Abstract
Acylglycerol kinase (AGK) is a mitochondrial lipid kinase that contributes to protein biogenesis as a subunit of the TIM22 complex at the inner mitochondrial membrane. Mutations in AGK cause Sengers syndrome, an autosomal recessive condition characterized by congenital cataracts, hypertrophic cardiomyopathy, skeletal myopathy, and lactic acidosis. We mapped the proteomic changes in Sengers patient fibroblasts and AGKKO cell lines to understand the effects of AGK dysfunction on mitochondria. This uncovered down-regulation of a number of proteins at the inner mitochondrial membrane, including many SLC25 carrier family proteins, which are predicted substrates of the complex. We also observed down-regulation of SFXN proteins, which contain five transmembrane domains, and show that they represent a novel class of TIM22 complex substrate. Perturbed biogenesis of SFXN proteins in cells lacking AGK reduces the proliferative capabilities of these cells in the absence of exogenous serine, suggesting that dysregulation of one-carbon metabolism is a molecular feature in the biology of Sengers syndrome.
INTRODUCTION
Mitochondria perform a diverse array of functions in mammalian cells, including production of ATP, induction of apoptosis, and calcium buffering (Anderson et al., 2019). Dysfunction of mitochondria is associated with many pathologies, including cancer, diabetes, and neurodegenerative disease. Mitochondrial diseases are genetic disorders that arise due to mutations in genes encoding proteins required for normal mitochondrial function (Jackson et al., 2018; Frazier et al., 2019). Mitochondria require 1200 nuclear-encoded proteins to function; these proteins are delivered to specific mitochondrial subcompartments (outer membrane; intermembrane space, inner membrane, and matrix) by translocation and sorting machineries (Wiedemann and Pfanner, 2017; Rath et al., 2020). Mutations in genes encoding various subunits of mitochondrial translocation machineries have been linked to a number of distinct mitochondrial diseases (Jackson et al., 2018).
The TIM22 complex is an inner membrane translocase that mediates the insertion of multipass transmembrane proteins into the mitochondrial inner membrane (Rehling et al., 2003). The TIM22 complex has been extensively studied in yeast; however, recent analyses in human cells have revealed substantial divergence of the complex in higher eukaryotes. The human TIM22 complex consists of 1) Tim22, the core pore-forming subunit; 2) the intermembrane space chaperones Tim9, Tim10, and Tim10b; and 3) Tim29 (Callegari et al., 2016; Kang et al., 2016) and acylglycerol kinase (AGK) (Kang et al., 2017; Vukotic et al., 2017), which are metazoan-specific subunits of the complex. The main substrates of the TIM22 complex are members of the SLC25 family of metabolite carrier proteins, which possess six transmembrane domains (Palmieri, 2013). The TIM22 complex also mediates membrane insertion of “TIM substrates,” Tim17, Tim23, and Tim22, which possess four transmembrane domains (Káldi et al., 1998; Kurz et al., 1999). Based on the properties of these proteins, it was thought that all substrates of the TIM22 complex would contain an even number of transmembrane domains. Recently, subunits of the mitochondrial pyruvate carrier have been identified as TIM22 complex substrates in both yeast and humans (Gomkale et al., 2020; Rampelt et al., 2020). One subunit of this complex (MPC2) has three transmembrane domains, suggesting that the current model of TIM22 complex import may need revising and that further substrates with odd numbers of transmembrane domains may exist.
In human cells, mutations in the AGK encoded subunit of the TIM22 complex are linked to Sengers syndrome, a severe mitochondrial disease characterized by congenital cataracts, hypertrophic cardiomyopathy, exercise intolerance, and lactic acidosis (Calvo et al., 2012; Mayr et al., 2012). As well as contributing to protein import at the TIM22 complex, AGK also functions as a lipid kinase, able to phosphorylate monoacylglycerol and diacylglycerol to produce phosphatidic acid and lysophosphatidic acid, respectively (Waggoner et al., 2004; Bektas et al., 2005). The lipid kinase activity of AGK is dispensable for its function at the TIM22 complex (Kang et al., 2017; Vukotic et al., 2017). Despite advances in the understanding of AGK function, how the protein’s dysfunction contributes to the molecular pathogenesis underlying Sengers syndrome is unclear.
Using quantitative proteomics, we set out to identify which mitochondrial functions and/or pathways are dysregulated in Sengers syndrome, with a view to expand our understanding of AGK function and Sengers syndrome pathology in an unbiased manner. We mapped the mitochondrial proteome of Sengers syndrome patient fibroblasts (Calvo et al., 2012; Kang et al., 2017), in addition to MCF7 and HEK293 cell lines lacking AGK (Kang et al., 2017), and identified extensive remodeling of the mitochondrial proteome. Consistent with the function of AGK at TIM22 and the known features of Sengers syndrome pathology, we observed a down-regulation of mitochondrial carrier proteins and Complex I subunits, in addition to down-regulation of enzymes involved in mitochondrial one-carbon (1C) metabolism (Ducker and Rabinowitz, 2017). By analyzing proteomic data sets from multiple systems of AGK or TIM22 complex dysfunction, we identified a list of candidate TIM22 complex substrates. We focused on sideroflexins (SFXNs), a family of proteins that mediate transport of serine into mitochondria and are required for efficient 1C metabolism (Kory et al., 2018). The proteomic data from Sengers patient fibroblasts and AGKKO HEK293 and MCF7 cell lines indicated down-regulation of SFXN proteins in the absence of functional AGK, and we show that these proteins rely on the TIM22 complex for their biogenesis. Consequently, loss of AGK in HEK293 cells leads to dependency on exogenous serine for normal proliferation. These data add further evidence to recent developments that suggest that mitochondrial dysfunction leads to changes in mitochondrial 1C metabolism (Bao et al., 2016; Nikkanen et al., 2016; Khan et al., 2017), and also provide a comprehensive list of candidate substrates of the human TIM22 complex.
RESULTS
Remodeling of the mitochondrial proteome is observed in Sengers syndrome patient fibroblasts and cell models of disease
We previously showed that two unrelated patients with Sengers syndrome had defects in the TIM22 complex and pathway (Kang et al., 2017). Patient 41 (referred to as Patient 1 in this study) possessed a compound heterozygous nonsense variant (p.Y390X) and splice variant that caused a shortened transcript with a premature stop codon (c.297+2T>C, pK75QfsX12) (Calvo et al., 2012). Patient 42 (referred to as Patient 2 in this study), possessed a homozygous splice variant that caused a shortened transcript with a premature stop codon (c.1131+1G>T, p.S350EfsX19) (Calvo et al., 2012). Label-free quantitative mass spectrometry was undertaken on mitochondria isolated from: 1) an AGKKO HEK293 cell line previously described (Kang et al., 2017); 2) the AGKKO HEK293 cell line reexpressing wild-type (WT) AGK or a kinase-dead AGK variant (AGKG126E) (Kang et al., 2017); 3) patient 1 and 2 fibroblasts and three control fibroblast lines (Calvo et al., 2012; Kang et al., 2017); and 4) an AGKKO MCF7 cell line (created for this study).
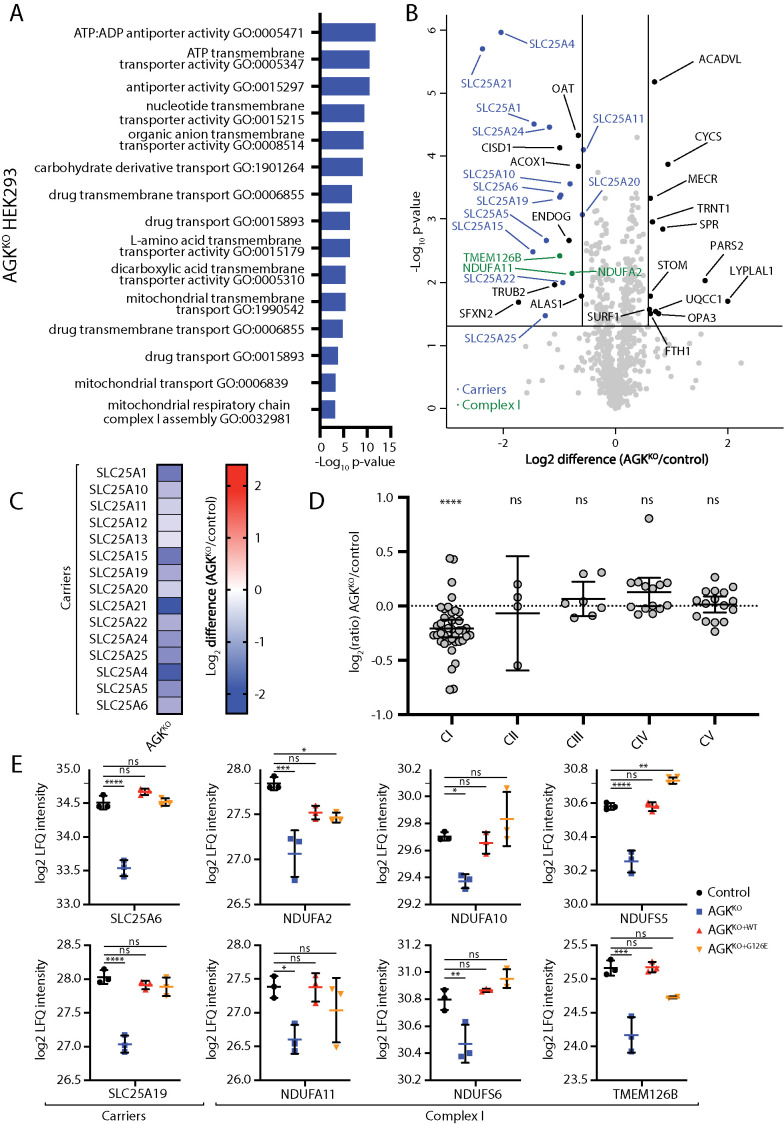
Across all cell lines, loss of AGK led to remodeling of the mitochondrial proteome. Gene ontology enrichment analysis performed on differentially expressed proteins in the AGKKO HEK293 cell line suggested that the two most significantly affected processes were mitochondrial transport and functioning of Complex I (NADH: ubiquinone oxidoreductase) of the mitochondrial electron transport chain (Figure 1, A and B; Supplemental Table S1). AGKKO HEK293 mitochondria had substantially reduced levels of SLC25 family members (13 members beyond the 1.5×-fold change cutoff) (Figure 1, B and C; Supplemental Table S1), confirming the role of AGK at the TIM22 complex and in the biogenesis of mitochondrial carrier proteins. The electron transport chain defect caused by AGKKO HEK293 cells seemed to be restricted to Complex I, as levels of proteins belonging to the other electron transport chain complexes were not significantly altered (Figure 1, B and D; Supplemental Figure 1, A and B; Supplemental Table S1). The observed defect in the assembly or stability of Complex I is consistent with observations of impaired mitochondrial respiration in tissues from Sengers syndrome patients (Haghighi et al., 2014).
FIGURE 1:
Proteomic characterization of an AGKKO HEK293 cell line. (A–D) Mitochondria were isolated from control and AGKKO HEK cells and subjected to label-free quantitative mass spectrometric analysis. (A) Gene Ontology (GO) enrichment analysis was performed for all proteins up-regulated or down-regulated >1.5-fold with p < 0.05. Significantly enriched GO terms are displayed, ranked by the p value associated with the term. Terms are associated with genes that have reduced abundance in AGKKO cells. (B) Volcano plot depicting the relative levels of proteins in AGKKO mitochondria compared with control HEK293. n = 3 biological replicates. The horizontal cutoff represents a p value of 0.05, while the vertical cutoffs represent 1.5-fold up- or down-regulation. SLC25 (carrier) family members (dark blue) and Complex I subunits (green) are indicated. (C) Log2 fold-change values (as compared with control) with p < 0.05 are depicted for SLC25 proteins in the AGKKO HEK293 cells. (D) Relative abundance of respiratory chain complexes (Complexes I–V) in AGKKO HEK293 cells as compared with control. Mean ± 95% CI is depicted. ****, p < 0.0001. (E) Log2-transformed LFQ values were determined for the indicated proteins from control, AGKKO, AGKKO+WT, and AGKKO+G126E HEK293 cells. Mean ± SD is depicted (n = 3). Statistical significance was determined using a one-way analysis of variance and Dunnett’s multiple comparisons test: *, p < 0.05, **, p < 0.01, ***, p < 0.001, ****, p < 0.0001.
