Abstract
The efficacy of chemotherapy for colon cancer is limited due to the development of chemoresistance. MicroRNA (miR)-188-5p is downregulated in various types of cancer. The aim of the present study was to explore the molecular role of miR-188 in oxaliplatin (OXA) resistance. An OXA-resistant colon cancer cell line, SW480/OXA, was used to examine the effects of miR-188-5p on the sensitivity of colon cancer cells to OXA. The target of miR-188-5p was identified using a luciferase assay. Cell cycle distribution was also assessed using flow cytometry. The measurement of p21 protein expression, Hoechst 33342 staining and Annexin V/propidium iodide staining was used to evaluate apoptosis. The expression of miR-188-5p significantly increased in SW480/OXA compared with wild-type SW480 cells. The luciferase assay demonstrated that miR-188-5p inhibited Ras GTPase-activating protein 1 (RASA1; also known as p120/RasGAP) luciferase activity by binding to the 3′-untranslated region of RASA1 mRNA, suggesting that miR-188-5p could target RASA1. In addition, miR-188-5p downregulation or RASA1 overexpression promoted the chemosensitivity of SW480/OXA, as evidenced by increased apoptosis and G1/S cell cycle arrest. Moreover, RASA1 silencing abrogated the increase in cell apoptosis induced by the miR-188-5p inhibitor. The findings of the present study suggested that miR-188-5p could enhance colon cancer cell chemosensitivity by promoting the expression of RASA1.
Keywords: chemoresistance, miR-188-5p, colon cancer, oxaliplatin, RASA1, apoptosis
Introduction
Colon cancer is the third most frequently diagnosed cancer and the second leading cause of cancer death in the United States (1). In the United States, 104,610 new cases of colon cancer were reported in 2020 (1). Among them, approximately 53,200 mortalities were reported annually in the United States (1). Overall, the incidence and death rates for colon cancer are on the decline (2). However, this trend is not observed in younger individuals from 15–35 years old (1–3). The two major causes of colon cancer are chromosomal instability and serrated neoplasia (3). Treatment options for colon cancer include endoscopic and surgical local excision, preoperative radiotherapy and chemotherapy (3). However, chemoresistance remains an obstacle to the treatment of colon cancer (4,5).
MicroRNA (miRNA/miR) plays a significant role in complex physiological and pathological processes through the regulation of target gene expression via binding to their 3′-untranslated region (UTR) (6). Usually, each miRNA has multiple target mRNA transcripts, resulting in various effects, such as cell proliferation, apoptosis, migration and invasion (7). Previous studies have highlighted the link between miRNA and the chemoresistant phenotype of different tumor types, which is mediated by abnormal regulation of apoptosis (8), cell cycle distribution (9) and activity of drug efflux transporters (10). Modulating the expression of miRNA using specific mimics or inhibitors normalizes gene regulatory networks and signaling pathways, thus sensitizing cancerous cells to chemotherapy (11). Therefore, miRNA-based gene therapy may represent an attractive approach for cancer therapy (11,12).
In various types of cancer, such as cervical cancer (13), non-small cell lung cancer (14), gastric cancer (15), glioma (16), colorectal cancer (CRC) (17), oral squamous cell carcinoma (18), hepatocellular carcinoma (19), prostate cancer (20), acute myeloid leukemia (21) and rectal cancer (22), miR-188-5p is downregulated. Moreover, transfection of miR-188-5p mimics reduces proliferation and migration while promoting apoptosis in A549 and H2126 cells (14). In in vivo xenograft models, miR-188-5p inhibits tumor growth of non-small cell lung cancer (14). Additionally, miR-188-3p regulates the expression of myeloid/lymphoid or mixed-lineage leukemia 4 (MLLT4) and promotes the migration of HCT116 and HRT18 cancer cells (17); it has also been proposed as a novel independent prognostic factor in patients with CRC (17). In locally advanced rectal cancer, miR-188-5p is associated with a complete pathological response to the neoadjuvant chemoradiotherapy (22). However, the role of miR-188 in drug-resistant cancer cells has yet to be examined.
Ras GTPase-activating protein 1 (RASA1; also known as p120/RasGAP) was the first identified RasGAP protein. RASA1 has been implicated in a number of biological processes, including actin filament polymerization, cell apoptosis and migration (23). RASA1 acts as a cancer suppressor gene in several types of tumor (24–32). RASA1 suppresses the effect of Ras proteins by enhancing their weak intrinsic GTPase activity, leading to an increase in the levels of the inactive, GDP-bound form of Ras (33). This causes aberrant intracellular signaling through the Ras-RAF-ERK pathway (33). A previous study has demonstrated that RASA1 protein levels are significantly decreased in colon cancer cells and that RASA1 is a target of miR-21, which promotes the malignancy of colon cancer cells (34). The upregulation of RASA1 inhibits cell proliferation and inhibits the RAS signaling pathway (34). Moreover, the expression levels of miR-223 and RASA1 are inversely correlated in tumor tissue from patients with CRC (35). RASA1 is also a validated target of miR-335, which is downregulated in CRC (36). Several studies have also indicated that onco-miR molecules, such as miR-21 and miR-182, promote tumor angiogenesis and lymph node metastasis by targeting RASA1 (37,38).
The present study aimed to explore the molecular role of miR-188-5p in oxaliplatin (OXA) resistance. miR-188-5p may enhance colon cancer cell chemosensitivity by promoting the expression of RASA1, providing a new insight into treatment options for OXA-resistant colon cancer.
Materials and methods
Cell culture and treatment
The human SW480 colon cancer cell line was purchased from American Type Culture Collection. To induce OXA resistance in SW480 cells (SW480/OXA), SW480 cells were seeded into a 24-well plate at a density of 1×105 cells/well and cultured in the presence of 0.5 mM OXA (Sigma-Aldrich; Merck KGaA; cat. no. O9512) for 24 h. Subsequently, the medium was replaced with OXA-free medium, and the cells were cultured for three days. From day 4, the drug concentration increase started. This procedure was continued for 6 months with a drug concentration increase at 0.4 µM/month, (0.5–2.5 mM) and the medium was changed at every other day. After appropriately increasing the concentration of OXA over a period of 6 months, a stable drug-resistant cell line SW480/OXA was obtained. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich; Merck KGaA) with 10% Fetal Bovine Serum (FBS; Gibco; Thermo Fisher Scientific Inc.) and 1% penicillin/streptomycin solution at 37°C with 5% CO2 saturated humidity.
