Abstract
Numerous studies have reported that abnormal cadherin 3 (CDH3) and microRNA (miRNA/miR)-665 expression can induce the progression of gastric cancer (GC). However, the mechanism of interaction between miR-665 and CDH3 in GC requires further investigation. The present study aimed to investigate the influence of miR-665 and CDH3 in GC development. The effect of miR-665 and CDH3 on GC tissues and cell lines was examined using reverse transcription-quantitative PCR. Subsequently, CDH3 protein expression in GC cell lines was detected using western blotting. To confirm the association between miR-665 and CDH3, a dual-luciferase reporter assay system was employed. Cell proliferation and adhesion were analyzed using BrdU ELISA, MTT and cell adhesion assays. Finally, caspase-3 activity assay kit and FITC apoptosis detection kit were used for the determination of apoptosis of GC cells. The current findings confirmed the upregulation of CDH3 expression in GC cell lines and tissues. Experimental results indicated that CDH3 overexpression increased cell proliferation and adhesion, but decreased the apoptosis level of the cells. Similarly, the miR-665 inhibitor enhanced cell proliferation and adhesion, but inhibited apoptosis of GC cells. Additionally, it was observed that CDH3 was a direct target of miR-665 in GC cells and that miR-665 inhibited CDH3 expression, thereby repressing the progression of GC. In conclusion, the present study suggested that by targeting CDH3, miR-665 suppressed the progression of GC. These findings may provide a significant theoretical basis for future GC clinical therapy.
Keywords: microRNA-665, cadherin 3, gastric cancer, proliferation, apoptosis, adhesion
Introduction
Gastric cancer (GC) refers to the deleterious inflammation of the stomach wall, and this type of tumor accounted for ~8.2% of tumor-associated deaths worldwide in 2018 (1). GC is a multifactorial disease that is associated with the environment, including Helicobacter pylori infection (2), smoking and alcohol consumption (3). To date, treatment options such as surgery, radiotherapy, chemotherapy and medication are the most frequently therapeutic strategies for patients with GC; however, these methods have not improved the recurrence and the poor prognosis of GC (4). Gene therapy has also been utilized in recent years to treat patients with GC (5). However, numerous target genes in GC remain unknown. By exploring the molecular mechanism of GC, the clinical outcomes of patients suffering from GC can be improved.
The cadherin 3 (CDH3) gene is located on chromosome 16q22.1 and consists of 18 exons. It is found in a six-cadherin bundle, and this calcium-dependent gene is comprised of five extracellular repeats, a cytoplasmic tail and a transmembrane region (6). The aberrant expression of CDH3 has been frequently observed in various types of cancer, including colorectal (7), pancreatic (8) and esophageal cancer (9). More specifically, CDH3 has been demonstrated to increase cell adhesion and invasion (10). However, only a few studies have been conducted to investigate the tumorigenic role of CDH3 in GC cells (11,12), revealing that CDH3 expression was upregulated in GC tissues. Therefore, it is noteworthy to investigate the impacts of CDH3 on GC.
MicroRNAs (miRNAs/miRs) can be referred to as non-coding protein sequences with the capability of regulating, decoding and controlling gene expression (13). Previous studies have revealed that miRNAs regulate cell proliferation, invasion and apoptosis, as well as serving a pivotal role in tumor progression (14,15). For instance, miR-665 has been associated with lung cancer progression by increasing cell proliferation, migration and invasion (16). Some studies have suggested that miR-665 may be used as a potential molecular target for various types of cancer, including pancreatic (17), breast (18) and ovarian carcinoma (19). Additionally, several studies on GC have suggested that by targeting different genes, miR-665 may act both as an oncogene and as a tumor inhibitor (20,21). However, the mechanism of interaction between miR-665 and CDH3 in GC requires further investigation.
The present study aimed to explore the effect of the miR-665/CDH3 interactome on the pathogenesis of GC. The objective included the investigation of potential bio-targets for GC treatment. It was hypothesized that miR-665 could suppress GC cells by targeting CDH3. The current study may be relevant in terms of providing a theoretical basis for GC clinical therapy.
Materials and methods
Bioinformatics analysis
The mRNA expression profiles GSE118916 (22) and GSE79973 (23) were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/?term=), a public database that stores gene expression microarrays. The upregulated differentially expressed genes (DEGs) were screened from GSE118916 and GSE79973 using log2-fold-change (logFC)>1 and adjusted (adj) P<0.05. STRING (https://string-db.org/) was used to analyze the Gene Ontology (GO) enrichment of the upregulated DEGs. The expression levels of the key genes in stomach adenocarcinoma (STAD) were analyzed using The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/).
Clinical samples and cell lines
All tissue samples, including tumor and non-tumor specimens, were obtained from 30 patients with GC (16 male and 14 female; median age, 60 years; age range 29–72 years) at Henan Provincial People's Hospital (Zhengzhou, China) between January 2019 and January 2020. All patients signed informed consent forms to participate in the study. The ethical protocols of the present study were approved by the Ethics Committee of Henan Provincial People's Hospital. The diagnosis of GC included a clinical examination, blood sampling, endoscopy (when clinically indicated) and computed tomography scanning of the chest, abdomen and pelvis. The tumor tissues were all confirmed by endoscopic biopsy or surgical specimens. Patients with a history of cancer or severe clinical symptoms and genetic diseases, and patients who received preoperative radiochemotherapy were excluded from the study. Non-tumor specimens were ≥1.5 cm from the tumor margins. All collected samples were stored at the recommended temperature (−80°C). The clinical features of patients who participated in the study are shown in Table I. The tumors were staged following the tumor-node-metastasis (TNM) staging system of the International Union Against Cancer (24). CDH3 was divided into high and low expression groups according to the median expression level of CDH3 (5.848). The association between CDH3 expression and clinical characteristics in patients with GC are shown in Table SI.
Table I.
Clinical characteristics of 30 patients with gastric cancer.
| Characteristics | N | Percentage, % |
|---|---|---|
| Age, years | ||
| >60 | 18 | 60.0 |
| ≤60 | 12 | 40.0 |
| Sex | ||
| Male | 16 | 53.3 |
| Female | 14 | 46.7 |
| Tumor size, cm | ||
| >5 | 19 | 63.3 |
| ≤5 | 11 | 36.7 |
| Histological grade | ||
| Low | 13 | 43.3 |
| High | 17 | 56.7 |
| TNM stage | ||
| I | 5 | 16.7 |
| II | 7 | 23.3 |
| III | 14 | 46.7 |
| IV | 4 | 13.3 |
| Lymph nodes status | ||
| Positive | 19 | 63.3 |
| Negative | 11 | 36.7 |
| Differentiation | ||
| Poor | 17 | 56.7 |
| Well/moderate | 13 | 43.3 |
All cell lines used were purchased from the American Type Culture Collection, including GC cell lines (HGC-27, GTL-16, MKN74 and AGS) and the gastric epithelial cell line (GES-1). MKN74 cells were maintained in F-12K medium, GES-1 cells were maintained in RPMI-1640 medium and AGS, HGC-27 and GTL-16 cells were kept in DMEM (Gibco; Thermo Fisher Scientific, Inc.). The media were supplemented with 100 U/ml penicillin and 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). Cells were incubated at 37°C in an atmosphere containing 5% CO2.