Previously, using biochemical approaches, including in vitro import assays, we and others established that the kinase activity of AGK was not required for its function in the import of mitochondria carrier proteins via the TIM22 complex (Kang et al., 2017; Vukotic et al., 2017). We set out to investigate this further using the above-outlined proteomic approach to determine whether any classes of proteins reduced in the AGKKO HEK293 cell line require the kinase activity of AGK for their biogenesis or stability. Label-free quantitative proteomics performed on mitochondria isolated from control HEK293 cells and AGKKO, AGKKO+WT, and AGKKO+G126E cells demonstrated that the reduced levels of carrier proteins and Complex I subunits could be rescued through expression of the kinase-dead AGK mutant (Figure 1E; Supplemental Table S1). SDS–PAGE and BN-PAGE analysis of these cell lines showed signs of a mild complex I defect in AGKKO cells, detectable via reduced levels of NDUFA10, a Complex I subunit (Supplemental Figure 1, A and B). The distribution of NDUFAF1, a Complex I assembly factor, across NDUFAF1-containing complexes on BN-PAGE was altered in AGKKO cells (Supplemental Figure 1C, lanes 21–24), supporting impaired Complex I assembly. The reduced levels of NDUFA10 and the altered distribution of NDUFAF1 observed in the KO were both reversed upon expression of WT or kinase-dead AGK. Consistent with a mild defect in Complex I biogenesis or stability, oxygen consumption rate measurements (Supplemental Figure 1D) indicated a functional respiration defect in AGKKO cells. Although both basal and maximal OCR rates were reduced in the AGKKO, only the reduced basal rate could be rescued by kinase-dead AGK (Supplemental Figure 1D). Together, these results suggest that the observed Complex I defect in AGKKO derives primarily from TIM22 complex dysfunction, rather than loss of lipid kinase activity. A role for the lipid kinase activity of AGK in direct or indirect regulation of Complex I cannot be excluded, however, as kinase-dead AGK was able to only partially rescue the oxygen consumption rate defect observed in AGKKO cells (Supplemental Figure 1D). However, the partial complementation may correlate with lower expression levels of the kinase-dead versus wild-type AGK (Kang et al., 2017). Additionally, a limited ability for kinase-dead AGK to rescue the OXPHOS defect observed in AGKKO cells is consistent with other published results (Vukotic et al., 2017). Notably, the reduced oxygen consumption rates did not affect the ability of the AGKKO cells to generate ATP via mitochondrial oxidative phosphorylation when provided with either Complex I or Complex II substrates (Supplemental Figure 1E).
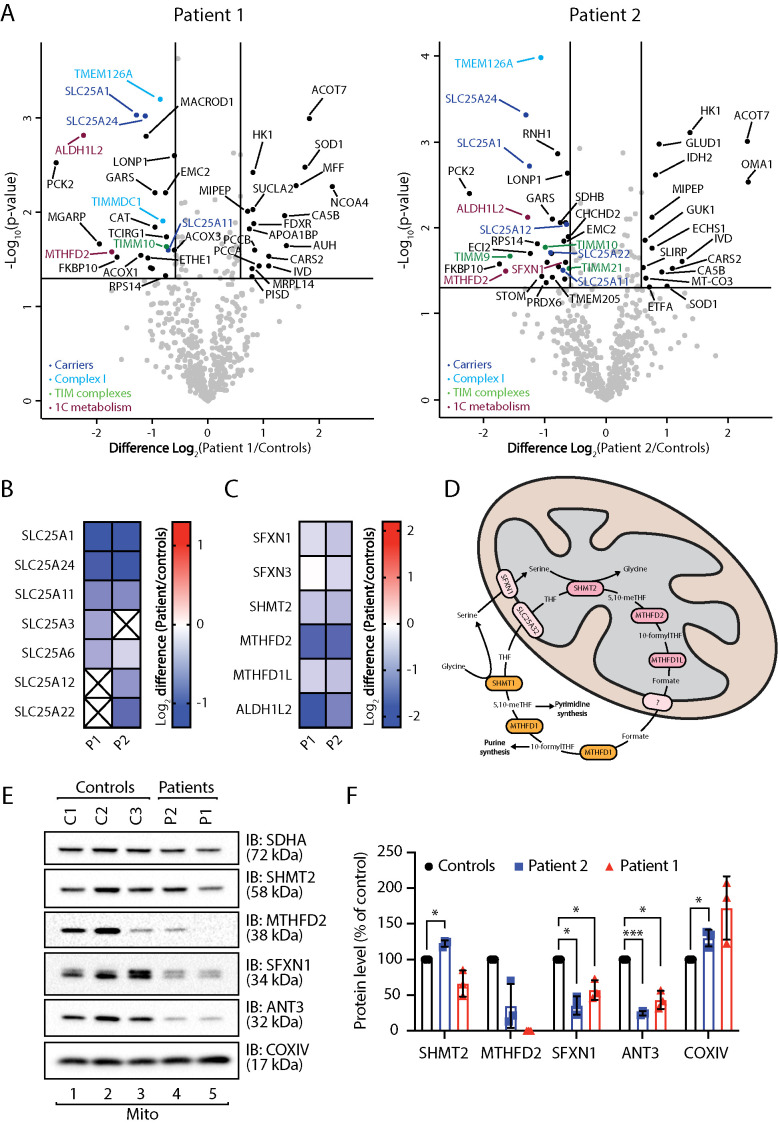
Proteomic changes observed in Sengers syndrome patient fibroblasts were mild compared with those observed in AGKKO cells (Figure 2A). Gene ontology enrichment analysis indicated up-regulation of a number of metabolic processes, including the TCA cycle, fatty acid oxidation, and amino acid catabolism (Supplemental Figure 2A), perhaps representing an attempt to compensate for TIM22 complex dysfunction and consequent bioenergetic impairment. However, these cells did not have an increase in mitochondrial mass as assessed by immunoblotting of whole-cell fractions (Supplemental Figure 2B) or up-regulation of mitochondrial biogenesis by transcriptional up-regulation of PGC1α (Supplemental Figure 2C) (Brown et al., 2010). Mitochondria from patient 1 and 2 fibroblasts showed a general reduction in the levels of SLC25 proteins (Figure 2, A and B; Supplemental Table S1), albeit not as significantly as in the AGKKO HEK293 cell line. Both patients also had lower amounts of Complex I related proteins relative to controls (Supplemental Figure 2D; Supplemental Table S1). This effect was less pronounced in the patient fibroblasts than in the AGKKO HEK293 cells, and the most significant changes in abundance were observed for proteins involved in Complex I assembly (TMEM126A, TIMMDC1) rather than proteins comprising the mature Complex I (Figure 2A; Supplemental Table S1). Oxygen consumption rate measurements exhibited a slight downward trend for only one of the patient fibroblast lines (Supplemental Figure 2E). The reduction fell within control range and was not significant, consistent with the absence of a reduction in the abundance of Complex I subunits (Supplemental Figure 2D). Interestingly, key proteins in the mitochondrial arm of 1C metabolism (SFXN1, SFXN3, SHMT2, MTHFD2, MTHFD1L, ALDH1L2) (Ducker and Rabinowitz, 2017) were down-regulated in both patient fibroblast lines (Figure 2, A, C, and D; Supplemental Table S1), hinting at dysregulation of this pathway in Sengers syndrome.
FIGURE 2:
Proteomic characterization of Sengers syndrome patient fibroblasts. (A, B) Mitochondria isolated from three independent control fibroblast cell lines and two Sengers patient fibroblast cell lines (Patient 1 [P1] and Patient 2 [P2]) were subjected to label-free mass spectrometric analysis. (A) Volcano plots depicting the relative levels of proteins in Sengers patient mitochondria compared with the averaged control data set. Significantly altered proteins are located outside the lines (p value: <0.05, fold change: >1.5× up or down). SLC25 members (dark blue), Complex I subunits or assembly factors (light blue), TIM complex subunits (green), and 1C metabolism proteins (plum) are indicated. (B) Log2 fold-change values (as compared with controls) with p < 0.05 are depicted for SLC25 family proteins in both patient fibroblast cell lines. (C) Log2 fold-change values (as compared with controls) are depicted for 1C metabolism proteins in both patient fibroblast cell lines. (D) Schematic depiction of 1C metabolism. 1C metabolism is a series of parallel and reversible reactions occurring in the mitochondria and the cytosol. In proliferating cells, the reaction proceeds such that formate is produced in the mitochondria and exported to the cytosol for use in biosynthetic reactions. (E) Mitochondrial lysates from control, Sengers syndrome Patient 1 and Patient 2 fibroblasts were analyzed by SDS–PAGE and Western blotting with the indicated antibodies. (F) The relative level of each protein was quantified and is represented as the mean ± SD (n = 3). One sample t test: *, p < 0.05, ***, p < 0.001.
1C metabolism generates one-carbon units required to synthesize many critical metabolites, including nucleotides, amino acids, and lipids (Ducker and Rabinowitz, 2017) (Figure 2D). Analysis of control and patient fibroblasts by Western blot was consistent with proteomic observations and demonstrated decreased abundance of SHMT2, MTHFD2, and SFXN1 (Figure 2, E and F). This was not likely due to an import defect of SHMT2 and MTHFD2 via TIM22 as the in vitro import of [35S]-SHMT2 and [35S]-MTHFD2 into mitochondria isolated from control and AGKKO cells presented normal import kinetics, unlike the known TIM22 substrate GC1 (Supplemental Figure 3A). Proteolytic processing of SHMT2 and MTHFD2 was evident, a typical feature of precursors with an N-terminal presequence that are imported via the TIM23 complex (Wiedemann and Pfanner, 2017). We could conclude that the reduction in the levels of these proteins was not due to import defects into mitochondria. quantitative–PCR (qPCR) measurement of mRNA levels of both SHMT2 and MTHFD2 in Sengers patient fibroblasts uncovered down-regulation of both genes, suggesting that their reduced abundance is due to a transcriptional response in patient fibroblasts (Supplemental Figure 3B). In line with our observation, other studies have observed changes in expression of mitochondrial 1C metabolism enzymes following mitochondrial dysfunction (Bao et al., 2016; Nikkanen et al., 2016; Khan et al., 2017).