Hoechst 33342 staining
SW480 and SW480/OXA cells (2×105/well) were seeded into a fibronectin-coated 12-well plate. After cell transfection, 5 µg/ml Hoechst 33342 staining solution (Beijing Solarbio Science & Technology Co., Ltd.; cat. no. C0031) was added to the cells, which were incubated for 20 min at room temperature. The cells were then washed three times with PBS. Coverslips were sealed and visualized under a confocal microscope (T1-SAM microscope; Nikon Corporation).
Reverse transcription-quantitative (RT-q) PCR
Total RNA was isolated from SW480 and SW480/OXA cells (5×105 cells/well in a 6-well plate) using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 15596026) according to the manufacturer's instructions. First-strand cDNA was synthesized using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.; cat. no. 4368814) according to the manufacturer's protocol. SYBR Green PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.; cat. no. A25741) was used to perform RT-qPCR. The qPCR thermocycling conditions were as follows: Initial denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 15 sec, annealing at 60°C for 25 sec and extension at 72°C for 30 sec. U6 and GAPDH were used as internal references for normalization. The 2−ΔΔCq method was applied to calculate relative mRNA levels (36). The primer sequences used in the study were: i) GAPDH forward, 5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse, 5′-ATGGCATGGACTGTGGTCAT-3′; ii) RASA1 forward, 5′-CAGTGGACGAAGGTGACTCT-3′ and reverse, 5′-AGGCGTTCTTCTGCTATCGT-3′; iii) U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and iv) miR-188-5p forward, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCTCCAC-3′ and reverse, 5′-ACACTCCAGCTGGGCATCCCTTGCATGGTGG-3′.
Cell transfection
The miR-188-5p inhibitor (5′-CCCUCCACCAUGCAAGGGAUG-3′; 2′-OMe-modified), negative control (NC) NC inhibitor (5′-CAGUACUUUUGUGUAGUACAA-3′), miR-188-5p mimics (sense, 5′-CAUCCCUUGCAUGGUGGAGGG-3′; antisense, 5′-CUCCACCAUGCAAGGGAUGUU-3′), NC mimics (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; antisense, 5′-ACGUGACACGUUCGGAGAATT-3′). The mimic and inhibitor NCs were scrambled sequences. RASA1 small interfering (si)RNA (5′-CAGTTTATGATGGGAGGCCGGTATT-3′), siRNA negative control (siNC, 5′-TTCTCCGAACGTTTCACGTTT-3′), RASA1 overexpression plasmid (pcDNA3.1-RASA1) and empty vector control (pcDNA3.1) were purchased from Shanghai GenePharma Co. Ltd. Cells were seeded in a 12-well plate (4×104 cells/well) and incubated overnight at 37°C with 5% CO2. At 70% confluency, cells were transfected with 100 pmol miR-188-5p mimics or NC mimics at room temperature for 48 h using Lipofectamine® 2000 (cat. no. 11668-500; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. SW480 and SW480/OXA cells were transfected with 50 pmol si-RASA1 or co-transfected with 50 pmol miR-188-5p inhibitor and si-RASA1. After 48 h, cells were collected for subsequent experiments.
Bioinformatics analysis
For the determination of potential target genes of miR-188-5p, various bioinformatics tools, including TargetScan (http://www.targetscan.org), miRDB (http://www.mirdb.org/) and PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html) databases were used. TargetScan was used to predict the binding mRNAs (218 mRNAs), similarly, 340 mRNAs and 229 mRNAs were predicted in miRDB and PITA databases, respectively. When the common mRNAs from the three databases were assessed, 6 mRNAs were obtained. RASA1 was one of the predicted target genes of miR-188-5p.
Luciferase activity assay
Luciferase reporter constructs were obtained by ligating the wild-type (WT) or mutant (MUT) version of the RASA1 3′-untranslated region (UTR) to the psiECHECK-2 vector containing a luciferase reporter gene (Promega Corporation; cat. no. C8021). SW480 and SW480/OXA cells (5×105/well) were seeded into 6-well plates and co-transfected with RASA1 WT or MUT and with miR-188-5p mimics or NC mimics using Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 11668-500). A Luciferase Reporter Gene Detection kit (Sigma-Aldrich; Merck KGaA; LUC1-1KT) was used to detect luciferase activity 48 h after transfection. Luciferase activity was normalized to the Renilla luciferase activity. Experiments were set up in triplicate and performed independently three times.
Western blot analysis
SW480 and SW480/OXA cells (1×106 cells/6-cm dish) were collected and lysed in lysis buffer containing phosphorylase inhibitor cocktail (Roche; Diagnostics; cat. no. 04693124001) and phenylmethanesulfonyl fluoride. Protein concentration was measured using the bicinchoninic acid (BCA) Protein Concentration Determination kit (Beyotime Institute of Biotechnology) following the manufacturer's instructions. An equal amount (30 µg) of each protein sample was resolved by SDS-PAGE on 8–12% gels, then transferred to PVDF membranes. Subsequently, the membranes were blocked using 5% skimmed milk powder for 1 h at room temperature, then incubated with primary antibodies at 4°C overnight. The primary antibodies were specific for p21 (Abcam; cat. no. ab227443; 1:1,000), RASA1 (Abcam; cat. no. ab2922; 1:1,000), Rs-GTP (Abcam; cat. no. ab69747; 1:2,000) and GAPDH (Abcam; cat. no. ab9485; 1:5,000). Goat anti-rabbit (Sigma-Aldrich; Merck KGaA; cat. no. 1:5,000) or goat anti-mouse IgG-HRP (Sigma-Aldrich; Merck KGaA; cat. no. AP-308P; 1:5,000) secondary antibodies were then added at room temperature for 1 h. The expression of the target proteins was normalized to the GAPDH internal control. The protein bands were visualized using an enhanced chemiluminescence kit (Beyotime Institute of Biotechnology) according to the manufacturer's instructions and quantified using the Gel-Pro-Analyzer v.4.0 software (Media Cybernetics, Inc.)
