotherapy. Furthermore, the plasma miR-33a-5p expression levels were associated with chemo-resistance in patients showing a good response to FP+OX+BEV chemotherapy. In the recent years, several studies have shown an association between miRNA and cancer chemo-resistance, not only in cancer tissues (12–14) but also in the plasma or serum samples (12,14). The use of CRC cell lines revealed the involvement of miR-215-3p and miR-203 in 5-fluorouracil (5-FU) and oxaliplatin sensitivity, respectively (12,14). The high expression of miR-17-5p in CRC tissues revealed FOLFOX resistance in mCRC patients (13). The expression levels of serum miR-19a were associated with chemo-resistance in advanced CRC patients treated with the first-line FOLFOX chemotherapy regimen (15). It is well known that microRNA is stable in the blood and, assuming clinical application, microRNAs in the plasma/serum are easier to measure and can be monitored repeatedly. Therefore, we focused on plasma miRNAs and performed an array analysis by the Toray 3D-gene, which was also employed when miR-1229-3p was identified as a chemo-resistant biomarker for gastric cancer (17). Regarding the relationship between plasma miR-33a-5p and chemotherapy resistance, the plasma of the same patient was collected over different time points from before the start of the treatment until disease progression or recurrence, suggesting that plasma miR-33a-5p can act as a potential molecular biomarker for monitoring resistance. This is the first study of its kind to report plasma miRNA as a biomarker for FP+OX+BV chemotherapy.
miR-33a-5p has been reported to be associated with arteriosclerosis in several studies and has also been considered as a candidate biomarker of atherosclerosis (33). In the recent years, it was reported that miR-33a-5p is involved in malignancy of cancer via peroxisome proliferator-activated receptor α (PPARα) and UV radiation resistance-associated gene (UVRAG). PPARα is a miR-33a-5p target genes (31) that belongs to the nuclear hormone receptor superfamily and plays physiological roles in energy homeostasis via modulating glucose and lipid metabolisms and transport (34). The PPARα expression has a major impact on the maintenance of mitochondrial β-oxidation (35). PPARα is also a tumor suppressor (36). In HCC, the high expression levels of miR-33a-5p were associated with larger tumor size, higher tumor stage, and poorer prognosis and high expression levels of miR-33a-5p being related with PPARα suppression (37). In CRC tissues, it was demonstrated that PPARα suppresses cyclooxygenase2 (COX2) and acts as a tumor suppressor agent to suppress inflammation, tumor growth, and metastasis (38). The expression of miR-33a-5p was not analyzed in the present study, but it may be involved in malignancy of CRC via PPARα suppression. UVRAG was initially identified for its complementary effect on UV sensitivity in xeroderma pigmentosum cells. Genetic association studies have demonstrated that human chromosomal region containing UVRAG is closely associated with the pathogenesis of various human cancers (39). UVRAG is a miR-33a-5p target gene and the suppression of UVRAG can activate the Notch pathway (40). The Notch pathway has been shown to be activated in multiple tumors, including in CRC (41). In CRC tissues, the activation of Notch pathway has been reported to promote tumor growth, invasion, and metastasis (42). In glioblastoma cells, miR-33a-5p activated the Notch pathway via suppression of UVRAG and was involved in the malignancy of glioblastoma (40). In CRC tissues, miR-33a-5p may activate the Notch pathway by suppressing UVRAG and thereby contributing to malignancy.
miR-33a-5p may also be involved in chemotherapy resistance via the action of the target genes PPARα, UVRAG, and TWIST. The overexpression of miR-33a-5p can downregulate PPARα (37). In CRC tissues, it has demonstrated that PPARα suppresses Vascular Endothelial Growth Factor (VEGF) (38). We can suggest that miR-33a-5p may reduce PPARα expression and promote VEGF, resulting in increasing the resistance against bevacizumab in CRC tissues (43). It is already known that the overexpression of miR-33a-5p downregulates UVRAG, which in turn activates the Notch pathway (40). It has also been reported that the activation of the Notch pathway is associated with resistance to chemotherapy (44). One of the downstream gene of the NOTCH pathway, hairy and enhancer of split 1 (HES1), is upregulated by the activation of the NOTCH pathway in CRC (42); HES1 can promote chemo-resistance to 5-FU for CRC patients (44). miR-33a-5p may upregulate the HES1 expression via activation of the Notch pathway and can promote the chemo-resistance to 5-FU. The miR-33a level was negatively correlated with the TWIST protein level in osteosarcoma cells, and miR-33a was upregulated in chemo-resistant osteosarcoma cell (32). TWIST has been suggested as a pivotal negative regulator of osteosarcoma chemo-resistance (45). In addition, miR-33a-5p has been reported to be involved in the chemo-resistance of cisplatin by downregulating TWIST in osteosarcoma cells (32). As oxaliplatin is a platinum antitumor agent similar to cisplatin, miR-33a-5p may be involved in the chemo-resistance of OX in CRC patients through the same mechanism. The expression of miR-33a in the plasma may influence chemo-resistance due to various mechanisms. The elucidation of the chemo-resistance mechanism is extremely important considering that it may contribute to the development of new drugs.