RNA extraction and reverse transcription-quantitative (RT-q)PCR
RNA extraction from tissues and cells was performed using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was obtained using the PrimeScript First Strand cDNA Synthesis kit (Takara Bio, Inc.) according to the manufacturer's protocol. Subsequently, qPCR was performed using SYBR® Premix Ex Taq™ (Takara Bio, Inc.) according to the manufacturer's protocol. The thermocycling conditions were as follows: Denaturation at 95°C for 20 sec, followed by 40 cycles of denaturation at 95°C for 1 min and annealing/extension at 60°C 20 sec. The expression levels of miR-665, CDH3, collagen type XVIII α1 chain (COL18A1) and fibroblast activation protein (FAP) were measured using the 2−ΔΔCq method, and U6 or GAPDH were used as an endogenous reference for miRNA or mRNA, respectively. The relative expression levels of miR-665, CDH3, COL18A1 or FAP were reported in relation to an internal control gene called ΔCt. The ΔCT and ΔΔCT values were calculated using the following mathematical formulas: ΔCt=CtmiRNA-126/CDH3-CtU6/GAPDH and ΔΔCt=ΔCtcase-ΔCtcontrol (25). All the primer sequences used in the study are listed in Table II.
Table II.
Primer sequences used for reverse transcription-quantitative PCR.
| Gene | Primer sequences (5′-3′) |
|---|---|
| miR-665 | Forward: GCCGAGACCAGGAGGCUGA |
| Reverse: CTCAACTGGTGTCGTGGA | |
| U6 | Forward: ATTGGAACGATACAGAGAAGATT |
| Reverse: GGA ACGCTTCACGAATTTG | |
| CDH3 | Forward: CAGGTGCTGAACATCACGGACA |
| Reverse: CTTCAGGGACAAGACCACTGTG | |
| COL18A1 | Forward: GGAGAGATTGGCTTTCCTGGAC |
| Reverse: CCTCATGCCAAATCCAAGGCTG | |
| FAP | Forward: GGAAGTGCCTGTTCCAGCAATG |
| Reverse: TGTCTGCCAGTCTTCCCTGAAG | |
| GAPDH | Forward: AGCCACATCGCTCAGACAC |
| Reverse: GCCCAATACGACCAAATCC |
miR, microRNA; CDH3, cadherin 3; COL18A1, collagen type XVIII α1 chain; FAP, fibroblast activation protein.
Western blotting
GC cells were lysed using RIPA lysis buffer (Sangon Biotech Co., Ltd.). Total protein concentration was quantified using the BCA method. Proteins (30 µg/lane) were separated via 10% SDS-PAGE and then transferred to PVDF membranes, which were blocked with 5% BSA (Gibco; Thermo Fisher Scientific, Inc.) for 2 h at 25°C, and washed three times with TBS-0.1% Tween-20. After blocking the membranes, primary antibodies against CDH3 (1:1,000; cat. no. ab242060; Abcam) and GAPDH (1:2,000; cat. no. ab181602; Abcam) were used to incubate the membranes overnight at 4°C. Subsequently, HRP-conjugated goat anti-rabbit IgG (1:5,000; cat. no. ab97051; Abcam) secondary antibody was incubated for 1 h at 25°C. The protein signal was subsequently visualized using an ECL system (Bio-Rad Laboratories, Inc.). Finally, the quantification of the density of each band was performed using Gel-Pro Analyzer 4.0 (Media Cybernetics, Inc.).
Cell transfection
The small interfering RNA (siRNA) used as a non-targeting negative control (si-NC) and the siRNA against CDH3 (si-CDH3) were obtained from Shanghai Tuoran Biological Technology Co., Ltd. For CDH3 overexpression, full-length CDH3 was synthesized and transformed into pcDNA3.1 (OE-CDH3), provided by Shanghai Tuoran Biological Technology Co., Ltd., and the corresponding empty vector was used as a NC (OE-NC). Using Lipofectamine® 3000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), the transfection of AGS and HGC-27 cells was performed using 50 nM siRNA or 2 µg/ml pcDNA3.1 in 24-well plates at 37°C for 48 h. Subsequent experiments were performed 48 h after transfection. The next procedure involved the selection of stable cell lines with puromycin (4 µg/ml). Additionally, the miR-665 inhibitor, miR-665 mimic and the corresponding non-targeting NCs (mimic-NC and inhibitor-NC) were obtained from Genewiz, Inc. The sequences of si-CDH3, miR-665 inhibitor, miR-665 mimic and their corresponding NCs are listed in Table III. HGC-27 and AGS cells were seeded into 24-well plates (2×105 cells/well). After the density of the cells was 50%, they were transfected with the miR-665 mimic, miR-665 inhibitor or corresponding negative controls at a concentration of 50 nM using Lipofectamine 3000 Transfection Reagent for 48 h at 37°C. Subsequent experiments were performed 48 h after transfection.
Table III.
Oligonucleotide sequences used for cell transfection.
| Oligonucleotide | Primer sequences (5′-3′) |
|---|---|
| si-CDH3 | Forward: UUCAACAGCAACCAGCCUGUUUCCU |
| Reverse: AGGAAACAGGCUGGUUGCUGUUGAA | |
| si-NC | Forward: CGGAAGGCCUAAUGCCGAAdTdT |
| Reverse: UUCGGCAUUAGGCCUUCCGdTdG | |
| miR-665 inhibitor | AGGGGCCUCAGCCUCCUGGU |
| inhibitor-NC | CAGUACUUUUGUGUAGUACAA |
| miR-665 mimic | Forward: CAGCAGCACACUGUGGUUUGU |
| Reverse: AGGGACCUCAGCCUCCUGGUUU | |
| mimic-NC | Forward: UUCUCCGAACGUGUCACGUTT |
| Reverse: ACGUGACACGUUCGGAGAATT |
si, small interfering RNA; CDH3, cadherin 3; NC, negative control; miR, microRNA.
MTT assay
MTT colorimetric assay was used to detect cell viability. HGC-27 and AGS cells were cultured into 96-well plates at a density of 5×103 cells/well. MTT (5 mg/ml) was then added to each well at four time points (0, 24, 48 and 72 h). After 4 h incubation at 37°C, acid isopropanol was added to each well. The cell samples were then mixed until the dark blue crystals dissolved completely. A microplate reader was used to read the absorbance at 570 nm. The triplicate repeated readings for each sample were averaged, and the medium background was subtracted from the analytical readings to correct the absorbance.
BrdU ELISA assay
The BrdU Cell Proliferation Assay kit (cat. no. 6813; Cell Signaling Technology, Inc.) was used to detect cell proliferation according to the manufacturer's protocol. First, 2×104 HGC-27 and AGS cells were cultured into each well of a 96-well plate. The next day, the cells were serum-starved overnight. Subsequently, the serum was added to the cells for 8 h. After that, cells were incubated for 8–12 h without removing the treatment media. The absorbance value was subsequently recorded at 450 nm using a microplate reader. BrdU was incorporated into the DNA of dividing cells, and the intensity of the luminescence recorded using a microplate reader was proportional to the amount of incorporated BrdU in the cells to evaluate cell proliferation.
Apoptosis assay
The FITC Annexin V Apoptosis Detection kit (BD Biosciences) was used for apoptosis analysis according to the manufacturer's protocol. After the cell lines were collected in 100 µl ice-cold PBS (6×104 cells), they were suspended with the binding buffer (100 µl) and treated with Annexin V-FITC (5 µl) and propidium iodide (5 µl) in the dark at 20–22°C for 15 min. The next procedure was the addition of 400 µl of the same binding buffer to the aforementioned binding buffer (100 µl). The cells were detected and characterized by flow cytometry (FACSCalibur; Becton, Dickinson and Company), and cells were analyzed using FlowJo v10.6.2 (FlowJo LLC). Early apoptosis (Annexin V-FITC+/PI−; Q4) and late apoptosis (Annexin V-FITC+/PI+; Q2) were considered as the apoptosis rate.