Immunofluorescence assay
SW480 and SW480/OXA cells (2×105/well) were seeded on a fibronectin-coated 12-well plate. After transfection with indicated mimics or inhibitors, the cells were fixed in 4% paraformaldehyde at room temperature for 10 min, then blocked with 1% BSA in PBS. The slides were incubated with a RASA1-specific primary antibody (1:1,000; Abcam; cat. no. ab2922) at room temperature for 1 h, then washed three times with PBS. An Alexa Fluor 555-conjugated goat anti-mouse secondary antibody (1:2,000; Abcam; cat. no. ab150114) was then added at room temperature for 40 min. The slides were washed with PBS three times and then stained with DAPI at room temperature for 10 min. For fluorescence visualisation, the Nikon C1 confocal microscope was used (Nikon Corporation; magnification, ×100).
Apoptosis assay
SW480 and SW480/OXA cells (2×105/well) were seeded onto a 6-well plate and transfected with siRNA or plasmid as indicated. After 48 h, the Annexin V-FITC apoptosis detection kit (BD Biosciences; cat. no. 556547) was used to detect apoptotic cells with Annexin V-FITC and propidium iodide (PI) double staining according to the manufacturer's instructions. Gated Annexin V+/PI+ cells as late stage apoptosis and Annexin V+/PI− cells as early stage apoptosis were assessed. The analysis was performed using a BD FACScan® II flow cytometer (BD Biosciences), and the data were analyzed using CellQuest Pro software version 5.1 (Becton, Dickinson and Company).
Cell cycle analysis
Cells (2×105 cells/well) were seeded onto a 6-well plate and transfected 12 h later. The cells were harvested 48 h after transfection, fixed in ice-cold 70% ethanol for at 4°C for 12 h, washed with PBS, then stained with 5 mg/ml propidium iodide (Sigma-Aldrich; Merck KGaA, cat. no. P4170) or isotype control antibody IgG2A (10 µl/106 cells; R&D Systems; cat. no. IC003A) in PBS supplemented with RNase A (Roche Diagnostics; cat. no. 10154105103) for 30 min at room temperature. The flow cytometry data were acquired using a FACSCalibur® (BD Biosciences) flow cytometer. Flow cytometry data were analyzed using Summit v.5.3 (DAT/EM Systems International). A histogram was plotted according to the distribution of nuclear DNA content and the frequency of cells in each phase of the cell cycle was determined based on their DNA ploidy profile.
Statistical analysis
The data are presented as the mean ± SD and were analyzed using SPSS 15.0 (SPSS, Inc.) All results are representative of at least three independent experiments unless stated otherwise. Unpaired two-tailed Student's t-test was used for comparisons between two groups. For multi-group comparisons, one-way ANOVA followed by Tukey's post hoc test was used. P<0.05 was considered to indicate a statistically significant difference.
Results
Inhibition of miR-188-5p upregulates RASA1 expression
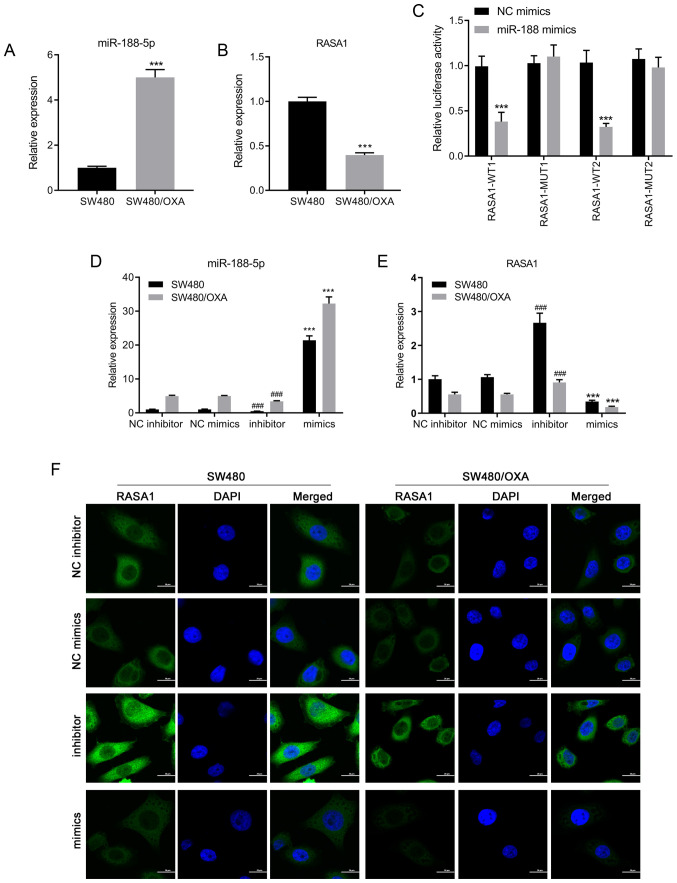
RT-qPCR was performed using SW480 and SW480/OXA cells to determine the expression levels of miR-188-5p. As shown in Fig. 1A, the expression of miR-188-5p was significantly upregulated in SW480/OXA cells compared with that in SW480 cells (P<0.001), suggesting that the expression of miR-188-5p may be associated with drug resistance in colon cancer cells. Bioinformatics analysis predicted that miR-188-5p potentially bound to RASA1 mRNA. There was a targeted binding association between miR-188-5p and RASA1. Moreover, mRNA expression levels of RASA1 were decreased in SW480/OXA cells compared with those in SW480 cells (Fig. 1B; P<0.001). To confirm whether RASA1 was a direct target of miR-188-5p, a luciferase assay was performed. Co-transfection with miR-188-5p mimics significantly reduced the luciferase activity of WT RASA1, but not MUT RASA1, compared with NC mimics (Fig. 1C; P<0.001). These results indicated that RASA1 was a target of miR-188-5p.
Figure 1.