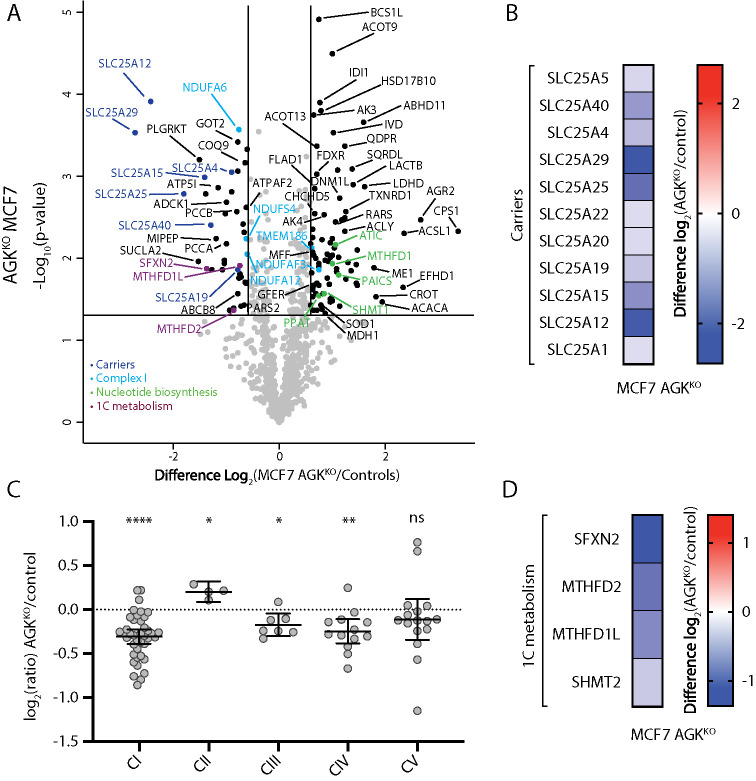
To understand the effect of AGK deletion in a different cellular context, we created an AGKKO MCF7 breast cancer cell line (Supplemental Figure 4). Overexpression of AGK in MCF7 cells and other cancer cell lines has been shown to increase cell proliferation and other tumorigenic properties (Wang et al., 2014). The ability of AGK to promote tumorigenesis has been thought to involve the production of LPA, a potent signaling molecule tthat drives cell growth and proliferation (Mills and Moolenaar, 2003). In light of the more recently discovered role for AGK as a subunit of the TIM22 complex, it also seems possible that overexpression of AGK serves to meet an increased requirement for mitochondrial metabolism. Indeed, mitochondrial proteomics from AGKKO MCF7 cells revealed a general down-regulation of SLC25 carrier proteins (Figure 3, A and B; Supplemental Table S1), confirming that the function of AGK as a component of the TIM22 complex is conserved in this cell type. Consistent with previous observations, a mild reduction in the level of Complex I (Figure 3C; Supplemental Table S1) and a significant down-regulation of mitochondrial 1C metabolism enzymes was observed (Figure 3D; Supplemental Table S1), again linking loss of AGK to remodeling of mitochondrial 1C metabolism.
FIGURE 3:
Proteomic characterization of an AGKKO MCF7 cell line. (A) Mitochondria isolated from control and AGKKO MCF7 cells were subjected to label-free mass spectrometric analysis (n = 3). The volcano plot depicts the relative levels of proteins in AGKKO MCF7 mitochondria compared with control. Significantly altered proteins are located outside the lines (p value: <0.05, fold change: >1.5× up or down). SLC25 members (dark blue), Complex I subunits or assembly factors (light blue), nucleotide biosynthesis enzymes (green), and 1C metabolism proteins (plum) are indicated. (B) Log2 fold-change values (as compared with control) are depicted for SLC25 proteins (p < 0.05) identified in the AGKKO MCF7 cells. (C) Relative abundance of respiratory chain complexes (complexes I–V) in AGKKO MCF7 cells as compared with control. Mean ± 95% CI is depicted. Ratio paired t test: *, p < 0.05, **, p < 0.05, ***, p < 0.001, ****, p < 0.0001 (n = 3). (D) Log2 fold-change values (as compared with control) for 1C metabolism proteins (p < 0.05) identified in the AGKKO MCF7 cells.
Sideroflexins are novel substrates of the TIM22 complex
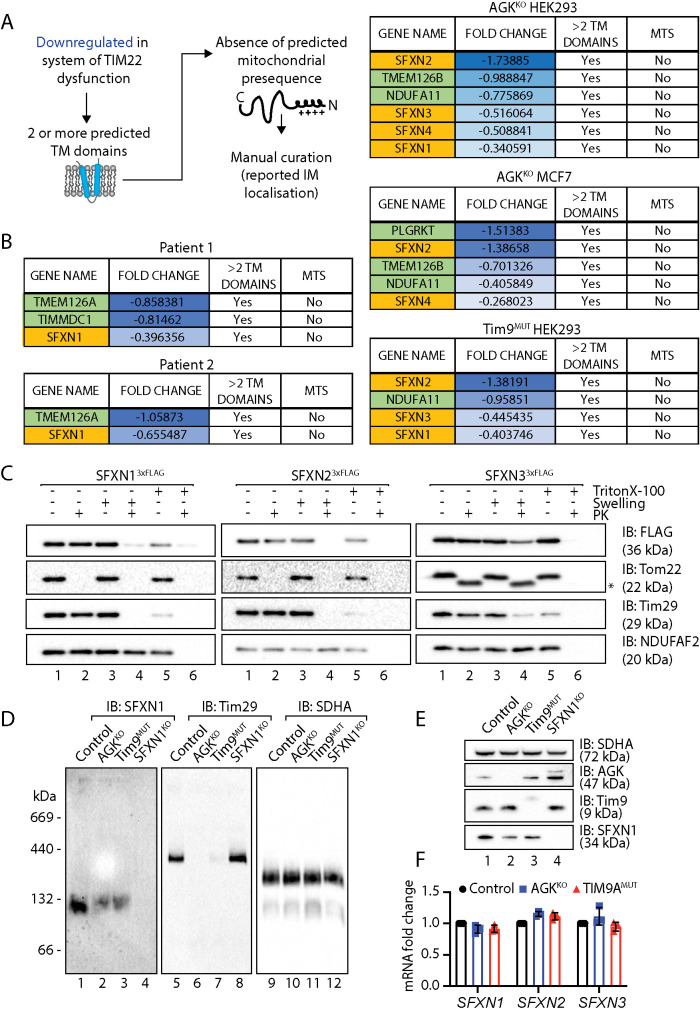
To date, two main classes of the TIM22 complex are known: 1) SLC25 proteins, which function as inner membrane metabolite transporters and possess six predicted transmembrane domains; and 2) TIM proteins (Tim17, Tim23, Tim22), which possess four predicted transmembrane domains and function as components of the inner membrane protein translocase complexes (Káldi et al., 1998; Kurz et al., 1999). As the primary function of AGK is in the import of polytopic transmembrane proteins at the TIM22 complex, the pathogenesis of Sengers syndrome is likely to arise due to the depletion of TIM22 complex substrates, which play diverse roles in mitochondrial metabolism. To try to identify other TIM22 complex substrates that could contribute to the pathology of Sengers syndrome, we analyzed a list of down-regulated proteins from patient fibroblast and AGKKO mitochondrial proteomics data sets. We hypothesized that a TIM22 substrate would 1) possess at least two predicted transmembrane domains, 2) be down-regulated in at least one of the proteomics data sets, and 3) lack a predicted mitochondrial targeting signal (MTS), as these proteins typically utilize the TIM23 complex (Figure 4A).
FIGURE 4:
Identification of TIM22 complex substrates. (A) Schematic representation of the parameters used to filter proteins from proteomic data sets to screen for novel TIM22 substrates. (B) Candidate TIM22 substrates identified following the pipeline in A in the indicated cell lines. SFXNs are highlighted in yellow. (C) Mitochondria were isolated from HEK293 cells stably expressing SFXN13xFLAG or SFXN23xFLAG or from HEK293 cells transiently transfected with SFXN33xFLAG. Intact mitochondria, mitoplasts (generated by hypoosmotic swelling of the outer membrane) or solubilized mitochondria were treated with or without proteinase K and analyzed by SDS–PAGE and Western blotting with the indicated antibodies. * indicates a proteolytic fragment of Tom22 sometimes detected due to incomplete proteolytic processing. (D) Mitochondria isolated from control, AGKKO, Tim9MUT, and SFXN1KO HEK293 cells were solubilized in 1% digitonin containing buffer and analyzed by BN-PAGE and immunoblotting with the indicated antibodies. (E) Mitochondrial lysates from control, AGKKO, Tim9MUT, and SFXN1KO HEK293 cells were analyzed by SDS–PAGE and Western blotting with the indicated antibodies. (F) Fold changes in mRNA expression for SFXN1, SFXN2, and SFXN3 in AGKKO and Tim9MUT HEK293 compared with control HEK293 cells were quantified by RT-qPCR and are expressed as mean ± SD (n = 3).
Down-regulated proteins were filtered to exclude those with fewer than two predicted transmembrane domains or those that contained a predicted MTS. A list of candidate TIM22 complex substrates was generated for Sengers patient fibroblasts, HEK293 and MCF7 AGKKO cell lines, and a Tim9MUT HEK293 cell line (Kang et al., 2019) (Figure 4, A and B; Supplemental Figure 5; Supplemental Table S2). Tim9 functions as a chaperone in the intermembrane space and at the TIM22 complex. Most proteins identified using this approach belonged to the SLC25 gene family of predicted TIM22 substrates (Supplemental Figure 5; Supplemental Table S2), which aligns with the known client portfolio of TIM22. Other candidate substrates included the sideroflexin (SFXN) family, of which at least one member was identified in data sets for all analyzed cell lines (Figure 4B; Supplemental Table S2). Interestingly, the remaining candidate substrates that emerged from this analysis were Complex I subunits (NDUFA11), Complex I assembly factors (TIMMDC1 and TMEM126B) (Formosa et al., 2018), and TMEM126A, a homologue of TMEM126B (Figure 4B; Supplemental Table S2). NDUFA11 contains four transmembrane domains and is a member of the Tim17 protein family (Žárský and Doležal, 2016) that also includes Tim22, Tim17, and Tim23, all of which are established substrates of the TIM22 complex. TIMMDC1 and TMEM126B are Complex I assembly factors with four predicted transmembrane domains (Formosa et al., 2018). TMEM126A is a paralogue of TMEM126B that also causes disease when mutated (Hanein et al., 2009; Meyer et al., 2010) and is likely to play a role in Complex I biogenesis. Like NDUFA11, TIMMDC1 is also a member of the Tim17 protein family (Žárský and Doležal, 2016).
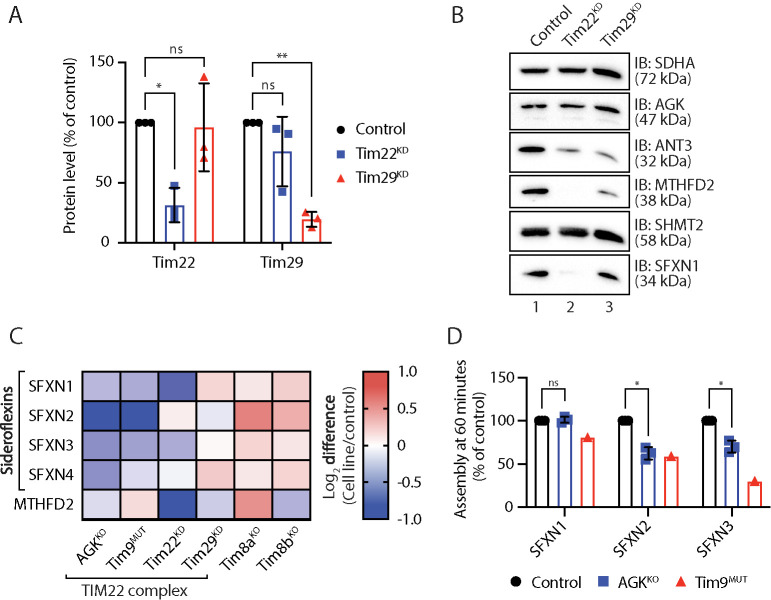
We were drawn to the SFXN protein family, as the proteomic profiling data revealed changes to enzymes involved in mitochondrial 1C metabolism and SFXN1, SFXN2, and SFXN3 were recently identified as serine transporters required for mitochondrial 1C metabolism (Kory et al., 2018). Reports on the localization and topology of the SFXN proteins vary (Kory et al., 2018; Mon et al., 2019). If they are genuine TIM22 complex substrates, we would expect the SFXN proteins to be localized to the inner mitochondrial membrane. We selected SFXN1, SFXN2, and SFXN3 as representatives of the family and determined their submitochondrial location using mitochondrial subfractionation in HEK293 cells stably expressing C-terminally FLAG-tagged versions of the proteins (Figure 4C). For SFXN1, SFXN2, and SFXN3 the C-terminal FLAG tag was only accessible to externally added proteinase K following hypo-osmotic disruption of the outer membrane (Figure 4C, lane 4) (note the inefficient rupturing of OM in the SFXN3 panel as indicated by incomplete digestion of Tim29, hence incomplete removal of FLAG signal), confirming that SFXN1, SFXN2, and SFXN3 localize to the inner mitochondrial membrane with their C-termini facing the intermembrane space. BN-PAGE analysis indicated that SFXN1 assembles into a complex migrating at ∼132 kDa (Figure 4D, lane 1), suggesting that the protein may exist in the inner membrane as a dimer or tetramer. The size of this complex on BN-PAGE is strikingly similar to that formed by SLC25 proteins, the major class of the TIM22 complex substrate (Kang et al., 2016, 2017, 2019).; The abundance of the SFXN1 complex was reduced in mitochondria isolated from AGKKO and Tim9MUT cells (Figure 4D, lanes 2 and 3) where the TIM22 complex is dysfunctional (Figure 4D, lanes 6 and 7). The complex was absent in a SFXN1 CRISPR/Cas9 genome-edited cell line (Figure 4D, lane 4; Supplemental Figure 6A), confirming the specificity of the antibody. Consistent with the reduced abundance of the complex on BN-PAGE, SFXN1 levels were also reduced on SDS–PAGE in AGKKO and Tim9MUT HEK293 cells (Figure 4E, lanes 2 and 3).