Caspase activity assay
This assay was performed using the caspase-3 activity assay kit (cat. no. 5723; Cell Signaling Technology, Inc.) according to the manufacturer's protocol. The first step was seeding AGS and HGC-27 cells into the 96-well plates (2×104 cells/well) and culturing the cells at the suggested operating conditions (37°C and 80% density). The caspase-3 assay loading solution was then prepared by adding N-Acetyl-Asp-Glu-Val-Asp-p-Nitroanilide (DEVD-pNA) substrates and DL-Dithiothreitol (DTT) reagents. This preparation was performed according to the manufacturer's instructions. After the cells were collected, they were suspended with ice-cold cell lysis buffer included in the aforementioned kit for 10 min and then incubated on ice. Then, the caspase-3 assay loading solution (100 µl/well) was added to the cells, and the mixture was incubated at 37°C for 2 h. The absorbance at 405 nm was measured using a microplate reader.
Cell adhesion assay
HGC-27 and AGS cells were transferred to a DMEM containing 10% FBS and seeded into 96-well plates (2×104 cells/well). A total of ~30 µl/well of Collagen I solution (40 µg/ml) (Chrono-log Corporation) was transferred to the 96-well plate and incubated at 4°C for 12 h. The next step was the removal of the solution, followed by air-drying the plate at 20–22°C. The cells were then washed with serum-free DMEM and cultured for 8 h at 37°C in serum-free DMEM. To dissociate them, 10 mM EDTA was added to the cells for 10 min at 25°C. The cells were then washed three times with PBS to remove EDTA and were suspended in DMEM with 0.1% BSA (2×105 cells/ml; Gibco; Thermo Fisher Scientific, Inc.). Subsequently, 100 µl cell suspension was added to the 96-well plate with air-dried Collagen I, and the cells were incubated at 37°C for 60 min. After the cells adhered to the plate surface, DMEM (100 µl) was added to eliminate other non-adherent cells. Next, the cells were incubated at 37°C for 1 h with DMEM containing 10% FBS. Subsequently, the MTT substrate (10 µl) was added to each well, and the cells were incubated for 2 h at 30°C. DMSO (100 µl) was then added to lyse the cells. Finally, the absorbance was measured at 570 nm using a microplate reader. The absorbance was positively associated with the number of adherent cells, so the optical density value was used to evaluate the adhesion ability of cells.
Luciferase assay
The miRNA upstream of CDH3 was predicted using bioinformatics softwares, including GSE93415 (26) from Gene Expression Omnibus DataSets (https://www.ncbi.nlm.nih.gov/gds/?term=), TargetScan (http://www.targetscan.org/), starBase (http://starbase.sysu.edu.cn/panCancer.php) and Venny 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html). The pmiRGLO plasmid vector (Promega Corporation) used in the present study contained both the firefly luciferase reporter gene and the internal control gene, Renilla luciferase. The pmiRGLO CDH3 3′-untranslated regions (3′-UTRs) wild-type (WT) vectors and pmiRGLO CDH3 3′-UTR mutated (MUT) vectors with the miR-665 binding sites replaced by a random nucleotide sequence were constructed. A 24-well plate was then used to seed AGS and HGC-27 cells (2×105 cells/well). The next step was the co-transfection of the cells with pmiRGLO CDH3-MUT or pmiRGLO CDH3-WT and with either miR-665 mimic or mimic-NC using Lipofectamine 3000 Transfection Reagent. After collecting the cells transfected for 48 h, Renilla luciferase and firefly luciferase activities were detected using the Dual Luciferase Reporter Assay System (Thermo Fisher Scientific, Inc.). Firefly luciferase activity was normalized to that of Renilla luciferase activity.
Statistical analysis
GraphPad Prism 7.0 (GraphPad Software, Inc.) was used for data analysis. To compare two variables in tissues, paired Student's t-test was utilized. To compare two groups in cell experiments, unpaired Student's t-test was used. To compare multiple groups, one-way ANOVA with Dunnett's or Tukey's post-hoc test was employed. Pearson correlation analysis was applied to assess the correlation between CDH3 and miR-665 expression. Fisher's exact test was used to determine the association between CDH3 expression and clinicopathological variables. Each experiment was performed thrice, and results were presented as the mean ± standard error. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of key genes involved in GC
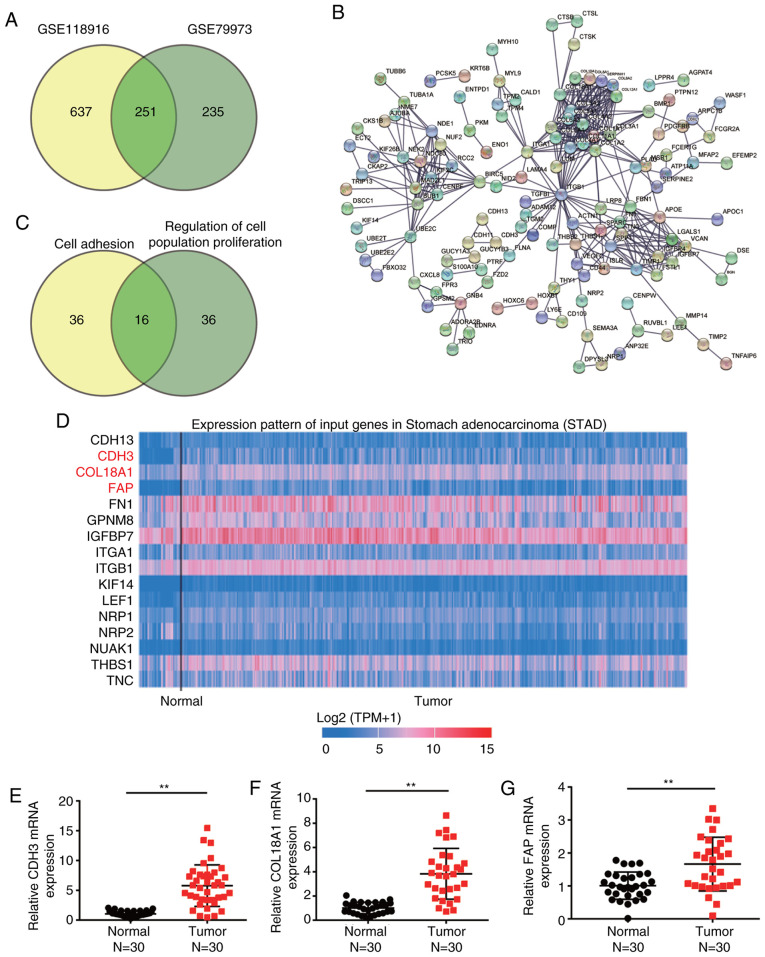
Two mRNA expression profiles (GSE118916 and GSE79973) were downloaded from the GEO database. The profiles were then applied to filter the DEGs involved in GC. A total of 251 upregulated DEGs were overlapped between GSE79973 and GSE118916, with logFC values >1 and adj. P<0.05 (Fig. 1A). The 251 overlapped DEGs were uploaded to STRING for GO enrichment analysis (Fig. 1B). After performing GO enrichment analysis, ‘cell adhesion’ and ‘regulation of cell population proliferation’ were significantly enriched following GO analysis, and 16 genes were involved in these two biological processes (Fig. 1C). By using TCGA analysis, CDH3, COL18A1 and FAP were found to be significantly upregulated in GC samples compared with in non-tumor samples (Fig. 1D). To identify and investigate these key genes, the expression levels of CDH3, COL18A1 and FAP were further detected in clinical tissues, revealing that they were all upregulated in tumor tissues compared with in normal tissues (Fig. 1E-G). Since CDH3 exhibited the highest upregulation, it was selected to be further explored in subsequent experiments.