Downregulation of miR-188-5p upregulates RASA1 expression in colon cancer. (A and B) mRNA expression of (A) miR-188-5p and (B) and RASA1 in SW480 or SW480/OXA cells. ***P<0.001 vs. SW480. (C) Luciferase activity in SW480 cells co-transfected with indicated plasmids and miR-188-5p or NC mimics. ***P<0.001 vs. NC mimics. (D and E) mRNA expression of (D) miR-188-5p and (E) RASA1 in SW480 or SW480/OXA cells transfected with miR-188-5p mimics or inhibitor. ***P<0.001 vs. NC mimics; ###P<0.001 vs. NC inhibitor. (F) RASA1 expression in SW480 or SW480/OXA cells transfected with miR-188-5p mimics or inhibitor. Scale bar, 20 µm. miR, microRNA; oxa, oxaliplatin; RASA1, Ras GTPase-activating protein 1; WT, wild-type; MUT, mutant; NC, negative control.
SW480 or SW480/OXA cells were transfected with miR-188-5p mimics or inhibitor to examine the regulatory effects of miR-188-5p on RASA1 expression. Transfection with miR-188-5p mimics significantly increased miR-188-5p expression and reduced RASA1 expression, compared with the respective negative controls. By contrast, miR-188-5p inhibitor transfection reduced miR-188-5p expression and increased RASA1 expression (Fig. 1D and E; all P<0.001). Moreover, immunofluorescence was used to detect the expression of RASA1, and the results suggested that transfection with miR-188-5p mimics decreased the expression of RASA1 at the protein level (Fig. 1F). Thus, inhibition of miR-188-5p increased the expression of its target gene, RASA1, both in SW480 and in SW480/OXA cells.
Targeting miR-188-5p promotes apoptosis in SW480/OXA cells
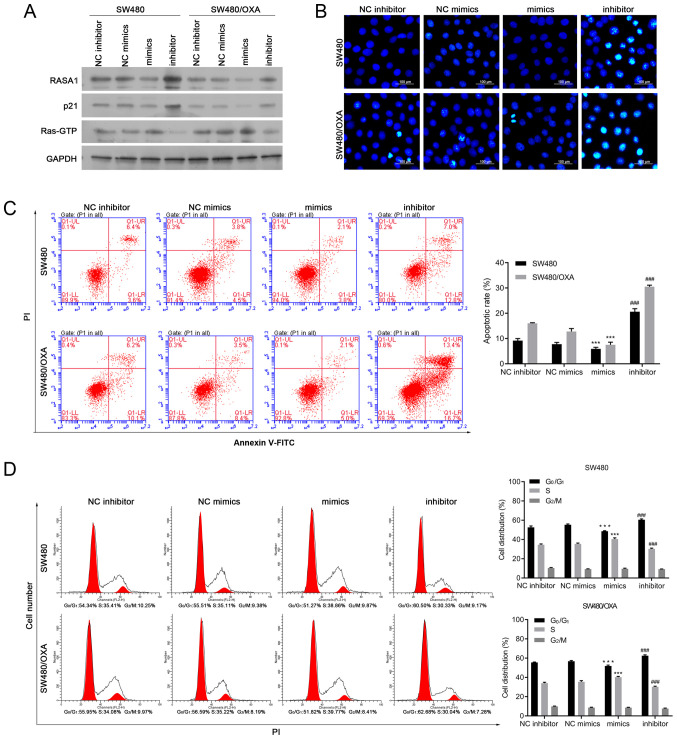
Transfections with miR-188-5p mimics or inhibitor were performed in SW480 or SW480/OXA cells to further examine the role of miR-188-5p on the apoptosis of colon cancer cells. The miR-188-5p mimics significantly decreased the protein expression levels of RASA1, compared with NC mimics; whereas miR-188-5p inhibitor significantly increased the expression of RASA1, compared with NC inhibitor (P<0.001; Figs. 2A and S1A). In addition, the expression of p21 decreased in miR-188-5p mimic-transfected cells and increased in miR-188-5p inhibitor-transfected cells, compared with that in NC mimics and NC inhibitor, respectively (Fig. 2A). Moreover, the expression of p21 was lower in miR-188-5p mimic-transfected SW480/OXA cells compared with that in mimic-transfected SW480 cells (Figs. 2A and S2A). As RASA1 suppresses the function of Ras and enhances the weak intrinsic GTPase activity of Ras proteins (34), the protein expression levels of Ras-GTP were then examined. The results demonstrated that miR-188-5p mimics significantly promoted the protein expression levels of Ras-GTP compared with NC mimics; whereas miR-188-5p inhibitor significantly decreased the expression of Ras-GTP compared with NC inhibitor (P<0.001 and P<0.01, respectively; Figs. 2A and S1A).The expression of Ras-GTP changed following miR-188-5p mimic or inhibitor transfection (Figs. 2A and S2A).
Figure 2.
Downregulation of miR-188-5p induces cell apoptosis in SW480/OXA cells. (A) Protein expression levels of RASA1, p21, and Ras-GTP in SW480 or SW480/OXA cells transfected with miR-188-5p mimics or inhibitors. (B) Hoechst 33342 staining of SW480 or SW480/OXA cells transfected with miR-188-5p mimics or inhibitor. Scale bar, 100 µm. (C) Apoptosis of SW480 or SW480/OXA cells transfected with miR-188-5p mimics or inhibitor. ***P<0.001 vs. NC mimics; ###P<0.001 vs. NC inhibitor. (D) Cell cycle distribution was analyzed by flow cytometry. ***P<0.001 vs. NC mimics, ###P<0.001 vs. NC inhibitor. miR, microRNA; oxa, oxaliplatin; RASA1, Ras GTPase-activating protein 1; PI, propidium iodide; FITC, fluorescein isothiocyanate; NC, negative control.
Moreover, in the Hoechst 33342 staining and Annexin V/PI assays, transfection with miR-188-5p mimics inhibited cell apoptosis compared with transfection with NC mimics, whereas miR-188-5p inhibitor promoted cell apoptosis compared with NC inhibitor in SW480 and SW480/OXA cells (P<0.01 and P<0.001, respectively; Figs. 2B and C and S2A). These data suggested that targeting miR-188-5p promoted apoptosis in both SW480 and SW480/OXA cells.