To confirm that the observed effect in the AGKKO cell line was related to TIM22 complex dysfunction rather than a loss of AGK lipid kinase activity, the levels of SFXN proteins were analyzed in proteomics data obtained from AGKKO HEK cells reexpressing WT AGK or kinase-dead AGK (AGKG126E). Similar to SLC25 proteins (Figure 1E), the reduced levels of SFXNs in AGKKO cells could be rescued through reexpression of both WT and kinase-dead AGK (Supplemental Figure 6B; Supplemental Table S1). To exclude the possibility that changes in the abundance of SFXN proteins were occurring due to alterations in gene expression, we measured mRNA abundance for SFXN1, SFXN2, and SFXN3 in AGKKO and Tim9MUT HEK293 cells, and these remained unchanged (Figure 4F), suggesting that the observed changes to the SFXN proteins were posttranscriptional. Taken together, these findings suggest that the SFXN family represents a novel class of TIM22 substrate and that their perturbed import in Sengers syndrome might contribute to the pathology of the disease.
To interrogate the requirement of the TIM22 complex for SFXN biogenesis, we analyzed mitochondria depleted of either Tim22 or Tim29 by small interfering RNA (siRNA) (Figure 5, A–C; Supplemental Table S1). Samples were retained for Western blot analysis (Figure 5B) and label-free quantitative mass spectrometric analysis (Figure 5C; Supplemental Table 1). Tim22 is the central channel-forming unit of the TIM22 complex, while Tim29 maintains TIM22 complex integrity and is required for the import of TIM substrates (Kang et al., 2016). In line with these different roles at the TIM22 complex, depletion of either Tim22 or Tim29 had different effects (Figure 5, A–C; Supplemental Table S1). Consistent with a requirement for the TIM22 complex in SFXN biogenesis, Tim22 knockdown substantially reduced the levels of SFXN1 (Figure 5, B, lane 2, and C; Supplemental Table S1), but had only a minimal impact on the levels of SFXN2 and SFXN4 (Figure 5C; Supplemental Table S1), perhaps due to the relatively short siRNA KD time course (72 h). Interestingly, MTHFD2 was undetectable by Western blot in the Tim22 KD (Figure 5B, lane 2), again suggesting that TIM22 complex dysfunction can induce remodeling of the mitochondrial 1C metabolism pathway. In contrast, depletion of Tim29 had no effect on the abundance of SFXN proteins (Figure 5, B, lane 3, and C; Supplemental Table S1), consistent with its proposed role as a subunit specifically required for the import of TIM proteins (Kang et al., 2016).
FIGURE 5:
Sideroflexins require the TIM22 complex for their biogenesis. (A) Mitochondrial lysates from triplicate sets of control, Tim22 knockdown (KD), and Tim29 KD HEK293 cells were analyzed by SDS–PAGE and Western blotting with antibodies specific for Tim22, Tim29, and SDHA (loading control). Levels of Tim22 and Tim29 were quantified and tabulated as mean ± SD (n = 3). One-sample t test: *, p < 0.05, **, p < 0.01. (B) Mitochondrial lysates from control, Tim22 KD, and Tim29 KD HEK293 cells were analyzed by SDS–PAGE and Western blotting. (C) Log2 fold-change values (as compared with respective controls) are depicted for selected proteins in the indicated cell lines. (D) [35S]-SFXN1, [35S]-SFXN2, and [35S]-SFXN3 were incubated with mitochondria isolated from control, AGKKO, and Tim9MUT HEK293 cells for 60 min before proteinase K (PK) treatment. Samples were solubilized in 1% digitonin containing buffer and analyzed by BN-PAGE and autoradiography. Assembled protein at 60 min in control, AGKKO, and Tim9MUT mitochondria was quantified. Graph depicts mean ± SD (n = 3 for AGKKO, n = 1 for Tim9MUT). One-sample t test: *, p < 0.05.
As an additional biochemical approach to investigate whether SFXN proteins are substrates of the TIM22 complex, we performed in vitro import of [35S]-SFXN1, [35S]-SFXN2, [35S]-SFXN3, and [35S]-Tim23 (as a control) into mitochondria isolated from control, AGKKO and hTim9MUT cells (Figure 5D). Import was performed for 60 min, after which mitochondria were isolated and separated by BN-PAGE and radioactive signal quantified. At 60 min the assembly of [35S]-SFXN2 and [35S]-SFXN3 was moderately compromised in AGKKO HEK293 mitochondria (Figure 5D), although SFXN1 assembly seemed normal (Figure 5D). The SFXN2 and SFXN3 import defects correspond with their reduced steady state levels in AGKKO mitochondria (Figure 1B; Supplemental Table S1). The normal assembly of SFXN1 was surprising considering that the endogenous SFXN1 complex was reduced in AGKKO cells (Figure 4D, lane 2). However, we reasoned that this discrepancy could be due to the fact that AGK is only a peripheral subunit of the TIM22 complex and that the essential Tim22 pore is still present and functional in the absence of AGK. In support of a role for the TIM22 complex, a clear assembly defect was apparent for SFXN1, SFXN2, and SFXN3 following import into Tim9MUT cells (Figure 5D). Together, these results suggest that SFXN proteins are substrates of the TIM22 complex and the import of these proteins requires Tim9 and AGK, as well as Tim22 itself, while Tim29 appears to be dispensable. Although the requirement of the TIM22 complex for the import of SFXN1 is less clear, the depletion of SFXN1 from cells lacking AGK may occur due to another yet unclear mechanism. The changes to SFXN proteins are specific to the TIM22 complex rather than a nonspecific response to stress, as Tim8aKO and Tim8bKO HEK293 mitochondria, which exhibit general mitochondrial and electron transport chain dysfunction but no TIM22 complex impairment (Kang et al., 2019), show no changes in abundance of SFXN proteins (Figure 5C).
Loss of AGK impairs growth in the absence of exogenous serine
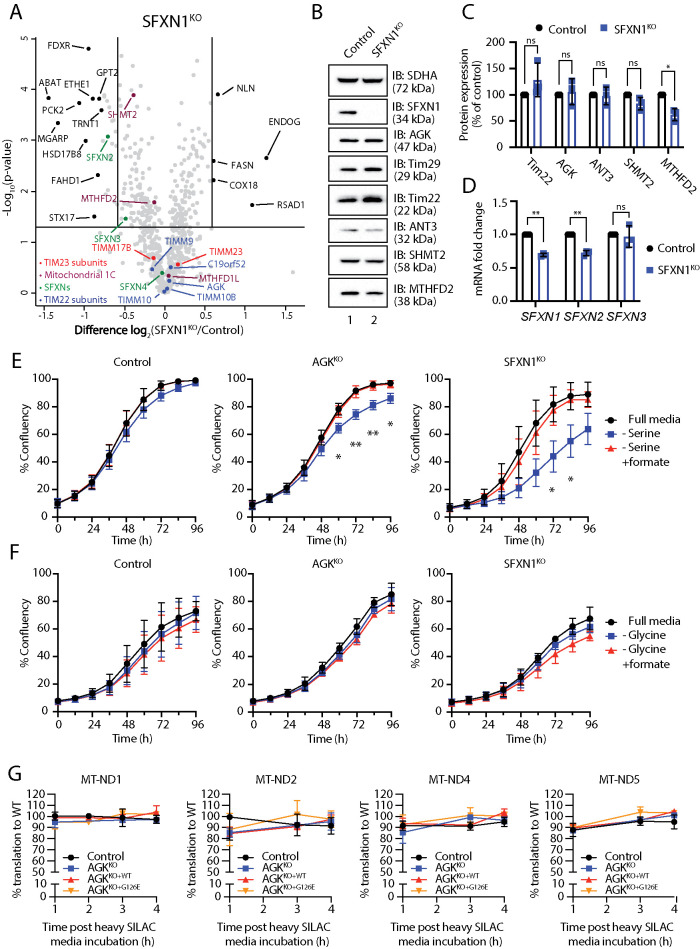
The lower abundance of mitochondrial 1C metabolism enzymes (MTHFD2, SHMT2, MTHFD1L, SLC25A32, SFXNs, ALDH1L2) in Sengers syndrome patient and AGKKO cell lines was of interest as 1C metabolism has surfaced as a pathway of importance in mitochondrial disease (Bao et al., 2016; Nikkanen et al., 2016; Khan et al., 2017). As SFXNs mediate a crucial step in mitochondrial 1C metabolism, we reasoned that depletion of SFXNs in Sengers syndrome fibroblasts or AGKKO cells could be responsible for the remodeling of this pathway. Analysis of SFXN1KO mitochondria using label-free quantitative proteomics revealed only small reductions in the levels of SHMT2 and MTHFD2 (Figure 6, A–C; Supplemental Table S1), suggesting that the metabolic remodeling observed in our model systems arises due to dysfunction induced by more than one single event (reduction in SFXN proteins). Knockout of SFXN1 had no reciprocal effect on the abundance of TIM22 complex subunits (Figure 6, A–C; Supplemental Table S1). Interestingly, depletion of SFXN1 also led to a reduction in the levels of SFXN2 and SFXN3 (Figure 6A; Supplemental Table S1). For SFXN2, but not SFXN3, this change was accompanied by a reduction in mRNA abundance (Figure 6D). The dependence of SFXN3 on SFXN1 for stability suggests that the two proteins may exist within a common complex.
FIGURE 6:
Loss of AGK limits cell proliferation in the absence of exogenous serine. (A) Mitochondria isolated from control and SFXN1KO HEK293 cells were subjected to label-free quantitative mass spectrometric analysis. Volcano plots depict the relative levels of mitochondrial proteins in each sample compared with control. n = 3 biological replicates. Horizontal cutoff represents p = 0.05 and vertical cutoffs represent – and + 1.5× fold change. TIM23 complex subunits (red), TIM22 complex subunits (blue), SFXN proteins (green), and 1C metabolism enzymes (plum) are indicated. (B) Mitochondrial lysates from control and SFXN1KO HEK293 cells were analyzed by SDS–PAGE and Western blotting. (C) Relative protein levels of selected proteins were quantified and are represented as mean ± SD (n = 3). One-sample t test: * p < 0.05. (D) Relative fold changes of mRNA expression for SFXN1, SFXN2, and SFXN3 in control and SFXN1KO HEK293 cells were determined using RT-qPCR and are represented as the mean ± SD (n = 3). One-sample t test: *, p < 0.05, **, p < 0.01. (E) Proliferation of control, AGKKO, and SFXN1KO HEK293 cells was monitored in complete media, serine-free media, and serine-free media supplemented with 1 mM formate. Confluency was measured at 12-h intervals and is depicted as mean ± SD (n = 4). Unpaired t test, * p < 0.05, ** p < 0.01. (F) Proliferation of control, AGKKO, and SFXN1KO HEK293 cells was monitored in complete media, glycine-free media, and glycine-free media supplemented with 1 mM formate. Confluency was measured at 12-h intervals and is depicted as mean ± SD (n = 3). (G) Pulse SILAC analysis of newly translated mtDNA-encoded OXPHOS subunits. SILAC media was added following 24 h treatment with chloramphenicol and analysis was performed at 1, 3, and 4 h post-SILAC media incubation. Log2-transformed heavy-peptide derived intensities are plotted relative to control. Statistical analysis was performed on each time point (n = 3) compared with control WT using a t test and FDR-1% with no significance recorded. Data are depicted as mean ± SD.