Figure 1.
CDH3 is a key gene in GC. (A) A total of 251 upregulated differentially expressed genes overlapped between GSE118916 and GSE79973 mRNA expression profiles. (B) Gene Ontology enrichment analysis of the 251 upregulated genes was performed using STRING. (C) A total of 16 genes were involved in ‘cell adhesion’ and ‘regulation of cell population proliferation’ according to STRING analysis. (D) High CDH3, COL18A1 and FAP expression was observed in tumor tissues based on TCGA database. The blue and red colors in TCGA database represent low and high expression, respectively. Reverse transcription-quantitative PCR detection of the expression levels of (E) CDH3, (F) COL18A1 and (G) FAP in collected GC and adjacent normal tissues (n=30). Data are presented as the mean ± SD, and at least three independent tests were performed per experiment. **P<0.001. GC, gastric cancer; CDH3, cadherin 3; COL18A1, collagen type XVIII α1 chain; FAP, fibroblast activation protein; TCGA, The Cancer Genome Atlas.
CDH3 expression is increased in GC cells
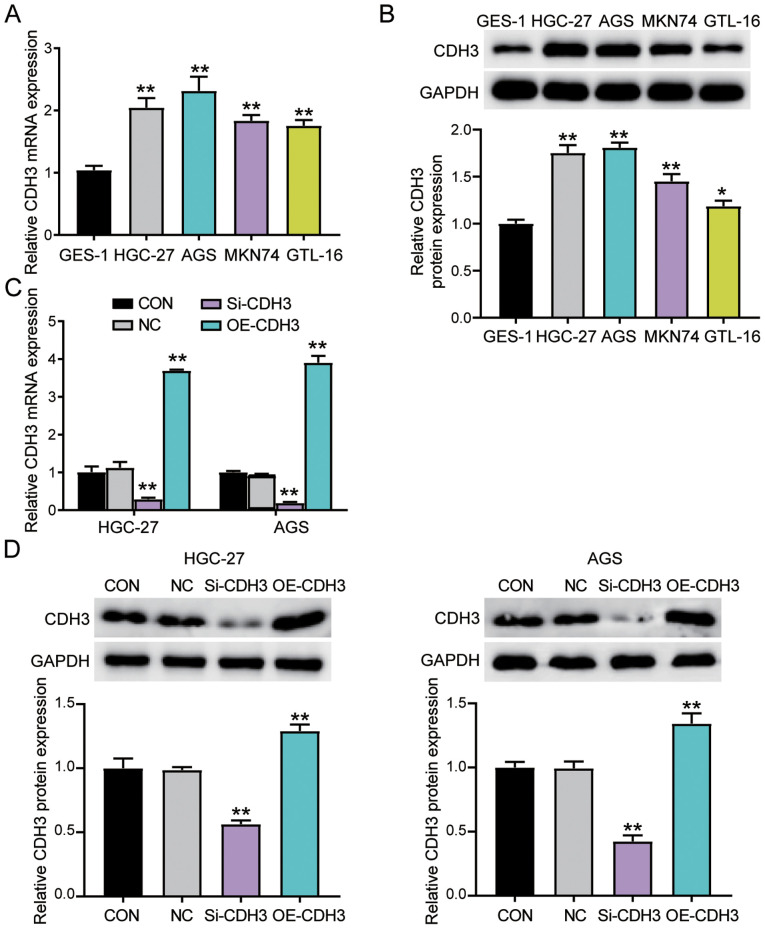
The association between CDH3 expression and clinical features in patients with GC was then analyzed. CDH3 expression was associated with tumor size, histological grade, TNM stage and advanced lymph nodes status (Table SI). However, CDH3 expression was not associated with other clinicopathological features, such as age, sex, differentiation, smoking, drinking alcohol and H. pylori infection. Furthermore, the expression levels of CDH3 were confirmed in the gastric epithelial cells (GES-1) and the gastric adenocarcinoma cells (AGS, MKN74, GTL-16 and HGC-27). CDH3 expression was significantly increased in all GC cells compared with in GES-1 cells (Fig. 2A). Western blotting was employed to analyze CDH3 protein expression. Consistent with gene expression, CDH3 protein expression in GC cells was significantly increased compared with in GES-1 cells (Fig. 2B). Since HGC-27 and AGS cell lines exhibited the highest CDH3 expression, they were selected to perform subsequent experiments. The transfection of si-CDH3 and OE-CDH3 in AGS and HGC-27 cells was performed to assess the function of CDH3 expression in GC cells. The transfection efficiency was confirmed using RT-qPCR and western blotting. The results revealed that overexpression of CDH3 led to a significant upregulation of CDH3 mRNA and protein expression, while si-CDH3 led to a significant downregulation of CDH3 mRNA and protein expression (Figs. S1A, 2C and D).
Figure 2.
Upregulation of CDH3 expression in GC cells. (A) RT-qPCR detection of CDH3 mRNA expression in gastric epithelial cells (GES-1) and gastric adenocarcinoma cell lines (AGS, MKN74, GTL-16 and HGC-27). **P<0.001 vs. GES-1 cells. (B) Measurement of CDH3 protein expression in GES-1, AGS, MKN74, GTL-16 and HGC-27 cell lines via western blotting. *P<0.05 and **P<0.001 vs. GES-1 cells. (C) CDH3 mRNA expression was identified using RT-qPCR in HGC-27 and AGS cells transfected with Si-CDH3 or OE-CDH3. **P<0.001 vs. NC (D) CDH3 protein expression was confirmed via western blotting in HGC-27 and AGS cells transfected with Si-CDH3 or OE-CDH3. Data are presented as the mean ± SD (n=3), and at least three independent tests were performed for every experiment. **P<0.001 vs. NC. CDH3, cadherin 3; Si-CDH3, small interfering RNA-CDH3; OE-CDH3, overexpression-CDH3; CON, blank control; NC, negative control of Si-CDH3 co-transfected with negative control of OE-CDH3; GC, gastric cancer; RT-qPCR, reverse transcription-quantitative PCR.