However, this hypothesis-generating study poses some limitations that need to be considered, particularly with respect to the sample size. In this cohort, only 15 patients who experienced PD were included. Future studies with a larger sample size are warranted to confirm these associations. In addition, it remains unclear as to which drug from FP, OX, and BEV induced the miR-33a-5p expression. To clarify which drug is unique to miR-33a-5p, further functional studies on the underlying biological mechanisms of miR-33a-5p are warranted. It is also important to analyze the relationship between the expression level of miR-33a-5p in the plasma and PPARα, UVRAG, and TWIST. Considering the clinical application, if the expression of miR-33a-5p in plasma is high before the start of the chemotherapy, we can prioritize other regimens as well as estimate the timing of the regimen change.
In conclusion, the miR-33a-5p expression level in the plasma can serve as a predictive marker of efficacy and as a biomarker of chemo-resistance in mCRC patients who receive the FP+OX+BEV regime. Our cumulative results suggest that miR-33a-5p may be involved in acquired resistance, which may contribute to the development of new mCRC therapeutics.
Acknowledgements
The authors would like to thank Mrs. Yoko Takagi and Mrs. Junko Inoue (Department of Specialized Surgeries, Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University) for their technical assistance.
Glossary
Abbreviations
- CRC
colorectal cancer
- mCRC
metastatic colorectal cancer
- FP
fluoropyrimidine
- OX
oxaliplatin
- ESMO
European Society for Medical Oncology
- miRNA
microRNA
- BEV
bevacizumab
- RT-qPCR
reverse transcription-quantitative PCR
- PR
partial response
- SD
stable disease
- PD
progression disease
- RQ
relative quantities
- PFS
progression free survival
- OS
overall survival
- HCC
hepatocellular carcinoma
- ROC
receiver operating characteristic
- CI
confidence interval
- HR
hazard ratio
- 5-FU
5-fluorouracil
- PPARα
peroxisome proliferator-activated receptor α
- UVRAG
UV radiation resistance-associated gene
- HES1
hairy and enhancer of split 1.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
MS, TI, MI, SO, SY, TM, and HU were involved in the conception and design of the study and development of the methodology. MS performed the experiments and collected laboratory data. MS, TI, SO and MI analyzed the results. MS, TI and SO edited the manuscript. TI, MI, SO, AK, SY, TM, KK, MT, HU, and YK assisted with all assays and analyses and in manuscript preparation. TI, MI, SO, AK, SY, TM, KK, MT, HU, and YK supervised the study. TI and SO confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was conducted in accordance with the Declaration of Helsinki and its later amendments or comparable ethical standards. The study protocol was approved by the Institutional Review Board of Tokyo Medical and Dental University, and written informed consent was obtained from all patients before enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Global Burden of Disease Cancer Collaboration. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schirripa M, Lenz HJ. Biomarker in CRC. Cancer J. 2016;22:156–164. doi: 10.1097/PPO.0000000000000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 4.Yamazaki K, Nagase M, Tamagawa H, Ueda S, Tamura T, Murata K, Eguchi Nakajima T, Baba E, Tsuda M, Moriwaki T, et al. Randomized phase III study of bevacizumab plus FOLFIRI and bevacizumab plus mFOLFOX6 as first-line treatment for patients with metastatic colorectal cancer (WJOG4407G) Ann Oncol. 2016;27:1539–1546. doi: 10.1016/S0923-7534(21)00374-4. [DOI] [PubMed] [Google Scholar]
- 5.Gustavsson B, Carlsson G, Machover D, Petrelli N, Roth A, Schmoll HJ, Tveit KM, Gibson F. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin Colorectal Cancer. 2015;14:1–10. doi: 10.1016/j.clcc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson A, Bodoky G, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 7.Kjersem JB, Ikdahl T, Lingjaerde OC, Guren T, Tveit KM, Kure EH. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol Oncol. 2014;8:59–67. doi: 10.1016/j.molonc.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X, Yu L, Wang L, Wang J, Wu Y, et al. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer. 2015;136:152–161. doi: 10.1002/ijc.28136. [DOI] [PubMed] [Google Scholar]