We noted that changes to 1C metabolism enzymes in our studied systems generally involved a reduction in protein abundance. This was unexpected, as mitochondrial stress typically induces up-regulation of mitochondrial 1C metabolism enzymes as a part of the integrated stress response (ISR) (Nikkanen et al., 2016; Khan et al., 2017; Quirós et al., 2017). HEK293 AGKKO cells showed no increase in sensitivity to actinonin (Supplemental Figure 7A), an inhibitor of mitochondrial translation and robust ISR inducer, indicating that the observed changes to 1C enzymes did not affect the ability of cells to cope with mitochondrial stress. Surprisingly, the ISR appeared to be intact in the AGKKO HEK cells, which were able to transcriptionally up-regulate SHMT2, MTHFD2, SLC7A11, and DDIT3 to the same extent as control cells (Supplemental Figure 7B), although this did not correlate with an increase in the protein abundance of SHMT2 and MTHFD2 (Supplemental Figure 7C). In fact, for MTHFD2, protein levels in both control and AGKKO HEK cells were reduced following treatment with actinonin (Supplemental Figure 7C). Although most studies of mitochondrial stress signaling show agreement between changes in transcript and protein abundance for MTHFD2, it appears that in some contexts this does not strictly apply. As we previously confirmed that MTHFD2 and SHMT2 do not require the TIM22 complex for their import (Supplemental Figure 3A), their reduced abundance following actinonin treatment or in various systems of TIM22 complex dysfunction suggests an additional mechanism, such as turnover within mitochondria. In support of this, studies of MTHFD2 have confirmed that it has a particularly short half-life and is subject to rapid regulation (Koufaris and Nilsson, 2018).
To confirm that reduced SFXN levels downstream of AGK/TIM22 complex dysfunction result in a functional defect in serine utilization at the mitochondrion, we monitored cell proliferation of control and AGKKO cells in serine-free media (Figure 6E). Under these conditions, intracellular serine levels are reduced and can be supplied only in limited quantities through the de novo serine biosynthesis pathway. Normal proliferation under these conditions requires efficient mitochondrial 1C metabolism, and any defects in this pathway, including reduced import of serine into the mitochondria, are likely to limit nucleotide synthesis and manifest as a growth defect. Indeed, while AGKKO HEK293 cells have no proliferation defect under standard culturing conditions (Kang et al., 2017), growth in serine-free media resulted in a mild but significant proliferation defect (Figure 6E, middle panel). This defect could be rescued through supplementation with 1 mM formate, a key product of the mitochondrial 1C cycle. Consistent with published observations (Kory et al., 2018), SFXN1KO cells, which lack the main mitochondrial serine transporter, showed a larger defect that was also rescued by supplementation with formate (Figure 6E, right panel). We also wondered whether a defect in glycine import could underpin the observed remodeling of 1C metabolism. Glycine can be utilized as a substrate for 1C metabolism through the glycine cleavage system (Kikuchi et al., 2008; Ducker and Rabinowitz, 2017), and the mitochondrial glycine transporter, SLC25A38, is a presumed substrate of the TIM22 complex. In media lacking glycine, neither control, AGKKO, nor SFXN1KO cells displayed a proliferation defect (Figure 6F). Together, these results suggest that AGK/TIM22 complex dysfunction reduces the efficiency of formate production through mitochondrial 1C metabolism, most likely by reducing the efficiency of SFXN import and assembly into the inner membrane.
Given our observation of Complex I changes in cells lacking AGK, we tested whether depletion of mitochondrial serine or 1C metabolism intermediates could be contributing to this defect by impairing mitochondrial translation, as serine catabolism is required to generate formyl-methionyl-tRNA for translation initiation of mitochondrial DNA (mtDNA)-encoded proteins (Minton et al., 2018). Additionally, serine is required as a proteinogenic amino acid for synthesis of mtDNA-encoded subunits. Pulse-SILAC labeling to track the expression of mtDNA-encoded subunits of the respiratory chain in control, AGKKO, AGKKO+WT, and AGKKO+G126E cells revealed no dysfunction in mitochondrial translation of mtDNA-encoded Complex I subunits (Figure 6G; Supplemental Table S3), or any other mtDNA-encoded OXPHOS subunits (Supplemental Figure 7D; Supplemental Table S3), indicating that the observed changes to Complex I were not driven by a mtDNA translation defect.
DISCUSSION
We characterized the mitochondrial proteomes of several cell lines lacking AGK, a subunit of the metazoan TIM22 complex. Our analyses included two fibroblast lines from patients with Sengers syndrome, a mitochondrial disease caused by loss of function mutations in AGK, as well as HEK293 and MCF7 AGKKO cell lines. Extensive proteome remodeling was observed in all cell models, and we analyzed the data with caution, understanding that it could represent a combination of primary and secondary effects given the diverse substrate spectrum of TIM22. Considering that the primary role for AGK as a component of the TIM22 complex is to facilitate the import of hydrophobic inner membranes, primary effects are likely to represent depletion of TIM22 substrates. Until recently, only two classes of TIM22 substrates were known: SLC25 carrier proteins, which mediate transport of metabolites across the inner membrane, and TIM proteins (Tim17/Tim23/Tim22), which are subunits of the inner membrane translocase complexes. These substrates with six and four transmembrane domains, respectively, have shaped dogma in the field as to how the TIM22 complex mediates insertion of substrates in a hairpin loop conformation (Rehling et al., 2003; Rehling et al., 2004). Based on this, the SFXN proteins with their five transmembrane domains do not fit with these previous models of TIM22 membrane insertion. However, subunits of the mitochondrial pyruvate carrier were recently identified as noncanonical substrates of the TIM22 complex (Gomkale et al., 2020; Rampelt et al., 2020), suggesting that substrates with odd numbers of transmembrane segments likely do utilize TIM22. It is also important to highlight that these models of TIM22 membrane insertion were conceived exclusively on biochemical work performed with yeast mitochondria, and in the past 5 yr the human TIM22 complex has emerged as the most divergent of the mitochondrial import translocases (Callegari et al., 2016; Kang et al., 2016, 2017; Vukotic et al., 2017). Thus, the true mechanism of membrane insertion via the human TIM22 complex awaits exploration.
Bioinformatic analysis of down-regulated proteins in various systems of AGK and TIM22 complex dysfunction allowed generation of a comprehensive list of candidate TIM22 complex substrates, including several novel TIM22 complex substrates. These included sideroflexins (SFXNs), of which SFXN1, SFXN2, and SFXN3 are thought to function as serine transporters (Kory et al., 2018), as well as several proteins involved in Complex I biogenesis and assembly. On the basis of our analysis showing reduced steady state levels and assembly of SFXNs in mitochondria lacking AGK or Tim9, we suggest that this family of proteins is indeed a novel substrate class of the TIM22 complex. Additionally, independent mass spectrometric analysis of AGKKO HEK293 demonstrated a striking down-regulation of SFXN proteins and identified SFXN2 as a high-confidence client protein of AGK (Vukotic et al., 2017), while SFXN1 has been enriched following Tim22 immunoprecipitation (Callegari et al., 2016).
Proteomic analysis of Sengers syndrome patient fibroblast mitochondria also revealed extensive metabolic remodeling, presumably representing a secondary effect arising from the depletion of inner membrane metabolite transport proteins. Many pathways involved in catabolism and ATP synthesis were up-regulated, while pathways including gluconeogenesis and mitochondrial 1C metabolism were repressed. A similar response was observed in AGKKO MCF7 cells, which exhibited down-regulation of mitochondrial 1C metabolism. The observation that mitochondrial 1C enzymes such as MTHFD2, MTHFD1L, and SHMT2 were down-regulated in systems of TIM22 complex dysfunction was surprising, as classical mitochondrial dysfunction caused by mtDNA lesions or acute OXPHOS inhibition typically results in activation of an ISR (Quirós et al., 2017; Fessler et al., 2020; Guo et al., 2020), which has been observed to lead to induction of MTHFD2, MTHFD1L, and SHMT2 both in vitro and in vivo (Bao et al., 2016; Nikkanen et al., 2016; Khan et al., 2017). Although we have not elucidated the exact mechanism for the changes in 1C metabolism, we have established that the changes in the levels of these enzymes are not due to reduced import capacity via the TIM22 complex and that they are not due to an ISR, which typically leads to an up-regulation of these enzymes. The extent to which dysfunction of mitochondrial 1C metabolism could contribute to mitochondrial disease is not clear. Mitochondrial 1C metabolism is known to play a crucial role in development (Momb et al., 2013), but its function in adult tissues is not as clear. Considering that the pathway plays a crucial role in redox balance through generation of glycine for glutathione synthesis and NADPH for glutathione cycling (Fan et al., 2014), it is possible that down-regulation of 1C metabolism limits the ability of cells in adult tissues to combat oxidative stress that occurs during mitochondrial dysfunction.
The comprehensive proteomic characterization performed in this study also provides mechanistic insight into other important characteristics of Sengers syndrome. One notable characteristic of Sengers syndrome is OXPHOS impairment, which is observed to varying degrees in muscle and heart in almost all cases of Sengers syndrome (Haghighi et al., 2014). In this study, a reduction in the abundance of Complex I was observed in both patient fibroblast cell lines and the HEK293 and MCF7 AGKKO cell lines. There are multiple potential explanations for this phenotype. Many individual SLC25 carrier proteins have been individually linked to disease (Palmieri et al., 2020). SLC25A4 mutations are able to cause a form of mitochondrial disease associated with mtDNA depletion and subsequent OXPHOS dysfunction (Kaukonen et al., 2000). It is possible that in Sengers syndrome, depletion of SLC25A4 and other carriers required for mitochondrial homeostasis leads to nucleotide imbalances or increased reactive oxygen species, resulting in mtDNA damage and subsequent depletion. However, mtDNA depletion is not observed in all Sengers syndrome cases (Haghighi et al., 2014), suggesting that it is not strictly necessary to induce the OXPHOS dysfunction observed in the disease. Additionally, a single case of mitochondrial disease caused by mutations in Tim22 itself resulted in impaired OXPHOS activity in muscle without mtDNA rearrangement or copy number reduction (Pacheu-Grau et al., 2018). An alternate explanation is provided by the identification of potential novel TIM22 substrates in this study. The TIM22 complex has not previously been linked to Complex I biogenesis; however, our analysis suggested that several proteins with direct roles in Complex I biogenesis may be substrates of the complex. NDUFA11, a member of the Tim17 protein family, is an accessory subunit of the complex (Stroud et al., 2016), while TMEM126B and TIMMDC1 are assembly factors (Formosa et al., 2018) (TIMMDC1 is also a Tim17 family member). This observation raises the interesting possibility that the Complex I defect arises at least in part by insufficient import of proteins that form part of the mature complex or contribute to its assembly. Understanding the biogenesis of these proteins in detail requires further biochemical investigation.