CDH3 acts as an oncogene in GC cells
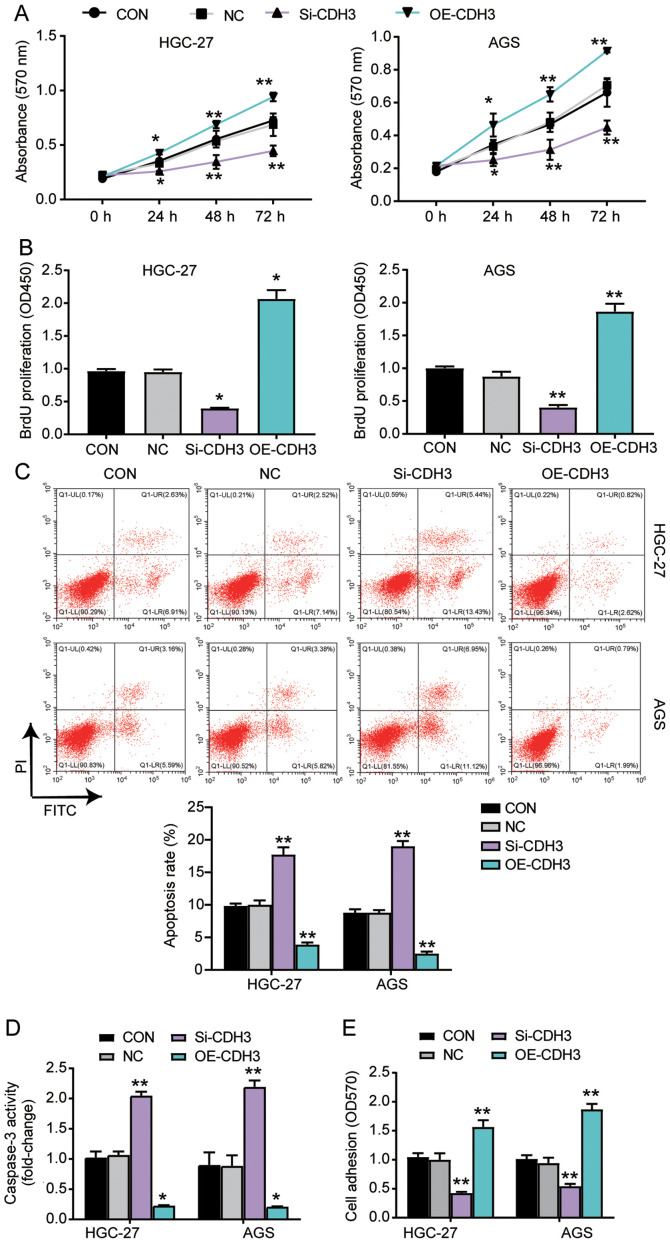
Several experiments were performed to explore the function of CDH3 in GC cells. The MTT assay results indicated that the cells transfected with OE-CDH3 presented higher cell viability than control cells, while the cells transfected with si-CDH3 exhibited lower cell viability than control cells (Fig. 3A). The results of the BrdU ELISA assay revealed that the proliferation of cells transfected with OE-CDH3 increased by 2-fold, while that of the cells transfected with si-CDH3 declined by 50% compared with control cells (Fig. 3B). The apoptosis level was ascertained using the Caspase-3 Activity Assay kit and Annexin V-FITC Apoptosis Detection kit (Fig. 3C and D). The results demonstrated that the apoptosis level and caspase 3 activity in cells transfected with si-CDH3 significantly increased compared with that in control cells, while there was a decrease in the apoptosis and caspase 3 activity of cells transfected with OE-CDH3 (Fig. 3C and D). Additionally, the adhesion ability of the OE-CDH3 group exhibited a >1.5-fold increase, while that of cells transfected with si-CDH3 decreased by 50% compared with control cells (Fig. 3E). Overall, the present results suggested that the overexpression of CDH3 enhanced the adhesion and proliferation of GC cells, but inhibited the apoptosis of GC cells.
Figure 3.
Overexpression of CDH3 enhances the viability, proliferation and adhesion, and represses the apoptosis of gastric cancer cells. (A) Cell viability was detected in AGS and HGC-27 cells transfected with OE-CDH3 or Si-CDH3 using the MTT assay. (B) Cell proliferation was ascertained in HGC-27 and AGS cells transfected with Si-CDH3 or OE-CDH3 by BrdU ELISA assay. (C) Apoptosis was determined in AGS and HGC-27 cells transfected with Si-CDH3 or OE-CDH3 using the FITC apoptosis detection kit. (D) Apoptosis was established in HGC-27 and AGS cells transfected with Si-CDH3 or OE-CDH3 by caspase activity assay kit. (E) Cell adhesion was detected in HGC-27 and AGS cells transfected with Si-CDH3 or OE-CDH3 using a cell adhesion assay kit. Data are presented as the mean ± SD (n=3), and at least three independent tests were performed for each experiment. *P<0.05 and **P<0.001 vs. NC. CDH3, cadherin 3; Si-CDH3, small interfering RNA-CDH3; OE-CDH3, overexpression-CDH3; CON, blank control; NC, negative control of Si-CDH3 co-transfected with negative control of OE-CDH3; OD, optical density.
miR-665 targets the CDH3 3′-UTR in GC
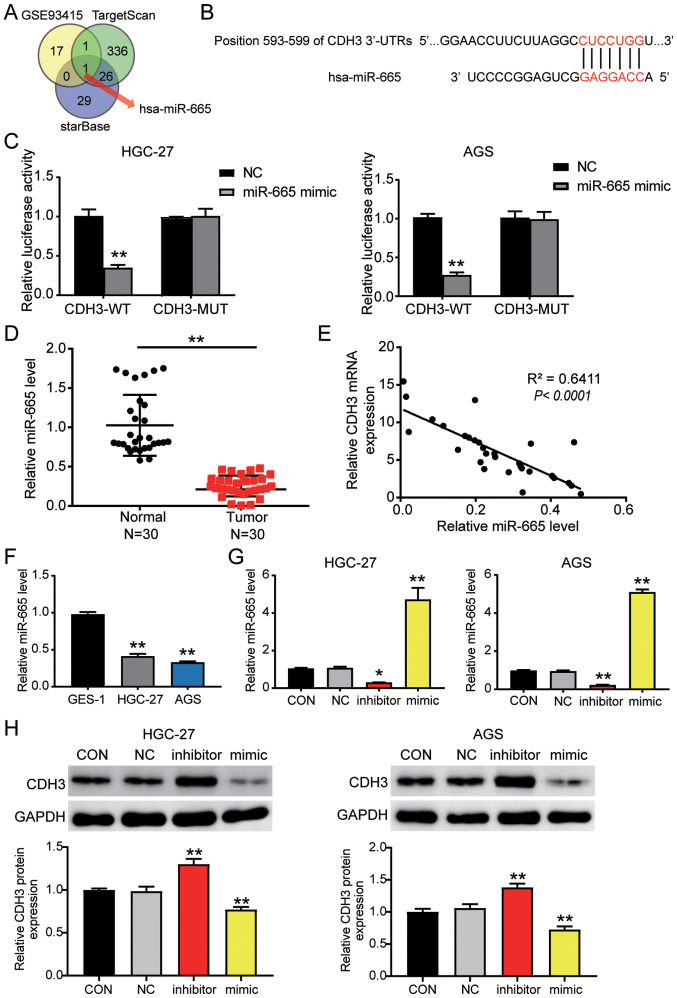
The GSE93415 profile was obtained from the GEO database and was used to screen the downregulated differentially expressed miRNAs with logFC <-1 and adj. P<0.05. TargetScan and starBase were used to predict the miRNAs binding to the CDH3 3′-UTR. Venny 2.1.0 analysis indicated an overlap of hsa-miR-665 (Fig. 4A). The TargetScan results revealed that miR-665 matched the position 593–599 of the CDH3 3′-UTR (Fig. 4B). Next, the luciferase activity detected in AGS and HGC-27 cells confirmed that the miR-665 mimic in the CDH3-WT group downregulated the luciferase activity by 60% compared with cells transfected with the mimic-NC; in contrast to the CDH3-WT cells, miR-665 mimic had no significant effect on the cells in the CDH3-MUT group (Fig. 4C). Additionally, miR-665 expression was decreased by 60% in tumor tissues compared with in adjacent non-tumor tissues (Fig. 4D). Furthermore, an inverse correlation was detected between CDH3 and miR-665 expression in GC tissues (Fig. 4E). miR-665 expression was also analyzed in GC cells, revealing that miR-665 expression in AGS and HGC-27 GC cells was decreased by 50% compared with that in GES-1 cells (Fig. 4F). After transfection, miR-665 expression in the cells transfected with the miR-665 mimic increased by 4-fold, while it decreased by 80% in the cells transfected with the miR-665 inhibitor compared with in the cells in the control group (Figs. S1B and 4G). Western blot analysis indicated that CDH3 protein expression in the miR-665 inhibitor-transfected cells was significantly increased, while it was significantly decreased in the cells transfected with the miR-665 mimic (Fig. 4H). Overall, these results indicated that by targeting CDH3 mRNA, miR-665 regulated GC cells.