- 11.Rasmussen MH, Jensen NF, Tarpgaard LS, Qvortrup C, Rømer MU, Stenvang J, Hansen TP, Christensen LL, Lindebjerg J, Hansen F, et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol Oncol. 2013;7:637–646. doi: 10.1016/j.molonc.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li XW, Qiu SJ, Zhang X. Overexpression of miR-215-3p sensitizes colorectal cancer to 5-fluorouracil induced apoptosis through regulating CXCR1. Eur Rev Med Pharmacol Sci. 2018;22:7240–7250. doi: 10.26355/eurrev_201811_16258. [DOI] [PubMed] [Google Scholar]
- 13.Fang L, Li H, Wang L, Hu J, Jin T, Wang J, Yang BB. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5:2974–2987. doi: 10.18632/oncotarget.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Wan G, Spizzo R, Ivan C, Mathur R, Hu X, Ye X, Lu J, Fan F, Xia L, et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol. 2014;8:83–92. doi: 10.1016/j.molonc.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q, Xia HW, Ge XJ, Zhang YC, Tang QL, Bi F. Serum miR-19a predicts resistance to FOLFOX chemotherapy in advanced colorectal cancer cases. Asian Pac J Cancer Prev. 2013;14:7421–7426. doi: 10.7314/APJCP.2013.14.12.7421. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, Zhou J, Li Y, Ye F, Wan X, Lu W, Xie X, Cheng X. miR-375 mediated acquired chemo-resistance in cervical cancer by facilitating EMT. PLoS One. 2014;9:e109299. doi: 10.1371/journal.pone.0109299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishibeppu K, Komatsu S, Imamura T, Kiuchi J, Kishimoto T, Arita T, Kosuga T, Konishi H, Kubota T, Shiozaki A, et al. Plasma microRNA profiles: Identification of miR-1229-3p as a novel chemoresistant and prognostic biomarker in gastric cancer. Sci Rep. 2020;10:3161. doi: 10.1038/s41598-020-59939-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu S, Ichikawa D, Kawaguchi T, Takeshita H, Miyamae M, Ohashi T, Okajima W, Imamura T, Kiuchi J, Arita T, et al. Plasma microRNA profiles: Identification of miR-23a as a novel biomarker for chemoresistance in esophageal squamous cell carcinoma. Oncotarget. 2016;7:62034–62048. doi: 10.18632/oncotarget.11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konishi H, Ichikawa D, Komatsu S, Shiozaki A, Tsujiura M, Takeshita H, Morimura R, Nagata H, Arita T, Kawaguchi T, et al. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer. 2012;106:740–747. doi: 10.1038/bjc.2011.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu S, Ichikawa D, Hirajima S, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Arita T, Konishi H, Shiozaki A, et al. Plasma microRNA profiles: Identification of miR-25 as a novel diagnostic and monitoring biomarker in oesophageal squamous cell carcinoma. Br J Cancer. 2014;111:1614–1624. doi: 10.1038/bjc.2014.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Wang SY, Shiboski S, Belair CD, Cooperberg MR, Simko JP, Stoppler H, Cowan J, Carroll PR, Blelloch R. miR-19, miR-345, miR-519c-5p serum levels predict adverse pathology in prostate cancer patients eligible for active surveillance. PLoS One. 2014;9:e98597. doi: 10.1371/journal.pone.0098597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng S, Xie L, Wu Y, Ding M, Zhang T, Wang X. MiR-144 inhibits growth and metastasis in colon cancer by down-regulating SMAD4. Biosci Rep. 2019;39:39. doi: 10.1042/BSR20181895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Ruan J, Liu X, Xiao C, Xiong J. MicroRNA-301a-3p suppressed the progression of hepatocellular carcinoma via targeting VGLL4. Pathol Res Pract. 2018;214:2039–2045. doi: 10.1016/j.prp.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, Ma SP, Yang D, Liu Y, Wang YP, Lin T, Li YX, Yang SH, Zhang WC, Wang XL. miR-142-3p suppresses cell growth by targeting CDK4 in colorectal Cancer. Cell Physiol Biochem. 2018;51:1969–1981. doi: 10.1159/000495721. [DOI] [PubMed] [Google Scholar]