The results presented provide a resource for developing a detailed understanding of the mitochondrial dysfunction that occurs following loss of AGK. They provide a catalogue on which future cases of Sengers syndrome can be diagnosed using proteomic profiling. In addition, we identified novel TIM22 complex substrates and provide preliminary biochemical evidence for a requirement of the TIM22 complex in the biogenesis of sideroflexin proteins, an emerging class of inner membrane proteins central to mitochondrial metabolism. Loss of TIM22 substrates, including the SLC25 proteins and sideroflexins, leads to extensive remodeling of the mitochondrial proteome and remodeling of metabolic pathways, including the mitochondrial arm of 1C metabolism. 1C metabolism has been implicated in mitochondrial disease, albeit as an up-regulated rather than a down-regulated pathway (Bao et al., 2016; Nikkanen et al., 2016; Khan et al., 2017). But mitochondrial diseases are clinically and genetically heterogeneous, and this heterogeneity is likely to also be a key player at the molecular level. Indeed, AGKKO cells exhibited a proliferation defect in the absence of serine that was rescuable with formate, suggesting that 1C metabolism is a key pathway that should be considered in the pathogenesis of mitochondrial disease.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell lines, cell culture, and siRNA transfection
Flp-In T-Rex 293 (Thermo Fisher Scientific), primary patient fibroblasts (Calvo et al., 2012), and MCF7 cells were cultured in DMEM (Thermo Fisher Scientific) containing 1% (vol/vol) penicillin–streptomycin (Thermo Fisher Scientific) and supplemented with 5% (vol/vol) fetal bovine serum. siRNA transfection was performed in cells plated overnight using scrambled siRNA (Sigma) or siRNA targeting Tim22 (5′ CCAUUGUGGGAGCCAUGUU 3′) (Sigma) or Tim29 (5′ GGCUCUUCGAUGAGAAGUA 3′) (Sigma). Briefly, siRNA was transfected at 10 nM using DharmaFECT (Dharmacon) according to the manufacturer’s instructions. Cells were transfected a second time 48 h after the first transfection and harvested 72 h after the first transfection.
Gene editing and screening
Editing of the SFXN1 and AGK genes was carried out using the pSpCas9(BB)-2A-GFP CRISPR-Cas9 construct (a gift from F. Zhang; Addgene, Broad Institute) (Ran et al., 2013). To edit SFXN1, guide RNAs targeting exon 7 (coding exon 6) of the SFXN1 gene were designed using CHOPCHOP. An oligonucleotide duplex formed from (5′ CACCGCGTTCGCCGACTCCCCCAAG 3′ and 5′ AAACCTTGGGGGAGTCGGCGAACGC 3′) was ligated into pSpCas9(BB)-2A-GFP and transfected into Flp-In T-Rex 293 cells, and single cells were obtained via fluorescence activated cell sorting based on green fluorescent protein fluorescence. Sorted cells were allowed to expand before screening. Screening was performed through Western blotting with a SFXN1 antibody, and clones were genetically verified using sanger sequencing of genomic DNA. AGKKO cells were generated as described previously, except using MCF7 cells rather than HEK293 (Kang et al., 2017). Clones were genetically verified by Sanger sequencing of genomic DNA.
Quantitative reverse transcription–PCR (RT-PCR)
Following RNA extraction using a NucleoSpin RNA kit (Macherey-Nagel), cDNA was synthesized with the Transcriptor First Strand cDNA Synthesis kit (Roche). Gene expression was determined using SYBR-green qPCR on the Lightcycler 480 (Roche). Gene expression was normalized to GAPDH and ACTB and determined using the ∆∆Ct method. Primer sequences used in this study are shown in Table 1.
TABLE 1:
Primers used.
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
| SFXN1 | TTGGCTTCTGTTTGGTGTTTG | TCCCTTATTGAAGTACACGCG |
| SFXN2 | CAGTTCTACAGGACGATGCC | TGGTTGTGGCTGTGAAGTAG |
| SFXN3 | ACTGTTACTGATCCTCGAAATCTG | CATACACATACTTGGCCCTCC |
| SLC7A11 | ATGCAGTGGCAGTGACCTTT | GGCAACAAAGATCGGAACTG |
| MTHFD2 | CTGCGACTTCTCTAATGTCTGC | CTCGCCAACCAGGATCACA |
| SHMT2 | CGAGTTGCGATGCTGTACTT | CTGCGTTGCTGTGCTGAG |
| DDIT3 | AGCCAAAATCAGAGCTGGAA | TGGATCAGTCTGGAAAAGCA |
| PGC1α | ACCAAACCCACAGAGAACAG | GGGTCAGAGGAAGAGATAAAGTTG |
| ACTB | AGAAAATCTGGCACCACACC | GGGGTGTTGAAGGTCTCAAA |
| GAPDH | GGTGTGAACCATGAGAAG | CCACAGTTTCCCGGAG |
Mitochondrial isolation, gel electrophoresis, and immunoblot analysis
Mitochondria were isolated from cultured mammalian cells through differential centrifugation (Kang et al., 2017). Cultured cells were harvested in phosphate-buffered saline (PBS) and isolated by centrifugation at 500 × g. Cells were homogenized in isolation buffer (20 mM HEPES-KOH [pH 7.6], 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 2 mg/ml bovine serum albumin) and the lysate was centrifuged at 800 × g to remove nuclear debris and intact cells. The supernatant containing mitochondria was centrifuged at 12,000 × g to obtain a crude mitochondrial pellet. Protein concentration in the mitochondrial pellet was determined using the Pierce BCA protein assay kit (Thermo Fisher Scientific).
Tris-tricine SDS–PAGE was performed as described previously (Kang et al., 2017). Solutions containing 10 or 16% (vol/vol) acrylamide solution (49.5% acrylamide, 1.5% bis-acrylamide) were made up in tricine gel buffer (1 M Tris-Cl, 0.1% [wt/vol] SDS, pH 8.45; 13% [vol/vol] glycerol included in the 16% mix). These solutions were used to pour 10–16% gels using a gradient mixer. Following polymerization, a stacking gel (4% [vol/vol] acrylamide solution in tricine gel buffer) was overlaid onto the gradient gel. Polymerization of both the gradient and stacking gels was achieved through sequential addition of TEMED (Sigma) and 10% APS (Sigma). Electrophoresis was performed using Tris-tricine SDS–PAGE anode buffer (50 mM Bis-Tris, pH 7.0) and cathode buffer (0.1 M Tris, 0.1 M tricine, 0.1% [wt/vol] SDS, pH 8.45). Pelleted mitochondria to be analyzed were resuspended in SDS loading dye (50 mM Tris-Cl [pH 6.8], 0.1 M dithiothreitol [DTT], 2% [wt/vol] SDS, 10 % [vol/vol] glycerol, 0.1% [wt/vol] bromophenol blue) and boiled.
Blue-Native (BN) PAGE was performed as described previously (Kang et al., 2017). Solutions containing 4 or 16% (vol/vol) acrylamide solution in BN gel buffer (66 mM ε-amino n-caproic acid, 50 mM Bis-Tris, pH 7.0) were used to pour 4–16% gradient gels. Following polymerization, a stacking gel (4% [vol/vol] acrylamide solution in BN gel buffer) was overlaid onto the gradient gel. Polymerization was initiated through sequential addition of TEMED and 10% APS. Electrophoresis was carried out overnight at 4°C using BN anode (50 mM Bis-Tris, pH 7.0) and cathode buffer (50 mM tricine, 15 mM Bis-Tris, 0.02% [wt/vol] Coomassie blue G250). Pelleted mitochondria to be analyzed were solubilized in digitonin solubilization buffer (20 mM Bis-Tris, 50 mM NaCl, 10% [vol/vol] glycerol, pH 7.4, 1% [wt/vol] digitonin). BN loading dye (0.5% [wt/vol] Coomassie blue G250, 50 mM ε-amino n-caproic acid, 10 mM Bis-Tris, pH 7.0) was added to the clarified supernatant before loading.
Gels were transferred onto polyvinylidene fluoride membranes (0.45 μM Immobilon-P; Merck) using the Owl HEP-1 Semidry Electroblotting system (Thermo Fisher Scientific). Following incubation with primary antibodies, horseradish peroxidase–coupled secondary antibodies (Sigma) and Clarity Western ECL Substrate (BioRad) were used for detection. Images were obtained using the ChemiDoc XRS+ imaging machine (BioRad). Quantification of Western blot signal was performed using the Image Lab software (BioRad) following the manufacturer’s instructions.
Cellular respiration measurements
Oxygen consumption rates (OCR) were measured in a Seahorse Bioscience XF24-3 Analyzer as previously described (Kang et al., 2019). Cells (50,000 [HEK293] or 25,000 [fibroblast]) were plated per well in XF24-3 culture plates (pretreated with poly-d-lysine for HEK293 cells) and grown overnight under standard culture conditions. Rates were measured in nonbuffered DMEM containing 25 mM glucose. HEK293 measurements consisted of three cycles each of 2 min mix, 2 min wait, and 3 min measure using the following inhibitors: 0.5 μM oligomycin, 0.1 μM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), 0.5 μM rotenone, and 0.3 μM antimycin A. Fibroblast cell measurements consisted of four cycles each of 2 min mix, 2 min wait, and 4 min measure using the following inhibitors: 1μM oligomycin, 0.7 μM FCCP, 0.5 μM rotenone, and 0.3 μM antimycin A. Cell numbers were normalized using CyQuant (Life Technologies) with four to five replicate wells per cell line measured in multiple plates (n = 4 for HEK293, n = 2 for fibroblasts). Basal OCR and nonmitochondrial respiration (following rotenone and antimycin A addition) were calculated from the average of all measurements. To calculate maximal respiration, either the initial measurement following FCCP addition (HEK293) or an average of all measurements (fibroblasts) was used.
ATP synthesis assay
ATP synthesis assays were performed essentially as described (Bird et al., 2014). Briefly, 10 μg of cultured HEK293 cells in technical duplicates were permeabilized with 50 μg/ml digitonin in ATP assay buffer (25 mM Tris, 150 mM KCl, 2 mM EDTA, 10 mM K2HPO4, pH 7.4) containing 1 mM ADP and the indicated substrate/inhibitor concentrations (succinate [1 mM], glutamate [10 mM], malate [10 mM], pyruvate [10 mM], rotenone [2.5 μM], malonate [1 mM]). Samples were incubated at 37 °C for 20 min and then transferred to ice. Reactions were stopped with the addition of 0.6 M perchloric acid and neutralized using a 2 M KOH/0.6 M MOPS solution. The ATP concentrations in each reaction were measured in a microplate reader (BMG Labtech, FLUOstar Omega) using the ATP Bioluminescence Assay Kit CLS II (Roche, 11699695001). ATP synthesis rates (nmol/mg protein/min) of each samples were averaged from four biological replicates (n = 4).
In vitro protein import and autoradiography
Open reading frames encoding SFXN1, SFXN2, SFXN3, MTHFD2, SHMT2, or GC1 were cloned into pGEM4z and used for transcription with the mMESSAGE mMACHINE SP6 kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Radiolabeled protein was translated from mRNA using the rabbit reticulocyte lysate system (Promega) and 35S-labeled methionine, according to the manufacturer’s instructions. Isolated mitochondria were resuspended in mitochondrial import buffer (250 mM sucrose, 5 mM magnesium acetate, 80 mM potassium acetate, 10 mM sodium succinate, 1 mM DTT, 5 mM ATP, 20 mM HEPES-KOH, pH 7.4), and imports were performed through incubation with radiolabeled proteins for the desired incubation time at 37°C and in the presence or absence of 10 μM FCCP (to dissipate membrane potential). Following import, each reaction was treated consecutively with proteinase K (50 μg/mL for 10 min on ice) and PMSF (1 mM for 5 min on ice) before reisolation for SDS–PAGE and BN-PAGE analysis. Radioactive signals were detected using a Typhoon phosphorimager (GE Healthcare). Analysis of autoradiography was performed using ImageJ software to calculate the intensity of each band. Background intensities were calculated by averaging the intensity of multiple areas of the gel away from the bands.