Figure 4.
CDH3 is a candidate target gene of miR-665. (A) miR-665 was identified by Venny 2.1.0 analysis. TargetScan and starBase were employed to ascertain the miRNAs binding to CDH3. GSE93415 was used to identify the miRNAs with low expression in GC. (B) TargetScan revealed the predicted binding sequences of CDH3 3′-UTR. (C) Dual luciferase assay kit was utilized in CDH3-WT and CDH3-MUT cells transfected with NC or miR-665 mimic. **P<0.001 vs. NC. (D) RT-qPCR was used to identify miR-665 expression in GC and adjacent normal tissues (n=30). **P<0.001. (E) Pearson's correlation analysis revealed the negative correlation between CDH3 and miR-665 expression in GC samples. RT-qPCR was used to detect the expression levels of miR-665 in (F) GES-1, HGC-27 and AGS cells, and (G) AGS and HGC-27 cells transfected with miR-665 inhibitor or miR-665 mimic. (H) Measurement of CDH3 protein expression in HGC-27 and AGS cells transfected with miR-665 inhibitor or miR-665 mimic. Data are presented as the mean ± SD (n=3), and at least three independent tests were performed for every experiment. *P<0.05 and **P<0.001 vs. NC. CDH3, cadherin 3; miR/miRNA, microRNA; WT, wild-type; MUT, mutant; CON, blank control; NC, negative control of miR-665 inhibitor co-transfected with negative control of miR-665 mimic; UTR, untranslated region; RT-qPCR, reverse transcription-quantitative PCR; GC, gastric cancer.
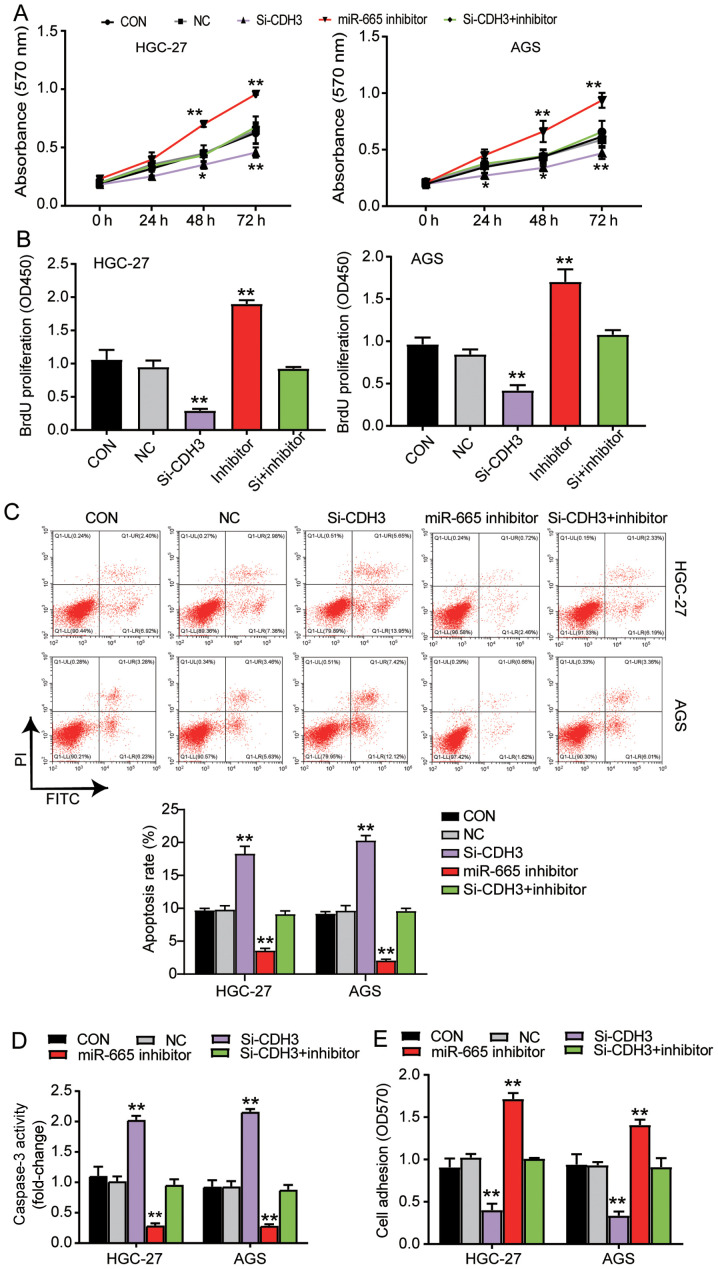
miR-665 targeting CDH3 inhibits GC progression
MTT assay were performed to study the function of miR-665 in GC cells. The results indicated that the cells transfected with the miR-665 inhibitor exhibited enhanced cell viability; additionally, the miR-665 inhibitor could reverse the suppressive effect of si-CDH3 on cell viability (Fig. 5A). Additionally, the results of the BrdU ELISA assay revealed a 2-fold increase in the proliferation of the cells transfected with the miR-665 inhibitor, and the miR-665 inhibitor reversed the suppressive effect of si-CDH3 on cell proliferation (Fig. 5B). The apoptosis level was identified using the two aforementioned kits, and the results indicated that apoptosis was suppressed by the miR-665 inhibitor, while the apoptosis in the group co-transfected with si-CDH3 and miR-665 inhibitor was similar to that in the control group (Fig. 5C and D). The cells transfected with the miR-665 inhibitor also exhibited a 1.5-fold increase in cell adhesion, and the negative effect of si-CDH3 on cell adhesion was relieved by the miR-665 inhibitor (Fig. 5E). Overall, the current results suggested that by targeting CDH3, miR-665 could inhibit cell proliferation and adhesion, but promote the apoptosis of GC cells.
Figure 5.
miR-665 targeting CDH3 inhibits proliferation and adhesion, and promotes apoptosis of gastric cancer cells. (A) Cell viability was ascertained in AGS and HGC-27 cells transfected with Si-CDH3, miR-665 inhibitor or Si-CDH3+miR-665 inhibitor by MTT assay. (B) Cell proliferation was analyzed in AGS and HGC-27 cells transfected with Si-CDH3, miR-665 inhibitor or Si-CDH3+miR-665 inhibitor by BrdU ELISA assay. Apoptosis was confirmed in AGS and HGC-27 cells transfected with Si-CDH3, miR-665 inhibitor or Si-CDH3+miR-665 inhibitor by (C) FITC apoptosis detection kit and (D) caspase-3 activity assay kit. (E) Cell adhesion was detected in HGC-27 and AGS cells transfected with miR-665 inhibitor, Si-CDH3 or Si-CDH3+miR-665 inhibitor using a cell adhesion assay kit. Data are presented as the mean ± SD, and at least three independent tests were performed for every experiment. *P<0.05 and **P<0.001 vs. NC. CDH3, cadherin 3; miR, microRNA; Si-CDH3, small interfering RNA-CDH3; CON, blank control; NC, negative control of miR-665 inhibitor co-transfected with negative control of Si-CDH3; OD, optical density.