- 27.Jiang M, Xu B, Li X, Shang Y, Chu Y, Wang W, Chen D, Wu N, Hu S, Zhang S, et al. O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit. Oncogene. 2019;38:301–316. doi: 10.1038/s41388-019-0834-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng H, Xue J, Yang S, Chen Y, Wang Y, Zhu Y, Wang X, Kuang D, Ruan Q, Duan Y, et al. Co-targeting of IGF1R/mTOR pathway by miR-497 and miR-99a impairs hepatocellular carcinoma development. Oncotarget. 2017;8:47984–47997. doi: 10.18632/oncotarget.18207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen Y, Qi L, Li Y, Zhang Y, Gao X, Zhu Y, Wang K. The downregulation of lncRNA PGM5-AS1 inhibits the proliferation and metastasis via increasing miR-484 expression in colorectal cancer. Cancer Biother Radiopharm. 2021;36:220–229. doi: 10.1089/cbr.2019.3059. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Men WL, Yan KM, Tie J, Nie YZ, Xiao HJ. MiR-340-5p is a potential prognostic indicator of colorectal cancer and modulates ANXA3. Eur Rev Med Pharmacol Sci. 2018;22:4837–4845. doi: 10.26355/eurrev_201808_15619. [DOI] [PubMed] [Google Scholar]
- 31.Chang W, Zhang L, Xian Y, Yu Z. MicroRNA-33a promotes cell proliferation and inhibits apoptosis by targeting PPARα in human hepatocellular carcinoma. Exp Ther Med. 2017;13:2507–2514. doi: 10.3892/etm.2017.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Huang Z, Wu S, Zang X, Liu M, Shi J. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. J Exp Clin Cancer Res. 2014;33:12. doi: 10.1186/1756-9966-33-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SH, Kim GJ, Umemura T, Lee SG, Cho KJ. Aberrant expression of plasma microRNA-33a in an atherosclerosis-risk group. Mol Biol Rep. 2017;44:79–88. doi: 10.1007/s11033-016-4082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR α in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoyama T, Peters JM, Iritani N, Nakajima T, Furihata K, Hashimoto T, Gonzalez FJ. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha) J Biol Chem. 1998;273:5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 36.Panigrahy D, Kaipainen A, Huang S, Butterfield CE, Barnés CM, Fannon M, Laforme AM, Chaponis DM, Folkman J, Kieran MW. PPARalpha agonist fenofibrate suppresses tumor growth through direct and indirect angiogenesis inhibition. Proc Natl Acad Sci USA. 2008;105:985–990. doi: 10.1073/pnas.0711281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang N, Chu ES, Zhang J, Li X, Liang Q, Chen J, Chen M, Teoh N, Farrell G, Sung JJ, et al. Peroxisome proliferator activated receptor alpha inhibits hepatocarcinogenesis through mediating NF-κB signaling pathway. Oncotarget. 2014;5:8330–8340. doi: 10.18632/oncotarget.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grau R, Punzón C, Fresno M, Iñiguez MA. Peroxisome-proliferator-activated receptor α agonists inhibit cyclo-oxygenase 2 and vascular endothelial growth factor transcriptional activation in human colorectal carcinoma cells via inhibition of activator protein-1. Biochem J. 2006;395:81–88. doi: 10.1042/BJ20050964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee G, Liang C, Park G, Jang C, Jung JU, Chung J. UVRAG is required for organ rotation by regulating Notch endocytosis in Drosophila. Dev Biol. 2011;356:588–597. doi: 10.1016/j.ydbio.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Sun T, Hu J, Zhang R, Rao Y, Wang S, Chen R, McLendon RE, Friedman AH, Keir ST, et al. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J Clin Invest. 2014;124:4489–4502. doi: 10.1172/JCI75284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh M, Katoh M. Notch signaling in gastrointestinal tract (Review) Int J Oncol. 2007;30:247–251. [PubMed] [Google Scholar]
- 42.Meng RD, Shelton CC, Li Y-M, Qin LX, Notterman D, Paty PB, Schwartz GK. gamma-Secretase inhibitors abrogate oxaliplatin-induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer Res. 2009;69:573–582. doi: 10.1158/0008-5472.CAN-08-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao L, Zhang D, Ma H, Jin M, Huang F, Zhang T. High VEGF-A level at baseline predicts poor treatment effect of bevacizumab-based chemotherapy in metastatic colorectal cancer: A meta-analysis. Panminerva Med. 2016;58:48–58. [PubMed] [Google Scholar]
- 44.Sun L, Ke J, He Z, Chen Z, Huang Q, Ai W, Wang G, Wei Y, Zou X, Zhang S, et al. HES1 promotes colorectal cancer cell resistance to 5 Fu by inducing of EMT and ABC transporter proteins. J Cancer. 2017;8:2802–2808. doi: 10.7150/jca.19142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y, Zang X, Huang Z, Zhang C. TWIST interacts with endothelin-1/endothelin A receptor signaling in osteosarcoma cell survival against cisplatin. Oncol Lett. 2013;5:857–861. doi: 10.3892/ol.2013.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.