Quantitative mass spectrometry and data analysis
A total of 50 ug pellets of whole-cells or isolated mitochondria (protein concentration determined using a Pierce BCA Assay Kit (Thermo Fisher Scientific)) were solubilized in solubilization buffer (1% [wt/vol] sodium deoxycholate, 100 mM Tris pH 8.1, 40 mM chloroacetamide [Sigma], and 10 mM Tris(2-carboxyethyl)phosphine hydrochloride [TCEP; BondBreaker; Thermo Fisher Scientific]) for 5 min at 99°C with 1500 rpm shaking followed by 15 min sonication in a water bath sonicator. Proteins were digested with trypsin (Thermo Fisher Scientific) at a 1:50 trypsin:protein ratio at 37°C overnight. The supernatant was transferred to stagetips containing 3 × 14 G plugs of 3M Empore SDB-RPS substrate (Sigma) as described previously (Kulak et al., 2014; Stroud et al., 2016). Ethyl acetate or isopropanol 99% (vol/vol) and 1% trifluoroacetic acid (TFA) [vol/vol] was added to the tip before centrifugation at 3000 × g at room temperature. Stagetips were washed first with ethyl acetate or isopropanol (99% [vol/vol]) and TFA (1% [vol/vol]) solution and then subjected to a second wash containing 0.2% (vol/vol) TFA. Peptides were eluted in 80% (vol/vol) acetonitrile and 1% (wt/vol) NH4OH and acidified to a final concentration of 1% (vol/vol) TFA before drying in a CentriVap Benchtop Vacuum Concentrator (Labconco). Peptides were reconstituted in 0.1% TFA and 2% ACN for analysis by liquid chromatography (LC)–MS/MS.
LC MS/MS was carried out on a QExactive plus Orbitrap mass spectrometer (Thermo Fisher Scientific) with a nanoESI interface in conjunction with an Ultimate 3000 RSLC nanoHPLC (Dionex Ultimate 3000. The LC system was equipped with an Acclaim Prepmap nano-trap column (Dionex C18; 100 Å, 75 μM × 50 cm). The tryptic peptides were injected into the enrichment column at an isocratic flow of 5 μl/min of 2% (vol/vol) CH3CN containing 0.1% (vol/vol) formic acid for 5 min applied before the enrichment column was switched in-line with the analytical column. The eluents were 5% dimethyl sulfoxide (DMSO) in 0.1% (vol/vol) formic acid (solvent A) and 5% DMSO in 100% (vol/vol) CH3CN and 0.1% (vol/vol) formic acid (solvent B). The flow gradient was 1) 0–6 min at 3% B, 2) 6–95 min at 3–22% B, 3) 95–105 min at 22–40% B, 4) 105–110 min at 40–80% B, 5) 110–115 min at 80% B, 6) 115–117 min at 80–3% B. Equilibration was performed with 3% B for 10 min before the next sample injection. The QExactive plus mass spectrometer was operated in the data-dependent mode. Full MS1 spectra were acquired in positive mode, 70,000 resolution, AGC target of 3e6, and maximum IT time of 50 ms. A loop count of 15 on the most intense targeted peptide was isolated for MS/MS. The isolation window was set to 1.2 m/z and precursors fragmented using stepped normalized collision energy of 28, 30, and 32. MS2 resolution was at 17,500, AGC target at 2e6, and maximum IT time of 50 ms. Dynamic exclusion was set to be 30 s.
Raw files were processed using the MaxQuant platform (version 1.6.5.0) (Cox and Mann, 2008) and searched against the UniProt human database (June 2019) using default settings for an label-free quantitation (LFQ) experiment with match between runs enabled. The proteinGroups.txt output from the search was processed in Perseus (version 1.6.2.2) (Tyanova et al., 2016). Briefly, entries “Only identified by site,” “Reverse,” and “Potential contaminant” were removed from the data sets. Log2-transformed LFQ intensities were grouped (control, knockout, patient) according to each experiment and filtered to have two out of three valid values in each group. Isolated mitochondria experiments were annotated for proteins present in the Mitocarta2.0 (Calvo et al., 2016) through matching by gene name. Mitocarta2.0-positive rows were filtered to include only mitochondrial entries and normalized using the “Subtract row cluster” function with “Known mitochondrial” entries from the IMPI (2017) (Smith and Robinson, 2016) database as reference. Two-sample t tests were performed between groups using p value for truncation (threshold p value < 0.05). Volcano plots were generated via scatter plots by selecting “Student’s T-test difference” and “-Log Student’s T-test p-value.”
OXPHOS plots were generated as described previously (Lake et al., 2017), with some modifications. Briefly, raw LFQ intensity values for OXPHOS proteins that contained at least two valid values in each experimental group were imported into Prism 8 software. Mean values for each gene in each group were obtained, and a ratio paired t test was performed on the Log2-transformed means.
For gene ontology (GO) enrichment analysis, proteins up-regulated or down-regulated >1.5-fold (>1.25-fold for fibroblasts) with p < 0.05 were submitted to the ClueGO plug-in (version 2.5.6) (Bindea et al., 2009) in Cytoscape (version 3.8.0) for identification of enriched GO biological processes. The GO term fusion function was used to reduce the complexity of the resulting networks. All other settings were left as default.
For prediction of TIM22 complex substrates, proteins down-regulated >1.25-fold in the indicated cell lines were submitted to the HMMTOP online tool for prediction of transmembrane domains (Tusnady and Simon, 2001), as well as the MitoFates online tool for prediction of mitochondrial presequences (Fukasawa et al., 2015). A protein was considered a candidate substrate if it contained two or more predicted transmembrane domains and lacked a predicted presequence.
Cell proliferation and viability measurements
Confluency of cells in 96-well plates was tracked over 96 h using IncuCyte FLR (Essen BioSciences) following the manufacturer’s guidelines. Five thousand cells were plated in the indicated media and allowed to adhere 2 h before the first reading. For dose response experiments, 10,000 cells were plated overnight and then dosed with actinonin (Sigma) at the indicated doses. Seventy-two hours following exposure, cell viability was determined by alamarBlue assay (Life Technologies) using Cytation 3 (Biotek).
Tissue culture, acquisition, and analysis of pulse-SILAC mass spectrometry data
Pulse-SILAC experiment was performed as previously described (Hock et al., 2020) with modifications. Cells were cultured in triplicate in DMEM high glucose) supplemented with 10% (vol/vol) fetal calf serum, penicillin–streptomycin, 50 μg/ml uridine, and 1 μg/ml tetracycline at 37 °C under an atmosphere of 5% CO2. Chloramphenicol (CAP; Sigma) was added at 50 µg/ ml for 24 h before pulse to inhibit mitochondrial translation. The next day, media was replaced with DMEM for SILAC (Thermo Fisher Scientific) supplemented with 10% (vol/vol) dialyzed fetal calf serum (dFCS; Thermo Fisher Scientific), penicillin/streptomycin (Life Technologies), 1 mM sodium pyruvate (Life Technologies), 1X Glutamax (Life Technologies), 3.5 g/l glucose, 50 µg/ml uridine, 1 µg/ml tetracycline, 600 mg/ml l-proline (Merck), 146 mg/ml -13C615N2-l-lysine-HCl, and 42 mg/l -13C615N2-l-arginine-HCl (Silantes). Time points were collected at 1, 3, and 4 h post–heavy SILAC media incubation. Cells were washed twice with PBS, pelleted at 500 × g, and frozen at −80°C until use.
Mitochondrial isolation was performed as previously described (Acín-Pérez et al., 2008) with modifications. Briefly, cell pellets were resuspended in buffer A (83 mM sucrose, 10 mM HEPES, pH 7.2) and homogenized with 10 strokes using a KIMBLE glass Dounce homogenizer (Sigma). An equal amount of buffer B (250 mM sucrose, 30 mM HEPES, pH 7.2) was added and centrifuged at 1000 × g for 5 min to remove cell debris and unbroken cells. Mitochondria were collected from the supernatant by centrifuging at 10,000 × g for 2 min. Protein concentration was determined using the Pierce BCA protein assay kit (Thermo Fisher Scientific) and normalized to 50 μg for quantitative proteomics preparation. Mitochondrial pellets were prepared for mass spectrometry using the same method as described under Quantitative mass spectrometry and data analysis.
Peptides were analyzed on an Orbitrap Eclipse Tribid mass spectrometer (Thermo Fisher Scientific). LC coupled MS/MS was carried out with a nanoESI interface in conjunction with an Ultimate 3000 RSLC nanoHPLC (Dionex Ultimate 3000). The LC system was equipped with an Acclaim Pepmap nano-trap column (Dionex-C18; 100 Å, 75 μ × 2 cm) and an Acclaim Pepmap RSLC analytical column (Dionex-C18; 100 Å, 75 μM × 50 cm). The tryptic peptides were injected to the trap column at an isocratic flow of 5 μl/min of 2% ACN containing 0.1% (vol/vol) formic acid for 5 min applied before the trap column was switched in-line with the analytical column. The eluents were 5% DMSO in 0.1% vol/vol formic acid (solvent A) and 5% DMSO in 100% vol/vol ACN and 0.1% vol/vol formic acid (solvent B). The flow gradient was 1) 0–6 min at 3% B, 2) 6–95 min, 3–23% B, 3) 95–105 min, 23–40% B, 4) 105–110 min, 40–80% B, 5) 110–115 min, 80–80% B, 6) 115–117 min, 80–3% B and equilibrated at 3% B for 10 min before the next sample injection.
For this experiment, the Orbitrap Eclipse mass spectrometer was operated in the data-dependent mode with a targeted inclusion list containing predicted peptides from the 13 mitochondrial DNA-encoded proteins. The inclusion list consists of mass/charge (m/z) and charge (z) or tryptic peptides (endogenous and SILAC labeled) predicted from in silico digest of target proteins using the Skyline software (MacLean et al., 2010). In addition, the inclusion list also contained peptides that have been previously observed in public data depositories through the PeptideAtlas site (Deutsch et al., 2008), reference peptides from ProteomicsDB (Schmidt et al., 2018), and the present study.
The acquisition method was created with the Orbitrap Tribid Tune version 3.3 acquisition software. The full MS1 spectra from 375 to 1500 m/z was acquired in positive mode at 120,000 resolution. Two scan priorities were created post–MS1 scans. The first priority was given to precursors that fulfill the mass and charge criteria. Precursors were then isolated using isolation window of 1.6 m/z and precursors fragmented using fixed normalized collision energy of 30. MS2 resolution was at 15,000, AGC target at 5e5, and maximum IT time of 100 ms. The second scan priority is activated when no matching mass from the target list is identified. Precursors were isolated with the same parameters with the difference of maximum IT of 22 ms. Dynamic exclusion was set to be 30 s.
Raw files were processed using the MaxQuant platform (version 1.6.10.43) (Cox and Mann, 2008) and searched against the UniProt human database (42,434 entries, June 2019) using default settings for a SILAC experiment with “label min. ratio count” set to 1 and match between runs enabled. From the proteinGroups.txt output file, mtDNA-encoded proteins were filtered by gene name and identified by at least two peptides. Heavy intensities were log2 transformed in Prism (version 8.4.3; GraphPad) and normalized to the maximum value detected in the WT line at 4 h pulse. The means from three experiments were plotted over time using Prism along with the SD. Statistical significance using t test was performed in Prism using the two-stage step-up method of Benjamini et al. (2006) and false discovery rate (FDR) of 1%.
Supplementary Material
Acknowledgments
T.D.J. and K.M.F. are supported by Australian Government Research Training Program (RTP) scholarships. D.H.H. is supported by a Melbourne International Research Scholarship. T.D.J., D.H.H., and Y.C.L are supported by Australian Mito Foundation PhD Top-Up scholarships. D.S. is supported by a Research Fellowship from the Mito Foundation. We acknowledge funding from the Australian Research Council (DP170101249 to D.S.), the National Health and Medical Research Council (NHMRC Project Grant 1140906 to D.A.S.; NHMRC Fellowship 1140851 to D.A.S; and NHMRC Fellowship 1155244 to D.R.T), the Australian Mito Foundation (D.R.T., A.E.F., and D.S.); and the Victorian Government’s Operational Infrastructure Support Program (D.R.T). Nicholas J. Clemons is supported by a Fellowship (MCRF16002) from the Victorian Government Department of Health and Human Services acting through the Victorian Cancer Agency. We thank Kristin Brown (Peter MacCallum Cancer Centre) for providing us with serine and glycine free DMEM.