Discussion
The present study confirmed the upregulation of CDH3 expression and the downregulation of miR-665 expression in GC tissues and cell lines. The overexpression of CDH3 increased cell proliferation and adhesion levels, and decreased the apoptosis level. On the other hand, the miR-665 mimic restrained the proliferation and adhesion of GC cells, and promoted their apoptosis. Moreover, CDH3 seemed to be a direct target of miR-665 in GC cells, meaning that miR-665 may inhibit CDH3 expression to suppress the progression of GC.
In the last three decades, several studies have reported the ability of CDH3 to accelerate the growth of multiple types of cancer, including lung (27), esophageal (9), colorectal (7) and breast cancer (28). A previous study has also suggested the involvement of CDH3 in promoting the motility of pancreatic cancer cells by activating Rho-family GTPases (8). Another study revealed that CDH3 upregulation associated with CDH3 promoter hypomethylation significantly influenced the proliferation of breast cancer cells (29). Moreover, in some studies, CDH3 expression was upregulated in GC cells (11,12). The detection of the CDH3 demethylation level in 36 primary gastric carcinoma samples revealed that 25/36 (69%) samples had an abnormal demethylation level, which may result in the promotion of gastric carcinoma (11). Another study reported the upregulation of CDH3 expression in 20/28 (71%) GC tissues based on a cDNA microarray, and immunohistochemical analyses further confirmed an increase in CDH3 protein expression in GC tissues (12). Consistent with the aforementioned studies, the present study demonstrated that upregulated CDH3 expression in GC tissues and cells augmented cell proliferation and adhesion, and decreased apoptosis.
Furthermore, miR-665 has been associated with numerous human malignancies, including pancreatic (17), breast (18), cervical (30) and ovarian cancer (31). Previous studies indicated that this miRNA could promote the occurrence of cancer, as well as suppress cancer. For instance, by targeting Homeobox Protein Hox-A10, miR-665 inhibited the progression of ovarian cancer (19). Additionally, miR-665 sponged by RHPN1 Antisense RNA 1 suppressed tumor development by preventing the activation of AKT3 (32). Nonetheless, miR-665 could facilitate the tumorigenesis of ovarian cancer cells by targeting SRC Kinase Signaling Inhibitor 1 (31). In addition, miR-665 overexpression enhanced the proliferation, migration and invasion of non-small cell lung cancer cells (16). Regarding GC, miR-665 has been documented in several studies to suppress the progression of GC (20,33). After binding to Protein Phosphatase 2 Regulatory Subunit Bα, this miRNA inhibited the proliferation, invasion and epithelial-mesenchymal transition (EMT) of GC cells (20). In addition, miR-665 attached to AKT3 suppressed GC cells by decreasing proliferation and increasing apoptosis. Additionally, miR-665 was observed to be a target of LINC00565, which accelerated the development of GC by upregulating AKT3 expression (33). However, by inhibiting its target gene suppressor of cytokine signaling 3 (SOCS3), miR-665 promoted the EMT of gastric adenocarcinoma cells (21). In the present study, miR-665 binding to CDH3 inhibited cell proliferation and adhesion, but enhanced apoptosis in GC cells.
A single miRNA may have a number of targets (34). More specifically, miR-665 is involved in numerous pathways involved in GC, since it can target AKT3 (33), Yes-associated protein 1 (YAP1) (35), cysteine-rich transmembrane bone morphogenetic protein regulator 1 (CRIM1) (36) and SOCS3 (21). However, in the current study, the effect of miR-665 on AKT3, YAP1, CRIM1 and SOCS3 was not explored. While the current study has investigated the proliferation, adhesion and apoptosis of GC cells, it is important to note that cancer occurrence is a complicated pathway interaction. The present study only confirmed the association between miR-665 and CDH3 in GC cells by analyzing the enrichment of target genes, so the lack of miRNA enrichment analysis and of survival analysis on the target genes and associated miRNAs are the other major limitations of the study. In the future, the potential signaling pathways and survival analysis of the target genes and associated miRNAs in GC should be further explored. Additionally, the current study only observed tumor characteristics at the cellular level. However, further studies are required to verify the current findings in animal models.
In conclusion, to the best of our knowledge, the present study was the first to demonstrate an interaction between miR-665 and CDH3, and to reveal that miR-665 may suppress the progression of GC by targeting CDH3. The present findings may offer further insight into the prognosis and treatment of GC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by Basic and Frontier projects of Science and Technology Department of Henan Province (grant no. 142300410050), Joint project of Henan Provincial Health and Family Planning Commission (grant no. 20170123), Henan Health and Family Planning Research and Innovation Talent Project (grant no. 51282) and Joint project of the National Natural Science Foundation of China (grant no. U1604174).
Funding
The present study was supported by Basic and Frontier projects of Science and Technology Department of Henan Province (grant no. 142300410050), Joint project of Henan Provincial Health and Family Planning Commission (grant no. 20170123), Henan Health and Family Planning Research and Innovation Talent Project (grant no. 51282) and Joint project of the National Natural Science Foundation of China (grant no. U1604174).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/gds/?term=).
Authors' contributions
XF and SD performed the experiments and data analysis. YB and LZ conceived and designed the study. LZ and SD assessed the raw data to ensure its legitimacy. XF reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Henan Provincial People's Hospital (Zhengzhou, China; approval no. 2019-04). All patients signed written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014;345:196–202. doi: 10.1016/j.canlet.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Dong J, Thrift AP. Alcohol, smoking and risk of oesophago-gastric cancer. Best Pract Res Clin Gastroenterol. 2017;31:509–517. doi: 10.1016/j.bpg.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Marin JJ, Al-Abdulla R, Lozano E, Briz O, Bujanda L, Banales JM, Macias RI. Mechanisms of resistance to chemotherapy in gastric cancer. Anticancer Agents Med Chem. 2016;16:318–334. doi: 10.2174/1871520615666150803125121. [DOI] [PubMed] [Google Scholar]
- 5.Biagioni A, Skalamera I, Peri S, Schiavone N, Cianchi F, Giommoni E, Magnelli L, Papucci L. Update on gastric cancer treatments and gene therapies. Cancer Metastasis Rev. 2019;38:537–548. doi: 10.1007/s10555-019-09803-7. [DOI] [PubMed] [Google Scholar]
- 6.Sprecher E, Bergman R, Richard G, Lurie R, Shalev S, Petronius D, Shalata A, Anbinder Y, Leibu R, Perlman I, et al. Hypotrichosis with juvenile macular dystrophy is caused by a mutation in CDH3, encoding P-cadherin. Nat Genet. 2001;29:134–136. doi: 10.1038/ng716. [DOI] [PubMed] [Google Scholar]