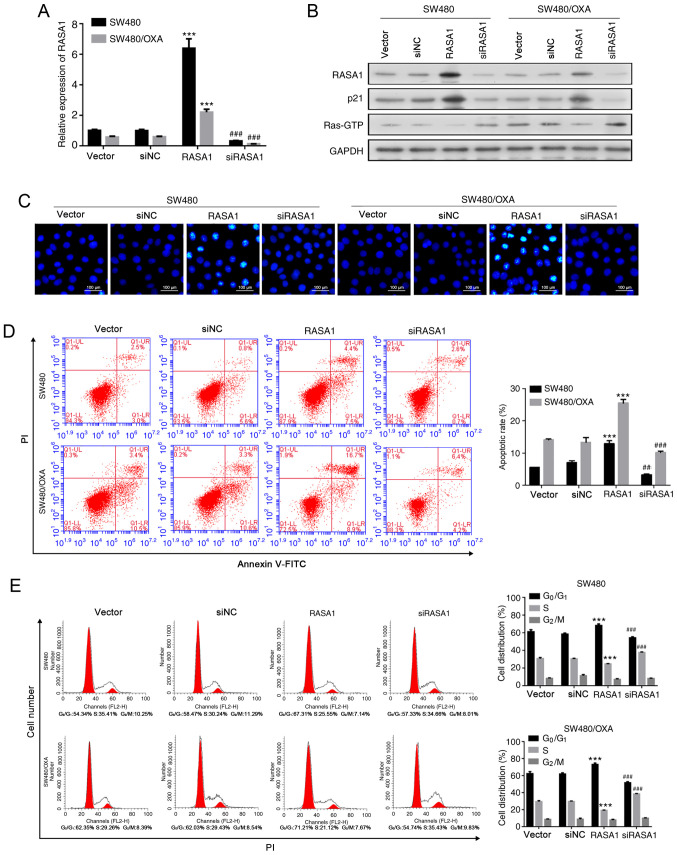
The effect of miR-188-5p on cell cycle progression was also determined. Transfection with miR-188-5p mimics reduced the G0G1-phase cell population and increased the proportion of cells in the S phase, while miR-188-5p inhibitor arrested cells at the G1 phase (Fig. 2D; all P<0.001). These data suggested that targeting miR-188-5p delays the cell cycle.S1B), compared with the respective negative controls. In addition, the overexpression of RASA1 decreased the expression of Ras-GTP, whereas siRASA1 transfection increased it, both in SW480 and SW480/OXA cells (Figs. 3B and S2B). and significantly Conversely, the expression of p21 decreased (Figs. 3B and S1B) following RASA1 silencing increased in RASA1-overexpressing SW480 and SW480/OXA cells (P<0.01and P<0.001, respectively; Figs. 3B and S1B).
Figure 3.
Overexpression of RASA1 accelerates apoptosis and inhibits the cell cycle. (A) Relative mRNA expression of RASA1 in SW480 or SW480/OXA cells transfected with RASA1 overexpression plasmid or RASA1 siRNA. ***P<0.001 vs. vector, ###P<0.001 vs. siNC group. (B) RASA1, p21, and Ras-GTP protein expression in SW480 or SW480/OXA cells transfected with RASA1 plasmid or RASA1 siRNA. (C) Hoechst 33342 staining in SW480 or SW480/OXA cells transfected with RASA1 plasmid or RASA1 siRNA. Scale bar, 100 µm. (D) Apoptosis rate in SW480 or SW480/OXA cells following transfection with RASA1 plasmid or RASA1 siRNA. ***P<0.001 vs. vector, ###P<0.001 vs. siNC group. (E) Cell cycle distribution was analyzed by flow cytometry. ***P<0.001 vs. vector; ##P<0.01, ###P<0.001 vs. siNC. Oxa, oxaliplatin; RASA1, Ras GTPase-activating protein 1; siNC, siRNA negative control; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Furthermore, apoptosis was decreased following RASA1 silencing (Figs. 3C and D and S2B; P<0.005). However, RASA1 overexpression promoted apoptosis, both in SW480 and SW480/OXA cells (Figs. 3C and D and S2B; P<0.001). In addition, SW480 and SW480/OXA cells accumulated in the G0G1 phase of the cell cycle following RASA1 overexpression, and the frequency of S-phase cells was reduced (Fig. 3E; P<0.001). By contrast, the frequency of G0G1 cells decreased, whereas the frequency of S-phase cells increased following RASA1 silencing (Fig. 3E; P<0.001). These findings further confirmed that RASA1 induced the cell apoptosis of colon cancer cells.
Suppression of miR-188-5p promotes SW480/OXA cell apoptosis by increasing RASA1 expression
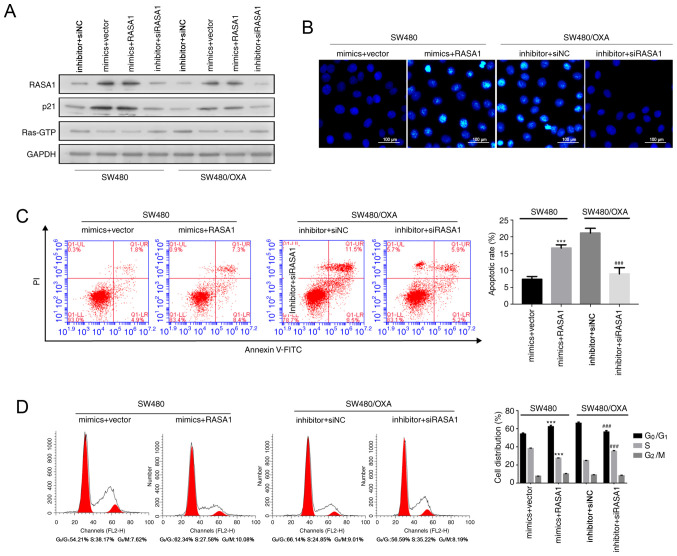
Co-transfection experiments were performed to determine whether the increase in apoptosis induced by the miR-188-5p inhibitor was dependent on RASA1. As shown in Figs. 4A and S1C RASA1 and p21 were upregulated following miR-188-5p mimic transfection, while the transfection with miR-188-5p inhibitor inhibited the expression of RASA1 and p21, compared with cells transfected with the mimics and empty vector control (P<0.001).
Figure 4.