Abbreviations used:
- AGK
acylglycerol kinase
- BN-PAGE
Blue Native PAGE
- KO
knockout
- mtDNA
mitochondrial DNA
- MTHFD2
bifunctional methylenetetrahydrofolate dehydrogenase, mitochondrial
- OXPHOS
oxidative phosphorylation
- SFXN
sideroflexin
- SHMT2
serine hydroxymethyltransferase, mitochondrial
- SILAC
stable isotope labeling with amino acids in cell culture
- TIM22
translocase of the inner membrane 22
- TIM23
translocase of the inner membrane 23
- 1C
one-carbon (metabolism).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-06-0390) on January 21, 2021.
REFERENCES
- Acín-Pérez R, Fernández-Silva P, Peleato ML, Pérez-Martos A, Enriquez JA (2008). Respiratory active mitochondrial supercomplexes. Mol Cell 32, 529–539. [DOI] [PubMed] [Google Scholar]
- Anderson AJ, Jackson TD, Stroud DA, Stojanovski D (2019). Mitochondria—hubs for regulating cellular biochemistry: emerging concepts and networks. Open Biol 9, 190126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et al. (2016). Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 6, e10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bektas M, Payne SG, Liu H, Goparaju S, Milstien S, Spiegel S (2005). A novel acylglycerol kinase that produces lysophosphatidic acid modulates cross talk with EGFR in prostate cancer cells. J Cell Biol 169, 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Abba MK, Daniel Y (2006). Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507. [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird MJ, Wijeyeratne XW, Komen JC, Laskowski A, Ryan MT, Thorburn DR, Frazier AE (2014). Neuronal and astrocyte dysfunction diverges from embryonic fibroblasts in the Ndufs4fky/fky mouse. Biosci Rep 34, e00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GC, Murphy MP, Jornayvaz FR, Shulman GI (2010). Regulation of mitochondrial biogenesis. Essays Biochem 47, 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callegari S, Richter F, Chojnacka K, Jans DC, Lorenzi I, Pacheu-Grau D, Jakobs S, Lenz C, Urlaub H, Dudek J, et al. (2016). TIM29 is a subunit of the human carrier translocase required for protein transport. FEBS Lett 590, 4147–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK (2016). MitoCarta2. 0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44, D1251–D1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, Laskowski A, Garone C, Liu S, Jaffe DB, et al. (2012). Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med 4, 118ra110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008). MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Lam H, Aebersold R (2008). PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep 9, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Rabinowitz JD (2017). One-carbon metabolism in health and disease. Cell Metab 25, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD (2014). Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler E, Eckl E-M, Schmitt S, Mancilla IA, Meyer-Bender MF, Hanf M, Philippou-Massier J, Krebs S, Zischka H, Jae LT (2020). A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 579, 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa LE, Dibley MG, Stroud DA, Ryan MT (2018). Building a complex complex: assembly of mitochondrial respiratory chain complex I. Semin Cell Dev Biol 76, 154–162. [DOI] [PubMed] [Google Scholar]
- Frazier AE, Thorburn DR, Compton AG (2019). Mitochondrial energy generation disorders: genes, mechanisms, and clues to pathology. J Biol Chem 294, 5386–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa Y, Tsuji J, Fu S-C, Tomii K, Horton P, Imai K (2015). MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics 14, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomkale R, Cruz-Zaragoza LD, Suppanz I, Guiard B, Montoya J, Callegari S, Pacheu-Grau D, Warscheid B, Rehling P (2020). Defining the substrate spectrum of the TIM22 complex identifies pyruvate carrier subunits as unconventional cargos. Curr Biol 30, 1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin Y-HT, Wiita AP, Xu K, Correia MA, Kampmann M (2020). Mitochondrial stress is relayed to the cytosol by an OMA1–DELE1–HRI pathway. Nature 579, 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi A, Haack TB, Atiq M, Mottaghi H, Haghighi-Kakhki H, Bashir RA, Ahting U, Feichtinger RG, Mayr JA, Rötig A, et al. (2014). Sengers syndrome: six novel AGK mutations in seven new families and review of the phenotypic and mutational spectrum of 29 patients. Orphanet J Rare Dis 9, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein S, Perrault I, Roche O, Gerber S, Khadom N, Rio M, Boddaert N, Jean-Pierre M, Brahimi N, Serre V, et al. (2009). TMEM126A, encoding a mitochondrial protein, is mutated in autosomal-recessive nonsyndromic optic atrophy. Am J Hum Genet 84, 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock DH, Reljic B, Ang C-S, Muellner-Wong L, Mountford HS, Compton AG, Ryan MT, Thorburn DR, Stroud DA (2020). HIGD2A is required for assembly of the COX3 module of human mitochondrial complex IV. Mol Cell Proteomics 19, 1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson TD, Palmer CS, Stojanovski D (2018). Mitochondrial diseases caused by dysfunctional mitochondrial protein import. Biochem Soc Trans 46, 1225–1238. [DOI] [PubMed] [Google Scholar]
- Káldi K, Bauer MF, Sirrenberg C, Neupert W, Brunner M (1998). Biogenesis of Tim23 and Tim17, integral components of the TIM machinery for matrix-targeted preproteins. EMBO J 17, 1569–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Anderson AJ, Jackson TD, Palmer CS, De Souza DP, Fujihara KM, Stait T, Frazier AE, Clemons NJ, Tull D, et al. (2019). Function of hTim8a in complex IV assembly in neuronal cells provides insight into pathomechanism underlying Mohr-Tranebjærg syndrome. eLife 8, e48828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Baker MJ, Liem M, Louber J, McKenzie M, Atukorala I, Ang CS, Keerthikumar S, Mathivanan S, Stojanovski D (2016). Tim29 is a novel subunit of the human TIM22 translocase and is involved in complex assembly and stability. eLife 5, e17463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Stroud DA, Baker MJ, Souza DDP, Frazier AE, Liem M, Tull D, Mathivanan S, McConville MJ, Thorburn DR, et al. (2017). Sengers syndrome-associated mitochondrial acylglycerol kinase is a subunit of the human TIM22 protein import complex. Mol Cell 67, 457–470. [DOI] [PubMed] [Google Scholar]
- Kaukonen J, Juselius JK, Tiranti V, Kyttälä A, Zeviani M, Comi GP, Keränen S, Peltonen L, Suomalainen A (2000). Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science 289, 782–785. [DOI] [PubMed] [Google Scholar]
- Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivelä R, Pessia A, Velagapudi V, Suomalainen A (2017). mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab 26, 419–428. [DOI] [PubMed] [Google Scholar]
- Kikuchi G, Motokawa Y, Yoshida T, Hiraga K (2008). Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad, Ser B 84, 246–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N, Wyant GA, Prakash G, uit de Bos J, Bottanelli F, Pacold ME, Chan SH, Lewis CA, Wang T, Keys HR, et al. (2018). SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science 362, eaat9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koufaris C, Nilsson R (2018). Protein interaction and functional data indicate MTHFD2 involvement in RNA processing and translation. Cancer Metab 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M (2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11, 319–324. [DOI] [PubMed] [Google Scholar]
- Kurz M, Martin H, Rassow J, Pfanner N, Ryan MT (1999). Biogenesis of Tim proteins of the mitochondrial carrier import pathway: differential targeting mechanisms and crossing over with the main import pathway. Mol Biol Cell 10, 2461–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake NJ, Webb BD, Stroud DA, Richman TR, Ruzzenente B, Compton AG, Mountford HS, Pulman J, Zangarelli C, Rio M, et al. (2017). Biallelic mutations in MRPS34 lead to instability of the small mitoribosomal subunit and Leigh syndrome. Am J Hum Genet 101, 239–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr JA, Haack TB, Graf E, Zimmermann FA, Wieland T, Haberberger B, Superti-Furga A, Kirschner J, Steinmann B, Baumgartner MR, et al. (2012). Lack of the mitochondrial protein acylglycerol kinase causes Sengers syndrome. Am J Hum Genet 90, 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Michaelides M, Tee LJ, Robson AG, Rahman F, Pasha S, Luxon LM, Moore AT, Maher ER (2010). Nonsense mutation in TMEM126A causing autosomal recessive optic atrophy and auditory neuropathy. Mol Vis 16, 650–664 [PMC free article] [PubMed] [Google Scholar]
- Mills GB, Moolenaar WH (2003). The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer 3, 582–591. [DOI] [PubMed] [Google Scholar]
- Minton DR, Nam M, McLaughlin DJ, Shin J, Bayraktar EC, Alvarez SW, Sviderskiy VO, Papagiannakopoulos T, Sabatini DM, Birsoy K, et al. (2018). Serine catabolism by SHMT2 is required for proper mitochondrial translation initiation and maintenance of formylmethionyl-tRNAs. Mol Cell 69, 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momb J, Lewandowski JP, Bryant JD, Fitch R, Surman DR, Vokes SA, Appling DR (2013). Deletion of Mthfd1l causes embryonic lethality and neural tube and craniofacial defects in mice. Proc Natl Acad Sci USA 110, 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mon EE, Wei F-Y, Ahmad RNR, Yamamoto T, Moroishi T, Tomizawa K (2019). Regulation of mitochondrial iron homeostasis by sideroflexin 2. J Physiol Sci 69, 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkanen J, Forsström S, Euro L, Paetau I, Kohnz RA, Wang L, Chilov D, Viinamäki J, Roivainen A, Marjamäki P, et al. (2016). Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metab 23, 635–648. [DOI] [PubMed] [Google Scholar]
- Pacheu-Grau D, Callegari S, Emperador S, Thompson K, Aich A, Topol SE, Spencer EG, McFarland R, Ruiz-Pesini E, Torkamani A, et al. (2018). Mutations of the mitochondrial carrier translocase channel subunit TIM22 cause early-onset mitochondrial myopathy. Hum Mol Genet 27, 4135–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F (2013). The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Aspects Med 34, 465–484. [DOI] [PubMed] [Google Scholar]
- Palmieri F, Scarcia P, Monné M (2020). Diseases caused by mutations in mitochondrial carrier genes SLC25: a review. Biomolecules 10, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, Auwerx J (2017). Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 216, 2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H, Sucec I, Bersch B, Horten P, Perschil I, Martinou J-C, van der Laan M, Wiedemann N, Schanda P, Pfanner N (2020). The mitochondrial carrier pathway transports non-canonical substrates with an odd number of transmembrane segments. BMC Biol 18, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S, Sharma R, Gupta R, Ast T, Chan C, Durham TJ, Goodman RP, Grabarek Z, Haas ME, Hung WH, et al. (2020). MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res 49, D1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N (2004). Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol 5, 519–530. [DOI] [PubMed] [Google Scholar]
- Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kühlbrandt W, Wagner R, Truscott KN, Pfanner N (2003). Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299, 1747–1751. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Samaras P, Frejno M, Gessulat S, Barnert M, Kienegger H, Krcmar H, Schlegl J, Ehrlich H-C, Aiche S, et al. (2018). ProteomicsDB. Nucleic Acids Res 46, D1271–D1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Robinson AJ (2016). MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res 44, D1258–D1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, Osellame LD, Stait T, Beilharz TH, Thorburn DR, et al. (2016). Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538, 123–126. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016). The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Vukotic M, Nolte H, Konig T, Saita S, Ananjew M, Kruger M, Tatsuta T, Langer T (2017). Acylglycerol kinase mutated in Sengers syndrome is a subunit of the TIM22 protein translocase in mitochondria. Mol Cell 67, 471–483. [DOI] [PubMed] [Google Scholar]
- Waggoner DW, Johnson LB, Mann PC, Morris V, Guastella J, Bajjalieh SM (2004). MuLK, a eukaryotic multi-substrate lipid kinase. J Biol Chem 279, 38228–38235. [DOI] [PubMed] [Google Scholar]
- Wang X, Lin C, Zhao X, Liu A, Zhu J, Li X, Song L (2014). Acylglycerol kinase promotes cell proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Mol Cancer 13, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N (2017). Mitochondrial machineries for protein import and assembly. Annu Rev Biochem 86, 685–714. [DOI] [PubMed] [Google Scholar]
- Žárský V, Doležal P (2016). Evolution of the Tim17 protein family. Biol Direct 11, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.