- 7.Kumara HMCS, Bellini GA, Caballero OL, Herath SAC, Su T, Ahmed A, Njoh L, Cekic V, Whelan RL. P-Cadherin (CDH3) is overexpressed in colorectal tumors and has potential as a serum marker for colorectal cancer monitoring. Oncoscience. 2017;4:139–147. doi: 10.18632/oncoscience.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniuchi K, Nakagawa H, Hosokawa M, Nakamura T, Eguchi H, Ohigashi H, Ishikawa O, Katagiri T, Nakamura Y. Overexpressed P-cadherin/CDH3 promotes motility of pancreatic cancer cells by interacting with p120ctn and activating rho-family GTPases. Cancer Res. 2005;65:3092–3099. doi: 10.1158/0008.5472.CAN-04-3646. [DOI] [PubMed] [Google Scholar]
- 9.Liu D, Wu K, Yang Y, Zhu D, Zhang C, Zhao S. Long noncoding RNA ADAMTS9-AS2 suppresses the progression of esophageal cancer by mediating CDH3 promoter methylation. Mol Carcinog. 2020;59:32–44. doi: 10.1002/mc.23126. [DOI] [PubMed] [Google Scholar]
- 10.Albergaria A, Ribeiro AS, Pinho S, Milanezi F, Carneiro V, Sousa B, Sousa S, Oliveira C, Machado JC, Seruca R, et al. ICI 182,780 induces P-cadherin overexpression in breast cancer cells through chromatin remodelling at the promoter level: A role for C/EBPbeta in CDH3 gene activation. Hum Mol Genet. 2010;19:2554–2566. doi: 10.1093/hmg/ddq134. [DOI] [PubMed] [Google Scholar]
- 11.Hibi K, Kitamura YH, Mizukami H, Goto T, Sakuraba K, Sakata M, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y. Frequent CDH3 demethylation in advanced gastric carcinoma. Anticancer Res. 2009;29:3945–3947. [PubMed] [Google Scholar]
- 12.Imai K, Hirata S, Irie A, Senju S, Ikuta Y, Yokomine K, Harao M, Inoue M, Tsunoda T, Nakatsuru S, et al. Identification of a novel tumor-associated antigen, cadherin 3/P-cadherin, as a possible target for immunotherapy of pancreatic, gastric, and colorectal cancers. Clin Cancer Res. 2008;14:6487–6495. doi: 10.1158/1078-0432.CCR-08-1086. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 15.Boehm M, Slack FJ. MicroRNA control of lifespan and metabolism. Cell Cycle. 2006;5:837–840. doi: 10.4161/cc.5.8.2688. [DOI] [PubMed] [Google Scholar]
- 16.Xia J, Li D, Zhu X, Xia W, Qi Z, Li G, Xu Q. Upregulated miR-665 expression independently predicts poor prognosis of lung cancer and facilitates tumor cell proliferation, migration and invasion. Oncol Lett. 2020;19:3578–3586. doi: 10.3892/ol.2020.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou B, Guo W, Sun C, Zhang B, Zheng F. Linc00462 promotes pancreatic cancer invasiveness through the miR-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell Death Dis. 2018;9:706. doi: 10.1038/s41419-018-0724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao XG, Hu JY, Tang J, Yi W, Zhang MY, Deng R, Mai SJ, Weng NQ, Wang RQ, Liu J, et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis. 2019;10:479. doi: 10.1038/s41419-019-1705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Jiang Y, Wan Y, Zhou S, Thapa S, Cheng W. MicroRNA-665 suppresses the growth and migration of ovarian cancer cells by targeting HOXA10. Mol Med Rep. 2018;18:2661–2668. doi: 10.3892/mmr.2018.9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M, Wang S, Yi A, Qiao Y. microRNA-665 is down-regulated in gastric cancer and inhibits proliferation, invasion, and EMT by targeting PPP2R2A. Cell Biochem Funct. 2020;38:409–418. doi: 10.1002/cbf.3485. [DOI] [PubMed] [Google Scholar]
- 21.Tang H, Long Q, Zhuang K, Yan Y, Han K, Guo H, Lu X. miR-665 promotes the progression of gastric adenocarcinoma via elevating FAK activation through targeting SOCS3 and is negatively regulated by lncRNA MEG3. J Cell Physiol. 2020;235:4709–4719. doi: 10.1002/jcp.29349. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Zhu Z, Zhao Y, Zhang Q, Wu X, Miao B, Cao J, Fei S. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci Rep. 2019;9:7827. doi: 10.1038/s41598-019-43924-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin Y, He J, Du J, Zhang RX, Yao HB, Shao QS. Overexpression of HS6ST2 is associated with poor prognosis in patients with gastric cancer. Oncol Lett. 2017;14:6191–6197. doi: 10.3892/ol.2017.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobin LH, Compton CC. TNM seventh edition: What's new, what's changed: Communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer. 2010;116:5336–5339. doi: 10.1002/cncr.25537. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Sierzega M, Kaczor M, Kolodziejczyk P, Kulig J, Sanak M, Richter P. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: The importance of miR-21 and miR-331. Br J Cancer. 2017;117:266–273. doi: 10.1038/bjc.2017.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsiao TF, Wang CL, Wu YC, Feng HP, Chiu YC, Lin HY, Liu KJ, Chang GC, Chien KY, Yu JS, Yu CJ. Integrative omics analysis reveals soluble cadherin-3 as a survival predictor and an early monitoring marker of EGFR tyrosine kinase inhibitor therapy in lung cancer. Clin Cancer Res. 2020;26:3220–3229. doi: 10.1158/1078-0432.CCR-19-3972. [DOI] [PubMed] [Google Scholar]
- 28.Albergaria A, Resende C, Nobre AR, Ribeiro AS, Sousa B, Machado JC, Seruca R, Paredes J, Schmitt F. CCAAT/enhancer binding protein β (C/EBPβ) isoforms as transcriptional regulators of the pro-invasive CDH3/P-cadherin gene in human breast cancer cells. PLoS One. 2013;8:e55749. doi: 10.1371/journal.pone.0055749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paredes J, Albergaria A, Oliveira JT, Jerónimo C, Milanezi F, Schmitt FC. P-cadherin overexpression is an indicator of clinical outcome in invasive breast carcinomas and is associated with CDH3 promoter hypomethylation. Clin Cancer Res. 2005;11:5869–5877. doi: 10.1158/1078-0432.CCR-05-0059. [DOI] [PubMed] [Google Scholar]
- 30.Cao L, Jin H, Zheng Y, Mao Y, Fu Z, Li X, Dong L. DANCR-mediated microRNA-665 regulates proliferation and metastasis of cervical cancer through the ERK/SMAD pathway. Cancer Sci. 2019;110:913–925. doi: 10.1111/cas.13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou P, Xiong T, Yao L, Yuan J. MicroRNA-665 promotes the proliferation of ovarian cancer cells by targeting SRCIN1. Exp Ther Med. 2020;19:1112–1120. doi: 10.3892/etm.2019.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, Yang T, Ji J, Zhao F, Li C, Han X. RHPN1-AS1 promotes cell proliferation and migration via miR-665/Akt3 in ovarian cancer. Cancer Gene Ther. 2020;28:33–41. doi: 10.1038/s41417-020-0180-0. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, Ni G, Mao L, Xue X, Zhang J, Wu W, Zhang S, Zhao H, Ding L, Wang L. LINC00565 promotes proliferation and inhibits apoptosis of gastric cancer by targeting miR-665/AKT3 axis. Onco Targets Ther. 2019;12:7865–7875. doi: 10.2147/OTT.S189471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2013;133:768–775. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]
- 35.Lin X, Huang C, Chen Z, Wang H, Zeng Y. CircRNA_100876 is upregulated in gastric cancer (GC) and promotes the GC cells' growth, migration and invasion via miR-665/YAP1 signaling. Front Genet. 2020;11:546275. doi: 10.3389/fgene.2020.546275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu KZ, Zhang CD, Zhang C, Pei JP, Dai DQ. miR-665 suppresses the epithelial-mesenchymal transition and progression of gastric cancer by targeting CRIM1. Cancer Manag Res. 2020;12:3489–3501. doi: 10.2147/CMAR.S241795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/gds/?term=).