Downregulation of miR-188-5p promotes SW480/OXA cell apoptosis by increasing RASA1 expression. (A) RASA1, p21, and Ras-GTP protein expression in SW480 or SW480/OXA cells following co-transfection with RASA1 plasmid or RASA1 siRNA and with miR-188-5p or inhibitor. (B) Hoechst 33342 staining in the different transfection groups. Scale bar, 100 µm. (C) Apoptosis rate in the different transfection groups. (D) Cell cycle distribution was analyzed by flow cytometry. ***P<0.001 vs. mimics + vector group; ###P<0.001 vs. inhibitor + siNC group. miR, microRNA; oxa, oxaliplatin; RASA1, Ras GTPase-activating protein 1; NC, negative control siNC, siRNA negative control; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Hoechst 33342 staining and Annexin V assays showed that overexpression of RASA1 enhanced apoptosis in miR-188-5p mimic-treated SW480 cells, compared with cells transfected with the mimics and empty vector control, whereas RASA1 silencing combined with miR-188-5p inhibitor transfection reduced the apoptosis rate (Figs. S2C, 4B and C; P<0.001). Moreover, RASA1 overexpression increased the frequency of cells in the G0G1 phase while decreasing the frequency of cells in the S phase of the cell cycle in miR-188-5p mimic-transfected SW480 cells (Fig. 4D). By contrast, siRASA1 and miR-188-5p inhibitor transfection resulted in the opposite effect (Fig. 4D; P<0.001). These results indicated that the inhibition of miR-188-5p induced SW480/OXA cell apoptosis by enhancing RASA1 expression.
Discussion
miR-188-5p was significantly upregulated in both SW480 and SW480/OXA cells. Transfection with miR-188-5p resulted in significant inhibition of SW480 and OXA-induced cell apoptosis, whereas both SW480 and SW480/OXA apoptosis significantly increased following miR-188-5p inhibition. Moreover, miR-188-5p transfection downregulated RASA1 expression. Moreover, the overexpression of miR-188-5p conferred OXA resistance, while downregulation of miR-188-5p sensitized resistant cells to OXA, which was blocked by inhibition of RASA1. In summary, the present study suggested that miR-188-5p could enhance colon cancer cell chemosensitivity by promoting the expression of RASA1.
Accumulating evidence has demonstrated the role of miR-188-5p in cancer development. However, the expression of miR-188-5p in OXA-resistant colon cancer cells has not been characterized. In the present study, the expression of miR-188-5p significantly increased in OXA-resistant colon cancer cells. A previous miRNA sequencing study by Zhao et al (14) involving 228 patients, six miRNA candidates, including miR-188-3p, were identified as strong predictors of patient survival. In addition, miR-188-3p promotes CRC cell migration both in vitro and in vivo partly by regulating MLLT4 expression, and high miR-188-3p expression is considered an independent prognostic factor of colon cancer (17). Moreover, a previous study reported the role of miR-188-5p in rectal cancer and response to neoadjuvant radio- and chemotherapy (22). The results of the present study were partly consistent with previous studies showing that miR-188-5p promoted colon cancer progression (22,39,40). In the present study, miR-188-5p inhibition promoted apoptosis, whereas the upregulation of miR-188-5p inhibited apoptosis in SW480/OXA cells, suggesting that targeting miR-188-5p promoted apoptosis in both SW480 and SW480/OXA cells. Moreover, cell cycle arrest in the G0G1 phase was induced, while apoptosis was increased following miR-188-5p inhibition. These results were consistent with previous studies showing that miR-188-5p controlled cell cycle progression via downregulation of multiple cyclins and cyclin-dependent kinase complexes involved in the G1/S transition (20,41). Although previous studies have indicated that miR-188-5p serves as a tumor suppressor and is downregulated in multiple types of cancer (15,42), miR-188-5p was upregulated in OXA-resistant colon cancer and RASA1 silencing abrogated the increase in cell apoptosis induced by the miR-188-5p inhibitor in the present study. Hence, miR-188-5p may enhance colon cancer cell chemosensitivity by promoting the expression of RASA1.
Of the several potential target genes for miR-188-5p predicted by TargetScan, PITA and miRDB Human database, RASA1 was selected for the present study. RASA1 acts as a suppressor of Ras function and enhances the weak intrinsic GTPase activity of Ras proteins, resulting in the inactive GDP-bound form of Ras (23). Previous studies have suggested that RASA1 acts as a tumor suppressor in hepatocellular carcinoma (42), human bronchial epithelial cells (43), breast cancer (44) and melanoma (45). In colon cancer cells, RASA1 is significantly downregulated, and has been identified as a target gene of miR-21, miR-223 and miR-335 (34,36). Moreover, onco-miRNA molecules, such as microRNA-21 and microRNA-182 promote tumor angiogenesis or lymph node metastasis by targeting RASA1 (37,38). p21 is a tumor suppresser gene that acts as negative regulator of the G1/S transition (46). Downregulation of p21 is observed in various human cancer types, including melanoma, gastric, ovarian and colon cancers (47). In the present study, the expression of miR-188-5p was negatively associated with that of RASA1 in OXA-resistant colon cancer cells. The overexpression of RASA1 promoted apoptosis, induced the expression of p21 and delayed cell cycle progression. Moreover, apoptosis in SW480 cells was reduced following miR-188-5p mimic transfection, while overexpression of RASA1 enhanced cell apoptosis in miR-188-5p mimic-transfected SW480 cells. By contrast, suppression of RASA1 abrogated cell apoptosis in miR-188-5p inhibitor-transfected SW480/OXA cells. These results suggested that miR-188-5p inhibition induced SW480/OXA cell apoptosis by enhancing RASA1 expression.
In conclusion, the present findings suggest that targeting miR-188-5p promoted apoptosis in SW480/OXA cells and suppression of miR-188-5p promoted SW480/OXA cell apoptosis by increasing RASA1 expression, thus providing new insight into treatment options for OXA-resistant colon cancer.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CRC
colorectal cancer
- miR/miRNA
microRNA
- OXA
oxaliplatin
- RASA1
Ras GTPase-activating protein 1
- UTR
untranslated region
Funding Statement
This study was supported by The Guilin Scientific Research and Technology Development Plan Project (grant no. 20170109-50).
Funding
This study was supported by The Guilin Scientific Research and Technology Development Plan Project (grant no. 20170109-50).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HW conceived and designed the study, and drafted and revised the manuscript. XZ, XSL, ZS, SJ and XG performed the experiments. XKL, XX, and LZ analyzed data and prepared the figures. XZ and XSL edited and revised the manuscript. HW and XZ confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.The Lancet Gastroenterology Hepatology, corp-author. Colorectal cancer screening: Is earlier better? Lancet Gastroenterol Hepatol. 2018;3:519. doi: 10.1016/S2468-1253(18)30205-X. [DOI] [PubMed] [Google Scholar]
- 3.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 4.Lai M, Liu G, Li R, Bai H, Zhao J, Xiao P, Mei J. Hsa_circ_0079662 induces the resistance mechanism of the chemotherapy drug oxaliplatin through the TNF-α pathway in human colon cancer. J Cell Mol Med. 2020;24:5021–5027. doi: 10.1111/jcmm.15122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y, Zhang J, Wang K, Han W, Wang X, Gao M, Wang Z, Sun Y, Yan H, Zhang H, et al. Quercetin overcomes colon cancer cells resistance to chemotherapy by inhibiting solute carrier family 1, member 5 transporter. Eur J Pharmacol. 2020;881:173185. doi: 10.1016/j.ejphar.2020.173185. [DOI] [PubMed] [Google Scholar]
- 6.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 7.Garofalo M, Croce CM. MicroRNAs: Master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 8.Xie Y, Tobin LA, Camps J, Wangsa D, Yang J, Rao M, Witasp E, Awad KS, Yoo N, Ried T, Kwong KF. MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells. Oncogene. 2013;32:2442–2451. doi: 10.1038/onc.2012.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamanaka S, Campbell NR, An F, Kuo SC, Potter JJ, Mezey E, Maitra A, Selaru FM. Coordinated effects of microRNA-494 induce G2/M arrest in human cholangiocarcinoma. Cell Cycle. 2012;11:2729–2738. doi: 10.4161/cc.21105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582–588. doi: 10.1016/j.bcp.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Angelo E, Vicentini C, Agostini M, Kiss A, Baffa R, Scarpa A, Fassan M. MicroRNAs as tools and effectors for patient treatment in gastrointestinal carcinogenesis. Curr Drug Targets. 2015;16:383–392. doi: 10.2174/1389450116666141210091454. [DOI] [PubMed] [Google Scholar]
- 12.D'Angelo E, Zanon C, Sensi F, Digito M, Rugge M, Fassan M, Scarpa M, Pucciarelli S, Nitti D, Agostini M. MiR-194 as predictive biomarker of responsiveness to neoadjuvant chemoradiotherapy in patients with locally advanced rectal adenocarcinoma. J Clin Pathol. 2018;71:344–350. doi: 10.1136/jclinpath-2017-204690. [DOI] [PubMed] [Google Scholar]
- 13.Gao C, Zhou C, Zhuang J, Liu L, Liu C, Li H, Liu G, Wei J, Sun C. MicroRNA expression in cervical cancer: Novel diagnostic and prognostic biomarkers. J Cell Biochem. 2018;119:7080–7090. doi: 10.1002/jcb.27029. [DOI] [PubMed] [Google Scholar]
- 14.Zhao L, Ni X, Zhao L, Zhang Y, Jin D, Yin W, Wang D, Zhang W. MiroRNA-188 Acts as tumor suppressor in Non-Small-Cell Lung cancer by Targeting MAP3K3. Mol Pharm. 2018;15:1682–1689. doi: 10.1021/acs.molpharmaceut.8b00071. [DOI] [PubMed] [Google Scholar]
- 15.Peng Y, Shen X, Jiang H, Chen Z, Wu J, Zhu Y, Zhou Y, Li J. MiR-188-5p suppresses gastric cancer cell proliferation and invasion via Targeting ZFP91. Oncol Res. 2018;27:65–71. doi: 10.3727/096504018X15191223015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N, Shi H, Zhang L, Li X, Gao L, Zhang G, Shi Y, Guo S. MiR-188 inhibits glioma cell proliferation and cell cycle progression through targeting β-Catenin. Oncol Res. 2018;26:785–794. doi: 10.3727/096504017X15127309628257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pichler M, Stiegelbauer V, Vychytilova-Faltejskova P, Ivan C, Ling H, Winter E, Zhang X, Goblirsch M, Wulf-Goldenberg A, Ohtsuka M, et al. Genome-Wide miRNA Analysis Identifies miR-188-3p as a novel prognostic marker and molecular factor involved in colorectal carcinogenesis. Clin Cancer Res. 2017;23:1323–1333. doi: 10.1158/1078-0432.CCR-16-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Liu H. MicroRNA-188 is downregulated in oral squamous cell carcinoma and inhibits proliferation and invasion by targeting SIX1. Tumour Biol. 2016;37:4105–4113. doi: 10.1007/s13277-015-4246-9. [DOI] [PubMed] [Google Scholar]
- 19.Fang F, Chang RM, Yu L, Lei X, Xiao S, Yang H, Yang LY. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J Hepatol. 2015;63:874–885. doi: 10.1016/j.jhep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Qi S, Zhang T, Wang A, Liu R, Guo J, Wang Y, Xu Y. MiR-188-5p inhibits tumour growth and metastasis in prostate cancer by repressing LAPTM4B expression. Oncotarget. 2015;6:6092–6104. doi: 10.18632/oncotarget.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinlong S, Lin F, Yonghui L, Li Y, Weidong W. Identification of let-7a-2-3p or/and miR-188-5p as prognostic biomarkers in cytogenetically normal acute myeloid leukemia. PLoS One. 2015;10:e0118099. doi: 10.1371/journal.pone.0118099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Della Vittoria Scarpati G, Falcetta F, Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK, D'Incalci M, De Placido S, Pepe S. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2012;83:1113–1119. doi: 10.1016/j.ijrobp.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 23.Pamonsinlapatham P, Hadj-Slimane R, Lepelletier Y, Allain B, Toccafondi M, Garbay C, Raynaud F. p120-Ras GTPase activating protein (RasGAP): A multi-interacting protein in downstream signaling. Biochimie. 2009;91:320–328. doi: 10.1016/j.biochi.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Yang XY, Guan M, Vigil D, Der CJ, Lowy DR, Popescu NC. p120Ras-GAP binds the DLC1 Rho-GAP tumor suppressor protein and inhibits its RhoA GTPase and growth-suppressing activities. Oncogene. 2009;28:1401–1409. doi: 10.1038/onc.2008.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvisi DF, Ladu S, Conner EA, Seo D, Hsieh JT, Factor VM, Thorgeirsson SS. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J Hepatol. 2011;54:311–319. doi: 10.1016/j.jhep.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mai A, Veltel S, Pellinen T, Padzik A, Coffey E, Marjomäki V, Ivaska J. Competitive binding of Rab21 and p120RasGAP to integrins regulates receptor traffic and migration. J Cell Biol. 2011;194:291–306. doi: 10.1083/jcb.201012126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, Zhang CY, Chen J, Zhang J. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1) J Biol Chem. 2013;288:9508–9518. doi: 10.1074/jbc.M112.367763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Organ SL, Hai J, Radulovich N, Marshall CB, Leung L, Sasazuki T, Shirasawa S, Zhu CQ, Navab R, Ikura M, Tsao MS. p120RasGAP is a mediator of rho pathway activation and tumorigenicity in the DLD1 colorectal cancer cell line. PLoS One. 2014;9:e86103. doi: 10.1371/journal.pone.0086103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma SB, Lin CC, Farrugia MK, McLaughlin SL, Ellis EJ, Brundage KM, Salkeni MA, Ruppert JM. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol Cell Biol. 2014;34:4143–4164. doi: 10.1128/MCB.00480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Liu T, Sun Q, Niu M, Jiang Y, Pang D. Downregulation of Ras GTPaseactivating protein 1 is associated with poor survival of breast invasive ductal carcinoma patients. Oncol Rep. 2015;33:119–124. doi: 10.3892/or.2014.3604. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, Cui Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42:D1070–D1074. doi: 10.1093/nar/gkt1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong B, Liu WW, Nie WJ, Li DF, Xie ZJ, Liu C, Liu YH, Mei P, Li ZJ. MiR-21/RASA1 axis affects malignancy of colon cancer cells via RAS pathways. World J Gastroenterol. 2015;21:1488–1497. doi: 10.3748/wjg.v21.i5.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun D, Wang C, Long S, Ma Y, Guo Y, Huang Z, Chen X, Zhang C, Chen J, Zhang J. C/EBP-β-activated microRNA-223 promotes tumour growth through targeting RASA1 in human colorectal cancer. Br J Cancer. 2015;112:1491–1500. doi: 10.1038/bjc.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Yang H, Yuan L, Liu G, Zhang C, Hong M, Liu Y, Zhou M, Chen F, Li X. Overexpression of miR-335 confers cell proliferation and tumour growth to colorectal carcinoma cells. Mol Cell Biochem. 2016;412:235–245. doi: 10.1007/s11010-015-2630-9. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Zhan X, Yan D, Wang Z. Circulating MicroRNA-21 is involved in lymph node metastasis in cervical cancer by targeting RASA1. Int J Gynecol Cancer. 2016;26:810–816. doi: 10.1097/IGC.0000000000000694. [DOI] [PubMed] [Google Scholar]
- 38.Zhu YJ, Xu B, Xia W. Hsa-mir-182 downregulates RASA1 and suppresses lung squamous cell carcinoma cell proliferation. Clin Lab. 2014;60:155–159. doi: 10.7754/clin.lab.2013.121131. [DOI] [PubMed] [Google Scholar]
- 39.Yan S, Yue Y, Wang J, Li W, Sun M, Gu C, Zeng L. LINC00668 promotes tumorigenesis and progression through sponging miR-188-5p and regulating USP47 in colorectal cancer. Eur J Pharmacol. 2019;858:172464. doi: 10.1016/j.ejphar.2019.172464. [DOI] [PubMed] [Google Scholar]
- 40.Wu J, Chen Z, Liu W, Zhang Y, Feng W, Yuan Y, Ye J, Wang L, Cai S, He Y, et al. MicroRNA-188-5p targeting Forkhead Box L1 promotes colorectal cancer progression via activating Wnt/β-catenin signaling. Oncol Res. 2020 Nov 23; doi: 10.3727/096504020X16061317886881. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 41.Wu J, Lv Q, He J, Zhang H, Mei X, Cui K, Huang N, Xie W, Xu N, Zhang Y. MicroRNA-188 suppresses G1/S transition by targeting multiple cyclin/CDK complexes. Cell Commun Signal. 2014;12:66. doi: 10.1186/s12964-014-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen YL, Huang WC, Yao HL, Chen PM, Lin PY, Feng FY, Chu PY. Down-regulation of RASA1 is associated with poor prognosis in human hepatocellular carcinoma. Anticancer Res. 2017;37:781–785. doi: 10.21873/anticanres.11377. [DOI] [PubMed] [Google Scholar]
- 43.Hayashi T, Desmeules P, Smith RS, Drilon A, Somwar R, Ladanyi M. RASA1 and NF1 are preferentially Co-mutated and define a distinct genetic subset of smoking-associated non-small cell lung carcinomas sensitive to MEK inhibition. Clin Cancer Res. 2018;24:1436–1447. doi: 10.1158/1078-0432.CCR-17-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suarez-Cabrera C, Quintana RM, Bravo A, Casanova ML, Page A, Alameda JP, Paramio JM, Maroto A, Salamanca J, Dupuy AJ, et al. A Transposon-based analysis reveals RASA1 Is involved in triple-negative breast cancer. Cancer Res. 2017;77:1357–1368. doi: 10.1158/0008-5472.CAN-16-1586. [DOI] [PubMed] [Google Scholar]
- 45.Sung H, Kanchi KL, Wang X, Hill KS, Messina JL, Lee JH, Kim Y, Dees ND, Ding L, Teer JK, et al. Inactivation of RASA1 promotes melanoma tumorigenesis via R-Ras activation. Oncotarget. 2016;7:23885–23896. doi: 10.18632/oncotarget.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pines J, Rieder CL. Re-staging mitosis: A contemporary view of mitotic progression. Nat Cell Biol. 2001;3:E3–E6. doi: 10.1038/35050676. [DOI] [PubMed] [Google Scholar]
- 47.Cai K, Dynlacht BD. Activity and nature of p21(WAF1) complexes during the cell cycle. Proc Natl Acad Sci USA. 1998;95:12254–12259. doi: 10.1073/pnas.95.21.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.